Abstract
RNA triphosphatase catalyzes the first step in mRNA cap formation, hydrolysis of the terminal phosphate from the nascent mRNA transcript. The RNA triphosphatase from the protozoan parasite Trypanosoma cruzi, TcCet1, belongs to the family of triphosphate tunnel metalloenzymes (TTMs). TcCet1 is a promising antiprotozoal drug target because the mechanism and structure of the protozoan RNA triphosphatases are completely different from those of the RNA triphosphatases found in mammalian and arthropod hosts. Here, we report several crystal structures of the catalytically active form of TcCet1 complexed with a divalent cation and an inorganic tripolyphosphate in the active-site tunnel at 2.20–2.51 Å resolutions. The structures revealed that the overall structure, the architecture of the tunnel, and the arrangement of the metal-binding site in TcCet1 are similar to those in other TTM proteins. On the basis of the position of three sulfate ions that cocrystallized on the positively charged surface of the protein and results obtained from mutational analysis, we identified an RNA-binding site in TcCet1. We conclude that the 5′-end of the triphosphate RNA substrate enters the active-site tunnel directionally. The structural information reported here provides valuable insight into designing inhibitors that could specifically block the entry of the triphosphate RNA substrate into the TTM-type RNA triphosphatases of T. cruzi and related pathogens.
Keywords: RNA triphosphatase, mRNA cap, capping enzyme, Trypanosoma cruzi, crystal structure, triphosphate tunnel metalloenzyme (TTM), protozoan parasite, TcCet1, RNA processing, RNA modification, enzyme structure
Introduction
Trypanosomatid parasites belong to the order kinetoplastida that causes several neglected diseases of humans and animals affecting close to 100 million people worldwide, including African sleeping sickness caused by Trypanosoma brucei, Chagas disease caused by Trypanosoma cruzi, and a spectrum of diseases caused by various Leishmania species (1). New therapeutics against trypanosomatids are needed because current medications are generally ineffective at late stages of infection or have severe side effects (2).
The 5′-cap (m7Gp) is an essential feature in eukaryotic cellular and viral mRNAs that functions to protect mRNA from degradation and to promote translation initiation. RNA triphosphatase, which catalyzes the first step in mRNA capping, is a promising target for antiprotozoal drug development because the mechanism of protozoan RNA triphosphatase is completely different from mammalian or arthropod host (3, 4). Metazoan and plant RNA triphosphatases belong to the cysteine–phosphatase enzyme superfamily that catalyzes a two-step phosphoryl transfer reaction in which the active site cysteine in the phosphate-binding loop attacks the γ-phosphate of triphosphate-terminated RNA (pppRNA) to form a covalent protein-cysteinyl-S-phosphate intermediate and release the diphosphate RNA product in the absence of metal cofactor (5, 6). The RNA triphosphatases of fungi, protozoa, and several DNA viruses belong to a TTM family that hydrolyzes γ-phosphorus of pppRNA in the presence of magnesium and NTP in the presence of either manganese or cobalt (7–17). The crystal structure of Saccharomyces cerevisiae RNA triphosphatase (Cet1) revealed that the catalytic core is located in a topologically closed hydrophilic tunnel composed of eight antiparallel β-strands (18). The Cet1 active site constitutes of 15 essential amino acid side chains that either stabilize the topology of the tunnel or make contacts to the divalent cation or sulfate ion (7, 19–21). The position of the sulfate ion was proposed to reflect the position of γ-phosphate of pppRNA and NTP or the leaving group phosphate (18). Two glutamate-containing motifs (designated as motifs A and C) comprise the metal-binding site and are conserved among all TTM family members (Fig. 1) (22–24).
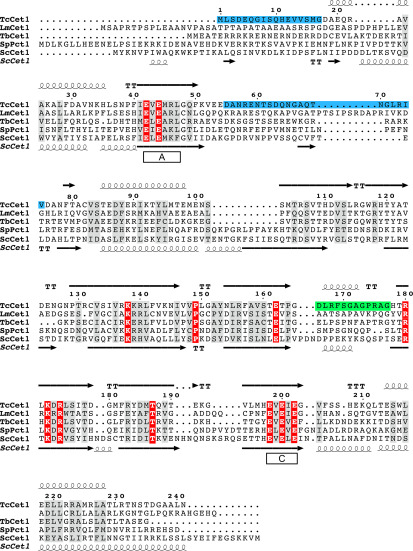
Figure 1.
Structure-based alignment of T. cruzi RNA triphosphatase. The secondary structure of T. cruzi RNA triphosphatase (TcCet1) is shown above the amino acid sequence. TcCet1 amino acids shaded in blue correspond to the segments that are removed in TcCet1(18–243 Δ55–75) protein. The TcCet1 sequence shaded in green (amino acid residues 166–177) corresponds to the segment that interacts with C13H13NO2 and C10H14N4O2 but was disordered in TcCet1(18–243 Δ55–75) manganese and Mn·PPPi–bound structures. The amino acid sequences of TcCet1 is aligned with Leishmania major (LmCet1), T. brucei (TbCet1), Schizosaccharomyces pombe (SpPct1), and S. cerevisiae (ScCet1) RNA triphosphatases. The secondary structure of ScCet1 is indicated below the aligned sequences. Identical side chains found in all polypeptides are highlighted in red. Amino acids with similar side chains are highlighted in gray. The positions of conserved motifs A and C, located within the catalytic domain of the metal-dependent RNA triphosphatases, are indicated. The alignment was prepared by ESPript. The secondary structure assignment was based on the DSSP program.
RNA triphosphatases from kinetoplastids have been characterized in T. brucei (TbCet1) and T. cruzi (TcCet1) (10, 11). Based on the primary structures of TbCet1 and TcCet1, the kinetoplastid enzymes contain all the putative counterparts of β-strands that comprise the yeast Cet1 triphosphate tunnel but lack extra domains appended to the N-terminal region that are essential for homodimerization and interaction with Ceg1 guanylyltransferase (25–27). TbCet1 is essential for procyclic cell growth (28) and can also complement the function of yeast Cet1 in S. cerevisiae as a cap-forming enzyme (11). Several classes of small molecular weight compounds have been reported to inhibit the triphosphatase activity of TbCet1, and mutagenesis studies have illuminated functional groups that are important for catalytic activity (28–30). What remains obscure is the structural basis of inhibition and the selectivity for the RNA substrate.
In the present study, we produced a catalytically active form of TcCet1 and crystallized the enzyme in complex with divalent cation and tripolyphosphate (PPPi) bound at the active site. We propose that the 5′-triphosphate terminus of RNA substrate directionally enters the active-site tunnel through a positively charged surface based on the three sulfate ions that cocrystallized with TcCet1. The structural information could provide valuable insight into designing inhibitors that could specifically block the entry of triphosphate RNA substrate in TTM-type RNA triphosphatases.
Results
Defining the minimal catalytic unit of Trypanosoma RNA triphosphatase
Deletion and limited proteolysis analyses of TbCet1 suggest that the hydrophilic N-terminal ∼20-amino acid segment is connected by protease-sensitive site and is dispensable for triphosphatase activity (30). To define the minimal catalytic core for structural analysis, we deleted several amino acids from N and C termini in TcCet1 (TcCet1(18–243)) and further made an internal deletion to remove a putative protease-sensitive loop (TcCet1(18–243 Δ55–75); Fig. 1 and Fig. S1). Similar deletions were made for TbCet1 (TbCet1(29–253) and TbCet1(29–253 Δ62–90)). The truncated TcCet1 and TbCet1 proteins were expressed in bacteria and purified from a soluble extract by nickel–agarose chromatography, and proteins were assayed for a release of 32Pi from [γ-32P]ATP in the presence of 2 mm manganese. Removal of N-terminal region apparently increased the ATPase activity of TcCet1 and TbCet1 1.3- and 2.6-fold, respectively, compared with their full-length enzymes (Fig. S1B). TcCet1(18–243 Δ55–75) and TbCet1(29-253 Δ62–90) showed modest reductions in activity: 90 and 60% of full-length enzymes, respectively. TcCet1(18–243 Δ55–75) had similar activity to the full-length TcCet1 in hydrolyzing pppRNA (Fig. S1C). We conclude that removal of putative protease-sensitive loop region does not significantly affect the triphosphatase activity.
Crystallization and structure determination of TcCet1(18–243 Δ55–75)
PPPi is a potent competitive inhibitor of TbCet1, suggesting that PPPi binds tightly to the active site of the enzyme (30). Although TbCet1 is capable of hydrolyzing PPPi, tripolyphosphatase activity is only 0.5% of its NTPase activity. We therefore cocrystallized TcCet1(18–243 Δ55–75) in the presence of PPPi and manganese (see “Experimental procedures”). The crystal was further soaked with potassium iodine to improve the mosaicity to obtain experimental phase information. The structure was then refined at 2.20 Å resolution with R/Rfree of 22.6%/26.1% (Fig. S2 and Table S1). The electron density corresponding to PPPi was evident, and a density for manganese was modeled adjacent to PPPi. We subsequently solved the structure of TcCet1(18–243 Δ55–75) in complex with manganese alone by means of molecular replacement phasing method using the TcCet1(18–243 Δ55–75) Mn·PPPi structure as a search model. The root-mean-square deviations between pairs of structures was 0.9 Å for the main chain atoms of residues 37–242, suggesting that binding of PPPi may not induce conformational change. Efforts to crystallize full-length TcCet1 and TbCet1 and other derivatives of the proteins were unsuccessful.
Two unique interfaces are formed in the structure of TcCet1(18–243 Δ55–75) (Fig. S2). The dimer-forming interface in the manganese-bound and Mn·PPPi–bound forms differ in the position of the N-terminal α-helix (from residues Asp18 to Leu36). Analysis with the PDBePISA server (32) reveals that the manganese-bound dimer interface buries 663.9 Å2 with a Complex Formation Significance Score of 0.000, suggesting that this interaction occurs only in the crystal. In the Mn·PPPi–bound form, the N-terminal α-helix is extended and protrudes from one protomer to the other, with an interface area of 654.1 Å2 and the calculated Complex Formation Significance Score of 0.440, suggesting that the interface plays an auxiliary role in complex formation. However, identical contact is formed by the monomeric enzyme, in which the N-terminal α-helix folds back by a connecting loop (Fig. S2C). Indeed, TcCet1 behaves as a monomer in solution at micromolar concentration, as judged by gel-filtration analysis and as previously noted (10). Thus, the two independent molecules of TcCet1 found in the asymmetric unit in Mn·PPPi–bound form can be described as a dimer formed by a crystal packing.
Structure of TcCet1 with tripolyphosphate in the active site
The overall structure of TcCet1(18–243 Δ55–75) is similar to other TTM proteins, composed of eight-stranded anti-parallel β-barrel with three α-helices surrounding the active-site tunnel (Fig. 2A). Structural comparison with yeast RNA triphosphatase Cet1 reveals that the diameter of the tunnel and the metal-binding site is nearly identical between the two enzymes (Fig. 2, C–E). Positions of 15 side chains in Cet1 active-site tunnel are conserved in TcCet1 (Fig. 2, B and E), of which 14 side chains were shown to be essential for Cet1 (7, 19, 21) and TbCet1 (30) triphosphatase activity. Three glutamates (Glu42, Glu44, and Glu210) from motifs A and C from the tunnel floor coordinate the Mn2+ and allows Mn2+ to interact with the PPPi at P1 and P2 positions (Fig. 2F). The basic side chains (Arg118 and Lys138) extended from the other side of the tunnel likely stabilize the interaction with PPPi. Lys182 and Arg184 interact with central phosphate (P2 position), and Lys138 interacts with phosphate at the P3 position of PPPi (Fig. 2F). A similar mode of PPPi binding and the metal arrangement was observed in the structures of the Arabidopsis protein AtTTM3·Mn·PPPi complex (24, 33). In TcCet1, Arg46 also forms water-mediated contact to Glu44 and Glu210.
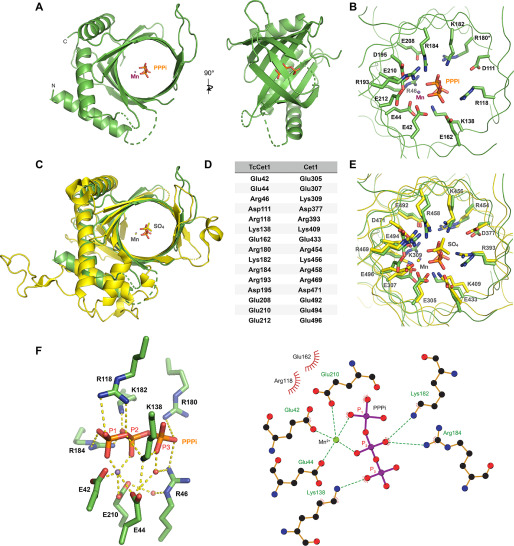
Figure 2.
Structure of TcCet1(18–243 Δ55–75) in complex with tripolyphosphate in the active site. A, crystal structure of TcCet1(18–243 Δ55–75) is depicted as a cartoon model. PPPi is shown as sticks, and manganese is shown as a magenta sphere. B, cross-section of the triphosphate tunnel of TcCet1(18–243 Δ55–75). C, the structure of TcCet1(18–243 Δ55–75) colored in green was superimposed on a yeast Cet1(210-549) colored in yellow (18) (Protein Data Bank entry 1D8H). The sulfate and manganese in Cet1(210-549) are depicted as yellow sticks and a yellow sphere, respectively. D, the amino acids of TcCet1 that correspond to active site residues of Cet1, as judged from the structural comparison. The Arg180 side chain was not visible in Mn·PPPi–bound structure and therefore was modeled from the manganese-bound structure. E, superimposition of triphosphate tunnel of TcCet1(18–243 Δ55–75) and Cet1(210-549). The image was prepared with PyMOL. F, the figure highlights the network of bonding interactions that coordinate PPPi, manganese, and two water molecules (red sphere) in TcCet1(18–243 Δ55–75). The images were prepared with PyMOL and LigPlus.
Based on the configuration between Mn2+ and PPPi, Mn2+ could facilitate the hydrolysis of phosphate at the P1 position. Hence, we predict that γ-phosphate of pppRNA and NTP will be situated at the position of P1 phosphate. In support of this view, superimposition of TcCet1 and yeast Cet1 structures reveal that P1 phosphate of PPPi is in close proximity to the sulfate ion present in the Cet1 structure (Fig. 2E), which has been proposed to mimic the position of γ-phosphate of pppRNA and NTP (18).
Screen for TcCet1 ligand by fluorescent-based protein thermal stability shift assay
We performed a fluorescent-based protein thermal stability shift assay to identify a potential ligand that can interact to stabilize the TcCet1 protein. In this procedure, TcCet1(18–243 Δ55–75) was incubated with CYPRO Orange dye, which preferentially binds to the hydrophobic core of the protein and emits fluorescence when the protein is unfolded by temperature shift. We determined that the Tm of TcCet1(18–243 Δ55–75) was 44.6 °C, and the addition of 2 mm ATP increased the Tm by 1.4 °C to 46.0 °C (Fig. 3A).
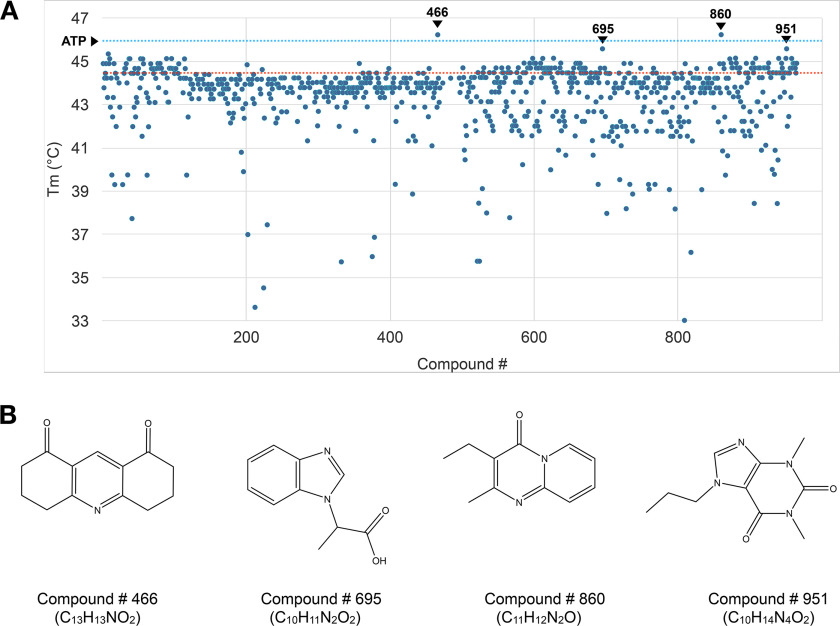
Figure 3.
Screening of small molecule that stabilizes TcCet1. A, melting temperatures (Tm) of TcCet1(18–243 Δ55–75) in the presence of 2 mm compounds were determined by thermal shift assay. Tm of TcCet1(18–243 Δ55–75) with DMSO (control) is indicated by the red dotted line (44.6 °C), and that with ATP is indicated by the blue dotted line (46.0 °C). Four compounds (compounds 466, 695, 860, and 951) that significantly increase the Tm are indicated. B, structures of compound 466 (C13H13NO2: 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione), compound 695 (C10H11N2O2: 2-(1H-benzimidazol-1-yl)propanoic acid), compound 860 (C11H12N2O: 3-ethyl-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one), and compound 951 (C10H14N4O2: 1,3-dimethyl-7-propyl-purine-2,6-dione).
We screened for compounds that can bind and increase the Tm of TcCet1(18–243 Δ55–75) using an in-house chemical compound library consists of 965 commercially available low-molecular-weight compounds (Fig. 3A). We identified four compounds that increase the Tm of TcCet1(18–243 Δ55–75) more than 1.0 °C when present at 2 mm concentration (compound 466 (C13H13NO2; Tm = 46.23 °C), compound 695 (C10H11N2O2; Tm = 45.58 °C), compound 860 (C11H12N2O; Tm = 46.23 °C), and compound 951 (C10H14N4O2; Tm = 45.58 °C)). The chemical structures of these compounds are shown in Fig. 3B. In addition, we identified 10 compounds that lower the Tm of TcCet1(18–243 Δ55–75) to below 37 °C. The structures of these compounds are shown in Fig. S3.
Structure of TcCet1 in complex with ligands
We attempted to cocrystallize TcCet1(18–243, Δ55–75) with the four compounds identified from the above screen that stabilize the enzyme. Only the complex formed with compound 951 (C10H14N4O2) crystallized with satisfactory quality for X-ray crystal diffraction analysis. By soaking 2 mm of compound 466 (C13H13NO2) in to C10H14N4O2-bound crystal, we also obtained the structure of TcCet1(18–243, Δ55–75)·C13H13NO2. The structures of TcCet1(18–243, Δ55–75) with the ligands were solved at 2.39 Å (complex with C10H14N4O2) or 2.51 Å (complexed with C13H13NO2) resolution using the TcCet1(18–243 Δ55–75) Mn·PPPi–bound structure as a search model (Fig. 4, A and B). The bound ligands were well-defined in the electron-density maps (Fig. S4) and bound on the same surface of the protein that forms a helical loop between amino acid residues 166 and 177, a region that was disordered in both manganese and Mn·PPPi–bound structures. Neither manganese nor PPPi is evident in both ligand-bound structures, and the tunnel cavities are slightly narrower compared with the manganese- or Mn·PPPi–bound structures (Fig. 4).
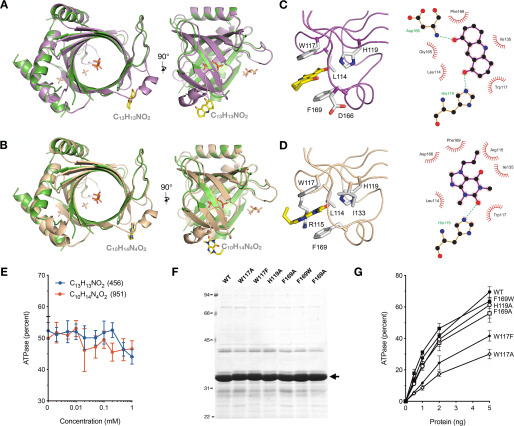
Figure 4.
Structures of TcCet1 in complex with C13H13NO2 and C10H14N4O2. A and B, crystal structures of TcCet1(18–243 Δ55–75) with C13H13NO2 (A, colored in violet) and C10H14N4O2 (B, colored in wheat) were superimposed with the TcCet1(18–243 Δ55–75) Mn·PPPi–bound complex (colored in green). Two segments, amino acids 35–39 and 166–177, were ordered in both C13H13NO2- and C10H14N4O2-bound structures but were disordered in the Mn·PPPi–bound structure. C and D, close-up view of C13H13NO2 (C) and C10H14N4O2 (D) binding sites on TcCet1(18–243 Δ55–75). The images were prepared with PyMOL and LigPlus. E, effect of C13H13NO2 or C10H14N4O2 on triphosphatase activity. Reaction mixtures (10 μl) containing 50 mm Tris-HCl, pH 7.5, 5 mm DTT, 1 mm MnCl2, 0.2 mm [γ-32P]ATP, 4 ng of TbCet1, and 0, 0.002, 0.005, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, or 1.0 mm of either C13H13NO2 (blue circle) or C10H14N4O2 (orange circle) were incubated for 15 min at 30 °C. The extents of ATP hydrolysis were plotted as a function of ligand concentration. The data shown represent the averages of three separate experiments with S.E. bars. F, aliquots (5 µg) of native WT TcCet1 and mutant proteins were analyzed by SDS-PAGE. Polypeptides were visualized by staining with Coomassie Blue dye. The position of WT and mutant TcCet1 polypeptide is denoted by an arrow. G, ATPase activity. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 1 mm MnCl2, 0.2 mm [γ-32P]ATP, and the indicated amounts of WT and mutant TcCet1 proteins were incubated for 15 min at 30 °C. The extents of ATP hydrolysis were plotted as a function of input protein. The data shown represent the averages of three separate experiments with S.E. bars.
Two aromatic residues, Phe169 from the loop and Trp117 from β4-strand, interact with C13H13NO2 and C10H14N4O2 through π–π stacking. The C13H13NO2 oxo-groups are hydrogen-bonded by His119 and Asp165 side chains (Fig. 4C). Similarly, one of the oxo-groups in C10H14N4O2 forms a hydrogen bond with the His119 side chain, and the other oxo-group is stabilized by Arg115 (Fig. 4D). Neither compound had a substantial effect on the triphosphatase activity of TcCet1, up to 1 mm concentration (Fig. 4E). Point mutations in Phe169 and His119 did not have significant impact on the triphosphatase activity, although Trp117 mutation showed a moderate effect with 20–50% of WT activity (Fig. 4, F and G). These results indicate that the ligand-binding site is not essential for TcCet1 activity and that ligand-bound structures constitute an active form of the enzyme.
Sulfate ions bound on the surface of TcCet1 reveal the entry site for triphosphate RNA
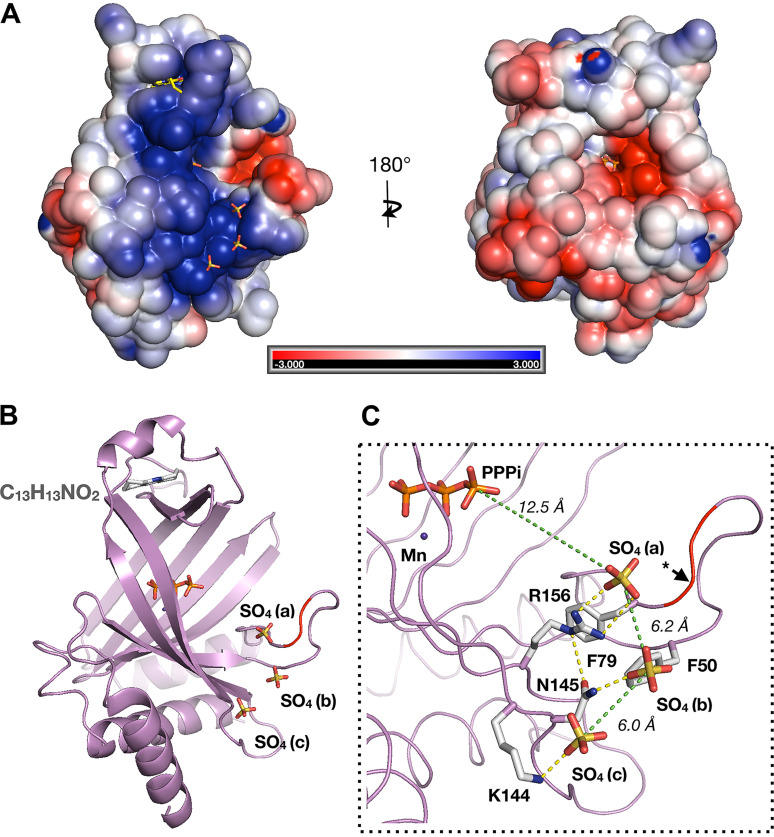
The striking feature of C13H13NO2- and C10H14N4O2-bound structures is that three sulfate ions, designated as SO4-a, SO4-b, and SO4-c, were bound on the same positively charged surface of the protein (Fig. 5, A and B). The distance between the three consecutive sulfur atoms is ∼6 Å each (S–S distance), which is similar to the distance between the two phosphorus atoms on a single-stranded RNA (Fig. 5C). We predict that three consecutive sulfate ions reflect the position of backbone phosphate on the RNA chain. Superposition of the ligand-bound and the PPPi-bound structures reveals that SO4-a is situated 12.6 Å (S-P distance) from the nearest phosphate on the PPPi in the active-site tunnel, suggesting that SO4-a likely occupies the position of phosphate between the second and third nucleotides of the RNA substrate. SO4-b and SO4-c may occupy the positions between the third and fourth and between the fourth and fifth nucleotides on the RNA, respectively.
Figure 5.
Sulfate ions bound on the positive surface of TcCet1. A, the space-filling surface images of TcCet1(18–243 Δ55–75) bound to C13H13NO2 were prepared with the program GRASP. The positive surface charge potential is shown in blue, and the negative potential is in red. C13H13NO2 and sulfate ions are shown as stick models. B, cartoon model of A. PPPi and manganese were modeled on the TcCet1(18–243 Δ55–75)·C13H13NO2 structure from the structure of PPPi·Mn–bound complex. C, close-up view of SO4-binding site. Distance between the three sulfur atoms and the distance between sulfur on SO4-a to the nearest phosphorous of PPPi are indicated by green dotted lines. Amino acid residues 54–76 are colored in red to indicate the region that was deleted in TcCet1, and the asterisk denotes a probable position of Arg58.
We hypothesized that three sulfate ions found in the structure of TcCet1 reflect the position of phosphodiester bonds of the RNA substrate; mutation in the residues surrounding the bound sulfate ions may affect the RNA triphosphatase activity but retains its ability to hydrolyze NTP. Alanine substitution was introduced at two positively charged residues, Arg156 and Lys144, that coordinate SO4-a and SO4-c, respectively (Fig. 5C). Phe50 and Phe79 were also selected, because these aromatic residues may participate in stacking interaction with the RNA bases. In addition, Arg58, situated within the internal deletion of TcCet1(18–243 Δ55–75), was substituted to alanine as a positive control.
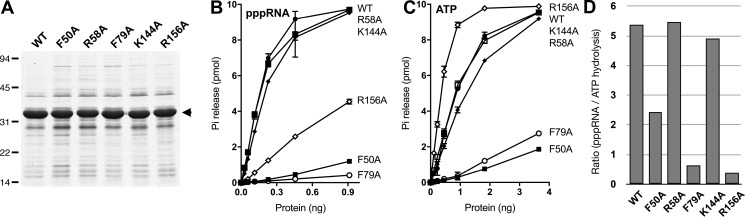
The full-length WT and Ala-substituted TcCet1 proteins were produced in bacteria (Fig. 6A). The RNA triphosphatase and ATPase activities were assayed for the release of 32Pi from 1 μm [γ-32P]pppRNA and [γ-32P]ATP, respectively (Fig. 6, B and C). The specific activities were calculated from the averages of the slopes of the titration curves in the linear range of enzyme dependence. Under this condition, TcCet1 can preferentially hydrolyze pppRNA, 5.4-fold higher than that for ATP (Fig. 6D). The R156A mutation showed significantly reduced RNA triphosphatase activity compared with the WT enzyme. The ratio of pppRNA hydrolysis/ATP hydrolysis was 0.4, suggesting that R156A is selectively impaired for hydrolyzing triphosphate RNA (13.5-fold lower compared with the WT enzyme). F50A and F79A display reduced pppRNA and ATP hydrolysis compared with the WT enzyme. The ratio of pppRNA/ATP hydrolysis by F50A and F79A was 2.4 and 0.6, respectively, implying that F79A was less active in hydrolyzing pppRNA than the ATP. K117A and R58A displayed near WT activity in hydrolyzing pppRNA and ATP. Consistent with our findings, F50A, F79A, and R156A mutations show significant reduction in binding to the nucleic acid, whereas R58A and K144A mutants maintained WT binding (Fig. S5). Taken together, these results support the notion that three sulfate ions found in the structure of TcCet1 reflect the position of phosphodiester bonds on the RNA, and the 5′-end of the RNA likely enters the active-site tunnel directionally through the positively charged nucleic acid–binding surface.
Figure 6.
Mutational analysis of putative RNA-binding surface of TcCet1. A, aliquots of the nickel–agarose preparations of the WT and mutant TcCet1 proteins (5 μg) indicated were analyzed by SDS-PAGE and visualized by staining with Coomassie Blue dye. B, RNA triphosphatase activity. The reaction mixtures contained 1 μm of [γ-32P]triphosphate-terminated RNA, 2 mm MgCl2 with WT, or mutant proteins as specified. The data shown represent the averages of three separate experiments with S.E. bars. C, ATPase activity. The reaction mixtures contained 1 μm of [γ-32P]ATP, 2 mm MnCl2 with WT, or mutant proteins as specified. The extent of Pi release is plotted as a function of input enzyme. The data shown represent the averages of three separate experiments with S.E. bars. D, efficiency of RNA hydrolysis versus ATP hydrolysis. The specific activities of WT and mutant TcCet1 RNA triphosphatase and ATPase activates were calculated from the data presented in B and C and are expressed as ratios of RTPase/ATPase activities and plotted for each protein indicated.
Discussion
The present study provides new insights into the structure and mechanism of TTM–RNA triphosphatase by capturing structures of catalytically active TcCet1 in complex with manganese and PPPi. The catalytic domain of TcCet1 adopts a characteristic TTM–enzyme fold with eight-stranded antiparallel β-barrel, and the arrangement of the metal-binding site is similar to other TTM–enzymes (18, 23). The RNA must bind near one of the tunnel openings to allow the 5′-triphosphate terminus to enter the active site. We further cocrystallized TcCet1 with the two phenolic compounds that were identified to stabilize the TcCet1. The crystals were grown in the presence of ammonium sulfate, and the TcCet1–ligand complexes contained three sulfate ions on an electrostatically positive surface near the tunnel entrance. Each sulfate ions are separated by ∼6 Å, similar to the distance between consecutive phosphates in single-stranded RNA. The distance between the PPPi found in the active-site tunnel and the closest sulfur atom was 12 Å. We predict that sulfate ions reflect positions of the second (pppN1pN2pN3pN4p), third (pppN1pN2pN3pN4p), and fourth (pppN1pN2pN3pN4p) backbone phosphates on the pppRNA substrate.
Two metal-binding sites were identified in the structures of other TTM members, including AtTTM3 polyphosphatase (24), ygiF polyphosphatase (24), and adenylate cyclase CyaB (34). The second metal ion participates in binding of the substrate, coordinating the triphosphate, or stabilizing the diphosphate leaving group for optimal catalysis. TcCet1 may employ two metal catalytic mechanisms, because previous studies suggest that TbCet1 and other RNA triphosphatases exhibit synergistic activation by magnesium and manganese (30, 35). However, only a single metal was found in the structure of TcCet1(18–243 Δ55–75). We speculate that the second metal is absent in the structure because PPPi lacks the nucleoside moiety found in NTP and pppRNA. Alternatively, an extra negative charge on the PPPi, which is not present in NTP or pppRNA, may cause interference with the binding of the second metal.
Mutational analysis of selected amino acids on the sulfate-binding surface identified Arg156 to be important for the RNA triphosphatase activity. Alanine substitution of Arg156 severely reduced RNA triphosphatase activity, but the mutant protein resulted in a 2-fold increase in NTPase activity. The Arg156 residue, located at the entrance of the tunnel, is conserved as arginine or lysine in cellular TTM-type RNA triphosphatases characterized to date, but is not present in other TTM–enzymes (13, 15, 23). The TcCet1 Arg156 counterpart for Cet1 is Lys427. Substitution of Cet1 Lys427 with Glu exhibits a cold-sensitive growth arrest phenotype in yeast, suggesting that a positive-charge residue at this position could be important for the Cet1 function in vivo (36). Two aromatic residues, Phe50 and Phe79, in the vicinity of sulfate ions were also crucial for TcCet1 RNA triphosphatase activity, suggesting that these aromatic residues may participate in stacking interaction with the nucleoside bases on the RNA. The effect of F79A substitution appeared to be much more severe than F50A, because Phe79 is positioned closer to the tunnel entrance than Phe50 and may have greater impact on stabilizing the 5′-end of the RNA.
The TTM–enzyme fold and the active site architecture are evolutionarily conserved but have different substrate specificity to hydrolyze phosphate. We propose that 5′-end of nascent RNA enters the triphosphate tunnel directionally through the positively charged RNA-binding surface to allow the γ-phosphate to be aligned in the active-site tunnel. This view is supported by cryo-EM and CX-MS analysis of yeast capping enzyme in complex with RNA polymerase II (37). In their model, the 5′-end of the newly synthesized RNA enters the Cet1 tunnel cavity in the same direction as we proposed for TcCet1. The directionality of substrate binding has been proposed to dictate the cleavage specificity of TTM (22, 24). Thiamine triphosphatase hydrolyzes thiamine triphosphate into thiamine diphosphate and Pi but is unable to hydrolyze pppRNA. The α-helix at the C-terminal end protrudes into the tunnel to interact with thiamine to orient the terminal phosphate for hydrolysis reaction, in the same direction as we proposed for TcCet1 (24). Similar C-terminal plug-in helix is also present in AtTTM3 tripolyphosphatase (24, 33) but is absent in TcCet1. The C-terminal plug-in helix may also function to block the unwanted substrate from entering the tunnel cavity, which may explain why thiamine triphosphatase and AtTTM3 cannot hydrolyze pppRNA. In the crystal structure of class IV adenylate cyclase, ATP is bound in a reverse orientation in the active-site tunnel, compared with the thiamine triphosphate of the thiamine triphosphatase (24, 34). This conformation of ATP allows the α-phosphate to position in a suitable orientation for an in-line nucleophilic attack by ribose O3′ at the cleavage site. Because TcCet1 can also hydrolyze γ-phosphate from NTP, we predict that triphosphate on the NTP will enter the tunnel in the same direction as pppRNA. In contrast, symmetric molecule such as PPPi may enter from either side of the tunnel. This may partly explain why PPPi is a potent inhibitor for the triphosphatase activity (30, 31).
In summary, we solved the structures of TcCet1 and illuminate the mechanism on how RNA substrate is being recognized and how the triphosphate end enters the active-site tunnel. The structural information of TcCet1 could be exploited in the development of effective inhibitor that could specifically block the entry of RNA substrate in the TTM-type RNA triphosphatase.
Experimental procedures
Expression plasmids for TcCet1 and TbCet1
The bacterial expression plasmid pET44-TcCet1 encodes T. cruzi RNA triphosphatase fused in-frame to an N-terminal His tag with a custom HRV3C protease recognition site. This was accomplished by amplifying the TcCet1 ORF from T. cruzi (Tulahuen strain) genome by PCR and inserting it into modified pET44, in which the plasmid segments encoding for Nus·Tag, S·Tag, and thrombin cleavage segments were replaced with a His tag and the HRV3C protease recognition site between NdeI and BamHI sites. The truncated allele TcCet1(18–243) was generated by PCR amplification using a mutagenic sense primer that replaces Gly17 with methionine and an antisense-strand primer that introduced stop codon in lieu of the codons for Ser244, and the fragment was inserted between NdeI and Sal1 restriction sites from the pET-TcCet1 template. TcCet1(18–243 Δ55–75) allele spanning from residues 18 to 243 with an internal deletion from 55 to 75 amino acids and point mutations in the TcCet1 gene were generated by synthetic oligonucleotides using the two-stage PCR-based overlap extension strategy. T. brucei RNA triphosphatase was amplified by PCR from pET–His/Smt3–TbCet1 (11) and inserted into pET44a plasmid between the NdeI and SalI restriction sites. The plasmids for expression of TbCet1(29–253) and TbCet1(29–253 Δ62–90) were generated by similar strategies described for TcCet1(18–243) and TcCet1(18–243 Δ55–75). The presence of the desired mutation was confirmed in every case by sequencing the entire insert. For deletion and mutational analysis, the plasmids were transformed into E. coli BL21 (DE3), and the proteins were purified from the soluble lysates by a Ni–NTA column as described (11).
Large-scale purification of TcCet1(18–243 Δ55–75)
E. coli BL21 (DE3) transformed with pET-TcCet1(18–243 Δ55–75) was grown in LB medium (6 liters) with 0.1 mg/ml ampicillin at 37 °C until the absorbance at 600 nm reached 0.6. The expression of TcCet1(18–243 Δ55–75) was induced by addition of isopropyl β-k-1-thiogalactopyranoside to a final concentration of 0.5 mm. The temperature was reduced to 16 °C following induction, and the cells were incubated for another 12–16 h. The bacteria were harvested by centrifugation and resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 0.3 M NaCl, 10 mm imidazole, 2 mm MnCl2 and 1 mm DTT), sonicated on ice, and centrifuged. The supernatant was mixed with Ni–NTA–Sepharose resin that had been equilibrated with lysis buffer, and the suspension was mixed by continuous rotation at 4 °C for 1 h. The nickel–Sepharose resin was poured into a column. The packed column was washed with lysis buffer containing 20 mm imidazole, and the N-terminal His tag was cleaved by GST-fused HRV3C protease on the column at 4 °C for 16 h. Tag-less TcCet1(18–243 Δ55–75) was recovered from the Ni–NTA column by elution buffer (50 mm Tris-HCl, pH 8.0, 0.1 m NaCl, 20 mm imidazole, 2 mm MnCl2, and 1 mm DTT). The eluate was passed through a Q-Sepharose (3 ml, GE Healthcare) column and then to a GS4B (3 ml, GE Healthcare) column, and the flow-through fractions were collected for each step. The GS4B flow-through fraction was concentrated by Amicon-Ultra (molecular mass cutoff, 10 kDa) to 20–30 mg/ml, and ∼15 mg of protein was applied to Superdex 75 gel filtration column (GE Healthcare) equilibrated with 10 mm HEPES (pH 7.5), 2 mm MnCl2, and 100 mm NaCl. Peak fractions containing the recombinant protein were collected, concentrated to ∼15 mg/ml, and stored at −80 °C. Purified TcCet1(18–243 Δ55–75) was used for crystallization and thermal shift assay.
Crystallization, data collection, and structure determination
Crystallization was performed by the hanging-drop vapor-diffusion method at 20 °C. Purified TcCet1(18–243 Δ55–75) (7.5 mg/ml) with a drop size of 1 μl was mixed with 1 μl of reservoir solution. The Manganese-bound form of crystal was obtained in 0.1 m HEPES-NaOH (pH 7.0–7.5), 14–20% PEG 20000. PPPi was added to a protein sample at 2.5 mm concentration prior to mixing with reservoir solution. The Mn·PPPi–bound form was crystalized in 0.1 m sodium acetate (pH 5.0) and 14–18% 1,4-butandiol. Compound 951 (C10H14N4O2: 1,3-dimethyl-7-propyl-purine-2,6-dione) was purchased from ChemBridge Corp., was added to a protein sample at 2 mm concentration with 2% DMSO, and then crystallized in 0.1 m HEPES (pH 7.5) and 1.4–1.6 m ammonium sulfate. Compound 466 (C13H13NO2: 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione) was purchased from Maybridge, Fisher Scientific. TcCet1(18–243 Δ55–75)·C10H14N4O2 crystal was soaked with 2 mm C13H13NO2 twice in 0.1 m HEPES (pH 7.5), 1.6 m ammonium sulfate, 2% DMSO, and 2 mm MnCl2 for 1 h to obtain the C13H13NO2-bound form structure. Prior to data collection, the crystals were transferred to 35% xylitol-containing reservoir solution for cryo-protection.
The diffraction data were collected at BL15A in National Synchrotron Radiation Research Center (Hsinchu, Taiwan) or BL-1A in the Photon Factory (Tsukuba, Japan). The data were integrated by XDS (38) and scaled by aimless in the CCP4 software package (39). For I-SAD data collection, the crystal was soaked with 0.25 m potassium iodine for 30 min at 293 K. Phenix AutoSol was used for the phase determination (40). In the Mn·PPPi complex, seven iodine sites in the asymmetric unit were located and used for phase determination and improvement, which yielded a traceable electron density map. The initial figure of merit was 0.327. Initial model building was performed by Phenix AutoBuild. Iterative rounds of manual adjustment in Coot and refinement using Phenix.refine were performed for structure refinement. The B-factors of phosphate atoms (α, β, and γ) were 89.21, 88.30, and 99.52, respectively.
Other crystal structures were solved by molecular replacement using Phaser in Phenix. Iodine and Mn·PPPi–bound form structures were used as a search model for the molecular replacement. The structures were refined to the indicated statistics using iterated rounds of manual adjustments in Coot (41, 42), followed by refinement using Phenix (40). Final coordinates and structure factors were submitted to the Protein Data Bank under accession codes 6L7W (manganese complex), 6L7V (Mn·PPPi complex), 6L7Y (C13H13NO2 complex), and 6L7X (C10H14N4O2 complex).
Thermal shift assay
A Library of 965 chemical compounds was assembled in-house from commercially available low-molecular-weight compounds based on the “rule of three” (molecular weight < 300, number of hydrogen bond donors ≤ 3, number of hydrogen bond acceptors ≤ 3, and ClogP ≤ 3), as described (43). Compounds were stored desiccated and were resuspended with DMSO to 100 mm prior to the assay. Thermal shift assay was carried out with 1 μg of TcCet1-(18–243 Δ55–75) and CYPRO Orange fluorescent dye (Thermo Fisher Scientific) (1:1000 dilution) in 96-well plates. The reaction mixture contained 10 mm HEPES (pH 7.5), 2 mm MnCl2, and 1 mm DTT, and 2 mm of each compound. The temperature of the plate was raised from 25 to 70 °C by an increment of 0.5 °C/5 s using the StepOne Plus real-time PCR system (Thermo Fisher Scientific). The resulting melting curves were analyzed by Protein Thermal Shift Software, version 1.1 (Thermo Fisher Scientific) to determine ΔTm.
Preparation of triphosphate terminated RNA
Substrate for RNA triphosphatase assay was prepared by in vitro transcription with T7 RNA polymerase from a partially duplexed oligonucleotide DNA (44). Oligonucleotides corresponding to the T7 promoter sequence (5′-TAATACGACTCACTATA-3′) and the complementary sequence with poly(dT) stretch (5′-TTTTTTTTTTTTTTTTTTTTTATAGTGAGTCGTATTA-3′) were annealed to produce a template for T7 RNA polymerase. The reaction mixture contained 1 mm of [γ-32P]ATP, 20 μm of duplex oligonucleotide DNA, 1 unit/μl of RNase inhibitor (Toyobo Co., Ltd.), and 2.5 units/μl of T7 RNA polymerase (New England Biolabs Inc.) in the provided reaction buffer. The reaction was carried out for 3 h at 37 °C. DNase I (Nippon Gene Co., Ltd.) was then added to digest the template oligonucleotide DNAs, according to the manufacturer's instructions. The product was extracted with phenol/chloroform/isoamyl alcohol, precipitated with ethanol, and separated on 20% denaturing polyacrylamide gel. The radioactive product that migrates between 7 and 9 nucleotides was excised from the gel and eluted by 10 mm Tris-HCl (pH8), 1 mm EDTA. The eluate was passed through a spin column to remove gel debris. The RNA was precipitated with ethanol and resuspended in water.
Triphosphatase assay
Reaction mixtures containing 50 mm Tris-HCl (pH 7.5), 2 mm DTT, 2 mm of divalent cation (MgCl2 or MnCl2) with either [γ-32P]ATP or [γ-32P]pppRNA, and the indicated amount of protein were incubated for 15 min at 30 °C. The reaction was quenched by the addition of EDTA or formic acid. The reaction products were separated by PEI-cellulose TLC with 0.45 m ammonium sulfate. The TLC plate was exposed to a PhosphorImager plate, scanned by BAS-2000 (FujiFilm, Japan), and quantitated by Image Gauge software.
Data availability
The atomic coordinates and structure factors have been deposited into the Protein Data Bank as entries 6L7V, 6L7W, 6L7X, and 6L7Y. All other information and data are available from the authors upon request.
Supplementary Material
Acknowledgments
We thank Osamu Tani from the National Institute of Advanced Industrial Science and Technology for performing thermal shift assay, Yanchang Liu and Moe Goto from University of Tsukuba for help with experiments, the beamline staffs of the Photon Factory (Tsukuba, Japan) and National Synchrotron Radiation Research Center (Shinchu, Taiwan) for X-ray crystallographic analysis, Dr. Toshiya Senda from the High Energy Accelerator Research Organization and Dr. Hitoshi Sakashita from the National Institute of Advanced Industrial Science and Technology for valuable discussion. This research was partially supported by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP20am0101071.
This article contains supporting information.
Author contributions—Y. T., N. K., K. F., and C. K. H. conceptualization; Y. T., N. K., and T. T. D. data curation; Y. T., N. K., T. T. D., and C. K. H. formal analysis; Y. T. and C. K. H. funding acquisition; Y. T., N. K., and C. K. H. investigation; Y. T., N. K., and C. K. H. methodology; Y. T., N. K., and C. K. H. writing-original draft; Y. T. and C. K. H. project administration; N. K. and C. K. H. resources; N. K. software; N. K. and T. T. D. validation; K. F. and C. K. H. writing-review and editing; C. K. H. supervision.
Funding and additional information—This material is based upon work supported by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research KAKENHI 16H05180 (to C. K. H.) and 18K15141 (to Y. T.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TTM
- triphosphate tunnel metalloenzyme
- pppRNA
- triphosphate-terminated RNA
- PPPi
- tripolyphosphate
- Ni–NTA
- nickel–nitrilotriacetic acid.
References
- 1. McCall L.-I., and McKerrow J. H. (2014) Determinants of disease phenotype in trypanosomatid parasites. Trends Parasitol. 30, 342–349 10.1016/j.pt.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 2. Field M. C., Horn D., Fairlamb A. H., Ferguson M. A., Gray D. W., Read K. D., De Rycker M., Torrie L. S., Wyatt P. G., Wyllie S., and Gilbert I. H. (2017) Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nat. Rev. Microbiol. 15, 217–231 10.1038/nrmicro.2016.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shuman S. (2001) The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb. Symp. Quant. Biol. 66, 301–312 10.1101/sqb.2001.66.301 [DOI] [PubMed] [Google Scholar]
- 4. Shuman S. (2002) What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3, 619–625 10.1038/nrm880 [DOI] [PubMed] [Google Scholar]
- 5. Takagi T., Moore C. R., Diehn F., and Buratowski S. (1997) An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell 89, 867–873 10.1016/S0092-8674(00)80272-X [DOI] [PubMed] [Google Scholar]
- 6. Changela A., Ho C. K., Martins A., Shuman S., and Mondragón A. (2001) Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 20, 2575–2586 10.1093/emboj/20.10.2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho C. K., Pei Y., and Shuman S. (1998) Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J. Biol. Chem. 273, 34151–34156 10.1074/jbc.273.51.34151 [DOI] [PubMed] [Google Scholar]
- 8. Gross C. H., and Shuman S. (1998) RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J. Virol. 72, 10020–10028 10.1128/JVI.72.12.10020-10028.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pei Y., Lehman K., Tian L., and Shuman S. (2000) Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 28, 1885–1892 10.1093/nar/28.9.1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Massayuki Kikuti C., Tersariol I. L., and Schenkman S. (2006) Divalent metal requirements for catalysis and stability of the RNA triphosphatase from Trypanosoma cruzi. Mol Biochem. Parasitol. 150, 83–95 10.1016/j.molbiopara.2006.06.012 [DOI] [PubMed] [Google Scholar]
- 11. Ho C. K., and Shuman S. (2001) Trypanosoma brucei RNA triphosphatase: antiprotozoal drug target and guide to eukaryotic phylogeny. J. Biol. Chem. 276, 46182–46186 10.1074/jbc.M108706200 [DOI] [PubMed] [Google Scholar]
- 12. Ho C. K., Gong C., and Shuman S. (2001) RNA triphosphatase component of the mRNA capping apparatus of Paramecium bursaria Chlorella virus 1. J. Virol. 75, 1744–1750 10.1128/JVI.75.4.1744-1750.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hausmann S., Vivarès C. P., and Shuman S. (2002) Characterization of the mRNA capping apparatus of the microsporidian parasite Encephalitozoon cuniculi. J. Biol. Chem. 277, 96–103 10.1074/jbc.M109649200 [DOI] [PubMed] [Google Scholar]
- 14. Ho C. K., and Shuman S. (2001) A yeast-like mRNA capping apparatus in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 98, 3050–3055 10.1073/pnas.061636198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kyrieleis O. J., Chang J., de la Peña M., Shuman S., and Cusack S. (2014) Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure. 22, 452–465 10.1016/j.str.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benarroch D., Smith P., and Shuman S. (2008) Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure 16, 501–512 10.1016/j.str.2008.01.009 [DOI] [PubMed] [Google Scholar]
- 17. Hausmann S., Altura M. A., Witmer M., Singer S. M., Elmendorf H. G., and Shuman S. (2005) Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1). J. Biol. Chem. 280, 12077–12086 10.1074/jbc.M412063200 [DOI] [PubMed] [Google Scholar]
- 18. Lima C. D., Wang L. K., and Shuman S. (1999) Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99, 533–543 10.1016/S0092-8674(00)81541-X [DOI] [PubMed] [Google Scholar]
- 19. Pei Y., Ho C. K., Schwer B., and Shuman S. (1999) Mutational analyses of yeast RNA triphosphatases highlight a common mechanism of metal-dependent NTP hydrolysis and a means of targeting enzymes to pre-mRNAs in vivo by fusion to the guanylyltransferase component of the capping apparatus. J. Biol. Chem. 274, 28865–28874 10.1074/jbc.274.41.28865 [DOI] [PubMed] [Google Scholar]
- 20. Bisaillon M., and Shuman S. (2001) Functional groups required for the stability of yeast RNA triphosphatase in vitro and in vivo. J. Biol. Chem. 276, 30514–30520 10.1074/jbc.M104936200 [DOI] [PubMed] [Google Scholar]
- 21. Bisaillon M., and Shuman S. (2001) Structure–function analysis of the active site tunnel of yeast RNA triphosphatase. J. Biol. Chem. 276, 17261–17266 10.1074/jbc.M100980200 [DOI] [PubMed] [Google Scholar]
- 22. Jain R., and Shuman S. (2008) Polyphosphatase activity of CthTTM, a bacterial triphosphate tunnel metalloenzyme. J. Biol. Chem. 283, 31047–31057 10.1074/jbc.M805392200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bettendorff L., and Wins P. (2013) Thiamine triphosphatase and the CYTH superfamily of proteins. FEBS J. 280, 6443–6455 10.1111/febs.12498 [DOI] [PubMed] [Google Scholar]
- 24. Martinez J., Truffault V., and Hothorn M. (2015) Structural determinants for substrate binding and catalysis in triphosphate tunnel metalloenzymes. J. Biol. Chem. 290, 23348–23360 10.1074/jbc.M115.674473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gu M., Rajashankar K. R., and Lima C. D. (2010) Structure of the Saccharomyces cerevisiae Cet1–Ceg1 mRNA capping apparatus. Structure 18, 216–227 10.1016/j.str.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ho C. K., Lehman K., and Shuman S. (1999) An essential surface motif (WAQKW) of yeast RNA triphosphatase mediates formation of the mRNA capping enzyme complex with RNA guanylyltransferase. Nucleic Acids Res. 27, 4671–4678 10.1093/nar/27.24.4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lehman K., Schwer B., Ho C. K., Rouzankina I., and Shuman S. (1999) A conserved domain of yeast RNA triphosphatase flanking the catalytic core regulates self-association and interaction with the guanylyltransferase component of the mRNA capping apparatus. J. Biol. Chem. 274, 22668–22678 10.1074/jbc.274.32.22668 [DOI] [PubMed] [Google Scholar]
- 28. Smith P., Ho C. K., Takagi Y., Djaballah H., and Shuman S. (2016) Nanomolar inhibitors of Trypanosoma brucei RNA triphosphatase. MBio. 7, e00058–16 10.1128/mBio.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Antczak C., Shum D., Radu C., Seshan V. E., and Djaballah H. (2009) Development and validation of a high-density fluorescence polarization-based assay for the trypanosoma RNA triphosphatase TbCet1. Comb. Chem. High Throughput Screen. 12, 258–268 10.2174/138620709787581729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gong C., Martins A., and Shuman S. (2003) Structure–function analysis of Trypanosoma brucei RNA triphosphatase and evidence for a two-metal mechanism. J. Biol. Chem. 278, 50843–50852 10.1074/jbc.M309188200 [DOI] [PubMed] [Google Scholar]
- 31. Gong C., and Shuman S. (2002) Chlorella virus RNA triphosphatase: mutational analysis and mechanism of inhibition by tripolyphosphate. J. Biol. Chem. 277, 15317–15324 10.1074/jbc.M200532200 [DOI] [PubMed] [Google Scholar]
- 32. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 33. Moeder W., Garcia-Petit C., Ung H., Fucile G., Samuel M. A., Christendat D., and Yoshioka K. (2013) Crystal structure and biochemical analyses reveal that the Arabidopsis triphosphate tunnel metalloenzyme AtTTM3 is a tripolyphosphatase involved in root development. Plant J. 76, 615–626 10.1111/tpj.12325 [DOI] [PubMed] [Google Scholar]
- 34. Gallagher D. T., Kim S.-K., Robinson H., and Reddy P. T. (2011) Active-site structure of class IV adenylyl cyclase and transphyletic mechanism. J. Mol. Biol. 405, 787–803 10.1016/j.jmb.2010.11.026 [DOI] [PubMed] [Google Scholar]
- 35. Martins A., and Shuman S. (2003) Mapping the triphosphatase active site of baculovirus mRNA capping enzyme LEF4 and evidence for a two-metal mechanism. Nucleic Acids Res. 31, 1455–1463 10.1093/nar/gkg244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takase Y., Takagi T., Komarnitsky P. B., and Buratowski S. (2000) The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol. Cell Biol. 20, 9307–9316 10.1128/MCB.20.24.9307-9316.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martinez-Rucobo F. W., Kohler R., van de Waterbeemd M., Heck A. J., Hemann M., Herzog F., Stark H., and Cramer P. (2015) Molecular basis of transcription-coupled pre-mRNA capping. Mol. Cell 58, 1079–1089 10.1016/j.molcel.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 38. Kabsch W. (2010) XDS. Acta Crystallogr. D 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 40. Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., et al. (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 43. Congreve M., Carr R., Murray C., and Jhoti H. (2003) A “rule of three” for fragment-based lead discovery? Drug Discov. Today 8, 876–877 10.1016/S1359-6446(03)02831-9 [DOI] [PubMed] [Google Scholar]
- 44. Ogino T., and Banerjee A. K. (2007) Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97 10.1016/j.molcel.2006.11.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited into the Protein Data Bank as entries 6L7V, 6L7W, 6L7X, and 6L7Y. All other information and data are available from the authors upon request.