Abstract
Background
Pin2/TRF1‐interacting protein, PinX1, was previously identified as a tumor suppressor. Here, we discovered a novel transcript variant of mPinX1 (mouse PinX1), mPinX1t (mouse PinX1t), in embryonic stem cells (ESCs). The aims of this investigation were (1) to detect the presence of mPinX1 and mPinX1t in ESCs and their differentiation derivatives; (2) to investigate the role of mPinX1 and mPinX1t on regulating the characteristics of undifferentiated ESCs and the cardiac differentiation of ESCs; (3) to elucidate the molecular mechanisms of how mPinX1 and mPinX1t regulate the cardiac differentiation of ESCs.
Methods and Results
By 5′ rapid amplification of cDNA ends, 3′ rapid amplification of cDNA ends, and polysome fractionation followed by reverse transcription–polymerase chain reaction, mPinX1t transcript was confirmed to be an intact mRNA that is actively translated. Western blot confirmed the existence of mPinX1t protein. Overexpression or knockdown of mPinX1 (both decreased mPinX1t expression) both decreased while overexpression of mPinX1t increased the cardiac differentiation of ESCs. Although both mPinX1 and mPinX1t proteins were found to bind to cardiac transcription factor mRNAs, only mPinX1t protein but not mPinX1 protein was found to bind to nucleoporin 133 protein, a nuclear pore complex component. In addition, mPinX1t‐containing cells were found to have a higher cytosol‐to‐nucleus ratio of cardiac transcription factor mRNAs when compared with that in the control cells. Our data suggested that mPinX1t may positively regulate cardiac differentiation by enhancing export of cardiac transcription factor mRNAs through interacting with nucleoporin 133.
Conclusions
We discovered a novel transcript variant of mPinX1, the mPinX1t, which positively regulates the cardiac differentiation of ESCs.
Keywords: cardiac development, cardiac differentiation, embryonic stem cell, PinX1, transcript variants
Subject Categories: Basic Science Research, Cell Biology/Structural Biology, Developmental biology, Stem Cells
Clinical Perspective
What Is New?
A new transcript variant (PinX1t) of a previously known tumor suppressor Pin2/TRF1‐interacting protein (PinX1) has been discovered.
Expression of PinX1t was found to be positively correlated to the cardiac differentiation of embryonic stem cells.
PinX1 protein was found to bind to nucleoporin 133 protein, a component of nuclear pore complex; PinX1t‐containing cells were found to have a higher cytosol‐to‐nucleus ratio of cardiac transcription factor mRNAs when compared with control cells.
Our data suggested that PinX1t may positively regulate cardiac differentiation by enhancing export of cardiac transcription factor mRNAs through interacting with Nucleoporin 133.
What Are the Clinical Implications?
Altering PinX1t expression/activity may be used as a way to enhance cardiac differentiation of embryonic stem cells (or pluripotent stem cells) so as to increase the generation of cardiomyocytes for future transplantation/regenerative medicine, drug screening, etc.
Introduction
Myocardial infarction has become the leading cause of death around the globe and is characterized by the loss of cardiomyocytes.1 Cardiomyocytes cannot be regenerated after myocardial infarction; therefore, embryonic stem cells (ESCs), which have unlimited self‐renewal potential and can differentiate into a variety of cells, including cardiomyocytes, have become a promising cell source for generating cardiomyocytes for transplantation therapy.2
Several crucial transcription factors, including Nkx2.5, Mef2C, Gata4, and Tbx5, have been identified to be important for ESCs to differentiate into cardiac lineage.3, 4, 5, 6 Knockdown of Gata4 leads to the decrease in expressions of cardiac structural genes and transcription factors including cardiac actin, myosin heavy chain, Nkx2.5 and Mef2C.7 Gata4 has also been reported to regulate Nkx2.5 transcription and interact with Tbx5 for cardiac differentiation.8, 9, 10, 11 Tbx5 overexpression promotes cardiac differentiation and increases the beating frequency of cardiomyocytes.12 Similarly, overexpression of Gata4 promotes cardiac differentiation, increases the cardiac‐specific gene expression, and increases the number of beating clusters.13 After the differentiation of ESCs into cardiomyocytes, actin and myosin filaments are arranged into a highly organized structure known as sarcomere.14, 15 Sarcomere structure is the basic unit of cardiomyocytes for force generation. A mature cardiomyocyte expresses different cardiac structural genes, including cardiac actin, myosin heavy chain, cardiac troponin T, and cardiac troponin I4; these cardiac structural genes together form the major constituents of sarcomere.
Pin2/TRF1 binding protein, PinX1, was first identified as a Pin2 binding protein by yeast‐2 hybrid screening using the cDNA library from human HeLa cells.16 PinX1 is characterized by having 2 special domains at its N‐ and C‐termini. At the N‐terminal, it contains a glycine‐rich region known as G‐Patch.16 Many proteins have been reported to be RNA‐binding proteins by having the G‐Patch domain,17 suggesting that PinX1 may be a potential RNA‐binding protein. At the C‐terminal, PinX1 contains a telomerase inhibitory domain, which allows PinX1 to directly bind to the catalytic subunit of telomerase, telomeric reverse transcriptase, and downregulate its activity.16 As such, PinX1 was reported to be tumor suppressor gene in various cancer cells through downregulating the activity of telomerase.18
In our present study, we discovered that there is a new transcript variant of PinX1 existed in mouse ESCs (mESCs). This novel transcript variant encodes a protein containing only the N‐terminal G‐Patch domain but not the C‐terminal telomerase inhibitory domain. As a result, this novel transcript variant is named as PinX1t, where “t” stands for “truncated.”
How mPinX1 and mPinX1t regulate pluripotency and differentiation in mESCs is unknown. On the other hand, the naturally occurring mPinX1t allowed us to have an in‐depth investigation on the cellular functions of the N‐terminal of mPinX1. This current study has investigated the expressions of mPinX1 and mPinX1t in undifferentiated and differentiated mESCs and the roles of mPinX1 and mPinX1t in maintaining the pluripotency of mESCs and in regulating the cardiac differentiation. This study has also elucidated the molecular mechanisms of how mPinX1/mPinX1t regulate the cardiac differentiation.
Materials and Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Detailed experimental procedures can be found in Data S1.
Cell Culture
Institutional review board approval was obtained for the use of animal cells in this study. All the procedures followed were in accordance with guidelines set by the Chinese University of Hong Kong. mESC line D3 was cultured and cardiac differentiation was induced as we previously described.19, 20, 21, 22
Polymerase Chain Reaction, Subcloning, and Sequencing for the First Identification of mPinX1t
Forward primer franking 23 bp upstream of mPinX1 (5′‐TAAGGGAATTCATCAGCGTTCGACAAACTTGAG‐3′; italic and bold sequence indicates sequence that is complementary to mPinX1 gene) and reverse primer franking 26 bp downstream of mPinX1 (5′‐TAAGGGCGGCCGCACAGTTGAGTGGTTGGAGGC‐3′; italic and bold sequence indicates sequence that is complementary to mPinX1 gene) coding sequence (accession number: NM_028228) were used in polymerase chain reaction (PCR) using mESCs and their differentiation derivatives as the template.
Polysome Fractionation Assay
Cycloheximide was added to mESCs and cells were pelleted and lysed on ice with hypotonic lysis buffer (50 mmol/L Tris‐HCl [pH 7.5], 2.5 mmol/L MgCl2, 1.5 mmol/L KCl, diethylpyrocarbonate water) supplemented with 0.59 mmol/L phenylmethylsulfonyl fluoride, 0.12 mg/mL cycloheximide, 2.35 mmol/L dithiothreitol and 0.09 U/μL ribonuclease inhibitor for 15 minutes. Then 0.59% Triton X‐100 and 0.59% Na deoxycholate were added to the lysate and vortexed. The lysate was centrifuged at 16 000g for 7 minutes at 4°C. A sucrose gradient density was set up by layering high‐density sucrose gradient buffer (20 mmol/L Tris HCl [pH 7.5], 140 mmol/L KCl, 5 mmol/L MgCl2, 50% sucrose, 0.5 mmol/L dithiothreitol, 100 μg/mL cycloheximide, diethylpyrocarbonate water) with low‐density sucrose gradient buffer (20 mmol/L Tris HCl [pH 7.5], 140 mmol/L KCl, 5 mmol/L MgCl2, 7% sucrose, 0.5 mmol/L dithiothreitol, 100 μg/mL cycloheximide, diethylpyrocarbonate water) in 1:1 ratio; 800 ng cell lysate was loaded on top of the sucrose gradient density and subjected to ultracentrifugation at 217 874g (35 000 rpm) for 3 hours. Fractions were collected to microcentrifuge tubes and Trizol LS solution (Life Technologies, Waltham, MA) was used for RNA extraction. The extracted RNA was reverse transcribed and mPinX1t expression was determined by quantitative PCR (qPCR) using mPinX1t specific primers. mPinX1t forward primer: 5′‐AAAGGGAAGGATCTGTCCTC‐3′ mPinX1t reverse primer: 5′‐CAATTTAATCCGAGGAGCCTGAG‐3′.
Full‐Length RNA Ligase‐Mediated Rapid Amplification of 5′ cDNA Ends Assay and RNA Ligase‐Mediated Rapid Amplification of 3′ cDNA Ends Assay
The rapid amplification of 5′ cDNA ends and rapid amplification of 5′ cDNA ends assays were carried according to the manufacturer's instructions (L1502‐01, Life Technologies, Rockville, MD). The 5′ untranslated region (UTR) sequence was amplified using 5′ GeneRacer‐specific forward primer: 5′‐CGACTGGAGCACGAGGACACTGA‐3′ and mPinX1t‐specific reverse primer: 5′‐CACAATTTAATCCGAGGAGCCTGAGCAA‐3′. A second‐round PCR using 1 μL of PCR product from the first‐round PCR was performed by changing the forward primer to 5′ GeneRacer nested primer: 5′‐GGACACTGACATGGACTGAAGGAGTA‐3′. For rapid amplification of 3′ cDNA ends (3′ RACE), the 3′ UTR sequence was amplified using 3′ GeneRacer‐specific reverse primer: 5′‐GCTGTCAACGATACGCTACGTAACG‐3′ and mPinX1t‐specific forward primer: 5′‐TTGCTCAGGCTCCTCGGATTAAATTGTG‐3′. A second‐round PCR using 1 μL of PCR product from the first‐round PCR was performed by changing the forward primer to 5′ nested primer: 5′‐GTGAAGGCAGAGCAACTTTGCCAGA‐3′ and reverse primer to 3′ GeneRacer nested primer: 5′‐CGCTACGTAACGGCATGACAGTG–3′.
Lentiviral Vector Subcloning and Packaging
cDNA encoding myc‐tag fused with the full coding sequence of mPinX1 and cDNA encoding myc‐tag fused with the full coding sequence of mPinX1t were subcloned into pWPI (Addgene, Cambridge, MA) to make the lentiviral backbone DNA plasmids pWPI‐myc‐mPinX1 and pWPI‐myc‐mPinX1t, respectively. Small hairpin RNA against mPinX1 (sense sequence: 5′‐AAGAAGAAAGTTTCCAGATAA‐3′) and scrambled small hairpin RNA sequence (sense sequence: 5′‐GAAACAGATAATAGACAGTTA‐3′) were subcloned into pLVTHM (Addgene) to make the lentiviral backbone DNA plasmids pLVTHM‐shmPinX1 and pLVTHM‐shscrambled. Lentiviral backbone DNA constructs and the packaging plasmids (psPAX2 and pMD2.G) were cotransfected into the human embryonic kidney (HEK)293FT cells in OPTI‐MEM I (Invitrogen, Waltham, MA) using Lipofectamine 2000 (Invitrogen). OPTI‐MEM I medium with the complexes was changed back to normal medium 6 hours after transfection and lentiviruses were collected at 24, 48, and 72 hours after transfection.
Lentiviral Vector‐Mediated Gene Transfer to mESCs
Stable mESC lines were made using lentiviral vector‐mediated gene transfer. Detailed experimental procedures can be found in Data S1.
Telomere Repeat Amplification Protocol Assay
The telomere repeat amplification protocol assay was done with TeloTAGGG telomerase PCR ELISA (Roche, Basel, Switzerland) according to the manufacturer's instructions.
Quantitative PCR
SYBR GREEN PCR Master Mix (Applied BioSystems, Foster City, CA) was used for qPCR, and the qPCR reaction was performed using ABI 7500 Fast Real Time PCR System. Detailed experimental procedures can be found in Data S1.
SDS PAGE and Western Blotting and Immunocytochemistry
Detailed experimental procedures can be found in Data S1.
RNA Immunoprecipitation Assay
Pwpi‐myc‐mPinX1 or Pwpi‐myc‐mPinX1t plasmids together with plasmids harboring the cDNAs of cardiac transcription factors, including pCI‐Gata4 (kindly provided by Prof. Qiangrong Liang in College of Osteopathic Medicine in New York Institute of Technology) or pAC‐CMV‐Tbx5 (kindly provided by Prof. Katherine Yutzey in the Division of Molecular Cardiovascular Biology at the Heart Institute, Cincinnati Children's Hospital Medical Center) were transfected into HEK293FT using Lipofectamine 2000 (Invitrogen). The details of RNA immunoprecipitation can be found in Data S1. Briefly, anti‐myc antibody (9B11; Cell Signaling Technology, Inc, Danvers, MA) at a dilution ratio of 1:250 and isotype antibody IgG2a (Abcam, Cambridge, UK) were used for the immunoprecipitation. RNA of the immunoprecipitates was extracted by Trizol LS. RNA was reverse transcribed into cDNA and subjected to qPCR analysis.
Yeast‐2 Hybrid
The yeast‐2 hybrid assay was performed using Matchmaker Gold Yeast Two‐Hybrid System (Clontech, Mountain View, CA) according to the instruction manual from the manufacturer.
Coimmunoprecipitation Assay
pWPI‐myc‐mPinX1t or pWPI‐myc‐mPinX1 together with CMV‐HA‐Nup133 were transfected into HEK293FT cells using Lipofectamine 2000. Detailed experimental procedures of coimmunoprecipitation can be found in Data S1.
Fractionation to Separate Cytosolic and Nuclear RNAs
The RNA fractionation protocol was modified from 2 publications23, 24 and detailed experimental procedures can be found in Data S1. For both the cytosolic and nuclear fractions resuspended in Trizol solution, spike in RNA (TATAA Universal RNA Spike I, TATAA Biocenter, Göteborg, Sweden) was added as loading control for normalization in later qPCR.25
Statistical Analysis
Data were presented as mean±SEM. Data between 2 groups were compared by unpaired Student t test. Data among 3 or more groups were compared by ANOVA followed by Tukey's multiple comparison tests. P<0.05 was considered to be statistically significant.
Results
mPinX1 and its Novel Transcript Variant Were Expressed in mESCs and Their Differentiation Derivatives
To investigate the expression of mPinX1 in mESCs and their differentiated derivatives, a pair of primers flanking the coding region of mPinX1 was designed and a routine PCR reaction was performed. As expected, a distinct band at 1074 bp, which corresponded to mPinX1, was observed in both the undifferentiated and differentiation derivatives of mESCs (Figure 1A). Interestingly, an additional band at 1185 bp was also observed in both the undifferentiated and differentiation derivatives (Figure 1A). This additional band was excised and sent for DNA sequencing. The result showed that the additional band contained an extra 111 nucleotides in between exon 6 and exon 7 of mPinX1 gene (Figure 1B). This additional 111 bp was blast‐searched and was found to be located in Locus NT_039606.7 of chromosome 14 of mouse genome where the mPinX1 gene is located. The position and the sequence of the extra sequence was annotated in Figure 1B. The result suggested a novel transcript variant of mPinX1 existed in both the undifferentiated and differentiated derivatives.
Figure 1.
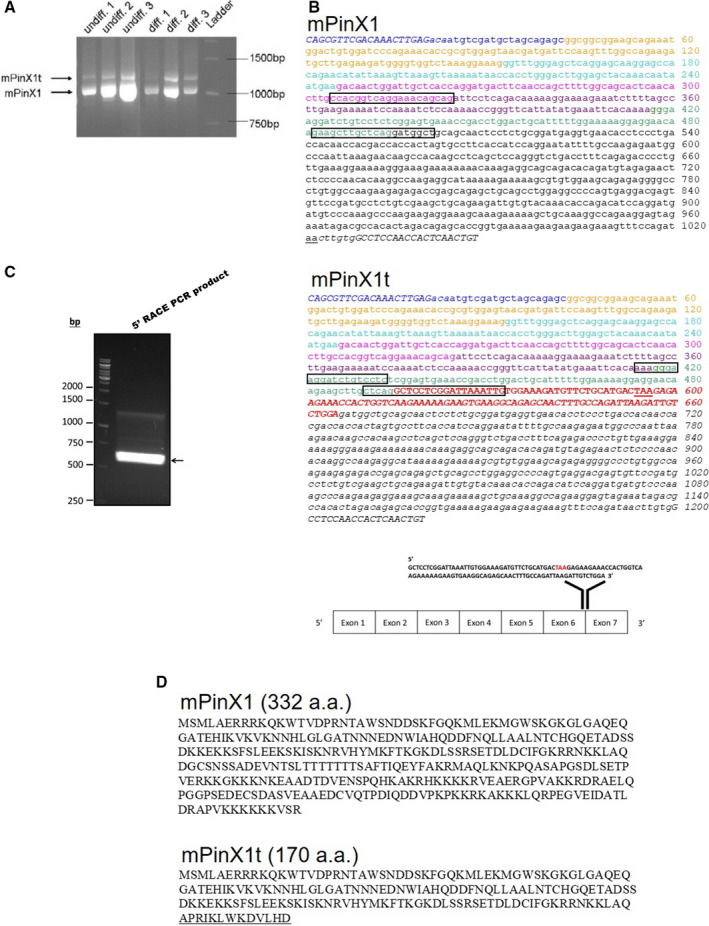
Discovery of mPinX1t, the novel transcript variant of mPinX1. A, Presence of 2 splice variants, mPinX1 and mPinX1t, as revealed by RT‐PCR. In the PCR, primers flanking the coding sequence of mPinX1 were used. Two splice variants were detected in both undifferentiated mESCs (undiff) and their differentiation derivatives (diff). B, cDNA sequences of (upper panel) mPinX1 and (middle panel) mPinX1t. Different colors represent different exons. Note that there is an extra exon in mPinX1t as represented by red and bold nucleotides. Start codon and stop codon are underlined. Italic nucleotides represent nucleotides at the untranslated regions. Italic and bold nucleotides located at the beginning and the end of the sequences represent PCR primer sequences used in (A). Boxed regions indicate the location of primers used in qPCR reactions. (Lower panel) Extra 111 nucleotides of mPinX1t was found to locate between exon 6 and exon 7 of mPinX1 sequence. Stop codon “TAA” is highlighted in red. C, Result of 5′RACE PCR reaction using primer specific for the 5′ added sequence and primer complementary to the mPinX1t sequence. The result indicated the presence of mPinX1t mRNA with intact 5′UTR. D, Amino acid sequence of (upper panel) mPinX1 and (lower panel) mPinX1t. mPinX1t contains the N‐terminal of mPinX1 (1–157 amino acids) but lacks the C‐terminal of mPinX1. Amino acids that are underlined represent the sequence that is present only in mPinX1t. 5′RACE indicates 5′ rapid amplification of cDNA ends; 5′UTR, 5′ untranslated region; mESCs, mouse embryonic stem cells; PCR, polymerase chain reaction; RT‐PCR, reverse transcription polymerase chain reaction.
mPinX1t Transcript Contained Intact 3′ Poly(A) Tail and 5′ Cap
Aforementioned PCR was conducted using cDNA reverse‐transcribed from whole‐cell RNA using oligo‐dT as primers. Therefore, those cDNAs should represent reverse‐transcribed product from RNA with poly(A) tail. To further confirm that the mPinX1t transcript was a functional mRNA (ie, with 5′ cap and 3′ poly(A) tail), RNA ligase‐mediated rapid amplification (RLM) 5′ rapid amplification of cDNA ends assay was carried out using total RNA from undifferentiated mESC lysate. Calf intestine alkaline phosphatase was used to selectively eliminate all 5′ truncated RNAs and non‐mRNAs that lack an intact 5′ cap. The RNA was then treated with tobacco acid pyrophosphatase to remove the cap structure from full‐length mRNAs. RNA oligo of known sequence was ligated to the 5′ end of the mRNA population. The resultant mRNA was reverse transcribed into cDNA by oligo‐dT primer and amplified using specific 5′ primer targeting the RNA oligo sequence and 3′ primer targeting mPinX1t‐specific sequence. A product between 500 and 750 bp was obtained after PCR (Figure 1C). The band was excised and subcloned into TOPO T‐vector. After restriction digestion, 3 different clones of the recombinant plasmids, representing 3 different 5′ UTR sequences of mPinX1t, were found, as revealed by 3 slightly different digestion patterns (eg, clone 2, clone 3, and clone 6 were 3 different clones; Figure S1). The 3 different 5′ UTRs were further analyzed by DNA sequencing and were found to be of different sequences, yet correspond to the known 5′ UTR of mPinX1 (data not shown).
In addition, RLM 3′ rapid amplification of 5′ cDNA ends assay was carried out using total RNA from undifferentiated mESC lysate. Our results indicated that mPinX1t transcript contains 3′ poly(A) tail; subsequent sequencing of the 3′ UTR confirmed the existence of an extra exon and the connection of this extra exon to the exon 7 of mPinX1. Altogether, our results indicated that mPinX1t exists as an intact mRNA with 5′ cap and 3′ poly(A) tail.
Sequence analysis revealed that the additional exon in mPinX1t contained a stop codon, making mPinX1t encodes for a protein containing only the N‐terminal but not the C‐terminal of mPinX1 (Figure 1D). Therefore, we named this novel transcript variant mPinX1t where “t” stands for “truncated.”
mPinX1t mRNA Was Actively Translating Into Protein
High‐sucrose density gradient polysome fractionation assay was performed and was found to successfully fractionate different RNAs into free ribonucleoprotein fractions, 40S and 60S (which contain 18S rRNA and 28S rRNA, respectively) fractions, monosome fractions, and polysome fractions (Figure 2A). Different RNA fractions (fractions 1, 2, 5, 6, 10, 11, 16, and 17; Figure 2A) were selected, reverse transcribed into cDNAs and amplified using mPinX1t‐specific primers by PCR reaction. Both the monosome and polysome fractions, but not the free ribonucleoprotein fractions, gave rise to mPinX1t‐specific band at 200 bp. These results suggested that mPinX1t mRNA was incorporated into ribosomes and actively translated into proteins (Figure 2B).
Figure 2.
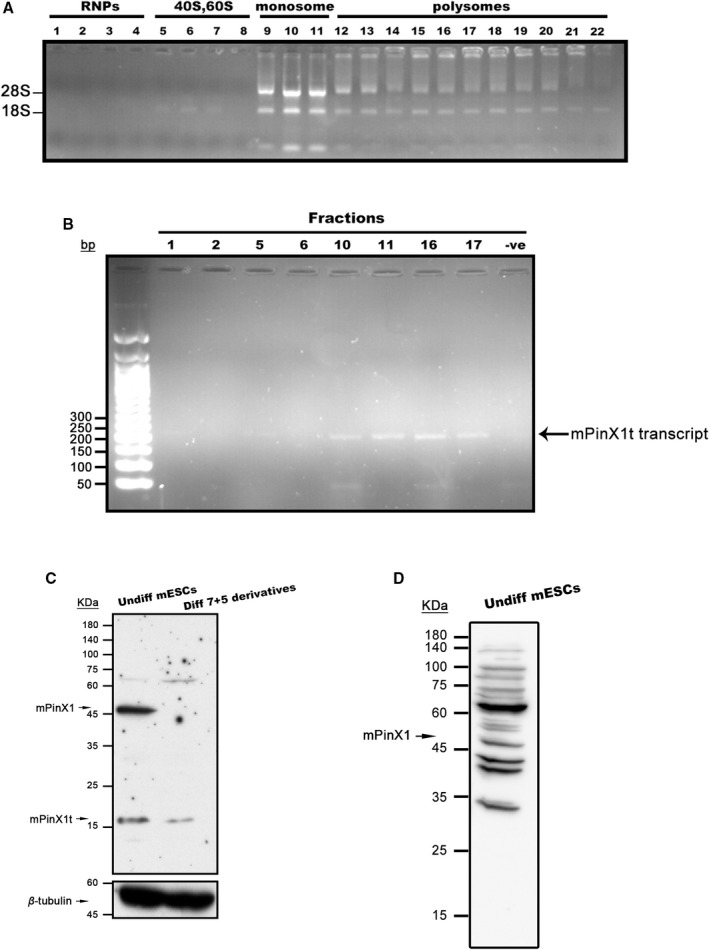
mPinX1t is a protein encoding gene. A and B, High‐density sucrose gradient polysome fractionation was performed using mESC lysate. A, Nondenaturing RNA agarose gel showing the presence and the integrity of 28S and 18S rRNAs in different fractions. Fractions 1 to 4 represent free ribonucleoprotein fractions; fractions 5 to 8 represent 40S, 60S fractions; fractions 9 to 11 represent monosome fractions; fractions 12 to 22 represent polysome fractions. B, PCR reactions of different fractions showing the presence of mPinX1t transcripts in monosome and polysome fractions. Equal volume of PCR product was loaded onto each lane. “–ve” represents negative control. C, Representative western blot showing the expressions of mPinX1 and mPinX1t proteins in undifferentiated mESCs and mESC differentiation derivatives at day 7+5. β‐Tubulin was used as the loading control. mPinX1 was observed at around 45 kDa and mPinX1t was observed between 15 and 25 kDa. D, Representative western blot showing the expression of mPinX1 proteins in undifferentiated mESCs using an antibody targeting the C‐terminal of mPinX1 (Orb47163). mPinX1 was observed at around 45 kDa; mPinX1t, which lacks the C‐terminal of mPinX1, was not detected. mESC indicates mouse embryonic stem cell; RNPs, ribonucleoproteins; PCR, polymerase chain reaction.
To validate the expression of mPinX1t at protein level, western blot analysis was performed using an anti‐PinX1, which has an epitope at the N‐terminal of PinX1. mPinX1 was detected at its expected size at ≈45 kDa, while mPinX1t was detected between 15 and 25 kDa (Figure 2C). On the other hand, using an antibody having an epitope at the C‐terminal of PinX1, mPinX1 but not mPinX1t (which lacks the C‐terminal of mPinX1) was detected (Figure 2D).
Establishment of Stable mESC Lines With Altered Expression of mPinX1 and mPinX1t
The expressions of mRNAs of mPinX1 and mPinX1t for mESCs at the undifferentiated stage and mESCs during the early stage of differentiation were plotted (Figure S2). Our results showed that the expression of mPinX1 and mPinX1t increased at the beginning of differentiation and peaked at around differentiation day 3 to 4, a time point where cardiac differentiation starts.26 The expression of mPinX1 and mPinX1t started to fall after the peak. These data hinted at the potential role of mPinX1 and mPinX1t and their interaction in regulating cardiogenesis. To study the effects of mPinX1 and mPinX1t on the cardiac commitment of mESCs, cell lines that were stably overexpressed or knocked down with mPinX1 or mPinX1t were established (Figure S3). While an attempt was made to knock down mPinX1t, we found that the unique 111 nucleotides in the mPinX1t transcript was too short to allow us to design effective small hairpin RNA (note that as revealed by RLM rapid amplification of 3′ cDNA ends, for mPinX1t transcript, exon 7 sequence of mPinX1 immediately followed the unique 111 nucleotide exon of mPinX1t. Therefore, the 111 nucleotides were the only unique sequence of mPinX1t that distinguished itself from mPinX1). As such, there was no specific small hairpin RNA to effectively knock down mPinX1t.
Altered Expression of mPinX1 and/or mPinX1t Did Not Affect the Characteristics of mESCs at the Undifferentiated State
First, whether altering the expression of mPinX1 and/or mPinX1t would affect the characteristics of mESCs at the undifferentiated state was examined. To study the long‐term effect on cell proliferation, different stable cell line clones were kept on being passaged for 38 to 39 passages starting from the day of their establishment. The number of cells was counted by trypan blue exclusion assays every time when the cells were passaged. Both overexpression of mPinX1 or mPinX1t or the knockdown of mPinX1 did not cause a significant change in cell proliferation in the long run as determined by trypan blue exclusion assays (Figure S4A and S4B). MTT assay also revealed that there was no significant difference in the proliferation of these stable overexpression/knockdown cell line clones when compared with their respective control cell lines (Figure S4C and S4D). In addition, the telomere repeat amplification protocol assay was done to examine the telomerase activity of the stable cell line clones. Our results showed that there was no significant change of the telomerase activity in these cell lines (Figure S4E and S4F).
Western blot was also done to examine the expression of pluripotent markers in these stable cell line clones. The expression of pluripotent markers, including octamer‐binding transcription factor 4 and SRY‐box transcription factor 2, were not consistently altered among these clones (Figure S4G and S4H).
Altogether, altering the expression of mPinX1 and/or mPinX1t did not affect the proliferation, telomerase activity, and pluripotency of mESCs.
Overexpression and Knockdown of mPinX1 Inhibited Cardiac Differentiation While Overexpression of mPinX1t Promoted Cardiac Differentiation
Upon differentiation, the overexpression and/or the knockdown of mPinX1/mPinX1t of different stable cell line persisted as expected (Figure S5). qPCR analysis showed that both overexpression and knockdown of mPinX1 decreased the expressions of cardiac structural markers including cardiac actin, cardiac troponin I, cardiac troponin T, and myosin heavy chain (Figure 3A and 3B). In contrast, overexpression of mPinX1t increased the expressions of these cardiac structural markers (Figure 3A).
Figure 3.
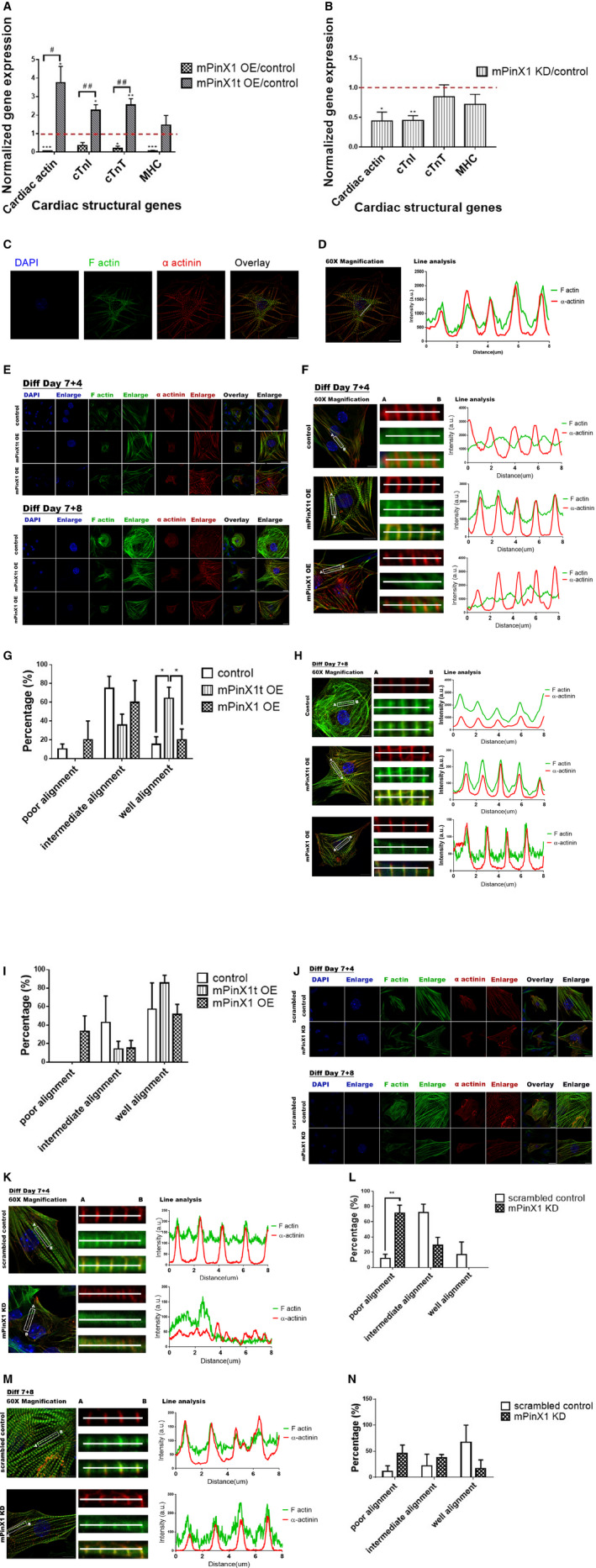
Effects of the overexpression and knockdown of mPinX1/mPinX1t on cardiac differentiation and sarcomere assembly. A, Bar chart showing the change in expressions of cardiac actin, cTnI, cTnT), and myosin heavy chain (MHC) of cells on differentiation day 7+25 in mPinX1 and mPinX1t overexpression lines normalized to that of control. Dotted line indicates the expression level of the control line. Overexpression of mPinX1 (mPinX1 OE) decreased while overexpression of mPinX1t (mPinX1t OE) increased the expressions of cardiac structural genes. Data were presented as mean±SEM (mPinX1 OE group, n=3; mPinX1t OE group, n=5; control group, n=5; where n represents data from each independent differentiations). *P<0.05, ***P<0.001 vs control line. # P<0.05, ## P<0.01 vs mPinX1 overexpression line. B, Bar chart showing the change in expressions of cardiac actin, cTnI, cTnT, and MHC of cells on differentiated day 7+25 in mPinX1 knockdown line normalized to that of control. Dotted line indicates the expression level of the control line. Knockdown of mPinX1 (mPinX1 KD) decreased the expression of cardiac structural genes. Data were presented as mean±SEM (n=3; where n represents data from each independent differentiations). *P<0.05, **P<0.01 vs control line. C, Immunostaining of α‐actinin and F‐actin of differentiation day 7+12 cardiomyocyte reveals proper sarcomere formation when cardiomyocytes are mature. Scale bar: 10 μm. D, Line analysis showing α‐actinin and F‐actin patterns of differentiation day 7+12 cardiomyocyte in (C). Mature cardiomyocytes have overlapping pattern of α‐actinin and F‐actin. Scale bar: 10 μm. E, Representative immunostaining results of α‐actinin and F‐actin of cardiomyocytes at early differentiation time point (day 7+4) and intermediate differentiation time point (day 7+8) in control line, mPinX1t overexpression line and mPinX1 overexpression line. Scale bar: 20 μm for photos of overlay views and 10 μm for photos with enlarged views. F, Line analysis of cardiomyocytes at early differentiation time point (day 7+4) in control line, mPinX1t overexpression line and mPinX1 overexpression line shown in (E). Scale bar: 10 μm. G, Bar graph summarizing the effects of mPinX1 and mPinX1t overexpression on sarcomere assembly at early differentiation time point (day 7+4). mPinX1t overexpression cell line showed a higher percentage of cardiomyocytes with a well‐aligned α‐actinin and F‐actin pattern, while it did not have cardiomyocytes with poor alignment. Data were presented as mean±SEM (15 cardiomyocytes from mPinX1 OE group, 19 cardiomyocytes from mPinX1t OE group, and 19 cardiomyocytes from control group were analyzed. Cells were obtained from 3 independent differentiations. The unit of statistical analysis was the number of independent differentiations). *P<0.05. H, Line analysis of cardiomyocytes at intermediate differentiation time point (day 7+8) in control line, mPinX1t overexpression line and mPinX1 overexpression line shown in (E). Scale bar: 10 μm. I, Bar graph summarizing the effects of mPinX1 and mPinX1t overexpression on sarcomere assembly at intermediate differentiation time point (day 7+8). Control, mPinX1 overexpression and mPinX1t overexpression cell lines all had >50% cardiomyocytes showing a well‐aligned α‐actinin and F‐actin pattern. The mPinX1t overexpression cell line had a trend of having higher percentage of cardiomyocytes with a well‐aligned α‐actinin and F‐actin pattern. The mPinX1 overexpression cell line had cardiomyocytes with poorly aligned α‐actinin and F‐actin pattern, while mPinX1t overexpression cell line or the control line did not have cardiomyocyte with poor alignment. Data were presented as mean±SEM (19 cardiomyocytes from mPinX1 OE group, 20 cardiomyocytes from mPinX1t OE group, and 19 cardiomyocytes from control group were analyzed. Cell were obtained from 3 independent differentiations. The unit of statistical analysis was the number of independent differentiations.). J, Representative immunostaining results of α‐actinin and F‐actin of cardiomyocytes at early differentiation time point (day 7+4) and intermediate differentiation time point (day 7+8) in scrambled control line and mPinX1 knockdown line. Scale bar: 20 μm for photos of overlay views and 10 μm for photos with enlarged views. K, Line analysis of cardiomyocytes at early differentiation time point (day 7+4) in scrambled control line and mPinX1 knockdown line shown in (J). Scale bar: 10 μm. L, Bar graph summarizing the effects of mPinX1 knockdown on sarcomere assembly at early differentiation time point (day 7+4). mPinX1 knockdown cell line showed a higher percentage of cardiomyocytes with poorly aligned α‐actinin and F‐actin pattern, while it did not have cardiomyocytes with good alignment. Data were presented as mean±SEM (17 cardiomyocytes from the mPinX1 KD group and 18 cardiomyocytes from the scrambled control group were analyzed. Cells were obtained from 3 independent differentiations. The unit of statistical analysis was the number of independent differentiations.). **P<0.01. M, Line analysis of cardiomyocytes at intermediate differentiation time point (day 7+8) in scrambled control line and mPinX1 knockdown line shown in (J). Scale bar: 10 μm. N, Bar graph summarizing the effects of mPinX1 knockdown on sarcomere assembly at intermediate differentiation time point (day 7+8). mPinX1 knockdown cell line had a trend of having higher percentage of cardiomyocytes with poorly aligned α‐actinin and F‐actin pattern and had a trend of having lower percentage of cardiomyocytes with a well‐aligned α‐actinin and F‐actin pattern. Data were presented as mean±SEM (17 cardiomyocytes from the mPinX1 KD group and 15 cardiomyocytes from the scrambled control group were analyzed. Cells were obtained from 3 independent differentiations. The unit of statistical analysis was the number of independent differentiations.) cTnI indicates cardiac troponin I; cTnT, cardiac troponin T; DAPI 4′,6‐diamidino‐2‐phenylindole.
Sarcomere assembly allows actin and myosin filaments to align in proper orientations for force contraction of cardiomyocytes and set the basis for heart contraction. α‐Actinin is one of the components of Z‐disc in sarcomere. α‐Actinin crosslinks with F‐actin to maintain the integrity of the Z‐disc structure.27 To study the role of mPinX1 and mPinX1t on the sarcomere assembly of cardiomyocytes, immunocytochemical staining on 2 sarcomere assembly‐related markers α‐actinin and F‐actin was done at different differentiation time points (differentiation day [7+4] and differentiation day [7+8]) among all the overexpression and knockdown cell lines of mPinX1 and mPinX1t. Proper alignment of α‐actinin and F‐actin in terminally differentiated cardiomyocytes would appear as a clear striated pattern of α‐actinin and F‐actin in which α‐actinin signal would lie within the peak of F‐actin signal when a line scan is performed (Figure 3C and 3D). For most cardiomyocytes in mPinX1 overexpression group, α‐actinin and F‐actin were not well aligned at the early differentiation time point (Figure 3E through 3G). On the other hand, a higher percentage of cardiomyocytes in the mPinX1t overexpression group had a well‐aligned α‐actinin and F‐actin pattern, and no cardiomyocyte in this group had a poorly aligned pattern (Figure 3G). The trend of difference among mPinX1 overexpression, mPinX1t overexpression, and control lines persisted at the immediate differentiation time point but was less apparent (Figure 3H and 3I).
A similar phenotype as that in the mPinX1 overexpression group could be observed in the mPinX1 knockdown cell line at the early differentiation time point (Figure 3J through 3L). Knockdown of mPinX1 led to an irregular α‐actinin and F‐actin pattern. A higher percentage of cardiomyocytes in the mPinX1 knockdown group had a poorly aligned α‐actinin and F‐actin pattern, and no cardiomyocyte in this group had a well‐aligned pattern (Figure 3L). The trend of difference between the mPinX1 knockdown line and control line persisted at the immediate differentiation time point but was less apparent (Figure 3M and 3N).
To further confirm the difference in the cardiomyocytes when mPinX1 and mPinX1t were differentially expressed, imaging on calcium transients (CaTs) was performed. When mPinX1t was overexpressed, the amplitude of the CaTs increased while the time to peak of CaTs decreased at both the early and late differentiation stages when compared with control (Figure S6). In addition, mPinX1t overexpression led to an increase in CaT frequency and basal calcium level in late differentiation stage. On the other hand, when mPinX1 was overexpressed, the basal calcium level decreased in early differentiation stage, while the time to peak of CaTs increased in late differentiation stage (Figure S6). In summary, CaT data support the notion that mPinX1t overexpression positively regulates cardiac differentiation while mPinX1 overexpression negatively regulates cardiac differentiation.
Taken together, overexpression and knockdown of mPinX1 decreased cardiac differentiation and led to a delay in proper sarcomere formation, while overexpression of mPinX1t increased cardiac differentiation and led to an earlier sarcomere formation.
mPinX1 Protein and mPinX1t Protein Bound to the mRNAs of Cardiac Transcription Factors Gata4 and Tbx5 While Only mPinX1t Protein but Not mPinX1 Protein Bound to the Nucleoporin 133
Sequence analysis revealed that both mPinX1 and mPinX1t contain the putative RNA‐binding domain G‐Patch at the N‐terminal. We hypothesized that mPinX1 and mPinX1t regulate cardiac differentiation through binding to the mRNAs of key cardiac transcription factors. By RNA immunoprecipitation assay, both mPinX1 and mPinX1t were found to bind to the mRNAs of the core cardiac transcription factors Gata4 and Tbx5 (Figure 4A through 4D). In HEK cells coexpressed with myc‐mPinX1 and mRNAs of cardiac transcription factors, when compared with the isotype control group, the pulldown fraction by anti‐myc was found to contain 7.47±2.02‐fold of Gata4 mRNA and 4.91±0.96‐fold of Tbx5 mRNA (Figure 4A and 4C). In HEK cells coexpressed with myc‐mPinX1t and mRNAs of cardiac transcription factors, when compared with the isotype control group, the pulldown fraction by anti‐myc was found to contain 2.80±0.70‐fold of Gata4 mRNA and 3.86±1.07‐fold of Tbx5 mRNA (Figure 4B and 4D). On the other hand, myc‐mPinX1t and myc‐mPinX1 were not found to bind to Rpl13A mRNA (Figure S7).
Figure 4.
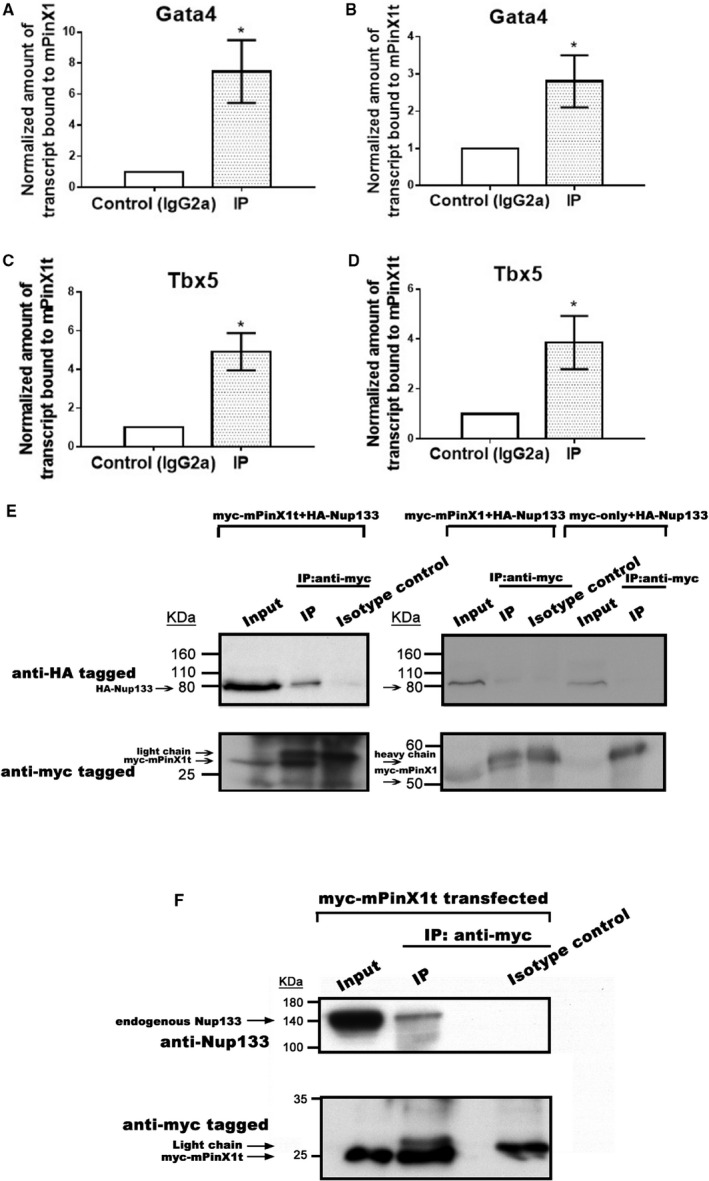
Both mPinX1 and mPinX1t proteins bound cardiac transcription factor mRNAs, while only mPinX1t protein but not mPinX1 protein‐bound Nucleoporin 133 (Nup133). A through D, RNA immunoprecipitation of HEK293FT cells overexpressed with (A and C) myc‐mPinX1 or (B and D) myc‐mPinX1t and (A and B) Gata4 or (C and D) Tbx5. Anti‐myc was used to perform the immunoprecipitation. IgG2a was used in isotype control experiment. The presence of (A and B) Gata4 mRNA or (C and D) Tbx5 mRNA in the immunoprecipitant was quantitated by subsequent quantitative polymerase chain reaction. The results showed that both mPinX1 and mPinX1t proteins could bind to Gata4 mRNA and Tbx5 mRNA. Data were presented as mean±SEM (n=3; where n represents independent RNA immunoprecipitation experiments). *P<0.05 vs control. E, Coimmunoprecipitation assay of HEK293FT cells overexpressed with myc‐mPinX1t (left panel) or myc‐mPinX1 and HA‐Nup133 (right panel). (Left panel) In the immunoprecipitant of anti‐myc (which contained myc‐mPinX1t), HA‐Nup133 was detected. No HA‐Nup133 was detected in the isotype control group. (Right panel) In the immunoprecipitant of anti‐myc (which contained myc‐mPinX1), HA‐Nup133 was not enhanced when compared with isotype control group. Another control experiment was also performed in which myc only and HA‐Nup133 were overexpressed in HE293FT cells. Using anti‐myc for immunoprecipitation, as expected, no HA‐Nup133 was detected in the immunoprecipitant. F, Coimmunoprecipitation assay of HEK293FT cells overexpressed with myc‐mPinX1t. As shown in the input lane, HEK293FT expressed endogenous Nup133. In the immunoprecipitant of anti‐myc (which contained myc‐mPinX1t), endogenous Nup133 was detected. No Nup133 was detected in the isotype control group. DAPI indicates 4′,6‐diamidino‐2‐phenylindole.
A yeast‐2 hybrid assay was carried out to screen out the mPinX1t‐interacting partners in mESCs. More than 100 interacting partners were identified and classified according to Gene Ontology terms classification (Figure S8). One of the interacting proteins was known as Nucleoporin 133 (Nup133). Nup133 is one of the components of nuclear pore complex, which is responsible for transport of macromolecules across the nuclear envelope from nucleus to cytoplasm.28, 29 We hypothesized that mPinX1t promotes cardiac differentiation through facilitating the transport of Gata4 and Tbx5 mRNAs from the nucleus to the cytoplasm for translation. To further validate the interaction between mPinX1t and Nup133, a coimmunoprecipitation assay was performed using HEK293FT cells coexpressed with myc‐mPinX1t and HA‐Nup133. Our result showed that myc‐mPinX1t could successfully pull down HA‐Nup133 (Figure 4E). Similar coimmunoprecipitation assay was performed in myc‐mPinX1 and HA‐Nup133 overexpressed HEK293FT. The result showed that myc‐mPinX1 failed to pull down HA‐Nup133 (Figure 4E).
To test if mPinX1t would interact with endogenous Nup133, a further coimmunoprecipitation assay was performed. In HEK293FT overexpressed with myc‐mPinX1t, endogenous Nup133 was present in the pulldown fraction by anti‐myc but not in its isotype control (Figure 4F). The result indicated that mPinX1t is able to interact with endogenous Nup133.
mPinX1t Increased the Ratio of Cytosolic Gata4 mRNAs to Nuclear Gata4 mRNAs; Knockdown of Nup133 Abolished the Effect
To investigate whether mPinX1t facilitates the export of cardiac transcription factor mRNAs from the nucleus to the cytoplasm, Gata4 and mPinX1/mPinX1t were coexpressed in HEK293FT, followed by fractionation of cytosolic and nuclear mRNAs. Reverse transcriptase qPCR on the Gata4 mRNAs was then performed using the cytosolic and nuclear fractions. In the mPinX1t and Gata4 cotransfected group, a higher ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs was detected when compared with that in the control group (empty vector and Gata4 cotransfected group) (Figure 5A). On the other hand, the mPinX1 and Gata4 cotransfected group tended to have a lower ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs (Figure 5A). Since interaction between mPinX1t and Nup133 was observed, to determine the importance of this interaction in the export of mRNA, the same experiment as above but with knockdown of Nup133 by small interfering RNA was performed. Interestingly, knockdown of Nup133 abolished the differences between different groups in term of the ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs (Figure 5B), suggesting the importance of interaction of mPinX1t and Nup133 in helping with the export of Gata4 mRNAs.
Figure 5.
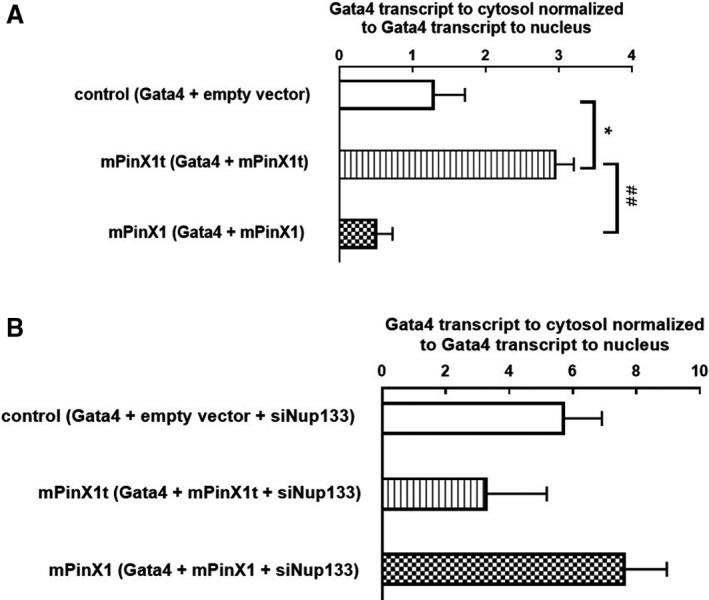
mPinX1t increased the ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs; knockdown of Nup133 abolished the effect. A, Gata4 and empty vector/mPinX1/mPinX1t were overexpressed in HEK293FT, followed by fractionation of cytosolic and nuclear mRNAs. RT‐qPCR on the Gata4 mRNAs was then performed using the cytosolic and the nuclear fractions. The amount of Gata4 transcripts in cytosol was normalized to that in nucleus. Our results showed that, in mPinX1t and Gata4 co‐transfected group, a higher ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs was detected when compared with that in control group (empty vector and Gata4 co‐transfected group). On the other hand, mPinX1 and Gata4 co‐transfected group tended to have a lower ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs. Data were presented as mean±SEM (n=3; where n represents independent experiments). *P<0.05 vs control group. ## P<0.01 vs mPinX1 group. B, Gata4, empty vector/mPinX1/mPinX1t and siRNA against Nup133 (siNup133) were overexpressed in HEK293FT, followed by fractionation of cytosolic and nuclear mRNAs. RT‐qPCR on the Gata4 mRNAs was then performed using the cytosolic and the nuclear fractions. The amount of Gata4 transcripts in cytosol was normalized to that in nucleus. Our results showed that there was no statistical difference in the ratio of cytosolic Gata4 mRNAs to nuclear Gata4 mRNAs between different groups. Data were presented as mean±SEM (n=3; where n represents independent experiments).
Discussion
The Discovery of a Novel Transcript Variant of mPinX1, mPinX1t
A new transcript variant of mPinX1, the mPinX1t, was detected in mESCs and their differentiated derivatives in a PCR reaction trying to detect the endogenous mPinX1 expression. Sequencing results revealed that this new transcript variant harbors an additional exon of 111 nucleotides located between exon 6 and exon 7 of mPinX1. This additional exon was found to be located in Locus NT_039606.7 of chromosome 14 of the mouse genome where the mPinX1 gene is located. Intriguingly, there is a stop codon within this extra 111 nucleotide sequence; rendering this new transcript encodes a protein that has the N‐terminal but lacks the C‐terminal of mPinX1. There is also an extra sequence that exists only in mPinX1t but not mPinX1 (see Figure 1D). RLM rapid amplification of 5′ cDNA ends followed by subsequent sequencing of the 5′ UTR revealed that there are at least 3 different sequences in the 5′ UTR of the mRNA of mPinX1t. These 3 different 5′ UTR sequences fall within the same locus of mPinX1 gene and completely aligned with the known 5′ UTR sequence of mPinX1. The result provided further evidence that mPinX1t is a transcript variant of mPinX1.
In fact, a search in the National Center for Biotechnology Information database indicated that human tissues also express a transcript variant similar to the transcript of mPinX1t; that is, this human PinX1 transcript variant is also predicted to encode a protein that lacks the C‐terminal of human PinX1. The result indicated that both mouse and human express at least 2 isoforms of PinX1 protein, and that the N‐terminal of PinX1 may have a unique function independent of the function of the C‐terminal of PinX1.
PinX1 is a well‐known tumor suppressor protein that works by negatively regulating the telomerase activity at its C‐terminal telomerase inhibitory domain.16 Many recent publications focused on how PinX1 regulates tumor progression and metastasis,18, 30, 31 while little has been done on the functions of the N‐terminal of mammalian PinX1. The newly discovered transcript variant mPinX1t provides a good opportunity for us to investigate the role and the function of the N‐terminal of mPinX1 in mESCs and their differentiated derivatives.
mPinX1t Encodes for a Functional Intact mRNA That Is Actively Translated Into Protein
Our RLM rapid amplification of 5′ cDNA ends assay and RLM rapid amplification of 3′ cDNA ends assay confirmed that mPinX1t encodes for a functional mRNA with intact 5′ cap and poly(A) tail. Consistently, polysome fractionation followed by PCR showed that mPinX1t mRNA was present in the monosome and polysome fractions, clearly suggesting that mPinX1t transcript encodes for an actively translating mRNA. Using an antibody having an epitope at the N‐terminal of PinX1 also revealed the existence of mPinX1 and mPinX1t proteins in both undifferentiated mESCs and differentiated derivatives. To our knowledge, our study is the first study to document the existence of a truncated isoform of PinX1 protein.
mPinX1 and mPinX1t Affect mESC Differentiation Into Cardiac Lineage
Various mESC lines with stable overexpression or knockdown of mPinX1 and/or mPinX1t were established to study the effect of mPinX1 and mPinX1t in determining the self‐renewal and pluripotent characteristics of mESCs and in regulating the cardiac differentiation of mESCs. Altering the expression level of neither mPinX1 nor mPinX1t could affect the proliferation, telomerase activity, and the pluripotent characteristics of mESCs.
ESCs have high levels of telomerase expression and activity, both of which are rapidly downregulated upon differentiation.32, 33 This high telomerase activity in ESCs is involved in maintaining the indefinite proliferation ability of ESCs.32 While PinX1 was first identified as a tumor suppressor protein and overexpression of it inhibited telomerase activity and shortened telomere,16 later studies also reported that PinX1 was associated with telomeres primarily at mitosis and that knockdown of PinX1 caused delayed mitotic entry.34 Therefore, knockdown of PinX1 may not necessarily lead to a change in proliferation at the cellular level because of its dual functions. In addition, many pathways are known to regulate telomerase activity in mESCs35, 36 and mPinX1 might not be the rate‐limiting physiological regulator of telomerase at the undifferentiated stage; therefore, its effect on telomerase activity and/or telomere length would be minor. Moreover, previous studies have shown that murine telomerase reverse transcriptase and RNA subunit of telomerase are not immediately essential for normal development and cell viability but is required for telomere‐length maintenance in vivo.37, 38 However, interestingly, mESCs can be obtained from telomeric reverse transcriptase knockout mice, and these cells can be maintained in culture.39 In addition, mice are known to have unusually long telomeres, and this may not be generalized to other species. This suggests that even if there is a change in telomerase activity/telomere length, mESCs may not be affected for many passages. All these may help explain why an alteration in mPinX1 expression level may not affect mESCs at undifferentiated state.
On the other hand, upon differentiation of mESCs, the effect of mPinX1 and mPinX1t could be observed. Stable overexpression and knockdown of mPinX1, which were also found to decrease mPinX1t expression, decreased cardiac differentiation has revealed the expression of cardiac structural markers and the sarcomere assembly. In contrast, stable overexpression of mPinX1t was found to promote cardiac differentiation. The result suggested that mPinX1t might be a potential positive regulator for cardiac differentiation. Previous studies reported that switching expression level of splice variants of some genes could be associated with stem cell differentiation.40, 41 In our study, the expression of mPinX1 was found to decrease to a low level during mESC differentiation, while the expression of mPinX1t was not found to decrease during mESC differentiation. This switch in the relative expression level of mPinX1 and mPinX1t is in line with the positive role of mPinX1t in the differentiation process of mESCs.
Both mPinX1 and mPinX1t Could Bind to mRNAs of Cardiac Transcription Factors While Only mPinX1t but Not mPinX1 Could Bind to Nup133
The amino acid sequence of mPinX1 and mPinX1t suggest that they both contain a G‐patch domain at their N‐terminal. Many previous reports showed that proteins containing a G‐patch domain are associated with RNA‐binding proteins or are able to interact with RNA themselves.17 We speculated that mPinX1 and mPinX1t proteins may regulate cardiac differentiation by interacting with the mRNAs of cardiac transcription factors. In fact, our RNA immunoprecipitation assay results indicated that both mPinX1 and mPinX1t proteins could interact with the mRNAs of cardiac transcription factors Gata4 and Tbx5.
A previous study has shown that human PinX1 could bind to the RNA‐binding domain of the human telomeric reverse transcriptase and the RNA subunit of human telomerase in vitro.42 In vivo study revealed that human PinX1 binds to the assembled human telomeric reverse transcriptase –human telomerase complex.41 This study suggested that mammalian PinX1 is capable of binding to RNA. Interestingly, the yeast homolog of mammalian PinX1, which bears the G‐patch, was found to be involved in preribosomal RNA processing and in the final 3′‐end trimming of U18 and U24 small nucleolar RNAs.43 A mutational analysis showed that the G‐patch is essential for these functions. Importantly, human PinX1 can complement the mutation of the G‐patch in yeast PinX1, suggesting that human PinX1 has a dual function in telomere length regulation and ribosomal RNA maturation. Our current study revealed a novel function of the mPinX1 and mPinX1t, which bear the G‐patch: They can bind to the mRNAs of cardiac transcription factors.
In our yeast‐2 hybrid assay, mPinX1t protein was found to interact with Nup133, a subunit of a protein complex that forms the nuclear pore at the nuclear membrane.28, 29 Our further coimmunoprecipitation assay validated the interaction between mPinX1t and Nup133. Interestingly, this interaction was not found between mPinX1 and Nup133.
mPinX1t Positively Regulates Cardiac Differentiation Possibly by Facilitating the Export of Cardiac Transcription Factor mRNAs From the Nucleus to the Cytosol
From the result of the RNA fractionation assay on Gata4 and mPinX1/mPinX1t cotransfected HEK293FT cells, cytosolic Gata4 mRNA to nuclear Gata4 mRNA ratio was found to be higher in mPinX1t‐containing cells. Interestingly, when Nup133 was knocked down, the difference between the effect of mPinX1t and mPinX1 in affecting the Gata4 mRNA transport was abolished. Previous study showed that yeast Nup133 assisted in mRNA export, as revealed by accumulation of poly(A)‐containing RNA in nucleus in yeast Nup133 negative cells.44 While previous studies showed the importance of mammalian Nup133 in the assembly of the nuclear pore complex and in anchoring the nuclear pore complex to the nuclear envelope,28, 29 the role of mammalian Nup133 in mediating the mRNA transport from nucleus to cytosol is unclear. Since we observed an increase in Gata4 mRNA transport from nucleus to cytosol when Nup133 was knocked down, we speculate that mammalian Nup133 negatively regulates mRNA export in this cellular context. Combined with our molecular data showing the interaction between mPinX1t and Nup133, we speculate that mPinX1t binds to Nup133 and relieves Nup133's inhibitory effect on mRNA transport; therefore, when mPinX1t is overexpressed, Gata4 mRNA transport from nucleus to cytosol increases. The cardiac transcription factor mRNAs in the cytoplasm can then be later translated to facilitate cardiac differentiation. However, when Nup133 is knocked down, the inhibitory effect of Nup133 no longer exists, and therefore the differences between mPinX1t‐overexpressed cells and mPinX1‐overexpressed cells is abolished.
Proposed Model on How mPinX1 and mPinX1t Regulated Cardiogenesis
In this study, mPinX1t was found to promote cardiac differentiation in early differentiation. A model is proposed to explain the observed phenotypes on how mPinX1t regulates cardiogenesis (Figure 6).
Figure 6.
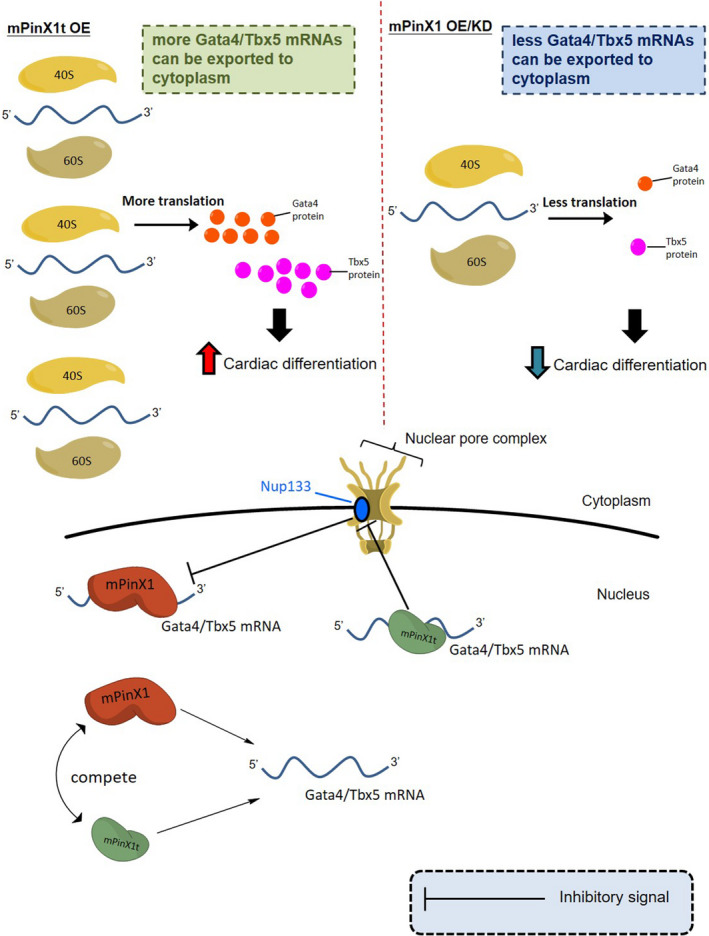
Schematic diagram showing a model of how mPinX1t regulates cardiac differentiation. mPinX1 and mPinX1t contain the RNA‐binding domain G‐Patch. This domain allows mPinX1 and mPinX1t to compete with each other for binding to the mRNAs of the cardiac transcription factors Gata4 and Tbx5. Only mPinX1t would interact with Nucleoporin 133 (Nup133), a component of the nuclear pore complex. We speculate that mammalian Nup133 negatively mediates the transport of mRNAs from nucleus to cytoplasm; mPinX1t interacts with Nup133 and relieves its inhibitory effect on the transport of Gata4 and Tbx5 mRNAs from nucleus to cytoplasm. Therefore, in the presence of mPinX1t, more Gata4 and Tbx5 mRNAs can be exported to the cytoplasm for later translation. This would result in larger amount of cardiac transcription factor proteins produced for cardiac differentiation. In the upper part of the figure, what happens in the cytoplasm under different scenarios are indicated. When mPinX1t is overexpressed, mPinX1t protein would bind to the mRNAs of Gata4 and Tbx5; these mPinX1t proteins would also interact with Nup133 to relieve its inhibitor effect on the transport the mRNAs, so more mRNAs are exported to the cytoplasm for translation. The translated Gata4 and Tbx5 proteins would then function as cardiac transcription factors for cardiac differentiation. When mPinX1 is overexpressed, it competes with mPinX1t and binds to Gata4 and Tbx5 mRNAs. Therefore, less mPinX1t protein binds to Gata4 and Tbx5 mRNAs. On the other hand, when mPinX1 is knocked down, mPinX1t is also knocked down; therefore, there is less mPinX1t protein to interact with Nup133. In both cases, mRNA transport inhibitor effect of Nup133 still exists; export of mRNAs of Gata4 and Tbx5 to the cytoplasm would decrease, leading to a decrease in cardiac differentiation.
From the result of RNA immunoprecipitation assay, both mPinX1 and mPinX1t were found to interact with the mRNAs of the key cardiac transcription factors Gata4 and Tbx5. In the yeast‐2 hybrid assay, mPinX1t but not mPinX1 was found to interact with Nup133. We speculate that mammalian Nup133 negatively mediates the transport of mRNAs from nucleus to cytoplasm; mPinX1t interacts with Nup133 and relieves its inhibitory effect on the transport of Gata4 and Tbx5 mRNAs from nucleus to cytoplasm for later translation.
Because of the high similarity between the N‐terminal of mPinX1 protein and mPinX1t protein, it is believed that mPinX1 and mPinX1t proteins might act as competitors to each other for binding to the mRNAs of Gata4 and Tbx5. When mPinX1 is overexpressed, mPinX1 competes with mPinX1t and binds to Gata4 and Tbx5 mRNAs. Therefore, less mPinX1t protein binds to Gata4 and Tbx5 mRNAs. On the other hand, when mPinX1 is knocked down, mPinX1t is also knocked down; therefore, there is less mPinX1t protein to interact with Nup133. In both cases, the mRNA transport inhibitor effect of Nup133 still exists; export of mRNAs of Gata4 and Tbx5 to the cytoplasm would decrease, leading to a decrease in cardiac differentiation. In contrast, when mPinX1t is overexpressed, mPinX1t protein binds to the mRNAs of Gata4 and Tbx5; these mPinX1t proteins also interact with Nup133 to relieve its inhibitor effect on the transport the mRNAs, so more mRNAs are exported to the cytoplasm for translation. The translated Gata4 and Tbx5 proteins would then function as cardiac transcription factors for cardiac differentiation.
In conclusion, we discovered a novel transcript variant of mPinX1, the mPinX1t, to be present in both the undifferentiated and differentiated mESCs. This mPinX1t exists as an intact mRNA and protein. Importantly, mPinX1t positively regulates cardiac differentiation of mESCs. While both mPinX1 protein and mPinX1t protein interact with the mRNAs of cardiac transcription factors, only mPinX1t protein but not mPinX1 protein interacts with Nup133, a component of the nuclear pore complex. Interestingly, mPinX1t‐containing cells was found to have a higher cytosol to nucleus ratio of cardiac transcription factor mRNAs when compared with control cells. The present study reports the presence of this novel mPinX1t protein and reveals its novel role in promoting cardiogenesis. This study provides important information for developmental biology and also provides novel insights on the possibility of increasing the yield of cardiomyocytes from ESCs, a hurdle that has to be overcome if ESC‐derived cardiomyocytes are applied for drug screening or for future therapeutic uses.
Sources of Funding
This work was supported by the General Research Fund (474711) from the University Grants Committee (UGC) of the Hong Kong SAR, and the Innovative Technology Fund of Innovation Technology Commission: Funding Support to Partner State Key Laboratories in Hong Kong. Drs Chan, Lau, and Ding were supported by the postgraduate studentships from the Chinese University of Hong Kong.
Disclosures
None.
Supporting information
Data S1. Supplemental materials and method.
Figure S1. Restriction digestion pattern of ligation products from RNA ligase‐mediated rapid amplification of cDNA ends (RLM 5′ RACE).
Figure S2. Expressions of mPinX1 and mPinX1t at mRNA level during early stage of mESC differentiation.
Figure S3. Establishment of mPinX1 overexpression (mPinX1 OE), mPinX1t overexpression (mPinX1t OE) and mPinX1 knockdown (mPinX1 KD) cell lines.
Figure S4. Lack of effects of mPinX1/mPinX1t overexpression and knockdown on the proliferation, pluripotency, and telomerase activity of mESCs.
Figure S5. Expression profile of mPinX1 and mPinX1t during early differentiation in different mPinX1/mPinX1t overexpression and knockdown cell lines.
Figure S6. Effects of overexpression of mPinX1 or mPinX1t on the calcium transients (CaTs) of mESC‐CMs.
Figure S7. mPinX1 and mPinX1t proteins did not universally bind to all mRNAs.
Figure S8. Gene Ontology analysis of mPinX1t interacting partners screened by the yeast‐2 hybrid assay.
(J Am Heart Assoc. 2020;9:e010240 DOI: 10.1161/JAHA.118.010240.)
References
- 1. Pagidipati NJ, Gaziano TA. Estimating deaths from cardiovascular disease: a review of global methodologies of mortality measurement. Circulation. 2013;127:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ali SR, Hippenmeyer S, Saadat LV, Luo L, Weissman IL, Ardehali R. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci USA. 2014;111:8850–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott IC. Life before Nkx2. 5: cardiovascular progenitor cells: embryonic origins and development. Curr Top Dev Biol. 2012;100:1–31. [DOI] [PubMed] [Google Scholar]
- 4. Rajala K, Pekkanen‐Mattila M, Aalto‐Setala K. Cardiac differentiation of pluripotent stem cells. Stem Cells Int. 2011;2011:383709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. Chamber‐specific cardiac expression of Tbx5 and heart defects in Holt‐Oram syndrome. Dev Biol. 1999;211:100–108. [DOI] [PubMed] [Google Scholar]
- 6. Benson DW, Silberbach GM, Kavanaugh‐McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, Garg V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326:368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang Y, Drysdale TA, Evans T. A role for GATA‐4/5/6 in the regulation of Nkx2.5 expression with implications for patterning of the precardiac field. Dev Biol. 1999;216:57–71. [DOI] [PubMed] [Google Scholar]
- 10. Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG, Zorn AM, Harvey RP. Cardiac T‐box factor Tbx20 directly interacts with Nkx2‐5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. [DOI] [PubMed] [Google Scholar]
- 11. Luna‐Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM, Mileikovsky M, Nagy A, Holloway AK, Pollard KS, Muller CW, Bruneau BG. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell. 2016;164:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fijnvandraat AC, Lekanne Deprez RH, Christoffels VM, Ruijter JM, Moorman AF. TBX5 overexpression stimulates differentiation of chamber myocardium in P19C16 embryonic carcinoma cells. J Muscle Res Cell Motil. 2003;24:211–218. [DOI] [PubMed] [Google Scholar]
- 13. Grepin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA‐4 transcription factor. Development. 1997;124:2387–2395. [DOI] [PubMed] [Google Scholar]
- 14. Rui Y, Bai J, Perrimon N. Sarcomere formation occurs by the assembly of multiple latent protein complexes. PLoS Genet. 2010;6:e1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boateng SY, Goldspink PH. Assembly and maintenance of the sarcomere night and day. Cardiovasc Res. 2008;77:667–675. [DOI] [PubMed] [Google Scholar]
- 16. Zhou XZ, Lu KP. The Pin2/TRF1‐interacting protein PinX1 is a potent telomerase inhibitor. Cell. 2001;107:347–359. [DOI] [PubMed] [Google Scholar]
- 17. Aravind L, Koonin EV. G‐patch: a new conserved domain in eukaryotic RNA‐processing proteins and type D retroviral polyproteins. Trends Biochem Sci. 1999;24:342–344. [DOI] [PubMed] [Google Scholar]
- 18. Zhou XZ. PinX1: a sought‐after major tumor suppressor at human chromosome 8p23. Oncotarget. 2011;2:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi Y, Qi Z, Li Z, Wong CK, So C, Lo IC, Huang Y, Yao X, Tsang SY. Role of TRPV1 in the differentiation of mouse embryonic stem cells into cardiomyocytes. PLoS One. 2015;10:e0133211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lo IC, Chan HC, Qi Z, Ng KL, So C, Tsang SY. TRPV3 channel negatively regulates cell cycle progression and safeguards the pluripotency of embryonic stem cells. J Cell Physiol. 2016;231:403–413. [DOI] [PubMed] [Google Scholar]
- 21. Law SK, Leung CS, Yau KL, Tse CL, Wong CK, Leung FP, Mascheck L, Huang Y, Sauer H, Tsang SY. Regulation of multiple transcription factors by reactive oxygen species and effects of pro‐inflammatory cytokines released during myocardial infarction on cardiac differentiation of embryonic stem cells. Int J Cardiol. 2013;168:3458–3472. [DOI] [PubMed] [Google Scholar]
- 22. Qi Z, Wong CK, Suen CH, Wang J, Long C, Sauer H, Yao X, Tsang SY. TRPC3 regulates the automaticity of embryonic stem cell‐derived cardiomyocytes. Int J Cardiol. 2016;203:169–181. [DOI] [PubMed] [Google Scholar]
- 23. Nabbi A, Riabowol K. Isolation of pure nuclei using a sucrose method. Cold Spring Harb Protoc. 2015;2015:773–776. [DOI] [PubMed] [Google Scholar]
- 24. Gagnon KT, Li L, Janowski BA, Corey DR. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc. 2014;9:2045–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bower NI, Moser RJ, Hill JR, Lehnert SA. Universal reference method for real‐time PCR gene expression analysis of preimplantation embryos. Biotechniques. 2007;42:199–206. [DOI] [PubMed] [Google Scholar]
- 26. Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. [DOI] [PubMed] [Google Scholar]
- 27. Sequeira V, Nijenkamp LL, Regan JA, van der Velden J. The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim Biophys Acta. 2014;1838:700–722. [DOI] [PubMed] [Google Scholar]
- 28. Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim SJ, Fernandez‐Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina‐Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Integrative structure‐function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol Cell Proteomics. 2014;13:2911–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai MY, Zhang B, He WP, Yang GF, Rao HL, Rao ZY, Wu QL, Guan XY, Kung HF, Zeng YX, Xie D. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci. 2010;101:1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng YZ, Zhang QY, Fu MT, Zhang ZF, Wei M, Zhou JY, Shi R. Low expression of PinX1 is associated with malignant behavior in basal‐like breast cancer. Oncol Rep. 2017;38:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hiyama E, Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96:1020–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Armstrong L, Saretzki G, Peters H, Wappler I, Evans J, Hole N, von Zglinicki T, Lako M. Overexpression of telomerase confers growth advantage, stress resistance, and enhanced differentiation of ESCs toward the hematopoietic lineage. Stem Cells. 2005;23:516–529. [DOI] [PubMed] [Google Scholar]
- 34. Yonekawa T, Yang S, Counter CM. PinX1 localizes to telomeres and stabilizes TRF1 at mitosis. Mol Cell Biol. 2012;32:1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gardano L, Pucci F, Christian L, Le Bihan T, Harrington L. Telomeres, a busy platform for cell signaling. Front Oncol. 2013;3:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang J, Li L. BMP signaling and stem cell regulation. Dev Biol. 2005;284:1–11. [DOI] [PubMed] [Google Scholar]
- 37. Niida H, Matsumoto T, Satoh H, Shiwa M, Tokutake Y, Furuichi Y, Shinkai Y. Severe growth defect in mouse cells lacking the telomerase RNA component. Nat Genet. 1998;19:203–206. [DOI] [PubMed] [Google Scholar]
- 38. Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. [DOI] [PubMed] [Google Scholar]
- 39. Cheong C, Hong KU, Lee HW. Mouse models for telomere and telomerase biology. Exp Mol Med. 2003;35:141–153. [DOI] [PubMed] [Google Scholar]
- 40. Yang Z, Sui Y, Xiong S, Liour SS, Phillips AC, Ko L. Switched alternative splicing of oncogene CoAA during embryonal carcinoma stem cell differentiation. Nucleic Acids Res. 2007;35:1919–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, Ta L, Mercola M, Conklin BR. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2009;5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Banik SS, Counter CM. Characterization of interactions between PinX1 and human telomerase subunits hTERT and hTR. J Biol Chem. 2004;279:51745–51748. [DOI] [PubMed] [Google Scholar]
- 43. Guglielmi B, Werner M. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J Biol Chem. 2002;277:35712–35719. [DOI] [PubMed] [Google Scholar]
- 44. Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplemental materials and method.
Figure S1. Restriction digestion pattern of ligation products from RNA ligase‐mediated rapid amplification of cDNA ends (RLM 5′ RACE).
Figure S2. Expressions of mPinX1 and mPinX1t at mRNA level during early stage of mESC differentiation.
Figure S3. Establishment of mPinX1 overexpression (mPinX1 OE), mPinX1t overexpression (mPinX1t OE) and mPinX1 knockdown (mPinX1 KD) cell lines.
Figure S4. Lack of effects of mPinX1/mPinX1t overexpression and knockdown on the proliferation, pluripotency, and telomerase activity of mESCs.
Figure S5. Expression profile of mPinX1 and mPinX1t during early differentiation in different mPinX1/mPinX1t overexpression and knockdown cell lines.
Figure S6. Effects of overexpression of mPinX1 or mPinX1t on the calcium transients (CaTs) of mESC‐CMs.
Figure S7. mPinX1 and mPinX1t proteins did not universally bind to all mRNAs.
Figure S8. Gene Ontology analysis of mPinX1t interacting partners screened by the yeast‐2 hybrid assay.


