Abstract
Rapid whole genome sequencing (rWGS) of peripheral blood has been used to detect microbial DNA in acute infections. Cytomegalovirus (CMV) is a herpesvirus capable of causing severe disease in neonates and immunocompromised patients. We identified CMV in patients undergoing diagnostic rWGS by matching reads that did not align to the human reference genome to a database of microbial genomes. rWGS was conducted on peripheral blood obtained from ill pediatric patients (age 1 day to 18 years). Reads not aligning to the human genome were analyzed using an in-house pipeline to identify DNA consistent with CMV infection. Of 669 patients who received rWGS from July 2016 through July 2019, we identified 28 patients (4.2%) with reads that aligned to the CMV reference genome. Six of these patients had clinical findings consistent with symptomatic CMV infection. Positive results were highly correlated (R2 > 0.99, p < 0.001) to a CMV-qPCR assay conducted on DNA isolated from whole blood samples. In acutely ill children receiving rWGS for diagnosis of genetic disease, we propose analysis of patient genetic data to identify CMV, which could impact treatment of up to 4% of children in the intensive care unit.
Subject terms: Clinical genetics, Microbial genetics, Paediatrics, Genetics, Medical research, Diseases, Infectious diseases
Introduction
Rapid whole genome sequencing (rWGS) of peripheral blood can rapidly diagnose genetic diseases and impact clinical care within 24 h1,2. A recent study showed a diagnosis rate of 43% in critically ill patients with a resultant change in management in 33% of patients1. rWGS allows clinicians to make actionable diagnoses in critically ill patients who may have an underpinning genetic or metabolic disease and is increasingly becoming a part of standard care. The concept of applying rWGS to detect infectious pathogens has also been utilized3–6. While plasma polymerase chain reaction (PCR) is often used for detection of infectious organisms, we theorized that an advantage of sequencing whole blood is that organisms that reside intracellularly within white blood cells are included in rWGS reads. Cytomegalovirus (CMV) is human herpesvirus 5 (HHV-5) and capable of causing disease in critically ill patients7. It can be found both intracellularly and extracellularly in humans. CMV can cause hepatitis, marrow suppression (including thrombocytopenia), encephalitis, retinitis, and pneumonitis. In neonates, CMV may also manifest as growth restriction, microcephaly, neurologic defects (including sensorineural hearing loss), and intellectual disability. Prevalence of congenital CMV ranges from 0.2 to 2.0% of pregnancies. Although most infected infants will have only latent infection without any sequelae, up to 12%—representing approximately 9,600 children per year in the United States—will experience neurological sequelae related to congenital CMV infection8. Recent studies have demonstrated improved outcomes in neonates with congenitally acquired CMV infection if antiviral treatment is started within the first 30 days of life9. Identifying CMV infection in critically ill children who are undergoing rWGS has the potential to significantly influence their outcomes. We sought to retrospectively detect the presence of CMV in patients undergoing rWGS and to identify those in whom CMV may have contributed to the severity of their illness.
Methods
Peripheral blood was obtained from acutely ill pediatric patients (age 1 day to 18 years) after parental consent and patient assent, where indicated, were obtained. Informed consent was obtained from all subjects or, if subjects were under 18 years, from a parent and/or legal guardian, including permission for publication. Research was conducted according to the principles of the Declaration of Helsinki and as approved by the University of California San Diego Institutional Review Board. All experimental protocols were approved by the University of California San Diego Institutional Review Board. Genomic DNA was extracted, paired-end genomic libraries were prepared without PCR, and sequenced (2 × 100 bp) on Illumina (San Diego, CA, USA) HiSeq 2,500, HiSeq 4,000 or NovaSeq 6,000 instruments, as previously described, to a whole genome redundant depth of ~ 40-fold1. Reads were aligned to the human genome (hg19) as previously described1. Unaligned reads were analyzed using an in-house built pipeline. Reads were first extracted from the whole genome binary alignment map (BAM) and realigned to the human genome with Burrows-Wheeler Aligner-Maximal Exact Match (BWA-MEM) using less stringent parameters (− c 10,000) to allow repetitive reads derived from the human genome to be aligned10. Reads with low quality (> 1/8th of the read length with a phred-like score of 1) or low complexity (> 25% of the read has the same dinucleotide) were removed. The remaining reads were aligned to a database of viral and bacterial genomes retrieved from the Microbial Genome Database (https://mbgd.genome.ad.jp/, accessed May 2017) and National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/genome/viruses/) (accessed Apr 2017)11. Duplicate alignments were then marked and removed using Sambamba (https://github.com/biod/sambamba)12. Only reads of at least 100 consecutive nucleotides were counted for each microbial species in each patient, this value was chosen to be stricter than the suggested metagenomic sequencing guideline (75 bp) to ensure accuracy in a clinical setting13. Read counts were normalized based on total rWGS coverage. We abstracted electronic health record data of all patients who had rWGS performed at Rady Children’s Institute for Genomic Medicine (RCIGM) between July 2016 and July 2019. In patients for whom charts were available in the electronic medical record, a manual review was performed by a pediatric infectious disease fellow (NR) to determine if the patients had clinical features consistent with CMV infection and/or positive clinical plasma CMV PCR results (DiaSorin Molecular CMV Primer Pair and the LIAISON MDX Thermocycler, Cypress, CA, USA). In false negative patients who received rWGS testing and were positive for CMV by plasma CMV PCR, but were negative by realignment of unaligned rWGS reads, we performed CMV PCR on the sample used for rWGS. We also performed CMV PCR testing on the saved DNA specimens used for rWGS testing on the patients in whom two or more reads of CMV were detected. Each qPCR run was performed with a high titer and low titer external positive control (Exact Diagnostics, Fort Worth, TX) in addition to a negative control. We performed Sanger sequencing PCR reactions as earlier described using previously validated primers on these DNA specimens to exclude the presence of a PCR inhibitor1.
Ethics approval
Peripheral blood was obtained from ill pediatric patients (age 1 day to 18 years) after parental consent and patient assent or consent where possible according to University of California San Diego Institutional Review Board. Research was conducted according to the principles of the Declaration of Helsinki.
Consent for publication
All patients were consented. When possible patient assent was obtained as well.
Results
From July 2016 through July 2019, 669 patients received rWGS at RCIGM. Of these, 164 were found to have at least one read that did not align to the reference human genome and that completely aligned to a human herpesvirus (HHV) genome: one subject with reads aligning to HHV-4 (Epstein-Barr virus, NC_009334.1), 28 with reads aligning to HHV-5 (CMV, NC_006273.2), 69 with HHV-6 (NC_001664.2 or NC_000898.1), and 66 with HHV-7 (NC_001716.2). The 28 patients in whom we detected two or more reads of HHV-5 (CMV) represented 4.2% of the total population evaluated. Through chart review we identified clinical characteristics suggestive of symptomatic CMV infection in six of these 28 patients (21%) (See Table 1, case summaries are found in the Supplement).
Table 1.
Symptomatic patients with CMV identified by rWGS pipeline.
| Patient ID | CMV reads (#) | Age | Genetic diagnosis | Patient description | Standard care testing for CMV | TCP | Hep | AVT | Hearing test |
|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | 3 | 8 d | Dursun syndrome | Profound neutropenia and cerebral infarct | NT | Yes | No | No | Passed |
| Patient 2 | 1,654 | 6 m | None found | Kaposiform hemangio-endothelioma with pneumocystis pneumonia | CMV plasma PCR negative. CMV serology positive. CMV in lungs on histopathology | Yes | Yes | No | NT |
| Patient 3 | 2 | 9 m | None found | Transient pancytopenia, megaloblastic | NT | Yes | No | No | NT |
| Patient 4 | 2 | 4 d | ASLD | Liver failure | NT | Yes | Yes | No | Passed |
| Patient 5 | 634 | 2 m | Ebstein anomaly | Ebstein anomaly status post heart transplant | Positive plasma PCR (1,892 IU/mL) | Yes | Yes | Yes | Passed |
| Patient 6 | 8 | 13 y | GPA | GPA with disseminated mucormycosis infection | Positive qualitative plasma PCR and bronchoalveolar lavage | Yes | Yes | No | NT |
ASLD arginosuccinate lyase deficiency, D days, M months, Y years, GPA granulomatous polyangiitis, TCP thrombocytopenia, Hep. Hepatitis, AVT antiviral treatment, NT not tested.
Of these six patients, three had never been evaluated for CMV infection. Only one patient had been started on antiviral treatment. To assess for false-negative results associated with our detection methods, we performed a chart review of all children who received rWGS testing at RCIGM to identify patients found to have CMV infection by standard-of-care testing. Given the difficulty of interpreting serology, we limited our search to patients with positive CMV PCR tests14. We identified five patients who were CMV quantitative PCR (qPCR) positive but not identified by rWGS read re-alignment (Table 1 in Supplementary material). Recognizing that CMV testing and rWGS were often done at different times during the patient’s hospital course, we performed CMV qPCR testing on the same DNA sample (dedicated CMV qPCR) that had been used for rWGS. PCR testing detected CMV in only one of five patients tested. In that case, CMV levels were too low (< 227 IU/mL) to be quantifiable, indicating a very low viral load. We additionally performed a dedicated CMV qPCR on the saved DNA specimen used for rWGS for the 28 patients with two counts or more of reads consistent with CMV (Table 2).
Table 2.
Patients with 2 reads or more reads of CMV detected in rWGS. CMV PCR was performed on the same DNA sample used for rWGS testing.
| Unaligned reads from rWGS | PCR | Sample age (months) |
|---|---|---|
| 2 | Not detected | 24 |
| 2 | Not detected | 19 |
| 2 | Not detected | 23 |
| 2 | Not detected | 17 |
| 2 | Not detected | 18 |
| 2 | Not detected | 15 |
| 2 | Not detected | 15 |
| 2 | Not detected | 15 |
| 2 | Not detected | 12 |
| 2 | Not detected | 19 |
| 2 | Not detected | 14 |
| 2 | Not detected | 17 |
| 2 | Not detected | 33 |
| 2 | Detected, but < 227 IU/mL detection limit | 14 |
| 3 | Not detected | 25 |
| 5 | Detected, but < 227 IU/mL detection limit | 14 |
| 8 | Detected, but < 227 IU/mL detection limit | 20 |
| 9 | Detected, but < 227 IU/mL detection limit | 19 |
| 11 | Detected, but < 227 IU/mL detection limit | 16 |
| 12 | 795 IU/mL, 2.9 LOG IU/mL | 14 |
| 19 | Not detected | 17 |
| 31 | 132 IU/mL, 2.12 LOG/mL | 1 |
| 36 | 345 IU/mL, 2.54 LOG IU/mL | 14 |
| 45 | 2,417 IU/mL, 3.38 LOG IU/mL | 27 |
| 69 | 1513 IU/mL, 3.18 LOG IU/mL | 23 |
| 85 | 1,071 IU/mL, 3.03 LOG IU/mL | 15 |
| 634 | 14,581 IU/mL, 4.16 LOG IU/mL | 20 |
| 1654 | 45,532 IU/mL, 4.66 LOG IU/mL | 29 |
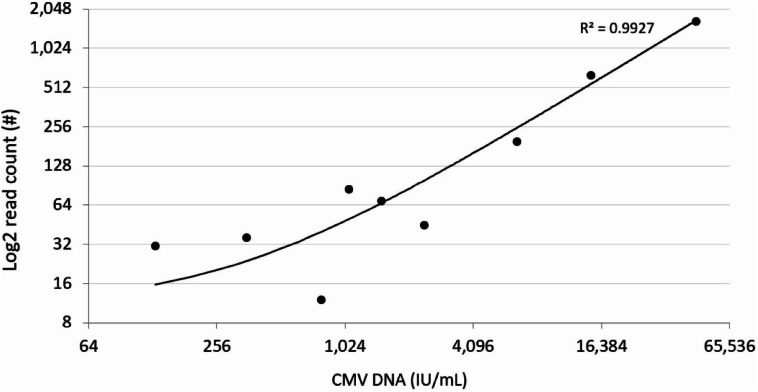
To assess for the presence of a PCR inhibitor, we used Sanger PCR reaction on all 28 samples tested. All of the samples underwent testing with two individual validated oligo sets. Each oligo set included one internal amplification control and one negative control. In 13 of the 14 patients with only two unaligned reads consistent with CMV, PCR reaction did not detect CMV. In a patient with three reads of CMV in WGS data and another with 19 reads in the rWGS data, CMV was not detected by PCR. Above a read count of 19, CMV was consistently detected by PCR reaction and quantifiable. The infectious unit count detected by PCR was highly correlated with the read count from rWGS (R2 = 0.9942; p < 0.0001; see Fig. 1.).
Figure 1.
Log2 CMV read count from rWGS versus viral load from CMV-quantitative PCR assay for 8 samples where rWGS detected CMV DNA and quantitative PCR could quantify viral load. rWGS read results are highly correlated to infectious unit determination by gold-standard quantitative PCR assay.
Discussion
Of 669 subjects who underwent rWGS at RCIGM from July 2016 through July 2019, we identified 28 patients with two or more DNA sequence reads consistent with CMV DNA. We also identified 164 patient genomes with unaligned reads consistent with DNA from any herpesvirus. Chart review identified clinical findings such as hepatitis, seizures, and thrombocytopenia that could be consistent with symptomatic CMV infection in 6 of the 28 in whom the presence of CMV was detected (see Table 1). Moreover, in these 6 case histories, even if the primary diagnosis may overlap with symptomology of CMV infection, CMV may have still have contributed to the symptoms experienced by these patients (and possibly even been the major cause of illness in case number 6). Of these, three patients had not previously been evaluated for CMV, and only one patient had been started on antiviral therapy. Two of the patients (cases 1 and 4) were neonates with possible congenital CMV infection and would have met inclusion criteria for treatment based on Kimberlin et al.9 In cases 2, 3, 5, and 6, though it is not possible to distinguish congenitally versus postnatally acquired infection, all of these patients were relatively immunosuppressed as a result of their primary diagnosis or from their treatment regimen.
In 5 cases with a positive clinical qPCR, we did not identify a CMV infection by WGS. In 4 of these cases, a confirmatory qPCR conducted on the plasma obtained for WGS was negative, likely due to a waning viremia. In the remaining case, CMV was detected, but below the detection limit for quantitation. In one case, a substantial HHV7 viremia was detected, and it may be possible that the initial CMV positive was due to cross reactivity with one of the qPCR primers for HHV7. Although qPCR can be highly specific, it is generally tested in the background of subjects with no viremia rather than a different HHV infection15. Future studies will need to be conducted to test if WGS can also be more sensitive than qPCR in some cases.
The clinical spectrum of CMV is highly varied and overlaps with many other disease processes. The ability of clinicians to identify active CMV viremia clinically can therefore be challenging16. In neonates in particular, even patients with mild disease initially may have significant morbidity, including sensorineural hearing loss17. A land mark study by Kimberlin and colleagues in 2015 demonstrated that morbidity from symptomatic congenital CMV infection could be reduced or averted by early initiation of antiviral therapy9. Recent studies have explored the utility of hearing screening to identify congenital CMV, though with a sensitivity of just 53%, this strategy will miss many affected patients8,18,19. Current diagnostic approaches are inadequate to identify CMV infection in vulnerable patients in a timely manner.
The use of unaligned reads from rWGS testing offers a cost-effective approach to identifying infectious pathogens while simultaneously testing for pathogenic variants in the host genome20. Given that most patients undergoing rWGS are critically ill, it is likely that many of these patients have an infectious process superimposed on an underlying medical condition that led to a severe disease state7,21,22. CMV, which is associated with significant morbidity in neonates and immunodeficient hosts, is a pathogen that may be implicated in many such cases. Analyzing rWGS for microbial DNA enables the testing laboratory to alert clinicians in a timely manner to the presence of this pathogen while simultaneously evaluating the host genome for other pathogenic conditions. The results of this assay are highly correlated to orthogonal testing with CMV-specific qPCR. Moreover, the additional cost of testing to confirm CMV infection following rWGS is low. Whole genome data may also play a role in understanding the natural history of congenital CMV beyond the identification of apparently symptomatic children. This is especially relevant given the calls for population-based whole genome sequencing of neonates and for population-level screening for CMV, which will identify many more children infected with CMV whose clinical course is uncertain16. Additional research using rWGS may help clarify which CMV positive neonates would benefit from potentially toxic antiviral therapy16.
This study has several limitations. We conducted our analysis retrospectively, and the patients we identified were therefore not systematically assessed for CMV infection by the treating team during their hospital stay. Furthermore, even in patients who were evaluated for CMV, the rWGS sample was often not obtained at the same time as the evaluation for CMV, which is significant given that CMV viremia may wane over time or resolve altogether23. Our chart review was also limited by the wide clinical variation in what may be attributable to CMV infection (hepatitis, cytopenia, etc.), and in most of our patients, their underlying metabolic or genetic disease process was also associated with many of the classic laboratory findings associated with CMV infection. PCR testing on the 28 samples from patients in whom two or more reads of CMV were detected by rWGS was done on DNA extracted from whole blood samples rather than on plasma as is traditionally done when CMV viremia is suspected. However, the common practice of obtaining plasma CMV PCR in the clinical setting is not necessarily due to a limitation in detection of CMV from whole blood24,25. Additionally, given the negative qPCR results for the majority of patients with only 2 reads of CMV and the positive correlation between read count and higher PCR quantitation, there is the intriguing possibility that rWGS may be more sensitive than qPCR for CMV, an observation that has been made for other organsims26. Further studies are required to prospectively compare and correlate CMV, and potentially other microbe, read count from rWGS with standard care testing, define the specificity and sensitivity of this approach, and ultimately to demonstrate clinical utility.
rWGS may be a rapid, extremely sensitive method to detect some microbial infections. We propose that, in critically ill children under going rWGS, unaligned reads be used to assess the presence of microbial DNA and that this analysis should be conducted regardless of prior clinical diagnosis. This approach has the potential to identify clinically relevant pathogens such as CMV, which may cause significant disease in certain vulnerable patient populations. Moreover, for CMV infections in particular, early initiation of antivirals may positively modify patient outcomes.
Supplementary information
Acknowledgments
Medical editing services were provided by Kevin Lomangino and Karen Doyle of KWF Consulting, Baltimore, MD, USA. This study was supported in part by grant U19HD077693 from NICHD and NHGRI to S.F.K., and gifts from the Liguori Family, John Motter and Effie Simanikas, Ernest and Evelyn Rady, and Rady Children’s Hospital San Diego. All authors had full access to all data, and MB had final responsibility for the decision to submit for publication.
Author contributions
Authors M.B., L.F., D.D., C.H., S.K., and N.R. conceived the study design. N.R. performed the chart review and drafted the initial manuscript. Author Y.D. performed sample processing. Authors M.B., L.F., D.D., S.K., C.H., and Y.D. reviewed and contributed to the manuscript.
Competing interests
All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: no financial relationships with any organizations that might have an interest in the submitted work in the previous three years and no other relationships or activities that could appear to have influenced the submitted work.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-67656-5.
References
- 1.Farnaes L, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genomic Med. 2018;3:10. doi: 10.1038/s41525-018-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark MM, et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med. 2019;11:eaat6177. doi: 10.1126/scitranslmed.aat6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blauwkamp TA, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019;4:663. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 4.Schlaberg R, et al. Viral pathogen detection by metagenomics and Pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J. Infect. Dis. 2017;215:1407–1415. doi: 10.1093/infdis/jix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thoendel MJ, et al. Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin. Infect. Dis. 2018;67:1333–1338. doi: 10.1093/cid/ciy303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leggett RM, et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat. Microbiol. 2019;5:1–13. doi: 10.1038/s41564-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davila S, et al. Viral DNAemia and immune suppression in pediatric sepsis. Pediatr. Crit. Care Med J. Soc. Crit. Care Med. World Fed. Pediatr. Intensive Crit. Care Soc. 2018;19:e14–e22. doi: 10.1097/PCC.0000000000001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig A, Hengel H. Epidemiological impact and disease burden of congenital cytomegalovirus infection in Europe. Eurosurveillance. 2009;14:19140. [PubMed] [Google Scholar]
- 9.Kimberlin DW, et al. Valganciclovir for symptomatic congenital cytomegalovirus disease. N. Engl. J. Med. 2015;372:933–943. doi: 10.1056/NEJMoa1404599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Q-Bio, ArXiv arXiv:1303.3997 (2013).
- 11.Uchiyama I. MBGD: microbial genome database for comparative analysis. Nucleic Acids Res. 2003;31:58–62. doi: 10.1093/nar/gkg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boppana SB, et al. Saliva polymerase-chain-reaction assay for cytomegalovirus screening in newborns. N. Engl. J. Med. 2011;364:2111–2118. doi: 10.1056/NEJMoa1006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross SA, Novak Z, Pati S, Boppana SB. Diagnosis of cytomegalovirus infections. Infect. Disord. Drug Targets. 2011;11:466–474. doi: 10.2174/187152611797636703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawlinson WD, et al. Congenital cytomegalovirus infection in pregnancy and the neonate: consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect. Dis. 2017;17:e177–e188. doi: 10.1016/S1473-3099(17)30143-3. [DOI] [PubMed] [Google Scholar]
- 17.Lanzieri TM, et al. Hearing loss in children with asymptomatic congenital cytomegalovirus infection. Pediatrics. 2017;139:e20162610. doi: 10.1542/peds.2016-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fowler KB, et al. A targeted approach for congenital cytomegalovirus screening within newborn hearing screening. Pediatrics. 2017;139:e20162128. doi: 10.1542/peds.2016-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park AH, et al. A diagnostic paradigm including cytomegalovirus testing for idiopathic pediatric sensorineural hearing loss. The Laryngoscope. 2014;124:2624–2629. doi: 10.1002/lary.24752. [DOI] [PubMed] [Google Scholar]
- 20.Sanford E, et al. Concomitant diagnosis of immune deficiency and Pseudomonas sepsis in a 19 month old with ecthyma gangrenosum by host whole-genome sequencing. Mol. Case Stud. 2018;4:a003244. doi: 10.1101/mcs.a003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnaes L, Coufal NG, Spector SA. Vaccine strain varicella infection in an infant with previously undiagnosed perinatal human immunodeficiency type-1 infection. Pediatr. Infect. Dis. J. 2019;38:413–415. doi: 10.1097/INF.0000000000002183. [DOI] [PubMed] [Google Scholar]
- 22.Walton AH, et al. Reactivation of multiple viruses in patients with sepsis. PLoS ONE. 2014;9:e98819. doi: 10.1371/journal.pone.0098819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng AP, et al. Cytomegalovirus DNAemia and disease: incidence, natural history and management in settings other than allogeneic stem cell transplantation. Haematologica. 2005;90:1672–1679. [PubMed] [Google Scholar]
- 24.Lisboa LF, et al. The clinical utility of whole blood versus plasma cytomegalovirus viral load assays for monitoring therapeutic response. Transplantation. 2011;91:231–236. doi: 10.1097/TP.0b013e3181ff8719. [DOI] [PubMed] [Google Scholar]
- 25.Chen XY, Hou PF, Bi J, Ying CM. Detection of human cytomegalovirus DNA in various blood components after liver transplantation. Braz. J. Med. Biol. Res. 2014;47:340–344. doi: 10.1590/1414-431X20133353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen SC, Fachmann MSR, Kiil K, Møller Nielsen E, Hoorfar J. Gene-based pathogen detection: can we use qPCR to predict the outcome of diagnostic metagenomics? Genes. 2017;8:332. doi: 10.3390/genes8110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.