Abstract
When faced with increased osmolarity in the environment, many bacterial cells accumulate the compatible solute ectoine and its derivative 5-hydroxyectoine. Both compounds are not only potent osmostress protectants, but also serve as effective chemical chaperones stabilizing protein functionality. Ectoines are energy-rich nitrogen and carbon sources that have an ecological impact that shapes microbial communities. Although the biochemistry of ectoine and 5-hydroxyectoine biosynthesis is well understood, our understanding of their catabolism is only rudimentary. Here, we combined biochemical and structural approaches to unravel the core of ectoine and 5-hydroxy-ectoine catabolisms. We show that a conserved enzyme bimodule consisting of the EutD ectoine/5-hydroxyectoine hydrolase and the EutE deacetylase degrades both ectoines. We determined the high-resolution crystal structures of both enzymes, derived from the salt-tolerant bacteria Ruegeria pomeroyi and Halomonas elongata. These structures, either in their apo-forms or in forms capturing substrates or intermediates, provided detailed insights into the catalytic cores of the EutD and EutE enzymes. The combined biochemical and structural results indicate that the EutD homodimer opens the pyrimidine ring of ectoine through an unusual covalent intermediate, N-α-2 acetyl-l-2,4-diaminobutyrate (α-ADABA). We found that α-ADABA is then deacetylated by the zinc-dependent EutE monomer into diaminobutyric acid (DABA), which is further catabolized to l-aspartate. We observed that the EutD–EutE bimodule synthesizes exclusively the α-, but not the γ-isomers of ADABA or hydroxy–ADABA. Of note, α-ADABA is known to induce the MocR/GabR-type repressor EnuR, which controls the expression of many ectoine catabolic genes clusters. We conclude that hydroxy–α-ADABA might serve a similar function.
Keywords: ectoine, chemical chaperone, salt stress, microbial community, structural biology, EutD, EutE, hydrolase, catalytic mechanism, deacetylase, metalloenzyme, pyrimidine, X-ray crystallography, enzyme catalysis, enzyme structure
Introduction
Many members of the three domains of life use compatible solutes as cytoprotectants (1–4). These highly water-soluble organic compounds are compliant with the physiology of both prokaryotic and eukaryotic cells and can thus be accumulated to very high intracellular concentrations. The function-preserving attributes of compatible solutes for proteins and other cellular components are reflected in their description as chemical chaperones (5–7). Microorganisms have widely adopted these solutes as stress protectants against the detrimental effects of high environmental osmolarity and salinity (4, 8). The degree of osmotic stress imposed onto the bacterial cell sensitively determines the size of the cytoplasmic compatible solute pool. Consequently, high osmolarity instigated water efflux, drop of vital turgor, and an undue increase in molecular crowding are counteracted by the accumulation of these organic osmolytes (9, 10). Microorganisms exploit compatible solutes not only as osmostress protectants but also in their cellular defense against extremes in both high and low growth temperatures (11–13), desiccation (14, 15), and oxidative stress (16). Bacteria and Archaea can synthesize a considerable variety of compatible solutes (e.g. glycine betaine, l-proline, and trehalose) (2, 4, 8). Also, they can import many of them via osmotically regulated transport systems (9, 10). By these means, severely osmotically stressed cells can accumulate compatible solutes in concentrations of up to several hundred millimolars (1).
The tetrahydropyrimidines ectoine (17) and its derivative 5-hydroxyectoine (18) (Fig. 1A) are prominent members of compatible solutes synthesized or imported by microorganisms (19). Like other compatible solutes, ectoines are preferentially excluded from the immediate hydration shell of proteins (20), a property hindering protein aggregation and preserving macromolecular functionality (5, 6, 21). These attributes of ectoines, and their water-binding and anti-inflammatory nature, led to various commercial applications, in particular in the area of skin care. Using Halomonas elongata as a cell factory, an industrial scale biotechnological production process for ectoines can deliver these valuable natural products on the scale of tons annually (21, 22).
Figure 1.
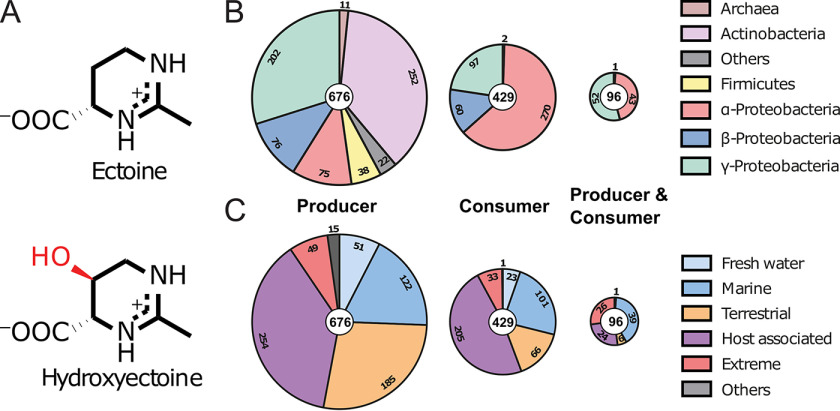
Eco-physiology of the ectoines. A, chemical structure of ectoine ((S)-2-methyl-1,4,5,6-tetrahydropyrimidine-4-carboxylic acid) and its derivative 5-hydroxyectoine ((4S,5S)-5-hydroxy-2-methyl-1,4,5,6-tetrahydropyrimidine-4-carboxylic acid). The 5-hydroxyl moiety of 5-hydroxyectoine is shown in red. B, distribution of microorganisms capable of ectoine production and/or degradation among different phyla. C, distribution of organisms capable of ectoine production and/or degradation among different habitats.
Reflecting the function of ectoines as osmostress protectants, transcription of their biosynthetic genes (ectABC/ectD) is up-regulated in response to increases in environmental osmolarity (23). Biosynthesis of ectoines is now a rather well biochemically and structurally understood process (23–26). Biosynthesis begins with the central microbial metabolite l-aspartate-β-semialdehyde, and involves the ectoine biosynthetic enzymes l-2,4-diaminobutyrate transaminase EctB (EC 2.6.1.76) (27, 28), l-2,4-diaminobutyrate acetyltransferase EctA (EC 2.3.1.178) (25), and ectoine synthase EctC (EC 4.2.1.108) (29), with l-2,4-diaminobutyrate (DABA) and N-γ-acetyl-l-2,4-diaminobutyrate (γ-ADABA) as the respective intermediates (24, 29). In a substantial subgroup of ectoine producers, ectoine can be further transformed into 5-hydroxyectoine through a regio- and stereospecific reaction catalyzed by the ectoine hydroxylase EctD (EC 1.14.11.55) (30, 31). Like compatible solute production in general (32), de novo synthesis of ectoine is costly. When the entire biosynthetic process is considered, cells growing heterotrophically in a minimal medium with glucose as the sole carbon source need to spend ∼40 ATP equivalents to produce just a single ectoine molecule (32).
In addition to their stress-protective and function-preserving properties, the nitrogen- and carbon-rich ectoine and 5-hydroxyectoine molecules (Fig. 1A) have found uses as nutrients for microorganisms (33–37). Ectoines are released into the environment from microbial producer cells through the transient opening of mechano-sensitive channels upon sudden osmotic down-shock (38), or simply when cells disintegrate. Environmental ectoines can then be re-captured by members of the microbial community either for re-use as osmostress protectants or recycled as nutrients (10, 23). Ectoine catabolism might also be triggered when cells capable of both ectoine synthesis and consumption are slowly shifted from high to lower environmental osmolarities (37).
A pioneering proteomics study by Jebbar et al. (35) focusing on the plant root-associated bacterium Sinorhizobium meliloti identified a substrate-inducible ABC-type transport system for the import of ectoines and several enzymes required for their catabolism. This study promoted the identification of orthologous genes in other species such as the highly salt-tolerant bacterium H. elongata (37) and the marine proteobacterium Ruegeria pomeroyi (39). Although R. pomeroyi is only an ectoine consumer (36), H. elongata can both synthesize and degrade ectoine (37). In contrast to the genes for ectoine/5-hydroxyectoine biosynthesis that are mostly contained in an evolutionarily conserved operon structure (23), the operons for the catabolism of ectoines are more varied, both with respect to gene order and content (36, 37). However, a common denominator of the catabolic gene cluster is the presence of a EutD/EutE (Eut: ectoine utilization) enzyme bi-module, regarded as central for ectoine utilization as a nutrient.
It is thought that ectoine degradation begins with the opening of the pyrimidine ring by the ectoine hydrolase EutD (DoeA) (EC 3.5.4.44) to form N-acetyl-diaminobutyrate (ADABA), which is then catabolized by the N-acetly-l-2,4-diaminobutyrate deacetylase EutE (DoeB) (EC 3.5.1.125) to acetate and diaminobutyrate (DABA) (36, 37). The Atf aminotransferase and the Ssd dehydrogenase subsequently transform DABA into l-aspartate with l-aspartate-β-semialdehyde as the intermediate (36, 37).
An initial in vivo characterization of the H. elongata DoeA (EutD) protein heterologously produced in Escherichia coli by Schwibbert et al. (37) suggests that this enzyme can form both the α- and γ-ADABA isomers from imported ectoine. Mutant studies in H. elongata also revealed that DoeB (EutE) specifically de-acetylates the α-ADABA isomer, whereas the DoeA (EutD)-formed γ-ADABA molecule was suggested to be re-cycled into ectoine via the EctC ectoine synthase (37). Moreover, the EutABC enzymes were speculated by Schulz et al. (36) to initially convert 5-hydroxyectoine into ectoine for further metabolism by the EutD/EutE bi-module. This proposal was primarily based on bioinformatics and the observation that a eutABC deletion mutant of R. pomeroyi was unable to metabolize 5-hydroxyectoine but retained the ability to utilze ectoine as a nutrient (23, 36).
Overall, most of the steps of the proposed ectoine and 5-hydroxyectoine degradation routes have not been experimentally challenged to date at any level of detail. Here, we clarify the catalytic mechanisms underlying the central steps in the catabolism of ectoine and 5-hydroxyectoine through an in-depth biochemical and structural analysis of the conserved EutD/EutE enzyme bi-module. Our study shows that the EutD and EutE enzymes from R. pomeroyi act in concert. The EutD hydrolase opens the pyrimidine ring of ectoine via an unusual covalent intermediate involving a conserved glutamate to generate α-ADABA, but not γ-ADABA. This is an important finding, as α-ADABA, but not γ-ADABA, serves as the internal inducer of the MocR/GabR-type repressor EnuR controlling the expression of the majority of ectoine/5-hydroxyectoine catabolic gene clusters (39, 40). Following α-ADABA synthesis by EutD, the zinc-dependent diaminobutyrate deacetylase EutE hydrolyzes this intermediate to produce acetate and the l-aspartate precursor DABA. Contrary to the previous proposal by Schulz et al. (36), we observed that the EutD protein also hydrolyzes 5-hydroxyectoine to form hydroxy–α-ADABA, a finding that requires a rethinking of the overall organization of the ectoine/5-hydroxyectoine degradation route.
Results
Phylogenomics and ecology of ectoine production and consumption
To study the phylogenomics of ectoine synthesis and metabolism, we analyzed all fully sequenced and annotated bacterial and archaeal genomes available in the IMG/M database (41). We used the amino acid sequence of the EctC ectoine synthase from Paenibacillus lautus (29), and that of the EutD ectoine hydrolase from R. pomeroyi (36) as search queries as markers for ectoine production and consumption, respectively. At the time of the search (November 2019) 8,850 fully sequenced prokaryotic genome sequences were represented in the IMG/M database. We found 676 predicted ectoine producers in these data set (7.5%) (665 originate from Bacteria and 11 from Archaea). 429 Ectoine consumers were found (4.8%); none was a member of the Archaea (Fig. 1B; Fig. S1). 96 Microbial species possessed both ectoine synthesis and degradation genes (less than 1%) (Fig. 1B); with respect to the total number of ectoine producers, this group of microorganisms rises to 14.2%.
Ectoine producers and consumers are found in most ecological niches ranging from freshwater, saltwater, and terrestrial, to extreme environments. They are also present in host-associated species including pathogens of animals, humans, and plants (Fig. 1C). Ectoine producers are found among the α-, β-, and γ-proteobacteria, actinobacteria, firmicutes, and some archaea (overall 12 phyla), whereas ectoine consumers are primarily found among the α-, β-, and γ-proteobacteria (Fig. 1B). In particular, both catabolic and anabolic ectoine genes were present in a restricted group of α- and γ-proteobacteria (Fig. 1B). These are primarily found in members of the Oceanospirallales, Vibrionales, and Rhodobacteriales (Fig. S1).
EutD and EutE cooperate to degrade ectoine
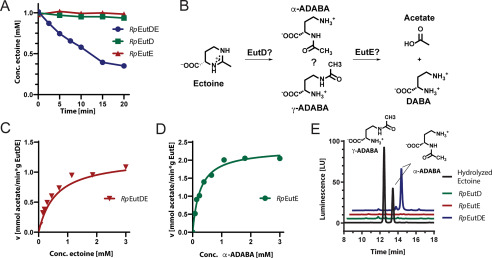
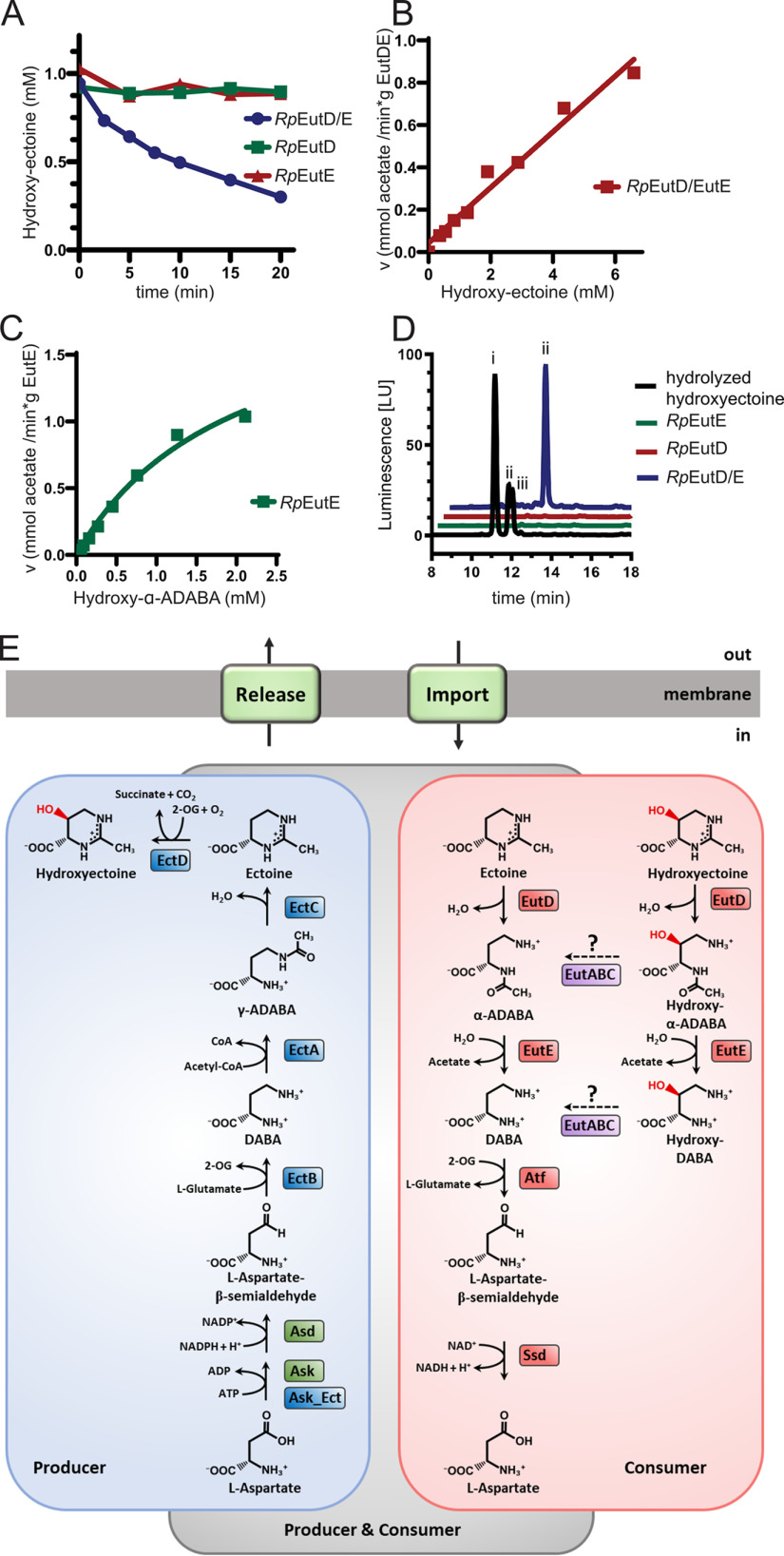
The wide distribution of bacteria able to consume ectoine prompted us to investigate the degradation route in mechanistic detail. We focused on the EutD/EutE enzyme bi-module as it is considered central for the catabolism of ectoine and 5-hydroxyectoine (36, 37). To dissect the molecular mechanism of ectoine degradation, we chose the marine bacterium R. Pomeroyi (Rp) as our model system, a bacterium that can only consume ectoine and in which the genetics and physiology of ectoine/5-hydroxyectoine degradation has already been studied to some extent (36, 39). Recombinantly produced in E. coli BL21(DE3), the RpEutD and RpEutE proteins were purified by a nickel-ion-affinity followed by size exclusion chromatography (Fig. S2, A and B). With the purified enzymes in hand, we qualitatively analyzed their ability to degrade 1 mm ectoine within 20 min by HPLC. Neither RpEutD nor RpEutE were capable of degrading ectoine (Fig. 2A). However, when both enzymes were present in the assay, substantial ectoine degradation was observed (Fig. 2A). These findings suggest that both enzymes cooperate for ectoine degradation. Next, we aimed at a better enzyme-kinetic understanding of the RpEutD/RpEutE enzyme pair. To this end, we employed a commercially available kit allowing the online detection of acetate, which together with DABA is a plausible reaction product of the EutD/EutE-catalyzed ectoine degradation pathway (Fig. 2B). Only when EutD and EutE were present together, did efficient acetate production occur (Fig. S2C), yielding apparent kM and Vmax values of 0.6 mm and 1.2 mmol/min/g, respectively (Fig. 2C). Our data indicate that the degradation of ectoine requires a complex of the EutD and EutE enzymes, which we tried to detect by size exclusion chromatography, isothermal calorimetry, and pull-down experiments. We were not able to visualize the complex by these methods arguing for its short-lived nature. Taken together, EutD and EutE cooperate for efficient ectoine degradation likely through a transient complex.
Figure 2.
Ectoine degradation by EutD and EutE. A, degradation of ectoine by RpEutD (green), RpEutE (red), and both enzymes together (blue), as monitored by HPLC. B, proposed pathway of ectoine degradation. EutD opens the pyrimidine ring of ectoine to generate ADABA, which can exist in the α- and γ-ADABA isomers. In the next step, EutE deacetylates ADABA into DABA and acetate. C, Michaelis-Menten plot of the enzyme pair RpEutD/EutE degrading ectoine. The product acetate was quantified via a commercially available absorption-based assay. kM = 0.6 mm; Vmax = 1.2 mmol/min/g. D, Michaelis-Menten plot of RpEutE degrading α-ADABA into DABA and acetate. kM = 0.2 mm; Vmax = 2.2 mmol/min/g. E, analysis of α-ADABA and γ-ADABA in E. coli cells expressing EutD, EutE, and both grown on ectoine containing minimal medium with glucose as the sole carbon source. HPLC analysis detects α-ADABA when both enzymes are present (blue). ADABA is not detected in cells expressing either RpEutD or RpEutE (green and red, respectively). The α-ADABA and γ-ADABA standards are shown in black.
Degradation of ectoine proceeds via α-ADABA
EutD catalyzed opening of the ectoine pyrimidine ring should yield ADABA, which can exist in the constitutional isomers α-ADABA and γ-ADABA (Fig. 2B). We prepared both ADABAs by alkaline hydrolysis of ectoine and subsequent separation by anion chromatography and then analyzed their use as substrates in the enzyme assays (Fig. S3). We analyzed the ability of RpEutE to degrade α- and γ-ADABA (Fig. 2B). RpEutE exclusively degraded α-ADABA with kM and Vmax values of 0.1 mm and 2.2 mmol/min/g, respectively (Fig. 2D). Thus, ectoine degradation by the R. pomeroyi enzymes exclusively proceeds via α-ADABA, a finding that is consistent with data obtained for the corresponding H. elongata enzymes (37). Our kinetic analysis also suggests that the EutD-mediated generation of α-ADABA is slower than the EutE-catalyzed degradation of α-ADABA, possibly making EutD the rate-limiting step (Fig. 2, C and D).
To show that ectoine degradation proceeds via α-ADABA also in vivo, we produced RpEutD, RpEutE, and both proteins together in E. coli strain MC4100 grown on minimal medium supplemented with 1 mm ectoine and 0.3 m NaCl to trigger ectoine import by the ProP and ProU transporters (42). Consistent with our in vitro analysis, neither α- nor γ-ADABA was detected when only either EutD or EutE was present (Fig. 2E). Surprisingly, when both enzymes were produced together exclusively α-ADABA was detected (Fig. 2E) but we could not find the expected reaction product of the EutD/EutE bi-module, DABA, by HPLC analysis. We have no explanation why α-ADABA is formed when EutD/EutE are co-produced, but EutE is unable to convert α-ADABA to DABA. It is important to keep in mind that both proteins are being produced in a recombinant background that might allow the EutD/EutE-dependent conversion of ectoine into α-ADABA, but somehow impairs the conversion of the latter into DABA. It might also be that EutE is inhibited by or limited for a yet unknown compound of the E. coli cytoplasm (e.g. zinc; see below). Whatever the precise reason might be, the co-expression of eutD/eutE allowed us to conclude that (i) the hydrolysis of ectoine by EutD exclusively proceeds via α-ADABA as the intermediate and that (ii) both EutD and EutE need to be present to achieve hydrolysis of ectoine. We note, however, that for the corresponding H. elongata enzymes, somewhat different data were reported since production of solitary recombinant DoeA (EutD) protein in E. coli yielded both ADABA isomers. However, these were produced at very low amounts despite the fact that 1 mm ectoine was fed to the recombinant cells (37).
EutD lacks the metal-binding site of M24-type aminopeptidases
Next, we wanted to determine the crystal structure of RpEutD. Unfortunately, the obtained RpEutD crystals were only of poor diffraction quality. However, the closely related EutD (DoeA) protein from H. elongata (He), which shares 62.5% sequence identity with RpEutD, produced well diffracting crystals (Table 1). The structure of the apo-state of HeEutD was determined to a resolution of 2.15 Å by molecular replacement employing the structure of E. coli (Ec) PepP as search model. EcPepP and HeEutD share 25.6% sequence identity.
Table 1.
Data collection and refinement statistics
Data were collected on ID30A-1 (MASSIF-1, ESRF), ID23-2 (ESRF), and MX14.2 (BESSY).
| HeEutD | HeEutD ectoine | RpEutE | RpEutE ADABA | HeEutD ADABA | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2 | P41212 | P6322 | P21 | P 41212 |
| Cell dimensions | |||||
| a, b, c (Å) | 120.37, 123.14, 61.49 | 158.2, 158.2, 122.32 | 99.19, 99.19, 134.62 | 76.667, 145.949, 164.116 | 157.073, 157.073, 124.072 |
| α, β, γ (°) | 90 97.184 90 | 90 90 90 | 90 90 120 | 90 92.30 90 | 90 90 90 |
| Wavelength (Å) | 0.966 | 0.873 | 0.972 | 0.918 | 0.9814 |
| Resolution (Å)a | 43.34–2.15 (2.22–2.15) | 43.88–2.25 (2.33–2.25) | 46.54–1.99 (2.06–1.99) | 46.46–2.5 (2.59–2.5) | 19.39–2.4 (2.48–2.4) |
| Rmerge | 0.0869 (0.621) | 0.1474 (1.243) | 0.0626 (4.804) | 0.213 (1.27) | 0.1901 (1.852) |
| I/σI | 8.96 (1.58) | 10.69 (1.96) | 23.00 (0.60) | 5.69 (0.97) | 12.O2 (1.38) |
| Completeness (%) | 98.26 (96.67) | 99.96 (99.97) | 99.48 (98.86) | 98.59 (99.67) | 99.14 (95.57) |
| Redundancy | 3.0 (2.9) | 8.9 (9.3) | 21.0 (20.2) | 3.9 (4.0) | 14.6 (14.6) |
| CC1/2 | 0.995 (0.659) | 0.997 (0.657) | 1 (0.551) | 0.984 (0.421) | 0.998 (0.766) |
| Refinement | |||||
| Resolution (Å) | 43.34–2.15 | 43.88–2.25 | 42.95–2.00 | 46.46–2.5 | 19.39–2.4 |
| No. reflections | 47,539 (4,593) | 73,757 (7,290) | 27,066 (2,612) | 122,999 (12,342) | 60,636 (5,766) |
| Rwork/Rfree | 0.23/0.26 | 0.16/0.19 | 0.24/0.29 | 0.20/0.26 | 0.18/0.21 |
| No. atoms | 6,432 | 6,847 | 2,331 | 30,865 | 6,658 |
| Protein | 6,158 | 6,273 | 2,327 | 29,396 | 6,291 |
| Ligand/ion | 0 | 30 | 0 | 22 | 29 |
| Water | 274 | 544 | 14 | 1447 | 338 |
| B-factors | 41.42 | 40.O2 | 80.66 | 49.99 | 52.27 |
| Protein | 41.30 | 39.29 | 80.76 | 50.46 | 52.25 |
| Ligand/ion | 0 | 72.17 | 0 | 97.10 | 63.62 |
| Water | 44.10 | 46.66 | 64.35 | 39.67 | 51.73 |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.005 | 0.003 | 0.031 | 0.003 | 0.008 |
| Bond angles (°) | 0.77 | 0.60 | 1.67 | 0.75 | 0.94 |
| Ramachandran | |||||
| Favored (%) | 97.66 | 97.71 | 98.36 | 97.92 | 97.22 |
| Allowed (%) | 2.34 | 2.16 | 1.32 | 2.08 | 2.53 |
| Outliers (%) | 0.00 | 0.13 | 0.00 | 0.00 | 0.25 |
a Values in parentheses are for highest-resolution shell.
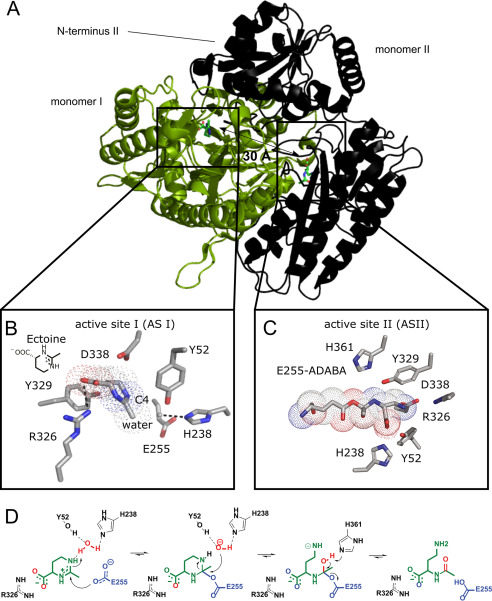
The crystal structure of HeEutD shows a highly intertwined homodimer (Fig. 3A). Homodimerization of HeEutD in solution was supported by multiangle light scattering experiments (Fig. S2A). EutD is closely related in structure and amino acid sequence to the M24-family of aminopeptidases, which usually require two metal ions for activity (Fig. S4, A and B). This could suggest that EutD operates in a metal-dependent manner. However, closer inspection of the active site architecture reveals that this cannot be the case. Aminopeptidases, such as EcPepP, typically coordinate two metal ions (e.g. manganese or zinc) within their active sites that are critical for catalyzing cleavage of the peptide bond (reviewed in Ref. 43). In these zinc-dependent proteases, both metals are coordinated by five amino acids (i.e. His-354, Asp-271, Asp-260, Glu-406, and Glu-383 in the case of EcPepP) (Fig. S4C). In EutD, however, the majority of the amino acids required for the metal ion coordination are not present, and instead were replaced by amino acids unfavorable for metal ion coordination (i.e. Pro-266, Tyr-329, Met-363, and Thr-376) (Fig. S4C). The only exception is Glu-255 of HeEutD, which is conserved between EutD enzymes and the M24-type aminopeptidases (Fig. S4C). These findings suggest that EutD operates in a metal-independent manner.
Figure 3.
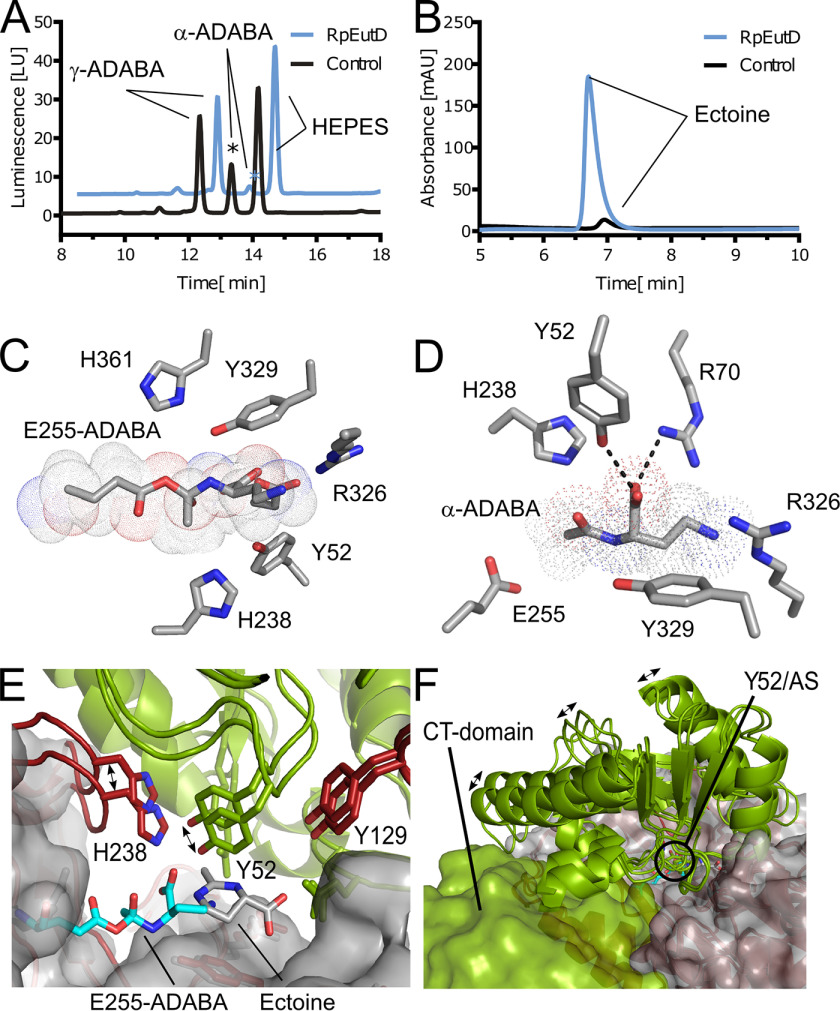
Crystal structure and mechanism of EutD. A, crystal structure of the EutD homodimer with one monomer shown as green cartoon and surface and the other as black cartoon indicating secondary structure. The localization of both actives sites is depicted. B, coordination of ectoine (surrounded by a dotted outline) and the putative catalytic water in the active site of HeEutD. Ectoine is coordinated by hydrogen bonds to Arg-326. The attacking water is hydrogen-bonded by His-238. Glu-255 and Tyr-52 can be observed in proximity. C, HeEutD active site with the α-ADABA intermediate covalently bound to Glu-255 (surrounded by a dotted outline). Further hydrogen bonds by His-238, Tyr-52, and Asp-338 keep the molecule in place. D, catalytic mechanism of ectoine hydrolysis by EutD. Ectoine, the catalytic water and Glu-255 are shown in green, red, and blue, respectively.
Ectoine ring cleavage proceeds via a covalent α-ADABA–Glu-255 adduct
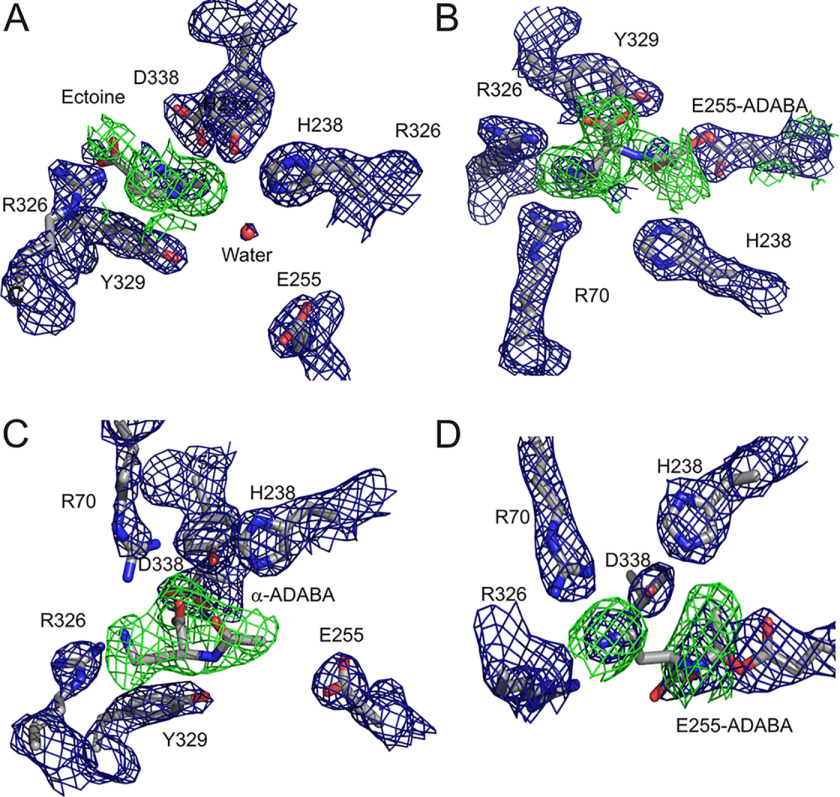
To gain further insights into the catalytic mechanism of the ectoine hydrolase, we determined the structure of HeEutD in the presence of ectoine at a resolution of 2.25 Å (Table 1). Inspection of one active site (AS I) of the EutD homodimer revealed electron densities that could be unambiguously attributed to ectoine (Fig. 4A, Fig. S5A). The ectoine is kept in place by a hydrogen bond between Arg-326 and the carboxyl moiety of the ligand (Fig. 3B). Moreover, a water molecule (with a B-factor of 45) is hydrogen-bonded by His-238 (Figs. 3B and 4A). Approximately 4 Å away from this water, Glu-255, the N5-nitrogen of the pyrimidine ring of ectoine, and Tyr-52 from the N-terminal domain of the opposing EutD chain can be found (Figs. 3B and 4A). Being 4.0 Å away from the C4 carbon of ectoine, this water might be ideally suited to provide the attacking hydroxyl anion for nucleophilic cleavage of the pyrimidine ring (Fig. 3B). Changing Glu-255 into aspartate, and Tyr-52 and His-238 into alanine residues yielded catalytically inactive EutD variants in our acetate assay, supporting their catalytic relevance (Table S1).
Figure 4.
Unbiased electron densities of EutD ligands. A, unbiased Fobs − Fcalc difference electron density of ectoine at 3σ shown as a green mesh. Bias was removed by refinement prior to incorporation of the ligand. Applies to all following unbiased densities (PDB ID 6TWK). B, unbiased Fobs − Fcalc difference electron density of α-ADABA covalently linked to glutamate 255 at 3σ shown as a green mesh. The refined 2Fobs − Fcalc electron density around glutamate 255 is shown as a blue mesh at 1.5 σ (PDB ID 6TWK). C, unbiased Fobs − Fcalc difference electron density of α-ADABA at 3σ shown as a green mesh (PDB ID 6YO9). D, unbiased Fobs − Fcalc difference electron density of α-ADABA covalently linked to glutamate 255 at 3σ shown as a green mesh. The refined 2Fobs − Fcalc electron density around glutamate 255 is shown as a blue mesh at 1.5σ (PDB ID 6YO9).
In the other active site (AS II) of the EutD homodimer, however, we did not observe electron densities corresponding to ectoine. Instead, a tubular density connected to Glu-255 was visible, which we assigned as α-ADABA with its C4 carbon linked to the carboxyl moiety of Glu-255 (Figs. 3C and 4B, and Fig. S5B). Thus, Glu-255 serves as electron donor forming a covalent adduct with α-ADABA upon opening of the pyrimidine ring (Fig. 3C). The correct distance of the Glu-255 carboxyl moiety and the ectoine substrate is essential. A E255D variant is unable to perform the catalytic reaction as monitored in the in vitro acetate assay, presumably because the side chain of aspartate is one methylene group shorter than that of glutamate (Table S1). Thus, the opening of the ectoine ring proceeds via a covalent α-ADABA-EutD adduct, which relies on Glu-255 (Fig. 3D).
Release of α-ADABA from EutD requires cleavage of its orthoester-like bond with Glu-255 accompanied with the abstraction of a proton. Glu-374 and His-361 localize in reasonable proximity to serve as possible proton-acceptors (Fig. S5E). The E374D and H361S variants decreased EutD activity to 20 and 5%, respectively (Table S1). Thus, His-361 could be the proton acceptor allowing α-ADABA release from Glu-255. However, His-361 might also be part of a proton exchange chain together with Glu-374. Taken together, we show that EutD opens the pyrimidine ring of ectoine in a metal-independent manner through the α-ADABA intermediate, which is transiently bound to Glu-255 (Fig. 3D).
EutD allows back-cyclization of α-ADABA into ectoine
Our data suggest that EutD only cleaves the ectoine ring in an efficient manner when EutE is present. Moreover, our structural data suggest that the covalently bound α-ADABA intermediate could be able to cycle back into the ectoine ring structure. To test whether a back-cyclization would indeed be possible, we exposed purified RpEutD to mixtures of α- and γ-ADABA and analyzed whether any of the two isomers would be converted back into ectoine. Our experiments clearly show that exclusively α-ADABA disappeared and ectoine was formed from it (Fig. 5, A and B). Thus, we conclude that RpEutD is able to perform the backward reaction from α-ADABA to ectoine.
Figure 5.
Reverse reaction and half-site cooperativity of EutD. A, ADABA isomers present in ectoine hydrolysate (black) and upon the addition of RpEutD (blue). In the absence of RpEutD three distinct peaks for α-ADABA, γ-ADABA, and a peak derived from the HEPES buffer were detected in this order. The size of the α-ADABA peak differing upon addition of RpEutD is marked with an asterisk. B, the remaining ectoine present in the ectoine hydrolysate (black) and formed ectoine upon addition of RpEutD (blue). C, HeEutD active site (AS) with the α-ADABA intermediate covalently bound to Glu-255 (surrounded by a dotted outline), as obtained from co-crystallization with α-ADABA. D, AS of RpEutD containing the product α-ADABA. The molecule is held in place by hydrogen bonds to Tyr-52, Asp-338, and Arg-326. E, superimposition of AS I and AS II, revealing distinct movements by His-238 and Tyr-52. In the intermediate bound state, this leads to a significant narrowing of the active site. F, comparison of the N-terminal architecture between AS I and AS II. Originating from conformational changes of Tyr-52, two helices and a loop region reposition themselves, potentially providing a topological signal.
To consolidate these findings at the structural level, we co-crystallized HeEutD in the presence of 1 mm α-ADABA to a resolution of 2.4 Å (Table 1). The crystal structure shows the HeEutD homodimer. A closer inspection of the two active sites revealed the α-ADABA product in one and the covalently bound α-ADABA–Glu-255 intermediate in the other active site (Figs. 4, C and D, 5, C and D; Fig. S5, C and D). The carboxyl moiety of the α-ADABA product interacts via hydrogen bonds with Arg-70 and Tyr-52 from the N terminus of the opposing monomer (Figs. 5D and 4C). The α-ADABA–Glu-255 intermediate in the other active site appears identical to its counterpart observed in the EutD structure obtained in the presence of 1 mm ectoine (see above) (Fig. 5C and 4D). Notably, the flexible amine group of the intermediate now points more toward Arg-326 rather than Asp-338, which could be explained by the flexible nature of the α-ADABA moiety. Taken together, our structural and biochemical analysis shows that EutD is able to form the covalent α-ADABA–Glu-255 intermediate also from the α-ADABA product, and allow the back-cyclization of α-ADABA into ectoine. Thus, the EutD-mediated cleavage of ectoine is reversible.
Cooperation between the active sites in the EutD homodimer
Our crystal structure shows a homodimer of EutD in which the ectoine substrate is present in one and the α-ADABA intermediate is found in the other active site. This indicates that both active sites, which are ∼30 Å away from each other (Fig. 3A), are able to cooperate with each other so that one active site “knows” the catalytic state of the other active site. Closer inspection of our structure shows that Tyr-52 of one monomer reaches into the other active site of the other monomer and vice versa. This tyrosine either locates in close proximity (4 Å) to the putative catalytic water in AS I or the α-ADABA–Glu-255 intermediate in AS II (Fig. 3). Moreover, it forms a hydrogen bond with the carboxyl moiety of the α-ADABA product (Fig. 5D). Therefore, Tyr-52 is an ideal candidate to sense the catalytic status of one active site and communicate to the other or vice versa. Superimposition of ASs I and II further reveals a significant movement of the loop harboring Tyr-52 when the reaction proceeds (Fig. 5E). This in turn leads to three major conformation changes in the N-terminal domain by moving two helices closer to the C-terminal domain as well as opening a loop region in the backside of the N-terminal domain (Fig. 5F). These topological changes directly affect the C-terminal domain, which is harboring the respective other active site, establishing the before-mentioned communication. It can be imagined that in this way the catalytic sites communicate with each other to establish their cooperativity.
To solidify this idea, we also employed the pKa prediction tool H++ (44) to analyze pKa differences of Glu-255 and thus its potential protonation state between AS I and AS II (substrate- versus intermediate-bound site). In the ectoine-bound AS I, Glu-255 is predicted to be completely deprotonated (pKa < 0), allowing the nucleophilic attack required for ectoine ring cleavage (Fig. 3D). In the intermediate-bound AS II, however, the calculated pKa of Glu-255 in its not α-ADABA–bound state would be 5.1. This pKa value would in principle allow the protonation of the α-ADABA–Glu-255 adduct to enable release of the α-ADABA product. Unfortunately, the prediction of the pKa value of the α-ADABA–Glu-255 intermediate with the H++ program is not straightforward due to its uncommon chemical nature. Thus, further studies need to address this issue in greater detail. Moreover, our in silico analysis also suggests that the pKa differences of Glu-255 between AS I and AS II are not caused by a single residue, but seem to be the consequence of an overall narrowing of AS II compared with AS I. A notable consequence of the AS II narrowing is that the catalytically relevant histidines 238 and 361 (Table S1) appear in closer proximity to the α-ADABA–Glu-255 intermediate. Again, since prediction of the pKa value of the α-ADABA–Glu-255 in this context is difficult, a definite statement cannot be made at this point. Taken together, our structural analysis suggests that the two catalytic half-sites of EutD cooperate in a highly synchronous manner via an intricate network of interactions.
EutE deacetylates α-ADABA in a zinc-dependent manner
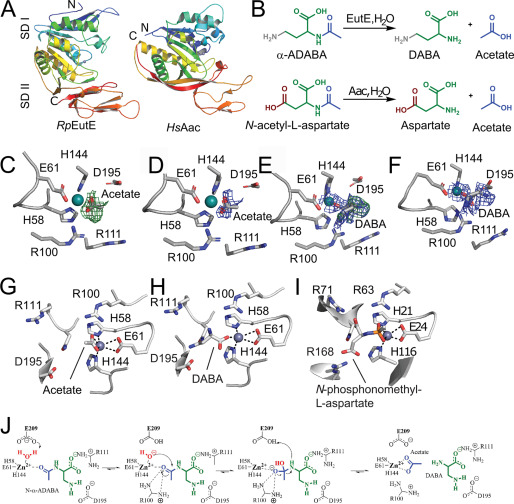
To complete the catalytic picture of ectoine degradation, we grew crystals of RpEutE in the presence of α-ADABA and determined the structure to a resolution of 2.5 Å by molecular replacement employing the structure of aspartoacylase (AAC) from Rhodobacter sphaeroides (PDB ID 3CDX; sequence identity 39.2%) (Table 1). RpEutE consists of two subdomains that appear in an extended crescent-like shape (Fig. 6A). It shares structural homology to the universally conserved AACs (reviewed in Ref. 45), including the zinc-binding site formed by His-58, His-144, and Glu-61 at the concave side of RpEutE (Fig. 6A). Alanine substitutions of His-58, His-144, and Glu-61 completely inactivated EutE proving its metal-dependence (Table S1). The RpEutE crystals grown in the presence of α-ADABA contained 12 EutE monomers within their asymmetric unit. Investigation of their active sites revealed unambiguous electron densities for three acetate/zinc products (Fig. 6, C and D) and seven DABA/zinc (Fig. 6, E and F), leaving two proteins within the asymmetric unit with empty active sites. Although acetate coordinates with zinc via its carboxyl moiety in a bi-dentate manner, DABA coordinates with the metal ion via one of its carboxyl oxygens in a uni-dentate way (Fig. 6, G and H).
Figure 6.
EutE deacetylates α-ADABA in a zinc-dependent manner. A, the crystal structure of RpEutE (left) shows that it is structurally related to AACs. Both structures are shown as cartoon in rainbow colors from their N to C termini. The two subdomains (SD) are indicated as SD I and SD II. B, deacetylation reactions of α-ADABA and N-acetyl-l-aspartate carried out by EutE and AACs, respectively. Note: α-ADABA and N-acetyl-l-aspartate only differ in their terminal amine and carboxyl groups, respectively. C, unbiased Fobs − Fcalc difference electron density of acetate at 3σ shown as a green mesh (PDB 6TWM). D, the 2Fobs − Fcalc after final refinement with acetate is shown as a blue mesh at 1.5 σ (PDB 6TWM). E, unbiased Fobs − Fcalc difference electron density of DABA at 3σ shown as a green mesh. 2 Fobs − Fcalc in the area is shown as a blue mesh at 1.5 σ (PDB 6TWM). F, the 2Fobs − Fcalc after final refinement with DABA is shown as a blue mesh at 1.5 σ (PDB 6TWM). G, coordination of the reaction product acetate. H, coordination of DABA in the active site of RpEutE. Both products remain coordinated to the catalytically active zinc ion, which is itself coordinated by His-58, His-144, and Glu-61. I, active site of a human AAC bound to an acetylaspartate mimic, depicting the likely substrate/transition state coordination in AACs (46). J, catalytic mechanism of EutE.
To detail the catalytic mechanism of EutE, we compared our DABA-product state of EutE with that of the human AAC bound to the substrate mimetic N-phosphonomethyl-l-aspartate (46) (Fig. 6I). AACs convert acetyl-aspartate (A-Asp) into aspartate and acetate, whereas EutE converts α-ADABA into DABA and acetate (Fig. 6B). The substrates of EutE and AAC only differ in their terminal moieties not involved in the deacetylation reaction (Fig. 6B). Comparing EutE with AAC identified Arg-111 of EutE to be involved in coordinating the carboxyl group of α-ADABA in an equivalent manner as in the AACs (Fig. 6, G–I; Table S1). Moreover, our comparison of EutE and AAC shows that Arg-100 coordinates the transition state of the EutE-catalyzed α-ADABA cleavage reaction similar to what has been observed for AACs (Fig. 6, G–I; Table S1).
The substrates of EutE and AAC differ in their terminal amino and carboxyl moieties, respectively (Fig. 6B). Thus, both enzymes must also differ to compensate these substrate specialties. Indeed, the terminal amine of the DABA product, which is also present in α-ADABA, hydrogen bonds with Asp-195 (Fig. 6H). An D195R variant is incapable of degrading α-ADABA, supporting a role of Asp-195 in substrate recognition (Table S1). Taken together, we show that EutE operates highly reminiscent to AACs (Fig. 6J). Substrate binding critically relies on Arg-111 and Asp-195, both forming hydrogen bonds to the carboxyl group and the amine of α-ADABA, respectively (Fig. 6J). The zinc ion further coordinates the carbonyl-oxygen of the α-ADABA peptide bond. The deacetylation reaction begins with water deprotonation promoted by the zinc ion and Glu-209 (Fig. 6J). The so-formed hydroxyl anion attacks the carbonyl-carbon of α-ADABA in a nucleophilic manner leading to the tetrahedral transition state, whose electron-negative character is compensated by the guanidinium moiety of Arg-100. Subsequently, Glu-209 promotes protonation and allows release of DABA and acetate (Fig. 6J).
RpEutD/EutE degrades 5-hydroxyectoine but less efficient than ectoine
Next, we wondered whether RpEutD/EutE would also be able to degrade 5-hydroxyectoine (Fig. 1A). As for ectoine (Fig. 2A), degradation of 5-hydroxyectoine was only observed by HPLC analysis when the RpEutD and RpEutE enzymes were present together in the in vitro assay (Fig. 7A; Fig. S2D). Using the acetate assay 5-hydroxectoine degradation by the RpEutD/EutE bi-module was also detected. However, no Michaelis-Menten–like behavior for the degradation of 5-hydroxyectoine could be observed (Fig. 7B). This finding suggests that one or both enzymes are not ideally suited for the degradation of 5-hydroxyectoine.
Figure 7.
Microbial ectoine degradation. A, degradation of 5-hydroxyectoine by RpEutD (green), RpEutE (red), and both enzymes together (blue), as monitored by HPLC. B, the velocity of acetate production as a function of 5-hydroxyectoine concentration was measured by the acetate assay. The enzymes do not exhibit a Michaelis-Menten–like behavior, suggesting that one or both enzymes are not ideally suited for degradation of 5-hydroxyectoine. C, Michaelis-Menten plot of RpEutE degrading α-ADABA into DABA and acetate. Estimated kM and Vmax values are 3 mm and 2 mmol/min/g, respectively. D, analysis of hydroxy–α-ADABA and hydroxy–γ-ADABA in E. coli cells producing EutD, EutE, or both proteins grown on hydroxyectoine containing minimal medium with glucose as the sole carbon source. HPLC analysis detects hydroxy–α-ADABA when both enzymes are present (blue). No hydroxy–ADABA is detected in cells expressing either RpEutD or RpEutE (green and red, respectively). A chemically hydrolyzed hydroxyectoine standard (black) shows three peaks for (i) hydroxy–γ-ADABA, (ii) hydroxy–α-ADABA, and (iii) a mixture of both isomers. E, ectoine degradation (red box) can be thought as opposite of its biosynthesis pathway (blue box). Both pathways separate at the respective ADABAs isomers.
Inspection of the EutD structure suggests the extra hydroxyl moiety of hydroxyectoine could well be accommodated within the active site of the enzyme, which was also true for the putative Glu-255–bound hydroxy–α-ADABA intermediate (Fig. S5G). However, analysis of the EutE structure suggests that Glu-187 provides steric hindrance for the hydroxyl moiety of the hydroxy–α-ADABA substrate (Fig. S5). To challenge this idea, we prepared both hydroxy–ADABA isomers with purities >95% by alkaline hydrolysis of 5-hydroxyectoine and subsequent separation by anion exchange chromatography (Fig. S3, lower panel). Although RpEutE was unable to degrade hydroxy–γ-ADABA, hydroxy–α-ADABA was converted into acetate albeit with significantly lower reaction velocity and a drastically increased kM value (Fig. 7C). To investigate the degradation of 5-hydroxyectoine in vivo, we produced RpEutD, RpEutE, and RpEutD/EutE in E. coli strain MC4100 grown on minimal medium supplemented with 1 mm 5-hydroxyectoine and 0.3 m NaCl (see above). Only when both enzymes were present was hydroxy–α-ADABA exclusively detected (Fig. 7D). The identity of this predicted reaction product of the RpEutD enzyme as hydroxy–α-ADABA was ascertained by NMR spectroscopy (Fig. S3). Collectively, we conclude from these in vitro and in vivo experiments that the EutD/EutE enzyme bi-module can degrade 5-hydroxyectoine via hydroxy–α-ADABA into acetate and hydroxy–DABA, albeit with lesser efficiency than ectoine.
As we were able to observe that RpEutD can catalyze the reverse reaction of ectoine degradation, we set out to test this for hydroxyectoine as well. To do so, we subjected a mixture of hydroxy–α- and hydroxy–γ-ADABA to RpEutD and monitored via HPLC analysis which of these compounds disappeared from the assay solution and if 5-hydroxyectoine would be formed. In this case, RpEutD used exclusively hydroxy–α-ADABA as the substrate for the backward reactions and formed the corresponding hydroxyectoine (Fig. S6).
Discussion
Ecophysiology of ectoine and 5-hydroxyectoine consumption
Like other compatible solutes, osmotically stimulated high-level production of ectoines requires the expenditure of considerable biosynthetic and energetic resources (32). It is therefore not surprising that microorganisms have found ways to exploit compatible solutes in general, and ectoine in particular, as nutrients, when they are no longer needed as osmostress protectants (47). A prime example for the utilization of compatible solutes as nutrients is the algal osmolyte dimethylsulfoniopropionate (DMSP) (48, 49). It is produced on a scale of millions of tons annually in marine habitats. Released into the water column, DMSP can be scavenged by microorganisms and exploited as carbon and sulfur source. Its metabolism drives the global sulfur cycle and the produced volatile dimethylsulfide (DMS) is considered as a contributor to global warming.
Another ecophysiological important process related to compatible solutes is the catabolism of the osmostress protectant choline in the gastrointestinal tracts of humans. This trimethylammonium compound can be catabolized by gut microbiota under anaerobic conditions to trimethylamine (TMA), a metabolite that can be further converted to trimethylamine-N-oxide (TMAO) in the human liver (50). Notably, several human diseases are associated with gut microbial production of TMA, and its derived metabolite TMAO (50).
Although certainly not as abundant in the biosphere as DMSP, choline, and glycine betaine (3), free ectoines have been detected in various ecosytems (23). Even if one considers only fully sequenced microbial genomes, ectoine biosynthetic genes can be found in about 7.5% of all species and strains currently represented in the IMG/M database. The widespread production of ectoine as an osmostress protectant likely provided an evolutionarily incentive to develop pathways for its catabolism. This event occurred primarily in the large phylum of the Proteobacteria (Fig. 1B; Fig. S1). Microorganisms populating marine, terrestrial, and plant-associated ecosystems are dominantly represented among the ectoine consumers (Fig. 1C). Most of the ectoine/5-hydroxyectoine using microorganisms can either exclusively synthesize or catabolize these nitrogen-rich compounds (Fig. 1B) but there is also an interesting group of bacteria that can do both. However, these are limited in number and their occurrence is restricted to particular subgroups of the α- and γ-Proteobacteria (Fig. 1B, Fig. S1).
The gene clusters for ectoine degradation often vary with respect to gene order and content (23, 36, 37). However, the vast majority encode the ectoine hydrolase EutD (EC 3.5.4.44) and the deacetylase EutE (EC 3.5.1.125) (Fig. S1). Here, we provide a comprehensive structural and biochemical analysis unraveling the molecular mechanism of the EutD/EutE enzymes, a bi-module positioned at the core of the ectoine/5-hydroxyectoine catabolic pathways, as it is required for opening of the pyrimidine ring of ectoines for their further degradation (Fig. 7E).
Catalytic mechanism for ectoine degradation
Our structural analysis shows that EutD enzymes share close structural and sequence conservation to the M24-family aminopeptidases, which rely on two highly coordinated metal ions for their activity (43). Despite this high overall structural similarity, EutD lacks most residues required for metal ion binding except for Glu-255. Instead, the metal ion site seen in the M24 aminopeptidase is changed in EutD to create a cavity well-suited to accommodate ectoine. Our study thus shows how the conserved enzyme fold of the M24-aminopeptidases was adapted in EutD to enable a new function in ectoine degradation. Generally spoken, the M24-aminopeptidase fold seems well-suited as a template to evolve new function. For instance, the ribosome-biogenesis factor Ebp1 (also Arx1) represents an ancient M24-aminopeptidase in which the substrate-binding pocket is maintained, but the zinc-ion–binding site was destroyed to evolve a ribosome-biogenesis factor (51) (Fig. S4C).
Only one of the metal-coordinating residues present in EcPepP remained preserved in EutDs, which is Glu-255. This residue plays several central roles in the catalytic reaction executed by EutD. Although it is not directly involved in ectoine binding, Glu-255 contributes to the coordination of a water molecule, which should be catalytically relevant by providing the attacking hydroxyl ion required for opening of the ectoine ring. Moreover, Glu-255 was found to be the “anchor” of the α-ADABA intermediate, which is covalently attached to this residue via an orthoester-like bond. Although covalently bonded reaction intermediates with cysteines or lysines are rather frequently described (52), this is to our knowledge, the first covalent intermediate involving a glutamate. Cleavage of this chemically labile orthoester-like bond requires the presence of a proton, which is in all likelihood provided by His-361, eventually being part of a proton exchange chain (Fig. 3D).
Both active sites of EutD homodimer operate in a concerted manner as clearly evidenced by the simultaneous presence of ectoine in one active site and the α-ADABA/Glu-255 adduct in the other. As efficient ectoine degradation only occurs when EutD and EutE are present, this suggests that EutE supports the α-ADABA release from EutD through transient interactions. But how could EutE then discriminate an α-ADABA–loaded active site of EutD from one that is either empty or has the substrate ectoine bound? Structural comparison of the EutD monomers harboring either ectoine or α-ADABA revealed a mobile loop (signal loop) adjacent to the center of the active sites (Fig. S5F). The position of this loop differs by 2.6 Å depending on whether substrate or intermediate is present. Therefore, we speculate that the signal loop provides a topological signal, guiding EutE to an α-ADABA–loaded AS of EutD. However, further experiments are required to challenge this idea.
EutE itself degrades α-ADABA into DABA and acetate. The enzyme shares high structural and sequence similarity to the AAC family, which is conserved across all kingdoms of life (45). Our study shows that EutE operates by a highly similar mechanism as AAC enzymes. Prior to the reaction, the substrate is coordinated by Arg-111, which is forming a hydrogen bond to the carboxyl group, as well as by Asp-195, which forms hydrogen bonds to the amine of α-ADABA (Fig. 6, G–I). The carbonyl-oxygen of the α-ADABA peptide bond coordinates the zinc ion, which contributes to further substrate binding. The deacetylation reaction starts with the deprotonation of a water molecule that is coordinated by the zinc ion and Glu-209. The newly formed hydroxyl anion performs a nucleophilic attack on the carbonyl-carbon of the α-ADABA, thereby forming the negatively charged, tetrahedral transition state, which is stabilized by the positive charge of the guanidinium moiety of the adjacent Arg-100. Release of the products of the EutE-catalyzed reaction, DABA and acetate, involves Glu-209, which promotes protonation of the transition state (Fig. 6J).
Rethinking the degradation scheme of 5-hydroxyectoine
Our study shows that EutD/EutE constitute the enzymatic core sufficient to degrade both ectoine and 5-hydroxyectoine (Fig. 7E). Whether further enzymes increase efficiency of 5-hydroxyectoine degradation remains to be shown (Fig. 7E). Our study also suggests that a previous proposal by Schulz et al. (36) in which 5-hydroxyectoine degradation would begin with the EutABC-mediated removal of its 5-hydroxyl moiety seems not to be valid (23). This proposal was primarily based on bioinformatics and on the observation that deletion of the eutABC genes abolishes growth of R. pomeroyi on 5-hydroxyectoine, whereas leaving growth on ectoine unaffected (36). In light of our data, the EutABC enzymes might contribute to degradation of 5-hydroxyectoine at the level of hydroxy–α-ADABA (Fig. 7E). Because EutB shares significant homology to the PLP-dependent group of threonine dehydratases, it is plausible that it removes the hydroxyl moiety of hydroxy–α-ADABA (Fig. 7E). Subsequently, EutC could reduce the resulting product into α-ADABA in a NADPH-dependent manner. The so-generated α-ADABA would then serve as a bona fide substrate of EutE.
Interestingly, a pair of functionally related enzymes operates in the widely distributed β-hydroxyasparte pathway (53). BhcB, the β-hydroxyasparte dehydratase, catalyzes a water elimination of its substrate to form iminosuccinate, an intermediate that is further reduced in an NADH-dependent reaction by the BhcD imminosuccinate reductase to produce l-aspartate (53). The role of EutA remains uncertain in such a scenario, because closer inspection of many genomes shows that eutA only sporadically appears in the context of the eutD/eutE gene cluster (Fig. S1). We cannot exclude that the EutABC enzymes could play a role in converting hydroxy–α-DABA to DABA for further catabolism to l-aspartate (Fig. 7E). Hence, future biochemical and structural studies are clearly required to challenge these ideas.
Regulation of bacterial ectoine metabolism
Ectoine degradation can be thought as the opposite of the ectoine biosynthetic route (Fig. 7E). We show that EutD/EutE-mediated ectoine degradation in R. pomeroyi proceeds via α-ADABA, but not γ-ADABA. This could nicely explain how producer/degrader cells can prevent futile ectoine production and consumption. Although the ectoine synthetase EctC primarily relies on γ-ADABA as the substrate (29), EutD selectively produces α-ADABA.
The ectoine biosynthetic genes are osmotically induced (19, 23), whereas those for ectoine/5-hydroxyectoine degradation are substrate inducible when ectoines are added to the growth medium (35, 36, 39). However, ectoine is not the true inducer; instead α-ADABA, and to a much lesser extend also the intermediate DABA, serve as internal inducers for the EnuR repressor (39, 40). This PLP-dependent MocR/GabR-type regulator (54) controls transcription of the ectoine/5-hydroxyectoine import and catabolic gene cluster of R. pomeroyi, and most likely, also in many other bacteria capable to degrade ectoines (23). Notably, γ-ADABA, the substrate of the ectoine synthase EctC does not serve as inducer for EnuR (39). In this way, ectoine biosynthesis and degradation are elegantly separated through the chemical uniqueness of the γ-ADABA and α-ADABA isomers. Since we found that EutD is able to convert 5-hydroxyectoine into hydroxy–α-ADABA, the possibility is raised that this compound also serves as an internal inducer for EnuR as well.
The genetic regulatory circuits controlling the expression of the genes for ectoine biosynthesis as an osmostress protectant and use of ectoine as a nutrient are clearly targeted for the physiological task at hand. However, for those microorganisms that can both synthesize and degrade ectoine, the situation can become complex due to intermediates shared in both pathways. For instance, Schwibbert et al. (37) reported that the EutD (DoeA) ectoine hydrolase from H. elongata, a microorganism capable of both ectoine production and catabolism, synthesizes both α-ADABA and γ-ADABA. Although α-ADABA is funneled to EutE (DoeB) for further catabolism, γ-ADABA is thought to serve as an intermediate in a new round of ectoine production (37). Conditions in natural settings that would require the simultaneous production and degradation of ectoine are not immediately obvious to us. Nevertheless, it should be noted that inactivation of the catabolic genes led to higher intracellular ectoine titers in the H. elongata industrial production host (37).
Concluding remarks
Use of compatible solutes as nutrients is an important aspect for the functioning of microbial ecosystems (47). Ectoine and 5-hydroxyectoine are among the most widely synthesized osmostress protectants in the microbial world. Severely osmotically stressed microorganisms synthesize and accumulate ectoines to cellular concentrations as high as hundreds of micromolar. The recent exiting discovery of ectoine production and uptake by halophilic protists and marine microalgae (55–57) raises the possibility for ectoines to function in microbial interactions with these eukaryotic cells. Similarly to the catabolism of the compatible solutes DMSP, choline, and glycine betaine, their environmental release can shape architecture and composition of microbial communities (57, 58).
Taken together, our study unravels the structure-based mechanism of the enzyme core essential for the catabolism of the stress protectants ectoine and 5-hydroxyectoine. The presented data clarify the key mechanism for ectoine consumption, an eco-physiologically relevant process preventing microbial ecosystems from losing valuable resources.
Materials and methods
Chemicals
Ectoine was kindly provided by bitop AG (Witten, Germany) and hydroxyectoine was purchased from Merck (Darmstadt, Germany). Ampicillin was purchased from Carl Roth GmbH (Karlsruhe, Germany). The γ-isomer of γ-ADABA was purchased from Chem-Impex International Inc. (Wood Dale, IL, USA). Ectoine and hydroxyectoine were hydrolyzed via alkaline hydrolysis as described for ectoine (59) and hydroxyectoine (60). The α-isomer of ADABA was separated from the hydrolysis product of ectoine by repeated chromatography on a silica gel column (Merck Silica Gel 60).
Database searches for ectoine producing and consuming microorganisms
For the phylogenomic analysis of ectoine biosynthetic and catabolic genes, we used the IMG/M database of the Joint Genome Institute and considered for our analysis only fully sequenced microbial genomes (41). At the time of the search (November 2019) 8,850 prokaryotic genomes were available; 8,557 were from members of the Bacteria, and 293 represented archaeal genomes. We used the amino acid sequence of the EctC ectoine synthase (29) from the Gram-positive bacterium Paenibacillus lautus as the search query for ectoine producers (61). EctC is the signature enzyme for the ectoine biosynthetic route, but a group of microorganisms exists that possesses a solitary EctC-type protein whose enzymatic function is unclear (23, 62). We therefore considered only EctC-type proteins as bona fide ectoine synthases when the inspected genome sequence also possessed genes for the two other enzymes (EctA–EctB) required for ectoine biosynthesis (23).
To identify ectoine consuming microorganisms, we used the amino acid sequence of the EutD ectoine hydrolase for the marine bacterium R. pomeroyi as the search query, as this enzyme is key for ectoine consumption. As the amino acid sequence of the EutD protein is related to other types of hydrolases (in particular to peptidases), we considered only EutD-type proteins as bona fide ectoine hydrolases when the eutD gene was part of a gene cluster containing other known ectoine-catabolizing genes. We then used the resulting 429 EutD-type proteins to construct a phylogenetic protein tree using software resources provided by the IMG/M website. Subsequently, the iTOL software suit (63) was used to visualize the EutD tree. Onto this tree, we projected information on the presence of ectoine biosynthetic genes in the microorganism under consideration derived from the above described EctC search. Furthermore, data on the habitat of ectoine-producing and consuming microorganisms were collected from information deposited in the IMG/M database or derived from literature searches. In the case of ectoine-consuming microorganisms, this information was incorporated into the iTOL-derived EutD protein tree. Similarly, information on the taxonomic affiliation of microorganisms was projected onto the tree; this information was derived from the IMG/M database. Amino acid sequence identities of EutD-type proteins relative to EutD protein from R. pomeroyi (used as the original search query) and that of the EutD (DoeA) protein from H. elongata were calculated using Clustal O (64). The amino acid sequence identity of ectoine hydrolases ranged between 41.88% (for Rhodobacter sphaeroides ATCC 17025) and 95.95% (for Leisingera sp. NJS204) when the R. pomeroyi EutD protein was used for the search and ranged between 39.56% (for Mycobacterium tuberculosis H37Ra) and 97.24% (for Halomonas beimenensis NTU-111) when the H. elongata EutD protein was used for the search.
Protein production, purification, and characterization
The genes encoding for the HeEutD, RpEutD, and RpEutE proteins were amplified by PCR and cloned into pET24d (Novagen) via the NcoI and XhoI restriction sites. All proteins contained an N-terminal His6 tag. Proteins were produced in E. coli BL21(DE3) (Novagen). Cells were lysed by a Microfluidizer (M110-L, Microfluidics). Cell debris after lysis was removed by high-speed centrifugation. All proteins were purified by nickel-ion affinity and size exclusion chromatography, as described previously (65). The SEC buffer consisted of 20 mm HEPES-Na (pH 7.5), 200 mm NaCl, 20 mm KCl, and 20 mm MgCl2.
For the analytical size exclusion chromatography (SEC) with multiangle light scattering (SEC-MALS), a sample (100 μl) of the purified EutD and EutE protein concentrations of 100 μm were injected at 4 °C on to a pre-equilibrated S200 300/10 GL analytical size exclusion column (GE Healthcare, München, Germany). The buffer at pH 7.5 contained 20 mm HEPES, 200 mm NaCl, 20 mm MgCl2, and 20 mm KCl. For the MALS-RI experiments, a multiangle light scattering and a differential refractive index detector (Postnova Analytics, Landsberg am Lech, Germany) was attached to the column.
Crystallization and structure determination
Crystallization was performed by the sitting-drop method at 20 °C in 600-nl drops consisting of equal parts of protein and precipitation solutions. Protein solutions of 300 μm (HeEutD) and 1 mm (RpEutE) were prepared in buffer described for SEC purification. For the crystals containing substrates, 1 mm (final concentration) ectoine or α-ADABA, respectively, were added and incubated at room temperature for 10 min. Crystallization conditions were: HeEutD with ectoine (0.2 m trisodium citrate, 20% (w/v) PEG 3350); RpEutE with α-ADABA (0.1 m Bicine (pH 9.0), 20% (v/v) PEG 6000). Prior to data collection, crystals were flash-frozen in liquid nitrogen using a cryo-solution that consisted of mother liquor supplemented with 20% (v/v) glycerol. Data were collected under cryogenic conditions at the European Synchrotron Radiation Facility (Grenoble, France) beamlines ID23-1 (66), ID23-2 and MASSIF-1 (67), and at beamline MX14.2 at BESSY II (Berlin, Germany) (68).
Data were processed with XDS and scaled with XSCALE (69). All structures were determined by molecular replacement with PHASER (70), manually built in COOT (71), and refined with PHENIX (72). The search model for the HeEutD-Apo structure was the E. coli aminopeptidase P (PDB ID 2BWS, sequence identity is 25.6%). The search model for all other EutD structures was the HeEutD-Apo structure (this study).
The search model for the RpEutE structure was the aspartoacylase from R. sphaeroides (PDB ID 3CDX, sequence identity is 39.2%). The search model for all other EutE structures was the RpEutE-Apo structure (this study).
For all ligand-bound structures of HeEutD and RpEutE, first the protein model was manually built to completeness and refined without placing waters or any other ligands. After completion of the protein model, the respective ligand was placed into the unbiased density (when present) and refined. In the last step, the water molecules were modeled. Final validation of the structures was carried out with the validation server of the PDB (73). Figures were prepared with PyMOL (RRID:SCR_000305).
Isolation of pure α-ADABA
A mixture of approximately 25% α-ADABA and 75% γ-ADABA made from chemically hydrolyzed ectoine was separated into the pure ADABA species by preparative anion exchange chromatography. A Metrohm Carb2 column (250 × 4 mm inner diameter) operated with a 90 mm (NH4)2CO3 eluent adjusted to pH 9.25 at 1 ml/min was used for the separation. Sample loading was 100 μl of a 50 mm solution of the ADABA mixture. The obtained separation factor for α-ADABA and γ-ADABA was 1.9 under these preparative conditions. The fractions of each ADABA species were collected, unified, and the eluent was removed using a vacuum drying oven at 40 °C overnight. The purity of the resulting α-ADABA sample was determined to 90% by HPLC.
HPLC-based assay
Ectoine and hydroxyectoine were detected by high pressure liquid chromatography (HPLC) using the 1260 Infinity system (Agilent Technologies, Walsbronn, Germany) with a GromSil Amino-1 PR, 3 μm column (125 × 4 mm) (Dr. Maisch GmbH, Ammerbruch, Germany) and a diode array detecting module (Agilent Technologies, Walsbronn, Germany) using a wavelength of 210 nm. The mobile phase consisted of 80% (v/v) acetonitrile (MeCN, HPLC grade) and 20% (v/v) H2O. The separation was performed, after injecting 10 μl of the sample, by an isocratic measurement over 15 min with a flow rate of 1 ml/min at 20 °C.
The ectoine degradation assay for HPLC-based analysis were performed in 200-μl assays containing 20 mm HEPES-Na (pH 7.5), 200 mm NaCl, 20 mm MgCl2, 20 mm KCl, 1 mm ectoine or hydroxyectoine, and 40 μg of purified EutD and/or EutE at a temperature of 30 °C. After 0, 5, 10, 15, and 20 min, a sample of 30 μl was taken and added to 30 μl of MeCN to stop the reaction. Before injecting 10 μl each into the HPLC system, the samples were centrifuged (13,300 rpm for 10 min at 4 °C) to remove the denatured enzymes.
Acetate assay
For the assays the commercially available kit from Sigma-Aldrich, named “Acetate Colorimetric Assay Kit” (catalog number MAK086) was used and applied according to the manufacturer's manual. In short, 1 nmol (final concentration = 10 μm) of the respective enzyme were mixed with acetate buffer from the kit to a total volume of 10 μl in the reaction well of a flat-bottom 96-well plate. The substrates, α-ADABA and ectoine, respectively, were prepared in a total of 15 μl of acetate buffer for final concentrations of 3, 1.8, 1.08, 0.65, 0.38, 0.23, 0.13, and 0.08 mm. The enzyme and substrate mixture of the kit were mixed accordingly to the instructions and added to the substrates. This mixture was then transferred to the prepared enzymes, in turn starting the reaction. The 450 nm absorption of each well was monitored for 15 min in 11-s intervals.
EutD-mediated synthesis of ectoines in its backward enzyme reaction
Assays were performed in 200-μl volumes containing 20 mm HEPES-Na (pH 7.5), 200 mm NaCl, 20 mm MgCl2, 20 mm KCl, and either 1 mm hydrolyzate of ectoine or hydroxyectoine. 40 μg of purified EutD were added to start the assay, which was performed at a temperature of 30 °C. After 1 h a sample of 30 μl was taken and added to 30 μl of MeCN to stop the reaction. The samples were centrifuged (13,300 rpm for 10 min at 4 °C) to remove the denatured enzymes before assessing the content of ectoines and ADABA-isomers in the samples, employing the HPLC-based detection methods described above.
Cultivation of recombinant E. coli strains for the hydrolysis of ectoine and hydroxyectoine
To determine the hydrolytic activity of EutD, EutE, and EutD/EutE on ectoine and hydroxyectoine, E. coli strain MC4100 (74) harboring plasmids pLH48, pLH49, pLH50, or the empty expression vector pTrc99a were grown in MMA (75) with 0.5% (w/v) glucose as a carbon source and ampicillin (100 μg ml−1) as an antibiotic. Growth took place in 100-ml Erlenmeyer flasks in a water bath set to 220 rpm at 37 °C. MMA pre-cultures were used to inoculate MMA supplemented with 0.3 m NaCl and 1 mm ectoine or hydroxyectoine to an OD578 of 0.1, and after growth to an OD578 of 0.5, 1 mm isopropyl 1-thio-β-d-galactopyranoside was added to the media to induce expression. 2-ml probes were sampled by centrifugation (5 min, 13,000 rpm, room temperature) directly before induction, 2 h, and 4 h after induction and the cell pellets and supernatants were separated and stored at −20 °C until further analysis.
In vivo detection of ectoine and hydroxyectoine degradation
To extract ectoine degradation products from E. coli, 1 ml of 20% (v/v) ethanol was added to the cell pellets and the probes were rigorously shaken for 1 h and subsequently centrifuged (30 min, 13,000 rpm, 4 °C). The supernatant was dried at 50 °C for 20 h. 100 μl of distilled water was added to the samples, which were then centrifuged for 30 min. 2 μl of the supernatant was derivatized using 3 μl of 9-fluorenylmethoxy carbonyl (FMOC) (25 mg/ml in acetonitrile), excessive FMOC was removed by addition of 6 μl of ADAM (7.6 mg/ml in 50% (w/v) borate buffer, 0.5 mol liter−1 (pH 7.7), and 50% (v/v) acetone). 489 μl of distilled water was added to the solution and the samples were centrifuged (15 min, 13,000 rpm at room temperature), before they were analyzed by isocratic HPLC. These measurements were performed with an Agilent 1260 Infinity LC system (Agilent, Waldbronn, Germany) and a Gemini 5-μm C18 110- Å column (Phenomenex, Torrance, CA, USA). The flow rate of the system was set to 1 ml min−1 and the Gemini column was operated at 40 °C. The following gradient of solvent A (80% (w/v) acetate buffer, 50 mmol liter−1, pH 4.2, 20% (v/v) acetonitrile, 0.5% (w/v) THF) and solvent B (20% acetate buffer, 50 mmol liter−1, pH 4.2, 80% (v/v) acetonitrile) was applied: 0 min, 0% solvent B; 2 min, 0% solvent B; 8 min, 20% solvent B; 16 min, 27% solvent B; 18 min, 54% solvent B; 20 min, 100% solvent B; 25 min, 100% solvent B; 26 min, 0% solvent B; 29 min, 0% solvent B. Samples were analyzed with the OpenLAB software suite (Agilent) and visualized with GraphPad Prism (GraphPad Software, Inc.).
Data availability
The atomic coordinates have been deposited in the Protein Data Bank under accession codes 6TWJ (Apo-HeEutD), 6TWK (HeEutD bound to ectoine and α-ADABA–Glu-255), 6YO9 (HeEutD bound to α-ADABA and α-ADABA–Glu-255), 6TWL (Apo-RpEutE) and 6TWM (product-bound state of RpEutE). The authors declare that all other data supporting the findings of this study are available within the article and its supplementary information files.
Supplementary Material
Acknowledgments
We thank the BESSYII (Berlin, Germany) and the European Synchrotron Radiation Facility (ESRF, Grenoble, France) for excellent support.
This article contains supporting information.
Author contributions—C.-N. M., L. H., F. A., and G. B. formal analysis; C.-N. M., L. H., F. A., A. A. R., I. W., L. C., and E. B. investigation; C.-N. M., L. H., F. A., L. C., E. B., and G. B. visualization; C.-N. M., L. H., and A. S. methodology; C.-N. M., L. H., E. B., and G. B. writing-original draft; F. A., E. B., and G. B. supervision; A. S. and G. B. resources; E. B. and G. B. conceptualization; E. B. and G. B. project administration; G. B. funding acquisition; G. B. validation; G. B. writing-review and editing.
Funding and additional information—This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) Collaborative Research Center (CRC) 987 “Microbial Diversity in Environmental Signal Response” (to G. B. and E. B.) and a Ph.D. fellowship from the International Max Planck Research School for Environmental, Cellular and Molecular Microbiology (IMPRS-Mic; Marburg) (to L. C.).
Conflict of interest—The authors declare no conflict of interest.
- EctA
- l-2,4-diaminobutyrate acetyltransferase
- EctB
- l-2,4-diaminobutyrate transaminase
- EctC
- ectoine synthase
- EctD
- ectoine hydroxylase
- Eut
- ectoine utilization
- Doe
- degradation of ectoines
- N-γ-ADABA
- N-γ-acetyl-l-2,4-diaminobutyrate
- N-α-ADABA
- N-α-acetyl-l-2,4-diaminobutyrate
- DABA
- l-2,4-diaminobutyrate
- DMSP
- dimethylsulfoniopropionate
- DMS
- dimethyl-sulfide
- TMA
- trimethlyamine
- TMAO
- trimethlyamine-N-oxide
- FMOC
- fluorenylmethyloxycarbonyl
- SEC
- size exclusion chromatography
- MALS
- multiangle light scattering
- PDB
- Protein Data Bank
- AC
- active site
- AAC
- aspartoacylase
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- ADAM
- 1-adamantylamine hydrochloride
- MMA
- Minimal Medium A.
References
- 1. Gunde-Cimerman N., Plemenitaš A., and Oren A. (2018) Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42, 353–375 10.1093/femsre/fuy009 [DOI] [PubMed] [Google Scholar]
- 2. Roesser M., and Müller V. (2001) Osmoadaptation in bacteria and archaea: common principles and differences. Environ. Microbiol. 3, 743–754 10.1046/j.1462-2920.2001.00252.x [DOI] [PubMed] [Google Scholar]
- 3. Yancey P. H. (2005) Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830 10.1242/jeb.01730 [DOI] [PubMed] [Google Scholar]
- 4. Kempf B., and Bremer E. (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170, 319–330 10.1007/s002030050649 [DOI] [PubMed] [Google Scholar]
- 5. Bolen D. W., and Baskakov I. V. (2001) The osmophobic effect: natural selection of a thermodynamic force in protein folding. J. Mol. Biol. 310, 955–963 10.1006/jmbi.2001.4819 [DOI] [PubMed] [Google Scholar]
- 6. Stadmiller S. S., Gorensek-Benitez A. H., Guseman A. J., and Pielak G. J. (2017) Osmotic shock induced protein destabilization in living cells and its reversal by glycine betaine. J. Mol. Biol. 429, 1155–1161 10.1016/j.jmb.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diamant S., Eliahu N., Rosenthal D., and Goloubinoff P. (2001) Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J. Biol. Chem. 276, 39586–39591 10.1074/jbc.M103081200 [DOI] [PubMed] [Google Scholar]
- 8. da Costa M. S., Santos H., and Galinski E. A. (1998) An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 61, 117–153 10.1007/BFb0102291 [DOI] [PubMed] [Google Scholar]
- 9. Wood J. M. (2011) Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 65, 215–238 10.1146/annurev-micro-090110-102815 [DOI] [PubMed] [Google Scholar]
- 10. Bremer E., and Krämer R. (2019) Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 73, 313–334 10.1146/annurev-micro-020518-115504 [DOI] [PubMed] [Google Scholar]
- 11. García-Estepa R., Argandoña M., Reina-Bueno M., Capote N., Iglesias-Guerra F., Nieto J. J., and Vargas C. (2006) The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J. Bacteriol. 188, 3774–3784 10.1128/JB.00136-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuhlmann A. U., Hoffmann T., Bursy J., Jebbar M., and Bremer E. (2011) Ectoine and hydroxyectoine as protectants against osmotic and cold stress: uptake through the SigB-controlled betaine-choline- carnitine transporter-type carrier EctT from Virgibacillus pantothenticus. J. Bacteriol. 193, 4699–4708 10.1128/JB.05270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann T., and Bremer E. (2011) Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193, 1552–1562 10.1128/JB.01319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanne C., Golovina E. A., Hoekstra F. A., Meffert A., and Galinski E. A. (2014) Glass-forming property of hydroxyectoine is the cause of its superior function as a desiccation protectant. Front. Microbiol. 5, 150 10.3389/fmicb.2014.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manzanera M., García de Castro A., Tøndervik A., Rayner-Brandes M., Strøm A. R., and Tunnacliffe A. (2002) Hydroxyectoine is superior to trehalose for anhydrobiotic engineering of Pseudomonas putida KT2440. Appl. Environ. Microbiol. 68, 4328–4333 10.1128/AEM.68.9.4328-4333.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brands S., Schein P., Castro-Ochoa K. F., and Galinski E. A. (2019) Hydroxyl radical scavenging of the compatible solute ectoine generates two N-acetimides. Arch. Biochem. Biophys. 674, 108097 10.1016/j.abb.2019.108097 [DOI] [PubMed] [Google Scholar]
- 17. Galinski E. A., Pfeiffer H. P., and Truper H. G. (1985) 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid: a novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 149, 135–139 10.1111/j.1432-1033.1985.tb08903.x [DOI] [PubMed] [Google Scholar]
- 18. Inbar L., and Lapidot A. (1988) The structure and biosynthesis of new tetrahydropyrimidine derivatives in actinomycin D producer Streptomyces parvulus: use of 13C- and 15N-labeled l-glutamate and 13C and 15N NMR spectroscopy. J. Biol. Chem. 263, 16014–16022 [PubMed] [Google Scholar]
- 19. Pastor J. M., Salvador M., Argandoña M., Bernal V., Reina-Bueno M., Csonka L. N., Iborra J. L., Vargas C., Nieto J. J., and Cánovas M. (2010) Ectoines in cell stress protection: uses and biotechnological production. Biotechnol. Adv. 28, 782–801 10.1016/j.biotechadv.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 20. Zaccai G., Bagyan I., Combet J., Cuello G. J., Demé B., Fichou Y., Gallat F. X., Galvan Josa V. M., von Gronau S., Haertlein M., Martel A., Moulin M., Neumann M., Weik M., and Oesterhelt D. (2016) Neutrons describe ectoine effects on water H-bonding and hydration around a soluble protein and a cell membrane. Sci. Rep. 6, 31434 10.1038/srep31434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kunte H., Lentzen G., and Galinski E. (2014) Industrial production of the cell protectant ectoine: protection mechanisms, processes, and products. Curr. Biotechnol. 3, 10–25 10.2174/22115501113026660037 [DOI] [Google Scholar]
- 22. Becker J., and Wittmann C. (2020) Microbial production of extremolytes: high-value active ingredients for nutrition, health care, and well-being. Curr. Opin. Biotechnol. 65, 118–128 10.1016/j.copbio.2020.02.010 [DOI] [PubMed] [Google Scholar]
- 23. Czech L., Hermann L., Stoveken N., Richter A. A., Hoppner A., Smits S. H. J., Heider J., and Bremer E. (2018) Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: genetics, phylogenomics, biochemistry, and structural analysis. Genes (Basel) 9, 177 10.3390/genes9040177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ono H., Sawada K., Khunajakr N., Tao T., Yamamoto M., Hiramoto M., Shinmyo A., Takano M., and Murooka Y. (1999) Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic eubacterium, Halomonas elongata. J. Bacteriol. 181, 91–99 10.1128/JB.181.1.91-99.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richter A. A., Kobus S., Czech L., Hoeppner A., Zarzycki J., Erb T. J., Lauterbach L., Dickschat J. S., Bremer E., and Smits S. H. J. (2020) The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine. J. Biol. Chem. 295, 2822–2838 10.1074/jbc.RA119.011277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peters P., Galinski E. A., and Trüper H. G. (1990) The biosynthesis of ectoine. FEMS Microbiol. Lett. 71, 157–162 10.1111/j.1574-6968.1990.tb03815.x 1601286 [DOI] [Google Scholar]
- 27. Richter A. A., Mais C. N., Czech L., Geyer K., Hoeppner A., Smits S. H. J., Erb T. J., Bange G., and Bremer E. (2019) Biosynthesis of the stress-protectant and chemical chaperon ectoine: biochemistry of the transaminase EctB. Front. Microbiol. 10, 2811 10.3389/fmicb.2019.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hillier H. T., Altermark B., and Leiros I. (2020) The crystal structure of the tetrameric DABA-aminotransferase EctB, a rate-limiting enzyme in the ectoine biosynthesis pathway. FEBS J. doi: 10.1111/febs.15265 10.1111/febs.15265 [DOI] [PubMed] [Google Scholar]
- 29. Czech L., Höppner A., Kobus S., Seubert A., Riclea R., Dickschat J. S., Heider J., Smits S. H. J., and Bremer E. (2019) Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis. Sci. Rep. 9, 364 10.1038/s41598-018-36247-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bursy J., Pierik A. J., Pica N., and Bremer E. (2007) Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J. Biol. Chem. 282, 31147–31155 10.1074/jbc.M704023200 [DOI] [PubMed] [Google Scholar]
- 31. Hoppner A., Widderich N., Lenders M., Bremer E., and Smits S. H. (2014) Crystal structure of the ectoine hydroxylase, a snapshot of the active site. J. Biol. Chem. 289, 29570–29583 10.1074/jbc.M114.576769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oren A. (1999) Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63, 334–348 10.1128/MMBR.63.2.334-348.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grammann K., Volke A., and Kunte H. J. (2002) New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J. Bacteriol. 184, 3078–3085 10.1128/JB.184.11.3078-3085.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vargas C., Jebbar M., Carrasco R., Blanco C., Calderon M. I., Iglesias-Guerra F., and Nieto J. J. (2006) Ectoines as compatible solutes and carbon and energy sources for the halophilic bacterium Chromohalobacter salexigens. J. Appl. Microbiol. 100, 98–107 10.1111/j.1365-2672.2005.02757.x [DOI] [PubMed] [Google Scholar]
- 35. Jebbar M., Sohn-Bösser L., Bremer E., Bernard T., and Blanco C. (2005) Ectoine-induced proteins in Sinorhizobium meliloti include an ectoine ABC-type transporter involved in osmoprotection and ectoine catabolism. J. Bacteriol. 187, 1293–1304 10.1128/JB.187.4.1293-1304.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schulz A., Stöveken N., Binzen I. M., Hoffmann T., Heider J., and Bremer E. (2017) Feeding on compatible solutes: a substrate-induced pathway for uptake and catabolism of ectoines and its genetic control by EnuR. Environ. Microbiol. 19, 926–946 10.1111/1462-2920.13414 [DOI] [PubMed] [Google Scholar]
- 37. Schwibbert K., Marin-Sanguino A., Bagyan I., Heidrich G., Lentzen G., Seitz H., Rampp M., Schuster S. C., Klenk H. P., Pfeiffer F., Oesterhelt D., and Kunte H. J. (2011) A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581 T. Environ. Microbiol. 13, 1973–1994 10.1111/j.1462-2920.2010.02336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Booth I. R., and Blount P. (2012) The MscS and MscL families of mechanosensitive channels act as microbial emergency release valves. J. Bacteriol. 194, 4802–4809 10.1128/JB.00576-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schulz A., Hermann L., Freibert S. A., Bönig T., Hoffmann T., Riclea R., Dickschat J. S., Heider J., and Bremer E. (2017) Transcriptional regulation of ectoine catabolism in response to multiple metabolic and environmental cues. Environ. Microbiol. 19, 4599–4619 10.1111/1462-2920.13924 [DOI] [PubMed] [Google Scholar]
- 40. Yu Q., Cai H., Zhang Y., He Y., Chen L., Merritt J., Zhang S., and Dong Z. (2017) Negative regulation of ectoine uptake and catabolism in Sinorhizobium meliloti: characterization of the EhuR gene. J. Bacteriol. 199, e00119–16 10.1128/JB.00119-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen I. A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J. R., Seshadri R., Smirnova T., Kirton E., Jungbluth S. P., Woyke T., Eloe-Fadrosh E. A., et al. (2019) IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47, D666–D677 10.1093/nar/gky901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jebbar M., Talibart R., Gloux K., Bernard T., and Blanco C. (1992) Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J. Bacteriol. 174, 5027–5035 10.1128/JB.174.15.5027-5035.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rawlings N. D., and Barrett A. J. (1993) Evolutionary families of peptidases. Biochem. J. 290, 205–218 10.1042/bj2900205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., and Onufriev A. (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371 10.1093/nar/gki464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Makarova K. S., Aravind L., and Koonin E. V. (1999) A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 8, 1714–1719 10.1110/ps.8.8.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le Coq J., Pavlovsky A., Malik R., Sanishvili R., Xu C., and Viola R. E. (2008) Examination of the mechanism of human brain aspartoacylase through the binding of an intermediate analogue. Biochemistry 47, 3484–3492 10.1021/bi702400x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Welsh D. T. (2000) Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24, 263–290 10.1111/j.1574-6976.2000.tb00542.x [DOI] [PubMed] [Google Scholar]
- 48. Curson A. R., Todd J. D., Sullivan M. J., and Johnston A. W. (2011) Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–859 10.1038/nrmicro2653 [DOI] [PubMed] [Google Scholar]
- 49. Moran M. A., Reisch C. R., Kiene R. P., and Whitman W. B. (2012) Genomic insights into bacterial DMSP transformations. Ann. Rev. Mar. Sci. 4, 523–542 10.1146/annurev-marine-120710-100827 [DOI] [PubMed] [Google Scholar]
- 50. Xu H., Wang X., Feng W., Liu Q., Zhou S., Liu Q., and Cai L. (2020) The gut microbiota and its interactions with cardiovascular disease. Microb. Biotechnol. 13, 637–656 10.1111/1751-7915.13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kowalinski E., Bange G., Bradatsch B., Hurt E., Wild K., and Sinning I. (2007) The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 581, 4450–4454 10.1016/j.febslet.2007.08.024 [DOI] [PubMed] [Google Scholar]
- 52. Heine A., DeSantis G., Luz J. G., Mitchell M., Wong C. H., and Wilson I. A. (2001) Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science 294, 369–374 10.1126/science.1063601 [DOI] [PubMed] [Google Scholar]
- 53. Schada von Borzyskowski L., Severi F., Krüger K., Hermann L., Gilardet A., Sippel F., Pommerenke B., Claus P., Cortina N. S., Glatter T., Zauner S., Zarzycki J., Fuchs B. M., Bremer E., Maier U. G., et al. (2019) Marine Proteobacteria metabolize glycolate via the β-hydroxyaspartate cycle. Nature 575, 500–504 10.1038/s41586-019-1748-4 [DOI] [PubMed] [Google Scholar]
- 54. Tramonti A., Nardella C., di Salvo M. L., Pascarella S., and Contestabile R. (2018) The MocR-like transcription factors: pyridoxal 5'-phosphate-dependent regulators of bacterial metabolism. FEBS J. 285, 3925–3944 10.1111/febs.14599 [DOI] [PubMed] [Google Scholar]
- 55. Harding T., Brown M. W., Simpson A. G., and Roger A. J. (2016) Osmoadaptative strategy and its molecular signature in obligately halophilic heterotrophic protists. Genome Biol. Evol. 8, 2241–2258 10.1093/gbe/evw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weinisch L., Kirchner I., Grimm M., Kuhner S., Pierik A. J., Rosselló-Móra R., and Filker S. (2019) Glycine betaine and ectoine are the major compatible solutes used by four different halophilic heterotrophic ciliates. Microb. Ecol. 77, 317–331 10.1007/s00248-018-1230-0 [DOI] [PubMed] [Google Scholar]
- 57. Fenizia S., Thume K., Wirgenings M., and Pohnert G. (2020) Ectoine from bacterial and algal origin is a compatible solute in microalgae. Mar. Drugs 18, 42 10.3390/md18010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Landa M., Burns A. S., Roth S. J., and Moran M. A. (2017) Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677–2690 10.1038/ismej.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kunte H. J., Galinski E. A., and Trüper H. G. (1993) A modified FMOC-method for the detection of amino acid-type osmolytes and tetrahydropyrimidines (ectoines). J. Microbiol. Meth. 17, 129–136 10.1016/0167-7012(93)90006-4 [DOI] [Google Scholar]
- 60. Sarciaux M., Pantel L., Midrier C., Serri M., Gerber C., Marcia de Figueiredo R., Campagne J. M., Villain-Guillot P., Gualtieri M., and Racine E. (2018) Total synthesis and structure-activity relationships study of odilorhabdins, a new class of peptides showing potent antibacterial activity. J. Med. Chem. 61, 7814–7826 10.1021/acs.jmedchem.8b00790 [DOI] [PubMed] [Google Scholar]
- 61. Mead D. A., Lucas S., Copeland A., Lapidus A., Cheng J. F., Bruce D. C., Goodwin L. A., Pitluck S., Chertkov O., Zhang X., Detter J. C., Han C. S., Tapia R., Land M., Hauser L. J., et al. (2012) Complete genome sequence of Paenibacillus strain Y4.12MC10, a novel Paenibacillus lautus strain isolated from Obsidian Hot Spring in Yellowstone National Park. Stand. Genomic Sci. 6, 381–400 10.4056/sigs.2605792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kurz M., Burch A. Y., Seip B., Lindow S. E., and Gross H. (2010) Genome-driven investigation of compatible solute biosynthesis pathways of Pseudomonas syringae pv. syringae and their contribution to water stress tolerance. Appl. Environ. Microbiol. 76, 5452–5462 10.1128/AEM.00686-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Letunic I., and Bork P. (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Han X., Altegoer F., Steinchen W., Binnebesel L., Schuhmacher J., Glatter T., Giammarinaro P. I., Djamei A., Rensing S. A., Reissmann S., Kahmann R., and Bange G. (2019) A kiwellin disarms the metabolic activity of a secreted fungal virulence factor. Nature 565, 650–653 10.1038/s41586-018-0857-9 [DOI] [PubMed] [Google Scholar]
- 66. Nurizzo D., Mairs T., Guijarro M., Rey V., Meyer J., Fajardo P., Chavanne J., Biasci J. C., McSweeney S., and Mitchell E. (2006) The ID23-1 structural biology beamline at the ESRF. J Synchrotron Radiat. 13, 227–238 10.1107/S0909049506004341 [DOI] [PubMed] [Google Scholar]
- 67. Svensson O., Malbet-Monaco S., Popov A., Nurizzo D., and Bowler M. W. (2015) Fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 71, 1757–1767 10.1107/S1399004715011918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mueller U., Darowski N., Fuchs M. R., Förster R., Hellmig M., Paithankar K. S., Pühringer S., Steffien M., Zocher G., and Weiss M. S. (2012) Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Radiat. 19, 442–449 10.1107/S0909049512006395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 72. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gore S., Sanz Garcia E., Hendrickx P. M. S., Gutmanas A., Westbrook J. D., Yang H., Feng Z., Baskaran K., Berrisford J. M., Hudson B. P., Ikegawa Y., Kobayashi N., Lawson C. L., Mading S., Mak L., et al. (2017) Validation of structures in the Protein Data Bank. Structure 25, 1916–1927 10.1016/j.str.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J. Mol. Biol. 104, 541–555 10.1016/0022-2836(76)90119-4 [DOI] [PubMed] [Google Scholar]
- 75. Miller J. H. (1972) Experiments in molecular genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited in the Protein Data Bank under accession codes 6TWJ (Apo-HeEutD), 6TWK (HeEutD bound to ectoine and α-ADABA–Glu-255), 6YO9 (HeEutD bound to α-ADABA and α-ADABA–Glu-255), 6TWL (Apo-RpEutE) and 6TWM (product-bound state of RpEutE). The authors declare that all other data supporting the findings of this study are available within the article and its supplementary information files.