Abstract
The Symbol Nomenclature for Glycans (SNFG) is a community-curated standard for the depiction of monosaccharides and complex glycans using various colored-coded, geometric shapes, along with defined text additions. It is hosted by the National Center for Biotechnology Information (NCBI) at the NCBI-Glycans Page (www.ncbi.nlm.nih.gov/glycans/snfg.html). Several changes have been made to the SNFG page in the past year to update the rules for depicting glycans using the SNFG, to include more examples of use, particularly for non-mammalian organisms, and to provide guidelines for the depiction of ambiguous glycan structures. This Glycoforum article summarizes these recent changes.
Keywords: database, glycobiology, glycoscience, software, symbol nomenclature
SNFG
The Symbol Nomenclature for Glycans (SNFG) is a community-curated, broadly utilized standard for the depiction of glycans using various colored, geometric shapes (Varki et al. 2015). It has been adopted by various publications including the journal Glycobiology (Haltiwanger 2016) and others listed at https://www.ncbi.nlm.nih.gov/glycans/snfgorg.html, and by major glycoscience databases (Table I). The historical background of the SNFG, which now spans nearly 40 years, has been previously reviewed (Varki et al. 2015). Currently, the SNFG is hosted by the National Center for Biotechnology Information (NCBI) at the NCBI Glycans Page (www.ncbi.nlm.nih.gov/glycans/snfg.html), and it is curated by an international group of researchers in the field (see SNFG Discussion Group list). Additionally, various software programs for sketching glycans have been developed to support the implementation of this nomenclature (Table II).
Table I.
SNFG compliant and actively maintained glycan databasesa
| Name | Link | Contents | Reference |
|---|---|---|---|
| Asian Community of Glycoscience and Glycotechnology | acgg.asia/db/ | Includes glycan disease, glycogene, lectin frontier and curated protocols developed by the Japanese Consortium for Glycobiology and Glycotechnology and other collaborators | Maeda et al. (2015) |
| Consortium for Functional Glycomics Gateway | www.functionalglycomics.org | Glycan array, Glycan mass spectrometry profiling, glycogene microarray and mouse phenotyping data repository | Raman et al. (2006) |
| Carbohydrate Structure Database (CSDB) | csdb.glycoscience.ru/database | Manually curated bacterial, plant and fungal carbohydrate structure database | Toukach and Egorova (2016) |
| CSDB_GT (CSDB Glycosyltransferases) | csdb.glycoscience.ru/gt.html | Manually curated database of glycosyltrasferases with confirmed activity (in 2019, of Escherichia coli and Arabidopsis thaliana) | Egorova et al. (2019) |
| GLYCAM-Web | Glycam.org | Tools for predicting 3D structures of carbohydrates, glycoproteins and carbohydrate–protein structures | Kirschner et al. (2008) |
| GLYCOSCIENCES.de | glycosciences.de | Various tools and databases for glycobiology, with focus on 3D structures and references to related PDB entries | Bohm et al. (2019) |
| Glycostore | glycostore.org | Glycan chromatographic retention properties | Zhao et al. (2018) |
| Glyco3D | glyco3d.cermav.cnrs.fr/home.php | Family of databases covering the 3D features of mono-, poly- and oligo-saccharides, glycosyltransferases and lectins and glycosaminoglycan-binding proteins | Perez et al. (2015) |
| Glycomics@ExPASy | www.expasy.org/glycomics | Various glycomics tools developed at the Swiss Institute of Bioinformatics and links to other sites | Mariethoz et al. (2018) |
| GlyCosmos | glycosmos.org | Portal for glycoscience data resources and repositories | … |
| GlyGen | www.glygen.org | Computational and Informatics Resources for Glycoscience research | … |
| GlyTouCan | glytoucan.org | International glycan structure repository | Tiemeyer et al. (2017) |
| KEGG GLYCANb | www.genome.jp/kegg/glycan/ | Glycan structures, relevant reactions and pathways | Kanehisa (2017) |
| MatrixDB | matrixdb.univ-lyon1.fr/ | Database of extracellular matrix protein, proteoglycan and polysaccharide interactions | Clerc et al. (2019) |
| MonosaccharideDB | monosaccharidedb.org | Monosaccharide database | |
| SugarBindDB | sugarbind.expasy.org | Pathogen Sugar-Binding Database | Mariethoz et al. (2016) |
| UniCarb-DB | unicarb-db.expasy.org | Glycomics mass spectrometry database | Campbell et al. (2014) |
Note: aTable only lists databases that follow a majority of the SNFG nomenclature guidelines, not all glycoscience resources.
bSNFG adoption in progress.
Table II.
SNFG drawing software
| Name | Purpose | Reference |
|---|---|---|
| 3D-SNFG | 3D adaptation of SNFG to visualize glycans in pdb structures (web based, implemented in VMD software) | Thieker et al. (2016) |
| 3D-SNFG in LiteMol | Implementation of 3D-SNFG in LiteMol for 3D display of glycans | Sehnal and Grant (2019) |
| DrawGlycan-SNFG | Converts IUPAC string to SNFG drawings (web based, stand-alone) | Cheng et al. (2017) |
| SugarSketcher | Drag and drop tool to sketch glycans (web based) | Alocci et al. (2018) |
| GlycanBuilder2 | GlycanBuilder extended for rendering SNFG sketches (stand-alone) | Tsuchiya et al. (2017) |
The overall goal of the SNFG is (1) to facilitate communications and presentations of monosaccharides and glycans for researchers in the glycosciences and for scientists and students less familiar with the field; (2) to ensure uniform usage of the nomenclature in the literature, thus helping to ensure scientific accuracy in journal and online publications; and (3) to continue to develop the SNFG and its applications to aid wider use by the scientific community. With these goals in mind, updates have been made this year to the NCBI Glycans page that hosts the SNFG. Specifically, the footnotes to Table I at the SNFG page that include the rules for depicting glycans have been modified. No changes were made to the table. The footnotes are now organized into 10 themes in order to help streamline their use. These footnotes provide guidelines for (i) general usage of the SNFG, (ii) CMYK/RGB color codes, (iii) symbol colors and shapes, (iv) ring configurations, (v) bond linkage presentation, (vi) sialic acids, (vii) glycan modifications, (viii) amino substitutions, (ix) handling ambiguous or partially defined glycans and (x) depicting non-glycan entities using SNFG renderings.
More examples have been included for non-mammalian species (Figure 1), with the realization that there is much greater diversity in monosaccharide composition and modifications in these organisms compared to vertebrates. This necessitates the need to engender greater flexibility with respect to SNFG use. To address this issue, several changes have been made.
Fig. 1.
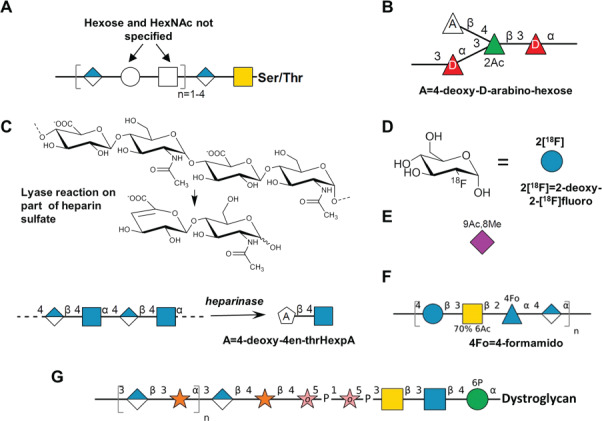
Footnotes, white symbols and variable modifications in the SNFG. (A) Drosophila O-glycan with generic Hexose and HexNAc (Tiemeyer et al. 2015). (B) Bacterial glycan from Citrobacter braakii including D-Fucose, 2-O-acetylation and 4-deoxy-D-arabinohexose (a deoxyhexose variant; Katzenellenbogen et al. 2003). All rings are pyranose. (C) Use of pentagon with single non-italicized letter to depict product not defined by the SNFG (lyase reaction shown above sketch, adopted from Shaya et al. 2010). (D) Positron emission tomography tracer 2-deoxy-2[18F]fluoro-α-D-glucopyranose. (E) 9-O-acetyl-8-O-methyl-N-acetylneuraminic acid (Neu5,9Ac28Me). Purple diamond, Neu5Ac, already includes 5Ac modification. (F) O-polysaccharide from Providencia alcalifaciens with variable modification on GalNAc (Ovchinnikova et al. 2007). (G) Glycan chain of α-dystroglycan containing ribitol-5-phosphate and phosphate at 6-position of mannose (Kanagawa et al. 2016). The “o” in the pink star represents the alditol of the monosaccharide.
First, white symbols of various shapes were previously used to define monosaccharides of unknown/undefined stereochemistry (white circle for hexose, type undefined, white diamond for deoxynonulosonic acids etc.). This usage of white symbols remains in the current revision. For example, Figure 1A depicts a Drosophila O-glycan that contains generic hexose and N-acetylhexosamine. However, while the white, flat hexagon was previously used to denote both heptoses and unknown monosaccharides, its usage is now restricted to depicting only unknown or partially defined monosaccharides.
Second, a single, non-italicized letter (A…Z) is now permitted inside white symbols to provide additional information about a monosaccharide that is not defined by a colored symbol. Footnotes/figure legends associated with these single letters convey structural details regarding this annotated symbol. When using the white symbol with a single letter, it is necessary to ensure that the chemical composition (and thus mass) of the entity corresponds to the composition of the selected white generic type. For example, the bacterial glycan in Figure 1B contains a deoxyhexose variant with known composition. A white triangle with letter “A” is used since this deoxyhexose is not defined by any colored SNFG symbol. A footnote is provided to explain the exact usage. In another case, if a monosaccharide has no equivalent representation in the SNFG table as in Figure 1C, a pentagon is used. In this case, heparinase action on a glycosaminoglycan results in a chemically defined product, but the generic type of the entity does not correspond to any of the standard white symbols in the SNFG table. Thus, a white pentagon is used with the letter “A” and an accompanying footnote.
Third, the rules for describing substituents have been reviewed and more examples are provided. For example, Figure 1D shows a commonly used fluoro-glucose analog, with a footnote providing a full explanation of the modification. Figure 1E illustrates a multiply-modified sialic acid, where the substituents are presented as a sequential/concatenated string. To the extent possible, abbreviations used to describe substituents should follow guidelines described in footnote 7 of the updated SNFG page. The presence of variable amounts of substituents may be indicated using the ± symbol or by indicating % presence if known, e.g. “70% 6Ac” to indicate presence of 6Ac on 70% of a residue or repeating unit (Figure 1F). Less common substituents like “4Fo”, which are not among the standard monosaccharide modifications listed on the SNFG page, may also be indicated using footnote as illustrated. The final example illustrates the use of italicized “o” within a pink star to depict ribitol in an O-glycan that is found on mammalian dystroglycan (Figure 1G).
Please note that the meaning of single letter annotations in colored SNFG symbols is different from those in the white symbols. In the colored symbols, such annotations can only be used to denote either “D”/“L” configuration or ring closure information (“p”, “f” or “o” for pyranose, furanose and alditol, respectively). In white symbols, they are used to annotate details regarding generic monosaccharide types as discussed in the above examples. More exhaustive examples of SNFG usage can be found at the NCBI-SNFG page, Essentials of Glycobiology textbook (https://www.ncbi.nlm.nih.gov/books/NBK310274/) and the database resources listed in Table I.
Overall, the goal of the SNFG is to make the field of glycobiology more visual and readily accessible, especially for new users and the larger biomedical community. As we make progress in this endeavor, the group anticipates working with the Glycan Informatics Advisory Group (GlyAg, www.ncbi.nlm.nih.gov/glycans/glyag.html) to populate more glycan-related pages at the NCBI and to establish links between these pages and other database resources (Table I). Comments from the scientific community are welcome, including examples of SNFG implementation in databases and software resources.
Author contributions
14Other members of the SNFG Discussion group: Alan Darvill, University of Georgia, USA; Anne Dell, Imperial College London, UK; Bernard Henrissat, Centre National de la Recherche Scientifique (CNRS), France; Carolyn Bertozzi, Stanford University, USA; Gerald Hart, University of Georgia, USA; Hisashi Narimatsu, Research Center of Medical Glycoscience, Japan; Hudson Freeze, Sanford-Burnham-Prebys Research Institute, USA; Issaku Yamada, The Noguchi Institute, Japan; James Paulson, Scripps Research Institute, USA; James Prestegard, University of Georgia, USA; Jamey Marth, University of California Santa Barbara, USA; JFG Vliegenthart, Bijvoet Center, Netherlands; Marilynn Etzler, UC Davis, USA; Markus Aebi, ETH Zürich, Switzerland; Minoru Kanehisa, Kyoto University, Japan; Naoyuki Taniguchi, Riken Global Research Cluster, Japan; Nathan Edwards, Georgetown University, USA; Pauline Rudd, National Institute for Bioprocessing Research & Training, UK; Peter Seeberger, Max-Planck-Institute of Colloids and Interfaces, Germany; Raja Mazumder, The George Washington University, USA; Rene Ranzinger, University of Georgia, USA; Richard Cummings, Harvard Medical School, USA; Ronald Schnaar, Johns Hopkins University School of Medicine, USA; Serge Perez, French National Centre for Scientific Research, France; Stuart Kornfeld, Washington University in St. Louis, USA; Taroh Kinoshita, Osaka University, Japan; William York, University of Georgia, USA; Yuriy Knirel, Russian Academy of Sciences, Russia
Acknowledgements
This is a global community-driven project. Some of the individuals who volunteer their time as members of the SNFG Discussion Group have received grant support from national funding agencies including the US National Institutes of Health, Russian Science Foundation, Swiss National Science Foundation, Research Data Australia, Japan Science and Technology Agency and the National Bioscience Database Center of Japan.
Other members of the SNFG Discussion Group: Alan Darvill, University of Georgia, USA; Anne Dell, Imperial College London, UK; Bernard Henrissat, Centre National de la Recherche Scientifique (CNRS), France; Carolyn Bertozzi, Stanford University, USA; Gerald Hart, University of Georgia, USA; Hisashi Narimatsu, Research Center of Medical Glycoscience, Japan; Hudson Freeze, Sanford-Burnham-Prebys Research Institute, USA; Issaku Yamada, The Noguchi Institute, Japan; James Paulson, Scripps Research Institute, USA; James Prestegard, University of Georgia, USA; Jamey Marth, , University of California Santa Barbara, USA; JFG Vliegenthart, Bijvoet Center, Netherlands; Marilynn Etzler, UC Davis, USA; Markus Aebi, ETH Zürich, Switzerland; Minoru Kanehisa, Kyoto University, Japan; Naoyuki Taniguchi, Riken Global Research Cluster, Japan; Nathan Edwards, Georgetown University, USA; Pauline Rudd, National Institute for Bioprocessing Research & Training, UK; Peter Seeberger, Max-Planck-Institute of Colloids and Interfaces, Germany; Raja Mazumder, The George Washington University, USA; Rene Ranzinger, University of Georgia, USA; Richard Cummings, Harvard Medical School, USA; Ronald Schnaar, Johns Hopkins University School of Medicine, USA; Serge Perez, French National Centre for Scientific Research, France; Stuart Kornfeld, Washington University in St. Louis, USA; Taroh Kinoshita, Osaka University, Japan; William York, University of Georgia, USA; Yuriy Knirel, Russian Academy of Sciences, Russia
Contributor Information
The SNFG Discussion Group:
Alan Darvill, Anne Dell, Bernard Henrissat, Carolyn Bertozzi, Gerald Hart, Hisashi Narimatsu, Hudson Freeze, Issaku Yamada, James Paulson, James Prestegard, Jamey Marth, J F G Vliegenthart, Marilynn Etzler, Markus Aebi, Minoru Kanehisa, Naoyuki Taniguchi, Nathan Edwards, Pauline Rudd, Peter Seeberger, Raja Mazumder, Rene Ranzinger, Richard Cummings, Ronald Schnaar, Serge Perez, Stuart Kornfeld, Taroh Kinoshita, William York, and Yuriy Knirel
Conflict of interest statement
None declared.
References
- Alocci D, Suchankova P, Costa R, Hory N, Mariethoz J, Varekova RS, Toukach P, Lisacek F. 2018. SugarSketcher: Quick and intuitive online glycan drawing. Molecules. 23, e3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm M, Bohne-Lang A, Frank M, Loss A, Rojas-Macias MA, Lutteke T. 2019. Glycosciences.DB: An annotated data collection linking glycomics and proteomics data (2018 update). Nucleic Acids Res. 47:D1195–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Nguyen-Khuong T, Hayes CA, Flowers SA, Alagesan K, Kolarich D, Packer NH, Karlsson NG. 2014. Validation of the curation pipeline of UniCarb-DB: Building a global glycan reference MS/MS repository. Biochim Biophys Acta. 1844:108–116. [DOI] [PubMed] [Google Scholar]
- Cheng K, Zhou Y, Neelamegham S. 2017. DrawGlycan-SNFG: A robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology. 27:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc O, Deniaud M, Vallet SD, Naba A, Rivet A, Perez S, Thierry-Mieg N, Ricard-Blum S. 2019. MatrixDB: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 47:D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova KS, Knirel YA, Toukach PV. 2019. Expanding CSDB_GT glycosyltransferase database with Escherichia coli. Glycobiology. 29:285–287. [DOI] [PubMed] [Google Scholar]
- Haltiwanger RS. 2016. Symbol Nomenclature for Glycans. Glycobiology. 26:217. [Google Scholar]
- Kanagawa M, Kobayashi K, Tajiri M, Manya H, Kuga A, Yamaguchi Y, Akasaka-Manya K, Furukawa JI, Mizuno M, Kawakami H et al. 2016. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 14:2209–2223. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. 2017. KEGG GLYCAN. In: Aoki-Kinoshita KF, editor A Practical Guide to Using Glycomics Databases. Japan: Springer; p. 177–193. [Google Scholar]
- Katzenellenbogen E, Kocharova NA, Zatonsky GV, Witkowska D, Bogulska M, Shashkov AS, Gamian A, Knirel YA. 2003. Structural and serological studies on a new 4-deoxy-d-arabino-hexose-containing O-specific polysaccharide from the lipopolysaccharide of Citrobacter braakii PCM 1531 (serogroup O6). Eur J Biochem. 270:2732–2738. [DOI] [PubMed] [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. 2008. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 29:622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Fujita N, Suzuki Y, Sawaki H, Shikanai T, Narimatsu H. 2015. JCGGDB: Japan Consortium for Glycobiology and Glycotechnology Database. Methods Mol Biol. 1273:161–179. [DOI] [PubMed] [Google Scholar]
- Mariethoz J, Alocci D, Gastaldello A, Horlacher O, Gasteiger E, Rojas-Macias M, Karlsson NG, Packer NH, Lisacek F. 2018. Glycomics@ExPASy: Bridging the gap. Mol Cell Proteomics. 17:2164–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariethoz J, Khatib K, Alocci D, Campbell MP, Karlsson NG, Packer NH, Mullen EH, Lisacek F. 2016. SugarBindDB, a resource of glycan-mediated host–pathogen interactions. Nucleic Acids Res. 44:D1243–D1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova OG, Bushmarinov IS, Kocharova NA, Toukach FV, Wykrota M, Shashkov AS, Knirel YA, Rozalski A. 2007. New structure for the O-polysaccharide of Providencia alcalifaciens O27 and revised structure for the O-polysaccharide of Providencia stuartii O43. Carbohydr Res. 342:1116–1121. [DOI] [PubMed] [Google Scholar]
- Perez S, Sarkar A, Rivet A, Breton C, Imberty A. 2015. Glyco3D: A portal for structural glycosciences. Methods Mol Biol. 1273:241–258. [DOI] [PubMed] [Google Scholar]
- Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. 2006. Advancing glycomics: Implementation strategies at the consortium for functional glycomics. Glycobiology. 16:82R–90R. [DOI] [PubMed] [Google Scholar]
- Sehnal D, Grant OC. 2019. Rapidly display glycan symbols in 3D structures: 3D-SNFG in LiteMol. J Proteome Res. 18:770–774. [DOI] [PubMed] [Google Scholar]
- Shaya D, Zhao W, Garron ML, Xiao Z, Cui Q, Zhang Z, Sulea T, Linhardt RJ, Cygler M. 2010. Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate. J Biol Chem. 285:20051–20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieker DF, Hadden JA, Schulten K, Woods RJ. 2016. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 26:786–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeyer M, Aoki K, Paulson J, Cummings RD, York WS, Karlsson NG, Lisacek F, Packer NH, Campbell MP, Aoki NP et al. 2017. GlyTouCan: An accessible glycan structure repository. Glycobiology. 27:915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeyer M, Nakato H, Esko JD. 2015. Arthropoda. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al., editors. Essentials of Glycobiology. New York: Cold Spring Harbor; p. 335–349. [Google Scholar]
- Toukach PV, Egorova KS. 2016. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res. 44:D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Aoki NP, Shinmachi D, Matsubara M, Yamada I, Aoki-Kinoshita KF, Narimatsu H. 2017. Implementation of GlycanBuilder to draw a wide variety of ambiguous glycans. Carbohydr Res. 445:104–116. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Walsh I, Abrahams JL, Royle L, Nguyen-Khuong T, Spencer D, Fernandes DL, Packer NH, Rudd PM, Campbell MP. 2018. GlycoStore: A database of retention properties for glycan analysis. Bioinformatics. 34:3231–3232. [DOI] [PubMed] [Google Scholar]


