Abstract
Background
There are various phenotypic methods for identifying class B and class A β-lactamase enzymes in Pseudomonas aeruginosa. The purpose of this study was to compare the sensitivity and specificity of different phenotypic methods with HRMA assay to detect β-lactamase-producing P. aeruginosa strains.
Methods
Eighty-eight of P. aeruginosa isolates were collected from different specimens. Conventional double-disk test (DDT) and EDTA-imipenem microbiological (EIM) were performed to detect ESBL and MBL-producing strains, respectively. Meanwhile, the Modified Hodge test and Carba-NP test were performed on all carbapenem-resistant strains. HRMA method and sensitivity and specificity of primers were determined based on the melt curve temperature range. In all comparisons, PCR was considered as the gold standard.
Results
Of the 402 isolates collected from different clinical specimens, 88 isolates of P. aeruginosa were identified. However, 43 strains were (48.88%) ESBL-producing, and 7 strains (7.95%) were MBL-producing. Also, using the Modified Hodge test and Carba-NP method, 11 (12.5%) and 19 (21.59%) strains were carbapenemase-producing, respectively. The results of the HRMA test revealed that genes coding for blaSHV, blaTEM, blaKPC, blaIMP, blaVIM, and blaGES were detected in 44.31%, 22.72%, 13.63%, 14.7%, 5.6%, and 2.27% of P. aeruginosa isolates. Nonetheless, for blaKPC and blaGES genes, sensitivity and specificity of the Carba-NP test were 90.47%, 94.87%, and 83.36%, 94.80%, respectively. However, sensitivity and specificity of MHT was 91.66%, 98.70%, and 77.77%, 96.42%, respectively. For blaSHV and blaTEM genes, sensitivity and specificity of DDT were 95.55%, 95.55%, and 86%, 83.50%, respectively. However, sensitivity and specificity of EMI were 77.77%, 97.59%, and 91.66%, 97.43% for blaVIM and blaIMP, respectively.
Conclusion
The HRMA is a powerful, accurate, closed-tube, rapid method for detecting β-lactamase genes in P. aeruginosa. The high sensitivity and specificity of this method, along with phenotypic tests, play a useful role in increasing the predictive value of clinical reports.
Keywords: Pseudomonas aeruginosa, high-resolution melting curve analysis, HRMA, β-lactamases, drug resistance
Background
Pseudomonas aeruginosa is one of the leading nosocomial pathogens worldwide.1 Nosocomial infections caused by this organism are often hard to treat because of both the intrinsic resistance of the species and its remarkable ability to acquire further mechanisms of β-lactamases enzymes to multiple groups of antimicrobial agents.2,3 However, in 1980 the β-lactamases were classified into four classes (A, B, C, and D).4,5 Class A and D include the classic and extended-spectrum β-lactamases (ESBLs) and are mostly composed of the blaTEM, blaSHV, CTX-M, and OXA enzymes, class B comprises the Metallo-β-lactamases (MBL), and finally, class C contains the AmpC β-lactamases (ACBL).6,7 Except for class B metalloenzymes, β-lactamases belong to the family of serine-reactive hydrolases.4,8,9 Carbapenemases, enzymes possessing the ability to inhibit almost all β-lactam antibiotics, including carbapenems, have been mainly detected in mentioned species.10 Until the early 1990s, carbapenemases were considered as species-specific, chromosomally encoded β-lactamases. Identification of genes encoding for carbapenemases on mobile genetic elements emerged the possible horizontal spread of these enzymes.4,11,12
There are various phenotypic methods for identifying β-lactamases enzyme-producing of P. aeruginosa, each with disadvantages and benefits.3 Double-disk synergy test for detection of strains belongs to class A β-lactamases, Modified Hodge test to detect class A β-lactamases and meropenem E-test strip to identify class B β-lactamases is common tests.13,14 One of the most critical advantages of phenotypic methods is their cost-effectiveness. Of course, the fatality of these methods and the low speed of diagnosis should be mentioned from its main disadvantages, and the need to use sensitive and rapid molecular methods along with these phenotypic methods is essential.10,15
High-Resolution Melting Analysis (HRMA) is an emerging technique used to discriminate DNA sequence variants.8 HRMA is based on accurate determination of the relationship between temperature and the extent of dissociation of a PCR amplicon. HRMA is almost performed universally in real-time PCR thermocyclers post-PCR.16 In HRMA, the fluorescence produced by a DNA intercalating dye (eg, SYBERGreen) is monitored during strand dissociation events during the melt phase. While phenotype-based detection methods have many advantages, molecular real-time amplification techniques have gained significant acceptance because they may provide more rapid detection, increased sensitivity, and specificity, and lack the risk of carry-over contamination associated with earlier methods.17,19 However, normalized and difference graphs were generated to assess the ability of the HRMA method to differentiate between bacterial strains.18
This study aimed to investigate different methods in diagnosing different strains of P. aeruginosa. Besides, by optimizing the HRMA assay, the errors and shortcomings of this method were investigated based on local and experimental conditions. Also, by comparing the sensitivity and specificity of phenotypic methods and the HRMA assay, the advantages, and disadvantages of different methods were discussed.
Methods
Study Design and Sampling
During 9 months (Dec 2017 to Sep 2018), 88 of P. aeruginosa strains were collected from various clinical isolates, including blood, sputum, urine, ulcers, and secretions. All isolates were transferred to the Microbiology Laboratory of Hamedan University of Medical Sciences for differential tests.
P. aeruginosa Isolates
Isolates suspected to be P. aeruginosa were identified by using Pseudomonas Cetrimide agar (Merck, Germany), and plates were incubated for 48 hours at 42ºC under aerobic conditions. Then, colonies grown on the Cetrimide agar were examined by various biochemical tests. Biochemical tests were performed to confirm P. aeruginosa using methyl-red and Voges – Proskauer (MRVP) tests, Oxidase, Catalase, hydrolysis of citrate utilization, Indole production, and fermentation of various sugars (All from Sigma-Aldrich, USA). Bacterial strains were stored at −20°C in Brain-Heart Infusion (BHI) broth (Merck, Germany) supplemented with 25% v/v glycerol.
Antibiotic Disc Susceptibility Testing
Antibiotic susceptibility testing was done as per the disc diffusion method (Bauer et al, 1966) following the guidelines of the Clinical Laboratory Standard Institute (CLSI) 2018 against 13 antibiotics of different classes. However, disc diffusion based on using antibiotic discs (MAST, UK); as ceftazidime (30 µg), doripenem (10 µg), meropenem (10 µg), imipenem (10 µg), cefepime (30 µg), ciprofloxacin (5 µg), gentamicin (10 µg), amikacin (30 µg), norfloxacin (10 µg), aztreonam (30 µg), piperacillin/tazobactam (100/10µg), Tobramycin (10 µg), and piperacillin (100 µg).
Detection of ESBL Strains by Double-Disk Confirmatory Test (DDCT)
Conventional DDCT was performed to detect ESBLs in a representative P. aeruginosa strain using cefotaxime + clavulanate (30/10 µg) with cefotaxime (30 µg) alone and ceftazidime + clavulanate (30/10 µg) with ceftazidime (30 µg) alone extended-spectrum cephalosporins (Mast, UK). Modifications of DDCT were applied according to Sahni et al,20 following the guidelines of Clinical Laboratory Standard Institute (CLSI) 2018, ESBL positive isolates show an increase of 5 mm zone of inhibition with clavulanic acid as compared to the zone size for CTX and CZD alone. K. pneumonia ATCC700603 and P. aeruginosa PAO-1 were applied as a positive and negative control, respectively.
Detection of MBL Strains by EIM Assay
EDTA-imipenem microbiological (EIM) assay was performed, as described by Marchiaro et al21 following the guidelines of the Clinical Laboratory Standard Institute (CLSI) 2018. MBL combined disc test was considered positive if the zone diameter difference between imipenem + EDTA and imipenem discs (MAST, UK) was larger than 7 mm. P. aeruginosa PAO-1 and P. aeruginosa NCTC 13359 were used as a positive and negative control, respectively.
Detection of Carbapenem-Resistant Strains by Modified Hodge Test and Carba-NP Test
The Modified Hodge test (MHT) was performed on all carbapenem-resistant isolates as described by Lee et al using E. coli ATCC 25922 as an organism sensitive to carbapenems, P. aeruginosa 18524928.2 (negative control) and Enterobacter cloacae 91419421 strain as a positive control. Two imipenem and ceftazidime sensitive strains were also chosen as additional negative controls. The surface of a Mueller Hinton agar plate was inoculated using a cotton swab with an overnight culture suspension of the E. coli 0.5 McFarland Standard. After several minutes of drying, 10 µg meropenem (Mast, UK) disk was placed at the center of the Muller Hinton (MH) plate, and suspected isolates from the overnight cultures were streaked heavily from the drug disk to the edge of the agar plate. Results were recorded after the overnight incubation at 36°C±1°C in an ambient-air incubator. The true positive results were regarded as the appearance of the “cloverleaf” inhibition zone due to enhanced growth of indicator strain toward the meropenem disk alongside the test strain.22
Preparation of DNA Extraction
Plasmid and DNA were extracted with a plasmid DNA purification kit (Qiagen, Germany). Extraction was performed based on the kit protocol.
Evaluation of Sensitivity and Specificity of Primers and HRMA Assay
Standard strains, including P. aeruginosa NCTC 13359 and P. aeruginosa ATCC 27853, were used to optimize HRMA assay. To the examination of the sensitivity of primers, a standard bacteria with a concentration of 0.5 McFarland (1.5×108 CFU/mL) was provided into serial dilutions of 107 to 10° CFU/mL. A real-time PCR test was performed for all the dilutions. The melting temperature of each product was determined in Singleplex PCR with SYBRGreen. To ensure that the difference in the Tm values between the primers was at least 2°C in order to avoid overlapping of peaks, different combinations of primers for each of the three genes were chosen.
Optimize of HRMA Assay
Singleplex HRMA assay and amplification were performed using a real-time PCR (ABI step one plus, USA). Reactions were carried out in a total volume of 20 µL included: HRMA Master Mix (HOT FIREPol® EvaGreen HRMA Mix) 4 µL, 1 µL of each primer (20pmol), 1 µL of bacterial DNA, and DEPS water. The cycling conditions were as follows: denaturing at 95°C for 15 min, followed by 40 cycles of 15 sec at 95°C, 59°C for one 30sec. Also, the melting curve was obtained following the melting steps: DNA double strands opening step – 95°C, 15 s, gradually heating step – 55°C, 15 s to 95°C with a temperature increase of 2% of the maximum setting of the machine. A ramping rate of 1% was also tested to see whether slower speed helps show more information about the melting curve.
Statically Analysis of Data
All statistical analyses were performed using the SPSS program, version 16.0 (SPSS Inc., Chicago, USA). The chi-squared test and Fisher’s exact test (two-sided) were used in order to analyze the qualitative data. The normal distribution of the quantitative data was tested for using the Shapiro–Wilk test. Also, melt curve profiles were assessed and analyzed using ABI Step one software version 2.3 (ABI Thermo Fisher Scientific, Inc., USA). HRMA data analyzed by ABI Step one High-Resolution Melt (HRMA) software v3.01 (ABI Thermo Fisher Scientific, Inc., USA). Data analysis was performed using a bivariate random-effects model to estimate pooled sensitivity, specificity, positive (PPV), and negative predictive values (NPV). PCR method was used as a gold-standard test.
Sanger Sequencing PCR and Phylogenic Tree
Sequencing was performed on standard PCR products on both the forward and reversed strands. PCR amplification was performed in a 25 ul mixture containing 12.5 µL master mix (Amplicon, Denmark), 1 µL forward primers and 1 µL reverse primers (according to Table 1), 1 µL of DNA and sterile DW (Sigma-Aldrich, St Louis, MO). Amplification of the target regions was performed in 35 cycles consisting of initial heat activation at 95°C for 10 min, denaturation at 95°C for 45 s, and elongation at 71°C for 1 min, with a final elongation at 72°C for 5 min. Annealing Tm and DNA amplification were carried out (Eppendorf thermocycler, Germany) with thermal cycling conditions consisting of Table 1. Electropherograms of generated sequences were inspected with MEGA6 software and Chromas software version 2.8 (Technelysium Pty. Ltd., Helensvale, Australia). Obtained DNA sequences were analyzed with the BLAST program, available from URL: http://www.ncbi.nlm.nih.gov. Primers used for sequencing are listed in Table 1.
Table 1.
Oligonucleotide Sequences Used in This Study
| Gene | Primer Name | Sequence of Primers | Melting Tm | Annealing Tm/Time | Product Size (bp) | References |
|---|---|---|---|---|---|---|
| blaSHV | SHV | F: TCCCATGATGAGCACCTTTAAA R: TCCTGCTGGCGATAGTGGAT |
88.57±0.5°C | 59 | 105 | [5] |
| blaTEM | TEM | F: GCATCTTACGGATGGCATGA R: GTCCTCCGATCGTTGTCAGAA |
82.4± 0.5°C | 58 | 101 | [5] |
| blaKPC | KPC | F: GATACCACGTTCCGTCTGG R: GCAGGTTCCGGTTTTGTCTC |
83.55±0.5°C | 60 | 254 | [8] |
| blaIMP | IMP | F: GGCTTAATTCTCGATCTATCCC R: CTAGCCAATAGTTAACTCCGC |
80.16±0.5°C | 61 | 114 | [3] |
| blaVIM | VIM | F: TCTCCACGCACTTTCATGAC R: GTGGGAATCTCGTTCCCCTC |
88.57±0.5°C | 60 | 124 | [3] |
| blaGIM | GES | F: GTTTTGCAATGTGCTCAACG R: TGCCATAGCAATAGGCGTAG |
86.86±0.5°C | 61 | 387 | 12 |
Results
In total, 88 P. aeruginosa isolates were collected. Of these isolates, 19 (21.59%) of the isolates were collected from blood, 26 (29.54%) from the wound, 10 (11.36%) from urine, and 11 (12.5%) from catheters. Tony-tow (25%) isolates were collected from other samples. However, 53 isolates (60.22%) were isolated from female patients, and 35 isolates (39.77%) from male patients.
Antibiotic Sensitivity Profile
Antibiotic susceptibility profiles were obtained after testing by the Kirby Bauer disk diffusion method according to CLSI guidelines 2018. The overall antibiotic resistance of clinical isolates is summarized in Table 2, which also illustrates the increased resistance level for ciprofloxacin (76.13%), followed by gentamicin (67.04%), ceftazidime (44.31%), imipenem (35.22%), piperacillin (32.94%), and doripenem and cefepime (30.68%). Additionally, the levels of non-susceptible to at least one agent in three or more antimicrobial categories hence considered as MDR, were unexpectedly high, affecting 42.04% of P. aeruginosa clinical isolates included in this study.
Table 2.
Antibiotic Susceptibility Profiles in Clinical Isolates of P. aeruginosa
| Antibiotics Resistance | β-lactamase genes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total(%) | blaSHV(%) | blaTEM(%) | blaKPC(%) | blaIMP(%) | blaVIM(%) | blaGES(%) | |||||||||
| S | I | R | S | R | S | R | S | R | S | R | S | R | S | R | |
| Ceftazidime | 33(37.5%) | 16(18.1%) | 39(44.3%) | 4(12.1%) | 33(84.6%) | 0(0%) | 19(48.7%) | 0(0%) | 11(28.2%) | 0(0%) | 13(33.3%) | 0(0%) | 5(12.8%) | 0(0%) | 2(5.1%) |
| Doripenem | 46(52.2%) | 15(17.0%) | 27(30.6%) | 0(0%) | 25(92.5%) | 0(0%) | 20(74.0%) | 0(0%) | 12(44.4%) | 0(0%) | 12(44.4%) | 0(0%) | 5(18.5%) | 0(0%) | 2(7.4%) |
| Meropenem | 54(61.3%) | 13(14.7%) | 21(23.8%) | 1(1.8%) | 17(80.9%) | 0(0%) | 20(95.2%) | 0(0%) | 12(57.1%) | 0(0%) | 13(61.9%) | 0(0%) | 5(23.8%) | 0(0%) | 2(9.5%) |
| Imipenem | 40(45.4%) | 17(19.3%) | 31(35.2%) | 3(7.5%) | 25(80.6%) | 0(0%) | 20(64.5%) | 0(0%) | 12(38.7%) | 0(0%) | 13(41.9%) | 0(0%) | 5(16.1%) | 0(0%) | 2(6.4%) |
| Cefepime | 50(56.8%) | 11(12.5%) | 27(27.2%) | 1(2.0%) | 26(96.2%) | 0(0%) | 19(70.3%) | 0(0%) | 12(44.4%) | 0(0%) | 10(37.0%) | 0(0%) | 5(18.5%) | 0(0%) | 2(7.4%) |
| Gentamycin | 18(20.4%) | 11(12.5%) | 59(67.0%) | 0(0%) | 39(66.1%) | 1(5.5%) | 18(31.0%) | 0(0%) | 12(20.3%) | 0(0%) | 13(22.0%) | 0(0%) | 5(8.4%) | 0(0%) | 2(3.3%) |
| Ciprofloxacin | 12(13.6%) | 9(10.2%) | 67(76.1%) | 0(0%) | 39(58.2%) | 0(0%) | 20(29.8%) | 0(0%) | 12(17.9%) | 0(0%) | 13(19.4%) | 0(0%) | 5(7.4%) | 0(0%) | 2(2.9%) |
| Amikacin | 52(59.1%) | 17(19.3%) | 19(21.5%) | 0(0%) | 15(78.9%) | 1(1.9%) | 15(78.9%) | 0(0%) | 12(63.1%) | 0(0%) | 13(68.4%) | 0(0%) | 5(26.3%) | 0(0%) | 2(10.5%) |
| Norfloxacin | 50(56.8%) | 18(20.4%) | 20(22.7%) | 7(14.0%) | 11(55.0%) | 2(4.0%) | 13(65.0%) | 2(4.0%) | 9(45.0%) | 5(1.0%) | 7(35.0%) | 0(0%) | 2(10.0%) | 0(0%) | 2(1.0%) |
| Aztreonam | 67(76.1%) | 9(10.2%) | 12(13.6%) | 0(0%) | 11(91.6%) | 0(0%) | 11(91.6%) | 5(7.1%) | 5(41.6%) | 0(0%) | 11(91.6%) | 0(0%) | 5(41.6%) | 0(0%) | 2(16.6%) |
| Tobramycin | 68(77.2%) | 9(10.2%) | 11(12.5%) | 0(0%) | 11(100%) | 0(0%) | 11(100%) | 0(0%) | 11(100%) | 0(0%) | 11(100%) | 0(0%) | 3(27.2%) | 0(0%) | 2(18.1%) |
| Piperacillin | 48(54.5%) | 11(12.5%) | 29(28.4%) | 3(5.1%) | 25(86.2%) | 3(5.1%) | 14(48.2%) | 2(3.4%) | 6(20.6%) | 2(3.4%) | 8(27.5%) | 0(0%) | 2(6.8%) | 0(0%) | 2(6.8%) |
| Piperacillin/tazobactam | 48(54.4%) | 16(18.1%) | 24(27.2%) | 2(4.1%) | 19(79.1%) | 3(6.2%) | 11(45.8%) | 3(6.2%) | 7(29.1%) | 3(6.2%) | 6(25.0%) | 0(0%) | 2(8.3%) | 0(0%) | 2(8.3%) |
| MDR | 37(22.4%) | 35(94.5%) | 17(45.9%) | 12(32.4%) | 12(32.4%) | 5(13.5%) | 2(5.4%) | ||||||||
| β-lactamase-producer strain | |||||||||||||||
| class A β-lactamases (ESBL) | 43(22.4%) | 39(90.6%) | 20(46.5%) | 10(23.2%) | 8(18.6%) | 5(11.6%) | 2(4.6%) | ||||||||
| class B β-lactamases (MBL) | 7(22.4%) | 5(71.4%) | 5(71.4%) | 7(100%) | 7(100%) | 5(71.4%) | 2(28.5%) | ||||||||
| class D β-lactamases (KPC) | 11 (12.5%) (Hodge test)19 (21.5%) (Carba-NP) | 11 (100%)17 (89.7%) | 10 (90.9%)14 (73.6%) | 11 (100%)12 (63.1%) | 11 (100%)9 (47.3%) | 5 (45.4%)4 (21.0%) | 2 (18.1%)2 (10.5%) | ||||||||
| Clinical isolates | |||||||||||||||
| Wound | 26(29.5%) | 17(65.3%) | 14(53.8%) | 8(30.6%) | 11(42.3%) | 11(42.3%) | 2(7.6%) | ||||||||
| Blood | 19(21.5%) | 13(68.4%) | 5(26.3%) | 2(10.5%) | 2(10.5%) | 2(10.5%) | 0(0%) | ||||||||
| Urine | 10(11.3%) | 5(50.0%) | 1(1.0%) | 1(1.0%) | 1(0%) | 1(0%) | 0(0%) | ||||||||
| Catheter | 11(12.5%) | 3(27.2%) | 0(0%) | 0(0%) | 0(0%) | 0(0%) | 0(0%) | ||||||||
| Other case | 22(25%) | 0(0%) | 0(0%) | 0(0%) | 0(0%) | 0(0%) | 0(0%) | ||||||||
Prevalence of Class B and Class A β-Lactamases in P. aeruginosa Isolates by Phenotypic Methods
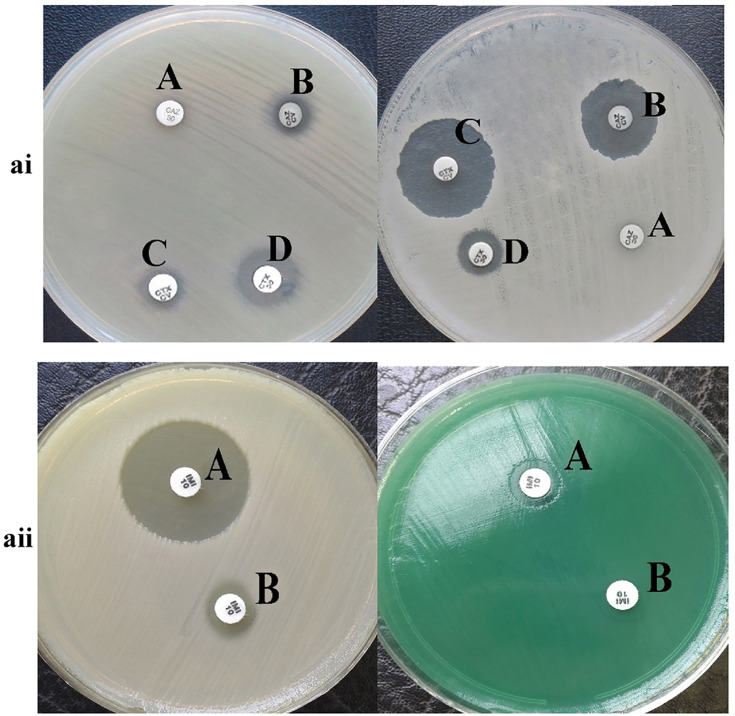
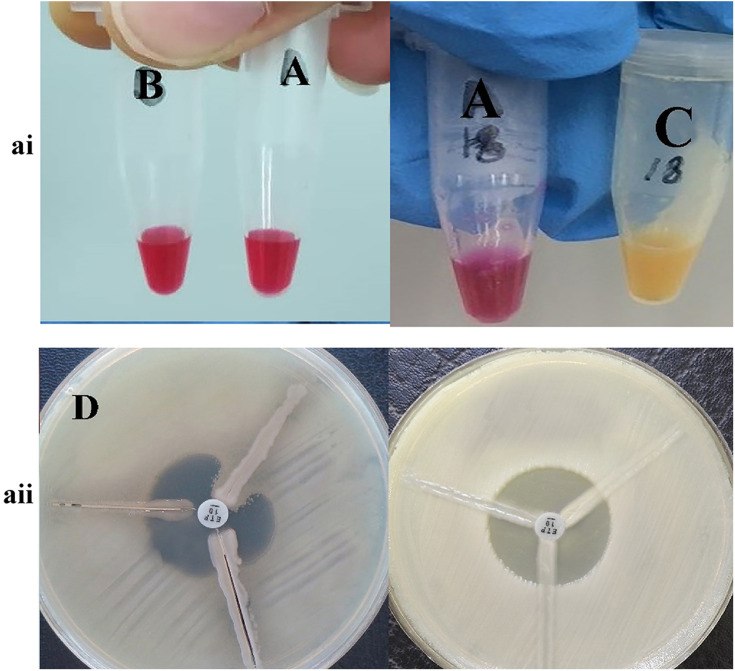
Results of phenotypic tests for the detection of different classes of β-lactamase enzymes for P. aeruginosa are shown in Table 2. In this case, 43 strains (48.88%) of P. aeruginosa isolates were ESBL-producing (Figure 1ai). Also, in class B β-lactamases, seven isolates (7.94%) of P. aeruginosa were MBL-producing (Figure 1aii). Carbapenemase-producing P. aeruginosa was identified in 11 (12.5%), and 19 (21.5%) isolates, by Modified Hodge test (Figure 2ai) and Carba-NP test (Figure 2aii) methods, respectively.
Figure 1.
ai: The result of phenotypic detection of ESBL-producing Pseudomonas aeruginosa by Double-Disk Confirmatory Test (DDCT) in Pseudomonas aeruginosa. A: Ceftazidime (30 µg); B: ceftazidime + clavulanate (30/10µg); C: cefotaxime + clavulanate (30/10µg); D: cefotaxime (30 µg). Top: ESBL negative strain; Bottom: ESBL positive strain; ESBL positive isolates show an increase of 5 mm zone of inhibition with clavulanic acid as compared to the zone size for CTX and CZD alone. aii: The result of phenotypic detection of MBL-producing Pseudomonas aeruginosa by EDTA-imipenem microbiological (EIM) test in Pseudomonas aeruginosa. A: EDTA + imipenem; B: imipenem. Top: MBL positive strain; Bottom: MBL negative strain; MBL considered positive when zone diameter difference between imipenem + EDTA and imipenem discs was larger than 7 mm.
Figure 2.
The result of phenotypic detection of carbapenemase-producing Pseudomonas aeruginosa by Modified Hodge test (right) and Carba-NP test test (left) in Pseudomonas aeruginosa. ai A: control negative strain; B: KPC negative strain by Carba-NP test test; C: KPC positive strain by Carba-NP test test. aii D: KPC positive strain by Modified Hodge test; (E) KPC negative strain by Modified Hodge test.
Prevalence of Class B and Class A β-Lactamases in P. aeruginosa Isolates by PCR Assay
Thirty-nine isolates (44.31%) with blaSHV gene, 20 isolates (22.7%) with blaTEM gene, 12 isolates (13.63%) with blaKPC gene, 13 isolates (14.77%) with blaIMP gene, 5 isolates (5.6%) with blaVIM gene, and 2 isolates (2.2%) with blaGES gene were detected. Details of the frequency of β-lactamase genes on different strains and clinical samples are shown in Table 2.
Phylogenetic Relationships of β-Lactamase Genes
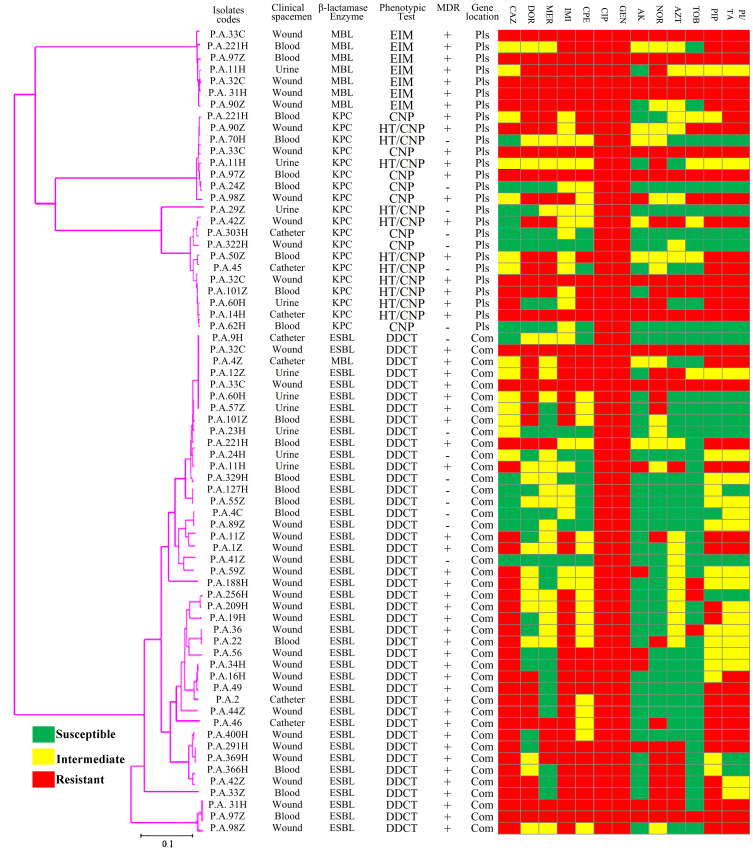
According to Figure 3, phylogenetic trees were constructed using the Neighbor-Joining technique, where evolutionary history and taxa were taken from 500 replicates to prepare the bootstrap consensus tree confidence limits. The phylogenetic tree was designed by determining the nucleotides sequence of blaSHV, blaTEM, blaKPC, blaIMP, blaVIM, and blaGES genes by sanger method. Indistinct and ambiguous sites were removed for each sequence pair. However, phylogenetic classification showed a high percentage of class A β-lactamases in P. aeruginosa strains (44.31%). At lower frequencies, percentages of 5.68% for the class B β-lactamases. Details of the different strains topping based on β-lactamase genes are illustrated in Figure 3.
Figure 3.
Phylogenetic tree of β-lactamases genes based on the gene sequencing of the class A and class B β-lactamases in P. aeruginosa isolates.
Abbreviations: MDR, multidrug-resistant; Com, chromosomal genes; Pls, plasmid genes; CAZ, ceftazidime; DOR, doripenem; MER, meropenem; IMI, imipenem; CPE, cefepime; GEN, gentamycin; CIP, ciprofloxacin; AK, amikacin; NOR, norfloxacin; AZT, aztreonam; TOB, tobramycin; PIP, piperacillin; PI/TA, piperacillin/tazobactam.
Analytical Sensitivity and Specificity of Primers by Real-Time PCR
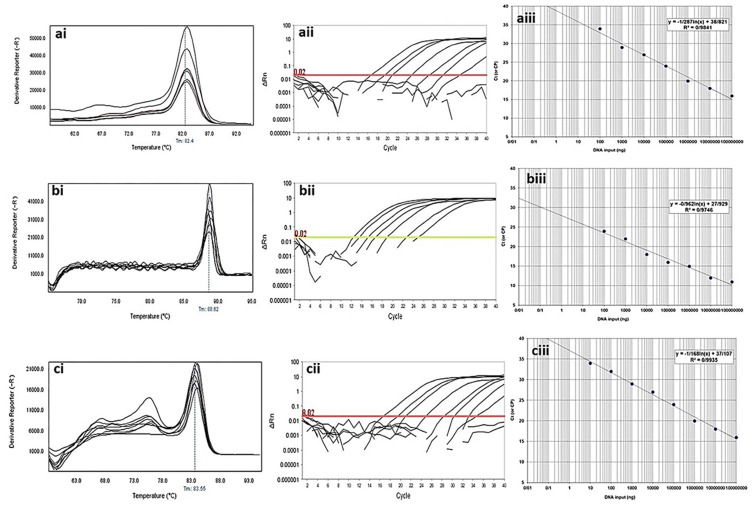
The observed Tm shown in the melting curves of gene amplification was equal to 83.5±0.5°C for blaKPC gene, 80.0±0.5°C for blaIMP gene, 84.8±0.5°C for blaVIM gene, 86.9±0.5°C for blaGES gene, 88.5±0.5°C for blaSHV gene and 82.2±0.5°C for blaTEM gene. The analysis of the threshold melting curves of the intended genes indicated a successful onset of gene replication in all prepared dilutions in different cycles. The sensitivity of the real-time PCR calculated from eight dilutions of the positive control monitored by the use of ABI Software V 3.2 and shown in Figures 4 and 5.
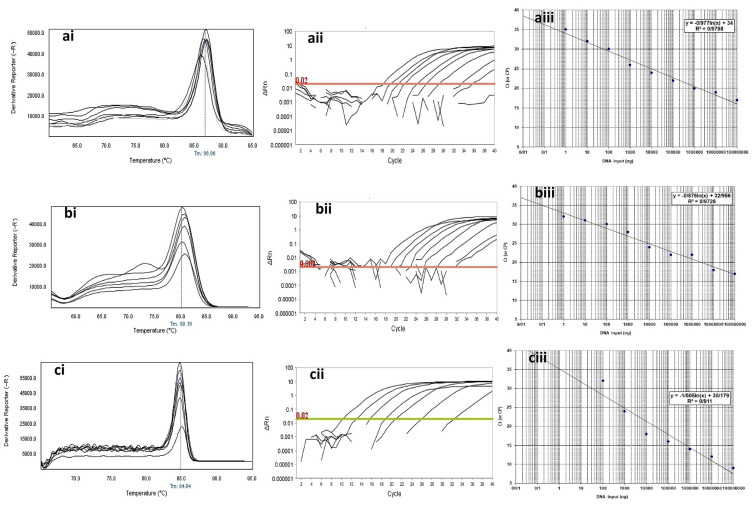
Figure 4.
Analytical sensitivity of Real-time PCR for primers used to detect class B β-lactamases genes in clinical isolates of P. aeruginosa. ai to aiii: blaGES; ai: Melting curve in 86.86°C; aii: Amplification curve; and aiii: Standard curve with efficiency=1.808. bi to biii: blaIMP gene; bi: Melting curve in 80.16°C; bii: Amplification curve; and biii: Standard curve with efficiency=1.742. ci to ciii: blaVIM gene; ci: Melting curve in 84.84°C; cii: Amplification curve; and ciii: Standard curve with efficiency=1.575. The mean of a: 108; b: 107; c: 106; d: 105; e: 104; f: 103; g: 102; h: 101 and i: 10° CFU/mL of DNA dilutions. Horizontal lines represent cycle threshold of Real-time PCR. One peak with a shoulder corresponds to genomic DNA amplification; no peak corresponds to no amplification. SYBR Green I Dye and single-tube reaction were used in this test. Also, Real-Time PCR was performed as single-step.
Figure 5.
Analytical sensitivity of real-time PCR for primers used to detect class A β-lactamases genes in clinical isolates of P. aeruginosa. ai to aiii: blaTEM gene; ai: Melting curve in 82.4°C; aii: Amplification curve; and aiii: Standard curve with efficiency=1.429. bi to biii: blaSHV gene; bi: Melting curve in 88.52°C; bii: Amplification curve; and biii: Standard curve with efficiency=1.742. ci to ciii: blaKPC gene; ci: Melting curve in 83.55°C; cii: Amplification curve; and ciii: Standard curve with efficiency=1.575. The mean of a: 108; b: 107; c: 106; d: 105; e: 104; f: 103; g: 102; h: 101 and i: 10° CFU/mL of DNA dilutions. Horizontal lines represent cycle threshold of Real-time PCR. One peak with a shoulder corresponds to genomic DNA amplification; no peak corresponds to no amplification. SYBR Green I Dye and single-tube reaction were used in this test. Also, Real-Time PCR was performed as single-step.
Detection of Class B and Class A β-Lactamases in P. aeruginosa Isolates by HRMA Assay
The results of the HRMA test revealed that out of 88 clinical isolates of P. aeruginosa, 39 isolates (44.31%) were blaSHV gene, 20 isolates (22.7%) were blaTEM gene. Also, 12 isolates were blaKPC gene (13.6%) and 14 isolates (15.9%) were blaIMP gene. Also, five isolates (5.6%) with blaVIM gene and two isolates (2.2%) with blaGES gene. The result of HRMA from DNA amplification of the positive strains monitored by the use of ABI Step one High-Resolution Melt (HRMA) software v3.01 and shown in Figures 6 and 7.
Figure 6.
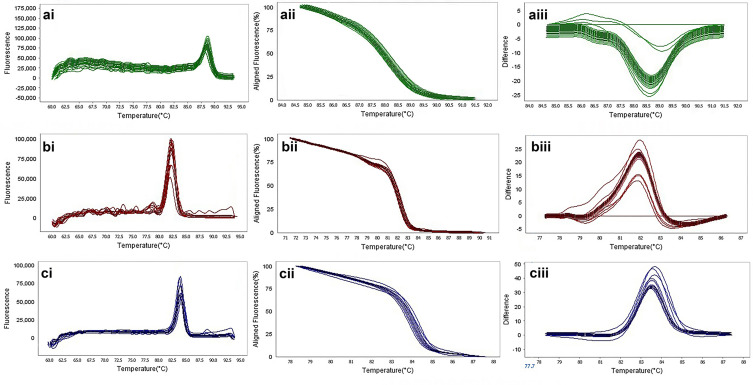
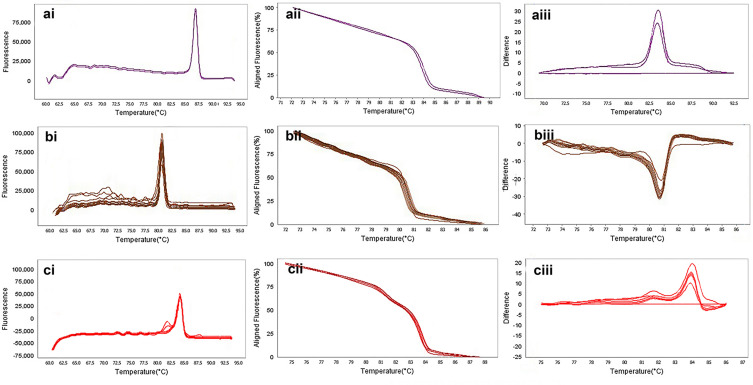
The result of molecular detection of class A β-lactamases genes by HRMA assay. a: blaSHV with 245 bp length and a melting temperature of 88.57±0/5°C; b: blaTEM with 101 bp length and a melting temperature of 82.4±0/5°C; c: blaKPC with 245 bp length and a melting temperature of 83.55±0/5°C. β-lactamases producing genes were amplified successfully using the EvaGreen Dye in the ABI Step-OnePlus machine by one-sept protocol. Primers specific melting peaks were obtained via HRMA analysis, allowing the differentiation of all investigated β-lactamase enzymes. Due to the highly saturating EvaGreen dye and the HRMA analysis, the accuracy of the resolution was ±0. 1–0.5 °C. (ai, bi, and ci) Melting curves; (aii, bii, and cii) normalized plot; and (aiii, biii, and ciii) difference plot.
Figure 7.
The result of molecular detection of class B β-lactamases genes by HRMA assay. a: blaGES with 72 bp length and a melting temperature of 86.86±0/5°C; b: blaIMP with 114 bp length and a melting temperature of 80.16±0/5°C; c: blaVIM with 105 bp length and a melting temperature of 84.80±0/5°C. β-lactamases producing genes were amplified successfully using the EvaGreen Dye in the ABI Step-OnePlus machine. Primers specific melting peaks were obtained via HRMA analysis, allowing the differentiation of all investigated β-lactamase enzymes. Due to the highly saturating EvaGreen dye and the HRMA analysis, the accuracy of the resolution was ±0. 1–0.5 °C. (ai, bi, and ci) Melting curves; (aii, bii, and cii) normalized plot; and (aiii, biii, and ciii) difference plot.
Sensitivity and Specificity of Methods
Based on Table 3 and statistical analysis of the results of the HRMA test for detection of class A and class B β-lactamases in P. aeruginosa strains, sensitivity and specificity of the HRMA method was 100%. Nonetheless, for the detection of carbapenemase-producing strains, sensitivity and specificity of Carba-NP test was 90.47% and 94.87%, respectively. However, sensitivity and specificity of MHT were 91.66% and 98.70%, respectively (All reported for blaKPC gene). For the detection of ESBL-producing strains, the sensitivity and specificity of the DDT were 95.55% and 95.55% (for blaSHV gene). Besides, for MBL-producing strains, the sensitivity and specificity of the EIM were 77.77% and 97.59%, respectively (for blaVIM gene).
Table 3.
Sensitivities, Specificities, Positive Predictive and Negative Predictive Values for Phenotypic and HRMA (blaSHV Gene for ESBL-Producing, blaKPC Gene for Carbapenemase-Producing and blaVIM Gene for MBL-Producing) Methods in β-Lactamase-Producing Strain of P. aeruginosa
| Methods | No. of P. aeruginosa Strains (Total n = 88) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC Strains (Total n = 12) | MBL Strains (Total n = 5) | ESBL Strains (Total n = 39) | Sen (%) | Sp (%) | PPV (%) | NPV (%) | ||||||||||
| True Positive | False Positive | False Negative | True Negative | True Positive | False Positive | False Negative | True Negative | True Positive | False Positive | False Negative | True Negative | |||||
| DDT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 43 | 4 | 2 | 86 | 95.55 | 95.55 | 91.48 | 97.72 |
| EIM assay | 0 | 0 | 0 | 0 | 7 | 2 | 2 | 81 | 0 | 0 | 0 | 0 | 77.77 | 97.59 | 97.59 | 77.77 |
| Hodge test | 11 | 1 | 1 | 76 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 91.66 | 98.70 | 91.66 | 98.70 |
| Carba-NP | 19 | 4 | 2 | 74 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 90.47 | 94.87 | 97.36 | 82.60 |
| HRM | 12 | 0 | 0 | 76 | 5 | 0 | 0 | 83 | 39 | 0 | 0 | 69 | 100 | 100 | 100 | 100 |
| Gold Standard | 12 | 0 | 0 | 76 | 5 | 0 | 0 | 83 | 39 | 0 | 0 | 69 | 100 | 100 | 100 | 100 |
Abbreviations: Sen, sensitivity; Sp, speciation; PPV, positive predictive values; NPV, negative predictive values.
As shown in Table 4, the sensitivity and specificity of the HRMA method was 100%. Also, the sensitivity and specificity of the Carba-NP test were 83.36% and 94.80%, respectively. However, sensitivity and specificity of MHT were 77.77% and 96.42%, respectively (All reported for blaGES gene). For the detection of ESBL-producing strains, the sensitivity and specificity of the DDT were 86% and 83.50%, respectively (for the blaTEM gene). In addition, for MBL strains, the sensitivity and specificity of the EIM were 91.66% and 97.43%, respectively (All reported for blaIMP gene).
Table 4.
Sensitivities, Specificities, Positive Predictive and Negative Predictive Values for Phenotypic and HRMA (blaTEM Gene for ESBL-Producing, blaIMP Gene for Carbapenemase-Producing and blaGES Gene for MBL-Producing) Methods in β-Lactamase-Producing Strain of P. aeruginosa
| Methods | No. of P. aeruginosa Strains (Total n = 88) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MBL Strains (Total n = 13) | KPC Strains (Total n = 2) | ESBL Strains (Total n = 20) | Sen (%) | Sp (%) | PPV (%) | NPV (%) | ||||||||||
| True Positive | False Positive | False Negative | True Negative | True Positive | False Positive | False Negative | True Negative | True Positive | False Positive | False Negative | True Negative | |||||
| DDT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 43 | 16 | 7 | 81 | 86 | 83.50 | 72.88 | 92.04 |
| EIM assay | 7 | 3 | 2 | 81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 77.77 | 96.42 | 70 | 91.01 |
| Hodge test | 0 | 0 | 0 | 0 | 11 | 2 | 1 | 76 | 0 | 0 | 0 | 0 | 91.66 | 97.43 | 84.61 | 98.70 |
| Carba-NP | 0 | 0 | 0 | 0 | 19 | 4 | 3 | 73 | 0 | 0 | 0 | 0 | 83.36 | 94.80 | 82.60 | 96.05 |
| HRM | 2 | 0 | 0 | 85 | 13 | 0 | 0 | 76 | 20 | 0 | 0 | 68 | 100 | 100 | 100 | 100 |
| Gold Standard | 2 | 0 | 0 | 85 | 13 | 0 | 0 | 76 | 20 | 0 | 0 | 68 | 100 | 100 | 100 | 100 |
Abbreviations: Sen, sensitivity; Sp, speciation; PPV, positive predictive values; NPV, negative predictive values.
According to Table 5, a significant relationship was observed between antibiotic resistance patterns, different classes of β-lactamase, and sensitivity/specificity of phenotypic methods and HRMA (p≤0.05). Thus, strains that had multiple antibiotic resistance played an essential role in enhancing the sensitivity and specificity of phenotypic and HRMA methods.
Table 5.
Relationship of Antibiotic Susceptibility Profiles, β-Lactamase Class and Sensitivity/Specificity of Phenotypic and HRMA in Clinical Isolates of P. aeruginosa
| Antibiotics Resistance | Methods | |||||
|---|---|---|---|---|---|---|
| DDT | EIM | Hodge test | Carba-NP | HRMA | Gold Standard | |
| Ceftazidime | 0.071 | 0.040 | 0.832 | 0.097 | 0.033 | 0.077 |
| Doripenem | 0.097 | 0.020 | 0.065 | 0.009 | 0.832 | 0.089 |
| Meropenem | 0.193 | 0.080 | 0.073 | 0.043 | 0.193 | 0.193 |
| Imipenem | 0.169 | 0.061 | 0.340 | 0.032 | 0.169 | 0.169 |
| Cefepime | 0.071 | 0.040 | 0.832 | 0.097 | 0.033 | 0.056 |
| Gentamycin | 0.097 | 0.020 | 0.065 | 0.009 | 0.832 | 0.117 |
| Ciprofloxacin | 0.193 | 0.080 | 0.073 | 0.043 | 0.193 | 0.146 |
| Amikacin | 0.169 | 0.061 | 0.340 | 0.032 | 0.169 | 0.088 |
| Norfloxacin | 0.071 | 0.040 | 0.832 | 0.097 | 0.033 | 0.077 |
| Aztreonam | 0.097 | 0.020 | 0.065 | 0.009 | 0.832 | 0.089 |
| Tobramycin | 0.193 | 0.080 | 0.073 | 0.043 | 0.193 | 0.193 |
| Piperacillin | 0.169 | 0.061 | 0.340 | 0.032 | 0.169 | 0.169 |
| Piperacillin/tazobactam | 0.071 | 0.040 | 0.832 | 0.097 | 0.033 | 0.056 |
| MDR | 0.218 | 0.824 | 0.343 | 0.383 | 0.647 | 0.052 |
| β-lactamase-producer strain | ||||||
| class A β-lactamases (ESBL) | 0.077 | 0.252 | 0.005 | 0.050 | 0.027 | 0.079 |
| class B β-lactamases (MBL) | 0.089 | 0.085 | 0.063 | 0.006 | 0.045 | 0.035 |
| class A β-lactamases (KPC) | 0.193 | 0.080 | 0.073 | 0.043 | 0.025 | 0.044 |
Discussion
One of the many concerning characteristics of P. aeruginosa is its low antibiotic susceptibility. It has a propensity to develop resistance during therapy, even evolving an MDR phenotype. Based on Table 2, a very high percent of P. aeruginosa isolates were resistant to ciprofloxacin (76.13%) and gentamycin, which (67.04%) is in agreement with the report of Kotwal et al,23 who found 82% and 77% resistance, respectively. This finding differs from an earlier report of Choudhary et al,24 who recorded 40% ciprofloxacin resistance among P. aeruginosa isolates; however, resistance to gentamycin found was 37%. Besides, the prevalence of carbapenemase-producing, ESBL-producing (both belong to class A β-lactamase), and MBL-producing (belong to class b β-lactamase) strains were 48.86%, 12.5%, and 7.95%, respectively. Under the present results, previous studies have demonstrated that the prevalence of MBL, ESBL, and carbapenemase-producing in P. aeruginosa isolates were more than 10%.23 The levels observed in this investigation are far below those observed by Farhan et al,25 who report 54% of ESBL-producing, 21% of carbapenemase-producing, and 52% of MBL-producing in P. aeruginosa isolates.
In the current study, based on Figure 3, phylogenetic diagrams of different P. aeruginosa strains were described. In this classification based on β-lactamase genes, the class A β-lactamase was the most abundant, and the blaTEM and blaSHV genes showed the most variation. These results reflect those of Karami et al,26 who also found that class A β-lactamase was the most prevalent.
In determining the specificity and sensitivity of the diagnostic method, the serial dilution plays a significant role. In some studies, Svec et al27 showed that the serial dilution of the DNA for the identification of different bacteria based on real-time PCR was of particular importance in increasing the sensitivity and specificity of the method. In Figures 4 and 5, the specificity/sensitivity of the method for class A and class B β-lactamase by real-time melting curve was described. The straight line was built through nine-fold serial dilutions, with a range from108 to 10°. Further, the temperature and length of the primer also play an essential role in the sensitivity of the real-time PCR reaction and efficiency of primers in the current study above 140%. The resulting plot showed an excellent correlation between the temperature and length of the primer and the method efficiency. This finding corroborates the ideas of Author links open overlay panel Wu et al,28 who suggested that the temperature and length of the primer for the identification of different bacteria based on real-time PCR was of particular importance in increasing the sensitivity and specificity of the method.
Based on Table 4 and a statistical analysis of the results of the HRMA test for the detection of carbapenemase-producing strains in P. aeruginosa, sensitivity, and specificity of the HRMA method was 100%. Also, the sensitivity and specificity of Carba-NP test was 90.47% and 94.87%, respectively. Also, sensitivity and specificity of MHT was 91.66% and 98.70%, respectively (All reported for blaKPC gene). For the detection of ESBL-producing strains, the sensitivity and specificity of the DDT were 95.55% and 95.55%, respectively (for blaSHV gene). In addition, for the detection of MBL-producing strains, the sensitivity and specificity of the EIM test were 77.77% and 97.59%, respectively. Given these results, we determined that the molecular identification of antibiotic-resistant bacteria is directly related to the resistance gene. Some genes have higher sensitivity and specificity to detect β-lactamase enzymes in P. aeruginosa. Peter et al,29 and Dortet et al,30 showed that phenotypic detection methods for class A and class B β-lactamase had been developed and used globally. While combined disk tests for MBL-producing (class B β-lactamase) P. aeruginosa, MHT for carbapenemase-producing, and double-disk test for ESBL-producing strains (class A β-lactamase) are frequently used due to cost-effectiveness and user-friendly manner. The specificity and sensitivity of the abovementioned methods are reported variously, 70–84% sensitivity, and 86–90% specificity.29,31,32
In our study, sensitivity and specificity of the Carba-NP test and MHT tests for the identification of carbapenemase-producing strains (class A β-lactamase) were 91.66%, 98.70% and 91.66%, 98.70%, respectively, Dortet et al,32 reported 100% sensitivity and specificity for the Carba-NP method. However, we showed that the sensitivity and specificity of the Carba-NP test were higher than the HTM test, which was reported by Bayramoğlu et al.22 They showed that phenotypic tests have high susceptibility and specificity to detect strains produced by P. aeruginosa. Also, for ESBL-producing strains, the sensitivity and specificity of the DD method were 95.55% and 95.55%, respectively, while for MBL-producing strains by EDTA-imipenem disk method, the sensitivity was 77.77% and specificity was 97.59% while Marchiaro et al21 and Bogiel et al33 reported that the sensitivity and specificity of the diagnostic method for MBL-producing strains is less than 85%, which is not consistent with our observations.
Our observations showed that the sensitivity and specificity of the HRMA method for the detection of ESBL-producing P. aeruginosa was 97.9% and 99.56%, respectively. For the identification of MBL-producing strains, the susceptibility and specificity of the HRMA method were 99.2% and 99.7%, respectively, and were 100% reported for the detection of carbapenemase-producing. Studies in UK,19 Denmark,34 and USA35 have shown that the sensitivity and specificity of HRMA to detect class A and class B β-lactamase are more than all phenotypic methods. However, in these studies, some phenotypic methods were used to detect strains producing ESBL and blaKPC with false positives and false negatives. It should be noted that phenotypic methods, although easy to access, but have many limitations. One of the most critical limitations of phenotypic methods is the inability to detect strains producing several enzymes. For example, routine phenotypic methods can not detect class A and class B β-lactamase enzymes simultaneously because phenotypic methods have limitations on implementation.
Table 5 shows that a relationship between antibiotic resistance and sensitivity and specificity of phenotypic tests. Elhariri et al36 reported that the frequency of the blaSHV gene is higher than that of the blaTEM gene. Rezai et al37 reported a high frequency of blaSHV genes in the study of the frequency of ESBL strains in P. aeruginosa. In the studies of Malkoçoğlu et al,38 and Giani et al,39 frequency of blaVIM gene was higher in comparison to the blaGES gene in P. aeruginosa strains.
The present study showed that the high sensitivity and specificity of the HRMA method in the detection of antibiotic-resistant strains in Pseudomonas aeruginosa clinical isolates compared to phenotypic methods explains its cost-effectiveness. Of course, optimizing real-time PCR in many cases can reduce the cost of response. One significant advantage of real-time chemistry platforms is that they do not require post-amplification processing to verify the specificity of the amplification products. Tong and Giffard40 demonstrated that real-time chemistries with exponential measurements clearly provide quantitative advantages of fast, precise, and accurate results. Besides, the number of reactions required to perform HRMA also plays an essential role in reducing costs. Murai et al41 found that the cost of real-time PCR reaction for a large number of bacterial isolates was more economical than for a small number.
However, based on our results, other advantages of an HRMA assay and real-time PCR technology are its ability to detect non-viable, fastidious, and unculturable organisms that would otherwise be missed by culture. Sirous et al,42 in other bacterias, showed that HRMA assay is a rapid, accurate, and cost-effective method possessing high sensitivity and specificity for the determination of antibiotic resistance. However, A PCR-positive, culture-negative specimen may reflect a real pathogen, yet detecting them would lead to a biased lower sensitivity and specificity value of the HRMA test. It should be noted that false positives could also be due to cell-free pathogen DNA circulating in the blood, from an old or controlled infection or contamination. Moreover, the quality and accuracy of these methods can be significantly enhanced by examining the distribution of genes responsible for resistance to β-lactamases and comparing these genes with the results of phenotypic methods. Because in some strains, it is possible that the resistance appears in a phenotypic form but does not have a resistance gene.
Limitations
There are several limitations to our study. First, the present study was conducted on a small number of clinical isolates of P. aeruginosa, while in order to reach a definitive and complete result, the population of the population should be increased and the number of strains with other resistance was also examined. Secondly, the focus on MDR and XDR strains is especially felt so that strains with broad resistance to different antibiotic groups can be detected in the shortest possible time. Hence, the detection of other strains, such as New Delhi Metallo-β-lactamase (NDM) strains, AmpC, and resistance to colistin, can also increase the strength of the study. Thirdly, the use of other molecular methods along with the HRMA method and the determination of their sensitivity and specificity can be helpful in choosing different molecular methods by researchers.
Conclusions
Our knowledge from this study suggests that some phenotypic methods are not suitable for identifying ESBL and MBL-producing P. aeruginosa strains. Therefore, cost-effectiveness (for a high number of samples), high speed and accuracy, and excellent sensitivity of the HRMA method can play an essential role in increasing the accuracy of clinical reporting. Moreover, methods such as HRMA can be used to reduce the diagnostic error in phenotypic methods with high speed and precision in identifying antibiotic-resistant bacteria. Hence, it can be concluded that the HRMA approach in identifying clinical isolates of P. aeruginosa carrying carbapenemase, MBL, and ESBL enzymes high efficiency.
Acknowledgments
The authors of this article are grateful to Hamadan University of Medical Sciences for their financial support in conducting the research.
Funding Statement
This article was conducted on the financial support of vice-chancellor for research of Hamadan University of Medical Sciences. This work was supported by a research grant from Hamadan University of Medical Sciences (Grant/Award Number: 9808145924).
Abbreviations
PPV, positive predictive values; TEM, temoneira; VEB, Verona integron-encoded metallo-β-lactamase; OXA, oxacillin hydrolyzing capabilities; MBL, metallo-β-lactamase; blaGES, Guiana extended-spectrum; blaIMP, imipenem; blaKPC, Klebsiella pneumoniae carbapenemase; PER, Pseudomonas extended resistant; ESBL, extended-spectrum beta-lactamase; PSE, Pseudomonas-specific enzymes; HRMA, high-Resolution Melt.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of Hamadan University of Medical Sciences (CodeNo: IR.UMSHA.REC.1398.573).
Consent for Publication
All the authors agree to publish the manuscript in Infection and Drug Resistance journal.
Author Contributions
HT and SD performed microbiological and molecular tests and wrote the manuscript. MYA and FK play a role in Project Administration. MA supervised all of the stages of designing the study, conducting the research, and writing the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Ullah W, Qasim M, Rahman H, Jie Y, Muhammad N. Beta-lactamase-producing Pseudomonas aeruginosa: phenotypic characteristics and molecular identification of virulence genes. J Chin Med Assoc. 2017;80(3):173–177. doi: 10.1016/j.jcma.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 2.Tahmasebi H, Yousef Alikhani M, Dehbashi S, Arabestani MR. Investigation of the relationship between the presence of chromosomal and plasmid-encoded ampc genes and type of clinical specimen in pseudomonas aeruginosa. J Babol Univ Med Sci. 2018;20(3):36–43. doi: 10.18869/acadpub.jbums.20.3.36 [DOI] [Google Scholar]
- 3.Kosykowska E, Dzieciątkowski T, Mlynarczyk G. Rapid Detection of NDM, VIM, KPC and IMP Carbapenemases by real-time PCR. J Bacteriol Parasitol. 2016;07(06). doi: 10.4172/2155-9597.1000299. [DOI] [Google Scholar]
- 4.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roschanski N, Fischer J, Guerra B, Roesler U. Development of a Multiplex real-time PCR for the Rapid Detection of the Predominant Beta-Lactamase Genes CTX-M, SHV, TEM and CIT-Type AmpCs in Enterobacteriaceae. PLoS One. 2014;9(7):e100956. doi: 10.1371/journal.pone.0100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansouri S, Kalantar Neyestanaki D, Shokoohi M, et al. Characterization of AmpC, CTX-M and MBLs types of β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli producing Extended Spectrum β-lactamases in Kerman, Iran. Jundishapur j Microbio. 2014;7(2):e8756–e. doi: 10.5812/jjm.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golshani Z, Ahadi AM, Sharifzadeh A. Occurrence of ambler class B metallo-β-lactamase gene in imipenem-resistant pseudomonas aeruginosa strains isolated from clinical samples. Zahedan J Res Med Sci. 2014;16(2):6–9. [Google Scholar]
- 8.Tahmasebi H, Dehbashi S, Arabestani MR. High resolution melting curve analysis method for detecting of carbapenemases producing pseudomonas aeruginosa. JKIMSU. 2018;7(4):70–77. [Google Scholar]
- 9.Haghi F, Zeighami H, Monazami A, Toutouchi F, Nazaralian S, Naderi G. Diversity of virulence genes in multidrug resistant Pseudomonas aeruginosa isolated from burn wound infections. Microb Pathog. 2018;115:251–256. doi: 10.1016/j.micpath.2017.12.052. [DOI] [PubMed] [Google Scholar]
- 10.van Dijk K, Voets GM, Scharringa J, et al. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin Microbiol Infect. 2014;20(4):345–349. doi: 10.1111/1469-0691.12322. [DOI] [PubMed] [Google Scholar]
- 11.Mlynarcik P, Roderova M, Kolar M. Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae. Jundishapur J Microbiol. 2016;9(1):e29314. doi: 10.5812/jjm.29314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gheorghe I, Czobor I, Chifiriuc MC, et al. Molecular screening of carbapenemase-producing Gram-negative strains in Romanian intensive care units during a one year survey. J Med Microbiol. 2014;63(Pt 10):1303–1310. doi: 10.1099/jmm.0.074039-0. [DOI] [PubMed] [Google Scholar]
- 13.de Lima-morales D, Ávila H, Soldi T, et al. Rapid detection of carbapenemase production directly from blood culture by colorimetric methods: evaluation in a routine microbiology laboratory. J Clin Microbiol. 2018. doi: 10.1128/jcm.00325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavelkovich A, Balode A, Edquist P, et al. Detection of carbapenemase-producing enterobacteriaceae in the baltic countries and st. petersburg area. Biomed Res Int. 2014;2014:7. doi: 10.1155/2014/548960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulou A, Grivakou E, Vrioni G, et al. Modified CLSI Extended-Spectrum β-lactamase (ESBL) Confirmatory test for phenotypic detection of ESBLs among Enterobacteriaceae producing various β-Lactamases. J Clin Microbiol. 2014;52(5):1483–1489. doi: 10.1128/JCM.03361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahmasebi H, Dehbashi S, Arabestani MR. Identification of gene mutation patterns obtained from resistance to mupirocin in methicillin-resistant staphylococcus aureus clinical strains, using high-resolution melting (HRMA) method. J Isfahan Med Sch. 2018;36(476):403–410. doi: 10.22122/jims.v36i476.9995. [DOI] [Google Scholar]
- 17.Chatzidimopoulos M, Ganopoulos I, Vellios E, Madesis P, Tsaftaris A, Pappas AC. Development of a two-step high-resolution melting (HRMA) analysis for screening sequence variants associated with resistance to the QoIs, benzimidazoles and dicarboximides in airborne inoculum of Botrytis cinerea. FEMS Microbiol Lett. 2014;360(2):126–131. doi: 10.1111/1574-6968.12594. [DOI] [PubMed] [Google Scholar]
- 18.Roth AL, Hanson ND. Rapid detection and statistical differentiation of KPC gene variants in gram-negative pathogens using high resolution melting and screenclust Analysis. J Clin Microbiol. 2012. doi: 10.1128/jcm.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards T, Williams C, Teethaisong Y, et al. A highly multiplexed melt-curve assay for detecting the most prevalent carbapenemase, ESBL and AmpC genes. bioRxiv. 2019:842963. doi: 10.1101/842963.. [DOI] [PubMed] [Google Scholar]
- 20.Sahni R, Mathai D, Sudarsanam T, et al. Extended-spectrum beta-lactamase producers: detection for the diagnostic laboratory. J Glob Infect Dis. 2018;10(3):140–146. doi: 10.4103/jgid.jgid_49_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchiaro P, Mussi MA, Ballerini V, et al. Sensitive EDTA-based microbiological assays for detection of metallo-β-lactamases in nonfermentative gram-negative bacteria. J Clin Microbiol. 2005;43(11):5648–5652. doi: 10.1128/jcm.43.11.5648-5652.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayramoglu G, Ulucam G, Gencoglu Ozgur C, Kilic AO, Aydin F. [Comparison of the modified Hodge test and the Carba NP test for detection of carbapenemases in Enterobacteriaceae isolates]. Mikrobiyol Bul. 2016;50(1):1–10. Turkish. doi: 10.5578/mb.10861 [DOI] [PubMed] [Google Scholar]
- 23.Kotwal A, Biswas D, Kakati B, Singh M. ESBL and MBL in cefepime resistant pseudomonas aeruginosa: an update from a rural area in northern India. J Clin Diagn Res. 2016;10(4):DC09–DC11. doi: 10.7860/JCDR/2016/18016.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhary V, Pal N, Hooja S. Prevalence and antibiotic resistance pattern of Metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from clinical specimens in a tertiary care hospital. J Mahatma Gandhi Inst Med Sci. 2019;24(1):19–22. doi: 10.4103/jmgims.jmgims_23_18. [DOI] [Google Scholar]
- 25.Farhan SM, Ibrahim RA, Mahran KM, Hetta HF, Abd El-Baky RM. Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect Drug Resist. 2019;12:2125–2133. doi: 10.2147/IDR.S198373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karami P, Mohajeri P, Yousefi Mashouf R, et al. Molecular characterization of clinical and environmental Pseudomonas aeruginosa isolated in a burn center. Saudi J Biol Sci. 2019;26(7):1731–1736. doi: 10.1016/j.sjbs.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. BDQ. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu DY, Ugozzoli L, Pal BK, Qian J, Wallace RB. The effect of temperature and oligonucleotide primer length on the specificity and efficiency of amplification by the polymerase chain reaction. DNA Cell Biol. 1991;10(3):233–238. doi: 10.1089/dna.1991.10.233. [DOI] [PubMed] [Google Scholar]
- 29.Peter S, Lacher A, Marschal M, et al. Evaluation of phenotypic detection methods for metallo-beta-lactamases (MBLs) in clinical isolates of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2014;33(7):1133–1141. doi: 10.1007/s10096-014-2059-1. [DOI] [PubMed] [Google Scholar]
- 30.Dortet L, Poirel L, Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clin Microbiol. 2012;50(11):3773–3776. doi: 10.1128/jcm.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manoharan A, Chatterjee S, Mathai D. Detection and characterization of metallo beta lactamases producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2010;28(3):241–244. doi: 10.4103/0255-0857.66486. [DOI] [PubMed] [Google Scholar]
- 32.Dortet L, Poirel L, Nordmann P. Rapid Detection of Carbapenemase-Producing Pseudomonas Spp. J Clin Microbiol. 2012;50(11):3773–3776. doi: 10.1128/jcm.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogiel T, Deptula A, Gospodarek E. Evaluation of different methods for detection of metallo-beta-lactamases in Pseudomonas aeruginosa clinical isolates. Pol J Microbiol. 2010;59(1):45–48. doi: 10.33073/pjm-2010-006 [DOI] [PubMed] [Google Scholar]
- 34.Njage PMK, Buys E, High Resolution A. DNA melting curve analysis for the rapid and efficient molecular diagnostics of extended spectrum β-lactamase determinants from foodborne. Escherichia Coli Microorganisms. 2020;8(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth AL, Hanson ND. Rapid detection and statistical differentiation of KPC gene variants in Gram-negative pathogens by use of high-resolution melting and ScreenClust analyses. J Clin Microbiol. 2013;51(1):61–65. doi: 10.1128/JCM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elhariri M, Hamza D, Elhelw R, Dorgham SM. Extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa in camel in Egypt: potential human hazard. Ann Clin Microbiol Antimicrob. 2017;16(1):21.d. doi: 10.1186/s12941-017-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezai MS, Ahangarkani F, Rafiei A, Hajalibeig A, Bagheri-Nesami M. Extended-spectrum beta-lactamases producing pseudomonas aeruginosa isolated from patients with ventilator associated nosocomial infection. Arch Clin Infect Dis. 2018;13(4):e13974. doi: 10.5812/archcid.13974. [DOI] [Google Scholar]
- 38.Gülşah M, Elif A, Banu B, Bariş O, Emin BM. VIM-1, VIM-2, and GES-5 Carbapenemases Among Pseudomonas aeruginosa Isolates at a Tertiary Hospital in Istanbul, Turkey. Microbial Drug Resist. 2017;23(3):328–334. doi: 10.1089/mdr.2016.0012. [DOI] [PubMed] [Google Scholar]
- 39.Giani T, Arena F, Pollini S, et al. Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J Antimicrob Chemother. 2017;73(3):664–671. doi: 10.1093/jac/dkx453. [DOI] [PubMed] [Google Scholar]
- 40.Tong SYC, Giffard PM. Microbiological applications of high-resolution melting analysis. J Clin Microbiol. 2012;50(11):3418–3421. doi: 10.1128/JCM.01709-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murai K, Lehenbauer TW, Champagne JD, Glenn K, Aly SS. Cost-effectiveness of diagnostic strategies using quantitative real-time PCR and bacterial culture to identify contagious mastitis cases in large dairy herds. Prev Vet Med. 2014;113(4):522–535. doi: 10.1016/j.prevetmed.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Sirous M, Khosravi AD, Tabandeh MR, Salmanzadeh S, Ahmadkhosravi N, Amini S. Molecular detection of rifampin, isoniazid, and ofloxacin resistance in Iranian isolates of Mycobacterium tuberculosis by high-resolution melting analysis. Infect Drug Resist. 2018;11:1819–1829. doi: 10.2147/IDR.S178831. [DOI] [PMC free article] [PubMed] [Google Scholar]