Abstract
Beilong virus (BeiV), a member of the newly recognized genus Jeilongvirus of family Paramyxoviridae, has been reported with limited geographic and host scopes, only in Hongkong, China and from two rat species. Here, by next-generation sequencing (NGS) on dominant wild small animal species in 4 provinces in China, we obtained a complete sequence of BeiV strain from Rattus norvegicus in Guangdong, neighboring HongKong, China. We then made an expanded epidemiological investigation in 11 provinces to obtain the geographic distribution and genetic features of this virus. Altogether 7168 samples from 2005 animals (1903 rodents, 100 shrews, 2 mustelidaes) that belonged to 33 species of Cricetidae, Muridae, Sciuridae and Dipodidae family of Rodentia, 3 species of Soricidae family of Soricomorpha, 2 species of Mustelidae family of Carnivora were examined by RT-PCR and sequencing. A positive rate of 3.7% (266/7168) was obtained that was detected from 22 animal species, including 5 species of Cricetidae family, 12 species of Muridae family, 2 species of Sciuridae family and 3 species of Soricidae family. Phylogenetic analyses based on 154 partial Large gene sequences grouped the current BeiV into two lineages, that were related to their geographic regions and animal hosts. Our study showed the wide distribution of BeiV in common species of wild rodents and shrews in China, highlighting the necessity of epidemiological study in wider regions.
Keywords: Jeilong virus, Beilong virus, Epidemiology, Small mammals, China
1. Introduction
The family Paramyxoviridae currently contains 78 species, organized in 17 genera (Metaavulavirus, Orthoavulavirus, Paraavulavirus, Synodonvirus, Aquaparamyxovirus, Ferlavirus, Henipavirus, Jeilongvirus, Morbillivirus, Narmovirus, Respirovirus, Salemvirus, Orthorubulavirus, Pararubulavirus,Cynoglossusvirus, Hoplichthysvirus, and Scoliodonvirus) (https://talk.ictvonline.org/). During the last decade, the discovery of many new viruses has revealed a much greater genetic diversity within family Paramyxoviridae than was previously recognized. Of special interest is the newly recognized genus Jeilongvirus, which contains 7 recognized species, namely Jun jeilongvirus (J-virus, JV) (Jack et al., 2005), Beilong jeilongvirus (Beilong virus, BeiV) (Li et al., 2006), Tailam jeilongvirus (Tailam virus, TaiV) (Woo et al., 2011), Lophuromys jeilongvirus 1 (Mount Mabu Lophuromys virus 1, MMLV-1), Lophuromys jeilongvirus 2 (Mount Mabu Lophuromys virus 2, MMLV-2), Miniopteran jeilongvirus (Shaan virus, ShaV), and Mydes jeilongvirus (Pohorje Myodes paramyxovirus 1, PMPV-1) (Vanmechelen et al., 2018), which were all identified from rodents.
Among this variety of Jeilongvirus members, BeiV was postulated to be originated from rodent, due to the previous amplification of BeiV from a rat kidney mesangial cell line (Li et al., 2006) and its close relationship to J virus and Tailam virus, both were of rodent sources. However, the epidemiological distribution and molecular evolution of BeiV remained to be rarely investigated, with epidemiology evidence only obtained from Hong Kong, China (Woo et al., 2012). In a territory-wide molecular survey that was performed in 2009 (Woo et al., 2012), BeiV was detected in 40 kidney and 9 spleen samples from 40 brown rats (Rattus norvegicus) and 3 black rats (R. rattus). In 2016, the same study group obtained the complete genome of the naturally occurring BeiV in brown and black rats, confirming their potential role as natural reservoirs of BeiV by phylogenetic analysis (Woo et al., 2016). Still it remained obscure whether BeiV was exclusively distributed in Hong Kong, China with limited host ranges. In this study, we made an expanded epidemiological investigation on various species of small mammals, including rodents, shrews and mustelidaes in eleven provinces across mainland China, to explore the epidemiological distribution and genetic features of BeiV by performing Next-generation sequencing (NGS) and RT-PCR test.
2. Materials and methods
2.1. Sample collection
From September 2013 to May 2019, wild small mammals, including rodents, shrews and mustelidaes were captured in 11 provinces (Beijing, Guangdong, Henan, Heilongjiang, Jilin, Liaoning, Inner Mongolia, Shandong, Xinjiang, Yunnan and Zhejiang) in China. The small animals were captured using snap traps and then identified by morphological features to the species level, which were further confirmed by sequencing of mitochondrial cytochrome b (mt-cyt b) gene (Nicolas et al., 2006). For each animal, at least one of the following 6 sample types was collected: heart, liver, spleen, lung, kidney and intestine content. All samples were stored at −80 °C until use.
2.2. NGS and whole genome sequencing of BeiV
The tissue samples from 5 to 10 animals of the same species and same sampling sites were pooled for metagenomic analysis by NGS. Briefly, the total RNA was extracted using an AllPrep DNA/RNA Mini Kit (Qiagen, Germany), from which rRNA was removed using MGIEasy rRNA Depletion Kit (BGI, China). A high-throughput sequencing library was constructed using an MGIEasy RNA Library Prep Kit (BGI). Viral gene libraries were then sequenced using the MGI2000 platform (BGI), sequencer with pair-end (150-bp) reads. After processing the original data by filtering adapter contamination, cutting low-quality and complexity reads, we mapped the clean reads to a host genome sequence using BWA (Version: 0.7.15). Then the remaining reads were aligned to the non-redundant bacterial, virus, fungal, and parasite databases using BWA. The MEGAHIT (v1.1.2) software was used to assemble the reads to obtain the scaffold sequence. Blast (version 2.5.0+) software was used to compare the scaffold sequence obtained after assembly to the non-redundant nucleotide database (NT) and non-redundant protein database (NR) database sequence of NCBI and extract the valid virus sequence. Specific primers were designed based on partial viral genomic sequences obtained by metagenomic analyses for PCR confirmation and whole genome sequencing of BeiV.
2.3. Reverse transcription-PCR (RT-PCR) sequencing for BeiV
Total nucleic acid was extracted using AllPrep DNA/RNA Mini Kit (Qiagen) from tissue samples and using the QIAamp viral RNA minikit (Qiagen) from intestine contents following manufacturer's instructions. The BeiV screening was performed by PCR amplification of a 440-bp fragment of the large (L) gene, located at the 3′ end of the BeiV genome and used modified primers LPW9739-F: 5′-GGAGGATTCCCTCATAGRGAA-3′ and LPW9741-R: 5′-CTCATATGTATTTACATTTAAACCA-3′ (Woo et al., 2012). PrimeScript™ One Step RT-PCR Kit was used according to the manufacturer's instructions for BeiV detection. Briefly, the PCR mix was in a volume of 25 μl containing 12.5 μl of one Step Buffer (2×), 1 μl of PrimeScript one Step Enzyme Mix, 1 μl of PCR primer mix (10 μM of sense and antisense each), total RNA 2 μl and RNase free dH2O (8.5 μl). RT-PCR was carried with one cycle of 50 °C for 30 min and 94 °C for 2 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s in PCR System 9700 (Applied Biosystems, USA). Modified primers (5′-CTTACGARATTCGRGACCT-3′ and 5′-CCCRCTGTCAKWACTCACTA-3′) (Woo et al., 2016) targeting the 521-bp fragment of the attachment glycoprotein (G) gene were used to reconfirm the positive results. The specific PCR products were all sequenced by Sanger method.
2.4. Phylogenetic analysis
Phylogenetic analysis involving all the species from family Paramyxoviridae revealed genetic diversity, which was correlated with the phylogenetic relationship between the BeiV sequences and other Paramyxoviridae species members (Table S1). The amino acid sequences of L protein from the currently detected BeiV and other representative species from family Paramyxoviridae that were downloaded from GenBank were aligned by ClustalW method using MEGA 7 (Kumar et al., 2016) with a gap opening penalty of 5 and a gap extension penalty of 1. Phylogenetic analyses based on L protein were performed using RAxML v8.2.12 (Stamatakis, 2014) with JTT as the amino acid substitution model and 1000 bootstrap replicates. Scale bars indicate protein substitutions per site, vertical bars represent the genera, and GenBank accession numbers are shown for the reference virus sequences.
To investigate the genetic feature of BeiV in relation with its host species and geographical regions, a phylogenetic tree was constructed using partial sequences (440-bp) of RdRp region of BeiV. The tree was built using the maximum likelihood method with T92 + G as nucleotide substitution model and bootstrapped with 1000 replicates. Amino acid sequences of G gene were aligned by ClustalW, and phylogenetic trees of the 42 sequences were constructed with MEGA 7.0 software using the Neighbor-Joining statistical method based on the JTT model.
2.5. Ethical issue
The study protocol was approved by the institutional review boards and the ethics committees of the Institute of the Academy of Military Medical Sciences. All animals were treated in accordance with the guidelines in the Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People's Republic of China, 1988).
2.6. Statistical analysis
The multivariate logistic regression analysis was performed to compare the detection rate of BeiV regarding the animal family, sample type, and geographic region, with 95% confidence intervals (CIs) and odds ratio (OR) estimated. The chi-square or Fisher's exact test were used for comparison of categorical variables. Statistical analyses were performed using R (version 3.5.3). All statistical tests were 2-tailed, and a significance level (P) of 0.05 was used.
2.7. Nucleotide sequence accession numbers
The BeiV sequences generated in this study were submitted to GenBank under the accession numbers MT085491 (full-length genome sequence), MT123352-MT123505 (partial L gene, Table S2) and MT649415-MT649456 (partial G gene, Table S3).
3. Results
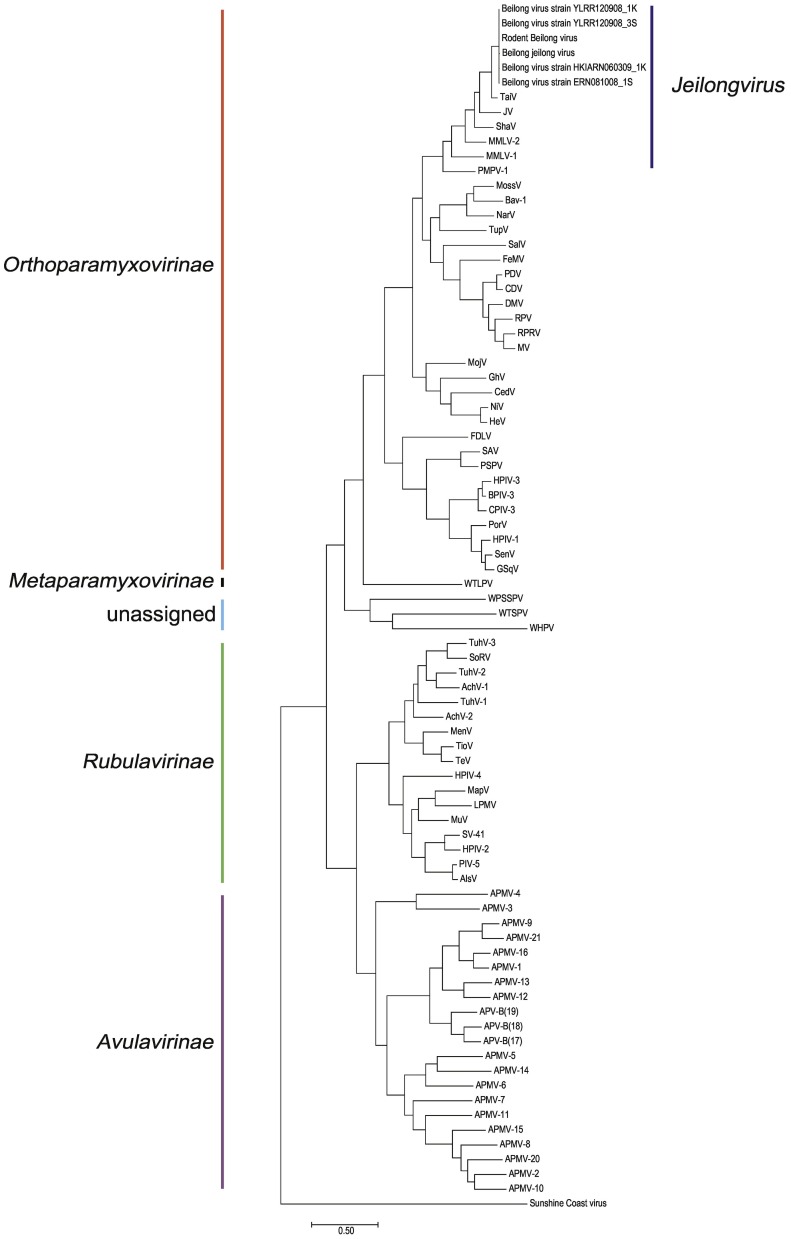
Four pooled samples that were prepared from R. norvegicus in Guangdong, R. norvegicus in Henan, Apodemus agrarius in Beijing and A. agrarius in Shandong, were subjected to metagenomic analysis by NGS. BeiV specific sequences were determined from pools in Guangdong (1043 reads), Beijing (115 reads) and Shandong (18 reads). A nearly complete sequence of BeiV was composed based on 1043 reads from the R. norvegicus samples in Guangdong. A set of 7 specific primers (Table S4) were designed based on parital viral genomic sequences obtained by NSG to fill in the gaps. Finally, we obtained the full-length genome sequence of BeiV genome (named Rodent Beilong virus) from R. norvegicus captured in Guangdong. To confirm the genome sequence of Rodent Beilong virus, we desinged 28 primer pairs (Table S5) for Sanger sequencing. The obtained consensus gene sequences were consistent with those from NGS, which were deposited in GenBank (accession number MT085491). The genome of the our rodents BeiV has 19,212 nucleotides with a G + C% content of 42.61%. Sequence comparison revealed that the current Rodent Beilong virus was highly similar to previously described BeiV that was isolated from the MHC line (GenBank accession number NC007803, with 95.21% similarity) (Li et al., 2006) and other four rodents BeiV (GenBank accession number KX940961, KX940962, KX940963 and KX940964, 97.66–98.14% similarity) that were described in HongKong, China (Woo et al., 2016). According to the phylogenetic trees based on L protein, the current rodents BeiV was clustered with previously desribed BeiV, and had a close relationship with other 6 species in genus Jeilongvirus while distinct from other members of family Paramyxoviridae (Fig. 1; Table S1). The multiple alignment showed 74.09% (TaiV), 68.93% (ShaV), 65.73% (PMPV-1), 65.34% (JV), 64.18% (MMLV-2), 60.21% (MMLV-1) similarity with other 6 species in genus Jeilongvirus.
Fig. 1.
The phylogenetic analysis of Beilong virus (BeiV) based on the complete large protein (L) sequences. Altogether 81 strains were used for the phylogenetic analysis the current BeiV, 5 previously published BeiV and 76 other Paramyxoviridae members. The tree was constructed by the Jones-Taylor-Thornton (JTT) method with 1000 bootstrap replicates and the evolutionary distance was indicated by the scale.
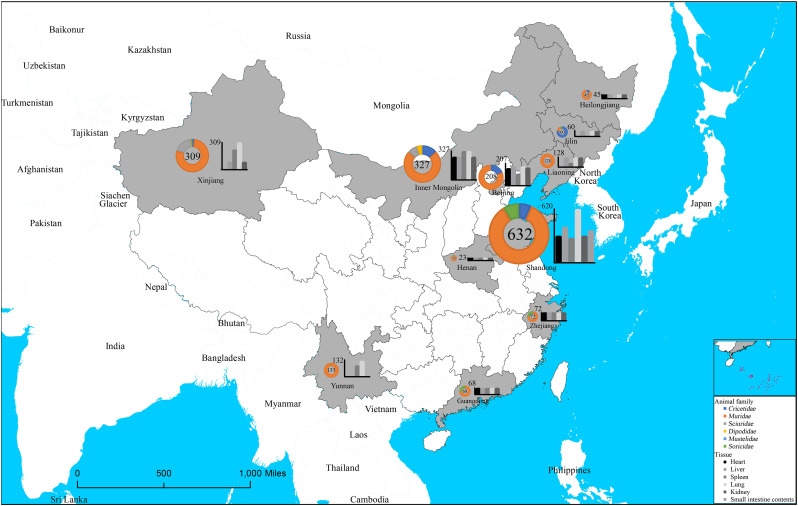
A total of 7168 samples from 2005 wild animals (1903 rodents, 100 shrews, 2 mustelidaes) that belonged to 33 species of Cricetidae, Muridae, Sciuridae and Dipodidae family of Rodentia, 3 species of Soricidae family of Soricomorpha, 2 species of Mustelidae family of Carnivora from 11 provinces across China were examined (Fig. 2 and Table S6). Six sample types were collected, including heart (n = 914), liver (n = 1356), spleen (n = 1303), lung (n = 1989), 1236 kidney (n = 1236) and small intestine content (n = 370), with two sample types tested from763 animals and three sample collected from 732 animals. An overall positive rate of 3.71% (266/7168) was obtained from the detected samples. The highest detection rate was determined from kidney (6.15%,76/1236), followed by that in spleen (5.76%, 75/1303), and much lower detection rates from liver (2.95%, 40/1356), lung (2.82%, 56/1989), heart (2.08%,19/914). No positive detection was obtained from the 370 intestinal content.
Fig. 2.
Geographical distribution of wild animals screened for Beilong virus (BeiV) in People's Republic of China, December 2003 to May 2019. Wild animals sampling (indicated in pies) and tissue (indicated in histograms) in China. The distribution map of specimens collected from eleven representative regions of China was generated by Arcgis 10.2. The size of the pie chart is in proportion with the total number of specimens collected from each region.
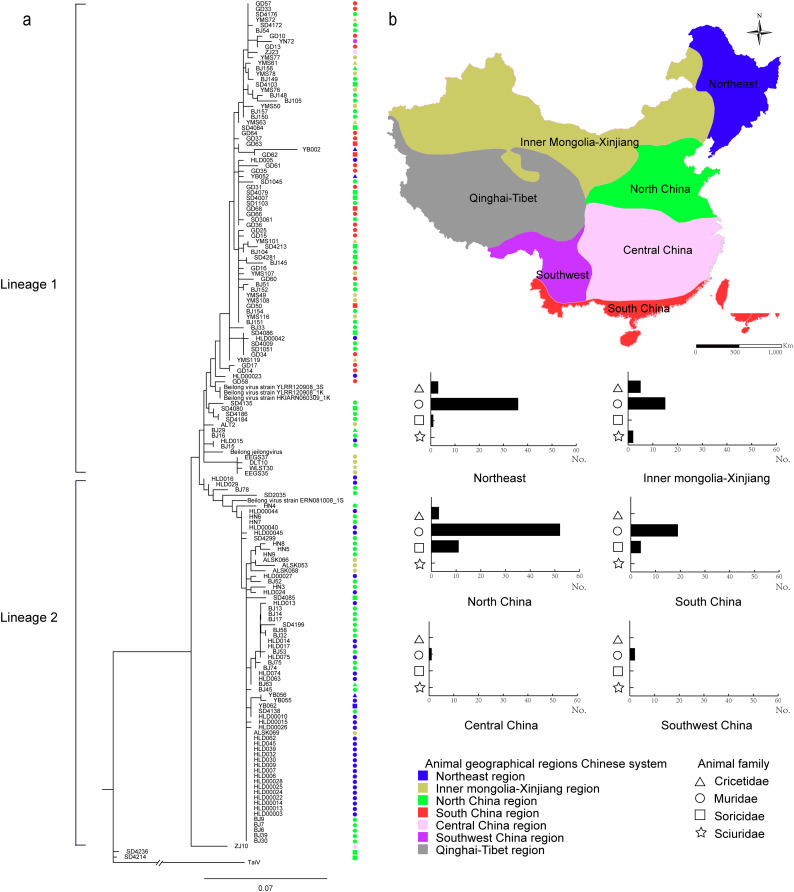
For each animal, the positive PCR result from any of the sample types was considered as positive. Thus the screening demonstrated a PCR positive rate of 7.68% (154/2005) for BeiV from the animals as a whole. Among all 38 animal species tested, 22 were positive for BeiV, including 12 Muridae, 5 Cricetidae, 2 Sciuridae and 3 Soricidae (Table S7). Four species of rodents and two sepcies of shrew had detection rate of higher than 15%, including 33.33% (1/3) in Myodes rufocanus, 28.57% (2/7) in Crocidura shantungensis, 20.83% (10/48) in Apodemus peninsulae, 17.57% (13/74) in Suncus murinus, 16.67% (1/6) in Allocricetulus eversmanni, and 15.52% (52/335) in R. norvegicus (Table S7). Both of 2 tested species of Mustelidae family were negative for BeiV. The currently surveyed 11 provinces were grouped into 6 zoogeographic regions which are defined by climatic and ecological characteristics (Xing et al., 2008), including Northeastern China (Liaoning, Jilin and Heilongjiang), Inner Mongolia-Xinjiang region, Northern China (Beijing, Henan and Shandong), Southern China (Guangdong), Central China (Zhejiang), and Southwest China (Yunnan) (Fig. 3b). The highest detection rate was observed in Southern China (33.82%, 23/68), followed by those obtained in Northeastern China (17.17%, 40/233), Northern China (7.65%, 66/863), Inner Mongolia-Xinjiang (3.46%, 22/636), Central China (2.78%, 2/72) and Southwestern China (0.76%, 1/133) (Table S5). The multivariate logistic regression analysis demonstrated that the samples obtained from Southern China, Northeastern, Inner Mongolia-Xinjiang and Northern China had significantly higher BeiV positive rate than those from the Southwestern China. The OR (95% CIs) were estimated to be 30.53 (4.12–226.28), 16.66 (2.27–122.16), 8.90 (1.23–64.68), and 7.81 (1.08–56.63), respectively. The Cricetidae species was associated with significantly higher detection rate than the other species, with OR (95% CIs) estimated to be 2.64 (1.08–6.46). The spleen and kidney had significantly higher detection rate than the other sample types (OR = 3.08, 95% CIs: 1.83–5.18, and OR = 2.73, 95% CIs: 1.62–4.60, respectively) (Table 1).
Fig. 3.
(a) Phylogenetic tree based on the partial L gene sequences of Beilong virus (BeiV). Phylogenetic tree of partial (440-bp) of L gene sequence from 154 BeiV sequence was reconstructed using the maximum-likelihood method. The bootstrap values of 1000 replicates were on each node; (b) Zoogeographic regions in China and sample host family distribution in each zoogeographic region.
Table 1.
Multiple logistic regression analysis on the Beilong virus detection rates.
| Variable | OR | 95% CIs | P value |
|---|---|---|---|
| Geographic region | |||
| Southwest China | Reference | Reference | |
| Southern China | 30.53 | 4.12–226.28 | < 0.001 |
| Northeastern China | 16.66 | 2.27–122.16 | 0.006 |
| Inner Mongolia-Xinjiang | 8.90 | 1.23–64.68 | 0.031 |
| Northern China | 7.81 | 1.08–56.63 | 0.042 |
| Central China | 1.88 | 0.19–18.31 | 0.589 |
| Animal family | |||
| Sciuridae | Reference | Reference | |
| Soricidae | 2.45 | 0.91–6.55 | 0.075 |
| Cricetidae | 2.64 | 1.08–6.46 | 0.034 |
| Muridae | 1.76 | 0.76–4.10 | 0.189 |
| Dipodidae | 0.00 | 0–7.50 | 0.970 |
| Mustelidae | 0.00 | 0.00 | 0.991 |
| Tissue type | |||
| Heart | Reference | Reference | |
| Spleen | 3.08 | 1.83–5.18 | < 0.001 |
| Kidney | 2.73 | 1.62–4.60 | < 0.001 |
| Liver | 1.28 | 0.73–2.25 | 0.385 |
| Lung | 1.40 | 0.82–2.39 | 0.219 |
OR, odds ratio; CIs, confidence intervals.
For each of the 154 BeiV-positive animals, partial sequences of L gene (440-bp) were obtained for the phylogenetic analyses. The current BeiVs were clustered in two major lineages, with mean(± standard deviation) of the amino acid sequence identity between two lineages as 95.78 ± 0.1%, lower than the identity within each lineage (98.28 ± 0.1% for Lineage 1 and 99.02 ± 0.1% for Lineage 2) (Fig. 3a, Table S8, Table S9). The clustering of lineage was largely related to their geographic regions. For instance, strains from the Southern China were clustered into Lineage 1, strains from the Northeastern China were clustered into Lineage 2, and sequences from Northern China were observed in both Lineage 1 and Lineage 2, with their surveyed locations geographically lying between Northeast China and Southern China. The animal hosts of Lineage 1 strains were mainly from the family Muridae, Cricetidae, Sciuridae and Soricidae, while the animal hosts of Lineage 2 strains were from family Cricetidae, Muridae, and Soricidae (Fig. 3b). This resulted in higher frequency of strains from Northeastern China and from Cricetidae and Soricidae species in the Lineage 1 (Table S9).
From the 154 BeiV-positive animals, 42 partial sequences of G gene (521-bp) were obtained for the phylogenetic analyses, which was grouped into four distinct clusters that were related to geographic origins (Fig. S1). Sequences obtained from Northeast, North and South region were identical to those from the BeiV strain ERN081008_1S and clustered in Lineage I. Sequences obtained from Northeast, North and Inner mongolia-Xinjiang region were clustered in Lineage II, together with sequences of BeiV strain HKIARN060309_1K, YLRR120908_1K and BeiV strain YLRR120908_3S. The animal hosts of Lineage I strains were mainly from the family Muridae and Soricidae, while the animal hosts of Lineage II strains were from family Muridae.
4. Discussion
Wild animals are extremely important natural hosts for the natural circulation of viral zoonosis (Bengis et al., 2004), posing increased global public health concerns, with the very recent example of the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) that were hypothesized to be originated from bats or from Pangolin (Zhou et al., 2020). During the past few decades, many new viruses from the Paramyxoviridae family have been isolated either from disease outbreaks or surveillance programs or simply by chance (Wang et al., 2008). Of special interest are the viruses originating from a wide range of animal hosts, such as bats, snakes, tree shrews, and rodents (Li et al., 2006). In recent years, the continuous discovery and characterization of Paramyxoviruses from rodents further expanded the genome diversity of these viruses, such as Mossman virus (Miller et al., 2003), Nariva virus (Lambeth et al., 2009), Mojiang paramyxovirus (Wu et al., 2014) and seven recognized species of genus Jeilongvirus (Jack et al., 2005; Li et al., 2006; Vanmechelen et al., 2018; Woo et al., 2011; Wu et al., 2018). In many of these reservoir host species, emerging viruses appear to be well adapted, with little or no evidence of clinical disease. However, the effects can sometimes be devastating when these viruses spill over into humans. This possibility underscored the need for persistent surveillance of these viral communities and the evolutionary processes that drive the emergence and adaptation of zoonotic viruses.
Here we provided evidence that BeiV, a rarely investigated Paramyxovirus, was widely distributed in a great variety of wide rodents and shrews in wide-range regions in China. The discovery and characterization of BeiV from 19 common wild rodents and 3 common wild shrews like M. rutilus and S. murinus expanded the host range of BeiV, indicating other possible animal hosts than just R.norvegicus and R. rattus. In our study, 3 out of 4 families of Rodentia, including Cricetidae, Muridae, Sciuridae are positive, and the rest are Soricomorpha, indicating the natural occurrence and wide distribution of BeiV in Rodentia in China. In addition, we found that shrew was one of the predominant animals harboring BeiV, while neither of the two tested species of Mustelidae family was positive for BeiV. This result is limited by small sample size of Mustelidae family, which warrants further study to validate. Positive detection of BeiV from high variety of organs, including heart, liver, spleen, kidney and lung samples suggested that BeiV can cause systemic infection in animal hosts. Despite of no direct evidence of detecting BeiV from blood sample, it is likely that the virus can be present in the bloodstream and might be transmitted among rodents through aggressive contacts, or to vectors by blood-sucking. These results were also consistent with the previous findings suggesting that BeiV can be detected in kidney and spleen samples, but not from respiratory or anal swabs (Woo et al., 2016). In another survey to characterize the virome of rodents in China, no BeiV was detected either, when the pharyngeal and anal swabs were used for NGS metagenomic analysis (Wu et al., 2018).
Our study indicated that BeiVs of small animal sources have previously unrecognized genome divergence. Paramyxoviruses are pleomorphic enveloped viruses possessing a linear, non-segmented, negative-sense, single-stranded RNA (NNS) genome. The entire genome length of the BeiV is 19,212 nucleotides, listed as the fourth largest among all known members of the Paramyxoviridae family Our findings indicate that BeiV contain well-defined lineages that circulate widely throughout the Mainland China within particular taxonomic groups of hosts. Furthermore, phylogenetic analyses revealed genetic diversity was correlated with geographic origins and animal hosts, possibly indicating a long-term co-evolution between virus and hosts. Although having the largest sample size for the detection of BeiV, the study is not designed as a nation-wide surveillance, thus not all regions were surveyed in the current research.
In summary, we found presence of BeiV in various species of wild small animals with wide geographic distribution in mainland China, indicating several well-defined BeiV lineages circulate widely within particular taxonomic groups of hosts. These findings extend the current knowledge on the known host and biogeographic range of BeiVs that had previously characterized. Although no evidence of human infection with BeiV can be determined at the current stage, the potential risks to humans and livestock cannot be ignored. Therefore further investigation is warranted to attain a more comprehensive understanding of BeiV, as well as other members in the family Paramyxoviridae.
The following are the supplementary data related to this article.
Phylogenetic analysis of 42 partial G (521-bp) gene sequences of Beilong virus (BeiV). The tree was constructed using the Jones-Taylor-Thornton (JTT) method by MEGA 7.0 with 1000 bootstrap replicates.
Nucleotide sequence accession numbers for RNA-directed RNA polymerase (Rdrp) phylogenetic tree.
Nucleotide sequence accession numbers for 154 partial L gene (440-bp).
Nucleotide sequence accession numbers for 43 partial G gene (521-bp).
Beilong virus specific primer pairs were designed to fill in the gaps.
Beilong virus specific primer pairs for the whole genome sequencing.
Samples of the 38 animal species used in this study and the provinces and dates of collection.
Detection rate of Beilong virus in wild rodents and small mammals of six families and from 10 locations in China.
Detection rate of Beilong virus among different animal families in six zoogeographic regions in China
The distribution of two lineage strains in and animal families
Crdit author statment
J.C., X.Z. and Y.W., X.Z., L.F., H.Z. and W.L. designed this study; J.C., X.Z. and P.Z. performed experiments; H.F., X.Z., X.L., H.L., W.S. and W.L. analyzed the data and wrote the manuscript; J.C., Y.W., K.D., F.J., M.J., Z.Y. and N.W. collected samples; J.C., X.Z., N.W., Y.W. and P.Z. contributed to the virus detection;
All of the authors reviewed the manuscript and approved the final version.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This work was funded by grants from China Mega-Project on Infectious Disease Prevention (No. 2018ZX10101003, 2017ZX10303401, 2018ZX10713002, 2018ZX10201001 and 2018ZX10301401), Qingdao people's livelihood science and technology program (14-2-3-29nsh) and the National Science Fund for Distinguished Young Scholars (No. 81825019).
References
- Bengis R.G., Leighton F.A., Fischer J.R., Artois M., Mörner T., Tate C.M. Revue Scientifique et Technique (International Office of Epizootics) Vol. 23. 2004. The role of wildlife in emerging and re-emerging zoonoses; pp. 497–511. [PubMed] [Google Scholar]
- Jack P.J.M., Boyle D.B., Eaton B.T., Wang L.-F. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J. Virol. 2005;79:10690–10700. doi: 10.1128/JVI.79.16.10690-10700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth L.S., Yu M., Anderson D.E., Crameri G., Eaton B.T., Wang L.F. Complete genome sequence of Nariva virus, a rodent paramyxovirus. Arch. Virol. 2009;154:199–207. doi: 10.1007/s00705-008-0287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yu M., Zhang H., Magoffin D.E., Jack P.J., Hyatt A., Wang H.Y., Wang L.F. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology. 2006;346:219–228. doi: 10.1016/j.virol.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Miller P.J., Boyle D.B., Eaton B.T., Wang L.-F. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology. 2003;317:330–344. doi: 10.1016/j.virol.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Nicolas V., Quérouil S., Verheyen E., Verheyen W., Mboumba J.F., Dillen M., Colyn M. Mitochondrial phylogeny of African wood mice, genus Hylomyscus (Rodentia, Muridae): implications for their taxonomy and biogeography. Mol. Phylogenet. Evol. 2006;38:779–793. doi: 10.1016/j.ympev.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanmechelen B., Bletsa M., Laenen L., Lopes A.R., Vergote V., Beller L., Deboutte W., Korva M., Avsic Zupanc T., Gouy de Bellocq J., Gryseels S., Leirs H., Lemey P., Vrancken B., Maes P. Discovery and genome characterization of three new Jeilongviruses, a lineage of paramyxoviruses characterized by their unique membrane proteins. BMC Genomics. 2018;19 doi: 10.1186/s12864-018-4995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Mackenzie J.S., Eaton B.T. 2008. Disease outbreaks caused by emerging paramyxoviruses of bat origin. [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Wong B.H.L., Wong A.Y.P., Poon R.W.S., Yuen K.-Y. Complete genome sequence of a novel paramyxovirus, Tailam virus, discovered in Sikkim rats. J. Virol. 2011;85:13473–13474. doi: 10.1128/JVI.06356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Wu Y., Lam C.S., Yuen K.Y. Novel variant of Beilong Paramyxovirus in rats, China. Emerg. Infect. Dis. 2012;18:1022–1024. doi: 10.3201/eid1806.111901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Wong A.Y.P., Wong B.H.L., Lam C.S.F., Fan R.Y.Y., Lau S.K.P., Yuen K.Y. Comparative genome and evolutionary analysis of naturally occurring Beilong virus in brown and black rats. Infect. Genet. Evol. 2016;45:311–319. doi: 10.1016/j.meegid.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Wu Z., Yang L., Yang F., Ren X., Jiang J., Dong J., Sun L., Zhu Y., Zhou H., Jin Q. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg. Infect. Dis. 2014;20:1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Lu L., Du J., Yang L., Ren X., Liu B., Jiang J., Yang J., Dong J., Sun L., Zhu Y., Li Y., Zheng D., Zhang C., Su H., Zheng Y., Zhou H., Zhu G., Li H., Chmura A., Yang F., Daszak P., Wang J., Liu Q., Jin Q. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 2018;6 doi: 10.1186/s40168-018-0554-9. 178–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zhou L., Zhang Y., Wang X. Geographical patterns based on faunal types of breeding birds and mammals in China. Integr. Zool. 2008;3:280–289. doi: 10.1111/j.1749-4877.2008.00107.x. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-41020-42012-41587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of 42 partial G (521-bp) gene sequences of Beilong virus (BeiV). The tree was constructed using the Jones-Taylor-Thornton (JTT) method by MEGA 7.0 with 1000 bootstrap replicates.
Nucleotide sequence accession numbers for RNA-directed RNA polymerase (Rdrp) phylogenetic tree.
Nucleotide sequence accession numbers for 154 partial L gene (440-bp).
Nucleotide sequence accession numbers for 43 partial G gene (521-bp).
Beilong virus specific primer pairs were designed to fill in the gaps.
Beilong virus specific primer pairs for the whole genome sequencing.
Samples of the 38 animal species used in this study and the provinces and dates of collection.
Detection rate of Beilong virus in wild rodents and small mammals of six families and from 10 locations in China.
Detection rate of Beilong virus among different animal families in six zoogeographic regions in China
The distribution of two lineage strains in and animal families