Abstract
Many proteobacteria, such as Escherichia coli, contain two main types of quinones: benzoquinones, represented by ubiquinone (UQ) and naphthoquinones, such as menaquinone (MK), and dimethyl-menaquinone (DMK). MK and DMK function predominantly in anaerobic respiratory chains, whereas UQ is the major electron carrier in the reduction of dioxygen. However, this division of labor is probably not very strict. Indeed, a pathway that produces UQ under anaerobic conditions in an UbiU-, UbiV-, and UbiT-dependent manner has been discovered recently in E. coli. Its physiological relevance is not yet understood, because MK and DMK are also present in E. coli. Here, we established that UQ9 is the major quinone of Pseudomonas aeruginosa and is required for growth under anaerobic respiration (i.e. denitrification). We demonstrate that the ORFs PA3911, PA3912, and PA3913, which are homologs of the E. coli ubiT, ubiV, and ubiU genes, respectively, are essential for UQ9 biosynthesis and, thus, for denitrification in P. aeruginosa. These three genes here are called ubiTPa, ubiVPa, and ubiUPa. We show that UbiVPa accommodates an iron–sulfur [4Fe-4S] cluster. Moreover, we report that UbiUPa and UbiTPa can bind UQ and that the isoprenoid tail of UQ is the structural determinant required for recognition by these two Ubi proteins. Since the denitrification metabolism of P. aeruginosa is believed to be important for the pathogenicity of this bacterium in individuals with cystic fibrosis, our results highlight that the O2-independent UQ biosynthetic pathway may represent a target for antibiotics development to manage P. aeruginosa infections.
Keywords: ubiquinone biosynthesis, coenzyme Q, anaerobic respiration, denitrification, Pseudomonas aeruginosa, iron-sulfur protein, UbiV, UbiU, UbiT, bacterial metabolism, hydroxylation, oxygen, quinone, ubiquinone, respiratory chain, coenzyme Q10 (CoQ10), metalloprotein, anaerobic metabolism
Introduction
The opportunistic pathogen Pseudomonas aeruginosa has a remarkable ability to grow under a variety of environmental conditions, such as soil and water as well as animal-, human-, and plant-host-associated environments. P. aeruginosa is responsible for numerous acute and chronic infections and poses a major health risk for patients with severe burns and cystic fibrosis (CF) or in severely immunocompromised states (1, 2).
The utilization of various carbon sources and energy metabolism (respiration or fermentation) might contribute to the environmental adaptation of P. aeruginosa (3). Its main energy-producing system is respiration, which requires a proton-motive force used for ATP synthesis. The proton-motive force is produced by the transfer of electrons and protons from reduced donors to oxidized acceptors via the quinone pool. Whereas the dehydrogenases and reductases involved in respiratory metabolism have been well described and annotated in the genome of P. aeruginosa PAO1 (4, 5), the composition of its quinone pool has not yet been fully established. Studies in the 1960s suggested ubiquinone 9 (UQ9) is a major quinone of aerobically grown P. aeruginosa (6); therefore, UQ9 is believed to be essential for aerobic respiration (7).
Proteobacteria contain two main types of quinones, benzoquinones and naphthoquinones, represented by UQ (or coenzyme Q) and menaquinone (MK)/demethylmenaquinone (DMK), respectively (8). Typically, MK and DMK function predominantly in anaerobic respiratory chains, whereas UQ is the major electron carrier used for the reduction of dioxygen by various cytochrome oxidases (8). Recent data indicated that the metabolic use of various quinone species according to environmental dioxygen availability is more complex than initially thought. Indeed, using E. coli as a model, we highlighted a pathway conserved across many bacterial species and able to produce UQ under anaerobic conditions (9). The classical UQ biosynthetic pathway requires O2 for three hydroxylation steps (10). Obviously, the flavin-dependent monooxygenases UbiI, UbiF, and UbiH, which catalyze the O2-dependent hydroxylation steps, are not involved in the anaerobic pathway, and the accessory UbiK and UbiJ proteins are not implicated in the assembly and/or stability of the aerobic Ubi complex (11). Seven proteins (UbiA, UbiB, UbiC, UbiD, UbiE, UbiG, and UbiX) catalyzing the prenylation, decarboxylation, and methylation of the phenyl ring of the 4-hydroxybenzoate precursor are common to both pathways (9). In addition, the anaerobic pathway requires UbiT, UbiU, and UbiV proteins. UbiT is homologous to UbiJ, and UbiU and UbiV are expected to be involved in O2-independent hydroxylations (9). However, as explained above, the metabolic relevance of the O2-independent UQ pathway is not yet clearly understood.
In the absence of O2, P. aeruginosa is able to carry out anaerobic respiration with nitrate and nitrite as terminal electron acceptors of the respiratory chain. This process, called denitrification, allows the reduction of soluble nitrate (NO3−) and nitrite (NO2−) to gaseous nitrous oxide (N2O) or molecular nitrogen (N2) (12). Because P. aeruginosa-infected mucus in CF airways is depleted of oxygen and enriched in nitrate and nitrite, the anaerobic metabolism of P. aeruginosa via the denitrification pathway is believed to be important for its pathogenicity (13). Four sequential reactions involving metalloenzymes are needed to reduce nitrate to N2, i.e. nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase. P. aeruginosa was considered a paradigm of the denitrification pathway, and all the reductases involved in this metabolism have been widely studied, as has the regulatory network controlling the denitrification genes (3, 4, 14). However, the anaerobic quinone pool of P. aeruginosa has not been characterized so far.
In the present study, we discovered that UQ9 is essential for the growth of the P. aeruginosa PAO1 strain in denitrification medium. We identified in this bacterium the ORFs PA3911, PA3912, and PA3913 as homologs to E. coli ubiT, ubiV, and ubiU, respectively. Our results showed that these three genes, here called ubiTPa, ubiVPa, and ubiUPa, are essential components of the O2-independent UQ9 biosynthetic pathway of P. aeruginosa. We demonstrated that (i) UbiVPa binds a [4Fe-4S] cluster and (ii) UbiUPa and UbiTPa copurify with UQ by recognizing the isoprenoid tail. Such a molecular pathway for UQ production was found only in proteobacteria (9), where it can exert an essential role under anaerobic conditions, as demonstrated here. Taken together, our results highlight that this pathway could be an interesting lead for the development of antibiotics targeting the denitrification metabolism.
Results
UQ9 is the major quinone of P. aeruginosa
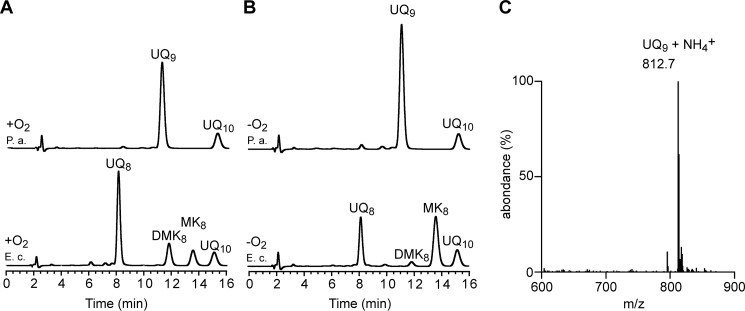
The quinone content of P. aeruginosa PAO1, grown under ambient air or anaerobic conditions (denitrification), was determined using electrochemical detection of lipid extracts separated by HPLC and compared with those obtained from E. coli. Whatever the conditions of growth, a major quinone species eluting at 11.5 min was present in the analyses of P. aeruginosa lipid extracts, with UQ10 being used as the standard (Fig. 1, A and B). MS analysis showed a predominant ammonium adduct (M+ NH4+) with an m/z ratio of 812.7 (Fig. 1C), together with minor adducts, such as Na+ (817.6) and H+ (795.7) (Fig. S1). These masses identify UQ9 (monoisotopic mass, 794.6) as the major quinone produced by P. aeruginosa. Membranes of E. coli contain UQ8 and naphthoquinones (DMK8 and MK8). The absence of detectable levels of naphthoquinones in P. aeruginosa lipid extracts, with or without oxygen (Fig. 1, A and B), is in agreement with the absence of their biosynthetic pathways (MK or futalosine pathways) in P. aeruginosa genomes. It is also interesting that the UQ content of E. coli was higher under aerobic compared with anaerobic conditions (97 ± 4 versus 42 ± 6 pmol of UQ8 per mg of cells), whereas we found the opposite for P. aeruginosa (95 ± 4 versus 126 ± 4 pmol of UQ9 per mg of cells). Together, our results establish that UQ9 is the major quinone of P. aeruginosa PAO1 and suggest that UQ9 is used under denitrification conditions.
Figure 1.
UQ9 is the major quinone used by P. aeruginosa under aerobic and anaerobic conditions. HPLC-ECD analysis of lipid extracts from 1 mg of cells after growth of E. coli MG1655 (E. c.) and P. aeruginosa PAO1 (P. a.) aerobically (+O2) in LB medium (A) or anaerobically (−O2) in denitrification medium (B). The chromatograms are representative of three independent experiments. The peaks corresponding to UQ8, UQ9, DMK8, MK8, and the UQ10, as a standard, are indicated. C, Mass spectrum of the quinone eluting at 11.5 min from extracts of P. aeruginosa grown in aerobic and anaerobic cultures.
Identification of ubi genes in the genome of P. aeruginosa PAO1
To identify the Ubi proteins of P. aeruginosa, IspB, UbiX, and UbiA to UbiK from E. coli MG1655 were first screened for homologs in the P. aeruginosa PAO1 protein sequence data set, available at RRID:SCR_006590, using the BLASTP software. As listed in Table S1, the analysis disclosed the presence of 11 homologous proteins (IspB, UbiA to UbiE, UbiG to UbiJ, and UbiX). As reported previously, the functional homolog of UbiF is a Coq7-like hydroxylase (15), and the corresponding PA0655 protein was shown to be essential for aerobic UQ9 biosynthesis (16). Overall, we propose that the O2-dependent UQ biosynthetic pathways in P. aeruginosa and E. coli share a similar pattern (Fig. S2).
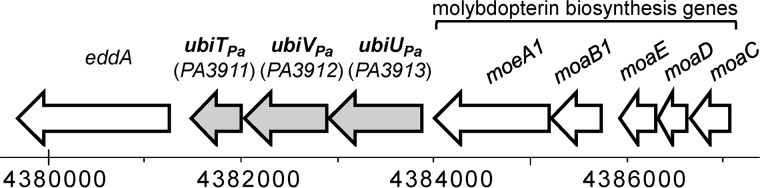
Under anaerobic conditions, E. coli still synthesizes UQ, and we recently identified three genes, which we called ubiT, ubiV, and ubiU, as essential for this process (9). Homologues of ubiT, ubiV, and ubiU were also identified in P. aeruginosa PAO1 and correspond to ORFs PA3911, PA3912, and PA3913, respectively (Table S1 and Fig. S2). These genes are called ubiTPa, ubiVPa, and ubiUPa here. The three genes are predicted to form an operon, ubiUVT (RRID:SCR_006590). Interestingly, this operon is located downstream of the genes moeA1, moaB1, moaE, moaD, and moaC, involved in the biosynthesis of the molybdopterin cofactor (MoCo) (Fig. 2), which is essential for nitrate reductase activity (17). Next, we evaluated the metabolic relevance of the O2-independent UQ biosynthetic pathway in P. aeruginosa by studying mutants of ubiTPa, ubiUPa, and ubiVPa genes.
Figure 2.
Genomic localization of the ubiUVT operon in P. aeruginosa PAO1. ORFs of the genes ubiTPa, ubiUPa, and ubiVPa are represented by gray arrows.
Tn mutants of ubiVPa and ubiUPa present a growth defect for denitrification and an impaired UQ9 content
The physiological importance of the proteins UbiTPa, UbiUPa, and UbiVPa was first investigated using transposon (Tn) mutants PW7609 (ubiTPa), PW7610 (ubiVPa), PW7611 (ubiVPa), PW7612 (ubiUPa), and PW7613 (ubiUPa) and the isogenic parental strain MPAO1 (WT strain from the Manoil collection) as a control (Table S2). Aerobic growth in LB medium was similar between the Tn mutants and the WT strain MPAO1 (Fig. S3A), and the Tn mutants presented a UQ9 level comparable to that of the WT (Fig. S3B). Thus, UbiTPa, UbiUPa, and UbiVPa are not involved in the O2-dependent UQ biosynthetic pathway of P. aeruginosa. In contrast, the growth of the ubiVPa and ubiUPa mutants was severely impaired under denitrification conditions (Fig. S3C), and their UQ9 content was strongly lowered (Fig. S3B). These results suggest the overall requirement of ubiUPa and ubiVPa for denitrification in P. aeruginosa, supposedly via their involvement in O2-independent UQ biosynthesis. Surprisingly, the growth of the ubiTPa Tn mutant PW7609 was not affected (Fig. S3C), and it showed around 40% UQ9 of the WT level in anaerobic cultures (Fig. S3B). In this mutant, the Tn is inserted at the fifth base of the ubiTPa gene, potentially leading to only partial inactivation of the gene (Table S2). We note that previous studies with E. coli ubi mutants showed that only 20% UQ was sufficient to maintain a WT growth phenotype (18, 19).
Denitrification is dependent on ubiTPa, ubiUPa, and ubiVPa genes via their involvement in O2-independent UQ biosynthesis
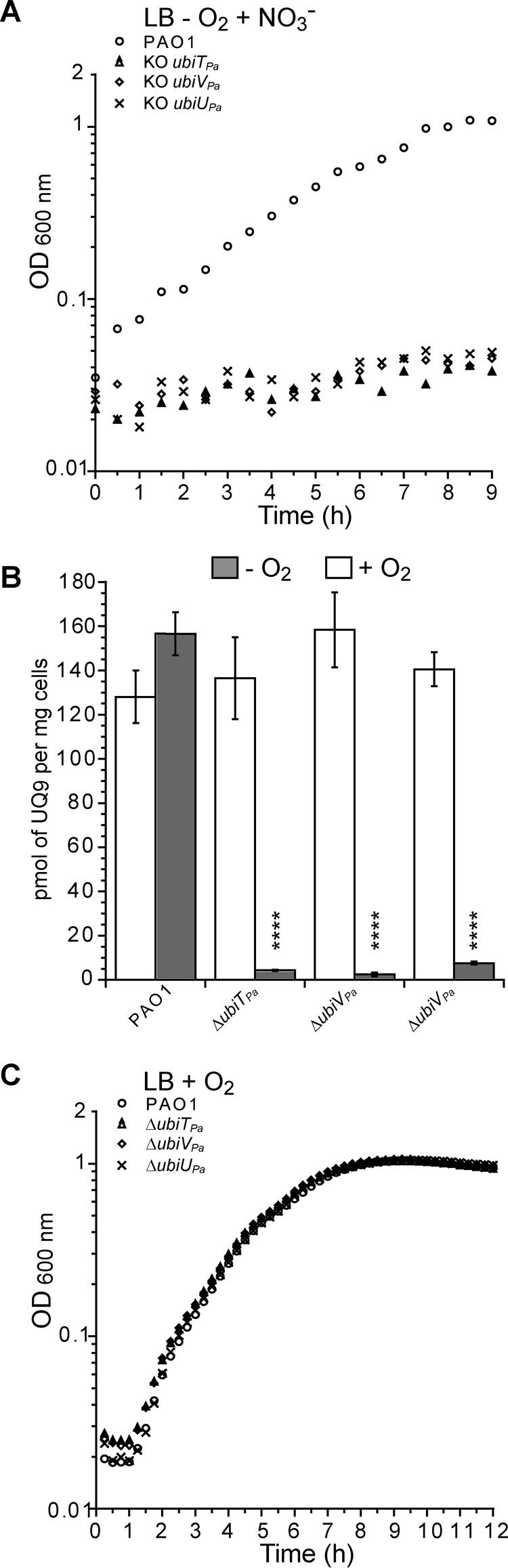
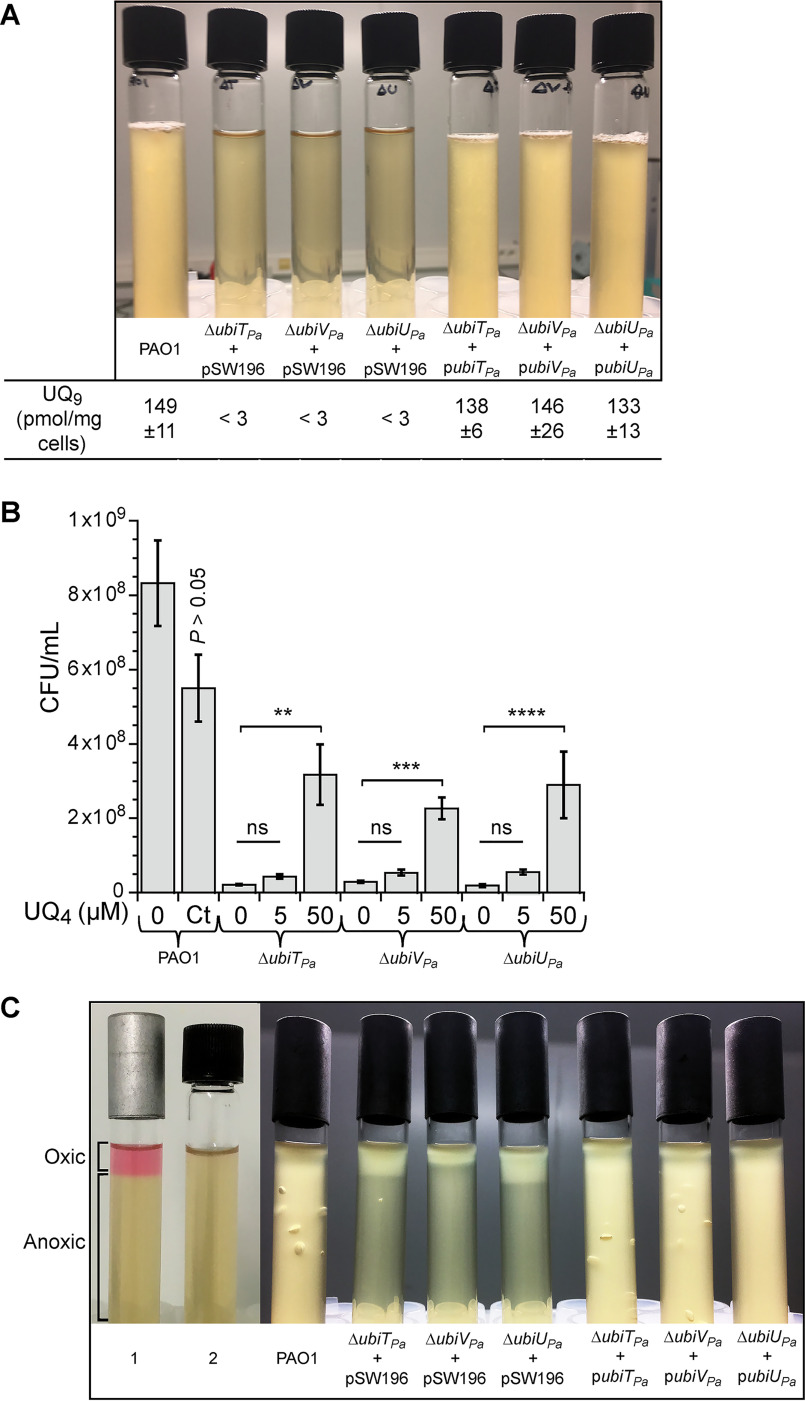
As ubiTPa, ubiVPa, and ubiUPa are localized next to each other in the genome of PAO1, the transposon inserted in the mutants previously studied might impact the expression of the neighboring genes. In addition, it is likely that the Tn mutant PW7609 is not properly disrupting the ubiTPa gene. Thus, for each of the three genes, we constructed knockouts (KO) as well as complementation mutants in the parental strain PAO1. All deletion mutant strains (here called ubiTUV-KO) shared a growth defect under denitrification coupled to a strong decrease of UQ9 content compared with the WT (Fig. 3, A and B), whereas UQ9 content and growth were normal under aerobic conditions (Fig. 3, B and C). Under anaerobic conditions, ubiTUV-KO strains accumulated an early UQ biosynthetic intermediate corresponding to nonaprenylphenol (NPP) (Fig. S2 and Table S3). This result suggests that the O2-independent UQ biosynthetic pathway is blocked downstream of NPP in these three mutants. Interestingly, upon complementation, bacterial growth and UQ9 levels were restored to those of the PAO1 strain used as a control (Fig. 4A).
Figure 3.
ubiUPa, ubiVPa, and ubiTPa are essential genes for anaerobic UQ9 biosynthesis and for denitrification. Shown are representative growth curves of WT PAO1 and ubiTUV-KO strains grown in denitrification medium (A) or aerobically in LB medium (C). B, Quantification of cellular UQ9 content (n = 3) in lipid extracts from WT PAO1 and KO cells grown aerobically in LB medium (white bars) (P > 0.05 by unpaired Student's t test) or in denitrification medium (gray bars) (****, P < 0.0001 by unpaired Student's t test). Error bars represent S.D.
Figure 4.
Complementation of ubiTUV-KO strains restores bacterial growth over the entire O2 range in a UQ-dependent manner. A, Photographs of culture tubes after overnight growth under anaerobic conditions in denitrification medium of ubiTUV-KO strains transformed with the empty vector pSW196 or the same vector carrying the corresponding WT allele (ubiTPa, ubiUPa, and ubiVPa). The parental strain PAO1 was used as a control (Ct), and the UQ9 content of WT and ubiTUV-KO strains cultured anaerobically was assayed (n = 3). B, ubiTUV-KO strains were cultured in denitrification medium supplemented with methanol-solubilized UQ4 at 5 or 50 μm final concentration. After 24 h of incubation, the numbers of CFU per ml (CFU/ml) of each KO strain were estimated and compared with those of the same strain grown without UQ4. As a control (P > 0.05 by unpaired Student's t test), the toxicity of methanol was tested on the parental strain grown in the same medium but supplemented with 4.5% (v/v) methanol (Ct), which is the final concentration of methanol corresponding to the adding of 50 μm UQ4. Data are representative of three independent experiments (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001; by unpaired Student's t test compared with condition without addition of UQ4; ns, not significant). C, As described for panel A, but in soft-agar tubes after overnight culture. All the strains studied were inoculated into anaerobic tubes and then exposed to ambient air to create an oxygen gradient. The controls correspond to soft-agar tubes supplemented with 2.5 µg/ml resazurin and then incubated with (1) or without (2) air. Oxic and anoxic parts of the agar are indicated. For all strains containing pSW196 vectors, denitrification medium was also supplemented with a 0.1% (w/v) final concentration of arabinose to induce the PBAD promoter.
To confirm that UQ was directly involved in the restoration of the anaerobic growth of the ubiTUV-KO strains, UQ4 solubilized in methanol was added to the denitrification medium at 5 and 50 μm final concentrations. After 24 h of anaerobic incubation, numbers of CFU per ml (CFU/ml) of each KO strain were estimated and compared with the same strain cultivated without UQ4. The most significant results were obtained with 50 μm UQ4, which increased the number of CFU of ubiTUV-KO strains by 8- to 15-fold (Fig. 4B). We noted a substantial toxicity of methanol on the WT strain (Fig. 4B, Ct lane), suggesting that the positive effect of UQ4 on the ubiTUV-KO strains is underestimated. Taken together, our results show unequivocally that UbiTPa, UbiUPa, or UbiVPa is needed for denitrification via its involvement in UQ biosynthesis.
UbiTPa, UbiUPa, and UbiVPa are needed for the process of denitrification
We used soft-agar experiments to examine dioxygen and nitrate requirements of ubiTUV-KO strains with or without the WT allele. Soft agar was prepared anaerobically in LB medium containing KNO3 at 100 mm final concentration and then exposed to ambient air. Oxygen diffuses through the agar to form a gradient, the highest concentration being at the top of the agar (Fig. 4C, lane 1). As shown in Fig. 4C, parental strain PAO1 and complemented ubiTUV-KO strains grew throughout the tube, because they were able to use aerobic respiration as well as denitrification. In contrast, the growth of the ubiTUV-KO strains harboring the empty vector was restricted to the oxygenated part of the medium, whereas the presence of the respective genes on the plasmids allowed growth in the anaerobic medium (Fig. 4C). The bubbles observed in the soft agar correspond to gas evolution of N2O and/or N2 (12), suggesting a restoration of the denitrification process in the lower part of the tube. Taken together, these results point to the requirement for ubiTPa, ubiUPa, and ubiVPa beyond the nitrate reduction step and support that UQ is probably essential for the entire denitrification process in P. aeruginosa.
Molybdopterin cofactors are not involved in anaerobic UQ9 biosynthesis
As mentioned previously, the ubiUVT operon is located downstream of the genes moeA1, moaB1, moaE, moaD, and moaC, involved in MoCo biosynthesis. Currently, MoCo-containing hydroxylases constitute the only family known to catalyze O2-independent hydroxylation reactions (20). Since three O2-independent hydroxylation reactions are needed to synthesize UQ anaerobically and UbiU and UbiV are suspected to be involved in these reactions (9), we reasoned that MoCo might be involved in this process, providing a rationale for the colocalization of the ubi and moa/moe genes. To test this hypothesis, we evaluated the ability of Tn mutants PW7614, PW7615/PW7616, PW7618/PW2470, PW7619/PW1920, and PW7621/PW7622 (Table S2), corresponding to Tn insertions in the ORFs moeA1, moaB1 (two mutants), moaE (two mutants), moaD (two mutants), and moaC (two mutants), respectively, to synthesize UQ9 without O2. However, MoCo is also essential for nitrate reductase activity and, thus, for denitrification (17). To overcome this problem, WT and Tn mutants were grown in LB medium using arginine as a fermentable energy source in rich medium. As expected, all the Tn mutants exhibited a growth defect in denitrification. However, anaerobic growth was rescued by the addition of arginine, as previously described (21); therefore, we were able to measure the UQ content of the cells under these conditions (Fig. S4). The UQ9 content of the MoCo Tn mutants was comparable to that of the WT strain (Fig. S4), suggesting that MoCo is not involved in anaerobic UQ9 biosynthesis.
Recombinant UbiVPa is an air-sensitive [4Fe-4S] cluster-containing protein
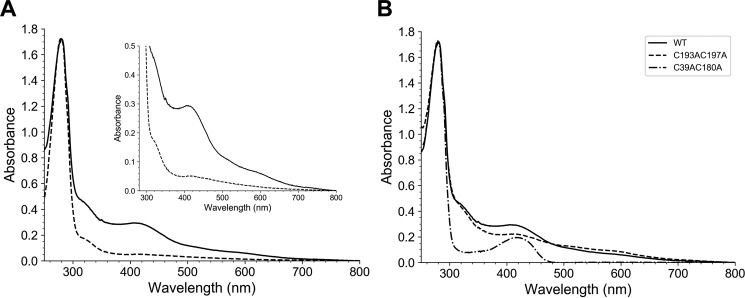
To gain insights into their biochemical properties, we produced and purified the three proteins in E. coli, with UbiVPa being the most soluble. First, we showed that UbiVPa, purified by size exclusion chromatography (SEC), behaved as a monomer (Fig. S5, A and B). Moreover, we noticed that the fraction containing the purified protein was slightly pink-colored with a UV-visible absorption spectrum characteristic of the presence of iron-sulfur species (22), with a band at 410 nm and broad and low-intensity shoulders between 450 and 600 nm (Fig. 5A, dotted line) (23). However, the amount of iron and sulfur (0.22 iron and 0.22 sulfur/monomer) was largely substoichiometric, suggesting a degradation of the [Fe-S] cluster during the purification of the protein under aerobic conditions, as already observed for many other Fe-S proteins. Consistent with this hypothesis, anaerobic reconstitution of the [Fe-S] cluster allowed us to obtain a brown-colored protein with a UV-visible spectrum displaying one broad absorption band at 410 nm, which is characteristic of a [4Fe-4S]2+ cluster (Fig. 5A, solid line) (24). The iron and sulfide determination yielded 3.90 ± 0.03 iron and 3.40 ± 0.20 sulfur/monomer of UbiVPa, consistent with the presence of one [4Fe-4S] cluster/protein (Table 1). As shown in Fig. S5C, the [Fe-S] cluster of UbiVPa was sensitive to air.
Figure 5.
Recombinant UbiVPa is a [4Fe-4S] cluster-containing protein. A, UV-visible absorption of metalloproteins UbiVPa (dotted line, 32.6 μm) and reconstituted holo-UbiVPa (solid line, 22.7 μm). The inset is an enlargement of the 300- to 800-nm region. The molar extinction coefficient, ε410, was determined to be 12.95 ± 0.5 mm−1 cm−1 for holo-UbiVPa. B, Comparative UV-visible absorption spectra of the WT and different Cys-to-Ala mutants of UbiVPa after metal cluster reconstitution. Proteins were analyzed at the following concentrations: 22.7 μm WT, 34.8 μm C39AC180A, and 15.9 μm C193AC197A. Proteins were suspended in buffer containing 50 mm Tris-HCl, 25 mm NaCl, 15% (v/v) glycerol, 1 mm DTT, pH 8.5.
Table 1.
Spectral characterization of UbiVPa and its variants
| Protein | A280/A410 | Contenta (nmol/nmol protein) |
|
|---|---|---|---|
| Iron | Sulfur | ||
| UbiVPa WT | 5.8 | 3.90 ± 0.03 | 3.40 ± 0.20 |
| UbiVPa C39AC180A | 9.0 | 1.10 ± 0.16 | 1.60 ± 0.19 |
| UbiVPa C193AC197A | 7.8 | 2.90 ± 0.05 | 3.00 ± 0.12 |
| UbiVPa C39AC193AC197A | ND | ND | ND |
a Iron and sulfur quantification of UbiVPa and its variants. Shown are the metal content and UV-visible properties after anaerobic reconstitution of their [Fe-S] clusters for WT and variants. ND, not determined.
Four strictly conserved cysteines (C39, C180, C193, and C197) arranged in a CXnCX12CX3C motif (where X represents any amino acid) are found in UbiVPa (9). To test if these four cysteines are important for the chelation of the [4Fe-4S] cluster present in UbiVPa, we generated two double mutants (C39AC180A and C193AC197A) and a triple mutant (C39AC193AC197A). All these mutants were colorless after purification under aerobic conditions and did not show any absorption band in the 350- to 550-nm region of their UV-visible spectra (Fig. S5D), suggesting that they were impaired in their capacity to accommodate a [Fe-S] cluster. After reconstitution under anaerobic conditions, UbiVPa C39AC193AC197A precipitated and its UV-visible spectrum could not be recorded. Although they also had a tendency to aggregate, 10% of the double mutants behaved as monomers, permitting us to perform some assays. Overall, their absorbance at 410 nm (Fig. 5B) and their iron and sulfur contents (Table 1) were largely decreased compared with those of the WT protein, suggesting that the four conserved cysteines are good candidates as ligands of the [4Fe-4S] cluster present in UbiVPa.
Recombinant UbiUPa and UbiTPa copurify with UQ8 in E. coli
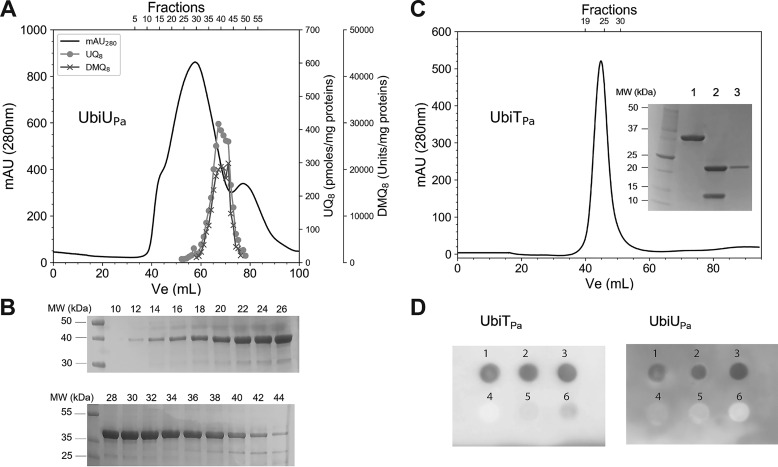
We have recently demonstrated that isoprenoid quinones were able to coelute with the Ubi proteins, such as UbiJ (11), and that UbiT exhibits a sterol carrier protein 2 (SCP2) domain, which is able to bind lipids (9). To that end, we performed lipid content analysis of the UbiTPa, UbiUPa, and UbiVPa fractions purified from E. coli extracts. No isoprenoid quinones were detected coeluting with UbiVPa (Table S4). In contrast, UQ8 and DMQ8 (2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone) were shown to copurify with UbiUPa. This protein was purified only in the presence of detergent, as it was insoluble without it. After a two-step purification protocol, including nickel-nitrilotriacetic acid (Ni-NTA) chromatography and SEC, the solubilized protein still had a tendency to form different oligomeric states, covering the fractions 14–44, as shown in Fig. 6, A and B. UQ8 and DMQ8 were mainly detected in the elution fractions 33–45 (Fig. 6A), corresponding to only a portion of the purified UbiUPa (Fig. 6B). The highest contents, i.e. 488.17 pmol UQ8 per mg of protein and 19,932 AU of DMQ8 per mg of protein, were assayed in fractions 39 and 40, respectively (Table S4). This corresponds to a UQ8/protein ratio of 1.5%. Taken together, these results show that UQ8 and DMQ8 copurify with UbiUPa depending on its oligomerization state.
Figure 6.
Recombinant UbiUPa and UbiTPa bind UQ8. A, Elution profile of UbiUPa. 70 mg of protein was loaded on a Superdex 200 16/60 chromatography column. Quantification of UQ8 and DMQ8 in each fraction was performed by HPLC-ECD MS. Recovery of 73 and 76%, respectively, for UQ8 and DMQ8 was calculated from the total content of all fractions compared with content of the UbiUPa-purified fraction deposited in the Superdex 200 column. B, Fractions 10–44, analyzed by SDS-PAGE for purity. C, Elution profile of UbiTPa on a Superdex 200 16/60 column. Inset, SDS-PAGE. Lane 1, 32-kDa TrxA-UbiTPa fusion protein; lane 2, after digestion with thrombin (UbiTPa, 19.6 kDa; TrxA, 12.1 kDa); lane 3, pooled fractions 20–30 of UbiTPa. Quantification of UQ8 (pool of fractions 20–30) was performed by HPLC-ECD MS. D, Protein-lipid overlay assay between UbiUPa and UbiTPa and different lipid ligands. 2 μl of six different lipid/compound potential candidates (1, UQ8; 2, UQ10; 3, solanesol; 4, 3-methylcatechol; 5, cholesterol; 6, POPE) at 20 mm final concentration were spotted on a PVDF membrane and then incubated with UbiTPa or UbiUPa (both proteins at 0.2 µg/ml final concentration). Detection of bound proteins was performed by chemiluminescence, as described in Experimental procedures. MW, molecular weights.
Due to a lack of solubility, UbiTPa was overproduced with E. coli thioredoxin (TrxA) as a gene fusion partner, as previously described (25). After the first step of the purification process (Ni-NTA chromatography), the 32-kDa TrxA-UbiTPa fusion protein was digested with thrombin. To remove the TrxA His-tagged protein, UbiTPa then was purified, by Ni-NTA chromatography coupled to SEC, as a high oligomeric form (Fig. 6C). The purified UbiTPa (pool of fractions 20–30) contained 9.75 ± 4.57 pmol UQ8 per mg of protein (Table S4), which corresponds to a UQ8/protein ratio of about 0.03%.
Recombinant UbiUPa and UbiTPa bind the isoprenoid tail of UQ
To further confirm the ability of UbiTPa and UbiUPa to bind UQ, a protein-lipid overlay assay was performed (Fig. 6D). We checked the possibility of these proteins recognizing UQ10, UQ8, solanesol, 3-methylcatechol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and cholesterol. Solanesol is a noncyclic terpene alcohol that consists of nine isoprene units, as found in UQ9. 3-Methylcatechol was chosen to mimic the head group of UQ. POPE is the major lipid component of the inner membrane of E. coli (26). Finally, cholesterol was used as a sterol standard. Figure 6D shows that UbiTPa and UbiUPa did not interact with 3-methylcatechol, POPE, or cholesterol under our experimental conditions. In contrast, both proteins were able to recognize UQ10, UQ8, and solanesol. We established the ability of UbiTPa to bind phosphatidic acid (PA), as previously demonstrated by Groenewold et al. (25) (Fig. S6). Together, we show that UbiTPa and UbiUPa are able to bind the isoprenoid tail of UQ, in agreement with their involvement in the O2-independent biosynthetic pathway of UQ.
Discussion
UQ acts as a membrane-embedded electron and proton shuttle and is a key molecule in the respiratory metabolism of proteobacteria. The biosynthesis of UQ under aerobic conditions has been widely studied and includes a series of enzymatic reactions in which a benzene ring undergoes a series of modifications involving a prenylation, a decarboxylation, three methylations, and three hydroxylations (27). Our chemical analysis identified UQ9 as the major quinone in the membranes of aerobically grown P. aeruginosa cells, which is in agreement with the literature (7, 16). We found in P. aeruginosa homologues of the genes known to be involved in UQ biosynthesis in E. coli, except ubiF (Table S1). Indeed, as already published, P. aeruginosa exhibits a yeast COQ7 protein homolog, which catalyzes the same reaction as UbiF from E. coli (15, 16). Therefore, both bacteria share a similar UQ biosynthetic pathway involving three hydroxylases, UbiI, UbiH, and UbiF, or COQ7, using O2 as a cosubstrate (Fig. S2). As no other isoprenoid quinone was detected in lipid extracts of P. aeruginosa, we suppose that UQ9 is essential for aerobic growth of this bacterium.
In the absence of oxygen, P. aeruginosa can grow by dissimilatory nitrate respiration by using nitrate or nitrite as alternative terminal electron acceptors of the respiratory chain. This metabolic process, known as denitrification, has been widely studied (3). However, the component acting to transfer electrons from primary dehydrogenases to nitrate or nitrite reductases was not clearly identified to date (3). In the present study, we identified UQ9 as the major quinone synthesized under anaerobic conditions. By LC-MS analysis, we also identified two related redox molecules, UQ8 and DMQ9, present in small amounts (see Fig. 1B, the peaks around 8 and 9.5 min) and nonaprenylphenol (NPP; see Fig. S2). NPP and DMQ9 are UQ9 biosynthetic intermediates (Fig. S2), and UQ8 was already detected in Pseudomonas lipid extracts under aerobic conditions (16). Taken together, these results suggest that P. aeruginosa possesses an O2-independent UQ biosynthetic pathway, which produces the major quinone species observed under anaerobic conditions. We recently identified such a pathway in E. coli (9). Here, we characterized three genes, ubiTPa (PA3911), ubiUPa (PA3913), and ubiVPa (PA3912), as essential for the anaerobic UQ9 biosynthesis in P. aeruginosa and dispensable for the aerobic one. These genes are homologs to the ubiT, ubiU, and ubiV genes previously identified in E. coli, which grows normally under anaerobic conditions in a UQ-independent manner because of the presence of naphthoquinones (9, 28, 29). In contrast, we demonstrated that these three genes were essential to anaerobic denitrification metabolism of P. aeruginosa, which is in agreement with the presence of a single quinone corresponding to UQ. In line with our results, ubiVPa (PA3912) and ubiUPa (PA3913) are expressed in response to anaerobic conditions (30), and the abundance of UbiUPa protein was increased during anaerobic growth (31). Moreover, using random transposon mutagenesis, ubiVPa (PA3912) and ubiUPa (PA3913) were already reported as essential for anaerobic growth of P. aeruginosa on nitrate and nitrite as alternative terminal electron acceptors of the respiratory chain (21).
Although its contribution is still poorly understood for within-host growth, anaerobic respiration of P. aeruginosa is likely to be significant for promoting virulence mechanisms in chronic lung infections (13). Indeed, the infected endobronchial mucus of CF patients is subject to severe hypoxia or even anoxia (32). A likely hypothesis is that accelerated O2 consumption in the biofilm results from activated polymorphonuclear leukocytes that produce superoxide (33) and nitric oxide (34). Indeed, high levels of nitrate and nitrite have been measured in sputum from CF patients (35). From all these observations, and as UQ is an essential component of the denitrification metabolism in P. aeruginosa, we propose that UbiTPa, UbiVPa, and UbiUPa contribute to the CF lung infection in patients (work in progress in our laboratory). This hypothesis is supported by a recent quantitative proteomics approach revealing the increased abundance of the three proteins in anaerobic biofilms grown under conditions of the cystic fibrosis lung (25). Moreover, as deduced from a high-throughput sequencing of Tn libraries from P. aeruginosa strain PA14, it appears that the ubiT gene was found to be essential for this bacterium to colonize the murine gastrointestinal tract (36), which suggests that O2-independent UQ biosynthesis is essential for bacterial virulence. This hypothesis is also supported by the essential contribution of UbiU and UbiV homologs to Yersinia ruckeri virulence (37).
As already suggested, homologs of UbiU and UbiV would belong to a new family of O2-independent hydroxylases (9). To date, only the MoCo-containing hydroxylases using water-derived oxygen are known to catalyze hydroxylations under anaerobic conditions (20). In our study, we have demonstrated that MoCo is not essential to the anaerobic UQ pathway, strengthening the hypothesis that hydroxylation reactions performed in a UbiU- and UbiV-dependent manner do not involve MoCo.
To better understand their functions, we decided to overproduce in E. coli, purify, and biochemically characterize UbiTPa, UbiUPa, and UbiVPa. Our results showed that recombinant UbiVPa is an air-sensitive Fe-S–containing protein, as UbiV from E. coli (9), and we demonstrated that cysteines 39, 180, 193, and 197 were ligands to the [4Fe-4S] cluster found in UbiVPa. These results confirm the conservation of a four-cysteine pattern coordinating an Fe-S cluster across homologs of UbiV. This pattern is also found in RlhA and TrhP (38, 39), two proteins that belong to the same protease U32 family as UbiU and UbiV, and are also involved in O2-independent hydroxylation reactions in E. coli (38, 39). However, the function of the iron-sulfur centers in the hydroxylation mechanism remains to be understood.
As a member of the U32 protease family, UbiUPa also presents four conserved cysteines (C169, C176, C193, and C232). Unfortunately, we failed to reconstitute an Fe-S cluster and instead obtained protein precipitation. Indeed, UbiUPa is an unstable protein. Unlike UbiU from E. coli, which forms a stable heterodimer UbiU-UbiV complex (9), we were not able to solubilize UbiUPa by coproducing it with its potential partner, UbiVPa. The fact that we produced P. aeruginosa proteins in E. coli could explain this difference in behavior. Nevertheless, a UbiU-UbiV complex in P. aeruginosa remains a reasonable hypothesis that needs further investigation. We were able to purify UbiUPa alone with significant quantities of UQ8 and DMQ8, whereas purified UbiVPa contained no quinones. This result was confirmed by a protein-lipid overlay assay, which showed that the isoprenoid tail of UQ was the structural determinant for the recognition by UbiUPa. From these results, we propose that UbiU would bind UQ and reaction intermediates of the anaerobic UQ pathway.
Homologs of UbiT and UbiJ contain an SCP2 domain (9, 40), involved in protein-lipid interactions, and UbiJ from E. coli copurified with UQ8 (11). Moreover, a previous study showed that PA3911 (UbiTPa) was able to bind specifically PA, the central hub of phospholipid metabolism (25). Here, we showed that UbiTPa binds to UQ8 and shares with UbiUPa the recognition of the isoprenoid tail of UQ. Taken together, these results support the hypothesis that UbiT is the counterpart of UbiJ under anaerobic conditions. We propose that UbiT binds UQ intermediates and stabilizes a putative anaerobic Ubi complex that has yet to be demonstrated.
Experimental procedures
Bacterial strains and growth conditions
P. aeruginosa and E. coli strains used in this study are listed in Table S2. We obtained the collection of transposon (Tn) mutants in the P. aeruginosa MPAO1 strain from the Manoil Laboratory, Department of Genome Science, University of Washington (41, 42). The Tn insertion sites of the mutant strains were verified by sequencing (GATC Biotech, Konstanz, Germany) using PCR primers recommended by the library curators. P. aeruginosa strains were aerobically maintained at 37 °C on lysogeny broth (LB) agar plates. For quinone assay, aerobic cultures (5 ml) were performed in LB medium at 37 °C with rotary shaking at 200 rpm. Anaerobic growth of P. aeruginosa was performed in 12-ml Hungate tubes containing LB medium supplemented with KNO3 as an electron acceptor (100 mm final concentration) (43), here called denitrification medium, and deoxygenated for 30 min by argon bubbling (O2 at <0.1 ppm) prior to autoclaving. In some experiments, LB medium was supplemented with arginine at a final concentration of 40 mm instead of KNO3 (44). Hungate tubes were inoculated through the septum with 100 μl of overnight cultures taken with disposable syringe and needles from closed Eppendorf tubes filled to the top. Cultures in Hungate tubes were used for measuring the quinone contents. For aerobic growth studies, aerobic overnight cultures were used to inoculate a 96-well plate to obtain a starting optical density at 600 nm (OD600) of 0.05 and further incubated with shaking at 37 °C. Changes in OD600 were monitored every 10 min for 12 h using the Infinite 200 PRO microplate reader (Tecan, Lyon, France). For anaerobic growth curve studies, overnight cultures in 50-ml closed tubes of non-degassed denitrification medium were used to inoculate 400-ml bottles to obtain a starting OD600 of 0.05. The bacteria then were grown anaerobically by sparging argon (O2 at <0.1 ppm), and bacterial cultures were monitored spectrophotometrically (OD600) at 30-min intervals for 9 h. E. coli MG1655 and DH5α were grown on LB agar or in LB liquid. When required, the medium was supplemented with ampicillin at 100 µg/ml for E. coli, carbenicillin at 250 µg/ml for P. aeruginosa, tetracycline at 60 µg/ml for E. coli and 100 µg/ml for P. aeruginosa, or gentamicin at 200 µg/ml for P. aeruginosa (Table S2). When necessary, UQ4 or 0.1% (w/v) arabinose, final concentration, was added to the medium to enhance bacterial anaerobic growth or to induce PBAD expression of pSW196-derived plasmids, respectively. Pseudomonas isolation agar medium (from DB) containing irgasan (25 µg/liter) was used for triparental mating to counterselect E. coli. For CFU counting, bacteria were suspended in PBS and cell suspensions were serially diluted in PBS. For each sample, 100 µl of at least four different dilutions were plated on LB plates and incubated for 24 h at 37 °C, and CFU were counted using a Scan 100 Interscience.
Plasmids and genetic manipulations
The plasmids and the primers used in this study are listed in Tables S2 and S4. All the plasmids produced in this work were checked using DNA sequencing (GATC Biotech, Konstanz, Germany). To generate P. aeruginosa deletion mutants, overlapping upstream and downstream flanking regions of ubiTPa, ubiUPa, and UbiVPa genes were obtained by PCR amplification using the PAO1 genome as the template and the oligonucleotides described in Table S5. The resulting fragments were then cloned into SmaI-cut pEXG2 plasmid by sequence- and ligation-independent cloning (45). To complement the mutants, the ubiTPa, ubiUPa, and ubiVPa fragments were generated by PCR amplification using the oligonucleotide pairs ubiT-PA-F/ubiT-PA-R, ubiU-PA-F/ubiU-PA-R, and ubiV-PA-F/ubiV-PA-R, respectively, and PAO1 genome as the template (Table S5). The fragments were EcoRI-SacI digested and inserted into the PBAD-harboring pSW196 plasmid, yielding the pubiTPa, pubiUPa, and pubiVPa plasmids, respectively (Table S2). The pEXG2- and pSW196-derived vectors were transferred into the P. aeruginosa PAO1 strain by triparental mating using pRK2013 as a helper plasmid (46). For allelic exchange using the pEXG2 plasmids, cointegration events were selected on Pseudomonas isolation agar plates containing gentamicin. Single colonies then were cultured on NaCl-free LB agar plates containing 10% (w/v) sucrose to select for the loss of the plasmid, and the resulting sucrose-resistant colonies were checked for mutant genotypes by PCR. To overproduce C-terminally His-tagged UbiVPa, the ubiVPa gene was cloned into the pET22b(+) vector. The ubiVPa insert was obtained by PCR amplification using the oligonucleotide pair pET22-UbiV-F and pET22-UbiV-R and the ubiVPa ORF as a template (Table S5). NdeI-XhoI-digested amplicon was ligated to NdeI-XhoI-digested pET22b(+) vector to obtain pET22-UbiVPa (Table S2). Variants of UbiVPa were obtained using the Q5 site-directed mutagenesis kit (New England Biolabs) (UbiVPa C193AC197A and UbiVPa C39AC193AC197A) and the QuikChange II XL site-directed mutagenesis kit (Agilent) (UbiVPa C39AC180A) according to the manufacturer's specifications, using pET22b-UbiVPa as the template (Table S2 and S5). The ubiUPa gene was cloned into pET-22b(+) by following the same protocol as that for the ubiVPa gene. The ubiTPa gene was synthesized by Eurofins with E. coli codon optimization. The synthetic gene then was cloned into the EcoRI/NotI sites of vector pET32a(+) (Novagen), resulting in plasmid pET32-TrxA-UbiTPa (Table S2).
Soft-agar study to evaluate the O2 dependence of growth
Soft-agar studies were performed in denitrification medium supplemented with agar at a 0.7% (w/v) final concentration. After argon bubbling (O2 at <0.1 ppm) for 30 min, the suspension (13 ml) was autoclaved in Hungate tubes. They were then placed in a 40 °C incubator and inoculated through the septum with 100 μl of overnight cultures taken with disposable syringe and needles from Eppendorf tubes filled to the top, mixed by inverting, and incubated at room temperature for 30 min to allow the agar to solidify. The tubes then were incubated under aerobic conditions with caps loosened at 37 °C for 24 h. A control experiment was performed with resazurin (0.25 µg/ml final concentration), used as an indicator of medium oxygenation. When required, the medium was supplemented with antibiotics.
Lipid extractions and quinone analysis
Cultures (5 ml under ambient air and 13 ml under anaerobic conditions) were cooled down on ice before centrifugation at 3200 × g at 4 °C for 10 min. Cell pellets were washed in 1 ml ice-cold PBS and transferred to preweighted 1.5-ml Eppendorf tubes. After centrifugation at 12,000 × g at 4 °C for 1 min and elimination of supernatant, the cell wet weight was determined (∼5–30 mg) and pellets were stored at −20 °C. Quinone extraction from cell pellets was performed as previously described (18). Lipid extracts corresponding to 1 mg of cell wet weight were analyzed by HPLC electrochemical detection-MS (ECD-MS) with a BetaBasic-18 column at a flow rate of 1 ml/min with mobile phases composed of methanol, ethanol, and a mix of 90% isopropanol, 10% ammonium acetate (1 M), and 0.1% TFA with mobile phase 1 (50% methanol, 40% ethanol, and 10% mix). When necessary, MS detection was performed on an MSQ spectrometer (Thermo Scientific) with electrospray ionization in positive mode (probe temperature, 400 °C; cone voltage, 80 V). Single-ion monitoring detected the following compounds: UQ8 (M+ NH4+), m/z 744–745, 6–10 min, scan time of 0.2 s; UQ9 (M+ NH4+), m/z 812–813, 9–14 min, scan time of 0.2 s; UQ10 (M+ NH4+), m/z 880.2–881.2, 10–17 min, scan time of 0.2 s; DMQ8 (M+ NH4+), m/z 714–715, 5–10 min, scan time of 0.2 s; and NPP (M+ NH4+), m/z 724–725, 8–13 min, scan time of 0.2 s. MS spectra were recorded between m/z 600 and 900 with a scan time of 0.3 s. ECD and MS peak areas were corrected for sample loss during extraction on the basis of the recovery of the UQ10 internal standard and then were normalized to cell wet weight. The peaks of UQ8 and UQ9 obtained with electrochemical detection were quantified with a standard curve of UQ10 as previously described (18).
Overproduction and purification of UbiV, UbiU, and UbiT from P. aeruginosa in E. coli
WT and UbiVPa variants were expressed and purified as previously described for E. coli proteins (9). Briefly, the pET-22b(+) plasmid, encoding WT or UbiVPa variants, were cotransformed with pGro7 plasmid (Takara Bio Inc.) into E. coli BL21 (DE3) ΔubiUV competent cells grown at 37 °C in LB medium, which was supplemented with ampicillin (50 µg/ml), kanamycin (50 µg/ml), and chloramphenicol (12.5 µg/ml). At an OD600 of 1.2, d-arabinose was added to the cultures at a final concentration of 2 mg/ml. At an OD600 of 1.8, cultures were cooled down on ice for 20 min, and IPTG was added at a final concentration of 0.1 mm. Cells then were allowed to grow further at 16 °C overnight. WT UbiVPa and the different variants were purified by Ni-NTA chromatography followed by SEC in buffer A (50 mm Tris-HCl, 25 mm NaCl, 15% [v/v] glycerol, pH 8.5) containing 1 mm DTT. The purified proteins were concentrated to 30–40 mg/ml using Amicon concentrators (30-kDa cutoff; Millipore).
The overproduction of WT UbiUPa was performed in E. coli BL21(DE3) ΔubiUV cells by following the same protocol as that for UbiVPa, except that UbiUPa overexpression was induced with 0.05 mm of IPTG, and the cell pellets were resuspended in buffer B (50 mm Tris-HCl, 500 mm NaCl, 15% [v/v] glycerol, pH 8.5) containing 0.2% (w/v) N-lauroylsarcosine sodium salt. After cell disruption by sonication, the clarified cell-free extracts were loaded onto a His-Trap FF crude column (GE Healthcare) pre-equilibrated with buffer B containing 0.1% (w/v) N-lauroylsarcosine sodium salt. The column was washed with 10 column volumes of buffer C (50 mm Tris-HCl, 500 mm NaCl, 15% [v/v] glycerol, 10 mm imidazole, pH 8.5) containing 6 mm N,N-dimethyldodecylamine N-oxide (LDAO) to remove nonspecifically bound E. coli proteins and then eluted with a linear gradient of 10 column volumes of buffer C containing 500 mm imidazole and 6 mm LDAO. Fractions containing UbiUPa were pooled and then loaded on a HiLoad 16/600 Superdex 200 pg (GE Healthcare) pre-equilibrated in buffer D (50 mm Tris-HCl, 150 mm NaCl, 15% [v/v] glycerol, pH 8.5) containing 3 mm LDAO. The purified proteins were concentrated using Amicon concentrators (100-kDa cutoff; Millipore), aliquoted, frozen in liquid nitrogen, and stored at −80 °C. For protein–lipid overlay, fractions 34–43 were pooled.
Overproduction of UbiTPa fused with the thioredoxin (TrxA-UbiTPa) in E. coli BL21(DE3) ΔubiUV cells was performed by following the same protocol as that for UbiUPa, except that overexpression of the chimeric gene was induced at an OD600 of 0.5 and the cell pellet was resuspended in buffer B containing 5% (w/v) sodium cholate. TrxA-UbiTPa was first purified by following the same protocol as that for UbiUPa by Ni-NTA chromatography, except that the HisTrap FF crude column was preequilibrated with buffer B containing 0.5% (w/v) sodium cholate and then eluted with buffer C containing 500 mm imidazole and 0.5% (w/v) sodium cholate. Fractions containing TrxA-UbiTPa were pooled and detergent was removed using a Hiprep 26/10 desalting column (GE Healthcare) preequilibrated with buffer D. The fusion protein was digested with thrombin (10 units/mg of TrxA-UbiTPa) at room temperature and then loaded on a HiLoad 16/600 Superdex 200 pg (GE Healthcare) coupled with a HisTrap FF crude column (GE Healthcare) preequilibrated with buffer D. The purified proteins were concentrated using Amicon concentrators (100-kDa cutoff; Millipore), aliquoted, frozen in liquid nitrogen, and stored at −80 °C.
[Fe-S] cluster reconstitution
The [Fe-S] cluster(s) of holo-UbiVPa and holo-variants was reconstituted as previously described (9). Briefly, a solution containing 100 μm of metalloproteins was treated with 5 mm DTT for 15 min at 20 °C and then incubated for 1 h with a 5-fold molar excess of both ferrous ammonium sulfate and l-cysteine. The reaction was initiated by the addition of a catalytic amount of the E. coli cysteine desulfurase CsdA (1–2% molar equivalent) and monitored by UV-visible absorption spectroscopy. After 1 h of incubation, the holo-proteins were then loaded onto a Superdex 75 Increase 10/300 GL column (GE Healthcare) preequilibrated with buffer A. The fractions containing the holo-proteins were pooled and concentrated to 20–30 mg/ml on a Vivaspin concentrator (30-kDa cutoff).
Protein–lipid overlay
To assess the lipid-binding properties of UbiTPa and UbiUPa, a protein–lipid overlay was performed as previously described (47). Briefly, 2 μl of 20 mm lipids in dichloromethane was spotted onto PVDF membrane and allowed to dry at room temperature for 1 h. The membranes were blocked in 3% (w/v) fatty acid-free BSA in TBST (50 mm Tris-HCl, 150 mm NaCl, and 0.1% [v/v] Tween-20, pH 7.5) for 1 h. The membranes were then incubated overnight at 4 °C with gentle stirring in the same solution containing 0.2 μg/ml of the indicated proteins. After washing six times for 30 min in TBST buffer, the membranes were incubated for 1 h with a 1/1000 dilution of anti-polyHis mAb (Sigma) and then for 1 h with a 1/10,000 dilution of anti-mouse–horseradish peroxidase conjugate (Thermo Fisher Scientific). His-tagged proteins bound to the membrane by virtue of its interaction with lipid were detected by enhanced chemiluminescence using Clarity Max Western ECL substrate (Bio-Rad).
Quantification methods
Protein concentrations were determined using the method of Bradford (Bio-Rad), with BSA as the standard. The iron and acid-labile sulfide were determined according to the method of Fish (48) and Beinert (49), respectively, before and after [4Fe-4S] cluster reconstitution.
UV-visible spectroscopy
UV-visible spectra were recorded in 1-cm-optic-path quartz cuvettes under aerobic conditions on a Cary 100 UV-visible spectrophotometer (Agilent) and under anaerobic conditions in a glove box on an XL-100 Uvikon spectrophotometer equipped with optical fibers.
Data availability
All data are contained within the manuscript and supporting material.
Supplementary Material
This article contains supporting information.
Author contributions—C.-D. T. V., J. M., S. E., B. F., and E. B. data curation; C.-D. T. V., J. M., S. E., B. F., E. B., F. P., M. L., and L. P. formal analysis; C.-D. T. V., M. L., and L. P. investigation; J. M., S. E., B. F., E. B., M. L., and L. P. methodology; F. B., M. F., M. L., and L. P. supervision; F. B., M. F., F. P., M. L., and L. P. funding acquisition; F. B., M. F., M. L., and L. P. validation; F. P. and L. P. visualization; M. L. and L. P. conceptualization; M. L. and L. P. writing-original draft; L. P. writing-review and editing.
Funding and additional information—This work was supported by the Agence Nationale de la Recherche (ANR), projects (An)aeroUbi ANR-15-CE11-0001-02, O2-taboo ANR-19-CE44-0014, DYNAMO ANR-11-LABX-0011-01 and ANR-10-LABX-62-IBEID, the University Grenoble Alpes (UGA), the French Centre National de la Recherche Scientifique (CNRS), and the Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA).
Conflict of interest—The authors declare that they have no competing interests.
- MK
- menaquinone
- UQ
- ubiquinone
- DMK
- dimethyl-menaquinone
- DMQ
- C6-demethoxy-ubiquinone
- NPP
- nonaprenylphenol
- SEC
- size exclusion chromatography
- SCP2
- sterol carrier protein 2
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- ECD
- electrochemical detection
- PA
- phosphatidic acid
- Tn
- transposon
- Ni-NTA
- nickel-nitrilotriacetic acid
- MoCo
- molybdopterin cofactor
- OD600
- optical density at 600 nm
- LDAO
- N,N-dimethyldodecylamine N-oxide
- POPE
- 3-methylcatechol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine.
References
- 1. Crull M. R., Somayaji R., Ramos K. J., Caldwell E., Mayer-Hamblett N., Aitken M. L., Nichols D. P., Rowhani-Rahbar A., and Goss C. H. (2018) Changing rates of chronic Pseudomonas aeruginosa infections in cystic fibrosis: a population-based cohort study. Clin. Infect. Dis. 67, 1089–1095 10.1093/cid/ciy215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Magill S. S., Edwards J. R., Bamberg W., Beldavs Z. G., Dumyati G., Kainer M. A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., Ray S. M., Thompson D. L., Wilson L. E., and Fridkin S. K. (2014) Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370, 1198–1208 10.1056/NEJMoa1306801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arai H. (2011) Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2, 103 10.3389/fmicb.2011.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams H. D., Zlosnik J. E., and Ryall B. (2007) Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71 10.1016/S0065-2911(06)52001-6 [DOI] [PubMed] [Google Scholar]
- 5. Torres A., Kasturiarachi N., DuPont M., Cooper V. S., Bomberger J., and Zemke A. (2019) NADH dehydrogenases in Pseudomonas aeruginosa growth and virulence. Front. Microbiol. 10, 75 10.3389/fmicb.2019.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Page A. C. Jr., Gale P., Wallick H., Walton R. B., Mc D. L., Woodruff H. B., Folkers K. (1960) Isolation of coenzyme Q10 from bacterial fermentation. Arch. Biochem. Biophys. 89, 318–321 10.1016/0003-9861(60)90062-X [DOI] [PubMed] [Google Scholar]
- 7. Matsushita K., Yamada M., Shinagawa E., Adachi O., and Ameyama M. (1980) Function of ubiquinone in the electron transport system of Pseudomonas aeruginosa grown aerobically. J. Biochem. 88, 757–764 10.1093/oxfordjournals.jbchem.a133028 [DOI] [PubMed] [Google Scholar]
- 8. Nowicka B., and Kruk J. (2010) Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys. Acta 1797, 1587–1605 10.1016/j.bbabio.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 9. Pelosi L., Vo C. D., Abby S. S., Loiseau L., Rascalou B., Hajj Chehade M., Faivre B., Gousse M., Chenal C., Touati N., Binet L., Cornu D., Fyfe C. D., Fontecave M., Barras F., et al. (2019) Ubiquinone biosynthesis over the entire O2 range: characterization of a conserved O2-independent pathway. mBio 10, e01319–19 10.1128/mBio.01319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alexander K., and Young I. G. (1978) Alternative hydroxylases for the aerobic and anaerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry 17, 4750–4755 10.1021/bi00615a024 [DOI] [PubMed] [Google Scholar]
- 11. Hajj Chehade M., Pelosi L., Fyfe C. D., Loiseau L., Rascalou B., Brugiere S., Kazemzadeh K., Vo C. D., Ciccone L., Aussel L., Coute Y., Fontecave M., Barras F., Lombard M., and Pierrel F. (2019) A soluble metabolon synthesizes the isoprenoid lipid ubiquinone. Cell Chem. Biol. 26, 482–492e487 10.1016/j.chembiol.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 12. Zumft W. G. (1997) Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 10.1128/.61.4.533-616.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen P. O., Kolpen M., Kragh K. N., and Kuhl M. (2017) Microenvironmental characteristics and physiology of biofilms in chronic infections of CF patients are strongly affected by the host immune response. APMIS 125, 276–288 10.1111/apm.12668 [DOI] [PubMed] [Google Scholar]
- 14. Borrero-de Acuna J. M., Timmis K. N., Jahn M., and Jahn D. (2017) Protein complex formation during denitrification by Pseudomonas aeruginosa. Microb. Biotechnol. 10, 1523–1534 10.1111/1751-7915.12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stenmark P., Grunler J., Mattsson J., Sindelar P. J., Nordlund P., and Berthold D. A. (2001) A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane-bound hydroxylase involved in ubiquinone biosynthesis. J. Biol. Chem. 276, 33297–33300 10.1074/jbc.C100346200 [DOI] [PubMed] [Google Scholar]
- 16. Jiang H. X., Wang J., Zhou L., Jin Z. J., Cao X. Q., Liu H., Chen H. F., and He Y. W. (2019) Coenzyme Q biosynthesis in the biopesticide Shenqinmycin-producing Pseudomonas aeruginosa strain M18. J. Ind. Microbiol. Biotechnol. 46, 1025–1038 10.1007/s10295-019-02179-1] [DOI] [PubMed] [Google Scholar]
- 17. Coelho C., and Romao M. J. (2015) Structural and mechanistic insights on nitrate reductases. Protein Sci. 24, 1901–1911 10.1002/pro.2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hajj Chehade M., Loiseau L., Lombard M., Pecqueur L., Ismail A., Smadja M., Golinelli-Pimpaneau B., Mellot-Draznieks C., Hamelin O., Aussel L., Kieffer-Jaquinod S., Labessan N., Barras F., Fontecave M., and Pierrel F. (2013) ubiI, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J. Biol. Chem. 288, 20085–20092 10.1074/jbc.M113.480368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loiseau L., Fyfe C., Aussel L., Hajj Chehade M., Hernandez S. B., Faivre B., Hamdane D., Mellot-Draznieks C., Rascalou B., Pelosi L., Velours C., Cornu D., Lombard M., Casadesus J., Pierrel F., et al. (2017) The UbiK protein is an accessory factor necessary for bacterial ubiquinone (UQ) biosynthesis and forms a complex with the UQ biogenesis factor UbiJ. J. Biol. Chem. 292, 11937–11950 10.1074/jbc.M117.789164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hille R. (2005) Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 433, 107–116 10.1016/j.abb.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 21. Filiatrault M. J., Picardo K. F., Ngai H., Passador L., and Iglewski B. H. (2006) Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74, 4237–4245 10.1128/IAI.02014-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imlay J. A. (2006) Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 10.1111/j.1365-2958.2006.05028.x [DOI] [PubMed] [Google Scholar]
- 23. Ollagnier de Choudens S., and Barras F. (2017) Genetic, biochemical, and biophysical methods for studying FeS proteins and their assembly. Methods Enzymol. 595, 1–32 10.1016/bs.mie.2017.07.015 [DOI] [PubMed] [Google Scholar]
- 24. Ollagnier-de Choudens S., Loiseau L., Sanakis Y., Barras F., and Fontecave M. (2005) Quinolinate synthetase, an iron-sulfur enzyme in NAD biosynthesis. FEBS Lett. 579, 3737–3743 10.1016/j.febslet.2005.05.065 [DOI] [PubMed] [Google Scholar]
- 25. Groenewold M. K., Massmig M., Hebecker S., Danne L., Magnowska Z., Nimtz M., Narberhaus F., Jahn D., Heinz D. W., Jansch L., and Moser J. (2018) A phosphatidic acid-binding protein is important for lipid homeostasis and adaptation to anaerobic biofilm conditions in Pseudomonas aeruginosa. Biochem. J. 475, 1885–1907 10.1042/BCJ20180257 [DOI] [PubMed] [Google Scholar]
- 26. Dowhan W. (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66, 199–232 10.1146/annurev.biochem.66.1.199 [DOI] [PubMed] [Google Scholar]
- 27. Aussel L., Pierrel F., Loiseau L., Lombard M., Fontecave M., and Barras F. (2014) Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys. Acta 1837, 1004–1011 10.1016/j.bbabio.2014.01.015 [DOI] [PubMed] [Google Scholar]
- 28. Sharma P., Teixeira de Mattos M. J., Hellingwerf K. J., and Bekker M. (2012) On the function of the various quinone species in Escherichia coli. FEBS J. 279, 3364–3373 10.1111/j.1742-4658.2012.08608.x [DOI] [PubMed] [Google Scholar]
- 29. Nitzschke A., and Bettenbrock K. (2018) All three quinone species play distinct roles in ensuring optimal growth under aerobic and fermentative conditions in E. coli K12. PLoS ONE 13, e0194699 10.1371/journal.pone.0194699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Filiatrault M. J., Wagner V. E., Bushnell D., Haidaris C. G., Iglewski B. H., and Passador L. (2005) Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect. Immun. 73, 3764–3772 10.1128/IAI.73.6.3764-3772.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu M., Guina T., Brittnacher M., Nguyen H., Eng J., and Miller S. I. (2005) The Pseudomonas aeruginosa proteome during anaerobic growth. J. Bacteriol. 187, 8185–8190 10.1128/JB.187.23.8185-8190.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K. C., Birrer P., Bellon G., Berger J., Weiss T., Botzenhart K., Yankaskas J. R., Randell S., Boucher R. C., and Doring G. (2002) Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109, 317–325 10.1172/JCI0213870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kolpen M., Hansen C. R., Bjarnsholt T., Moser C., Christensen L. D., van Gennip M., Ciofu O., Mandsberg L., Kharazmi A., Doring G., Givskov M., Hoiby N., and Jensen P. O. (2010) Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65, 57–62 10.1136/thx.2009.114512 [DOI] [PubMed] [Google Scholar]
- 34. Kolpen M., Bjarnsholt T., Moser C., Hansen C. R., Rickelt L. F., Kuhl M., Hempel C., Pressler T., Hoiby N., and Jensen P. O. (2014) Nitric oxide production by polymorphonuclear leucocytes in infected cystic fibrosis sputum consumes oxygen. Clin. Exp. Immunol. 177, 310–319 10.1111/cei.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Linnane S. J., Keatings V. M., Costello C. M., Moynihan J. B., O'Connor C. M., Fitzgerald M. X., and McLoughlin P. (1998) Total sputum nitrate plus nitrite is raised during acute pulmonary infection in cystic fibrosis. Am. J. Respir. Crit. Care Med. 158, 207–212 10.1164/ajrccm.158.1.9707096 [DOI] [PubMed] [Google Scholar]
- 36. Skurnik D., Roux D., Aschard H., Cattoir V., Yoder-Himes D., Lory S., and Pier G. B. (2013) A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 9, e1003582 10.1371/journal.ppat.1003582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Navais R., Mendez J., Perez-Pascual D., Cascales D., and Guijarro J. A. (2014) The yrpAB operon of Yersinia ruckeri encoding two putative U32 peptidases is involved in virulence and induced under microaerobic conditions. Virulence 5, 619–624 10.4161/viru.29363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakai Y., Kimura S., and Suzuki T. (2019) Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability. Nat. Commun. 10, 2858 10.1038/s41467-019-10750-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lauhon C. T. (2019) Identification and characterization of genes required for 5-hydroxyuridine synthesis in Bacillus subtilis and Escherichia coli tRNA. J. Bacteriol. 201, e00433–19 10.1128/JB.00433-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aussel L., Loiseau L., Hajj Chehade M., Pocachard B., Fontecave M., Pierrel F., and Barras F. (2014) ubiJ, a new gene required for aerobic growth and proliferation in macrophage, is involved in coenzyme Q biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium. J. Bacteriol. 196, 70–79 10.1128/JB.01065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Held K., Ramage E., Jacobs M., Gallagher L., and Manoil C. (2012) Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194, 6387–6389 10.1128/JB.01479-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V., and Manoil C. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A 100, 14339–14344 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wagner V. E., Bushnell D., Passador L., Brooks A. I., and Iglewski B. H. (2003) Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095 10.1128/JB.185.7.2080-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hassett D. J. (1996) Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J. Bacteriol. 178, 7322–7325 10.1128/JB.178.24.7322-7325.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jeong J. Y., Yim H. S., Ryu J. Y., Lee H. S., Lee J. H., Seen D. S., and Kang S. G. (2012) One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 78, 5440–5443 10.1128/AEM.00844-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Figurski D. H., and Helinski D. R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U S A 76, 1648–1652 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y., and Lemmon M. A. (1998) Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273, 30497–30508 10.1074/jbc.273.46.30497 [DOI] [PubMed] [Google Scholar]
- 48. Fish W. W. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158, 357–364 10.1016/0076-6879(88)58067-9 [DOI] [PubMed] [Google Scholar]
- 49. Beinert H. (1983) Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131, 373–378 10.1016/0003-2697(83)90186-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript and supporting material.