Abstract
Ribosome profiling (RIBO-Seq) has improved our understanding of bacterial translation, including finding many unannotated genes. However, protocols for RIBO-Seq and corresponding data analysis are not yet standardized. Here, we analyzed 48 RIBO-Seq samples from nine studies of Escherichia coli K12 grown in lysogeny broth medium and particularly focused on the size-selection step. We show that for conventional expression analysis, a size range between 22 and 30 nucleotides is sufficient to obtain protein-coding fragments, which has the advantage of removing many unwanted rRNA and tRNA reads. More specific analyses may require longer reads and a corresponding improvement in rRNA/tRNA depletion. There is no consensus about the appropriate sequencing depth for RIBO-Seq experiments in prokaryotes, and studies vary significantly in total read number. Our analysis suggests that 20 million reads that are not mapping to rRNA/tRNA are required for global detection of translated annotated genes. We also highlight the influence of drug-induced ribosome stalling, which causes bias at translation start sites. The resulting accumulation of reads at the start site may be especially useful for detecting weakly expressed genes. As different methods suit different questions, it may not be possible to produce a “one-size-fits-all” ribosome profiling data set. Therefore, experiments should be carefully designed in light of the scientific questions of interest. We propose some basic characteristics that should be reported with any new RIBO-Seq data sets. Careful attention to the factors discussed should improve prokaryotic gene detection and the comparability of ribosome profiling data sets.
Keywords: mRNA, bacterial genetics, bacterial transcription, translation, gene mapping, computational biology, polyribosome, ribosome profiling (RIBO-Seq), size selection, transcriptomics
Introduction
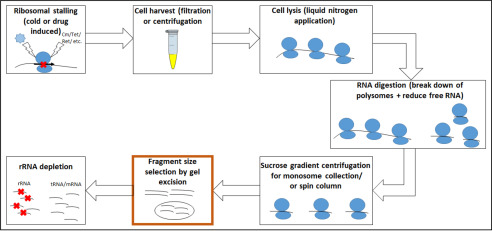
Ribosome profiling (RIBO-Seq) is a specialized form of RNA-Seq. In this method, translating ribosomes of a bacterial culture are isolated and treated with RNases in vitro. Ribosome-protected mRNA fragments, the “footprints,” are then isolated and sequenced in high throughput. This allows taking a snapshot of translation, hence the “translatome,” as compared with the transcriptome obtained in typical RNA-Seq (1–3). New discoveries from this approach include previously undetected “intergenic” (4) and overlapping genes (5), a deeper understanding of translational regulation (6, 7), and the finding that different ribosome types may be used in translating different genes (8). Important experimental steps include RNA digestion, monosome purification, RNA extraction, and footprint size selection (1, 9) (Fig. 1). However, each of these steps can be performed using a number of variations. It has been shown that different steps, such as RNA extraction, size selection, or rRNA depletion play an important role in RNA sequencing–based transcriptome analysis (10–12). In such experiments, RNA species of interest are enriched by depleting high-abundance rRNA (13, 14). To reduce the amount of rRNA present in an RNA-Seq experiment, size selection has been performed as well (15). However, a major concern is the size range to choose for RNA-Seq (11), as genes of interest can have different lengths. For RIBO-Seq, the length of protected footprints is given by the ribosomal occupancy, whereas for RNA-Seq, the fragment size is more dependent on the whole gene lengths. Therefore, choosing an overly narrow cut-off in RNA-Seq may exclude potentially interesting targets, such as long noncoding RNAs (16, 17). Here, we particularly focus on ribosomal profiling approaches and introduce each critical step (Fig. 1) and the available methods.
Figure 1.
Overview of crucial experimental steps having an influence on the RNA fragments obtained for ribosome profiling. The step we primarily want to focus on is highlighted in orange.
Ribosome stalling in general and for start site detection
Unwanted changes in the translatome during processing can be minimized through ribosome stalling (1, 18), induced either through application of drugs or rapid cooling (1, 9). Chloramphenicol (Cm) has been the most commonly used antibiotic for inducing stalling; however, recent studies have shown that it does not fully inhibit prokaryotic translation. Whereas Cm stops the elongation of most ribosomes, initiation still progresses, leading to an accumulation of ribosomes at the start codon (19). To avoid this bias, rapid filtration for harvest can be combined with immediate freezing in liquid nitrogen (19, 20). With this method, no ribosomal accumulation at the translational start site is observed (19, 21). Three studies included in our survey used filtration and flash freezing (22–24). We examine some of the effects of Cm further below.
Rather than seeking to avoid bias from stalling, some studies intentionally use drug-related bias in translation start sites because of the improved detection of a gene's start codon. For instance, in bacteria, tetracycline (Tet) stops protein elongation by preventing tRNA binding to the ribosomal A-site, preventing binding of the anticodon (25). Thus, Tet has been used for start site mapping. However, as this blocking is a reversible process, trapping at the start site is only semi-specific (25, 26). A study performed by Meydan et al. (27) tested the antibiotic retapamulin (Ret), from the class of pleuromutilins, as an elongation blocker (28). The antibacterial peptide Onc112 was shown by Weaver et al. (29) to be suitable in stalling ribosomes precisely at the initiation site by destabilizing the initiation complex such that subsequent elongation is prevented (30). Although ribosome profiling data sets produced with Ret and Onc112 show differences in the distance between read ends and the upstream start codon, both allow precise start site detection (29).
Cell harvesting and lysis
Either rapid filtration or centrifugation can be used for harvesting ribosomes. For rapid filtration, the culture is quickly transferred to a membrane and filtered by applying vacuum pressure (20). Cells are scraped from the membrane and flash-frozen in liquid nitrogen. The alternative is centrifugation of cooled cell suspensions (21, 31). For both, proceeding quickly is necessary to guarantee ribosomal stalling before any cold-shock response alters the translatome (31, 32). Rapid filtration is the method of choice as it seems to result in less variation, possibly because of faster ribosomal stalling (18, 24, 31).
Before freezing the cells in liquid nitrogen, lysis buffer should be added (18, 31). The buffers used vary in their composition but should guarantee the stabilization of the ribosome-bound mRNA complex (31). Negative effects may occur if excess amounts of some salts such as magnesium are used (21, 31, 33, 34). These either destabilize ribosomes, thereby releasing the mRNA, or conversely increase folding of unbound mRNAs, preventing digestion and leading to these fragments being mistaken as footprints (31). The frozen cells are pulverized in a grinder mill or using a mortar and pestle to release the ribosomes (21, 31, 35).
Ribosomal footprint generation
The prepared ribosomes are incubated with endo- and/or exonucleases to digest any unprotected mRNA, aiming to retrieve monosomal ribosomes with a footprint inside (9, 31). Polysomes will be separated into monosomes as unprotected mRNA is digested (9, 31). The length of remaining protected fragments inside the ribosomes is given by the size of the ribosome. For eukaryotes, the ribosomal footprints are about ∼ 28–30 nucleotides (nt) (2, 9, 18). In contrast, the ribosome footprint length is still highly debated for prokaryotes. Some studies hypothesize a length of 23 or 24 nt due to this being the most common fragment length (19, 36, 37). In contrast, other studies, including a few of those examined here, consider “ribosomal footprints” to span a much larger range of up to 42 nt (22, 23, 38).
Now the question of the RNases useful for ribosome footprinting arises. In eukaryotes, the endonuclease RNase I is used in many studies. RNase I has no cleaving bias for specific nucleotides, and the unprotected mRNA appears to be trimmed right to the edge of the ribosome (9, 18). RNase I is claimed to not work in bacteria and especially Escherichia coli, as it is bound by the 30S ribosome subunit and therefore inhibited (19, 39). Evidence from our group suggests that RNase I degrades the ribosome in E. coli LF82. Nevertheless, ribosome profiling experiments have been conducted successfully using RNase I alone (4, 40) or in combination with a mix of other enzymes (41), producing footprints of a size of ∼23 nt. Because of the assumption of RNase I unsuitability, the enzyme micrococcal nuclease (MNase) is normally used for prokaryotic experiments instead, despite this enzyme having some sequence specificity (42), likely contributing to greater variability in ribosome footprint lengths. The best enzyme or mix of enzymes to use in bacterial ribosome profiling deserves further empirical study and may differ across species.
In eukaryotes, it is possible to detect reading frames for single proteins in RIBO-Seq. The mRNA progresses stepwise one codon at a time through the ribosome, with RIBO-Seq reads showing a clear corresponding periodicity. Thus, a single-codon resolution is achievable (2, 19). In contrast, for prokaryotes, reading frames can be detected in a sum signal, but the resolution of standard ribosome footprinting is insufficient for individual genes (43). One approach for increased precision uses the mRNA interferase toxin RelE in addition to MNase. RelE only cleaves the mRNA in the A-site of the ribosome if activated in a stress condition, and it normally cleaves between the second and third base of a codon (44–46). Therefore, RelE is suitable for reading frame determination (44). However, because RelE cleaves the footprint within the ribosomes and MNase cleaves the unprotected mRNA outside the ribosomes (47, 48), the fragments resulting from combining them are even shorter, which can be harder to map accurately (44). Consequently, careful size selection or special mapping approaches may be important if using RelE.
Performing size selection and rRNA depletion
To enrich for ribosomes from the crude cell lysate after RNase digestion, sucrose gradient centrifugation is performed (18). There are, however, limitations in detecting and obtaining the layer containing the monosomes. Enrichment of ribosomes can also be performed using gel filtration (9, 49). Nevertheless, gradient centrifugation is still the most common method (24, 44, 50, 51).
From the enriched ribosome fraction, the total RNA is isolated. Putative ribosomal footprints are separated from other RNAs (e.g. rRNA and tRNA) by performing a size selection using gel electrophoresis. Samples are loaded onto, for example, a TBE-urea gel, and certain fragment sizes are excised after comparison with a corresponding DNA or RNA ladder (18, 43). Another possibility for fragment size selection is to use a size-exclusion spin column, but this has rarely been used (52). Size selection is a crucial step because it seems to have a major impact on the sequencing results. There is no clear consensus on which read lengths to choose when conducting bacterial ribosome profiling. In the beginning of ribosome footprinting of bacteria, several groups isolated reads ranging from 28 to 40 nt (24, 53), whereas others chose fragments of 23 ± 3 nt (37) or used a range starting with fragments as short as 15 nt (54, 55). In a recent study, Mohammad et al. (19) claimed that the diversity in length of bacterial ribosomal footprints depends on the characteristics of prokaryotic ribosomes and suggested that a broad range of read lengths (15-40 nt) should be taken for analysis. They claim that sampling this full range of read lengths yields the most informative output, with a peak at around 24 nt (19, 36). Several studies have used this range (19, 54, 55). Unfortunately, the RNases used for mRNA clipping also degrade the rRNA to some extent, and reads in the footprint range are not all ribosomal footprints. Therefore, rRNA depletion is necessary, helping to ensure that fewer contaminating rRNA reads are sequenced. Ingolia (18) provided a method for depletion in the first RIBO-Seq protocol published, and since then different kits have become available for prokaryotic rRNA removal, such as RiboZero (Illumina), RiboMinus (Thermo Fisher Scientific), MICROBExpress (Thermo Fisher Scientific), and others. The efficacy of different rRNA removal methods has been discussed elsewhere and is an area of continuing development (14, 56–59).
Evaluation of RIBO-Seq data
To discriminate between noncoding RNA and protein-coding mRNA, a direct comparison between RIBO-Seq and RNA-Seq results of the same culture can be conducted. Evidence for translation of ORFs is given by dividing the reads per kilobase per million sequenced reads (RPKM) values obtained in RIBO-Seq by the values from RNA-Seq experiments. This ratio, designated ribosomal coverage value (RCV), is a measure of the extent to which an mRNA is translated. It has been suggested that an RNA should be considered as expressing a protein if the RCV is at least 0.355 (40). The appropriate threshold will vary somewhat between strains and samples, a point that may deserve further study.
The diversity of methods that have been employed and the increasing use of RIBO-Seq prompted us to examine and compare available bacterial ribosome profiling data sets to assess the methods used and their influence on the output. As other studies have already investigated the effect of ribosomal stalling (19, 25, 29, 30) and the RNases used for ribosome footprinting (9, 18, 41, 42), we particularly focus on the size-selection step and the resulting read length distribution to shed light on its influence on the outcome. Further, we also make recommendations for future prokaryotic experiments.
Results
Size selection
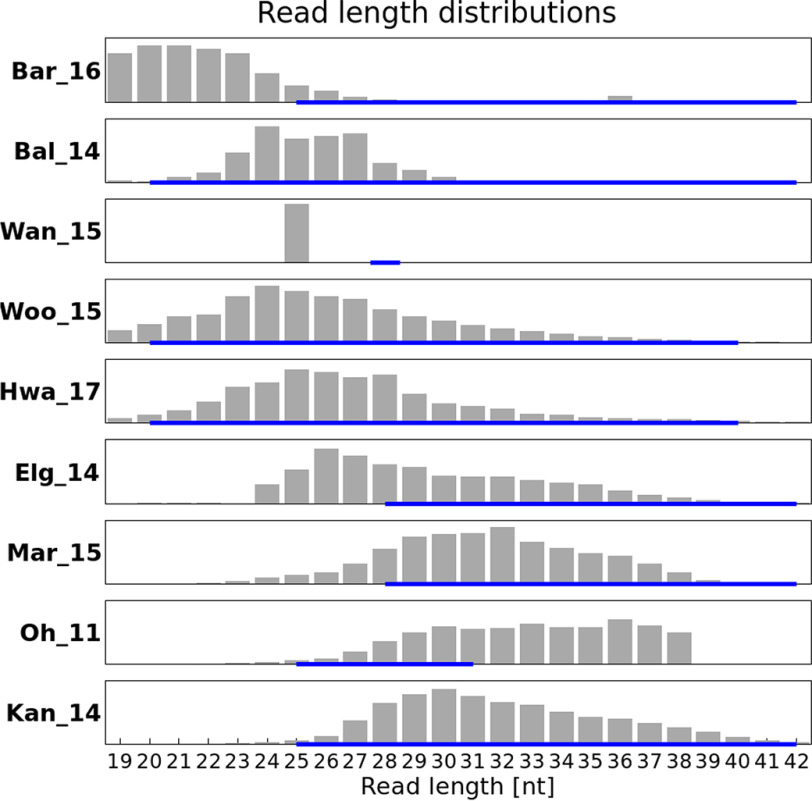
For eight experiments, the gel size selection performed was reported (Table S1). We were interested in reads between 19 to 42 nt, the approximate range for possible ribosome footprints. For several samples (Oh_11, Woo_15, Mar_16, Kan_14, Hwa_17, and Bal_14), reads longer than 42 nt (not shown) were detected, possibly because of insufficient trimming. These were not taken into consideration as, given their length, they are unlikely to represent ribosomal footprints (at least for monosomes). In general, analyzing the trimmed and aligned reads mapping to mRNA and omitting reads mapping to either rRNA or tRNA, we expect to find a footprint distribution pattern matching the reported size-selection thresholds in each publication. These patterns are representing the protected mRNA fragments; thus, the expectation is that a predominant length will be detected surrounded by shorter and longer fragments because of conformity issues of the ribosome. In reality, we find that for some samples, the distribution patterns differ significantly from the reported size selection. Fig. 2 shows the actual distributions for one representative sample per experiment.
Figure 2.
Length distribution of reads after trimming of different samples an exclusion of rRNA and tRNA (one example data set per project) with reported size selection (blue line below).
Here, we examine the results for each set, shown in Fig. 2. For set Bar_16, we were not able to find any information about the performance of size selection. The two samples belonging to this experiment had a peak at 20-21 nt (i.e. the shortest fragments) (Fig. 2, first panel). In Fig. 2, the third panel shows the discrepancy for experiment Wan_15 between a size selection performed around 28 nt and the actual read length of 25 nt. For these samples, trimming had already been performed for the data released by the group. Reads were digitally trimmed to 25 nucleotides (personal communication)3. In set Elg_14 (Fig. 2, sixth panel), size selection was performed between 28 and 42 nt. The peak is found at 26 nt, which should have been excluded by their intended size selection. Both of these examples illustrate the difficulties in choosing the margin for the gel cut. For four experiments, namely Bal_14, Hwa_17, Kan_14, and Woo_15, the observed read length distribution is situated within their performed size selection (Fig. 2, second, fourth, fifth, and ninth panels). The chosen ranges varied between 20 and 30 or 40 nt, with peak values between 20 and 30 nt. For sample Oh_11 (Fig. 2, eighth panel), a size selection from 25 to 31 nt was performed. The intended range was obtained, but the upper limit was not successfully implemented. Results from this analysis overall imply that fragments of 24–27 nt in length are the most frequently protected. Therefore, they might be the most informative regarding gene positions; thus, one should aim for these sizes at selection. To test this hypothesis, we performed an analysis focusing on the length of reads aligned to annotated protein-coding genes compared with rRNA and tRNA genes.
Specific read lengths correspond to different RNA types
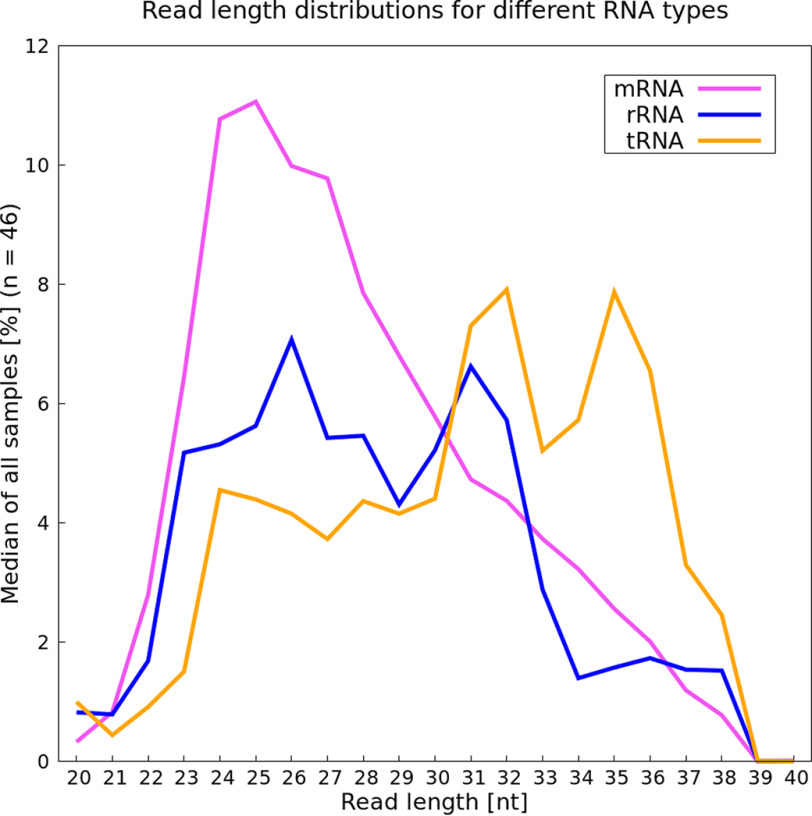
To test whether different RNA types (mRNA, rRNA, and tRNA) vary in their read length, we analyzed reads of lengths 20–40 nt in each sample. The two samples from experiment Wan_15 were excluded from this analysis, because these samples were missing a distribution range because of a digital size selection after sequencing (Fig. 2, third panel). The percentage of the fragment length distribution within each RNA type was calculated per sample. Based on these 46 percentage values, a median value was calculated for each specific length (Fig. 3). A peak at around 24–27 nt was observed for mRNA, with tRNA and rRNA more likely to have longer reads, especially tRNA. Additional calculations were performed regarding the mean values for either the read length distributions within an RNA type (i.e. mean calculation mentioned above) or the mean value per type at a specific length and can be found in the supporting information (Figs. S1 and S2). For the second mean value analysis, the read amount corresponding to a distinct RNA type was analyzed. Numbers of reads corresponding to, for example, rRNA with a specific length were divided by the total amount of reads with the same length.
Figure 3.
Read length analysis for three RNA categories. The median calculation is based on the fragment length distribution (in percent) of the different RNA types within each sample (n = 46). Pink, mRNA; blue, rRNA; orange, tRNA.
In our analysis we detected a peak at 24 and 25 nt with a slightly lower number of reads with a length of 26-27 nt for mRNA (Fig. 3, pink line). This supports the hypothesis that reads with these lengths are the most informative ones regarding protein-coding genes. Thus, in cases where rRNA/tRNA depletion is imperfect and the special cases of long reads are not of interest, we recommend choosing a narrower cut-off. Size selection is not completely precise, but a range between 22 and 30 nt can be targeted to obtain the most informative reads. For reads mapping to rRNA, a peak can be detected at 26 nt of length (Fig. 3, blue line). Located fully within our suggested range, these reads cannot be excluded because of a narrower selection. Therefore, additional rRNA depletion during the experiment is advised to minimize the amount of sequenced reads that are wasted. The second peak of 31 nt in rRNA read lengths would be excluded with our suggested size selection. This corresponds particularly to 5S rRNA (Fig. S3), which is not targeted in some standard depletion methods (14), another reason to consider size selection as part of an rRNA depletion strategy. Reads mapping specifically to tRNA are predominantly longer, having two peak values at a length of 32 and 35 nt (Fig. 3, orange line). Again, with our suggested size selection, these reads could mostly be excluded.
Longer reads in 5'-UTR region
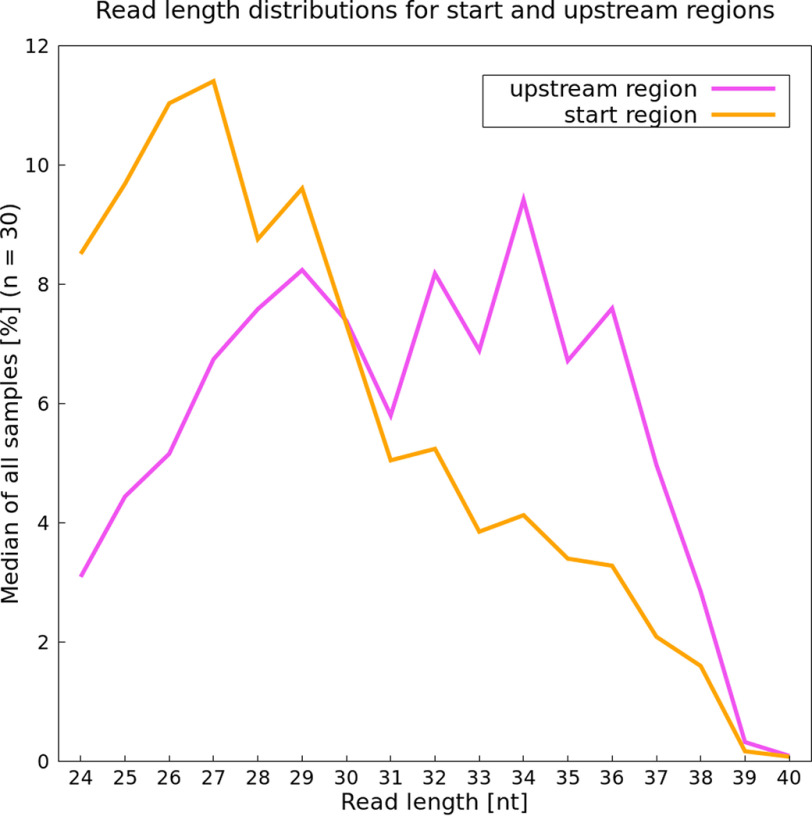
There is some evidence that reads in the range of 28–40 nt are associated with incorporated Shine–Dalgarno (SD) motifs (53, 54). We thus expect that reads mapping upstream of a start codon should also be longer because of SD sequences in this region. For this comparison, we chose to analyze all reads ranging from 24 to 40 nt. However, because of their narrow size selection ranging on average from 20 to 30 nt, experiments Wan_15 and Bal_14 were excluded from this analysis. Further, Bar_16 was also excluded because no size selection was reported and the upper part of the examined size range of reads was missing. Thus, 30 samples remained for this analysis.
A clear trend for read lengths in start and stop regions can be seen. Reads mapping directly in the start region tend to be shorter, with 27 nt being the most frequent length (Fig. 4). However, reads mapping in the 5′-UTR region of genes are substantially longer, with 34 nt as the most common length (Fig. 4). The SD motif, often located in the upstream region (60), potentially results in longer reads because of the interaction between the anti-SD sequence and the mRNA. Additionally, boxplots showing the number of reads for each length over all samples were created (Fig. S4, A–D). Results for the analysis of the stop region and over the whole gene length are also included. The most prominent read length for these two regions again is 27 nt (Fig. S4, C and D). These results also support our hypothesis that this length is particularly associated with protein-coding characteristics.
Figure 4.
Prevalence of specific read lengths in the start (orange) and 5′-UTR, potentially including a Shine–Dalgarno sequence (pink), based on their median (in percent).
Potential influence of chloramphenicol on read length
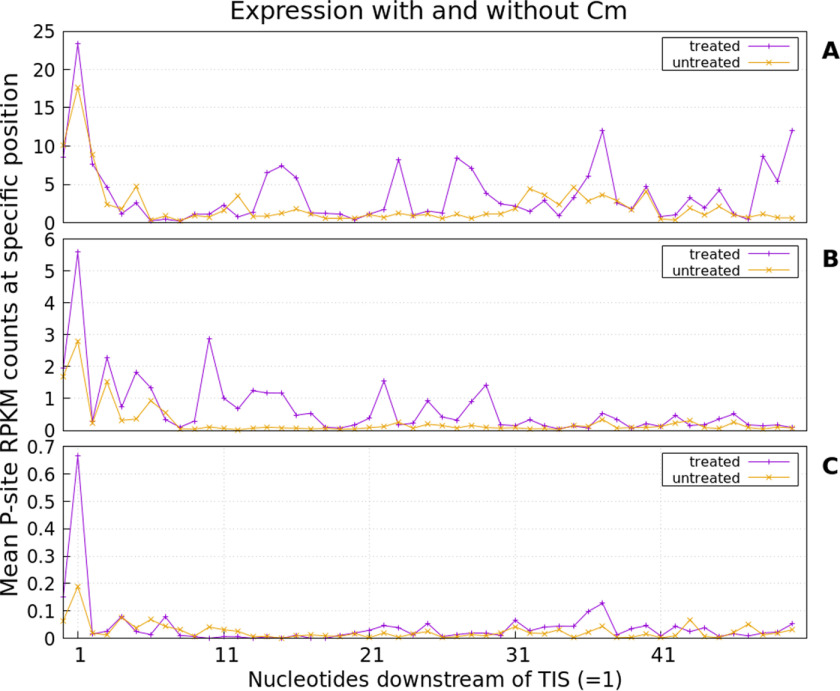
Drug-induced stalling of ribosomes at the translation initiation site (TIS) was examined for Cm-treated samples. We can confirm this phenomenon (i.e. in drug-treated samples, ribosome footprints in the start region of genes are enriched). However, results depend on gene expression. In Fig. 5, the average read accumulation in the start region is shown. For highly expressed genes (Fig. 5A), drug-induced stalling results in an only slightly higher accumulation of reads at the TIS compared with untreated samples. Despite stalling, translation does not cease completely, resulting in several additional accumulations along the mRNA even in treated samples. However, drug application leads to a visibly increased number of reads located at the TIS for genes expressed at an intermediary level (Fig. 5B). Interestingly, the largest effect of stalling is observed for weakly expressed genes (Fig. 5C).
Figure 5.
Average read accumulation for all analyzed genes being highly (A), medium (B), and weakly expressed (C). Cm-treated samples (purple) were compared with untreated samples (orange). In general, the drug treatment appears to promote read accumulation at the TIS.
Sufficient coverage depth for ribosome profiling
Genome coverage is defined as how often a base is sequenced on average during an experiment. In RNA-Seq experiments, expression values vary greatly between genes, depending on their expression in a particular growth stage or environment. To “catch” all genes being transcribed, the number of reads being sequenced needs to be sufficient to also detect weakly expressed genes (61). Increasing the number of reads sequenced beyond a certain point will likely lead to saturation of the proportion of genes detectable. Haas et al. (61) claimed that about 5–10 million reads covering mRNA in RNA-Seq approaches is enough to also detect weakly expressed genes while minimizing false positives. The appropriate threshold for “false positives” could well be discussed. In any case, in RNA-Seq, a number of 2 million reads already seems to be sufficient for medium to highly expressed genes (62). However, the required number of reads depends on the specifics of each experiment. If the particular interest lies in using ribosome profiling to detect weakly expressed genes (5, 41, 43, 63), a certain read number matching a gene is needed to confidently confirm expression (i.e. an RPKM above a certain threshold). We adapted the calculation from Haas et al. (61) for RIBO-Seq. Prediction of ORFs was conducted as described under “Experimental procedures.” Fig. 6 provides an overview.
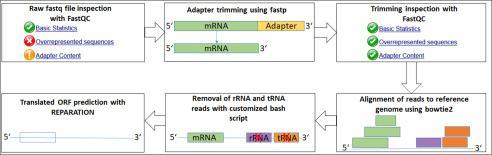
Figure 6.
Analysis pipeline overview for prediction of translated open reading frames.
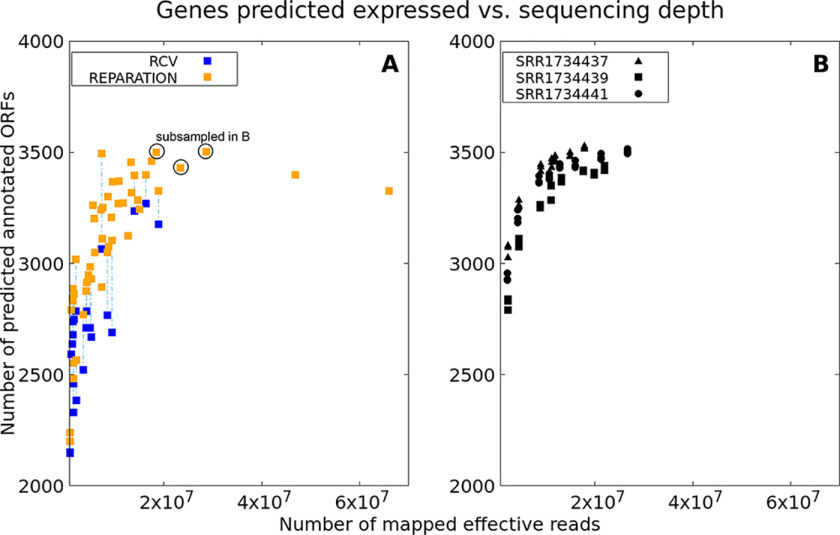
The total numbers of reads after excluding rRNA or tRNA were compared with the number of predicted open reading frames by REPARATION, which represents detectable annotated genes (Fig. 7A). Haas et al. (61) were able to detect “all but 2 of 4149 ORFs annotated” with a threshold of at least one read mapping in an ORF for RNA-Seq data. Mapping of just a single read is a very lenient criterion, and with our analysis, we cannot reach the same detection level. For predictions by REPARATION, we required at least three reads mapping to a predicted ORF. With this threshold, the number of annotated genes detected in a RIBO-Seq experiment reaches saturation with 20 million reads per sample after rRNA/tRNA removal, detecting ∼3500 genes (82% of 4242 known annotated genes, NC_000913.3). At least 20 million reads mapping to annotated genes seem to be an appropriate amount for ribosome profiling data and does not exceed the number of reads predicted to be excessive for RNA-Seq (i.e. 50 million) (61). However, the appropriate thresholds for weakly expressed unannotated genes require further research.
Figure 7.
A, plot showing relation between the numbers of effective reads sequenced (i.e. without rRNA/tRNA) and the number of annotated genes predicted to be potentially translated based on REPARATION (≥3 reads, orange) or REPARATION prediction plus an RCV threshold (≥0.355, blue). Results for data with both estimations (i.e. RNA-Seq available) are connected via dashed lines. B, relation between numbers of annotated genes predicted to be translated for subsampled data sets having a high sequencing depth originally (samples from Woo_15; each in triplicate; SRR1734437; SRR1734439; SRR1734441). Gene number estimations are based on REPARATION predictions as before.
If RNA-Seq data were available, RCVs were also calculated (Fig. 7A). Predictions of genes from REPARATION were considered true positives if having an RCV ≥ 0.355 (27). For samples for which both REPARATION-based predictions and RCV values were available, data points were connected with dashed lines in Fig. 7A Overall, this analysis supports the finding that 20 million reads are sufficient to detect most of the annotated genes in RIBO-Seq experiments. To check this conclusion, the analysis was repeated through subsampling of three deeply sequenced samples (Woo_15; samples SRR1734437, SRR1734439, and SRR1734441). Reducing the number of reads is accompanied by a loss of gene predictions possible (Fig. 7B). The subsampling confirms that a sequencing depth of around 20 million reads is appropriate for estimating expressed annotated genes. Above 20 million reads, the number of genes predicted increases no further. Future work on this question could include recent improvements in gene predictions from RIBO-Seq data (64, 65) and should take into account the many previously unrecognized small proteins (29, 43, 60, 63, 66).
Discussion
Size selection
Careful size selection should help to ensure that only ribosome-protected footprints are sequenced (18). We find that a discrepancy between the purported selected size and the actual read length distribution sequenced is common. This discrepancy was expected to some extent, as gel excision does not have absolute precision. It has recently been suggested that a range from 15 to 40 nt should be taken into consideration for the experimental size selection and data analysis (19). We argue that a narrower selection range has some significant benefits, as longer fragments have a higher probability of mapping to either tRNA or rRNA, and mapping shorter reads is more error-prone. Using a better size selection, even though size selection can never be absolute, unwanted RNA species can potentially be excluded from the sample before sequencing, resulting in a higher coverage of protein-coding mRNA reads. The relative interest of longer reads associated for instance with upstream regions, and those in our suggested range, has to be weighed carefully in each case however.
Four data sets show a read distribution within their selection (Bal_14, Hwa_17, Kan_14, and Woo_15). The other four sets also had reads of length outside their chosen selection. The actual size range obtained is highly dependent on the precision with which gel excision is conducted. The typical range of between 20 and 40 nt will normally result in a spectrum of various fragment lengths with a maximum number of reads at around 24 nt (19, 44, 67). This same peak value can also be achieved when using a size selection from 20 to 30 nt (51) or, even more narrowly, of 23 ± 3 nt, as used for a different E. coli strain (37). For the purposes of gene expression analysis, we suggest choosing the narrower range (i.e. aiming for peak values around 24–27 nt) because our results indicated that the vast majority of useful reads have these lengths. These lengths predominantly map to protein-coding mRNA (Fig. 3; see below). Choosing fragments of 25–42 nt in length resulted in a peak value at around 30 nt (Kan_14) (38). It could be argued that shifting the lower boundary value up (around 5 nt) results in a distribution shift as a whole. However, the size selection should have little correlation with the fragment length peak obtained, because this primarily depends on the protection by the ribosome from nuclease digestion (1, 2, 19). The reasons for this shift are not fully clear; an insufficient digestion may have led to longer fragments in this one study.
Specific read lengths correspond to different RNA types
rRNA and tRNA make up around 95-97% of the total RNA in bacterial cells (61, 68); hence, their removal, by fragment size selection or rRNA depletion before library preparation, increases the proportion of reads mapping to mRNA. Reads ranging in length from 24 up to 27 nt seem to be the dominant ones for mRNA (Fig. 3). tRNA reads appear to be longer than mRNA reads. If an aim is to exclude excessive tRNA reads, size selection should again be below 30 nt. To further consider whether a narrower size selection does in fact exclude tRNAs, the percentage of remaining reads mapping to tRNA was compared (Table S4). The samples with a size selection of 20–30 nt contained less than 1% tRNA (Table S4), and size selection spanning from 25 to 31 nt resulted in a remaining tRNA percentage of 11.5% on average, less than with a range from 20 to 40 for size selection (33.5%). Choosing a cut-off for size selection at around 30 nt should ensure exclusion of most of the tRNA. The lowest numbers of reads mapping to rRNA are also found in experiments with a size selection of 20–30 nt. In contrast, the second lowest remaining rRNA amount (15.2%) can be found with a size selection performed at 25–42 nt. Even though the selection is in the higher range, the amount of rRNA is fairly low compared with the other data sets. These results suggest that for rRNA removal, a properly performed depletion step is as important as size selection.
Reads belonging to rRNA show two prominent peaks at 26 and 31 nt. The shorter peak is primarily because of 16S rRNA (Fig. S3). The longer fragments at 31 nt could be excluded in higher proportion if the size selection threshold did not exceed 30 nt. To exclude remaining rRNA fragments, several options for depletion are currently available. Three experiments specified using commercial kits, namely RiboZero (Illumina; Woo_15 and Hwa_17) or MICROBExpress (Thermo Fisher Scientific; Elg_14) for rRNA depletion. The remaining experiments performed standard protocol rRNA depletion based on biotin-labeled rRNA hybridization. The average percentage of rRNA remaining in kit-treated samples is not significantly lower than in samples where no commercial kit was used (Table S4). The lowest rRNA amount remaining can be found in a set for which no kit-based depletion method was mentioned (Bal_14). This suggests that size selection might be contributing more effectively to rRNA depletion. However, rRNA fragments with a length of 26 nt cannot be excluded by size selection. Thus, any size selection should ideally be performed in conjunction with best practices in rRNA depletion to ensure low amounts of rRNA reads.
Longer reads in the 5′-UTR region
Generally, for E. coli, the main SD sequence is located around 8 nt upstream of a start codon and aids in translation initiation because of binding a 16S rRNA of the 30S small subunit, allowing the ribosome to assemble at the coding sequence (69–75). The interaction of the SD sequence and its counterpart anti-SD results in the assembly of the ribosome complex. This mechanism might cause protection of longer fragments than expected normally (76, 77). As already mentioned under “Results,” in cases where reads with a Shine–Dalgarno-like motif are of interest, a size range exceeding 30 nt should be taken into consideration, but this will decrease the overall percentage of useful reads unless very effective rRNA depletion is performed.
Further experimental improvements
In the early RIBO-Seq protocols, a DNA ladder was used as a sizing standard (18). Experiments performed by our group, however, show that using single-strand RNA molecules as a ladder is more suitable. Therefore, orienting gel excision of RNA fragments based on a DNA ladder leads to an incorrect size range and probably larger fragments than intended. We recommend using a single-stranded RNA ladder with a mixture of random sequences of the intended size to ensure a more precise size selection. Besides this, excision accuracy and fragment separation are dependent on the gel resolution achieved and thus are also factors contributing to the performance of size selections.
Sufficient coverage depth for ribosome profiling
For RIBO-Seq experiments, a higher read amount is necessary to detect annotated genes independent of their expression levels than has been reported for RNA-Seq experiments. We recommend sequencing to at least 20 million mapped effective reads (rRNA/tRNA excluded). This should lead to a broad range of strongly and weakly expressed genes being detectable.
Adjustments for detecting new genes
To improve detection of unannotated translated ORFs, the use of drug-induced ribosomal stalling by elongation inhibitors such as Cm, Onc112, or Ret is currently the most promising strategy. Because of read accumulation bias at the TIS (19, 29), the start site is more clearly evidenced with these methods, which is very useful when trying to select the correct ORF among candidates in the same region, particularly important for short and weakly expressed ORFs. We can confirm an accumulation bias around the start codon because of Cm application. However, we found that Cm stalls ribosomes of highly and weakly expressed genes differently. For highly expressed genes, Cm makes hardly any difference to read coverage at the start site (Fig. 5A). However, these genes are usually of lesser interest with regard to novelties, because they are typically well-known and well-analyzed. In contrast, for weakly expressed genes, Cm causes increased ribosomal stalling at the start site compared with standard RIBO-Seq, thereby improving detection of such genes (Fig. 5C). We also find that the application of Cm is associated with high “periodicity” or reading frame specificity in alignments of length 31 in the start site region (Fig. S5), further confirming the usefulness of Cm in detecting the start site and reading frame. Careful analysis of read length and periodicity following the use of different drugs for ribosome stalling has the potential to dramatically improve start site and reading frame prediction for weakly expressed ORFs. More recent studies using Ret or Onc112 showed that these substances are particularly suitable for detecting translation initiation sites, including those potentially representing varying lengths of the same protein (proteoforms) (27, 29, 78, 79). The accumulation bias also allows the detection between two ORFs situated in different reading frames in the same region (29). A similar result has been found in the model archaeal species Haloferax volcanii using the drug harringtonine (80). Clearly, the use of elongation inhibitors is useful for detecting yet-unrecognized translated ORFs, especially with regard to detection of the reading frame and translation initiation site.
As all samples used for our analysis were grown in LB medium, the expression of novel genes (i.e. genes unknown to science) is expected to be limited. The expression of novel intergenic or overlapping genes is expected to be low, for instance, if they typically function in stress response or other atypical culture conditions (a question that remains unanswered). Their detection might be improved via the detection of translation start sites because of Cm application. We suggest minimizing the time between adding antibiotics and finally harvesting the bacteria; otherwise, the translatome is likely to also change in response to the drug (19) (Fig. S6). Because of this effect, we recommend combining data from drug-treated samples with normal ribosome profiling experiments. Some researchers use rapid filtration followed by cell flash freezing to stall the ribosomes, avoiding the potential translatome changes associated with chloramphenicol use (19). However, as noted, the read accumulation bias potentially aids in detecting new ORFs.
Increasing the sequencing depth also aids in detecting weakly expressed unannotated genes present in a sample. However, some studies have claimed that ORFs that are antisense to annotated genes detectable by increasing the read depth mostly result from transcription initiation or termination inaccuracies and are, therefore, presumably nonfunctional sequences (61, 81–83). In contrast, a number of other researchers have proposed that many antisense RNA sequences are in fact functional (84–86), even potentially for protein coding (87). Several studies have shown that there are still a number of unannotated genes present in even the well-studied E. coli K12 (88–90). Phenotypes for overlapping genes in enterohemorrhagic E. coli (EHEC) recently detected through ribosome profiling have been found in different environmental conditions (e.g. salt stress (5), anaerobiosis (41), or arginine supplementation (63)). This may imply that mRNAs of such genes are present in a relatively low amount when cultivated in standard LB medium. Increasing read depth and sampling from other growth conditions should improve the ability to find and predict such low-abundance proteins. We considered the number of putatively translated ORFs that are located partially overlapping an annotated gene or fully embedded antiparallel to an annotated gene. Surprisingly, for both categories, the threshold of 20 million effective reads also seems sufficient to reach saturation in the number of such putative genes expressed (Fig. S7, A and B); however, the extent to which this is an artifact of the prediction tools used remains to be determined.
The introduction of ribosome profiling by Ingolia et al. (2) in 2009 was the first stepping stone for directly investigating entire “translatomes,” beginning in eukaryotes. Since then, investigation of the bacterial translatome has led to improvements in the experimental protocols for prokaryotes, including the first analysis of an archaeal organism's translatome. On some topics that have been disputed we can make suggestions from our analysis. There still is no consensus about which size selection to choose for bacteria to obtain the most informative ribosomal footprints. Our recommendation for a basic analysis of gene expression is to choose a size selection between 22 and 30 nt, to not only exclude longer fragments associated with tRNAs and rRNAs during size selection but also to enrich for the fragments that are most likely to be protein-coding. However, if researchers are interested in the 5′-UTR region of genes potentially including, for instance, the Shine–Dalgarno sequence or are using modified methods such as ribosome stalling with chloramphenicol, size selection should be adapted according to the expected fragment length. The total number of mappable reads required for RIBO-Seq appears to be around 20 million reads after excluding those mapping to rRNA/tRNA regions. This is much higher than available for some experiments. With this number of reads, most annotated genes that are translated at the point of harvesting are detected. However, this result may be limited by the prediction tool used in this study (REPARATION). The RCV threshold provides an additional criterion, which can be used to ensure translation of candidate ORFs. This value is also interesting in studying translation response.
After surveying the literature, we suggest that a few questions deserve further study in optimizing protocols for ribosome profiling studies. On the experimental side, appropriate depletion of rRNA will be important for improving the affordability of high-depth ribosome profiling; without good protocols here, much more than half of the data sequenced is not useful for the intended purposes. The appropriate sequencing depth to detect all translated genes under different conditions and in different bacteria also requires attention, part of the general need to develop minimal expectations for new ribosome profiling data. The appropriate enzyme for degradation of mRNA not protected by a ribosome also deserves further study, given the diversity used in bacterial experiments. In analysis of ribosome profiling data, the “ribosome coverage value” has not been studied in depth but is potentially very useful for distinguishing noncoding from protein-coding RNA.
Finally, in light of variation in the detail of reporting in existing studies, we emphasize the importance of publishing the complete protocol with all details for future reference and analyses (listed in Table S5). Only with exact information provided (e.g. concerning the adapter sequence used, ribosome stalling methods, harvesting, and size selection) can subsequent comparative analyses be performed accurately. Alongside the experimental protocols chosen, we recommend that reports of new data sets should include the number of reads mapping to regions outside of rRNA and tRNA, and the length distribution of these reads, as an indicator of quality and comprehensiveness. It is likely that there is yet much to discover regarding the complexity of the bacterial translatome, and the further development of ribosome profiling will be a key contributor to this advance.
Experimental procedures
Sample selection
For our analysis, we compared 48 available RIBO-Seq E. coli K12 samples from nine different experiments (Table S1) (7, 22–24, 38, 44, 50, 51, 67) with all samples grown in LB medium. These were all of the experiments available at the time of analysis that used E. coli K12 grown in LB medium. One additional set was not included, as the paper was retracted (91). Only substrains of K12 (i.e. BW25113/BWDK, MG1655, and MC4100) were considered here because of their close phylogenetic relationship (23 samples of substrain MG1655, 15 samples of BW25113, and 10 samples of MC4100) (92–95). Between the samples assessed, there are differences in the experimental procedures for ribosome stalling (if applied), cell harvest, and size selection, allowing comparison of the outcomes. Abbreviations of each data set were created for further use (Fig. 2). Original Gene Expression Omnibus database identifiers, study abbreviations, and the experimental variations are listed in Table S1. We were particularly interested in the effect of the size-selection step on the read length distributions of trimmed and mapped reads. One important scientific question related to this concerns whether a particular length is most suitable for detecting protein-coding ORFs.
Bioinformatic data analysis
First, raw fastq files were inspected using FastQC v0.11.4 (RRID:SCR_014583). From this, adapter contaminations and overrepresented sequences can be inferred. Before performing the analysis, another frequently used adapter (5′-CTG TAG GCA CCA TCA AT-3′) (24, 51, 67) was added to the existing FastQC adapter list. Two experiments each used unique adapters, which were defined as input settings for the trimming as well (7, 23). For the remaining four experiments, the adapter sequences used were not stated (22, 38, 44, 50). While inspecting all samples with FastQC, adapters or other contaminants (overrepresented sequences) were identified. If available, we used the published or detected adapter sequences for trimming; otherwise, we chose the most overrepresented sequences. This includes poly(A) sequences, which were used by Wan_15 as adapters. Special care is necessary if random barcoding is applied to avoid PCR duplicates (96, 97). These short sequences would not be detected automatically, because of their random sequences. With, for instance, “end-to-end alignment” their location at the end of a read could subsequently prevent mapping because of mismatches. However, with “local alignment” the read sequence should be “soft-clipped” from the end, allowing mapping. In any case, none of the studies used reported using random barcoding in their experiments.
The software used during processing data is summarized in Table S2 (either default settings or as specified in the table). The custom pipelines connecting these tools are available upon request. Trimming was performed using fastp version 0.14.2 (98). First, the identified adapter sequence was trimmed, and second, if present, overrepresented sequences >3% were chosen for trimming as well. For some samples from the set Bal_14, for example, a second trimming step was performed, as a single overrepresented sequence constituted over 30% of the remaining reads. Automatic detection of adapters by fastp, followed by their trimming, was not successful, although it should be possible according to the program description. Therefore, we specified the sequences for trimming. Subsequent mapping of reads was performed using Bowtie2 version 2.2.6 with local alignment (99). Reads mapping to rRNA or tRNA were directed into a separate file. Prediction of translated open reading frames was performed with REPARATION, a ribosomal profiling–assisted reannotation tool (100). For compatibility with our system, the REPARATION workflow was adjusted by replacing UBLAST with DIAMOND for choosing the training set, after ORF prediction with Prodigal. Predictions matching annotated genes were then used to analyze the total number of mappable reads in a sample (after removal of rRNA/tRNA) necessary for detection of the genes. For a second verification of the translation status of these genes, the RCV was calculated, and genes with RCV ≥ 0.355 were considered to be translated. Besides ribosome profiling, RNA-Seq data are also necessary for this evaluation and were available for 22 of our 48 chosen samples. Additionally, three samples sequenced at greater depth (from Woo_15: SRR1734437, SRR1734439, SRR1734441) were used to compare predicted genes and sequencing depth. Each sample was analyzed at various coverage depths, with random subsampling at different depths repeated in triplicate.
The length distributions of trimmed reads mapping to annotated protein-coding mRNA were compared with the published experimental size selection used. Lengths of reads mapping to either tRNA or rRNA were also analyzed to test whether different read lengths correlate with specific types of RNA. Similar to the previous analysis, potential differences in length corresponding to a specific type of rRNA (5S, 16S, or 23S) were analyzed. We further investigated potential differences in read lengths mapping upstream of or directly within the start region of genes. The length distribution patterns in either the start region (from the start codon of an annotated gene to 25 nt downstream of the start) or in the 5′-UTR (in our case, 25 nt upstream of the start codon of each annotated gene) for all annotated genes were calculated. Additionally, read length distribution in the stop region (25 nt upstream of a stop codon) and the distribution for the remainder of the gene (between start +25 and stop −25) were calculated. Similarly, the median number of reads, based on the read distribution in the region of interest, was calculated for all annotated genes per sample.
Claims that Cm induces ribosomal stalling and causes a read accumulation at the start region were investigated. All samples from set Oh_11 were used for this comparison, as four were treated with Cm, whereas the remaining four were not. However, harvesting methods also differed between the two sample subsets; Cm samples were centrifuged, whereas untreated samples were harvested by rapid filtration. Nevertheless, in this set we analyzed the ribosome footprint RPKMs of annotated genes and created sets of highly, medium, and weakly expressed genes (Table S3). For each expression category, 10 genes were chosen that showed approximately the same expression status throughout all eight samples. Approximate P-site locations were inferred as 15 nt upstream of the 3′ end of each read. By counting this specific position, each read will only be considered once for further analysis. Thus, the P-site locations of each read near the start region were calculated, investigating a potential accumulation of reads at the start position. Within each expression category, the mean read depth values at each genome coordinate were calculated. These values were compared between treated (i.e. Cm) and untreated sample sets, with values for each sample normalized by sample sequence depth. In addition, a similar comparison based on the median read amount at each unique position per subset was performed to verify the obtained results (data not shown).
Data availability
Sources of all original data are listed in Table S1. All processed data and bash scripts used for analyses are available upon request from Alina Glaub (alina.glaub@tum.de).
Supplementary Material
Acknowledgments
We thank Michaela Kreitmeier for helpful comments on RIBO-Seq and Joseph T. Wade for information on digital trimming in their study.
This article contains supporting information.
Author contributions—A. G. data curation; A. G. and Z. A. formal analysis; A. G. visualization; A. G. writing-original draft; C. H. validation; K. N. and Z. A. conceptualization; K. N. and Z. A. writing-review and editing; Z. A. supervision.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
J. T. Wade, personal communication.
- RIBO-Seq
- ribosome profiling
- Cm
- chloramphenicol
- Tet
- tetracycline
- Ret
- retapamulin
- nt
- nucleotides
- MNase
- micrococcal nuclease
- RPKM
- reads per kilobase per million sequenced reads
- RCV
- ribosomal coverage value
- SD
- Shine–Dalgarno
- TIS
- translation initiation site.
References
- 1. Ingolia N. T. (2016) Ribosome footprint profiling of translation throughout the genome. Cell 165, 22–33 10.1016/j.cell.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingolia N. T., Ghaemmaghami S., Newman J. R., and Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Popa A., Lebrigand K., Paquet A., Nottet N., Robbe-Sermesant K., Waldmann R., and Barbry P. (2016) RiboProfiling: a Bioconductor package for standard Ribo-seq pipeline processing. F1000Res 5, 1309 10.12688/f1000research.8964.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neuhaus K., Landstorfer R., Fellner L., Simon S., Schafferhans A., Goldberg T., Marx H., Ozoline O. N., Rost B., Kuster B., Keim D. A., and Scherer S. (2016) Translatomics combined with transcriptomics and proteomics reveals novel functional, recently evolved orphan genes in Escherichia coli O157:H7 (EHEC). BMC Genomics 17, 133 10.1186/s12864-016-2456-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vanderhaeghen S., Zehentner B., Scherer S., Neuhaus K., and Ardern Z. (2018) The novel EHEC gene asa overlaps the TEGT transporter gene in antisense and is regulated by NaCl and growth phase. Sci. Rep. 8, 17875 10.1038/s41598-018-35756-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brar G. A., and Weissman J. S. (2015) Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol. Cell Biol. 16, 651–664 10.1038/nrm4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J., Rennie W., Liu C., Carmack C. S., Prévost K., Caron M. P., Massé E., Ding Y., and Wade J. T. (2015) Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res. 43, 12 10.1093/nar/gkv1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y.-X., Xu Z., Wang B.-W., Ge X., Zhu J.-H., Sanyal S., Lu Z. J., and Javid B. (2019) Selective translation by alternative bacterial ribosomes. bioRxiv 10.1101/605931 [DOI] [PMC free article] [PubMed]
- 9. Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., and Weissman J. S. (2012) The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui P., Lin Q., Ding F., Xin C., Gong W., Zhang L., Geng J., Zhang B., Yu X., Yang J., Hu S., and Yu J. (2010) A comparison between ribo-minus RNA-sequencing and polyA-selected RNA-sequencing. Genomics 96, 259–265 10.1016/j.ygeno.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 11. van Vliet A. H. (2010) Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 302, 1–7 10.1111/j.1574-6968.2009.01767.x [DOI] [PubMed] [Google Scholar]
- 12. Sultan M., Amstislavskiy V., Risch T., Schuette M., Dökel S., Ralser M., Balzereit D., Lehrach H., and Yaspo M. L. (2014) Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics 15, 675 10.1186/1471-2164-15-675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z., and Duan X. (2011) Ribosomal RNA depletion for massively parallel bacterial RNA-sequencing applications. Methods Mol Biol 733, 93–103 10.1007/978-1-61779-089-8_7 [DOI] [PubMed] [Google Scholar]
- 14. Petrova O. E., Garcia-Alcalde F., Zampaloni C., and Sauer K. (2017) Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterial biofilm and mixed pathogen culture transcriptomes. Sci. Rep. 7, 41114 10.1038/srep41114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu J. M., Livny J., Lawrence M. S., Kimball M. D., Waldor M. K., and Camilli A. (2009) Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res 37, e46 10.1093/nar/gkp080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., Loh E., Gripenland J., Tiensuu T., Vaitkevicius K., Barthelemy M., Vergassola M., Nahori M. A., Soubigou G., Régnault B., et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 17. Rasmussen S., Nielsen H. B., and Jarmer H. (2009) The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 73, 1043–1057 10.1111/j.1365-2958.2009.06830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ingolia N. T. (2010) Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 470, 119–142 10.1016/s0076-6879(10)70006-9 [DOI] [PubMed] [Google Scholar]
- 19. Mohammad F., Green R., and Buskirk A. R. (2019) A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife 8, 10.7554/eLife.42591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGlincy N. J., and Ingolia N. T. (2017) Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 10.1016/j.ymeth.2017.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becker A. H., Oh E., Weissman J. S., Kramer G., and Bukau B. (2013) Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat. Protoc. 8, 2212–2239 10.1038/nprot.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marks J., Kannan K., Roncase E. J., Klepacki D., Kefi A., Orelle C., Vázgues-Laslop N., and Mankin A. S. (2016) Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl. Acad. Sci. U.S.A. 113, 12150–12155 10.1073/pnas.1613055113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elgamal S., Katz A., Hersch S. J., Newsom D., White P., Navarre W. W., and Ibba M. (2014) EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 10, e1004553 10.1371/journal.pgen.1004553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oh E., Becker A. H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R. J., Typas A., Gross C. A., Kramer G., Weissman J. S., and Bukau B. (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 10.1016/j.cell.2011.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakahigashi K., Takai Y., Kimura M., Abe N., Nakayashiki T., Shiwa Y., Yoshikawa H., Wanner B. L., Ishihama Y., and Mori H. (2016) Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 23, 193–201 10.1093/dnares/dsw008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chopra I., and Roberts M. (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260, table of contents 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meydan S., Marks J., Klepacki D., Sharma V., Baranov P. V., Firth A. E., Margus T., Kefi A., Vazquez-Laslop N., and Mankin A. S. (2019) Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell 74, 481–493 10.1016/j.molcel.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dornhelm P., and Högenauer G. (1978) The effects of tiamulin, a semisynthetic pleuromutilin derivative, on bacterial polypeptide chain initiation. Eur. J. Biochem. 91, 465–473 10.1111/j.1432-1033.1978.tb12699.x [DOI] [PubMed] [Google Scholar]
- 29. Weaver J., Mohammad F., Buskirk A. R., and Storz G. (2019) Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10, 10.1128/mBio.02819-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seefeldt A. C., Nguyen F., Antunes S., Pérébaskine N., Graf M., Arenz S., Inampudi K. K., Douat C., Guichard G., Wilson D. N., and Innis C. A. (2015) The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 22, 470–475 10.1038/nsmb.3034 [DOI] [PubMed] [Google Scholar]
- 31. Bartholomäus A., Del Campo C., and Ignatova Z. (2016) Mapping the non-standardized biases of ribosome profiling. Biol. Chem. 397, 23–35 10.1515/hsz-2015-0197 [DOI] [PubMed] [Google Scholar]
- 32. Giuliodori A. M., Fabbretti A., and Gualerzi C. (2019) Cold-responsive regions of paradigm cold-shock and non-cold-shock mRNAs responsible for cold shock translational bias. Int. J. Mol. Sci. 20, 457 10.3390/ijms20030457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blobel G., and Sabatini D. (1971) Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U.S.A. 68, 390–394 10.1073/pnas.68.2.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ron E. Z., Kohler R. E., and Davis B. D. (1968) Magnesium ion dependence of free and polysomal ribosomes from Escherichia coli. J. Mol. Biol. 36, 83–89 10.1016/0022-2836(68)90221-0 [DOI] [PubMed] [Google Scholar]
- 35. Chew G. L., Pauli A., Rinn J. L., Regev A., Schier A. F., and Valen E. (2013) Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development 140, 2828–2834 10.1242/dev.098343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li G.-W., Burkhardt D., Gross C., and Weissman J. S. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hücker S. M., Simon S., Scherer S., and Neuhaus K. (2017) Transcriptional and translational regulation by RNA thermometers, riboswitches and the sRNA DsrA in Escherichia coli O157:H7 Sakai under combined cold and osmotic stress adaptation. FEMS Microbiol. Lett. 364, fnw262 10.1093/femsle/fnw262 [DOI] [PubMed] [Google Scholar]
- 38. Kannan K., Kanabar P., Schryer D., Florin T., Oh E., Bahroos N., Tenson T., Weissman J. S., and Mankin A. S. (2014) The general mode of translation inhibition by macrolide antibiotics. Proc. Natl. Acad. Sci. U.S.A. 111, 15958–15963 10.1073/pnas.1417334111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kitahara K., and Miyazaki K. (2011) Specific inhibition of bacterial RNase T2 by helix 41 of 16S ribosomal RNA. Nat. Commun. 2, 549 10.1038/ncomms1553 [DOI] [PubMed] [Google Scholar]
- 40. Neuhaus K., Landstorfer R., Simon S., Schober S., Wright P. R., Smith C., Backofen R., Wecko R., Keim D. A., and Scherer S. (2017) Differentiation of ncRNAs from small mRNAs in Escherichia coli O157:H7 EDL933 (EHEC) by combined RNAseq and RIBOseq - ryhB encodes the regulatory RNA RyhB and a peptide, RyhP. BMC Genomics 18, 216 10.1186/s12864-017-3586-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hücker S. M., Vanderhaeghen S., Abellan-Schneyder I., Scherer S., and Neuhaus K. (2018) The novel anaerobiosis-responsive overlapping gene ano is overlapping antisense to the annotated gene ECs2385 of Escherichia coli O157:H7 Sakai. Front. Microbiol. 9, 931 10.3389/fmicb.2018.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dingwall C., Lomonossoff G. P., and Laskey R. A. (1981) High sequence specificity of micrococcal nuclease. Nucleic Acid Res. 12, 2659–2673 10.1093/nar/9.12.2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hücker S. M., Ardern Z., Goldberg T., Schafferhans A., Bernhofer M., Vestergaard G., Nelson C. W., Schloter M., Rost B., Scherer S., and Neuhaus K. (2017) Discovery of numerous novel small genes in the intergenic regions of the Escherichia coli O157:H7 Sakai genome. PLoS ONE 12, e0184119. 10.1371/journal.pone.0184119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hwang J. Y., and Buskirk A. R. (2017) A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res 45, 327–336 10.1093/nar/gkw944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hurley J. M., Cruz J. W., Ouyang M., and Woychik N. A. (2011) Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5′ end of coding regions in vivo. J. Biol. Chem. 286, 14770–14778 10.1074/jbc.M110.108969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., and Ehrenberg M. (2003) The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 10.1016/S0092-8674(02)01248-5 [DOI] [PubMed] [Google Scholar]
- 47. Calviello L., and Ohler U. (2017) Beyond read-counts: Ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet. 33, 728–744 10.1016/j.tig.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 48. Gerashchenko M. V., and Gladyshev V. N. (2017) Ribonuclease selection for ribosome profiling. Nucleic Acids Res 45, e6 10.1093/nar/gkw822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jelenc P. C. (1980) Rapid purification of highly active ribosomes from Escherichia coli. Anal. Biochem. 105, 369–374 10.1016/0003-2697(80)90472-8 [DOI] [PubMed] [Google Scholar]
- 50. Bartholomäus A., Fedyunin I., Feist P., Sin C., Zhang G., Valleriani A., and Ignatova Z. (2016) Bacteria differently regulate mRNA abundance to specifically respond to various stresses. Philos. Trans. A Math. Phys. Eng. Sci. 374, 20150069 10.1098/rsta.2015.0069 [DOI] [PubMed] [Google Scholar]
- 51. Balakrishnan R., Oman K., Shoji S., Bundschuh R., and Fredrick K. (2014) The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 42, 13370–13383 10.1093/nar/gku1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Latif H., Szubin R., Tan J., Brunk E., Lechner A., Zengler K., and Palsson B. O. (2015) A streamlined ribosome profiling protocol for the characterization of microorganisms. BioTechniques 58, 329–332 10.2144/000114302 [DOI] [PubMed] [Google Scholar]
- 53. Li G.-W., Oh E., and Weissman J. S. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541, 10.1038/nature10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buskirk A. R., and Green R. (2017) Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160183 10.1098/rstb.2016.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burkhardt D. H., Rouskin S., Zhang Y., Li G. W., Weissman J. S., and Gross C. A. (2017) Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 6, e22037 10.7554/eLife.22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herbert Z. T., Kershner J. P., Butty V. L., Thimmapuram J., Choudhari S., Alekseyev Y. O., Fan J., Podnar J. W., Wilcox E., Gipson J., Gillaspy A., Jepsen K., BonDurant S. S., Morris K., Berkeley M., et al. (2018) Cross-site comparison of ribosomal depletion kits for Illumina RNAseq library construction. BMC Genomics 19, 199 10.1186/s12864-018-4585-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Culviner P. H., Guegler C. K., and Laub M. T. (2020) A simple, cost-effective, and robust method for rRNA depletion in RNA-sequencing studies. bioRxiv 10.1101/2020.01.06.896837 [DOI] [PMC free article] [PubMed]
- 58. Huang Y., Sheth R. U., Kaufman A., and Wang H. H. (2020) Scalable and cost-effective ribonuclease-based rRNA depletion for transcriptomics. Nucleic Acids Res. 48, e20 10.1093/nar/gkz1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kraus A. J., Brink B. G., and Siegel T. N. (2019) Efficient and specific oligo-based depletion of rRNA. Sci. Rep. 9, 12281 10.1038/s41598-019-48692-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. VanOrsdel C. E., Kelly J. P., Burke B. N., Lein C. D., Oufiero C. E., Sanchez J. F., Wimmers L. E., Hearn D. J., Abuikhdair F. J., Barnhart K. R., Duley M. L., Ernst S. E. G., Kenerson B. A., Serafin A. J., and Hemm M. R. (2018) Identifying new small proteins in Escherichia coli. Proteomics 18, e1700064 10.1002/pmic.201700064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Haas B. J., Chin M., Nusbaum C., Birren B. W., and Livny J. (2012) How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes?. BMC Genomics 13, 734 10.1186/1471-2164-13-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Landstorfer R., Simon S., Schober S., Keim D., Scherer S., and Neuhaus K. (2014) Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 15, 353 10.1186/1471-2164-15-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hücker S. M., Vanderhaeghen S., Abellan-Schneyder I., Wecko R., Simon S., Scherer S., and Neuhaus K. (2018) A novel short l-arginine responsive protein-coding gene (laoB) antiparallel overlapping to a CadC-like transcriptional regulator in Escherichia coli O157:H7 Sakai originated by overprinting. BMC Evol. Biol. 18, 21 10.1186/s12862-018-1134-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Clauwaert J., Menschaert G., and Waegeman W. (2019) DeepRibo: a neural network for precise gene annotation of prokaryotes by combining ribosome profiling signal and binding site patterns. Nucleic Acids Res. 47, e36 10.1093/nar/gkz061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Giess A., Jonckheere V., Ndah E., Chyżyńska K., Van Damme P., and Valen E. (2017) Ribosome signatures aid bacterial translation initiation site identification. BMC Biol 15, 76 10.1186/s12915-017-0416-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miravet-Verde S., Ferrar T., Espadas-García G., Mazzolini R., Gharrab A., Sabido E., Serrano L., and Lluch-Senar M. (2019) Unraveling the hidden universe of small proteins in bacterial genomes. Mol. Syst. Biol. 15, e8290 10.15252/msb.20188290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Woolstenhulme C. J., Guydosh N. R., Green R., and Buskirk A. R. (2015) High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21 10.1016/j.celrep.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rosenow C., Saxena R. M., Durts M., and Gingeras T. R. (2001) Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29, E112 10.1093/nar/29.22.e112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Amin M. R., Yurovsky A., Chen Y., Skiena S., and Futcher B. (2018) Re-annotation of 12,495 prokaryotic 16S rRNA 3′ ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences. PLoS ONE 13, e0202767 10.1371/journal.pone.0202767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ma J., Campbell A., and Karlin S. (2002) Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184, 5733–5745 10.1128/JB.184.20.5733-5745.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shultzaberger R. K., Bucheimer R. E., Rudd K. E., and Schneider T. D. (2001) Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 313, 215–228 10.1006/jmbi.2001.5040 [DOI] [PubMed] [Google Scholar]
- 72. Hui A., and de Boer H. A. (1987) Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 84, 4762–4766 10.1073/pnas.84.14.4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shine J., and Dalgarno L. (1975) Determinant of cistron specificity in bacterial ribosomes. Nature 254, 34–38 10.1038/254034a0 [DOI] [PubMed] [Google Scholar]
- 74. Shine J., and Dalgarno L. (1974) The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 71, 1342–1346 10.1073/pnas.71.4.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Steitz J. A., and Jakes K. (1975) How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 72, 4734–4738 10.1073/pnas.72.12.4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ingolia N. T. (2014) Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 10.1038/nrg3645 [DOI] [PubMed] [Google Scholar]
- 77. Lareau L. F., Hite D. H., Hogan G. J., and Brown P. O. (2014) Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife 3, e01257 10.7554/eLife.01257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Smith C., Canestrari J. G., Wang J., Derbyshire K. M., Gray T. A., and Wade J. T. (2019) Pervasive translation in Mycobacterium tuberculosis. bioRxiv 10.1101/665208 [DOI] [PMC free article] [PubMed]
- 79. Saito K., Green R., and Buskirk A. R. (2020) Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. eLife 9, 10.7554/eLife.55002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gelsinger D. R., Dallon E., Reddy R., Mohammad F., Buskirk A. R., and DiRuggiero J. (2020) Ribosome profiling in Archaea reveals leaderless translation, novel translational initiation sites, and ribosome pausing at single codon resolution. bioRxiv 10.1101/2020.02.04.934349 [DOI] [PMC free article] [PubMed]
- 81. Lloréns-Rico V., Cano J., Kamminga T., Gil R., Latorre A., Chen W. H., Bork P., Glass J. I., Serrano L., and Lluch-Senar M. (2016) Bacterial antisense RNAs are mainly the product of transcriptional noise. Sci. Adv. 2, e1501363 10.1126/sciadv.1501363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S., Becher D., Bisicchia P., Botella E., Delumeau O., Doherty G., et al. (2012) Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- 83. Raghavan R., Sloan D. B., and Ochman H. (2012) Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio 3, 10.1128/mBio.00156-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lejars M., Kobayashi A., and Hajnsdorf E. (2019) Physiological roles of antisense RNAs in prokaryotes. Biochimie 164, 3–16 10.1016/j.biochi.2019.04.015 [DOI] [PubMed] [Google Scholar]
- 85. Grainger D. C. (2016) The unexpected complexity of bacterial genomes. Microbiology 162, 1167–1172 10.1099/mic.0.000309 [DOI] [PubMed] [Google Scholar]
- 86. Passalacqua K. D., Varadarajan A., Weist C., Ondov B. D., Byrd B., Read T. D., and Bergman N. H. (2012) Strand-specific RNA-seq reveals ordered patterns of sense and antisense transcription in Bacillus anthracis. PLoS ONE 7, e43350 10.1371/journal.pone.0043350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ardern Z., Neuhaus K., and Scherer S. (2020) Are antisense proteins in prokaryotes functional? bioRxiv 10.1101/2020.02.20.958058 [DOI] [PMC free article] [PubMed]
- 88. Fellner L., Bechtel N., Witting M. A., Simon S., Schmitt-Kopplin P., Keim D., Scherer S., and Neuhaus K. (2014) Phenotype of htgA (mbiA), a recently evolved orphan gene of Escherichia coli and Shigella, completely overlapping in antisense to yaaW. FEMS Microbiol. Lett. 350, 57–64 10.1111/1574-6968.12288 [DOI] [PubMed] [Google Scholar]
- 89. Kurata T., Katayama A., Hiramatsu M., Kiguchi Y., Takeuchi M., Watanabe T., Ogasawara H., Ishihama A., and Yamamoto K. (2013) Identification of the set of genes, including nonannotated morA, under the direct control of ModE in Escherichia coli. J. Bacteriol. 195, 4496–4505 10.1128/JB.00304-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Delaye L., Deluna A., Lazcano A., and Becerra A. (2008) The origin of a novel gene through overprinting in Escherichia coli. BMC Evol. Biol. 8, 31. 10.1186/1471-2148-8-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu B., and Chen C. (2018) Translation Elongation Factor 4 (LepA) Contributes to tetracycline susceptibility by stalling elongating ribosomes. Antimicrob. Agents Chemother. 62, 10.1128/AAC.02356-17 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92. Goodall E. C. A., Robinson A., Johnston I. G., Jabbari S., Turner K. A., Cunningham A. F., Lund P. A., Cole J. A., and Henderson I. R. (2018) The essential genome of Escherichia coli K-12. mBio 9, 10.1128/mBio.02096-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Grenier F., Matteau D., Baby V., and Rodrigue S. (2014) Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2, e01038–14 10.1128/genomea.01038-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Peters J. E., Thate T. E., and Craig N. L. (2003) Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185, 2017–2021 10.1128/JB.185.6.2017-2021.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lau B. T., and Ji H. P. (2017) Single molecule counting and assessment of random molecular tagging errors with transposable giga-scale error-correcting barcodes. BMC Genomics 18, 745 10.1186/s12864-017-4141-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Fu Y., Wu P. H., Beane T., Zamore P. D., and Weng Z. (2018) Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics 19, 531 10.1186/s12864-018-4933-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen S., Zhou Y., Chen Y., and Gu J. (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ndah E., Jonckheere V., Giess A., Valen E., Menschaert G., and Van Damme P. (2017) REPARATION: ribosome profiling assisted (re-)annotation of bacterial genomes. Nucleic Acids Res. 45, e168 10.1093/nar/gkx758 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sources of all original data are listed in Table S1. All processed data and bash scripts used for analyses are available upon request from Alina Glaub (alina.glaub@tum.de).