Abstract
Trichosporon asahii is a pathogenic fungus that causes deep mycosis in patients with neutropenia. Establishing an experimental animal model for quantitatively evaluating pathogenicity and developing a genetic recombination technology will help to elucidate the infection mechanism of T. asahii and promote the development of antifungal drugs. Here we established a silkworm infection model with a transgenic T. asahii strain expressing eGFP. Injecting T. asahii into silkworms eventually killed the silkworms. Moreover, the administration of antifungal agents, such as amphotericin B, fluconazole, and voriconazole, prolonged the survival time of silkworms infected with T. asahii. A transgenic T. asahii strain expressing eGFP was obtained using a gene recombination method with Agrobacterium tumefaciens. The T. asahii strain expressing eGFP showed hyphal formation in the silkworm hemolymph. Both hyphal growth and the inhibition of hyphal growth by the administration of antifungal agents were quantitatively estimated by monitoring fluorescence. Our findings suggest that a silkworm infection model using T. asahii expressing eGFP is useful for evaluating both the pathogenicity of T. asahii and the efficacy of antifungal drugs.
Subject terms: Animal disease models, Small molecules, Fungal infection
Introduction
Trichosporon asahii is a basidiomycetous yeast widely distributed in the environment1–4, and commonly isolated from human blood, sputum, skin, feces, and urine5–8. T. asahii is a pathogenic fungus that causes severe deep mycosis in patients with neutropenia8–10. Whereas the mortality rate of deep mycosis caused by Candida albicans is approximately 40%, that caused by T. asahii is approximately 80%, and the prognosis is poor11. T. asahii is resistant to echinocandin antifungal drugs and causes severe infections in patients treated with micafungin12. Moreover, strains resistant to amphotericin B and azole antifungal drugs such as fluconazole have been isolated from patients13,14. Therefore, T. asahii has become a serious clinical problem as a pathogenic fungus that causes systemic infection in immunocompromised hosts8.
T. asahii forms hyphae, which are branching filamentous structures14. In pathogenic fungi that form hyphae such as Candida albicans, hyphal formation is crucial for the host epithelial cell damage and biofilm formation that are involved in infections15. As with C. albicans, T. asahii makes treatment difficult by forming a biofilm on devices such as catheters14,16. Hyphal growth of T. asahii in blood vessels causes necrotic thrombi, and may contribute to infection17. The molecular mechanisms of infection caused by T. asahii, however, remain unclear. Establishing a simple animal experimental system for systemic infection with hyphal formation of T. asahii and developing genetic recombination technology in T. asahii will contribute to elucidate the infection mechanism.
The silkworm, an invertebrate, is a useful experimental animal for evaluating the pathogenicity of pathogenic microorganisms and the therapeutic effects of antibacterial, antifungal, and antiviral drugs18–22. Silkworms have advantages for conducting infection experiments that require a large number of individuals23–25 as they are much less expensive to rear and maintain than mammals and their experimental use partly avoids the ethical problems associated with that of mammalian models23,24. These advantages allow for the use of a large number of silkworms to calculate the dose of a pathogen required to kill half of the silkworms (LD50) and the administration dose of an antifungal drug required to promote survival in half of the silkworms (ED50) for quantitative evaluation of the pathogenicity of microorganisms and the efficacy of therapeutic agents26–28. Furthermore, silkworm infection models can be used for in vivo screening experiments, and allow for the identification of virulence-related genes of microorganisms and therapeutically effective drugs. Virulence-related genes of pathogenic microorganisms such as Staphylococcus aureus, Candida albicans, and Candida glabrata have been discovered using silkworm infection models and gene-deficient mutant libraries19,29,30. Avirulent mutants that exhibit lower pathogenicity against silkworms also exhibit lower pathogenicity in infection experiments in mice. Many studies using antimicrobial drugs, toxic compounds, and natural products demonstrate that the therapeutic effects, pharmacokinetic parameters, and toxicity are similar between silkworms and mammals31–35. Moreover, lysocin E36, nosokomycin37, ASP239738, and GPI036334 were identified by exploratory research using a silkworm infection model from microbial culture broths and chemical libraries as novel antimicrobial compounds that show therapeutic effects in mouse infection experiments. Therefore, the silkworm infection model is useful for elucidating pathogenic mechanisms and evaluating the efficacy of antifungal drugs24,39.
Fungal hyphae comprise multiple cells, making it difficult to quantitatively estimate hyphal formation by counting colony-forming units40. Fluorescence imaging using enhanced green fluorescent protein (eGFP) is a simple and effective method for quantitatively evaluating cell proliferation and cell death in vivo41,42. We previously established a silkworm infection model using transgenic dermatophytes expressing eGFP, and developed a method for quantifying hyphal formation by fluorescence imaging43.
In the present study, we demonstrated that T. asahii can kill silkworms and successfully established a transgenic T. asahii strain expressing eGFP by genetic recombination using Agrobacterium tumefaciens. The therapeutic effects of antifungal drugs can be evaluated using a silkworm infection model with the transgenic T. asahii strain expressing eGFP. The silkworm infection model and gene recombination technology established in this study were effective toward elucidating the infection mechanism of T. asahii.
Material and methods
Culture of T. asahii
T. asahii JCM2466 was grown on Sabouraud dextrose agar plates and incubated at 27 °C for 2 days.
Silkworm rearing
Eggs of silkworms were purchased from Ehime-Sanshu Co., Ltd. (Ehime, Japan), disinfected, and hatched at 25–27 °C. The silkworms were fed an artificial diet, Silkmate 2S, containing antibiotics purchased from Ehime-Sanshu Co., Ltd. (Ehime, Japan). Fifth instar larvae were used in the infection experiments.
Silkworm infection experiments
Silkworm fifth instar larvae were fed an artificial diet (Silkmate 2S; Ehime-Sanshu Co., Ltd., Ehime, Japan) overnight. T. asahii grown on Sabouraud agar plates was suspended in physiological saline solution (0.9% w/v NaCl) and filtered through a 40-µm cell strainer (Corning, NY, USA). A suspension (50 μl) of the T. asahii cells was injected into the silkworm hemolymph using a 1-ml tuberculin syringe (Terumo Medical Corporation, Tokyo, Japan). Silkworms injected with T. asahii cells were placed in an incubator and their survival was monitored.
LD50 measurement
T. asahii JCM2466 (2.9 × 103 to 1.8 × 107 cells) was injected into the silkworm hemolymph. Survival of the silkworms at 48 h was monitored. The LD50, which is the dose of T. asahii required to kill half of the silkworms, was determined from combined data of 6 independent experiments by simple logistic regression model using Prism 8 (GraphPad Software, LLC, San Diego, CA, USA, https://www.graphpad.com/scientific-software/prism/).
Antifungal agents
Amphotericin B (Wako, Osaka, Japan), fluconazole (Wako, Osaka, Japan), and voriconazole (Tocris Bioscience, Bristol, UK) were dissolved in dimethyl sulfoxide (Wako, Osaka, Japan) and stored at − 80 °C until use.
Minimum inhibitory concentration determination
The minimum inhibitory concentration (MIC) was determined using a drug sensitivity test kit, yeast-like fungi DP (Eiken Chemical, Tokyo, Japan), according to the Clinical and Laboratory Standards Institute (CLSI) M27-A3 method. Briefly, T. asahii cells (2 × 103 cells per well of a 96 well plate) were incubated with twofold serial dilutions of antifungal agents at 37 °C for 2 days, and the MIC values were determined.
ED50 measurement
To evaluate the therapeutic effects of antifungal agents, T. asahii cells (1–5 × 106 cells) were injected into the silkworm hemolymph, and various concentrations of the antifungal agents (50 μl) dissolved in saline were injected immediately afterwards into the silkworm hemolymph. The doses were created by fourfold serial dilutions. To determine the ED50 values, five or six silkworms were injected with each dose of the antifungal agents. Survival of the silkworms at 48 h was monitored. The ED50 values were calculated from combined data of 4–5 independent experiments by simple logistic regression model using Prism 8 (GraphPad Software, LLC, San Diego, CA, USA, https://www.graphpad.com/scientific-software/prism/).
Construction of T. asahii expressing eGFP
The plasmid for expressing eGFP in T. asahii was constructed according to a previous report43. The eGFP gene was introduced into a pAg1-NAT1 vector44. The promoter sequence of the actin gene of Cryptococcus neoformans (CnPactin) was incorporated upstream of the eGFP gene, and the termination sequence of the Aspergillus nidulans trpC gene was incorporated downstream of the eGFP gene45. Cloning was performed using the infusion method according to the general method (In-Fusion HD Cloning Kit, Takara, Shiga, Japan). The primers used for polymerase chain reaction (PCR) amplification of each DNA region are shown in Table 1.
Table 1.
Primers used in the study.
| Primers | Nucleic acid sequence |
|---|---|
| pAg1-NAT for infusion cloning | |
| FpAg1-NAT | AGGATCCTTGCGCGCCTAGGC |
| RpAg1-NAT | GAAGAGATGTAGAAACTAGCT |
| CnPactin for infusion cloning | |
| FCnPactin(-pAg1-NAT) | TTTCTACATCTCTTCGCTGCGAGGATGTGAGCTGGA |
| RCnPactin(-eGFP) | CTCGCCCTTGCTCACCATGCCTCGATGGCCTCGGCGTCC |
| eGFPTtrpC for infusion cloning | |
| FeGFPTtrpC(-CnPactin) | TTTCTACATCTCTTCATGGTGAGCAAGGGCGAGGAG |
| ReGFPTtrpC(-pAg1-NAT) | GCGCGCAAGGATCCTAAAGAAGGATTACCTCTAAACAAGTGTACC |
The pAg1-NAT1-eGFP was introduced into the T. asahii JCM2466 strain using the A. tumefaciens-mediated transformation method described previously46,47. The pAg1-NAT-eGFP was introduced into A. tumefaciens EHA105 strain by electroporation and transformants were grown on 2 × YT agar containing rifampicin (50 µg/ml), chloramphenicol (25 µg/ml), and kanamycin (50 µg/ml). The transformant was co-cultured with the T. asahii JCM2466 strain at 27 °C for 2 days. The candidate transgenic T. asahii expressing eGFP were isolated as colonies grown on Sabouraud dextrose agar containing nourseothricin (50 µg/ml) and cefotaxime (100 µg/ml). Introduction of the eGFP gene into genome of the candidate strains was confirmed by PCR using the primers described in Table 1.
Imaging of eGFP-expressing T. asahii in silkworms
Fluorescence imaging using eGFP-expressing fungi in silkworms was performed according to a previous report43. eGFP-expressing T. asahii cells (2 x 106 cells) were injected into silkworms. The silkworms were reared at 37 °C and their hemolymph was collected after 24 h. The hemolymph was placed on glass slides and covered by a glass coverslip. The samples were examined with bright light or ultraviolet light under a microscope equipped with a fluorescence lens (DP-74; Olympus, Tokyo, Japan). The pictures were randomly obtained and the image fluorescence was determined by Image J software (ImageJ 1.47t; National Institutes of Health, Bethesda, MD, https://imagej.nih.gov/ij/).
Statistical analysis
The significance of differences between groups was calculated using the Tukey–Kramer method. P < 0.05 was considered significant.
Results
Killing of silkworms by injection of Trichosporon asahii cells
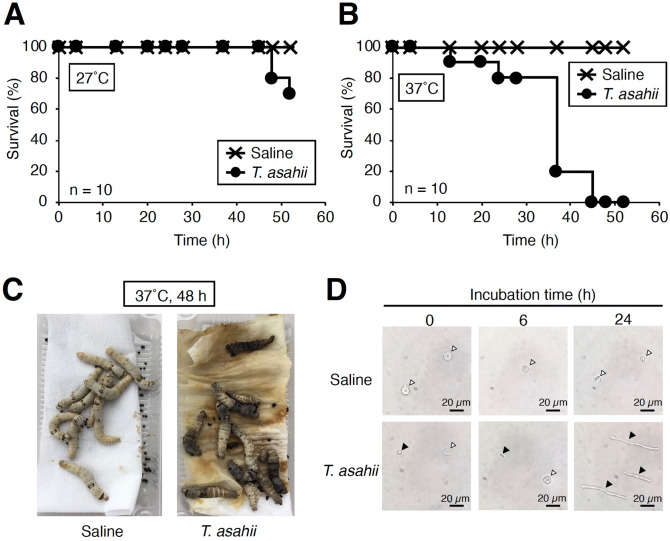
To establish a silkworm infection model with fungi, the silkworm rearing temperature after infection should be determined24,48. In Cryptococcus neoformans (H99 strain), by injection of 107 cells into silkworm hemolymph, the silkworms survived for 2 days rearing at 27 °C, but all silkworms died within 2 days at 37 °C27. Therefore, we first determined the rearing temperature at which T. asahii would kill silkworms. When silkworms injected with 1.9 × 106 cells of T. asahii were reared at 27 °C, most of silkworms survived at 48 h, but when reared at 37 °C, all of the silkworms died within 48 h (Fig. 1A–C). Hyphal formation of T. asahii was observed in the silkworm hemolymph after T. asahii injection under 37 °C rearing conditions (Fig. 1D). Silkworms died in a dose-dependent manner after administering T. asahii (Fig. 2A). The LD50 was 3.9 × 105 cells (Fig. 2B and Supplementary Fig. 1). On the other hand, silkworms injected with autoclave-treated T. asahii cells did not die (Fig. 3A,B). We previously reported the LD50 value of C. neoformans after 48 h at 37 °C27, which are the same experimental conditions we used to determine the LD50 value of T. asahii. The LD50 value of the C. neoformans H99 strain was 6 × 106 cells per larva. The LD50 value of the T. asahii JCM2466 strain determined in this study is lower than that of the C. neoformans H99 strain, indicating that the pathogenicity of T. asahii JCM2466 strain is higher than that of the C. neoformans H99 strain under these experimental conditions.
Figure 1.
Killing of silkworms by injection of T. asahii. Saline or T. asahii JCM2466 (2 × 106 cells) was injected into the silkworm hemolymph. Survival of the silkworms at 27 °C (A) and 37 °C (B) was monitored. n = 10/group. (C) Silkworms after injection at 48 h under 37 °C incubation are shown. (D) Saline or T. asahii JCM2466 (2 × 106 cells) was injected into the silkworm hemolymph. Silkworm hemolymph was collected at 0, 6, and 24 h after injection and observed with a microscope. White arrow indicates hemocytes of silkworm. Black arrowheads indicate T. asahii cells.
Figure 2.
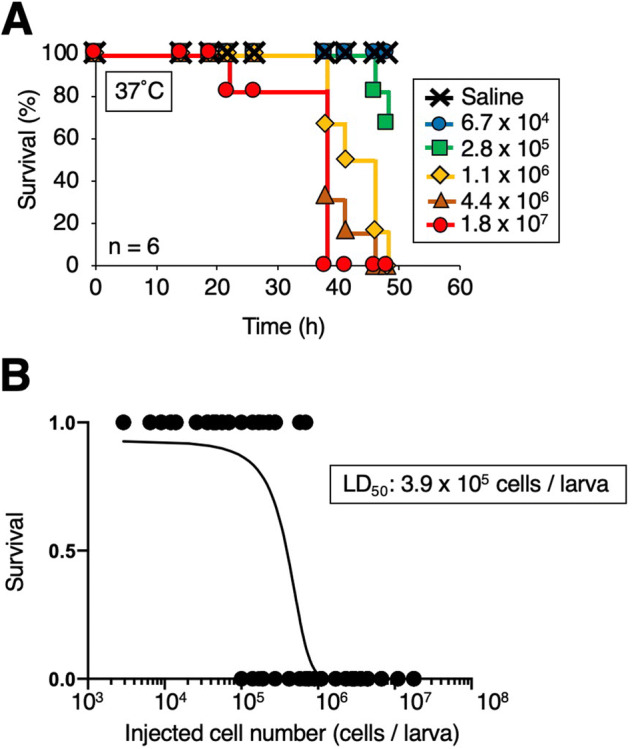
Death of silkworms according to the number of T. asahii cells administered. (A) Saline or T. asahii JCM2466 (6.7 × 104 – 1.8 × 107 cells) was injected into the silkworm hemolymph. Survival of the silkworms at 37 °C was monitored. n = 6/group. (B) Survival of the silkworms at 48 h was monitored. The LD50, which is the dose of T. asahii required to kill half of the silkworms, was determined from combined data of six independent experiments.
Figure 3.
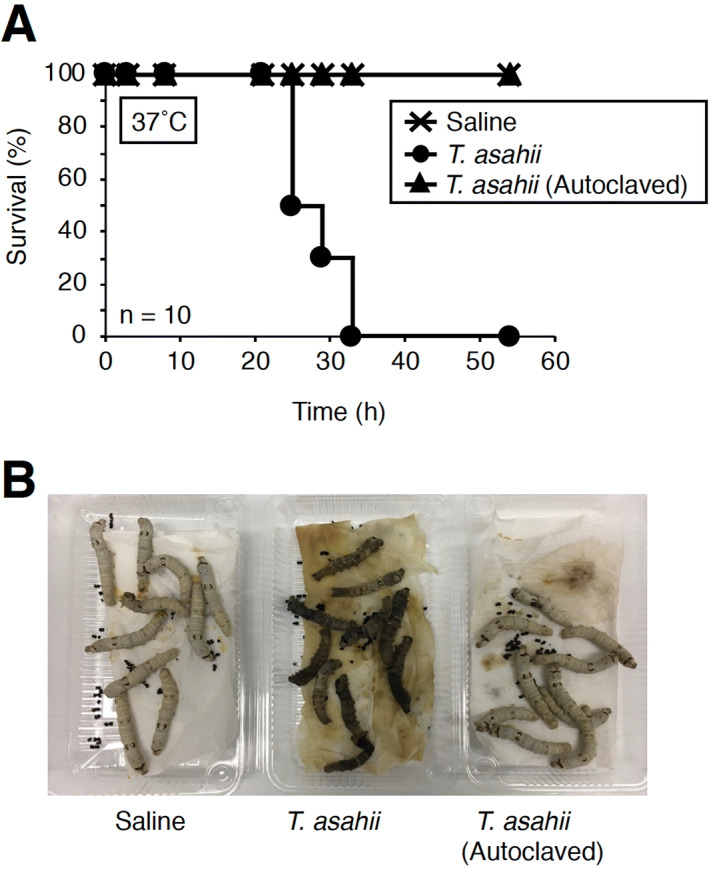
Effect of heat-treatment of T. asahii cells on the silkworm killing ability. (A) Saline, T. asahii JCM2466 (3 × 106 cells), or autoclaved T. asahii JCM2466 (autoclaved 3 × 106 cells) was injected into the silkworm hemolymph. Survival of the silkworms at 37 °C was monitored. n = 10/group. (B) Silkworms after injection at 48 h under 37 °C incubation are shown.
Evaluation of the therapeutic effects of antifungal drugs using the silkworm infection model
We previously found that the therapeutic effect of antifungal drugs could be evaluated using a silkworm infection model with C. neoformans27. Administration of the antifungal drugs amphotericin B, fluconazole, and voriconazole prolonged the silkworm survival time after injection of T. asahii (Fig. 4A). The ED50 value of amphotericin B, fluconazole, and voriconazole was 1.3, 3.9, and 0.4 µg g−1 of larva, respectively (Table 2 and Supplementary Fig. 2). Hyphal formation of T. asahii in the silkworm hemolymph at 24 h after injection was inhibited by administering amphotericin B, fluconazole, and voriconazole (Fig. 4B). These findings suggest that the therapeutic effect of antifungal drugs can be quantitatively evaluated using the silkworm infection model with T. asahii.
Figure 4.
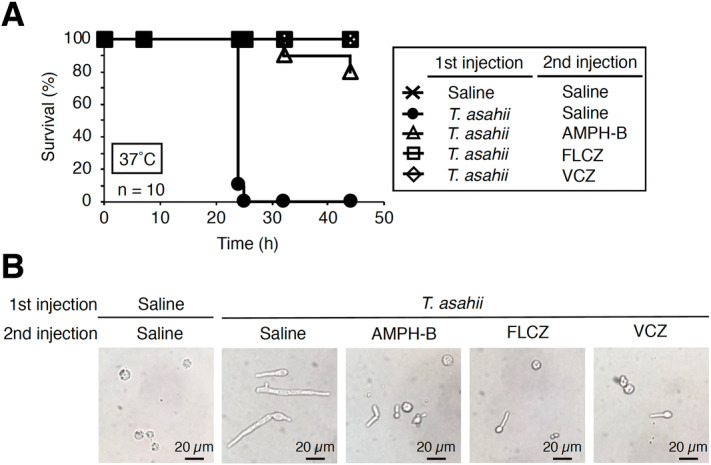
Therapeutic effects of antifungal drugs in silkworms infected with T. asahii. (A) Saline or T. asahii JCM2466 (3 × 106 cells) was injected into the silkworm hemolymph, followed by injection of 50 µl of amphotericin B (AMPH-B, 100 µg/ml), fluconazole (FLCZ, 100 µg/ml), or voriconazole (VCZ, 100 µg/ml) into the hemolymph. Silkworms were reared at 37 °C and survival of the silkworms was monitored. (B) Silkworm hemolymph was collected at 24 h after injection and observed with a microscope.
Table 2.
ED50 and MIC values of amphotericin B, fluconazole, and voriconazole against T. asahii JCM2466.
| ED50 (µg g−1 of larva) | MIC (µg ml−1) | |
|---|---|---|
| Amphotericin B | 1.3 | 1 |
| Fluconazole | 3.9 | 8 |
| Voriconazole | 0.4 | 0.25 |
Establishment of a transgenic T. asahii strain expressing eGFP
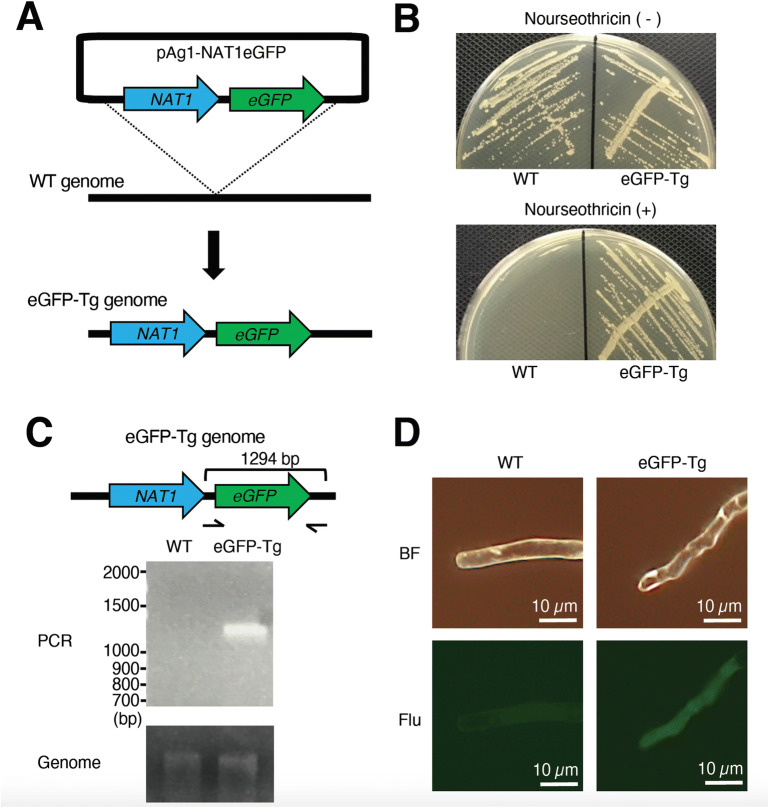
By gene recombination using Agrobacterium tumefaciens, we successfully introduced a gene into dermatophytes, which cause a superficial cutaneous fungal infection46. Using the Agrobacterium gene transfer method, a plasmid inserted with a nourseothricin resistance gene and a gene encoding eGFP was introduced into T. asahii to obtain a strain that grew on agar medium containing nourseothricin (Fig. 5A,B). When the genome of the nourseothricin-resistant strain was used as a template, a band of the expected size of the eGFP gene was amplified by PCR, but when the wild strain genome was used as a template, no amplification was observed (Fig. 5C). Moreover, the nourseothricin-resistant strain emitted green fluorescence when irradiated with excitation light (Fig. 5D). We named the strain eGFP-Tg. These results suggest that a nourseothricin-resistance gene and gene encoding eGFP could be introduced into T. asahii by gene recombination using A. tumefaciens.
Figure 5.
Establishment of transgenic T. asahii expressing eGFP. (A) Schematic illustration of gene transfer of the nourseothricin resistance gene NAT1 and the gene encoding eGFP to the T. asahii JCM2466 strain. (B) JCM2466 strain and JCM2466 eGFP-Tg strains were spread on Sabouraud agar medium containing nourseothricin (50 µg/ml) and incubated at 27 °C for 2 days. (C) Detection of gene encoding eGFP in the chromosome of the T. asahii strain by PCR. Full-length gels are presented in Supplementary Fig. 5. (D) The JCM2466 strain and the T. asahii eGFP-Tg strain were observed with a fluorescence microscope. BF: bright field. Flu: fluorescent, which is field of view under conditions of irradiation with excitation light for fluorescence detection.
Fluorescence imaging analysis of T. asahii infection
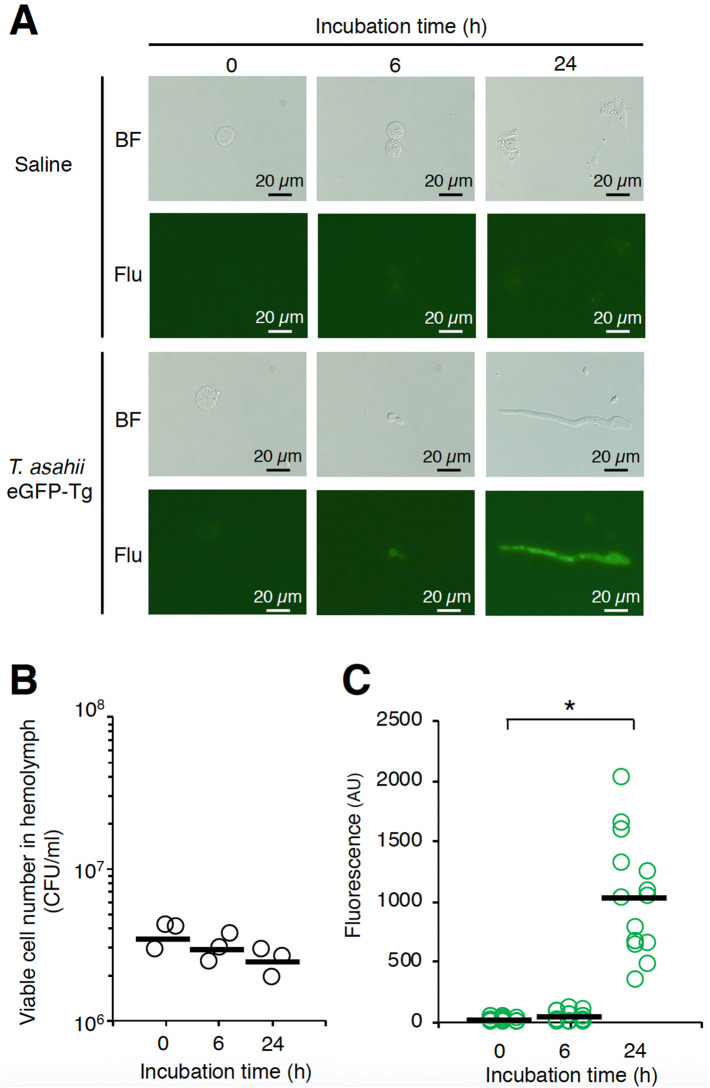
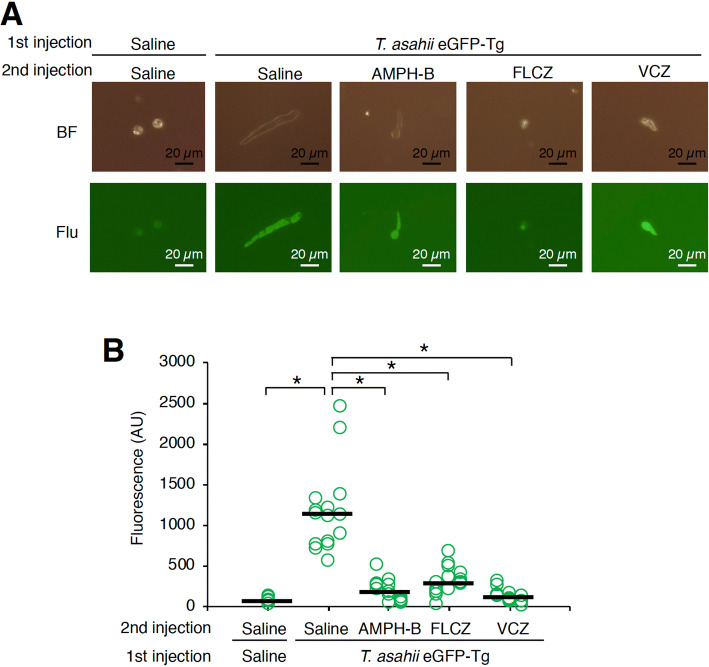
We examined whether T. asahii grew in silkworms by microscopic observation of the hemolymph of silkworms infected with T. asahii. Hyphal formation was observed in the silkworm hemolymph at 24 h after injection of the T. asahii eGFP-Tg strain (Fig. 6A). On the other hand, we observed no increase in the number of colony-forming units of T. asahii in the hemolymph (Fig. 6B). We quantified the hyphal formation of dermatophytes in silkworms by measuring fluorescence intensity using transgenic dermatophytes expressing eGFP43. The fluorescence intensity increased in hemolymph of silkworms at 24 h after injection of T. asahii eGFP-Tg strain (Fig. 6C). The increased fluorescence intensity was attenuated by administering the antifungal drugs amphotericin B, fluconazole, and voriconazole (Fig. 7A,B). We also confirmed that the pathogenicity of the T. asahii eGFP-Tg strain in the silkworm infection model was similar to that of the wild-type strain (Supplementary Figs. 3 and 4). These results suggest that fluorescence imaging using the transgenic T. asahii strain expressing eGFP is effective for quantitatively evaluating hyphal formation in the host.
Figure 6.
Growth of transgenic T. asahii expressing eGFP in silkworm hemolymph. Saline or T. asahii JCM2466 eGFP-Tg strain (1 × 106 cells) was injected into the silkworm hemolymph. Silkworm hemolymph was collected at 0, 6, and 24 h after injection and observed with a fluorescence microscope (A). The colony forming units (B) and fluorescence (C) were calculated. BF: bright field. Flu: fluorescent, in which the field of view was irradiated with excitation light for fluorescence detection. Scale bar, 20 µm. Statistically significant differences between groups were evaluated using Tukey–Kramer method. *P < 0.05.
Figure 7.
Antifungal drug-induced inhibition of growth of transgenic T. asahii expressing eGFP in silkworm hemolymph. (A) Saline or T. asahii JCM2466 eGFP-Tg strain (3 × 106 cells) was injected into the silkworm hemolymph, followed by injection of 50 µl of amphotericin B (AMPH-B, 100 µg/ml), fluconazole (FLCZ, 100 µg/ml), or voriconazole (VCZ, 100 µg/ml) into the hemolymph. Silkworm hemolymph was collected at 24 h after injection and observed with a fluorescence microscope (A). The fluorescence was calculated (B). Statistically significant differences between groups were evaluated using Tukey–Kramer method. *P < 0.05.
Discussion
We established a silkworm infection model with T. asahii and evaluated the therapeutic effects of antifungal drugs using the silkworm infection model. Moreover, we generated a transgenic T. asahii strain expressing eGFP by gene recombination using A. tumefaciens. The silkworm infection model and the T. asahii gene recombination technology will be useful for elucidating the infection mechanisms of T. asahii and developing antifungal drugs.
Silkworm systemic infection models caused by pathogenic fungi, such as C. albicans, C. tropicalis, C. glabrata, C. neoformans, Aspergillus fumigatus, and dermatophytes, are reported26,27,29,30,38,43. T. asahii infection models using mice with neutropenia and Galleria mellonella, an insect, have been reported4. In these infection models, the number of viable T. asahii cells in the host organs is measured by colony-forming units and therefore quantifying the actual cell proliferation of T. asahii may not be accurate. The fluorescence quantification method using the transgenic T. asahii expressing eGFP can be used to quantify cell proliferation by considering hyphal formation in the infection models using mice and Galleria mellonella.
We demonstrated that the death of silkworms caused by T. asahii was abolished by autoclaving the fungal cells, and that antifungal drugs inhibited the hyphal formation in silkworms and effectively treated silkworm infections. T. asahii also forms arthroconidia that are segmented conidia. The arthroconidia of T. asahii were observed in multinucleated giant cells present in the blood of a patient with Job’s syndrome49. To our knowledge, however, the molecular mechanisms involved in hyphal and arthroconidia formation in T. asahii are unknown. Genetic evidence from experiments using a mutant that cannot form hyphae or arthroconidia is needed to clarify the contribution of hyphal and arthroconidia formation to the pathogenicity of T. asahii. Silkworms are useful for identifying virulence genes of pathogenic microorganisms24. Novel pathogenesis-related genes of Staphylococcus aureus, Pseudomonas aeruginosa, Bacillus cereus, C. albicans, and C. glabrata have been identified by screening mutants with reduced pathogenicity in silkworms from genetic-disrupted mutant libraries19,29,30,50,51. We previously reported in generating gene-deficient strains of dermatophytes by gene recombination using A. tumefaciens46. In the present study, we revealed that gene recombination using A. tumefaciens is a useful transgenic technique in T. asahii. Future studies will establish gene-deficient strains and screening for virulence genes in T. asahii. On the other hand, in the experimental system established in this study, multiple experiments are required to determine LD50 value by the simple logistic regression model. Moreover, there are the difference in LD50 values between each experiment (Supplementary Fig. 1). Optimizing the experimental condition to enable more reproducible measurement of LD50 values is a future subject.
Silkworms are useful for evaluating the pharmacokinetics and toxicity of compounds31–35. In mammalian animals, organs such as the intestinal tract, liver, and kidney govern the pharmacokinetics of drugs. Recent studies revealed that silkworms have functionally similar organs that affect drug pharmacokinetics and toxicity32–35. Many in vitro and in vivo analyses revealed that the absorption of compounds from the silkworm intestinal tract is similar to that in mammals26,27,31,43.
The ED50 values of antibiotics in silkworms are similar to those in mice26. The difference in the total clearance, volume of distribution, and half-life values of anti-microbial agents such as chloramphenicol, tetracycline, vancomycin, rifampicin, micafungin, and fluconazole is less than 10-fold between silkworms and mammals35. The LD50 values for compounds are also similar between silkworms and mammals28,32. Lysocin E, nosokomycin, ASP2397, and GPI0363 were identified by exploratory research using a silkworm infection model from microbial culture broths and chemical libraries as novel antimicrobial compounds that show therapeutic effects in mouse infection experiments34,36–38. Furthermore, we demonstrated that substances identified by in vivo screening using a silkworm disease model are effective in humans52. Therefore, silkworms are useful for in vivo screening of compounds that are candidate antibacterial and antifungal agents.
The ED50 values of amphotericin B and voriconazole in the silkworm infection model with T. asahii were 1.3 mg/kg and 0.4 mg/kg, respectively. A previous study using a guinea pig infection model with T. asahii reported amphotericin B and voriconazole ED50 values greater than 1.5 mg/kg and 5–10 mg/kg, respectively53. Therefore, the ED50 value of voriconazole in the silkworm model is lower than that in the guinea pig model. In the previous report using the guinea pig model, antifungal drugs were administered 24 h after infection. On the other hand, in the silkworm infection model, antifungal drugs were administered immediately after infection. The difference in the timing of the administration may account for the difference in ED50 values. T. asahii exhibits natural resistance to candin antifungals such as micafungin, and strains resistant to azole antifungals such as fluconazole and amphotericin B have also been clinically isolated12–14. New antifungal drugs might be obtained by in vivo screening using a silkworm infection model with a multidrug-resistant T. asahii strain.
In conclusion, the silkworm T. asahii infection model is useful for quantitatively evaluating the pathogenicity of T. asahii and the therapeutic effects of antifungal drugs. We also revealed that T. asahii can be genetically modified using Agrobacterium. The transgenic T. asahii expressing eGFP might be useful for in vivo fluorescence imaging in various infection models.
Supplementary information
Acknowledgements
We thank Saki Azami, Haruka Shiga, Tae Nagamachi, Hikari Moriyama, Yuki Yamashita, and Asami Yoshikawa (Meiji Pharmaceutical University) for their technical assistance rearing the silkworms. This project was supported by JSPS KAKENHI Grant number JP17K08288 and JP20K07022 (Scientific Research (C) to Y.M.). This study was supported in part by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development, AMED (Grant to T.S.).
Author contributions
Y.M. conceived of the project, designed the experimental approach, performed experiments, analyzed the data, and prepared the draft manuscript. H.Y., Y.Y., and Y.T. performed experiments to determine the LD50 and ED50 values. T.Y. designed the binary plasmid for establishing the transgenic T. asahii strain expressing eGFP. T.S. supervised the project.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-67841-6.
References
- 1.Sugita T, Nishikawa A, Ichikawa T, Ikeda R, Shinoda T. Isolation of Trichosporon asahii from environmental materials. Med. Mycol. 2000;38:27–30. doi: 10.1080/mmy.38.1.27.30. [DOI] [PubMed] [Google Scholar]
- 2.Sugita T, et al. Genetic diversity and biochemical characteristics of Trichosporon asahii isolated from clinical specimens, houses of patients with summer-type-hypersensitivity pneumonitis, and environmental materials. J. Clin. Microbiol. 2001;39:2405–2411. doi: 10.1128/JCM.39.7.2405-2411.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo, A. L., Padovan, A. C. B. & Chaves, G. M. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin. Microbiol. Rev.24, 682–700 (2011). [DOI] [PMC free article] [PubMed]
- 4.Mariné M, et al. The development of animal infection models and antifungal efficacy assays against clinical isolates of Trichosporon asahii, T. asteroides, and T. inkin. Virulence. 2015;6:476–486. doi: 10.1080/21505594.2015.1020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang E, Sugita T, Tsuboi R, Yamazaki T, Makimura K. The opportunistic yeast pathogen Trichosporon asahii colonizes the skin of healthy individuals: analysis of 380 healthy individuals by age and gender using a nested polymerase chain reaction assay. Microbiol. Immunol. 2011;55:483–488. doi: 10.1111/j.1348-0421.2011.00341.x. [DOI] [PubMed] [Google Scholar]
- 6.Gouba N, Raoult D, Drancourt M. Eukaryote culturomics of the gut reveals new species. PLoS ONE. 2014;9:e106994. doi: 10.1371/journal.pone.0106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho O, Matsukura M, Sugita T. Molecular evidence that the opportunistic fungal pathogen Trichosporon asahii is part of the normal fungal microbiota of the human gut based on rRNA genotyping. Int. J. Infect. Dis. 2015;39:87–88. doi: 10.1016/j.ijid.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Duarte-Oliveira C, et al. The Cell Biology of the Trichosporon-Host Interaction. Front. Cell. Infect. Microbiol. 2017;7:118. doi: 10.3389/fcimb.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh TJ, et al. Experimental Trichosporon infection in persistently granulocytopenic rabbits: implications for pathogenesis, diagnosis, and treatment of an emerging opportunistic mycosis. J. Infect. Dis. 1992;166:121–133. doi: 10.1093/infdis/166.1.121. [DOI] [PubMed] [Google Scholar]
- 10.Walsh TJ, Melcher GP, Lee JW, Pizzo PA. Infections due to Trichosporon species: new concepts in mycology, pathogenesis, diagnosis and treatment. Curr. Top. Med. Mycol. 1993;5:79–113. [PubMed] [Google Scholar]
- 11.Krcmery V, et al. Hematogenous trichosporonosis in cancer patients: report of 12 cases including 5 during prophylaxis with itraconazol. Support Care Cancer. 1999;7:39–43. doi: 10.1007/s005200050221. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M, et al. Micafungin Breakthrough Fungemia in Patients with Hematological Disorders. Antimicrob. Agents Chemother. 2018;62:324. doi: 10.1128/AAC.02183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toriumi Y, Sugita T, Nakajima M, Matsushima T, Shinoda T. Antifungal pharmacodynamic characteristics of amphotericin B against Trichosporon asahii, using time-kill methodology. Microbiol. Immunol. 2002;46:89–93. doi: 10.1111/j.1348-0421.2002.tb02663.x. [DOI] [PubMed] [Google Scholar]
- 14.Iturrieta-González IA, Padovan ACB, Bizerra FC, Hahn RC, Colombo AL. Multiple species of Trichosporon produce biofilms highly resistant to triazoles and amphotericin B. PLoS ONE. 2014;9:e109553. doi: 10.1371/journal.pone.0109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naglik JR, König A, Hube B, Gaffen SL. Candida albicans-epithelial interactions and induction of mucosal innate immunity. Curr. Opin. Microbiol. 2017;40:104–112. doi: 10.1016/j.mib.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Bonaventura G, et al. Biofilm formation by the emerging fungal pathogen Trichosporon asahii: development, architecture, and antifungal resistance. Antimicrob. Agents Chemother. 2006;50:3269–3276. doi: 10.1128/AAC.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mada, P. K., Ayoade, F., Li, A. & Todd, J. Trichosporon asahii septic thrombophlebitis following lower extremity amputation in an immunocompetent host. BMJ Case Rep.2018, bcr–2017–221441 (2018). [DOI] [PMC free article] [PubMed]
- 18.Kaito C, Akimitsu N, Watanabe H, Sekimizu K. Silkworm larvae as an animal model of bacterial infection pathogenic to humans. Microb. Pathog. 2002;32:183–190. doi: 10.1006/mpat.2002.0494. [DOI] [PubMed] [Google Scholar]
- 19.Kaito C, et al. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 2005;56:934–944. doi: 10.1111/j.1365-2958.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki S, Matsumoto Y, Sekimizu K, Kaito C. Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol. Lett. 2012;326:116–124. doi: 10.1111/j.1574-6968.2011.02439.x. [DOI] [PubMed] [Google Scholar]
- 21.Miyashita A, et al. Lipopolysaccharide O-antigen of enterohemorrhagic Escherichia coli O157:H7 is required for killing both insects and mammals. FEMS Microbiol. Lett. 2012;333:59–68. doi: 10.1111/j.1574-6968.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 22.Castillo Y, Suzuki J, Watanabe K, Shimizu T, Watarai M. Effect of vitamin A on Listeria monocytogenes infection in a silkworm model. PLoS ONE. 2016;11:e0163747. doi: 10.1371/journal.pone.0163747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii M, Matsumoto Y, Sekimizu K. Usefulness of silkworm as a host animal for understanding pathogenicity of Cryptococcus neoformans. Drug Discov. Ther. 2016;10:9–13. doi: 10.5582/ddt.2016.01015. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Sekimizu K. Silkworm as an experimental animal to research for fungal infections. Microbiol. Immunol. 2019;63:41–50. doi: 10.1111/1348-0421.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto Y. Facilitating drug discovery in human disease models using insects. Biol. Pharm. Bull. 2020;43:216–220. doi: 10.1248/bpb.b19-00834. [DOI] [PubMed] [Google Scholar]
- 26.Hamamoto H, et al. Quantitative evaluation of the therapeutic effects of antibiotics using silkworms infected with human pathogenic microorganisms. Antimicrob. Agents Chemother. 2004;48:774–779. doi: 10.1128/AAC.48.3.774-779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto Y, et al. Quantitative evaluation of cryptococcal pathogenesis and antifungal drugs using a silkworm infection model with Cryptococcus neoformans. J. Appl. Microbiol. 2012;112:138–146. doi: 10.1111/j.1365-2672.2011.05186.x. [DOI] [PubMed] [Google Scholar]
- 28.Usui K, et al. Acute oral toxicity test of chemical compounds in silkworms. Drug Discov. Ther. 2016;10:57–61. doi: 10.5582/ddt.2016.01025. [DOI] [PubMed] [Google Scholar]
- 29.Hanaoka N, et al. Identification of the putative protein phosphatase gene PTC1 as a virulence-related gene using a silkworm model of Candida albicans infection. Eukaryot. Cell. 2008;7:1640–1648. doi: 10.1128/EC.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueno K, et al. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS ONE. 2011;6:e24759. doi: 10.1371/journal.pone.0024759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamamoto H, et al. Effects of molecular mass and hydrophobicity on transport rates through non-specific pathways of the silkworm larva midgut. Int. J. Antimicrob. Agents. 2005;26:38–42. doi: 10.1016/j.ijantimicag.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Hamamoto H, Tonoike A, Narushima K, Horie R, Sekimizu K. Silkworm as a model animal to evaluate drug candidate toxicity and metabolism. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009;149:334–339. doi: 10.1016/j.cbpc.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Asami Y, Horie R, Hamamoto H, Sekimizu K. Use of silkworms for identification of drug candidates having appropriate pharmacokinetics from plant sources. BMC Pharmacol. 2010;10:7–6. doi: 10.1186/1471-2210-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paudel A, et al. Pharmacokinetic parameters explain the therapeutic activity of antimicrobial agents in a silkworm infection model. Sci. Rep. 2018;8:1578–1588. doi: 10.1038/s41598-018-19867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamamoto H, Horie R, Sekimizu K. Pharmacokinetics of anti-infectious reagents in silkworms. Sci. Rep. 2019;9:9451–9458. doi: 10.1038/s41598-019-46013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamamoto H, et al. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat. Chem. Biol. 2015;11:127–133. doi: 10.1038/nchembio.1710. [DOI] [PubMed] [Google Scholar]
- 37.Uchida R, et al. Nosokomycins, new antibiotics discovered in an in vivo-mimic infection model using silkworm larvae. I: Fermentation, isolation and biological properties. J. Antibiot. 2010;63:151–155. doi: 10.1038/ja.2010.9. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura I, et al. Discovery of a new antifungal agent ASP2397 using a silkworm model of Aspergillus fumigatus infection. J. Antibiot. 2017;70:41–44. doi: 10.1038/ja.2016.106. [DOI] [PubMed] [Google Scholar]
- 39.Kaito C. Understanding of bacterial virulence using the silkworm infection model. Drug Discov. Ther. 2016;10:30–33. doi: 10.5582/ddt.2016.01020. [DOI] [PubMed] [Google Scholar]
- 40.Clemons KV, Stevens DA. Conventional or molecular measurement of Aspergillus load. Med. Mycol. 2009;47(Suppl 1):S132–S137. doi: 10.1080/13693780802213340. [DOI] [PubMed] [Google Scholar]
- 41.Zimmer M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem. Rev. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 42.Hoffman RM. Strategies for In Vivo Imaging Using Fluorescent Proteins. J. Cell. Biochem. 2017;118:2571–2580. doi: 10.1002/jcb.25677. [DOI] [PubMed] [Google Scholar]
- 43.Ishii M, Matsumoto Y, Yamada T, Abe S, Sekimizu K. An invertebrate infection model for evaluating anti-fungal agents against dermatophytosis. Sci. Rep. 2017;7:12289. doi: 10.1038/s41598-017-12523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alshahni MM, Makimura K, Yamada T, Takatori K, Sawada T. Nourseothricin acetyltransferase: a new dominant selectable marker for the dermatophyte Trichophyton mentagrophytes. Med. Mycol. 2010;48:665–668. doi: 10.3109/13693780903330555. [DOI] [PubMed] [Google Scholar]
- 45.Yamada T, Makimura K, Uchida K, Yamaguchi H. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes. Med. Mycol. 2005;43:533–544. doi: 10.1080/13693780500057619. [DOI] [PubMed] [Google Scholar]
- 46.Yamada T, et al. Agrobacterium tumefaciens-mediated transformation of the dermatophyte, Trichophyton mentagrophytes: an efficient tool for gene transfer. Med. Mycol. 2009;47:485–494. doi: 10.1080/13693780802322240. [DOI] [PubMed] [Google Scholar]
- 47.Iwata A, et al. Development of a tightly regulatable copper-mediated gene switch system in dermatophytes. Appl. Environ. Microbiol. 2012;78:5204–5211. doi: 10.1128/AEM.00464-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishii M, Matsumoto Y, Nakamura I, Sekimizu K. Silkworm fungal infection model for identification of virulence genes in pathogenic fungus and screening of novel antifungal drugs. Drug Discov. Ther. 2017;11:1–5. doi: 10.5582/ddt.2016.01080. [DOI] [PubMed] [Google Scholar]
- 49.Chakrabarti A, et al. Generalized lymphadenopathy caused by Trichosporon asahii in a patient with Job's syndrome. Med. Mycol. 2002;40:83–86. doi: 10.1080/mmy.40.1.83.86. [DOI] [PubMed] [Google Scholar]
- 50.Chieda Y, et al. Pathogenicity of gacA mutant of Pseudomonas aeruginosa PA01 in the silkworm. Bombyx mori. FEMS Microbiol. Lett. 2005;244:181–186. doi: 10.1016/j.femsle.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 51.Tran S-L, et al. Haemolysin II is a Bacillus cereus virulence factor that induces apoptosis of macrophages. Cell. Microbiol. 2011;13:92–108. doi: 10.1111/j.1462-5822.2010.01522.x. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto Y, Ishii M, Hasegawa S, Sekimizu K. Enterococcus faecalis YM0831 suppresses sucrose-induced hyperglycemia in a silkworm model and in humans. Commun. Biol. 2019;2:157. doi: 10.1038/s42003-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serena C, Gilgado F, Mariné M, Pastor FJ, Guarro J. Efficacy of voriconazole in a guinea pig model of invasive trichosporonosis. Antimicrob. Agents Chemother. 2006;50:2240–2243. doi: 10.1128/AAC.00045-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.