Abstract
C-type lectins that contain collagen-like domains are known as collectins. These proteins are present both in the circulation and in extravascular compartments and are central players of the innate immune system, contributing to first-line defenses against viral, bacterial, and fungal pathogens. The collectins mannose-binding lectin (MBL) and surfactant protein D (SP-D) are regulated by tissue fibroblasts at extravascular sites via an endocytic mechanism governed by urokinase plasminogen activator receptor–associated protein (uPARAP or Endo180), which is also a collagen receptor. Here, we investigated the molecular mechanisms that drive the uPARAP-mediated cellular uptake of MBL and SP-D. We found that the uptake depends on residues within a protruding loop in the fibronectin type-II (FNII) domain of uPARAP that are also critical for collagen uptake. Importantly, however, we also identified FNII domain residues having an exclusive role in collectin uptake. We noted that these residues are absent in the related collagen receptor, the mannose receptor (MR or CD206), which consistently does not interact with collectins. We also show that the second C-type lectin-like domain (CTLD2) is critical for the uptake of SP-D, but not MBL, indicating an additional level of complexity in the interactions between collectins and uPARAP. Finally, we demonstrate that the same molecular mechanisms enable uPARAP to engage MBL immobilized on the surface of pathogens, thereby expanding the potential biological implications of this interaction. Our study reveals molecular details of the receptor-mediated cellular regulation of collectins and offers critical clues for future investigations into collectin biology and pathology.
Keywords: Receptor endocytosis, innate immunity, receptor structure-function, protein turnover, collagen, collectin, immune regulation, surfactant protein, mannose binding lectin, Endo180, mannose receptor, CD206
Introduction
Collectins are a group of proteins that make up a central part of the innate immune system. These proteins, which include mannose-binding lectin (MBL) and the surfactant proteins (SP) -A and -D, are pattern recognition molecules that interact with conserved structures on the surface of pathogens as well as apop-totic cells. The binding of collectins to these biological surfaces, a process mediated by a C-type calcium-dependent lectin-like domain (CTLD), leads to activation of complement, enhanced phagocytosis, or pathogen inactivating agglutination. Importantly, collectins also contain a short collagen-like triple-helical domain, which provides critical structural properties and mediates downstream functions via interactions with host effector proteins (1–3).
In homeostasis, MBL is confined to the circulation, whereas lung collectins SP-D and SP-A mainly cover the luminal side of the alveolar walls (4, 5). Upon tissue injury and initiation of inflammation, MBL may enter extravascular sites such as dermis and the peritoneum to exert anti-pathogenic functions (6–8). Similarly, the alveolar levels of SP-D and SP-A may increase dramatically following acute lung injury (9–14). Once the injury becomes resolved, the collectin levels quickly return to that of homeostatic conditions (10, 15), suggesting that potent catabolic mechanisms exist. Accordingly, we have recently shown that MBL and SP-D can be efficiently cleared from extravascular sites by tissue fibroblasts in a process that involves the cellular uptake and routing of these collectins to lysosomes. This process is undertaken by an endocytic collagen receptor known as urokinase plasminogen activator receptor-associated protein (uPARAP or Endo180). We have also shown that this pathway regulates endogenous levels of SP-D after lung injury (16).
uPARAP is part of the mannose receptor (MR or CD206) protein family, and the family member MR is also a known collagen receptor. The MR protein family also includes the non-collagen-binding receptors, phospholipase A2 receptor (PLA2R1), and DEC-205 (also known as CD205). All four receptors are constitutively recycling endocytic receptors that transport ligands bound in the extracellular space to the endosomal or lysosomal compartments (17). The collagen-binding properties of uPARAP and MR appear to be very similar, with binding mechanisms centered around their fibronectin type-II (FNII) domain (18). Although this might suggest a common binding site, also for other proteins with collagen-like motifs, it turns out that MR does not bind to collectins (16). This observation implies that uPARAP utilizes distinct mechanisms for interacting with collectins and collagens, despite the molecular similarities between these ligands, imposed by their triple-helical regions.
Although this distinction may seem unexpected, it is less surprising from the perspective of biological regulation. In addition to the fact that collagens and collectins are very different protein families with respect to function, they also differ strongly with respect to regulation dynamics. The maintenance of collagen structures is characterized by a rather constant turnover followed by reconstruction (19), and this pattern stands in contrast to the rapid secretion or transport of collectins to damaged or infected tissues, which is followed by a rapid clearance, when the injury is resolved (10–12, 15). Despite these differences, the turnover of both collectins and collagens is facilitated by the same cell types, including fibroblasts and macrophages (16, 20–23). However, if these cells can utilze a receptor that exclusively mediates the uptake of collagens and not collectins, i.e. MR, or a receptor that can mediate either collagen or collectin uptake, i.e. uPARAP, this will allow a differential regulatory system.
These considerations make it important to learn about the structure-function relationship that dictates the differential binding of structurally related ligands by these receptors. In the present study, we take advantage of established assays for quantifying the cellular uptake of relevant ligands, mediated by endocytic receptors, to investigate the molecular mechanisms governing the uptake of collectins and solubilized collagens by uPARAP. We find that the uptake of collectins and collagens depend on some of the same features in uPARAP's FNII domain, but that this domain also contains elements with a selective role in collectin uptake. We also find that the uptake of individual collectins require specific elements in uPARAP, because the active lectin domain in uPARAP is required for the uptake of SP-D only. Finally, we find that uPARAP can engage collectins in complex with pathogens, which suggests a downstream effector role of uPARAP in addition to the regulation of soluble collectin levels.
Results
The FNII domain is critical for uPARAP-mediated uptake of collectins
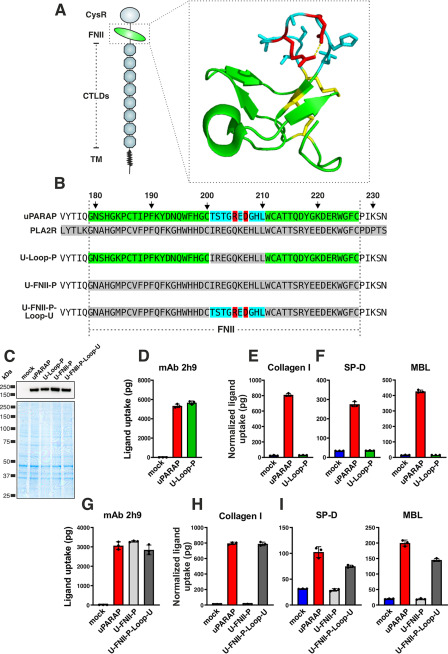
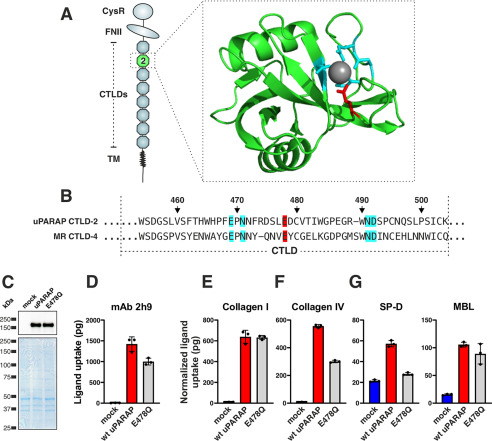
The well-conserved FNII domain is the 2nd of 10 N-terminal domains that constitute the extracellular region of uPARAP (Fig. 1A). Even though the FNII domain consists of only 49 residues (Fig. 1B, residues Gly-179–Cys-227 in uPARAP), it contains a number of elements essential for uPARAP's ability to bind structural collagens. These elements include a loop of 10 residues protruding from the central core of the domain (Fig. 1, A and B, residues Thr-201–Leu-210, highlighted in blue or red) (18, 24, 25). The protruding loop is flanked by two cysteines that are invariant for this type of domain (Fig. 1B, Cys-200 and Cys-212). These cysteines both form disulfide bonds (to separate cysteines in the domain), and the resulting structural constraints ensure the status of the protruding loop as a well-defined domain feature.
Figure 1.
A protruding loop in the FNII domain is essential for uPARAP-mediated uptake of collectins and collagen. A, overview of domains in uPARAP and the crystal structure of uPARAP's FNII domain (24). CysR, cysteine rich; TM, transmembrane region. The protruding loop is shown with side chains as sticks (residues in blue). A salt bridge within the loop, formed between Arg-205 and Asp-207 (residues in red), is represented by a dotted yellow line. Cysteines and disulfide bonds are highlighted (residues in yellow). B, alignment of FNII domain sequences from uPARAP, PLA2R1, and three uPARAP mutant receptors: U-loop-P, uPARAP with residues Thr-201–Leu-210 replaced by the corresponding residues from PLA2R1; U-FNII-P, uPARAP with complete FNII domain (Gly-179–Cys-227) replaced by FNII residues from PLA2R1; U-FNII-P-loop-U, the U-FNII-P mutant receptor with uPARAP residues Thr-201–Leu-210 reintroduced. Amino acid numbering follows the uPARAP sequence (accession no. NP_032652.3). C, western blotting analysis for the detection of uPARAP in mock transfected CHO-K1 cells and cells transfected to express uPARAP, U-loop-P, U-FNII-P, or U-FNII-P-loop-U (top panel). Nonreducing SDS-PAGE and Coomassie Brilliant Blue staining of protein lysates was included as loading controls for the western blotting (bottom panel). D–I, assay for uptake of radiolabeled mAb 2h9 (D and G), collagen type I (E and H), and SP-D and MBL (F and I) by mock-transfected CHO-K1 cells and cells transfected to express uPARAP or the indicated mutants. Analysis was performed in triplicate. The uptake of collagen type I, SP-D, and MBL was normalized to the uptake of anti-uPARAP antibody 2h9 to adjust for minor differences in the receptors' expression levels (see “Experimental procedures”). Data are presented as mean ± S.D. with individual data points also shown. Analysis was performed in triplicate.
The structural similarities between collagens and collectins suggest that these ligands could share some of the critical binding determinants in uPARAP. Therefore, to study any similarities in binding, we started out investigating engineered uPARAP variants with modifications known to affect collagen binding. We first utilized a uPARAP FNII-domain mutant construct (Fig. 1B, U-loop-P) in which the protruding loop was replaced with the counterpart from the related PLA2R1, a receptor known to facilitate the endocytosis of soluble PLA2 (26), but which does not interact with collagens (18). In transfected cells, U-loop-P was expressed at the same level as WT uPARAP (Fig. 1C) and retained complete endocytic capacity, as demonstrated by its ability to mediate the cellular uptake of the anti-uPARAP mAb 2h9 (27) (Fig. 1D). In contrast to mAb 2h9, the cellular uptake of collagen was completely abolished with the U-loop-P mutant (Fig. 1E), thereby recapitulating the role of the protruding loop in this function of uPARAP (18). We then examined the uptake of MBL and SP-D by WT uPARAP and U-loop-P and found that all endocytic activity toward these ligands was lost in the mutant (Fig. 1F). This observation demonstrated a crucial role of the FNII domain and its protruding loop in the cellular uptake of collectins. To further explore the binding requirements, we utilized two additional uPARAP mutants: U-FNII-P, in which all 49 FNII domain residues of uPARAP (Gly-179–Cys-227) were replaced with residues from the FNII domain of PLA2R1, and U-FNII-P-loop-U, wherein the protruding loop (Thr-201–Leu-210) from uPARAP was reintroduced in place of the PLA2R1 loop in the U-FNII-P mutant construct (Fig. 1, B, C, and G). As expected, the U-FNII-P mutant was devoid of collagen uptake activity, but the reintroduction of the protruding loop from uPARAP was sufficient to achieve a complete collagen uptake rescue in U-FNII-P-loop-U, when compared with WT (Fig. 1H). When examining the uptake of SP-D and MBL, a similar pattern was observed for the U-FNII-P mutant and a partial rescue was also achieved for these ligands in the U-FNII-P-loop-U construct (Fig. 1I). Taken together, these observations establish that the protruding loop not only constitutes a central element required for uPARAP to bind SP-D and MBL, but also that this loop is sufficient for introducing collectin-binding capacity within the framework of the FNII domain of PLA2R1.
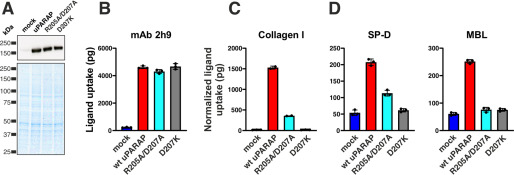
Within the protruding loop of the FNII domain in uPARAP, a salt bridge is formed between residues Arg-205 and Asp-207 (Fig. 1, A and B, residues in red), and mutations disrupting this salt bridge are sufficient to completely abolish collagen uptake by uPARAP without compromising overall receptor folding or endocytic recycling properties (24, 25). To study a potential role of the salt bridge in MBL and SP-D binding, we utilized two mutants with amino acid substitutions introduced to disrupt the salt bridge and destabilize the loop region: one mutant, in which two alanines replaced the positively and negatively charged residues (R205A/D207A), and another mutant, in which a more aggressive destabilization was achieved by replacing the aspartic acid with a positively charged lysine (D207K), thereby creating a construct with two positive charges facing each other. R205A/D207A and D207K mutants were expressed at the same level as WT uPARAP (Fig. 2A), retained endocytic activity toward mAb 2h9 (Fig. 2B) and lost all or most of the ability to facilitate collagen uptake (Fig. 2C), with just a low residual activity remaining in R205A/D207A. We then analyzed the potential of these mutants to bind SP-D and MBL (Fig. 2D). The uptake of SP-D closely mimicked the situation observed for collagen with a low residual uptake remaining in the R205A/D207A mutant and a complete loss of any uptake activity recorded for D207K. For MBL, both mutants had lost all cellular uptake activity. Evidently, the Arg-205 and Asp-207 residues are critical for uPARAP's ability to bind collectins, most likely due to the structural stability imposed by the salt bridge in the protruding loop in the FNII domain.
Figure 2.
Importance of a salt bridge stabilizing the protruding loop in uPARAP's FNII domain. A, western blotting analysis for the detection of uPARAP in HEK-293T cells transfected with the indicated constructs (top panel). Loading controls were included as described in the legend to Fig. 1 (bottom panel). B–D, assay for uptake of radiolabeled mAb 2h9 (B), collagen type I (C), and SP-D and MBL (D) by mock HEK-293T cells and cells expressing uPARAP or the indicated mutants. The uptake of collagen type I, SP-D, and MBL was normalized to the uptake of anti-uPARAP antibody 2h9 (see “Experimental procedures”). Data are presented as mean ± S.D. with individual data points shown. Analysis was performed in triplicate with the exception of collagen uptake by WT uPARAP and R205A/D207A, which was performed in duplicates.
Key residues in the FNII domains of uPARAP and MR are discriminators governing collectin binding
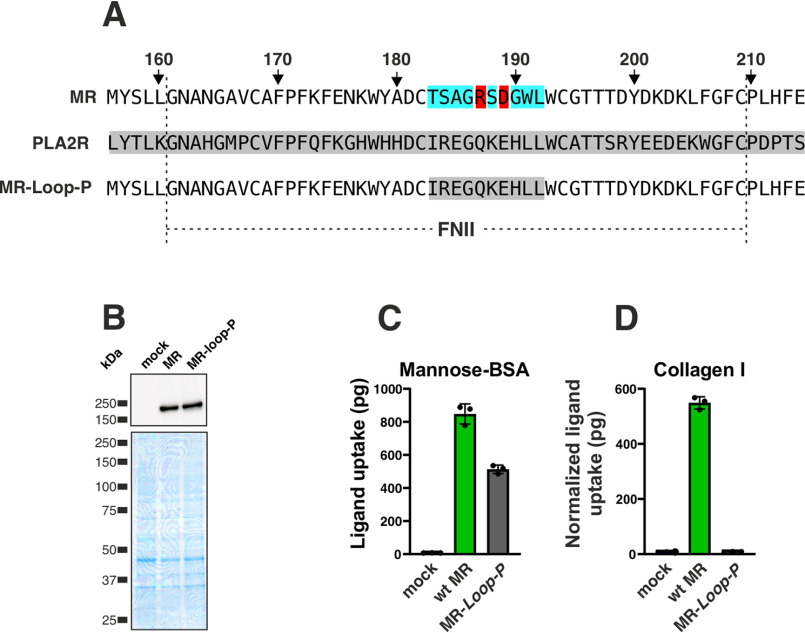
The experiments described above position the FNII domain at the center of uPARAP's function as a receptor for SP-D and MBL and suggest that one or more receptor-binding sites are located within the collagen-like triple-helical regions of these ligands. On this background, the intriguing fact that the related collagen receptor, MR, is not a receptor for collectins, becomes even more interesting from a molecular perspective. In particular, we have shown that the FNII domain from MR can replace the counterpart in uPARAP without any consequence for collagen binding (18). We therefore set out to identify potential molecular entities in the FNII domain that distinguish uPARAP from MR with respect to the function as a collectin receptor. First, it was necessary to establish if the protruding loop in the FNII domain of MR plays the same central role in collagen uptake as it does for uPARAP. To this end, we constructed an MR mutant, MR-loop-P, in which the protruding loop in the FNII domain (Thr-183–Leu-192) was replaced with the counterpart from PLA2R1 (Fig. 3A). MR-loop-P was expressed at a level similar to WT MR (Fig. 3B) and efficiently undertook the cellular uptake of a positive control ligand, mannose-BSA, a well-established MR ligand, which is independent of the FNII domain (28) (Fig. 3C). However, the insertion of the PLA2R1 loop into MR resulted in a complete loss of collagen uptake activity (Fig. 3D), thereby establishing the protruding loop as a key feature in FNII domain-mediated functions for both uPARAP and MR.
Figure 3.
MR binding to collagen depends on the FNII domain protruding loop. A, alignment of FNII domain sequences from MR, PLA2R1, and MR-loop-P (MR-loop-P, MR mutant receptor with residues Thr-183–Leu-192 replaced by the corresponding residues from PLA2R1). The amino acid numbering here follows the MR sequence (accession no. NP_032651). B, western blotting analysis for the detection of MR in CHO-K1 cells transfected with the indicated constructs (top panel). Loading controls were included as described in the legend to Fig. 1 (bottom panel). C and D, assay for uptake of radiolabeled mannose-BSA (C) and collagen type I (D) by mock CHO-K1 cells and cells expressing MR or MR-loop-P. The uptake of collagen type I was normalized to the uptake of mannose-BSA to adjust for differences in the receptors' expression levels (see “Experimental procedures”). Data are presented as mean ± S.D. with individual data points also shown. Analysis was performed in triplicate.
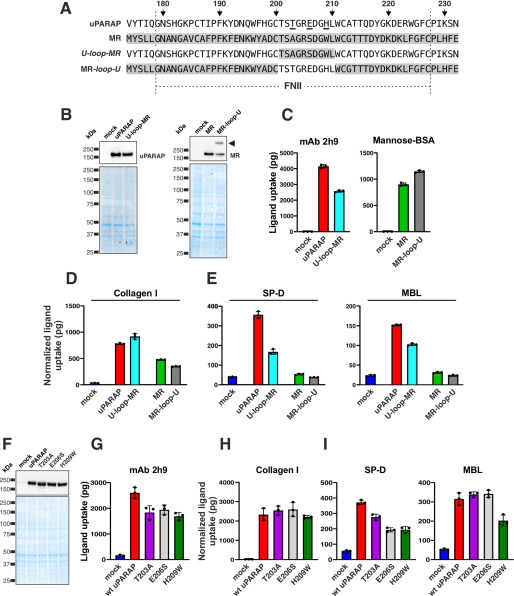
We then moved on to address if differences between the critical collagen-binding loops in the two receptors could explain the collectin binding by uPARAP, but not MR. For this purpose, two uPARAP and MR mutants were designed: U-loop-MR and MR-loop-U (Fig. 4A). In these mutants, the protruding loops in the FNII domains were replaced with the loop from the other receptor. Both mutants could be expressed at a level comparable with the WT receptors and retained their ability to internalize control ligands (Fig. 4, B and C). Confirming the highly similar collagen uptake mechanisms for uPARAP and MR, both loop mutants also retained all collagen uptake activity (Fig. 4D). When examining the cellular uptake of SP-D and MBL mediated by these mutants, two observations were made: 1) a reduction in collectin uptake activity, but not a complete loss, was recorded for U-loop-MR, and 2) no gain of activity was obtained with the MR-loop-U mutant (Fig. 4E). To further detail the differences between the two receptors' FNII domains, we expressed three single-amino acid uPARAP mutants: T203A, E206S, and H209W (Fig. 4, F and G). Each of these mutants represented one of the three amino acid substitutions found when comparing the sequences of the protruding loops from the FNII domains of uPARAP and MR, respectively (Fig. 4A, residues underlined in uPARAP sequence). None of these mutations had any consequence for the collagen uptake activity (Fig. 4H), consistent with the results obtained with the U-loop-MR mutant. A partial effect on collectin uptake was, however, observed: SP-D uptake was reduced for all three mutants (mainly for E206S and H209W), whereas only H209W affected MBL uptake (Fig. 4I). Note that minor differences in the expression levels for WT uPARAP and the T203A, E206S, and H209W mutants were observed (mAb 2h9 uptake values, Fig. 4G). This was adjusted for by normalization of collagen, SP-D, and MBL uptake values to the mAb 2h9 uptake (see “Experimental procedures”). In summary, these experiments identify key residues in uPARAP's FNII domain that are not found in MR and that are important for uPARAP's ability to bind collectins.
Figure 4.
Amino acid residues Thr-203, Glu-206, and His-209 are critical for the interaction of uPARAP with collectins. A, alignment of FNII domain sequences from uPARAP, MR, U-Loop-MR, and MR-Loop-U (u-Loop-MR, uPARAP mutant receptor with residues Thr-201–Leu-210 replaced by the corresponding residues from MR (Thr-183–Leu-192); MR-Loop-U, MR mutant receptor with residues Thr-183–Leu-192 replaced by the corresponding residues from uPARAP (Thr-201–Leu-210)). The amino acid numbering indicated at the top of the panel follows the uPARAP sequence. B, western blotting analysis for the detection of uPARAP (top left) or MR (top right) in CHO-K1 cells transfected with the indicated constructs (top panel). Loading controls were included as described in the legend to Fig. 1 (bottom panel). Note, the presence of an extra band in the MR-loop-U mutant lane (arrowhead; possibly representing an MR dimer) was considered of minor functional importance, judged from the uptake of mannose-BSA and collagen mediated by this mutant receptor (C and D). C–E, assay for uptake of radiolabeled mAb 2h9 and mannose-BSA (C), collagen type I (D), and SP-D and MBL (E) by mock CHO-K1 cells and cells expressing uPARAP, MR, or the indicated mutants. F, western blotting analysis for the detection of uPARAP in HEK-293T cells transfected with the indicated constructs (top panel). Loading controls were included as described in the legend to Fig. 1 (bottom panel). G–I, assay for uptake of radiolabeled mAb 2h9 (G), collagen type I (H), and SP-D and MBL (I) by mock HEK-293T cells and cells expressing WT uPARAP or indicated mutants. The uptake of collagen type I, SP-D, and MBL was normalized to the uptake of anti-uPARAP antibody 2h9 (uPARAP, U-loop-MR, T203A, E206S, and H209W) or mannose-BSA (MR and MR-loop-U) to adjust for differences in the receptors' expression levels (see “Experimental procedures”). Data are presented as mean ± S.D. with individual data points also shown. Analysis was performed in triplicate (C–E and G–I).
uPARAP's active lectin domain is critical for the cellular uptake of SP-D
Although critical for collectin binding in uPARAP itself, the FNII loop from uPARAP was not sufficient to confer MR with collectin binding and conversely, the loss of this feature in uPARAP did not lead to a complete loss of binding in this receptor (Fig. 4E). Consequently, additional molecular features distinguish the two receptors when it comes to the collectin-binding function. We speculated that one such feature could be uPARAP's active lectin domain, CTLD2 (Fig. 5A), because we have previously shown that this domain contributes to the uptake of glycosylated collagen subtypes, including collagen type IV (29). The five amino acid residues, known to be intimately involved in the Ca2+-dependent sugar-binding mediated by CTLDs (30), are conserved in CTLD2 (Fig. 5, A and B, uPARAP CTLD2, residues highlighted in blue or red) and this domain has indeed been shown to bind sugars with the expected structural requirements (31). MR also contains at least one active CTLD domain with the same five residues conserved (Fig. 5B, MR CTLD4), and it has been shown that mutating a specific glutamic acid residue to a glutamine in MR CTLD4 (Fig. 5B, residue highlighted in red) disrupts the sugar-binding activity without affecting the Ca2+ binding or the structural integrity of the domain (32). To test a role of the lectin activity of uPARAP's CTLD2 in collectin uptake, we constructed a uPARAP E478Q mutant (Fig. 5, C and D), which mimicked the MR CTLD4 mutation (Fig. 5B). The lectin mutant was not affected in its ability to mediate the cellular uptake of collagen type I (Fig. 5E), a collagen with a low level of glycosylation, but as expected, a partial loss of collagen type IV uptake activity was observed (Fig. 5F). When then examining the lectin mutant's ability to facilitate collectin uptake, we observed an almost complete loss of activity toward SP-D but, surprisingly, no effect on the cellular uptake of MBL (Fig. 5G). As above, differences in expression levels (mAb 2h9 uptake values, Fig. 5D) for WT uPARAP and E478Q were adjusted for by normalization of uptake values recorded for the other ligands (collagen I, collagen IV, SP-D, and MBL). Thus, these experiments clearly demonstrated a role of CTLD2 in the uptake of SP-D, whereas the lack of effect toward MBL pointed to an additional layer of complexity in the structural requirements of uPARAP as a collectin receptor.
Figure 5.
The lectin activity in CTLD-2 is critical for uPARAP's interaction with SP-D. A and B, overview of domains in uPARAP and the crystal structure of uPARAP's CTLD2 (24) (A) and alignment of sequences from uPARAP CTLD2 and MR CTLD4 (B). The sequence alignments include residues that coordinate to a Ca2+-ion (gray sphere in A), which is critical for the domain's ability to bind sugars like mannose or glucose (highlighted in blue or red) (24, 31). The Glu residue in MR CTLD4, which aligns with uPARAP Glu-478 (red), can be mutated to a Gln residue to disrupt lectin activity (32). Amino acid numbering follows the uPARAP sequence. C, western blotting analysis for the detection of uPARAP in CHO-K1 cells transfected with the indicated constructs (top panel). Loading controls are included as described in the legend to Fig. 1 (bottom panel). D–G, assay for uptake of radiolabeled mAb 2h9 (D), collagen type I (E), collagen type IV (F), and SP-D and MBL (G) by mock CHO-K1 cells and cells expressing WT uPARAP or E478Q mutant. The uptake of collagen type I, collagen type IV, SP-D, and MBL was normalized to the uptake of anti-uPARAP antibody 2h9 (see “Experimental procedures”). Data are presented as mean ± S.D. with individual data points also shown. Analysis was performed in triplicate.
uPARAP engages MBL immobilized on the surface of pathogens
When taking part in an innate immune response following injury, collectins become deposited on the surface of apoptotic cells or different types of pathogens, unlike under homeostatic conditions, where these proteins are mainly present as soluble components (2, 3, 33). Therefore, having identified various elements in uPARAP with a role in the uptake of free collectins, we next focused on the physical state of the collectin ligands.
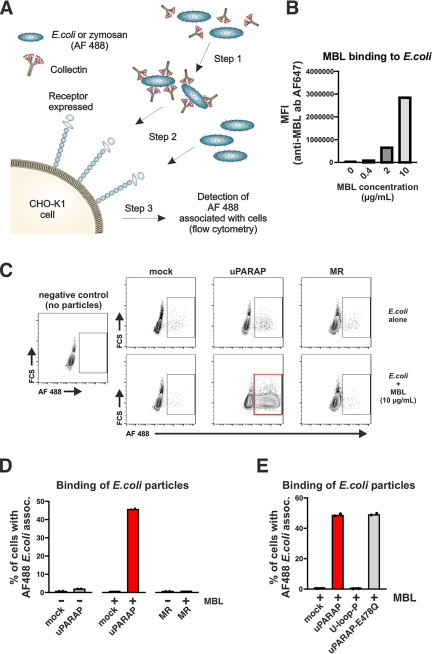
To investigate if uPARAP can function as a receptor for surface-immobilized collectins, we set up a flow cytometry-based assay for measuring a potential association between uPARAP-expressing cells and collectins deposited on bioparticles. Alexa FluorTM (AF) 488-labeled, heat-inactivated Escherichia coli, and yeast (zymosan) bioparticles were preincubated with recombinant collectins before being presented to cells expressing the receptors to be analyzed (Fig. 6A). To establish the assay conditions, we first showed that the recombinant collectins could be immobilized on the surface of the bioparticles. To this end, we used an AF 647-labeled antibody against MBL to demonstrate binding of the collectin to the bioparticles, exploiting that the bioparticles can be detected in a flow cytometer. In this experiment, a large concentration-dependent immobilization of MBL on both E. coli (Fig. 6B) and zymosan (Fig. S1A) was recorded.
Figure 6.
Interaction of uPARAP with MBL immobilized on the surface of E. coli bioparticles. A, assay for the evaluation of the interaction between cells and surface-bound collectins. Step 1: incubation of AF 488-conjugated bioparticles with collectins. Step 2: incubation of CHO-K1 cells expressing receptor of interest (uPARAP or MR) or mock CHO-K1 cells with bioparticles pre-incubated with collectin. Bioparticles incubated without collectins are also added as a control. Step 3: analysis of fluorescence associated with CHO-K1 cells that were incubated with AF 488-conjugated bioparticles. B, quantification of MBL binding to the surface of E. coli bioparticles. Immobilized MBL was detected using a primary anti-MBL antibody followed by an AF 647-conjugated secondary antibody. The AF 647 signal associated with the bioparticles was quantified using flow cytometry. MFI, mean fluorescence intensity. C and D, assay for the association between AF 488-conjugated E. coli bioparticles and mock-transfected CHO-K1 cells or cells transfected to express uPARAP or MR. Examples of flow charts showing AF 488 fluorescence associated with cells exposed to bioparticles that were preincubated with MBL (C, bottom panels) or incubated without collectin (C, top panels). A negative control sample, in which no bioparticles were added to the cells, is shown in C (left panel). The percentages of cells scoring positive for an association with bioparticles are presented in bar charts (D). E, assay for the association between E. coli bioparticles and mock-transfected CHO-K1 cells or cells transfected to express uPARAP, U-loop-P, or E478Q uPARAP mutant. E. coli bioparticles were preincubated with 10 µg/ml of MBL. Presentation of results in bar charts as in D. Analysis was performed in duplicate. Data are presented as mean with individual data points also shown (D and E).
We then investigated if the deposition of MBL on the surface of E. coli could induce a uPARAP-dependent association with cells in culture. AF 488-labeled E. coli particles, with or without MBL immobilized on the surface, were incubated with uPARAP-expressing cells, after which the percentage of cells associated with the fluorescent particles was determined (Fig. 6, C and D). Strikingly, uPARAP expression combined with MBL immobilization resulted in an association of particles with a large fraction (∼50%) of the cells, whereas less than 2% of the cells became associated with the particles in the absence of MBL. A negligible association with E. coli particles was also found in parallel measurements with MR-expressing cells or mock-transfected cells, whether or not MBL was present.
A similar effect of uPARAP expression was obtained when using MBL deposited on AF 488-labeled zymosan particles, although the association between cells and the zymosan particles was lower (Fig. S1, B and C). In this case, however, a stringent comparison with MR-expressing cells could not be performed. Thus, when using zymosan particles, a large association of particles with MR-expressing cells was observed, both in the presence and absence of MBL, precluding a concise evaluation of any contribution from the collectin (Fig. S1C). This is in line with previous reports, in which MR has been demonstrated to be a receptor for glycoproteins and carbohydrate moieties with terminal mannose or GlcNAc residues, found, e.g. on zymosan (34, 35).
We next analyzed the performance of the U-loop-P and the E478Q uPARAP mutants in this assay to test the role of the FNII and the CTLD2 domains, respectively. Matching our results obtained using soluble MBL (Figs. 1F and 5G), the association of cells with the MBL-coated E. coli particles was abolished in the U-loop-P mutant, whereas the E478Q substitution in CTLD2 had no effect (Fig. 6E).
Finally, we investigated if immobilized SP-D could induce a similar association between cells and E. coli or zymosan particles. However, even though SP-D binding to the surface of both particle types was observed (Fig. S2A), no association was recorded between the particles and mock or uPARAP-expressing cells after SP-D immobilization (Fig. S2, B and C). This observation adds further to the notion that the interaction of uPARAP with MBL and SP-D, respectively, involves both common and distinct molecular mechanisms.
Discussion
The levels of extravascular collectins change dramatically during acute infections, tissue damage, and resolution of injury, but very little is known about collectin regulation and turnover. We recently demonstrated that collectins share a cellular uptake mechanism with structural collagens in the form of the endocytic cell surface receptor uPARAP, and that uPARAP facilitates a cellular clearance of SP-D and MBL from injured tissues (16). Here we show that the uptake of collectins involves many of the same molecular features in uPARAP as those involved in the uptake of collagen, but that collectin uptake also depends on unique receptor elements. These elements constitute critical determinants that distinguish uPARAP from the related collagen receptor MR, and, importantly, may dictate a preference of uPARAP for collectin or collagen uptake in a given tissue and/or injury state. Given that the physiological conditions governing the turnover of collectins and structural collagens are very different, it is not surprising that specific structural elements in otherwise similar receptors serve a discriminatory function toward these ligands.
In addition to the specificity distinguishing uPARAP from MR, we show that the uptake of MBL is governed by uPARAP's FNII domain, whereas the uptake of SP-D requires both uPARAP's FNII domain and the lectin activity of CTLD2. FNII domains from multiple proteins, including fibronectin, matrix metalloproteinases-2 and -9, MR, and uPARAP, facilitate the interaction with native, cleaved, or partially denatured collagens. Although the exact binding site has not been identified in every case, these interactions with all likelihood involve the triple-helical regions of collagens (36–41). Similarly, the importance of the FNII domain for collectin uptake by uPARAP clearly suggests that the binding site for this receptor is located in the collagen-like domains of SP-D and MBL.
Certain other proteins have also been suggested to have a binding site in this part of MBL. Of particular importance, the binding site for the MBL-associated serine proteases (MASPs), which are essential for complement activation through the lectin pathway, has been mapped to residues just downstream of the hinge region in the collagen-like domain (42, 43). Because the collagen-like domain in MBL is short, spanning only 59 amino acids, it is possible that uPARAP and MASPs have overlapping binding sites. Consequently, uPARAP may preferentially interact with MBL molecules not associated with MASPs. In relationship to the uPARAP-mediated turnover of MBL in dermal fibroblasts (16), we therefore propose that MBL is more efficiently targeted for intracellular degradation when not in a complex with MASPs. This may constitute a mechanism that regulates MBL and complement activation in the dermis. Interestingly, such a competition phenomenon has already been demonstrated for the cell surface-associated proteins calreticulin and CD91, because their interactions with MBL are potently inhibited by MASPs (44, 45). The biological implications of these observations, however, remain to be addressed.
Although collectin and collagen uptake mechanisms share many common features within the FNII domain, including the Arg-205–Asp-207 salt bridge, these mechanisms are not identical. uPARAP residues Thr-203, Glu-206, and His-209 are exclusively important for collectin uptake and this observation also provides a partial molecular explanation for the lack of collectin uptake activity in MR. However, the findings that the T203A, E206S, and H209W mutants, as well as the uPARAP mutant carrying the entire MR loop (U-loop-MR), did retain some uptake activity toward SP-D and MBL, and that the MR mutant carrying the uPARAP loop (MR-loop-U) did not gain any activity, provided a clear indication that additional molecular features distinguish the two receptors with respect to collectin interactions. The results obtained using the E478Q, lectin-deficient uPARAP mutant (Fig. 5) suggest that CTLD2 constitutes one such feature, at least for the interaction with SP-D, because MR does not have lectin activity in the corresponding domain. In this connection, we note that the functional interplay between the FNII domain and CTLD2 in uPARAP is not completely understood. The two domains may be working in parallel as two separate ligand-binding entities, a hypothesis consistent with our previous study on the uPARAP-mediated uptake of collagen IV (29), or CTLD2 may instead indirectly affect ligand binding mediated by FNII through various suggested mechanisms (24, 25, 46).
The uPARAP mutant carrying the PLA2R1 FNII domain with the uPARAP protruding loop reintroduced (U-FNII-P-loop-U) showed a complete functional rescue for collagen uptake, but only a partial rescue for collectin uptake (Fig. 1). This observation strongly suggests that even inside the FNII domain, certain elements in addition to the protruding loop are involved in collectin uptake specifically.
Using bioparticles, we demonstrate that uPARAP, in addition to soluble collectins, can interact with collectins immobilized on the surface of E.coli and zymosan (Fig. 6 and Fig. S1). This finding suggests that uPARAP may be more involved in MBL biology than previously recognized. For instance, it is possible that uPARAP is involved in the phagocytosis of pathogens and apoptotic cells promoted by MBL (2, 47–49). Although uPARAP is predominantly expressed by fibroblasts and related mesenchymal cells and does not engage in phagocytic processes undertaken by these cells (50), it should be noted that some expression of uPARAP has also been demonstrated on phagocytic immune cells, including subpopulations of macrophages and immature dendritic cells (20, 51, 52).1
In contrast to MBL, surface-immobilized SP-D appears not to interact with uPARAP, because SP-D failed to promote the association of pathogen particles with uPARAP-expressing cells (Fig. S2). It is possible that the collagen-like region of SP-D is not exposed to host-binding partners when the protein is immobilized on the surface of pathogens. This could be a direct physical consequence of the assembly of SP-D into an oligomeric cruciform-like structure (53), which places the collagen-like domains of the individual SP-D trimeric subunits at the center of the supramolecular structure. MBL oligomers, on the other hand, form a more flower bouquet-like structure (33, 54), which leaves the collagen-like domain more directly exposed to the solvent phase, also when MBL is immobilized on the surface of a pathogen, and this orientation of the collagen-like domain could be more favorable for a simultaneous interaction with other host proteins, including uPARAP. An additional difference may arise due to the pathogen-binding properties of the two collectins. Unlike MBL, SP-D can mediate aggregation of pathogen particles (3), which could result in additional shielding of the collagen-like domains. Although we cannot provide a molecular explanation at this time, the different structures of MBL and SP-D oligomers may also be the reason why uPARAP's active lectin domain is only required for the uptake of the latter collectin.
The difference between pathogen-associated MBL and SP-D with respect to interactions with uPARAP may also be important in the innate immune response to viral lung infections. SP-D has been demonstrated to bind the surface of corona-type and certain other viral particles, and this enables SP-D to block the infection of cultured human bronchial epithelial cells with coronavirus (55, 56). This specific role of SP-D in the innate immune response to viral lung infection is further supported by the demonstrations of a large increase in the level of alveolar SP-D, but not SP-A, in corona-infected rats (15) and significantly elevated serum SP-D levels in patients suffering from severe acute respiratory syndrome coronavirus, when compared with healthy control subjects (57). Accordingly, by a selective targeting of soluble SP-D for cellular clearance and not SP-D in complex with pathogens, uPARAP may modulate the SP-D–mediated innate immune response to viral infections. Furthermore, the inability of pathogen-associated SP-D to interact with uPARAP likely excludes uPARAP as a possible novel mechanism by which viral particles might gain entrance into cells, even during lung infection conditions where SP-D levels are very high.
In conclusion, our study identifies molecular elements governing uPARAP-mediated uptake of collectins and demonstrates differences between the structural requirements for collagen and collectin uptake. For the first time, we also demonstrate interactions between uPARAP and pathogen-associated MBL, and this discovery could be essential for the understanding of the functional relationship between collectins and uPARAP in biology and disease.
Experimental procedures
Reagents and cells
The following reagents were purchased from commercial sources: recombinant human SP-D, recombinant human and recombinant mouse MBL, and mouse anti-human SP-D antibody (Bio-Techne), collagen type I from rat tail (BD Biosciences), 125I (PerkinElmer), collagen type IV from human placenta (Sigma-Aldrich), E. coli, zymosan AF 488-conjugated bioparticles, and goat anti-mouse AF 647-conjugated antibody (Thermo Fisher Scientific), and d-mannose-BSA (14 atom spacer, Dextra Laboratories). Mouse anti-human MBL antibodies Hyb-131-10 and Hyb-131-11 were kindly provided by Professor Peter Garred, Rigshospitalet, Denmark. In-house produced mouse anti-uPARAP mAb 2h9 was described previously (27). The epitope for 2h9 in uPARAP is located outside the FNII and CTLD2 domains and this feature allows for the utilization of 2h9 as primary antibody for the detection of uPARAP and all studied mutant constructs in western blotting analysis, as well as the utilization of 2h9 as a positive control ligand in receptor uptake assay.
Flp-In CHO-K1 cells (Thermo Fischer Scientific, RRID:CVCL_U424) were maintained in Dulbecco's modified Eagle's medium/F-12 supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin. Selection reagents Zeocin (100 μg/ml, Thermo Fisher Scientific) and hygromycin B (400 μg/ml, Thermo Fisher Scientific), respectively, were also added to the growth media before and after transfection. HEK-293T cells (Thermo Fisher Scientific, RRID:CVCL_0063) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Generation of receptor expression plasmids
cDNAs encoding murine uPARAP (accession no. NM_008626) and MR (accession no. NM_008625) were cloned into the pcDNA5/FRT/TO expression vector as described previously (18) and this expression vector system was used for stable expression of receptors in CHO-K1 cells or transient expression of receptors in HEK-293T cells (see below). Receptor mutant constructs were generated using a Q5 site-directed mutagenesis kit following the manufacturer's instructions (New England Biolabs). All constructs generated are listed in Table 1 along with the corresponding mutagenic primers used in the PCR. The U-FNII-P mutant receptor was generated in a previous study (18).
Table 1.
List of receptor mutants and primers used for generating the expression constructs
| Receptor mutant construct | Description | Mutagenesis primer sequences |
|---|---|---|
| R205A/D207A | Point mutations in FNII from uPARAP | 5'-AGCTGGGCACCTGTGGTGTGCC-'3 |
| 5'-TCCGCGCCAGTGCTGGTGCAGCC-'3 | ||
| D207K | Point mutation in FNII from uPARAP | 5'-TGGCAGAGAAAAGGGGCACCTGTG-'3 |
| 5'-GTGCTGGTGCAGCCGTGG-'3 | ||
| T203A | Point mutation in FNII from uPARAP | 5'-CTGCACCAGCGCTGGCAGAGA-'3 |
| 5'-CCGTGGAACCACTGGTTG-'3 | ||
| E206S | Point mutation in FNII from uPARAP | 5'-CACTGGCAGATCAGATGGGCACCTG-'3 |
| 5'-CTGGTGCAGCCGTGGAAC-'3 | ||
| H209W | Point mutation in FNII from uPARAP | 5'-AGAAGATGGGTGGCTGTGGTGTGCCACC-'3 |
| 5'-CTGCCAGTGCTGGTGCAG-'3 | ||
| E478Q | Point mutation in FNII from uPARAP | 5'-TGACAGCCTGCAAGACTGTGTCAC-'3 |
| 5'-CGAAAGTTGTTGGGCTCAAAG-'3 | ||
| U-loop-MR | uPARAP with FNII residues Thr-201-Leu-210 replaced by Thr-183–Leu-192 from MR FNII | 5'-TCGGACGGATGGCTCTGGTGTGCCACCACCCAG-'3 |
| 5'-GCGCCCGGCAGAGGTGCAGCCGTGGAACCACTG-'3 | ||
| MR-loop-P | MR with FNII residues Thr-183–Leu-192 replaced by Ile-196–Leu-205 from PLA2R FNII | 5'-GAAGATGGGCACCTGTGGTGTGGAACCACCACT-'3 |
| 5'-TCTGCCAGTGCTGGTGCAGTCTGCATACCACTTG-'3 | ||
| MR-loop-U | MR with FNII residues Thr-183–Leu-192 replaced by Thr-201–Leu-210 from uPARAP FNII | 5'-GAAAGAGCACCTGCTCTGGTGTGGAACCACC-'3 |
| 5'-TGCCCTTCTCGGATGCAGTCTGCATACCACTTG-'3 | ||
| U-loop-P | uPARAP with FNII residues Thr-201–Leu-210 replaced by Ile-196–Leu-205 from PLA2R FNII | 5'-AAAGAGCACCTGCTGTGGTGTGCCACCACCCAG-'3 |
| 5'-CTGCCCTTCTCGGATGCAGCCGTGGAACCACTG-'3 | ||
| U-FNII-P | uPARAP with FNII residues Gly-179–Cys-227 replaced by Gly-174–Cys-222 from PLA2R FNII | (18) |
| U-FNII-P-loop-U | U-FNII-P residues Ile-201–Leu-210 replaced by Thr-201–Leu-210 from uPARAP FNII | 5'-GAAGATGGGCACCTGTGGTGTGCCACCACAAGC-'3 |
| 5'-TCTGCCAGTGCTGGTACAGTCATGGTGCCAGTG-'3 |
Cell transfections
CHO-K1 cell monoclones with a stable expression of WT uPARAP, MR, or mutant constructs were generated using the Flp-In system according to the manufacturer's instructions in combination with a Lipofectamine 3000 Transfection Reagent kit (Thermo Fisher Scientific). A mock cell line was also generated by transfecting cells with an empty pcDNA5/FRT/TO expression vector. HEK-293T cells were transfected to transiently express WT uPARAP or mutant constructs using either Lipofectamine 2000 or Lipofectamine 3000 Reagent kits (Thermo Fisher Scientific). One μg of expression plasmid DNA was used per 75,000 cells seeded. 24 h after transfection, cells were harvested to generate cell lysates for use in western blotting or were used in radioligand uptake assays (see below). Due to a much superior transient transfection efficiency when compared with CHO-K1 cells, HEK-293T cells were used to express R205A/D207A, D207K, T203A, E206S, and H209W uPARAP mutants, which were not included in the initial study design. This was done to circumvent the time-consuming need to generate CHO-K1 monoclones with stable expression of these constructs.
Western blotting
Transfected CHO-K1 and HEK-293T cells were washed twice in PBS and then detached using PBS containing 5 mm EDTA. Cells were then pelleted by centrifugation at 200 × g and lysed in lysis buffer (1% Triton X-100, 50 mm Tris, and 100 mm NaCl, pH 7.4) containing protease inhibitor cocktail set III (1:200, Calbiochem). The protein concentration in the whole cell lysates was determined using a BCA protein assay kit (ThermoFisher Scientific). Proteins in the cell lysates were separated by SDS-PAGE (NuPAGE 4–12% BisTris gels, Invitrogen) before blotting onto a polyvinylidene difluoride membrane (iBlot transfer stack, Invitrogen). The membranes were blocked using a 2% BSA in PBS solution followed by probing with either mouse anti-uPARAP 2h9 (1 μg/ml) (27) or rat anti-MR (1 μg/ml, clone MR5D3, Bio-Rad) primary antibodies overnight at 4 °C. Primary antibodies were detected using the relevant horseradish peroxidase-coupled secondary antibodies (1:6000, Dako). Primary and secondary antibodies were diluted in PBS containing 0.1% Tween 20 detergent. When detecting uPARAP or MR in CHO-K1 cell lysates, respectively, 1.5 or 7.5 μg of total protein was analyzed. When detecting uPARAP in HEK-293T cell lysates, 2 μg of total protein was analyzed. As a loading control, 10 μg of total protein from each lysate was separated by SDS-PAGE and stained using Coomasssie Brilliant Blue.
Internalization of radiolabeled proteins
Human SP-D, human or mouse MBL, anti-uPARAP antibody 2h9, mannose-BSA, collagen type I from rat tail, and collagen type IV (20 μg of each) were labeled with 125I using a previously established procedure (58). The assays used to determine the uptake of radiolabeled ligands by transfected CHO-K1 or HEK-293T cells were described previously (CHO-K1 (16) and HEK-293T (18)). For CHO-K1 cells, triplicates of 100,000 cells seeded in a 24-well plate format were assayed for each condition. For HEK-293T cells, triplicates of 150,000 cells seeded in a 12-well plate format were assayed. The fraction of internalized radiolabeled ligand (125I radioactivity) was measured on a Wizard2 γ-counter (PerkinElmer Life Scineces). The uptake of anti-uPARAP antibody 2h9 was used as a positive control and measure of the expression level for WT uPARAP and all uPARAP-based mutant receptors. The uptake of mannose-BSA served the same purpose for WT MR and all MR-based mutant receptors. To adjust for any differences in expression levels between WT uPARAP and uPARAP mutant receptors, the uptake of each ligand was normalized to the uptake of antibody 2h9. The uptake of ligands by each MR mutant receptor was normalized in the same manner using mannose-BSA uptake.
Binding of collectins to bioparticles and association of bioparticles with transfected cells
Collectin binding to heat-inactivated AF 488-conjugated E. coli or zymosan bioparticles (Thermo Fisher Scientific) was detected using the following procedure. Human MBL or SP-D (0-10 μg/ml) was incubated with 0.25 mg/ml of bioparticles for 1 h at 37 °C. SP-D and MBL binding to the bioparticles was then detected by incubation with primary antibody solutions (2 μg/ml of mouse anti-human SP-D antibody, Bio-Techne, or a mixture of Hyb-131-10 and Hyb-131-11 mouse anti-human MBL antibodies at a concentration of 1 μg/ml each) for 30-60 min at room temperature. Primary antibodies were then detected using incubation with AF 647-conjugated goat anti-mouse secondary antibody at 2 μg/ml (Thermo Fisher Scientific) for 30-60 min at room temperature. All incubations steps involving bioparticles, collectins, and antibodies were performed in a TBS buffer supplemented with 2 mm CaCl2 and 0.1% Tween 20 and with gentle mixing. In between each step, the bioparticles were washed twice in fresh buffer. Buffer change was enabled by pelleting bioparticles by centrifugation at 1000 × g for 5 min The level of AF 647 fluorescence associated with the bioparticles was determined using an Accuri C6 Flow Cytometer (BD Biosciences).
To assay for the association of bioparticles with transfected CHO-K1 cells, the bioparticles were initially incubated with 10 μg/ml of hMBL or hSP-D, or incubated in the absence of collectins as described above. Subsequently, the bioparticles were added to CHO-K1 cells at a final concentration of 20 μg/ml for 3 h at 37 °C. The adherent CHO-K1 cells, with any associated bioparticles, were then harvested using a 0.25% trypsin/EDTA solution supplemented with 50 μg/ml of proteinase K (Roche) and filtered using a 50-μm mesh. Finally, the percentage of CHO-K1 cells associated with the AF 488-conjugated bioparticles was determined using an Accuri C6 Flow Cytometer (BD Biosciences).
Data availability
All the presented data are contained within the article.
Supplementary Material
Acknowledgments
We thank Katharina Hassing, Britt Kongstofte, and Mia Kristine Grønning Høg for expert technical assistance.
This article contains supporting information.
Author contributions—K. S. N., N. B., L. H. E., and H. J. J. conceptualization; K. S. N. and H. J. J. data curation; K. S. N. and H. J. J. formal analysis; K. S. N., O. K., and H. J. J. investigation; K. S. N., O. K., and H. J. J. methodology; K. S. N., N. B., and H. J. J. writing-original draft; K. S. N., N. B., L. H. E., and H. J. J. writing-review and editing; N. B. and L. H. E. resources; N. B., L. H. E., and H. J. J. supervision; N. B. and L. H. E. funding acquisition; N. B., L. H. E., and H. J. J. visualization; N. B. and L. H. E. project administration; H. J. J. validation.
Funding and additional information—This work was supported by the Danish Medical Research Council Grants 4092-00387B and DFF-4004-00340 (to N. B. and H. J. J.), Danish Cancer Society Grants R90-A5989, R146-A9326-16-S2, R231-A13832, and R222-A13103 (to N. B., L. H. E., and H. J. J.), the Simon Fougner Hartmanns Family Foundation, Region Hovedstadens forskningsfond, The Novo Nordisk Foundation, and Dansk Kræftforskningsfond.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
K. Norregaard, O. Krigslund, N. Behrendt, L. H. Engelholm, and H. J. J̈urgensen, unpublished results.
- MBL
- mannose-binding lectin
- SP
- surfactant protein
- CTLD
- C-type calcium-dependent lectin-like domain
- uPARAP
- urokinase plasminogen activator receptor-associated protein
- PLA2
- phospholipase A2
- MR
- mannose receptor
- FNII
- fibronectin type-II
- MASP
- MBL-associated serine protease
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- AF
- alexa fluor
- PLA2R1
- PLA2 receptor.
References
- 1. Fraser D. A., and Tenner A. J. (2008) Directing an appropriate immune response: the role of defense collagens and other soluble pattern recognition molecules. Curr. Drug Targets 9, 113–122 10.2174/138945008783502476 [DOI] [PubMed] [Google Scholar]
- 2. Litvack M. L., and Palaniyar N. (2010) Review: soluble innate immune pattern-recognition proteins for clearing dying cells and cellular components: implications on exacerbating or resolving inflammation. Innate Immun. 16, 191–200 10.1177/1753425910369271 [DOI] [PubMed] [Google Scholar]
- 3. Casals C., Garcia-Fojeda B., and Minutti C. M. (2019) Soluble defense collagens: sweeping up immune threats. Mol. Immunol. 112, 291–304 10.1016/j.molimm.2019.06.007 [DOI] [PubMed] [Google Scholar]
- 4. Garred P., Larsen F., Madsen H. O., and Koch C. (2003) Mannose-binding lectin deficiency–revisited. Mol. Immunol. 40, 73–84 10.1016/S0161-5890(03)00104-4 [DOI] [PubMed] [Google Scholar]
- 5. Ordonez S. R., Veldhuizen E. J. A., van Eijk M., and Haagsman H. P. (2017) Role of soluble innate effector molecules in pulmonary defense against fungal pathogens. Front. Microbiol. 8, 2098 10.3389/fmicb.2017.02098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Messias-Reason I. J., Nisihara R. M., and Mocelin V. (2011) Mannan-binding lectin and ficolin deposition in skin lesions of pemphigus. Arch. Dermatol. Res. 303, 521–525 10.1007/s00403-011-1132-1 [DOI] [PubMed] [Google Scholar]
- 7. Lokitz M. L., Zhang W., Bashir M., Sullivan K. E., Ang G., Kwon E. J., Lin J. H., and Werth V. P. (2005) Ultraviolet-B recruits mannose-binding lectin into skin from non-cutaneous sources. J. Investig. Dermatol. 125, 166–173 10.1111/j.0022-202X.2005.23794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stuart L. M., Takahashi K., Shi L., Savill J., and Ezekowitz R. A. (2005) Mannose-binding lectin-deficient mice display defective apoptotic cell clearance but no autoimmune phenotype. J. Immunol. 174, 3220–3226 10.4049/jimmunol.174.6.3220 [DOI] [PubMed] [Google Scholar]
- 9. Aono Y., Ledford J. G., Mukherjee S., Ogawa H., Nishioka Y., Sone S., Beers M. F., Noble P. W., and Wright J. R. (2012) Surfactant protein-D regulates effector cell function and fibrotic lung remodeling in response to bleomycin injury. Am. J. Respir. Crit. Care Med. 185, 525–536 10.1164/rccm.201103-0561OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaunsbaek M. Q., Rasmussen K. J., Beers M. F., Atochina-Vasserman E. N., and Hansen S. (2013) Lung surfactant protein D (SP-D) response and regulation during acute and chronic lung injury. Lung 191, 295–303 10.1007/s00408-013-9452-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McIntosh J. C., Swyers A. H., Fisher J. H., and Wright J. R. (1996) Surfactant proteins A and D increase in response to intratracheal lipopolysaccharide. Am. J. Respir. Cell Mol. Biol. 15, 509–519 10.1165/ajrcmb.15.4.8879185 [DOI] [PubMed] [Google Scholar]
- 12. Forbes L. R., and Haczku A. (2010) SP-D and regulation of the pulmonary innate immune system in allergic airway changes. Clin. Exp. Allergy 40, 547–562 10.1111/j.1365-2222.2010.03483.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Helden H. P., Kuijpers W. C., Steenvoorden D., Go C., Bruijnzeel P. L., van Eijk M., and Haagsman H. P. (1997) Intratracheal aerosolization of endotoxin (LPS) in the rat: a comprehensive animal model to study adult (acute) respiratory distress syndrome. Exp. Lung Res. 23, 297–316 10.3109/01902149709039228 [DOI] [PubMed] [Google Scholar]
- 14. Savani R. C., Godinez R. I., Godinez M. H., Wentz E., Zaman A., Cui Z., Pooler P. M., Guttentag S. H., Beers M. F., Gonzales L. W., and Ballard P. L. (2001) Respiratory distress after intratracheal bleomycin: selective deficiency of surfactant proteins B and C. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L685–696 10.1152/ajplung.2001.281.3.L685 [DOI] [PubMed] [Google Scholar]
- 15. Funk C. J., Manzer R., Miura T. A., Groshong S. D., Ito Y., Travanty E. A., Leete J., Holmes K. V., and Mason R. J. (2009) Rat respiratory coronavirus infection: replication in airway and alveolar epithelial cells and the innate immune response. J. Gen. Virol. 90, 2956–2964 10.1099/vir.0.014282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jürgensen H. J., Norregaard K. S., Sibree M. M., Santoni-Rugiu E., Madsen D. H., Wassilew K., Krustrup D., Garred P., Bugge T. H., Engelholm L. H., and Behrendt N. (2019) Immune regulation by fibroblasts in tissue injury depends on uPARAP-mediated uptake of collectins. J. Cell Biol. 218, 333–349 10.1083/jcb.201802148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. East L., and Isacke C. M. (2002) The mannose receptor family. Biochim. Biophys. Acta 1572, 364–386 10.1016/S0304-4165(02)00319-7 [DOI] [PubMed] [Google Scholar]
- 18. Jürgensen H. J., Johansson K., Madsen D. H., Porse A., Melander M. C., Sørensen K. R., Nielsen C., Bugge T. H., Behrendt N., and Engelholm L. H. (2014) Complex determinants in specific members of the mannose receptor family govern collagen endocytosis. J. Biol. Chem. 289, 7935–7947 10.1074/jbc.M113.512780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonnans C., Chou J., and Werb Z. (2014) Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 10.1038/nrm3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Madsen D. H., Leonard D., Masedunskas A., Moyer A., Jürgensen H. J., Peters D. E., Amornphimoltham P., Selvaraj A., Yamada S. S., Brenner D. A., Burgdorf S., Engelholm L. H., Behrendt N., Holmbeck K., Weigert R., et al. (2013) M2-like macrophages are responsible for collagen degradation through a mannose receptor-mediated pathway. J. Cell Biol. 202, 951–966 10.1083/jcb.201301081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greenhill S. R., and Kotton D. N. (2009) Pulmonary alveolar proteinosis: a bench-to-bedside story of granulocyte-macrophage colony-stimulating factor dysfunction. Chest 136, 571–577 10.1378/chest.08-2943 [DOI] [PubMed] [Google Scholar]
- 22. Jürgensen H. J., van Putten S., Norregaard K. S., Bugge T. H., Engelholm L. H., Behrendt N., and Madsen D. H. (2020) Cellular uptake of collagens and implications for immune cell regulation in disease. Cell. Mol. Life Sci. 10.1007/s00018-020-03481-3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herbein J. F., and Wright J. R. (2001) Enhanced clearance of surfactant protein D during LPS-induced acute inflammation in rat lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L268–L277 10.1152/ajplung.2001.281.1.L268 [DOI] [PubMed] [Google Scholar]
- 24. Yuan C., Jürgensen H. J., Engelholm L. H., Li R., Liu M., Jiang L., Luo Z., Behrendt N., and Huang M. (2016) Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding. Biochem. J. 473, 2359–2368 10.1042/BCJ20160276 [DOI] [PubMed] [Google Scholar]
- 25. Paracuellos P., Briggs D. C., Carafoli F., Lončar T., and Hohenester E. (2015) Insights into collagen uptake by C-type mannose receptors from the crystal structure of Endo180 domains 1-4. Structure 23, 2133–2142 10.1016/j.str.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zvaritch E., Lambeau G., and Lazdunski M. (1996) Endocytic properties of the M-type 180-kDa receptor for secretory phospholipases A2. J. Biol. Chem. 271, 250–257 10.1074/jbc.271.1.250 [DOI] [PubMed] [Google Scholar]
- 27. Sulek J., Wagenaar-Miller R. A., Shireman J., Molinolo A., Madsen D. H., Engelholm L. H., Behrendt N., and Bugge T. H. (2007) Increased expression of the collagen internalization receptor uPARAP/Endo180 in the stroma of head and neck cancer. J. Histochem. Cytochem. 55, 347–353 10.1369/jhc.6A7133.2006 [DOI] [PubMed] [Google Scholar]
- 28. Taylor M. E., Bezouska K., and Drickamer K. (1992) Contribution to ligand binding by multiple carbohydrate-recognition domains in the macrophage mannose receptor. J. Biol. Chem. 267, 1719–1726 [PubMed] [Google Scholar]
- 29. Jürgensen H. J., Madsen D. H., Ingvarsen S., Melander M. C., Gårdsvoll H., Patthy L., Engelholm L. H., and Behrendt N. (2011) A novel functional role of collagen glycosylation: interaction with the endocytic collagen receptor uparap/ENDO180. J. Biol. Chem. 286, 32736–32748 10.1074/jbc.M111.266692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weis W. I., Drickamer K., and Hendrickson W. A. (1992) Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature 360, 127–134 10.1038/360127a0 [DOI] [PubMed] [Google Scholar]
- 31. East L., Rushton S., Taylor M. E., and Isacke C. M. (2002) Characterization of sugar binding by the mannose receptor family member, Endo180. J. Biol. Chem. 277, 50469–50475 10.1074/jbc.M208985200 [DOI] [PubMed] [Google Scholar]
- 32. Mullin N. P., Hitchen P. G., and Taylor M. E. (1997) Mechanism of Ca2+ and monosaccharide binding to a C-type carbohydrate-recognition domain of the macrophage mannose receptor. J. Biol. Chem. 272, 5668–5681 10.1074/jbc.272.9.5668 [DOI] [PubMed] [Google Scholar]
- 33. Holmskov U., Thiel S., and Jensenius J. C. (2003) Collections and ficolins: humoral lectins of the innate immune defense. Annu. Rev. Immunol. 21, 547–578 10.1146/annurev.immunol.21.120601.140954 [DOI] [PubMed] [Google Scholar]
- 34. Ezekowitz R. A., Sastry K., Bailly P., and Warner A. (1990) Molecular characterization of the human macrophage mannose receptor: demonstration of multiple carbohydrate recognition-like domains and phagocytosis of yeasts in Cos-1 cells. J. Exp. Med. 172, 1785–1794 10.1084/jem.172.6.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor M. E., and Drickamer K. (1993) Structural requirements for high affinity binding of complex ligands by the macrophage mannose receptor. J. Biol. Chem. 268, 399–404 [PubMed] [Google Scholar]
- 36. Briknarová K., Grishaev A., Bányai L., Tordai H., Patthy L., and Llinás M. (1999) The second type II module from human matrix metalloproteinase 2: structure, function and dynamics. Structure 7, 1235–1245 10.1016/S0969-2126(00)80057-X [DOI] [PubMed] [Google Scholar]
- 37. Dzamba B. J., Wu H., Jaenisch R., and Peters D. M. (1993) Fibronectin binding site in type I collagen regulates fibronectin fibril formation. J. Cell Biol. 121, 1165–1172 10.1083/jcb.121.5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pickford A. R., Smith S. P., Staunton D., Boyd J., and Campbell I. D. (2001) The hairpin structure of the (6)F1(1)F2(2)F2 fragment from human fibronectin enhances gelatin binding. EMBO J. 20, 1519–1529 10.1093/emboj/20.7.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu X., Chen Z., Wang Y., Yamada Y., and Steffensen B. (2005) Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and -2. Biochem. J. 392, 127–134 10.1042/BJ20050650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Madsen D. H., Engelholm L. H., Ingvarsen S., Hillig T., Wagenaar-Miller R. A., Kjøller L., Gårdsvoll H., Hoyer-Hansen G., Holmbeck K., Bugge T. H., and Behrendt N. (2007) Extracellular collagenases and the endocytic receptor, urokinase plasminogen activator receptor-associated protein/Endo180, cooperate in fibroblast-mediated collagen degradation. J. Biol. Chem. 282, 27037–27045 10.1074/jbc.M701088200 [DOI] [PubMed] [Google Scholar]
- 41. Leitinger B., and Hohenester E. (2007) Mammalian collagen receptors. Matrix Biol. 26, 146–155 10.1016/j.matbio.2006.10.007 [DOI] [PubMed] [Google Scholar]
- 42. Wallis R., Shaw J. M., Uitdehaag J., Chen C. B., Torgersen D., and Drickamer K. (2004) Localization of the serine protease-binding sites in the collagen-like domain of mannose-binding protein: indirect effects of naturally occurring mutations on protease binding and activation. J. Biol. Chem. 279, 14065–14073 10.1074/jbc.M400171200 [DOI] [PubMed] [Google Scholar]
- 43. Teillet F., Lacroix M., Thiel S., Weilguny D., Agger T., Arlaud G. J., and Thielens N. M. (2007) Identification of the site of human mannan-binding lectin involved in the interaction with its partner serine proteases: the essential role of Lys55. J. Immunol. 178, 5710–5716 10.4049/jimmunol.178.9.5710 [DOI] [PubMed] [Google Scholar]
- 44. Pagh R., Duus K., Laursen I., Hansen P. R., Mangor J., Thielens N., Arlaud G. J., Kongerslev L., Højrup P., and Houen G. (2008) The chaperone and potential mannan-binding lectin (MBL) co-receptor calreticulin interacts with MBL through the binding site for MBL-associated serine proteases. FEBS J. 275, 515–526 10.1111/j.1742-4658.2007.06218.x [DOI] [PubMed] [Google Scholar]
- 45. Duus K., Thielens N. M., Lacroix M., Tacnet P., Frachet P., Holmskov U., and Houen G. (2010) CD91 interacts with mannan-binding lectin (MBL) through the MBL-associated serine protease-binding site. FEBS J. 277, 4956–4964 10.1111/j.1742-4658.2010.07901.x [DOI] [PubMed] [Google Scholar]
- 46. Boskovic J., Arnold J. N., Stilion R., Gordon S., Sim R. B., Rivera-Calzada A., Wienke D., Isacke C. M., Martinez-Pomares L., and Llorca O. (2006) Structural model for the mannose receptor family uncovered by electron microscopy of Endo180 and the mannose receptor. J. Biol. Chem. 281, 8780–8787 10.1074/jbc.M513277200 [DOI] [PubMed] [Google Scholar]
- 47. Brouwer N., Dolman K. M., van Zwieten R., Nieuwenhuys E., Hart M., Aarden L. A., Roos D., and Kuijpers T. W. (2006) Mannan-binding lectin (MBL)-mediated opsonization is enhanced by the alternative pathway amplification loop. Mol. Immunol. 43, 2051–2060 10.1016/j.molimm.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 48. Ogden C. A., deCathelineau A., Hoffmann P. R., Bratton D., Ghebrehiwet B., Fadok V. A., and Henson P. M. (2001) C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194, 781–795 10.1084/jem.194.6.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jack D. L., Lee M. E., Turner M. W., Klein N. J., and Read R. C. (2005) Mannose-binding lectin enhances phagocytosis and killing of Neisseria meningitidis by human macrophages. J. Leukocyte Biol. 77, 328–336 10.1189/jlb.0604342 [DOI] [PubMed] [Google Scholar]
- 50. Sprangers S., Behrendt N., Engelholm L., Cao Y., and Everts V. (2017) Phagocytosis of collagen fibrils by fibroblasts in vivo is independent of the uPARAP/Endo180 receptor. J. Cell. Biochem. 118, 1590–1595 10.1002/jcb.25821 [DOI] [PubMed] [Google Scholar]
- 51. Jürgensen H. J., Silva L. M., Krigslund O., van Putten S. M., Madsen D. H., Behrendt N., Engelholm L. H., and Bugge T. H. (2019) CCL2/MCP-1 signaling drives extracellular matrix turnover by diverse macrophage subsets. Matrix Biol. Plus 1, 100003 10.1016/j.mbplus.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sheikh H., Yarwood H., Ashworth A., and Isacke C. M. (2000) Endo180, an endocytic recycling glycoprotein related to the macrophage mannose receptor is expressed on fibroblasts, endothelial cells and macrophages and functions as a lectin receptor. J. Cell Sci. 113, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 53. Crouch E., Persson A., Chang D., and Heuser J. (1994) Molecular structure of pulmonary surfactant protein D (SP-D). J. Biol. Chem. 269, 17311–17319 [PubMed] [Google Scholar]
- 54. Miller A., Phillips A., Gor J., Wallis R., and Perkins S. J. (2012) Near-planar solution structures of mannose-binding lectin oligomers provide insight on activation of lectin pathway of complement. J. Biol. Chem. 287, 3930–3945 10.1074/jbc.M111.320341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leth-Larsen R., Zhong F., Chow V. T., Holmskov U., and Lu J. (2007) The SARS coronavirus spike glycoprotein is selectively recognized by lung surfactant protein D and activates macrophages. Immunobiology 212, 201–211 10.1016/j.imbio.2006.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Funk C. J., Wang J., Ito Y., Travanty E. A., Voelker D. R., Holmes K. V., and Mason R. J. (2012) Infection of human alveolar macrophages by human coronavirus strain 229E. J. Gen. Virol. 93, 494–503 10.1099/vir.0.038414-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu Y. P., Liu Z. H., Wei R., Pan S. D., Mao N. Y., Chen B., Han J. J., Zhang F. S., Holmskov U., Xia Z. L., de Groot P. G., Reid K. B., Xu W. B., and Sorensen G. L. (2009) Elevated plasma surfactant protein D (SP-D) levels and a direct correlation with anti-severe acute respiratory syndrome coronavirus-specific IgG antibody in SARS patients. Scand. J. Immunol. 69, 508–515 10.1111/j.1365-3083.2009.02245.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Behrendt N., Ronne E., and Dano K. (1996) Domain interplay in the urokinase receptor. Requirement for the third domain in high affinity ligand binding and demonstration of ligand contact sites in distinct receptor domains. J. Biol. Chem. 271, 22885–22894 10.1074/jbc.271.37.22885 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the presented data are contained within the article.