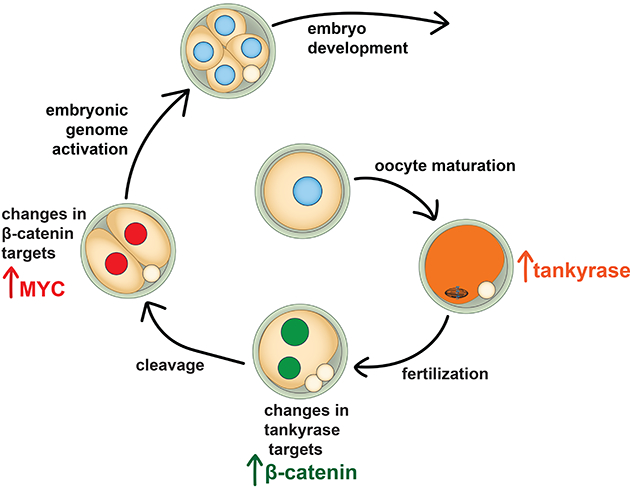
Summary
Embryonic genome activation (EGA) is orchestrated by an intrinsic developmental program initiated during oocyte maturation with translation of stored maternal mRNAs. Here we show that tankyrase, a poly(ADP-ribosyl) polymerase that regulates β-catenin levels, undergoes programmed translation during oocyte maturation and serves an essential role in mouse EGA. Newly translated TNKS triggers proteasomal degradation of axin, reducing targeted destruction of β-catenin and promoting β-catenin-mediated transcription of target genes, including Myc. MYC mediates ribosomal RNA transcription in 2-cell embryos, supporting global protein synthesis. Suppression of tankyrase activity using knockdown or chemical inhibition causes loss of nuclear β-catenin and global reductions in transcription and histone H3 acetylation. Chromatin and transcriptional profiling indicate that development arrests prior to the mid-2-cell stage, mediated in part by reductions in β-catenin and MYC. These findings indicate that post-transcriptional regulation of tankyrase serves as a ligand-independent developmental mechanism for post-translational β-catenin activation and is required to complete EGA.
Keywords: Tankyrase, β-catenin, axin, embryonic genome activation, mouse, preimplantation embryo, ATAC-seq, WNT signaling pathway, post-transcriptional regulation
Graphical Abstract
eTOC Blurb
β-catenin signaling during development is normally mediated by Wnt ligands. Gambini, Stein et al. show that ligand-independent, post-transcriptional regulation of tankyrase levels during mouse oocyte maturation leads to β-catenin activity necessary for embryonic genome activation.
Introduction
The “oocyte-to-embryo transition” encompasses the series of developmental events that gives rise to the totipotent cleavage stage preimplantation embryo. Critical aspects of this transition include meiotic maturation-associated recruitment for translation of specific maternally stored mRNAs, widespread chromatin remodeling, an increase in protein synthesis, and initiation of transcription from the embryonic genome, or “embryonic genome activation” (EGA) (Schultz et al., 1979; Schultz et al., 2018). Because full-grown oocytes are transcriptionally quiescent, programming of EGA depends on translation of dormant maternal mRNAs during maturation and following fertilization. Many dormant maternal mRNAs encode proteins essential for critical processes in 1-cell (1C) and 2-cell (2C) embryos, such as DNA replication, chromatin remodeling and RNA degradation (Schultz et al., 2018). What has not yet been determined is how specific transcription factors are activated to direct the process of EGA.
EGA occurs embryo-autonomously in simple culture media in vitro, indicating that secreted ligands from somatic tissues are not needed; instead, an intrinsic developmental program drives this process. In mice, a minor wave of embryonic transcription occurs during the 1C stage that is essential for embryo development (Abe et al., 2018). This minor wave is somewhat promiscuous, generally independent of core promoter elements, and is associated with transcription of numerous retrotransposons (Abe et al., 2015). However, some key transcripts are generated at the 1C stage. An important example is Dux, which encodes a homeobox transcription factor that synchronizes the major wave of embryonic transcription during the 2C stage (Chen and Zhang, 2019; De Iaco et al., 2017; De Iaco et al., 2020; Hendrickson et al., 2017). DUX is subsequently repressed following initiation of major EGA by a protein-RNA complex comprised of the retrotransposon long interspersed element 1 (LINE1) RNA, nucleolin and tripartite motif-containing 28 (TRIM28); DUX repression is necessary for embryos to progress beyond the 2C stage (Hendrickson et al., 2017; Percharde et al., 2018). β-catenin (encoded by Ctnnb1) could participate in EGA based on its expression and localization patterns. It is a protein with dual functions: regulation of cell adhesion and transcriptional activation in the canonical WNT signaling pathway. β-catenin’s cell adhesion function is required for compaction at the 8-cell embryo stage and its role as a transcription factor is required for gastrulation (De Vries et al., 2004; Haegel et al., 1995; Huelsken et al., 2000; Rudloff and Kemler, 2012). However, β-catenin is detected in nuclei of oocytes, 1C and 2C embryos (De Vries et al., 2004; Xie et al., 2008). Furthermore, both Ctnnb1 mRNA and tankyrase 1 (Tnks) mRNA, which encodes a protein that upregulates β-catenin transcriptional activity, have all the hallmarks of dormant maternal mRNAs (Kemler et al., 2004; Pan et al., 2008). These hallmarks include relatively high mRNA levels in oocytes and the presence of “cytoplasmic polyadenylation elements” in the 3′-untranslated region that direct polyadenylation during maturation (Mendez and Richter, 2001). Polyadenylation causes an apparent increase in mRNA levels during oocyte maturation, despite the lack of transcription at this time, along with recruitment to polysomes for translation. The observations that both β-catenin and a protein that upregulates β-catenin transcriptional activity appear to be encoded by dormant maternal mRNAs, combined with β-catenin nuclear localization at the 1C and 2C stages, suggest that β-catenin has a critical transcription function during EGA. Of note, embryos reported to lack maternal and zygotic β-catenin are abnormally small and misshapen by the blastocyst stage, indicating a failure earlier in preimplantation development (Messerschmidt et al., 2016).
Stringent control of β-catenin activity is accomplished via post-translational regulation by a complex pathway (MacDonald et al., 2009). The β-catenin “destruction complex” is comprised of a scaffolding protein, axin (AXIN1 or AXIN2), glycogen synthase kinase 3 (GSK3), casein kinase 1 (CK1), adenoma polyposis coli (APC), and an E3-ubiquitin ligase. In the absence of WNT signaling, β-catenin is phosphorylated by the combined actions of GSK3 and CK1; these phosphorylation events lead to its ubiquitination and rapid proteasomal degradation. Activation of the WNT signaling pathway via ligand-induced activation of frizzled receptors leads to suppression of β-catenin ubiquitination and the accumulation of newly synthesized, non-phosphorylated β-catenin that translocates to the nucleus to carry out transcription functions. A second layer of β-catenin regulation is via post-translational control of axin. Axin proteins undergo poly(ADP-ribosyl)ation by tankyrase proteins (TNKS or TNKS2); this modification promotes axin ubiquitination and proteasomal degradation. In the absence of tankyrase activity, axin levels increase, leading to more efficient destruction of β-catenin and inhibition of WNT signaling (Huang et al., 2009). Finally, E-cadherin at the cell surface competes with the destruction complex for β-catenin binding, and thus can serve as a reservoir of active β-catenin (De Vries et al., 2004; Hulsken et al., 1994).
Here we tested the hypothesis that tankyrase generated during oocyte maturation activates β-catenin-mediated transcription, which in turn is essential for EGA. Tankyrase protein levels increased significantly during maturation, confirming that Tnks is a dormant maternal mRNA. Inhibition of tankyrase activity in 1C embryos increased AXIN1 and AXIN2, reduced nuclear active β-catenin, and caused developmental arrest prior to the mid-2C stage. The arrested embryos had altered chromatin status, persistent DNA double strand breaks, significantly reduced transcription levels, particularly of ribosome components, and loss of nuclear MYC (myelocytomatosis oncogene) associated with a global failure of new protein translation. We conclude that developmentally regulated translation of a β-catenin post-translational regulator, tankyrase, serves as a ligand-independent mechanism to activate β-catenin-mediated transcription required for completion of EGA.
Results
Tankyrase activity is required for preimplantation embryo development
We first attempted to test directly whether β-catenin had a critical role in EGA using a knockdown approach with both siRNAs and morpholino oligonucleotides targeting Ctnnb1. Despite multiple attempts, we were unable to detectably decrease β-catenin levels in 2C embryos (Figure S1A). This finding was not completely unexpected because, as outlined above, β-catenin is tightly regulated at the protein level and the E-cadherin already present in oocytes and early embryos could serve as an additional source of β-catenin. We next attempted to generate β-catenin-null embryos by crossing β-catenin-floxed mice (Ctnnb1-f/f) with mice carrying the ZP3-cre transgene as previously reported (Messerschmidt et al., 2016). Surprisingly, 2C embryos derived by parthenogenetic activation of eggs from Ctnnb1-f/f; ZP3-cre females had only a 26% decrease in β-catenin relative to controls (Figure S1B). Blastocyst stage parthenotes had a normal appearance and most (11/14; 79%) had β-catenin levels similar to controls, but a few (3/13; 21%) had no or minimally detectable protein (Figure S1C). Oocytes from Ctnnb1-f/f; ZP3-cre females had only a ~50% decrease in Ctnnb1 mRNA, including exons that should have been excised (Figure S1D). These findings suggested that deletion efficiency of the floxed allele was low, resulting in residual stored mRNA or expressed protein.
The above results led us to test whether we could knock down β-catenin levels by increasing activity of the destruction complex. An in silico examination of microarray data on the destruction complex components and its modulators revealed that Tnks and Tnks2 transcripts were present in oocytes, though Tnks was more abundant (Pan et al., 2008). Furthermore, Tnks, like Ctnnb1, was apparently upregulated during oocyte maturation, suggesting it was a dormant maternal mRNA (Pan et al., 2008). Of note, mice singly deficient in either TNKS or TNKS2 are viable and fertile, but deficiency in both TNKS and TNKS2 results in embryonic lethality by E10 (Chiang et al., 2008). However, based on the use of EIIa-Cre to delete the floxed alleles, these double knockout embryos would have had normal levels of maternally-derived TNKS protein.
Tnks and Tnks2 mRNAs were detected in oocytes and were either stable or apparently increased in 1C embryos (Figures S2A and S2B). To determine if either tankyrase was transcribed during EGA we tested whether inhibition of transcription using α-amanitin impacted mRNA levels. This treatment did not affect Tnks, but reduced Tnks2 mRNA by ~50% (Figures S2C and S2D), indicating that Tnks is solely maternally derived whereas Tnks2 is also transcribed from the embryonic genome. Immunoblot analysis using an antibody that recognizes both tankyrase proteins (Smith et al., 1998) showed that TNKS was present at low levels in germinal vesicle-intact (GV) oocytes but was recruited for translation and/or stabilized during oocyte maturation such that the amount in metaphase II (MII) eggs was increased by ~2.5-fold (Figures 1A and S2E). TNKS levels remained relatively stable after MII up to the 2C stage, whereas TNKS2 was not detected until the late 2C stage. Interestingly, the band detected for TNKS at the MII stage was slightly higher in apparent molecular weight compared to that in 2C embryos. Consistent with previous observations in somatic cells (Ha et al., 2012), this size shift was due to TNKS phosphorylation (Figure S2F). Because TNKS phosphorylation is associated with increased stability and poly-ADP ribosylation activity (Ha et al., 2012), these findings are consistent with the hypothesis that TNKS activity is important in 1C embryos.
Figure 1. Tankyrase activity is essential for preimplantation embryo development.
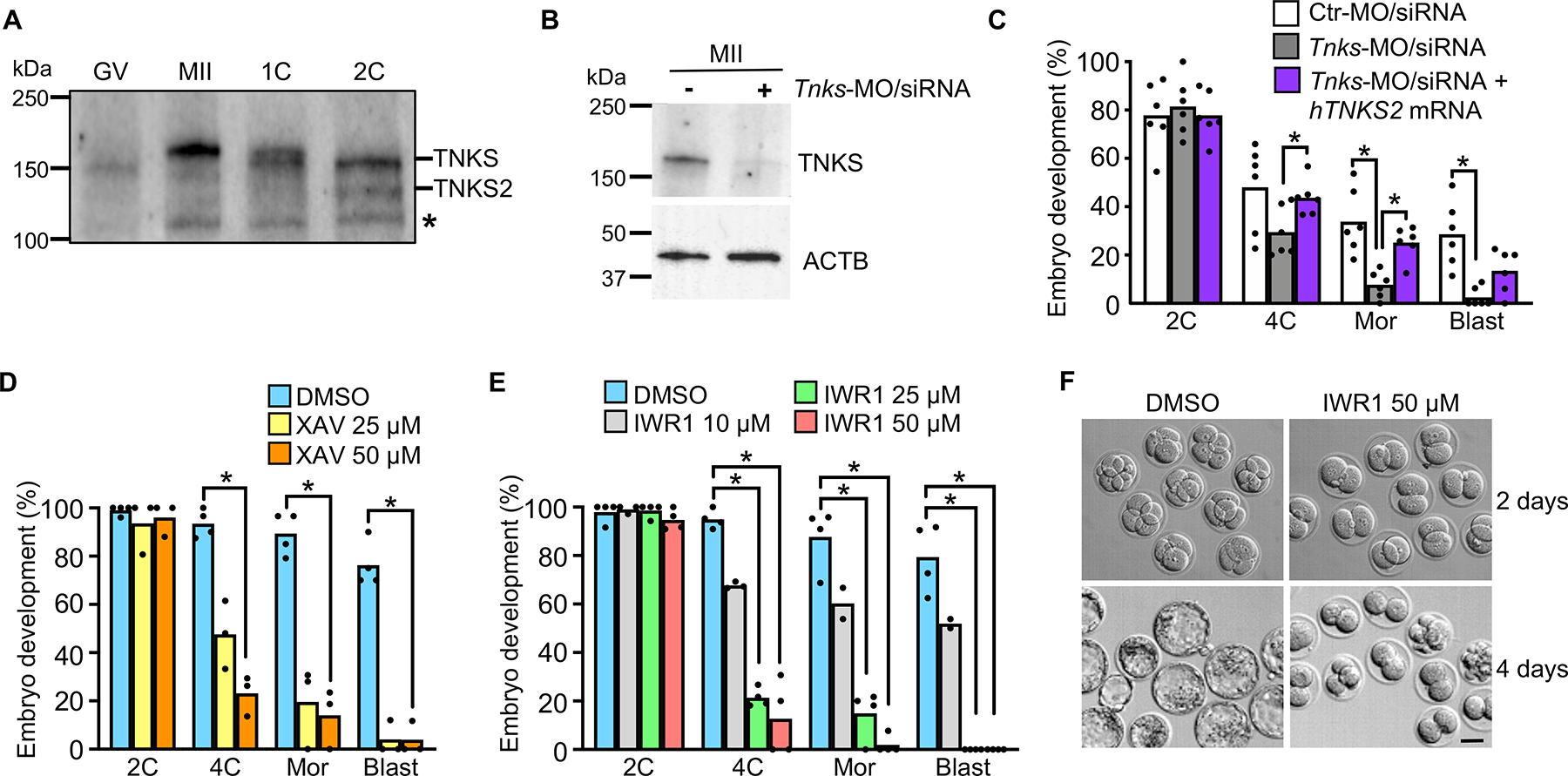
(A) Immunoblot of TNKS and TNKS2. N=3; 50 oocytes/eggs/embryos per lane. Asterisk indicates non-specific band. (B) Immunoblot analysis of TNKS in eggs following microinjection at the GV stage using the indicated combination of morpholino (MO) and siRNA (upper panel). β-actin, loading control (lower panel). N=2; 58 eggs/lane in representative blot shown. (C) Percentage of fertilized embryos to reach specified stages following microinjection at the GV stage with the indicated morpholino/siRNA/mRNA combination. N=6, 21–67 1C embryos/group/replicate; *p<0.05, ANOVA with Dunnett’s. (D) Percentage of 1C embryos to reach specified stages following culture with XAV939. N=4; 16–38 1C embryos/group/replicate; *p<0.05, Kruskal-Wallis with Dunn’s. (E) Percentage of 1C embryos to reach specified stages following culture with IWR1. N=3; 16–33 1C embryos/group/replicate; *p<0.05, Kruskal-Wallis with Dunn’s. (F) Embryos at 2 days and 4 days following culture from 1C stage in DMSO or IWR1. Scale bar=40 μm. Median or mean, as appropriate, and all values shown in graphs. Abbreviations in this and all subsequent figures: GV, GV-intact oocyte; MII, MII-arrested egg; 1C, one-cell embryo; 2C, two-cell embryo; 4C, four-cell embryo; 8C, eight-cell embryo; Mor, morula; Blast, blastocyst.
See also Figures S1 and S2.
To elucidate the role of TNKS during early preimplantation embryo development we used a knockdown approach to target Tnks mRNA prior to its recruitment for translation during oocyte maturation. GV oocytes were microinjected with both siRNA and morpholino oligonucleotides targeting Tnks to improve knockdown efficiency. This approach resulted in a dramatic although not complete loss of TNKS protein by the MII stage (Figure 1B). Following in vitro fertilization, less than 30% of the TNKS-knockdown embryos cleaved to the 4C stage, <10% developed into morulae, and only rare blastocysts were observed (Figure 1C). This development success was significantly lower at the morula and blastocyst stages than that of embryos injected with Ctr-MO/siRNA. Importantly, Tnks knockdown was rescued to a large extent by co-injection of human TNKS2 mRNA, which would not be targeted by the Tnks-MO/siRNA (Figure 1C).
To confirm these results, we used two different small molecule inhibitors that are selective for tankyrase relative to most other poly(ADP-ribose) polymerases (PARPs). XAV939 inhibits tankyrase activity by interfering with the nicotinamide-binding active site, but it also inhibits other PARPs (Karlberg et al., 2010; Thorsell et al., 2017). IWR1-endo (IWR1) induces structural changes in tankyrases without binding the active site and lacks inhibitory activity toward other PARPs (Karlberg et al., 2010; Narwal et al., 2012; Thorsell et al., 2017). As expected, 1C stage embryos cultured in either inhibitor had increased TNKS levels by the late 2C stage due to a lack of auto-ADP(ribosyl)ation by TNKS (Figure S2G). XAV939-treated embryos arrested beginning at the 2C stage, and the degree of arrest was proportional to the inhibitor concentration (Figure 1D). Similar to XAV939, IWR1 had a concentration-dependent effect on embryo development (Figure 1E). Culture of embryos in 50 μM IWR1-exo, an IWR1 diastereomer with ~10-fold lower activity (Chen et al., 2009; Huang et al., 2009), had no impact on embryo development (Figure S2H). Other than arrest in development, neither inhibitor caused a morphological abnormality in embryo appearance until the embryos had been arrested for a prolonged period (Figure 1F). Because of the specificity of IWR1 for tankyrase inhibition, further experiments were done with IWR1 and not XAV939. Inhibition of tankyrase activity beginning in the late 2C stage did not impact embryo development until the morula stage, and later initiation of treatment with IWR1 had even less impact on development (Figure S2I). Because TNKS2 is not present until the 2C stage, the inhibitor-induced arrest in development at the 2C stage observed following inhibitor treatment beginning at the 1C stage must be mediated by TNKS, not TNKS2. Taken together, these findings indicate that TNKS function is essential during early phases of EGA and that TNKS and/or TNKS2 activities are essential following midpreimplantation gene activation at the 4C-stage.
WNT signaling pathway components mediate effects of tankyrase inhibition
Both total and active (non-phosphorylated) β-catenin are detected in mouse 1C and 2C-stage embryos (De Vries et al., 2004; Xie et al., 2008), but there was no detailed analysis of active β-catenin in the different pronuclear (PN) stages (Adenot et al., 1997). We reasoned that if β-catenin transcriptional activity were involved in the first (minor) wave of EGA, then localization of active β-catenin in the PNs should correlate with transcriptional activation. Active β-catenin localized to both male and female PN beginning at pronuclear stage PN2 (Figure 2A). There was a subsequent accumulation of active β-catenin in both PN over time, but the male PN had significantly higher levels of β-catenin relative to the female PN beginning at PN3. This finding is consistent with the higher transcription activity in the male PN relative to the female PN (Aoki et al., 1997), and supports the idea that β-catenin could mediate transcriptional activation of genes required during minor EGA.
Figure 2. WNT signaling components mediate tankyrase inhibition effects.
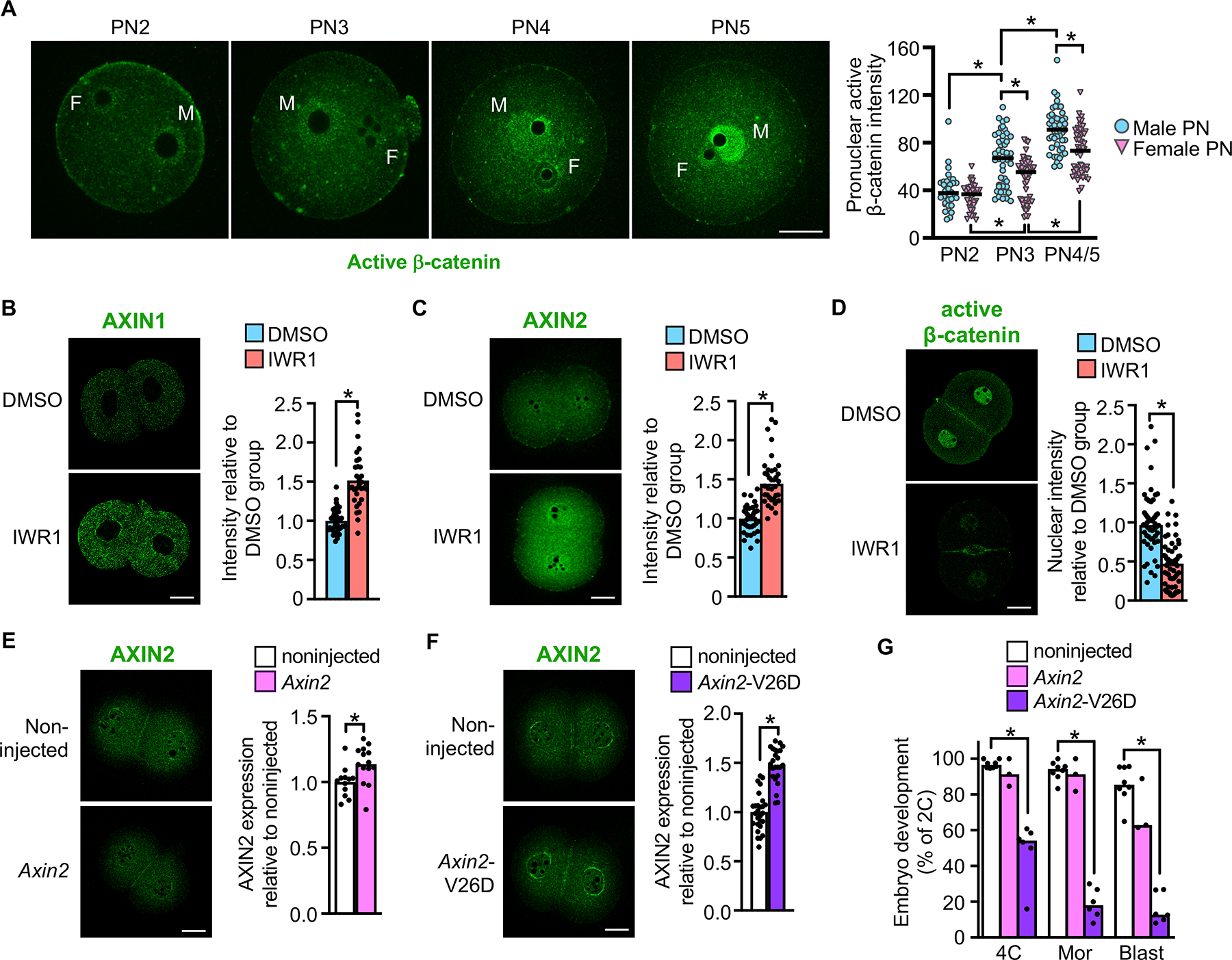
(A) Active β-catenin staining across pronuclear (PN) stages and quantification. F, female PN; M, male PN. N=3; 9–24 embryos/group/replicate; *p<0.05, Kruskal-Wallis test with Dunn’s. (B) AXIN1 in late 2C embryos and quantification. N=3; 32–33 embryos/group; *p<0.05, t-test. (C) AXIN2 in late 2C embryos and quantification. N=3; 36–38 embryos/group; *p<0.05, Mann Whitney test. (D) Active β-catenin in late 2C embryos and quantification. N=4; 52–59 embryos/group; *p<0.05, Mann Whitney test. (E) AXIN2 in late 2C embryos following Axin2 mRNA microinjection at 1C-stage and quantification. N=3; 12–14 embryos/group. p<0.05, t-test. (F) AXIN2 in late 2C embryos following Axin2-V26D mRNA microinjection at the 1C-stage and quantification. N=3; 24–27 embryos/group. p<0.05, t-test. (G) Percentage of embryos to reach the various preimplantation embryo stages following microinjection at the 1C-stage with the indicated mRNA. N=4; 30–54 1C embryos/group/replicate; *p<0.05, ANOVA with Dunnett’s. Scale bars=20 μm. Median or mean, as appropriate, and all values shown in graphs.
Tankyrase inhibitors impair β-catenin signaling by preventing tankyrase-mediated poly(ADP-ribosyl)ation of axin proteins. Stabilization of these key components of the β-catenin destruction complex causes a reduction in total β-catenin protein levels and, consequently, a decrease in active β-catenin translocation into the nucleus. To determine if axin stabilization could explain our results, we tested whether tankyrase inhibition using IWR1 altered axin and β-catenin levels in 2C embryos. AXIN1 was detected in the cytoplasm and AXIN2 was detected in both the nucleus and cytoplasm of 2C embryos (Figures 2B and 2C). The levels of both proteins increased in response to IWR1 treatment. Active β-catenin was detected near the plasma membrane and in nuclei of 2C embryos, but its level in nuclei was reduced by ~60% in IWR1-treated embryos (Figure 2D).
We next tested whether axin upregulation could explain the observed embryo arrest phenotype. Early 1C embryos were microinjected with mRNA encoding AXIN2, but AXIN2 protein levels in 2C embryos could barely be distinguished from non-injected controls (Figure 2E). AXIN2 can be stabilized by mutating valine 26 of the tankyrase-binding domain to an aspartate residue, which prevents tankyrase-mediated poly(ADP-ribosyl)ation and thereby slows protein turnover (Qian et al., 2011). Microinjection of mRNA encoding AXIN2-V26D into early 1C embryos caused a small but significant increase in AXIN2 expression at the 2C stage (Figure 2F). When these embryos were cultured, there was a ~50% reduction in the success of cleavage to the 4C stage, and relatively few embryos formed morulae or blastocysts (Figure 2G). In contrast, microinjection of mRNA encoding wild type AXIN2 did not impact embryo development. These data indicate that AXIN2 overexpression impairs preimplantation development, and that tankyrase activity is required to strictly regulate axin protein levels to allow progression of early embryo development.
Tankyrase inhibition is associated with alterations in chromatin accessibility
Extensive chromatin remodeling occurs during the oocyte-to-embryo transition, including global DNA demethylation, alterations in histone modifications, and replacement of specific histones with variant forms that together alter the open chromatin landscape (Lin et al., 2014; Schultz et al., 2018). To determine if tankyrase inhibition impaired embryo development due to a failure of chromatin reprogramming we used ATAC-seq (Buenrostro et al., 2013) to profile open chromatin in late 2C embryos treated with IWR1 or vehicle control. Correlation between replicates using genome-wide bins was 0.87–0.90, indicating high reproducibility (Figure S3A). To identify differentially open chromatin regions, we used DESeq2 to analyze the set of 65,535 high confidence regions using a stringent cutoff of false discovery rate (FDR)<0.05. This analysis identified 1409 regions that were significantly less open in IWR1 vs. control (IWR1-low) and 2345 regions that were significantly more open in IWR1 vs. control (IWR1-high) (Figure 3A). These differential regions (n=3754) corresponded to only 5.7% of all high-confidence ATAC-seq peaks (Figure 3B), indicating that the majority of the open chromatin landscape in IWR1-treated and control embryos was similar. Given the large reduction in β-catenin in nuclei of IWR1-treated embryos (Figure 2D), these findings suggest that the global open chromatin state is driven mainly by mechanisms unrelated to β-catenin.
Figure 3. Tankyrase activity is necessary to generate the appropriate chromatin status for EGA.
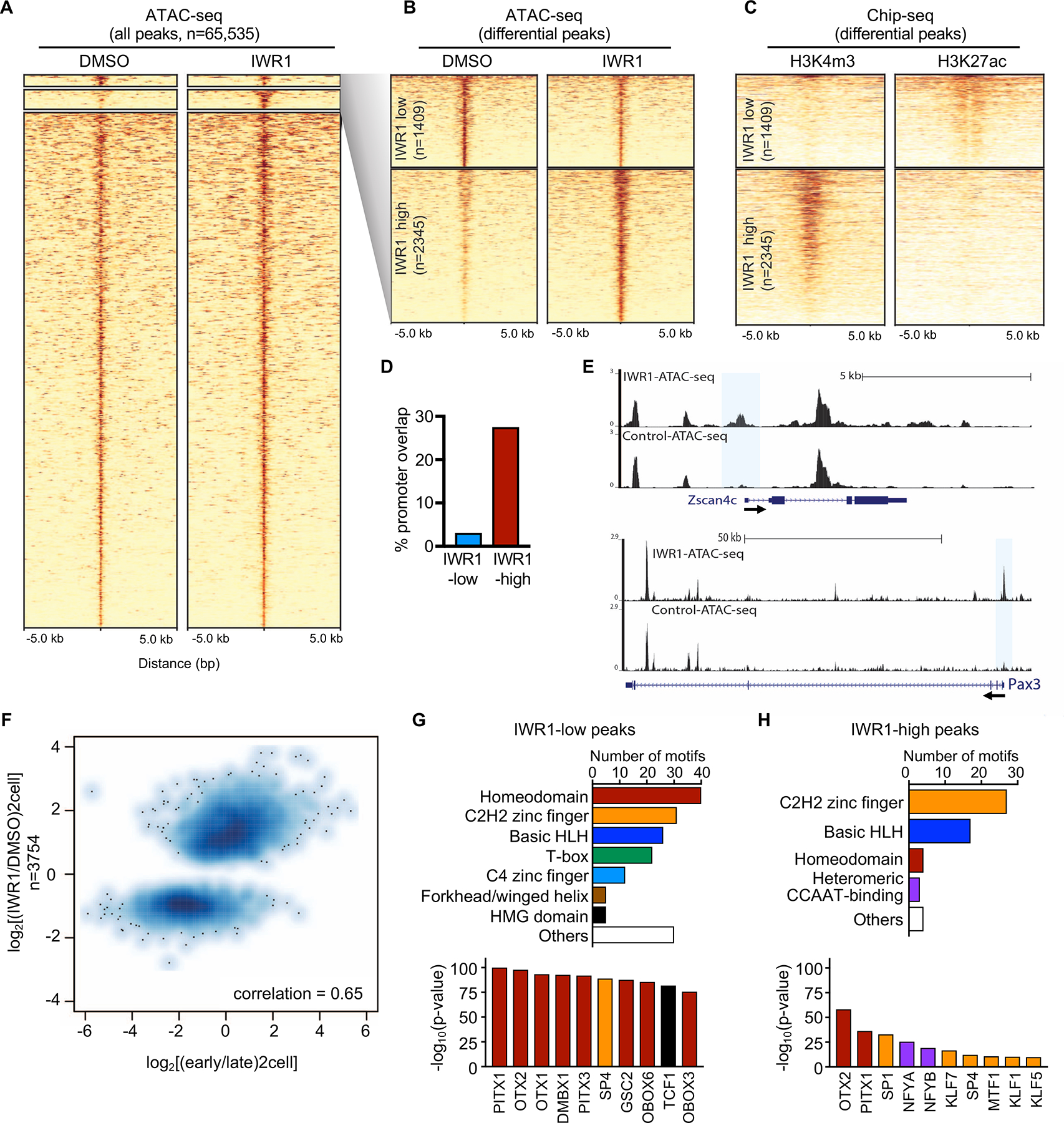
(A) Heatmap of ATAC-seq of 2C stage embryos treated with IWR1 or DMSO beginning at the 1C stage (22–23 hrs post-hCG) and cultured until the late 2C stage (48 hrs post-hCG). High confidence peaks (n=65,535) filtered for differential ATAC-seq signal. (B) Heatmap of differential ATAC-seq peaks (expanded from A) showing IWR1-low and IWR1-high peaks at FDR<0.05 using DESeq2. (C) Heatmap of ChIP-seq data from (Dahl et al., 2016) using late 2C-stage mouse embryos. Left, H3K4me3 ChIP-seq; Right: H3K27ac ChIP-seq. (D) Percentage of differential ATAC-seq peaks that overlap with promoter regions. (E) Genome browser snapshots showing ATAC-seq peaks in IWR1 or DMSO treated 2C embryos for EGA genes Zscan4c and Pax3. Differential open promoters highlighted by blue boxes. (F) Scatterplot of ATAC-seq IWR1 vs. DMSO differential peaks compared to early vs. late 2C ATAC-seq from (Wu et al., 2016). Strong positive correlation = 0.65. (G,H) Motif analysis of IWR1-low (G) and IWR1-high (H) peak regions. Upper panels: Total number of significantly enriched motifs in the indicated transcription factor family. Lower panels: Most significantly enriched motifs. Bars color-coded to match motif families in upper panels.
See also Figure S3.
To better characterize the IWR1 differential open chromatin regions, we compared our ATAC-seq data to H3K27ac and H3K4me3 ChIP-seq data from late 2C embryos (Dahl et al., 2016). IWR1-low sites co-occurred with H3K27ac+/H3K4me3- regions whereas IWR1-high sites co-occurred at H3K4me3+/H3K27ac- regions (Figure 3C). Based on these two histone post-translational modification datasets, we predicted that IWR1-low sites were non-promoter active regulatory regions (putative distal regulatory elements), whereas the IWR1-high sites were promoters. Consistent with these predictions, IWR1-low regions showed only ~3% overlap with an annotated TSS whereas IWR1-high regions showed ~28% overlap with an annotated TSS (Figures 3D and S3B). Inspection of open chromatin maps revealed promoters at genes such as Zscan4c and Pax3, which exhibit IWR1-high promoter open chromatin (Figure 3E). These loci can be contrasted with the Myc gene locus, which has no significant differences in open chromatin (Figure S3C).
Early 2C (G1 or early S) mouse embryo chromatin is characterized by an initial EGA heavily biased to retrotransposons of the MERVL long terminal repeat (LTR) class and other genes such as Dux and Zscan4 (Hendrickson et al., 2017; Wu et al., 2016). To test if there was a relationship between differential open chromatin regions in IWR1-treated embryos and those in early or late 2C embryos, we compared the ATAC-seq data to published open chromatin maps from these embryo stages (Wu et al., 2016). Interestingly, there was a moderate positive correlation (0.65) between IWR1-differential open chromatin regions and early vs. late 2C stage chromatin (Figure 3F). Specifically, open chromatin regions enriched in early vs. late 2C embryos was also classified as IWR1-high chromatin whereas IWR1-low chromatin had higher late/early open chromatin ratios. These comparisons indicate that IWR1-treated embryos retain a subset of early 2C open chromatin (biased towards proximal promoter regions) but fail to activate a subset of late 2C open chromatin at distal putative enhancer regions.
To test if IWR1-low chromatin was driven by DNA sequence-specific transcription factors that could explain the failure to undergo EGA properly, we performed a motif analysis. Several high-scoring motifs were recovered from IWR1-low chromatin that corresponded to a diverse set of transcription factors strongly enriched for homeodomain motifs (Figure 3G and Table S1). In addition, high mobility group (HMG)-box motifs including TCF and LEF were highly enriched, consistent with a role for β-catenin in EGA. Motif analysis of IWR1-high peaks also showed enrichment for homeodomains, but in addition there was high enrichment for SP1 and NFYA/B (Figure 3H and Table S1). These motifs are consistent with the presence of SP1 in pronuclei of late 1C and 2C embryos (Worrad et al., 1994; Worrad and Schultz, 1997) and the role for NFYA in EGA (Lu et al., 2016).
Tankyrase inhibition arrests development prior to the mid-2C stage
To determine how tankyrase inhibition altered embryo gene expression profiles we performed RNA-seq on control and IWR1-treated embryos (Table S2). There was high reproducibility between biological replicates (Figure 4A). After restricting the analysis to transcripts with RPKM>1 in at least one sample and adjusted p≤0.01, there were 2779 down- and 2918 upregulated transcripts. Altered transcript expression by RNA-seq was highly and directly correlated with the altered peaks identified by ATAC-seq (Figure 4B and Table S3). We took advantage of an existing RNA-seq dataset that included early and late 2C embryos (Wu et al., 2016) to further characterize the IWR1-treated embryos. Transcripts were sorted into three groups: significantly different only in the early vs. late 2C embryos, significantly different only in the IWR1 vs. DMSO 2C embryos, or significantly different in both datasets. Hierarchical clustering clearly demonstrated that the IWR1-treated 2C embryos were far more similar to early 2C embryos; as expected, the DMSO-treated embryos were very similar to late 2C embryos (Figure 4C).
Figure 4. Tankyrase activity is required for EGA to progress to the mid-2C stage.
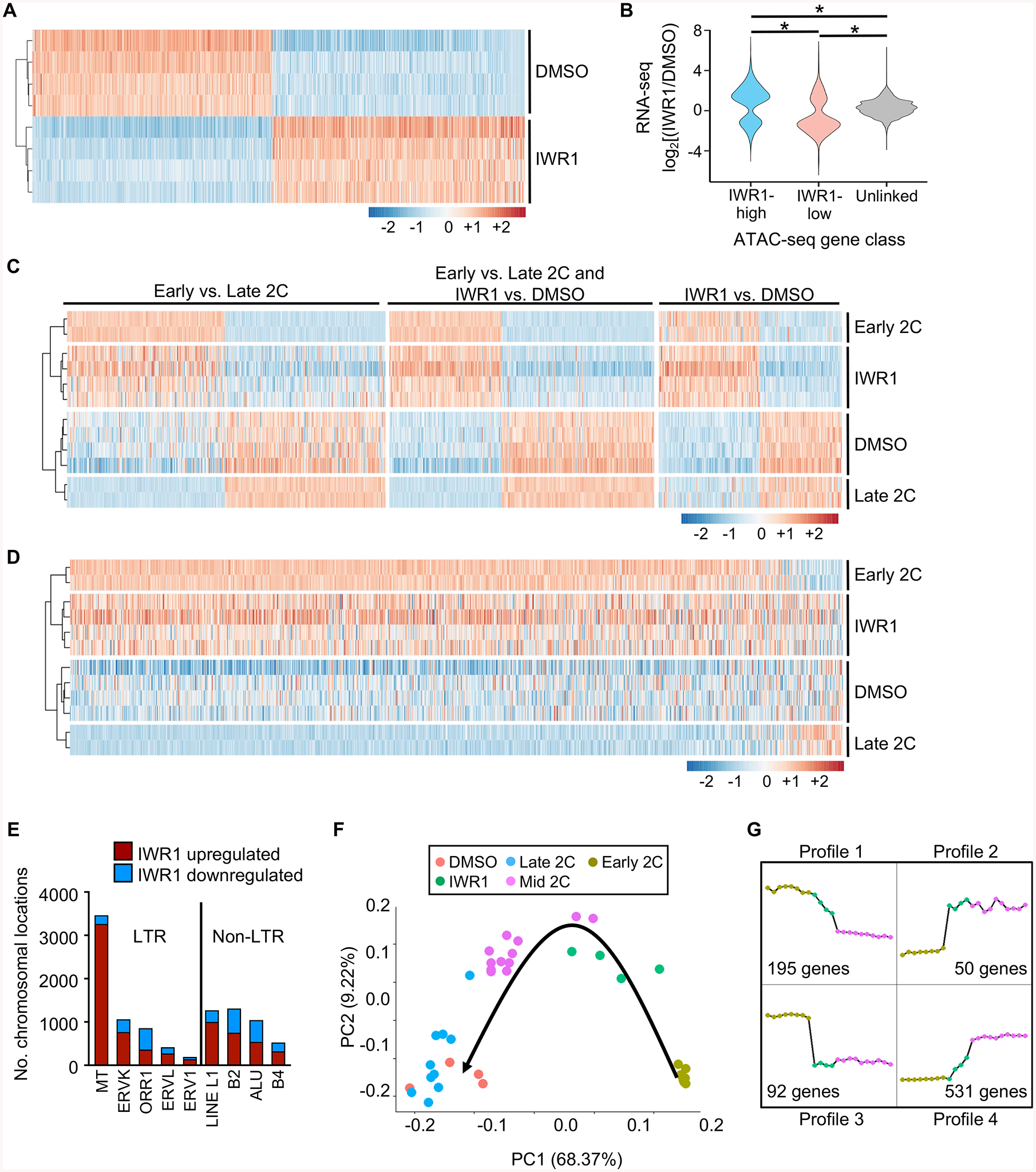
(A) Differentially expressed (DE) genes sorted by log2fc between DMSO and IWR1, clustered by Euclidean distance. 5697 DE genes; p_adj<0.01 and |log2fc|>1. Z-scores plotted. (B) Violin plot showing correlation between ATAC-seq gene class and RNA-seq gene expression levels. Expression ratio (IWR1/DMSO) of genes within 100 kb of a differential ATAC-seq peak (DESeq2 FDR<0.05) compared to each other and to unlinked genes. *p≤2.2E-16, one-sided Wilcoxon-Mann-Whitney test. (C) Comparison of gene expression differences between earlyand late 2C embryos and IWR1 and DMSO treated embryos. Left set DE (p_adj<0.01, |log2fc|>1) only between early and late 2C embryos. Middle set DE in both early/late and IWR1/DMSO comparisons. Right set DE only between IWR1/DMSO. Early/late 2C data sets from (Wu et al., 2016). Clustered by Euclidean distance prior to conversion to z-scores. Plotted values are z-scores that were computed separately for the two experiments – IWR1/DMSO and early/late 2C. (D) Heatmap of gene expression differences between the early and late 2C embryos and IWR1 and DMSO treated embryos of transcripts degraded ≥8-fold between oocyte and 2C stages (Xue et al., 2013). Genes ordered by log2fc in comparison between early and late 2C data sets. Clustered by Euclidean distance prior to z-score conversion. Plotted values are z-scores, which were computed separately for the two experiments – IWR1/DMSO and early/late. (E) Number of chromosomal regions up- or downregulated for each repetitive element. (F) Principal component analysis of single cell RNA-seq data from mouse embryos in the early, mid, and late 2C stages (Deng et al., 2014) and DMSO/IWR1-treated 2C embryos. Black arrow, progression of time in development. (G) Gene expression profiles generated from RNA-seq datasets from early and mid 2C embryos (Deng et al., 2014), and IWR1-treated 2C embryos. Profiles derived from all genes common to all 3 datasets. Number of genes represents the number present in the set of genes differentially expressed between early and mid 2C embryos that displayed the indicated profile. Graph legend shown in (F).
A critical process in early embryo development is targeted degradation of maternal transcripts (Ma et al., 2013). We used a published dataset (Xue et al., 2013) to identify maternal transcripts that are normally degraded at least 8-fold between the oocyte and 2C stages. Comparison of these transcripts in the early and late 2C embryos (Wu et al., 2016) with IWR1-treated and control 2C embryos demonstrated that, like the early 2C embryos, IWR1-treated embryos had not completed degradation of maternal transcripts (Figure 4D). Consistent with this finding was that the highly abundant class of mouse transcript (MT) retrotransposons, which are actively degraded during and following oocyte maturation, were highly upregulated in the IWR1-treated 2C embryos relative to controls (Figure 4E). Most remaining categories of retrotransposons were also more often upregulated than downregulated, but the difference was not as dramatic. These findings indicate that the IWR1-treated embryos have an open chromatin state and transcript expression pattern strongly resembling that of early 2C-stage embryos.
To determine more accurately the timing of developmental arrest of the IWR1-treated embryos, we compared our IWR1 and DMSO RNA-seq data to published single cell RNA-seq data from mouse embryos in the early, mid, and late 2C stages (Deng et al., 2014). All genes with differential expression detected in one of the three comparisons between early, mid, and late 2C stages were selected, and then principal component (PC) analysis was employed to compare the five data sets. As expected, this analysis demonstrated that the DMSO controls clustered tightly with late 2C embryos (Figure 4F). In contrast, the IWR1-treated 2C embryos clustered in between the early and mid 2C embryos, except for the presence of two cells from a single mid 2C embryo that clustered with the IWR1 group. Because PC1 differences correlated with time, it is likely that these two outliers were from an embryo at a developmentally earlier stage than the remainder of the mid 2C embryos. Consistent with the idea that major EGA was initiated to some degree in the IWR1 embryos was that Dux expression was repressed, similar to the DMSO embryos, yet its downstream target Zscan4 was upregulated (Table S2). Dux repression occurred despite a significant decrease in expression of mRNAs encoding both nucleolin and TRIM28 (Table S2), which together with LINE1 RNA suppress Dux expression (Percharde et al., 2018). Taken together, these findings indicate that IWR1-treated embryos arrest soon after the initiation of major EGA at the 2C stage. To determine the transcripts that were the primary drivers of the differences between early, IWR1-treated, and mid 2C embryos we performed an analysis of transcript expression patterns of all genes identified in both early and mid 2C embryos, regardless of differential expression status, using EPIG software (Table S4) (Chou et al., 2007). This analysis revealed six distinct profiles, of which only four contained genes that were also differentially expressed between the early and mid 2C embryos (Figure 4G). Profiles 1 and 3 would be expected of maternal transcripts or 1C embryo transcripts that are degradedin the early to mid 2C stage. Indeed, over 50% of the differentially expressed transcripts in Profiles 1 and 3 were also identified as degraded maternal transcripts in (Xue et al., 2013). Profiles 2 and 4 reflect transcripts whose expression was activated in the very early 2C stage. These transcripts represent a unique dataset of early 2C EGA transcripts.
To determine the overall cause of embryo developmental arrest following tankyrase inhibition, we assessed DNA replication, transcriptional activity, and histone modifications (Figure 5A). Following S-phase of the second cell cycle, there was no detectable difference in DNA synthesis between IWR1-treated and control 2C-stage embryos. However, there was a large reduction in transcription, suggesting an almost complete failure of EGA. Furthermore, although histone H4 acetylation was no different in the treated embryos, histone H3 acetylation was reduced by ~40%. A large reduction in acetylation of H3 lysine 27 (H3K27ac) was responsible in part for the overall decrease in H3 acetylation. Because H3K27ac is tightly associated with transcriptional activation and active enhancers, this finding is consistent with EGA failure.
Figure 5. Tankyrase activity is necessary for embryonic transcription, chromatin modifications and DNA repair.
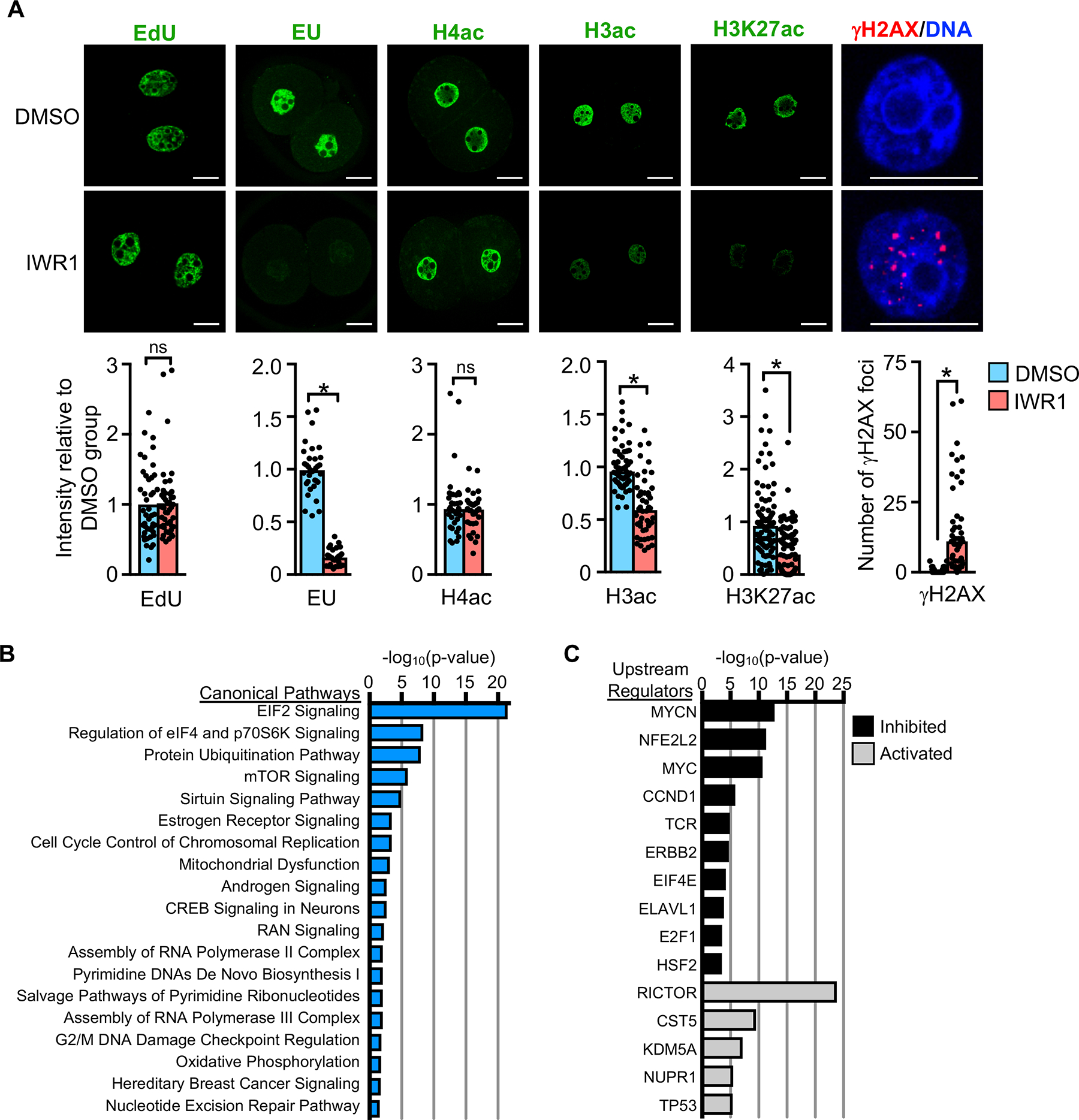
(A) Nuclear staining in 2C embryos following culture in DMSO or IWR1. EdU, indicator of DNA replication levels; N=3, 23–24 embryos/group. EU, indicator of global transcription activity; N=3, 28–34 embryos/group; *p<0.05, t-test. H4ac, histone H4 acetylation; N=2, 18–20 embryos/group. H3ac, histone H3 acetylation; N=3, 24–27 embryos/group; *p<0.05, t-test. H3K27ac, histone H3 lysine 27 acetylation; N=4, 30–35 embryos/group; *p<0.05, t-test. γH2AX, histone H2AX phosphorylation as measure of DNA double-strand breaks. DNA counterstained with DAPI, merged image shown. N=3, 20–25 embryos/group; *p<0.05, t-test. Graphs show mean and all values. Scale bars=20 μm. (B,C) Ingenuity® Pathway Analysis. (B) Highly significant canonical pathways of differentially expressed genes in DMSO vs. IWR1 embryos. (C) Highly significant upstream regulators of differentially expressed genes in DMSO vs. IWR1 embryos.
See also Figure S4.
Previous studies have implicated poly(ADP-ribosyl)ation via PARP family proteins, possibly including tankyrase, as an essential step in DNA repair (De Vos et al., 2012). For this reason, we also examined the 2C embryos for evidence of double strand DNA breaks using γH2AX staining. IWR1-treated embryos had a dramatic increase in nuclear γH2AX foci relative to controls, in which γH2AX foci were rarely detected (Figure 5A). Taken together, these results indicate that embryos lacking tankyrase activity have normal DNA replication but do not effectively repair DNA damage, and initiate but largely fail to undergo major EGA.
Ingenuity Pathway Analysis on the differentially expressed genes in the IWR1 and DMSO embryo RNA-seq data revealed highly significant enrichment for several canonical pathways regulating protein translation, including EIF2 signaling, regulation of eIF4 and p70S6K signaling, protein ubiquitination, and mTOR signaling (Figure 5B and Table S5). There was also enrichment for DNA repair pathways, which was not surprising given the persistent DNA double strand breaks. Upstream regulator analysis, which identifies molecules upstream of the dataset genes that could explain the observed expression changes, identified activation of RICTOR and inhibition of MYCN and MYC as highly significant (Figure 5C and Table S6). Consistent with these findings, genome set enrichment analysis (GSEA) (Subramanian et al., 2005), indicated that the hallmark gene sets significantly (FDR<0.05) downregulated in the IWR1 group relative to DMSO included unfolded protein response, mTORC1 signaling, and MYC targets (Figure S4 and Table S7). Both IWR1-high and IWR1-low chromatin from our ATAC-seq analysis were enriched for basic helix-loop-helix motifs, which include the consensus motifs for MYC and its binding partner MAX (Figure 3G and 3H and Table S1). However, the motif enrichment for these factors was far more significant for the IWR1-low chromatin (Table S1). These findings suggest that tankyrase activity is essential for proper induction of transcriptional responses required to initiate MYC-mediated ribosomal biogenesis and assembly and protein translation in the 2C embryo.
Tankyrase activity is required for new protein translation
Based on the RNA-seq analysis, we next tested whether overall protein synthesis was affected in the IWR1-treated embryos. Newly translated proteins were detected by incorporation of O-propargyl-puromycin (OPP) (Liu et al., 2012). Control embryos had small cytoplasmic puncta and large amounts of protein accumulation in nuclei, particularly in nucleoli, likely representing newly translated ribosomal proteins (Figure 6A). In contrast, protein synthesis was barely detectable in IWR1-treated embryos. A central mechanism for translational control involves phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α), which represses the initiation phase of protein synthesis. We found that eIF2α-P was much more abundant in IWR1-treated 2C embryos (increased 2.9-fold ± 0.4), indicating that protein synthesis was repressed in response to tankyrase inhibition (Figure 6B).
Figure 6. Tankyrase and β-catenin support generation of MYC and global protein translation necessary for embryo development.
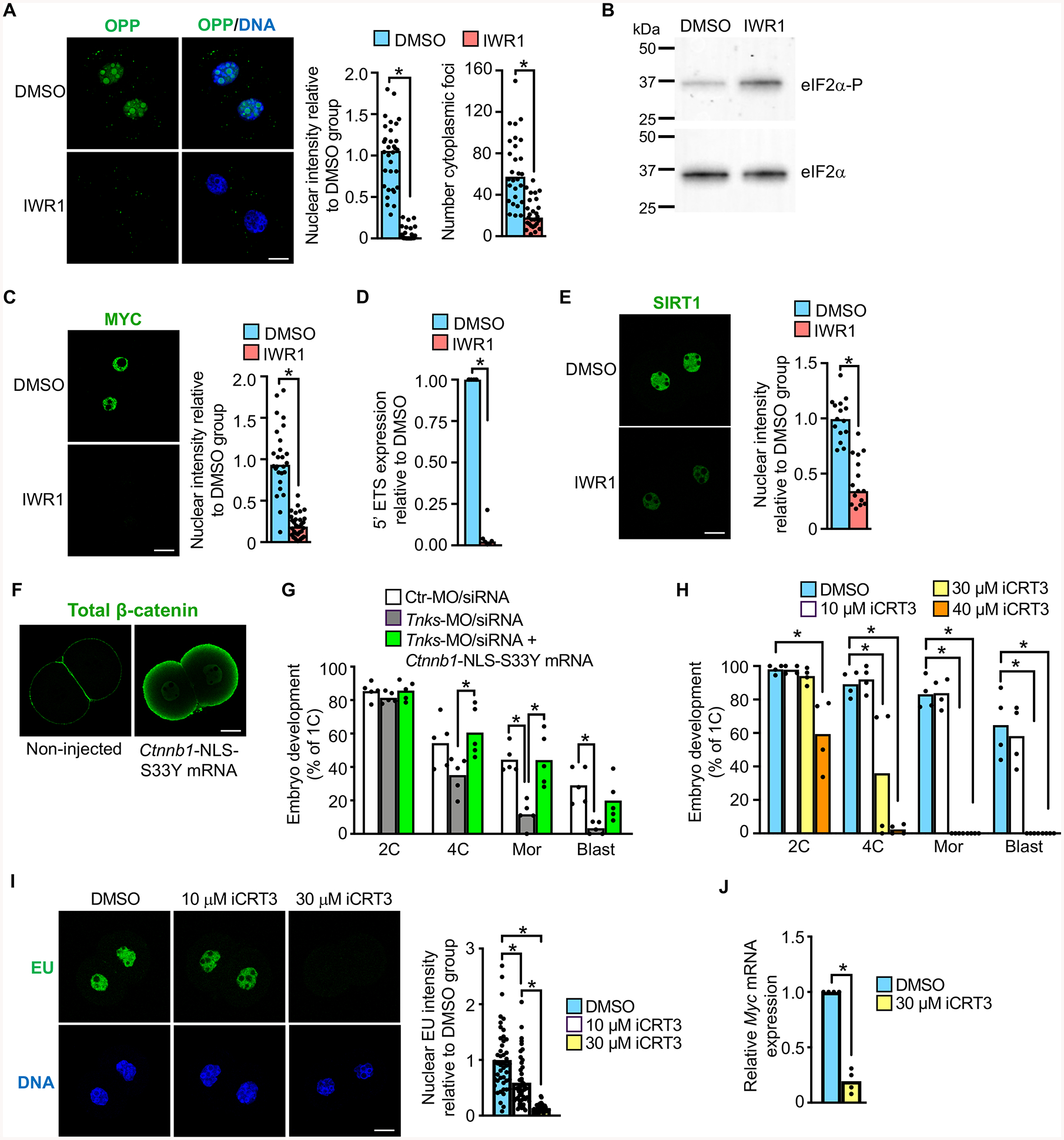
(A) OPP staining as indicator of newly translated protein in DMSO or IWR1-treated late 2C embryos. Graphs show nuclear intensity and total number of cytoplasmic foci/group. *p<0.05, t-test; N=3, 28–32 embryos/group. (B) Immunoblot of eIF2α and eIF2α-P in DMSO or IWR1-treated late 2C embryos; 100 embryos/lane. Blot representative of N=3 replicates. (C) MYC immunofluorescence and relative intensity in DMSO or IWR1-treated late 2C embryos. *p<0.05, t-test; N=3, 26–33 embryos/group. (D) Relative expression of 5′ external transcribed spacer (5′ ETS) in DMSO or IWR1-treated late 2C embryos. *p<0.05, Mann-Whitney test; N=6. (E) SIRT1 immunofluorescence and relative intensity in DMSO or IWR1-treated late 2C embryos. *p<0.05, t-test; N=3, 15 embryos/group. (F) Total β-catenin immunofluorescence in late 2C embryos that were either not injected or microinjected at the 1C-stage with Ctnnb1-NLS-S33Y mRNA. (G) Percentage of 1C embryos to reach specified stages following microinjection at the GV-stage with the indicated MO/siRNA ± mRNA. N=5, 19–47 1C embryos/group/replicate; *p<0.05, ANOVA with Dunnett’s. (H) Percentage of 1C embryos to reach specified stages following culture with DMSO or iCRT3. N=4; 21–44 1C embryos/group/; *p<0.05, Kruskal-Wallis with Dunn’s. (I) EU staining and quantification in 2C embryos cultured in DMSO or iCRT3. Upper panels, EU; lower panels, DNA. N=3, 46–47 embryos/group; *p<0.05, Kruskal-Wallis with Dunn’s. (J) Relative Myc expression in 2C embryos cultured in DMSO or iCRT3. N=4, *p<0.05, Kruskal Wallis with Dunn’s. Median or mean, as appropriate, and all values shown in graphs.
See also Figures S5 and S6.
There were three obvious candidate mediators of the observed transcription and translation arrest in response to tankyrase inhibition. First, PI3K (phosphatidylinositol 3-kinase) signaling via PDK1 (pyruvate dehydrogenase kinase, isoenzyme 1) has an essential role in supporting EGA (Halet et al., 2008; Zheng et al., 2010). PI3K signaling can be inhibited by tankyrase through post-translational modulation of PTEN (phosphatase and tensin homolog deleted on chromosome ten) levels (Li et al., 2015). However, PTEN levels were unchanged in IWR1-treated 2C embryos (Figure S5A). Second, the transcription factor YAP1 (yes-associated protein 1) is essential for EGA in the mouse because it promotes transcription of genes necessary for protein translation (Yu et al., 2016). TNKS alters nuclear localization of YAP1 by promoting degradation of angiomotin family proteins, which sequester YAP1 in the cytoplasm (Wang et al., 2015). However, there was no difference in the total amount of YAP1 protein or in nuclear localized YAP1 in IWR1-treated 2C embryos (Figure S5B), indicating that altered YAP1 localization does not contribute to the impaired protein translation. This finding is not surprising because published microarray data indicates that none of the angiomotin family proteins are expressed in mouse embryos until after the 2C stage (Zeng and Schultz, 2005). Third, MYC was previously suggested to be a critical regulator of genes essential for development beyond the 2C stage, particularly due to its role in supporting protein synthesis, and it is detected in nuclei of early preimplantation embryos (Suzuki et al., 2009; Zeng and Schultz, 2005). Furthermore, Myc is a direct target of transcriptional activation by β-catenin and, hence, a likely target for tankyrase inhibition (He et al., 1998). Myc was downregulated ~4.5-fold in IWR1-treated 2C embryos relative to controls in our RNA-seq data set (Table S2). MYC nuclear protein levels were also dramatically reduced following IWR1 treatment (Figure 6C). Consistent with this finding, transcription of ribosomal DNA to generate the 45S rRNA precursor needed for ribosome synthesis was barely detectable as indicated by relative expression of the 5′ external transcribed spacer (Eichler and Craig, 1994) (Figure 6D). MYC also regulates transcription of the NAD+-dependent protein deacetylase SIRT1, which is highly expressed in 2C embryos (Tang et al., 2017; Yuan et al., 2009). Sirt1 mRNA was downregulated ~4.6-fold and nuclear SIRT1 protein was significantly reduced in IWR1-treated 2C embryos (Table S2 and Figure 6E). Taken together, these findings suggest that loss of MYC is a major contributor to the observed arrest of protein translation in response to tankyrase inhibition.
β-catenin functions downstream of tankyrase to promote EGA
To determine if tankyrase activity was required for embryo development solely because of its impact on axin/β-catenin signaling, we tested whether overexpression of nuclear active β-catenin could rescue tankyrase inhibition by IWR1. We first generated Ctnnb1-S33Y mRNA, which encodes a degradation-resistant form of β-catenin (Morin et al., 1997), but microinjection of this mRNA into 1C embryos did not increase nuclear active β-catenin in 2C embryos. We next added a canonical nuclear localization signal (NLS) to the mutant construct. Microinjection of Ctnnb1-NLS-S33Y mRNA into 1C embryos resulted in an observable increase in nuclear β-catenin at the 2C stage, although much of the protein localized to the cortex (Figure 6F). However, overexpression of nuclear β-catenin did not overcome embryo developmental arrest in response to IWR1 treatment, suggesting that tankyrase had roles in addition to modulating β-catenin signaling.
To determine if β-catenin was, instead, a significant downstream modulator of tankyrase action, we tested whether overexpression of nuclear active β-catenin could rescue embryo development after tankyrase knockdown using the MO/siRNA approach, which does not completely eliminate tankyrase protein (Figure 1B). GV oocytes were microinjected with Ctr-MO/siRNA, Tnks-MO/siRNA, or Tnks-MO/siRNA + Ctnnb1-NLS-S33Y mRNA. The oocytes were matured, fertilized in vitro, and evaluated for embryo development. Impaired development to the 4C stage and beyond in Tnks KD embryos was completely rescued to control levels by overexpression of nuclear active β-catenin (Figure 6G).
To test the importance of nuclear β-catenin in EGA and embryo development using alternative methods, we used two different inhibitors that block direct interactions between β-catenin and TCF transcription factors, iCRT14 and iCRT3 (Gonsalves et al., 2011). iCRT14 completely prevented embryo development beyond the PN stage and caused significant decreases in both Myc expression and new protein synthesis (Figure S6). Although iCRT14 effectively blocks β-catenin-TCF interactions, it may also directly impact TCF-DNA binding, an activity not observed for iCRT3 (Gonsalves et al., 2011). One-cell embryos cultured in iCRT3 had a concentration-dependent failure of development beyond the 2C stage (Figure 6H). These embryos did not undergo EGA as indicated by EU staining, and had minimal Myc transcripts (Figure 6I and 6J). Together, these findings indicate that β-catenin is a critical downstream target of tankyrase during early preimplantation embryo development and that β-catenin has an essential role in EGA.
Discussion
The data reported here demonstrate that TNKS activity is essential for driving completion of EGA in the mouse. Maternally generated Tnks mRNA is translated and the protein stabilized through phosphorylation during oocyte maturation. TNKS-mediated ADP-ribosylation of axin leads to its degradation, increasing nuclear active β-catenin and promoting β-catenin-dependent Myc transcription, as well as transcription of additional genes required for key aspects of the oocyte-to-embryo transition. β-catenin may have additional chromatin-regulatory activities independent of transcriptional activation, such as binding chromatin at non-promoter sites as is observed in Xenopus embryos (Nakamura et al., 2016). Chromatin profiling shows that inhibiting TNKS activity does not completely block early aspects of major EGA, but remarkably prevents decommissioning of early 2C chromatin and opening of late 2C chromatin, which is reflected in the transcriptome (Figures 3 and 4). Furthermore, MYC stimulates ribosome biogenesis and thereby synthesis of proteins required to carry out the developmental program (summarized in Figure 7). Our data, combined with a previous report that tankyrase function is essential for mouse post-implantation embryonic development (Chiang et al., 2008), indicate that tankyrase activity is essential both immediately following fertilization and following implantation.
Figure 7. Working model of tankyrase-mediated ligand-independent β-catenin activation resulting in MYC production to support embryo development.
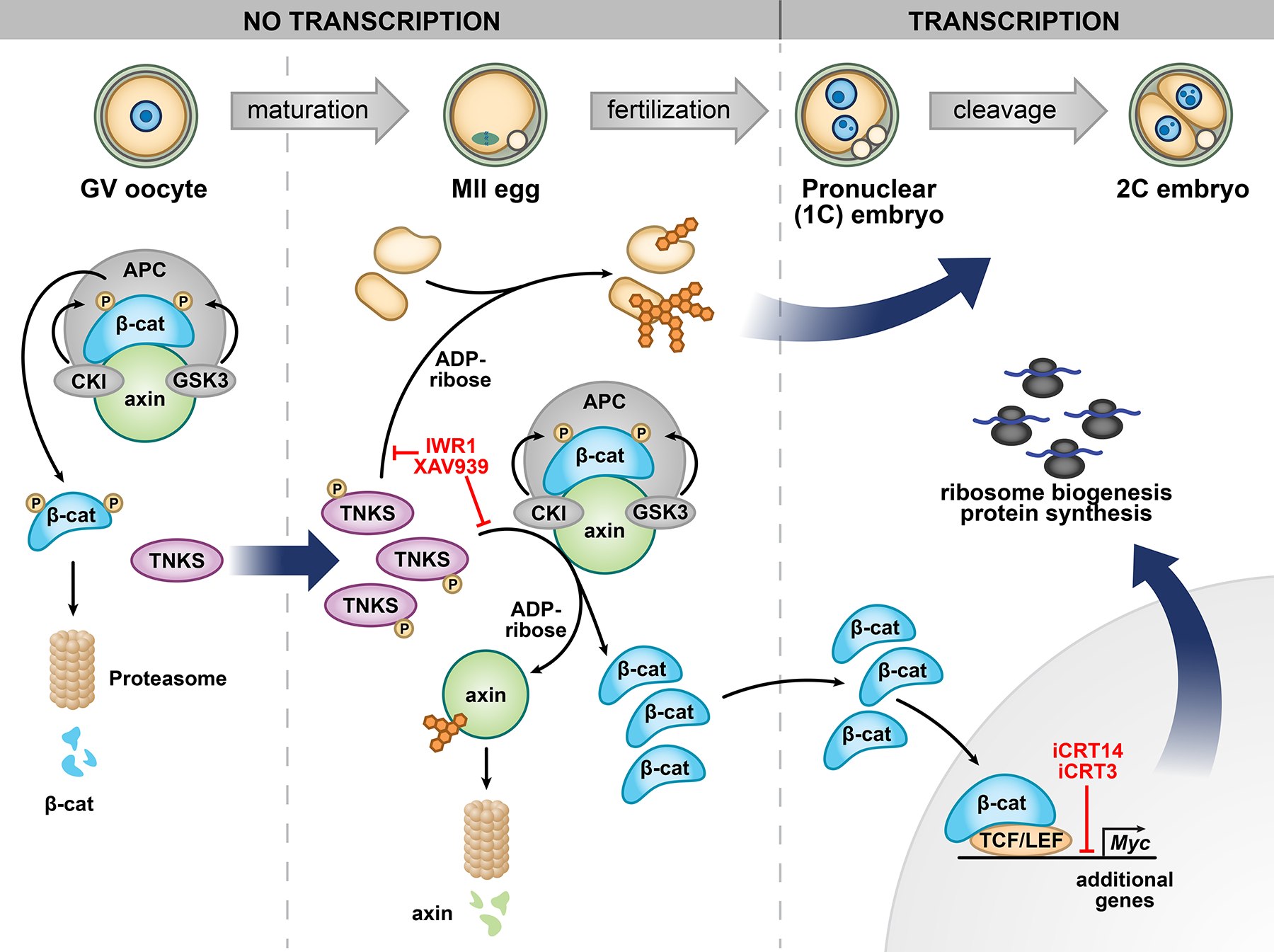
See Discussion for full model description. Sites of action of inhibitors are indicated. TNKS, tankyrase 1; APC, adenomatosis polyposis coli; β-cat, β-catenin; CKI, casein kinase I; GSK3, glycogen synthase kinase 3; TCF/LEF, T-cell factor/lymphoid enhancer-binding factor proteins.
Determination of the role of canonical WNT signaling in mammalian preimplantation embryo development has been challenging. The abnormal development of blastocyst stage mouse embryos having reduced levels of β-catenin indicates, however, that it does have an essential role (Messerschmidt et al., 2016). Our findings that (1) TNKS inhibition increased axin, reduced nuclear active β-catenin, and impaired EGA and embryo development and (2) inhibition of β-catenin-TCF interactions similarly affected EGA and embryo development provide compelling evidence that sufficient nuclear β-catenin activity is essential at the 2C stage. Rescue of the TNKS knockdown phenotype by overexpression of β-catenin indicates that the Wnt signaling pathway is a major target of TNKS action.
TNKS inhibition could also impact β-catenin-independent pathways necessary for embryo development. For example, TNKS inhibition could contribute to the embryo arrest phenotype by causing direct axin-mediated suppression of Myc transcription or MYC degradation (Arnold et al., 2009; Rennoll et al., 2014). Our observation that DUX repression occurred despite decreased expression of nucleolin and TRIM28, which are required to repress DUX (Percharde et al., 2018), suggests that tankyrase could regulate additional factors that influence DUX function. Other roles have been reported for TNKS that could in theory impact preimplantation embryo development, including regulation of YAP1 localization, PTEN levels, spindle function, telomere elongation, and telomere cohesion (Chang et al., 2005; Cook et al., 2002; Kim and Smith, 2014; Li et al., 2015; Smith et al., 1998; Troilo et al., 2016). We found that PTEN levels and YAP1 levels and localization were unchanged following TNKS inhibition, and it was shown previously that embryos lacking both TNKS and TNKS2 have normal telomere length (Chiang et al., 2008). The TNKS inhibition-mediated 2C stage embryo arrest phenotype occurs prior to formation of the mitotic spindle, and TNKS inhibition beginning at the 4C stage did not impact cleavage to morula (Figure S2I), indicating that abnormal spindle function or cell cycle misregulation cannot explain the phenotype. However, TNKS ADP(ribosyl)ates telomeric repeat binding factor 1; this modification interferes with its ability to bind telomeric DNA, promoting telomere resolution (Kim and Smith, 2014; Smith et al., 1998). Persistent telomere cohesion that eventually resolved but led to significant DNA damage could prevent future cell divisions; this idea is consistent with the DNA double strand breaks observed in 2C stage embryos following IWR1 treatment (Figure 5A). Taken together, these data suggest that a primary essential function of TNKS during early embryo development is to reduce axin levels, consequently activating β-catenin-mediated gene transcription, increasing MYC levels, and supporting translation of proteins essential for completion of the oocyte-to-embryo transition.
The WNT signaling pathway is a key regulator of embryo development via its impact on cell proliferation and differentiation (MacDonald et al., 2009). In zebrafish and Xenopus, WNT target genes are transcribed prior to widespread genome activation; expression of these genes from the embryonic genome is essential for embryo development (Leung et al., 2003; Skirkanich et al., 2011; Takahashi et al., 2000; Yang et al., 2002). WNT signaling is tightly regulated at multiple levels, from WNT ligand-receptor interactions to transcriptional activation/repression and post-translational modifications that alter protein localization and protein degradation. The predominant mode of activating WNT signaling during normal development is via secreted WNT ligands that bind plasma membrane receptors to initiate downstream cellular responses via β-catenin (canonical WNT signaling) or via other effectors (non-canonical WNT signaling) (Angers and Moon, 2009). However, a recent study demonstrated that in zebrafish and Xenopus, ligand-independent activation of β-catenin signaling by huluwa, which promotes tankyrase-mediated axin degradation, is required for proper embryonic patterning (Yan et al., 2018). In an interesting parallel to this work, here we identified a mechanism of WNT signaling activation during normal mammalian development that is independent of ligand-receptor interaction: endogenously programmed, post-transcriptional regulation of tankyrase protein levels during oocyte maturation that leads to nuclear localization of active β-catenin and activation of canonical WNT signaling pathways required for EGA.
STAR ★ METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carmen Williams (williamsc5@niehs.nih.gov).
Materials Availability
Plasmids generated in this study have been or will be deposited to Addgene: pGEMHE-NLS-Beta-catenin-S33Y, ID #138537; pGEMHE-TNKS-2, ID #pending; pIVT2-Axin2, ID #pending; pIVT2-Axin2-V26D, ID #pending.
Data and Code Availability
The sequencing data generated during this study are available at GEO DataSets, accession #GSE123815.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal procedures complied with National Institutes of Health animal care guidelines under an approved protocol. The following mice were used: 6–10-week old NSA (CF-1) females were obtained from Envigo and housed 5 females per cage. 2–8-month old B6SJLF1/J males were obtained from the Jackson Laboratory, were single-housed, and were used for breeding and in vitro fertilization. Ctnnb1f/f mice were purchased from the Jackson Laboratory (stock # 022775). Ctnnb1f/f females were crossed to Zp3-cre males (JAX, stock # 003651) to generate oocyte-specific Ctnnb1 conditional knockout (cKO) mice. Mice were housed in a temperature-controlled environment under a 12 h light:12 h dark cycle.
METHOD DETAILS
Egg and embryo collection
Metaphase II (MII)-arrested eggs and preimplantation embryos were collected from 6- to 10-week-old CF1 female mice. Females were injected with 7.5 IU eCG, followed 48 h later by 7.5 IU hCG. MII eggs were obtained from the oviducts 13–15 h after hCG injection. For embryo collection, after hCG administration, females were mated to B6SJLF1/J males. One-cell, two-cell, eight-cell, and blastocyst stage embryos were flushed from the oviducts/uteri of superovulated and mated mice 16–20, 44, 68 and 96-h post-hCG, respectively. Females were sacrificed by CO2 asphyxiation. The collection medium was MEM, Hepes supplemented with 0.1% PVA (MEM/PVA).
Inhibitor treatment
One-cell embryos were cultured in a humidified atmosphere of 5% CO2, 5% O2 and 90% N2 in 4-well dishes containing 500 μl of KSOM with 10, 25 or 50 μM of the tankyrase inhibitor IWR1-endo (IWR1); 50 μM IWR1-exo; 25 or 50 μM of tankyrase inhibitor XAV939; 1, 2.5, 5 or 10 μM of the β-catenin inhibitor iCRT14;; 10, 30 or 40 μM of the β-catenin inhibitor iCRT3 or the respective v/v% of vehicle (DMSO). Note that inhibitor treatments were performed in the absence of mineral oil to prevent loss of inhibitors into the oil phase. Embryos were monitored daily for 5 days and the stages of development as indicated by cell number and morphological appearance were recorded. To block zygotic transcription in late 2C embryos, 1C embryos were cultured in KSOM containing 24 μg/ml α-amanitin.
Oocyte collection, microinjection, and in vitro maturation
GV-intact oocytes were collected from female mice 46–48 h following injection with 7.5 IU eCG. Females were sacrificed by CO2 asphyxiation. Ovaries were placed in MEM/PVA containing 2.5 μM milrinone, and antral follicles were punctured using 27G needles. Cumulus cells were removed by gentle pipetting through a narrow bore capillary. Oocytes were held in MEMα containing 5% calf serum and 2.5 μM milrinone at 37°C in a humidified atmosphere of 5% CO2 in drops of pre-equilibrated medium under light mineral oil. Oocyte microinjections were performed using a Leica DMI 6000B inverted microscope equipped with a XenoWorks micromanipulator system (Sutter Instruments, Novato, CA). Oocytes were injected into the cytoplasm with 5–10 pl of morpholino oligonucleotides, siRNAs, or mRNAs. For knockdown experiments, a combination of 1 mM Tnks or control morpholino oligonucleotide and 50 μM Tnks or control siRNA pipette concentration was used. Rescue groups were injected with 1 mM Tnks morpholino, 50 μM Tnks siRNA and 1 μg/μl hTNKS2 or Ctnnb1 mRNAs. Morpholinos and siRNAs are listed in the Key Resources Table. Following microinjection, oocytes were placed back in the incubator in MEMα containing 5% calf serum and 2.5 μM milrinone for 4 h and then washed free of milrinone and in vitro matured for 16 h in MEMα containing 5% fetal calf serum.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 647 mouse anti-γH2AX (pS139) | BD Biosciences | 560447; RRID:AB_1645414 |
| Rabbit monoclonal anti-AXIN2 | Abcam | ab109307; RRID:AB_10862550 |
| Rabbit polyclonal anti-AXIN1 | Thermo Fisher | 34–5900; RRID:AB_2533178 |
| Rabbit monoclonal anti-c-MYC [Y69] | Abcam | ab32072; RRID:AB_731658 |
| Rabbit anti-TNKS1 465 antiserum | (Smith et al., 1998) | N/A |
| Rabbit polyclonal anti-histone H3K27Ac | Active Motif | 39133; RRID:AB_2561016 |
| Rabbit polyclonal anti-acetyl-histone H3 | Millipore | 06–599; RRID:AB_2115283 |
| Rabbit Anti-acetyl-histone H4 serum | Millipore | 06–866; RRID:AB_310270 |
| Mouse monoclonal anti-active-β-catenin, clone 8E7 | Millipore | 05–665; RRID:AB_309887 |
| Mouse anti-β-catenin, IgG1 | BD Biosciences | 610153; RRID:AB_397554 |
| eIF2α-S1(phospho S51) | Abcam | ab32157; RRID:AB_732117 |
| Mouse monoclonal anti-YAP1 | Santa Cruz Biotechnology Inc | sc-101199; RRID: AB_1131430 |
| Rabbit monoclonal anti-eIF2α | Cell Signaling | 5324; RRID:AB_10692650 |
| Rabbit polyclonal anti-SIRT1 | Cell Signaling | 2028; RRID:AB_1196631 |
| PTEN (D4.3) XP Rabbit mAb antibody | Cell Signaling Technology | 9188; RRID:AB_2253290 |
| Mouse anti-beta-actin monoclonal antibody, unconjugated, clone AC-15 | Sigma | A1978; RRID:AB_476692 |
| Alexa 488 goat anti-rabbit IgG | Thermo-Fisher | A-11034; RRID:AB_2576217 |
| Alexa 488 donkey anti-mouse IgG | Thermo-Fisher | A-21202; RRID:AB_141607 |
| Peroxidase-conjugated donkey anti-rabbit IgG | Jackson ImmunoResearch | 711-035-152; RRID:AB_10015282 |
| Peroxidase-conjugated goat anti-mouse IgG | Jackson ImmunoResearch | 115-035-174; RRID:AB_2338512 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| IWR1-endo (IWR1) | Sigma | 10161 |
| IWR1-exo | Cayman Chemical | 13598 |
| XAV939 | Sigma | X3004 |
| iCRT3 | Sigma | SML0211 |
| iCRT14 | Tocris | 4299 |
| DMSO | Thermo-Fisher | D12345 |
| Lambda Protein Phosphatase | NEB | P0753S |
| Cytochalasin D | Sigma | C8273 |
| α-amanitin | Sigma | A2263 |
| RNase A | Thermo-Fisher | EN0531 |
| Hyaluronidase | Sigma-Aldrich | H3884 |
| Calf serum | Atlanta Biologicals | S11495 |
| Minimal essential medium α (MEMα) | Thermo-Fisher | 3251 |
| MEM, Hepes (MEM) | Thermo-Fisher | 12360038 |
| EmbryoMax® KSOM Medium (KSOM) | MilliporeSigma | MR-106-D |
| EmbryoMax® Human Tubal Fluid (HTF) | MilliporeSigma | MR-070-D |
| AlbuMAX™ I Lipid-Rich BSA | Thermo-Fisher | 11020021 |
| Poly (vinyl alcohol), average MW 30-70,000 (PVA) | Sigma-Aldrich | P8136 |
| Equine chorionic gonadotropin (eCG) | Lee Biosolutions | 493-10 |
| Human chorionic gonadotropin (hCG) | Sigma-Aldrich | C1063 |
| IGEPAL® CA-630 | Sigma-Aldrich | I8896 |
| Dimethylformamide | Sigma-Aldrich | PHR1553 |
| Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Milrinone | Sigma | M4659 |
| Critical Commercial Assays | ||
| Click-iT™ Plus OPP Alexa Fluor™ 488 Protein Synthesis Assay Kit | Thermo-Fisher | C10456 |
| Click-iT™ EdU Alexa Fluor™ 488 Imaging Kit | Thermo-Fisher | C10337 |
| Click-iT™ RNA Alexa Fluor™ 488 Imaging Kit | Thermo-Fisher | C10329 |
| SuperSignal™ West Femto Maximum Sensitivity Substrate | Thermo-Fisher | 34095 |
| PicoPure™ RNA Isolation Kit | Applied Biosystems™ | KIT0204 |
| SYBR™ Green PCR Master Mix | Applied Biosystems™ | 4309155 |
| SMART-Seq® v4 Ultra® Low Input RNA Kit for Sequencing | Takara Bio USA, Inc. | 634888 |
| QuikChange II XL Site-Directed Mutagenesis Kit | Agilent | 200521 |
| Nextera DNA Library Preparation Kit | Illumina | FC-121-1030 |
| Nextera Index Kit (24 indexes, 96 samples) | Illumina | FC-121-1011 |
| MinElute PCR Purification Kit | Qiagen | 28004 |
| QIAprep Spin Miniprep Kit | Qiagen | 27104 |
| AmpliCap-MaxTM T 7 High Yield Message Maker Kit | CELLSCRIPT ™ | C-ACM04037 |
| Phusion High-Fidelity PCR Master Mix | NEB | M0531S |
| MEGAscript™ T7 Transcription Kit | Thermo-Fisher | AM1334 |
| MEGAclear™ Transcription Clean-Up Kit | Thermo-Fisher | AM1908 |
| Deposited Data | ||
| Raw and processed ATAC-seq and RNA-seq data | This paper | GEO: GSE123815 |
| Experimental Models: Organisms/Strains | ||
| Mice: Hsd:NSA(CF-1) females | Envigo | 033 |
| Mice: B6SJLF1/J males | Jackson Laboratory | 100012 |
| Mice: B6(Cg)- Ctnnb1<tm1Knw>/J | Jackson Laboratory | 022775 |
| Mice: C57BL/6-Tg(Zp3-cre)93Knw/J | Jackson Laboratory | 003651 |
| Oligonucleotides | ||
| TNKS-siRNA | Thermo-Fisher Silencer Select siRNAs | 4390771 Assay ID: s75316 |
| β-catenin siRNA | Thermo-Fisher Silencer Select siRNAs | 4390771 Assay ID: s63418 |
| Silencer Select Negative Control No. 1 | Thermo-Fisher Silencer Select siRNAs | 4390843 |
| TNKS morpholino (TNKS-MO): 5′-GGTCTGCTTTGCAGTGAATATCCAT-3′ | Gene Tools | N/A |
| β-catenin morpholino: CTTGAGTAGCCATTGTCCACGCAGC | Gene Tools | N/A |
| Standard Control morpholino (Ctr-MO): 5′-CCTCTTACCTCAGTTACAATTTATA-3′ | Gene Tools | N/A |
| Primers: see Supplementary Table S8 | ||
| Recombinant DNA | ||
| pIVT2-Axin2 | This paper | N/A |
| pIVT2-Axin2-V26D | This paper | N/A |
| pcDNA3-S33Y Beta-catenin | Addgene (Kolligs et al., 1999) | Cat#19286 |
| pGEMHE-mCherry-mTrim21 | Addgene (Clift et al., 2017) | Cat#105522 |
| pFLAG-TNKS-2 | Addgene (Sbodio et al., 2002) | Cat#34691 |
| pGEMHE-TNKS-2 | This paper | N/A |
| pGEMHE-NLS-Beta-catenin-S33Y | This paper | N/A |
| Software and Algorithms | ||
| Prism 7 | GraphPad Software | RRID:SCR_002798 URL: https://www.graphpad.com/ |
| ImageJ | (Schneider et al., 2012) | RRID:SCR_003070 URL: https://imagej.net/ |
| Trim Galore! | Babraham Bioinformatics | RRID:SCR_011847 URL: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Bowtie 2 (v2.2.6) | (Langmead and Salzberg, 2012) | RRID:SCR_016368 URL: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Cutadapt (v1.11) | (Martin, 2011) | RRID:SCR_011841 URL: http://code.google.com/p/cutadapt/ |
| STAR Aligner (v2.5.1b) | (Dobin et al., 2013) | https://intranet.birmingham.ac.uk/it/teams/infrastructure/research/bear/bluebear/applications/star-aligner-v2-5.1b.aspx |
| featureCounts (v1.5.0-p1) | (Liao et al., 2014) | RRID:SCR_012919 URL: http://bioinf.wehi.edu.au/featureCounts/ |
| DESeq2 | (Love et al., 2014) | RRID:SCR_015687 URL: https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| MACS2 | (Feng et al., 2012) | http://liulab.dfci.harvard.edu/MACS/ |
| Gene Set Enrichment Analysis (GSEA) | (Subramanian et al., 2005) | RRID:SCR_003199 URL: http://www.broadinstitute.org/gsea/ |
| Ingenuity® Pathway Analysis version 01-07 | Qiagen Inc. | N/A |
| Other | ||
Parthenogenetic activation
Metaphase II eggs were subjected to parthenogenetic activation by culture in Ca2+/Mg2+-free CZB medium (85.35 mM NaCl, 4.83 mM KCl, 1.18 mM KH2PO4, 0.11 mM EDTA, 25.12 mM NaHCO3, 31.3 mM Na lactate, 0.27 mM Na pyruvate, 10 μg/ml gentamicin, 10 μg/ml phenol red, 0.1% PVA) containing 10 mM SrCl2 and 2 μg/mL cytochalasin D for 4 h in a humidified atmosphere of 5% CO2, 5% O2 and 90% N2. They were then washed and cultured in KSOM in a humidified atmosphere of 5% CO2, 5% O2 and 90% N2.
In vitro fertilization (IVF)
Caudae epididymides and upper part of vas deferens were dissected from a 2–8-month-old B6SJLF1/J male, placed in the mineral oil phase of a dish containing a 500-μl drop of HTF with 4 mg/ml BSA (HTF/BSA; 3 mg/ml of AlbuMAX™ Lipid-Rich BSA was added to commercial HTF), covered in mineral oil. Using a dissecting needle, the tissue was gently pressed to release sperm, that were dragged into the HTF/BSA medium. The dish was placed in an incubator with a humidified atmosphere of 5% CO2 in air for 15 minutes. The 500-μL sperm suspension was pipetted into a clean round-bottom culture tube and 150 μL were transferred to the bottom of a round-bottom culture tube containing 850 μL of HTF/BSA to allow sperm to swim up and capacitate for 1 h. 500 μL from the top part of the suspension was removed and sperm were counted using a hemocytometer.
In vitro matured eggs were washed through 5 drops of HTF/BSA, then placed in a 250-μL drop of HTF/BSA containing 106 sperm/ml in a 60-mm petri dish covered with oil. Insemination was carried out for 4 h at 37°C in an atmosphere of 5% CO2, 5% O2 and 90% N2. Then, eggs were washed through 3 drops of HTF/BSA and 3 drops of KSOM and transferred to KSOM at 37°C in an atmosphere of 5% CO2, 5% O2 and 90% N2 for further culture.
One-cell embryo microinjection
One-cell embryo microinjections were performed using a Leica DMI 6000B inverted microscope equipped with a XenoWorks micromanipulator system and PrimeTech PMM-150FU piezo drill. One-cell embryo injections with mRNAs encoding Axin2 or Axin2V26D were completed prior to 18 h following hCG injection. Axin2 and Axin2V26D were injected at a final pipette concentration of 1.5 μg/μl. Embryos were cultured for 5 days in KSOM in a humidified atmosphere of 5% CO2, 5% O2 and 90% N2.
Cloning and in vitro mRNA production
Mouse Axin2 cDNA was made by RT-PCR of total RNA isolated from mouse vagina tissue and was cloned into pIVT vector (Igarashi et al., 2007) between the SalI and XbaI sites. Axin2V26D mutant construct was made using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). pcDNA3-S33Y β-catenin was purchased from Addgene (Kolligs et al., 1999). A FLAG tag was added to the C-terminus, a 2X nuclear localization signal was added to the N-terminus, an AgeI restriction site was added to the 5’ end, and an XbaI restriction site was added to the 3’ end of β-catenin-S33Y via PCR. pGEMHE-mCherry-Trim21 purchased from Addgene (Clift et al., 2017) was digested with AgeI and XbaI to remove the mCherry-Trim21 sequences and β-catenin-S33Y was cloned into the pGEMHE vector between the AgeI and XbaI sites. pFLAG-TNKS-2 was purchased from Addgene (Sbodio et al., 2002). An AgeI restriction site was added to the 5’ and an Xba1 site to the 3’ end of TNKS-2 via PCR. TNKS-2 was cloned into the pGEMHE vector between the AgeI and XbaI sites. For microinjection, cRNAs were made by in vitro transcription using the AmpliCap-Max T7 High Yield Message Maker Kit (CELLSCRIPT, Madison, WI).
RNA isolation and real-time RT-PCR
Fifty oocytes/eggs/embryos at different stages of development were pipetted in a microcentrifuge tube and stored at −80°C until RNA extraction. Total RNA was isolated from oocytes, eggs, or embryos using the PicoPure RNA isolation kit (KIT0204, Thermo Fisher). For the developmental profile, enhanced Gfp (eGfp) cRNA was transcribed in vitro from pIVT-eGFP (Igarashi et al., 2007) and 1 ng was added to each sample prior to RNA isolation to serve as an internal control. Real-time RT-PCR was performed as previously described (Jefferson et al., 2013), using cDNA from two oocytes/embryos per reaction. All transcripts were amplified from the same three sets of oocytes/embryos to enable comparison of CT values. Relative gene expression was calculated using the ΔCT method (Pfaffl, 2001) using either eGfp (developmental profile) or Gapdh (experiments to assay Myc and Ctnnb1 mRNA) for normalization. Amplification was carried out using an ABI Prism 7900 sequence detection system and analysis software (Applied Biosystems). Primer sequences are in the Key Resources Table.
Immunofluorescent staining, microscopy and quantification
Zona pellucidae were removed immediately prior to fixation using acid Tyrode’s solution (137 mM NaCl, 2.68 mM KCl, 1.63 mM CaCl2, 0.49 mM MgCl2, 5.55 mM glucose, 0.01 mM polyvinyl pyrrolidone, pH 2.5). Embryos were fixed for 30 min in 2.5% paraformaldehyde in PBS and then permeabilized for 20 min in 0.1% Triton X-100 in PBS. Embryos were then incubated at 4°C overnight in primary antibody (diluted 1:100 for H3K27Ac, H3ac, YAP1, H4ac, c-MYC, total β-catenin, active-β-catenin, AXIN1, and AXIN2; diluted 1:10 for anti-γH2AX; diluted 1:500 for SIRT1), washed, then incubated in Alexa Fluor® 488 anti-rabbit or anti-mouse IgG (Thermo Fisher; 1:500) for 1 h at room temperature, except for anti-γH2AX that is a conjugated antibody. All embryos were mounted in Vectashield containing 1.5 μg/mL DAPI (Vector Laboratories, Burlingame, CA) and slides were scanned using a Zeiss LSM 510 UV confocal microscope. All immunofluorescence experiments were repeated a minimum of three different times using at least 10 embryos per group.
Immunoblotting
GV oocytes, eggs, 1C and 2C embryos were collected in 5 μL of 2X Tris-glycine-SDS sample buffer including protease inhibitors, immediately frozen on dry ice and stored at −80°C. Numbers of cells run per lane are indicated in the Figure Legends. For detection of TNKS phosphorylation, half of the eggs were treated with 2000 U λ phosphatase (New England BioLabs) and incubated at 30°C for 2 h before gel loading. Samples were heated at 99°C for 5 min and loaded on a BioRad 4–15% gradient Tris-glycine gel, then transferred to low fluorescence PDVF membrane using a Trans-Blot®Transfer System™ (BioRad). Blots were briefly rinsed in TBS, blocked in 5% non-fat milk in TBST for 1 h, and incubated overnight at 4°C in primary antibody (diluted 1:1000 in blocking solution for anti-TNKS1 465 serum, EIF2α, actin; diluted 1:500 for eIF2α-S1, PTEN). The anti-TNKS1 465 serum was raised against the TNKS SAM domain, but it cross-reacts with purified TNKS2 (Susan Smith, NYU, personal communication). Blots were washed in TBST, incubated for 1 h at room temperature in peroxidase-conjugated anti-rabbit or anti-mouse IgG (Jackson ImmunoResearch) diluted 1:10,000 in 1% non-fat milk in TBST, then washed in TBST. The signal was detected with SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Scientific) and imaged using a BioRad ChemiDoc™ Touch Imaging system.
Analysis of protein synthesis, transcription, and DNA replication
Embryos were cultured in KSOM containing 10 μM 5-ethynyl-2′-deoxyuridine (EdU) or 1 mM ethynyl uridine (EU) for 1 h or 20 μM O-propargylpuromycin (OPP) for 3 h and fixed in 4% paraformaldehyde in PBS for 25 min at room temperature. Time of fixation in all cases was at 48 h after hCG administration. EdU, EU or OPP incorporation into DNA, RNA or newly translated proteins, respectively, was detected using Click-iT® EdU Alexa Fluor® 488 Imaging Kit, Click-iT® RNA Alexa Fluor® 488 Imaging Kit, or Click-iT® Plus OPP Alexa Fluor® 488 Protein Synthesis Assay Kit (Thermo-Fisher), according to the manufacturer’s protocols. Fluorescence was detected on a Zeiss LSM 510 UV confocal microscope.
RNA-sequencing and analysis
Four independent collections of 10 late 2-cell embryos/group were performed. Embryos were placed in 1 μL of PBS in a 1.5 ml tube and frozen at −80°C until library preparation. mRNA libraries were constructed using the SMART-Seq® v4 Ultra® Low Input RNA Kit (TaKaRa) following the manufacturer’s instructions. Unique barcode adapters were applied to each library. Libraries were pooled in equimolar ratio and sequenced together on an Illumina NextSeq with HighOutput flow cell and sequencing reagents. At least 45 million 76-base read pairs were generated for each individual library. Adapter and low-quality tails were removed from sequenced reads using Cutadapt (v1.11) (Martin, 2011) prior to alignment to the mm10 reference genome using STAR (v2.5.1b) (Dobin et al., 2013). RNA abundance was quantified using uniquely-mapped reads only with featureCounts (v1.5.0-p1) (Liao et al., 2014) and the RefSeq annotation downloaded from the UCSC Genome Browser database (Karolchik et al., 2014). Differential expression between experimental groups was carried out using DESeq2 (Love et al., 2014). Comparisons were restricted to DMSO treated (n=4) vs IWR treated (n=4), early 2-cell (early, n=2) vs late 2-cell (late, n=2) using data from (Wu et al., 2016), and oocyte (n=2) and 2-cell data (n=3) from (Xue et al., 2013). In each case, genes with a false discovery rate below 0.01 and a fold-change over 2 were considered as significant. Hierarchical clustering was performed using the R function hclust with Euclidean distances between variance stabilizing transformed count data. Visualization was carried out by transforming this data to z-scores within comparison sets prior to heatmap generation. Ingenuity Pathways Analysis was carried out on genes identified using DESeq2 with FPKM>1, padj<0.01 and |log fold change|≥1.
Several steps were needed to compare the current IWR1 and DMSO RNA-seq data sets to previously published 2-cell stage single-cell data (Deng et al., 2014). Z-scores used for the heatmap visualizations were also used for the IWR1 and DMSO RNA-seq data, whereas the single-cell RNA-seq data was first log-transformed and depth normalized before independently being converted to z-scores. The R function ‘prcomp’ was then used to perform a principal component analysis on these combined z-score data sets. Subsequent to this analysis, two mid-2-cell stage cells were removed from the single-cell data as outliers. The remaining z-scores were analyzed using the software EPIG (Extracting Patterns and Identifying co-expressed Genes) version 1.2 to extract patterns of co-expressed genes. EPIG was carried out with parameter settings “Min cluster size = 100; Cluster resolution (correlation) = 0.8” for pattern extraction and “SNR p-value = 0.001; r-value = 0.8” for gene categorization.
Low-Input Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq)
Two independent collections of 100 late 2-cell embryos/group were performed. Zona pellucidae were removed using acid Tyrode’s solution followed by mechanical removal of polar bodies by pipetting embryos up and down in Ca2+/Mg2+-free CZB medium. Embryos were placed in a 1.5-ml tube containing 4 μl of ice-cold 1x lysis buffer (10mM Tris-HCl, pH 7.4; 3 mM MgCl2, 0.1% IGEPAL CA630 for 15 min, followed by addition of 6 μL of 2x Tris-DMF-tagmentation buffer (20 mM Tris-HCl, pH 8.0; 10 mM MgCl2; 20% dimethylformamide). Transposase reaction was started by the addition of 2 μL of Nextera Tn5 transposase (Illumina) and incubation at 37°C for 30 min. To stop the reaction, 0.5 μL of 10% SDS was added. DNA was then purified using the Qiagen MinElute PCR purification Kit, and eluted in 22 μL of elution buffer. Amplification by 15 cycles of PCR was performed using Phusion High-Fidelity PCR Master Mix with HF Buffer and Nextera universal primer and bar code primers (Key Resources Table). PCR products were purified using columns, and fragment size was analyzed on a bioanalyzer.
To deplete mitochondrial fragments from the ATAC-seq libraries, we used the CRISPR/Cas9-assisted removal of mitochondrial DNA (CARM) procedure from (Wu et al., 2016). gRNA sequences were designed to target the entire mouse mitochondrial genome and were flanked by the following sequences: TAATACGACTCACTATAGGG – target – GGTTTTAGAGCTAGAAATA (Table S11). We then extended these oligos with CARM forward (T7_primer) and CARM reverse (gRNA_long_R_oligo) primers (Key Resources Table) to incorporate T7 primer binding sites and tacrRNA sequence. PCR products were pooled and in vitro transcribed with MEGAscript T7 Transcription kit (Thermo Fisher #AM1334) at 42°C for 18 h. In vitro transcribed products were purified using the MEGAclear Transcription Clean-Up kit (Thermo Fisher # AM1908). ATAC-seq libraries were subjected to CARM by incubation for 2 h at 37°C. The mix consisted of: 1 μg gRNA, 2 μg CAS9, ~500 ng DNA (10 μL), 10X buffer and RNase-free water. RNase A was added and the mix was incubated for 15 min at 37°C, followed by addition of stop buffer (30% glycerol, 1.2% SDS, 25 mM EDTA, pH 8 in water) and incubation at 37°C for 15 min.
ATAC-seq libraries were sequenced on an Illumina NextSeq 500 with HighOutput flow cell. At least 95 million 51-base read pairs were generated for each library. Paired-end, raw read files were first processed by Trim Galore (Babraham Institute) to trim low-quality reads and remove adapters. Processed reads were then aligned to mm10 with Bowtie2 v2.2.6 (Langmead and Salzberg, 2012) with the following parameters: (--t --q --N1 --L 25 --X 2000 --no-mixed --no-discordant). Remaining reads that mapped to mitochondria were removed from the BAM file, and reads were filtered to remove PCR duplicates. ATAC-seq peaks were called with MACS2 (Feng et al., 2012) ‘callpeak’ (--B –nomodel –nolambda --shift −100 --extsize 200 --SPMR), generating bedgraph and bigwig files. Peaks from replicates were merged into a union set. Reads from each replicate that overlapped peaks were counted and differential peaks were identified using DESeq2 with FDR < 0.05. Peaks were connected to linked genes using GREAT (McLean et al., 2010).
Differential motif enrichment analysis
IWR-low and IWR-high ATAC-seq differential regions (500 bp regions centered around the summit of the peak) were identified using DESeq2 (FDR<0.05) and were analyzed using MEME-suite (http://meme-suite.org/tools/meme-chip) to identify underlying motifs (Bailey et al., 2009). Significant transcription factors for both groups were classified according to (Wingender et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of immunofluorescence intensity was performed using ImageJ software (Schneider et al., 2012). A region of interest was drawn around each area (nucleus or whole embryo) and the average pixel intensity was determined. The values for each area were then averaged to generate an intensity level for each embryo. Intensity was expressed relative to the average intensity of the control embryos for each independent experiment. Quantification of foci for γH2AX and OPP images was performed manually. Densitometry analysis of immunoblots was performed using ImageJ software (Schneider et al., 2012). All others statistical analyses were performed using GraphPad Prism software except genome-wide data analyses which were performed using R plug-in (DESeq2). Comparisons of two groups were performed using two-tailed t-tests for normally distributed data, with Welch’s correction in case of unequal standard deviations, or Mann Whitney tests for non-parametric data. Multiple group results were analyzed using ANOVA for normally distributed data or Kruskal-Wallis test for non-parametric data. Detailed information is included in the figure legends.
Supplementary Material
Supplemental tables not included in main supplemental PDF
Table S1. Motif Analysis of ATAC-seq Peaks, Related to Figure 3.
Table S2. IWR1 vs DMSO Differential Expression Analysis, Related to Figure 4.
Table S4. Genes differentially expressed between early- and mid-2C embryos with expression patterns in EPIG profiles 1–4, Related to Figure 4.
Table S5. IPA Canonical Pathways, Related to Figure 5.
Table S6. Upstream Regulators of Gene Expression Differences in IWR1- and DMSO-treated 2C embryos, Related to Figure 5.
Table S7. Enriched HALLMARK gene sets in DMSO group relative to IWR1 group. Related to Figure 5.
Table S8. Primers used in this study. Related to STAR Methods.
Highlights.
Tankyrase protein levels increase during mouse oocyte maturation
Tankyrase promotes β-catenin activity in a ligand-independent fashion
β-catenin activates MYC-mediated ribosomal biogenesis
Tankyrase activity is essential for embryonic genome activation during the 2C stage
Acknowledgements
We thank Susan Smith (NYU) for tankyrase antibodies and Jurrien Dean, Richard Schultz, Paul Wade, and Guang Hu for manuscript critiques. This work was supported by the Intramural Research Program of the NIH, National Institutes of Environmental Health Sciences, 1ZIAES102985 (CJW) and the Howard Hughes Medical Institute (BJC). EJG was supported by the Lalor Foundation Postdoctoral Fellowship and the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the NIH (Award Number F32HD094500). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, and Aoki F (2015). The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3’ processing. EMBO J 34, 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe KI, Funaya S, Tsukioka D, Kawamura M, Suzuki Y, Suzuki MG, Schultz RM, and Aoki F (2018). Minor zygotic gene activation is essential for mouse preimplantation development. Proc Natl Acad Sci U S A 115, E6780–E6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot PG, Mercier Y, Renard JP, and Thompson EM (1997). Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124, 4615–4625. [DOI] [PubMed] [Google Scholar]
- Angers S, and Moon RT (2009). Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol 10, 468–477. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, and Schultz RM (1997). Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181, 296–307. [DOI] [PubMed] [Google Scholar]
- Arnold HK, Zhang X, Daniel CJ, Tibbitts D, Escamilla-Powers J, Farrell A, Tokarz S, Morgan C, and Sears RC (2009). The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J 28, 500–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Coughlin M, and Mitchison TJ (2005). Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol 7, 1133–1139. [DOI] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 5, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, and Zhang Y (2019). Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet 51, 947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S, and Hodes RJ (2008). Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One 3, e2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JW, Zhou T, Kaufmann WK, Paules RS, and Bushel PR (2007). Extracting gene expression patterns and identifying co-expressed genes from microarray data reveals biologically responsive processes. BMC Bioinformatics 8, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, and Schuh M (2017). A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell 171, 1692–1706 e1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, and Smith S (2002). Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Verp S, Offner S, Grun D, and Trono D (2020). DUX is a non-essential synchronizer of zygotic genome activation. Development 147, dev177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Schreiber V, and Dantzer F (2012). The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol 84, 137–146. [DOI] [PubMed] [Google Scholar]
- De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, and Knowles BB (2004). Maternal beta-catenin and E-cadherin in mouse development. Development 131, 4435–4445. [DOI] [PubMed] [Google Scholar]
- Deng Q, Ramskold D, Reinius B, and Sandberg R (2014). Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler DC, and Craig N (1994). Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 49, 197–239. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, and Liu XS (2012). Identifying ChIP-seq enrichment using MACS. Nat Protoc 7, 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S, Nagourney R, Cardozo T, Brown AM, and DasGupta R (2011). An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci U S A 108, 5954–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha GH, Kim HS, Go H, Lee H, Seimiya H, Chung DH, and Lee CW (2012). Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell Death Differ 19, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, and Kemler R (1995). Lack of beta-catenin affects mouse development at gastrulation. Development 121, 3529–3537. [DOI] [PubMed] [Google Scholar]
- Halet G, Viard P, and Carroll J (2008). Constitutive PtdIns(3,4,5)P3 synthesis promotes the development and survival of early mammalian embryos. Development 135, 425–429. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, and Kinzler KW (1998). Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, and Birchmeier W (2000). Requirement for beta-catenin in anterior-posterior axis formation in mice. J Cell Biol 148, 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsken J, Birchmeier W, and Behrens J (1994). E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J Cell Biol 127, 2061–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Knott JG, Schultz RM, and Williams CJ (2007). Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol 312, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson WN, Chevalier DM, Phelps JY, Cantor AM, Padilla-Banks E, Newbold RR, Archer TK, Kinyamu HK, and Williams CJ (2013). Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Mol Endocrinol 27, 1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg T, Markova N, Johansson I, Hammarstrom M, Schutz P, Weigelt J, and Schuler H (2010). Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor. J Med Chem 53, 5352–5355. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. (2014). The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42, D764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler R, Hierholzer A, Kanzler B, Kuppig S, Hansen K, Taketo MM, de Vries WN, Knowles BB, and Solter D (2004). Stabilization of beta-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development 131, 5817–5824. [DOI] [PubMed] [Google Scholar]
- Kim MK, and Smith S (2014). Persistent telomere cohesion triggers a prolonged anaphase. Mol Biol Cell 25, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs FT, Hu G, Dang CV, and Fearon ER (1999). Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol 19, 5696–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Bischof J, Soll I, Niessing D, Zhang D, Ma J, Jackle H, and Driever W (2003). bozozok directly represses bmp2b transcription and mediates the earliest dorsoventral asymmetry of bmp2b expression in zebrafish. Development 130, 3639–3649. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, Wang W, Songyang Z, Lin C, Yang L, et al. (2015). Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev 29, 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, and Ramalho-Santos M (2014). Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell 30, 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, and Salic A (2012). Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A 109, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, and Zhang Y (2016). Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 165, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Strnad H, Svoboda P, and Schultz RM (2013). Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod 88, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, and He X (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17, 10–12. [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, and Richter JD (2001). Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2, 521–529. [DOI] [PubMed] [Google Scholar]
- Messerschmidt D, de Vries WN, Lorthongpanich C, Balu S, Solter D, and Knowles BB (2016). beta-catenin-mediated adhesion is required for successful preimplantation mouse embryo development. Development 143, 1993–1999. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, and Kinzler KW (1997). Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275, 1787–1790. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, de Paiva Alves E, Veenstra GJ, and Hoppler S (2016). Tissue- and stage-specific Wnt target gene expression is controlled subsequent to beta-catenin recruitment to cis-regulatory modules. Development 143, 1914–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narwal M, Venkannagari H, and Lehtio L (2012). Structural basis of selective inhibition of human tankyrases. J Med Chem 55, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Pan H, Ma P, Zhu W, and Schultz RM (2008). Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol 316, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, and Ramalho-Santos M (2018). A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 174, 391–405.e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Mahaffey JP, Alcorn HL, and Anderson KV (2011). Tissue-specific roles of Axin2 in the inhibition and activation of Wnt signaling in the mouse embryo. Proc Natl Acad Sci U S A 108, 8692–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennoll SA, Konsavage WM Jr., and Yochum GS (2014). Nuclear AXIN2 represses MYC gene expression. Biochem Biophys Res Commun 443, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudloff S, and Kemler R (2012). Differential requirements for beta-catenin during mouse development. Development 139, 3711–3721. [DOI] [PubMed] [Google Scholar]
- Sbodio JI, Lodish HF, and Chi NW (2002). Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem J 361, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM, Letourneau GE, and Wassarman PM (1979). Program of early development in the mammal: changes in patterns and absolute rates of tubulin and total protein synthesis during oogenesis and early embryogenesis in the mouse. Dev Biol 68, 341–359. [DOI] [PubMed] [Google Scholar]
- Schultz RM, Stein P, and Svoboda P (2018). The oocyte-to-embryo transition in mouse: past, present, and future. Biol Reprod 99, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, and Klein PS (2011). An essential role for transcription before the MBT in Xenopus laevis. Dev Biol 357, 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, and de Lange T (1998). Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Abe K, Inoue A, and Aoki F (2009). Expression of c-MYC in nuclear speckles during mouse oocyte growth and preimplantation development. J Reprod Dev 55, 491–495. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto J, and Asashima M (2000). Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development 127, 5319–5329. [DOI] [PubMed] [Google Scholar]
- Tang S, Fang Y, Huang G, Xu X, Padilla-Banks E, Fan W, Xu Q, Sanderson SM, Foley JF, Dowdy S, et al. (2017). Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. EMBO J 36, 3175–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsell AG, Ekblad T, Karlberg T, Low M, Pinto AF, Tresaugues L, Moche M, Cohen MS, and Schuler H (2017). Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. J Med Chem 60, 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troilo A, Benson EK, Esposito D, Garibsingh RA, Reddy EP, Mungamuri SK, and Aaronson SA (2016). Angiomotin stabilization by tankyrase inhibitors antagonizes constitutive TEAD-dependent transcription and proliferation of human tumor cells with Hippo pathway core component mutations. Oncotarget 7, 28765–28782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li N, Li X, Tran MK, Han X, and Chen J (2015). Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins. Cell Rep 13, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Haubrock M, and Donitz J (2015). TFClass: a classification of human transcription factors and their rodent orthologs. Nucleic Acids Res 43, D97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrad DM, Ram PT, and Schultz RM (1994). Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development 120, 2347–2357. [DOI] [PubMed] [Google Scholar]
- Worrad DM, and Schultz RM (1997). Regulation of gene expression in the preimplantation mouse embryo: temporal and spatial patterns of expression of the transcription factor Sp1. Mol Reprod Dev 46, 268–277. [DOI] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. [DOI] [PubMed] [Google Scholar]
- Xie H, Tranguch S, Jia X, Zhang H, Das SK, Dey SK, Kuo CJ, and Wang H (2008). Inactivation of nuclear Wnt-beta-catenin signaling limits blastocyst competency for implantation. Development 135, 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Chen J, Zhu X, Sun J, Wu X, Shen W, Zhang W, Tao Q, and Meng A (2018). Maternal Huluwa dictates the embryonic body axis through beta-catenin in vertebrates. Science 362. [DOI] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, and Klein PS (2002). Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development 129, 5743–5752. [DOI] [PubMed] [Google Scholar]
- Yu C, Ji SY, Dang YJ, Sha QQ, Yuan YF, Zhou JJ, Yan LY, Qiao J, Tang F, and Fan HY (2016). Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res 26, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Minter-Dykhouse K, and Lou Z (2009). A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol 185, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, and Schultz RM (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 283, 40–57. [DOI] [PubMed] [Google Scholar]
- Zheng W, Gorre N, Shen Y, Noda T, Ogawa W, Lundin E, and Liu K (2010). Maternal phosphatidylinositol 3-kinase signalling is crucial for embryonic genome activation and preimplantation embryogenesis. EMBO Rep 11, 890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables not included in main supplemental PDF
Table S1. Motif Analysis of ATAC-seq Peaks, Related to Figure 3.
Table S2. IWR1 vs DMSO Differential Expression Analysis, Related to Figure 4.
Table S4. Genes differentially expressed between early- and mid-2C embryos with expression patterns in EPIG profiles 1–4, Related to Figure 4.
Table S5. IPA Canonical Pathways, Related to Figure 5.
Table S6. Upstream Regulators of Gene Expression Differences in IWR1- and DMSO-treated 2C embryos, Related to Figure 5.
Table S7. Enriched HALLMARK gene sets in DMSO group relative to IWR1 group. Related to Figure 5.
Table S8. Primers used in this study. Related to STAR Methods.
Data Availability Statement
The sequencing data generated during this study are available at GEO DataSets, accession #GSE123815.


