Abstract
Background
The aberrant expression of circular RNAs (circRNAs) has been identified as a novel trait of cancers. However, the role of circRNAs in colorectal cancer (CRC) remains to be elucidated.
Methods
Informatic analysis was performed to identify circRNAs in CRC tissues and adjacent tissues. Gain- and loss-of-function experiments were constructed to analyze hsa_circ_001806 roles in CRC cell stemness by sphere-formation, ALDH activity, stemness marker expression and tumor-initiating ability assays. CCK8 cell viability was carried out to evaluate hsa_circ_0001806 roles in CRC cell viability. Luciferase reporter and pull-down assays were used to reveal the underlying mechanisms.
Results
Hsa_circ_0001806 was significantly upregulated in CRC tissues and correlated with TNM stage, depth of invasion, lymphatic metastasis and distant metastasis. Hsa_circ_0001806 promoted the stemness of CRC cells, as evident by increasing sphere-formation ability, ALDH1 activity and stemness marker expression while had no effect on cell viability. Mechanistically, the same miR-193-5p-binding sites are shared between hsa_circ_0001806 and COL1A1. Hsa_circ_0001806 upregulates COL1A1 expression in a miR-193-5p-dependent manner, which is essential for hsa_circ_0001806-mediated regulation on CRC cell stemness.
Conclusion
CircRNA hsa_circ_0001806 may act as a promising therapeutic target by facilitating the stemness of CRC cells via activating the hsa_circ_0001806/miR-193a-5p/COL1A1 axis.
Keywords: hsa_circ_0001806, miR-193a-5p, COL1A1, ceRNA, stemness, colorectal cancer
Introduction
Colorectal carcinoma (CRC) is the third most common cancer, with 1.8 million newly diagnosed cases and 0.88 million deaths annually.1 Exploring the underlying mechanisms of CRC progression is crucial to developing more effective treatment options. Plenty of evidences have shown that cancer stem cell (CSC) is the origin of tumor progression, and there are also CSCs existing in CRC.2 Therefore, it is very important to elucidate the underlying mechanisms contributing to colorectal CSC progression.
Accumulating evidence indicates that colorectal CSC progression results from multiple complex factors that include genetic, molecular and epigenetic alterations.3 Circular RNAs (circRNAs) are defined as a class of non-coding transcripts and characterized by the closed-loop structures, and their regulatory roles have attracted enormous interest recently.4 The mechanisms by which circRNAs function as competing endogenous RNAs (ceRNAs) have been mostly studied in cancer research. In addition, the other cellular RNA mechanisms, including regulation of PTMs and protein translation, have also been revealed.5 For example, circNSUN2, which can form an RNA-protein ternary complex with IGF2BP2 and HMGA2, promotes CRC liver metastasis by enhancing the mRNA stability of HMGA2;6 Overexpression of ciRS-7 abrogates the miR-7-mediated suppression on gastric cancer progression via antagonizing miR-7-mediated PTEN/PI3K/AKT pathway;5 CircRNA hsa_circRNA_102958 promotes the CRC progression via sponging miR-585.7 However, the roles of circRNAs in colorectal CRC progression are still unclear.
There are many members of collagen family, including COL1A1, COL1A2, etc., among which, COL1A1 has been reported to be involved in the progression of several cancers.8 COL1A1 supports the matrix structure and forms the stromal part of most solid tumors, and mainly affects cell proliferation, differentiation, adhesion and metastasis through extracellular matrix receptor interaction pathway and local adhesion pathway, which is mainly related to tumor invasion and metastasis.9 Since CSC contribute to tumor metastasis, COL1A1 might be involved in CSC progression.10 And the underlying mechanisms by which COL1A1 is regulated are still unclear in CRC. In the present study, we initially explored the expression profile of circRNA, miRNA and mRNA in CRC and paired adjacent tissues in TCGA, GEO datasets and clinical tissues. Then, hsa_circ_0001806 was identified to be upregulated in CRC patients and to promote CRC cell stemness by functioning as a ceRNA of COL1A1. This study indicates that hsa_circ_0001806 exerts an oncogenic role and may be a potential checkpoint in CRC diagnosis and therapy.
Materials and Methods
Tissues and Cell Lines
Thirty-one pairs of CRC tissues and corresponding adjacent tissues were collected from CRC patients who underwent surgical resection without preoperative chemoradiotherapy at the Affiliated Suzhou Science&Technology Town Hospital of Nanjing Medical University between October 2017 and October 2019. This study was approved by the Ethics Committee of the Affiliated Suzhou Science&Technology Town Hospital of Nanjing Medical University. All patients provided written informed consent, and that this experiment was conducted in accordance with the Declaration of Helsinki. The human CRC cells HCT-116, DLD-1, SW480, CT-26, SW620, SW1116, HT29, LOVO and the human normal colon epithelial cells NCM460 were purchased from the Chinese Academy of Science. All cell lines were cultured with DMEM supplemented with 10% fetal bovine serum and penicillin-streptomycin solution.
Computational Analysis
GSE126095 is the Agilent gene chip used to examine circRNA expression changes in 10 paired CRC and adjacent mucosal tissues, and used to explore the correlation between circRNA expression dysregulation and CRC tumourigenesis and progression. It was obtained from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database. The “fold change” (ie, the ratio of the group averages) between the groups for each circRNA was computed. Statistically significant difference was evaluated by t-test. CircRNAs with 2 fold changes and P-values of <0.05 were considered to be significantly different. The expression profiles of miRNA and mRNA in “fragments per kilobase of transcript per million mapped reads” for patients with CRC were downloaded from the TCGA database. circRNA expression profile data in raw read counts were downloaded for the GEO database. These raw data were processed by background correction and normalization using the “affy” package of R/Bioconductor. The “limma” package was used to identify the differentially expressed circRNAs (DEcircRNAs), while the edgeR package was used to identify the differentially expressed miRNAs (DEmiRNAs) and mRNAs (DEmRNAs) between normal samples and tumor samples. The cut-off values of circRNA were set at the adjusted P-value of <0.01 and |log2 fold change (FC)| >3.0, while the cut-off values of miRNA and mRNA were set at the adjusted P-value of <0.01 and |log2 FC| >2.0. We used GSEA to obtain enriched gene sets in the KEGG data set, and 17,631 mRNA expression grouped by the high- or low-expression of hsa_circ_001806 was used as input.
RNA Isolation and qRT-PCR
Total RNA was isolated from tissue and cells using TRIzol reagent (Vazyme, Nanjing, China). cDNA was reverse transcribed from total RNA using HiScript II Q RT SuperMix for qPCR (Vazyme, Nanjing, China). The transcripts were amplified and detected using AceQ SYBR Green PCR Master Mix (Vazyme, Nanjing, China), and the results were obtained by qRT-PCR. The circRNA and mRNA levels were normalized to the GAPDH levels, while the miRNA levels were normalized to the U6 levels.
RNA-Fluorescence in situ Hybridization (RNA-FISH)
RNA-FISH for hsa_circ_0001806 was performed in SW620 and HT-29 cells. The detailed procedure was referred to the previous study.11 The 5ʹ FAM-labeled Locked Nucleic Acid (LNA) probe directed against hsa_circ_0001806 and 4, 6-diamidino-2-phenylindole (DAPI) were purchased from the Biomics (Nantong, China).
Plasmid Construction, Lentivirus Package and Transfection
For upregulating and downregulating the expression of hsa_circ_0001806, full-length hsa_circ_0001806 (Len-circ_0001806) and the shRNA sequence targeting hsa_circ_0001806 (sh-circ_0001806) were synthesized by GenePharma (Shanghai, China) and cloned into the lentivirus vectors. CRC cells infected with shRNAs targeting hsa_circ_0001806 or negative control hairpin shRNA, termed shRNA 1, shRNA2 and shControl, were verified by qRT-PCR. The infected cells were then treated with puromycin to select the stably infected cells. We determined that shRNA 1 held the highest knockdown efficiency and was used for the mechanistic studies. To construct the luciferase reporter vector, the sequences of hsa_circ_0001806 and COL1A1 3′UTR were cloned downstream of the pMIR-Reporter vector. A Mutagenesis Kit (Vazyme, Nanjing, China) was used to generate mutations in the miR-193-5p-binding sites in hsa_circ_0001806 and the COL1A1 3′UTR sequence. The overexpression and knockdown lentivirus of miR-193-5p was purchased from GenePharma, designated lenti-miR-193-oe and lenti-miR-193-kd, respectively. For decreasing the expression of COL1A1 in CRC cells, the siRNA (si-COL1A1) and negative control sequences were synthesized by GenePharma. Si-COL1A1 was transfected into CRC cells with hsa_circ_0001806 overexpression.
Tumor Sphere Formation Analysis
Cells were cultured in 12-well low-adherent plates with a density of 1000/mL using sphere-culturing medium (DMEM/F12 without serum with 1 × B27 reagent, EGF (20 ng/mL), bFGF (20 ng/mL), insulinolin (5 g/mL), hydrocortisone (1 g/mL) and 1% penicillin and streptomycin), and the fresh medium were added every 3 days. After 10 days of culture, sphere formation (>50 μm) was observed under the microscope and counted.
ALDH Activity Assay
Since CSCs hold significantly higher level of ALDH activity than the parental cancer cells, ALDH activity was measured to evaluate the stemness of NPC cells. The detection was performed referring to the protocols of ALDH Activity Assay Kit (Colorimetric) (Taipei, China).
CCK8 Cell Viability Assay
Cells with or without ectopic expression of hsa_circ_0001806 were seeded into the 96-well plates. After the cells were attached (0 h), CCK8 was added and incubated for another 1 h at 24 h, 48 h and 72 h. The absorbance value was measured at 450 nm and used to evaluate the cell viability which is positively correlated with the absorbance value.
Pull-Down Assay
Briefly, a total of 1 × 107 CRC cells were lysed and incubated with C-1 magnetic beads (Thermo Fisher Scientific, USA) bound to a hsa_circ_0001806 probe or oligo probe at 4°C overnight. We extracted RNA from the magnetic beads after washing the beads three times. The isolated RNA mixtures were then analyzed by qRT-PCR.
Dual-Luciferase Reporter Assay
The wild-type (Wt) or mut hsa_circ_0001806 plasmid or COL1A1 was co-transfected with miR-193a-4p mimics using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) into CRC cells. Finally, a Dual-Luciferase Reporter System Kit (Promega, Madison, WI, USA) was used to detect the firefly and Renilla luciferase activity.
Western Blot Assay
Immunoblot analysis was conducted as previously described [23]. Briefly, total proteins were lysed by Cell lysis buffer for Western and IP (Beyotime, China) containing protease inhibitors (Beyotime, China). Total cellular protein samples were separated by 10% SDS-PAGE gel and completely transferred onto a PVDF membrane (Millipore, USA). After incubating with the primary antibodies overnight, the membranes were then incubated with goat anti-rabbit IgG H&L (Abcam, USA) for 2 h. After washing the PVDF membrane three times, a chemiluminescence system was used to detect signals which were analyzed using FluorChem FC3 Software (ProteinSimple, USA). GAPDH was used to normalize protein loading. The original Western blots were indicated in Supplemental Figure S1.
Tumor-Initiating Ability Evaluation
All animal experiments were performed with the approval of Ethics Committee for Animal Experimentation of the Affiliated Suzhou Science & Technology Town Hospital of Nanjing Medical University, and this work followed institutional guidelines for the ethical and legal treatment of animals. Six-week athymic BALB/c nude mice were purchased from the Charles River (Beijing, China). For tumor-limiting dilution assays, tumor cells were mixed 1:1 with Matrigel matrix (BD Biosciences, San Jose, CA) and subcutaneously implanted in mice. After 15 days, all mice were killed and tumor tissues were collected and weighed. And the CSC rate in CRC cells was determined using the online analysis tool (ELDA: http://bioinf.wehi.edu.au/software/elda/).
Statistical Analysis
All the above experimental assays were repeated in triplicate. Data are represented as the means ± standard deviation (SD). Student’s t test or two-way ANOVA was used to determine the statistical significance of the difference between two or multiple groups, and the χ2 test was used to assess the correlation between hsa_circ_0001806 levels and clinical features. Data were analyzed and presented using Prism software 8.0 (GraphPad Software, USA). P < 0.05 was considered to be a statistically significant difference.
Results
Hsa_circ_0001806 Level Is Upregulated in CRC
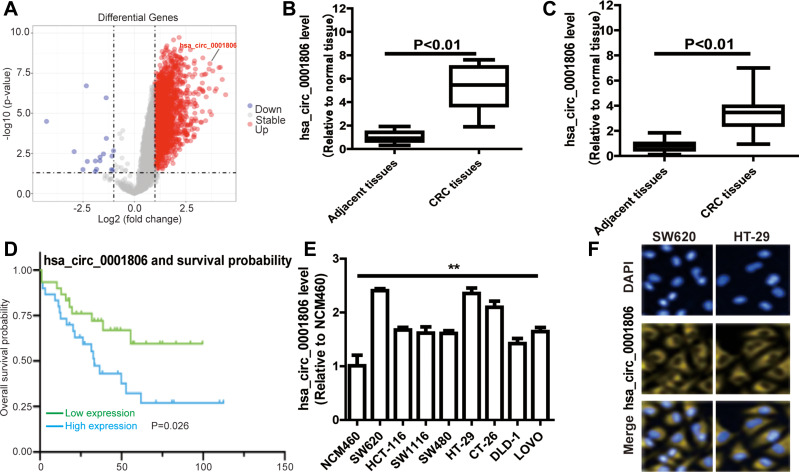
We used the expression profiles of circRNAs (GSE126095, which contains 3962 circRNA probes) as input to identify the differentially expressed circRNAs. A total of 1827 dysregulated circRNAs were identified in CRC tissues, of which 1808 circRNAs were upregulated and 19 circRNAs were downregulated (Figure 1A). Among these, a novel circRNA named hsa_circ_0001806 was differentially expressed in CRC tissues and corresponding adjacent tissues (Figure 1B). Then, the aberrant expression of hsa_circ_0001806 was confirmed using qRT-PCR assay in clinical samples (n = 31, P < 0.001) (Figure 1C). Kaplan–Meier survival curves of CRC patients (n = 31) demonstrated that the overall survival of CRC patients with a high hsa_circ_0001806 level was significantly lower than that of hsa_circ_0001806 low-expression CRC patients (*P = 0.026) (Figure 1D). Next, the 31 CRC patients were grouped into hsa_circ_0001806high and hsa_circ_0001806low groups by the median level of hsa_circ_0001806 to investigate the clinical significance of the upregulated expression of hsa_circ_0001806 in CRC. The statistical results showed that the expression of hsa_circ_0001806 was highly correlated with the TNM stage, depth of invasion, lymphatic metastasis and distant metastasis (Table 1). Additionally, hsa_circ_0001806 level was examined in different CRC cells and normal colon epithelial cells, and it was found that hsa_circ_0001806 exhibited a higher level in CRC cells, especially in SW620 and HT-29, which were used as the research subjects (Figure 1E). To investigate the cellular localization of hsa_circ_0001806, RNA-FISH assay was performed. The results revealed that hsa_circ_0001806 was preferentially located in the cytoplasm of SW620 and HT-29 cells (Figure 1F). Furthermore, we avoided trans-splicing or genomic rearrangements, including head-to-tail splicing, by employing several universal circRNA detection methods.12 We first designed convergent and divergent primers to amplify hsa_circ_0001806 (98 bp) and linear mRNA (238 bp). We used cDNA and genomic DNA (gDNA) from CRC cells as templates to amplify hsa_circ_0001806 from cDNA using only divergent primers, while no amplification product was observed from genomic DNA (Supplementary Figure S2A). We also used Sanger sequencing to confirm the head-to-tail splicing in the RT-PCR product of hsa_circ_0001806 with the expected size (Supplementary Figure S2B). RNase R is an exoribonuclease that can degrade RNA from its 3′ to 5′ end but does not act on circRNA.13 As shown in Supplementary Figure S2C, hsa_circ_0001806 was resistant to RNase R.
Figure 1.
Hsa_circ_0001806 is upregulated in CRC. (A) The volcano plot visualizes the expression of circRNAs in 10 paired samples of CRC tissues in GSE126095 dataset. (B) Hsa_circ_0001806 expression was detected in GSE126095 dataset. (C) Hsa_circ_0001806 expression was detected in 31 pairs of CRC and adjacent tissue samples. (D) Kaplan-Meier survival curve of overall survival in 31 patients with CRC according to hsa_circ_0001806 expression. Patients were stratified into high expression and low expression groups by the median expression. (E) Hsa_circ_0001806 expression was detected and was examined in different CRC cells and normal colon epithelial cells. (F) RNA-FISH assay was constructed to determine the localization of hsa_circ_0001806 in SW620 and HT-29 cells. Data were presented as mean ± s.d; **P < 0.01 vs control.
Table 1.
Relevance Analysis of Hsa_circ_0001806 Expression in CRC Patients
| Variables | All Patients | circ_0001806 | P value | |
|---|---|---|---|---|
| Low | High | |||
| All cases | 31 | 15 | 16 | |
| Age (years) | 0.621 | |||
| <60 | 15 | 7 | 8 | |
| ≥60 | 16 | 8 | 8 | |
| Gender | 0.239 | |||
| Male | 18 | 7 | 7 | |
| Female | 13 | 8 | 9 | |
| Tumour diameter (cm) | 0.578 | |||
| <5 | 12 | 6 | 7 | |
| ≥5 | 19 | 9 | 9 | |
| TNM Stage | 0.129 | |||
| I+II | 19 | 7 | 8 | |
| III+IV | 12 | 8 | 8 | |
| Depth of invasion | 0.372 | |||
| T1+T2 | 13 | 8 | 9 | |
| T3+T4 | 8 | 7 | 7 | |
| Lymphatic metastasis | 0.327 | |||
| Yes | 13 | 6 | 7 | |
| No | 18 | 9 | 9 | |
| Distant metastasis | 0.008 | |||
| Yes | 14 | 5 | 6 | |
| No | 17 | 10 | 10 | |
| CEA (ng/mL) | 0.632 | |||
| <5 | 18 | 11 | 9 | |
| ≥5 | 13 | 4 | 7 | |
| Tumor location | 0.591 | |||
| Colon | 13 | 10 | 10 | |
| Rectum | 18 | 5 | 6 | |
Hsa_circ_0001806 Promotes the Stemness of CRC Cells
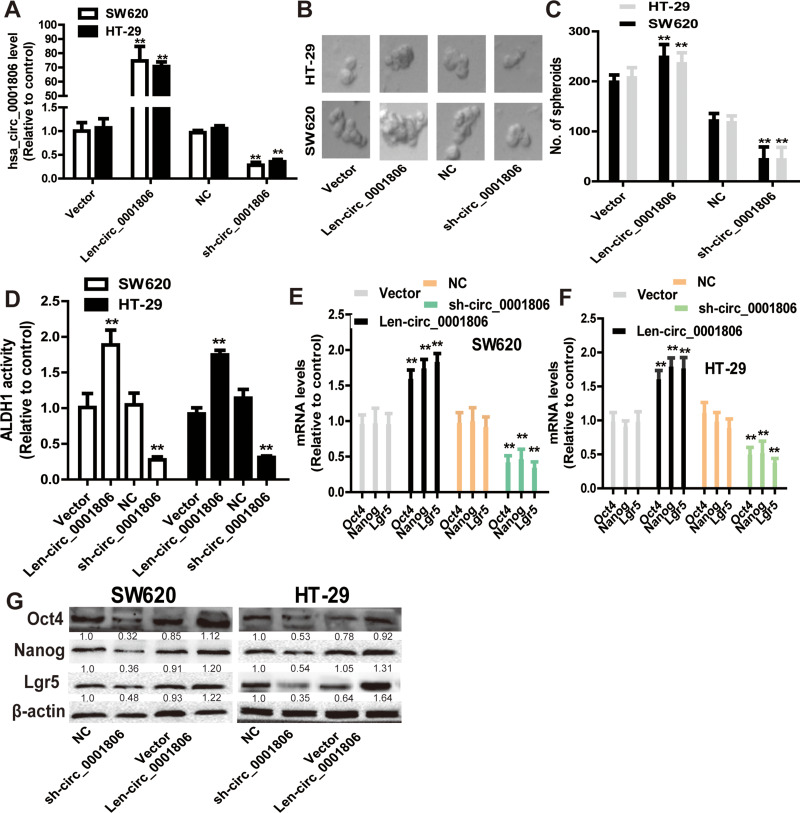
Since CSCs are regarded as the origin of tumor progression, we focused on the effects of hsa_circ_0001806 on CRC cell stemness. Hsa_circ_0001806 was stably overexpressed and knocked down in SW620 and HT-29 cells via lentivirus infection. The efficiency was confirmed by qRT-PCR assay (Figure 2A). Firstly, the effects of ectopic expression of hsa_circ_000180 on CRC sphere-formation ability were examined. As shown in Figure 2B and C, overexpression of hsa_circ_0001806 significantly increased the sphere size and number of CRC cells, while knockdown of hsa_circ_0001806 exerted an opposite effect. Furthermore, ALDH1 activity was increased in CRC cells with hsa_circ_0001806 overexpression, while reduced in CRC cells with hsa_circ_0001806 knockdown (Figure 2D). Additionally, hsa_circ_0001806 overexpression increased the expression of stemness critical regulators (Oct4, Nanog and Lgr5), while hsa_circ_0001806 knockdown decreased it (Figure 2E–G).
Figure 2.
Hsa_circ_0001806 promotes the stemness of CRC cells. (A) The infection efficiency of hsa_circ_0001806 overexpression or knockdown was confirmed by qRT-PCR. (B and C) The sphere size and number were measured in SW620 and HT-29 cells with ectopic expression of hsa_circ_0001806. (D) ALDH1 activity was examined in CRC cells with ectopic expression of hsa_circ_0001806. (E–G) The expression of stemness critical regulators (Oct4, Nanog and Lgr5) was detected in CRC cells with ectopic expression of hsa_circ_0001806. Data were presented as mean ± s.d; **P < 0.01 vs control.
Hsa_circ_0001806 Enhances the Tumor-Initiating Ability of CRC Cells
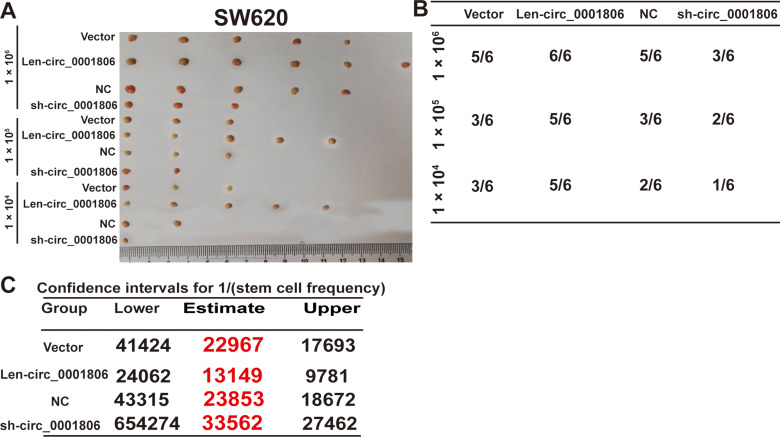
Next, we examined the effects of hsa_circ_0001806 on the tumor-initiating ability of CRC cells, which is positively correlated with the stemness of tumor cells. As shown in Figure 3A and B, the tumor-initiating ability was significantly increased in CRC cells with hsa_circ_0001806 overexpression, and decreased in CRC cells with hsa_circ_0001806 knockdown, which was evident by the change of tumor-formation ratio. Consistently, the CSC rate was analyzed using the ELDA tool and it was found that the CSC rate was increased in CRC cells with hsa_circ_0001806 overexpression, and decreased in CRC cells with hsa_circ_0001806 knockdown (Figure 3C). Thus, our results demonstrate that hsa_circ_0001806 can promote the stemness of CRC cells.
Figure 3.
Hsa_circ_0001806 enhances the tumor-initiating ability of CRC cells. (A) Tumor images derived from CRC cells with ectopic expression of hsa_circ_0001806 at the different cell number. (B) The tumor formation rate was evaluated in CRC cells depicted in (A). (C) ELDA assay was used to analyze the CSC rate in the tumors described in (A).
Hsa_circ_0001806 Acts as an miR-193-5p Sponge to Increase COL1A1 Expression
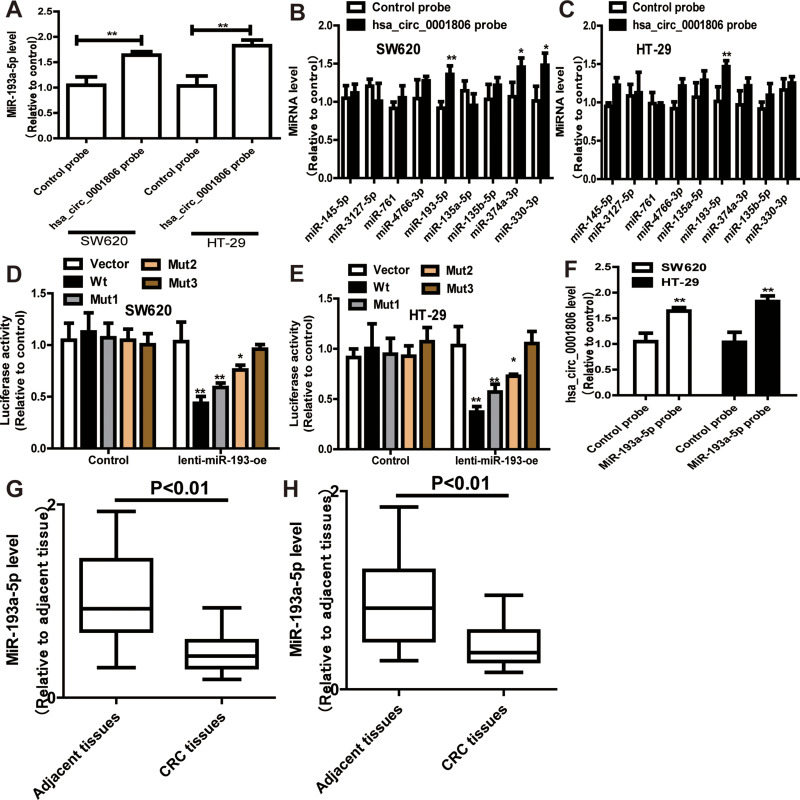
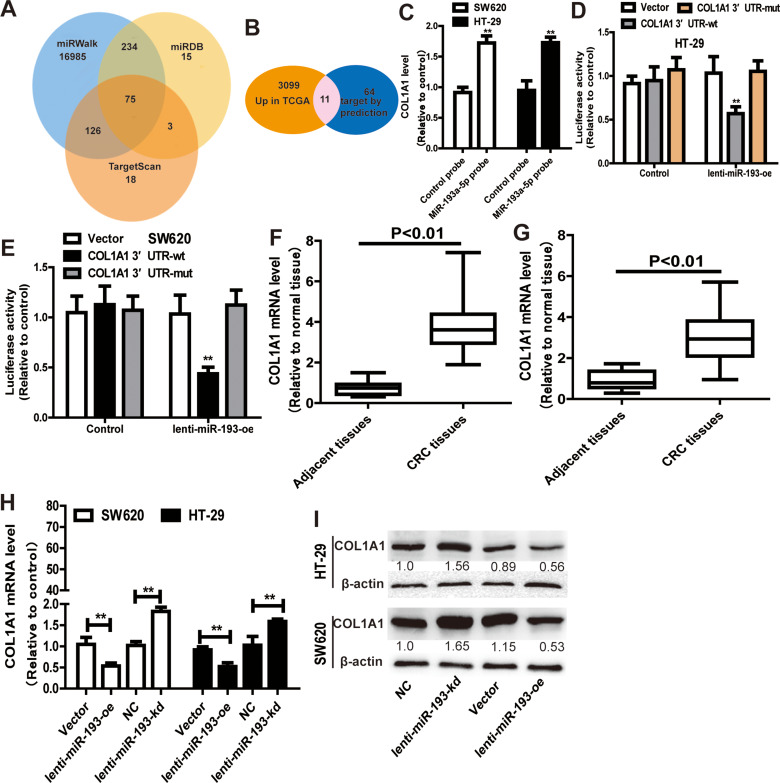
To identify the potential hsa_circ_0001806-sponging miRNA, we selected the top 10 candidate miRNAs according to the presence of miRNA recognition elements in the hsa_circ_0001806 sequence (Table 2). An RNA pull-down assay was performed with a biotinylated hsa_circ_0001806 probe to investigate the potential hsa_circ_0001806-sponging miRNA to study the interaction between hsa_circ_0001806 and miR-193a-5p (Figure 4A). The results indicated that hsa_circ_0001806 could specifically bind to miR-193a-5p in both SW620 and HT-29 cell lines but not the other miRNAs predicted by miRanda (Figure 4B and C). The luciferase reporter assay was used to further demonstrate the interaction between hsa_circ_0001806 and miR-193a-5p, and the results showed that miR-193a-5p significantly reduced the luciferase activity of the hsa_circ_0001806-WT reporter but had weaker effects on the activities of the mutated miR-193a-5p binding site reporters, including hsa_circ_0001806 MUT1, hsa_circ_0001806 MUT2 and hsa_circ_0001806 MUT3, especially that of the hsa_circ_0001806 MUT3 (Figure 4D and E). In addition, we also verified the interaction of miR-193a-5p and hsa_circ_0001806 using a biotinylated miR-193a-5p probe to conversely successfully capture hsa_circ_0001806 (Figure 4F). On the basis of these findings, we also found downregulated expression of miR-193a-5p in a TCGA data set, indicating that miR-193a-5p was significantly downregulated in CRC tissues and upregulated in adjacent tissues (Figure 4G). Then, the aberrant expression of miR-193a-5p was shown using qRT-PCR (n = 31, P < 0.001) (Figure 4H).
Table 2.
Top 10 Targets Predicted by miRanda
| Seq1 | Seq2 | Tot Score | Tot Energy | Max Score | Max Energy | Strand | Len1 | Len2 | Positions |
|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-193a-5p | hsa_circ_0001806 | 308 | −47.1 | 158 | −24.55 | 824 | 22 | 432 | 273194 |
| hsa-miR-4510 | hsa_circ_0001806 | 308 | −51.93 | 164 | −26.69 | 19197 | 22 | 432 | 302352 |
| hsa-miR-6809-3p | hsa_circ_0001806 | 298 | −32.11 | 156 | −17.11 | 25909 | 21 | 432 | 92150 |
| hsa-miR-520g-3p | hsa_circ_0001806 | 292 | −38.3 | 147 | −20.66 | 4011 | 24 | 432 | 23849 |
| hsa-miR-3168 | hsa_circ_0001806 | 291 | −24.35 | 147 | −14.19 | 15881 | 17 | 432 | 169113 |
| hsa-miR-520h | hsa_circ_0001806 | 290 | −29.89 | 146 | −19.59 | 4021 | 22 | 432 | 24551 |
| hsa-miR-1231 | hsa_circ_0001806 | 286 | −36.74 | 144 | −20.86 | 6962 | 20 | 432 | 2921 |
| hsa-miR-508-5p | hsa_circ_0001806 | 284 | −46.58 | 144 | −26.43 | 4050 | 23 | 432 | 53183 |
| hsa-miR-6830-3p | hsa_circ_0001806 | 281 | −24.93 | 141 | −14.49 | 25951 | 23 | 432 | 87269 |
| hsa-miR-6770-5p | hsa_circ_0001806 | 168 | −24.38 | 168 | −24.38 | 25830 | 24 | 432 | 352 |
Figure 4.
Hsa_circ_0001806 specifically binds to miR-193a-5p. (A) RNA pull down was performed to determine the miR-193a-5p level in RNA pulled down by hsa_circ_0001806 probe in CRC cells. (B and C) The levels of miRNAs as indicated were determined in RNA pulled down by hsa_circ_0001806 probe in CRC cells. (D and E) Dual-luciferase reporter assay was performed to determine the direct binding between hsa_circ_0001806 and miR-193a-5p based on their complementary sequences. (F) Hsa_circ_0001806 level was detected in RNA pulled down by miR-193a-5p probe in CRC cells. (G and H) MiR-193a-5p level was examined in GSE126095 dataset (G) and clinical samples (H). Data were presented as mean ± s.d; *P < 0.05, **P < 0.01 vs control.
We predicted the potential targets of miR-193a-5p by three online tools (miRanda, TargetScan and miRDB), and 75 candidate mRNAs were selected (Figure 5A). Then, the upregulated mRNAs in the TCGA data set were compared to the candidate targets, and among these, COL1A1 was chosen for further study (Figure 5B). To investigate the interaction between miR-193a-5p and COL1A1, we measured the expression of COL1A1 in the RNA mixture captured by the biotinylated miR-193a-5p probe. The positive results validated the predicted relationship (Figure 5C). Moreover, the luciferase reporter assay was used to further confirm the interaction between COL1A1 and miR-193a-5p, and the result showed that miR-193a-5p expression significantly reduced the luciferase activity of the COL1A1 WT reporter but not that of the COL1A1 MUT reporter (Figure 5D and E). In addition, we also tested the dysregulation of COL1A1 in the TCGA COAD and READ data sets, revealing that COL1A1 was significantly upregulated in CRC tissues and downregulated in adjacent tissues (Figure 5F). Then, the aberrant expression of COL1A1 was confirmed using qRT-PCR (n = 31, P < 0.001) (Figure 5G). Next, the mRNA and protein levels of COL1A1 were found to be altered in SW620 and HT-29 cells with overexpression or knockdown of miR-193a-5p and could be restored by knockdown or overexpression of COL1A1 (Figure 5H and I).
Figure 5.
MiR-193a-5p specifically binds to COL1A1. (A) Venn diagram showing 75 genes that are putative miR-193a-5p targets computationally predicted by three algorithms (miRWalk, miRDB and TargetScan). (B) These computed results were compared with the upregulated genes in GSE126095 dataset. (C) COL1A1 level was detected in RNA pulled down by miR-193a-5p probe in CRC cells. (D and E) Dual-luciferase reporter assay was performed to determine the direct binding between COL1A1 and miR-193a-5p based on their complementary sequences. (F and G) COL1A1 level was examined in GSE126095 dataset (F) and clinical samples (G). (H and I) The expression of COL1A1 was measured in CRC cells with ectopic expression of miR-193a-5p. Data were presented as mean ± s.d; **P < 0.01 vs control.
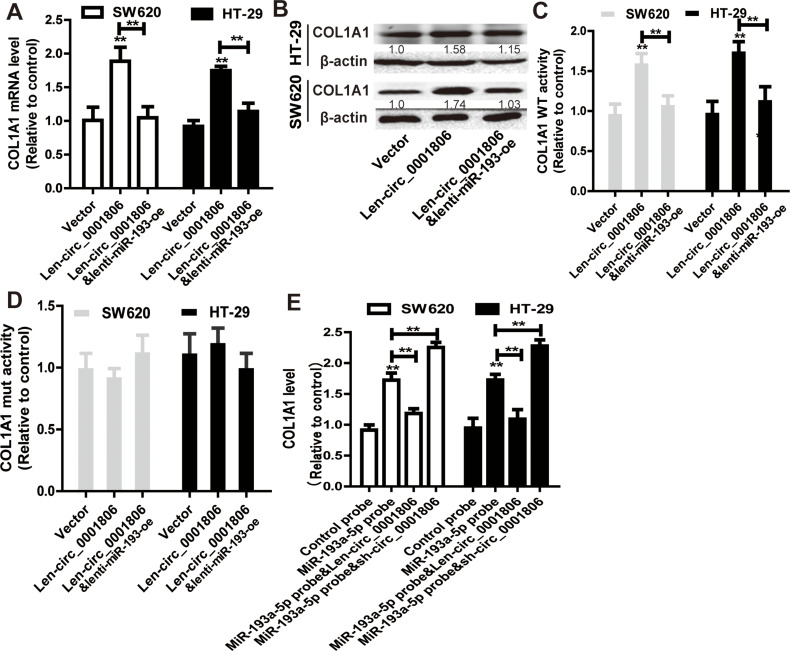
Moreover, it was found that hsa_circ_0001806 could positively regulate the expression of COL1A1 in a miR-193a-5p-dependent manner (Figure 6A and B). In consistent, hsa_circ_0001806 positively regulated the activity of COL1A1 WT reporter in a miR-193a-5p-dependent manner, but had no effect on the activity of COL1A1 mut reporter (Figure 6C and D). Additionally, the abundance of COL1A1 in the RNA mixture captured by the biotinylated miR-193a-5p probe was reduced by hsa_circ_0001806 overexpression, and was increased by hsa_circ_0001806 knockdown (Figure 6E). Therefore, these results indicate that hsa_circ_0001806 overexpression acts as a ceRNA for COL1A1 via sponging miR-193a-5p in CRC cells.
Figure 6.
Hsa_circ_0001806 acts as a ceRNA for COL1A1 via competitively binding to miR-193a-5p. (A and B) COL1A1 expression was examined in CRC cells with hsa_circ_0001806 overexpression as well as miR-193a-5p overexpression or not. (C) Dual-luciferase reporter assay was performed to determine the activity of COL1A1 WT in the cells described in (A). (D) Dual-luciferase reporter assay was performed to determine the activity of COL1A1 MUT in the cells described in (A). (E) COL1A1 level was detected in RNA pulled down by miR-193a-5p probe in the cells depicted in (A). Data were presented as mean ± s.d; **P < 0.01 vs control.
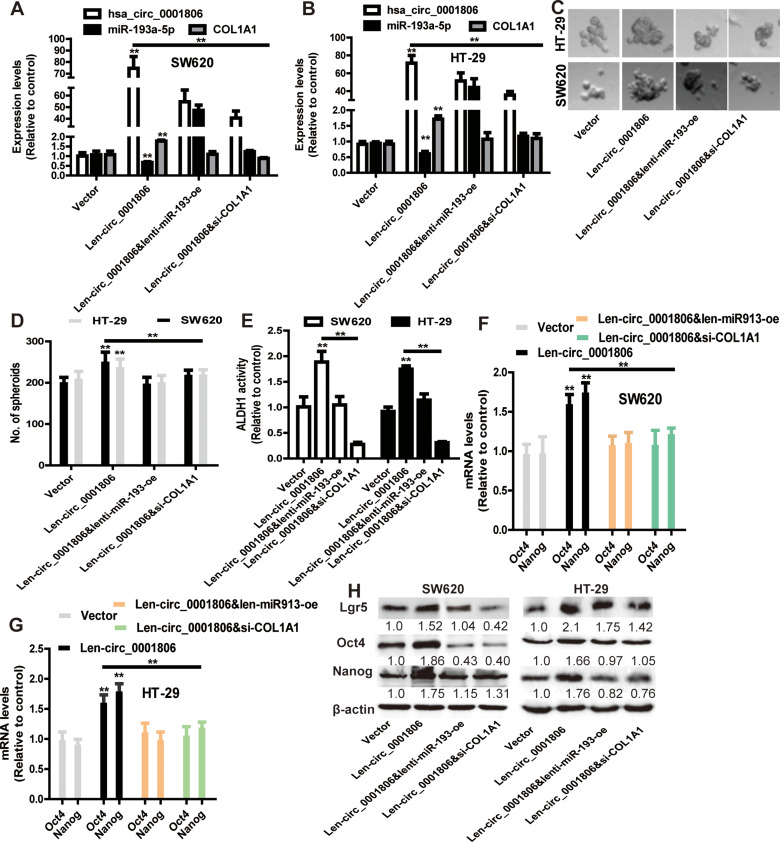
Hsa_circ_0001806 Promotes the Stemness of CRC Cells Dependent on miR-193-5p/COL1A1 Axis
Finally, we explored whether hsa_circ_0001806 exerts its effects on CRC cell stemness through the miR-193-5p/COL1A1 axis. MiR-193a-5p was overexpressed, and COL1A1 was knocked down in CRC cells with hsa_circ_0001806 overexpression. The overexpressed efficiency of miR-193a-5p and knockdown efficiency of COL1A1 was confirmed (Figure 7A and B). As shown in Figure 7C and D, the promoting effects of hsa_circ_0001806 overexpression on sphere-formation capacity were rescued by miR-193a-5p overexpression and COL1A1 knockdown in CRC cells, as evident by the change of sphere size and number. Additionally, hsa_circ_0001806 overexpression-induced ALDH1 activity was attenuated by miR-193a-5p overexpression and COL1A1 knockdown (Figure 7E). Consistently, the promoting effects of hsa_circ_0001806 overexpression on stemness marker expression were attenuated by miR-193a-5p overexpression and COL1A1 knockdown (Figure 7F–H). Thus, these results demonstrate that hsa_circ_0001806 positively regulates the stemness of CRC cells through the miR-193-5p/COL1A1 axis.
Figure 7.
Hsa_circ_0001806 promotes the stemness of CRC cells dependent on miR-193-5p/COL1A1 axis. (A and B) The expression of hsa_circ_0001806, miR-193a-5p and COL1A1 was detected in CRC cells with hsa_circ_0001806 overexpression plus miR-193a-5p overexpression or COL1A1 knockdown. (C and D) The sphere size and number were examined in the cells described in (A). (E) ALDH1 activity was measured in the cells depicted in (A). (F–H) The expression of stemness critical regulators was determined in the cells described in (A). Data were presented as mean ± s.d; **P < 0.01 vs control.
Discussion
In this study, we identified hsa_circ_0001806 as a markedly dysregulated circRNA in CRC tissues, which was elevated with the increase in TNM stage, depth of invasion, lymphatic metastasis and distant metastasis. Gain- and loss-of-function experiments revealed that hsa_circ_0001806 promoted the stemness of CRC cell in vitro and in vivo. Hsa_circ_0001806, functioning as a ceRNA by competitively binding to miR-193a-5p, abrogated the suppressive effect of miR-193a-5p on its target mRNA COL1A1. Knockdown of COL1A1 or miR-193-5p overexpression partially reversed the promoting effects of hsa_circ_0001806 overexpression on the stemness of CRC cells, indicating that hsa_circ_0001806 promotes CRC cell stemness via the ceRNA mechanism.
The roles of circRNAs in colorectal CSC progression are still unclear. To identify potential stemness regulators, we used bioinformatics analysis to filter upregulated and downregulated circRNAs and their candidate targets. Among these findings, we determined that hsa_circ_0001806 was downregulated in adjacent colorectal tissues but upregulated in CRC tissues and could promote the stemness of CRC cells in vitro and in vivo, indicating that hsa_circ_0001806 is a potential target in CRC. In searching the targets of hsa_circ_0001806, COL1A1, a major component of collagen type I, attracted our attention. We found that COL1A1 expression is increased and associated with tumour metastasis in various cancers, such as breast cancer, hepatocellular carcinoma, gastric cancer, colorectal carcinoma, non-small cell lung cancer, prostate cancer and cervical cancer.14 Here, we found that COL1A1 level was upregulated in CRC tissues, and bioinformatic analysis revealed that COL1A1 was highly correlated with the OS and DFS of CRC patients. Remarkably, hsa_circ_0001806 could upregulate the mRNA level of COL1A1, and the promoting role of hsa_circ_0001806 in CRC cell stemness could be almost offset by downregulating COL1A1. Moreover, increased expression of COL1A1 in CRC cells was significantly associated with a lower survival rate in vivo. This result suggests that hsa_circ_0001806-mediated effects are linked with COL1A1 expression.
Notably, the ceRNA network theory presumes that circRNA, lncRNA and mRNA share the same MREs (miRNA response elements), binding to miRNAs, and enhancing the expression of each others.15 In the current work, various bioinformatic softwares were used to filter target miRNAs that shared the same MREs in hsa_circ_0001806 and COL1A1. In addition, we performed biotinylated RNA/miRNA pull-down assays to evaluate the competitive binding activities of hsa_circ_0001806 and COL1A1 to miR-193a-5p. Further luciferase reporter assays indicated that miR-193a-5p significantly decreased the luciferase activity of hsa_circ_0001806-wt. Notably, our results showed that hsa_circ_0001806 was preferentially located in the cytoplasm of CRC cells, where the ceRNA network can happen.16 All of these positive results revealed that hsa_circ_0001806 and COL1A1 could directly bind to miR-193a-5p. Rescue experiments showed that hsa_circ_0001806 significantly suppressed the effects of miR-193a-5p on COL1A1, suggesting that hsa_circ_0001806 may regulate COL1A1 expression in CRC by functioning as a ceRNA. Notably, our results showed that knockdown of COL1A1 rescued the upregulation of hsa_circ_0001806 in CRC cells with hsa_circ_0001806 overexpression (Figure 7A and B), which means that COL1A1 might act as a ceRNA for hsa_circ_0001806 conversely, this should be investigated in the future. MiR-193a-5p has been studied in several tumours.17,18 For example, miR-193a-5p suppressed cell metastasis and EMT (epithelial-to-mesenchymal transition) by targeting E-cadherin in non-small cell lung cancers.19 However, the roles of miR-193a-5p in CRC cell stemness remain under investigation. In this study, we found that miR-193a-5p was sponged by hsa_circ_0001806, resulting in less miR-193a-5p targeting COL1A1 and enhancing CRC cell stemness. The existence of the hsa_circ_0001806/miR-193a-5p/COL1A1 axis improves our understanding of the underlying mechanism of colorectal CSC progression. However, it is noteworthy that COL1A1 are primarily produced by extracellular matrix components, such as cancer-associated fibroblasts (CAFs). Thus, the contribution of CRC cells to changes in collagen deposition may be led by the COL1A1 in CAFs. For example, the expression of T-lymphoma invasion and metastasis-inducing protein-1 (TIAM1) is upregulated in CAFs and contribute to CRC cell stemness;20 And CAFs can produce Netrin-1 to control cancer cell stemness.21 Therefore, further studies should be performed to compare the expression level of COL1A1 in CRC and CAFs and explore the roles of CAFs in CRC cell stemness.
Conclusively, our study determined that hsa_circ_0001806, competitively binding to miR-193a-5p, offsets the inhibition of miR-193a-5p on COL1A1 and then promotes CRC cell stemness. These findings provide a novel insight into exploring the molecular mechanism of development and progression of CRC and a potential therapeutic target for CRC.
Acknowledgments
This work was supported by the Suzhou High-tech Zone Youth Science and Technology Fund Project (2017Q0042) and Suzhou Science and Technology Town Hospital Pre-research Fund Project (2019D05).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.van Zelm R, Coeckelberghs E, Sermeus W, Aeyels D, Panella M, Vanhaecht K. Protocol for process evaluation of evidence-based care pathways: the case of colorectal cancer surgery. Int J Evid Based Healthc. 2018;16(3):145–153. doi: 10.1097/XEB.0000000000000149 [DOI] [PubMed] [Google Scholar]
- 2.Shirmohamadi M, Eghbali E, Najjary S, et al. Regulatory mechanisms of microRNAs in colorectal cancer and colorectal cancer stem cells. J Cell Physiol. 2019. [DOI] [PubMed] [Google Scholar]
- 3.Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247(5):708–718. doi: 10.1002/path.5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2017. [DOI] [PubMed] [Google Scholar]
- 6.Chen RX, Chen X, Xia LP, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. doi: 10.1038/s41467-019-12651-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, Wu B, Xia J, Ye L, Yang X. Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res. 2019;11:6887–6893. doi: 10.2147/CMAR.S212180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MB, Liu YY, Xing ZY, et al. Itraconazole-induced inhibition on human esophageal cancer cell growth requires AMPK activation. Mol Cancer Ther. 2018;17(6):1229–1239. doi: 10.1158/1535-7163.MCT-17-1094 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Wang Y, Zhang J, Zhong J, Yang R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Rep. 2018;17(4):5037–5042. doi: 10.3892/mmr.2018.8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L, Xiang C, Li X, et al. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11(1):72. doi: 10.1186/s13045-018-0613-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Luo X, Wu W, et al. The long non-coding RNA MACC1-AS1 promotes nasopharyngeal carcinoma cell stemness via suppressing miR-145-mediated inhibition on SMAD2/MACC1-AS1 axis. Biomed Pharmacother. 2020;125:109986. doi: 10.1016/j.biopha.2020.109986 [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78(17):4812–4825. doi: 10.1158/0008-5472.CAN-18-0532 [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. doi: 10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan K, Yan R, Zhu K, et al. Itraconazole inhibits the proliferation of gastric cancer cells in vitro and improves patient survival. Oncol Lett. 2018;16(3):3651–3657. doi: 10.3892/ol.2018.9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan YJ, Ma JY, Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19:183. doi: 10.1186/s12935-019-0905-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Zhou L, Lu L, et al. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013;32(29):3432–3442. doi: 10.1038/onc.2012.360 [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Qu CB, Zhang Y, et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38(14):2516–2532. doi: 10.1038/s41388-018-0602-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Y, Xie W, Song DG, Powell DJ Jr. Control of triple-negative breast cancer using ex vivo self-enriched, costimulated NKG2D CAR T cells. J Hematol Oncol. 2018;11(1):92. doi: 10.1186/s13045-018-0635-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumi D, Toden S, Ureta E, Ishimoto T, Baba H, Goel A. TIAM1 promotes chemoresistance and tumor invasiveness in colorectal cancer. Cell Death Dis. 2019;10(4):267. doi: 10.1038/s41419-019-1493-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung PJ, Rama N, Imbach J, et al. Cancer-associated fibroblasts produce netrin-1 to control cancer cell plasticity. Cancer Res. 2019;79(14):3651–3661. doi: 10.1158/0008-5472.CAN-18-2952 [DOI] [PubMed] [Google Scholar]