Abstract
We aimed to develop machine learning models to accurately predict bronchiolitis severity, and to compare their predictive performance with a conventional scoring (reference) model. In a 17-center prospective study of infants (aged < 1 year) hospitalized for bronchiolitis, by using routinely-available pre-hospitalization data as predictors, we developed four machine learning models: Lasso regression, elastic net regression, random forest, and gradient boosted decision tree. We compared their predictive performance—e.g., area-under-the-curve (AUC), sensitivity, specificity, and net benefit (decision curves)—using a cross-validation method, with that of the reference model. The outcomes were positive pressure ventilation use and intensive treatment (admission to intensive care unit and/or positive pressure ventilation use). Of 1,016 infants, 5.4% underwent positive pressure ventilation and 16.0% had intensive treatment. For the positive pressure ventilation outcome, machine learning models outperformed reference model (e.g., AUC 0.88 [95% CI 0.84–0.93] in gradient boosted decision tree vs 0.62 [95% CI 0.53–0.70] in reference model), with higher sensitivity (0.89 [95% CI 0.80–0.96] vs. 0.62 [95% CI 0.49–0.75]) and specificity (0.77 [95% CI 0.75–0.80] vs. 0.57 [95% CI 0.54–0.60]). The machine learning models also achieved a greater net benefit over ranges of clinical thresholds. Machine learning models consistently demonstrated a superior ability to predict acute severity and achieved greater net benefit.
Subject terms: Diseases, Health care, Medical research, Signs and symptoms
Introduction
Bronchiolitis is the leading cause of infant hospitalization in the US, accounting for 107,000 infant hospitalizations each year with direct cost of 734 million US dollars1. Even among hospitalized infants, the severity of bronchiolitis can range from moderate severity (which requires observation and supportive therapies, such as supplemental oxygen, fluid, and nutrition) to near-fatal and fatal infections. Previous studies have identified individual risk factors for higher severity of bronchiolitis (e.g., young age, prematurity, viral etiology)2–5 and developed prediction scoring models (e.g., logistic regression models)6–9. However, identifying the subgroup of infants with bronchiolitis who require higher acuity care (e.g., positive pressure ventilation, intensive care unit [ICU] admission) remains an important challenge. The difficulty and uncertainty of predicting acute severity—and, consequently, the appropriate level of care for infants with bronchiolitis—are reflected by the well-documented variability in inpatient management across the nation1,10–12.
Machine learning models have gained increasing attention because of their advantages, such as the ability to incorporate high-order, nonlinear interactions between predictors and to yield more accurate and stable predictions. Indeed, recent studies have reported that the use of machine learning models provide a high predictive ability in various conditions and settings—e.g., sepsis13,14, asthma exacerbation15, emergency department (ED) triage16,17, and unplanned transfers to ICU18. Despite the clinical and research promise, no study has yet examined the utility of modern machine learning models in predicting outcomes in infants hospitalized for bronchiolitis—a large population with high morbidity and health resource use.
In this context, we aimed to develop machine learning models that accurately predict acute severity in infants hospitalized with bronchiolitis, and compare their predictive performance with that of conventional scoring approaches6.
Results
During 2011–2014, 1,016 infants with bronchiolitis were enrolled into a 17-center prospective cohort study. The median age at the enrolment was 3.2 months (IQR 1.6–6.0), 40% were female, and 42% were non-Hispanic white. The length-of-hospital stay varied widely from 0 to 60 days (median, 2 days) (Table 1). Clinical data had a small proportion of missingness; most had < 1% missingness (e.g., missingness on oxygen saturation with the use of supplemental oxygen, 0.1%) while the maximum proportion of missing was 4.8% (eTable 3 in Additional file 1). Overall, 55 infants (5.4%) underwent positive pressure ventilation and 163 infants (16.0%) had intensive treatment outcome.
Table 1.
Patient characteristics and clinical outcomes in 1,016 infants hospitalized for bronchiolitis.
| Variables | n = 1,016 |
|---|---|
| Demographics | |
| Age (month), median (IQR) | 3.2 (1.6–6.0) |
| Female sex | 406 (40.0) |
| Race/ethnicity | |
| Non-Hispanic white | 430 (42.0) |
| Non-Hispanic black | 239 (23.5) |
| Others | 347 (34.2) |
| Medical history | |
| Prenatal maternal smoking | 147 (14.7) |
| Gestational age (week) | |
| 32–33 | 35 (3.4) |
| 34–36 | 151 (14.9) |
| 37–39 | 417 (41.0) |
| 40–41 | 391 (38.5) |
| ≥ 42 | 22 (2.2) |
| Birth weight (kg) | |
| 0–1.3 | 3 (0.3) |
| 1.4–2.2 | 61 (6.0) |
| 2.3–3.1 | 343 (33.9) |
| ≥ 3.2 | 604 (59.7) |
| Postnatal ICU admission | 167 (16.4) |
| Previous hospital admission | 162 (16.0) |
| Previous ICU admission | 17 (1.7) |
| Previous breathing problems (count) | 32 (3.2) |
| 0 | 810 (79.7) |
| 1 | 160 (15.7) |
| 2 | 46 (4.5) |
| History of eczema | 149 (14.7) |
| Parent-reported symptoms at home | |
| Poor feeding | 32 (3.2) |
| Cyanosis within 24 h | 92 (9.1) |
| Apnea | 131 (12.9) |
| Apnea within 24 h | 86 (8.5) |
| Duration of symptom ( 24 h) | 53 (5.2) |
| Signs and symptom at ED | |
| Vital signs at presentation | |
| Temperature (F), median (IQR) | 99.4 (98.8–100) |
| Pulse rate (bpm), median (IQR) | 162 (150–176) |
| Respiratory rate (per min), median(IQR) | 48 (40–60) |
| Use of supplemental oxygen (%) | 51 (5) |
| Oxygen saturation level (%) at room air (IQR) | 96 (94–98) |
| Oxygen saturation level (%) with the use of supplemental oxygen (IQR) | 98 (95–100) |
| Wheeze | 602 (62.3) |
| Severity of retraction | |
| None | 192 (19.6) |
| Mild | 431 (43.9) |
| Moderate/severe | 358 (36.5) |
| Apnea | 56 (5.5) |
| Dehydration | 392 (39.5) |
| Virology | |
| RSV | 821 (80.8) |
| Length of hospital stay (days), range | 0–60 |
| Clinical outcomes | |
| Positive pressure ventilation usea | 55 (5.4) |
| Intensive treatment useb | 163 (16.0) |
Data are no. (%) of infants unless otherwise indicated. Percentages may not equal 100, because of rounding and missingness.
bpm beats per minute, IQR interquartile range, ICU intensive care unit, RSV respiratory syncytial virus.
aInfants with bronchiolitis who underwent continuous positive airway ventilation and/or mechanical ventilation.
bInfants with bronchiolitis who were admitted to ICU and/or who underwent positive pressure ventilation.
Predicting positive pressure ventilation outcome
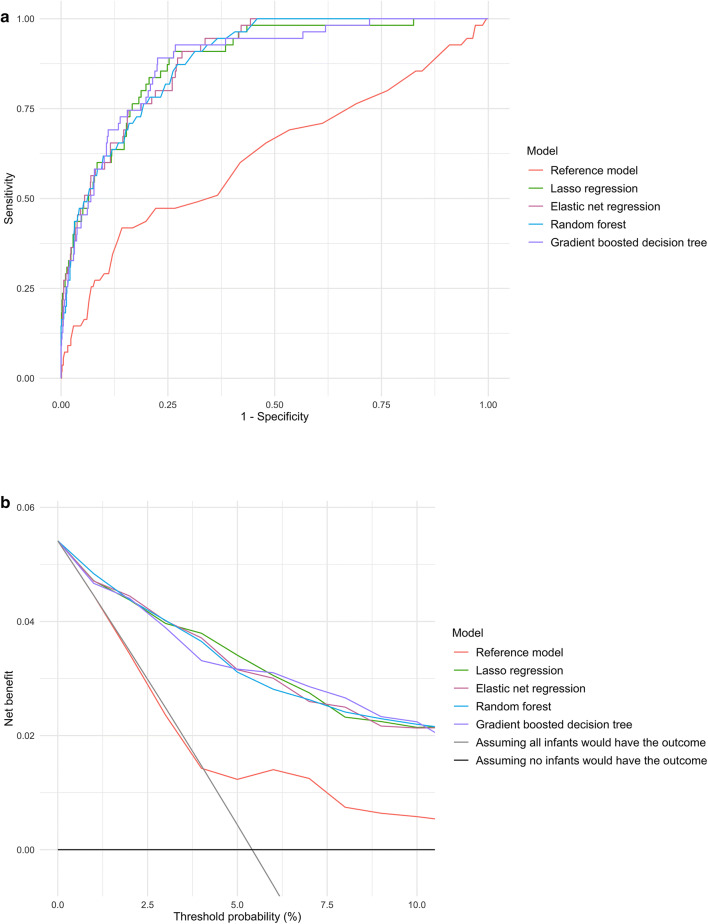
In the prediction of positive pressure ventilation outcome, the discriminatory abilities of all models are summarized in Fig. 1A and Table 2. All four machine learning models demonstrated significantly superior AUCs (all P < 0.001). For example, compared with the reference model (AUC 0.62 [95% CI 0.53–0.70]), the AUC was higher in the elastic net regression (AUC 0.89 [95% CI 0.85–0.92]) and gradient boosted decision tree (AUC 0.88 [95% CI 0.84–0.93]) models. Similarly, compared with the reference model, all machine learning models also achieved a significant net reclassification improvement (all P < 0.001).
Figure 1.
Prediction ability of the reference and machine learning models for positive pressure ventilation outcome in the overall cross-validation dataset. (A) Receiver-operating-characteristics (ROC) curves. The corresponding value of the area under the receiver-operating-characteristics curve (AUC) for each model are presented in Table 2. (B) Decision curve analysis. X-axis indicates the threshold probability for positive pressure ventilation outcome; Y-axis indicates the net benefit. Compared to the reference model, the net benefit of all machine learning models was larger over the range of clinical threshold.
Table 2.
Prediction performance of the reference, and machine learning models in infants hospitalized for bronchiolitis.
| Outcomes and models | AUC | P-valuea | NRIb | P-valueb | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Positive pressure ventilation outcome | ||||||||
| Reference model | 0.62 (0.53–0.70) | Reference | Reference | Reference | 0.62 (0.49–0.75) | 0.57 (0.54–0.60) | 0.075 (0.054–0.097) | 0.96 (0.95–0.97) |
| Logistic regression with Lasso regularization | 0.88 (0.84–0.93) | < 0.001 | 1.09 (0.87–1.32) | < 0.001 | 0.84 (0.73–0.93) | 0.79 (0.77–0.82) | 0.19 (0.14–0.24) | 0.99 (0.99–0.99) |
| Logistic regression with elastic net regularization | 0.89 (0.85–0.92) | < 0.001 | 1.05 (0.82–1.28) | < 0.001 | 0.89 (0.80–0.96) | 0.73 (0.70–0.75) | 0.15 (0.11–0.18) | 0.99 (0.99–0.99) |
| Random forest | 0.89 (0.85–0.92) | < 0.001 | 1.17 (0.96–1.38) | < 0.001 | 0.85 (0.75–0.95) | 0.74 (0.71–0.76) | 0.15 (0.12–0.21) | 0.99 (0.99–0.99) |
| Gradient boosted decision tree | 0.88 (0.84–0.93) | < 0.001 | 1.08 (0.84–1.33) | < 0.001 | 0.89 (0.80–0.96) | 0.77 (0.75–0.80) | 0.17 (0.08–0.21) | 0.99 (0.99–0.99) |
| Intensive treatment outcome | ||||||||
| Reference model | 0.62 (0.57–0.67) | Reference | Reference | Reference | 0.58 (0.55–0.62) | 0.58 (0.50–0.66) | 0.21 (0.18–0.24) | 0.88 (0.86–0.89) |
| Logistic regression with Lasso regularization | 0.79 (0.76–0.83) | < 0.001 | 0.68 (0.52–0.84) | < 0.001 | 0.75 (0.69–0.82) | 0.70 (0.66–0.73) | 0.31 (0.26–0.38) | 0.94 (0.93–0.94) |
| Logistic regression with elastic net regularization | 0.80 (0.76–0.83) | < 0.001 | 0.58 (0.42–0.74) | < 0.001 | 0.72 (0.64–0.79) | 0.74 (0.71–0.77) | 0.33 (0.28–0.41) | 0.93 (0.92–0.94) |
| Random forest | 0.79 (0.75–0.84) | < 0.001 | 0.70 (0.55–0.86) | < 0.001 | 0.70 (0.63–0.77) | 0.78 (0.76–0.81) | 0.37 (0.29–0.45) | 0.93 (0.92–0.94) |
| Gradient boosted decision tree | 0.79 (0.75–0.84) | < 0.001 | 0.72 (0.57–0.87) | < 0.001 | 0.74 (0.67–0.80) | 0.74 (0.71–0.77) | 0.33 (0.26–0.42) | 0.93 (0.92–0.94) |
AUC area under the receiver-operating-characteristic curve, NRI net reclassification improvement, PPV positive predictive value, NPV negative predictive value.
aP-value was calculated to compare area-under-the-curve of the reference model with that of each machine model.
bWe used continuous NRI and its P-value.
Additionally, compared with the reference model, all machine learning models also demonstrated a higher sensitivity (e.g., 0.62 [95% CI 0.49–0.75] in the reference model vs. 0.89 [95% CI 0.80–0.96] in the elastic net regression; Table 2) and specificity (e.g., 0.57 [95% CI 0.54–0.60] in the reference model vs. 0.79 [95% CI 0.77–0.82] in the Lasso regression model). More specifically, all machine learning models correctly predicted a larger number of infants who underwent positive pressure ventilation (true-positives) with a fewer number of predicted outcomes (Table 3). For example, the reference scoring system categorized most infants (n = 629, 62%) into the prediction score groups of 2–3. The reference model correctly identified 16 out of 25 infants who underwent positive pressure ventilation, while predicting that 265 infants would have undergone positive pressure ventilation. In contrast, the gradient boosted decision tree model correctly identified 23 (of 25) patients, while predicting that 135 infants would have undergone positive pressure ventilation in the same patient groups. Considering the low prevalence of the positive pressure ventilation outcome, all models had a high negative predictive value (e.g., 0.96 [95% CI 0.95–0.97] in the reference model vs. 0.99 [95% CI 0.99–0.99] in the Lasso regression model; Table 2).
Table 3.
The number of actual and predicted outcomes of prediction models, according to the score of the reference model.
| Reference model (score) | Positive pressure ventilation use n (%) |
Reference model | Lasso regression | Elastic net regression | Random forest | Gradient boosted tree | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
||
| 0: (n = 41) | 1 (2.4) | 0 | 6 | 0 | 7 | 0 | 8 | 1 | 9 | 0 | 8 |
| 1: (n = 64) | 3 (4.7) | 1 | 15 | 3 | 11 | 2 | 14 | 2 | 11 | 2 | 11 |
| 2: (n = 359) | 13 (3.6) | 9 | 156 | 12 | 80 | 11 | 106 | 11 | 78 | 12 | 79 |
| 3: (n = 270) | 12 (4.4) | 7 | 109 | 10 | 52 | 12 | 65 | 10 | 58 | 11 | 56 |
| 4: (n = 122) | 3 (0.8) | 2 | 46 | 1 | 20 | 1 | 30 | 2 | 41 | 1 | 24 |
| 5: (n = 58) | 8 (13.8) | 3 | 22 | 7 | 21 | 8 | 24 | 7 | 31 | 8 | 24 |
| 6: (n = 15) | 0 (0.0) | 0 | 3 | 0 | 6 | 0 | 6 | 0 | 8 | 0 | 6 |
| 7: (n = 41) | 5 (12.5) | 3 | 22 | 3 | 22 | 5 | 30 | 4 | 28 | 5 | 29 |
| 8: (n = 24) | 4 (16.7) | 2 | 8 | 4 | 12 | 4 | 14 | 4 | 17 | 4 | 15 |
| 9: (n = 11) | 0 (0.0) | 0 | 2 | 0 | 4 | 0 | 4 | 0 | 7 | 0 | 5 |
| 10: (n = 8) | 5 (62.5) | 3 | 3 | 5 | 7 | 5 | 7 | 5 | 8 | 5 | 7 |
| 11: (n = 3) | 1 (33.3) | 0 | 2 | 1 | 2 | 1 | 3 | 1 | 3 | 1 | 2 |
| 12: (n = 0) | 0 (0) | ||||||||||
| Overall (n = 1,016) | 55 (5.4) | 30 (55) | 394 | 46 (84) | 244 | 49 (89) | 311 | 47 (85) | 299 | 49 (89) | 266 |
| Reference model (Score) | Intensive treatment n (%) |
Reference model | Lasso regression | Elastic net regression | Random forest | Gradient boosted tree | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
Correctly identified outcome n (%) |
Predicted outcome n |
||
| 0: (n = 41) | 2 (4.9) | 1 | 10 | 1 | 8 | 0 | 5 | 0 | 4 | 0 | 4 |
| 1: (n = 64) | 8 (12.5) | 1 | 11 | 5 | 17 | 5 | 11 | 5 | 12 | 5 | 9 |
| 2: (n = 359) | 44 (12.2) | 21 | 157 | 27 | 133 | 23 | 111 | 26 | 77 | 24 | 87 |
| 3: (n = 270) | 36 (13.3) | 18 | 118 | 28 | 75 | 28 | 71 | 23 | 64 | 28 | 75 |
| 4: (n = 122) | 17 (13.8) | 8 | 53 | 9 | 36 | 9 | 33 | 7 | 40 | 10 | 53 |
| 5: (n = 58) | 17 (29.3) | 6 | 26 | 14 | 30 | 14 | 30 | 15 | 28 | 15 | 29 |
| 6: (n = 15) | 3 (20.0) | 0 | 3 | 3 | 7 | 2 | 7 | 3 | 9 | 3 | 12 |
| 7: (n = 41) | 19 (47.5) | 15 | 28 | 19 | 38 | 19 | 36 | 18 | 32 | 18 | 35 |
| 8: (n = 24) | 7 (29.2) | 4 | 8 | 7 | 18 | 7 | 17 | 7 | 16 | 7 | 17 |
| 9: (n = 11) | 1 (9.1) | 0 | 3 | 1 | 10 | 1 | 9 | 1 | 6 | 1 | 7 |
| 10: (n = 8) | 7 (87.5) | 3 | 4 | 7 | 7 | 7 | 7 | 7 | 8 | 7 | 8 |
| 11: (n = 3) | 2 (66.7) | 0 | 1 | 2 | 3 | 2 | 3 | 2 | 3 | 2 | 3 |
| 12: (n = 0) | 0 (0) | ||||||||||
| Overall (n = 1,016) | 163 (16.0) | 77 (47) | 422 | 123 (75) | 382 | 117 (72) | 340 | 114 (70) | 299 | 120 (74) | 339 |
Likewise, in the decision curve analysis (Fig. 1B), all four machine learning models outperformed the reference model, demonstrating a greater net benefit throughout the range of clinical thresholds, indicating that the machine learning prediction would more accurately identify high-risk infants (true-positives) while taking the trade-off with false-positives into consideration.
Predicting intensive treatment outcome
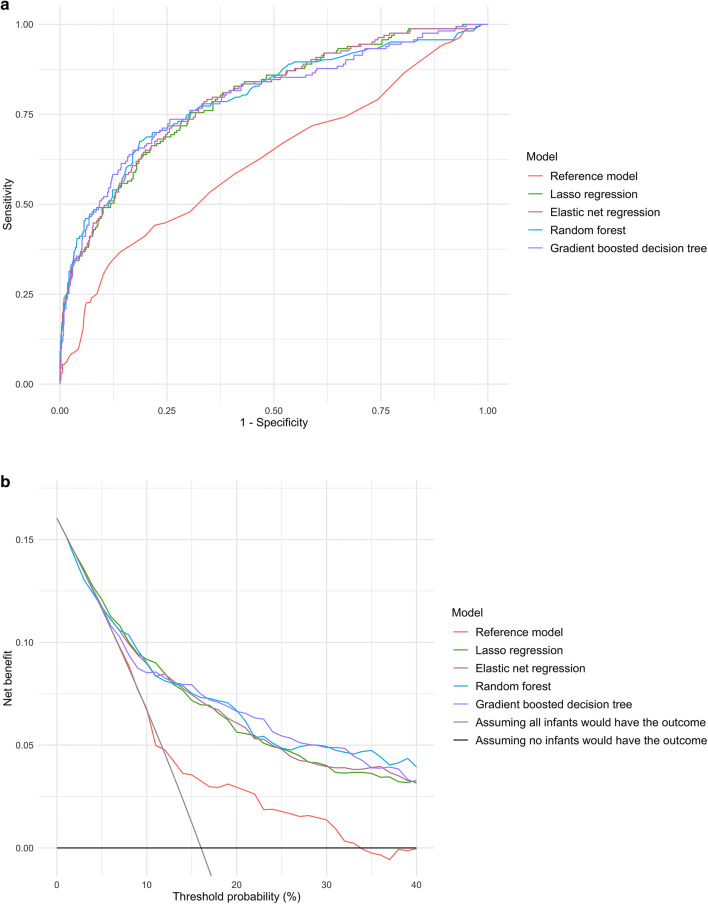
In the prediction of intensive treatment outcome, the discriminatory abilities of all models are shown in Fig. 2A and Table 2. All four machine learning models demonstrated a significantly higher AUC (all P < 0.001). For example, compared with the reference model (AUC 0.62 [95% CI 0.57–0.67]), the AUC was higher in the elastic net regression (AUC 0.80 [95% CI 0.76–0.83]) and random forest (AUC 0.79 [95% CI 0.75–0.84]) models. Similarly, compared with the reference model, all machine learning models also achieved significant net reclassification improvement (all P < 0.001).
Figure 2.
Prediction ability of the reference and machine learning models for intensive treatment outcome in the overall cross-validated dataset. (A) Receiver-operating-characteristics (ROC) curves. The corresponding values of the area under the receiver-operating-characteristics curve (AUC) for each model are presented in Table 2. (B) Decision curve analysis. X-axis indicates the threshold probability for intensive treatment outcome; Y-axis indicates the net benefit. Compared to the reference model, the net benefit of all machine learning models was larger over the range of clinical threshold.
Additionally, all machine learning models demonstrated a higher sensitivity (e.g., 0.58 [95% CI 0.49–0.75] in the reference model vs. 0.75 [95% CI 0.69–0.82] in the Lasso regression; Table 2) and specificity (e.g., 0.58 [95% CI 0.50–0.66] in the reference model vs. 0.78 [95% CI 0.76–0.81] in the random forest model). For example, among the infants categorized into the reference score groups of 2–3 (62% of cohort infants), the reference model correctly identified 39 out of 80 infants who had intensive treatment, while predicting that 275 infants would have had intensive treatment (Table 3). In contrast, the gradient boosted decision tree correctly identified 52 (out of 80) infants with the outcome, while predicting that 162 infants would have had intensive treatment. Likewise, in the decision curve analysis (Fig. 2B), all four machine learning models outperformed the reference model, demonstrating a greater net benefit throughout the range of clinical thresholds.
Variable importance
To yield insights into the relevance of each predictor, eFigures 1 and 2 (Additional file 1) summarized the 15 most important predictors of random forest and gradient boosted decision tree models for each outcome. In the prediction of positive pressure ventilation outcome, age, oxygen saturation level with the use of supplemental oxygen, and other vital signs [at the presentation] were the most important predictors in both models (eFigures 1A and 2A). Likewise, in the prediction of intensive treatment outcome, similar predictors were considered important in the both models (eFigures 1B and 2B).
Discussion
In this analysis of multicenter prospective cohort data from 1,016 infants, we applied four modern machine learning approaches (i.e., Lasso regression, elastic net regression, random forest, and gradient boosted decision tree) to the prediction of acute severity outcomes of bronchiolitis. Compared to the reference model that was derived in an ED sample6, these machine learning models consistently demonstrated a superior performance in predicting positive pressure ventilation and intensive treatment outcomes, including AUC and net reclassification. Additionally, the machine learning models achieved a higher sensitivity and specificity for the two outcomes, in both the overall cohort and the majority of cohort infants that were categorized into the reference score groups of 2–3. Furthermore, the decision curve analysis also demonstrated the net benefit of machine learning models was also greater—i.e., a larger number of true-positives considering a trade-off with false-positives—across a range of clinical thresholds. To the best of our knowledge, this is the first study that has investigated the performance of modern machine learning models in predicting severity in infants with bronchiolitis.
One of the main objectives in the risk stratification of infants with bronchiolitis is to promptly identify infants at risk for higher severity and efficiently utilize finite healthcare resources. The American Academy of Pediatrics bronchiolitis guideline2 highlights the importance of assessing the risk in infants with bronchiolitis. However, optimal risk stratification and prediction remains a challenge as the clinical course in this population (even in infants hospitalized for bronchiolitis) is highly variable10–12. Previous studies, by using conventional modeling (e.g., logistic regression models), have reported a moderate ability to predict severity outcomes (e.g., ED-to-hospital admission, hospital length-of-stay, ICU admission, positive pressure ventilation use) of infants with bronchiolitis6–9,19. Although the use of an expanded set of predictors—e.g., repeated examinations and invasive monitoring during hospital course—may yield better predictive performance, it is often impractical in the real-world acute care settings with an aim to promptly risk-stratify these infants. Alternatively, the use of advanced machine learning models may improve the clinician’s decision-making ability. Indeed, machine learning models have recently been applied to the prediction of various disease conditions and clinical settings, such as early identification of mortality risk in patients with sepsis13, rehospitalization in patients with heart failure20, intensive treatment outcomes in patients with asthma exacerbation15, unplanned transfer to ICU18, and escalated care at pediatric ED triage16. Our multicenter study builds on these earlier reports, and extends them by demonstrating that the modern machine learning models outperform conventional approaches in predicting higher severity of infants with bronchiolitis. While external validation is warranted, these machine learning models using routinely-available predictors can be implemented to clinical practice (e.g., online risk calculators or build-in risk assessment systems)—similar to existent clinical scoring rules.
Clinical prediction systems strive for an appropriate balance between sensitivity and specificity because of the trade-off relationship between these two factors in the context of prevalence of clinical outcomes. In the present study, we observed that the reference score model did not effectively categorize most infants (i.e., 62% of cohort were categorized into the two score groups) or appropriately predicted infants who developed the outcomes. By contrast, the machine learning models correctly identified a larger number of true-positives (i.e., higher sensitivity). This finding supports the utility of these models in the target population, for which the one of the major priorities is to reduce “missed” high-risk cases. Additionally, the machine learning models also had a fewer number of false-positives (i.e., higher specificity) in predicting both outcomes while they were imperfect in the setting of relatively-smaller prevalence of outcome (5.4% for positive pressure ventilation use). This may mitigate excessive resource use in this large population. These findings are further supported by the decision curve analysis that demonstrated a greater net benefit of the machine learning models incorporating the trade-offs between true-positives and false-positives across the wide ranges of clinical thresholds.
There are several potential explanations for the observed gains in the predictive abilities of machine learning models. For example, machine learning models incorporate high-order interactions between predictors and nonlinear relationships with outcomes. Additionally, machine learning models are able to mitigate potential overfitting by adopting several methods, such as regularization, out-of-bagging estimation, and cross-validation. Furthermore, the use of large multicenter data with rigorous quality assurance might have contributed to low bias and variance in the machine models. Although the machine learning models achieved superior predictive ability, their performance remained imperfect. This may be explained, at least partially, by the limited set of predictors, subjectivity of some data elements (e.g., parent-reported symptoms at home), variable clinical factors after prehospitalization assessment (e.g., ED management and patient responses), difference in clinician’s practice patterns, and availability of intensive care resources. Notwithstanding the complexity and challenges of clinical prediction in infants with bronchiolitis, machine learning models have scalable advantages in the era of health information technology, such as automated sophistication of models through the sequential extraction of electronic health records, continuous non-invasive physiological monitoring, natural language processing, and reinforcement learning21–24. In the past, this scalability had not been attainable with the use of conventional approaches. Taken together, our findings and recent developments support cautious optimism that modern machine learning may enhance the clinician’s ability as an assistive technology.
Our study has several potential limitations. Firstly, the data may be subject to measurement bias and missingness. However, the study was conducted by trained investigators using a standardized protocol, which led to the low proportion of missingness in the predictors (eTable 3 in Additional file 1). Secondly, the clinical thresholds for these outcomes may depend on local resources and vary between clinicians and hospitals (e.g., different criteria for admission to the ICU). Yet, the decision curve analysis demonstrated the greater benefit of the machine learning models across the wide range of clinical thresholds. Lastly, the study cohort consisted of a racially/ethnically- and geographically-diverse US sample of infants hospitalized with bronchiolitis. While the severity of this population was highly variable and the model used pre-hospitalization data, our models might not be generalizable to infants in ambulatory settings. External validation of the models in different populations and settings is necessary. Nonetheless, our data remain highly relevant for the 107,000 infants hospitalized yearly in the US1.
Conclusion
Based on data from a multicenter prospective cohort of 1,016 infants with bronchiolitis, we developed four machine learning models to predict severity of illness. By using prehospitalization data as predictors, these models consistently yielded superior performance—a higher AUC, net reclassification, sensitivity, and specificity—in predicting positive pressure ventilation and intensive treatment outcomes over the reference model6. Specifically, these advanced machine learning models correctly predicted a larger number of infants with higher severity—with a fewer number of false-positives—who would not be appropriately predicted by the conventional models. Moreover, the machine learning models also achieved a greater net benefit across wide ranges of clinical thresholds. Although an external validation is warranted, the current study lends support to the application of machine learning models to the prediction of acute severity in infants with bronchiolitis. Machine learning models have a potential to enhance clinicians’ decision-making ability and hence to improve clinical care and optimize resource utilization in this high morbidity population.
Methods
Study design, setting and participants
The current study aimed to develop machine learning models that accurately predict acute severity in infants with bronchiolitis, by using the data from a multicenter prospective cohort study of 1,016 infants hospitalized for bronchiolitis—the 35th Multicenter Airway Research Collaboration (MARC-35) study25,26. MARC-35 is coordinated by the Emergency Medicine Network (EMNet, https://www.emnet-usa.org27) an international research collaboration with 246 participating hospitals. Briefly, at 17 sites across 14 U.S. states (eTable 1 in Additional file 1), MARC-35 enrolled infants (aged < 1 year) who were hospitalized with an attending physician diagnosis of bronchiolitis during three consecutive bronchiolitis seasons (November 1 to April 30) during 2011–2014. The diagnosis of bronchiolitis was made according to the American Academy of Pediatrics bronchiolitis guidelines2, defined as acute respiratory illness with a combination of rhinitis, cough, tachypnea, wheezing, crackles, and retractions. We excluded infants who were transferred to a participating hospital > 24 h after initial hospitalization or with a preexisting heart and lung disease, immunodeficiency, immunosuppression or gestational age of < 32 weeks.
We followed the Standards for Reporting Diagnostic Accuracy statement guideline for the reporting of prediction models28. The institutional review board of the 17 participating hospitals (Alfred I. duPont Hospital for Children, Arnold Palmer Hospital for Children, Boston Children's Hospital, Children's Hospital of Los Angeles, Children's Hospital of Philadelphia, Children's Hospital of Pittsburgh, The Children's Hospital at St. Francis, The Children's Mercy Hospital & Clinics, Children's National Medical Center, Cincinnati Children's Hospital and Medical Center, Connecticut Children's Medical Center, Dell Children's Medical Center of Central Texas, Norton Children's Hospital, Massachusetts General Hospital, Phoenix Children's Hospital, Seattle Children's Hospital, Texas Children's Hospital) approved the study. Written informed consent was obtained from the parent or guardian.
Predictors
For predictors in the machine learning models, we selected variables based on clinical plausibility and a priori knowledge3,6–9,29–31. These predictors—which are available in most prehospitalization settings—included demographics (age, sex, and race/ethnicity), medical history (prenatal maternal smoking, gestational age, birth weight, postnatal ICU admission, history of hospital and ICU admission, history of breathing problems, and history of eczema), parent-reporting symptoms (poor feeding, cyanosis, apnea, and duration of symptoms), ED presentation (vital signs [temperature, pulse rate, respiratory rate, oxygen saturation], interaction between oxygen saturation and supplemental oxygen use, wheezing, retractions, apnea, and dehydration), and detection of respiratory syncytial virus (RSV) by PCR25. These clinical data were obtained through a structured interview and medical record review by trained physicians and investigators using a standardized protocol26. All data were reviewed at the EMNet Coordinating Center at Massachusetts General Hospital (Boston, MA), and site investigators were queried about missing data and discrepancies identified by manual data checks.
Outcomes
The primary outcome was the use of positive pressure ventilation—continuous positive airway pressure ventilation and/or intubation during inpatient stay32. The secondary outcome was intensive treatment defined as a composite of ICU admission and/or the use of positive pressure ventilation during the inpatient stay3, 31. In this observational study, patients were managed at the discretion of treating physicians. These two outcomes have been employed for outcomes in the MARC-35 study.
Statistical analysis
In the training sets (80% randomly-selected samples) in fivefold cross-validation, we developed five models: the reference model6 and four machine learning models for each outcome. As the reference model, we fit logistic regression models using the predictors of a previously-established clinical prediction score that was derived using an ED sample6. We selected this prediction score as the reference model since it was recently developed in a large sample and focused on similar clinical outcomes reflecting acute severity of bronchiolitis6,33. The predictors included age, poor feeding, oxygen saturation, retractions, apnea, and dehydration, excluding nasal flaring/grunting, based on the availability of data in the current study (eTable 2).
Next, using the prehospitalization predictors, we developed four machine learning models: (1) logistic regression with Lasso regularization (Lasso regression), (2) logistic regression with elastic net regularization (elastic net regression), (3) random forest, and (4) gradient boosted decision tree models. First, Lasso regression is an extension of regression-based models that has an ability to shrink (or regularize) the predictor coefficients toward zero, thereby effectively selecting important predictors and improving interpretability of the model34. Lasso regression computes the optimal regularization parameter (lambda) that minimizes the sum of least square plus L1-shrinkage penalty using a cross-validation method35. Second, elastic net regression is another regression-based model incorporating both Lasso-regularization and Ridge-regularization34,36. Elastic net regression calculates the optimal regularization parameter that minimizes the sum of least square plus weighted L1-shrinkage penalty and weighted L2-shrinkage penalty. We used R glmnet and caret packages for Lasso regression and elastic net regression models37,38. Third, random forest is an ensemble of decision trees generated by bootstrapped training samples with random predictor selection in tree induction34,39. We created a hyperparameter tuning grid to identify the best set of parameters using cross-validation methods. We used randomForest and caret packages to construct random forest models38,40. Lastly, gradient boosted decision tree is another ensemble method which constructs new simple tree models predicting the errors and residuals of the previous model. When adding a new tree, this model uses a gradient descent algorithm minimizes a loss function41. We performed hyperparameter tuning sequentially using a fivefold cross-validation method. We used R xgboost and caret packages to construct gradient boosted decision tree38,42. To minimize potential overfitting, we utilized several methods—e.g., regularizations (or penalizations) in Lasso and elastic net regression models, out-of-bag estimation in random forest models, and cross-validation in all models.
As for the predictor engineering methods of the machine learning models, we preprocessed predictors sequentially. First, we investigated non-linear relationships between the continuous predictors and outcomes and created quadric terms of age, respiratory rate, and temperature. These quadratic terms were used only for regression-based machine learning models (i.e., logistic regression models with Lasso regularization and those with elastic net regularization). Second, we also chose either of highly-correlated predictors (e.g., age and weight at hospitalization). Third, we imputed predictors with missing values (eTable 3) using bagged tree imputation. Fourth, we converted continuous predictors into normalized scales using Yeo-Johnson transformation. Categorical predictors were coded as dummy variables while birth weight, gestational age, previous breathing problem, and degree of retraction were coded as ordinal variables. Fifth, to incorporate clinically evident interaction between oxygen saturation level and use of supplemental oxygen, we created an interaction term between oxygen saturation and use of supplemental oxygen. Lastly, we removed predictors that are highly sparse in the dataset. We applied these preprocessing methods independently to the training sets and the test sets to avoid carrying the information from the training sets to the test sets. We used R recipe package for these predictor preprocessing43.
To examine the variable importance in the random forest, we used permutation-based variable importance—normalized average values of difference between the prediction accuracy of out-of-bag estimation and that of the same measure after permutating each predictor. In the gradient boosted model, we also computed the variable importance that is summed over iterations39. We graphically presented the rank of variable importance using unscaled values.
To measure the test performance of each model, we computed the overall cross-validation performance from the test sets (the remaining randomly-selected 20% samples). As the predictive performance, we used (1) the area under the receiver-operating-characteristic curve (AUC), (2) net reclassification improvement, (3) confusion matrix results (i.e., sensitivity, specificity, positive predictive value, and negative predictive value), and (4) net benefit from decision curve analysis. To compare the AUC between the models, we used Delong’s test44. To compute AUC and its confidential interval, we used pROC package45. We also used the net reclassification improvement to quantify whether a new model provides clinically relevant improvements in prediction when compared to the reference model46. To compute the net reclassification improvement, we used PredictABEL package47. To address the class imbalance in the both outcomes, we employed the value with the shortest distance to the top-left part of the AUC plot as the threshold for the confusion matrix39.The decision curve analysis incorporates the information on both the benefit of correctly predicting the outcome (true-positives) and the relative harm of incorrectly labelling patients as if they would have the outcome (false-positives)—i.e., the net benefit48–52. We made a graphical presentation of the net benefit for each model over a range of threshold probabilities (or clinical preferences) of the outcome as decision curves. We used decision curve analysis R source code from Memorial Sloan Kettering Cancer Center53 and plotted the graphs using ggplot2 package54. We performed all analysis with R version 3.5.1 (R Foundation for Statistical Computing, Vienna, Austria)55.
Supplementary information
Acknowledgements
We thank the MARC-35 study hospitals and research personnel for their ongoing dedication to bronchiolitis and asthma research (see eTable 1 in Additional file 1). We also thank Ashley Sullivan, MPH, MS and Janice Espinola, MPH (Massachusetts General Hospital, Boston, MA) as well as Alkis Togias, MD (National Institute of Allergy and Infectious Diseases) for their helpful contributions. This study was supported by Grants from the National Institutes of Health (Bethesda, MD): U01 AI-087881, R01 AI-114552, R01 AI-108588, R01 AI-134940, and UG3/UH3 OD-023253. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding organizations were not involved in the collection, management, or analysis of the data; preparation or approval of the manuscript; or decision to submit the manuscript for publication.
Abbreviations
- MARC-35
The 35th Multicenter Airway Research Collaboration study
- AUC
Area-under-the-curve
- CI
Confidential interval
- ICU
Intensive care unit
- ED
Emergency department
- EMNet
Emergency Medicine Network
- RSV
Respiratory syncytial virus
- IQR
Interquartile range
- NRI
Net reclassification improvement
- PPV
Positive predictive value
- NPV
Negative predictive value
Author contributions
Y.R. contributed to the design of the study, data analysis, and interpretation of results, and writing the manuscript. C.A.C. organized the study cohort and interpretation of results, and critically reviewed the manuscript. C.G.M., J.M.M., P.A.P., S.C.P., and S.J.T. contributed to the acquisition and maintaining of the data, and critically reviewed the manuscript. K.H. contributed to the analysis of data, design of the study, and interpretation of result, and critically reviewed the manuscript. All authors finally approved the final version of manuscript.
Data availability
The datasets generated and analysed during the current study are not publicly available because of the informed consent documents. Per the informed consent documents of the MARC research participants, the data sharing and use are limited to the severe bronchiolitis, recurrent wheezing, asthma and related concepts. Accordingly, the data are not publicly available but available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-67629-8.
References
- 1.Fujiogi, M. et al. Trends in bronchiolitis hospitalizations in the United States: 2000–2016. Pediatrics144(6), e20192614 (2019). 10.1542/peds.2019-2614. [DOI] [PMC free article] [PubMed]
- 2.Ralston SL, et al. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics. 2014;134:e1474–1502. doi: 10.1542/peds.2014-2742. [DOI] [PubMed] [Google Scholar]
- 3.Mansbach JM, et al. Prospective multicenter study of children with bronchiolitis requiring mechanical ventilation. Pediatrics. 2012;130:e492–500. doi: 10.1542/peds.2012-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasegawa K, Mansbach JM, Camargo CA. Infectious pathogens and bronchiolitis outcomes. Expert Rev. Anti Infect. Ther. 2014;12:817–828. doi: 10.1586/14787210.2014.906901. [DOI] [PubMed] [Google Scholar]
- 5.Mansbach JM, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch. Pediatr. Adolesc. Med. 2012;166:700–706. doi: 10.1001/archpediatrics.2011.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freire, G. et al. Predicting escalated care in infants with bronchiolitis. Pediatrics142(3), e20174253 (2018). 10.1542/peds.2017-4253. [DOI] [PubMed]
- 7.Walsh P, Rothenberg SJ, O’Doherty S, Hoey H, Healy R. A validated clinical model to predict the need for admission and length of stay in children with acute bronchiolitis. Eur. J. Emerg. Med. Off. J. Eur. Soc. Emerg. Med. 2004;11:265–272. doi: 10.1097/00063110-200410000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Voets S, van Berlaer G, Hachimi-Idrissi S. Clinical predictors of the severity of bronchiolitis. Eur. J. Emerg. Med. Off. J. Eur. Soc. Emerg. Med. 2006;13:134–138. doi: 10.1097/01.mej.0000206194.85072.33. [DOI] [PubMed] [Google Scholar]
- 9.Damore D, Mansbach JM, Clark S, Ramundo M, Camargo CA. Prospective multicenter bronchiolitis study: Predicting intensive care unit admissions. Acad. Emerg. Med. Off. J. Soc. Acad. Emerg. Med. 2008;15:887–894. doi: 10.1111/j.1553-2712.2008.00245.x. [DOI] [PubMed] [Google Scholar]
- 10.Christakis DA, et al. Variation in inpatient diagnostic testing and management of bronchiolitis. Pediatrics. 2005;115:878–884. doi: 10.1542/peds.2004-1299. [DOI] [PubMed] [Google Scholar]
- 11.Macias CG, et al. Variability in inpatient management of children hospitalized with bronchiolitis. Acad. Pediatr. 2015;15:69–76. doi: 10.1016/j.acap.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Mansbach JM, et al. Hospital course and discharge criteria for children hospitalized with bronchiolitis. J. Hosp. Med. 2015;10:205–211. doi: 10.1002/jhm.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor RA, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: A local big data-driven, machine learning approach. Acad. Emerg. Med. Off. J. Soc. Acad. Emerg. Med. 2016;23:269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemati S, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit. Care Med. 2018;46:547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto T, Camargo CAJ, Faridi MK, Yun BJ, Hasegawa K. Machine learning approaches for predicting disposition of asthma and COPD exacerbations in the ED. Am. J. Emerg. Med. 2018;36:1650–1654. doi: 10.1016/j.ajem.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 16.Goto T, Camargo CA, Faridi MK, Freishtat RJ, Hasegawa K. Machine learning-based prediction of clinical outcomes for children during emergency department triage. JAMA Netw. Open. 2019;2:e186937. doi: 10.1001/jamanetworkopen.2018.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raita Y, et al. Emergency department triage prediction of clinical outcomes using machine learning models. Crit. Care Lond. Engl. 2019;23:64. doi: 10.1186/s13054-019-2351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellner B, et al. Predicting unplanned transfers to the intensive care unit: A machine learning approach leveraging diverse clinical elements. JMIR Med. Inform. 2017;5:e45. doi: 10.2196/medinform.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansbach JM, et al. Prospective multicenter study of bronchiolitis: predicting safe discharges from the emergency department. Pediatrics. 2008;121:680–688. doi: 10.1542/peds.2007-1418. [DOI] [PubMed] [Google Scholar]
- 20.Mortazavi BJ, et al. Analysis of machine learning techniques for heart failure readmissions. Circ. Cardiovasc. Qual. Outcomes. 2016;9:629–640. doi: 10.1161/CIRCOUTCOMES.116.003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priesol AJ, Cao M, Brodley CE, Lewis RF. Clinical vestibular testing assessed with machine-learning algorithms. JAMA Otolaryngol. Head Neck Surg. 2015;141:364–372. doi: 10.1001/jamaoto.2014.3519. [DOI] [PubMed] [Google Scholar]
- 22.Bacchi S, et al. Deep Learning Natural language processing successfully predicts the cerebrovascular cause of transient ischemic attack-like presentations. Stroke. 2019 doi: 10.1161/STROKEAHA.118.024124. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Prediction of emergency department hospital admission based on natural language processing and neural networks. Methods Inf. Med. 2017;56:377–389. doi: 10.3414/ME17-01-0024. [DOI] [PubMed] [Google Scholar]
- 24.Kolachalama VB, et al. Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int. Rep. 2018;3:464–475. doi: 10.1016/j.ekir.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansbach JM, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J. Allergy Clin. Immunol. 2016;137:1909–1913.e4. doi: 10.1016/j.jaci.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa K, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur. Respir. J. 2016;48:1329–1339. doi: 10.1183/13993003.00152-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emergency Medicine Network. https://www.emnet-usa.org/. Accessed 17 June 2020.
- 28.Bossuyt PM, et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corneli HM, et al. Bronchiolitis: Clinical characteristics associated with hospitalization and length of stay. Pediatr. Emerg. Care. 2012;28:99–103. doi: 10.1097/PEC.0b013e3182539098. [DOI] [PubMed] [Google Scholar]
- 30.Yusuf S, Caviness AC, Adekunle-Ojo AO. Risk factors for admission in children with bronchiolitis from pediatric emergency department observation unit. Pediatr. Emerg. Care. 2012;28:1132–1135. doi: 10.1097/PEC.0b013e31827132ff. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa K, et al. Risk factors for requiring intensive care among children admitted to ward with bronchiolitis. Acad. Pediatr. 2015;15:77–81. doi: 10.1016/j.acap.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart CJ, et al. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. A multiomic analysis. Am. J. Respir. Crit. Care Med. 2017;196:882–891. doi: 10.1164/rccm.201701-0071OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo G, Nkoy FL, Gesteland PH, Glasgow TS, Stone BL. A systematic review of predictive modeling for bronchiolitis. Int. J. Med. Inf. 2014;83:691–714. doi: 10.1016/j.ijmedinf.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 34.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning: with Applications in R. New York: Springer-Verlag; 2013. [Google Scholar]
- 35.Tibshirani R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B Methodol. 1996;58:267–288. [Google Scholar]
- 36.Zou H, Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 37.Friedman, J. et al. glmnet: Lasso and Elastic-Net Regularized Generalized Linear Models. (2020). https://cran.rproject.org/web/packages/glmnet/glmnet.pdf. Accessed 17 June 2020.
- 38.Kuhn, M. The caret Package. http://topepo.github.io/caret/index.html. Accessed 17 June 2020.
- 39.Kuhn M, Johnson K. Applied Predictive Modeling. New York: Springer-Verlag; 2013. [Google Scholar]
- 40.Cutler, F. original by L. B. and A. & Wiener, R. port by A. L. and M. randomForest: Breiman and Cutler’s Random Forests for Classification and Regression. (2018). https://cran.r-project.org/web/packages/randomForest/randomForest.pdf. Accessed 17 June 2020.
- 41.Natekin, A. & Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobotics7, 21 (2013). [DOI] [PMC free article] [PubMed]
- 42.Chen, T. et al. xgboost: Extreme Gradient Boosting. (2018). https://cran.r-project.org/web/packages/xgboost/vignettes/xgboost.pdf. Accessed 17 June 2020.
- 43.Kuhn, M., Wickham, H. & RStudio. recipes: Preprocessing Tools to Create Design Matrices. (2018). https://cran.rproject.org/web/packages/recipes/recipes.pdf. Accessed 17 June 2020.
- 44.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 45.Robin, X. et al. pROC: Display and Analyze ROC Curves. (2020). https://cran.r-project.org/web/packages/pROC/pROC.pdf. Accessed 17 June 2020.
- 46.Kerr KF, et al. Net reclassification indices for evaluating risk prediction instruments: A critical review. Epidemiol. Camb. Mass. 2014;25:114–121. doi: 10.1097/EDE.0000000000000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundu, S., Aulchenko, Y. S. & Janssens, A. C. J. W. PredictABEL: Assessment of Risk Prediction Models. (2020). https://cran.rproject.org/web/packages/PredictABEL/PredictABEL.pdf. Accessed 17 June 2020.
- 48.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. JAMA. 2015;313:409–410. doi: 10.1001/jama.2015.37. [DOI] [PubMed] [Google Scholar]
- 50.Rousson V, Zumbrunn T. Decision curve analysis revisited: Overall net benefit, relationships to ROC curve analysis, and application to case–control studies. BMC Med. Inform. Decis. Mak. 2011;11:45. doi: 10.1186/1472-6947-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Calster B, et al. Reporting and interpreting decision curve analysis: A guide for investigators. Eur. Urol. 2018;74:796–804. doi: 10.1016/j.eururo.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biostatistics: Decision Curve Analysis|Memorial Sloan Kettering Cancer Center. https://www.mskcc.org/departments/epidemiology-biostatistics/biostatistics/decision-curve-analysis. Accessed 17 June 2020.
- 54.Wickham, H. et al. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. (2019). https://cran.rproject.org/web/packages/ggplot2/index.html. Accessed 17 June 2020.
- 55.R: The R Project for Statistical Computing. https://www.r-project.org/. Accessed 17 June 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are not publicly available because of the informed consent documents. Per the informed consent documents of the MARC research participants, the data sharing and use are limited to the severe bronchiolitis, recurrent wheezing, asthma and related concepts. Accordingly, the data are not publicly available but available from the corresponding author on reasonable request.