Abstract
CBFA2/RUNX1 partner transcriptional co-repressor 3 (CBFA2T3, also known as MTG16 or ETO2) is a myeloid translocation gene family protein that functions as a master transcriptional corepressor in hematopoiesis. Recently, it has been shown that CBFA2T3 maintains leukemia stem cell gene expression and promotes relapse in acute myeloid leukemia (AML). However, a role for CBFA2T3 in myeloid differentiation of AML has not been reported. Here, we show that CBFA2T3 represses retinoic acid receptor (RAR) target gene expression and inhibits all-trans-retinoic acid (ATRA)-induced myeloid differentiation of AML cells. ChIP-Seq revealed that CBFA2T3 targets the RARα/RXRα cistrome in U937 AML cells, predominantly at myeloid-specific enhancers associated with terminal differentiation. CRISPR/Cas9-mediated abrogation of CBFA2T3 resulted in spontaneous and ATRA-induced activation of myeloid-specific genes in a manner correlated with myeloid differentiation. Importantly, these effects were reversed by CBFA2T3 re-expression. Mechanistic studies showed that CBFA2T3 inhibits RAR target gene transcription by acting at an early step to regulate histone acetyltransferase recruitment, histone acetylation, and chromatin accessibility at RARα target sites, independently of the downstream, RAR-mediated steps of transcription. Finally, we validated the inhibitory effect of CBFA2T3 on RAR in multiple AML subtypes and patient samples. To our knowledge, this is the first study to show that CBFA2T3 down-regulation is both necessary and sufficient for enhancing ATRA-induced myeloid gene expression and differentiation of AML cells. Our findings suggest that CBFA2T3 can serve as a potential target for enhancing AML responsiveness to ATRA differentiation therapies.
Keywords: CBFA2/RUNX1 partner transcriptional co-repressor 3 (CBFA2T3), all-trans-retinoic acid (ATRA), acute myeloid leukemia (AML), cell differentiation, gene transcription, transcriptional corepressor, histone acetyltransferase, histone acetylation, chromatin accessibility, epigenetics, cancer biology, nuclear receptor, transcription corepressor, chromatin immunoprecipitation (ChIP), chromatin regulation
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous malignancies that arise from hematopoietic stem and progenitor cells (HSPCs) at various points in the hierarchy of myeloid development. A defining characteristic of AML is myeloid differentiation arrest, in which immature blasts suppress terminal maturation and thereby evade senescence and apoptosis. Under normal conditions, myeloid differentiation of HSPCs is facilitated by RARα/RXRα-induced gene transcription, which occurs upon binding of RARα to its ligand, all-trans-retinoic acid (ATRA), the predominant endogenous metabolite of vitamin A. ATRA binding promotes the release of transcriptional co-repressors and the recruitment of transcriptional activators, resulting in activation of genes involved in terminal myeloid differentiation (1).
Deregulated retinoid signaling has been extensively studied in the context of t(15;17) AML (also known as acute promyelocytic leukemia), which expresses a fusion protein between the promyelocytic leukemia protein (PML) and the retinoic acid receptor α (RARA), known as PML-RARα. The PML-RARα fusion protein is unresponsive to physiologic levels of ATRA and acts as a dominant inhibitor of retinoid signaling (2). A supraphysiological level of ATRA, however, is sufficient to activate PML-RARα, which renders t(15;17) AML highly sensitive to ATRA differentiation therapy. Other subtypes of AML maintain resistance to both physiologic and therapeutic concentrations of ATRA, and the mechanisms they use to evade normal RAR signaling remain unclear. There are several reports that non-t(15;17) AMLs develop indirect means of repressing retinoid signaling and differentiation, such as loss-of-function mutations and transcriptional repression of key myeloid transcription factors, including CEBPA, RUNX1, and PU.1 (3, 4). AML blasts can also up-regulate various cofactors and epigenetic enzymes that have been shown to directly or indirectly repress RARα/RXRα transcriptional activation, including LSD1, MN1, and PRAME (5–8). These discoveries have led to the promising use of LSD1 and HDAC inhibitors, which can sensitize non-t(15;17) AML to ATRA-mediated differentiation and apoptosis (9–11).
CBFA2T3 (also known as ETO2 or MTG16) is a hematopoietic corepressor that forms stoichiometric complexes with E-proteins (e.g. E2A and HEB) to facilitate exchange of coactivators including p300 and GCN5 histone acetyltransferases (HATs), with nuclear receptor corepressors (NCoR/SMRT) and HDACs (12, 13). Underscoring its important role in hematopoietic transcriptional regulation, CBFA2T3 is frequently involved in leukemogenic translocations producing CBFA2T3 fusion proteins, including RUNX1-CBFA2T3 in therapy-related AML (14) and CBFA2T3-GLIS2 in pediatric AMLs (15). WT CBFA2T3 plays important roles in both normal and malignant hematopoiesis, where it inhibits terminal erythro-megakaryocytic differentiation (16), maintains stemness of long-term hematopoietic stem cells (HSCs) (17), and promotes the expansion of leukemia stem cells (LSCs) and AML relapse (18). Intriguingly, CBFA2T3 knockout in murine HSPCs promotes differentiation along the granulo-monocytic lineage at the expense of erythro-megakaryocytic development, phenocopying the effect of ATRA treatment (19, 20).
We recently found that retinoic acid treatment rapidly down-regulates CBFA2T3 expression in NB-4 t(15;17) acute promyelocytic leukemia cells (18). Based on these results, we hypothesized that there is an antagonistic cross-talk between CBFA2T3 and RAR-driven transcription. Here, we demonstrate that CBFA2T3 is causally involved in inhibiting ATRA-mediated myeloid gene expression and AML differentiation. We also show that CBFA2T3 function is linked to decreased chromatin accessibility at RARα target genes via regulation of HAT recruitment and histone acetylation, independent of the downstream, RAR-involved steps of transcription. These results are supported by multiple complementary results at mechanistic, functional, and biological levels. In particular, our loss-of-function and gain-of-function assays have demonstrated a causal role for CBFA2T3 in regulating ATRA/RAR signaling. Finally, we validated these results in multiple AML cell lines representing different AML subtypes, and in AML patient samples, suggesting that CBFA2T3 is a general inhibitor of ATRA/RAR-induced myeloid differentiation. Thus, CBFA2T3 may serve as a new therapeutic target to overcome ATRA resistance in AML.
Results
CBFA2T3 targets the RAR/RXR cistrome
Given the reported role of CBFA2T3 in inhibiting hematopoietic differentiation, along with the documented function of RAR in promoting myeloid differentiation, we hypothesized that CBFA2T3 inhibits RAR-dependent transcription. Because this predicts that CBFA2T3 should bind to RAR sites, we performed ChIP-Seq assays and mapped the CBFA2T3 binding sites in U937 cells, an FAB M5 AML cell line minimally responsive to ATRA (21). CBFA2T3 ChIP-Seq was performed in both untreated (18) and CBFA2T3-overexpressed U937 cells to help identify the true binding sites of CBFA2T3. These binding sites were annotated to the nearest transcriptional start site (TSS) using HOMER (22). Next, to unbiasedly assess the function of the CBFA2T3-occupied genes, hypergeometric tests were performed to assess the enrichment of CBFA2T3-occupied genes across all known gene sets of the MSigDB database (Fig. S1). This analysis revealed highly significant enrichment of CBFA2T3 at (i) RARα/PML-RARα-bound genes, (ii) genes related to myeloid/leukocyte differentiation and activation, and (iii) genes depleted in hematopoietic/leukemic stem cells (HSCs/LSCs) (Fig. S1). Indeed, of the 22,596 gene sets tested, the “MARTENS_BOUND_BY_PML_RARA_FUSION” gene set was found to be the most significantly enriched set for CBFA2T3 binding. Together, these results suggest that CBFA2T3 is capable of targeting the RARα/PML-RARα cistrome.
To further validate these results, we analyzed public ChIP-Seq data sets of both PML-RARα/RXRα from PML-RARα–transduced U937 cells (23) and endogenous RARα from MV4-11 cells (an FAB M5 AML cell line) (24). Given that the above CBFA2T3 ChIP-Seq was performed in U937 cells not expressing PML-RARα, we limited the following analyses to the shared binding sites between PML-RARα in U937 cells and RARα in MV4-11 cells, thus excluding off-target peaks introduced by the PML moiety. Under these criteria, 21.1 and 23.5% of CBFA2T3 peaks contained RARα sites in untreated and CBFA2T3-overexpressing U937 cells, respectively, and 63.6% of RARα peaks were co-occupied by CBFA2T3 (Fig. 1A). Quantification of read densities from both experiments showed that ectopically expressed CBFA2T3 specifically localized to the RAR cistrome in a distribution matching that of RARα (Fig. 1B).
Figure 1.
CBFA2T3 is highly enriched at genomic loci occupied by RARα. A, peak overlap between CBFA2T3 peaks (endogenous and exogenously expressed) and RARα cistrome in U937 cells. Significance of overlap was assessed with hypergeometric tests. B, read coverage from α-CBFA2T3 ChIP-Seq in U937 cells within the RARα cistrome. Background regions were randomly selected with HOMER to match GC content of RARα cistromic sites. C, IGV tracks depicting CBFA2T3 and RARα binding in U937 and MV4-11 cells. α-H3K27ac ChIP-Seq, DNase-Seq, and RNA-Seq data from human CD34+ HSPCs (immature) and PBMCs (differentiated) were visualized to demarcate active enhancer regions related to myeloid differentiation. Tracks were normalized by 1,000,000/(total read count), and all tracks corresponding to the same ChIP or cell type (HSPCs versus PBMCs) were set to the same scaling factor.
Finally, we visualized gene loci with high degrees of overlap between CBFA2T3 and RARα (Fig. 1C). This revealed a number of myeloid-specific enhancers and promoters controlling the integrin CD11b (ITGAM), a marker of ATRA-mediated differentiation, PU.1 (SPI1), and CEBPA, master transcription factors required for terminal myeloid differentiation (25). Importantly, many of these regions have documented regulatory functions in myeloid differentiation, including the −14 kb SPI1 upstream regulatory element, of which heterozygous deletion is sufficient to cause AML in mice (4), and the +34/+42 kb CEBPA enhancers, which drive neutrophil- and monocyte-specific CEBPA expression (26). These data were further integrated with H3K27ac ChIP-Seq, DNase HS-Seq, and RNA-Seq data from primary CD34+ HSCs and differentiated PBMCs, showing that these regulatory loci are conserved in multiple hematopoietic cell types and have higher transcriptional activity upon terminal myeloid differentiation (Fig. 1C). In summary, the results presented above support a functional role for CBFA2T3 in regulating RARα-dependent transcription.
CBFA2T3 abrogation derepresses myeloid-specific transcriptional program
To further explore the functional involvement of CBFA2T3 in regulating RAR-mediated gene expression, we used CRISPR/Cas9-mediated gene editing to knock out CBFA2T3. To increase editing frequency while facilitating screening of the edited clones, a dual gRNA strategy (27) was used to target exons 8 and 10, a region that encodes the NHR2 domain necessary for the corepressor function of MTG corepressors including CBFA2T3 (28–30). A U937 clone (U937 C) was identified to contain a single-nucleotide insertion in exon 8, resulting in a premature stop codon shortly downstream of the intron/exon junction (Fig. 2A). This led to the loss of detectable CBFA2T3 protein expression and significantly reduced CBFA2T3 mRNA expression due to nonsense-mediated decay (31) of CBFA2T3 transcripts (Fig. 2, A and B) (data not shown).
Figure 2.
CBFA2T3 deletion enhances transcriptional response to ATRA in U937 cells. A, CBFA2T3 deletion strategy depicting targeted Sanger sequencing of clone C and Western blotting demonstrating loss of CBFA2T3 protein in U937 C cells. B, volcano plot depicting genes with significantly altered expression (red) between U937 C and U937 WT. Differential expression testing was performed with QL F-tests in edgeR. Ampli-Seq was performed in duplicate for each biological sample. C, gene set enrichment analysis shows significant enrichment of CBFA2T3 target genes, integrin pathway genes, and ATRA response genes among the genes up-regulated in U937 C. P values were empirically derived with permutation testing. D, heatmap of GROUP 1 and GROUP 2 genes among four sample groups (U937 WT/C ± ATRA), scaled across samples (rows). Among the genes that were highly/significantly up-regulated in U937 WT upon ATRA treatment (FDR < 0.005, n = 199), GROUP 1 genes were defined as those having higher expression in U937 C upon ATRA treatment (U937 C/WT > 1.2, n = 82) and GROUP 2 as those with lower expression (U937 C/WT < 0.8, n = 36). E, normalized expression (TPM) of GROUP 1 and GROUP 2 genes shows that GROUP 1 genes are more highly expressed in U937 C under basal conditions (DMSO). Significance was assessed with the Mann–Whitney U test. F, enrichment of gene sets within GROUP 1 and GROUP 2 gene categories assessed with the hypergeometric distribution and p values adjusted using the Benjamini–Hochberg method. Ratio represents the fraction of GROUP 1/2 genes also belonging to a given gene set. Significant results (FDR < 0.05) are labeled with the Q value. Error bars, S.D.
We assessed the global effect of CBFA2T3 loss on gene expression by performing whole-transcriptome Ampli-Seq assays in U937 WT and U937 C cells, treated with vehicle (DMSO) or ATRA (1 µm) (Table S2). Compared with U937 WT cells, U937 C cells expressed lower levels of CBFA2T3 expectedly and higher levels of genes related to granulocyte-specific integrin signaling and activation, including TYROBP, ITGAD, and CEACAM6 (32, 33). In total, in the absence of ATRA, 75 genes were significantly up-regulated, whereas 54 genes were down-regulated in U937 C cells.
Gene set enrichment analysis confirmed a genome-wide increase in the expression of CBFA2T3 target genes (Fig. 2C, top). The loss of CBFA2T3 also spontaneously increased the expression of integrin-related genes, which are highly expressed in differentiated leukocytes (Fig. 2C, middle). Finally, and in agreement with the ChIP-Seq data, abrogation of CBFA2T3 also significantly up-regulated ATRA-responsive genes (Fig. 2C, bottom). These results are consistent with an active role of CBFA2T3 in maintaining the normal repression of RAR target genes in U937 cells.
Next, we focused on a set of high-confidence ATRA-induced genes identified from U937 WT cells (n = 199, FDR < 0.005), many of which are bona fide ATRA-responsive genes, including NCF1, CD38, and CD11b/ITGAM (Fig. 2D). Of these 199 genes, 82 (GROUP 1) were more highly expressed in U937 C cells compared with U937 WT cells, both with and without ATRA treatment (Fig. 2E). This shows that CBFA2T3 is required for maintaining the repression of these genes to below-basal levels.
Interestingly, a smaller subset of genes (n = 32, GROUP 2) had a blunted response to ATRA in U937 C cells, prompting us to further characterize the functions of GROUP 1 and GROUP 2 genes. To this end, we compared gene sets enriched in each group, as well as in a control group of genes (OTHER, n = 81) showing no differential response to ATRA between U937 WT and U937 C. This analysis revealed that GROUP 1 genes were enriched for genes related to myeloid/granulocytic differentiation and genes down-regulated in normal and leukemic stem cells, which are known to express high levels of CBFA2T3 (18) (Fig. 2F). In contrast, GROUP 2 was enriched for genes that function as inhibitors of ATRA-mediated differentiation or antigens of LSCs, including LYN, CD93, and CD36 (34–36). These GROUP 2 genes may not be directly regulated by CBFA2T3 but perhaps serve as a downstream, counterregulatory response to promote resistance to myeloid differentiation.
Loss of CBFA2T3 sensitizes U937 cells to ATRA-induced myeloid differentiation
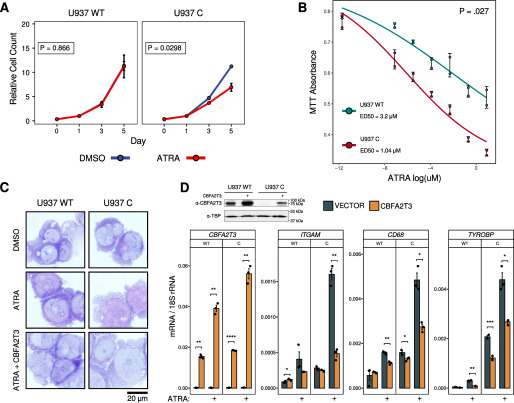
We next sought to explore the biological effects of CBFA2T3 ablation on myeloid differentiation. Treatment of U937 C cells with 1 µm ATRA decreased cell expansion compared with control U937 WT cells (Fig. 3A). Analyses of cell viability with MTT assays showed that U937 C cells had higher dose-dependent sensitivity to ATRA, with a nearly 3-fold reduction in ED50 (Fig. 3B). Further analyses of cell morphology with Wright–Giemsa staining showed that, whereas U937 WT had minimal changes upon ATRA treatment as reported (21), U937 C displayed evidence of increased differentiation, as shown by decreased nuclear/cytoplasmic ratios, decreased basophilia, and loss of nucleoli in cytospin cell preparations (Fig. 3C).
Figure 3.
U937 C cells exhibit increased sensitivity to ATRA-mediated differentiation. A. U937 WT and U937 C cells were treated with 1 μm ATRA over 5 days, and cell counts were recorded after trypan blue staining (n = 2/group). Generalized linear models were fit to assess cell count as a function of treatment and time point, and Wald tests were used to determine the significance of ATRA treatment as a predictive variable. B, the MTT assay was used to assess viable cell counts (n = 2) after 5 days of ATRA treatment over a range of concentrations. A four-parameter log-logistic model was used to fit curves and estimate ED50 concentrations. The p value represents the significance of cell line (WT/C) as a model variable obtained from the Wald test. C, Wright–Giemsa stains of U937 cells transduced with pCDH-MSCV-CBFA2T3 or an empty vector control and treated for 5 days with 1 μm ATRA. Slides were imaged at 100× magnification. D, top left, Western blotting analysis showing CBFA2T3 protein expression in empty vector and pCDH-MSCV-CBFA2T3 transduced U937 WT and U937 C cells, using TBP as a loading control. Top right and bottom, RT-qPCR was performed to measure CBFA2T3, ITGAM, CD68, and TYROBP gene expression after a 48-h treatment with 1 μm ATRA in U937 WT/C cells with or without pCDH-MSCV-CBFA2T3 (n = 3). In the Western blotting analysis, ectopic CBFA2T3 was expressed at lower levels in U937 C cells than its levels in U937 WT cells. This may conceivably be due to possible processing of CBFA2T3 transcripts by CAS9/sgRNA2 in the U937 C cells. In RT-qPCR assays, the primers used to detect CBFA2T3 expression were complementary to a 5′ region of the CBFA2T3 transcript, which may detect both unprocessed and processed CBFA2T3 transcripts, explaining why the result did not show reduced expression of the ectopic CBFA2T3 transcripts in the U937 C cells. Student's t test was used to assess significance (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001). Error bars, S.D.
To test whether the above effects were indeed due to the loss of CBFA2T3, we ectopically expressed CBFA2T3 in both U937 WT and U937 C cells. This not only increased CBFA2T3 protein expression in U937 WT cells but also restored its expression in U937 C cells (Fig. 3D, top). Functionally, the re-expression of CBFA2T3 prevented the morphological changes seen in ATRA-treated U937 C cells (Fig. 3C) and reversed spontaneous and ATRA-induced activation of GROUP 1 genes (ITGAM, CD68, and TYROBP) in the U937 C cells (Fig. 3D). Ectopic CBFA2T3 also blunted the low-level ligand-dependent response of U937 WT cells (Fig. 3D). Together, these results support a causal role for CBFA2T3 in inhibiting ATRA responses of RAR target gene transcription.
CBFA2T3 function is linked to decreased chromatin accessibility independent of RARα-involved steps of transcription
Activation of transcription may occur through mechanisms dependent on or independent of chromatin remodeling and accessibility. To provide a mechanistic insight into CBFA2T3 regulation of GROUP 1 genes, we focused on the CD11b gene (ITGAM), given its well-studied association with blast differentiation. We first assayed the effect of loss of CBFA2T3 expression on chromatin accessibility of the promoter and TSS regions of ITGAM by micrococcal nuclease (MNase) digestion assays (37). These regions include the −1 (UN1) and +1 (DN1) nucleosomes, the remodeling of which has been shown to be important for preinitiation complex assembly and transcriptional activation (38) (Fig. 4B). We first noted that chromatin derived from U937 C cells was more sensitive to MNase digestion compared with U937 WT cells (Fig. 4A). This supports a possible global chromatin compaction effect of CBFA2T3. Next, MNase-qPCR assays were performed to determine site-specific accessibility at the −1/+1 nucleosomes (UN1/DN1) as well as the nucleosome-free region (NFR)/ITGAM TSS (Fig. 4, B and C). To reveal gene-specific effects of CBFA2T3, we normalized the qPCR results to the housekeeping gene RPL30, whose expression was not affected by depletion of CBFA2T3 (data not shown), consistent with its lack of detectable binding by CBFA2T3 or RARα (Fig. 4B). The MNase-qPCR results showed that at all assayed ITGAM TSS-proximal sites, the chromatin was more accessible in U937 C cells compared with U937 WT cells (Fig. 4C).
Figure 4.
Loss of CBFA2T3 promotes increased accessibility at the CD11b (ITGAM) locus. A, chromatin isolated from U937 WT or U937 C was subject to digestion with increasing concentrations of MNase, and nucleosomal-length DNA distribution was visualized with gel electrophoresis. B, IGV tracks (from Fig. 1) showing the specific sites analyzed in our MNase- and ChIP-qPCR experiments. CBFA2T3 and RARα bind strongly to this region, and higher CBFA2T3 binding (indicated in CD34+ HSCs, which express high levels of CBFA2T3) is associated with lower DNase and H3K27ac signals. The RPL30 site (right) shows a lack of CBFA2T3/RARα binding. The cell type and track annotations apply to both ITGAM and RPL30 gene loci. C, relative (digested/undigested) nucleosomal protection of ITGAM sites was quantified via qPCR with specific primers (UN1, NFR, and DN1) to ensure a predominantly mononucleosomal distribution. The experiment was performed in technical triplicates and normalized to primers targeting a RPL30 nucleosomal site. D, ChIP-qPCR shows increased association of p300 and GCN5 at the NFR and DN1 sites, along with corresponding increases in H3K27ac/H3K9ac, in U937 C cells under basal conditions. The experiment was performed in technical triplicates, and a nonspecific IgG antibody was used as a negative control for immunoprecipitation efficiency. E, U937 WT and U937 C were transduced with pCDH-MSCV-FLAG-RARA or an empty vector control and treated with 1 μm ATRA for 24 h. RT-qPCR was performed to measure ITGAM expression. F, 293T cells were co-transduced with CBFA2T3, FLAG-HEB, and FLAG-RARα, and cell lysates were immunoprecipitated (IP) with the α-CBFA2T3 antibody. FLAG-RARα was detected by anti-FLAG M2 antibody. FLAG-HEB was detected by an anti-HEB antibody. Error bars, S.D.
As a corepressor of E-proteins, CBFA2T3 has been shown to competitively prevent p300 and GCN5 recruitment (13, 30, 39). Analyses of ChIP-Seq also showed that the E-protein HEB was present at the CBFA2T3-binding site at ITGAM NFR/TSS in multiple AML cells (GSM1122309 (40) and GSM585586 (41)). We thus asked whether CBFA2T3 inhibits chromatin accessibility by inhibiting HAT recruitment and histone acetylation. ChIP-qPCR assays were performed to measure p300 and GCN5 recruitment and H3K27 and H3K9 acetylation, which are catalyzed by p300 and GCN5, respectively, to mark active, accessible chromatin. As shown in Fig. 4D, the loss of CBFA2T3 expression in U937 C cells strongly increased p300 and GCN5 recruitment to ITGAM NFR and DN1 with minimal effects on their levels at the RPL30 site. Acetyl-H3K27 and acetyl-H3K9 were also increased at NFR, DN1, and, to a lesser extent, UN1 in U937 C cells compared with U937 WT cells (Fig. 4D).
Because these results were obtained in the absence of exogenous RAR ligands, we hypothesized that CBFA2T3 regulated an early chromatin step prior to RARα recruitment. This predicted that, by making chromatin accessible to RARα, the loss of CBFA2T3 should render transcription susceptible to enhanced regulation by RARα under both DMSO and ATRA conditions. To test this, we ectopically expressed RARα in U937 WT and U937 C cells. Consistent with the prediction, ectopic RARα in U937 C cells dramatically increased expression of CD11b upon ATRA treatment (Fig. 4E). Empty vector–transduced U937 WT cells did not respond to ATRA (Fig. 4E), consistent with the idea that the chromatin was inaccessible to RARα binding. However, by increasing RARα expression, ectopic RARα in WT cells manifested a ligand-dependent gene transcription comparable in magnitude with that seen in the empty vector–transduced U937 C cells (Fig. 4E). This shows that sufficiently high levels of RARα (which mirrors aberrant PML-RARα expression) may partially overcome the inaccessible state of CBFA2T3-bound RAR cistromic sites to augment transcriptional activation upon ATRA treatment. In U937 C cells, by increasing chromatin accessibility, loss of CBFA2T3 expression allows endogenous RARα to bind and induce target gene activation in response to ATRA.
Notably, in the absence of ATRA, ectopic RARα strongly reduced the high basal level transcription seen in U937 C cells (Fig. 4E, DMSO, VECTOR versus RARα). This shows that ectopic RARα can function both as a potent repressor and as a potent activator, in the absence of CBFA2T3, under DMSO and ATRA conditions, respectively, thus supporting the idea that CBFA2T3 is not directly involved in modulating RARα's transcriptional activities. This was further indicated by co-immunoprecipitation assays showing that CBFA2T3 was incapable of binding to RARα in 293T cells overexpressing both proteins (Fig. 4F). In contrast, CBFA2T3 strongly bound to the HEB E-protein expectedly (13).
Taken together, these results support the idea that CBFA2T3 and RARα function at separate, sequential steps in ATRA/RARα-dependent gene transcription. CBFA2T3 inhibits chromatin accessibility to limit RARα recruitment, thereby preventing ATRA-induced, RARα-mediated gene activation. Mechanistically, CBFA2T3 precludes p300 and GCN5 HAT recruitment and histone acetylation, consistent with its demonstrated corepressor function for E-proteins (13, 30, 39).
CBFA2T3 generally inhibits ATRA-mediated effects in various AML cell types
To extend these findings to other AML cell types, we first examined HL-60 cells, an M2 AML that is relatively less mature than U937 cells (42). Depletion of CBFA2T3 using two independent shRNAs (Fig. 5A) inhibited cell growth in a manner synergistic with ATRA treatment (Fig. 5B). Although CBFA2T3 knockdown alone can inhibit HL-60 cell proliferation, it was insufficient to prevent long-term cell expansion or induce maximal differentiation without concomitant ATRA treatment (Fig. 5C). Assessing the effects of loss and gain of CBFA2T3 on basal and ATRA-induced expression of ITGAM showed that CBFA2T3 knockdown resulted in high-level, constitutive expression of CD11b in HL-60 cells (Fig. 5D). In overexpression assays, whereas ATRA can modestly induce ITGAM expression in a time-dependent manner, these effects were almost completely blocked by ectopic expression of CBFA2T3 (Fig. 5E). Finally, we provided evidence that depletion of CBFA2T3 promotes ATRA responses in multiple other AML cell lines, including SKNO-1, a model of M2, t(8;21)+ AML, and THP-1, a model of M5 AML (Fig. 6A).
Figure 5.
CBFA2T3 inhibits ATRA response in more immature HL-60 AML. A, CBFA2T3 mRNA expression in HL-60 cells 72 h after transduction with SCR (scrambled shRNA control) or CBFA2T3-specific shRNAs (#164 or #165). Whole-cell lysates for immunoblotting were similarly collected 72 h after transduction. B, transduced HL-60 cells were grown with 1 μm ATRA until reaching confluence, and counts were recorded after trypan blue staining (n = 2/group). A Wald test was used to assess treatment effect on cell expansion. C, Wright–Giemsa stains of HL-60 cells transduced with shCBFA2T3#164 or SCR and treated for 5 days with 1 μm ATRA. Slides were imaged at ×100 magnification. D, RT-qPCR was performed to measure ITGAM gene expression after a 48-h treatment with 1 μm ATRA (n = 3). Student's t test was used to assess significance (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). E, stably transduced HL-60 cells were treated with DMSO or ATRA for 2 and 4 days (fresh ATRA/medium added daily), and ITGAM expression was measured (n = 3) via RT-qPCR. Error bars, S.D.
Figure 6.
CBFA2T3 suppresses RARα function in multiple AML cell lines and across AML patients. A, 1 week following transduction, SKNO.1, U937, and THP-1 cells were grown with 1 μm ATRA until reaching confluence, and counts were recorded after trypan blue staining (n = 2/group). A Wald test was used to assess treatment effect on cell expansion. B, Pearson correlation among AML patients between CBFA2T3 expression and “gene set scores” (see “Experimental procedures”). High CBFA2T3 levels are associated with low expression of “BROWN_MYELOID_DEVELOPMENT_UP” (genes increasing with myeloid differentiation) and “MARTENS_TRETINOIN_RESPONSE_UP” (genes up-regulated in ATRA-treated NB-4 cells) in GSE14468 (n = 526) and GSE13159 (n = 1066). The fill of each point represents the patient's “CHYLA_CBFA2T3_TARGETS_UP” score (genes that are increased in CBFA2T3-/- murine HSPCs). Error bars, S.D.
CBFA2T3 antagonizes RAR signaling in primary AML patients
Finally, we sought to document the antagonistic effects between CBFA2T3 and RARα in clinical AML samples. To this end, we examined two of the largest AML microarray studies, the GSE14468 AML patient cohort (n = 526) (43) and the multicenter MILE cohort (GSE13159, n = 1066) (44), both of which include patients spanning multiple cytogenetic categories, FAB morphology categories, and prognoses. As a surrogate for RARα transcriptional activity, we computed a “gene set score” for each patient, representing the mean expression of all genes belonging to a particular gene set, z-scored across patients to prevent skewing toward highly expressed genes (see “Experimental procedures”). Using this metric, we observed highly significant, negative correlations between CBFA2T3 expression and the “BROWN_MYELOID_CELL_DEVELOPMENT_UP” gene set (163 genes) as well as the “MARTENS_TRETINOIN_RESPONSE_UP” set (856 genes) (Fig. 6B).
Next, representing the “CHYLA_CBFA2T3_TARGETS_UP” gene set (n = 373) (containing CBFA2T3 target genes up-regulated in CBFA2T3 knockout murine HSPCs) as a third fill variable revealed that patients with low CBFA2T3 levels and high RARα target gene expression also expressed higher levels of CBFA2T3 target genes. This shows decreased functionality of CBFA2T3 (and not merely decreased CBFA2T3 expression) in these low-CBFA2T3 and high-RARα activity patients. These data suggest a functional antagonism between CBFA2T3 and RAR/myeloid differentiation pathways, thus extending CBFA2T3/RARα antagonism to diverse AML patient populations.
Discussion
Together, these results demonstrate the novel finding that CBFA2T3 is a general inhibitor of the RAR-dependent myeloid differentiation program in AML. Our ChIP-Seq analyses reveal a strong, genome-wide overlap between CBFA2T3 and the RARα cistrome in U937 cells, and our functional studies, encompassing both loss-of-function and gain-of-function assays, demonstrate a direct, repressive effect of CBFA2T3 at these genomic loci. Mechanistically, multiple lines of evidence support the idea that CBFA2T3 mediates its anti-ATRA/RARα effects by acting at an early step to regulate HAT recruitment, histone acetylation, and chromatin accessibility at RARα target sites. This restricts RARα recruitment, thus inhibiting ATRA-dependent, RARα-mediated target gene transcription. An RARα-independent role for CBFA2T3 in antagonizing HAT recruitment and histone acetylation is well-supported by its previously demonstrated corepressor function for E-proteins (13, 39), which are part of the multiprotein pioneer transcription factor complex important for lineage specification in hematopoiesis (12, 45). Thus, optimal binding of RARα to its myeloid-specific target genes may require the assistance from these pioneer factors and their associated chromatin-remodeling enzymes to increase chromatin accessibility. CBFA2T3 inhibits this step to preclude ATRA-induced, RARα-mediated transcription. Adding further support to this idea, we found that ATRA-sensitive AML cells, including NB-4 and MV-4-11, and U937 cells overexpressing PML-RARα all had detectable levels of apo-RARα binding at myeloid-specific genes, such as ITGAM, whereas no such binding of endogenous apo-RARα can be detected in ATRA-resistant AML cells, such as WT U937 cells (data not shown). Our Ampli-Seq studies showed that depletion of CBFA2T3 does not affect RARα expression (data not shown). Most AML cells, including those belonging to minimally differentiated AML (M0/M1), also express a functional RARα (46). Thus, it is possible that a preoccupied, poised RARα (or PML-RARα) on chromatin sites is a major determinant of the ATRA sensitivity of AML cells. Finally, given that CBFA2T3 also binds to HDACs, a role for CBFA2T3 in early chromatin-dependent steps of RARα target gene transcription also sheds new light on the synergism between HDAC inhibitors and ATRA in promoting AML differentiation (47).
We identified 82 genes (GROUP 1) that were more highly expressed in U937 C upon ATRA treatment. Most of these genes play roles in functionally mature granulocytes, and the enrichment analysis returned many neutrophilic gene ontologies related to phagocytosis, superoxide production, and chemotaxis. Notably, however, many GROUP 1 genes are specifically expressed in the monocytic lineage, such as CD68, EMR1, and C2 among others (48–50). These results align with the fact that whereas RARα signaling has most commonly been associated with granulocytic differentiation, there is significant cross-talk with monocytic-macrophage differentiation, especially in combination with vitamin D treatment (51). This bilineage gene expression may also represent a “promiscuous” phase of multilineage gene expression, which has been shown to immediately precede loss of multipotency and lineage commitment in hematopoietic progenitor cells (52).
Although not directly addressed in this study, a number of observations support the notion that ATRA sensitivity may be linked to changes in CBFA2T3 expression. For example, we previously found that NB-4 cells, which are highly susceptible to ATRA-mediated differentiation, rapidly terminate active CBFA2T3 transcription upon ATRA treatment (18). Additionally, CBFA2T3 levels are increased in relapsed AML, a phenomenon that is driven mostly by M4 and M5 subtypes of AML. M4/M5 blasts exhibit a high degree of myelomonocytic differentiation and relatively low CBFA2T3 expression, which may present a selective pressure, favoring the expansion of high-CBFA2T3 subclones that have a greater capacity to resist RARα signaling and terminal differentiation.
Interestingly, GCN5 functions as an important driver of this post-remission up-regulation of CBFA2T3 (18). Recently, GCN5 was also shown to promote ATRA resistance in multiple AML subtypes (53). Therefore, the degree to which this depends on CBFA2T3 up-regulation is an important direction of future studies. Although HDAC inhibitors have been shown to promote ATRA sensitivity in vitro, their off-target toxicities have limited their clinical efficacy (54). CBFA2T3 may therefore serve as a new target to allow specific induction of the RAR-driven myeloid differentiation pathway. Conceivably, this may be achieved by combining different approaches (e.g. by inhibiting GCN5 to down-regulate CBFA2T3 expression, by preventing CBFA2T3 interactions with transcription factors such as E-proteins (39), or by preventing CBFA2T3 oligomerization (15)). These methods offer the hope of extending the success of ATRA differentiation therapy for t(15;17) AML to a majority of AML patients.
Experimental procedures
Cell culture, chemicals, plasmids, primers, and antibodies
AML cells were maintained in RPMI 1640 with 10% fetal bovine serum. Kasumi-1, U937, SKNO-1, and THP-1 cells have been described previously (18), and the HL-60 cell line was a gift from Herman Sintim (55). Lentiviral particles were produced in HEK293T cells as described previously (39). ATRA and validated shRNAs targeting CBFA2T3 were obtained from Millipore Sigma. For CBFA2T3 overexpression experiments, cDNA of the full-length NM_005187 isoform was ordered from Dharmacon (Clone ID: 5227349, Lafayette, CO, USA) and cloned into the pCDH-MSCV-EF1α-Puro lentivector (System Biosciences, Palo Alto, CA, USA). For RARα overexpression experiments, FLAG-RARα (pcDNA-FLAG-RARα, Addgene plasmid 35555) was cloned into the pCDH-MSCV-EF1α-Puro lentivector. All vectors were Sanger-sequenced to verify correct inserts, and stably transduced clones were selected for 48 h following infection with 2 μg/µl puromycin. CBFA2T3 protein was probed with an in-house antibody raised against the NHR1 domain. β-Actin (A2228, Millipore Sigma) or TBP (AB28175, Abcam, Cambridge, MA, USA) was used as loading control. All shRNA, gRNA, and qPCR primer sequences are listed in Table S1.
CRISPR-Cas9 deletion of CBFA2T3
gRNA sequences targeting exon 8 and 10 of CBFA2T3 (NM_005187) were obtained from GenScript (Piscataway, NJ, USA), cloned into the dual-cassette pKLV2.2-h7SKgRNA5-hU6gRNA5-PGKpuro-W (56) lentivector (Addgene plasmid 72666), and transduced into U937 cells stably expressing Cas9 from the pKLV2-EF1aBsd2ACas9-W lentivector (Addgene plasmid 67978). Following puromycin selection, cells were diluted to 48 cells/10 ml in RPMI 1640 diluted 1:2 with 45-µm-filtered media collected from the bulk U937 population and seeded in 100-µl wells. Wells receiving a single cell were marked the following day and grown up to clonal populations. Clones were assessed for CBFA2T3 protein expression, and knockout clones were subject to PCR amplification of the targeted regions for sequencing. Control U937 WT cells were obtained in an identical process but initially transduced with the empty lentivector lacking gRNAs. FLAG-tagged HEB plasmid has been described previously (30).
Gene expression, pathway, and patient analyses
RT-qPCR was performed as described previously (39). 18S rRNA was used for normalization control. For all RT-qPCR results in figures, experiments were performed in triplicate, and bars represent mean values, whereas error bars represent S.D. Whole-genome AmpliSeq analysis was performed on duplicate samples using Ion AmpliSeq Kit for Chef DL8 and Ion 540 Kit-Chef (Thermo Fisher Scientific, Waltham, MA, USA), and edgeR (57) was used to assess differential expression. Gene set enrichment analysis (58) was performed using fgsea (59) against MSigDB 30 C2 V5.1 gene sets. P values were empirically derived using 100,000 permutations. AML patient microarray studies, GSE14468 (43) and GSE13159 (44), were downloaded from GEO using GEOquery (60) and analyzed in R. To calculate “gene set scores” for each patient, we averaged the z-score (relative to all patients) of every gene in the gene set as follows,
| (Eq. 1) |
where σ represents S.D., Pi represents the raw expression value of gene i in an individual patient, P represents the average expression of gene i across all patients, and n represents the number of genes in a given gene set. Ontological enrichment of genes was performed with the clusterProfiler package (61).
ChIP-Seq and analysis
ChIP-Seq of overexpressed CBFA2T3 in U937 cells was performed as described previously, and ChIP-Seq data from endogenous CBFA2T3 are available at GSE126953 (18). Additional ChIP data for PML-RARα/RARα in U937 and MV4-11 cells were downloaded from SRA studies SRP001549 (23) and SRP103029 (24), respectively. Raw FASTQ files were aligned to hg19 with STAR (62), and peak calling, peak overlaps, motif enrichment, and tag count distributions were assessed using HOMER (22). CBFA2T3 peak ontology enrichment was performed with the chipenrich package (63). CBFA2T3 antibody (SC-9741) was obtained from (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Sequencing was performed at the Genome Technology Access Center at Washington University in St. Louis. Peaks were visualized in IGV, and additional H3K27ac, DNase, and RNA-Seq signal tracks were imported directly from the DeepBlue epigenomic data server (64).
Chromatin accessibility and ChIP-qPCR assays
MNase digestion and qPCR assays were performed as described previously (37). Briefly, we cross-linked chromatin in U937 cells with 4% paraformaldehyde and lysed the cells using the SimpleChip Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA, USA). Nuclei were further lysed in hypotonic lysis buffer (20 mm HEPES (pH 7.5), 0.25 m sucrose, 3 mm MgCl2, 0.2% Nonidet P-40, 3 mm 2-mercaptoethanol), and extracted chromatin was subject to MNase digestion for 20 min at 37 °C with varying concentrations. Following reverse cross-linking, DNA was extracted by phenol/chloroform/isoamyl alcohol (25:24:1). qPCR was used to quantify specific sequences (UN1/NFR/DN1), and the RPL30 exon 3 locus was used as a normalization control. ChIP-qPCR was performed using the SimpleChip Enzymatic Chromatin IP Kit as described previously (18). GCN5 and p300 antibodies were from Santa Cruz Biotechnology (SC-20698 and SC-584, respectively). H3K9ac and H3K27ac antibodies were from Abcam (ab32129 and ab4729, respectively).
Assessment of ATRA sensitivity
Dose-response sensitivity to ATRA was assessed by seeding cells in duplicate 100-µl wells at an initial concentration of 60,000 cells/ml with increasing concentrations of ATRA (0, 0.0008, 0.004, 0.02, 0.1, 0.5, and 2.5 µm). Cell viability was assessed with the MTT assay 5 days later according to the manufacturer's instructions (Promega, Madison, WI). Log-logistic models were fit with the drc package (65). To assess cell morphology, cells were washed with PBS and diluted to a concentration of 5E5 cells/ml, and cytospins were prepared using a Thermo Shandon Cytospin 4 at 500 rpm for 5 min. Slides were fixed in methanol for 1 min and stained with Wright–Giemsa (WS16, Millipore Sigma) according to the manufacturer's instructions. Slides were imaged on a Leica DM6 B bright-field microscope. For cell expansion assays, cells were seeded in duplicate at 10E4 cells/ml in 200 µl of RPMI 1640 with 1 µm ATRA and counted with a hemocytometer. Fresh medium with ATRA was added every other day to maintain a constant volume.
Coimmunoprecipitation assay
293T cells were transfected using TurboFect Transfection Reagent (Thermo Fisher Scientific). Cells were lysed in lysis buffer (20 mm HEPES, pH 7.9, 1 mm EDTA, 20% glycerol, and protease inhibitors) containing 400 mm NaCl and 0.5% Nonidet P-40. Cell extracts were immunoprecipitated with CBFA2T3 antibody (SC-9741). Following washing with lysis buffer, bound proteins were subjected to Western blotting analysis. Input lanes show 1.5% of the total. Anti-FLAG M2 antibody was from Sigma. HEB antibody was from Santa Cruz Biotechnology (SC-357).
Data availability
The ChIP-Seq results from CBFA2T3-transduced U937 cells have been deposited to the National Institutes of Health GEO database under GSE126953.
Supplementary Material
This article contains supporting information.
Author contributions—N. S., C. G., and J. Z. conceptualization; N. S. and C. G. data curation; N. S. and J. Z. formal analysis; N. S., C. G., and J. Z. investigation; N. S. writing-original draft; N. S., C. G., and J. Z. writing-review and editing; J. Z. resources; J. Z. supervision; J. Z. funding acquisition; J. Z. project administration.
Funding and additional information—This work is supported by National Institutes of Health Grants R01HL093195 (to J. Z.) and R21CA178513 (to J. Z.), a fund from Saint Louis University (to J. Z.), a Brennan Summer Fellowship Award from Saint Louis University (to N. S.), and National Institutes of Health Grant T32GM008306-26A1 (to N. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- AML
- acute myeloid leukemia
- CBFA2T3
- CBFA2/RUNX1 partner transcriptional co-repressor 3, or core-binding factor, runt domain, α subunit 2, translocated to 3, also known as MTG16 or ETO2
- PML
- promyelocytic leukemia
- ATRA
- all-trans-retinoic acid
- RAR
- retinoic acid receptor
- RXR
- retinoid X receptor
- HSPCs
- hematopoietic stem and progenitor cells
- LSCs
- leukemia stem cells
- TSS
- transcriptional start site
- ITGAM
- integrin subunit αM, also known as CD11b
- HAT
- histone acetyltransferase
- GCN5
- general control non-depressible protein 5
- p300
- E1A-binding protein p300
- HDAC
- histone dacetylase
- HSCs
- hematopoietic stem cells
- PBMC
- peripheral blood mononuclear cell
- FDR
- false discovery rate
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MNase
- micrococcal nuclease
- qPCR
- quantitative PCR
- NFR
- nucleosome-free region
- H3K9
- histone H3 Lys-9
- H3K27
- histone H3 Lys-27
- ac
- acetylation
- gRNA
- guide RNA.
References
- 1. Collins S. J. (2002) The role of retinoids and retinoic acid receptors in normal hematopoiesis. Leukemia 16, 1896–1905 10.1038/sj.leu.2402718 [DOI] [PubMed] [Google Scholar]
- 2. Ablain J., and de Thé H. (2014) Retinoic acid signaling in cancer: the parable of acute promyelocytic leukemia. Int. J. Cancer 135, 2262–2272 10.1002/ijc.29081 [DOI] [PubMed] [Google Scholar]
- 3. Takei H., and Kobayashi S. S. (2019) Targeting transcription factors in acute myeloid leukemia. Int. J. Hematol. 109, 28–34 10.1007/s12185-018-2488-1 [DOI] [PubMed] [Google Scholar]
- 4. Rosenbauer F., Wagner K., Kutok J. L., Iwasaki H., Le Beau M. M., Okuno Y., Akashi K., Fiering S., and Tenen D. G. (2004) Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36, 624–630 10.1038/ng1361 [DOI] [PubMed] [Google Scholar]
- 5. Schenk T., Chen W. C., Göllner S., Howell L., Jin L., Hebestreit K., Klein H.-U., Popescu A. C., Burnett A., Mills K., Casero R. A. Jr., Marton L., Woster P., Minden M. D., Dugas M., et al. (2012) Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 18, 605–611 10.1038/nm.2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niebel D., Kirfel J., Janzen V., Höller T., Majores M., and Gütgemann I. (2014) Lysine-specific demethylase 1 (LSD1) in hematopoietic and lymphoid neoplasms. Blood 124, 151–152 10.1182/blood-2014-04-569525 [DOI] [PubMed] [Google Scholar]
- 7. Heuser M., Argiropoulos B., Kuchenbauer F., Yung E., Piper J., Fung S., Schlenk R. F., Dohner K., Hinrichsen T., Rudolph C., Schambach A., Baum C., Schlegelberger B., Dohner H., Ganser A., et al. (2007) MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in patients with AML. Blood 110, 1639–1647 10.1182/blood-2007-03-080523 [DOI] [PubMed] [Google Scholar]
- 8. Bullinger L., Schlenk R. F., Gotz M., Botzenhardt U., Hofmann S., Russ A. C., Babiak A., Zhang L., Schneider V., Dohner K., Schmitt M., Dohner H., and Greiner J. (2013) PRAME-induced inhibition of retinoic acid receptor signaling-mediated differentiation–a possible target for ATRA response in AML without t(15;17). Clin. Cancer Res. 19, 2562–2571 10.1158/1078-0432.CCR-11-2524 [DOI] [PubMed] [Google Scholar]
- 9. Sugino N., Kawahara M., Tatsumi G., Kanai A., Matsui H., Yamamoto R., Nagai Y., Fujii S., Shimazu Y., Hishizawa M., Inaba T., Andoh A., Suzuki T., and Takaori-Kondo A. (2017) A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia 31, 2303–2314 10.1038/leu.2017.59 [DOI] [PubMed] [Google Scholar]
- 10. Smitheman K. N., Severson T. M., Rajapurkar S. R., McCabe M. T., Karpinich N., Foley J., Pappalardi M. B., Hughes A., Halsey W., Thomas E., Traini C., Federowicz K. E., Laraio J., Mobegi F., Ferron-Brady G., et al. (2019) Lysine specific demethylase 1 inactivation enhances differentiation and promotes cytotoxic response when combined with all-trans retinoic acid in acute myeloid leukemia across subtypes. Haematologica 104, 1156–1167 10.3324/haematol.2018.199190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang Y., Liao G., and Yu B. (2019) LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J. Hematol. Oncol. 12, 129–129 10.1186/s13045-019-0811-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steinauer N., Guo C., and Zhang J. (2017) Emerging roles of MTG16 in cell-fate control of hematopoietic stem cells and cancer. Stem Cells Int. 2017, 6301385–6301385 10.1155/2017/6301385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Kalkum M., Yamamura S., Chait B. T., and Roeder R. G. (2004) E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305, 1286–1289 10.1126/science.1097937 [DOI] [PubMed] [Google Scholar]
- 14. Gamou T., Kitamura E., Hosoda F., Shimizu K., Shinohara K., Hayashi Y., Nagase T., Yokoyama Y., and Ohki M. (1998) The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood 91, 4028–4037 10.1182/blood.V91.11.4028.411a45_4028_4037 [DOI] [PubMed] [Google Scholar]
- 15. Thirant C., Ignacimouttou C., Lopez C. K., Diop M. B., Le Mouël L., Thiollier C., Siret A., Dessen P., Aid Z., Rivière J., Rameau P., Lefebvre C., Khaled M., Leverger G., Ballerini P., et al. (2017) ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell 31, 452–465 10.1016/j.ccell.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 16. Schuh A. H., Tipping A. J., Clark A. J., Hamlett I., Guyot B., Iborra F. J., Rodriguez P., Strouboulis J., Enver T., Vyas P., and Porcher C. (2005) ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol. Cell Biol. 25, 10235–10250 10.1128/MCB.25.23.10235-10250.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer M. A., Moreno-Miralles I., Hunt A., Chyla B. J., and Hiebert S. W. (2012) Myeloid translocation gene 16 is required for maintenance of haematopoietic stem cell quiescence. EMBO J. 31, 1494–1505 10.1038/emboj.2011.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steinauer N., Guo C., Huang C., Wong M., Tu Y., Freter C. E., and Zhang J. (2019) Myeloid translocation gene CBFA2T3 directs a relapse gene program and determines patient-specific outcomes in AML. Blood Adv. 3, 1379–1393 10.1182/bloodadvances.2018028514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chyla B. J., Moreno-Miralles I., Steapleton M. A., Thompson M. A., Bhaskara S., Engel M., and Hiebert S. W. (2008) Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol. Cell Biol. 28, 6234–6247 10.1128/MCB.00404-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tocci A., Parolini I., Gabbianelli M., Testa U., Luchetti L., Samoggia P., Masella B., Russo G., Valtieri M., and Peschle C. (1996) Dual action of retinoic acid on human embryonic/fetal hematopoiesis: blockade of primitive progenitor proliferation and shift from multipotent/erythroid/monocytic to granulocytic differentiation program. Blood 88, 2878–2888 10.1182/blood.V88.8.2878.bloodjournal8882878 [DOI] [PubMed] [Google Scholar]
- 21. Shi L., Weng X.-Q., Sheng Y., Wu J., Ding M., and Cai X. (2016) Staurosporine enhances ATRA-induced granulocytic differentiation in human leukemia U937 cells via the MEK/ERK signaling pathway. Oncol. Rep. 36, 3072–3080 10.3892/or.2016.5123 [DOI] [PubMed] [Google Scholar]
- 22. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martens J. H. A., Brinkman A. B., Simmer F., Francoijs K.-J., Nebbioso A., Ferrara F., Altucci L., and Stunnenberg H. G. (2010) PML-RARα/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 17, 173–185 10.1016/j.ccr.2009.12.042 [DOI] [PubMed] [Google Scholar]
- 24. McKeown M. R., Corces M. R., Eaton M. L., Fiore C., Lee E., Lopez J. T., Chen M. W., Smith D., Chan S. M., Koenig J. L., Austgen K., Guenther M. G., Orlando D. A., Lovén J., Fritz C. C., et al. (2017) Superenhancer analysis defines novel epigenomic subtypes of non-APL AML, including an RARα dependency targetable by SY-1425, a potent and selective RARα agonist. Cancer Discov. 7, 1136–1153 10.1158/2159-8290.CD-17-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pundhir S., Bratt Lauridsen F. K., Schuster M. B., Jakobsen J. S., Ge Y., Schoof E. M., Rapin N., Waage J., Hasemann M. S., and Porse B. T. (2018) Enhancer and transcription factor dynamics during myeloid differentiation reveal an early differentiation block in Cebpa null progenitors. Cell Rep. 23, 2744–2757 10.1016/j.celrep.2018.05.012 [DOI] [PubMed] [Google Scholar]
- 26. Avellino R., Havermans M., Erpelinck C., Sanders M. A., Hoogenboezem R., van de Werken H. J. G., Rombouts E., van Lom K., van Strien P. M. H., Gebhard C., Rehli M., Pimanda J., Beck D., Erkeland S., Kuiken T., et al. (2016) An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation. Blood 127, 2991–3003 10.1182/blood-2016-01-695759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bauer D. E., Canver M. C., and Orkin S. H. (2015) Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp. e52118 10.3791/52118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gelmetti V., Zhang J., Fanelli M., Minucci S., Pelicci P. G., and Lazar M. A. (1998) Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell Biol. 18, 7185–7191 10.1128/mcb.18.12.7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J., Hug B. A., Huang E. Y., Chen C. W., Gelmetti V., Maccarana M., Minucci S., Pelicci P. G., and Lazar M. A. (2001) Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell Biol. 21, 156–163 10.1128/MCB.21.1.156-163.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo C., Hu Q., Yan C., and Zhang J. (2009) Multivalent binding of the ETO corepressor to E proteins facilitates dual repression controls targeting chromatin and the basal transcription machinery. Mol. Cell Biol. 29, 2644–2657 10.1128/MCB.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Popp M. W., and Maquat L. E. (2016) Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell 165, 1319–1322 10.1016/j.cell.2016.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuijpers T. W., Hoogerwerf M., van der Laan L. J., Nagel G., van der Schoot C. E., Grunert F., and Roos D. (1992) CD66 nonspecific cross-reacting antigens are involved in neutrophil adherence to cytokine-activated endothelial cells. J. Cell Biol. 118, 457–466 10.1083/jcb.118.2.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pereira S., and Lowell C. (2003) The Lyn tyrosine kinase negatively regulates neutrophil integrin signaling. J. Immunol. 171, 1319–1327 10.4049/jimmunol.171.3.1319 [DOI] [PubMed] [Google Scholar]
- 34. Iriyama N., Yuan B., Hatta Y., Takagi N., and Takei M. (2016) Lyn, a tyrosine kinase closely linked to the differentiation status of primary acute myeloid leukemia blasts, associates with negative regulation of all-trans retinoic acid (ATRA) and dihydroxyvitamin D3 (VD3)-induced HL-60 cells differentiation. Cancer Cell Int. 16, 37–37 10.1186/s12935-016-0314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iwasaki M., Liedtke M., Gentles A. J., and Cleary M. L. (2015) CD93 marks a non-quiescent human leukemia stem cell population and is required for development of MLL-rearranged acute myeloid leukemia. Cell Stem Cell 17, 412–421 10.1016/j.stem.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farge T., Saland E., de Toni F., Aroua N., Hosseini M., Perry R., Bosc C., Sugita M., Stuani L., Fraisse M., Scotland S., Larrue C., Boutzen H., Féliu V., Nicolau-Travers M.-L., et al. (2017) Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 7, 716–735 10.1158/2159-8290.CD-16-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alenghat T., Yu J., and Lazar M. A. (2006) The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor. EMBO J. 25, 3966–3974 10.1038/sj.emboj.7601280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang C., and Pugh B. F. (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 10.1038/nrg2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gow C.-H., Guo C., Wang D., Hu Q., and Zhang J. (2014) Differential involvement of E2A-corepressor interactions in distinct leukemogenic pathways. Nucleic Acids Res. 42, 137–152 10.1093/nar/gkt855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mandoli A., Singh A. A., Jansen P. W., Wierenga A. T., Riahi H., Franci G., Prange K., Saeed S., Vellenga E., Vermeulen M., Stunnenberg H. G., and Martens J. H. (2014) CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 28, 770–778 10.1038/leu.2013.257 [DOI] [PubMed] [Google Scholar]
- 41. Martens J. H., Mandoli A., Simmer F., Wierenga B. J., Saeed S., Singh A. A., Altucci L., Vellenga E., and Stunnenberg H. G. (2012) ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood 120, 4038–4048 10.1182/blood-2012-05-429050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dalton W. T. Jr., Ahearn M. J., McCredie K. B., Freireich E. J., Stass S. A., and Trujillo J. M. (1988) HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood 71, 242–247 10.1182/blood.V71.1.242.bloodjournal711242 [DOI] [PubMed] [Google Scholar]
- 43. Wouters B. J., Löwenberg B., Erpelinck-Verschueren C. A. J., van Putten W. L. J., Valk P. J. M., and Delwel R. (2009) Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 113, 3088–3091 10.1182/blood-2008-09-179895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kohlmann A., Kipps T. J., Rassenti L. Z., Downing J. R., Shurtleff S. A., Mills K. I., Gilkes A. F., Hofmann W.-K., Basso G., Dell'orto M. C., Foà R., Chiaretti S., De Vos J., Rauhut S., Papenhausen P. R., et al. (2008) An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br. J. Haematol. 142, 802–807 10.1111/j.1365-2141.2008.07261.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li L., Jothi R., Cui K., Lee J. Y., Cohen T., Gorivodsky M., Tzchori I., Zhao Y., Hayes S. M., Bresnick E. H., Zhao K., Westphal H., and Love P. E. (2011) Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat. Immunol. 12, 129–136 10.1038/ni.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grande A., Montanari M., Manfredini R., Tagliafico E., Zanocco-Marani T., Trevisan F., Ligabue G., Siena M., Ferrari S., and Ferrari S. (2001) A functionally active RARα nuclear receptor is expressed in retinoic acid non responsive early myeloblastic cell lines. Cell Death Differ. 8, 70–82 10.1038/sj.cdd.4400771 [DOI] [PubMed] [Google Scholar]
- 47. Trus M. R., Yang L., Suarez Saiz F., Bordeleau L., Jurisica I., and Minden M. D. (2005) The histone deacetylase inhibitor valproic acid alters sensitivity towards all trans retinoic acid in acute myeloblastic leukemia cells. Leukemia 19, 1161–1168 10.1038/sj.leu.2403773 [DOI] [PubMed] [Google Scholar]
- 48. Lubbers R., van Essen M. F., van Kooten C., and Trouw L. A. (2017) Production of complement components by cells of the immune system. Clin. Exp. Immunol. 188, 183–194 10.1111/cei.12952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chistiakov D. A., Killingsworth M. C., Myasoedova V. A., Orekhov A. N., and Bobryshev Y. V. (2017) CD68/macrosialin: not just a histochemical marker. Lab. Invest. 97, 4–13 10.1038/labinvest.2016.116 [DOI] [PubMed] [Google Scholar]
- 50. Waddell L. A., Lefevre L., Bush S. J., Raper A., Young R., Lisowski Z. M., McCulloch M. E. B., Muriuki C., Sauter K. A., Clark E. L., Irvine K. M., Pridans C., Hope J. C., and Hume D. A. (2018) ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 9, 2246–2246 10.3389/fimmu.2018.02246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakajima H., Kizaki M., Ueno H., Muto A., Takayama N., Matsushita H., Sonoda A., and Ikeda Y. (1996) All-trans and 9-cis retinoic acid enhance 1,25-dihydroxyvitamin D3-induced monocytic differentiation of U937 cells. Leuk. Res. 20, 665–676 10.1016/0145-2126(96)00020-3 [DOI] [PubMed] [Google Scholar]
- 52. Hu M., Krause D., Greaves M., Sharkis S., Dexter M., Heyworth C., and Enver T. (1997) Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785 10.1101/gad.11.6.774 [DOI] [PubMed] [Google Scholar]
- 53. Kahl M., Brioli A., Bens M., Perner F., Kresinsky A., Schnetzke U., Hinze A., Sbirkov Y., Stengel S., Simonetti G., Martinelli G., Petrie K., Zelent A., Böhmer F.-D., Groth M., et al. (2019) The acetyltransferase GCN5 maintains ATRA-resistance in non-APL AML. Leukemia 33, 2628–2639 10.1038/s41375-019-0581-y [DOI] [PubMed] [Google Scholar]
- 54. Quintás-Cardama A., Santos F. P. S., and Garcia-Manero G. (2011) Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia 25, 226–235 10.1038/leu.2010.276 [DOI] [PubMed] [Google Scholar]
- 55. Dayal N., Opoku-Temeng C., Hernandez D. E., Sooreshjani M. A., Carter-Cooper B. A., Lapidus R. G., and Sintim H. O. (2018) Dual FLT3/TOPK inhibitor with activity against FLT3-ITD secondary mutations potently inhibits acute myeloid leukemia cell lines. Future Med. Chem. 10, 823–835 10.4155/fmc-2017-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tzelepis K., Koike-Yusa H., De Braekeleer E., Li Y., Metzakopian E., Dovey O. M., Mupo A., Grinkevich V., Li M., Mazan M., Gozdecka M., Ohnishi S., Cooper J., Patel M., McKerrell T., et al. (2016) A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 10.1016/j.celrep.2016.09.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sergushichev A. A. (2016) [Algorithm for cumulative calculation of gene set enrichment statistic]. Sci. Tech. J. Inform. Technol. Mech. Optics 956–959 10.17586/2226-1494-2016-16-5-956-959 [DOI] [Google Scholar]
- 60. Davis S., and Meltzer P. S. (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 10.1093/bioinformatics/btm254 [DOI] [PubMed] [Google Scholar]
- 61. Yu G., Wang L.-G., Han Y., and He Q.-Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Welch R. P., Lee C., Imbriano P. M., Patil S., Weymouth T. E., Smith R. A., Scott L. J., and Sartor M. A. (2014) ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res 42, e105 10.1093/nar/gku463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Albrecht F., List M., Bock C., and Lengauer T. (2016) DeepBlue epigenomic data server: programmatic data retrieval and analysis of epigenome region sets. Nucleic Acids Res 44, W581–W586 10.1093/nar/gkw211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ritz C., Baty F., Streibig J. C., and Gerhard D. (2015) Dose-response analysis using R. PLoS ONE 10, e0146021 10.1371/journal.pone.0146021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-Seq results from CBFA2T3-transduced U937 cells have been deposited to the National Institutes of Health GEO database under GSE126953.