Abstract
DEAD-box helicase 5 (DDX5) is a founding member of the DEAD-box RNA helicase family, a group of enzymes that regulate ribonucleoprotein formation and function in every aspect of RNA metabolism, ranging from synthesis to decay. Our laboratory previously found that DDX5 is involved in energy homeostasis, a process that is altered in many cancers. Small cell lung cancer (SCLC) is an understudied cancer type for which effective treatments are currently unavailable. Using an array of methods, including short hairpin RNA–mediated gene silencing, RNA and ChIP sequencing analyses, and metabolite profiling, we show here that DDX5 is overexpressed in SCLC cell lines and that its down-regulation results in various metabolic and cellular alterations. Depletion of DDX5 resulted in reduced growth and mitochondrial dysfunction in the chemoresistant SCLC cell line H69AR. The latter was evidenced by down-regulation of genes involved in oxidative phosphorylation and by impaired oxygen consumption. Interestingly, DDX5 depletion specifically reduced intracellular succinate, a TCA cycle intermediate that serves as a direct electron donor to mitochondrial complex II. We propose that the oncogenic role of DDX5, at least in part, manifests as up-regulation of respiration supporting the energy demands of cancer cells.
Keywords: DEAD-box helicase 5 (DDX5), RNA helicase, gene expression, mitochondrial metabolism, TCA cycle, small cell lung cancer, bioenergetics, energy homeostasis, cell proliferation, Krebs cycle, respiration
Introduction
The DEAD-box RNA helicase DDX5 is overexpressed in multiple types of cancers in which its expression is necessary for cell proliferation and tumor growth (1–4). Similar to other RNA helicases, DDX5 is involved in many steps of RNA metabolism, such as rRNA biogenesis, transcriptional regulation, alternative splicing, and microRNA processing (5). Our laboratory showed that DDX5 is an active RNA helicase in vitro that is functionally conserved with its Saccharomyces cerevisiae ortholog DBP2 (6). We also found that deleting DBP2 leads to global changes of mRNA secondary structures in S. cerevisiae that correlate with altered transcriptional termination (7). Similarly, other groups have shown that DDX5 controls alternative splicing in a manner that may involve secondary structure remodeling (8, 9). This suggests that DDX5 and Dbp2 remodel RNA secondary structures in nascent RNA during transcription and pre-mRNA maturation steps, likely altering gene expression in the process.
In addition to RNA remodeling, DDX5 also acts as cofactor for oncogenic transcription factors in several cancer types (2, 10). DDX5 expression activates the oncogenic Wnt and mammalian target of rapamycin signaling pathways involved in cell-fate determination and cell growth (3, 11, 12). Interestingly, depletion of DDX5 in mammalian cells or deletion of DBP2 in S. cerevisiae lead to differential expression of metabolic genes (13, 14) and alteration of glycolysis and mitochondrial respiration activity (6, 15). Moreover, a DDX5 S480A polymorphism has been connected to metabolic syndrome in humans, which is typified by abnormalities in primary metabolic pathways (16). This suggests that the DDX5 family is involved in energy metabolism, a process that is misregulated in many cancers (17).
Small cell lung cancer (SCLC) accounts for 15% of lung cancer cases and has a 5-year survival rate of less than 10% (18). SCLC is an aggressive disease because of its fast growth rate and early development of metastases. This cancer shows positive initial response to chemotherapies but develops resistance rapidly, resulting in relapse within 2 years. SCLC originates from neuroendocrine cells, with the classic subtype of SCLC defined by expression of the neuroendocrine-specific transcriptional factor achaete-scute homolog 1, a lineage-specific oncogene (18, 19). SCLC invariably inactivates the tumor suppressor genes TP53 and RB transcriptional corepressor 1. Factors such as the MYC family of oncogenes, the apoptosis regulator Bcl-2, and the tetraspanin cell-surface family member CD151 have been correlated with SCLC (20). However, SCLC still remains largely uncharacterized, and there are currently no effective pharmacological strategies to prevent recurrence of the disease.
Despite diverse genetic landscapes, a common theme of cellular transformation is up-regulation of metabolic pathways to support rapid growth and biomass production (21). The observation by Otto Warburg in the 1920's that cancer cells continue to convert glucose to lactate through lactic acid fermentation, despite the availability of oxygen to support respiration, has now been observed in multiple cancer cell lines and within solid tumors (22). It is now established that this type of “reprogramed” metabolism is not used for ATP production but rather to produce glycolytic intermediates that form building blocks for rapid cellular growth. Importantly, cancer cells still generate ATP through mitochondrial respiration to fuel biosynthesis (21). Thus, mitochondrial defects can be a barrier for malignancy, suggesting the necessity of mitochondrial functions in carcinogenesis. Herein, we show that DDX5 depletion leads to growth defects and global changes of gene expression in drug-resistant SCLC cells. Moreover, we find that DDX5 is necessary for mitochondrial function and production of the TCA cycle intermediate succinate. This suggests that DDX5 may be a novel drug target for small cell lung cancer in the future.
Results
DDX5 is required for the growth of SCLC cell lines
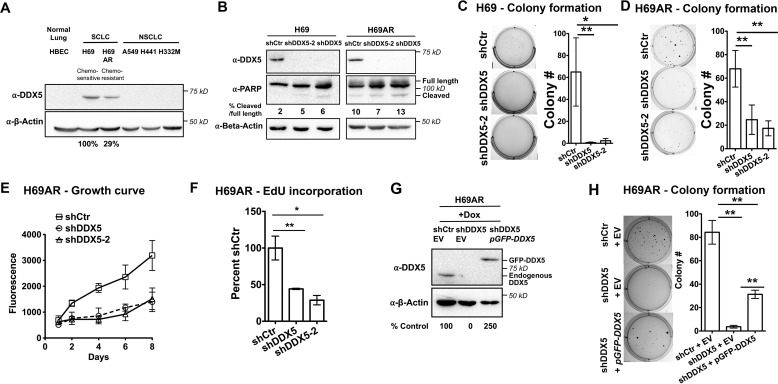
SCLC is an aggressive cancer that rapidly develops chemoresistance and currently lacks effective therapy options. DDX5 is an RNA helicase whose overexpression is correlated with multiple cancer types, including breast, colon, and prostate (23), and is associated with resistance to therapeutics in the non–small cell lung cancer cell line H1299 (24). To determine whether DDX5 is necessary for SCLC growth, we first measured DDX5 protein levels in transformed and nontransformed cell lines. Analysis of DDX5 levels by Western blotting of the noncancerous human bronchial epithelial cell line HBEC-3KT (HBEC); two SCLC cell lines, H69 (chemosensitive) and H69AR (chemoresistant); and three non–small cell lung cancer cell lines, A549, H441, and H332M, shows that DDX5 is expressed in both chemosensitive and chemoresistant SCLC cells but is below detection in the other cell lines (Fig. 1A). This is consistent with prior reports that DDX5 may be dispensible for some tissue functions in whole animals (13). To determine whether DDX5 expression supports the transformed properties of SCLC, such as cell invasion and aberrant growth, we knocked down DDX5 in H69 and H69AR cells using inducible, short hairpin RNAs (shRNAs). Two shRNAs (shDDX5 and shDDX5-2) with different target sites were used to knockdown DDX5. A Western blotting showed efficient knockdown of DDX5 using either shDDX5 construct (Fig. 1B). Thus, we used shDDX5 for simplicity in further experimentation.
Figure 1.
Knockdown of DDX5 reduces proliferation and soft agar colony formation of SCLC cells. A, Western blotting of DDX5 in HBEC, SCLC, and NSCLC cell lines. B, Western blotting of DDX5 and PARP in H69 and H69AR cells stably transfected with shRNAs targeting DDX5 or with a nontargeting control (shCtr). C and D, soft agar colony formation assays of H69 or H69AR cells, with or without DDX5 knockdown. 10,000 cells/well of H69 cells or 5000 cells/well of H69AR cells were seeded in a 0.3% agar layer in 6-well plates. H69 and H69AR cells were grown for 30 and 21 days, respectively, with growth medium added twice a week. The bar graphs in the right panels show colony counts from at least three replicates, whereas the left panels show representative pictures of wells. E, growth curve of H69AR cells with or without DDX5 knockdown. H69AR cells were seeded at 200 cells/well in 96-well plates on day 0, and the relative cell number was measured on the indicated days. Cell growth was measured using CyQuant fluorescent DNA dye to quantify cell number on the days indicated. The data show the means ± S.D. of four biological replicates. F, measurement of cell proliferation by quantifying newly synthesized DNA using the nucleotide analog EdU after 24 h of growth. Incorporated EdU was measured using the Click-iT EdU Alexa Fluor 647 Assay (Invitrogen, C10356). G, Western blotting of DDX5 in H69AR cells expressing shRNA-resistant GFP–DDX5. GFP–DDX5 is overexpressed as compared with endogenous DDX5. H, soft agar assays of cells expressing GFP–DDX5. Ectopic overexpression of GFP–DDX5 partially rescues the ability of H69AR cells to form colonies in soft agar. The bar graph shows colony counts from three biological replicates, and the left panel shows one representative picture for each cell line. Colonies were quantified using OpenCFU. The data show the means ± S.D. of three biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001. EV, empty vector; Dox, doxycycline.
To determine whether DDX5 promotes anchorage-independent growth in SCLC, a hallmark of carcinogenesis, we measured the abilities of H69 and H69AR cells to form colonies in soft agar. Although both H69 and H69AR cells readily form colonies, cells expressing either shDDX5 or shDDX5-2 showed decreased colony formation, with decreases of >90% in H69 cells and >60% in H69AR cells (Fig. 1, C and D). To validate that DDX5 is required for growth of H69AR in normal culture conditions, we measured the cell density of H69AR cells over 8 days and plotted the growth curves. This showed that cells expressing shDDX5 and shDDX5-2 grew slower than cells expressing shCtr but similar to each other (Fig. 1E). By the end of the 8th day, the density of DDX5-depleted cells was less than 50% of the control cells. Consistently, DDX5 depletion reduces proliferation of H69AR cells by more than 50%, as evidenced by measurement of EdU incorporation into newly synthesized DNA after 24 h (Fig. 1F). Negative regulators of tumorigenesis often inhibit tumor growth via inducing apoptosis. To test whether this is true for DDX5 in SCLC, we analyzed the cleavage of poly(ADP-ribose) polymerase (PARP) using Western blotting. Interestingly, an increase in PARP cleavage was not observed after knocking down DDX5 above levels of the shCtr control alone (Fig. 1B), suggesting that apoptosis is not triggered. To confirm that the growth defects are due to DDX5 knockdown, we ectopically expressed an shRNA-resistant, N-terminally GFP-tagged DDX5 construct in our DDX5 knockdown cells (Fig. 1G). Soft agar assays demonstrated that GFP–DDX5 expression partially rescues the anchorage-independent growth of cells expressing shDDX5 as compared with the control (shCtr) (Fig. 1H). Cells ectopically expressing DDX5 showed a 10-fold increase in colony count as compared with cells expressing the empty vector, but only to 40% of WT. This partial rescue may be attributed to reduced functionality of the N-terminally tagged DDX5. Regardless, rescue to 40% suggests that growth defects are attributed to DDX5 knockdown. Taken together, these data indicate that DDX5 depletion slows down growth of H69AR cells but does not trigger programed cell death.
Genes involved in respiration are down-regulated upon DDX5 depletion in SCLC cells
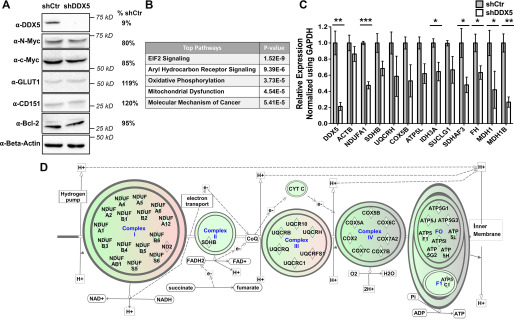
To explore the target of DDX5 in chemoresistant SCLC cells, we first used a Western blotting to analyze the levels of several proteins whose overabundance has been linked to SCLC (20). This includes the MYC oncogenes, the GLUT1 gene encoding the major glucose transporter, the tetraspanin CD151, and the apoptosis regulator BCL-2 (20). None of these factors showed altered protein levels upon DDX5 depletion (Fig. 2A).
Figure 2.
RNA-Seq reveals dysregulated mitochondrial function upon DDX5 depletion. A, Western blotting of factors known to be involved in SCLC tumorigenesis. H69AR cells with or without DDX5 knockdown were used. GLUT1, glucose transporter type 1; CD151, tetraspanin CD151 molecule; Bcl-2, apoptosis regulator. B, table of the top canonical pathways (from IPA) associated with DEGs upon DDX5 knockdown. Canonical pathways were identified using the Qiagen IPA toolkit. C, independent validation of misregulated mitochondrial function upon depletion of DDX5 using RT-qPCR. Primer sets spanning exon junctions were used to quantify mRNA levels, which were then normalized to the expression levels of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The relative expression levels of each gene in H69AR + shCtr cells were arbitrarily set to 1. The data represent the means ± S.D. of three biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001. D, schematic depicting down-regulation of genes involved in the OxPhos pathway. DEGs are shown as rhombuses. Green or pink fill in the rhombus indicates down-regulation or up-regulation, respectively. Green background indicates that the pathway is predicated to be down-regulated.
To globally identify genes that are affected by DDX5, RNA-Seq was performed with H69AR cells expressing either shCtr or shDDX5. We obtained ∼2 × 108 mapped reads for each of three biological replicates, with genome mapping rates from 90% to 95% (Table S4) and high correlation between replicates (Pearson correlation coefficient r ≥ 0.99 for shCtr and r ≥ 0.96 for shDDX5; Fig. S1). Statistical analysis of the data using DESeq2 and edgeR revealed 6727 differentially expressed genes (DEGs) upon DDX5 knockdown with a false discovery rate (FDR) ≤ 0.05 (Table S5). Five mitochondria-encoded transcripts (cytochrome c oxidase II, NADH:ubiquinone oxidoreductase core subunit 2, and three tRNA genes) were also identified as DEGs in the analysis. Pathway analysis of the DEGs was performed using the Ingenuity Pathway Analysis (IPA) toolkit, which uses a curated database to identify significantly altered gene expression pathways. This analysis revealed oxidative phosphorylation (OxPhos) and mitochondrial function among the top significantly altered pathways upon DDX5 knockdown (Fig. 2B). Most DEGs found in OxPhos and mitochondrial function pathways are down-regulated upon DDX5 depletion (Table S5) and as confirmed by RT-qPCR of select genes (Fig. 2C). Other top scoring pathways were molecular mechanism of cancer, EIF2 signaling pathway linked to cellular stress, and aryl hydrocarbon receptor signaling involved in response to environmental stimuli (Fig. 2B). The OxPhos pathway is predicted to be deactivated by IPA (z score = −6.03; Table S6), with down-regulation of 37 genes involved in the formation of complex I–V (Fig. 2D).
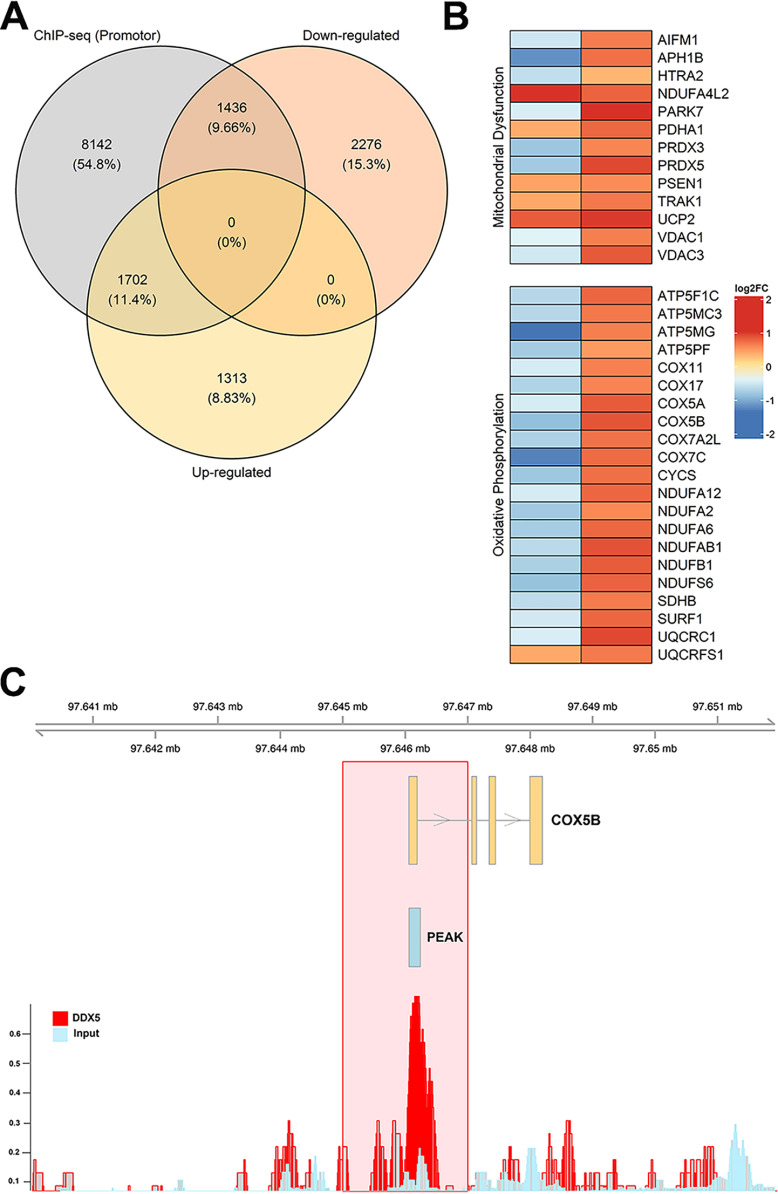
To determine whether DDX5 might directly regulate expression of genes involved in mitochondrial respiration, we remapped reads from published DDX5–ChIP-Seq from HeLa cells (25). This revealed that DDX5 binds 11,281 genes, consistent with the prior report. A total of 37,924 DDX5 peaks were detected, 25,996 of which were found within 1KB of the transcription start site (TSS). Approximately half (3138 of 6727 genes) of the DEGs identified through our RNA-Seq of SCLC lines have a corresponding DDX5 ChIP peak within 1KB of the TSS, with an equal distribution between down-regulated (1436) and up-regulated (1313) genes (59–62) (Fig. 3A). In addition to promoter binding, DDX5 is also found associated with terminators (Fig. S2A). Comparison of all DDX5 ChIP-Seq peaks (promoter + terminator) to our RNA-Seq also revealed a similar distribution between up- and down-regulated genes upon DDX5 knockdown (Fig. S2B). Interestingly, all of the DEGs linked to mitochondrial dysfunction or oxidative phosphorylation by IPA are bound by DDX5 at or near the promoter (within 1 kb of the TSS) with the vast majority (28 of 34 genes, or 82%) down-regulated upon DDX5 knockdown (Fig. 3, B and C), suggesting that DDX5 promotes expression of these genes.
Figure 3.
Differentially expressed genes upon DDX5 knockdown are also bound by DDX5 at promoters. ChIP-Seq data were downloaded from Gene Expression Omnibus (GSE24126), processed, and aligned to the genome. The significance of overlap between DDX5-bound (ChIP-Seq) and DDX5-misregulated (up-regulated or down-regulated) genes was determined using the R-Bioconductor package GeneOverlap (59). Venn diagram, heat map, and genomic visualization figures were generated using the R packages VennDiagram, ComplexHeatmap, and Gviz, respectively (60–62). A, overlap between DDX5-bound (within 1 kb of promoter) and differentially expressed genes from RNA-Seq. B, heat map of selected genes from oxidative phosphorylation and mitochondrial dysfunction pathways with log2 fold change values denoting the direction of gene expression (RNA-Seq, left column) and DDX5-binding strength (ChIP-Seq, right column). C, genomic visualization showing the DDX5-bound peak signal for the gene COX5B.
Cellular respiration is compromised upon DDX5 depletion
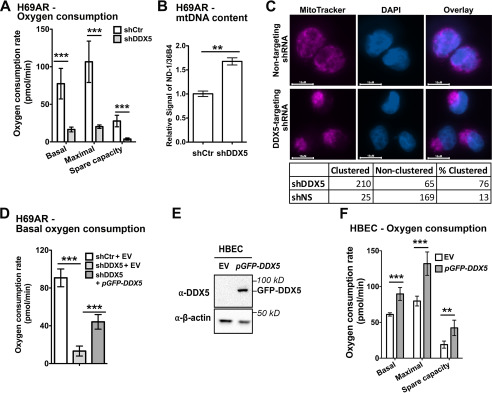
Gene expression analysis suggests that DDX5 facilitates cellular respiration in SCLC. To test this, we analyzed the rates of oxygen consumption using the Seahorse extracellular flux analyzer, which measures dissolved oxygen concentration in the culture medium in real time. Analysis of respiration using the Seahorse revealed that the basal and maximal oxygen consumption rates are decreased by ∼80% upon DDX5 depletion (Fig. 4A). The spare capacity is also decreased, revealing a reduced ability to respond to increased energy demand.
Figure 4.
DDX5 promotes mitochondrial function and respiration. A, oxygen consumption rates of H69AR cells with or without DDX5 knockdown were measured using the Seahorse XFe24 metabolic flux analyzer. The data show the means ± S.D. of four biological replicates. B, measurement of mitochondrial DNA content. The relative amount of the mitochondria-specific NADH-ubiquinone oxidoreductase chain 1 (ND-1) gene to the nuclear ribosomal protein lateral stalk subunit P0 (36B4) gene was measured by qPCR, with the ratio in H69AR + shCtr cells arbitrarily set to 1. C, mitochondrial staining in H69AR cells stably transfected with doxycycline-inducible shRNAs targeting DDX5 or with a nontargeting control as visualized by fluorescence microscopy. The cells were grown overnight on polylysine-treated coverslips and then treated with 500 nm MitoTracker Deep Red for mitochondrial visualization. The cells were then fixed in 0.6% formaldehyde, and DNA was stained with 300 nm DAPI and serves as a nuclear marker. The cells were manually counted and individually categorized as exhibiting a “clustered” or “nonclustered” morphological phenotype. D, oxygen consumption rates of H69AR cells in the presence of shDDX5-resistant DDX5 or empty vector. E, Western blotting of DDX5 in HBEC cells stably transfected with empty vector (EV) or pGFP–DDX5. F, oxygen consumption rates of HBEC cells ± GFP–DDX5. The data show the means ± S.D. of six biological replicates. **, P < 0.01; ***, P < 0.001.
Next, we reasoned that reduced oxygen consumption may be due to decreased mitochondria activity. To test this, we first measured the mitochondrial DNA (mtDNA) content using qPCR. Interestingly, we observed a 1.7-fold, statistically significant increase in mtDNA content, relative to the nuclear genome (Fig. 4B). This increase in mtDNA is consistent with mitochondrial dysfunctions observed in other cancer types (26). To determine whether there is a mitochondrial morphology defect upon knockdown of DDX5, we stained H69AR cells with MitoTracker to visualize mitochondria (Fig. 4C). This revealed a pronounced mitochondrial clustering defect, a phenotype that correlates with reduced inner membrane potential and mitochondrial function (27, 28). 76% of cells expressing the shDDX5 construct showed a clustering phenotype compared with 13% of the cells expressing the nontargeting shRNA control. Interestingly, we also observed a decrease in cell size of ∼30% upon DDX5 knockdown. We then asked if overexpression of shDDX5-resistant GFP–DDX5 in H69AR cells could rescue the respiration defects upon DDX5 knockdown. GFP–DDX5 expression partially rescued the reduced oxygen consumption rates upon DDX5 knockdown to ∼50% of the control (Fig. 4D), similar to rescue of colony formation activity (Fig. 1F). To determine whether DDX5 overexpression is sufficient to increase respiration in nontransformed cells, we ectopically overexpressed a GFP-tagged DDX5 in the normal lung HBEC cells (Fig. 4E). This showed that overexpression of DDX5 increases oxygen consumption rates by one-third in nontransformed lung cells (Fig. 4F). Taken together, our studies suggest that DDX5 expression facilitates invasive growth behavior of SCLC by promoting mitochondrial function.
DDX5 knockdown reduces intracellular succinate concentrations
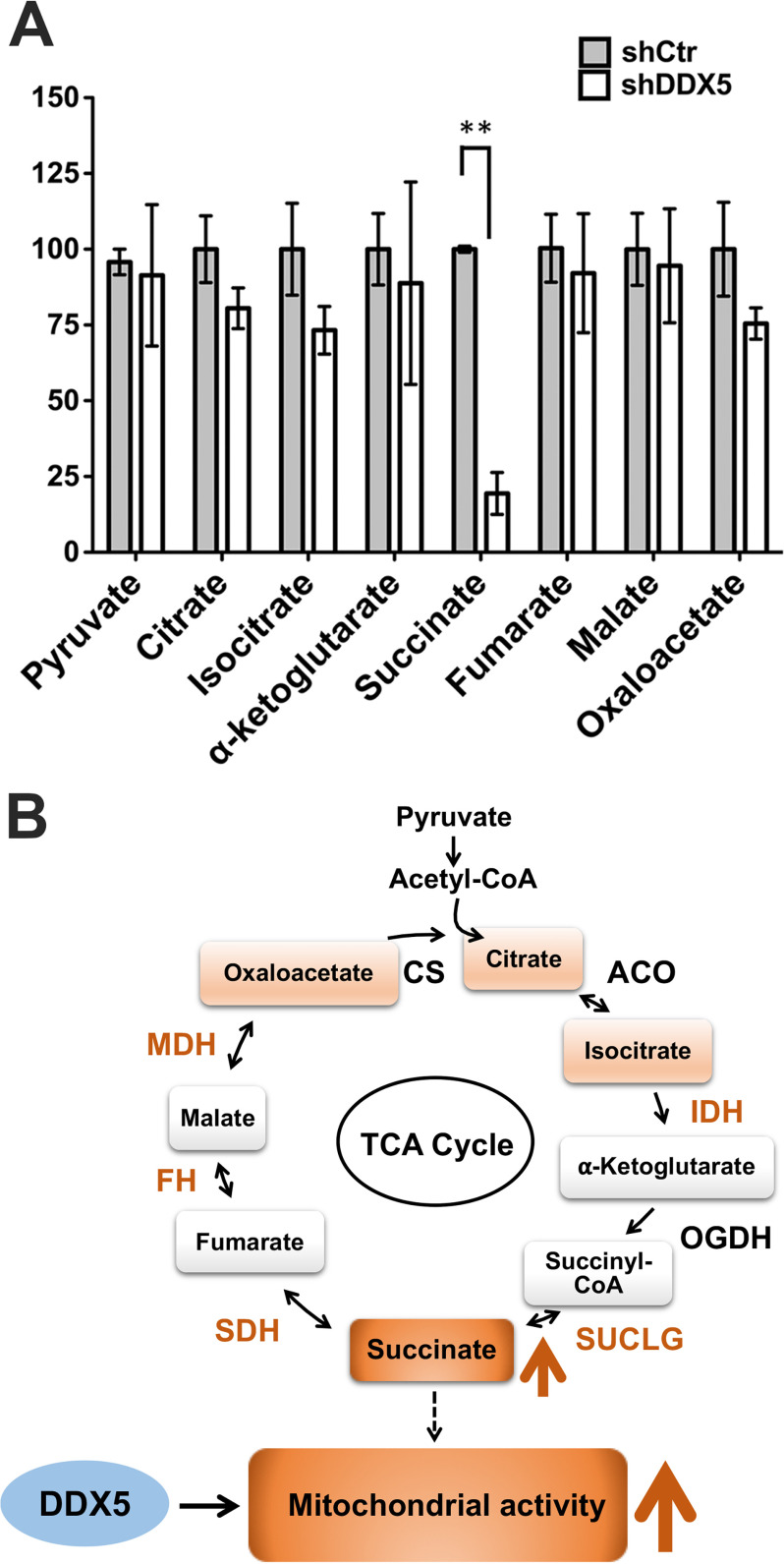
The almost complete loss of mitochondrial respiration upon DDX5 knockdown in SCLC seems inconsistent with the statistically significant but modest (30–60%) decrease in expression of OxPhos genes (Fig. 2C and Table S5). Thus, we hypothesized that DDX5 knockdown induces a respiratory bottleneck that abolishes respiration. To identify the bottleneck, we conducted metabolite profiling to quantify the relative amount of intracellular TCA cycle intermediates using HPLC–MS/MS (29). This revealed that succinate, a product of succinyl-CoA ligase, is specifically and significantly reduced (80%) upon depletion of DDX5 (Fig. 5A). This suggests that succinate accumulation/utilization is uniquely controlled and that insufficient succinate may be the bottleneck for mitochondria function in cells lacking DDX5.
Figure 5.
Depletion of DDX5 reduces intracellular succinate. A, TCA cycle metabolite profiling by MS (HPLC–MS/MS) of H69AR cells ± DDX5 knockdown. The relative amount of each metabolite was normalized to cell wet weight. B, model for DDX5 function in respiration. DDX5 is required for efficient respiration likely through promoting OxPhos gene expression (Fig. 2) and succinate production. Succinate is particularly important to the electron transport chain because it is a direct electron donor. Future work is needed to determine the precise molecular role of DDX5 in this process. Orange color indicates down-regulation of gene expression or decrease of intracellular TCA metabolite. CS, citrate synthase; ACO, aconitase; IDH, isocitrate dehydrogenase; OGDH, oxoglutarate dehydrogenase; SUCLG, succinyl-CoA ligase; SDH, succinate dehydrogenase; FH, fumarase; MDH, malate dehydrogenase. **, P < 0.01.
Succinate is not only an intermediate in the TCA cycle (Fig. 5B) but also a direct electron donor for the electron transport chain through succinate dehydrogenase (complex II) (30). Because lung cancers rely heavily on mitochondrial respiration for growth (31), these cancers may be particularly susceptible to inhibition of mitochondrial function. The findings herein implicate the RNA helicase DDX5 in this process, suggesting that DDX5 may be a new target for the development of lung cancer therapies.
Discussion
DEAD-box RNA helicases are the largest class of enzymes in the RNA helicase family, acting as nonprocessive, ATP-dependent RNA-binding proteins, whose activity can be coupled to remodeling of secondary structure and/or RNA–protein complexes (32). Somewhat paradoxically, up-regulation of members of the DEAD-box protein family, including DDX1, DDX2, DDX3, and DDX5, has been linked to cellular transformation and correlates with multiple cancer types including breast, colon, bone, and prostate (33, 34). Herein, we assessed the role of DEAD-box helicase DDX5 in drug-resistant SCLC, a highly aggressive, metastatic cancer with a high mortality rate. Our results show that DDX5 is required for anchorage-independent growth and normal respiration of SCLC cells, consistent with the notion that mitochondrial functions are necessary for carcinogenesis (35, 36). Moreover, we find that DDX5 expression is necessary for accumulation of succinate, an oncometabolite whose up-regulation correlates with chemoresistance (37), suggesting that up-regulation of DDX5 may be part of the underlying mechanism for drug resistance in SCLC.
DDX5 was one of the first members of the DEAD-box helicase family to be enzymatically characterized and, in the 30 years since, has now been implicated in multiple transcriptional and cotranscriptional processes suggestive of a role as an RNA chaperone for nascent transcripts (5). This raises the question of how a general chaperone that acts in a wide array of gene expression steps could promote uncontrolled cell growth. Although we do not know the precise molecular basis of this activity, two possibilities exist for a putative mechanism for DDX5 in supporting rapid cellular proliferation. First, a biochemically distinct pool of DDX5 may exist in vivo that regulates transcription of specific genes directly. DDX5 is both polyubiquitinated and phosphorylated, modifications that appear to correlate with distinct activities. Specifically, phosphorylation of Tyr-593 at the C terminus of DDX5 promotes cellular proliferation and expression of genes involved in cell growth such as c-MYC and Cyclin D1 (11, 23). Interestingly, DDX5 was recently shown to unwind a transcriptionally repressive G-quadraplex in the c-MYC promoter, illustrating an unexpected, DNA-dependent activity for this enzyme (38). Direct regulation of transcriptional activity is supported by ChIP profiles, which show enrichment of DDX5 at promoters of genes required for mitochondrial function (Fig. 3, B and C). Second, the secondary structure of a given pre-mRNA may determine the requirement for DDX5. In support of this, studies from our laboratory have shown that the budding yeast counterpart of DDX5, termed Dbp2, promotes cellular metabolism and expression of genes involved in energy homeostasis (15). Interestingly, Dbp2 appears to regulate gene expression at the level of transcription termination by remodeling mRNA structure, suggesting that the underlying secondary structure of RNA may confer Dbp2 dependence to the expression of specific genes (7). DDX5 is functionally equivalent to S. cerevisiae Dbp2 (6, 39), suggesting that DDX5 may function similarly, depending primarily on signatures that reside in the nascent gene transcript itself. A recent study showing that DDX5-binding sites are in close proximity to structured regions near exons suggests that a similar mechanism may be involved in alternative splicing (9), although is unknown whether DDX5 functions in termination in mammalian cells. Future studies are necessary to determine the precise mechanism by which DDX5 promotes respiration.
Our studies show that loss of DDX5 in SCLC cells impairs mitochondrial function, a defect that is consistent with up-regulation of mtDNA and, likely, accumulation of defective mitochondria (26). This is in contrast to our previous results in mouse liver cells whereby DDX5 knockdown leads to down-regulation of glycolysis and up-regulation of respiration (6). DDX5 expression is below detection by Western blotting in normal lung epithelial HBEC cells and tested NSCLC cells (Fig. 1A), suggesting that DDX5 is not required for the growth of these cell lines. Similarly, ablation of DDX5 expression in mESC or human MCF-7 cells does not impact cell proliferation, and knockdown in HeLa cells (to 5% of control siRNA) only slightly alters the proliferation rate (2, 40, 41). DDX5 expression is developmentally regulated and is not detectable in mouse embryos until embryonic day 11.5 (42). Moreover, DDX5 is dispensable for normal hematopoiesis and tissue functions in adult mice (13) and for the function of mature adipocytes, despite being required to initiate adipogenesis (43). These data suggest that DDX5 is dispensable in many, fully differentiated cell types, and we reason that the differences are likely due to cell type–/tissue-specific metabolism (i.e. mouse hepatocyte versus transformed human lung cell). For example, the hepatocyte-specific glucose transporter 2 and glucokinase regulator encode proteins that mediate glucose import and regulate glucose metabolism through inhibiting glucokinase, respectively (44, 45). We also observed down-regulation of glycolysis in S. cerevisiae cells upon deletion of DBP2 (15), consistent with the fact that budding yeast also rely predominantly on glycolysis over respiration when glucose is abundant (46).
In addition to mitochondrial dysfunction, we also observed a striking decrease in succinate accumulation in SCLC cells upon DDX5 knockdown. This decrease in necessary metabolites may account for loss of respiration and slowed growth because succinate is necessary for both oxidative phosphorylation and the TCA cycle (Fig. 5B). Interestingly, succinate has been described as an oncometabolite, along with fumarate and 2-hydroxyglutarate, that functions both in primary metabolism but also facilitates epigenetic changes by inhibiting α-ketoglutarate dioxygenases (47). Tumors with succinate dehydrogenase mutations that cause succinate accumulation are highly metastatic and aggressive, displaying decreased expression of genes needed for cellular differentiation and/or stabilizing HIF1α, which promotes cell growth and survival (48, 49). One possible explanation for the accumulation of succinate in SCLC cells involves tumor necrosis factor receptor–associated protein 1 (TRAP1), a member of the heat shock protein 90 family shown to be overexpressed in SCLC and associated with drug resistance in various cancers (47, 50, 51). TRAP1 down-regulates SDH activity and induces succinate accumulation and HIF1α stabilization (52). Regulation of TRAP1 expression by DDX5 could then help explain the decrease of succinate accumulation and proliferation observed upon DDX5 knockdown. Future studies are needed to determine both how DDX5 promotes succinate accumulation and whether reduction of succinate is both necessary and sufficient to inhibit SCLC growth and invasion.
SCLC is a highly aggressive lung cancer with a median survival of 9 months that encompasses ∼15% of all lung cancer cases worldwide (RRID:SCR_018570). There are currently no effective treatment options or early detection methods to date, making SCLC a major health challenge around the world. Studying SCLC has been a challenge for the community because of the dearth of patient samples, however, with characterization of cell lines and limited tissue samples providing the majority of mechanistic insight. Understanding the underlying mechanisms of cellular transformation and establishment of chemoresistance in SCLC requires defining key pathways that support this aggressive cancer with the goal of developing future targeted chemotherapies.
Experimental procedures
Plasmids and shRNAs
Plasmids and primers used for cloning and expression can be found in Tables S1 and S2, respectively. DDX5 cDNA (Gene ID: 1655, Dharmacon: 3528578) was subcloned into the PAcGFP1-Hyg-C1 vector to generate pAcGFP-Hyg-DDX5, using PCR primers HindIII DDX5 forward and SalI DDX5 reverse. The shDDX5 (sh2)–resistant pGFP–DDX5 was then generated using site-directed mutagenesis with primers shDDX5-resistant DDX5 forward and reverse.
The SMARTvector-inducible lentiviral shRNAs (Dharmacon) were used for depleting DDX5. A nonsilencing shRNA was used as a control (VSC11651), and two shRNAs were used to target DDX5 (V3SH11252): shDDX5 (ATCCAATCCACTTAGAGCA) and shDDX5-2 (TTAGAACCCAGTCACGCTC). We generated the lentiviral particles for transduction of the shRNAs using the Trans-lentiviral packaging system (Dharmacon, TLP5917).
Cell culture, transfection, and lentivirus transduction
H69 and H69AR cells were obtained from the American Type Culture Collection and cultured as instructed. The HBEC-3KT (HBEC) cell line was a gift from Dr. Andrea Kasinski (Purdue University) and was cultured with Keratinocyte-SFM medium (Gibco, 17005042). Transfection was performed with Lipofectamine 2000 reagent (Invitrogen, 11668027). For lentivirus transduction (TU), a multiplicity of infection of <0.3 TU/cell was used to prevent multiple genome insertion events. Briefly, the cells were mixed with 6 µg/ml Polybrene and virus in the absence of serum (fetal bovine serum). 10% or 20% fetal bovine serum was added to the mixture after a 6-h incubation in a humidified CO2 incubator for H69 or H69AR cells, respectively. After 24 h, the virus-containing medium was replaced with fresh medium. For H69 cells, the cell–virus mixture was centrifuged at 300 × g for 1 h at 25 °C before being put into the incubator.
Western blotting
The cells were lysed using 1× lysis buffer (Cell Signaling, 9803) with the addition of protease inhibitor (Roche, 11873580001). Protein levels were analyzed using the following antibodies: anti-DDX5 (Millipore, 05-850), anti–β-actin (Sigma, A5441), anti-PARP (Cell Signaling, 9532), anti–N-Myc (Abiocode, R1281-2), anti–c-Myc (Cell Signaling, D84C12), anti-Glut1 (Santa Cruz, SC377228), anti-CD151 (Santa Cruz, SC271216), and anti-Bcl-2 (Santa Cruz, SC7382).
Growth analysis
To measure growth in normal culture conditions, H69AR cells were treated with 1 µg/ml doxycycline for 3 days and subsequently seeded at 400 cells/well in 96-well plates (Corning, 3603). Live cells were labeled with DNA-binding fluorescent dye on indicated days using the CyQUANT® direct cell proliferation kit (Thermo Fisher, C35011) during an 8-day period. We used the relative fluorescence to infer the cell count. Background fluorescence from unlabeled cells was subtracted from the relative fluorescence. Doubling times were calculated using GraphPad Prism.
Soft agar assays were conducted as described (53). H69 (10,000 cells/well) or H69AR (5000 cells/well) were treated with 1 µg/ml doxycycline for 3 days and then seeded in 6-well plates. Assays with H69 cells were incubated for 30 days, and assays with H69AR cells were incubated for 21 days.
RNA-Seq and data analysis
Three biological replicates of the H69AR cells stably transfected with shCtr or shDDX5 were used for RNA-Seq. Total RNA was extracted using TRIzol (Thermo Fisher, 15596026), treated with DNase (Thermo Fisher, AM2238), and then subjected to library preparation using the TruSeq® stranded total RNA library prep kit (Illumina, 20020596). Unique dual index adapters (TruSeq RNA UD Indexes Illumina, 20022371) were substituted for the single index adapters. Libraries were clustered on Illumina NovaSeq S4 (300 cycle) cassettes for paired-end, 150-bp read sequencing.
Data quality control (minimum Phred score, 30; minimum read length, 50 bp) was performed using the TrimGalore toolkit (version 0.4.4) (RRID:SCR_011847). Quality trimmed reads were mapped to human reference genome (GRCh38) downloaded from the Ensembl database, using the STAR aligner (version 2.5.4b) (54). Reads aligned to each gene feature were counted via HTSeq package, followed by differential expression analysis using DESeq2 and edgeR methods (55, 56). Genes with FDR ≤ 0.05 and differential expression by both methods were denoted as significant. Functional and pathway analysis was performed with 6727 significant DEGs using IPA (57). Pathways with IPA assigned p values of <0.05 were denoted as significant, and pathway direction was determined by assigned z scores.
DDX5 ChIP-Seq reanalysis
The DDX5 ChIP-Seq data were derived from published data sets (25). Raw reads of DDX5 ChIP and input were downloaded from Gene Expression Omnibus with accession number GSE24126. Adaptor sequences were removed using Trimmomatic (version 0.36). Reads were aligned to the human genome (GRCh38) using bowtie (version 1.2.2). Duplications are marked and removed using Picard tools (version 1.9). The peaks were determined by MACS2 narrow peak calling (version 2.1.2) with FDR (q value) cutoff of 0.05. The figure of peaks and signal intensity was generated through Gviz R package (version 1.28.1).
RT-qPCR
Total RNA was extracted and subjected to reverse transcription using the QuantiTect kit (Qiagen, 205310). qPCR was conducted using the SYBR green master mix (Applied Biosystems, 4309155) to analyze the expression levels of specific mature mRNAs using primer pairs spanning exon-junctions (Table S3). The relative expression level is calculated using the ΔΔCq method using glyceraldehyde-3-phosphate dehydrogenase as the reference gene.
EdU incorporation
H69AR cells were seeded in 96-well plates at 40000 cells/well 24 h prior to the experiment. The cells were incubated with 10 μm EdU for 4 h, fixed with 3.7% formaldehyde, and permeabilized. The relative amount of incorporated EdU was then quantified using the Click-iTTM EdU Alexa FluorTM 647 HCS assay (Invitrogen, C10356).
Metabolic analysis
To measure extracellular flux, 20,000 cells/well of H69AR cells or 40,000 cells/well of HBEC cells were seeded in 24-well plates. H69AR cells were treated with 1 µg/ml doxycycline for 3 days before seeding. The respiration profiles of these cells were measured using the Seahorse XFe24 metabolic flux analyzer according to the Mito-stress test kit (Agilent, 103015-100) protocols. Basal glycolysis rates were measured at the same time, but before adding any drug from the kit. Data analysis was performed using the Seahorse report generator (Agilent). For assays with metformin treatment, metformin was added to the cells 24 h prior to the experiment. H69 cells could not be analyzed because they are nonadherent and form aggregates.
For lactate quantification, H69AR cells were treated with 1 µg/ml doxycycline for 3 days and then seeded in 6-well plates. After 40 h, the cells were washed using PBS and lysed. Intracellular lactate concentrations were measured according to the lactate assay kit (Sigma, MAK064).
Mitochondrial morphology characterization
H69AR cells stably transfected with doxycycline-inducible shRNAs targeting DDX5 or with a nontargeting control were treated with 1 µg/ml doxycycline for 48 h to induce shRNA expression. Both cell lines were seeded at 0.5 × 106 cells/well onto polylysine-treated coverslips in a 6-well plate and left to grow overnight with 1 µg/ml doxycycline. The cells were then treated with 500 nm MitoTracker Deep Red (Thermo Fisher Scientific, M22426) for mitochondrial visualization, according to the manual. The cells were fixed using 0.6% formaldehyde, and DNA was stained with 300 nm DAPI to serve as a nuclear marker. The cells were visualized using a Leica DM6 microscope with a 40× objective.
DAPI-stained nuclei were measured to compare cell size, because typical nuclei boundaries are more easily defined than the irregularly shaped outer boundaries of the cell. Leica LAS X software was utilized for nuclei measurement.
TCA metabolite profiling
Sample preparation
H69AR cells were grown in the presence of 1 µg/ml doxycycline for 3 days, and 100 mg of cells were pelleted and washed. A Bligh–Dyer extraction, a standard procedure used to isolate total lipid fractions from biological samples (58), was performed to lyse the cells and extract the TCA cycle intermediates. 300 µl of methanol and 150 µl of water was added to the microcentrifuge tube, followed by sonication for 5 min. 300 µl of chloroform and 150 µl water was added. The samples were vortexed and then centrifuged. The top layer was lyophilized and reconstituted in 100 µl of 50% acetonitrile and 50% water prior to HPLC–MS/MS analysis.
HPLC–MS/MS
TCA cycle intermediates were analyzed as described (29), using an Agilent 1290 Infinity II HPLC and 6470 triple quadrupole mass spectrometer. An Agilent HILIC-Z (2.1 × 150 mm, 2.7 μm) column was used at a flow rate of 0.3 ml/min. Mobile phase A was 90% acetonitrile, 5% isopropanol, and 5% 200 mm ammonium acetate in water at pH 9. Mobile phase B was 90% water, 5% isopropanol, and 5% 200 mm ammonium acetate in water at pH 9. A linear gradient elution was used as follows: initial conditions 5% B; 0–5 min: isocratic at 5% B; 5–15 min: gradient to 100% B; 15–16 min: isocratic at 100% B. Column re-equilibration was 16–17 min: gradient to 5% B; 17–20 min: isocratic at 5% B. Effluent was transferred to the mass spectrometer using negative electrospray ionization. Identification was based on retention time of authentic standards (pure TCA metabolites) and multiple reaction monitoring.
Data availability
All of the data described herein is within this article. RNA-Seq data have been deposited in the NCBI Gene Expression Omnibus database (GSE142024). The reagents, cell lines, plasmids, etc., are available upon request by contacting the corresponding author.
Supplementary Material
Acknowledgments
We thank Dr. Bruce Cooper at the Purdue Metabolite Profiling facility for running the HPLC–MS/MS experiments and analyzing the results. We also thank the Purdue Genomics Core facility and Dr. Nadia Atallah at the Collaborative Core for Cancer Bioinformatics, the Walther Cancer Foundation, Purdue University Center for Cancer Research, for helping with the RNA-Seq and data analysis and members of the Tran lab for critical reading and analysis of this manuscript. Special thanks goes to Kirsten Westerhouse for assisting with tissue culture work. The HBEC cell line and NSCLC cell lysates were gifts from Dr. Andrea Kasinski, Purdue University.
This article contains supporting information.
Author contributions—Z. X. and E. J. T. conceptualization; Z. X. and M. P. R. data curation; Z. X. validation; Z. X. and M. P. R. investigation; Z. X. and S. M. U. methodology; Z. X., M. P. R., and S. M. U. writing-original draft; M. P. R. and E. J. T. writing-review and editing; S. M. U. formal analysis; E. J. T. supervision; E. J. T. funding acquisition.
Funding and additional information—This work was supported by funds from the Department of Biochemistry and Purdue University Center for Cancer Research through National Institutes of Health Grant P30 CA023168 and by a kind gift from Mary Slevin in honor of her parents. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ChIP-Seq
- ChIP sequencing
- SCLC
- small cell lung cancer
- NSCLC
- non–small cell lung cancer
- HBEC
- HBEC-3KT
- shRNA
- short hairpin RNA
- shDDX5
- DDX5-targeting shRNA
- shCtr
- nontargeting shRNA
- siRNA
- small interfering RNA
- EdU
- 5-ethynyl-2′-deoxyuridine
- IPA
- Ingenuity Pathway Analysis
- OxPhos
- oxidative phosphorylation
- TSS
- transcription start site
- DEG
- differentially expressed gene
- mtDNA
- mitochondrial DNA
- TRAP1
- tumor necrosis factor receptor–associated protein 1
- c-Myc
- Myc proto-oncogene protein
- N-Myc
- neuroblastoma Myc
- PARP
- poly(ADP-ribose) polymerase
- GFP
- green fluorescent protein
- RNA-Seq
- RNA sequencing
- FDR
- false discovery rate
- qPCR
- quantitative PCR
- DAPI
- 4′,6′-diamino-2-phenylindole
- TCA
- tricarboxylic acid
References
- 1. Shin S., Rossow K. L., Grande J. P., and Janknecht R. (2007) Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 67, 7572–7578 10.1158/0008-5472.CAN-06-4652 [DOI] [PubMed] [Google Scholar]
- 2. Wortham N. C., Ahamed E., Nicol S. M., Thomas R. S., Periyasamy M., Jiang J., Ochocka A. M., Shousha S., Huson L., Bray S. E., Coombes R. C., Ali S., and Fuller-Pace F. V. (2009) The DEAD-box protein p72 regulates ERα-/oestrogen-dependent transcription and cell growth and is associated with improved survival in ERα-positive breast cancer. Oncogene 28, 4053–4064 10.1038/onc.2009.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Du C., Li D. Q., Li N., Chen L., Li S. S., Yang Y., Hou M. X., Xie M. J., and Zheng Z. D. (2017) DDX5 promotes gastric cancer cell proliferation in vitro and in vivo through mTOR signaling pathway. Sci. Rep. 7, 42876 10.1038/srep42876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clark E. L., Hadjimichael C., Temperley R., Barnard A., Fuller-Pace F. V., and Robson C. N. (2013) p68/Ddx5 supports β-catenin & RNAP II during androgen receptor mediated transcription in prostate cancer. PLoS One 8, e54150 10.1371/journal.pone.0054150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xing Z., Ma W. K., and Tran E. J. (2019) The DDX5/Dbp2 subfamily of DEAD-box RNA helicases. Wiley Interdiscip. Rev. RNA 10, e1519 10.1002/wrna.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xing Z., Wang S., and Tran E. J. (2017) Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with S. cerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism. RNA 23, 1125–1138 10.1261/rna.060335.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lai Y. H., Choudhary K., Cloutier S. C., Xing Z., Aviran S., and Tran E. J. (2019) Genome-wide discovery of DEAD-box RNA helicase targets reveals RNA structural remodeling in transcription termination. Genetics 212, 153–174 10.1534/genetics.119.302058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., and Wu J. Y. (2011) RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell Biol. 31, 1812–1821 10.1128/MCB.01149-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee Y. J., Wang Q., and Rio D. C. (2018) Coordinate regulation of alternative pre-mRNA splicing events by the human RNA chaperone proteins hnRNPA1 and DDX5. Genes Dev. 32, 1060–1074 10.1101/gad.316034.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clark E. L., Coulson A., Dalgliesh C., Rajan P., Nicol S. M., Fleming S., Heer R., Gaughan L., Leung H. Y., Elliott D. J., Fuller-Pace F. V., and Robson C. N. (2008) The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 68, 7938–7946 10.1158/0008-5472.CAN-08-0932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang L., Lin C., and Liu Z. R. (2006) P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from β-catenin. Cell 127, 139–155 10.1016/j.cell.2006.08.036 [DOI] [PubMed] [Google Scholar]
- 12. Arun G., Akhade V. S., Donakonda S., and Rao M. R. (2012) mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol. Cell Biol. 32, 3140–3152 10.1128/MCB.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mazurek A., Park Y., Miething C., Wilkinson J. E., Gillis J., Lowe S. W., Vakoc C. R., and Stillman B. (2014) Acquired dependence of acute myeloid leukemia on the DEAD-box RNA helicase DDX5. Cell Rep. 7, 1887–1899 10.1016/j.celrep.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beck Z. T., Cloutier S. C., Schipma M. J., Petell C. J., Ma W. K., and Tran E. J. (2014) Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics 198, 1001–1014 10.1534/genetics.114.170019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S., Xing Z., Pascuzzi P. E., and Tran E. J. (2017) Metabolic adaptation to nutrients involves coregulation of gene expression by the RNA helicase Dbp2 and the Cyc8 corepressor in Saccharomyces cerevisiae. G3 (Bethesda) 7, 2235–2247 10.1534/g3.117.041814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo J., Hong F., Loke J., Yea S., Lim C. L., Lee U., Mann D. A., Walsh M. J., Sninsky J. J., and Friedman S. L. (2010) A DDX5 S480A polymorphism is associated with increased transcription of fibrogenic genes in hepatic stellate cells. J. Biol. Chem. 285, 5428–5437 10.1074/jbc.M109.035295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pavlova N. N., and Thompson C. B. (2016) The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gazdar A. F., Bunn P. A., and Minna J. D. (2017) Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer 17, 765–765 10.1038/nrc.2017.106 [DOI] [PubMed] [Google Scholar]
- 19. Asai N., Ohkuni Y., Kaneko N., Yamaguchi E., and Kubo A. (2014) Relapsed small cell lung cancer: treatment options and latest developments. Ther. Adv. Med. Oncol. 6, 69–82 10.1177/1758834013517413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teicher B. A. (2014) Targets in small cell lung cancer. Biochem. Pharmacol. 87, 211–219 10.1016/j.bcp.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 21. DeBerardinis R. J., and Chandel N. S. (2016) Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koppenol W. H., Bounds P. L., and Dang C. V. (2011) Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 10.1038/nrc3038 [DOI] [PubMed] [Google Scholar]
- 23. Fuller-Pace F. V. (2013) DEAD box RNA helicase functions in cancer. RNA Biol. 10, 121–132 10.4161/rna.23312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cohen A. A., Geva-Zatorsky N., Eden E., Frenkel-Morgenstern M., Issaeva I., Sigal A., Milo R., Cohen-Saidon C., Liron Y., Kam Z., Cohen L., Danon T., Perzov N., and Alon U. (2008) Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 10.1126/science.1160165 [DOI] [PubMed] [Google Scholar]
- 25. Yao H., Brick K., Evrard Y., Xiao T., Camerini-Otero R. D., and Felsenfeld G. (2010) Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 10.1101/gad.1967810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shapovalov Y., Hoffman D., Zuch D., de Mesy Bentley K. L., and Eliseev R. A. (2011) Mitochondrial dysfunction in cancer cells due to aberrant mitochondrial replication. J. Biol. Chem. 286, 22331–22338 10.1074/jbc.M111.250092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Asins G., Serra D., and Hegardt F. G. (1991) Isolation and partial characterization of a protein with HMG-CoA reductase phosphatase activity associated with rat liver microsomal membranes. J. Lipid Res. 32, 1391–1401 [PubMed] [Google Scholar]
- 28. Thomas W. D., Zhang X. D., Franco A. V., Nguyen T., and Hersey P. (2000) TNF-related apoptosis-inducing ligand-induced apoptosis of melanoma is associated with changes in mitochondrial membrane potential and perinuclear clustering of mitochondria. J. Immunol. 165, 5612–5620 10.4049/jimmunol.165.10.5612 [DOI] [PubMed] [Google Scholar]
- 29. Al Kadhi O., Melchini A., Mithen R., and Saha S. (2017) Development of a LC-MS/MS method for the simultaneous detection of tricarboxylic acid cycle intermediates in a range of biological matrices. J. Anal. Methods Chem. 2017, 5391832 10.1155/2017/5391832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tretter L., Patocs A., and Chinopoulos C. (2016) Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim. Biophys. Acta 1857, 1086–1101 10.1016/j.bbabio.2016.03.012 [DOI] [PubMed] [Google Scholar]
- 31. Kalainayakan S. P., FitzGerald K. E., Konduri P. C., Vidal C., and Zhang L. (2018) Essential roles of mitochondrial and heme function in lung cancer bioenergetics and tumorigenesis. Cell Biosci. 8, 56 10.1186/s13578-018-0257-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jarmoskaite I., and Russell R. (2011) DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip. Rev. RNA 2, 135–152 10.1002/wrna.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fuller-Pace F. V. (2013) The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim. Biophys. Acta 1829, 756–763 10.1016/j.bbagrm.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 34. Mohibi S., Chen X., and Zhang J. (2019) Cancer the'RBP'eutics–RNA-binding proteins as therapeutic targets for cancer. Pharmacol. Ther. 203, 107390 10.1016/j.pharmthera.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joshi S., Tolkunov D., Aviv H., Hakimi A. A., Yao M., Hsieh J. J., Ganesan S., Chan C. S., and White E. (2015) The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 13, 1895–1908 10.1016/j.celrep.2015.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weinberg F., Hamanaka R., Wheaton W. W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G. M., Budinger G. R., and Chandel N. S. (2010) Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 107, 8788–8793 10.1073/pnas.1003428107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rahman M., and Hasan M. R. (2015) Cancer metabolism and drug resistance. Metabolites 5, 571–600 10.3390/metabo5040571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu G., Xing Z., Tran E. J., and Yang D. (2019) DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 116, 20453–20461 10.1073/pnas.1909047116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iggo R. D., Jamieson D. J., MacNeill S. A., Southgate J., McPheat J., and Lane D. P. (1991) p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol. Cell Biol. 11, 1326–1333 10.1128/MCB.11.3.1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H., Lai P., Jia J., Song Y., Xia Q., Huang K., He N., Ping W., Chen J., Yang Z., Li J., Yao M., Dong X., Zhao J., Hou C., et al. (2017) RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell 20, 571 10.1016/j.stem.2017.03.014 [DOI] [PubMed] [Google Scholar]
- 41. Jalal C., Uhlmann-Schiffler H., and Stahl H. (2007) Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 35, 3590–3601 10.1093/nar/gkm058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stevenson R. J., Hamilton S. J., MacCallum D. E., Hall P. A., and Fuller-Pace F. V. (1998) Expression of the “dead box” RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J. Pathol. 184, 351–359 [DOI] [PubMed] [Google Scholar]
- 43. Ramanathan N., Lim N., and Stewart C. L. (2015) DDX5/p68 RNA helicase expression is essential for initiating adipogenesis. Lipids Health Dis. 14, 160 10.1186/s12944-015-0163-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karim S., Adams D. H., and Lalor P. F. (2012) Hepatic expression and cellular distribution of the glucose transporter family. World J. Gastroenterol. 18, 6771–6781 10.3748/wjg.v18.i46.6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raimondo A., Rees M. G., and Gloyn A. L. (2015) Glucokinase regulatory protein: complexity at the crossroads of triglyceride and glucose metabolism. Curr. Opin. Lipidol. 26, 88–95 10.1097/MOL.0000000000000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Diaz-Ruiz R., Rigoulet M., and Devin A. (2011) The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta 1807, 568–576 10.1016/j.bbabio.2010.08.010 [DOI] [PubMed] [Google Scholar]
- 47. Dalla Pozza E., Dando I., Pacchiana R., Liboi E., Scupoli M. T., Donadelli M., and Palmieri M. (2020) Regulation of succinate dehydrogenase and role of succinate in cancer. Semin. Cell Dev. Biol. 98, 4–14 10.1016/j.semcdb.2019.04.013 [DOI] [PubMed] [Google Scholar]
- 48. Selak M. A., Armour S. M., MacKenzie E. D., Boulahbel H., Watson D. G., Mansfield K. D., Pan Y., Simon M. C., Thompson C. B., and Gottlieb E. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7, 77–85 10.1016/j.ccr.2004.11.022 [DOI] [PubMed] [Google Scholar]
- 49. Zhao T., Mu X., and You Q. (2017) Succinate: an initiator in tumorigenesis and progression. Oncotarget 8, 53819–53828 10.18632/oncotarget.17734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee J. H., Kang K. W., Kim J. E., Hwang S. W., Park J. H., Kim S. H., Ji J. H., Kim T. G., Nam H. Y., Roh M. S., Lee E. H., Park M. I., Kim M. S., and Lee H. W. (2015) Differential expression of heat shock protein 90 isoforms in small cell lung cancer. Int. J. Clin. Exp. Pathol. 8, 9487–9493 [PMC free article] [PubMed] [Google Scholar]
- 51. Matassa D. S., Agliarulo I., Avolio R., Landriscina M., and Esposito F. (2018) TRAP1 regulation of cancer metabolism: dual role as oncogene or tumor suppressor. Genes (Basel) 9, 195 10.3390/genes9040195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sciacovelli M., Guzzo G., Morello V., Frezza C., Zheng L., Nannini N., Calabrese F., Laudiero G., Esposito F., Landriscina M., Defilippi P., Bernardi P., and Rasola A. (2013) The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metab. 17, 988–999 10.1016/j.cmet.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Borowicz S., Van Scoyk M., Avasarala S., Karuppusamy Rathinam M. K., Tauler J., Bikkavilli R. K., and Winn R. A. (2014) The soft agar colony formation assay. J. Vis. Exp. e51998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Krämer A., Green J., Pollard J. Jr., and Tugendreich S. (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- 59. Shen L., and Sinai M. (2019) GeneOverlap: test and visualize gene overlaps. R package version 1.22.0, RRID:SCR_018419
- 60. Gu Z., Eils R., and Schlesner M. (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 61. Chen H. (2018) VennDiagram: generate high-resolution Venn and Euler plots. R package version 1.6.20, RRID:SCR_002414
- 62. Hahne F., and Ivanek R. (2016) Statistical genomics: methods and protocols. In Visualizing Genomic Data Using Gviz and Bioconductor (Mathé E., and Davis S., eds) pp. 335–351, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 63. Ramírez F., Dündar F., Diehl S., Grüning B. A., and Manke T. (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data described herein is within this article. RNA-Seq data have been deposited in the NCBI Gene Expression Omnibus database (GSE142024). The reagents, cell lines, plasmids, etc., are available upon request by contacting the corresponding author.