Abstract
Cytochrome P450 1A1 (CYP1A1) catalyzes the metabolic activation of polycyclic aromatic hydrocarbons (PAHs) such as benzo[a]pyrene (B[a]P) and is transcriptionally regulated by the aryl hydrocarbon receptor (AhR)/AhR nuclear translocator (ARNT) complex upon exposure to PAHs. Accordingly, inhibition of CYP1A1 expression reduces production of carcinogens from PAHs. Although transcription of the CYP1A1 gene is known to be repressed by transforming growth factor-β (TGF-β), how TGF-β signaling is involved in the suppression of CYP1A1 gene expression has yet to be clarified. In this study, using mammalian cell lines, along with shRNA-mediated gene silencing, CRISPR/Cas9-based genome editing, and reporter gene and quantitative RT-PCR assays, we found that TGF-β signaling dissociates the B[a]P-mediated AhR/ARNT heteromeric complex. Among the examined Smads, Smad family member 3 (Smad3) strongly interacted with both AhR and ARNT via its MH2 domain. Moreover, hypoxia-inducible factor 1α (HIF-1α), which is stabilized upon TGF-β stimulation, also inhibited AhR/ARNT complex formation in the presence of B[a]P. Thus, TGF-β signaling negatively regulated the transcription of the CYP1A1 gene in at least two different ways. Of note, TGF-β abrogated DNA damage in B[a]P-exposed cells. We therefore conclude that TGF-β may protect cells against carcinogenesis because it inhibits CYP1A1-mediated metabolic activation of PAHs as part of its anti-tumorigenic activities.
Keywords: aryl hydrocarbon receptor (AhR), AhR nuclear translocator (ARNT), benzo[a]pyrene, CYP1A1, hypoxia-inducible factor 1α (HIF-1α), Smad, transforming growth factor β (TGF-β), polycyclic aromatic hydrocarbon (PAH), xenobiotic metabolism, gene regulation, aryl hydrocarbon receptor (AhR), hypoxia-inducible factor (HIF), Smad transcription factor
Introduction
The cytochrome P450 (CYP) superfamily, known as a family of heme-containing enzymes, can catalyze the monooxygenation of not only endogenous substrates but also xenobiotics. The human body has 57 different CYPs, which contribute to assorted metabolic and biosynthetic pathways (1). Among them, the CYP1A family, consisting of CYP1A1 and CYP1A2, is known to activate a variety of compounds with carcinogenic properties. CYP1A1 catalyzes epoxidation of an aromatic ring with a vacant position to initiate carcinogenesis via the formation of a highly reactive moiety that can promote oncogenic mutation in the genome. On the other hand, CYP1A1 is implicated in the metabolization of ingested natural aryl hydrocarbon receptor (AhR) ligands in the intestine to fine-tune intestinal immunity (2).
Some of the environmental carcinogens that CYP1A1 can metabolize promote the activation of the CYP1A1 gene via xenobiotic-responsive elements (XREs) in its promoter as a cis-element (3). Both the AhR and the AhR nuclear translocator (ARNT), which possess an N-terminal bHLH (basic helix-loop-helix) motif flanking a PAS (Per, ARNT, Sim) domain in their structures, play an essential role in the activation of the CYP1A1 gene through their binding to XREs (4, 5). The inactive AhR that interacts with chaperon complex, including Hsp90, a co-chaperone p23, and hepatitis B virus X-associated protein 2 (XAP2), is located in cytosols. When AhR binds to polycyclic aromatic hydrocarbons (PAHs), AhR dissociates from the complex among Hsp90, p23, and XAP2 to translocate to the nucleus, where AhR forms a heteromeric complex with ARNT. Then the AhR/ARNT complex can bind to XREs within the CYP1A1 promoter to enhance its transcription, in cooperation with coactivators and chromatin-remodeling factors (6–8). On the other hand, the AhRR (AhR repressor), which is also induced by PAHs, forms a complex with ARNT to compete with AhR/ARNT for the binding to XRE, followed by recruitment of transcriptional corepressors such as ANKRA2, HDAC4, and HDAC5 to suppress the CYP1A1 gene (9). The activity of AhR can be regulated by interaction with other transcription factors or its phosphorylation. Estrogen receptor α or glucocorticoid receptor augments PAH-induced CYP1A1 mRNA expression through its physical interaction with AhR, whereas NF-κB or HIF-1α that is stabilized under hypoxia suppresses CYP1A1 expression (10, 11). Phosphorylation of AhR by protein kinase C or c-Src further up-regulates the CYP1A1 gene, whereas that by protein kinase A attenuates CYP1A1 induction by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (12). Besides, the classical mitogen-activating protein kinase pathway increases the TCDD-mediated induction of CYP1A1 via AhR and simultaneously sensitizes AhR for ubiquitin-dependent degradation (13, 14). In addition, AhR is rapidly degraded via cytoplasmic ubiquitin/proteasome pathway after AhR is exported from the nucleus (7, 15).
The canonical transforming growth factor (TGF)-β signaling is transmitted via the receptor complex between TGF-β type II receptor (TβRII) and TGF-β type I receptor (TβRI or activin receptor-like kinase 5 (ALK5)), followed by the phosphorylation of the receptor-regulated Smad (R-Smad) proteins (i.e. Smad2 and Smad3). Subsequently, the two phosphorylated R-Smads can form a complex with Smad4 to go to the nucleus, where the Smad complex regulates the transcription of TGF-β target genes together with other transcriptional factors, coactivators, and corepressors (16–25).
TGF-β is known to possess the inhibitory action of the enzymatic activity of CYP1A1, which is induced by TCDD. When the mRNA expression of the CYP1A1 gene was examined, TGF-β directly down-regulated the transcription of the CYP1A1 gene and other AhR-regulated genes upon TCDD (26). As one of its inhibitory mechanisms, TGF-β repressed the transcription of the AhR gene through the Smad4-dependent pathway (27). However, an inhibitory mechanism(s) other than the TGF-β–mediated suppression of AhR expression might also contribute to the negative regulation of CYP1A1 gene expression by TGF-β.
In this current study, we found that Smad3 activated upon TGF-β stimulation attenuates B[a]P-induced CYP1A1 gene expression via its interaction with both AhR and ARNT. Furthermore, TGF-β retained AhR in the cytosol. Besides, HIF-1α stabilized by TGF-β could compete with AhR for binding to ARNT, which was followed by dissociation of the AhR/ARNT heteromeric complex. Therefore, TGF-β signaling possesses a dual role of inhibiting the transcription of the CYP1A1 gene. Consistently, TGF-β could reduce the DNA damage provoked by B[a]P owing to the decreased expression of CYP1A1.
Results
Inhibition of CYP1A1 expression by TGF-β
B[a]P, one of the PAHs, is a well-known inducer of the CYP1A1 gene. When A549 cells were stimulated with B[a]P, we could see the induction of CYP1A1 mRNA with the maximum expression 6 h after the treatment with B[a]P (Fig. 1A). Like the CYP1A1 gene, B[a]P could also induce the expression of CYP1B1 mRNA (Fig. 1B), as described previously (26, 28). On the other hand, AhR mRNA was not affected by B[a]P (Fig. 1B). Pretreatment of the cells with different doses of TGF-β for 2 h exhibited suppression of the CYP1A1 and CYP1B1 mRNA expression induced by B[a]P, although the inhibitory effect of TGF-β on B[a]P-induced CYP1B1 mRNA expression was marginal (Fig. 1, C and D). Furthermore, we could observe TGF-β–mediated suppression of B[a]P-induced CYP1B1 protein in mouse hepatocytes as well (Fig. 1E), whereas we could not succeed in detecting endogenous CYP1A1 protein using several commercially available antibodies (data not shown).
Figure 1.
Inhibition of B[a]P-induced CYP1A1 and CYP1B1 expression by TGF-β. A, induction of CYP1A1 mRNA by B[a]P. A549 cells were stimulated with 10 µm B[a]P for 1, 2, 6, and 24 h. The expression of CYP1A1 mRNA was detected by qPCR. B, induction of CYP1B1 and AhR mRNAs by B[a]P. The experiments were performed according to A. The expressions of CYP1B1 (●) and AhR mRNAs (■) were detected by qPCR. C and D, inhibitory effect of TGF-β on B[a]P-induced expression of CYP1A1 and CYP1B1 mRNAs. A549 cells were pretreated with 1 or 5 ng/ml TGF-β 2 h before exposure to 10 µm B[a]P for 2 h. The expression of CYP1A1 and CYP1B1 mRNAs was detected by qPCR. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks. E, suppression of B[a]P-induced CYP1B1 protein expression by TGF-β. Hepatocytes from WT mice were stimulated with 5 ng/ml TGF-β 4 h before exposure to 1 µm B[a]P cells for 12 or 24 h. Top and bottom panels show the expression of CYP1B1 and β-actin, respectively. The intensity of the band for CYP1B1 was normalized using the intensity of the band corresponding to β-actin. Relative intensity was calculated relative to the value for 12-h treatment of cells with B[a]P alone.
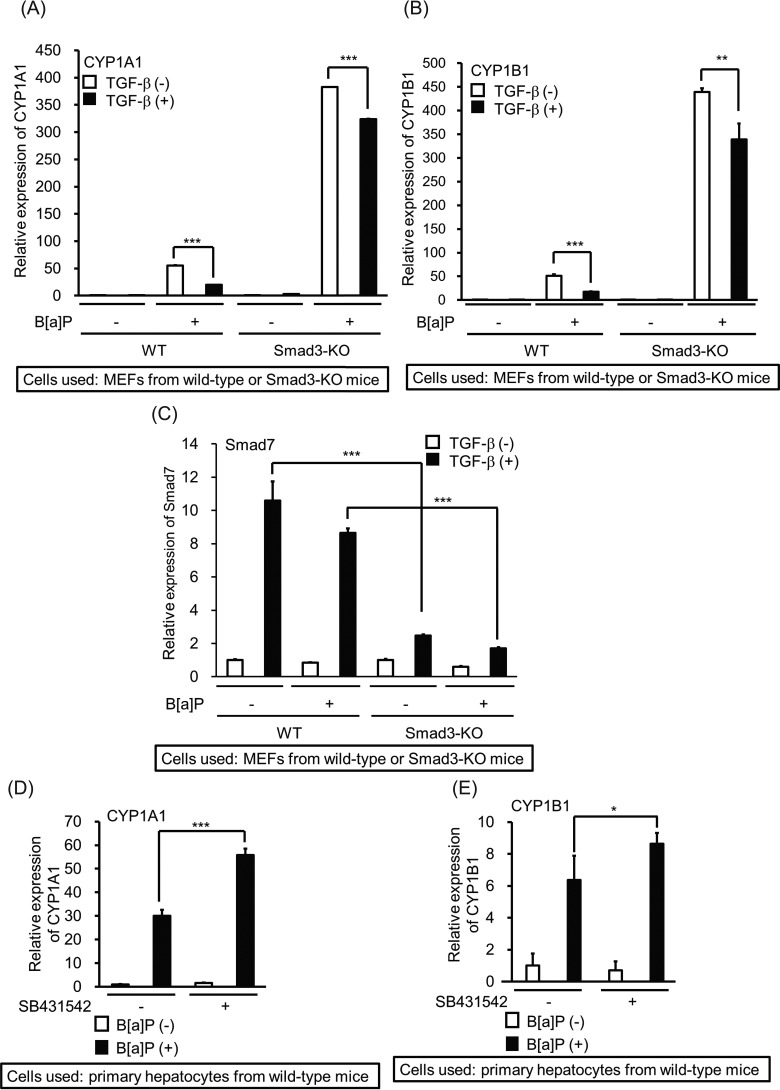
To determine whether the TGF-β/Smad pathway is implicated in the inhibitory action of B[a]P-mediated CYP1A1 expression, we prepared primary hepatocytes from both WT and Smad3-knockout (Smad3-KO) mice lacking TGF-β/Smad3 signaling (29, 30). Although TGF-β perturbed B[a]P-induced CYP1A1 transcription in hepatocytes from the WT mice, CYP1A1 mRNA induced by B[a]P was only weakly affected by TGF-β in the Smad3-deficient hepatocytes (Fig. 2A). Similarly, TGF-β did not reduce the B[a]P-induced transcription of the CYP1B1 gene in Smad3-deficient hepatocytes as much as in the WT hepatocytes (Fig. 2B). Curiously, high expression of both CYP1A1 and CYP1B1 mRNAs was observed without TGF-β stimulation in hepatocytes from Smad3-KO mice. We hypothesized that endogenous TGF-β secreted from hepatocytes might be involved in suppression of the basal expressions of CYP1A1 and CYP1B1 transcription in WT mice. To prove our hypothesis, we stimulated hepatocytes from WT mice with B[a]P 8 h after the addition of SB431542, a TGF-β type I receptor kinase inhibitor (Fig. 2, D and E). The B[a]P-induced mRNA expression of both CYP1A1 and CYP1B1 was further enhanced in the presence of SB431542. These results indicate that endogenous TGF-β might be partially implicated in inhibition of CYP1A1 and CYP1B1 expression. Of note, similar inhibitory effects by TGF-β were observed both in human and mouse cells even though the human AhR not only shares limited sequence identity to mouse AhR but also shows different affinity to TCDD or other AhR ligands to mouse AhR (31–34).
Figure 2.
Attenuation of TGF-β–mediated inhibition of B[a]P-induced CYP1A1 and CYP1B1 mRNA expressions. A–C, TGF-β–mediated alteration of B[a]P-induced mRNA expression in Smad3-deficient hepatocytes. After 4 h of starvation, hepatocytes from WT or Smad3-KO mice were stimulated with 5 ng/ml TGF-β 2 h before exposure to 1 µm B[a]P cells for 2 h. The expressions of CYP1A1 (A), CYP1B1 (B), and Smad7 (C) mRNAs were measured by qPCR. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks (A and B). To show whether hepatocytes from WT and Smad3-KO mice were responsive to TGF-β, the expression of Smad7 mRNA was used as the positive control. Significant differences between WT and Smad3-KO hepatocytes with TGF-β in the absence or presence of B[a]P are indicated with asterisks (C). D and E, hepatocytes from WT mice were stimulated with 1 µm B[a]P 2 h after exposure to 3 µm SB431542 for 8 h. The expressions of CYP1A1 (D) and CYP1B1 (E) mRNAs were measured by qPCR. Significant differences between B[a]P alone and combination of B[a]P with SB431542 are indicated with asterisks.
Implication of XREs in TGF-β–mediated repression of CYP1A1 promoter activity
The promoter activity of the CYP1A1 gene by PAHs is regulated via XREs as cis-elements, where the AhR/ARNT complex binds (3). To seek the negative cis-element(s) through which TGF-β represses the B[a]P-induced promoter activity of the CYP1A1 gene, we cloned the fragment from +19 to −4150 of the human CYP1A1 gene and inserted it into the pGL3-basic vector (CYP1A1(−4150)-luc). In parallel, we made one 5′ deletion mutant of the CYP1A1 promoter, termed CYP1A1(−1249)-luc (Fig. 3A). When we investigated whether the human CYP1A1 promoter includes a TGF-β–mediated inhibitory cis-element(s), the B[a]P-induced activities of both CYP1A1(−4150)-luc and CYP1A1(−1249)-luc were significantly perturbed by TGF-β (Fig. 3B). To narrow down the negative cis-element(s) in the CYP1A1 promoter, we made three luciferase reporter constructs deleted from the 5′ distal region of CYP1A1(−1249)-luc, termed CYP1A1(−931)-luc, CYP1A1(−620)-luc, and CYP1A1(−313)-luc (Fig. 3A). Then, after the cells were prestimulated with TGF-β, the B[a]P-induced activity of each reporter was evaluated (Fig. 3C). The TGF-β–mediated inhibition of the reporter activity by B[a]P declined, dependent on the decreased numbers of XREs in their promoter regions. In particular, the CYP1A1(−313)-luc reporter lacking a typical XRE in its promoter region was not affected by either B[a]P or TGF-β. Thus, we hypothesized that TGF-β−mediated repression of the CYP1A1 promoter activity induced by B[a]P involves XREs. To test this hypothesis, we inserted two, four, and six copies of the XRE in front of a minimal promoter (35), termed pGL3ti-(XRE)2-luc, pGL3ti-(XRE)4-luc, and pGL3ti-(XRE)6-luc, respectively (Fig. 3D), and tested the effect of TGF-β on the B[a]P-induced activity of these constructs in A549 cells. In all three reporter constructs, TGF-β inhibited the B[a]P-induced reporter activities, although the inducibility of the reporter activity upon B[a]P stimulation depended on the numbers of XREs (Fig. 3E). On the other hand, the luciferase reporter possessing four mutated XREs, termed pGL3ti-(mXRE)4-luc, could respond against B[a]P nor TGF-β (Fig. S1, A–C).
Figure 3.
TGF-β–mediated inhibition of B[a]P-induced CYP1A1 promoter activity via XRE. A, schematic presentation of deletion mutants for the human CYP1A1 promoter. luc gene, luciferase gene; XRE, xenobiotic-responsive element; SBE, Smad-binding element. B, inhibition of B[a]P-driven reporter activity by TGF-β. A549 cells were transfected with either CYP1A1(−4150)-luc or CYP1A1(−1249)-luc together with pCH110. Twenty-four hours later, the cells were pretreated with 5 ng/ml TGF-β 4 h before exposure to 1 µm B[a]P for 18 h. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks. C, inhibitory effect of TGF-β on B[a]P-induced CYP1A1 reporter activities. The experiments were performed according to the description in B. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks. D, illustration of luciferase reporters including XRE as an enhancer. The core XRE sequence is 5′-CACGCGA-3′/3′-GTGCGCT-5′. E, inhibition of B[a]P-driven XRE reporter activities by TGF-β. The experiments were performed according to the description in B. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks.
Requirement of the canonical Smad pathway to inhibit the CYP1A1 promoter activity induced by B[a]P
To investigate whether the Smad pathway is indispensable for the inhibition of the CYP1A1 promoter activity induced by B[a]P, we introduced pGL3ti-(XRE)2-luc into mouse embryonic fibroblasts (MEFs) from either Smad3 heterozygous (Smad3-HET) or Smad3-KO mice (29, 30), which was followed by pretreatment with TGF-β 2 h before the addition of B[a]P. As seen in Fig. 4A, deficiency of the Smad3 gene disabled TGF-β–mediated suppression of the CYP1A1 promoter activity induced by B[a]P. Although both Smad2 and Smad3 can transduce the intracellular canonical Smad pathway together with Smad4, Smad2 does not possess the ability to bind to the specific DNA element (termed SBE) because of the insertion of a 30-amino-acid length immediately behind the DNA-binding domain in the MH1 domain of Smad2 (36, 37). When either Smad2 or Smad3 was transfected into MEFs from Smad3-KO mice together with pGL3ti-(XRE)2-luc, Smad2 and Smad3 improved TGF-β–mediated repression of the reporter activity induced by B[a]P, although the effect of Smad2 was weaker than that of Smad3 (Fig. 4B). We also observed that the introduction of Smad2Δex3 (38) could also recover TGF-β–mediated inhibition of B[a]P-induced reporter activity, like Smad3 (data not shown). To further confirm that TGF-β/Smad3 signaling perturbs B[a]P-induced CYP1A1 gene activation, we generated A549 cells lacking the Smad3 gene by means of the CRISPR/Cas9 system (A549-S3KO cells) (Fig. S2). In addition to A549-S3KO cells, we established A549-S3KO cells overexpressing human Smad3, termed A549-S3KO(S3OE) cells (Fig. 4C). When each cell line was transfected with pGL3ti-(XRE)2-luc, enhancement of the B[a]P-induced reporter activity in A549-S3KO cells could be observed. Furthermore, TGF-β–mediated suppression of the B[a]P-induced reporter activity in A549-S3KO cells was lowered when compared with that in the control cells. On the other hand, introduction of human Smad3 into A549-S3KO cells improved the ability of TGF-β–mediated suppression of the B[a]P-induced reporter activity (Fig. 4D).
Figure 4.
Requirement of Smads for the inhibition of B[a]P-induced CYP1A1 promoter activity by TGF-β. A, effect of TGF-β on B[a]P-driven XRE reporter activities in Smad3-deficient MEFs. MEFs from Smad3-HET or Smad3-KO mice were transfected with pGL3ti-(XRE)2-luc and pCH110. The experiments were performed according to the description in the legend to Fig. 3B. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks. B, effect of Smad2 or Smad3 on TGF-β–mediated inhibition of B[a]P-driven XRE reporter activities in Smad3-KO MEFs. The experiments were performed according to the description in A except for the transfection of either Smad2 or Smad3 in Smad3-KO MEFs. Smad3-HET, MEFs from Smad3+/− mice; Smad3-KO, MEFs from Smad3−/− mice. Significant differences between B[a]P alone and combination of B[a]P with TGF-β are indicated with asterisks. C, expression of Smad3 protein in A549 cells lacking the Smad3 gene. The top and bottom panels show the expression of Smad3 and β-actin, respectively. Control, A549-P1 cells carrying the empty vector; A549-S3 KO, A549-P1 cells lacking the Smad3 gene; A549-S3 KO(S3OE), A549-S3 KO cells overexpressing human Smad3. D, effect of TGF-β on B[a]P-driven XRE reporter activities in A549-S3 KO. A549-P1, A549-S3 KO, or A549-S3 KO(S3OE) cells were transfected with pGL3ti-(XRE)2-luc and pCH110. The experiments were performed according to the description in the legend to Fig. 3B. Significant differences between control and A549-S3 KO cells or between control and A549-S3 KO(S3OE) cells are indicated with asterisks.
Because Smad proteins are known to interact with a number of transcriptional factors (37), we tried to examine their interaction with AhR and ARNT. As seen in Fig. 5 (A and B), Smad3 could associate with both AhR and ARNT most strikingly among the Smads examined. Thus, we tried to clarify the involvement of TGF-β/Smad3 signaling in the transcriptional control of the CYP1A1 gene in the following experiments. When we tested the endogenous interaction between Smad3 and AhR in A549 cells, we could see a TGF-β–dependent association between the two proteins (Fig. 5C). We also tried to show the endogenous interaction between Smad3 and ARNT. However, Smad3 was never coimmunoprecipitated with ARNT using the anti-ARNT antibody that can detect endogenous ARNT protein in A549 cells. Instead of coimmunoprecipitation, we adopted a proximity ligation assay (PLA) to detect the endogenous interaction between Smad3 and ARNT in cells. When both rabbit anti-ARNT and mouse anti-Smad3 antibodies were simultaneously added to the fixed sample in which cells were pretreated with TGF-β for 2 h, a large number of red spots could be observed (Fig. 5D). On the other hand, no dots were detected when mouse anti-Myc9E10 and rabbit anti-ARNT antibodies were used for the PLA (Fig. 5E).
Figure 5.
Interaction of Smads with AhR or ARNT. A, interaction of AhR with Smads. COS7 cells were transfected with the indicated plasmids and harvested for the co-IP experiments. The interaction between Smads and AhR is shown in the top panel. The total expressions of AhR and FLAG-Smads are indicated in the middle and bottom panels, respectively. The intensity of the band for immunoprecipitated AhR was normalized using the intensity of the band corresponding to total AhR. Relative intensity was calculated relative to the value for V5-AhR plus FLAG-Smad2. B, interaction of ARNT with Smads. COS7 cells were transfected with the indicated plasmids and harvested for the co-IP experiments. The interaction between Smads and ARNT is shown in the top panel. The total expressions of Myc-ARNT and FLAG-Smads are indicated in the middle and bottom panels, respectively. The intensity of the band for immunoprecipitated ARNT was normalized using the intensity of the band corresponding to total ARNT. Relative intensity was calculated relative to the value for Myc-ARNT plus FLAG-Smad2. C, endogenous interaction between Smad3 and AhR. A549 cells were stimulated with 5 ng/ml TGF-β for 2 h and harvested for the co-IP experiments. The anti-Myc9E10 antibody was used as the control antibody for immunoprecipitation. The interaction between Smad3 and AhR is shown in the top panel. The total expressions of AhR, Smad3, phosphorylated Smad2 (PS2), and β-actin are indicated in the second, third, fourth, and bottom panels, respectively. The bands corresponding to phosphorylated Smad2 indicate that the cells were responsive to TGF-β. The intensity of the band for immunoprecipitated AhR was normalized using the intensity of the band corresponding to total AhR. Relative intensity was calculated relative to the value for the band immunoprecipitated with anti-Smad3 antibody in the absence of TGF-β stimulation. D and E, endogenous interaction between Smad3 and ARNT. A549 cells were stimulated with 5 ng/ml TGF-β for 2 h. Then PLA was carried out. The red dots show the interaction between Smad3 and ARNT. The nuclei were stained with DAPI (D). As the negative control, anti-Myc9E10 antibody instead of anti-Smad3 antibody was used. No red dots were observed when the cells were stimulated with 5 ng/ml TGF-β for 2 h (E). Bar, 10 µm. ND, not determined.
Disruption of the AhR/ARNT complex by TGF-β/Smad3 signaling
Having shown that Smad3 can interact not only with AhR but also with ARNT, we hypothesized that TGF-β/Smad3 signaling can dissociate the complex between AhR and ARNT. The treatment of A549 cells with B[a]P promoted the complex formation between AhR and ARNT, whereas the introduction of Smad3 reduced it. TGF-β stimulation obviously further dissociated the AhR/ARNT complex in the presence of Smad3 (Fig. 6A).
Figure 6.
Disruption of the AhR/ARNT complex by TGF-β. A, dissociation between AhR and ARNT upon TGF-β stimulation. 911 cells were transfected with the indicated plasmids. Forty hours later, the cells were pretreated with 5 ng/ml TGF-β 4 h before exposure to 1 µm B[a]P for 2 h. Then the cells were harvested for the co-IP experiments using an anti-V5 antibody. The heterodimer formation between AhR and ARNT is shown in the top panel. The total expressions of ARNT, AhR, Smad3, and phosphorylated Smad3 are indicated in the second, third, fourth, and bottom panels, respectively. The band corresponding to phosphorylated Smad3 indicates that the cells were responsive to TGF-β. The intensity of the band for immunoprecipitated ARNT was normalized using the intensity of the band corresponding to total ARNT. Relative intensity was calculated relative to the value for Myc-ARNT plus V5-AhR in the presence of B[a]P. B, determination of the interacting domain of Smad3 with AhR. COS7 cells were transfected with the indicated plasmids. Then the cells were harvested for the co-IP experiments using an anti-V5 antibody. The interaction of Smad3 mutants with AhR is shown in the top panel. The total expressions of 6xMyc-Smad3 mutants and V5-AhR are indicated in the middle and bottom panels, respectively. The intensity of the band for each Smad3 mutant precipitated with AhR was normalized using the intensity of the band corresponding to each of the total Smad3 mutants. Relative intensity was calculated relative to the value for each Smad3 mutant alone. C, determination of the interacting domain of Smad3 with ARNT. COS7 cells were transfected with the indicated plasmids. Then the cells were harvested for the co-IP experiments using an anti-V5 antibody. The interaction of Smad3 mutants with ARNT is shown in the top panel. The total expressions of 6xMyc-Smad3 mutants and V5-ARNT are indicated in the middle and bottom panels, respectively. The intensity of the band for each Smad3 mutant precipitated with ARNT was normalized using the intensity of the band corresponding to each of the total Smad3 mutants. Relative intensity was calculated relative to the value for each Smad3 mutant alone. D, dissociation of the AhR/ARNT complex by the Smad3 MH2 domain. 911 cells were transfected with the indicated plasmids. Then the cells were harvested for the co-IP experiments using an anti-V5 antibody. The interaction of V5-AhR with Myc-ARNT is shown in the top panel. The total expression of V5-AhR was detected with an anti-V5 antibody after immunoprecipitation using total lysates by an anti-V5 antibody (second panel). The total expressions of Myc-ARNT and 6xMyc-Smad3 MH2 are indicated in the third and bottom panels, respectively. The intensity of the band for immunoprecipitated ARNT was normalized using the intensity of the band corresponding to total ARNT. Relative intensity was calculated relative to the value for Myc-ARNT plus V5-AhR.
Smad3 is composed of three domains: the Mad homology 1 (MH1), linker, and MH2 domains (36). To confirm which domain(s) is required for Smad3 to interact with AhR and ARNT, we carried out a coimmunoprecipitation (co-IP) assay. We observed that both AhR and ARNT interact with the MH2 domain of Smad3, whereas the Linker + MH2 domain of Smad3 only marginally associated with AhR and ARNT (Fig. 6, B and C). Consistently, overexpression of the Smad3 MH2 domain partially disrupted the B[a]P-induced complex between AhR and ARNT (Fig. 6D). The AhR/ARNT complex can bind to XREs in the presence of B[a]P. Overexpression of the MH2 domain of Smad3 indeed inhibited the binding of the AhR/ARNT complex to XREs (Fig. 7, A and B), although Smad3 by itself did not have any ability to bind to XREs directly (Fig. S3). We were also convinced that TGF-β interrupted the binding of the AhR/ARNT complex to XREs in the presence of B[a]P (Fig. 7C).
Figure 7.
Inhibitory effect of TGF-β/Smad3 signal on DNA binding of the AhR/ARNT complex. A, interference of the Smad3 MH2 domain in DNA binding of AhR. 911 cells were transfected with the indicated plasmids. Forty hours later, the cells were stimulated with 1 µm B[a]P for 2 h. Then the cell lysates were mixed with biotinylated (XRE)2. AhR that binds to DNA was detected by Western blotting analysis using an anti-V5 antibody (top). The expressions of Myc-ARNT, V5-AhR, and 6xMyc-Smad3 MH2 were evaluated using anti-Myc9E10 (second panel), anti-AhR (third panel), and anti-Myc9E10 (bottom panel) antibodies, respectively. The cells were pretreated with 10 µm MG-132 2 h before the addition of B[a]P to stabilize the proteins in the cells. The intensity of the band for AhR binding to DNA was normalized using the intensity of the band corresponding to total AhR. Relative intensity was calculated relative to the value for Myc-ARNT plus V5-AhR. B, inhibitory effect of the Smad3 MH2 domain on DNA binding of ARNT. 911 cells were transfected with the indicated plasmids. Forty hours later, the cells were stimulated with 1 µm B[a]P 2 h before preparation of the cell lysates. The cells were pretreated with 10 µm MG-132 2 h before the addition of B[a]P to stabilize the proteins in the cells. The experiments were carried out according to the description in A. ARNT that binds to DNA was detected by Western blotting analysis using an anti-Myc9E10 antibody (top panel). The expressions of Myc-ARNT, V5-AhR, and 6xMyc-Smad3 MH2 were evaluated using anti-Myc9E10 (second panel), anti-AhR (third panel), and anti-Myc9E10 (bottom panel) antibodies, respectively. The intensity of the band for ARNT binding to DNA was normalized using the intensity of the band corresponding to total ARNT. Relative intensity was calculated relative to the value for Myc-ARNT plus V5-AhR. C, dissociation of ARNT from XREs upon TGF-β stimulation. A549 cells were pretreated with 5 ng/ml TGF-β 2 h before exposure to 1 µm B[a]P for 2 h. Then the cell lysates were mixed with biotinylated (XRE)2. ARNT that binds to DNA was detected by Western blotting analysis using anti-ARNT antibody (top panel). The expressions of AhR, ARNT, phosphorylated Smad2, and β-actin were evaluated using anti-AhR (second panel), anti-ARNT (third panel), anti-phosphorylated Smad2 (PS2) (fourth panel), and anti-β-actin (bottom panel) antibodies, respectively. The bands corresponding to phosphorylated Smad2 indicate that the cells responded to TGF-β. The intensity of the band for ARNT binding to DNA was normalized using the intensity of the band corresponding to total ARNT. Relative intensity was calculated relative to the value for B[a]P stimulation alone. ND, not determined.
Contribution of HIF-1α to the inhibition of TGF-β–mediated transcriptional repression of the CYP1A1 gene induced by B[a]P
In addition to AhR, ARNT can form a heteromeric complex with HIF-1α, whose expression is known to be induced by TGF-β and contributes to the activation of hypoxia-related genes (39–41). Indeed, we could detect a prolonged t½ of HIF-1α protein in the presence of TGF-β when HIF-1α was expressed in 911 cells (Fig. 8A). Next, we investigated whether HIF-1α stabilized by TGF-β interrupts the B[a]P-induced transcription of the CYP1A1 gene. To address this possibility, we used constitutively active HIF-1α (caHIF-1α) in which two proline residues targeted by prolyl hydroxylase (PHD) were replaced with alanine and glycine at positions 402 and 564, respectively (39), because the WT HIF-1α protein was unstable in the cells. Then caHIF-1α was transfected with pGL3ti-(XRE)2-luc in the presence or absence of B[a]P. As seen in Fig. 8B, caHIF-1α dose-dependently interfered with the reporter activity induced by B[a]P. When we transfected the fusion gene between AhR and the Gal4 DNA-binding domain (Gal4 DBD) together with ARNT, we could observe transactivation of the reporter gene including the Gal4 DNA-binding sequence as an enhancer, although B[a]P marginally enhanced its transactivation. caHIF-1α obviously blocked the basal and B[a]P-induced reporter activities (Fig. 8C). To further confirm the above results, we also tested whether decreased expression of HIF-1α promoted TGF-β–mediated suppression of B[a]P-induced CYP1A1 transcription in A549 cells. Thus, we generated two independent HIF-1α–knocked-down cell lines, termed A549-shHIF-1α-2 and A549-shHIF-1α-3 (Fig. 8D). When these cells were transfected with pGL3ti-(XRE)2-luc, their TGF-β–mediated suppression of B[a]P-induced CYP1A1 transcription was reduced when compared with that of A549-shCont (Fig. 8E). Therefore, HIF-1α stabilized by TGF-β might contribute to inhibition of B[a]P-induced CYP1A1 expression to some extent. From these results, we speculated that HIF-1α disrupted the heteromeric complex between AhR and ARNT. Expectedly, HIF-1α promoted the dissociation between AhR and ARNT.
Figure 8.
Effect of HIF-1α on CYP1A1 promoter activity. A, prolongation of the t½ of HIF-1α upon TGF-β stimulation. 911 cells transfected with FLAG-HIF-1α were pretreated with 50 ng/ml cycloheximide (CHX) before stimulation of the cells with 5 ng/ml TGF-β. Then the cell lysates were prepared at the indicated times. The top and bottom panels indicate the expressions of HIF-1α and β-actin by anti-FLAG and anti-β-actin antibodies, respectively. Cont, without TGF-β stimulation. The intensity of the band for HIF-1α was normalized using the intensity of the band corresponding to β-actin. Relative intensity was calculated relative to the value for FLAG-HIF-1α without treatment with CHX. B, inhibition of B[a]P-induced reporter activity by HIF-1α. A549 cells were transfected with the indicated plasmids. Twenty-four hours later, the cells were exposed to 1 µm B[a]P for 18 h. Significant differences from the control with B[a]P in either the absence or the presence of AhR are indicated with asterisks. C, inhibition of B[a]P-induced reporter activity by HIF-1α using a Gal4 luciferase reporter. The expression plasmid for the fusion protein between Gal4 DBD and AhR, ARNT, Gal4-TATA-luc, and pCH110 was transfected with or without caHIF-1α in A549 cells. Twenty-four hours later, the cells were stimulated with or without 1 µm B[a]P for 18 h. Significant differences from the control without HIF-1α in either the absence or the presence of B[a]P are indicated with asterisks. D, expression of HIF-1α in A549 cells carrying the knocked-down vectors for HIF-1α. To detect endogenous HIF-1α protein, cells were cultured under hypoxia 28 h before cell lysis. The top and bottom panels indicate the expressions of HIF-1α and β-actin, respectively. A549-shCont, A549 cells carrying pLKO.1 alone; A549-shHIF-1α-2, A549 cells carrying pLKO.1-HIF-1α-sh2; A549-shHIF-1α-3, A549 cells carrying pLKO.1-HIF-1α-sh3. The intensity of the band for HIF-1α was normalized using the intensity of the band corresponding to β-actin. Relative intensity was calculated relative to the value for HIF-1α under hypoxia in A549-shCont. E, partial recovery of B[a]P-induced reporter activity in the presence of TGF-β. HIF-1α–knocked-down cells were transfected with pGL3ti-(XRE)2-luc and CH110. Twenty-four hours later, the cells were exposed to 1 µm B[a]P for 6 h after a 4-h treatment with 5 ng/ml TGF-β. Significant differences between A549-shCont and either A549-shHIF-1α-2 or A549-shHIF-1α-3 are indicated with asterisks. F, HIF-1α–mediated dissociation between AhR and ARNT. 293T cells were transfected with the indicated plasmids. The interaction between AhR and ARNT is shown in the top panel. The total expressions of Myc-ARNT, V5-AhR, and FLAG-HIF-1α are indicated in the second, third, and bottom panels, respectively. The intensity of the band for ARNT precipitated with AhR was normalized using the intensity of the band corresponding to total ARNT. Relative intensity was calculated relative to the value for Myc-ARNT plus V5-AhR. ND, not determined.
Effect of TGF-β on the genomic instability mediated by B[a]P
Because TGF-β inhibits the expression of CYP1A1 induced by B[a]P, the CYP1A1-mediated metabolic activation of B[a]P might decrease in cells exposed to TGF-β. To evince this possibility, we examined the expression of phosphorylated histone H2AX (γ-H2AX), which is an indicator of DNA double-strand breaks (42). Fluorescence microscopy showed that B[a]P-induced expression of γ-H2AX was reduced upon TGF-β stimulation (Fig. 9A). Consistently, Western blotting analysis also confirmed the reduction of B[a]P-induced γ-H2AX protein upon TGF-β stimulation (Fig. 9B). On the other hand, loss of TGF-β/Smad3 signaling in hepatocytes potentiated expression of B[a]P-mediated γ-H2AX protein even in the presence of TGF-β (Fig. 9C).
Figure 9.
TGF-β-mediated inhibition of B[a]P-induced double-strand DNA breaks. A, inhibition of B[a]P-induced γ-H2AX foci by TGF-β. A549 cells were pretreated with 5 ng/ml TGF-β 4 h before stimulation of the cells with 39.6 µm B[a]P for 24 h. Red color, nuclei showing double-stranded DNA breaks; blue color, nucleus with DAPI. Bar, 50 µm. B, suppression of γ-H2AX expression by TGF-β. The experiments were performed according to the description in A. Then the cell lysates were prepared for Western blotting analysis. Top panel, γ-H2AX expression; bottom panel, β-actin expression. The intensity of the band for γ-H2AX was normalized using the intensity of the band corresponding to β-actin. Relative intensity was calculated relative to the value for TGF-β stimulation alone. C, B[a]P-induced γ-H2AX expression by TGF-β in hepatocytes from Smad3 KO mice. Hepatocytes from either WT or Smad3 KO mice were pretreated with 5 ng/ml TGF-β 4 h before stimulation of the cells with 39.6 µm B[a]P for 24 h. The following experiments were carried out according to B. Top panel, γ-H2AX expression; bottom panel, β-actin expression. The intensity of the band for γ-H2AX was normalized using the intensity of the band corresponding to β-actin. Relative intensity was calculated relative to the value for no stimulus in hepatocytes from WT mice. ND, not determined.
Discussion
Integration of several diverse signal transduction pathways is implicated in tumor development. Among them, TGF-β is known to possess biphasic effects during tumorigenicity: in the early phase of tumorigenicity, TGF-β suppresses tumor growth via induction of cyclin-dependent kinase inhibitors and repression of c-Myc expression, whereas in the late stage, it promotes cell motility, the epithelial mesenchymal transition, angiogenesis, and suppression of cytotoxic T cells and NK cells (43). Thus, the TGF-β signaling pathway is the so-called “Jekyll and Hyde” of cancer because it possesses the contradictory roles of tumor suppression and tumor promotion (44). PAHs, including B[a]P and TCDD, are involved in the induction of CYP1A1 metabolizing them. Hence, PAHs become potential toxicants and chemical carcinogens with a concomitant formation of epoxide by epoxide hydroxylase. Thus, inhibition of CYP1A1 expression might protect the cells from the genotoxic action of CYP1A1-activated PAHs. It has been reported that CYP1A1 is down-regulated by TGF-β. However, the molecular mechanism by which TGF-β interferes in the transcription of the CYP1A1 gene is not fully understood except for reports that TGF-β perturbs the expression of AhR mRNA (26, 45). In this study, we hypothesized that TGF-β signaling possesses other inhibitory machinery to regulate transcription of the CYP1A1 gene besides repression of AhR expression. Interestingly, the inhibitory action of TGF-β depended on the presence of XREs in the CYP1A1 promoter because the luciferase activity of CYP1A1(-313)-luc, which has no XREs in its promoter, was not affected by TGF-β. To further confirm that TGF-β inhibits the transcription of the CYP1A1 promoter, we assessed the luciferase reporters in which the XREs are multimerized. Expectedly, TGF-β hampered B[a]P-induced luciferase reporters, including the XRE in the proximal region of the minimal promoter. Curiously, there were no inhibitory actions of TGF-β on B[a]P-induced activity of pGL3ti-(XRE)2-luc in Smad3-deficient MEFs. On the other hand, the introduction of either Smad2 or Smad3 in Smad3-deficient MEFs improved the responsiveness to TGF-β, although the introduction of Smad2 had a smaller effect than that of Smad3. This finding triggered the idea that Smad3 might interfere in the B[a]P-mediated interaction between AhR and ARNT. As expected, among the examined Smads, Smad3 associated with both AhR and ARNT most strongly, although Smad2 and Smad4 could interact with them as well. In particular, the MH2 domain of Smad3 played a crucial role in the dissociation of AhR and ARNT (Fig. 6D). Consistently, AhR and ARNT lost the ability to bind to the XREs when the MH2 domain of Smad3 was overexpressed (Fig. 7, A and B). This event might occur because heterodimerization between AhR and ARNT is necessary for the AhR/ARNT complex to bind to the XREs (3). These lines of evidence might support the notion that the MH2 domain of Smad2 is also required for Smad2 to suppress the B[a]P-induced CYP1A1 expression by TGF-β because the MH2 domains between Smad2 and Smad3 are highly homologous.
Because HIF-1α, which has been reported to be induced by TGF-β, interacts with ARNT (46), we hypothesized that HIF-1α prompts dissociation between AhR and ARNT to perturb B[a]P-mediated CYP1A1 transcription upon TGF-β stimulation. We could detect obvious prolongation of the t½ of HIF-1α protein when cells were stimulated with TGF-β. Furthermore, caHIF-1α inhibited B[a]P-induced reporter activities of both pGL3ti-(XRE)2-luc and Gal4-TATA-luc. Indeed, HIF-1α dissociated the complex formation between AhR and ARNT upon TGF-β stimulation, suggesting that stabilized HIF-1α might interfere with B[a]P-mediated association between AhR and ARNT to suppress the transcription of the CYP1A1 gene. Although McMahon et al. (39) revealed that TGF-β inhibits the expression of PHD2 to increase HIF-1α protein, we could not see any differences in mRNA expression among PHD1, PHD2, and PHD3 when cells were stimulated with TGF-β (data not shown). Thus, an unknown mechanism(s) other than decreased expression of PHD2 might lead HIF-1α to be stabilized in the presence of TGF-β. We plan to elucidate this unknown mechanism in the future.
CYP1A1 can oxidatively metabolize PAHs including B[a]P to convert them into mutagens that covalently bind to DNAs (3). Thus, we postulated that TGF-β suppresses B[a]P-induced genetic instability because of a diminishment of CYP1A1 expression. Using the expression of γ-H2AX as a biomarker for DNA double-strand breaks (47), we found that TGF-β obviously blocked B[a]P-induced genotoxicity. In the present study, we have not measured metabolites of B[a]P in cells using HPLC because we could evaluate TGF-β–mediated inhibition of B[a]P-induced genotoxicity by detection of γ-H2AX protein. To further investigate the effect of TGF-β on inhibition of CYP1A1 expression, the detection of metabolites for B[a]P using HPLC will be required.
Müller et al. (48) could also observe the inhibition of CYP1A1 enzymatic activity when cells were stimulated with TGF-β, although they did not know the exact mechanism by which TGF-β inhibited the expression of B[a]P-induced CYP1A1 transcription. Therefore, our current study proposes a novel mechanism in which TGF-β/Smad3 signaling hampers the B[a]P-induced transcription of the CYP1A1 gene. Additionally, TGF-β marginally maintained the cytoplasmic retention of AhR in the presence of B[a]P (data not shown), which was consistent with the results from Staršíchová et al. (27).
AhR has been reported to potentiate the TGF-β signaling pathway in glioma to promote its tumorigenicity (49, 50). Therefore, B[a]P-induced AhR activation might affect the TGF-β/Smad signaling pathway, although we did not examine the effect of the AhR/ARNT complex on TGF-β/Smad3 signaling. Thus, we must also investigate the involved intricate signaling circuit in the future.
In conclusion, the TGF-β/Smad3 pathway promotes dissociation between AhR and ARNT even in the presence of B[a]P. Additionally, HIF-1α stabilized by TGF-β competes with AhR for binding to ARNT. Consequently, the metabolic activation of B[a]P by CYP1A1 might be lowered owing to decreased expression of CYP1A1 (Fig. 10).
Figure 10.
Proposed model for TGF-β/Smad signaling to inhibit CYP1A1 promoter activity. Without TGF-β, after B[a]P binds to AhR, the complex between B[a]P and AhR enters the nucleus, where AhR interacts with ARNT. Then AhR/ARNT complex recognizes XREs on the CYP1A1 promoter to activate the CYP1A1 gene. Consequently, the CYP1A1 protein metabolically activates B[a]P to make it an ultimate carcinogen. With TGF-β, activated Smad3 (and maybe activated Smad2) upon TGF-β stimulation interacts with AhR and/or ARNT to interfere in the heteromeric complex between AhR and ARNT. Additionally, HIF-1α stabilized by TGF-β promotes dissociation between the AhR and ARNT complex. Therefore, enough of the AhR/ARNT complex cannot bind to XREs in the CYP1A1 promoter even when exposed to B[a]P. Thus, TGF-β signaling might be able to protect cells from increased carcinogenic risk via metabolic activation of B[a]P by CYP1A1.
Experimental procedures
Antibodies
Antibodies were obtained from the following sources: anti-FLAG M5 mouse mAb (F4042-1MG) from Sigma–Aldrich; anti-DYKDDDDK tag mAb (040-30953) corresponding to anti-FLAG antibody from FUJIFILM Wako; anti-Myc9E10 mAb (sc-40) from Santa Cruz Biotechnology, Inc.; anti-Myc tag rabbit polyclonal antibody (pAb) (06-549) from Merck; anti-V5 tag mouse mAb (011-23594) from FUJIFILM Wako; anti-β-actin mouse mAb (AC-15) (sc-69879) from Santa Cruz Biotechnology; anti-AhR rabbit pAb (ABS522) from Merck; anti-ALK5 (V-22) rabbit pAb (sc-398) from Santa Cruz Biotechnology; anti-ARNT1 mouse mAb (sc-17812) from Santa Cruz Biotechnology; anti-HIF-1α rabbit mAb (ab51608) from Abcam; anti-phospho-histone H2A.X (Ser-139) mouse mAb (05-636) corresponding to anti-γ-H2AX antibody from Merck; anti-Smad3 rabbit mAb (04-1035) from Merck; anti-phospho-Smad3 (Ser-423/425) (C25A9) rabbit mAb (9520T) (PS3) from Cell Signaling Technology; anti-mouse IgG HRP-linked sheep pAb (NA931-1ML) from GE Healthcare; and anti-rabbit IgG HRP-linked donkey F(ab)2 fragment (NA9340-1ML) from GE Healthcare. Anti-phosphorylated Smad2 rabbit pAb (PS2) antibody was homemade (51).
Cell culture
911, 293A, A549, and COS7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Nacalai Tesque) containing 10% fetal calf serum (FCS; Invitrogen). MEFs established from Smad3-HET and Smad3-KO mice (29, 30) were cultured in DMEM containing 10% FCS. Primary hepatocytes from WT and Smad3-KO mice were prepared and cultured according to the methods described by Nemoto et al. (52) and Sakuma et al. (53). Then the hepatocytes were cultured in collagen-coated dishes. All animal work was performed with institutional approval by the Animal Care and Use Committee of Showa Pharmaceutical University (approval P-2018-01-R1, dated April 19, 2018).
Expression plasmids
Human AhR, ARNT, and HIF-1α cDNAs were kindly provided by Prof. Y. Fujii-Kuriyama (University of Tsukuba, Japan) and Prof. S. Hara (Showa University, Japan) (54, 55). caHIF-1α, in which prolines are mutated into alanine and glycine at positions 402 and 564 (39), was made with PrimeStar HS DNA Polymerase (Takara). Then the fragment was inserted into a pcDEF3 vector (56). All constructs described above possessed the FLAG, Myc, or V5 epitope tag at their N terminus. For CYP1A1(−4150)-luc, the human CYP1A1 promoter from −4150 to +19 was amplified by PCR using the genome from human A549 cells as a template (referring to the sequence information of NC_000015.10 in the NCBI Reference Sequence) and cloned into pGL3-basic (Promega). For CYP1A1(−1249)-luc, CYP1A1(−931)-luc, CYP1A1(−620)-luc, and CYP1A1(−313)-luc, the fragments from −1249 to +19, −931 to +19, −620 to +19, and −313 to +19 were respectively isolated by PCR-based amplification using CYP1A1(−4150)-luc as a template and inserted into pGL3-basic. To generate pGL3ti-(XRE)2-luc, pGL3ti-(XRE)4-luc, and pGL3ti-(XRE)6-luc, two oligonucleotides (5′-TCGAGTCTCACGCGAGCCGGGACTCTCACGCGAGCCGGGAC-3′/3′-CAGAGTGGCTCGGCCCTGAGAGTGCGCTCGGCCCTGAGCT-5′; the underlined letters are the XREs) were multimerized and subcloned into pGL3ti-luc (35). The pGL3ti-(mXRE)4-luc reporter including mutated XRE was constructed using two oligonucleotides (5′-TCGAGTCTaAaaaaAGCCGGGACTCTaAaaaaAGCCGGGAC-3′/3′-CAGAtTttttTCGGCCCTGAGAtAttttTCGGCCCTGAGCT-5′; the lowercase letters indicate mutated nucleotides). Gal4-DBD-AhR was made by fusion between Gal4 DBD (57) and AhR according to Itoh et al. (58). The nucleotide sequences of all the plasmids were confirmed by sequencing. The other constructs have been described previously (59–61).
Lentiviral shRNAs for HIF-1α
The lentiviral vectors for HIF-1α shRNA were constructed using a pLKO.1 vector (Addgene) (62). The sequences for HIF-1α-sh2 and HIF-1α-sh3 were 5′-CCGGAACTGATGACCAGCAACTTGActcgagTCAAGTTGCTGGTCATCAGTTTTTTTG-3′/3′-TTGACTACTGGTCGTTGAACTgagctcAGTTCAACGACCAGTAGTCAAAAAAACTTAA-5′ and 5′-CCGGAAGCACAGTTACAGTATTCCActcgagTGGAATACTGTAACTGTGCTTTTTTTG-3′/3′-TTCGTGTCAATGTCATAAGGTgagctcACCTTATGACATTGACACGAAAAAAACTTAA-5′, respectively. Each lentiviral vector was transfected into 293T cells together with psPAX2 and pMD2.G. After 48 h of transfection, the media were collected as a source of lentiviruses. Each lentivirus was incubated in DMEM containing 8 µg/ml Polybrene (Sigma–Aldrich) for 2 h and then added to the A549 cell culture dishes. Twelve hours after infection, the cells were washed and cultured in medium. The infected A549 cells, which became puromycin-resistant, were used for the experiments.
Genome editing using the CRISPR/Cas9 system
A549 cells were cloned and used as a parental cell line (A549-P1), which was authenticated by short tandem repeat analysis. Knockout of SMAD3 was conducted using Double Nickase plasmid (catalog no. sc-4000069-NIC-2; Santa Cruz Biotechnology). Briefly, cells were grown until 70% confluence on a 6-well plate and then transfected with 1 μg of plasmid DNA using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). After 24 h of incubation, the cells were treated with 1 μg/ml puromycin (Sigma–Aldrich) to eliminate nontransfected cells within 48 h. Deletion/disruption of the target genes was confirmed by sequencing (Fig. S2). There are three SMAD3 loci in A549 cells.
Transcriptional reporter assays
One day before transfection, A549 cells and MEFs were seeded at 1.2 × 105 and 1.0 × 105 cells/well in 12- and 6-well plates, respectively. The cells were transfected with a reporter gene, pCH110 (GE Healthcare), and the indicated plasmids by use of polyethyleneimine (PEI; Polysciences). When 5 ng/ml TGF-β1 (Peprotech) was added to the wells 24 h after transfection, the cells were cultured in the absence of FCS for 18 h. In all the experiments, β-gal activity was measured to normalize for transfection efficiency. Each transfection was carried out in triplicate and repeated at least twice.
Immunoprecipitation and Western blotting analysis
To detect interactions among the proteins, plasmids were transfected into COS7 cells (5 × 105 cells/6-cm dish), 911 cells (1.5 × 106 cells/10-cm dish), or 293T cells (3.0 × 106 cells/10-cm dish) by use of PEI. Forty hours after the transfection, the cells were lysed in 500 µl of TNE buffer (10 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 100 units/ml aprotinin, 2 mm sodium vanadate, 40 mm NaF, and 20 mm β-glycerophosphate). The cell lysates were precleared with protein G–Sepharose beads (GE Healthcare) for 30 min at 4 °C and then incubated with the indicated antibodies for 2 h at 4 °C. The protein complexes were immunoprecipitated by incubation with protein G–Sepharose beads for 30 min at 4 °C and then washed three times with TNE buffer. The immunoprecipitated proteins and aliquots of the total cell lysates were boiled for 5 min in sample buffer, separated by SDS-PAGE, and transferred to UltraCruz Nitrocellulose Pure Transfer membranes (Santa Cruz Biotechnology). The membranes were probed with the primary antibodies, which were detected with horseradish peroxidase–conjugated secondary antibodies and a chemiluminescent substrate (Western BLoT Quant HRP Substrate, Takara). The protein expression in the total cell lysates was evaluated by Western blotting analysis.
PLA
A549 cells on cover glasses coated with 0.1% gelatin (Sigma–Aldrich) were cultured with DMEM. After TGF-β stimulation, the cover glasses were washed once with PBS, fixed for 10 min with 4% paraformaldehyde, washed three times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and again washed three times with PBS. The following procedures were performed according to the manufacturer's instructions (Olink Bioscience). To visualize the fluorescence, a confocal microscope (Nikon, A1R) was used.
RNA preparation and quantitative PCR (qPCR) analysis
Total RNAs from A549 cells and hepatocytes from WT or Smad3-KO mice were extracted using a ReliaPrep RNA Cell Miniprep System (Promega). Reverse transcription was performed with a High-Capacity RNA-to-cDNA kit (Thermo Fisher Scientific). qPCR was performed using a KAPA SYBR Fast qPCR kit (KAPA). All reactions were carried out on a Thermal Cycler Dice (TAKARA). Each sample was analyzed in triplicate at least twice for each PCR measurement. The melting curves were checked to ensure specificity. Relative quantification of mRNA expression was calculated using the standard curve method with the β-actin level. Before qPCR, we used agarose gel electrophoresis to confirm that the DNA fragment amplified using each primer set was a single band with the correct size. The primer sets are listed in Table 1.
Table 1.
Primer sets for qPCR
| Species | Gene | Sequence (5′ to 3′) |
|---|---|---|
| Human | CYP1A1 | GATGGTCAAGGAGCACTACA |
| TACAAAGACACAACGCCCCT | ||
| CYP1B1 | ACGTACCGGCCACTATCACT | |
| ATACAAGGCAGACGGTCCCT | ||
| AhR | GTCTAAGGTGTCTGCTGGAT | |
| TAGACTGGACCCAAGTCCAT | ||
| β-actin | CAAGAGATGGCCACGGCTGCT | |
| TCCTTCTGCATCCTGTCGGCA | ||
| Mouse | CYP1A1 | TTCATCCTTCGTCCCCTTCA |
| CCCAAACCAAAGAGAGTGAC | ||
| CYP1B1 | TTCTGGACAAGTTCCTGAGG | |
| ATGACATATGGCAGGTTGGG | ||
| Smad7 | GGAAGTCAAGAGGCTGTGTT | |
| GTCTTCTCCTCCCAGTATGC | ||
| β-actin | TGAACCCTAAGGCCAACCGTG | |
| GCTCATAGTCTTCTCCAGGG |
DNA affinity precipitation
911 cells were seeded at 1.5 × 106 cells/well in a 10-cm dish 1 day before transfection. The cells were transfected with the indicated plasmids by use of PEI. Forty hours after the transfection, the cells were pretreated with 50 µm MG-132 (FUJIFILM Wako) 2 h before stimulation of the cells with 1 µm B[a]P (Tokyo Chemical Industry) for 2 h. Then the cells were lysed in 1 ml of TNE buffer. The cell lysates were precleared with 12 µg/ml poly(dI·dC) (GE Healthcare) and streptavidin agarose (Sigma–Aldrich) for 30 min and incubated with 24 µm biotinylated (XRE)2 for 2 h at 4 °C. Subsequently, streptavidin agarose was added to the reaction mixture and incubated for 30 min at 4 °C. After the precipitates were washed with TNE buffer three times, the precipitates and aliquots of the total lysates were separated by SDS-PAGE. Then the proteins were transferred to the membrane for Western blotting analysis. The membrane was incubated with the indicated primary antibodies. The primary antibodies were detected as described above. The sequences of biotinylated (XRE)2 are as follows: 5′-biotinated-CTCTCACGCGAGCCGGGACTCTCACGCGAGCCGGGACG-3′/3'-CATGGAGAGTGCGCTCGGCCCTGAGAGTGCGCTCGGCCCTGCGATC-5′ (the underlined letters indicate the XREs). When the endogenous proteins were used for DNA affinity precipitation, A549 cells were prestimulated with 5 ng/ml TGF-β 2 h before treatment with 1 µm B[a]P for 2 h.
Measurement of protein t½
FLAG-HIF-1α was transfected into 911 cells by use of PEI. After 40 h, the cells were pretreated with 50 ng/ml cycloheximide (Sigma–Aldrich) 30 min before the addition of 5 ng/ml TGF-β for the indicated times. Then Western blotting analysis was performed as described above.
γ-H2AX staining
A549 cells were seeded at 3 × 104 cells/12-well plate on glass cover slides coated with 0.1% gelatin (in PBS). The cells were then pretreated with 5 ng/ml TGF-β 4 h before stimulation of the cells with 39.6 µm B[a]P for 24 h. Then the cover glasses were washed once with PBS, fixed for 10 min with 4% paraformaldehyde, washed three times with PBS, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and again washed three times with PBS. The cover glasses were blocked with 5% BSA in PBS at 37 °C for 1 h and incubated with 5% BSA (in PBS) containing γ-H2AX antibody at 4 °C overnight. The cover glasses were then washed three times with PBS, incubated with 5% BSA (in PBS) containing Alexa Fluor 555–conjugated goat anti-mouse IgG antibody (diluted 1:250) (A-21422, Thermo Fisher Scientific) at room temperature for 1 h, and then washed three times with PBS. The nuclei were stained with 4′,6-amino-2-phenylidole (DAPI). A confocal microscope was used to visualize the fluorescence.
Statistical analysis
Data are expressed as mean ± S.D. (n = 3) unless otherwise mentioned. Significance was assessed using the t test. Probability values below 0.05, 0.01, and 0.001 were considered significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Data availability
All data generated during this study except for data described as “data not shown” are included in this article. The data described as “data not shown” can be obtained from the corresponding author (Susumu Itoh, sitoh@ac.shoyaku.ac.jp) upon request.
Supplementary Material
Acknowledgments
Prof. T. Sakuma (Ohu University) kindly showed us how to prepare hepatocytes from mice. We thank F. Miyamasu for excellent English proofreading and K. Ohmura, M. Kikuma, M. Fukuta, and T. Numanyu for helpful technical support.
This article contains supporting information.
Author contributions—N. N., N. S., K. M., and S. Itoh conceptualization; N. N., N. S., Y. K., D. N., E. S., Y. T., S. Y., Y. H., S. Ikeno, M. M., K. S., K. Y., K. M., and S. Itoh data curation; N. N., N. S., Y. K., D. N., E. S., Y. T., S. Y., Y. H., S. Ikeno, M. M., K. S., and K. Y. formal analysis; N. N., N. S., S. Ikeno, M. M., K. S., K. Y., K. M., and S. Itoh supervision; N. N. and S. Itoh funding acquisition; N. N., N. S., Y. K., D. N., K. M., and S. Itoh validation; N. N., N. S., Y. K., D. N., E. S., Y. T., S. Y., Y. H., S. Ikeno, M. M., K. S., K. Y., and K. M. investigation; N. N., N. S., Y. K., D. N., E. S., Y. T., S. Y., Y. H., S. Ikeno, M. M., K. S., K. Y., and K. M. methodology; N. N. and S. Itoh writing-original draft; N. N., K. M., and S. Itoh project administration; N. N., N. S., Y. K., D. N., E. S., Y. T., S. Y., Y. H., S. Ikeno, M. M., K. S., K. Y., K. M., and S. Itoh writing-review and editing.
Funding and additional information—This research was supported by the Smoking Research Foundation (to S. Itoh). This work was also supported by the Japan Society for the Promotion of Science Core-to-Core Program, “Cooperative International Framework in TGF-β Family Signaling.”
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CYP
- cytochrome P450
- XRE
- xenobiotic-responsive element
- AhR
- aryl hydrocarbon receptor
- ARNT
- AhR nuclear translocator
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- TGF-β
- transforming growth factor-β
- R-Smad
- receptor-regulated Smad
- KO
- knockout
- MEF
- mouse embryo fibroblast
- IP
- immunoprecipitation
- PLA
- proximity ligation assay
- MH
- Mad homology
- caHIF-1α
- constitutively active HIF-1α
- PHD
- prolyl hydroxylase
- DBD
- DNA-binding domain
- pAb
- polyclonal antibody
- HRP
- horseradish peroxidase
- DMEM
- Dulbecco's modified Eagle's medium
- FCS
- fetal calf serum
- PEI
- polyethyleneimine
- qPCR
- quantitative PCR
- DAPI
- 4′,6-diamidino-2-phenylindole
- HIF-1α
- hypoxia-inducible factor 1α
- PAH
- polycyclic aromatic hydrocarbon
- B[a]P
- benzo[a]pyrene.
References
- 1. Guengerich F. P., Waterman M. R., and Egli M. (2016) Recent structural insights into cytochrome P450 function. Trends Pharmacol. Sci. 37, 625–640 10.1016/j.tips.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schiering C., Wincent E., Metidji A., Iseppon A., Li Y., Potocnik A. J., Omenetti S., Henderson C. J., Wolf C. R., Nebert D. W., and Stockinger B. (2017) Feedback control of AHR signalling regulates intestinal immunity. Nature 542, 242–245 10.1038/nature21080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whitlock J. P., Jr. (1999) Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39, 103–125 10.1146/annurev.pharmtox.39.1.103 [DOI] [PubMed] [Google Scholar]
- 4. Sogawa K., and Fujii-Kuriyama Y. (1997) Ah receptor, a novel ligand-activated transcription factor. J. Biochem. 122, 1075–1079 10.1093/oxfordjournals.jbchem.a021864 [DOI] [PubMed] [Google Scholar]
- 5. Fujii-Kuriyama Y., and Mimura J. (2005) Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 338, 311–317 10.1016/j.bbrc.2005.08.162 [DOI] [PubMed] [Google Scholar]
- 6. Endler A., Chen L., and Shibasaki F. (2014) Coactivator recruitment of AhR/ARNT1. Int. J. Mol. Sci. 15, 11100–11110 10.3390/ijms150611100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Larigot L., Juricek L., Dairou J., and Coumoul X. (2018) AhR signaling pathways and regulatory functions. Biochim. Open 7, 1–9 10.1016/j.biopen.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu K., Meng Q., Zhang Z., Yi T., He Y., Zheng J., and Lei W. (2019) Aryl hydrocarbon receptor pathway: role, regulation and intervention in atherosclerosis therapy (Review). Mol. Med. Rep. 20, 4763–4773 10.3892/mmr.2019.10748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hahn M. E., Allan L. L., and Sherr D. H. (2009) Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochem. Pharmacol. 77, 485–497 10.1016/j.bcp.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fradette C., Bleau A. M., Pichette V., Chauret N., and Du Souich P. (2002) Hypoxia-induced down-regulation of CYP1A1/1A2 and up-regulation of CYP3A6 involves serum mediators. Br. J. Pharmacol. 137, 881–891 10.1038/sj.bjp.0704933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tian Y. (2009) Ah receptor and NF-κB interplay on the stage of epigenome. Biochem. Pharmacol. 77, 670–680 10.1016/j.bcp.2008.10.023 [DOI] [PubMed] [Google Scholar]
- 12. Oesch-Bartlomowicz B., and Oesch F. (2009) Role of cAMP in mediating AHR signaling. Biochem. Pharmacol. 77, 627–641 10.1016/j.bcp.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 13. Chen S., Operaña T., Bonzo J., Nguyen N., and Tukey R. H. (2005) ERK kinase inhibition stabilizes the aryl hydrocarbon receptor: implications for transcriptional activation and protein degradation. J. Biol. Chem. 280, 4350–4359 10.1074/jbc.M411554200 [DOI] [PubMed] [Google Scholar]
- 14. Henklová P., Vrzal R., Ulrichová J., and Dvořák Z. (2008) Role of mitogen-activated protein kinases in aryl hydrocarbon receptor signaling. Chem. Biol. Interact. 172, 93–104 10.1016/j.cbi.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 15. Yin J., Sheng B., Qiu Y., Yang K., Xiao W., and Yang H. (2016) Role of AhR in positive regulation of cell proliferation and survival. Cell Prolif. 49, 554–560 10.1111/cpr.12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng X. H., and Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 10.1146/annurev.cellbio.21.022404.142018 [DOI] [PubMed] [Google Scholar]
- 17. Shi Y., and Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- 18. Derynck R., and Zhang Y. E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425, 577–584 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- 19. Derynck R., and Budi E. H. (2019) Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 12, eaav5183 10.1126/scisignal.aav5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Derynck R., and Akhurst R. J. (2007) Differentiation plasticity regulated by TGF-β family proteins in development and disease. Nat. Cell Biol. 9, 1000–1004 10.1038/ncb434 [DOI] [PubMed] [Google Scholar]
- 21. Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weiss A., and Attisano L. (2013) The TGFβ superfamily signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2, 47–63 10.1002/wdev.86 [DOI] [PubMed] [Google Scholar]
- 23. Heldin C. H., and Moustakas A. (2016) Signaling receptors for TGF-β family members. Cold Spring Harbor Perspect. Biol. 8, a022053 10.1101/cshperspect.a022053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morikawa M., Derynck R., and Miyazono K. (2016) TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 8, a021873 10.1101/cshperspect.a021873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. David C. J., and Massagué J. (2018) Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 10.1038/s41580-018-0007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Döhr O., Sinning R., Vogel C., Münzel P., and Abel J. (1997) Effect of transforming growth factor-β1 on expression of aryl hydrocarbon receptor and genes of Ah gene battery: clues for independent down-regulation in A549 cells. Mol. Pharmacol. 51, 703–710 10.1124/mol.51.5.703 [DOI] [PubMed] [Google Scholar]
- 27. Staršíchová A., Hrubá E., Slabáková E., Pernicová Z., Procházková J., Pěnčíková K., Šeda V., Kabátková M., Vondráček J., Kozubík A., Machala M., and Souček K. (2012) TGF-β1 signaling plays a dominant role in the crosstalk between TGF-β1 and the aryl hydrocarbon receptor ligand in prostate epithelial cells. Cell. Signal. 24, 1665–1676 10.1016/j.cellsig.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 28. Shimada T., Inoue K., Suzuki Y., Kawai T., Azuma E., Nakajima T., Shindo M., Kurose K., Sugie A., Yamagishi Y., Fujii-Kuriyama Y., and Hashimoto M. (2002) Arylhydrocarbon receptor-dependent induction of liver and lung cytochromes P450 1A1, 1A2, and 1B1 by polycyclic aromatic hydrocarbons and polychlorinated biphenyls in genetically engineered C57BL/6J mice. Carcinogenesis 23, 1199–1207 10.1093/carcin/23.7.1199 [DOI] [PubMed] [Google Scholar]
- 29. Ashcroft G. S., Yang X., Glick A. B., Weinstein M., Letterio J. L., Mizel D. E., Anzano M., Greenwell-Wild T., Wahl S. M., Deng C., and Roberts A. B. (1999) Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat. Cell Biol. 1, 260–266 10.1038/12971 [DOI] [PubMed] [Google Scholar]
- 30. Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., and Deng C. (1999) Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-β. EMBO J. 18, 1280–1291 10.1093/emboj/18.5.1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ema M., Ohe N., Suzuki M., Mimura J., Sogawa K., Ikawa S., and Fujii-Kuriyama Y. (1994) Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J. Biol. Chem. 269, 27337–27343 [PubMed] [Google Scholar]
- 32. Connor K. T., and Aylward L. L. (2006) Human response to dioxin: aryl hydrocarbon receptor (AhR) molecular structure, function, and dose-response data for enzyme induction indicate an impaired human AhR. J. Toxicol. Environ. Health B Crit. Rev. 9, 147–171 10.1080/15287390500196487 [DOI] [PubMed] [Google Scholar]
- 33. Flaveny C. A., Murray I. A., and Perdew G. H. (2010) Differential gene regulation by the human and mouse aryl hydrocarbon receptor. Toxicol. Sci. 114, 217–225 10.1093/toxsci/kfp308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramadoss P., and Perdew G. H. (2004) Use of 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin as a probe to determine the relative ligand affinity of human versus mouse aryl hydrocarbon receptor in cultured cells. Mol. Pharmacol. 66, 129–136 10.1124/mol.66.1.129 [DOI] [PubMed] [Google Scholar]
- 35. Jonk L. J., Itoh S., Heldin C. H., ten Dijke P., and Kruijer W. (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273, 21145–21152 10.1074/jbc.273.33.21145 [DOI] [PubMed] [Google Scholar]
- 36. Hata A., and Chen Y. G. (2016) TGF-β signaling from receptors to Smads. Cold Spring Harb. Perspect. Biol. 8, a022061 10.1101/cshperspect.a022061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hill C. S. (2016) Transcriptional control by the SMADs. Cold Spring Harb. Perspect. Biol. 8, a022079 10.1101/cshperspect.a022079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yagi K., Goto D., Hamamoto T., Takenoshita S., Kato M., and Miyazono K. (1999) Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 274, 703–709 10.1074/jbc.274.2.703 [DOI] [PubMed] [Google Scholar]
- 39. McMahon S., Charbonneau M., Grandmont S., Richard D. E., and Dubois C. M. (2006) Transforming growth factor β1 induces hypoxia-inducible factor-1 stabilization through selective inhibition of PHD2 expression. J. Biol. Chem. 281, 24171–24181 10.1074/jbc.M604507200 [DOI] [PubMed] [Google Scholar]
- 40. Basu R. K., Hubchak S., Hayashida T., Runyan C. E., Schumacker P. T., and Schnaper H. W. (2011) Interdependence of HIF-1α and TGF-β/Smad3 signaling in normoxic and hypoxic renal epithelial cell collagen expression. Am. J. Physiol. Renal Physiol. 300, F898–F905 10.1152/ajprenal.00335.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rozen-Zvi B., Hayashida T., Hubchak S. C., Hanna C., Platanias L. C., and Schnaper H. W. (2013) TGF-β/Smad3 activates mammalian target of rapamycin complex-1 to promote collagen production by increasing HIF-1α expression. Am. J. Physiol. Renal Physiol. 305, F485–F494 10.1152/ajprenal.00215.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yuan J., Adamski R., and Chen J. (2010) Focus on histone variant H2AX: to be or not to be. FEBS Lett. 584, 3717–3724 10.1016/j.febslet.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seoane J., and Gomis R. R. (2017) TGF-β family signaling in tumor suppression and cancer progression. Cold Spring Harb. Perspect. Biol. 9, a022277 10.1101/cshperspect.a022277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bierie B., and Moses H. L. (2006) Tumour microenvironment: TGFβ: the molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer 6, 506–520 10.1038/nrc1926 [DOI] [PubMed] [Google Scholar]
- 45. Wolff S., Harper P. A., Wong J. M., Mostert V., Wang Y., and Abel J. (2001) Cell-specific regulation of human aryl hydrocarbon receptor expression by transforming growth factor-β1. Mol. Pharmacol. 59, 716–724 10.1124/mol.59.4.716 [DOI] [PubMed] [Google Scholar]
- 46. Majmundar A. J., Wong W. J., and Simon M. C. (2010) Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309 10.1016/j.molcel.2010.C09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuo L. J., and Yang L. X. (2008) γ-H2AX—a novel biomarker for DNA double-strand breaks. In Vivo 22, 305–309 [PubMed] [Google Scholar]
- 48. Müller G. F., Döhr O., El-Bahay C., Kahl R., and Abel J. (2000) Effect of transforming growth factor-β1 on cytochrome P450 expression: inhibition of CYP1 mRNA and protein expression in primary rat hepatocytes. Arch. Toxicol. 74, 145–152 10.1007/s002040050667 [DOI] [PubMed] [Google Scholar]
- 49. Gramatzki D., Pantazis G., Schittenhelm J., Tabatabai G., Köhle C., Wick W., Schwarz M., Weller M., and Tritschler I. (2009) Aryl hydrocarbon receptor inhibition downregulates the TGF-β/Smad pathway in human glioblastoma cells. Oncogene 28, 2593–2605 10.1038/onc.2009.104 [DOI] [PubMed] [Google Scholar]
- 50. Silginer M., Burghardt I., Gramatzki D., Bunse L., Leske H., Rushing E. J., Hao N., Platten M., Weller M., and Roth P. (2016) The aryl hydrocarbon receptor links integrin signaling to the TGF-β pathway. Oncogene 35, 3260–3271 10.1038/onc.2015.387 [DOI] [PubMed] [Google Scholar]
- 51. Persson U., Izumi H., Souchelnytskyi S., Itoh S., Grimsby S., Engström U., Heldin C. H., Funa K., and ten Dijke P. (1998) The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 434, 83–87 10.1016/S0014-5793(98)00954-5 [DOI] [PubMed] [Google Scholar]
- 52. Nemoto N., Sakurai J., Tazawa A., and Ishikawa T. (1989) Proline-dependent expression of aryl hydrocarbon hydroxylase in C57BL/6 mouse hepatocytes in primary culture. Cancer Res. 49, 5863–5869 [PubMed] [Google Scholar]
- 53. Sakuma T., Bhadhprasit W., Hashita T., and Nemoto N. (2008) Synergism of glucocorticoid hormone with growth hormone for female-specific mouse Cyp3a44 gene expression. Drug Metab. Dispos. 36, 878–884 10.1124/dmd.107.019935 [DOI] [PubMed] [Google Scholar]
- 54. Hara S., Hamada J., Kobayashi C., Kondo Y., and Imura N. (2001) Expression and characterization of hypoxia-Inducible factor (HIF)-3α in human kidney: suppression of HIF-mediated gene expression by HIF-3α. Biochem. Biophys. Res. Commun. 287, 808–813 10.1006/bbrc.2001.5659 [DOI] [PubMed] [Google Scholar]
- 55. Horii K., Suzuki Y., Kondo Y., Akimoto M., Nishimura T., Yamabe Y., Sakaue M., Sano T., Kitagawa T., Himeno S., Imura N., and Hara S. (2007) Androgen-dependent gene expression of prostate-specific antigen is enhanced synergistically by hypoxia in human prostate cancer cells. Mol. Cancer Res. 5, 383–391 10.1158/1541-7786.MCR-06-0226 [DOI] [PubMed] [Google Scholar]
- 56. Goldman L. A., Cutrone E. C., Kotenko S. V., Krause C. D., and Langer J. A. (1996) Modifications of vectors pEF-BOS, pcDNA1 and pcDNA3 result in improved convenience and expression. BioTechniques 21, 1013–1015 10.2144/96216bm10 [DOI] [PubMed] [Google Scholar]
- 57. Sadowski I., and Ptashne M. (1989) A vector for expressing GAL4(1–147) fusions tn mammalian cells. Nucleic Acids Res. 17, 7539–7539 10.1093/nar/17.18.7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Itoh S., Ericsson J., Nishikawa J., Heldin C. H., and ten Dijke P. (2000) The transcriptional co-activator P/CAF potentiates TGF-β/Smad signaling. Nucleic Acids Res. 28, 4291–4298 10.1093/nar/28.21.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakano N., Itoh S., Watanabe Y., Maeyama K., Itoh F., and Kato M. (2010) Requirement of TCF7L2 for TGF-β-dependent transcriptional activation of the TMEPAI gene. J. Biol. Chem. 285, 38023–38033 10.1074/jbc.M110.132209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakano N., Maeyama K., Sakata N., Itoh F., Akatsu R., Nakata M., Katsu Y., Ikeno S., Togawa Y., Vo Nguyen T. T., Watanabe Y., Kato M., and Itoh S. (2014) C18 ORF1, a novel negative regulator of transforming growth factor-β signaling. J. Biol. Chem. 289, 12680–12692 10.1074/jbc.M114.558981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakano N., Kato M., and Itoh S. (2016) Regulation of the TMEPAI promoter by TCF7L2: the C-terminal tail of TCF7L2 is essential to activate the TMEPAI gene. J. Biochem. 159, 27–30 10.1093/jb/mvv117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., et al. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study except for data described as “data not shown” are included in this article. The data described as “data not shown” can be obtained from the corresponding author (Susumu Itoh, sitoh@ac.shoyaku.ac.jp) upon request.