Abstract
Isolated hypogonadotropic hypogonadism (IHH), combined pituitary hormone deficiency (CPHD), and septo-optic dysplasia (SOD) constitute a disease spectrum whose etiology remains largely unknown. This study aimed to clarify whether mutations in SMCHD1, an epigenetic regulator gene, might underlie this disease spectrum. SMCHD1 is a causative gene for Bosma arhinia microphthalmia syndrome characterized by arhinia, microphthalmia and IHH. We performed mutation screening of SMCHD1 in patients with etiology-unknown IHH (n = 31) or CPHD (n = 43, 19 of whom also satisfied the SOD diagnostic criteria). Rare variants were subjected to in silico analyses and classified according to the American College of Medical Genetics and Genomics guidelines. Consequently, a rare likely pathogenic variant, p.Asp398Asn, was identified in one patient. The patient with p.Asp398Asn exhibited CPHD, optic nerve hypoplasia, and a thin retinal nerve fiber layer, and therefore satisfied the criteria of SOD. This patient showed a relatively low DNA methylation level of the 52 SMCHD1-target CpG sites at the D4Z4 locus. Exome sequencing for the patient excluded additional variants in other IHH/CPHD-causative genes. In vitro assays suggested functional impairment of the p.Asp398Asn variant. These results provide the first indication that SMCHD1 mutations represent a rare genetic cause of the HH-related disease spectrum.
Subject terms: Genetics, Diseases, Endocrinology, Medical research, Molecular medicine
Introduction
Hypogonadotropic hypogonadism (HH) is a multifactorial disorder that occurs either as an isolated hormonal abnormality (isolated HH, IHH) or in combination with other pituitary hormone deficiencies (combined pituitary hormone deficiency, CPHD)1,2. HH is frequently accompanied by craniofacial and neurological abnormalities, such as microphthalmia, anosmia, and septum pellucidum/corpus callosum hypoplasia2–6. The current understanding is that IHH and CPHD belong to a disease spectrum, which includes septo-optic dysplasia (SOD) and holoprosencephaly at the most severe end and IHH at the mildest end1,7. This disease spectrum results from defective organogenesis from the cranial placodes1,8. To date, more than 40 genes have been implicated in this disease spectrum1,9. However, mutations in these genes account for only about 50% of IHH cases and less than 20% of CPHD cases, indicating that other causative genes remain to be identified1,9.
SMCHD1 encodes an epigenetic regulator that controls DNA methylation of multiple genomic loci10–12. Previous studies have shown that SMCHD1 is involved in the regulation of several monoallelically expressed genes11,13, and is particularly enriched in the nuclear territory of the inactive X chromosome (Xi)14, 15. Yet, the precise function of this protein remains to be clarified. Recently, heterozygous SMCHD1 mutations were identified in patients with Bosma arhinia microphthalmia syndrome (BAMS), an extremely rare syndrome whose triad is the absence of the nose, microphthalmia, and IHH16–18. In addition, SMCHD1 mutations are known to cause facioscapulohumeral muscular dystrophy type 2 (FSHD2), particularly when the mutations co-occurred with disease-susceptible alleles at the D4Z4 locus19,20. BAMS/FSHD2 patients with SMCHD1 mutations frequently exhibit DNA hypomethylation in the DUX4 promoter region at the D4Z4 locus, possibly reflecting impaired regulatory activity of the mutant SMCHD1 proteins16,19. Previous studies revealed that BAMS-causative SMCHD1 mutations consist solely of missense substitutions within or very close to the GHKL ATPase domain, while FSHD2-causative mutations include several missense, nonsense, and frameshift variants widely distributed in the 48 coding exons16–18. It has been proposed that gain-of-function and loss-of-function mutations in SMCHD1 result in BAMS and FSHD2, respectively, although four mutations have been associated with both conditions16,18,21, 22. More than 40 sporadic and familial cases with BAMS due to SMCHD1 mutations have been reported to date16–18. Clinical analysis of the familial cases suggested variable expressivity of the BAMS triad16. However, given that previous molecular analyses of SMCHD1 primarily focused on individuals with facial anomalies or muscular dystrophy, further studies are needed to clarify the phenotypic variations of SMCHD1 abnormalities. In particular, it remains to be elucidated whether SMCHD1 mutations can underlie IHH or CPHD cases without nasal malformations. To address this issue, we performed mutation screening of SMCHD1 in 74 patients with HH-related disorders.
Results
Mutation screening of SMCHD1 for 74 patients with HH-related disorders
The study was approved by the Institutional Review Board Committee at the National Center for Child and Development and performed after obtaining informed consent from the participants or their parents. We performed mutation screening of SMCHD1 in 74 patients, i.e., 31 patients clinically diagnosed with IHH and 43 patients with CPHD. Nineteen of the CPHD patients also satisfied the diagnostic criteria of SOD23. All 74 patients had normal nasal structures and harbored no pathogenic variants in any of the known causative genes of IHH or CPHD.
All 48 coding exons of SMCHD1 and their flanking intronic regions were amplified and subjected to next generation sequencing. The functional consequence of each variant was assessed by in silico analyses. The variants were classified according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines for interpretation of sequence variants24.
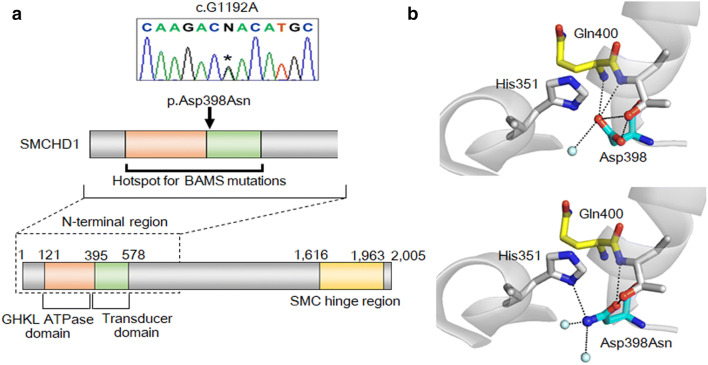
Consequently, two rare heterozygous variants, namely, c.G1192A (p.Asp398Asn) and c.G54C (p.Glu18Asp), were identified in patients 1 and 2, respectively (Table 1). Patients 1 and 2 were from the CPHD and IHH groups, respectively. Of the two variants, p.Asp398Asn in patient 1 was scored as deleterious by all of the five in silico programs used in this study. This variant has not been submitted to the ClinVar database. The variant resides in exon 10 within the hotspot for BAMS-causative mutations18, and affects a conserved amino acid adjacent to the ATPase domain (Fig. 1a). In silico protein structural analysis suggested that this substitution creates a new hydrogen bond between the 398th and 351st residues and disrupts a hydrogen bond between the 398th and 400th residues (Fig. 1b). The p.Asp398Asn variant was ranked as likely pathogenic according to the ACMG/AMP guidelines (Table 1). The variant was confirmed by Sanger sequencing (Fig. 1a). Although we attempted to obtain DNA samples from the patient’s parents, such samples were unavailable.
Table 1.
Characteristics of two rare SMCHD1 variants.
| Patient | Pheno-type | Variant | ClinVara | dbSNPb | Allele frequency | In silico analysis | ACMG/AMP guideline | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cDNA | Protein | 1000Gc | gnomADd | HGVDe | PolyPhen-2f | SIFTg | CADDh | M-CAPi | Mutation-Tasterj | |||||
| 1 | CPHD | c.G1192A | p.Asp398Asn | No data | No data | No data | No data | No data | 1.000 | 0.000 | 26.2 | 0.163 | Disease causing | Likely pathogenic |
| 2 | IHH | c.G54C | p.Glu18Asp | Likely benign | rs779165187 | No data | 20/7,642 | 5/2,402 | 0.004 | 0.246 | 9.9 | 0.087 | Polymorphism | Likely benign |
ACMG/AMP the American College of Medical Genetics and Genomics and the Association for Molecular Pathology, CPHD combined pituitary hormone deficiency, IHH Isolated hypogonadotropic hypogonadism. Scores indicative of a damaging variant are boldfaced.
aClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
bdbSNP (https://www.ncbi.nlm.nih.gov/snp/).
cThe 1000 Genome Database (https://www.internationalgenome.org/1000-genomes-browsers/).
dThe gnomAD browser (https://gnomad.broadinstitute.org/).
eThe Human Genetic Variation Database (http://www.hgvd.genome.med.kyoto-u.ac.jp/).
fPolymorphism Phenotyping v2 (http://genetics.bwh.harvard.edu/pph2/).
gSorting Intolerant From Tolerant (http://sift.jcvi.org/).
hCombined Annotation Dependent Depletion (https://cadd.gs.washington.edu/).
iMendelian Clinically Applicable Pathogenicity (http://bejerano.stanford.edu/mcap/).
jMutationTaster (http://www.mutationtaster.org/).
Figure 1.
The p.Asp398Asn variant of SMCHD1. (a) The genomic position of the c.G1192A variant. The asterisk in the chromatogram denotes the mutated nucleotide. This variant resided in exon 10, within the hotspot of Bosma arhinia microphthalmia syndrome (BAMS) mutations18. The orange, green, and yellow indicate the GHKL ATPase domain, the transducer domain, and SMC hinge region, respectively. (b) Protein structure prediction of the p.Asp398Asn variant. This variant was predicted to alter multiple hydrogen bonds.
The p.Glu18Asp variant in patient 2 was found in dbSNP as rs779165187 (Table 1). Reportedly, its allele frequency in the general population was 0.21–0.26%. This variant was assessed as benign/tolerate by most of the five in silico programs, and has been submitted to the ClinVar database as a likely benign variant. Furthermore, the variant resided in exon 1, outside of the hotspot for BAMS-causative mutations18. Altogether, the p.Glu18Asp variant was classified as likely benign according to the ACMG/AMP guidelines. Thus, we excluded this variant from further analyses.
Whole exome sequencing of patient 1
To exclude the possibility that CPHD of patient 1 was caused by a pathogenic variant in other genes, we performed exome sequencing for this individual. Consequently, we identified 64 heterozygous rare variants in 63 protein-coding genes, which were assessed as deleterious (Supplementary Table 1). However, none of the 63 genes, except for SMCHD1, was associated with IHH or CPHD.
Clinical analysis of patient 1
We examined the clinical features of patient 1 with the p.Asp398Asn variant. This patient was a Japanese girl born to non-consanguineous parents at 37 weeks of gestation. Her birth weight was 2,216 g (− 1.3 SD). No dysmorphic features were noted. She had episodes of oxygen desaturation from four hours after birth, and was diagnosed with combined deficiencies of adrenocorticotropic hormone (ACTH), thyroid stimulating hormone (TSH), and growth hormone (GH). She received hydrocortisone and levothyroxine supplementation from the neonatal period, and GH treatment from eight months of age. She showed mild mental retardation from early childhood and underwent a special education program. She lacked spontaneous pubertal development in her teens and was diagnosed with HH. From 13 years of age, she received estrogen replacement therapy.
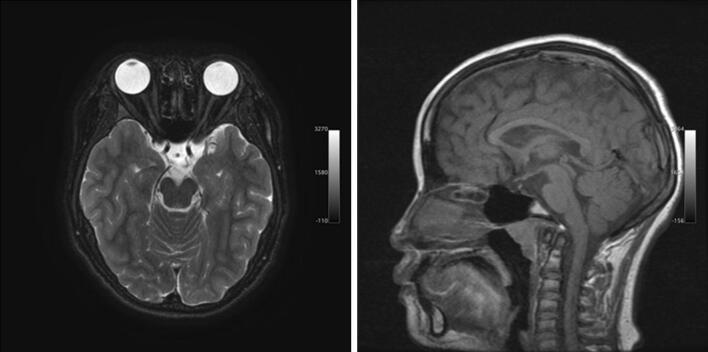
At 17 years of age, she measured 148 cm (− 1.9 SD) and weighted 60.2 kg (+ 1.0 SD). She retained normal visual activity with myopia. However, optical coherence tomography revealed decreased thickness of the retinal nerve fiber layer. Brain magnetic resonance imaging delineated ectopic posterior pituitary and optic nerve hypoplasia, together with normal eye and nose structure (Fig. 2). Endocrine evaluation demonstrated the lack of responses of ACTH (basal, < 0.2 pmol/L; stimulated, < 0.2 pmol/L), GH (basal, < 1.0 µg/L; stimulated, < 1.0 µg/L), and cortisol (basal, 8.3 nmol/L; stimulated, > 5.5 nmol/L) to the growth hormone releasing peptide 2 stimulation25. Thus, this patient satisfied the diagnostic criteria of SOD23. Physical evaluation detected no clinical features indicative of FSHD226,27, except for mild fatigability, mild mental retardation, and slight instability in walking on tiptoes and heels (Supplementary Table 2).
Figure 2.
Brain magnetic resonance imaging of patient 1. Patient 1 exhibited ectopic posterior pituitary and optic nerve hypoplasia. The anterior pituitary lobe was not visible. The structures of the eyes and nose were apparently normal.
The 55-year-old father, 51-year-old mother, and 23-year-old brother of patient 1 were phenotypically normal. Allegedly, the family had no history of congenital endocrine disorders, craniofacial anomalies, or muscular dystrophy.
DNA methylation analysis
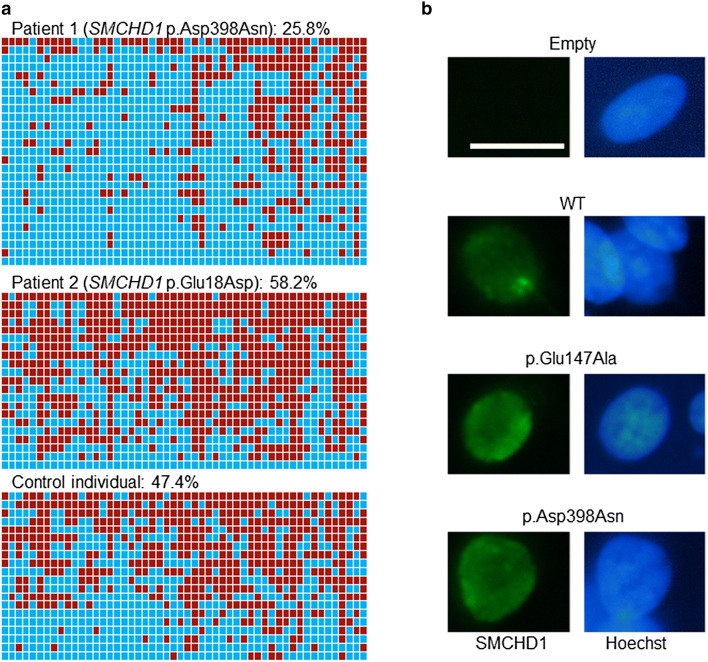
We analyzed the DNA methylation status of patient 1 by bisulfite sequencing. Samples from patient 2 and a control individual without SMCHD1 variants were also analyzed. The DUX4 promoter region containing 52 SMCHD1-target CpG sites was PCR-amplified and subcloned16,19. We found that the average methylation rate of patient 1 was 25.8% (range of 27 clones, 0–80.8%) (Fig. 3a), which was markedly lower than previously reported data of control individuals (56.5%)16, and close to the cutoff value of the diagnostic criteria of FSHD2 (25%)19. Average methylation rates of patient 2 and the control individual were 58.2% and 47.4% respectively (Fig. 3a), which were almost comparable to the reference data of previous studies16.
Figure 3.
Functional analyses of the p.Asp398Asn variant. (a) The results of DNA methylation analysis. DNA methylation statuses of 52 CpG sites in the DUX4 promoter region are shown16. The red and blue boxes depict methylated and unmethylated CpG sites, respectively. The average methylation rate of patient 1 was 25.8%, which is close to the cut off value of the diagnostic criteria of fascioscapulohumeral muscular dystrophy type 2 (25%)19. The average methylation rates of patient 2 and a control individual without SMCHD1 variants were 58.2% and 47.4%, respectively. (b) Representative results of in vitro functional assays. HEK293 cells transfected with the expression vectors of SMCHD1 or an empty vector were stained with anti-FLAG M2 for SMCHD1 (green) and Hoechst 33342 for DNA (blue). WT, full-length wildtype SMCHD1; p.Glu147Ala, an artificially created variant lacking the GHKL ATPase activity; p.Asp398Asn, a variant identified in patient 1. Scale bar, 20 μm.
Furthermore, we attempted to perform Southern blot analysis to examine the number of D4Z4 repeats28,29. However, the quality and amount of the patient’s DNA sample were not sufficient for Southern blotting. Thus, it remains unknown whether patient 1 retains a normal number of D4Z4 repeats.
In vitro functional assays for the p.Asp398Asn variant
To analyze the effect of the p.Asp398Asn variant on protein function, we performed in vitro assays. It is known that wildtype (WT) SMCHD1 accumulates to the territory of the Xi, whereas an artificially created variant lacking GHKL ATPase activity (p.Glu147Ala) shows a diffuse distribution in the nucleus14,15. Thus, we transfected HEK293 cells with an expression vector for the p.Asp398Asn variant, together with expression vectors for the WT SMCHD1 and the p.Glu147Ala variant. As reported previously14,15, WT SMCHD1 was observed as nuclear foci (Fig. 3b). In contrast, both the p.Asp398Asn and p.Glu147Ala variants were diffusely distributed in the nucleus (Fig. 3b).
Discussion
Mutation screening of SMCHD1 in 74 patients with etiology-unknown IHH/CPHD identified two rare variants each in one patient. According to the ACMG/AMP guidelines, the p.Asp398Asn and p.Glu18Asp variants were classified into the likely pathogenic and likely benign groups, respectively. Several findings of this study support the pathogenicity of the p.Asp398Asn variant. First, this variant resides within the hotspot for BAMS mutations in an evolutionally constrained genomic region18, and has not been identified in the general population. Second, this variant was predicted to be damaging by all in silico programs used in this study. In particular, the high Combined Annotation Dependent Depletion (CADD) score of 26.2 is remarkable, because Shaw et al. have shown that SMCHD1 variants with CADD scores of > 25.0 frequently result in BAMS16. Third, protein structural prediction suggested that this substitution alters hydrogen bonds. Specifically, it likely disrupts a hydrogen bond between the 398th and 400th residues, the latter being a nucleotide whose mutation results in BAMS16. Fourth, whole exome sequencing of patient 1 excluded other pathogenic variants in known IHH/CPHD-causative genes. Fifth, the relatively low DNA methylation rate of the SMCHD1-target CpG sites in patient 1 possibly reflects compromised activity of the mutant SMCHD1 protein as an epigenetic regulator16,19. Although the methylation rate was slightly above the cutoff value for the diagnostic criteria of FSHD219, this does not argue against the pathogenicity of the p.Asp398Asn variant, because a certain percentage of previously reported BAMS patients with pathogenic SMCHD1 mutations showed methylation rates higher than 30%16. Lastly, in vitro functional assays suggested functional impairment of the mutant SMCHD1. The p.Asp398Asn variant failed to accumulate in the Xi territory. The diffuse distribution pattern of the p.Asp398Asn variant was similar to that of an artificially created variant lacking GHKL ATPase activity14.
The phenotype of patient 1 with the p.Asp398Asn variant has both similarities and differences with that of previously reported BAMS patients. Patient 1 exhibited not only HH but also combined deficiencies of ACTH, TSH, and GH. On the other hand, she lacked ocular or nasal malformations, except for optic nerve hypoplasia and a thin retinal nerve fiber layer. These results indicate that the clinical consequences of SMCHD1 mutations are broader than currently recognized. Actually, there seems to be a considerable overlap in the phenotype between the SMCHD1 mutation-mediated syndrome and the HH-related disease spectrum, because septum pellucidum/corpus callosum hypoplasia, hearing loss, and cleft palate, HH, and ocular anomalies have been documented in both conditions2–6. However, considering that likely pathogenic SMCHD1 variants were absent from 73 of 74 patients in our cohort, such variants appear to play only a minor role in the etiology of HH and its related disorders.
Previous studies have suggested that IHH in BAMS patients results from defective nasal placode formation and/or impaired projection of the gonadotropin releasing hormone neuron16, 17. However, such mechanism cannot account for the development of CPHD. Since SMCHD1 is known to regulate monoallelically expressed genes11,13, CPHD in our patient may reflect aberrant expression of some SMCHD1 target genes in the brain. Indeed, some genes involved in hypothalamus/pituitary development, such as POU1F1 and MKRN3, were reported to show monoallelic expression30,31. Further studies are necessary to clarify the role of SMCHD1 in craniofacial and brain development.
Patient 1 exhibited no apparent clinical features of FSHD2. These results are consistent with previous findings that BAMS patients with SMCHD1 mutations rarely manifest FSHD216. The absence of muscle weakness in our patient and most BAMS patients may reflect the complex oligogenic or multifactorial nature of FSHD221. Previous studies have suggested that SMCHD1 mutations alone would not be sufficient to cause FSHD216,21. Moreover, we cannot exclude the possibility that our patient may develop FSHD2 in later life29.
In summary, we identified two rare heterozygous SMCHD1 variants in two of 74 patients with IHH and CPHD. Of the two variants, p.Asp398Asn was predicted to be a likely pathogenic variant. In vitro assay suggested functional impairment of the p.Asp398Asn protein. The results of this study provide the first indication that SMCHD1 variants represent a rare genetic cause of the HH-related disease spectrum.
Materials and methods
Patients
The study group consisted of 31 patients clinically diagnosed with IHH and 43 patients with CPHD. Nineteen of the CPHD patients satisfied the diagnostic criteria of SOD, i.e., pituitary hormone deficiency associated with optic nerve hypoplasia and/or midline brain defects23. One patient with IHH showed eye anomaly (microphthalmia). Twenty-one of the IHH and one of the CPHD patients had anosmia, and were therefore diagnosed with Kallmann syndrome. None of the 74 patients had nasal malformations.
Prior to the present study, all participants underwent sequence analysis for 43 known causative genes of IHH and CPHD (Supplementary Methods). Patients carrying possibly pathogenic variants in these genes were excluded from this study.
Mutation screening of SMCHD1 for 74 patients with HH-related disorders
Mutation screening of SMCHD1 was performed using genomic DNA samples of the patients. First, all 48 coding exons of SMCHD1 and their flanking intronic regions were amplified using the HaloPlex HS Target Enrichment System (Design ID 31368-1548826336; Agilent Technologies, Palo Alto, CA, USA), and sequenced as 150 bp paired-end reads on a NextSeq sequencer (Illumina, San Diego, CA, USA). The methods for read alignment and variant call are described in the Supplementary Information. Then, we examined the position of detected variants in the SMCHD1 protein. Also, we referred to dbSNP (https://www.ncbi.nlm.nih.gov/snp/), the 1000 genome database (https://www.internationalgenome.org/1000-genomes-browsers/), the gnomAD browser (https://gnomad.broadinstitute.org/), and the Human Genetic Variation Database (http://www.hgvd.genome.med.kyoto-u.ac.jp/), to examine the frequency of variants in the general population. Polymorphisms that account for ≥ 1% of the alleles in the general population were excluded from further analysis. Next, we examined whether the variants have been submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
Functional consequences of rare variants were predicted using five in silico programs, namely, Polymorphism Phenotyping v2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/), Sorting Intolerant From Tolerant (SIFT, http://sift.jcvi.org/), CADD (https://cadd.gs.washington.edu/), Mendelian Clinically Applicable Pathogenicity (M-CAP, http://bejerano.stanford.edu/mcap/), and MutationTaster (http://www.mutationtaster.org/). Variants with PolyPhen-2 scores of > 0.8, SIFT scores of < 0.05, CADD scores of > 20, M-CAP scores of > 0.025, or MutationTaster results of “disease causing” were assessed as damaging. The effects of the variants on the protein structure were predicted using PyMOL (version 2.3; Schrödinger). Lastly, the variants were classified according to the ACMG/AMP guideline for interpretation of sequence variants24. A variant scored as likely pathogenic was confirmed by Sanger sequencing. Primer sequences are available upon request.
Whole exome sequencing for patient 1
Whole exome sequencing for patient 1 was performed in Macrogen Japan (Kyoto, Japan) using SureSelect Human All Exon Kit v6 (Agilent Technologies). We searched for rare variants in protein-coding genes, whose frequency in the general population is less than 1%. Next, we selected nonsense, frameshift, and splice-site mutations, as well as missense variants that were assessed as deleterious by all of the five in silico programs. Variants which were reported as benign or likely_benign in the ClinVar database were excluded. Lastly, we investigated whether the affected genes have previously been associated with IHH or CPHD1,9.
DNA methylation analysis of patient 1
We examined DNA methylation of the 52 SMCHD1-target CpG sites in the DUX4 promoter region at the D4Z4 locus. We utilized the previously described methods with slight modifications16,19. In brief, the patient’s genomic DNA sample was treated with bisulfite using the EZ DNA Methylation-Gold Kit (ZYMO RESEARCH, Irvine, CA, USA). Then, the DUX4 promoter region containing 52 CpG sites was PCR-amplified and subcloned into a TOPO TA vector (Invitrogen, Carlsbad, CA, USA). Twelve clones were picked and sequenced. The methylation rates of the region were calculated from the average of 20 or more clones. The results of the patient were compared to previous data obtained from SMCHD1 mutation-negative control individuals16,19 and to the cutoff value for the diagnostic criteria of FSHD2 (25%)19.
In vitro functional assay for the p.Asp398Asn variant
The expression vector for full-length WT SMCHD1 was created by inserting SMCHD1 cDNA into a modified pcDNA5/FRT/TO vector (Invitrogen) with the FLAG-tag at the N-terminus15. Expression vectors for the p.Glu147Ala and p.Asp398Asn variants were created by site-directed mutagenesis. HEK293 cells were transiently transfected with WT or variant expression vectors or an empty vector using Lipofectamine 3000 (Invitrogen). After 48 h incubation, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.25% Triton X-100 for 15 min, blocked with 3% fetal bovine serum blocking buffer, and incubated with Alexa 488-conjugated anti-FLAG M2 (F1804, Sigma-Aldrich, St. Louis, MO, USA) at room temperature for 1 h. Hoechst 33342 (Sigma-Aldrich) was used for nuclear staining. Fluorescence images were acquired using Olympus IX71 (Olympus cellSens software, Olympus Co., Tokyo, Japan). These experiments were repeated three times.
Study approval
All methods were carried out in accordance with relevant guidelines and regulations. This study was approved by the Institutional Review Board Committee at the National Center for Child and Development and performed after obtaining informed consent from the participants or their parents.
Supplementary information
Acknowledgements
We thank Drs. T. Yoshida and S. Narumi and Ms. A. Ueda for their technical assistance. We also thank Drs. T. Aasada, Y. Ando, H. Arimura, H. Fukuoka, A. Hakoda, J. Hamada, Y. Hasegawa, R. Horikawa, Y. Hotta, J. Kannno, S. Kawano, H. Kihara, S. Kinjo, S. Kitagawa, K. Komiya, J. Kurimoto, H. Mabe, E. Makita, N. Matsuura, C. Miyagi, Y. Miyoshi, K. Mori, Y. Naiki, H. Narumi, N. Namba, C. Numakura, M. Ono, R. Hirano, T. Sasaki, H. Sawada, S. Suzuki, M. Tachibana, I. Tamada, T. Tanaka, T. Usui, T. Wada, Y. Wada, A. Yamane and S. Yamashita for providing DNA samples and clinical information of the patients. This work was supported by Grants from the Japan Society for the Promotion of Science (17H06428), the Japan Agency for Medical Research and Development (20ek0109464h0001), the National Center for Child Health and Development (2019A-1), and the Takeda Science Foundation. The founders have no role in study design, data collection, data analysis, manuscript preparation, or publication decisions.
Author contributions
M.F. contributed to the study conception and design. K.K., E.S., K.I., Ka.N., A.H. Ke.M. and M.M. participated in the acquisition of laboratory data. Ke.N. and Ko.M. participated in the acquisition of clinical samples and phenotypic information. K.K., Ko.N., R-S.N., C.O., T.O., M.F. and M.M. participated in analysis and interpretation of data. K.K., M.F. and M.M. drafted the manuscript. All authors have critically reviewed the manuscript and approved the final version.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kenichi Kinjo and Keisuke Nagasaki.
These authors jointly supervised: Maki Fukami and Mami Miyado.
Contributor Information
Maki Fukami, Email: fukami-m@ncchd.go.jp.
Mami Miyado, Email: miyado-m@ncchd.go.jp.
Supplementary information
is available for this paper at 10.1038/s41598-020-67715-x.
References
- 1.Fang Q, et al. Genetics of combined pituitary hormone deficiency: Roadmap into the genome era. Endocr. Rev. 2016;37:636–675. doi: 10.1210/er.2016-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maione L, et al. Genetics in endocrinology: Genetic counseling for congenital hypogonadotropic hypogonadism and Kallmann syndrome: New challenges in the era of oligogenism and next-generation sequencing. Eur. J. Endocrinol. 2018;178:R55–R80. doi: 10.1530/EJE-17-0749. [DOI] [PubMed] [Google Scholar]
- 3.Macchiaroli A, et al. A novel heterozygous SOX2 mutation causing congenital bilateral anophthalmia, hypogonadotropic hypogonadism and growth hormone deficiency. Gene. 2014;534:282–285. doi: 10.1016/j.gene.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 4.Manara R, et al. Brain changes in Kallmann syndrome. Am. J. Neuroradiol. 2014;35:1700–1706. doi: 10.3174/ajnr.A3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tajima T, Ishizu K, Nakamura A. Molecular and clinical findings in patients with LHX4 and OTX2 mutations. Clin. Pediatr. Endocrinol. 2013;22:15–23. doi: 10.1297/cpe.22.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tziaferi V, Kelberman D, Dattani MT. The role of SOX2 in hypogonadotropic hypogonadism. Sex. Dev. 2008;2:194–199. doi: 10.1159/000152035. [DOI] [PubMed] [Google Scholar]
- 7.Raivio T, et al. Genetic overlap in Kallmann syndrome, combined pituitary hormone deficiency, and septo-optic dysplasia. J. Clin. Endocrinol. Metab. 2012;97:E694–E699. doi: 10.1210/jc.2011-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr. Rev. 2009;30:790–829. doi: 10.1210/er.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topaloğlu AK. Update on the genetics of idiopathic hypogonadotropic hypogonadism. J. Clin. Res. Pediatr. Endocrinol. 2017;9:113–122. doi: 10.4274/jcrpe.2017.S010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansz N, Chen K, Murphy JM, Blewitt ME. The epigenetic regulator SMCHD1 in development and disease. Trends Genet. 2017;33:233–243. doi: 10.1016/j.tig.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Mould AW, et al. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin. 2013;6:19. doi: 10.1186/1756-8935-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkie AOM. Many faces of SMCHD1. Nat. Genet. 2017;49:176–178. doi: 10.1038/ng.3776. [DOI] [PubMed] [Google Scholar]
- 13.Gendrel AV, et al. Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol. 2013;33:3150–3165. doi: 10.1128/MCB.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brideau NJ, et al. Independent mechanisms target SMCHD1 to trimethylated histone H3 lysine 9-modified chromatin and the inactive X chromosome. Mol. Cell. Biol. 2015;35:4053–4068. doi: 10.1128/MCB.00432-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nozawa RS, et al. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struct. Mol. Biol. 2013;20:566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 16.Shaw ND, et al. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017;49:238–248. doi: 10.1038/ng.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon CT, et al. De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat. Genet. 2017;49:249–255. doi: 10.1038/ng.3765. [DOI] [PubMed] [Google Scholar]
- 18.Lemmers RJLF, et al. SMCHD1 mutation spectrum for facioscapulohumeral muscular dystrophy type 2 (FSHD2) and Bosma arhinia microphthalmia syndrome (BAMS) reveals disease-specific localisation of variants in the ATPase domain. J. Med. Genet. 2019;56:693–700. doi: 10.1136/jmedgenet-2019-106168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones TI, et al. Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing. Clin. Epigenetics. 2014;6:23. doi: 10.1186/1868-7083-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmers RJLF, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mul K, et al. FSHD type 2 and Bosma arhinia microphthalmia syndrome: Two faces of the same mutation. Neurology. 2018;91:e562–e570. doi: 10.1212/WNL.0000000000005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurzau AD, et al. FSHD2- and BAMS-associated mutations confer opposing effects on SMCHD1 function. J. Biol. Chem. 2018;293:9841–9853. doi: 10.1074/jbc.RA118.003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb EA, Dattani MT. Septo-optic dysplasia. Eur. J. Hum. Genet. 2010;18:393–397. doi: 10.1038/ejhg.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura T, et al. Concordant and discordant adrenocorticotropin (ACTH) responses induced by growth hormone-releasing peptide-2 (GHRP-2), corticotropin-releasing hormone (CRH) and insulin-induced hypoglycemia in patients with hypothalamopituitary disorders: Evidence for direct ACTH releasing activity of GHRP-2. Endocr. J. 2010;57:639–644. doi: 10.1507/endocrj.K10E-017. [DOI] [PubMed] [Google Scholar]
- 26.DeSimone AM, Pakula A, Lek A, Emerson CP. Facioscapulohumeral muscular dystrophy. Compr. Physiol. 2017;7:1229–1279. doi: 10.1002/cphy.c160039. [DOI] [PubMed] [Google Scholar]
- 27.Strafella C, et al. The variability of SMCHD1 gene in FSHD patients: Evidence of new mutations. Hum. Mol. Genet. 2019;28:3912–3920. doi: 10.1093/hmg/ddz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Deutekom JC, et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 29.Larsen M, et al. Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur. J. Hum. Genet. 2015;23:808–816. doi: 10.1038/ejhg.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abreu AP, Macedo DB, Brito VN, Kaiser UB, Latronico AC. A new pathway in the control of the initiation of puberty: The MKRN3 gene. J. Mol. Endocrinol. 2015;54:R131–R139. doi: 10.1530/JME-14-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto N, et al. Monoallelic expression of normal mRNA in the PIT1 mutation heterozygotes with normal phenotype and biallelic expression in the abnormal phenotype. Hum. Mol. Genet. 1994;3:1565–1568. doi: 10.1093/hmg/3.9.1565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.