Abstract
Skp1, a subunit of E3 Skp1/Cullin-1/F-box protein ubiquitin ligases, is modified by a prolyl hydroxylase that mediates O2 regulation of the social amoeba Dictyostelium and the parasite Toxoplasma gondii. The full effect of hydroxylation requires modification of the hydroxyproline by a pentasaccharide that, in Dictyostelium, influences Skp1 structure to favor assembly of Skp1/F-box protein subcomplexes. In Toxoplasma, the presence of a contrasting penultimate sugar assembled by a different glycosyltransferase enables testing of the conformational control model. To define the final sugar and its linkage, here we identified the glycosyltransferase that completes the glycan and found that it is closely related to glycogenin, an enzyme that may prime glycogen synthesis in yeast and animals. However, the Toxoplasma enzyme catalyzes formation of a Galα1,3Glcα linkage rather than the Glcα1,4Glcα linkage formed by glycogenin. Kinetic and crystallographic experiments showed that the glycosyltransferase Gat1 is specific for Skp1 in Toxoplasma and also in another protist, the crop pathogen Pythium ultimum. The fifth sugar is important for glycan function as indicated by the slow-growth phenotype of gat1Δ parasites. Computational analyses indicated that, despite the sequence difference, the Toxoplasma glycan still assumes an ordered conformation that controls Skp1 structure and revealed the importance of nonpolar packing interactions of the fifth sugar. The substitution of glycosyltransferases in Toxoplasma and Pythium by an unrelated bifunctional enzyme that assembles a distinct but structurally compatible glycan in Dictyostelium is a remarkable case of convergent evolution, which emphasizes the importance of the terminal α-galactose and establishes the phylogenetic breadth of Skp1 glycoregulation.
Keywords: s: X-ray crystallography, NMR, molecular dynamics simulation, Skp1, SCF, glycogenin, cytoplasmic glycosylation, post-translational modification, Toxoplasma, Pythium, X-ray crystallography, nuclear magnetic resonance (NMR), molecular dynamics, glycosyltransferase, Toxoplasma gondii, E3 ubiquitin ligase, post-translational modification (PTM)
Introduction
A prominent mechanism of O2 sensing in metazoa involves an O2-dependent prolyl 4-hydroxylase (PHD2) that generates degrons on hypoxia-inducible factor-α (HIFα) that are recognized by the von Hippel Lindau subunit of the E3(VBC) ubiquitin ligase, leading to its polyubiquitination and degradation in the 26S proteasome (1). Thus, low O2 inhibits PHD2 and stabilizes HIFα to dimerize with HIFβ and transcriptionally activate genes appropriate to respond to low O2 (2). Protists also have a PHD2-like gene that encodes the evolutionary predecessor and likely ortholog of PHD2 (3). However, protists lack HIFα, and protist PhyA enzymes instead hydroxylate Skp1, a subunit of the Skp1/Cullin-1/F-box protein (SCF) family of E3 ubiquitin ligases (4) that are related to the E3(VBC) ubiquitin ligases. In the social amoeba Dictyostelium discoideum, Skp1 prolyl hydroxylation is involved in mediating the O2 checkpoint for fruiting body formation (5). Thus, both prolyl hydroxylases contribute to regulation of the proteome, but transcriptionally in animals and likely via degradation in protists.
The mechanism by which Skp1 hydroxylation contributes to O2 sensing has been examined in D. discoideum (6). Remarkably, the hydroxyproline (Hyp) residue is sequentially modified by a series of five glycosyltransferase (GT) reactions leading to the assembly of a linear pentasaccharide of recently defined structure (7). Genetic studies reveal a strong Skp1-dependent role for the GTs in O2 sensing (8), and radiotracer and biochemical complementation studies indicate that Skp1 is the sole target of these GTs. Skp1 serves as an adaptor that links the F-box protein (FBP) and Cullin-1 subunits of the SCF complex through mostly independent binding events. Interactome studies indicate that glycosylation promotes accumulation of Skp1/FBP subcomplexes in vitro and in vivo (9, 10), thereby potentially activating the respective SCF complexes and contributing to the degradation of all of the substrates that are recognized by the cellular repertoire of FBPs. Current evidence indicates that the glycan assumes a relatively rigid structure that organizes the intrinsically disordered subsite-2 region involved in FBP recognition. By increasing the fraction of time that the C-terminal region is folded into α-helix-8 and extended to form subsite-2, the glycan is thought to promote the interaction of Skp1 with FBPs in the cell (7). However, the importance of the exact glycan sequence and the significance of the terminal (fifth) sugar deserve further attention because there is limited precedent for glycan-imposed order on local protein structure (6) and because the information will provide needed new approaches to probe its role in cellular O2 sensing.
Toxoplasma gondii is an apicomplexan parasite that latently infects a sizable fraction of many human populations (11). Although chronic infections are usually clinically benign, reactivation, as can occur in immunosuppressed individuals, can lead to serious diseases of the central nervous system and other organs. Moreover, human fetuses are subject to serious birth defects in the case of acute maternal infection. Previous studies documented that the T. gondii prolyl hydroxylase (phyA) can complement a disruption of phyA in D. discoideum and contribute to O2-dependent growth of T. gondii in a fibroblast monolayer growth assay (12). Furthermore, coding sequences for the first three GT activities are present, although, surprisingly, T. gondii still assembles a pentasaccharide glycan on its Skp1 (13). This led to the discovery of a novel enzyme that assembles the fourth sugar (14) and, as reported here, another enzyme that mediates addition of the final sugar. Analytical studies revealed that the fourth sugar is different from that of D. discoideum, but the fifth has remained unknown. These findings have raised important questions, including why are the GTs and glycans different between these two protists, what constrains their evolution, and are they compatible with the conformational regulation model posited for D. discoideum?
Pythium ultimum is an oomycete plant pathogen that resides in the stramenopile branch of the larger TSAR group of protozoans to which the apicomplexans also belong. P. ultimum is the agent for root rot disease in agriculturally important crops (15, 16) and is related to P. insidiosum, the agent for debilitating pythiosus in humans and other mammals (17). We recently showed P. ultimum PhyA and the first GT Gnt1 constitute a bifunctional protein that is active toward Skp1A but not the second Skp1B encoded by its genome (18). Thus O2-dependent post-translational regulation of Skp1 appears to be widespread, and P. ultimum and protists from other lineages have evidently evolved a second Skp1 that avoids O2-dependent glycoregulation. We show here that the terminal glycosylation of P. ultimum Skp1 is like that of T. gondii rather than that of D. discoideum.
By identifying the Gat1 GT that mediates the addition of the final sugar to the T. gondii, we have been able to determine the complete glycan sequence and to show that Gat1 is specific for Skp1, regulates Skp1, and contributes to optimal growth in cell culture. Despite a distinct structure, the glycan mediates a conserved conformational effect on Skp1. We propose that the selection pressure for a 5-sugar chain was so strong that, in lineages where the Gat1 progenitor was evolving to acquire a new function, the later-evolving amoebozoa acquired a novel GT mechanism to ensure assembly of a glycan that was structurally similar enough to still conformationally control Skp1.
Results
The fifth and final Skp1 sugar is an α-linked Gal and depends on TgGat1
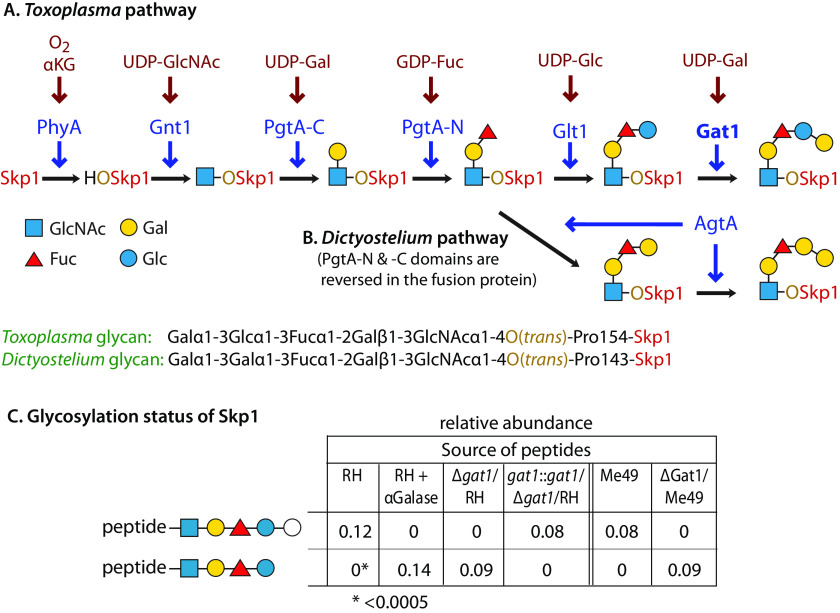
Previous studies described the mechanism of assembly of the first four sugars on TgSkp1-Pro-154 (Fig. 1A), but the left the identity of the final sugar (other than its being a hexose) unresolved (13, 14). The corresponding sugar in Dictyostelium (Fig. 1B) is a 3-linked αGal that is susceptible to removal with green coffee bean α-galactosidase. Similar treatment of tryptic peptides from TgSkp1, isolated from the standard type 1 strain RH by immunoprecipitation, resulted in complete conversion of the pentasaccharide form of the glycopeptide to the tetrasaccharide form, indicating that the terminal Hex is an αGal residue (Fig. 1C).
Figure 1.
Gat1 is required for terminal α-galactosylation of Skp1 in parasites. A and B, schematic of Skp1 glycosylation pathway in Toxoplasma (14) (this work) and Dictyostelium (3), which modifies a single hydroxyproline associated with its F-box–binding region, depicted using CFG glycan symbols (19). The identity of Gat1 as the final enzyme in the Toxoplasma pathway and the nature of the final sugar are reported here. C, dependence of the terminal sugar on Gat1 and its characterization. Skp1 was immunoprecipitated from type 1 RH and type 2 ME49 strains and their genetic derivatives, and peptides generated by trypsinization were analyzed by nano-LC-MS. In addition, Skp1 from RH was treated with green coffee bean α-gal after trypsinization. The values represent the levels of pentasaccharide-peptide and tetrasaccharide-peptide levels detected, after normalization to all detected modification states of the peptide; note that the values have only partial relative meaning because of the low and varied detection efficiency of glycopeptides. As indicated, the detection threshold was <0.0005. The open circle for the terminal sugar of the pentasaccharide indicates that it was only known as a hexose at the time of the experiment. See Table S2 and Fig. S5 for primary data. Similar results were obtained in independent samples from RH and Δgat1/RH (not shown).
Genomics studies predict the existence of four cytoplasmically localized GTs whose functions are not assigned (20), and one of these, referred to as Gat1, was predicted to be the missing Skp1 GT on account of the phylogenetic co-distribution of its gene in protists that possess Glt1-like genes (14). To test the dependence of Skp1 glycosylation on Tggat1, the gene was disrupted using CRISPR/Cas9 in the RH strain, yielding gat1Δ as described in Fig. S4A. PCR studies confirmed replacement with a dhfr cassette, and enzyme assays (see below) showed a loss of enzyme activity. The resulting clone produced a version of Skp1 in which the 5-sugar glycopeptide was no longer detectable, but ions corresponding to the 4-sugar glycopeptide were detected at similar abundance (Fig. 1C). Similar results were obtained in the type 2 ME49 strain (Fig. S4A and Fig. 1C) and in strain RHΔΔ in which gat1 was disrupted by homologous recombination using a different selection marker, referred to as Δgat1-1 (Table 1 and Fig. S3B; data not shown). The similar results obtained by different genetic methods in distinct genetic backgrounds indicate that Tggat1 is required for the addition of the terminal sugar, but whether this was a direct or indirect effect was unclear.
Table 1.
Strains employed in this study
| Description | Name | Parental strain | Genotype | Gene targeted | Selection marker | Reference |
|---|---|---|---|---|---|---|
| RH (type 1) | WT | 21 | ||||
| Δgat1/RH | MM12, cl.A8 | RH | Δgat1 | gat1 | DHFR | This report |
| gat1+/Δgat1/RH | MM21, cl.E12 | MM21 | Δuprt/gat1::gat1-Ty | uprt | Δuprt | This report |
| RHΔΔ | RH | Δku80/Δhxgprt | 22 | |||
| Δgat1-1/RHΔΔ | KR10, cl.A1 | RHΔΔ | Δku80/Δgat1 | gat1 | HXGPRT | This report |
| Δgat1-2/RHΔΔ | MM16, cl.B7 | RHΔΔ | Δku80/Δhxgprt/Δgat1 | gat1 | DHFR | This report |
| gat1+/Δgat1-2/ RHΔΔ | MM24, cl.G10 | MM16 | Δku80/Δhxgprt/Δuprt/tub::gat1-3×HA | uprt | Δuprt | This report |
| Me49-RFP (type 2) | MM8, cl.A10a | Me49 | rfp+ | 23 | ||
| Δgat1/Me49-RFP | MM14, cl.B5 | Me49-RFP | Δgat1 | gat1 | DHFR | This report |
a High RFP expression level clone from original source.
Toxoplasma growth depends partially on gat1
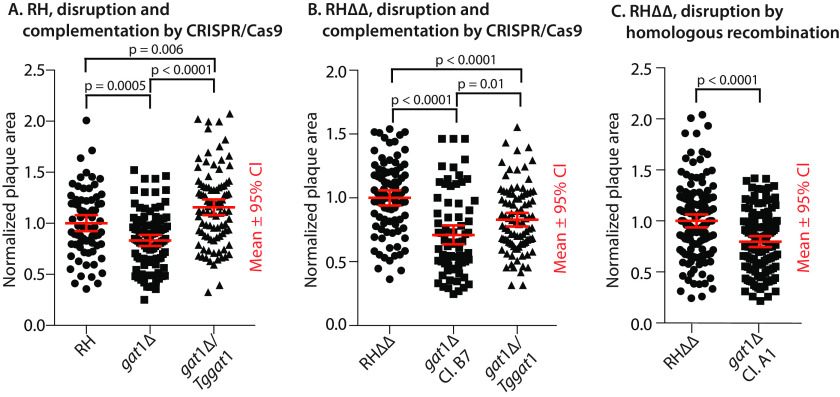
Parasites require invasion of mammalian host cells to establish a niche within an intracellular parasitophorous vacuole to proliferate (24, 25). In two-dimensional fibroblast monolayers, parasites lyse out and invade neighboring cells to repeat the cycle, resulting in plaques whose area is a measure of efficiency of these cellular processes. Past studies showed that plaque size growth is compromised by mutational blockade of Skp1 hydroxylation and earlier steps of the glycosylation pathway (12–14). Similarly, disruption of gat1 also resulted in modestly smaller plaques in both RH or RHΔΔ backgrounds (Fig. 2, A and B) and in RHΔΔ in which gat1 was disrupted using homologous recombination without CRISPR-Cas9 (Fig. 2C). To determine whether the effects were specific to the genetic lesion at the gat1 locus, the RH and RHΔΔ KO strains generated using CRISPR/Cas9 were modified again by CRISPR/Cas9 to introduce single copies of epitope-tagged versions of the gat1 coding locus, downstream of an endogenous gat1 promoter cassette or a tubulin promoter cassette, respectively, into the uprt locus. The expected insertions were confirmed using PCR (Figs. S3C and S4B). As a result, TgGat1-3×HA could be detected by Western blotting of tachyzoites in the RHΔΔ background (Fig. S4D), and, as discussed below, the complemented RHΔΔ strain restored Skp1 glycosylation according to a biochemical complementation test. Although TgGat1-Ty expressed under its own promoter cassette in the RH background was not detected (not shown), enzyme activity was partially restored in the TgGat1-Ty strain (Fig. S4C). Both strains exhibited larger plaque sizes than their respective KO parents (Fig. 2, A and B), confirming that the effect on growth was due to the original loss of Gat1.
Figure 2.
Parasite growth depends on Gat1. Parasites were plated at clonal density on two-dimensional monolayers of HFFs and allowed to invade, proliferate, lyse out, and reinfect neighboring fibroblasts. After 5.5 days, cultures were fixed, stained, and analyzed for the areas occupied by lysed fibroblasts (plaques). Data from each of three independent trials, which were each normalized to the parental strain, were merged for presentation. A, comparison of the type 1 RH strain before and after gat1 replacement using CRISPR/Cas9 and complementation with gat1 under control of its own promoter cassette in the uprt locus. B, comparison of RHΔΔ, gat1-2Δ/RHΔΔ, and the latter complemented with gat1 under control of a tubulin promoter in the uprt locus. C, comparison of gat1-1Δ/RHΔΔ, prepared by homologous recombination. Significance of differences in plaque areas between parasite strains was assessed by Student's t test. CI, confidence interval.
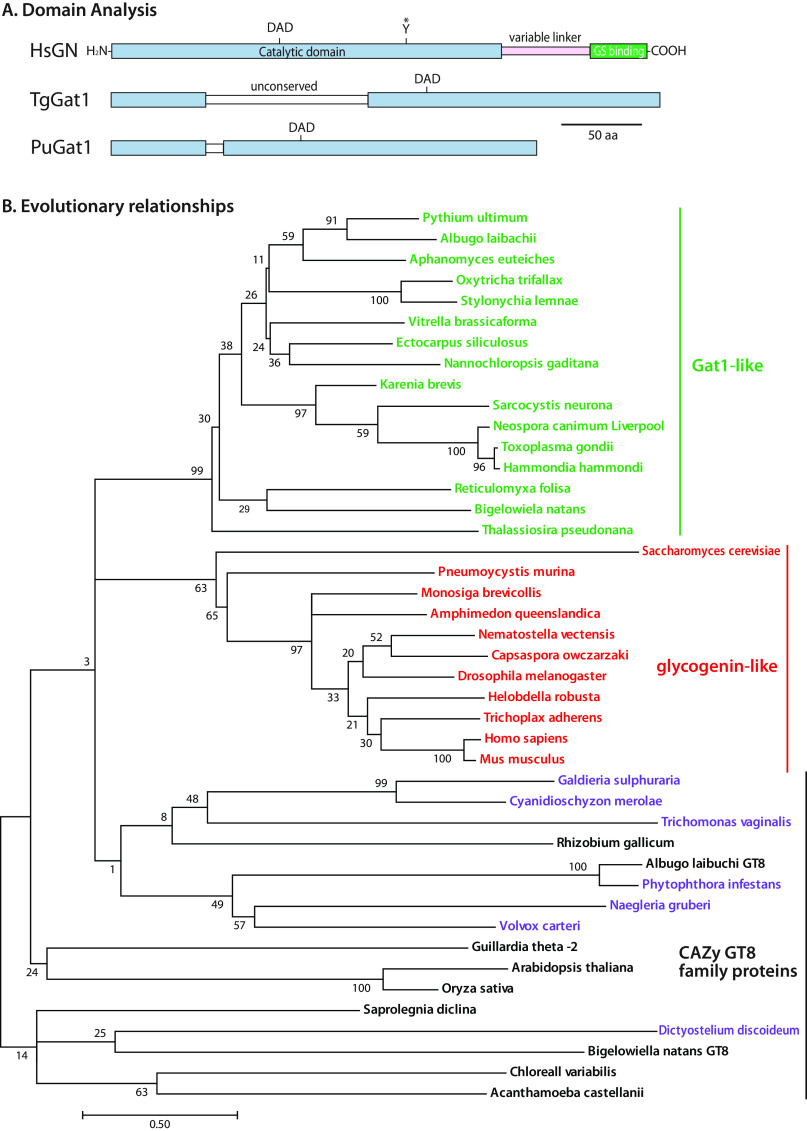
Gat1 is closely related to glycogenin
An evolutionary analysis was conducted to gain further insight into the function of TgGat1. Based on searches of genomic databases using BLASTP, TgGat1 belongs to the CAZy GT8 family. The top-scoring hits, with Expect values of <10−32, were found only in protists that contain Toxoplasma PgtA-like and Glt1-like sequences (Fig. S7) and lack D. discoideum AgtA-like sequences, suggesting a common function. The most similar sequences, in searches seeded with the putative catalytic domain, belong to glycogenin, with Expect values of ≥E−27. All other homologous sequences had Expect values of ≥10−22. Glycogenin is a dimeric α4-glucosyltransferase that can prime the synthesis of glycogen in the cytoplasm of yeast and animals by a mechanism that involves autoglucosylation. Glycogenin appears not to be involved in starch formation and is an evolutionarily recent addition to the glycogen biosynthesis pathways that occur in the absence of glycogenin activity in bacteria and many unicellular eukaryotes (26). The cyst-forming stage of T. gondii accumulates crystalline amylopectin (27), an α1,4Glc polymer with α1,6-linked branches that resembles glycogen, in its cytoplasmic compartment. T. gondii amylopectin is assembled by a UDP-Glc–based metabolism that is related to the floridean starch of the red alga Cyanidioschyzon merolae and, to a lesser extent, to that of glycogen storing animals and fungi. Homologs of glycogen synthase (27) and glycogen phosphorylase (28) regulate the accumulation of amylopectin in this parasite, and TgGat1 has been annotated as a glycogenin (28, 29). Related genes have been implicated in promoting starch formation in red algae (30, 31). Thus, Gat1 might have a function in regulating T. gondii amylopectin formation.
Glycogenin consists of a CAZy GT8 family catalytic domain plus a C-terminal glycogen synthase–binding domain separated by a linker, whereas Gat1 consists only of a single catalytic domain (Fig. 3A). TgGat1 is predicted to be a 345-amino acid protein encoded by a single exon gene in the Type I GT1 strain (TGGT1_310400). It is 34% identical to rabbit (Oryctolagus cuniculus) glycogenin over the catalytic domain but includes a poorly conserved 90-amino acid sequence that interrupts the catalytic domain (Fig. 3A). This region is likely unstructured based on secondary structure prediction by the XtalPred server (32). The Gat1-like sequence from P. ultimum (Uniprot K3WC47) lacks this sequence, so it was analyzed for comparison. PuGat1 is predicted to be a 266-amino acid protein encoded by a two-exon gene, annotated as PYU1_G002535-201 (transcript ID PYU1_T002538).
Figure 3.
Gat1 is closely related to glycogenin sequences. A, domain analysis of Gat1 from T. gondii and P. ultimum compared with human glycogenin-1. B, the evolutionary history of the sequence of the Gat1 catalytic domain was inferred by using a maximum likelihood method. The tree with the highest log likelihood (−13,279.56) is shown. Gat1 and Gat1-like sequences are colored green, glycogenin and glycogenin-like sequences are in red, and characterized and other selected other CAZy GT8 sequences are in black, or in purple if predicted to reside in the secretory pathway rather than the cytoplasm. The percentage of trees in which the associated taxa clustered together is shown at each branch. Branch lengths are measured by the number of substitutions per site. See Figs. S6–S8 for alignments.
To further evaluate the evolutionary relationship of these putative GTs, their catalytic domains and those of the most closely related or known sequences from the CAZy GT8 family (Fig. S7) were aligned (Fig. S8) and analyzed by a maximum likelihood method (Fig. 3B). The results suggest that Gat1 and glycogenin evolved separately from a common ancestor. Although the last common ancestor was not resolved, Gat1 was potentially the predecessor to glycogenin owing to its presence in other primitive unicellular eukaryotes that bear no evidence of glycogenin-like sequences and because there is currently no evidence for the existence of close homologs of Gat1 and glycogenin within the same clade that would suggest that they evolved as paralogs of a gene duplication. However, the possibility that either one or the other product of an ancestral gene duplication was always lost in every extant derivative cannot be excluded. Gat1 and glycogenin each possess unique conserved sequence motifs (Fig. S6) that potentially support functional differences.
Gat1 is a terminal Skp1 α-galactosyltransferase
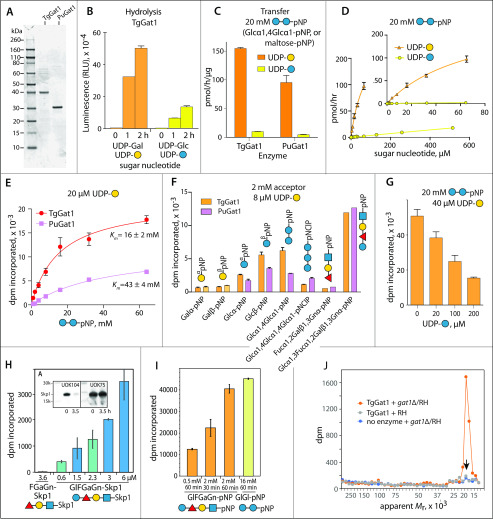
To determine whether TgGat1 can directly modify Skp1, the predicted full-length protein was expressed as a His6-tagged conjugate in Escherichia coli, purified on a TALON resin, and treated with TEV protease, leaving an N-terminal GlyHis-dipeptide stub before the start Met (Fig. 4A). The presumptive ortholog from P. ultimum was prepared similarly. A screen for UDP-sugar hydrolysis activity of TgGat1 yielded, after extended reaction times, only UDP-Gal and UDP-Glc as candidate substrates from a panel of six common UDP-sugars (Fig. S9A). A quantitative comparison showed ∼7-fold greater activity toward UDP-Gal than UDP-Glc (Fig. 4B).
Figure 4.
Gat1 preferentially galactosylates the Skp1 glycan in vitro. A, recombinantly expressed and purified preparations of TgGat1 and PuGat1 were analyzed by SDS-PAGE and staining with Coomassie blue. B, temporal dependence of UDP-Gal and UDP-Glc hydrolysis. The averages and S.D. of three technical replicates are shown. A similar profile was observed with a different enzyme concentration. See Fig. S9E for a trial with higher enzyme concentrations. C, transferase activity utilizing 8 μm UDP-Gal or UDP-Glc toward 20 mm Glcα1,4Glcα-pNP (maltose-pNP) for TgGat1 and PuGat1. The averages and S.D. of two technical replicates are shown; similar profiles were in two independent assays with a different TgGat1 preparation. D, UDP-Gal and UDP-Glc concentration dependence of TgGat1 transferase activity toward 20 mm maltose-pNP. The averages and S.D. of two technical replicates are shown, and an independent trial with TgGat1 and PuGat1 against UDP-Gal is shown in Fig. S9F. E, maltose-pNP concentration dependence of TgGat1 and PuGat1 transferase activity from 20 μm UDP-Gal. The averages and S.D. of two technical replicates are shown. F, relative Gal-transferase activity of TgGat1 and PuGat1 toward different acceptors. The averages and S.D. of three technical replicates are shown. Similar results were obtained in three independent trials. G, effect of UDP-Glc concentration on the Gal-transferase activity of TgGat1. Reactions were incubated for 1 h. The averages and S.D. of two technical replicates are shown. H, Gal-transferase activity of TgGaT1 toward varied concentrations of GlFGaGn-Skp1, in the presence of 40 μm UDP-Gal (1 µCi) after a 1-h incubation. Data from independent preparations of TgSkp1 are colored in different shades. FGaGn-Skp1 is included for comparison. Error bars, S.D. of duplicate measurements. Inset, Western blots of the Skp1 preparations used, where FGaGn-Skp1, which is recognized specifically by pAb UOK104, is largely converted in a 3.5-h reaction using Glt1 and UDP-Glc to GlFGaGn-Skp1, which is recognized only by the pan-specific pAb UOK75. I, reactions with synthetic oligosaccharides conjugated to pNP were conducted in parallel using the same conditions. J, biochemical complementation to detect Gat1 substrates. Desalted S100 extracts of RH and gat1Δ/RH were reacted with recombinant Gat1 in the presence of UDP-[3H]Gal, and the product of the reaction was separated on an SDS-polyacrylamide gel, which was sliced into 40 bands for liquid scintillation counting. The migration position of Skp1 is marked with an arrow. See Fig. S9 (H and I) for trials using different strains.
The ability of Gat1 to transfer Gal or Glc to another sugar, rather than water, was tested using a substrate analog for glycogenin, Glcα1,4Glcα1-pNP (maltose-pNP), which mimics the terminal disaccharide of glycogen and starch and has a terminal αGlc, as found on the Skp1 tetrasaccharide. Although Gat1 from either T. gondii or P. ultimum could modify maltose-pNP using either sugar nucleotide (Fig. 4C), the enzymes strongly preferred UDP-Gal (Fig. 4D). Furthermore, TgGat1 activity was not saturated by UDP-Glc at 0.5 mm (Fig. 4D), whereas TgGat1 and PuGat1 exhibited apparent Km values for UDP-Gal in the range of 30-15 μm (Fig. S9F) These values were greater than the 2-4 μm values reported for rabbit and yeast glycogenins for UDP-Glc (33, 34). TgGat1 and PuGat1 exhibited higher apparent Km values for maltose-pNP in the range of 16-43 mm (Fig. 4D), which were greater than the 4 mm value reported for rabbit glycogenin (35).
Extending the acceptor to three sugars or decreasing it to one resulted in less activity, but either anomer of Glc-pNP was acceptable (Fig. 4F). A similar pattern was observed for the P. ultimum and T. gondii enzymes. The enzymes were specific for terminal Glc acceptors as activity toward Gal-pNP was not detected. In comparison, pNP-Skp1 tetrasaccharide (GlFGaGn-pNP), which mimics the natural acceptor on Skp1, was a superior acceptor substrate (Fig. 4F) with a Km of 1.5 mm (Fig. S9G), and the truncated trisaccharide FGaGn-pNP was inactive, indicating that Glc was the position of attachment. Thus, the Gat1 enzymes preferred their native tetrasaccharide acceptor substrate and UDP-Gal as a donor but tolerated, with low efficiency, the preferred substrates of glycogenin, UDP-Glc, and α4-linked oligomers Glc.
The importance of Skp1 as context for the acceptor glycan was examined using GlFGaGn-Skp1, which was prepared by reaction of FGaGn-Skp1 with UDP-Glc and Glt1, resulting in loss of the trisaccharide epitope (Fig. 4H, inset). In a comparison of acceptor concentration dependence, TgGat1 was about 33 times more active toward GlFGaGn-Skp1 than free GlFGaGn-pNP (Fig. 4, H and I), based on ∼33 times less activity (dpm incorporated) at 0.001 times the substrate concentration (substrate concentrations were both in the linear response range). The reaction with GlFGaGn-Skp1 did not approach saturation at the highest concentration tested, 6 μm. Thus, the TgGat1 reaction was much more efficient when the tetrasaccharide was associated with its native substrate Skp1 and thus consistent with Gat1 being directly responsible for modifying Skp1 in the cell.
A characteristic of glycogenin is its ability to modify the HO group of a Tyr side chain near its active site with αGlc and then to repeatedly modify the 4-position of the Glc with another αGlc and repeat the process up to 8-12 sugars. Thus, when isolated as a recombinant protein expressed in UDP-Glc–positive E. coli, glycogenin is partly glucosylated (36). However, neither TgGat1 nor PuGat1 prepared in a similar manner were found to be glycosylated, based on an exact mass measurement using nLC/MS (Fig. S10, A–E). Furthermore, following incubation with either UDP-Gal or UDP-Glc, no change in SDS-PAGE mobility (Fig. S10D) or exact mass were observed (Fig. S10E). Thus, no evidence for autoglycosylation activity of Gat1 from either species could be detected.
Skp1 is the only detectable substrate of Gat1 in parasite extracts
GlFGaGn-Skp1 is a substrate for Gat1, but are there others? This was addressed by complementing extracts of gat1Δ parasites with recombinant TgGat1 in the presence of UDP-[3H]Gal and measuring incorporation of 3H after display of the proteome on a one-dimensional SDS-polyacrylamide gel. A high level of incorporation of 3H that depended on the addition of enzyme was observed at the position of Skp1 (Fig. 4J), as expected, but negligible dpm were detected elsewhere in the gel. Furthermore, negligible dpm were incorporated into Skp1 in RH parental cells, indicating that little GlFGaGn-Skp1 accumulates in WT cells. Similar results were observed in studies of gat1Δ clones in the RHΔΔ and Me49 backgrounds (Fig. S9, H and I). Finally, complementation of the gat1Δ clone in RHΔΔ with Gat1 expressed under the tubulin promoter resulted in the absence of measurable incorporation into Skp1, confirming specificity for Gat1 expression per se.
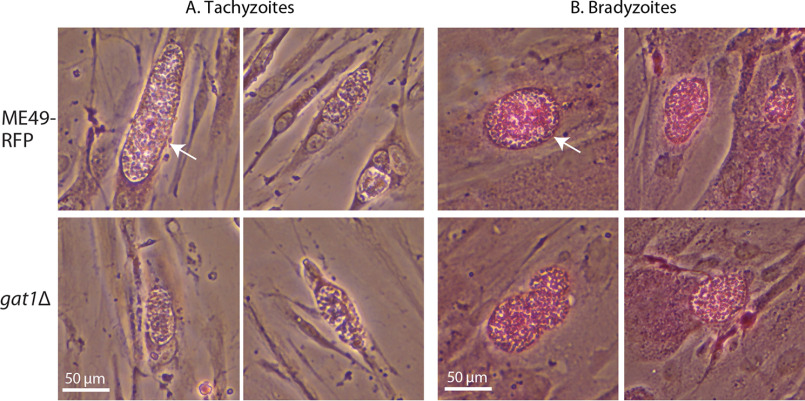
Although Gat1 was unable to serve as its own GT acceptor in the manner of glycogenin, its ability to modify α4Glc oligomers in vitro, albeit with low efficiency, raised the possibility that it may affect amylopectin (starch) biosynthesis in cells by applying αGal residues to its nonreducing termini. Starch normally accumulates to substantial levels in bradyzoites, a slow-growing form of the parasite that is induced by stress. Because induction of bradyzoite differentiation in cell culture is more efficient in type 2 strains, Me49 and gat1Δ/Me49 cells were induced by pH upshift and examined for differentiation by labeling of bradyzoite cyst walls with FITC-DBA-lectin and for starch with the periodic acid/Schiff's base reagent. As shown in Fig. 5, ME49 bradyzoites accumulated substantial levels of starch relative to tachyzoites, and no difference in the pattern or level was ascertained in gat1Δ cells using this qualitative assessment. Thus, Gat1 does not appear to affect starch synthesis, which is consistent with the absence of terminal Gal residues in a sugar composition analysis of T. gondii starch (29, 37) and the finding that gat1 is expressed equally in starch-poor tachyzoites and starch-rich bradyzoites based on transcript analysis (29).
Figure 5.
Starch appears unaffected in gat1Δ parasites. To promote normal starch accumulation, rapidly proliferating tachyzoites (A) of the type II strain Me49 (RFP-expressing) and its gat1Δ derivative were induced to differentiate as slow-proliferating bradyzoite cysts (B) in human foreskin fibroblasts. Cultures were fixed and stained with periodic acid/Schiff's base to reveal starch as a purple adduct. Arrow in (A), parasitophorous vacuole containing dozens to hundreds of tachyzoites within a fibroblast. Arrow in (B), cyst containing dozens of slow-growing bradyzoites, as confirmed by labeling of the cyst wall with FITC-DBA lectin (not shown). Scale bars, 50 μm. Two independent trials yielded similar results.
Gat1 generates a Galα1,3Glc linkage
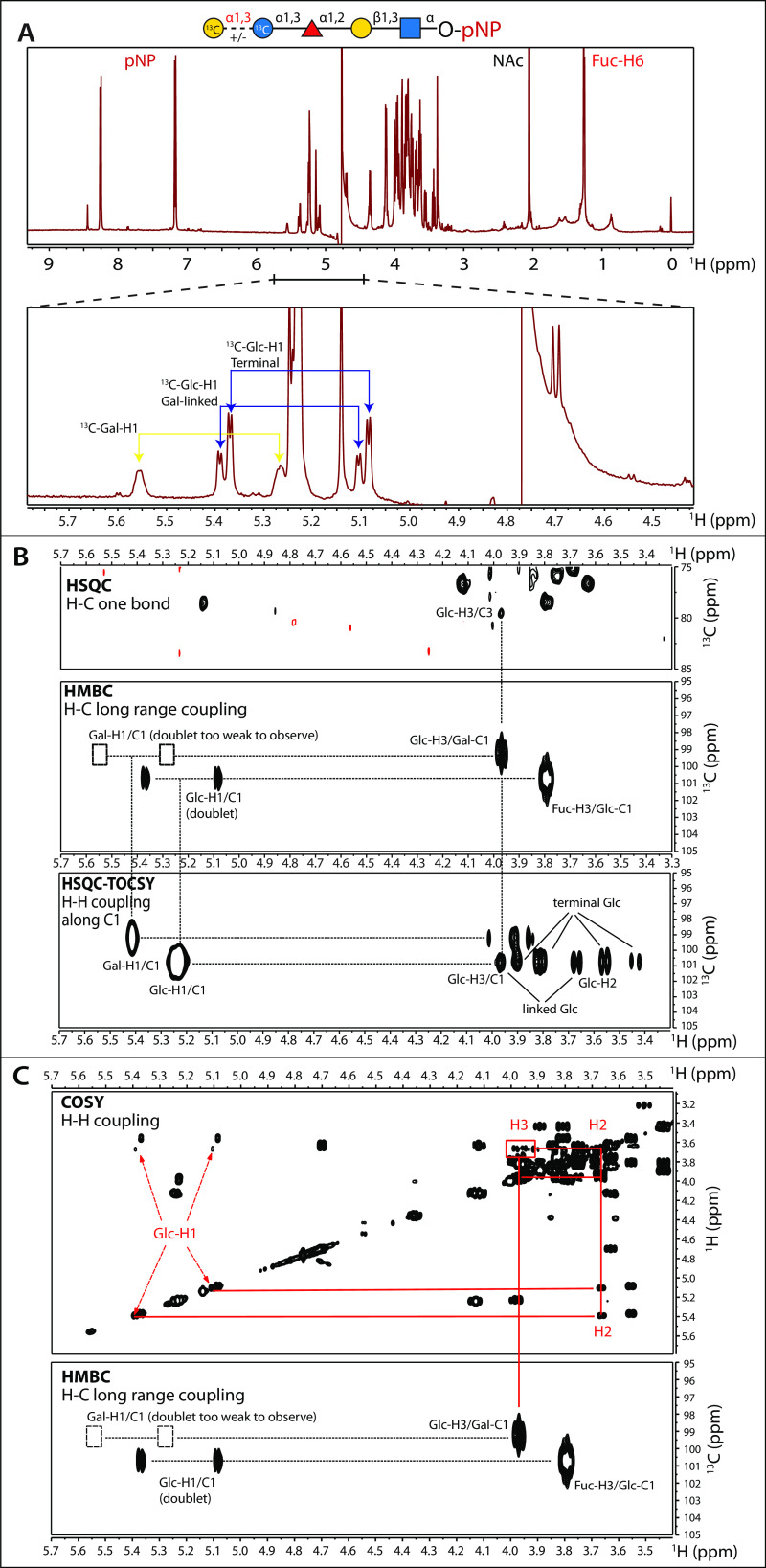
To determine the glycosidic linkage of the αGal residue transferred by TgGat1, the previously prepared (13C6)GlFGaGn-pNP (14) was modified with TgGat1 using UDP-[U-13C]Gal as the donor substrate. The pentasaccharide reaction product (∼30% conversion) was analyzed together with the tetrasaccharide starting material by NMR. The previous assignment of the chemical shifts of the tetrasaccharide (14) facilitated provisional assignment of the additional terminal Gal chemical shifts using the CASPER program (38) and confirmed by analysis of the two-dimensional COSY, TOCSY, and HMBC spectra (Fig. 6). One-dimensional 1H NMR spectra reveal the presence of the mixture of tetra- and pentasaccharides, followed by a downfield shift in the 13C-Glc-H1 peaks upon linkage to the terminal 13C-Gal (Fig. 6A). The HMBC 1H–13C correlation spectrum shows the (Fig. 6B, center panel) connection from Gal-H1 to Glc-H3, consistent with the downfield peak in the 1H–13C HSQC spectrum (Fig. 6B, top panel), and the proton resonances in the HSQC-TOCSY spectrum (Fig. 6B, bottom panel), establishing the glycosidic linkage between the terminal αGal and underlying αGlc as 1→3. Finally, a 1H–1H COSY spectrum confirms the assignments of the underlying Glc-H1, H2, and H3 (Fig. 6C). Consistent results were obtained when the tetrasaccharide was modified in the presence of PuGat1 (data not shown). Taken together, our NMR analyses are most consistent with the glycan structure, Galα1,3Glcα1,3Fucα1,2Galβ1,3GlcNAcα-, indicating that TgGat1 is a retaining UDP-Gal:glucoside α1,3-galactosyltransferase. Not only does Gat1 transfer the α-anomer of a different sugar compared with glycogenin, it attaches it to a different position (4- not 3-) of the acceptor αGlc residue.
Fig. 6.
Gat1 assembles a Galα1,3Glc linkage on the Skp1 tetrasaccharide. Shown is NMR analysis of the TgSkp1 pentasaccharide. [1-13C]Glcα1,3Fucα1,2Gal-β1,3GlcNAcα1-pNP was partially (30%) modified by TgGat1 or PuGat1 in the presence of UDP-[U-13C]Gal. A, one-dimensional 600-MHz 1H NMR spectrum. Magnification shows the region of 13C-anomeric carbons in the mixture of modified and unmodified tetrasaccharide. A cartoon diagram of the TgSkp1 pentasaccharide attached to pNP (Galα1,3Glcα1,3Fucα1,2Gal-β1,3GlcNAcα1-pNP) is shown at the top using CFG glycan symbols (19). B, 1H–13C HSQC, HMBC, and HSQC-TOCSY spectra demonstrating anomeric carbon to ring proton correlations. The Gal-H1/C1 doublet peaks were too weak to observe and are indicated by boxes. C, 1H–1H COSY and 1H–13C HMBC spectra. Identical results were obtained using [1-13C]GlFGaGn-pNP modified by PuGat1 (not shown). See Fig. S11 for a listing of chemical shift values.
Comparison of crystal structures explains the catalytic differences between Gat1 and glycogenin
To further probe the relationship between Gat1 and glycogenin, we compared their structures by X-ray crystallography. Attempts to crystallize TgGat1 were unsuccessful, even after deletion of its unconserved insert (Fig. 3A). However, PuGat1, which lacks this insert, was co-crystallized in the presence of Mn2+ and UDP.
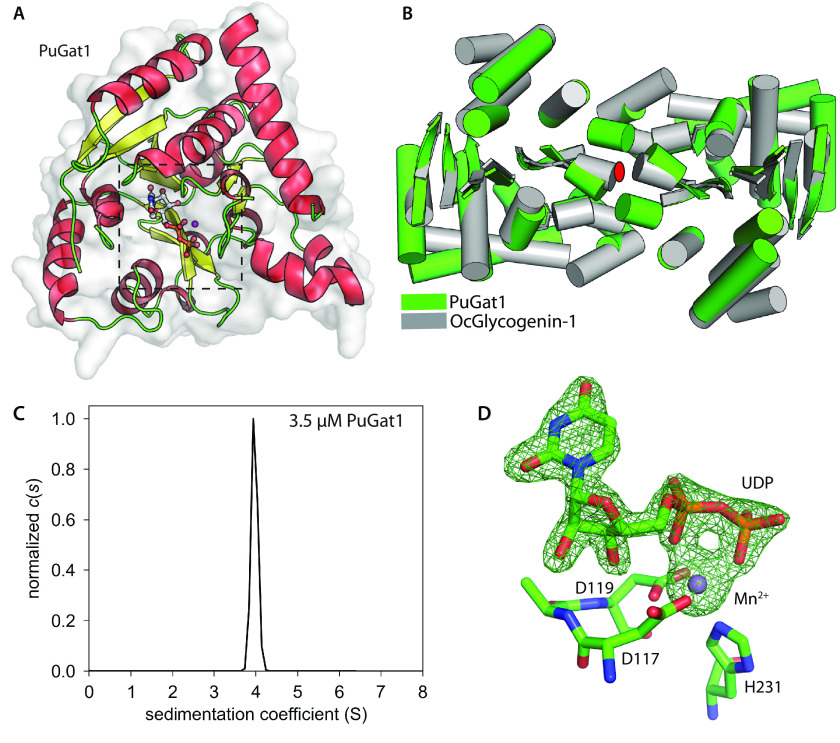
The crystal structure of PuGat1 in complex with Mn2+ and UDP was solved using single-wavelength anomalous dispersion phasing of a Pt2+ derivative, and the resolution was extended to 1.76 Å using a native data set (Table S3). The asymmetric unit contains a single chain of PuGat1 with unambiguous electron density for the nucleotide and Mn2+ ion (Fig. 7A). The overall structure of PuGat1 reveals a canonical GT-A fold (39) consisting of eight α-helices and eight β-strands. The N terminus (residues 1-8) and two loops (residues 80-96 and 242-244) are disordered and were not modeled. The structure is similar to glycogenin-1 from the rabbit O. cuniculus (Oc-glycogenin-1), which superimposes 213 corresponding Cα atoms with a root mean square deviation of 3.3 Å despite a sequence identity of only 34% (Fig. 7B). The application of crystallographic symmetry shows that PuGat1 forms the same dimer described (40) for the Oc-glycogenin-1 structure (Fig. 7B). According to PISA (41) analysis, the PuGat1 dimer interface buries 1,090 Å2 with a favorable p value of 0.107, which suggests that the dimer contact is stable. Sedimentation velocity analysis of 3.5 μm PuGat1 reveals a c(s) distribution consisting of single species at 4.0 S, which corresponds to the predicted value of 4.2 S for a dimer (Fig. 7C). The slightly slower sedimentation indicates that the enzyme in solution is less compact than that observed in the crystal structure. PuGat1 was dimeric even at 0.3 μm (Fig. S12), suggesting that it forms a dimer with an affinity >2-fold higher than that of Oc-glycogenin-1 (which has a reported Kd of 0.85 μm) (42). Based on gel filtration and preliminary sedimentation velocity experiments (not shown), TgGat1 is also a dimer.
Figure 7.
Gat1 is structurally related to glycogenin. A, asymmetric unit of crystallized Gat1 from P. ultimum in complex with UDP and Mn2+. α-Helices are shown in red, β-strands in yellow, and loops in green, and the active site with bound ligand is boxed. B, PuGat1 forms a homodimer that closely superimposes on Oc-glycogenin-1 (PDB entry 1LL2). Cylinders, α-helices; arrows, β-sheets; red ellipse, 2-fold symmetry axis perpendicular to the page. C, sedimentation velocity data modeled as a continuous c(s) distribution (normalized to 1.0) yields an S value for 3.5 μm PuGat1 that is close to the predicted value for a stable dimer in solution. Fig. S12 shows that the dimer is stable down to at least 0.3 μm. D, UDP is coordinated in nearly identical fashion to that of glycogenin-1, based on the difference density map (Fo − Fc) that was contoured at 5σ, calculated after omitting UDP and Mn2+ and subjecting the structure to simulated annealing. Octahedral coordination of Mn2+ is satisfied by the DXD motif, His-231, and UDP. The comparison with glycogenin-1 is illustrated in Fig. S13.
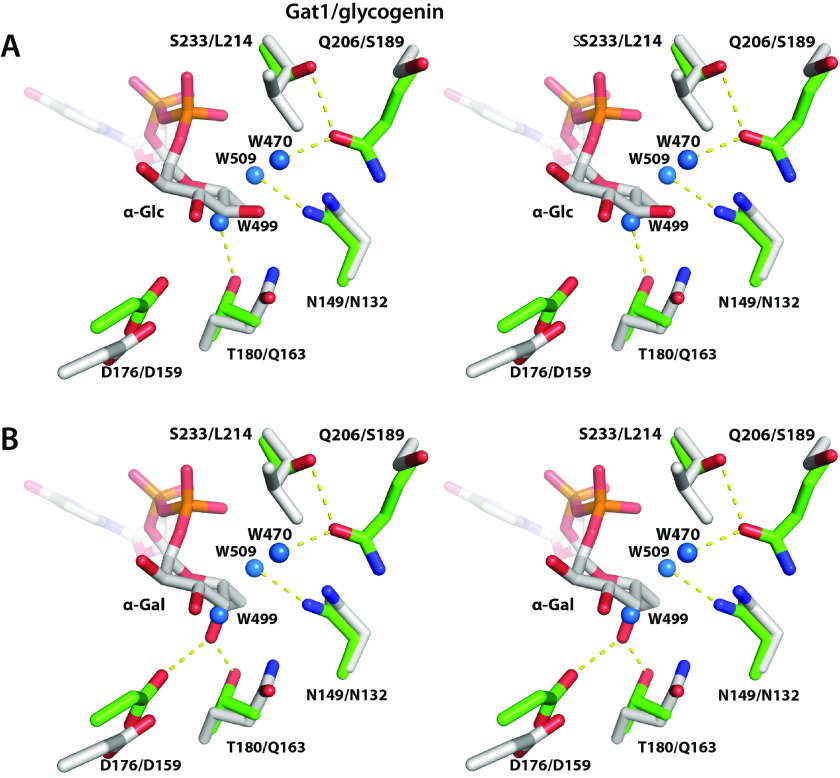
The PuGat1 active site shows that the conserved DXD motif (43) and a conserved His residue coordinate the Mn2+ ion using the Oδ2 atom of Asp-117, both the Oδ1 and Oδ2 atoms of Asp-119, and the Nε2 atom of His-231 (Fig. 7D). The Mn2+ ion is also coordinated by the oxygen atoms from the α- and β-phosphates of UDP. Comparing the PuGat1 and Oc-glycogenin-1 active sites shows that all of the interactions with the nucleotide are conserved, with the exception of the interactions with N3 and O4 of the uracil ring (Fig. S13). Other changes in Gat1 active site include a Leu to Ser substitution at residue 233, which can potentially remove a packing interaction with the donor sugar in PuGat1 (Fig. 8). The hydroxyl of the substituted Ser-233 forms a hydrogen bond with the adjacent side chain of Gln-206. A water molecule (W509) replaces the Leu side chain and forms a hydrogen bond with Asn-149.
Figure 8.
Active-site geometry explains Gat1's preference for UDP-Gal rather than UDP-Glc. Comparison of the sugar-binding pockets of PuGat1 and Oc-glycogenin-1 displayed as wall-eyed stereo view. A, the Glc moiety is modeled based on the Oc-glycogenin-1 crystal structure with its intact sugar nucleotide. B, the Gal moiety is modeled by flipping the stereochemistry of Glc at the C4′ position. PuGat1 and glycogenin-1 side chains are represented by green and gray sticks, respectively, the yellow dashes represent hydrogen bonds, and water molecules are represented by blue spheres.
Comparing the crystal structure of UDP-Glc bound Oc-glycogenin-1 with PuGat1 immediately suggests a reason for why these enzymes have different donor specificities (Fig. 8). Superimposing the Oc-glycogenin-1:UDP-Glc structure onto PuGat1 shows that UDP-Glc would displace water 499 coordinated by Thr-180 (Fig. 8A). This would leave the Thr-180 hydroxyl group unsatisfied, and the unfavorable burial of a polar group likely explains why UDP-Glc is a poor donor (Fig. 8A). In contrast, we modeled in UDP-Gal by flipping the stereochemistry at the C4 position. The O4 atom of Gal would be ideally positioned to satisfy the Thr-180 hydroxyl group. Concurrently, Asp-176, whose Cα atom underwent a 2.3-Å shift relative to its location in glycogenin, would be in position to receive a hydrogen bond from the O4 atom of the Gal (Fig. 8B). Altogether, Gat1's sugar donor preference for UDP-Gal is likely due to formation of favorable hydrogen bonds with the O4 atom of Gal in contrast to the burial of the Thr-180 hydroxyl group when binding UDP-Glc.
Computational modeling predicts the specificity of Gat1 toward the Skp1 tetrasaccharide
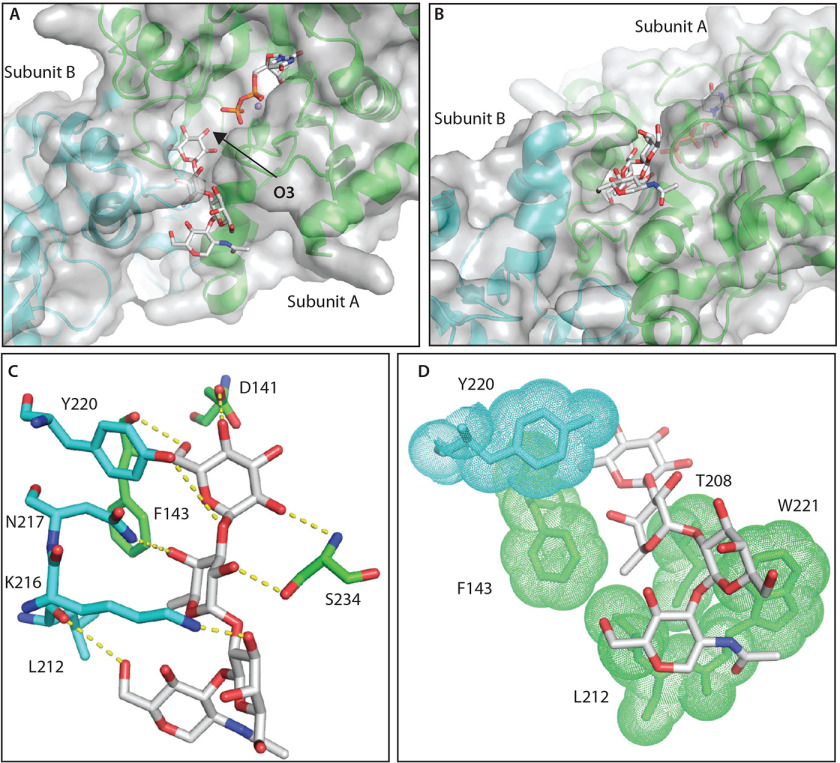
To address the basis of Gat1's preference for the GlFGaGn- glycan, the lowest-energy conformation of the reducing form of the glycan generated by GLYCAM-web (RRID:SCR_018260) was docked using AutoDock Vina. A plausible docking mode was selected based on the requirement that the C′3-hydroxyl group must be oriented toward the anomeric carbon of the donor sugar to serve as the nucleophile for the addition of the Gal and that the glycan does not clash with the other subunit of the dimer. Of 100 docking simulations, only the top-scoring pose with a binding energy score of −5.7 kcal/mol satisfied the selection requirement. In this pose, the glycan adopted an alignment in a groove formed by Gat1 dimerization (Fig. 9, A and B). The glycan is stabilized by hydrogen bond contributions from the side chains or peptide backbones of residues Asp-141, Phe-143, and Ser-234 from subunit A and residues Leu-212, Lys-216, Asn-217, and Tyr-220 from subunit B (Fig. 9C). In addition, nonpolar interactions against the faces of the sugar moieties are provided by residues Thr-208, Leu-212, Phe-143, and Trp-221 from subunit A and residue Tyr-220 from subunit B (Fig. 9D). Phe-143, Tyr-220, Trp-221, and Ser-234 are uniquely conserved in Gat1 proteins relative to glycogenins (Fig. S6). The packing interactions from conserved hydrophobic residues are likely the major contributors in terms of binding energy. The extensive electrostatic and packing complementarity, which could not be achieved using the same approach using the α4Glc-tetramer recognized by glycogenin, can explain the distinct preference of Gat1 for the Skp1 tetrasaccharide acceptor substrate.
Figure 9.
Computational docking explains specificity of Gat1 for the Skp1-tetrasaccharide. A, top docking pose of the Skp1-tetrasaccharide in the PuGat1 active site and a groove formed by the dimer. B, 90° turn of the image shown in A. C, hydrogen-bonding interactions of the glycan. D, hydrophobic packing of the sugar faces and fucose-methyl (in sticks) with nonpolar surfaces (sticks/dots) of Gat1 subunit A (green) and B (cyan).
The Toxoplasma glycan influences Skp1 helix-8 extension via sugar-protein contacts
The glycan-protein contacts described previously for D. discoideum Skp1 correlated with the extension of helix-8, which was interpreted to provide better access by FBPs. To address whether this mechanism is conserved for T. gondii, despite the difference of the fourth sugar, the computational studies were repeated on glycosylated TgSkp1. Energy-minimized structures of the glycans from DdSkp1 and TgSkp1 revealed only a difference in the position of O4 of the fourth sugar (due to the Glc/Gal configurational inversion) (Fig. S14A). Six all-atom MD simulations were performed without coordinate constraints for 250 ns each to allow greater sampling of conformational space. Three of the simulations began with a 50-ns pre-equilibration of the glycan with Cα atom constraints on the polypeptide, whereas the others proceeded directly. As before, the simulations did not converge on a common structure, so a combined time-resolved linear regression analysis of the six simulations was conducted to identify correlations between helix extension and calculated polar and nonpolar interaction energies with each moiety of the glycan, and the results are shown in Table 2.
Table 2.
MMGBSA-derived per-residue energies between the protein and glycan that exhibit a strong correlation with helix extension (distance in Fig. 10A) according to a linear regression analysis of 48 bins from the six MD simulations (Fig. S14E)
Polar energies represent a sum of the electrostatic and polar solvation energies, whereas the nonpolar energies are composed of the van der Waals and nonpolar solvation energies. Only interactions with an average polar/nonpolar energy less than −0.5 were considered.
| Glycan | Protein | Average energy | r2 | |
|---|---|---|---|---|
| kcal/mol | ||||
| Polar | αGal | Asn-147 | −0.6 | 0.73 |
| Glc | Asn-150 | −0.6 | 0.60 | |
| Fuc | Asn-150 | −1.2 | 0.50 | |
| Nonpolar | αGal | Asn-147 | −0.81 | 0.73 |
| GlcNAc | Phe-152 | −1.5 | 0.66 | |
| βGal | Phe-152 | −1.1 | 0.64 | |
| GlcNAc | Asn-150 | −0.65 | 0.54 | |
| αGal | Ile-148 | −0.67 | 0.52 |
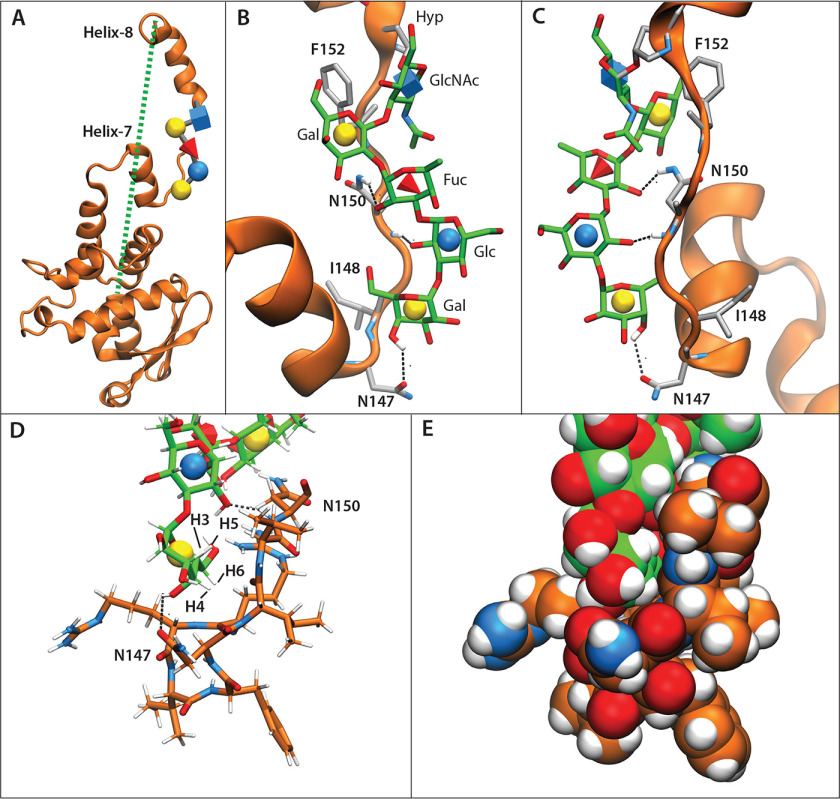
A representative frame from a simulation with a strong correlation between the extension of helix-8 (dotted green line) and glycan contacts with the polypeptide chain is shown in Fig. 10 (A–C). Glycan-protein contacts involve both polar hydrogen bonds and nonpolar van der Waals interactions between sugar and amino acid residues. The strongest correlated polar interactions (Table 2) correspond to three hydrogen bonds between sugars and amino acids within the unstructured peptide chain located between helix-7 and -8 (residues 147-152), as depicted in Figs. 9C and 10B. Notably, the 4-OH of αGlc (the fourth sugar), which is epimeric to the D. discoideum fourth sugar, is oriented toward the solvent, thus not affecting protein contacts. Nonpolar interactions also occur within this region, with the terminal αGal moiety showing the highest correlation (Table 2). The hydrophobic face of the terminal αGal can pack against a nonpolar pocket consisting of planar faces of peptide backbone (Figs. 9E and 10D). Specifically, its C3–C5 hydrogens can pack with the backbone of residues 147-149, and a hydrogen on C6 can be buried within residues 143 and 144 (Fig. 10D). Both the polar and nonpolar interactions identified here involve amino acids that are conserved across both organisms, with the exception of Val-149, which is Lys in DdSkp1 (Fig. S14D). This substitution would not be expected to affect Gal burial because the majority of interactions are with the protein backbone at this position (not shown). The previously noted hydrogen bonds 1 and 3 to helix-7, involving Glu-140 (Glu-129 in DdSkp1), were also observed in this study (Fig. S14C) but were poorly correlated (r2 = 0.0 versus 0.63 average for the other three) with helix-8 extension (Fig. S14E). The energetic analyses applied to the T. gondii variant confirm its ability to form an organized structure that can still similarly influence local Skp1 polypeptide organization. The new studies emphasize the interaction of the glycan with the interhelix region rather than helix-7 and contributions of packing of the outer sugars, including the terminal αGal. As previously hypothesized, these effects on the conformational ensemble of TgSkp1 have the potential to make it more receptive to FBP binding.
Figure 10.
Packing of the glycan with Skp1 can explain F-box–binding site conformation. T. gondii GaGlFGaGn-Skp1 was subjected to six 250-ns all-atom molecular dynamics simulations. A, frame representative of the glycan-protein interaction and associated helix-8 extension, from a simulation (Equil-1; see Fig. S14E) in which the glycan was pre-equilibrated for 50 ns prior to the start of the simulation. The dotted green line refers to the distance from the C terminus to the center of mass of residues 1-136 and ranged from 18 to 61 Å. B, zoom-in of A depicting the glycan (carbon atoms in green) and amino acids (carbon atoms in gray) described in Table 2. Dotted black lines depict hydrogen bonds contributing to the polar energies described in Table 2. C, the back side of B. D and E, packing of terminal sugars against the polypeptide. Carbon atoms of the peptide are in orange. D, glycan and peptide represented as sticks. E, as in D, with glycan and peptide represented by spheres.
Discussion
Glycosylation mediates much of the effect of O2-dependent prolyl hydroxylation of Skp1 in D. discoideum and T. gondii, and the present work emphasizes the importance of the fifth and final sugar of the glycan. The T. gondii glycan adopts a similar constrained conformation as described for D. discoideum despite the sequence difference (Fig. S14, A and B), and new all-atom molecular dynamics simulations showed a conserved cis-interaction with the back side of the intrinsically disordered region of Skp1 that comprises subsite-2 of the FBP-binding pocket (Fig. S14C). A deeper time-resolved energetics analysis of polar and nonpolar contacts showed a strong dynamic correlation between burial of the peripheral (especially the fifth) sugars in the turn from helix-7 to the loop connecting helix-8, and the extension of helix-8 which exposes subsite-2 for FBP docking (Fig. 10 and Table 2). This analysis de-emphasized the previous correlation of hydrogen bonds with helix-7 in favor of contacts with loop residues between helix-7 and helix-8 (Fig. S14E). This quantitative analysis of the trajectories extends critical features of the prior study in D. discoideum (7) by emphasizing the role of nonpolar packing of the glycan terminus in helix-8 extension.
The fifth sugar is attached by Gat1, an unusual GT that resides in the cytoplasmic compartment rather than the secretory pathway of the cell and appears to be dedicated to only Skp1. Although confined to the protist kingdom, it is nevertheless descended from a widely distributed lineage of sugar nucleotide–dependent CAZy GT8 GTs and may be the evolutionary predecessor of glycogenin, a GT that modulates glycogen formation in the cytoplasm of yeast, fungi, and animals. Although Gat1s from T. gondii and P. ultimum catalyze the transfer of αGal from UDP-Gal to nonreducing terminal Glc acceptors (Fig. 4 and Fig. S9), their ability to utilize UDP-Glc at low efficiency could be rationalized by the organization of the active site in the crystal structure (Fig. 8) and anticipates this evolutionary transition. The ability of UDP-Glc to inhibit Gat1's galactosyltransferase activity (Fig. 4G) might be regulatory in cells. The recently reported capability of glycogenin to transfer Gal at certain steps (44) lends further support to the evolutionary relationship. Although Gat1 is able to modify Glc in multiple contexts, it prefers Glc at the terminus of the native Skp1 tetrasaccharide over Glc in a native starch- or glycogen-like glycan (Fig. 4 and Fig. S9). Furthermore, Gat1 was substantially more reactive when the glycan was attached to Skp1 (Fig. 4, H and I), indicating that the apoprotein contributes to increased activity. The increase in activity may be due to the recognition of Skp1 protein by Gat1, but this hypothesis remains to be tested.
Computational docking showed how the Skp1 glycan alone in its calculated lowest-energy state can naturally fit within a groove along the dimer interface of PuGat1 (Fig. 9A), in a manner that cannot be achieved with an α4-glucan as in starch or glycogen in its lowest-energy state (not shown). The orientation of the acceptor Glc of the Skp1 glycan in this docking mode also supports the formation of the α1,3 linkage that was determined by NMR analysis of the product of the reaction of both PuGat1 and TgGat1 with a synthetic version of the Skp1 tetrasaccharide as an acceptor (Fig. 6). Furthermore, a high degree of conservation between the two enzymes is supported by their very similar activity and specificity characteristics (Fig. 4 and Fig. S9). The described binding mode is a possible explanation for the greater activity of the free tetrasaccharide as an acceptor substrate compared with the α4-linked glucans. Overall, the data suggest that recognition of Skp1 as a substrate includes both active-site preference for the specific glycan and separate determinants on the polypeptide. Owing to the limited computational scope of this study, however, further analysis is needed to fully define the basis for Skp1 recognition.
The significance of the terminal sugar is echoed by the slow growth phenotype of gat1Δ parasites in the monolayer plaque assay. This was attributed to the absence of Gat1 because similar results were obtained in independent knockouts in three different genetic backgrounds, and the defect was corrected by genetic complementation under its own promoter or a strong tubulin promoter in the uprt locus (Fig. 2). Because no other Gat1 targets were detected by biochemical complementation of gat1Δ extracts (Fig. 4J and Fig. S9 (H and I)), and no effect on starch formation was observed (Fig. 5), it is likely that failure to fully glycosylate Skp1 is responsible for the growth defect. This interpretation is consistent with similar effects of knocking out earlier GTs in the pathway and their selectivity for Skp1 in vitro (13, 14). Furthermore, in D. discoideum, genetic manipulations of Skp1 levels interact with its glycosylation with respect to O2 sensing (8). Skp1 interactome studies in D. discoideum indicated that five sugars were clearly more effective in promoting binding of FBPs than three sugars (10) but were, however, unable to resolve the roles of the fourth and fifth sugars owing to their being added by the same GT (45). Although we cannot completely exclude some other function of Gat1, we note that at 345 amino acids, nearly all of its sequence appears devoted to the enzymatic domain (Fig. 3A). Studies are under way to identify SCF substrates and how they are affected by Skp1 modification and influence parasite growth and fitness.
The importance of the fifth sugar is reinforced by the observation that the amoebozoa, which lack both Glt1 and Gat1 (Fig. 1, A and B), evolved a new unrelated enzyme, AgtA, to fulfill the role. AgtA is a dual-function GT that applies both the fourth and fifth sugars, each an α3-linked Gal. This raises the interesting possibility that AgtA evolved to compensate for the unexplained loss of Glt1 and Gat1, with recursive addition of the same sugar being an accessible evolutionary pathway to recover the pentasaccharide. Gat1 sequences are found in alveolates (including T. gondii), stramenopiles (including P. ultimum), and rhizaria (which together with telonemids are known as the TSAR group) and archaeplastids (which together with the TSAR group are known as bikonts), but not in the unikonts that include the amoebozoa, fungi, and animals (46, 47). Whereas the fate of Glt1 in the unicots is unknown, its disappearance potentially freed Gat1 to evolve into glycogenin, an α4-glucosyltransferase that, in vitro, can autoglucosylate itself to prime glycogen synthesis in fungi and metazoa. The enzymes share catalytic properties (see above) and perfectly conserve structural similarities, including the same dimer interface (Figs. 7 and 9), although further studies are needed to support the model of lineal descent. Thus, it is interesting to speculate whether the proposed transition conserved a cellular function. Although glycogenin acquired a C-terminal glycogen synthase–binding domain (Fig. 3A) (48) that seems consistent with its in vitro capacity to initiate glycogen assembly, data now exist indicating that glycogenin is not required for glycogen synthesis in yeast and mice (49, 50), although glycogen levels are affected by an unknown mechanism. The trans-glycosyltransferase activity for this enzyme lineage in bikonts protists raises the possibility of an unanticipated activity in unicots that could help explain the glycogenin-KO findings in yeast and mice.
Experimental procedures
Maintenance of host cells and parasite manipulations
Cultures of human foreskin fibroblast (HFF; ATCC SCRC-1041) or hTERT HFF (BJ-5ta, ATCC CRL-4001) were maintained Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin (Corning) at 37 °C in a humidified CO2 (5%) incubator. Type 1 RH (21), RHΔKu80ΔHXGPRT (RHΔΔ) (22), and type 2 ME49-RFP (23) strains of T. gondii were cultured on HFF or hTERT HFF monolayers in the same medium as described for host cells except that 1% (v/v) fetal bovine serum was used where stated and cloned by limiting dilution in 96-well plates. Parasites were kept in media without drug for plaque assays, which were performed as before (20). Transfections were conducted by electroporation using a Bio-Rad Gene Pulser Xcell at 1.5 kV and 25 microfarads with 2-mm electroporation cuvettes (VWR International) in Cytomix buffer (10 mm KH2PO4/K2HPO4 (pH 7.6), 120 mm KCl, 0.15 mm CaCl2, 5 mm MgCl2, 25 mm HEPES, 2 mm EDTA).
Disruption and complementation of gat1
TGGT1_310400 (ToxoDB, RRID:SCR_013453), referred to as gat1 (Fig. S1), was disrupted by two independent approaches. In the first method, a disruption DNA was prepared from the vector pmini.GFP.ht (a gift from Dr. Gustavo Arrizabalaga) in which the hxgprt gene is flanked by multiple cloning sites as described (13). 5′- and 3′-flanking targeting sequences were PCR-amplified from strain RHΔΔ with primer pairs Fa and Ra and pairs Fb and Rb, respectively (Table S1). The 5′-fragment was digested with ApaI and XhoI and cloned into similarly digested pmini.GFP.ht. The resulting plasmid was digested with XbaI and NotI and ligated to the similarly digested 3′-flanking DNA. The resulting vector was linearized with PacI and electroporated into strain RHΔΔ, selected under 25 μg/ml mycophenolic acid and 25 μg/ml xanthine, and cloned by limiting dilution. Genomic DNA was screened by PCR to identify Tggat1 disruption clones (Fig. S3A), using primers listed in Table S1.
The second approach was based on a double-CRISPR/Cas9 method as detailed previously (20), with minor modifications. To generate the dual guide (DG) plasmid, a fragment of p2 containing the guide RNA gat1-63 expression cassette was PCR-amplified using primers plasmid 3 FOR and plasmid 3 REV (Table S1), digested with NsiI, and ligated into the NsiI site of a dephosphorylated p3 containing guide RNA gat1-968. The type 1 RH and type 2 ME49 strains were co-transfected with pDG-Gat1 (10 μg) and a dihydroxyfolate reductase (dhfr) amplicon (1 μg) by electroporation (Fig. S4A). CRISPR/Cas9-mediated disruption in RHΔΔ parasites was done similarly except that pDG-Gat1 was co-transfected with a dhfr amplicon containing 45-bp homology arms targeting gat1 (Fig. S3B). Gat1Δ parasites were subsequently selected in 1 μm pyrimethamine (Sigma). The expected replacement of nucleotides 63-968 (relative to A of the ATG start codon) with the dhfr cassette was confirmed by PCR using primers listed in Table S1.
To complement gat1Δ RH parasites, a Tggat1 DNA fragment consisting of the coding sequence of gat1 plus ∼1.2 kb each of 5′- and 3′-flanking DNA sequences was generated by PCR from RH genomic DNA using primers Fa and Rb, which contained ApaI and NotI restriction sites, respectively. After treatment with ApaI and NotI, the PCR product was cloned into similarly digested pmini.GFP.ht plasmid in place of its hxgprt cassette, to generate pmini.Tggat1. The plasmid was transformed into E. coli Top10 cells and purified by using a Monarch miniprep kit (New England Biolabs). A Ty tag DNA sequence was inserted at the 3′-end of the gat1 coding sequence using a Q5 site-directed mutagenesis kit (New England Biolabs) and Fn and Rn primers, yielding pmini.Tggat1.Ty. The sequence was confirmed using primers Fl and Rl (Table S1). RH gat1Δ clone 8 was complemented by co-electroporation with a PCR amplicon from pmini.Tggat1.Ty (1 μg) and a sgUPRT CRISPR/Cas9 plasmid (10 μg) targeting the uprt locus using the guide DNA sequence 5′-ggcgtctcgattgtgagagc (51) (Fig. S4B). Transformants were selected with 10 μm fluorodeoxyuridine (Sigma), and drug-resistant clones were screened by PCR with primers listed in Table S1.
To complement gat1 in RHΔΔ parasites, the UPRT Vha1 cDNA shuttle vector containing TgVhaI cDNA (52) was modified to generate a Gat1-HA complementation plasmid using the New England Biolabs HiFi Builder method. The vector backbone, containing 5′- and 3′-flanking uprt targeting sequences and a Tg-tubulin promoter and 3×HA sequence, was PCR-amplified from the shuttle vector using primers Ft and Rt. The coding sequence of TgGat1 was PCR-amplified from pmini.Tggat1 plasmid using primers Fu and Ru, which had 18-21 nucleotides complementary to the terminal ends of the vector (Fig. S3C). The gel-purified PCR fragments were incubated with HiFi DNA assembly enzyme mix (New England Biolabs) and transformed into E. coli Top10 cells, yielding pUPRTgat13×HA. The Tggat1 sequence was confirmed using primers Fl and Rl. Complementation in a gat1Δ clone derived from RHΔΔ was done similarly except that a gat1-3×HA PCR amplicon with 5′ and 3′ uprt homology arms was used in the transfection.
Bradyzoite induction
ME49-RFP tachyzoites were differentiated to bradyzoites using alkaline pH (53). HFF monolayers were preincubated with sodium bicarbonate–free RPMI (Corning) containing 50 mm HEPES-NaOH (pH 8.1) for 24 h and then infected with tachyzoites and maintained at ambient atmosphere at 37 °C with medium replacement every 24 h. Differentiation was monitored by labeling with Dolichos biflorus agglutinin (DBA) (54). Infected HFF monolayers formed on 25-mm coverslips were washed with PBS (Corning), fixed with 4% paraformaldehyde in PBS for 10 min, washed with PBS, and permeabilized with 1% Triton X-100 (Bio-Rad) in PBS for 10 min, all at room temperature. Samples were blocked with 3% (w/v) BSA in PBS for 1 h, incubated for 2 h with 5 μg/ml FITC-DBA lectin (Vector Laboratories Inc.) in 1% BSA in PBS, and washed with PBS. The coverslips were mounted with ProLong Gold antifade reagent (Invitrogen) on glass slides. The slides were imaged by phase-contrast and fluorescence microscopy on a Zeiss Axioskop 2 Mot Plus.
Periodic acid staining
To assess amylopectin levels, parasite-infected HFF monolayers on 25-mm glass coverslips were washed with plain PBS, fixed with ice-cold MeOH for 5 min, washed with PBS, and incubated with 1% periodic acid in deionized H2O in the dark for 10 min (55). The coverslips were washed with deionized H2O, incubated with Schiff reagent for 15 min, washed once with deionized H2O, and finally rinsed with running tap water for 10 min. The stained coverslips were dehydrated by sequential immersion in 70% (v/v), 80, 90, and 100% EtOH, mounted on glass slides using Permount mounting medium (Fisher Scientific), and imaged on an EVOS XL Core microscope (Invitrogen).
Expression and purification of recombinant TgGat1 and PuGat1
The single exon-coding sequence of Tggat1 cDNA was amplified by PCR from RH genomic DNA using primers, Gat1 Fw and Gat1 Rv (Table S1), cloned into PCR4-TOPO TA (Invitrogen), and transformed into E. coli Top 10 cells. The plasmid was double-digested with BamHI and NheI to yield the gat1 fragment that was cloned into similarly digested pET15-TEVi plasmid (Invitrogen), resulting in the original 346-amino acid coding sequence of Gat1 extended at its N terminus with a His6 tag and TEV protease cleavage site (MGSSHHHHHHSSGRENLYFQGH-). A similar N-terminal modification of rabbit glycogenin did not significantly alter its enzymatic activity (35). The predicted coding sequence for Pugat1 was inferred from PYU1_G002535-201 (EnsemblProtists, RRID:SCR_013154, P. ultimum genome). The coding sequence was codon-optimized for E. coli expression (Fig. S2), chemically synthesized by Norclone Biotech (London, Ontario, Canada), and inserted in the NdeI and XhoI sites of pET15b-TEV. The expressed protein was extended at its N terminus with MGSSHHHHHHSSGENLYFQGH-.
TgGat1 or PuGat1 were expressed in and purified from E. coli BL21-Gold cells as described previously for TgGlt1 (14), through the purification on a 5-ml Co2+ TALON resin column. The eluted protein was dialyzed in 50 mm Tris-HCl (pH 8.0), 300 mm NaCl, 1 mm EDTA, and 2 mm β-mercaptoethanol, followed by 25 mm Tris-HCl (pH 8.0), 200 mm NaCl, 1 mm β-mercaptoethanol, and 5 mm MnCl2. The sample was treated with 2 μm His6-TEV protease, 5 μm TCEP in the same buffer overnight at 20 °C and reapplied to another Co2+ TALON column. The flow-through fraction was dialyzed in 25 mm Tris-HCl (pH 8.0), 50 mm NaCl, 1 mm β-mercaptoethanol, 2 mm MnCl2. The sample was concentrated by centrifugal ultrafiltration, and aliquots were stored at −80 °C. As indicated, the preparations were further purified by gel filtration on a Superdex200 column in the same buffer.
SDS-PAGE and Western blotting
Samples were suspended in diluted with Laemmli sample buffer and typically electrophoresed on a 4–12% gradient SDS-polyacrylamide gel (NuPAGE Novex, Invitrogen). Gels were either stained with Coomassie Blue or transferred to a nitrocellulose membrane using an iBlot system (Invitrogen). Blots were typically blocked in 5% nonfat dry milk in TBS and probed with a 1:1,000-fold dilution of the antibody of interest in the milk solution, followed by secondary probing with a 1:10,000-fold dilution of Alexa 680–labeled goat anti-rabbit IgG secondary antibody (Invitrogen). Blots were imaged on a LI-COR Odyssey IR scanner and analyzed in Adobe Photoshop with no contrast enhancement. For measuring incorporation of radioactivity, 1-mm thick 7–20% acrylamide gels were prepared manually, as detailed (13).
Preparation of Skp1 peptides
To monitor Skp1 glycosylation status, endogenous TgSkp1 was purified from parasite extracts essentially as described (13). Briefly, frozen pelleted tachyzoites (1 × 108) were resuspended in 8 m urea in 50 mm HEPES-NaOH (pH 7.4), incubated on ice for 1 h and at 50 °C for 5 min, and diluted 8-fold in IP buffer (0.2% Nonidet P-40 in 50 mm HEPES-NaOH, pH 7.4). The lysates were centrifuged at 21,000 × g for 20 min at 4 °C, and 100 μl of the supernatant (5 × 107 cells) were incubated with 5 μl of UOK75 rabbit antibody (affinity-purified anti-TgSkp1) coupled to protein A/G magnetic agarose beads (Pierce, 78609) for 1 h at 4 °C. Beads were captured in a DynaMag-2 magnet (Life Technologies) according to the manufacturer's directions and washed three times with 50 mm HEPES-NaOH (pH 7.4); three times with 10 mm Tris-HCl (pH 7.4), 50 mm NaCl; and once with 50 mm NaCl. Bound Skp1 was eluted twice with 60 μl of 133 mm triethylamine (sequencing grade; Pierce, 25108) by incubating for 15 min at RT and immediately neutralized with 40 μl of 0.2 m acetic acid. The eluted material was pooled and dried under vacuum, reconstituted in 8 m urea, 10 mm Tris-HCl (pH 7.4), reduced in 10 mm DTT for 40 min at RT, and alkylated in 50 mm chloroacetamide for 30 min at RT. Samples were then diluted to 2 m urea with 10 mm Tris-HCl (pH 7.4) and digested in 10 μg/ml Trypsin Gold (mass spectrometry grade; Promega, V5280) overnight at RT. Excess trypsin was quenched by the addition of 1% (v/v) TFA (Pierce, 28904) on ice for 15 min and centrifuged at 1,800 × g for 15 min at 4 °C. The supernatants were adsorbed to C18 pipette tips (Bond Elut OMIX C18, Agilent, A7003100) and eluted in 50% acetonitrile (ACN, OptimaTM LC–MS grade; Fisher, A955-4), 0.1% formic acid (FA, LC–MS grade; Pierce, 28905). Eluted peptides were vacuum-dried and reconstituted in 40 μl of 5% ACN, 0.05% TFA, and 4 μl were analyzed by nano-LC-MS/MS.
Treatment of TgSkp1 peptides with α-gal
Trypsinates from above were centrifuged at 1,800 × g for 15 min at 4 °C. The supernatants were dried under vacuum, resuspended in 100 mm sodium citrate phosphate buffer (pH 6.0), and treated with 3.6 milliunits of green coffee bean α-gal (Calbiochem) for 18 h at 37 °C. An additional 3.6 milliunits of α-gal was added for 8 h. After treatment, peptides were processed as above.
MS of TgSkp1 peptides
Reconstituted peptides were loaded onto an Acclaim PepMap C18 trap column (300 μm, 100 Å) in 2% (v/v) ACN, 0.05% (v/v) TFA at 5 μl/min, eluted onto and from an Acclaim PepMap RSLC C18 column (75 μm × 150 mm, 2 μm, 100 Å) with a linear gradient consisting of 4–90% solvent B (solvent A: 0.1% FA; solvent B: 90% ACN, 0.08% (v/v) FA) over 180 min, at a flow rate of 300 nl/min with an Ultimate 3000 RSLCnano UHPLC system, into the ion source of an Orbitrap QE+ mass spectrometer (Thermo Fisher Scientific). The spray voltage was set to 1.9 kV, and the heated capillary temperature was set to 280 °C. Full MS scans were acquired from m/z 350 to 2,000 at 70,000 resolution, and MS2 scans following higher energy collision-induced dissociation (30) were collected for the top 10 most intense ions, with a 30-s dynamic exclusion. The acquired raw spectra were analyzed using Sequest HT (Proteome Discoverer 2.2, Thermo Fisher Scientific) with a full MS peptide tolerance of 10 ppm and MS2 peptide fragment tolerance of 0.02 Da and filtered to generate a 1% target decoy peptide-spectrum match false discovery rate for protein assignments. All known glycoforms for TgSkp1-specific glycopeptides were manually searched for and verified. Searches were performed against the T. gondii (strain ATCC 50853/GT1) proteome (Uniprot proteome ID UP000005641, downloaded May 18, 2018; 8,450 entries) plus a list of common keratin, immunoglobulin-derived, and trypsin contaminants. Carbamidomethylation of Cys was set as a fixed/static modification, and oxidation of Met, deamidation of Asn and Gln residues, and acetylation of protein N termini were set as variable/dynamic modifications. Searches were performed with trypsin cleavage specificity, allowing two missed cleavage events and a minimum peptide length of 6 residues. All known glycoforms for TgSkp1-specific glycopeptides were manually searched for and verified. The raw files were uploaded to the Figshare server with ID 10.6084/m9.figshare.12272882 (Skp1 glycopeptides raw data, Fig. S5).
Enzyme assays
Sugar nucleotide hydrolysis
Recombinant TgGat1 (0.625-2.5 μm) was incubated with a given UDP-sugar (50 μm) (Promega) in 50 mm HEPES-NaOH (pH 7.4), 2 mm MnCl2, 5 mm DTT, in a final volume of 20 μl, for 1-16 h at 37 °C. The UDP generated was quantitated using the UDP-Glo assay (Promega) as described (56).
Glycosyltransferase activity toward small glycosides
In the standard reaction, TgGat1 or PuGat1 was incubated with 2 mm synthetic glycosides (pNP-α-galactoside, pNP-β-galactoside, pNP-α-glucoside, pNP-β-glucoside, pNP-α-maltoside, chloro-4-nitrophenyl-α-maltotrioside, FGaGn-pNP, GlFGaGn-pNP), 8 μm UDP-Gal (unlabeled), 0.17 μm UDP-[3H]Gal (15.6 μCi/nmol; American Radiolabeled Chemicals), 50 mm HEPES-NaOH (pH 7.0), 2 mm MnCl2, 5 mm DTT, in a final volume of 30 μl, for 1 h at 37 °C. Pilot studies indicated a pH optimum of 7.0, with 50% activity at pH 8.0 and 75% activity at pH 6.0 for TgGat1. Salt dependence studies showed maximal activity with no added NaCl or KCl and 35% activity at a 800 mm concentration of either salt. Activity showed a ∼6-fold preference for MnCl2 over MgCl2, with activity maximal at 2 mm MnCl2. The enzyme was essentially inactive in NiCl2, CoCl2, and CaCl2 (Fig. S9, A–D). For kinetic studies, concentrations and times were varied as indicated, and kinetic parameters were analyzed according to the Michaelis–Menten model based on the least squares fitting method in GraphPad Prism software. Reactions were stopped by the addition of 1 ml of 1 mm ice-cold Na-EDTA (pH 8.0), and incorporation of radioactivity into pNP-glycosides was analyzed by capture and release from a Sep-Pak C18 cartridge and scintillation counting (14).
Glycosyltransferase activity toward GlFGaGn-Skp1
To prepare Tg-GlFGaGn-Skp1, 2 nmol (40 μg) of recombinant TgSkp1 FGaGn-Skp1 (14) was incubated with 1.3 nmol (88 μg) of Tg-His6-Glt1, 4 nmol of UDP-Glc, 1.2 units of alkaline phosphatase (Promega), 50 mm HEPES-NaOH (pH 8.0), 5 mm DTT, 2 mm MnCl2, 2 mm MgCl2, 2 mg/ml BSA in a final volume of 121 μl for 3.5 h at 37 °C. The reaction was initiated by the addition of UDP-Glc and terminated by freezing at −80°C. Reaction progress was monitored by Western blotting with pAb UOK104, which is specific for FGGn-Skp1 from either D. discoideum or T. gondii (57), followed by probing with pAb UOK75, which is pan-specific for all TgSkp1 isoforms, for normalization. Approximately 85% of total Skp1 was modified. GlFGaGn-Skp1 was purified from Glt1 on a mini-QAE column using a Pharmacia Biotech SMART System. ∼19 μg of GlFGaGn-Skp1 was applied to a mini-QAE column pre-equilibrated with 50 mm Tris-HCl (pH 7.8), 5 mm MgCl2, 0.1 mm EDTA (buffer A) and eluted with a gradient from 0% A to 100% buffer B (50 mm Tris-HCl (pH 7.8), 5 mm MgCl2, 0.1 mm EDTA, 300 mm NaCl) in 40 min at a flow rate of 240 μl/min. GlFGaGn-Skp1 fractions were identified based on A280 and Western blotting probing with pAb UOK75.
TgGat1 (0.127 μm) was incubated with GlFGaGn-Skp1 or FGaGn-Skp1 (0.9-3.6 μm), 3.2 μm UDP-[3H]Gal (15.6 μCi/nmol), 40 μm UDP-Gal (unlabeled), 0.2% (v/v) Tween 20, 50 mm HEPES-NaOH (pH 7.0), 2 mm MnCl2, 5 mm DTT, in a final volume of 20 μl, for 1 h at 37 °C. The reaction was stopped by the addition of 4× Laemmli sample buffer, 1 m DTT (50 mm final concentration), and 2 μg of soybean trypsin inhibitor and boiled for 3 min SDS-PAGE, and incorporation of radioactivity into Skp1 was performed as described above.
To detect Gat1 activity in cells, a cytosolic extract was prepared by hypotonic lysis, ultracentrifugation at 100,000 × g for 1 h, and desalting as described previously (13). 25 μg of desalted S100 protein was incubated with 10-50 nmol of Tg-GlFGaGn-Skp1 and 1.0 μCi of UDP-[3H]Gal (15.6 μCi/nmol) for 5 h, and incorporation of radioactivity into protein was assayed by SDS-PAGE and scintillation counting as described above.
Glycosyltransferase activity toward parasite extracts
To search for Gat1 substrates, cytosolic S100 fractions (180 μg of protein) were incubated with 0.13 μm TgGat1, 2.0 μCi of UDP-[3H]Gal (15.6 μCi/nmol) in a final volume of 60 μl containing 50 mm HEPES-NaOH (pH 7.0), 2 mm MnCl2, 5 mm DTT, 30 mm NaF, 0.2% Tween 20, at 37 °C for 1 h, supplemented with 1.7 μg of Tg-GlFGaGN-Skp1 as indicated. Incorporation of radioactivity was monitored by the SDS-PAGE assay as described above.
MS of Skp1
To evaluate its glycosylation status, recombinant PuGat1 or TgGat1 (purified by gel filtration) was incubated with or without UDP-Gal or UDP-Glc in the absence of added acceptor substrate and diluted to 50 ng/μl Skp1 with 2% acetonitrile, 0.05% (v/v) TFA. 250-500 ng of protein (5-10 μl) was injected into an Acclaim PepMap C4 trap cartridge (300 μm × 5 mm) equilibrated with 0.05% TFA, 2% acetonitrile, ramped up with an increasing gradient to 0.1% formic acid, 25% acetonitrile, and introduced into an Acclaim PepMap analytical C4 column (75 μm × 15 cm, 5-μm pore size) maintained at 35 °C in an Ultimate 3000 RSLC system coupled to a QE+ Orbitrap mass spectrometer (Thermo Scientific). After equilibrating the analytical column in 98% LC–MS Buffer A (water, 0.1% formic acid) for 10 min and a 6-min ramp up to 27% LC–MS Buffer B (90% (v/v) acetonitrile, 0.1% formic acid), separation was achieved using a linear gradient from 27 to 98% Buffer B over 20 min at a flow rate of 300 nl/min. The column was regenerated after each run by maintaining it at 98% Buffer B for 5 min. The effluent was introduced into the mass spectrometer by nanospray ionization in positive ion mode via a stainless-steel emitter with spray voltage set to 1.9 k, capillary temperature set at 250 °C, and probe heater temperature set at 350 °C. The MS method consisted of collecting full ITMS (MS1) scans (400-2,000 m/z) at 140,000 resolution in intact protein mode (default gas P set to 0.2). PuGat1 species eluting between 17.5 and 21.5 min and TgGat1 species eluting between 18.5 and 22.5 min (∼60–80% acetonitrile) were processed with Xcalibur Xtract deconvolution software to generate monoisotopic masses from the multicharged, protonated ion series. Because TgGat1 MS spectra were not isotopically resolved, masses were extracted after MS spectra deconvolution using the ReSpect algorithm in the BioPharma Finder suite (Thermo Scientific), with a 20 ppm deconvolution mass tolerance and 25 ppm protein sequence matching mass tolerance. For consistency, PuGat1 was also deconvoluted and re-extracted using the ReSpect algorithm and the same conditions. The data raw files were uploaded to the Figshare server with ID 10.6084/m9.figshare.12272909 (recombinant Gat1 intact protein raw files, Fig. S10).
Structure determination of PuGat1
A PuGat1:UDP:Mn2+ complex in 50 mm HEPES-NaOH, pH 7.4, 75 mm NaCl, 2 mm DTT, 5 mm UDP, and 5 mm MnCl2 was crystallized at 20 °C using a hanging-drop vapor diffusion method over a reservoir containing 8–12% (w/v) PEG4000, 0.4 m ammonium sulfate, and 0.1 m sodium acetate at pH 4.0. Crystals were obtained overnight and were transferred to a reservoir solution containing 15% (v/v) of a cryoprotectant mixture (1:1:1, ethylene glycol/DMSO/glycerol) and flash-cooled with liquid N2. The complex crystallized in space group P42212 and diffracted to 1.76 Å (Table S3). X-ray data were collected remotely at the SER-CAT 22-BM beamline at the Argonne National Laboratory using a Fast Rayonix 300HS detector and processed using XDS (58), with 5% of the data omitted for cross-validation.
PuGat1:UDP:Mn2+ crystals were soaked with platinum cyanide for heavy-atom phasing, cryoprotected, and frozen as above. PuGat1:UDP:Pt2+ crystals were isomorphous to PuGat1:UDP:Mn2+ crystals and diffracted to 2.1 Å (Table S3). PuGat1:UDP:Mn2+ crystals were alternatively soaked in UO2 (not shown). The crystals, which diffracted to 2.4 Å in the same space group, lacked UDP density, suggesting displacement by UO2.
The crystal structure of PuGat1:Pt2+ was solved using single-wavelength anomalous dispersion. The data were obtained at a wavelength of 1.85 Å for maximum anomalous signal. A single Pt2+ site was located using PHENIX (59), and the resulting phases had an acceptable figure of merit of 0.31. The model was subjected to iterative cycles of refinement and yielded a final model with Rwork/Rfree of 0.21/0.24 (Table S3). The structure of PuGat1:UDP:Mn2+ was solved using rigid-body refinement of PuGat1:Pt2+. The resulting model was subjected to iterative cycles of refinement and yielded a final model with Rwork/Rfree of 0.18/0.21 (Table S3). Images were rendered in PyMOL (60), and secondary structures were assigned based on DSSP (61, 62).
Glycan docking
The lowest-energy conformation of the TgSkp1 tetrasaccharide (Glcα1,3Fucα1,2Galβ1,3GlcNAcα1-OH) was generated via GLYCAM (63). Hydrogen atoms were added, and the electrostatic surface was generated using AutoDockTools (64). A grid box with dimensions 26 × 26 × 34 Å was placed over the ligand-binding site based on where the acceptor is bound in the glycogenin/glucan complex. The ligand was kept rigid, because AutoDock Vina is not parameterized specific to glycan torsion angles. 100 binding modes were calculated with the lowest binding energy scored at −4.4 kcal/mol and the highest binding energy scored at −5.7 kcal/mol.
Sedimentation velocity studies
PuGat1 was further purified on a Superdex S200 gel filtration column (GE Healthcare) equilibrated with 20 mm potassium phosphate (pH 7.4), 50 mm KCl, 0.5 mm tris(2-carboxyethyl)phosphine. Protein concentration was calculated from A280 measured in an Agilent 8453 UV-visible spectrophotometer, based on a molar absorptivity (ε280) of 60,390 m−1 cm−1, which was calculated from the PuGat1 sequence using ProtParam (65). Samples were diluted to 0.3-11 μm, loaded into 12-mm double-sector Epon centerpieces equipped with quartz windows, and equilibrated for 2 h at 20 °C in an An60 Ti rotor. Sedimentation velocity data were collected using an Optima XLA analytical ultracentrifuge (Beckman Coulter) at a rotor speed of 50,000 rpm at 20 °C. Data were recorded at 280 nm for protein samples at 3.5–11 μm and at 230/220 nm for samples at 0.3–1.5 μm, in radial step sizes of 0.003 cm. SEDNTERP (66) was used to model the partial specific volume of PuGat1 (0.73818 ml/g) and the density (1.0034 g/ml) and viscosity (0.0100757 poise) of the buffer. Using SEDFIT (67), data were modeled as continuous c(s) distributions and were fit using baseline, meniscus, frictional coefficient, and systematic time-invariant and radial-invariant noise. Predicted sedimentation coefficient (s) values for the PuGat1 monomer (2.8 S) and dimer (4.2 S) were calculated using HYDROPRO (68). Data fit and c(s) plots were generated using GUSSI (69).
Molecular dynamics simulations
The model for TgSkp1 was built as described previously for DdSkp1 (7). Briefly, a homology model of TgSkp1 was generated with the SWISS-MODEL web server (70) based on the human Skp1 template from Protein Data Bank (PDB) entry 2ASS (71), and missing residues were appended with UCSF Chimera (72). Molecular dynamics simulations were performed as described previously. Briefly, MD simulations were performed with the pmemd.cuda version of AMBER14 (73). The amino acid and carbohydrate residues were parameterized with the FF12SB and GLYCAM06 (J-1) force fields, respectively (74, 75). The systems were neutralized with Na+ ions and solvated using the TIP3P water model (76) in a truncated octahedral box with 15-Å distance from the solute to the end of the unit cell. Electrostatic interactions were treated with the particle mesh–Ewald algorithm, and a cut-off for nonbonded interactions was set to 8 Å (77). SHAKE was employed to constrain hydrogen-containing bonds, enabling an integration time step of 2 fs. Restraints were imposed in specific situations and were enforced with a 10-kcal/mol Å2 energy barrier in each case. Each minimization step consisted of 1,000 cycles of the steepest descent method (1,000 cycles), followed by 24,000 cycles using the conjugate gradient approach. The systems were heated to 300 K under NVT conditions over 60 ps, employing the Berendsen thermostat with a coupling time constant of 1 ps. The subsequent simulations were performed under NPT conditions. A torsion term that corrects 4(trans)-hydroxyproline residue (Hyp) ring puckering was included in simulations of the O-linked residue type based on previous studies that indicate that the ring is primarily exo when glycosylated (78, 79). This torsion term has been adopted in GLYCAM06 (version K).
A 50-ns simulation of the protein was performed with Cα cartesian constraints on all amino acids except those generated by Chimera. The fully glycosylated isoform was created by adding the TgSkp1 pentasaccharide to the exo-pucker conformation of hydroxyproline (residue 154). Six independent simulations were performed. Three ran for 250 ns directly, whereas the other three began with an additional 50 ns in which the protein was restrained to allow the glycan time to adapt to the protein conformation.
Computational analysis
Structural images were created with Visual Molecular Dynamics (80) and the 3D-SNFG plugin (81). The structure depicted in Fig. 7 was created by identifying the frame from equil-1 that consisted of the lowest root mean square deviation to the average structure as calculated by cpptraj (82). The cpptraj program was also used to distribute the latter 200 ns of the six simulations into 48 bins containing 250 frames each for analysis. Per-residue MMGBSA energies were calculated with MMPBSA.py.MPI with igb = 2 and idecomp = 3. A Bash script was used to calculate the correlation coefficients (83).
Phylogenetic analysis of enzyme sequences
Proteins related to TgGat1 were searched for using a BLASTP (version 2.4.0) search seeded with the full-length TgGat1 protein sequence against the NCBI nonredundant database (December 2016). The evolutionary relationship of Gat1-like sequences was investigated by using a maximum likelihood method (84) and conducted in MEGA7 (85). Catalytic domains from 43 CAZy GT8 sequences were selected based on their relatedness to Gat1, glycogenin, or known function and consisted of 196 positions. Sequence alignments were manually curated in BioEdit (version 7.2.5). An initial tree(s) for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. A discrete γ distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.1608)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 1.02% of sites).
Data availability
All atomic coordinates and structure factors were deposited in the RCSB PDB under accession codes 6MW5 (PuGat1 Pt2+ derivative) and 6MW8 (PuGat1 native). The raw data files for MS were uploaded to the Figshare server with ID 10.6084/m9.figshare.12272882 (Skp1 glycopeptides raw data, Fig. S5) and ID 10.6084/m9.figshare.12272909 (recombinant Gat1 intact protein raw files, Fig. S10).
Supplementary Material
Acknowledgments
We thank Kentuan Hicks, Nathan Beattie, and Safal Shrestha for assistance.
This article contains supporting information.
Author contributions—M. M., H. W. K., D. T., K. R., H. v. d. W., R. J. W., Z. A. W., and C. M. W. conceptualization; M. M., H. W. K., D. T., M. O. S., E. G.-P., K. R., and C. M. W. formal analysis; M. M. validation; M. M., H. W. K., D. T., M. O. S., E. G.-P., K. R., P. Z., N. G. D., H. v. d. W., H. T. I., and J. N. G. investigation; M. M., H. W. K., D. T., M. O. S., E. G.-P., K. R., P. Z., N. G. D., H. v. d. W., H. T. I., J. N. G., L. W., R. J. W., Z. A. W., and C. M. W. methodology; M. M., H. W. K., D. T., M. O. S., E. G.-P., K. R., and C. M. W. writing-original draft; M. M., H. W. K., D. T., R. J. W., Z. A. W., and C. M. W. writing-review and editing; H. W. K., M. O. S., E. G.-P., P. Z., and C. M. W. data curation; D. T., E. G.-P., and C. M. W. visualization; L. W., R. J. W., Z. A. W., and C. M. W. supervision; C. M. W. resources; C. M. W. funding acquisition; C. M. W. project administration.
Funding and additional information—M. M. was supported in part by National Institutes of Health (NIH) Grant T32-AI060546; H. W. K. was supported in part by NIH Grant T32-GM107004; and K. H. was supported by National Science Foundation REU Grant DBI-1426834. This project was supported in part by NIH Grant RO1-GM084383 (to C. M. W. and Ira Blader), Grant #14-140 from the Mizutani Foundation for Glycoscience (to C. M. W. and Ira Blader), NIH Grant P41-GM103490 (to L. W., senior investigator), NIH Grant 8P41-GM103390 (Resource for Integrated Glycotechnology to J. H. Prestegard), NIH Grant R01-GM114298 (to Z. A. W.), and NIH Grant S10-OD021762 (to John Rose). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- SCF
- Skp1/Cullin-1/F-box protein subcomplex of E3 Cullin-RING-1 ubiquitin ligases
- DBA
- Dolichos biflorus lectin
- Dd
- Dictyostelium discoideum
- FBP
- F-box protein
- GaGlFGaGn–
- Galα1,3Glcα1,3Fucα1,2Galβ1,3GlcNAcα1–
- GT
- glycosyltransferase
- Hyp
- (2S,4R)-4-hydroxy-l-proline
- pAb
- polyclonal antibody
- pNP
- para-nitrophenol
- Pu
- Pythium ultimum
- RHΔΔ
- RHΔku80Δhxgprt
- Tg
- Toxoplasma gondii
- Oc
- Oryctolagus cuniculus
- TEV
- tobacco etch virus
- COSY
- correlation spectroscopy
- TOCSY
- total correlation spectroscopy
- HMBC
- heteronuclear multiple-bond correlation spectroscopy
- HSQC
- heteronuclear single-quantum correlation spectroscopy
- HFF
- human foreskin fibroblast
- RT
- room temperature
- ACN
- acetonitrile
- FA
- formic acid
- PDB
- Protein Data Bank.
References
- 1. Islam M. S., Leissing T. M., Chowdhury R., Hopkinson R. J., and Schofield C. J. (2018) 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 87, 585–620 10.1146/annurev-biochem-061516-044724 [DOI] [PubMed] [Google Scholar]
- 2. Pugh C. W., and Ratcliffe P. J. (2017) New horizons in hypoxia signaling pathways. Exp. Cell Res. 356, 116–121 10.1016/j.yexcr.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. West C. M., and Blader I. J. (2015) Oxygen sensing by protozoans: how they catch their breath. Curr. Opin. Microbiol. 26, 41–47 10.1016/j.mib.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Willems A. R., Schwab M., and Tyers M. (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695, 133–170 10.1016/j.bbamcr.2004.09.027 [DOI] [PubMed] [Google Scholar]
- 5. West C. M., van der Wel H., and Wang Z. A. (2007) Prolyl 4-hydroxylase-1 mediates O2 signaling during development of Dictyostelium. Development 134, 3349–3358 10.1242/dev.000893 [DOI] [PubMed] [Google Scholar]
- 6. West C. M., and Kim H. W. (2019) Nucleocytoplasmic O-glycosylation in protists. Curr. Opin. Struct. Biol. 56, 204–212 10.1016/j.sbi.2019.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheikh M. O., Thieker D., Chalmers G., Schafer C. M., Ishihara M., Azadi P., Woods R. J., Glushka J. N., Bendiak B., Prestegard J. H., and West C. M. (2017) O2 sensing-associated glycosylation exposes the F-box-combining site of the Dictyostelium Skp1 subunit in E3 ubiquitin ligases. J. Biol. Chem. 292, 18897–18915 10.1074/jbc.M117.809160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z. A., Singh D., van der Wel H., and West C. M. (2011) Prolyl hydroxylation- and glycosylation-dependent functions of Skp1 in O2-regulated development of Dictyostelium. Dev. Biol. 349, 283–295 10.1016/j.ydbio.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheikh M. O., Schafer C. M., Powell J. T., Rodgers K. K., Mooers B. H., and West C. M. (2014) Glycosylation of Skp1 affects its conformation and promotes binding to a model F-box protein. Biochemistry 53, 1657–1669 10.1021/bi401707y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sheikh M. O., Xu Y., van der Wel H., Walden P., Hartson S. D., and West C. M. (2015) Glycosylation of Skp1 promotes formation of Skp1/cullin-1/F-box protein complexes in Dictyostelium. Mol. Cell. Proteomics 14, 66–80 10.1074/mcp.M114.044560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halonen S. K., and Weiss L. M. (2013) Toxoplasmosis. Handb. Clin. Neurol. 114, 125–145 10.1016/B978-0-444-53490-3.00008-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y., Brown K. M., Wang Z. A., van der Wel H., Teygong C., Zhang D., Blader I. J., and West C. M. (2012) The Skp1 protein from Toxoplasma is modified by a cytoplasmic prolyl 4-hydroxylase associated with oxygen sensing in the social amoeba Dictyostelium. J. Biol. Chem. 287, 25098–25110 10.1074/jbc.M112.355446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rahman K., Zhao P., Mandalasi M., van der Wel H., Wells L., Blader I. J., and West C. M. (2016) The E3 ubiquitin ligase adaptor protein Skp1 Is glycosylated by an evolutionarily conserved pathway that regulates protist growth and development. J. Biol. Chem. 291, 4268–4280 10.1074/jbc.M115.703751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rahman K., Mandalasi M., Zhao P., Sheikh M. O., Taujale R., Kim H. W., van der Wel H., Matta K., Kannan N., Glushka J. N., Wells L., and West C. M. (2017) Characterization of a cytoplasmic glucosyltransferase that extends the core trisaccharide of the Toxoplasma Skp1 E3 ubiquitin ligase subunit. J. Biol. Chem. 292, 18644–18659 10.1074/jbc.M117.809301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lévesque C. A., Brouwer H., Cano L., Hamilton J. P., Holt C., Huitema E., Raffaele S., Robideau G. P., Thines M., Win J., Zerillo M. M., Beakes G. W., Boore J. L., Busam D., Dumas B., et al. (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, R73 10.1186/gb-2010-11-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kamoun S., Furzer O., Jones J. D., Judelson H. S., Ali G. S., Dalio R. J., Roy S. G., Schena L., Zambounis A., Panabières F., Cahill D., Ruocco M., Figueiredo A., Chen X. R., Hulvey J., et al. (2015) The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434 10.1111/mpp.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaastra W., Lipman L. J., De Cock A. W., Exel T. K., Pegge R. B., Scheurwater J., Vilela R., and Mendoza L. (2010) Pythium insidiosum: an overview. Vet. Microbiol. 146, 1–16 10.1016/j.vetmic.2010.07.019 [DOI] [PubMed] [Google Scholar]
- 18. van der Wel H., Gas-Pascual E., and West C. M. (2019) Skp1 isoforms are differentially modified by a dual function prolyl 4-hydroxylase/N-acetylglucosaminyltransferase in a plant pathogen. Glycobiology 29, 705–714 10.1093/glycob/cwz049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gas-Pascual E., Ichikawa H. T., Sheikh M. O., Serji M. I., Deng B., Mandalasi M., Bandini G., Samuelson J., Wells L., and West C. M. (2019) CRISPR/Cas9 and glycomics tools for Toxoplasma glycobiology. J. Biol. Chem. 294, 1104–1125 10.1074/jbc.RA118.006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sabin A. B. (1941) Toxoplasmic encephalitis in children. J. Am. Med. Assoc. 116, 801–807 10.1001/jama.1941.02820090001001 [DOI] [Google Scholar]
- 22. Fox B. A., Ristuccia J. G., Gigley J. P., and Bzik D. J. (2009) Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot. Cell 8, 520–529 10.1128/EC.00357-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grainger J. R., Wohlfert E. A., Fuss I. J., Bouladoux N., Askenase M. H., Legrand F., Koo L. Y., Brenchley J. M., Fraser I. D., and Belkaid Y. (2013) Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat. Med. 19, 713–721 10.1038/nm.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blader I. J., Coleman B. I., Chen C. T., and Gubbels M. J. (2015) Lytic cycle of Toxoplasma gondii: 15 years later. Annu. Rev. Microbiol. 69, 463–485 10.1146/annurev-micro-091014-104100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bisio H., and Soldati-Favre D. (2019) Signaling cascades governing entry into and exit from host cells by Toxoplasma gondii. Annu. Rev. Microbiol. 73, 579–599 10.1146/annurev-micro-020518-120235 [DOI] [PubMed] [Google Scholar]
- 26. Ball S., Colleoni C., Cenci U., Raj J. N., and Tirtiaux C. (2011) The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J. Exp. Bot. 62, 1775–1801 10.1093/jxb/erq411 [DOI] [PubMed] [Google Scholar]
- 27. Coppin A., Dzierszinski F., Legrand S., Mortuaire M., Ferguson D., and Tomavo S. (2003) Developmentally regulated biosynthesis of carbohydrate and storage polysaccharide during differentiation and tissue cyst formation in Toxoplasma gondii. Biochimie 85, 353–361 10.1016/S0300-9084(03)00076-2 [DOI] [PubMed] [Google Scholar]
- 28. Sugi T., Tu V., Ma Y., Tomita T., and Weiss L. M. (2017) Toxoplasma gondii requires glycogen phosphorylase for balancing amylopectin storage and for efficient production of brain cysts. mBio 8, 10.1128/mBio.01289-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coppin A., Varré J.-S. P., Lienard L., Dauvillée D., Guérardel Y., Soyer-Gobillard M.-O., Buléon A., Ball S., and Tomavo S. (2005) Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J. Mol. E 60, 257–267 10.1007/s00239-004-0185-6 [DOI] [PubMed] [Google Scholar]
- 30. Pancha I., Shima H., Higashitani N., Igarashi K., Higashitani A., Tanaka K., and Imamura S. (2019) Target of rapamycin-signaling modulates starch accumulation via glycogenin phosphorylation status in the unicellular red alga Cyanidioschyzon merolae. Plant J. 97, 485–499 10.1111/tpj.14136 [DOI] [PubMed] [Google Scholar]
- 31. Pancha I., Tanaka K., and Imamura S. (2019) Overexpression of a glycogenin, CmGLG2, enhances floridean starch accumulation in the red alga Cyanidioschyzon merolae. Plant. Signal. Behav. 14, 1596718 10.1080/15592324.2019.1596718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Slabinski L., Jaroszewski L., Rychlewski L., Wilson I. A., Lesley S. A., and Godzik A. (2007) XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23, 3403–3405 10.1093/bioinformatics/btm477 [DOI] [PubMed] [Google Scholar]
- 33. Lomako J., Lomako W. M., and Whelan W. J. (1988) A self-glucosylating protein is the primer for rabbit muscle glycogen biosynthesis. FASEB J. 2, 3097–3103 10.1096/fasebj.2.15.2973423 [DOI] [PubMed] [Google Scholar]
- 34. de Paula R. M., Wilson W. A., Roach P. J., Terenzi H. F., and Bertolini M. C. (2005) Biochemical characterization of Neurospora crassa glycogenin (GNN), the self-glucosylating initiator of glycogen synthesis. FEBS Lett. 579, 2208–2214 10.1016/j.febslet.2005.02.075 [DOI] [PubMed] [Google Scholar]
- 35. Cao Y., Steinrauf L. K., and Roach P. J. (1995) Mechanism of glycogenin self-glucosylation. Arch. Biochem. Biophys. 319, 293–298 10.1006/abbi.1995.1295 [DOI] [PubMed] [Google Scholar]
- 36. Alonso M. D., Lomako J., Lomako W. M., Whelan W. J., and Preiss J. (1994) Properties of carbohydrate-free recombinant glycogenin expressed in an Escherichia coli mutant lacking UDP-glucose pyrophosphorylase activity. FEBS Lett. 352, 222–226 10.1016/0014-5793(94)00962-7 [DOI] [PubMed] [Google Scholar]
- 37. Guérardel Y., Leleu D., Coppin A., Liénard L., Slomianny C., Strecker G., Ball S., and Tomavo S. (2005) Amylopectin biogenesis and characterization in the protozoan parasite Toxoplasma gondii, the intracellular development of which is restricted in the HepG2 cell line. Microbes Infect. 7, 41–48 10.1016/j.micinf.2004.09.007 [DOI] [PubMed] [Google Scholar]
- 38. Jansson P. E., Stenutz R., and Widmalm G. (2006) Sequence determination of oligosaccharides and regular polysaccharides using NMR spectroscopy and a novel Web-based version of the computer program CASPER. Carbohydr Res. 341, 1003–1010 10.1016/j.carres.2006.02.034 [DOI] [PubMed] [Google Scholar]
- 39. Moremen K. W., and Haltiwanger R. S. (2019) Emerging structural insights into glycosyltransferase-mediated synthesis of glycans. Nat. Chem. Biol. 15, 853–864 10.1038/s41589-019-0350-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gibbons B. J., Roach P. J., and Hurley T. D. (2002) Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J. Mol. Biol. 319, 463–477 10.1016/S0022-2836(02)00305-4 [DOI] [PubMed] [Google Scholar]
- 41. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 42. Bazán S., Issoglio F. M., Carrizo M. E., and Curtino J. A. (2008) The intramolecular autoglucosylation of monomeric glycogenin. Biochem. Biophys. Res. Comm. 371, 328–332 10.1016/j.bbrc.2008.04.076 [DOI] [PubMed] [Google Scholar]
- 43. Bourne Y., and Henrissat B. (2001) Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11, 593–600 10.1016/S0959-440X(00)00253-0 [DOI] [PubMed] [Google Scholar]
- 44. Bilyard M. K., Bailey H. J., Raich L., Gafitescu M. A., Machida T., Iglésias-Fernández J., Lee S. S., Spicer C. D., Rovira C., Yue W. W., and Davis B. G. (2018) Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis. Nature 563, 235–240 10.1038/s41586-018-0644-7 [DOI] [PubMed] [Google Scholar]
- 45. Schafer C. M., Sheikh M. O., Zhang D., and West C. M. (2014) Novel regulation of Skp1 by the Dictyostelium AgtA α-galactosyltransferase involves the Skp1-binding activity of its WD40 repeat domain. J. Biol. Chem. 289, 9076–9088 10.1074/jbc.M113.528679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burki F., Roger A. J., Brown M. W., and Simpson A. G. B. (2020) The new tree of eukaryotes. Trends Ecol. E 35, 43–55 10.1016/j.tree.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 47. Brown M. W., Heiss A. A., Kamikawa R., Inagaki Y., Yabuki A., Tice A. K., Shiratori T., Ishida K. I., Hashimoto T., Simpson A. G. B., and Roger A. J. (2018) Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biol. E 10, 427–433 10.1093/gbe/evy014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeqiraj E., Tang X., Hunter R. W., García-Rocha M., Judd A., Deak M., von Wilamowitz-Moellendorff A., Kurinov I., Guinovart J. J., Tyers M., Sakamoto K., and Sicheri F. (2014) Structural basis for the recruitment of glycogen synthase by glycogenin. Proc. Natl. Acad. Sci. U.S.A. 111, E2831–E2840 10.1073/pnas.1402926111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torija M. J., Novo M., Lemassu A., Wilson W., Roach P. J., François J., and Parrou J. L. (2005) Glycogen synthesis in the absence of glycogenin in the yeast Saccharomyces cerevisiae. FEBS Lett. 579, 3999–4004 10.1016/j.febslet.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 50. Testoni G., Duran J., García-Rocha M., Vilaplana F., Serrano A. L., Sebastián D., López-Soldado I., Sullivan M. A., Slebe F., Vilaseca M., Muñoz-Cánoves P., and Guinovart J. J. (2017) Lack of glycogenin causes glycogen accumulation and muscle function impairment. Cell Metab. 26, 256–266.e4 10.1016/j.cmet.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 51. Shen B., Brown K., Long S., and Sibley L. D. (2017) Development of CRISPR/Cas9 for efficient genome editing in Toxoplasma gondii. Methods Mol. Biol. 1498, 79–103 10.1007/978-1-4939-6472-7_6 [DOI] [PubMed] [Google Scholar]
- 52. Stasic A. J., Chasen N. M., Dykes E. J., Vella S. A., Asady B., Starai V. J., and Moreno S. N. J. (2019) The Toxoplasma vacuolar H+-ATPase regulates intracellular pH and impacts the maturation of essential secretory proteins. Cell Reports 27, 2132–2146.e7 10.1016/j.celrep.2019.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soête M., Camus D., and Dubrametz J. F. (1994) Experimental induction of bradyzoite-specific antigen expression and cyst formation by the RH strain of Toxoplasma gondii in vitro. Exp. Parasitol. 78, 361–370 10.1006/expr.1994.1039 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y. W., Halonen S. K., Ma Y. F., Wittner M., and Weiss L. M. (2001) Initial characterization of CST1, a Toxoplasma gondii cyst wall glycoprotein. Infect. Immun. 69, 501–507 10.1128/IAI.69.1.501-507.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dubey J. P., Lindsay D. S., and Speer C. A. (1998) Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 11, 267–299 10.1128/CMR.11.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheikh M. O., Halmo S. M., Patel S., Middleton D., Takeuchi H., Schafer C. M., West C. M., Haltiwanger R. S., Avci F. Y., Moremen K. W., and Wells L. (2017) Rapid screening of sugar-nucleotide donor specificities of putative glycosyltransferases. Glycobiology 27, 206–212 10.1093/glycob/cww114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chinoy Z. S., Schafer C. M., West C. M., and Boons G. J. (2015) Chemical synthesis of a glycopeptide derived from Skp1 for probing protein specific glycosylation. Chemistry 21, 11779–11787 10.1002/chem.201501598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. DeLano W. L. (2018) The PyMOL Molecular Graphics System, version 2.1, Schroedinger, LLC, New York [Google Scholar]
- 61. Touw W. G., Baakman C., Black J., Te Beek T. A. H., Krieger E., Joosten R. P., and Vriend G. (2015) A series of PDB related databases for everyday needs. Nucleic Acids Res. 43, D364–D368 10.1093/nar/gku1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kabsch W., and Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- 63. Woods R. J. (2014) GLYCAM Web, Complex Carbohydrate Research Center, University of Georgia, Athens, GA [Google Scholar]
- 64. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 30, 2785–2791 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., and Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. in The Proteomics Protocols Handbook (Walker J. M., ed) pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 66. Laue T. M., Shah B. D., Ridgeway T. M., and Pelletier S. L. (1992) Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., and Horton J., eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 67. Schuck P. (2000) Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ortega A., Amorós D., and García de la Torre J. (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101, 892–898 10.1016/j.bpj.2011.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brautigam C. A. (2015) Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 10.1016/bs.mie.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 70. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 71. Hao B., Zheng N., Schulman B. A., Wu G., Miller J. J., Pagano M., and Pavletich N. P. (2005) Structural basis of the Cks1-dependent recognition of p27Kip1 by the SCFSkp2 ubiquitin ligase. Mol. Cell 20, 9–19 10.1016/j.molcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 72. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 73. Götz A. W., Williamson M. J., Xu D., Poole D., Le Grand S., and Walker R. C. (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555 10.1021/ct200909j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Case D. A., Cheatham T. E. 3rd, Darden T., Gohlke H., Luo R., Merz K. M. Jr, Onufriev A., Simmerling C., Wang B., and Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., and Woods R. J. (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 10.1002/jcc.20820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 77. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Physics 98, 10089–10092 10.1063/1.464397 [DOI] [Google Scholar]
- 78. Park S., Radmer R. J., Klein T. E., and Pande V. S. (2005) A new set of molecular mechanics parameters for hydroxyproline and its use in molecular dynamics simulations of collagen‐like peptides. J. Comput. Chem. 26, 1612–1616 10.1002/jcc.20301 [DOI] [PubMed] [Google Scholar]
- 79. Owens N. W., Braun C., O'Neil J. D., Marat K., and Schweizer F. (2007) Effects of glycosylation of (2S,4R)-4-hydroxyproline on the conformation, kinetics, and thermodynamics of prolyl amide isomerization. J. Am. Chem. Soc. 129, 11670–11671 10.1021/ja073488d [DOI] [PubMed] [Google Scholar]
- 80. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 81. Thieker D. F., Hadden J. A., Schulten K., and Woods R. J. (2016) 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology 26, 786–787 10.1093/glycob/cww076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Roe D. R., and Cheatham T. E. 3rd (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- 83. Miller B. R. 3rd, McGee T. D. Jr., Swails J. M., Homeyer N., Gohlke H., and Roitberg A. E. (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- 84. Le S. Q., and Gascuel O. (2008) An improved general amino acid replacement matrix. Mol. Biol. E 25, 1307–1320 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 85. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. E 33, 1870–1874 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 88. West C. M., van der Wel H., and Gaucher E. A. (2002) Complex glycosylation of Skp1 in Dictyostelium: implications for the modification of other eukaryotic cytoplasmic and nuclear proteins. Glycobiology 12, 17R–27R 10.1093/glycob/12.2.17R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All atomic coordinates and structure factors were deposited in the RCSB PDB under accession codes 6MW5 (PuGat1 Pt2+ derivative) and 6MW8 (PuGat1 native). The raw data files for MS were uploaded to the Figshare server with ID 10.6084/m9.figshare.12272882 (Skp1 glycopeptides raw data, Fig. S5) and ID 10.6084/m9.figshare.12272909 (recombinant Gat1 intact protein raw files, Fig. S10).