Abstract
Cytosine base editors (CBEs) enable targeted C•G-to-T•A conversions in genomic DNA. Recent studies report that BE3, the original CBE, induces a low frequency of genome-wide Cas9-independent off-target C•G-to-T•A deamination in mouse embryos and in rice. Here we develop multiple rapid, cost-effective methods to screen the propensity of different CBEs to induce Cas9-independent deamination in E. coli and in human cells. We use these assays to identify CBEs with reduced Cas9-independent deamination and validate via whole-genome sequencing that YE1, a narrowed-window CBE variant, displays background levels of Cas9-independent off-target editing. We engineered YE1 variants that retain the substrate-targeting scope of high-activity CBEs while maintaining minimal Cas9-independent off-target editing. The suite of CBEs characterized and engineered in this study collectively offer ~10- to 100-fold lower average Cas9-independent off-target DNA editing while maintaining robust on-target editing at most positions targetable by canonical CBEs, and thus are especially promising for applications in which off-target editing must be minimized.
Editorial summary
Methods to efficiently detect Cas9-independent cytosine base editor off-target activity enables the identification and development of highly active variants with minimal off-target editing.
Cytosine base editors (CBEs) are genome editing agents consisting of a cytidine deaminase fused to a catalytically impaired Cas9 protein and one or more copies of a uracil glycosylase inhibitor (UGI)1,2. Deamination of cytosine within a base editing activity window (canonically, protospacer positions ~4–8, counting the PAM as positions 21–23) in the single-stranded DNA loop displaced by the Cas9 guide RNA generates uracil, which is partially protected from base excision by the UGI. Selective nicking of the opposite DNA strand biases cellular DNA repair to replace the non-edited strand, resulting in the conversion of a target C•G base pair to a T•A base pair1–3. CBEs have achieved high levels of single-nucleotide polymorphism (SNP) conversion with low levels of indels in numerous cell types and organisms, including animal models of human genetic diseases3–7.
Like other Cas9-directed genome editing tools, base editors can bind to off-target genomic loci that have high sequence homology to the target protospacer. A subset of these Cas9-dependent off-target binding events can lead to base editing1,8–11, which can be minimized by using Cas9 variants with higher DNA specificity, or by delivering base editors as transient protein:RNA complexes rather than expressing them from longer-lived DNA constructs11.
In addition to Cas9-dependent off-target base editing, deamination from Cas9-independent binding of a base editor’s deaminase domain to DNA represents a distinct type of off-target base editing. Yang, Gao, and their respective co-workers recently reported that when overexpressed in mouse embryos and rice, BE3, the original CBE, induces random genome-wide mutations at average frequencies of 5×10−8 per bp and 5.3×10−7 per bp, respectively12,13. These off-target edits likely arise from the intrinsic DNA affinity of BE3’s deaminase domain, independent of the guide RNA-programmed DNA binding of Cas912,13. Ye and co-workers subsequently demonstrated that CBEs can also induce Cas9-independent off-target mutations in human induced pluripotent stem cells14. Unlike Cas9-dependent off-target editing, Cas9-independent deamination occurs at different loci between cells, making it difficult to characterize by targeted high-throughput sequencing. Extensive whole-genome sequencing experiments such as those performed by Yang, Gao, Ye, and their respective co-workers are low-throughput, expensive, and time-intensive, limiting their use for evaluating and engineering CBE variants with decreased Cas9-independent deamination activity. Here, we describe the development of methods to efficiently evaluate the propensity of a base editor to cause Cas9-independent deamination, and the application of these methods to identify and engineer CBE variants that minimize Cas9-independent DNA editing.
Results
Bacterial rifampin resistance assay
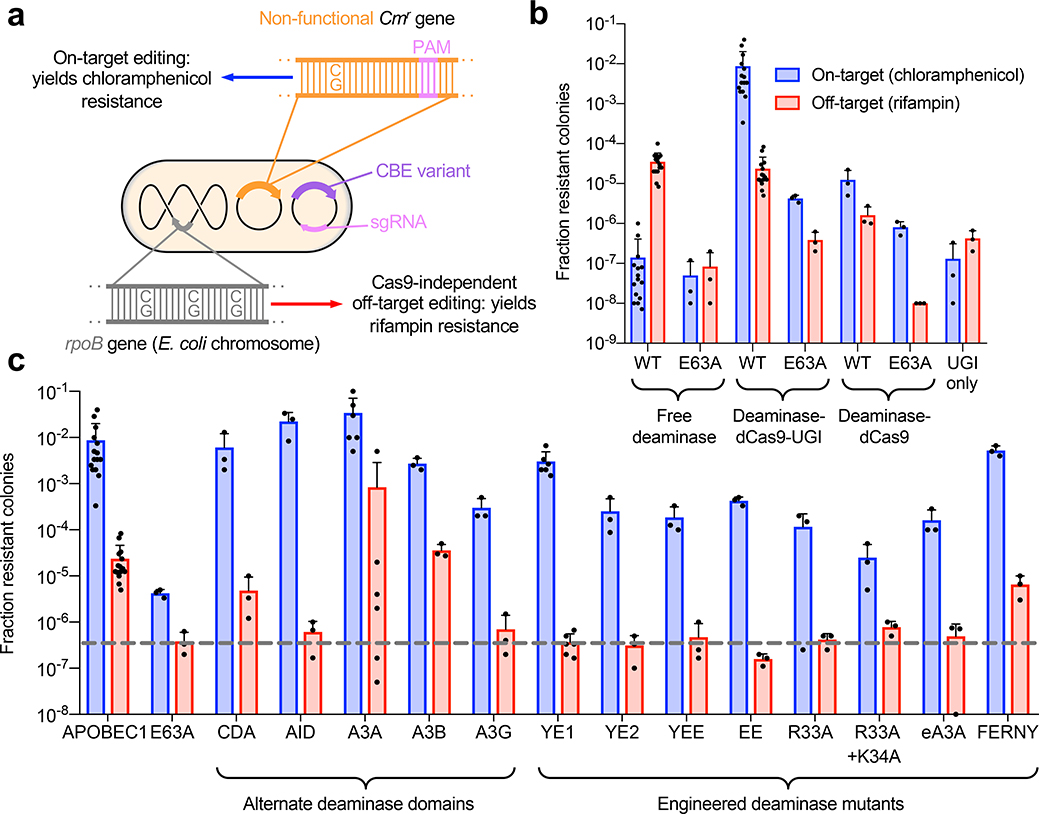
First, we assayed Cas9-independent deamination by CBEs in bacteria using a rifampin resistance assay. Measuring resistance to the antibiotic rifampin has previously been used to characterize the activity and mutagenicity of proteins expressed in E. coli15–19. Deaminase-catalyzed C•G-to-T•A mutations in the rpoB gene render E. coli resistant to rifampin. We hypothesized that cells transformed with a plasmid encoding a CBE with Cas9-independent deamination activity would become resistant to rifampin at a frequency that reflects the magnitude of this activity. To simultaneously assess the on-target activity of the base editor, we also transformed a second plasmid encoding a chloramphenicol acetyltransferase with an inactivating T•A-to-C•G point mutation, together with a guide RNA that directs the CBE to revert this point mutation. Base editors with higher on-target activity more effectively rescue chloramphenicol resistance8. Survival rates on chloramphenicol thus reflect on-target editing efficiency, while survival rates on rifampin reflect Cas9-independent deamination activity (Fig. 1a).
Figure 1. On-target and Cas9-independent off-target DNA editing in E. coli.
(a) Experimental design. (b) Assay validation. Fraction of resistant colonies was calculated relative to number of E. coli plated on maintenance antibiotics. Data are shown as individual data points and mean ± SEM for n=3 or n=15 bacterial colonies. E63A refers to a catalytically inactivated APOBEC1 E63A mutant. (c) Performance of CBEs that use alternative deaminases. All constructs are in the deaminase–dCas9–UGI (BE2) architecture. The grey dotted line indicates the background level of rifampin resistance of the inactive APOBEC1 E63A deaminase control. Data are shown as individual data points and mean ± SEM for n=3 or n=15 bacterial colonies.
To validate this assay, we measured the chloramphenicol and rifampin resistance of bacteria transformed with wild-type APOBEC1, the deaminase used in BE3, and the catalytically inactive E63A mutant of APOBEC1 in three different architectures: as free deaminases, as deaminase–dCas9–UGI fusions, or as deaminase–dCas9 fusions lacking the UGI domain (Fig. 1b). We used dCas9 instead of Cas9 nickase for bacterial assays because E. coli lack the nick-directed mismatch repair pathway that enables improved editing by Cas9 nickase CBEs in mammalian cells20. Compared to the background resistance levels of untethered inactive APOBEC1 E63A construct, untethered active APOBEC1 induced a 1,000-fold increase in rifampin resistance and a 10-fold increase in chloramphenicol resistance. The APOBEC1–dCas9–UGI base editor yielded the same level of rifampin resistance as that of untethered APOBEC1, but a 250-fold higher level of chloramphenicol resistance. These data are consistent with high on-target activity of CBEs and with Cas9-independent off-target mutagenesis.
To confirm that rifampin resistance was accompanied by CBE-induced mutations, we sequenced the rpoB gene of rifampin-resistant colonies and observed primarily C•G-to-T•A mutations (Supplementary Fig. 1). The inactive APOBEC1 E63A–dCas9 fusion resulted in rifampin resistance levels equivalent to the background of the assay, suggesting that dCas9 alone does not contribute to these off-target mutations. Compared to the APOBEC1–dCas9–UGI fusion, both the APOBEC1–dCas9 fusion and the APOBEC1 E63A–dCas9-UGI exhibited a substantial decrease in rifampin resistance. These results suggest that both the deaminase domain of the base editor and the UGI domain can contribute to Cas9-independent off-target mutagenesis.
To minimize Cas9-independent deamination, we focused on the deaminase domain for two reasons. First, the rifampin resistance frequency from expression of APOBEC1 alone was 100-fold higher than the average rifampin resistance from expression of UGI alone (Fig. 1b). Second, when we analyzed the off-target DNA sequencing data from Yang and coworkers12, we found a strong 5’ T preference among edited cytosines (Supplementary Fig. 2). This preference suggests that APOBEC1, which has a preference for deaminating 5’ TC substrates21, is primarily responsible, as opposed to UGI, which is not known to cause a sequence context bias among C•G-to-T•A mutations.
Many CBE variants with alternate deaminase domains have now been reported1,22–33. We measured the chloramphenicol and rifampin resistance of E. coli transformed with virtually all previously reported CBEs, starting with naturally occurring APOBEC1, AID, CDA, APOBEC3A, APOBEC3B, and APOBEC3G deaminases (Fig. 1c, Supplementary Fig. 3). E. coli transformed with CBEs that use CDA, APOBEC3A, and APOBEC3B exhibited rifampin resistance levels that were comparable to or higher than the rifampin resistance arising from the original APOBEC1 base editor, consistent with our recent characterization of high editing activity from CDA- and APOBEC3A-derived CBEs30. In contrast, APOBEC3G and AID base editors produced significantly lower levels of rifampin resistance, suggesting they generate less Cas9-independent deamination in bacteria.
Next, we expanded our panel of deaminases to include engineered deaminase variants that we and others previously developed for base editing applications. We created APOBEC1 variants W90Y+R126E (YE1), W90Y+R132E (YE2), R126E+R132E (EE), and W90Y+R126E+R132E (YEE) to narrow the on-target base editing window31. In addition, Joung and co-workers engineered APOBEC1 R33A and APOBEC1 R33A+K34A to have lower off-target RNA editing32. Joung and co-workers also designed an engineered APOBEC3A (eA3A) to have a strict 5’ T sequence context requirement33. Finally, we recently reported FERNY, a truncated, ancestrally reconstructed deaminase, which lacks an RNA-binding motif that could mediate nonspecific interactions with nucleic acids30. Promisingly, most of these engineered CBEs yielded substantially lower rifampin resistance levels. In particular, eA3A, YE1, YE2, EE, YEE, R33A, and R33A+K34A APOBEC1 variants all resulted in rifampin resistance frequencies equivalent to that of the inactive APOBEC1 E63A–dCas9–UGI control (Fig. 1c, Supplementary Fig. 3). These results indicate that several base editor variants have much lower Cas9-independent deamination in E. coli, consistent with their original design goals of lower deamination activity31,32 or increased requirements for deamination33.
Bacterial thymidine kinase toxicity assay
Numerous studies have shown that cytidine deaminases exhibit strong sequence context preferences; for example, while APOBEC1 prefers 5’-TC substrates, A3G prefers 5’-CC substrates, and AID prefers 5’-GC substrates23,24,29,30. To ensure that the results of the rifampin assay were not skewed by the sequence contexts of particular cytosines whose mutagenesis led to rifampin resistance, we sought to recapitulate the rifampin assay using a different selectable target gene with a different set of cytosines that yield resistance when deaminated, and a different set of 5’ base contexts. To do this, we inserted a single copy of the herpes simplex virus thymidine kinase (HSV-TK) gene into the E. coli chromosome. HSV-TK leads to toxicity in the presence of the nucleoside analogue dP34. We reasoned that off-target C•G-to-T•A mutations in the HSV-TK gene that inactive the enzyme would lead to survival on dP. Indeed, while the dynamic range of this assay was narrower than that of the rifampin assay, we observed the same trends: rAPOBEC1, A3A, A3B, and CDA induced more mutagenesis, whereas most other CBEs induced levels of dP resistance comparable to background. Sequencing the HSV-TK gene confirmed various resistant alleles caused by C•G-to-T•A mutations (Supplementary Fig. 4). The consistency between the rifampin and HSV-TK resistance assays suggests that sequence context bias plays a minimal role in the above results.
Human cell orthogonal R-loop assay
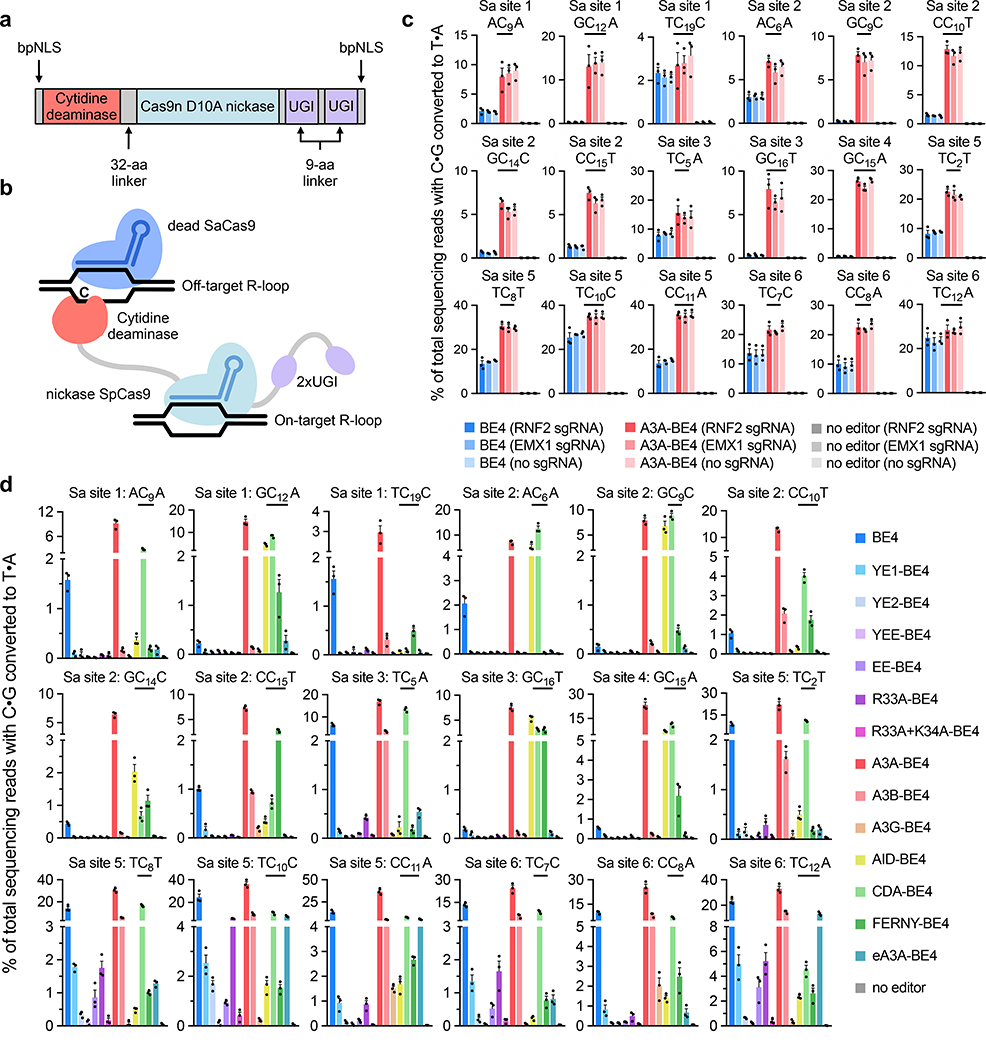
Next, we developed assays for Cas9-independent deamination by CBEs in human cells that are not dependent on whole-genome sequencing. Since the above results, as well as the findings of Yang, Gao, and their respective co-workers12,13, all suggest that the frequency of stochastic Cas9-independent deamination by BE3 is well below the ~0.1% detection limit of practical high-throughput DNA sequencing experiments, we developed an assay that magnifies Cas9-independent off-target deamination at specific loci that can be monitored by targeted high-throughput sequencing. All of the deaminases used in CBEs to date deaminate single-stranded DNA or RNA efficiently, but not double-stranded nucleic acids, and recent reports detailing Cas9-independent deamination noted that the observed mutations were enriched in transcribed regions of the genome12,13. We therefore reasoned that generating long-lived single-stranded DNA at specific positions would create artificially high Cas9-independent deamination levels that could be detected by targeted amplicon sequencing.
To evaluate the ability of different base editors to deaminate cytosines in single-stranded DNA regions unrelated to their on-target loci, we co-transfected HEK293T cells with plasmids encoding an SpCas9-based CBE, an SpCas9 on-target guide RNA, a catalytically inactive S. aureus Cas9 (dSaCas9), and an SaCas9 guide RNA targeting a genomic locus unrelated to the on-target site (Fig. 2a and b). We generated all editors for mammalian cell experiments using the current “BE4max” architecture, with optimized codon usage and the optimized structure of NLS–deaminase–Cas9 nickase–UGI–UGI–NLS (Supplementary Sequences)4,35. Deamination of cytosines in the R-loop formed by dSaCas9 should occur in a CBE-dependent, but SpCas9 guide RNA-independent, manner. Indeed, high-throughput sequencing of six dSaCas9 loci three days after plasmid cotransfection resulted in off-target deamination by APOBEC1-based BE44,35 that was easily detected by targeted DNA sequencing (0.4–25%), and were independent of the on-target SpCas9 guide RNA (Fig. 2C). Encouragingly, A3A-BE430, a CBE that uses APOBEC3A, demonstrated substantially higher off-target deamination of dSaCas9-generated R-loops relative to BE4 (Fig. 2b, Supplementary Fig. 5a), consistent with its higher frequency of generating resistant colonies in the bacterial rifampin assay (Fig. 1c), and with the previously reported high degree of mutagenicity of APOBEC3A in human cells36. These results collectively suggest that in trans deamination within R-loops generated by an orthogonal Cas9 homolog can be used to assess the propensities of SpCas9-derived CBEs to mediate Cas9-independent deamination.
Figure 2. Cas9-independent deamination by CBEs in HEK293T cells.
(a) BE4max-like architecture for all CBE constructs used in mammalian cell experiments. (b) Schematic of Cas9-independent deamination of cytosines within dSaCas9-induced R-loops by SpCas9 CBEs. (c) and (d) Cas9-independent off-target C•G-to-T•A editing frequencies detected by targeted high-throughput sequencing of six dSaCas9 loci following cotransfection with SpCas9-targeted CBEs. Each subplot shows the observed C•G-to-T•A conversion of a single underlined cytosine and its immediate sequence context. Transfections in (c) were performed with one of two SpCas9 sgRNAs (targeting the RNF2 or EMX1 genomic loci) or no SpCas9 sgRNA, with on-target editing controls shown in Supplementary Fig. 5a. Transfections in (d) were performed with one SpCas9 sgRNA targeting the RNF2 genomic locus, with on-target editing controls shown in Supplementary Fig. 5b. For (c) and (d), data are shown as individual data points and means ± SEM for n = 3 independent biological replicates performed on different days.
To identify base editor variants that exhibit reduced Cas9-independent deamination relative to BE4 in human cells, we evaluated the same panel of 14 deaminase domains (APOBEC1, CDA, AID, APOBEC3A, eA3A, APOBEC3B, APOBEC3G, and FERNY; and APOBEC1 mutants YE1, YE2, YEE, EE, R33A, and R33A+K34A)1,22–33 in the BE4max architecture for their ability to deaminate dSaCas9-induced R-loops in trans. Base editors with narrowed on-target DNA editing windows such as YE1-BE4, YE2-BE4, and EE-BE4, or with reduced RNA editing propensities such as R33A-BE4, again exhibited substantially reduced Cas9-independent DNA deamination compared to BE4 (Fig. 2d, Supplementary Fig. 5b). Indeed, YEE-BE4 and R33A+K34A-BE4 displayed nearly undetectable levels of Cas9-independent deamination in this assay. Nearly all of the other CBE variants assayed displayed comparable or higher levels of Cas9-independent deamination relative to BE4 for at least a subset of off-target cytosines within SaCas9-induced R-loops. Compared to BE4, CBEs derived from CDA, AID, and FERNY exhibited higher levels of Cas9-independent deamination at 5’-GC substrates, as expected given their higher activity on 5’-GC sequences than APOBEC123,24, 30. Likewise, eA3A-BE4 and A3G-BE4 displayed moderate to high levels of Cas9-independent deamination at 5’-TCR and 5’-CC substrates respectively, also consistent with their known sequence context preferences29,33. All transfected constructs had similar effects on cell viability (Supplementary Fig. 6), which indicates that cell viability is not a confounding factor in this assay. These trends agree with the results of the rifampin resistance and thymidine kinase assays, with the exception of AID-BE4: our results show higher amounts of off-target editing by AID-BE4 in mammalian cells compared to in E. coli. This observation is consistent with previous studies that show higher AID activity in human cells compared to bacteria, potentially due to protein/protein interaction partners or post-translational modifications to the enzyme17,37. These data suggest that R33A-BE4, YE1-BE4, YE2-BE4, EE-BE4, YEE-BE4, and R33A+K34A-BE4 are especially promising CBE variants for applications in which Cas9-independent off-target editing must be minimized.
In vitro kinetics assay
We hypothesized that a primary determinant of Cas9-independent deamination propensity is the catalytic efficiency of the enzyme. Ideal CBE deaminases should inefficiently catalyze deamination of substrates that are present at low concentrations (such as Cas9-independent off-target sites) but efficiently deaminate on-target substrates when presented at high effective local concentration due to DNA binding of the tethered Cas9 domain. To test this hypothesis, we purified three different CBE proteins and measured their kcat/Km values in vitro for a 5’-Cy3-labeled ssDNA oligonucleotide that contained a single cytosine and was unrelated to the sgRNA present in the reaction. To measure reaction velocities, we quantified uracil-containing product formation by gel densitometry following USER enzyme treatment38. YE1–dCas9–UGI and APOBEC3A–dCas9–UGI have kcat/Km values for ssDNA that are 69-fold lower and 1.3-fold higher, respectively, than that of APOBEC1–dCas9–UGI (Supplementary Fig. 7). These findings are consistent with the results of the orthogonal R-loop assays in Fig. 2, as well as the rifampin resistance assays in Fig. 1, and support a model in which CBEs with higher kcat/Km values for ssDNA have a greater propensity for Cas9-independent deamination in cells.
Human cell ssDNA deamination assay
As an additional independent assay of Cas9-independent deamination by CBEs in mammalian cells, we also measured intracellular deamination frequencies from BE4, A3A-BE4, YE1-BE4, YEE-BE4, and R33A+K34A-BE4 of a co-transfected 164-mer ssDNA oligonucleotide containing 35 cytosines in HEK293T cells, in light of previous reports that endogenous deaminases can induce mutagenesis in transfected ssDNA oligonucleotides (Supplementary Fig. 8)39. We observed that A3A-BE4 showed 4.4-fold higher Cas9-independent off-target editing compared to BE4, while YE1-BE4, YEE-BE4, and R33A+K34A-BE4 showed 1.7-, 3.2-, and 1.4-fold lower average Cas9-independent off-target editing relative to BE4 at the twelve 5’-TC cytosines present in the oligonucleotide that were deaminated above background (Supplementary Fig. 8), again concordant with findings from the other assays. Taken together, the results from the rifampin and HSV-TK resistance assays in bacteria, orthogonal R-loop assay in human cells, kinetic assay in vitro, and ssDNA deamination assay in human cells are consistent with a model in which CBEs with deaminases that have a low intrinsic catalytic efficiency (kcat/Km) for cytosine-containing ssDNA substrates exhibit lower Cas9-independent off-target deamination.
Comparison of adenine and cytosine base editors
Previous studies that detected Cas9-independent off-target DNA editing by CBEs did not detect off-target editing induced by the canonical adenine base editor, ABE12,13, so ABE should produce minimal off-target editing in our assays. Indeed, in the rifampin and HSV-TK resistance assays, ABE induced background levels of resistance, and in the orthogonal R-loop and intracellular ssDNA deamination assays, ABE induced only very low levels of off-target A•T-to-G•C editing (Supplementary Fig. 9). Therefore, low off-target activity as assessed by the methods developed in this study is consistent with low off-target activity as assessed by previous whole-genome sequencing studies12,13.
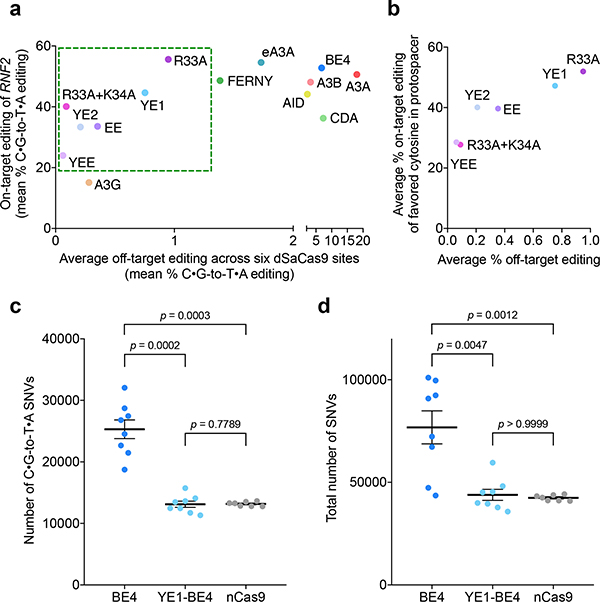
Each deaminase domain tested has a distinct on-target editing and off-target editing profile, which is shown in Fig. 3A. Of the CBEs that we identified as being especially promising for minimizing Cas9-independent editing, YE1-BE4 and R33A-BE4 offer the best balance between decreased off-target editing and robust on-target activity (Fig. 3B, Supplementary Fig. 10). Meanwhile, YE2-BE4, EE-BE4, R33A+K34A-BE4, and YEE-BE4 produce even lower off-target editing but with a significant decrease in average on-target activity tested across six sites (Fig. 3b and Supplementary Fig. 10).
Figure 3. YE1 balances efficient on-target editing with greatly decreased Cas9-independent editing as confirmed by whole-genome sequencing.
(a) On-target editing versus average off-target editing for all CBEs in this study. The y-axis reflects the mean on-target control editing at the on-target RNF2 locus used in the orthogonal R-loop assay, and the x-axis reflects the mean off-target editing for six orthogonal R loops. The green box indicates CBE variants that have substantially decreased Cas9-independent off-target editing but retain appreciable on-target activity. See Supplementary Fig. 5 and Fig. 2d for mean values and SEM at individual sites. (b) Average maximum on-target and average off-target editing for constructs with decreased Cas9-independent editing events. The y-axis reflects average editing across six on-target protospacers of the most highly edited cytosine within that protospacer. The x-axis reflects the average off-target editing in the orthogonal R-loop assay. See Supplementary Fig. 10 and Fig. 2d for mean values and SEM at individual sites (c) Number of C•G-to-T•A single-nucleotide variants (SNVs) relative to the initial parent sample detected by whole-genome sequencing (WGS). (d) Total number of SNVs relative to the initial parent sample detected by WGS. For both (c) and (d), each dot represents the number of SNVs called in a clonal population of cells relative to the parent sample. Each clonal population was derived from a single GFP-positive cell that was isolated after flow sorting HEK293T cells transfected with a CBE–P2A–GFP construct for the GFP-positive cells. Horizontal lines and error bars indicate mean number of SNVs ± SEM for n=8 (BE4 and YE1) or n=7 (nCas9). P values were calculated using the two-sided Mann-Whitney U test.
Whole-genome sequencing of treated human cells
To further validate that our methods are representative of genome-wide Cas9-independent off-targets, we performed whole-genome sequencing (WGS) of HEK293T cells treated with BE4, YE1-BE4, or a Cas9 D10A nickase control. Four days following transfection with an sgRNA plasmid and a plasmid encoding either BE4, YE1-BE4, or nCas9(D10A) co-translationally fused to GFP, we isolated the top ~25% of GFP-positive cells by flow cytometry, diluted single cells into individual wells, and grew them into clonal populations for 16 days before genomic DNA extraction. This approach ensured that CBE-derived off-target mutations would be present at high allele frequencies within the clonal samples derived from a single CBE-treated cell (Supplementary Fig. 11). We performed WGS at an average depth of 77x on all samples and determined all single-nucleotide variants (SNVs) present in each sample using the intersection of variants called by three algorithms (Supplementary Fig. 11, Supplementary Table 3, Supplementary Table 4). In order to restrict our analysis to SNVs that were generated following CBE treatment, we filtered out SNVs that were present in the original clonal population of cells prior to CBE treatment.
WGS results revealed that BE4, but not YE1, produced significantly more C•G-to-T•A SNVs than the Cas9 nickase-only negative control (Fig. 3c). These observations confirmed the findings of Yang, Gao, and their respective coworkers that CBEs containing wild type rAPOBEC1 produce off-target C•G-to-T•A SNVs in a Cas9-independent manner. We also found that BE4-treated samples contained more non-C•G-to-T•A SNVs than YE1 or nickase samples (Fig. 3d, Supplementary Fig. 12), consistent with previous reports that deaminase overexpression in HEK293 cells leads to overall increased SNVs of all types40. The frequency of BE4-mediated off-target edits that we observed (8.0×10−6/bp) was also much higher than either of the previously reported values (5×10−8/bp and 5.3×10−7/bp reported by Yang and Gao, respectively). This difference likely arises from different delivery methods, our sorting of cells to isolate those that express CBEs most highly, our clonal expansion approach to maximizing SNV detection sensitivity, and the different cell types used. Importantly, the above WGS results confirmed the findings of other assays: YE1 exhibits significantly reduced Cas9-independent off-target editing compared to BE4; indeed, YE1 treatment did not lead to statistically significant differences relative to the Cas9 nickase-only control (Fig. 3d, Supplementary Fig. 12).
Engineering CBE variants with minimal off-target activity and expanded targeting scope
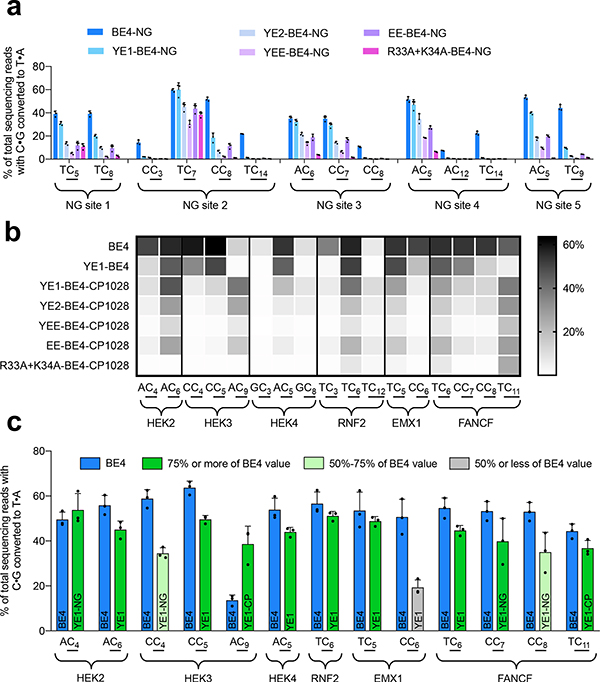
Because YE1 and the CBE variants that we assessed to have minimal Cas9-independent off-target activity all exhibit narrowed on-target DNA editing windows31 (YE1-BE4, YE2-BE4, YEE-BE4, and EE-BE4), or a specific DNA sequence context requirement32 (R33A+K34A-BE4), we sought to expand the targeting scope of these CBEs in order to increase their overall utility. We tested if these deaminases are compatible with SpCas9-NG, one of two recently reported Cas9 variants that recognize a broadened NG PAM41,42 and found that YE1, and to a lesser extent YE2, YEE, EE, and R33A+K34A, maintained compatibility with SpCas9-NG nickase (Fig. 4a). YE1-NG expands the targeting scope of CBEs while maintaining substantially decreased Cas9-independent off-target activity (Supplementary Fig. 13).
Figure 4. Expanding the utility of CBEs with decreased Cas9-independent off-targets through protein engineering.
(a) On-target base editing by different Cas9-NG-targeted CBEs across five different sites with NG PAMs (non-NGG) in HEK293T cells. Individual data points and mean values ± SEM are shown for n = 3 independent biological replicates. (b) On-target base editing by circularly permuted CBEs across six sites in HEK293T cells. Intensities reflect the mean editing value from three biological replicates, which are shown in Supplementary Fig. 14. (c) Summary of the on-target editing of BE4 and engineered YE1 variants across six test loci. Blue bars indicate BE4 values for particular cytosines, and green bars represent YE1 variants that can be used to target those cytosines. The name of each YE1 variant used is indicated on the bar. Shading reflects the efficiency that each cytosine can be edited by a YE1 variant when compared to BE4. Individual data points and mean values ± SEM for n = 3 independent biological replicates are shown.
Next, we replaced the SpCas9 nickase domain of YE1-BE4, YE2-BE4, YEE-BE4, EE-BE4, and R33A+K34A-BE4 with CP1028, a circularly permuted SpCas9 variant43. We recently reported that some circularly permutated Cas9 variants can widen or shift the on-target editing window of CBEs and ABEs4,43. Indeed, in HEK293T cells at a variety of endogenous loci, we observed that YE1-BE4-CP1028, YE2-BE4-CP1028, and EE-BE4-CP1028 exhibit base editing activity windows shifted towards the PAM compared to that of non-permuted YE1-BE4 (Fig. 4b, Supplementary Fig. 14). Collectively, YE1-BE4 and YE1-BE4-CP1028 enable targeting of nearly all cytosines present in the original base editing activity window of BE4, with the exception of sites that contain long multi-C repeats (Supplementary Fig. 15). In addition, YEE-BE4-CP1028 and R33A+K34A-BE4-CP1028 were also active at a subset of sites tested and showed shifted editing windows at those sites (Fig. 4b).
Variants such as YEE-BE4 and R33A+K34A-BE4 are intriguing in that they offer extremely low, if any, off-target deamination in our orthogonal R-loop assay, but they are only active at a subset of on-target sites. To further increase the target sequence compatibility of R33A+K34A-BE4, which exhibits a 5’-TC requirement for base editing, we incorporated H122L and D124N, two mutations that we recently found during the continuous evolution of APOBEC1 to enable efficient deamination of 5’-GC substrates30. The resulting R33A+K34A+H122L+D124N-BE4 variant (referred to as AALN-BE4) indeed changed the profile of targetable C’s relative to the original R33A+K34A variant, enabling editing of some positions that were not accessed by R33A+K34A-BE4 (Supplementary Fig. 16). Importantly, the AALN variant maintains the minimized levels of Cas9-independent deamination shown by R33A+K34A-BE4, and circularly permuted variants likewise displayed Cas9-independent deamination levels equivalent to or lower than their unpermuted counterparts (Supplementary Figs. 17 and 18). This result indicates that deaminases with the lowest number of off-target edits can be engineered to enhance their targeting scope without disrupting their minimal off-target editing profile.
Next, we assessed if the CBEs that exhibit minimal Cas9-independent deamination have altered propensities to generate other unwanted editing outcomes, such as indels and Cas9-dependent off-target DNA base editing. We observed that all of these variants (YE1-BE4, YE2-BE4, YEE-BE4, EE-BE4, R33A+K34A-BE4, R33A-BE4, AALN-BE4, the CP1028 variants of the first five of these variants, and the Cas9-NG variants of the same five CBEs), induce lower or comparable levels of indels relative to BE4 across all on-target genomic sites tested in this study (Supplementary Fig. 19). Moreover, all seven CBE variants (YE1-BE4, YE1-BE4-CP1028, YE2-BE4, EE-BE4, YEE-BE4, R33A+K34A-BE4, and AALN-BE4) showed much lower levels of Cas9-dependent off-target DNA editing than BE4 when tested at 20 genomic sites previously identified by GUIDE-seq44 to be the most highly edited off-target substrates of SpCas9 nuclease for three target loci (Supplementary Fig. 20). In addition, we note that YE1-BE3, R33A-BE3, and R33A+K34A-BE3 were recently found to exhibit substantially reduced levels of transcriptome-wide Cas9-independent RNA off-target editing compared to BE332,45. We confirmed that these variants exhibit decreased Cas9-independent off-target editing of three abundant RNA transcripts and found that YEE also shows decreased RNA off-target editing (Supplementary Fig. 21). These results collectively indicate that the CBEs that minimize Cas9-indepdendent off-target editing do not suffer from higher levels of other forms of unwanted editing; in general they give rise to fewer indels, less Cas9-dependent DNA off-target editing, and less RNA off-target editing.
Collectively, the expanded targeting capabilities of engineered YE1 variants (YE1-BE4, YE1-BE4-CP1028, and YE1-BE4-NG,) enable targeting, in principle, of 65% of pathogenic SNPs in ClinVar that can be corrected by a C•G-to-T•A edit, compared to the only 19% that can be targeted by SpCas9-YE1max alone (Supplementary Fig. 22). The known pathogenic SNPs that can be targeted by these engineered CBEs include the vast majority (~80%) of pathogenic SNPs that can be targeted with the most broadly targetable current-generation BE4max variants, and far outnumber the SNPs targetable by SpCas9-BE4max alone, the most widely-used CBE (Supplementary Fig. 22).
Analysis of expression levels and off-target editing by RNPs
Finally, we explored how base editor expression and exposure contribute to Cas9-independent off-target editing. Western blots of BE4, YE1-BE4, YE1-BE4-NG, and A3A-BE4 in HEK293T cells revealed that the expression levels of YE1-BE4 and YE1-BE4-NG were comparable to that of BE4. However, A3A-BE4 had drastically reduced expression compared to the other three editors, in stark contrast with its higher levels of off-target editing (Supplementary Fig. 23). We then transfected a variant of BE4 with only one, as opposed to two, nuclear localization signals. This construct should have lower levels of CBE trafficked to the nucleus, and therefore lower effective dosing. Indeed, we saw decreased off-target editing in the R-loop assay when we included only one NLS (Supplementary Fig. 24). This collection of experiments conveys that while expression of a base editor influences Cas9-independent off-target editing, it cannot fully explain the propensity of an editor to perform Cas9-independent deamination.
These results also suggested that limiting the time of exposure to the base editor through protein delivery might decrease Cas9-independent off-target editing. Therefore, we delivered a 1xNLS-BE4 construct into HEK293T cells as a protein:RNA complex and measured levels of orthogonal R-loop deamination: average Cas9-independent deamination decreased 21-fold relative to plasmid delivery, while retaining similar on-target editing efficiencies (Supplementary Fig. 23). Therefore, even if a specific target can only be edited to an acceptable level by a BE4-like CBE that uses a deaminase with a high kcat/Km, protein delivery may still provide a path forward to minimize Cas9-independent off target editing.
Discussion
The assays developed and applied in this study enable rapid and cost-effective profiling of base editors for Cas9-independent deamination of DNA in bacteria and mammalian cells, and complement in vivo methods such as those performed by Yang, Gao, and their respective coworkers12,13. The WGS data collected in this study validate that these assays are representative of genome-wide off-target DNA mutagenesis rates, and suggest that those CBEs that show low off-target editing in these assays should exhibit low levels of genome-wide off targets. We anticipate that the assays used here will provide a valuable means of evaluating many CBE variants much more efficiently and with much lower costs than experiments that require extensive whole-genome sequencing12,13.
The many deaminases and CBEs characterized and generated in this study collectively form a landscape of base editing options with different on-target and off-target editing characteristics, plotted in Fig. 3a. Given this landscape, and the fact that the 5×10−8/bp mutation rate attributed to Cas9-independent deamination by BE3 in mouse embryos12 is lower than the observed rate of spontaneous mutation in many mammalian somatic cell types in vivo46–50, the optimal choice of base editor depends strongly on a given application’s on-target sequence context, on-target PAM availability, target tissue type, and the extent to which minimizing low levels of Cas9-independent deamination is critical. For applications in which off-target editing must be strictly minimized, we recommend YE1-BE4, YE2-BE4, YEE-BE4, EE-BE4, R33A+K34A-BE4, YE1-BE4-CP1028, YE1-BE4-NG, and AALN-BE4 variants, each of which offer ~10- to 100-fold lower levels of Cas9-independent off-target DNA editing (Fig. 1–Fig. 3), ~5-to 50-fold lower levels of Cas9-dependent off-target DNA editing (Supplementary Fig. 20), and lower or similar levels of indel formation (Supplementary Fig. 19), while maintaining ~50–90% of average on-target DNA editing levels (Fig. 3a, Fig. 3b, Fig. 4c) relative to BE4max. Additionally, base editor exposure may also be limited to achieve lower off-target editing. Collectively, the diverse targeting capabilities of this suite of CBEs, especially those that utilize the YE1 deaminase domain, enable high-fidelity base editing at the vast majority of previously accessible target sites with efficiencies approaching those of BE4 (Supplementary Fig. 22, Fig. 4c).
Methods
Cloning
All plasmids for this study were created using either USER cloning or KLD cloning as described previously1. DNA was amplified using PhusionU Green Multiplex PCR Master Mix (Thermo Fisher Scientific). Mach1 (Invitrogen) or Turbo (New England BioLabs) chemically competent E. coli were used for plasmid construction.
Preparation and transformation of chemically competent E. coli
Commercially available chemically competent BL21 E. coli (New England BioLabs) were transformed with a plasmid harboring an inactivated chloramphenicol resistance gene. Transformed cells were plated on LB media + 1.5% agar supplemented with maintenance antibiotic (kanamycin, 30 μg/mL). The following day, a single colony was picked and grown overnight in 2xYT media supplemented with maintenance antibiotic. The overnight culture was diluted 100-fold into 50 mL of 2xYT media supplemented with maintenance antibiotic and grown at 37 °C with shaking at 230 rpm to OD600 ~0.4–0.6. Cells were collected by centrifugation at 3,400 g for 10 min at 4°C. The cell pellet was resuspended by gentle stirring in 2.5 mL of cold LB media followed by 2.5mL of 2x TSS (LB media supplemented with 5% v/v DMSO, 10% w/v PEG 3350, and 20 mM MgCl2). After thorough resuspension, cells were aliquoted, frozen on dry ice, and stored at −80 °C until use.
To transform cells, 100 mL of competent cells thawed on ice were added to a pre-chilled mixture of plasmid (2 μL) in 98 μL KCM solution (100 mM KCl, 30 mM CaCl2, and 50 mM MgCl2 in H2O). The mixture was incubated on ice for 20 min and heat shocked at 42 °C for 75 s followed by addition of 500 μL of SOC media (New England BioLabs). Cells were recovered at 37 °C with shaking at 230 rpm for 1 h, streaked on 2xYT media + 1.5% agar plates containing the appropriate antibiotics, and incubated at 37 °C for 14–16 h.
Rifampin assay
Chemically competent E. coli harboring a plasmid encoding an inactivated chloramphenicol resistance gene were transformed with a plasmid encoding a base editor + guide RNA. Transformed cells were plated on maintenance antibiotics (30 μg/mL kanamycin, 50 μg/mL spectinomycin, with no chloramphenicol). The following day, colonies were picked and grown overnight in Davis Rich Medium (DRM) and maintenance antibiotics. Overnight cultures were diluted 1:100 into DRM and maintenance antibiotics and grown at 37 °C with shaking at 230 rpm When cells reached OD600 = 0.5, 5 mM rhamnose was added to induce base editor expression. After 18 hours, 700 μL of each culture was centrifuged at 3,400 g for 10 min and the cell pellet was resuspended in 150 μL total DRM. Serial dilutions in H2O of each resuspended culture were plated on three different conditions in parallel: (1) 2xYT agar + 30 μg/mL kanamycin + 50 μg/mL spectinomycin + 20 mM glucose, (2) 2xYT agar + 30 μg/mL kanamycin + 50 μg/mL spectinomycin + 20 mM glucose + 100 μg/mL rifampin, or (3) 2xYT agar + 30 μg/mL kanamycin + 50 μg/mL spectinomycin + 20 mM glucose + 10 μg/mL chloramphenicol. Surviving colonies were counted following an incubation at 37 °C for 24 h after plating. To obtain survival rates, the number of colonies in the chloramphenicol or rifampin conditions were divided by the number of colonies counted on the maintenance antibiotic plate.
Sanger sequencing of rpoB mutations from rifampin-resistant colonies
Rifampin-resistant colonies were picked into 10 μL of H2O and heated at 95°C for 10 min, followed by PCR using primers AB1678 (5’-AATGTCAAATCCGTGGCGTGAC) and AB1682 (5’-TTCACCCGGATACATCTCGTCTTC). Each fragment was sequenced twice using primers AB1680 (5’-CGGAAGGCACCGTAAAAGACAT) and AB1683 (5’-CGTGTAGAGCGTGCGGTGAAA).
HSV thymidine kinase assay
Lambda red recombineering was performed as described previously51 in order to chromosomally integrate a single copy of the HSV thymidine kinase gene under a constitutive promoter and β-lactamase into the tonB locus of BL21 E. coli. The resulting strain was transformed with a plasmid encoding a base editor + guide RNA. Transformed cells were plated on plasmid maintenance antibiotics (50 μg/mL carbenicillin, 50 μg/mL spectinomycin). The following day, colonies were picked and grown overnight in DRM and maintenance antibiotics. Overnight cultures were diluted 1:100 into DRM and maintenance antibiotics and grown at 37 °C with shaking at 230 rpm. When cells reached OD600 = 0.5, 5 mM rhamnose was added to induce base editor expression. After 18 hours, 700 μL of each culture was centrifuged at 3,400 g for 10 min and the cell pellet was resuspended in 150 μL total DRM. Serial dilutions in H2O of each resuspended culture were plated on two different conditions in parallel: (1) 2xYT agar + 50 μg/mL carbenicillin + 50 μg/mL spectinomycin + 20 mM glucose or (2) 2xYT agar + 50 μg/mL carbenicillin + 50 μg/mL spectinomycin + 20 mM glucose + 10 μM 6-(β-D-2-Deoxyribofuranosyl)-3,4-dihydro-8H-pyrimido-[4,5-c][1,2]oxazin-7-one (dP). Surviving colonies were incubated at 37 °C for 24 h after plating, then counted. To obtain survival rates, the number of colonies in the dP condition was divided by the number of colonies counted on the maintenance antibiotic plate.
Sanger sequencing of HSV thymidine kinase mutations from dP-resistant colonies
dP-resistant colonies were picked into 10 μL of H2O and heated at 95 °C for 10 min, followed by PCR using primers AR393 (5’-AGGCAGTGGGATTGTGGTG) and AR394 (5’-CGGTCAGCATTAATATTGAAGTGTGG). Each fragment was sequenced three times using primers AB301 (5’-ATAAAGTTGCAGGACCACTTCT), AR341 (5’- GCAAGCAGCCCGTAAAC), and AR392 (5’-CGTACGTCGGTTGCTATG).
Analysis of BE3-induced point mutations in mouse embryos reported by Yang and coworkers12
Using the genomic locations of all C•G-to-T•A SNVs reported by Yang and coworkers in tables S6 and S7 of their recent work12 the flanking sequences (20 base pairs on either side) were extracted from the mouse mm10 reference genome [GCA_000001635.2]. These flanking sequences were aligned, fixing the mutant cytosine in each case at position 21, and the resulting alignment was used to produce a sequence logo using WebLogo 3.6.0 (ref. 52). The custom Python script used for this analysis is included in Supplementary Note 1.
Cell culture
HEK293T cells were maintained in DMEM + GlutaMAX (Life Technologies) supplemented with 10% (v/v) fetal bovine serum. Cells were cultured at 37 °C with 5% carbon dioxide and were confirmed to be negative for mycoplasma by testing with MycoAlert (Lonza Biologics).
Mammalian cell transfections
HEK293T cells were seeded in a 48-well, poly-D-lysine-coated plate (Corning) and transfected at 70% confluence. Plasmids were prepared for transfection using either a ZymoPURE II midi prep kit (Zymo Research Corporation) or a Qiagen midi prep kit (Qiagen). For on-target editing experiments, 750 ng of base editor plasmid and 250 ng of guide RNA plasmid were co-transfected into HEK293T cells using 1.5 μL of Lipofectamine 2000 (ThermoFisher Scientific) per well as directed by the manufacturer. 20 ng of pmaxGFP transfection control plasmid (Lonza Biologics) was used as a transfection control. For orthogonal R-loop assays to measure off-target editing, 200 ng of SpCas9 guide RNA plasmid, 200 ng of SaCas9 guide RNA plasmid, 300 ng of base editor plasmid, and 300 ng of dSaCas9 plasmid were co-transfected into HEK293T cells using 1.5 μL of Lipofectamine 2000. For controls involving no base editor or no sgRNA, pUC19 DNA was used to maintain the total quantity of transfected DNA at 1000 ng. For the intracellular oligonucleotide deamination experiment, 750 ng of base editor plasmid, 250 ng of guide RNA plasmid, and 1 pmol of ssDNA oligonucleotide (Integrated DNA Technologies) were co-transfected into HEK293T cells using 1.5 μL of Lipofectamine 2000.
High-throughput sequencing of genomic DNA
Genomic DNA was sequenced using methods previously described1. Briefly, genomic DNA was isolated from HEK293T cells three days after transfection. Cells were washed with PBS and then lysed with 150 μL of lysis buffer consisting of 10 mM Tris-HCl (pH 7), 0.05% SDS, and 25 μg/mL of Proteinase K (ThermoFisher Scientific) at 37 °C for 1 h and then heat inactivated at 80 °C for 30 min. Following lysis, 1 μL of the genomic DNA lysate was used as input for the first of two PCR reactions. Genomic loci were amplified using a PhusionU PCR kit (Life Technologies) PCR1 primers for genomic loci are listed in the Supplementary Materials. 30 cycles of PCR were performed for all loci with an annealing temperature of 61 °C and an extension time of 30 s. For sequencing of the cotransfected ssDNA oligonucleotide, 22 cycles of PCR1 were performed. PCR1 products were confirmed on a 2% agarose gel. 1 μL of PCR1 was used as an input for PCR2 to install Illumina barcodes. PCR2 was conducted using a Phusion HS II kit (Life Technologies). Following PCR2, samples were pooled and gel extracted in a 2% agarose gel using a Qiaquick Gel Extraction Kit (Qiagen). Library concentration was quantified using the Qubit High-Sensitivity Assay Kit (ThermoFisher Scientific). Samples were sequenced on an Illumina MiSeq instrument (paired-end read, R1: 200–280 cycles, R2: 0 cycles) using an Illumina 300 v2 Kit (Illumina).
High-throughput sequencing data analysis
Sequencing reads were demultiplexed using the MiSeq Reporter (Illumina) and fastq files were analyzed using Crispresso2 (ref. 53). Representative analysis input and usage are described in Supplementary Note 2. Prism 8 (GraphPad) was used to generate dot plots and bar plots of these data. Base-editing values are representative of n=3 independent biological replicates performed at different times, generally by different researchers, with the mean ± SEM shown.
Protein expression and purification for in vitro assays
Base editor purification was performed as described previously11, with a few modifications. BL21DE3* (ThermoFisher Scientific) chemically competent E. coli were transformed with a plasmid encoding N-terminally 6xHis-tagged base editor under control of an IPTG-induced T7 promoter. Individual colonies were picked and grown in 1 L of 2xYT media until OD600 ~0.7–0.8. Cells were cold shocked on ice for 1–2 h, then induced with 1 mM IPTG (isopropyl-β-D-thiogalactoside; Gold Biotechnology) and grown for a further 12–16 h at 16 °C with shaking at 220 rpm Cells were collected by centrifugation at 6,000 g for 20 min and the resulting cell pellet was resuspended in 25 mL high-salt buffer (100 mM Tris–Cl pH 8.0, 1 M NaCl, 5 mM tris(2-carboxyethyl)phosphine (TCEP; Sigma-Aldrich), 20% glycerol) supplemented with 0.4 mM phenylmethane sulfonyl fluoride (PMSF; Sigma-Aldrich) and EDTA-free protease inhibitor pellet (Roche, 1 pellet per 50mL lysis buffer used). Cells were lysed by sonication (6 min total, 3 s on, 3 s off) and the lysate was cleared by centrifugation at 22,000 g for 20 min. The cleared lysate was incubated with 1.5 mL of TALON Cobalt resin (Clontech) with rotation at 4 °C for 1–2 h. The resin was washed two times with 15 mL cold high-salt buffer and bound protein was eluted in medium-salt buffer (100 mM Tris–HCl pH 8.0, 0.5 M NaCl, 20% glycerol, 5 mM TCEP) supplemented with 200 mM imidazole. The isolated protein was then buffer-exchanged with low-salt buffer and concentrated using an Amicon Ultra-15 centrifugal filter unit (100,000 molecular weight cutoff). The isolated protein was further purified on a 5 mL Hi-Trap HP SP (GE Healthcare) cation exchange column using an Akta Pure FPLC. Protein-containing fractions were pooled and concentrated using an Amicon Ultra-15 centrifugal filter unit (100,000 molecular weight cutoff). Proteins were quantified using Quick Start Bradford reagent (Bio-Rad) using BSA standards (Bio-Rad) and stored short-term at 4 °C.
Protein purity was characterized by SDS-PAGE analysis. Briefly, proteins were denatured at 95 °C for 10 min in Laemmli sample loading buffer (Bio-Rad) supplemented with 2 mM dithiothreitol (DTT; Sigma-Aldrich) and separated by electrophoresis at 200 V for 40 min on a Bolt 4–12% Bis-Tris Plus (ThermoFisher Scientific) pre-cast gel in Bolt MES SDS running buffer (ThermoFisher Scientific). Gels were stained with InstantBlue reagent (Expedeon) for 1 h and washed several times with H2O before imaging with a G:Box Chemi XRQ (Syngene).
In vitro deamination assays. A 5’-Cy3-labeled ssDNA oligonucleotide
(5’-Cy3-ATTATTATTATTTCTATTTATTTATTTATTT) was purchased as an HPLC-purified oligonucleotide from Integrated DNA Technologies (IDT). All reactions were performed in reaction buffer11 (20 mM HEPES pH 7.5, 150 mM KCl, 0.5 mM dithiothreitol (DTT), 0.1 mM EDTA, 10 mM MgCl2) with concentrations of 5’-Cy3-labeled oligonucleotide varying from 0.2–100 μM and concentrations of each purified base editor protein that were >20-fold lower than the substrate concentration assayed in each case. Base editor proteins were incubated at room temperature for 5 min with a non-targeting sgRNA added in a 1:1 molar ratio. Subsequently, the 5’-Cy3-labeled oligonucleotide was added to the appropriate concentration and the reactions were incubated at 37 °C for 30 min. Reactions were stopped by the addition of buffer PB (100 μL, Qiagen) and isopropanol (25 μL) and purified on a MinElute spin column (Qiagen), eluting in 15 μL of CutSmart buffer (New England BioLabs). USER enzyme (1.5 U, New England BioLabs) was added to the purified ssDNA and incubated at 37 °C for 1 h. 10 μL of the resulting solution was combined with 10 μL of loading buffer (0.09 M tris(hydroxymethyl)aminomethane, 0.09 M sodium tetraborate, 10 mM EDTA pH 8.0, 10 M urea, 20% sucrose, 0.1% SDS) and loaded on a 10% TBE-urea gel (Bio-Rad) that was pre-run in 0.5x TBE buffer for 15 min at 180 V. The cleaved uracil-containing products were resolved from the uncleaved cytosine-containing starting material by electrophoresis for 30 min at 180 V, and the gel was imaged on a GE Typhoon FLA 7000 imager. The ratio of product to substrate bands was quantified by densitometry using ImageJ and used to calculate initial reaction velocities. Nonlinear regression was performed using Prism 8 (GraphPad) to fit these data to the Michaelis-Menten equation in order to determine kcat and Km values. Calculation of the propagated error in the kcat/Km ratio from the individual errors in the kcat and Km parameters estimated by the regression is described in Supplementary Note 3.
Transfection and fluorescence-activated cell sorting for WGS samples
HEK293T cells were transfected with 750 ng of CBE–P2A–GFP constructs and 250 ng of RNF2-targeting guide RNA as described above. Four wells were transfected for each tested CBE or control (Cas9 nickase instead of a CBE). Four days after transfection, cells were trypsinized with 50 μL of trypsin per well, and resuspended in 200 uL of DMEM (50% FBS (v/v), 100U/mL penicillin/streptomycin). Wells of the same editor were pooled, and cells were filtered through a cell strainer cap (VWR International). Flow sorting was performed on a FACS Aria II (BD Biosciences) sorter using BDFACS Diva software. Cells were gated on forward/side scatter and then gated for GFP signal compared to an untransfected negative control. Cells were then gated on fluorescence intensity. Intensity gates were set to contain the top 28% of GFP-positive YE1–P2A–GFP cell, which corresponded to the top 30% of GFP-positive BE4–P2A–GFP cells and the top 45% of GFP-positive Cas9 nickase–P2A–GFP positives cells (see Supplementary Note 5). Approximately 70,000 cells were collected for each sample in bulk. Of these, about 20,000 cells were sequenced for bulk on-target editing efficiency at the RNF2 locus. The remaining cells were diluted to a concentration of 6 cells/mL (equivalent to 0.9 cells/well) in DMEM (10% FBS (v/v), 100U/mL penicillin-streptomycin). 150 μL of this diluted mixture was pipetted into each well of a 96-well plate. Wells were monitored daily to ensure that each population of cells came from only a single cell. Cells were split into a 48-well, poly-D-lysine-coated plate (Corning) and grown for 16 days before harvesting.
Whole-genome sequencing sample preparation
Cells were lysed using a DNA Agencourt Advance (Beckman Coulter) according to manufacturer instruction. Briefly, 100 μL of lysis buffer (95 μL of Beckman lysis buffer, 2.5 μL of proteinase K (Thermo Fisher), and 2.5 μL of 1M DTT) were added to each well and incubated for 5 min at 37 °C. Lysate was then transferred to PCR strips and incubated at 55 °C for 1 hour. 50 μL of Beckman Binding Buffer 1 (Beckman Coulter) was added, and samples were incubated for 2 min before the addition of magnetic beads contained in Beckman Binding Buffer 2 (Beckman Coulter). Samples were incubated for 5 min and then placed on a magnetic plate for 10 min. Supernatant was removed, and beads were washed twice with 70% ethanol. DNA was then resuspended in 50 μL of elution buffer. Samples were placed on a magnetic plate, and the supernatant containing the purified DNA was removed and transferred to fresh tubes. DNA yields were quantified with a Nanodrop. Libraries were created using a Kapa HyperPrep Plus kit according to manufacturer instruction. 800 ng of purified DNA per sample was diluted to a total volume of 35 μL in 10 mM Tris-HCl (pH 8). 5 μL of KAPA frag buffer and 10 μL of Kapa frag enzyme were added to each reaction. Samples were placed in a pre-cooled PCR block and then heated to 37 °C for 12 min. Immediately after 12 min, samples were placed on ice, and 7 μL of End Repair and A-tailing buffer and 3 μL of End Repair and A-tailing enzyme mix were added immediately to each sample. Samples were mixed and then heated at 20 °C for 30 min and then 65 °C for 30 min in a thermocycler with the lid temperature set to 85 °C. Following this incubation, 10 μL of DNA ligase, 30 μL of DNA ligation buffer, and 10 μL of 15 μM KAPA Adapter primers were added. This mixture was then incubated at 20 °C for 15 min. A post-ligation cleanup was performed by adding 88 μL of Kapa Pure beads to the adapter mix. After a 10-min incubation, beads were collected on a magnetic plate, and the supernatant was discarded. Samples were washed twice with 200 μL of 80% ethanol. Beads were dried for 4 min, and 55 μL of elution buffer was added. After incubation, 50 μL of purified DNA was removed from beads.
WGS library size selection and quality control
A size selection was performed on the purified library. A 0.5x cut was performed to remove fragments greater than 1 kb: 25 μL of Kapa Pure beads were added to the eluted library, incubated, and placed on a magnet. The supernatant was collected and saved. 10 μL of fresh Kapa Pure beads were then added to the supernatant to perform a 0.7x second cut. After incubation, libraries were placed on a magnet and the supernatant was removed and discarded. Beads were washed twice with 200 μL of 80% ethanol and then dried for 4 min. 40 μL of 10 mM Tris-HCl (pH 8) was added to the beads to elute the final library. Each individual genome was quantified using the Kapa Quantification kit as described previously1. Library length was determined using an Agilent High Sensitivity DNA Kit and an Agilent 2100 Electrophoresis Bioanalyzer according to manufacturer instructions. Mean fragment length for final libraries was approximately 700 bp.
Whole-genome sequencing and data analysis
Sequencing was performed at the Broad Institute Genomics Platform on an Illumina NovaSeq 6000 using two S4 flow cells. Initial data processing and read alignment was performed by the Broad Institute Genomics Platform. Reads were demultiplexed and aligned to the hg19 (b37) reference genome using BWA-MEM (v0.7.7) (ref. 54). Aligned bams were sorted and optical duplicates were marked using Picard tools (v2.21). Base quality recalibration was performed using GATK (v3.4). All subsequent analyses were performed using the FAS RC Cannon high-performance computing cluster (Harvard University). Sequencing coverage was calculated using mosdepth (v0.2.6) (ref. 55). We conducted variant calling on every sample independently using three algorithms, GATK HaplotypeCaller (v4.1.3.0) (ref. 56), freebayes (v1.3.1) (ref. 57), and VarScan (v2.4.3) (ref. 58), assuming a ploidy of four and a minimum alternate allele read frequency of 0.1 to call an SNV. We called SNVs on the GATK-recommended genomic intervals56 that exclude highly repetitive regions such as centromeres and telomeres. We used bcftools (v1.9) to find the intersection of the variants called by all three algorithms in order to generate high-confidence variant calls. For all treated samples, we used bcftools to filter out variants in the treated sample that were present in the parent in order to retain only de novo variants that arose post treatment with base editors. We also used bcftools to filter out variants present at allele frequencies greater than 0.5 as previously reported40 in order to restrict analysis to variants that likely arose as a result of base editor treatment. Finally, we used bcftools to exclude variants that exhibited at least one of the following poor quality metrics based on the GATK vcf annotations: QD < 2, FS > 60, SOR > 3, MQ < 40, MQRankSum < −5. These final, high-quality variant calls for each treated sample were used for all downstream analyses.
RNA off-target editing analysis
HEK293T cells were transfected with 750 ng of plasmid encoding editors and 250 ng of guide RNA plasmid as described above. Cells were lysed 48 hours after transfection using the RNeasy kit (Qiagen) following manufacturer instructions. Briefly, media was aspirated, and cells were washed with ice cold PBS. To lyse, 350 μL of RLT buffer was added to each well. Cells were pipetted vigorously and then transferred to a DNA eliminator column. Columns were spun at 8000xg for 30 seconds, and 350 μL of 70% ethanol was added to the flow through, which was then applied to an RNeasy spin column. The mixture was centrifuged for 8000xg for 30 seconds. The column was then washed with 700 μL of RW1 buffer and then twice with 500 μL of RPE buffer. The membrane was dried by centrifuging at 8000xg for 1 min. Purified RNA was eluted with 40 μL of RNase-free water, and 2 μL of RNase-OUT (Fisher Scientific) was added. cDNA was generated using SuperScript IV (Thermo Fisher Scientific). 2 μL of purified RNA was combined with 1 μL of dNTPs, 1 μL of a poly T primer, and 9 μL of RNase-free water. The mixture was heated to 65 °C for 5 min and then placed on ice for 1 min. 4 μL of 5x superscript buffer, 1 μL of SSIV reverse transcriptase 1 μL of 0.1M DTT, and 1 μL of RNase OUT were then added. Two additional reactions were also performed, and reverse transcriptase was not added, as a control for gDNA contamination. Reverse transcription reactions were heated to 50 °C for 10 min, then to 80 °C for 10 min and then placed on ice. 1 μL of RNAse H was added, and the samples were heated to 37 °C for 20 min to degrade RNA. 1 uL of this reaction was used as a template for the first PCR of amplicon sequencing: the remaining protocol is identical to that used for gDNA sequencing (see above). Primers used for each cDNA amplicon and amplicon sequences are listed in the supplementary information. The no-RT controls were also subjected to Miseq prep, and it was ensured that there were negligible read counts for these samples.
Western blot analysis
HEK293T cells were transfected with 750 ng of plasmid encoding C-terminal 3xHA-tagged base editors and 250 ng of guide RNA plasmid as described above. Cells were lysed 48 h post transfection at 4 °C for 30 min in RIPA buffer (Thermo Fisher) supplemented with 1 mM phenylmethane sulfonyl fluoride (PMSF; Sigma-Aldrich) and EDTA-free protease inhibitor pellet (Roche, 1 pellet per 50mL lysis buffer used). Lysates were cleared by centrifugation at 12,000 rpm for 20 min. Total protein concentration was quantified using Quick Start Bradford reagent (Bio-Rad) using BSA standards (Bio-Rad). Protein extracts were denatured at 95 °C for 10 min in Laemmli sample loading buffer (Bio-Rad) supplemented with 2 mM dithiothreitol (DTT; Sigma-Aldrich) and were separated by electrophoresis at 180 V for 40 min on a Bolt 4–12% Bis-Tris Plus (ThermoFisher Scientific) pre-cast gel in Bolt MES SDS running buffer (ThermoFisher Scientific). 10 μg of total protein was loaded per well. Transfer to a PVDF membrane was performed using an iBlot 2 Gel Transfer Device (ThermoFisher Scientific) according to the manufacturer’s protocols. The membrane was cut in half at the 75 kDa marker and each half was blocked separately in Odyssey Blocking Buffer (LI-COR) in TBS for 1 h at room temperature with rocking. The high molecular weight half was incubated with rabbit anti-HA (Cell Signaling Technologies 3724S; 1:1000 dilution) in SuperBlock Blocking Buffer (ThermoFisher Scientific) at 4°C overnight with rocking. The low molecular weight half was incubated with rabbit anti-GAPDH (Cell Signaling Technologies 5174S; 1:1000 dilution) in SuperBlock Blocking Buffer (ThermoFisher Scientific) at 4 °C overnight with rocking. The membranes were washed 2x with TBST (TBS + 0.5% Tween-20) for 10 min each at room temperature, then incubated with goat anti-rabbit 680RD (LI-COR 926–68071) diluted 1:10,000 in SuperBlock for 1 h at room temperature. The membrane was washed as before and imaged using an Odyssey Imaging System (LI-COR).
Cell viability assay
HEK293T cells were seeded in a 96-well, clear-bottomed black plate (Corning) and transfected at 70% confluence with 200 ng of base editor plasmid, 40 ng of guide RNA plasmid, and 0.5 μL of Lipofectamine 2000 (ThermoFisher Scientific) per well. 48 or 72 h post transfection, cell viability was measured using the CellTiter-Glo Reagent (Promega) according to the manufacturer’s protocol. Luminescence was measured using an Infinite M1000 Pro microplate reader (Tecan).
Protein nucleofections
To compare on-target editing and off-target editing at orthogonal R-loops using DNA or ribonucleoprotein (RNP) delivery of base editors, cells were first lipofected as described above with the respective plasmids, supplemented to 1000 ng total with pUC19 plasmid if base editor plasmid was not included. 24 hours after lipofection, to allow time for expression of SaCas9 and formation of the R-loop, cells that were treated only with dSaCas9 and orthogonal guide RNA plasmids were trypsinized in 50 μL of TrypLE express (Life Technologies) per well for 5 min at 37 °C. Cells were suspended and trypsin was quenched with an equal volume of fresh media. Cells were counted in a Countess II cell counter (ThermoFisher Scientific) and 200,000 cells per protein nucleofection sample were apportioned into a single tube. These cells were centrifuged for 8 min at 100 g, the supernatant was discarded, and cells were resuspended in 10 μL per 200,000 cells of nucleofection solution supplemented as described by the manufacturer (Lonza, SF Cell Line 4D-Nucleofector X Kit S). RNP solutions were prepared by adding 100 pmol of chemically modified sgRNA (Synthego) to 10 μL of supplemented nucleofection solution per sample. 94 pmol of BE4 protein (expressed and purified by Aldevron, and provided as a generous gift from Prof. Mark Osborn) was added to a final volume of 12 μL, and RNP complexes were formed by incubation at room temperature for 5 min. 12 μL of RNP solution was mixed with 200,000 cells in 10 μL of nucleofection solution per sample, and added to a Nucleocuvette (Lonza, SF Cell Line 4D-Nucleofector X Kit S). Cells were nucleofected in a Lonza 4D Nucleofector using program CM-130 according to the manufacturer’s instructions. Immediately following nucleofection, cells were recovered for 5 min by adding 80 μL of pre-warmed media. 30 μL of recovered cells from each sample were diluted to a final volume of 250 μL in fresh media and incubated at 37 °C for 2 more days before extraction of genomic DNA from all samples, including those treated only with DNA. Three different splits of cells were used in triplicate samples for each treatment.
Data Availability
High-throughput sequencing and whole-genome sequencing data is deposited in the NCBI Sequence Read Archive (PRJNA553240). Plasmids used in this study are available from Addgene. Amino acid sequences of all base editors in this study are provided in the Supplementary Sequences.
Code Availability
The script used to analyze the SNVs reported by Yang and coworkers12 is provided in Supplementary Note 1. The script and parameters used for running CRISPResso2 analyses is provided in Supplementary Note 2. The script used for calculating the number of pathogenic SNPs targetable by CBEs is provided in Supplementary Note 4.
Supplementary Material
Acknowledgements
We thank Ben Thuronyi for helpful discussions, Kevin Zhao, Tony Huang, and Shannon Miller for providing guide RNA plasmids, and Beverly Mok for providing an sgRNA for in vitro experiments. Mark Osborn, Aldevron, and the Kidz1stFund provided the BE4 protein used for protein delivery experiments, and Kathryn Tian and Smriti Pandey provided assistance with experiments. We thank Donald Court for providing pSIM5, which was used for recombineering the HSV-TK gene onto the E. coli chromosome. We thank Tamara Mason and Erin LaRoche for assistance with WGS library preparation and FAS Research Computing at Harvard University for computational resources. This work was supported by U.S. NIH U01 AI142756, RM1 HG009490, R35 GM118062, HHMI, the Bill and Melinda Gates Foundation, and the St. Jude Collaborative Research Consortium. A.R. is an NSF Graduate Research Fellow and was supported by NIH Training Grant T32 GM095450. J.L.D is supported by the Hertz Foundation and the NSF GFRP fellowship. G.A.N. is a Howard Hughes Medical Institute Fellow of the Helen Hay Whitney Foundation. Flow cytometry was supported by Cancer Center Support (core) Grant P30-CA14051 from the NCI.
Footnotes
Competing interests
The authors declare competing financial interests. D.R.L. is a consultant and cofounder of Editas Medicine, Pairwise Plants, Beam Therapeutics, and Prime Medicine, companies that use genome editing. J.L.D., A.R., and D.R.L. through the Broad Institute have filed patent applications on aspects of this work.
References
- 1.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168, 20–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rees HA & Liu DR Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paz Zafra MS, E.M.; Katti A; Foronda M; Breinig M; Schweitzer AY; Simon A; Han T; Goswami S,; Montgomery E; Thibado J; Kastenhuber ER; Sanchez-Rivera FJ; Shi J; Vakoc CR; Lowe SW; Tschaharganeh DF; Dow LE Optimized base editors enable efficient editing in cells, organoids and mice. Nature Biotechnology 36, 888–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim K et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol 35, 435–437 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y et al. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat Commun 8, 118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zong Y et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35, 438–440 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol 35, 475–480 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Liang P et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nature Communications 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rees HA et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 8, 15790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo ES, Y.; Wei W; Yuan T; Ying W; Sun H; Yuan L; Steinmetz LM; Li Y; Yang H Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin SZ, Y.; Gao QG; Zhu Z; Wang Y; Qin P; Liang C; Wang D; Qiu J; Zhang F; Gao C Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364 (2019). [DOI] [PubMed] [Google Scholar]
- 14.McGrath E et al. Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing. Nat Commun 10, 5353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badran AH & Liu DR Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat Commun 6, 8425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garibyan L Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2, 593–608 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Harris RSP-M, S.K.; Neuberger MS RNA Editing Enzyme APOBEC1 and Some of Its Homologs Can Act as DNA Mutators. Molecular Cell 10, 1247–1253 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Kohli RM et al. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem 284, 22898–22904 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Popodi E, Tang H & Foster PL Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A 109, E2774–2783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukui K DNA mismatch repair in eukaryotes and bacteria. J Nucleic Acids 2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saraconi GS,.; Sala C; Mattiuz G; Conticello SG The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biology 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729-aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Ma Y et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 13, 1029–1035 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Hess GT et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 13, 1036–1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X et al. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat Biotechnol 36, 946–949 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Coelho MA et al. BE-FLARE: a fluorescent reporter of base editing activity reveals editing characteristics of APOBEC3A and APOBEC3B. BMC Biol 16, 150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St Martin A et al. A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC-Cas9 or cleavage by Cas9 in living cells. Nucleic Acids Res 46, e84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin AS et al. A panel of eGFP reporters for single base editing by APOBEC-Cas9 editosome complexes. Sci Rep 9, 497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z et al. Highly precise base editing with CC context-specificity using engineered human APOBEC3G-nCas9 fusions. Bioarchive (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thuronyi BWK, L.W.; Levy JM; Yeh W; Zheng C; Newby GA; Wilson C; Bhaumik M; Shubina-Oleinik O; Holt JR; Liu DR Continuous evolution of base editors with expanded target compatibility and improved activity. Nature Biotechnology in press (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunewald J et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gehrke JM et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nature Biotechnology 36, 977–982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tashiro Y, Fukutomi H, Terakubo K, Saito K & Umeno D A nucleoside kinase as a dual selector for genetic switches and circuits. Nucleic Acids Res 39, e12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koblan LW et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nature Biotechnology 36, 843–846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan K et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 47, 1067–1072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eto T, Kinoshita K, Yoshiwaka K, Muramatsu M & Honjo T RNA-editing cytidine deaminase Apobec-1 is unable to induce somatic hypermutation in mammalian cells. PNAS 100, 12895–12898 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter MA et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem 287, 34801–34808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei L et al. APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks. Nat Struct Mol Biol 25, 45–52 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Akre MK et al. Mutation Processes in 293-Based Clones Overexpressing the DNA Cytosine Deaminase APOBEC3B. PLoS One 11, e0155391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimasu HS, X.; Ishiguro S; Gao L; Hirano S; Okazaki S; Noda T; Abudayyeh O; Gootenberg JS; Mori H; Oura S; Holmes B; Tanaka M; Seki M; Hirano H; Aburatani H; Ishitani R; Ikawa M; Yachie N; Zhang F; Nureki O Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang TP et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nature Biotechnology 37, 626–631 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou C et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature (2019). [DOI] [PubMed] [Google Scholar]
- 46.Hazen JL et al. The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning. Neuron 89, 1223–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milholland B et al. Differences between germline and somatic mutation rates in humans and mice. Nat Commun 8, 15183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong X et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat Methods 14, 491–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynch M Evolution of the mutation rate. Trends Genet 26, 345–352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahbari R et al. Timing, rates and spectra of human germline mutation. Nat Genet 48, 126–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
(References below are for online methods)
- 51.Thomason LC et al. , Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 106, 1 16 11–39 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Crooks GE et al. , WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clement K et al. , CRISPResso2: Accurate and Rapid Analysis of Genome Editing Data from Nucleases and Base Editors. Nature Biotechnology 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedersen BS, Quinlan AR, Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Auwera GA et al. , From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garrison E, Marth G, Haplotype-based variant detection from short-read sequencing. arXiv preprint (2012). [Google Scholar]
- 58.Koboldt DC et al. , VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing and whole-genome sequencing data is deposited in the NCBI Sequence Read Archive (PRJNA553240). Plasmids used in this study are available from Addgene. Amino acid sequences of all base editors in this study are provided in the Supplementary Sequences.