Abstract
Findings from recent studies have indicated that enzymes containing more than one catalytic domain may be particularly powerful in the degradation of recalcitrant polysaccharides such as chitin and cellulose. Some known multicatalytic enzymes contain several glycoside hydrolase domains and one or more carbohydrate-binding modules (CBMs). Here, using bioinformatics and biochemical analyses, we identified an enzyme, Jd1381 from the actinobacterium Jonesia denitrificans, that uniquely combines two different polysaccharide-degrading activities. We found that Jd1381 contains an N-terminal family AA10 lytic polysaccharide monooxygenase (LPMO), a family 5 chitin-binding domain (CBM5), and a family 18 chitinase (Chi18) domain. The full-length enzyme, which seems to be the only chitinase produced by J. denitrificans, degraded both α- and β-chitin. Both the chitinase and the LPMO activities of Jd1381 were similar to those of other individual chitinases and LPMOs, and the overall efficiency of chitin degradation by full-length Jd1381 depended on its chitinase and LPMO activities. Of note, the chitin-degrading activity of Jd1381 was comparable with or exceeded the activities of combinations of well-known chitinases and an LPMO from Serratia marcescens. Importantly, comparison of the chitinolytic efficiency of Jd1381 with the efficiencies of combinations of truncated variants—JdLPMO10 and JdCBM5-Chi18 or JdLPMO10-CBM5 and JdChi18—indicated that optimal Jd1381 activity requires close spatial proximity of the LPMO10 and the Chi18 domains. The demonstration of intramolecular synergy between LPMOs and hydrolytic enzymes reported here opens new avenues toward the development of efficient catalysts for biomass conversion.
Keywords: ascorbic acid, carbohydrate-binding protein, carbohydrate metabolism, cellulose, chitin, chitinase, copper monooxygenase, LPMO, lytic polysaccharide monooxygenase, multimodular
Introduction
The enzymatic conversion of recalcitrant polysaccharides, such as chitin and cellulose, requires the combined action of several enzymes. For chitin conversion, the key enzymes involved are endochitinases, processive exochitinases, lytic polysaccharide monooxygenases (LPMOs), and β-glucosaminidases (1), and analogous enzymes are used for cellulose conversion (2). Each of these enzymes may contain one or more carbohydrate-binding modules (CBMs) that promote substrate binding. In recent decades, the enzymatic conversion of cellulose has been studied in much detail, at both the fundamental and the industrial levels (3), whereas less is known about chitin conversion. It is clear, however, that there are similarities between the challenges met by chitin- or cellulose-converting enzyme systems and between the enzymatic solutions that nature has developed (4, 5).
Recently, several reports have appeared showing that single enzymes containing more than one carbohydrate-active catalytic unit may be particularly powerful in degrading cellulose or chitin. Brunecky et al. (6, 7) studied a multidomain cellulase from the thermophilic bacterium Caldicellulosiruptor bescii comprising a GH9 and a GH48 cellulase and three cellulose-binding CBMs. They found that under certain conditions, this single enzyme performed as well as highly developed commercial cellulase cocktails. In another study, Larsbrink et al. (8) showed that the efficiency of a chitin-targeting enzyme system encoded by a polysaccharide utilization locus of Flavobacterium johnsoniae depends on one single secreted 155.5-kDa multidomain protein comprising two GH18 chitinases, one of which was proposed to have a predominantly endo-character, whereas the other was predicted to have an exo-processive character. The two chitinase domains are connected by a region of ∼700 residues that contains several domains predicted to be involved in substrate binding but whose precise functions remain unclear. Further studies with recombinantly produced individual GH18 domains showed synergy between the two domains but also showed that the combined individual domains were much (2–10-fold) less active than the full-length protein when acting on β-chitin or, even more so, α-chitin.
In 2005, it was shown that the activity of GHs on chitin was enhanced by addition of a protein that at the time was thought to be a substrate-disrupting CBM (9). Later it became clear that this protein, and similar proteins acting on cellulose, in fact are redox enzymes that break glycosidic bonds (10–13). Today these enzymes are referred to as lytic polysaccharide monooxygenases (LPMOs), and their discovery has revolutionized the field of enzymatic conversion of recalcitrant polysaccharides. LPMOs use oxidative chemistry to cleave polysaccharide chains while in their crystalline context. Their activity depends on the external supply of reducing power and an oxidant, which was originally thought to be oxygen (10) but which may also be hydrogen peroxide (14–16). By introducing chain breaks in crystalline parts of the substrate, LPMOs boost the activity of hydrolytic enzymes (9, 10, 17–19), and they are part of today's commercial enzyme cocktails for saccharification of cellulose (3, 20). Bacterial LPMOs are classified in family AA10 in the CAZy database (21) (AA stands for “auxiliary activity”). These LPMO10s may occur as single catalytic domains or may contain additional domains, in particular CBMs (22).
Interestingly, some LPMO10s are appended to glycoside hydrolase (GH) modules, thus potentially forming enzymes with the unprecedented ability to catalyze both hydrolytic and oxidative cleavage of polysaccharides. Concerning the known synergy between LPMOs and GHs, and the abovementioned observations concerning the beneficial effects of covalently linking GH domains, multidomain enzymes with both GH and LPMO domains could be particularly efficient. To assess this hypothesis, we have studied one such enzyme, named Jd1381, from the Gram-positive bacterium Jonesia denitrificans. Judged by its predicted amino acid sequence, this trimodular enzyme consists of a small N-terminal LPMO10 domain (JdLPMO10) followed by a CBM5, which belongs to a family of CBMs known to bind chitin (JdCBM5), and a C-terminal chitinase domain belonging to family GH18 (JdChi18). These three domains are connected by linkers of ∼26 and 28 residues with low sequence complexity (Fig. 1A; see below). Genomic and preliminary proteomic analysis (see below) indicated that Jd1381 might be the only enzyme with chitinolytic activity produced by J. denitrificans, which would make this enzyme the sole determinant of this bacterium's ability to grow on chitin.
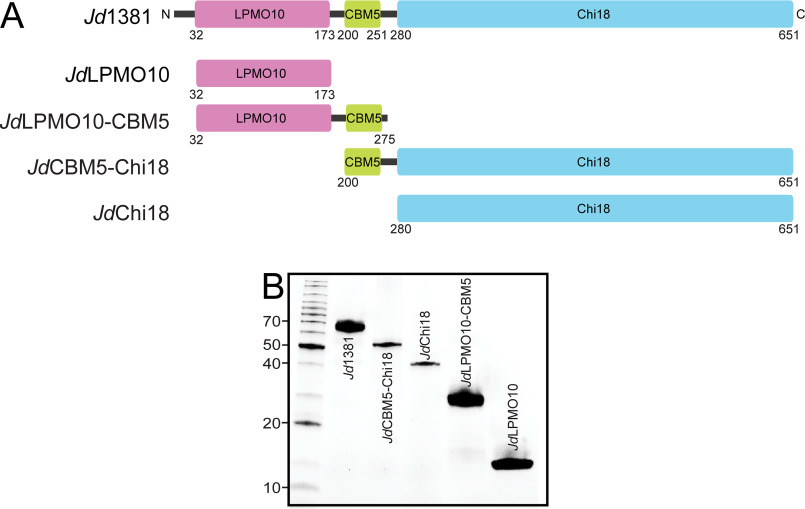
Figure 1.
Modular structure of Jd1381 and its truncated versions. A, the full-length enzyme Jd1381 contains an N-terminal signal peptide (residues 1–31; black line), an AA10 type LPMO (LPMO10, residues 32–173; purple), a CBM5 domain (residues 200–251; green), and a C-terminal GH18 type chitinase (Chi18, residues 280–651; light blue). Two linkers (black lines) connect the CBM5 domain to the LPMO10 and Chi18 domains, containing 26 and 28 residues, respectively. The four additional pictures show the domain organizations of truncated versions of Jd1381 produced in this study, named JdLPMO10, JdLPMO10-CBM5, JdCBM5-Chi18, and JdChi18. B, SDS-PAGE gel showing the recombinant enzymes produced and purified in this study. The left lane shows protein size markers with their sizes indicated in kDa.
Here we describe biochemical properties of recombinantly produced full-length Jd1381 and its truncated versions. Thus, we have analyzed how and to what extent the LPMO, the CBM5, and the Chi18 contribute to the chitin-degrading efficiency of this three-domain protein. Importantly, this approach allowed us to assess the importance of the spatial proximity of the LPMO and the GH18 in Jd1381. To understand the potential of Jd1381, we examined its degradative activity on α- and β-chitins, followed by an in-depth exploration of enzyme performance on α-chitin, the most crystalline chitin form.
Results
The genome of J. denitrificans contains only one gene annotated as chitinase, namely Jden_1381, encoding the protein here referred to as Jd1381. We observed that the bacterium grew better on β-chitin than on α-chitin. In α-chitin the polysaccharide chains are arranged an anti-parallel fashion, giving a recalcitrant material that is stabilized by a high number of hydrogen bonds, with tightly packed chains (23). In β-chitin the polysaccharide chains are arranged in a parallel fashion, which gives reduced packing tightness, fewer intrachain hydrogen bonds, and an increased number of hydrogen bonds with water (24). Because it was more easily degraded, we used β-chitin in a preliminary analysis of the secretome of J. denitrificans grown on chitin (Fig. S1 and Table S2), which showed that Jd1381 is one of the most abundantly secreted proteins.
To determine the function of each of the predicted carbohydrate active domains (LPMO, Chi18) and confirm that they are indeed free-standing domains, we recombinantly expressed the individual LPMO and Chi18 domains with and without the flanking carbohydrate binding domain (CBM5) by generating truncated versions of Jd1381 (Fig. 1A). The full-length Jd1381 and truncated variants were successfully expressed and purified as soluble proteins, suggesting that the predicted domains fold into functional catalytic units (Fig. 1B).
Three-dimensional model of the JdChi18 domain
In a previous study, the crystal structure of the LPMO domain, JdLPMO10, was determined (25, 26). Structural and functional data showed that this 142-residue LPMO domain, the smallest LPMO domain characterized so far, is a chitin-active LPMO with C1-oxidizing activity.
Using the crystal structure of the Streptomyces thermoviolaceus GH18 chitinase “chitinase 40” (Protein Data Bank entry 4W5U) as a template (68.7% sequence identity to JdChi18), a reliable three-dimensional model could be built of the GH18 domain of Jd1381 (Qmean= 0.58) (Fig. 2). The predicted structure of JdChi18 was compared with the structures of the three chitinases produced by the well-known chitinolytic bacterium Serratia marcescens, for which abundant functional data are available (1, 27–32). These chitinases are SmChi18A (exo-processive, acting from the reducing end), SmChi18B (exo-processive, acting from the nonreducing end), and SmChi18C (endo-acting, nonprocessive).
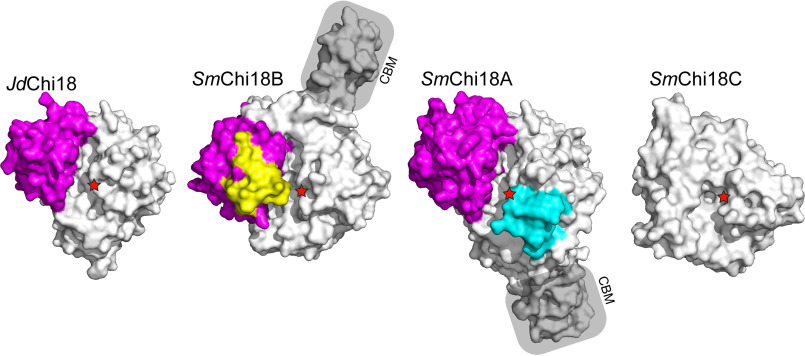
Figure 2.
Structural comparison of JdChi18 with well-studied chitinases from S. marcescens. The figure shows surface representations of the four enzymes, looking into the substrate-binding clefts of the enzymes from the top. The so-called α + β domain is colored magenta, whereas extra loops (relative to JdChi18) in SmChi18B and SmChi18A are shown in yellow and cyan, respectively. The catalytic acid is indicated by red stars in all four enzymes. CBMs are labeled and shaded. Note that for SmChi18C only the catalytic domain is shown and that this domain lacks the α + β insertion. SmChi18C has two additional domains likely involved in substrate binding that are not shown because the structure of the full-length protein is not known.
The GH18 domain of Jd1381 contains the so-called α + β insertion, which is often associated with processive exo-acting chitinases and which lacks in true endo-chitinases such as SmChi18C (27). Pairwise structural superposition of JdChi18 with SmChi18A showed that JdChi18 shares multiple surface-exposed conserved aromatic residues that are involved in substrate binding by SmChi18A and that endow this latter enzyme with processive properties (Fig. S2). Although the presence of such residues suggests strong substrate-binding and processive action, JdChi18 lacks loops that are typically found in processive exo-chitinases (Fig. 2) and that contribute to making the substrate-binding cleft “deep” and even “tunnel-like.” Thus, based on the structural model, it seems that JdChi18 is a “hybrid” between a (processive) exo-chitinase and an endo-chitinase.
Hydrolytic and oxidative degradation of chitin
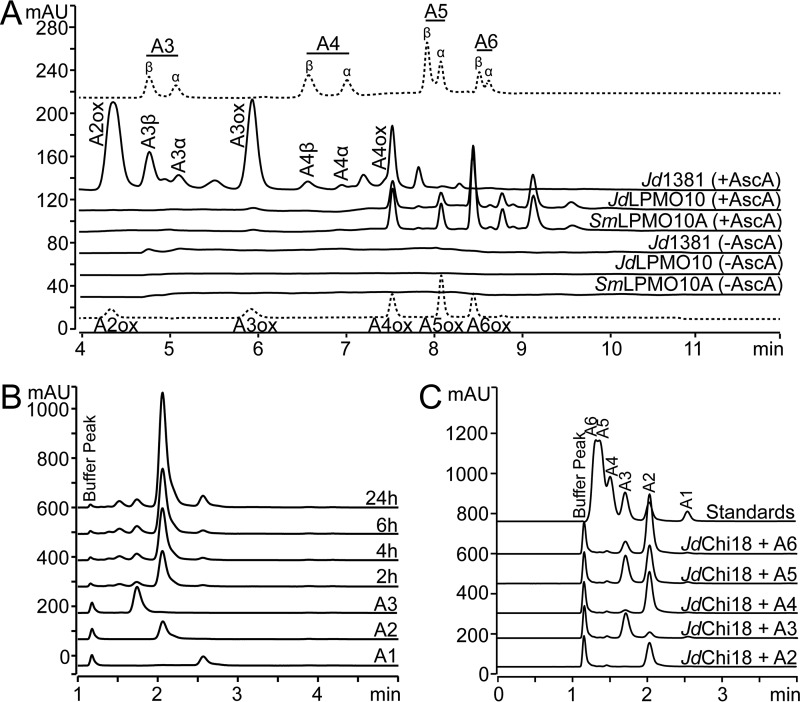
The chitinolytic activities of Jd1381 and its individual LPMO and GH18 domains were explored using α-chitin, β-chitin, and soluble chito-oligosaccharides (degree of polymerization in the range 2–6) as substrates (Fig. 3). To characterize oxidative cleavage by full-length Jd1381 and its LPMO only, purified proteins were assayed with β-chitin as substrate in the presence and absence of ascorbic acid (the reductant required for LPMO activity). The previously characterized chitin-active LPMO from S. marcescens (SmLPMO10A, also known as CBP21) was included as a positive control (Fig. 3A). The LPMO domain gave a product profile dominated by oxidized GlcNAc4–GlcNAc8, with even-numbered products being most abundant, similar to the product profile produced by S. marcescens, SmLPMO10A. Full-length Jd1381 yielded shorter products, in particular oxidized GlcNAc2, GlcNAc3, and GlcNAc4, which is likely due to the presence of the chitinase domain, which will cleave the longer oxidized products. As expected, oxidized products were only generated in reactions with added ascorbic acid. The most abundant soluble product by far in the reactions containing the chitinase, i.e. reactions with Jd1381, was chitobiose ((GlcNAc)2 or A2), as shown in Fig. 3B, discussed below. Unexpectedly, Fig. 3A shows that the accumulation of longer nonoxidized oligomeric products, which is limited relative to the accumulation of chitobiose, was higher in the reaction with ascorbic acid. This could indicate that, under the conditions used here, chitinase activity on short chito-oligosaccharides is inhibited by products of the LPMO reaction.
Figure 3.
Catalytic activity of Jd1381 and its individual domains. Chito-oligosaccharides are labeled as “An,” where n indicates the number of sugars; ox indicates oxidation. A, soluble oxidized products released by Jd1381 or JdLPMO10 or SmLPMO10A from β-chitin, analyzed by HPLC using a HILIC column. The reactions contained 1 μm Cu(II) saturated enzyme and 10 g/liter substrate in 20 mm BisTris, pH 6.0, with or without 1 mm AscA. The reactions were incubated for 24 h at 40 °C, with shaking at 1000 rpm. This experiment was repeated three times, with similar results. The upper and lower chromatograms (dotted lines) show standard mixtures containing native (A3–A6) and oxidized (A2ox–A6ox) chito-oligosaccharides, respectively. Native oligomers appear as double peaks because the α- and β-anomers are separated, as indicated. The small peaks eluting in between A3 and A3ox, between A4 and A4ox and after A5 are products of unknown nature. Note that by far the most dominant soluble product generated in the reactions with Jd1381 (regardless of the presence of AscA) was the nonoxidized dimer (chitobiose, A2, GlcNAc2), which is not visible in A and was analyzed using another chromatographic method, as shown in B. B, nonoxidized products generated from β-chitin by Jd1381, analyzed by ion-exclusion chromatography. The reaction conditions were as described above, with added ascorbic acid. The compounds eluting prior to GlcNAc2 are oxidized products. C, products generated by JdChi18 from soluble chito-oligosaccharides in reaction mixtures containing 0.5 mm substrate and 10 nm JdChi18 in 20 mm BisTris, pH 6.0, after incubation for 5 h at 40 °C. The standard mixture contained 50 μm of A1–A6. Fig. S3 shows that full-length Jd1381 and JdChi18 are equally active toward A5.
Analysis of nonoxidized products generated by Jd1381 from β-chitin showed production of large amounts of chitobiose (Fig. 3B), and a similar dominance of chitobiose was observed in reactions with α-chitin (not shown). Degradation reactions with oligomeric substrates showed that both the full-length enzyme and its recombinantly produced GH18 domain, JdChi18, readily cleave soluble chito-oligosaccharides with a degree of polymerization of four or higher, whereas GlcNAc3 is cleaved slowly (Fig. 3C). Of note, the dominance of chitobiose in reactions with β-chitin and GlcNAc6 is indicative of some degree of processivity (33). Activity assays with chitopentaose showed that full-length Jd1381 and JdChi18, i.e. the chitinase domain only, are equally active on this short oligomeric substrate and that product formation was not affected by the presence of ascorbic acid (Fig. S3). Under the conditions used, both enzyme variants displayed a rate of ∼10 s−1, which is comparable with rates determined for other GH18 chitinases using similar conditions (8, 29, 34).
The effect of the LPMO domain in chitin conversion
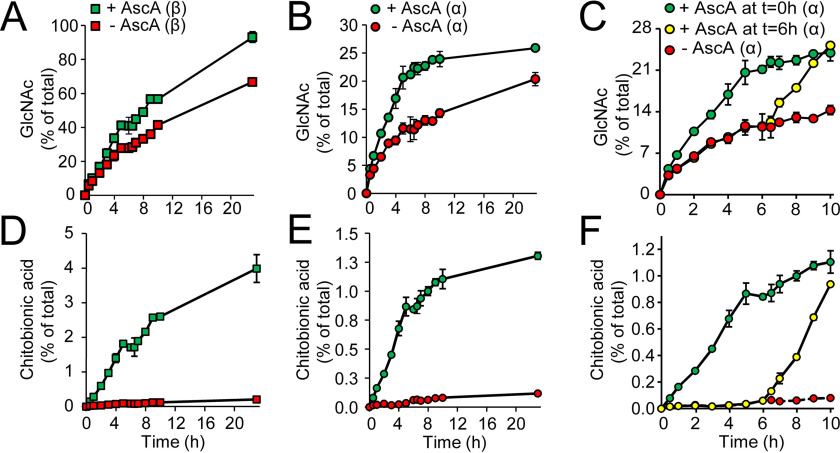
To verify that the LPMO domain in Jd1381 contributes to the efficiency of chitin conversion by Jd1381, the degradation of α- and β-chitins was assessed while controlling the activity of the LPMO domain by varying the addition of the reductant, ascorbic acid (AscA). Indeed, activation of the LPMO10 domain by addition of AscA increased the degradation of both α- and β-chitin. The increased saccharification yield upon LPMO activation (Fig. 4, A–C) was accompanied by the increased generation of oxidized products (Fig. 4, D–F). Of note, Fig. 4 (C and F) shows that addition of AscA later during the reaction (at t = 6 h instead of at t = 0 h) leads to an immediate boost in chitin saccharification (Fig. 4C) and generation of oxidized products (Fig. 4F). Reactions lacking AscA did show some LPMO activity (red symbols in Fig. 4, D–F), indicating the presence of some reducing power in the substrate, in line with previous observations (9). Thus, the LPMO was not totally inactive in the absence of AscA, which means that the real contribution of the LPMO to the overall saccharification efficiency may be larger than suggested by Fig. 4.
Figure 4.
The contribution of LPMO activity to the chitinolytic activity of Jd1381. Reaction mixtures contained 1 μm Cu(II) saturated Jd1381, 10 g/liter chitin (α or β, as indicated) and, when indicated in the Fig. 1 mm AscA, in 20 mm BisTris, pH 6.0, and were incubated at 40 °C with shaking at 1000 rpm. A–C show time courses for the solubilization of GlcNAc, and D–F show corresponding time courses for the solubilization of oxidized products. The legends shown in A–C also apply to the respective panels shown below (D–F). In the reactions shown in A, B, D, and E, AscA was added at 0 h, whereas C and F show reactions with AscA supplementation at either 0 or 6 h. Before analysis, soluble products generated by Jd1381 were converted to GlcNAc and chitobionic acid by overnight incubation with 1.5 μm SmCHB (chitobiase, an N-acetylhexosaminidase) at 37 °C (60). The data points represent the means of three independent experiments ± S.D. Incubation of these chitin substrates under the same conditions but in the absence of enzyme did not yield detectable levels of soluble products.
The progress curves in Fig. 4A show that Jd1381 with an activated LPMO domain catalyzes almost complete conversion of β-chitin after 23 h. Fig. 4D shows that 4% of the substrate solubilized after 23 h appears as chitobionic acid, which means that 2% of the glycosidic bonds has been cleaved by the LPMO domain. For α-chitin, complete conversion was not achieved, and the amount of oxidized products was lower (Fig. 4, B and E). The results show that the approximate initial conversion rate of α-chitin by Jd1381 without activation of the LPMO domain is 0.27 s−1, whereas this rate was 0.40 s−1 for fully activated Jd1381 (i.e. in the presence of AscA; see supporting text for calculations). For β-chitin, these values were 0.58 and 0.69 s−1, respectively.
The impact of spatial proximity of the LPMO and chitinase domains on catalytic efficiency of Jd1381
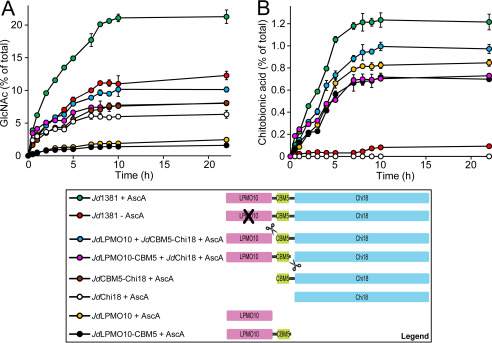
To evaluate the impact of the modular architecture of Jd1381 on the interplay between the LPMO10 and the Chi18 domains, the degradation of α-chitin by full-length Jd1381 was compared with the catalytic efficiency of mixtures of truncated versions of the protein. The results (Fig. 5) show that intact Jd1381 is more efficient in degrading α-chitin compared with mixtures of its separately produced domains. The data show that the boosting effect of the individually expressed LPMO10 domain on chitin solubilization by the individually expressed Chi18 domain is modest and that the full potential of Jd1381 is only harnessed when the two catalytic units are covalently linked.
Figure 5.
The effect of domain proximity on the efficiency of Jd1381. The panels show solubilization of GlcNAc (A) and oxidized products (B) during incubation of 10 g/liter α-chitin with 1 μm of Jd1381 with AscA (green circles) or JdCBM5-Chi18 (brown circles) or JdChi18 (white circles) or JdLPMO10 (orange circles) or JdLPMO10-CBM5 (black circles) or combinations of 1 μm JdLPMO10 and 1 μm JdCBM5-Chi18 (light blue circles) or 1 μm JdLPMO10-CBM5 and 1 μm JdChi18 (pink circles) in 20 mm BisTris (pH 6.0). The red circles show product formation in a reaction with Jd1381 without added AscA. Prior to use, all enzymes were copper-saturated, and the reactions were incubated at 40°C with shaking at 1000 rpm. Before analysis, soluble products generated by the enzymes were converted to GlcNAc and chitobionic acid by overnight incubation with 1.5 μm SmCHB (a chitobiase) at 37 °C (60). The data points represent the mean of three independent experiments ± S.D. In B, the curves representing JdCBM5-Chi18 and JdChi18 overlap; hence only one (for JdChi18) is visible.
Binding to chitin
Binding assays (Fig. S4) showed that all enzyme variants bind to α-chitin and β-chitin, albeit with different efficiencies. The chitinase domain alone, JdChi18, bound equally well to both chitin forms. For α-chitin, the LPMO domain alone showed similar binding as the chitinase domain, whereas binding toward β-chitin was somewhat weaker. Interestingly, variants containing the CBM showed considerably more binding to α-chitin, compared with the individual Chi18 and LPMO domains, whereas the CBM hardly affected binding to β-chitin. Thus, the full-length enzyme and truncated variants containing a CBM bind considerably better to α-chitin compared with β-chitin.
Comparison of Jd1381 with other chitinolytic enzyme systems
Fig. S5 shows data for hydrolysis of α-chitin by Jd1381 or by the well-known chitinases SmChi18A or SmChi18C in the absence or presence of SmLPMO10A. Importantly, the results show that Jd1381 is almost as efficient as a combination of SmChiA (reducing end exo-chitinase) and SmLPMO10A, whereas this single enzyme clearly outperforms a combination of SmChiC (endo-chitinase) and SmLPMO10A. Of note, the high performance of Jd1381 was only achieved when the LPMO domain was activated (Fig. S5).
Modification of α-chitin particles
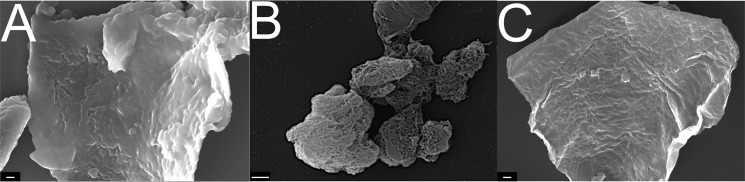
Morphological changes in cellulose upon treatment with LPMOs alone or in combination with a cellulase have been demonstrated in several studies using atomic force microscopy and scanning EM (SEM) (18, 19, 35–39). These studies have convincingly shown that LPMOs facilitate fibrillation of cellulose fibers. Here, we examined how the surface morphology of chitin particles was affected by Jd1381, using SEM. Fig. 6 shows SEM images of chitin particles before and after a 3.5-h treatment with Jd1381 in the presence and absence of AscA. The SEM images generally revealed high particle heterogeneity. The presence of AscA (i.e. a reaction in which the LPMO domain was active; Fig. 6B) led to an increased occurrence of smaller particles with pronounced indentations that were never observed in the other reactions (Fig. 6, A and C). Of note, most studies showing LPMO effects on cellulose were done with high doses of LPMO (10–20 mg/g DM) (35). Here, we used a lower enzyme concentration, namely 6.6 mg of Jd1381 per gram of DM, which corresponds to an LPMO dose of ∼1.5 mg/g DM.
Figure 6.
The impact of Jd1381 on α-chitin particles. A, SEM image showing an α-chitin particle before enzymatic treatment. B and C, SEM images showing α-chitin particles after treatment with Jd1381 in the presence (B) and absence (C) of AscA. Note that the particles shown in B were exposed to an active LPMO domain, whereas the particles shown in C were not. The reaction mixtures contained 1 μm Cu(II) saturated enzyme, 10 g/liter chitin, and, when indicated, 1 mm AscA, in 20 mm BisTris, pH 6.0, and were incubated at 40 °C with shaking at 1000 rpm for 3.5 h prior to imaging. The control reaction (A) did include AscA but lacked the enzyme. The scale bars indicate 1 μm in A and C and 2 μm in B.
Discussion
The synergistic effects of combining individually produced LPMOs and GHs during biomass conversion have been well-documented in the past (9, 10, 16–18, 20, 37, 40–43). Herein, we investigated chitin degradation by a unique trimodular enzyme, Jd1381. The enzyme contains two glycosidic bond-breaking catalytic domains (an LPMO10 and a Chi18) with distinct catalytic mechanisms. To our knowledge, this is the first study showing the degradation of carbohydrates by a natural multimodular and multicatalytic CAZyme that combines the redox chemistry catalyzed by an LPMO with the hydrolytic chemistry catalyzed by a GH.
The data above show that Jd1381 is optimized to work on polymeric, insoluble substrates, as suggested by the presence of an LPMO domain. Although it is not possible to determine the exact nature of the activity of the Chi18 domain (endo, exo, processive) based on the present data, its predicted structural features (Fig. 2 and Fig. S2) indicate that the enzyme is predominantly endo-acting. The presence of conserved tryptophan residues in the substrate-binding cleft (Fig. S2) and the strong dominance of chitobiose in the product mixtures (Fig. 3B) indicate that the chitinase binds strongly to the substrate (as confirmed by the binding studies depicted in Fig. S4) and may be processive. Indeed, JdChi18 possesses most of the conserved substrate-binding residues found in the processive chitinase SmChi18A (Fig. S2). These conserved aromatic residues include Trp293 in the −3 subsite (Trp167 in SmChi18A) that is thought to be important for steering the directionality of processive action in SmChi18A, which acts from the reducing end (28, 44). Of note, literature shows that GH18 chitinases may show combinations of functional features, like mixed endo/exo activity, and includes examples of endo-processive enzymes (32, 45). Considering its good performance in chitin degradation, it is tempting to speculate that the chitinase domain of Jd1381 combines features of multiple GH18 chitinases with complementary activities as they are encountered in the secretomes of chitin-degrading microbes such as S. marcescens (1, 31).
Fig. 4A shows that the initial rate for β-chitin degradation by Jd1381 hardly depends on activation of the LPMO but that LPMO activity becomes important for efficiency later during the reaction. However, in the case of the more recalcitrant chitin form, α-chitin, the reaction with ascorbic acid, and thus LPMO activity, performs better all the way from the start of the reaction (Fig. 4B). The role of CBMs in promoting the activity of chitinases, glycoside hydrolases in general, and LPMOs is well-established in the literature and was not investigated in detail in this study. Of note, the effect of CBM domains on LPMO performance is highly complex, because it may depend on the substrate concentration and because CBM-mediated substrate binding affects not only LPMO activity but also stability (see Ref. 46 for a detailed discussion). The chitin degradation experiments with truncated Jd1381 variants (Fig. 5) did not reveal clear effects of the presence of the CBM. Interestingly, the binding studies depicted in Fig. S4 indicate that the CBM5 has a larger effect on binding to α-chitin compared with β-chitin. It would thus seem that Jd1381 is optimized for interacting with its most likely natural substrate, which is α-chitin rather than β-chitin.
Comparison of the apparent initial enzyme rates for degradation of α-chitin, which can be estimated from data shown in Figs. 4 and 5 and Fig. S5, with initial rates derived from reactions with other chitinolytic enzyme cocktails described here (Fig. S5) or in the literature (8, 29, 31, 34, 47–49) shows that Jd1381 is an efficient enzyme. Data comparison is complicated because most published studies do not show kinetic constants, whereas the substrates (source of the chitin, particle size) and reaction conditions may vary. A rough assessment of apparent kinetic constants for individual chitinases from published progress curves with α-chitin typically yields values between 0.05 and 0.3 s−1. For example, the data published for the multiple chitinases from Cellvibrio japonicus (34) suggest that the single enzyme that is most effective on α-chitin has an apparent rate of ∼0.30 s−1. Data published for the allegedly powerful multidomain ChiA from F. johnsoniae (8) suggest an apparent initial rate below 0.1 s−1. The degradation reactions described in the present report suggest rates of ∼0.25 s−1 for SmChiA, 0.1 s−1 for SmChiC and 0.27 s−1 for Jd1381 in the absence of AscA.
The true activity of Jd1381 becomes apparent when comparing the apparent initial rate of Jd1381 in the presence of AscA, i.e. with an active LPMO domain, with comparable enzyme cocktails. The data in Fig. 4 show an apparent initial rate for fully activated Jd1381 on the order of 0.40 s−1, which corresponds to an appreciable one-third of the initial rate obtained when using an optimized mixture containing three S. marcescens chitinases, the S. marcescens LPMO, a chitobiase, and ascorbic acid, at a similar protein loading (47) (see supporting text for the underlying calculations). The data in Fig. S5 shows that activated Jd1381 is as efficient as the most efficient chitinase of S. marcescens, SmChiA, combined with SmLPMO10A, and outperforms the combination of SmChiC and SmLPMO10A. Activated Jd1381 also seems to outperform the combination SmChiC with CjLPMO10A, an LPMO that seems better suited to act on α-chitin than SmLPMO10A (Ref. 48; also see supporting text). Finally, the apparent initial rate of activated Jd1381 acting on α-chitin amounted to ∼80% of the apparent initial rate achieved by a combination of the three S. marcescens chitinases, SmChi18A, SmChi18B, and SmChi18C, with a particularly powerful four-domain chitin-active LPMO10 from Bacillus cereus named BcLPMO10A (Ref. 49; also see supporting text). Considering these observations, it is clear that trimodular Jd1381 is an efficient enzyme that, on its own, may provide J. denitrificans with the catalytic power needed to degrade and grow on chitin.
Reduced LPMOs may engage in off-pathway reactions that damage residues in the catalytic center and lead to enzyme inactivation (14, 50). The risk of inactivation is particularly high in the absence of substrate, when the reduced LPMO, in solution, may react with molecular oxygen or hydrogen peroxide to generate damaging reactive oxygen species (14, 46, 51–53). Thus, both the activity and the stability of an LPMO will benefit from the presence of a CBM, which keeps the LPMO in close proximity to the substrate, increasing the effective substrate concentration. Likewise, chitinase activity may promote LPMO activity because chitinase activity will “peel off” LPMO-oxidized chitin chains from the fibril, thus exposing new productive binding sites for the LPMO. Although not all combinations of truncated variants of Jd1381 show this latter effect, perhaps because of stability issues, this effect is indeed evident when comparing the full-length enzyme with truncated variants containing the LPMO domain only. In the presence of ascorbic acid, the full-length enzyme produced more oxidized products compared with truncated variants lacking the chitinase domain (Fig. 5B).
The present study of Jd1381 adds an important element to understanding the GH–LPMO interplay. Our findings indicate that the efficiency of Jd1381 is based on a “proximity-induced” synergy between two covalently coupled catalytic domains with totally different catalytic functions. Beneficial effects of spatial proximity of multiple GHs are known from the studies of multidomain enzymes alluded to above (6–8) and have also been observed in studies of cellulosomes, which are large multienzyme complexes used by some anaerobic bacteria to degrade cellulose (54, 55). Interestingly, Arfi et al. (43) showed that the combined efficiency of a cellulase and an LPMO was larger when the enzymes were combined in a designer cellulosome than when acting free in solution. It would be interesting to obtain further insights into the dynamics that underlie the cooperation between the covalently linked GH and LPMO domains of Jd1381. These two domains are both anchored to the substrate by the CBM5 domain in between them (Fig. 1), and one may expect that the length and nature of the two linkers have a significant impact on the overall efficiency of the trimodular enzyme. A first glimpse of such dynamics was recently provided by an NMR study of full-length ScLPMO10C, which is cellulose-active and comprises a CBM and an LPMO domain (46).
Despite the efficiency of combining hydrolytic and oxidative glycoside bond splitting activities in one protein, as in Jd1381, such an arrangement is not commonly identified in genes from bacterial genomes. On the other hand, several eukaryotic marine organisms contain genes encoding both LPMO10 and GH18 domains. Because substantially fewer eukaryotic genomes have been sequenced compared with prokaryotic genomes, it may be that such multicatalytic enzyme architectures are favored among more complex organisms. In conclusion, the challenges related to optimizing LPMO action, as discussed above and in, for example, Ref. 56, and the demonstrated efficiency of Jd1381 both indicate that multidomain enzymes comprising both oxidative and hydrolytic polysaccharide-degrading activities are among the most promising enzymes for biomass conversion.
Experimental procedures
Chemicals
BisTris buffer salt was purchased from VWR (Radnor, PA, USA); GlcNAc (purity > 99%) and ascorbic acid were purchased from Sigma–Aldrich. GlcNAc2 (purity > 95%), GlcNAc3 (purity > 95%), GlcNAc4 (purity > 95%), GlcNAc5 (purity > 95%), and GlcNAc6 (purity > 95%) were purchased from Megazyme (Bray, Ireland).
Enzymes
Enzymes from S. marcescens BJL200 encoded by chia (GenBank accession number Z36294; SmChi18A (57)), chic (AJ630582, SmChi18C (58)), cbp21 (AY665558, SmLPMO10A (59)), and chb (L43594, SmCHB (60)) were overexpressed in Escherichia coli and purified as described previously (47). The enzyme concentrations were determined using a Bradford protein assay kit (Bio-Rad).
Chitinous substrates
α-Chitin from shrimp (Pandalus borealis) shells was purchased from Chitinor AS (Senjahopen, Norway) and ball-milled using a planetary ball mill PM 100 (Retsch, Haan, Germany) with a stainless-steel jar and zirconium oxide balls. The chitin powder was ground at 450 rpm for 15 min with 5-min milling periods being interrupted by 2-min pauses, to avoid excess heating, to a final average particle size of ∼0.2 mm. β-Chitin from squid pen with an average particle size of 0.8 mm was purchased from France Chitin (Orange, France). Chito-oligosaccharides were purchased from Megazyme (Bray, Ireland).
Cloning, expression, and purification
For production of full-length Jd1381, the gene encoding mature Jd1381 (residues 32–651, UniProt ID; C7R4I0) was codon optimized for E. coli expression, de novo synthesized, and cloned behind an IPTG-inducible T5 promoter in the pD441-CH expression vector by ATUM (Newark, CA, USA) as a fusion construct with an N-terminal E. coli OmpA signal peptide and a C-terminal His6 motif (Gly-(His)6). The expression vector was transformed into chemically competent E. coli BL21 (New England Biolabs). Note that the natural signal peptide of Jd1381 was replaced by the E. coli OmpA signal peptide. For production of truncated versions of Jd1381, six primers (Table S1) incorporating a BamHI or a NotI restriction site (for JdLPMO10 and JdLPMO-CBM5) or a NdeI or a XhoI restriction site (for JdCBM5-Chi18 and JdChi18) were used. The four generated PCR fragments encoded JdLPMO10, the LPMO catalytic domain only (residues 32–173), JdLPMO10-CBM5, the LPMO and the CBM5 domain (residues 32–275), JdCBM5-Chi18, the CBM5 and the Chi18 domain (residues 200–651), and JdChi18, the Chi18 catalytic domain only (residues 280–651); see Fig. 1 for an overview.
For cloning JdLPMO10 and JdLPMO10-CBM5, the gene fragments were inserted into the pUCBB/eGFP expression vector (61). The pUCBB/eGFP vector was pre-cut with BamHI and NotI restriction endonucleases (New England Biolabs), to remove nucleotides encoding the eGFP protein, prior to ligating the gene fragments using the T4 DNA ligase kit (Invitrogen), following instructions provided by the supplier. The expression vector was transformed by heat shock into chemically competent One Shot® BL21 Star™ (DE3) cells (Invitrogen).
For cloning JdCBM5-Chi18 and JdChi18, the gene fragments were inserted behind an IPTG-inducible T7 promoter in the pET26b expression vector (Novagen, Merck Millipore). The pET26b vector was digested with the restriction endonucleases NdeI and XhoI (New England Biolabs), and the gene fragments were ligated using T4 DNA ligase (Roche), following instructions provided by the supplier. The expression vectors were transformed by heat shock into chemically competent E. coli BL21 (DE3) cells (New England Biolabs).
For production of full-length Jd1381, the expression strain was cultivated in a 1-liter fermenter (DASGIP® benchtop bioreactors for cell culture; Eppendorf, Hamburg, Germany) filled with 0.7 liter of mineral salt medium (2 g/liter yeast extract, 15 g/liter glycerol, 100 mm KH2PO4, 30 mm Na2HP-O4, 30 mm (NH4)2SO4, 9 mm citric acid, 5 mm MgSO4, 10 ml/liter trace element solution, 15 μm thiamine). Batch fermentation was carried out at 37 °C, 25% dissolved oxygen saturation, and pH 7.0 (regulated with 5 m ammonium hydroxide and 3 m phosphoric acid). Upon complete consumption of the initial carbon source, the temperature was switched to 22 °C, and gene expression was initiated by an exponential feed (750 g/liter glucose monohydrate, 20 g/liter magnesium sulfate heptahydrate, 2.5 mm IPTG, initial feed rate of 4.4 ml/h, µ = 0.04 h−1). After 16 h of fed-batch cultivation, the cells were harvested by centrifugation.
Jd1381 was purified in a stepwise manner using nickel–nitrilotriacetic acid affinity chromatography followed by anion-exchange chromatography. 30 g of cell pellet (wet weight) obtained from a 400-ml culture volume were resuspended in 120 ml of buffer (50 mm Tris/HCl, 300 mm NaCl, pH 8.0), and the cells were disrupted by two passages through a microfluidizer M-110P (Microfluidics, Westwood, MA, USA) at 1000 bar. After centrifugation for 30 min at 35,000 × g, the cleared lysate was sterilized by filtering using a 0.2 μm Nalgene Rapid-Flow sterile bottle-top filter unit (Thermo Scientific, Waltham, MA, USA). Hereafter, the filtered lysate was applied to a 20-ml nickel–nitrilotriacetic acid–Sepharose column connected to an ÄKTA express FPLC system (GE Healthcare Lifesciences), which was then washed with 10 column volumes buffer A containing 20 mm imidazole. Bound protein was eluted with buffer B containing 200 mm imidazole. Fractions containing the target protein were pooled, diluted 10-fold with a loading buffer used for anion-exchange chromatography (buffer A; 20 mm Tris/HCl, pH 7.5), and applied to a 5-ml HiTrap Q column (GE Healthcare Lifesciences). After washing the column with 10 column volumes of buffer A, the proteins were eluted with a linear salt gradient (0–1.0 m NaCl for 20 min at 5 ml/min). The peak fractions were pooled, frozen at −80 °C, and lyophilized (the remaining salt was removed as part of the gel-filtration steps related to the copper saturation step described below).
For production of JdLPMO10 and JdLPMO10-CBM5, fresh transformant colonies were inoculated in LB medium containing 50 µg/ml ampicillin and grown at 30 °C for 16 h at 170 rpm. The cells were harvested by centrifugation at 9,800 × g for 10 min, and a periplasmic fraction was prepared using a cold osmotic shock method (62). The resulting periplasmic extract was passed through a 0.2-μm sterile filter using Amicon Ultra centrifugal filters (Merck Millipore) prior to purification. JdLPMO10 and JdLPMO10-CBM5 were purified by loading periplasmic extracts adjusted to buffer A (20 mm Tris/HCl, pH 8.0) onto a 5-ml HiTrap DEAE FF anion exchanger (GE Healthcare Lifesciences) connected to an ÄKTA purifier FPLC system (GE Healthcare Lifesciences). The enzymes were eluted by applying a linear salt gradient (0–500 mm NaCl) over 60 min at a flow rate of 1 ml/min. Fractions containing the target proteins were pooled and concentrated using Amicon Ultra centrifugal filters (molecular mass cutoff, 10 kDa; Merck Millipore). For further purification, the samples were loaded onto a HiLoad 16/60 Superdex 75 size-exclusion column (GE Healthcare Lifesciences), with a running buffer consisting of 20 mm Tris/HCl, pH 8.0, and 250 mm NaCl, using a flow rate of 0.8 ml/min. Protein purity was analyzed by SDS-PAGE. Fractions containing pure protein were pooled and concentrated, and the buffer was exchanged to 20 mm Tris, pH 8.0, using Amicon Ultra centrifugal filters (molecular mass cutoff, 10 kDa; Merck Millipore), prior to storage at 4 °C. Protein concentrations were determined using the Bradford assay (Bio-Rad).
For purification of JdCBM5-Chi18 and JdChi18, precultures were prepared by growing the expression strains in 5 ml of LB medium supplemented with 50 µg/ml kanamycin at 37 °C for 16 h. Hereafter, the precultures were used to inoculate 500 ml of LB medium supplemented with 50 µg/ml kanamycin and grown at 37 °C for 4 h at 170 rpm. Expression was induced by supplementing with 0.2 mm IPTG, and the cultures were grown at 22 °C for 16 h. The cells were harvested by centrifugation at 9800 × g for 10 min, and cytoplasmic extracts were prepared by sonicating the cell pellets, after which the enzymes were purified by refolding from inclusion bodies following a protocol described elsewhere (34). Prior to purification, the refolded JdCBM5-Chi18 and JdChi18 were buffer-exchanged to 20 mm Tris, pH 8.0, 0.1 m NaCl and sterilized by filtration as described above. For purification, the enzymes were loaded onto a HisTrap HP 5-ml column (GE Healthcare Lifesciences) connected to an ÄKTA purifier FPLC system (GE Healthcare Lifesciences). The enzymes were eluted by using a linear gradient ending at 500 mm imidazole over 60 min at a flow rate of 1 ml/min. The fractions containing the enzymes were concentrated, and the buffer was exchanged to 20 mm Tris, pH 8.0, using Amicon Ultra centrifugation filters (molecular mass cutoff, 10 kDa; Merck Millipore). Of note, the specific activity of these refolded enzymes toward chitopentaose was identical to the specific activity of the full-length enzyme (Fig. S3).
The yields of the production and purification procedures described above were 4, 12, 40, 50, and 50 mg pure enzyme per liter of culture for JdChi18, JdCBM5-Chi18, JdLPMO10-CBM5, JdLPMO10, and Jd1381, respectively. Prior to use in enzyme reactions, Jd1381, JdLPMO10, JdLPMO10-CBM5, JdCBM5-Chi18, JdChi18, and SmLPMO10A (also called CBP21) were saturated with Cu(II) by incubation with CuSO4 in a 1:3 molar ratio (enzyme:Cu(II)) for 30 min at room temperature (so, all enzyme preparations, including those not containing an LPMO domain, were treated in exactly the same manner). Excess Cu(II) was removed using gravity flow separation through a PD MidiTrap G-25 column in 20 mm Tris/HCl, pH 8.0 (GE Healthcare Lifesciences).
Chitin degradation experiments
For qualitative analysis of activity and product profiles in reactions with chitin, reaction mixtures contained 1 μm of Jd1381 or JdLPMO10 or SmLPMO10A and 10 g/liter β-chitin and, when indicated, 1 mm AscA in 20 mm BisTris, pH 6.0. The reaction mixtures were incubated at 40 °C in a preheated Eppendorf Comfort Thermomixer with a thermostated lid, with shaking at 1000 rpm. For analysis of nonoxidized products generated by Jd1381, the reactions were stopped by mixing 50-µl samples with an equal volume of 50 mm H2SO4 and diluted 10 times with 5 mm H2SO4. Subsequently, the soluble products were separated by filtration using a 96-well filter plate operated with a Millipore vacuum manifold (Merck Millipore). The soluble nonoxidized products were then analyzed by HPLC using a Rezex column, as described below. For qualitative analysis of oxidized products, the reactions were stopped by directly separating the soluble products from the substrate by filtration using the same procedure as above. Hereafter, 26 µl of the sample was immediately mixed with 74 µl of 100% acetonitrile and analyzed by HPLC using a HILIC column as described below.
For assays with soluble substrates, reaction mixtures containing 100 μm chitobiose, chitotriose, chitotetraose, chitopentaose, or chitohexaose in 20 mm BisTris, pH 6.0, were prepared and placed in a thermomixer preheated to 40 °C. Subsequently, the enzyme was added to a final concentration of 5 nm, and the reactions were further incubated for 5 h. Samples (20 µl) were taken at appropriate time points and mixed with 20 µl of 50 mm H2SO4 to stop the reactions. Hereafter, the samples were immediately placed in the HPLC system and analyzed using a Rezex column, as described below.
For quantitative analysis of enzyme activity, the reactions were set up containing 10 g/liter of α or β-chitin in 20 mm BisTris, pH 6.0, with or without 1 mm AscA and incubated at 40 °C in a ThermoMixer, with shaking at 1000 rpm. Samples of reaction supernatants (10 µl) were taken at various time points after spinning the reaction tubes at 11,000 × g for 5 min. To convert the soluble products to GlcNAc and chitobionic acid (GlcNAcGlcNAc1A), as described previously (60), all samples were treated with 1.5 μm of the GH20 β-N-acetylhexosaminidase from S. marcescens (also known as chitobiase or SmCHB) overnight at 37 °C. The samples were diluted with 5 mm H2SO4 and analyzed by HPLC using a Rezex column, as described below. All reactions were performed in triplicate. Additional degradation reactions for studying enzyme synergy in the degradation of α-chitin (10 g/liter) were performed in exactly the same manner.
Binding assay
Binding of Jd1381 variants to α-chitin and β-chitin was assessed by mixing 2 μm of enzyme with 10 g/liter chitin, 1 mm AscA in 20 mm BisTris, pH 6.0, followed by incubation at 40 °C with shaking at 1000 rpm in an Eppendorf Comfort Thermomixer with a thermostated lid. At time points 60, 120, 240, and 360 min, samples were taken, and insoluble chitin with bound enzymes was separated from the soluble fraction by filtration using a 96-well filter plate operated with a Millipore vacuum manifold (Merck Millipore). The concentration of unbound enzymes in the filtrate was determined using a Bradford protein assay kit (Bio-Rad).
Product analysis and quantification
Nonoxidized hydrolysis products and chitobionic acid (GlcNAcGlcNAc1A or A2ox) were analyzed and quantified using a Dionex Ultimate 3000 ultra HPLC system (Dionex Corp., Sunnyvale, CA, USA), equipped with a Rezex RFQ-Fast acid H+ (8%) 7.8 × 100-mm column (Phenomenex, Torrance, CA) preheated to 85 °C, using 5 mm H2SO4 as mobile phase, at a flow rate of 1 ml/min. The solutes were separated isocratically and detected using UV absorption at 194 nm. The amounts of GlcNAc (GlcNAc or A1) and chitobiose (GlcNAc2 or A2) were quantified using GlcNAc and GlcNAc2 standards, which were regularly analyzed. For analysis and quantification of chitobionic acid, a standard stock solution (10 mm) of chitobionic acid was produced in-house by treating GlcNAc2 with a chito-oligosaccharide oxidase from Fusarium graminearum (63), as described previously (60).
Qualitative analysis of oxidized products was performed according to a previously described method (40). In short, oxidized products were analyzed by ultra HPLC using a HILIC column and the following gradient: 0–5 min (74% acetonitrile), 5–7 min (74–62% acetonitrile), 7–8 min (62% acetonitrile), 8–10 min (62–74% acetonitrile), and 10–12 min (74% acetonitrile). Standards (A2ox–A6ox) were prepared in-house by oxidizing a mixture of chito-oligosaccharides of different lengths with chito oligosaccharide oxidase from F. graminearum (63), as described previously (60).
Proteomics
A secretome from J. denitrificans 20603 grown on β-chitin was prepared by inoculating (4 ml) of a preculture grown in 5 ml of brain–heart infusion medium for 16 h at 37 °C and 170 rpm, into 500 ml of M9 minimal medium containing 2% β-chitin, 6 g/liter Na2HPO4, 3 g/liter KH2PO4, 1 g/liter NH4Cl, 0.5 g/liter NaCl, 1 mm thiamine HCl, 0.1 mm CaCl2, and 1 mm MgSO4. The culture was incubated at 37 °C for 5 days, until all chitin was consumed. The cells were removed by centrifugation at 32,816 × g for 15 min followed by sterilizing the supernatant by filtration using a 0.22-μm Sterilecup filter unit (Merck Millipore). The culture supernatant was concentrated ∼20 times by ultrafiltration using a Viva Flow 200 system equipped with a 10-kDa membrane and then subjected to SDS-PAGE. The gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad) and destained before bands of interest were cut out for in-gel digestion, essentially performed as described by Shevchenko et al. (64). Before trypsination, proteins were reduced with DTT and alkylated with iodoacetamide. The peptides were purified using ZipTip C18 pipette tips (Merck Millipore), dried under vacuum, and dissolved in 10 µl of 2% (v/v) acetonitrile, 0.1% (v/v) TFA.
For LC–MS/MS analysis, peptide samples were loaded onto a trap column (Acclaim PepMap100, C18, 5 μm, 100 Å, 300-μm inner diameter × 5 mm; Thermo Scientific) and backflushed onto a 50-cm analytical column (Acclaim PepMap RSLC C18, 2 μm, 100 Å, 75-μm inner diameter; Thermo Scientific). At the start, the columns were in 96% solution A (0.1% (v/v) formic acid), 4% solution B (80% (v/v) acetonitrile, 0.1% (v/v) formic acid), and a 70-min gradient was used to elute the peptides. The gradient was as follows: 4 to 13% solution B in 2 min, 13 to 45% in 52 min, and finally to 55% B in 3 min, before starting a wash phase at 90% B, all at a flow rate of 300 nl/min. To isolate and fragment the 10 most intense peptide precursor ions at any given time throughout the chromatographic elution, the Q-Exactive mass spectrometer (Thermo Scientific) was operated in data-dependent mode to switch automatically between Orbitrap-MS and higher-energy collisional dissociation Orbitrap-MS/MS acquisition. The selected precursor ions were then excluded for repeated fragmentation for 20 s. The resolution was set to r = 70,000 and r = 35,000 for MS and MS/MS, respectively. For optimal acquisition of MS/MS spectra, automatic-gain-control target values were set to 1,000,000 charges and a maximum injection time of 128 ms.
MS raw files were converted to Mascot Generic file format, and a database search was carried out using Mascot Deamon version 2.5.1 (Matrix Science Ltd., London, UK). The data were searched against a database containing the reference proteome of J. denitrificans 20603 (2495 sequences), downloaded from Uniprot, and supplemented with common contaminants such as keratins, trypsin, and BSA. Trypsin was used as digestion enzyme, and one missed cleavage was allowed. N-terminal acetylation, oxidation of methionine, and deamidation of asparagine and glutamine were set as variable modifications, whereas carbamidomethylation of cysteine residues was set as a fixed modification. Peptide tolerance was set to ±10 ppm (with #13C = 1), and the MS/MS tolerance was set to ±20 milli-mass units, whereas the peptide charge state was set to 2+, 3+ and 4+. A decoy database was generated by Mascot.
Homology modeling of JdChi18
The part of the Jd1381 protein sequence representing the GH18 domain was identified through annotation of the domain structure using the dbCAN server (RRID:SCR_013208) (65). This prediction was verified by aligning the predicted GH18 sequence with the amino acid sequences of structurally characterized GH18 chitinases. Based on these analyses, residues 280–651 were determined to represent the GH18 domain (named JdChi18). The 3D structure of JdChi18 was modeled using the Swiss-Model software (66) operated in automatic mode, using a S. thermoviolaceus GH18 chitinase called “chitinase 40” as a template (Protein Data Bank entry 4W5U).
Scanning electron microscopy
To detect modifications of chitin particles caused by LPMO and/or chitinase activity, scanning electron micrographs were taken. α-Chitin (10 g/liter) was treated with 1 μm Jd1381 in the presence or absence of 1.0 mm AscA as a reducing agent at 40 °C and 1000 rpm for 3.5 h. Control reactions without enzyme, but with otherwise identical composition and incubation conditions, were also performed. The reactions were stopped by removing the liquid fraction by centrifugation using a benchtop Eppendorf centrifuge 5418. Chitin particles (the pellet) were processed in a BAL-TEC critical point drying CPD 030 (BAL-TEC AG, Balzers, Germany) and coated with gold-palladium using a Polaron sputter coater SC 7640 (Quorum Technologies Ltd., East Sussex, UK). The coated samples were examined in a Zeiss Evo 50 EP scanning electron microscope (Carl Zeiss).
Data availability
Further details regarding our preliminary analysis of the secretome of J. denitrificans grown on chitin can be obtained from the corresponding author upon request. All other data are contained within the article and the supporting information.
Supplementary Material
This article contains supporting information.
Author contributions—S. M. and T. R. T. formal analysis; S. M. and V. G. H. E. validation; S. M., T. R. T., S. C., and A. N. investigation; S. M., F. A., and G. V.-K. visualization; S. M., T. R. T., F. A., S. C., and A. N. methodology; S. M., T. R. T., F. A., G. V.-K., and V. G. H. E. writing-original draft; S. M., T. R. T., F. A., S. C., C. S.-D., A. N., J. M., G. V.-K., and V. G. H. E. writing-review and editing; C. S.-D. and J. M. resources; C. S.-D., G. V.-K., and V. G. H. E. supervision; G. V.-K. and V. G. H. E. conceptualization; V. G. H. E. funding acquisition; V. G. H. E. project administration.
Funding and additional information—This work was supported by the Research Council of Norway through Project 221576 (to S. M., T. R. T., G. V.-K., and V. G. H. E.), called MarPol, and Project 247001 (to S. M., A. N., J. M., G. V.-K., and V. G. H. E.), which is Norway's contribution to a project in the European Research Area Industrial Biotechnology (ERA-IB) called ChitoTex: Development and production of new insect chitosan and chitosan based functional coatings for yarns and textile fabrics.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- LPMO
- lytic polysaccharide monooxygenase
- CBM
- carbohydrate-binding module
- DM
- dry matter
- GH
- glycoside hydrolase
- Jd
- J. dentrificans
- SEM
- scanning electron microscopy
- Sm
- S. marcescens
- AscA
- ascorbic acid
- IPTG
- isopropyl β-d-thiogalactopyranoside
- HILIC
- hydrophilic interaction liquid chromatography.
References
- 1. Vaaje-Kolstad G., Horn S. J., Sørlie M., and Eijsink V. G. (2013) The chitinolytic machinery of Serratia marcescens: a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 280, 3028–3049 10.1111/febs.12181 [DOI] [PubMed] [Google Scholar]
- 2. Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., Ståhlberg J., and Beckham G. T. (2015) Fungal cellulases. Chem. Rev. 115, 1308–1448 10.1021/cr500351c [DOI] [PubMed] [Google Scholar]
- 3. Harris P. V., Xu F., Kreel N. E., Kang C., and Fukuyama S. (2014) New enzyme insights drive advances in commercial ethanol production. Curr. Opin. Chem. Biol. 19, 162–170 10.1016/j.cbpa.2014.02.015 [DOI] [PubMed] [Google Scholar]
- 4. Eijsink V. G., Vaaje-Kolstad G., Vårum K. M., and Horn S. J. (2008) Towards new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol. 26, 228–235 10.1016/j.tibtech.2008.02.004 [DOI] [PubMed] [Google Scholar]
- 5. Beeson W. T., Vu V. V., Span E. A., Phillips C. M., and Marletta M. A. (2015) Cellulose degradation by polysaccharide monooxygenases. Annu. Rev. Biochem. 84, 923–946 10.1146/annurev-biochem-060614-034439 [DOI] [PubMed] [Google Scholar]
- 6. Brunecky R., Alahuhta M., Xu Q., Donohoe B. S., Crowley M. F., Kataeva I. A., Yang S. J., Resch M. G., Adams M. W., Lunin V. V., Himmel M. E., and Bomble Y. J. (2013) Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342, 1513–1516 10.1126/science.1244273 [DOI] [PubMed] [Google Scholar]
- 7. Brunecky R., Donohoe B. S., Yarbrough J. M., Mittal A., Scott B. R., Ding H., Taylor Ii L. E., Russell J. F., Chung D., Westpheling J., Teter S. A., Himmel M. E., and Bomble Y. J. (2017) The multi domain Caldicellulosiruptor bescii CelA cellulase excels at the hydrolysis of crystalline cellulose. Sci. Rep. 7, 9622 10.1038/s41598-017-08985-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larsbrink J., Zhu Y., Kharade S. S., Kwiatkowski K. J., Eijsink V. G. H., Koropatkin N. M., McBride M. J., and Pope P. B. (2016) A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotech. Biofuels 9, 260 10.1186/s13068-016-0674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaaje-Kolstad G., Horn S. J., van Aalten D. M., Synstad B., and Eijsink V. G. (2005) The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497 10.1074/jbc.M504468200 [DOI] [PubMed] [Google Scholar]
- 10. Vaaje-Kolstad G., Westereng B., Horn S. J., Liu Z., Zhai H., Sørlie M., and Eijsink V. G. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- 11. Quinlan R. J., Sweeney M. D., Lo Leggio L., Otten H., Poulsen J. C., Johansen K. S., Krogh K. B., Jørgensen C. I., Tovborg M., Anthonsen A., Tryfona T., Walter C. P., Dupree P., Xu F., Davies G. J., et al. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U.S.A. 108, 15079–15084 10.1073/pnas.1105776108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phillips C. M., Beeson W. T., Cate J. H., and Marletta M. A. (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 10.1021/cb200351y [DOI] [PubMed] [Google Scholar]
- 13. Forsberg Z., Vaaje-Kolstad G., Westereng B., Bunæaes A. C., Stenstrøm Y., MacKenzie A., Sørlie M., Horn S. J., and Eijsink V. G. (2011) Cleavage of cellulose by a CBM33 protein. Protein Sci. 20, 1479–1483 10.1002/pro.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bissaro B., Røhr Å. K., Müller G., Chylenski P., Skaugen M., Forsberg Z., Horn S. J., Vaaje-Kolstad G., and Eijsink V. G. H. (2017) Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13, 1123–1128 10.1038/nchembio.2470 [DOI] [PubMed] [Google Scholar]
- 15. Bissaro B., Várnai A., Røhr Å. K., and Eijsink V. G. H. (2018) Oxidoreductases and reactive oxygen species in conversion of lignocellulosic biomass. Microbiol. Mol. Biol. Rev. 82, e00029–18 10.1128/MMBR.00029-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Müller G., Chylenski P., Bissaro B., Eijsink V. G. H., and Horn S. J. (2018) The impact of hydrogen peroxide supply on LPMO activity and overall saccharification efficiency of a commercial cellulase cocktail. Biotechnol. Biofuels 11, 209 10.1186/s13068-018-1199-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris P. V., Welner D., McFarland K. C., Re E., Navarro Poulsen J. C., Brown K., Salbo R., Ding H., Vlasenko E., Merino S., Xu F., Cherry J., Larsen S., and Lo Leggio L. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 10.1021/bi100009p [DOI] [PubMed] [Google Scholar]
- 18. Eibinger M., Ganner T., Bubner P., Rošker S., Kracher D., Haltrich D., Ludwig R., Plank H., and Nidetzky B. (2014) Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J. Biol. Chem. 289, 35929–35938 10.1074/jbc.M114.602227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eibinger M., Sattelkow J., Ganner T., Plank H., and Nidetzky B. (2017) Single-molecule study of oxidative enzymatic deconstruction of cellulose. Nat. Commun. 8, 894 10.1038/s41467-017-01028-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Müller G., Várnai A., Johansen K. S., Eijsink V. G., and Horn S. J. (2015) Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol. Biofuels 8, 187 10.1186/s13068-015-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levasseur A., Drula E., Lombard V., Coutinho P. M., and Henrissat B. (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6, 41 10.1186/1754-6834-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horn S. J., Vaaje-Kolstad G., Westereng B., and Eijsink V. G. (2012) Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45 10.1186/1754-6834-5-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minke R., and Blackwell J. (1978) The structure of α-chitin. J. Mol. Biol. 120, 167–181 10.1016/0022-2836(78)90063-3 [DOI] [PubMed] [Google Scholar]
- 24. Gardner K. H., and Blackwell J. (1975) Refinement of the structure of β-chitin. Biopolymers 14, 1581–1595 10.1002/bip.1975.360140804 [DOI] [PubMed] [Google Scholar]
- 25. Mekasha S., Forsberg Z., Dalhus B., Bacik J. P., Choudhary S., Schmidt-Dannert C., Vaaje-Kolstad G., and Eijsink V. G. (2016) Structural and functional characterization of a small chitin-active lytic polysaccharide monooxygenase domain of a multi-modular chitinase from Jonesia denitrificans. FEBS Lett. 590, 34–42 10.1002/1873-3468.12025 [DOI] [PubMed] [Google Scholar]
- 26. Bacik J. P., Mekasha S., Forsberg Z., Kovalevsky A. Y., Vaaje-Kolstad G., Eijsink V. G. H., Nix J. C., Coates L., Cuneo M. J., Unkefer C. J., and Chen J. C. (2017) Neutron and atomic resolution x-ray structures of a lytic polysaccharide monooxygenase reveal copper-mediated dioxygen binding and evidence for N-terminal deprotonation. Biochemistry 56, 2529–2532 10.1021/acs.biochem.7b00019 [DOI] [PubMed] [Google Scholar]
- 27. Payne C. M., Baban J., Horn S. J., Backe P. H., Arvai A. S., Dalhus B., Bjørås M., Eijsink V. G., Sørlie M., Beckham G. T., and Vaaje-Kolstad G. (2012) Hallmarks of processivity in glycoside hydrolases from crystallographic and computational studies of the Serratia marcescens chitinases. J. Biol. Chem. 287, 36322–36330 10.1074/jbc.M112.402149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Igarashi K., Uchihashi T., Uchiyama T., Sugimoto H., Wada M., Suzuki K., Sakuda S., Ando T., Watanabe T., and Samejima M. (2014) Two-way traffic of glycoside hydrolase family 18 processive chitinases on crystalline chitin. Nat. Commun. 5, 3975 10.1038/ncomms4975 [DOI] [PubMed] [Google Scholar]
- 29. Horn S. J., Sørbotten A., Synstad B., Sikorski P., Sørlie M., Vårum K. M., and Eijsink V. G. (2006) Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 273, 491–503 10.1111/j.1742-4658.2005.05079.x [DOI] [PubMed] [Google Scholar]
- 30. Jana S., Hamre A. G., Wildberger P., Holen M. M., Eijsink V. G., Beckham G. T., Sørlie M., and Payne C. M. (2016) Aromatic-mediated carbohydrate recognition in processive Serratia marcescens chitinases. J. Phys. Chem. B 120, 1236–1249 10.1021/acs.jpcb.5b12610 [DOI] [PubMed] [Google Scholar]
- 31. Tuveng T. R., Hagen L. H., Mekasha S., Frank J., Arntzen M. Ø., Vaaje-Kolstad G., and Eijsink V. G. H. (2017) Genomic, proteomic and biochemical analysis of the chitinolytic machinery of Serratia marcescens BJL200. Biochim. Biophys Acta Proteins Proteom. 1865, 414–421 10.1016/j.bbapap.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 32. Sikorski P., Sørbotten A., Horn S. J., Eijsink V. G., and Vårum K. M. (2006) Serratia marcescens chitinases with tunnel-shaped substrate-binding grooves show endo activity and different degrees of processivity during enzymatic hydrolysis of chitosan. Biochemistry 45, 9566–9574 10.1021/bi060370l [DOI] [PubMed] [Google Scholar]
- 33. Horn S. J., Sørlie M., Vårum K. M., Väljamäe P., and Eijsink V. G. (2012) Measuring processivity. Methods Enzymol. 510, 69–95 10.1016/B978-0-12-415931-0.00005-7 [DOI] [PubMed] [Google Scholar]
- 34. Monge E. C., Tuveng T. R., Vaaje-Kolstad G., Eijsink V. G. H., and Gardner J. G. (2018) Systems analysis of the glycoside hydrolase family 18 enzymes from Cellvibrio japonicus characterizes essential chitin degradation functions. J. Biol. Chem. 293, 3849–3859 10.1074/jbc.RA117.000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Villares A., Moreau C., Bennati-Granier C., Garajova S., Foucat L., Falourd X., Saake B., Berrin J. G., and Cathala B. (2017) Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci. Rep. 7, 40262 10.1038/srep40262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song B., Li B., Wang X., Shen W., Park S., Collings C., Feng A., Smith S. J., Walton J. D., and Ding S. Y. (2018) Real-time imaging reveals that lytic polysaccharide monooxygenase promotes cellulase activity by increasing cellulose accessibility. Biotechnol. Biofuels 11, 41 10.1186/s13068-018-1023-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu J., Tian D., Renneckar S., and Saddler J. N. (2018) Enzyme mediated nanofibrillation of cellulose by the synergistic actions of an endoglucanase, lytic polysaccharide monooxygenase (LPMO) and xylanase. Sci. Rep. 8, 3195 10.1038/s41598-018-21016-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Valls C., Pastor F. I. J., Roncero M. B., Vidal T., Diaz P., Martínez J., and Valenzuela S. V. (2019) Assessing the enzymatic effects of cellulases and LPMO in improving mechanical fibrillation of cotton linters. Biotechnol. Biofuels 12, 161 10.1186/s13068-019-1502-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moreau C., Tapin-Lingua S., Grisel S., Gimbert I., Le Gall S., Meyer V., Petit-Conil M., Berrin J. G., Cathala B., and Villares A. (2019) Lytic polysaccharide monooxygenases (LPMOs) facilitate cellulose nanofibrils production. Biotechnol. Biofuels 12, 156 10.1186/s13068-019-1501-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakagawa Y. S., Kudo M., Loose J. S., Ishikawa T., Totani K., Eijsink V. G., and Vaaje-Kolstad G. (2015) A small lytic polysaccharide monooxygenase from Streptomyces griseus targeting α- and β-chitin. Febs J. 282, 1065–1079 10.1111/febs.13203 [DOI] [PubMed] [Google Scholar]
- 41. Paspaliari D. K., Loose J. S., Larsen M. H., and Vaaje-Kolstad G. (2015) Listeria monocytogenes has a functional chitinolytic system and an active lytic polysaccharide monooxygenase. FEBS J. 282, 921–936 10.1111/febs.13191 [DOI] [PubMed] [Google Scholar]
- 42. Karnaouri A., Muraleedharan M. N., Dimarogona M., Topakas E., Rova U., Sandgren M., and Christakopoulos P. (2017) Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol. Biofuels 10, 126 10.1186/s13068-017-0813-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arfi Y., Shamshoum M., Rogachev I., Peleg Y., and Bayer E. A. (2014) Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc. Natl. Acad. Sci. U.S.A. 111, 9109–9114 10.1073/pnas.1404148111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zakariassen H., Aam B. B., Horn S. J., Vårum K. M., Sørlie M., and Eijsink V. G. (2009) Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 284, 10610–10617 10.1074/jbc.M900092200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuusk S., Sørlie M., and Väljamäe P. (2017) Human chitotriosidase is an endo-processive enzyme. PLoS One 12, e0171042 10.1371/journal.pone.0171042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Courtade G., Forsberg Z., Heggset E. B., Eijsink V. G. H., and Aachmann F. L. (2018) The carbohydrate-binding module and linker of a modular lytic polysaccharide monooxygenase promote localized cellulose oxidation. J. Biol. Chem. 293, 13006–13015 10.1074/jbc.RA118.004269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mekasha S., Byman I. R., Lynch C., Toupalová H., Anděra L., Næs T., Vaaje-Kolstad G., and Eijsink V. G. H. (2017) Development of enzyme cocktails for complete saccharification of chitin using mono-component enzymes from Serratia marcescens. Process Biochem. 56, 132–138 10.1016/j.procbio.2017.02.021 [DOI] [Google Scholar]
- 48. Forsberg Z., Nelson C. E., Dalhus B., Mekasha S., Loose J. S., Crouch L. I., Røhr Å. K., Gardner J. G., Eijsink V. G., and Vaaje-Kolstad G. (2016) Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus. J. Biol. Chem. 291, 7300–7312 10.1074/jbc.M115.700161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mutahir Z., Mekasha S., Loose J. S. M., Abbas F., Vaaje-Kolstad G., Eijsink V. G. H., and Forsberg Z. (2018) Characterization and synergistic action of a tetra-modular lytic polysaccharide monooxygenase from Bacillus cereus. FEBS Lett. 592, 2562–2571 10.1002/1873-3468.13189 [DOI] [PubMed] [Google Scholar]
- 50. Loose J. S. M., Arntzen M. Ø., Bissaro B., Ludwig R., Eijsink V. G. H., and Vaaje-Kolstad G. (2018) Multipoint precision binding of substrate protects lytic polysaccharide monooxygenases from self-destructive off-pathway processes. Biochemistry 57, 4114–4124 10.1021/acs.biochem.8b00484 [DOI] [PubMed] [Google Scholar]
- 51. Forsberg Z., Bissaro B., Gullesen J., Dalhus B., Vaaje-Kolstad G., and Eijsink V. G. H. (2018) Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J. Biol. Chem. 293, 1397–1412 10.1074/jbc.M117.817130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eijsink V. G. H., Petrovic D., Forsberg Z., Mekasha S., Røhr Å. K., Várnai A., Bissaro B., and Vaaje-Kolstad G. (2019) On the functional characterization of lytic polysaccharide monooxygenases (LPMOs). Biotechnol. Biofuels 12, 58 10.1186/s13068-019-1392-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petrović D. M., Várnai A., Dimarogona M., Mathiesen G., Sandgren M., Westereng B., and Eijsink V. G. H. (2019) Comparison of three seemingly similar lytic polysaccharide monooxygenases from Neurospora crassa suggests different roles in plant biomass degradation. J. Biol. Chem. 294, 15068–15081 10.1074/jbc.RA119.008196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith S. P., Bayer E. A., and Czjzek M. (2017) Continually emerging mechanistic complexity of the multi-enzyme cellulosome complex. Curr. Opin. Struct. Biol. 44, 151–160 10.1016/j.sbi.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 55. Artzi L., Bayer E. A., and Moraïs S. (2017) Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 15, 83–95 10.1038/nrmicro.2016.164 [DOI] [PubMed] [Google Scholar]
- 56. Chylenski P., Bissaro B., Sørlie M., Røhr Å. K., Várnai A., Horn S. J., and Eijsink V. G. H. (2019) Lytic polysaccharide monooxygenases in enzymatic processing of lignocellulosic biomass. ACS Catalysis 9, 4970–4991 10.1021/acscatal.9b00246 [DOI] [Google Scholar]
- 57. Brurberg M. B., Eijsink V. G., and Nes I. F. (1994) Characterization of a chitinase gene (chiA) from Serratia marcescens BJL200 and one-step purification of the gene product. FEMS Microbiol. Lett. 124, 399–404 10.1111/j.1574-6968.1994.tb07315.x [DOI] [PubMed] [Google Scholar]
- 58. Synstad B., Vaaje-Kolstad G., Cederkvist F. H., Saua S. F., Horn S. J., Eijsink V. G., and Sørlie M. (2008) Expression and characterization of endochitinase C from Serratia marcescens BJL200 and its purification by a one-step general chitinase purification method. Biosci. Biotechnol. Biochem. 72, 715–723 10.1271/bbb.70594 [DOI] [PubMed] [Google Scholar]
- 59. Vaaje-Kolstad G., Houston D. R., Riemen A. H., Eijsink V. G., and van Aalten D. M. (2005) Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280, 11313–11319 10.1074/jbc.M407175200 [DOI] [PubMed] [Google Scholar]
- 60. Loose J. S., Forsberg Z., Fraaije M. W., Eijsink V. G., and Vaaje-Kolstad G. (2014) A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett. 588, 3435–3440 10.1016/j.febslet.2014.07.036 [DOI] [PubMed] [Google Scholar]
- 61. Vick J. E., Johnson E. T., Choudhary S., Bloch S. E., Lopez-Gallego F., Srivastava P., Tikh I. B., Wawrzyn G. T., and Schmidt-Dannert C. (2011) Optimized compatible set of BioBrick™ vectors for metabolic pathway engineering. Appl. Microbiol. Biotechnol. 92, 1275–1286 10.1007/s00253-011-3633-4 [DOI] [PubMed] [Google Scholar]
- 62. Manoil C., and Beckwith J. (1986) A genetic approach to analyzing membrane protein topology. Science 233, 1403–1408 10.1126/science.3529391 [DOI] [PubMed] [Google Scholar]
- 63. Heuts D. P., Winter R. T., Damsma G. E., Janssen D. B., and Fraaije M. W. (2008) The role of double covalent flavin binding in oligosaccharide oxidase from Fusarium graminearum. Biochem. J. 413, 175–183 10.1042/BJ20071591 [DOI] [PubMed] [Google Scholar]
- 64. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 65. Yin Y., Mao X., Yang J., Chen X., Mao F., and Xu Y. (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further details regarding our preliminary analysis of the secretome of J. denitrificans grown on chitin can be obtained from the corresponding author upon request. All other data are contained within the article and the supporting information.