Abstract
COVID-19 has ravaged the world and is the greatest of pandemics in human history, in the absence of treatment or vaccine the mortality and morbidity rates are very high. The present investigation was undertaken to screen and identify the potent leads from the Indian Ayurvedic herb, Asparagus racemosus (Willd.) against SARS-CoV-2 using molecular docking and dynamics studies. The docking analysis was performed on the Glide module of Schrödinger suite on two different proteins from SARS-CoV-2 viz. NSP15 Endoribonuclease and spike receptor-binding domain. Asparoside-C, Asparoside-D and Asparoside -F were found to be most effective against both the proteins as confirmed through their docking score and affinity. Further, the 100 ns molecular dynamics study also confirmed the potential of these compounds from reasonably lower root mean square deviations and better stabilization of Asparoside-C and Asparoside-F in spike receptor-binding domain and NSP15 Endoribonuclease respectively. MM-GBSA based binding free energy calculations also suggest the most favourable binding affinities of Asparoside-C and Asparoside-F with binding energies of −62.61 and −55.19 Kcal/mol respectively with spike receptor-binding domain and NSP15 Endoribonuclease.
Highlights
Asparagus racemosus have antiviral potential
Phytochemicals of Shatavari showed promising in-silico docking and MD results
Asparaoside-C and Asparoside-F has good binding with target proteins
Asparagus racemosus holds promise as SARS-COV-2 (S) and (N) proteins inhibitor
Communicated by Ramaswamy H. Sarma
Keywords: Asparagus racemosus, Shatavari; Molecular docking, nCov-2019, Antiviral, Indian Ayurveda, Pandemic disease
1. Introduction
During the month of December 2019, a global pandemic emerged from the Wuhan city of China, called as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and has become a major threat to the whole world due to the range of population it has infected throughout the world (Zhou et al., 2020). The pandemic COVID 19 has affected around 212 countries with almost half million death throughout the globe. The corona viruses are highly enveloped single stranded RNA with a largest genome size (26-32-kilobases) that plays critical role in initial RNA synthesis of the infectious cycle, template for replication and transcription and also act as a substrate for packaging into the progeny virus. In all types of corona viruses, two-thirds of the genome encodes a replicase polyproteins, pp1ab, that consists of two overlapping open reading frames (ORFs) i.e. ORF1a and ORF1b and are processed by viral proteases to undergo cleavage into 16 different non-structural proteins (NSPs), involved in transcription and replication (Boopathi et al., 2020; Cotten et al., 2013; Gupta et al., 2020). Among these the NSP15 Endoribonuclease is a nidoviral RNA uridylate-specific endoribonuclease (NendoU) which comprises of C-terminal catalytic domain, specific for uridine, acts on single- as well as double stranded RNA (Elfiky, 2020; Mittal et al., 2020). Studies have reported that the NendoU activity of NSP15 is mainly attributed to the observed protein interference with innate immune response and is therefore, a very essential segment for understanding the biological progression of coronavirus (Bhardwaj et al., 2008; Deng & Baker, 2018). Another NSP that helps 2019-nCoV to get entry into the host cells involves a densely glycosylated, homotrimeric class I fusion spike (S) protein (Sarma et al., 2020). This S protein present in a metastable prefusion conformation, undergoes structural rearrangement for the viral membrane to get fused with the membrane of host cell. The process is accelerated as a result of binding of S1 subunit to the host-cell receptor and transition of the S2 subunit to a highly stable postfusion conformation (Hasan et al., 2020). Further, recent studies have reported that, spike (S) glycoprotein possess a human angiotensin-converting enzyme 2 (ACE2) binding site and has 10 to 20-fold higher binding affinity towards 2019-nCoV S as compared to SARS-CoVs (Chikhale et al., 2020; Choudhury, 2020; Han & Král, 2020; Ou et al., 2020; Pillaiyar et al., 2020; Sinha et al., 2020a).
Even though, scientist throughout the world are trying their level best to identify or discover a new lead or repurposing of drugs which could be effective against SARS-CoV-2, it is a long way to go before we have a vaccine or drug molecule to treat COVID-19.The U.S. Food and Drug Administration has now issued an emergency use authorization for Remdesivir for the treatment of suspected or laboratory-confirmed adults and children hospitalized with severe symptoms of SARS-CoV-2 disease (FDA news release, 2020). The effectiveness of the treatment based on traditional medicinal plants has been reported during the 2003 SARS (Wen et al. 2011; Hoever et al,. 2005; Chen et al., 2008; Li et al., 2005; Lin et al., 2008; Ryu et al., 2010) . Therefore, studies on medicinal plants based on their history and traditional uses have already started among the scientific community (Al-Khafaji et al., 2020; Elmezayen et al., 2020; Enmozhi et al., 2020; Joshi et al., 2020; Sinha et al., 2020b; Wahedi et al., 2020; Zhang et al., 2020).
Asparagus racemosus (Willd.) is a well-known medicinal plant, grown in the tropical and subtropical regions of India, where its therapeutic importance is well documented in the Indian and British Pharmacopoeia along with various traditional system of medicines such as Ayurveda, Siddha and Unani (Gurav & Gurav, 2014; Singh & Geetanjali, 2016). Traditionally, this plant is popularly known as ‘Shatavari’, and described in Ayurveda as a potential rasayana which prevents ageing, provides immunity, increases longevity, improves mental function and also helps in treatment of diseases related to female reproductive system, inflammation, dysentery, biliousness, tumor and diseases of the blood and eyes (Alok et al., 2013; Singh & Geetanjali, 2016). The pharmacological studies carried out on the plant has revealed the potential adaptogenic, immunostimulant, anti-inflammatory, anti-microbial, anti-oxidant, anti-ulcer, galactogogue, phytoestrogenic, neuroprotective, aphrodisiac, anti-dyspepsia, anti-tussive and anti-cancerous activities (Alok et al., 2013; Boonsom et al., 2012; Singh & Geetanjali, 2016; Upadhyay et al., 2020). The major phytoconstituents reported in the plant includes steroidal saponins such as Shatavarin (I-XI mainly I, IV and X), Aasparacosides (A-F), Racemosol, Asparagamine, Racemoside A-B, Asparagoside F, Quercetin, Rutin, Kaempferol and many more (Alok et al., 2013; Lalert et al., 2018; Singh & Geetanjali, 2016).
Shatavari has been reported for its anti-bacterial, anti-candidal, anti-viral activities (Mandal et al., 2000; Raza et al., 2015; Shah et al., 2014; Uma et al., 2009) . Recent studies have reported that, the drug under investigation, if possess a potent immunomodulatory, anti-inflammatory and antioxidant potential could prove an effective agent in treatment of COVID 19 (Jayawardena et al., 2020; Patwardhan et al., 2020; Tillu et al., 2020). Further, all these suggested properties are well equipped by the phytoconstituents from Asparagus racemosus.
Thus, keeping the above view into consideration, the present investigation was undertaken to determine the efficacy of 32 different bioactive molecules from the plant Asparagus racemosus against two different protein targets i.e. NSP15 Endoribonuclease and spike receptor-binding domain from SARS-CoV-2 with the help of molecular docking and molecular dynamics studies.
2. Material and methods
2.1. Molecular docking studies
Molecular docking studies (Supplementary File) of 32 phytochemicals of A. racemosus were performed on the NSP15 Endoribonuclease and 2019-nCoV spike receptor-binding domain using the experimentally solved crystal structures of PDB IDs 6W01 and 6M0J respectively. The crystal structures have resolution of 1.9 and 2.45 Å respectively and are suitable for docking studies. Schrödinger Glide SP module was used for docking simulation (Sinha et al., 2020a; Verma et al., 2020).
2.2. Molecular dynamics (MD) simulation and molecular mechanics-generalized born solvent accessibility (MM-GBSA) analysis
AMBER18 software package was used for MD simulations and ligands were parameterized with ANTECHAMBER. The protein-ligand complexes prepared in xLeap were subjected to 100 ns MD simulations on Nvidia V100-SXM2-16GB Graphic Processing Unit using the PMEMD.CUDA module. Further, the 100 ns trajectories were subjected to MM-GBSA analysis using Amber18 and Amber18 tools on all the 10000 frames (Supplementary File).
3. Results and discussion
3.1. Molecular docking
To determine the potency and understand the insights into the possible mechanism of 32 different herbal-based ligands, molecular docking simulation study was carried out on two proteins i.e. spike receptor-binding domain and NSP15 Endoribonuclease from SARS-CoV-2.The spike receptor-binding domain from the virus gets entry into the host cell by combining with ACE-2 sreceptor, while NSP15 Endoribonuclease is said to be responsible for replication of virus genetic material. Docking simulation study was also performed on the remdesivir, a reference drug, on the same protein targets and the results were compared with the test ligands (Muralidharan et al., 2020). Remdesivir was chosen as a reference drug as it is a well-known RNA dependent RNA polymerase, a nsp protein, inhibitor. Further, remdesivir was chosen as reference drug because it is being used as a agent under clinical investigation for the treatment of ncov-19 (Sinha et al., 2020b). On the basis of the docking score (Table 1) and binding interaction (Tables 2 and 3), only the best five tested ligands along with the reference drug with respect to different proteins have been discussed in our study. The docking scores of top scorer A. racemosus bio-actives are given in Table 1 whereas the docking scores of other phytochemicals are given in supplementary information Table S1. The structures of all A. racemosus phytochemicals investigated in present investigation are given in supplementary information Figure S1.
Table 1.
List of bio-actives with docking scores with the PDB: 6W01 and PDB: 6M0J of SARS-CoV-2.
| Sr. No. | Pubchem CID | Name | Docking Score |
|
|---|---|---|---|---|
| 6W01 | 6M0J | |||
| 1 | 158598 | Asparoside-C | −7.542 | −7.165 |
| 2 | 158597 | Asparoside-D | −7.069 | −6.445 |
| 3 | 101406647 | Shatavarin-I (Asparaoside-B ) | −6.524 | −5.524 |
| 4 | 101847691 | Shatavarin-X | −6.431 | −5.621 |
| 5 | 102253062 | Racemoside-A | −6.238 | −5.993 |
| 6 | 101422489 | Asparoside-A | −6.03 | −5.396 |
| 7 | 101847687 | Shatavarin-VI | −5.747 | −3.961 |
| 8 | 102253063 | Racemoside-B | −5.713 | −5.409 |
| 9 | 129626614 | Asparagoside-F | −5.609 | −6.615 |
| 10 | 5280343 | Quercetin | −5.605 | −4.519 |
| 11 | 158595 | Asparanin-D | −5.551 | −5.001 |
| 12 | 44203607 | Shatavaroside-B | −5.477 | −5.348 |
| 13 | 5280863 | Kaempferol | −5.357 | −4.569 |
| 14 | 273773749 | Curillin-H | −5.254 | −5.952 |
| 15 | 121304016 | Remdesivir | −5.28 | −5.94 |
Table 2.
Binding interaction of different bioactive herbal ligands with the active site of SARS-CoV-2 spike RBD (PDB ID: 6M0J).
| S. No. | Ligand | Interaction (PDB-6M0J) |
||
|---|---|---|---|---|
| H-Bonding | No. of H-Bond |
|||
| HBD | HBA | |||
| 1 | Asparoside-C | Gly 496, Gln 414, Ser 494, Thr 415, Tyr 453 | 6 | 2 |
| 2 | Asparoside-D | Gly 502, Ser 494, Lys 417, Asp 420 Tyr 449, Gln 498 | 4 | 3 |
| 3 | Shatavarin-I | Gly 447, Lys 444, Glu 406, Arg 403, Gly 496 | 4 | 3 |
| 4 | Shatavarin-X | Gly 496, Lys 417, Glu 406,Gln 409,Tyr 453, Arg 403 | 2 | 5 |
| 5 | Racemoside-A | Glu 484, Gly 496, Ser 494 | 4 | 1 |
| 6 | Remdesivir | Arg 403, Glu 406, Tyr 453 | 1 | 2 |
H-Bond: Hydrogen Bonding, HBD: Hydrogen bond donor, HBA: Hydrogen bond acceptor in respect to ligand.
Table 3.
Binding interaction of different bioactive herbal ligands with the active site of SARS-CoV-2 NSP15 endoribonuclease (PDB ID: 6W01).
| S. No. | Ligand | Interaction (PDB-6M0J) |
||
|---|---|---|---|---|
| H-Bonding | No. of H-Bond |
|||
| HBD | HBA | |||
| 1 | Asparoside-C | Glu 234, Gly 230, Val 292, Hip 235, Asp 240 | 5 | 3 |
| 2 | Asparoside-F | Glu 234, Gly 230, Ala 232, Hip 235, Asp 240, Glu 340, Val 339 | 5 | 3 |
| 3 | Asparoside-D | Glu 340, His 243, Gln 245, Asp 240, Asn 278, Leu 346 | 6 | 1 |
| 4 | Rutin | Asp 240, Gln 245, Hip 235, Hip 250, Glu 340 | 4 | 3 |
| 5 | Racemoside-A | Thr 341, Hip 235, Glu 340, Hip 250 | 3 | 3 |
| 6 | Remdesivir | Val 292, Hip 235, Thr 341 | 1 | 2 |
H-Bond: Hydrogen Bonding, HBD: Hydrogen bond donor, HBA: Hydrogen bond acceptor in respect to ligand.
Figure 1.
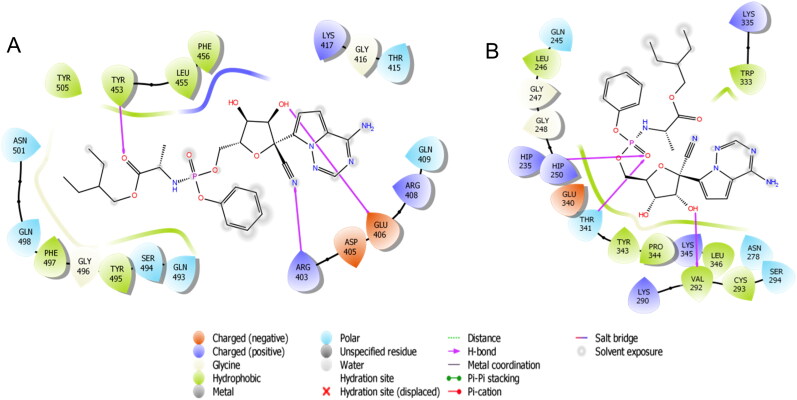
Binding-interaction analysis of remdesivir at the binding site of A. SARS-CoV-2 spike receptor-binding domain (PDB ID: 6M0J) and B. SARS-CoV-2 NSP15 Endoribonuclease.
3.1.1. Docking studies of the reference drug with SARS-CoV-2 spike receptor-binding domain (RBD) and NSP15 endoribonuclease
Remdesivir was properly positioned into the catalytic site of SARS-Cov-2 spike RBD assembled by polar Asn 501, Gln 409, Gln 493, Ser 494, Gln 498, Thr 415 charged Lys 417, Arg 403, Glu 406, Asp 405, Arg 408 hydrophobic Tyr 453, Tyr 505, Phe 497, Tyr 495, Leu 455, Phe 456 and Gly 416, Gly 496 amino acids with docking score -5.28 Kcal/mol. The carbonitrile group of remdesivir showed H-bonding with Arg 403, hydroxyl group of tetrahydrofuran ring exhibited H-bonding with Glu 406 and carboxyl group of remdesivir depicted H-bond interaction with Tyr 453 (Figure 1A).
The result outcomes from the docking of remdesivir with SARS-CoV-2 NSP15 Endoribonuclease are presented in (Table 1 and Figure 1B). Remdesivir was connected with the binding site mainly through H-bonding and showed docking score -5.94 Kcal/mol. The hydroxyl group of tetrahydrofuran ring exhibited H-bonding with Val 292 and the oxygen of phosphoramide exhibited bi-furcated H-bonding with (protonated His) Hip 235, Thr 341 (Figure 3A).
Figure 3.
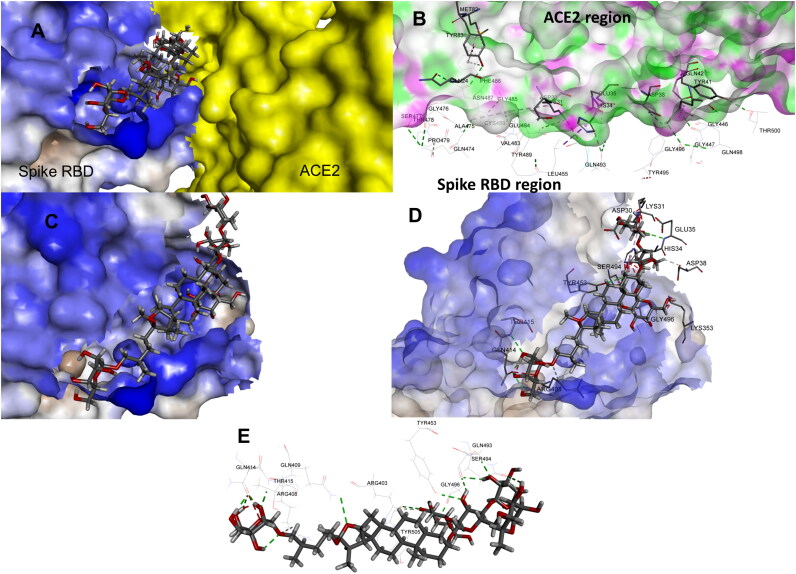
Binding pose of Asparoside-C at the SARS-CoV2 RBD and ACE2 interface A. Asparoside-C pose at the binding site, B. Interacting residues of SARS-COV2 RBD and ACE2, C. Binding pose of Asparoside-C on the surface of RBD, D. Binding pose of Asparoside-C along with the interacting residues, E. Binding interactions with various residues.
SARS-CoV-2 gets entry into the host cell through spike receptor-binding domain, which gets fused with ACE-2 receptor and its binding interaction depends on the binding of S1 subunit, having RBD. The RBD is consists of 5 twisted β sheets i.e. β1, β2, β3, β4 and β7 which are antiparallel to each other, where the extended inclusion is observed between β4 and β7 containing some α loops which designated as receptor binding motif (RBM). This RBM are said to contain almost all the residues that plays critical role in connectingn-COVID19 with ACE-2. Recently, it has been confirmed that, from all the residues between Arg 319 to Phe 541 only the 17 residues Lys 417, Gly 446, Tyr 449, Tyr 453, Leu 455, Phe 456, Ala 475, Phe 486, Asn 487, Tyr 489, Gln 493, Gly 496, Gln 498, Thr 500, Asn 501, Gly 502, Tyr 505 are considered to be critical for binding with ACE-2, where it gets connected with 20 residues of N-terminal peptidase domain of ACE-2. From these 17 residues Gln 493, Asn 501, Tyr 449, Tyr 489 and Tyr 505 are found to get strongly connected with the help of H-bonding and Lys 417 via a salt bridge interaction (Figure 1).
3.1.2. Docking studies of the natural bioactive ligand with SARS-CoV-2 spike RBD
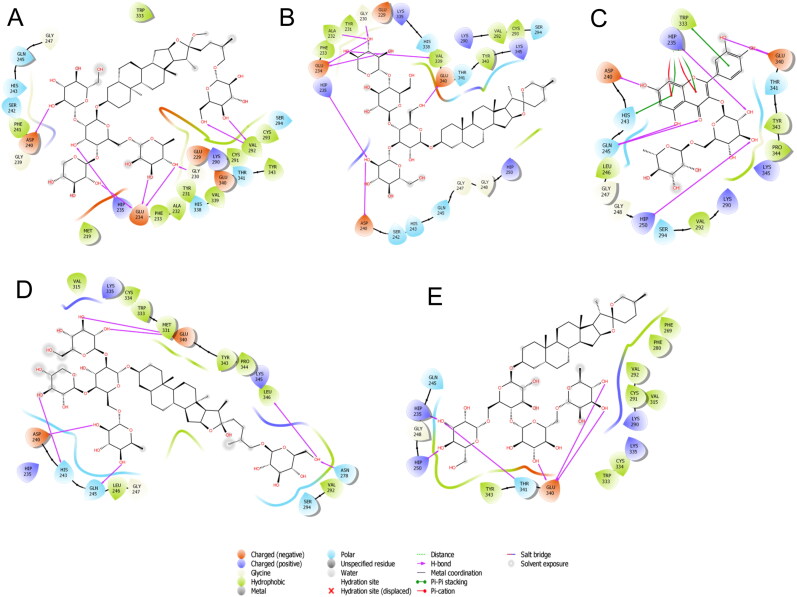
Asparoside-C was properly positioned into the binding pocket constructed by polar Gln 493, Ser 494, Gln 498, Asn 501, Gln 409, Gln 414, Thr 415 charged Glu 406, Arg 403, Lys 417, Asp 405, Glu 484 hydrophobic Tyr 453, Tyr 495, Tyr 505, Phe 497, Tyr 449, Tyr 489, Leu 455, Phe 456 and Gly 416, Gly 496 amino acids with docking score -7.542 Kcal/mol. Hydroxyl group of oxane ring of Asparoside-C exhibited H-bonding with Gly 496, Gln 414, Ser 494, hydroxyl group of other oxane ring showed bi-furcated H-bonding with hydrophilic acceptor Ser 494 and hydrophobic donor Tyr 453, hydroxyl group of terminal oxane ring also interacted by H-bonding with Gln 414, Thr 415 and hydroxymethyl group of terminal oxane ring depicted H-bond interaction with Gln 414 (Figure 2A). The bonding energy of Asparoside-C was found to be higher than all the reference drugs. All the oxane rings of was found to be properly buried into the outer concave surface of RBM and stabilized by 7 H-binding interaction. In accordance with our previous study (Sinha et al., 2020a), in which we explained the importance of oxane rings of saikosaponins during binding. Asparoside-D was properly positioned into the catalytic site assembled by polar Gln 493, Ser 494, Gln 498, Asn 501, Thr 415, Asn 460, Thr 500 charged Arg 403, Lys 417, Asp 420 hydrophobic Tyr 453, Tyr 495, Tyr 505, Phe 456, Tyr 449, Tyr 421, Leu 455 and Gly 416, Gly 496, Gly 502 amino acids with docking score -7.069 Kcal/mol. Hydroxyl group of oxane ring of Asparoside-D exhibited H-bonding with Gly 502, Ser 494, Lys 417, Asp 420 and hydroxyl group of other oxane ring showed bi-furcated H-bonding with Tyr 449, Gln 498 (Figure 2C). After structural elucidation of Asparoside-C and Asparoside-D we found that number of oxane ring was same in both the ligand, but aglycone part was differ. A hydroxyl group of aglycone part in Asparoside-C is naturally derivetized into methoxy in Asparoside-D, so we can say that Asparoside-D is the methoxy derivative of Asparoside-C and may be responsible for the lower binding affinity of Asparoside-D. Ser 494 residue was found to be common to form H-bond with both ligand and also found in the close proximity with other ligands in binding pocket.
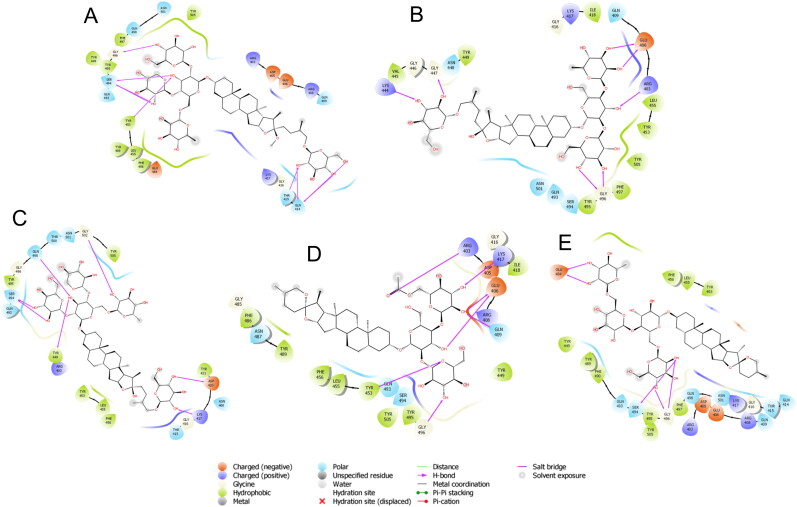
Figure 2.
Binding-interaction analysis of A. Asparoside-C, B. Shatavarin-I, C. Asparoside-D, D. Shatavarin-X, E. Racemoside-A with SARS-CoV-2 spike receptor-binding domain (PDB ID: 6M0J).
Shatavarin-I was substituted by 3 oxane rings on one side of aglycone and other side substituted by only one oxane ring which are properly fit into the binding pocket with docking score -6.524 Kcal/mol. The residue Glu 406 and Gly 496 showed bi-furcated H-bonding with the hydroxyl group of oxane ring (Figure 2B). Shatavarin-X have only 3 oxane rings attached to the aglycone part and was found to bind with the binding pocket constructed by polar Gln 493, Ser 494, Gln 409, Asn 487 charged Arg 403, Lys 417, Glu 406, Arg 408, Asp 405 hydrophobic Tyr 453, Tyr 495, Tyr 505, Phe 456, Tyr 449, Leu 455, Ile 418, Phe 486, Tyr 489 and Gly 416, Gly 496, Gly 485 amino acids with docking score -6.431 Kcal/mol. Hydroxyl group of oxane ring of Shatavarin-X exhibited H-bonding with Gly 496, Lys 417, Glu 406, and the total 6 H-bonding interaction was found with the backbone of protein (Figure 2D). The lesser number of oxane ring in Shatavarin-X might be the reason for its little bit lower docking score than Shatavarin-I. Racemoside-A was found to bind with the binding pocket constructed by polar Gln 493, Ser 494, Gln 409, Asn 501, Gln 498, Gln 414, Thr 415 charged Arg 403, Lys 417, Glu 484, Arg 408, Asp 405, Glu 406 hydrophobic Tyr 453, Tyr 495, Tyr 505, Phe 456, Tyr 449, Leu 455, Phe 490, Tyr 489, Phe 497 and Gly 416, Gly 496 amino acids with docking score -6.238 Kcal/mol. Hydroxyl group of oxane ring of Racemoside-A exhibited H-bonding with Gly 496, Glu 484 and hydroxyl group of oxane ring also showed bi-furcated H-bonding with Gly 496, Ser 494 (Figure 2E). The residues Gln 414, Tyr 415, Arg 408, Gln 409, Arg 403, Tyr 505, Tyr 453, Ser 494, Gly 496 and Gln 493 of spike RBD are involved in interaction with ACE2 surface residues Ile21, Gln 24, Leu29, Asp 30, Lys31, His 34, Glu 35, Tyr 41 and Gln 42. Asparoside-C blocks these interactions by forming hydrogen bond interaction with spike RBD residues Gln 414, Tyr 415, Arg 408, Gln 409, Arg 403, Tyr 505, Tyr 453, Ser 494, Gly 496 and Gln 493 residues (shown in Figure 3).
The NSP15, bears a catalytic C-terminal domain, which is constitutes of a set of trimers i.e. hexamers and responsible for cutting the double-stranded (ds) RNA substrates with uridine specificity via the Mn + 2-dependent endoribonuclease activity. Additionally, each monomeric unit is comprised of ∼345 amino acids which are folded into three domains viz. N-terminal, middle and nidoviral RNA uridylate-specific endoribonuclease (NendoU) C-terminal catalytic domain, where the NendoU C-terminal catalytic domain comprises of two β-sheets which are antiparallel. These β-sheets contain six key amino acids viz. His 235, His 250, Lys 290, Thr 341, Tyr 343, and Ser 294. Among them, His 235, His 250, Lys 290 constitutes the catalytic triad, His 235 serves as general acid, His 250 acts a base, while Ser 294 together with Tyr 343 have been found to govern U specificity.
3.1.3. Docking studies of the natural bioactive ligands with SARS-CoV-2 NSP15 endoribonuclease
Total 32 bioactive ligand was docked into the binding pocket of this protein and based on their docking score, Asparoside-C (-7.165 Kcal/mol), Asparoside-F (-6.615 Kcal/mol), Asparoside-D (-6.445 Kcal/mol), Rutin (-6.346 Kcal/mol) and Racemoside-A (-5.993 Kcal/mol) was selected as a potent lead. All the selected ligand was found to be tightly fit into the binding pocket and interaction with the catalytic triad. These ligands also showed higher docking score as compare to the reference drugs. Hydroxyl group of oxane ring of Asparoside-C stabilized through H-bonding with Glu 234, Gly 230, Val 292, Hip 235, Asp 240 and hydroxymethyl group showed H-bonding with Val 292 (Figure 4A). Asparoside-F was found to interact with the binding pocket through tri-furcated H-bonding with Glu 234, Gly 230, Ala 232, in which two are donor and one acceptor in nature, bi- furcated H-bonding with Hip 235, Asp 240 and conventional H-bonding with Val 339, Glu 234 (Figure 4B).
Figure 4.
Binding-interaction analysis of A. Asparoside-C, B. Asparoside-F, C. Rutin, D. Asparoside-D, E. Racemoside-A with NSP15 endoribonuclease (PDB ID: 6W01).
Rutin was found to be the least ranked ligand and get stabilized into the binding pocket through the chromenone and oxane ring. The hydroxyl group of chromenone ring exhibited H-bonding with Asp 240, Gln 245 and carboxyl group of chromenone ring showed H-bonding with Gln 245. Hydroxyl group of oxane ring exhibited H-bonding with Hip 235, Hip 250 and both hydroxyl group of phenyl ring showed H-bonding with Glu 340. The chromenone ring depicted π-π interactions with His 243, Hip 235, π-cation interaction with Hip 235 and phenyl ring also showed π-π interactions with Trp 333 (Figure 4C). Hydroxyl group of oxane ring of Asparoside-D exhibited H-bonding with Glu 340, His 243, Gln 245, Asp 240 and terminal hydroxymethyl group showed bi- furcated H-bonding with Asn 278, Leu 346 (Figure 4D).
3.1.4. Docking studies of the natural bioactive ligands with SARS-CoV-2 main protease, papain like protease and RNA dependent RNA polymerase
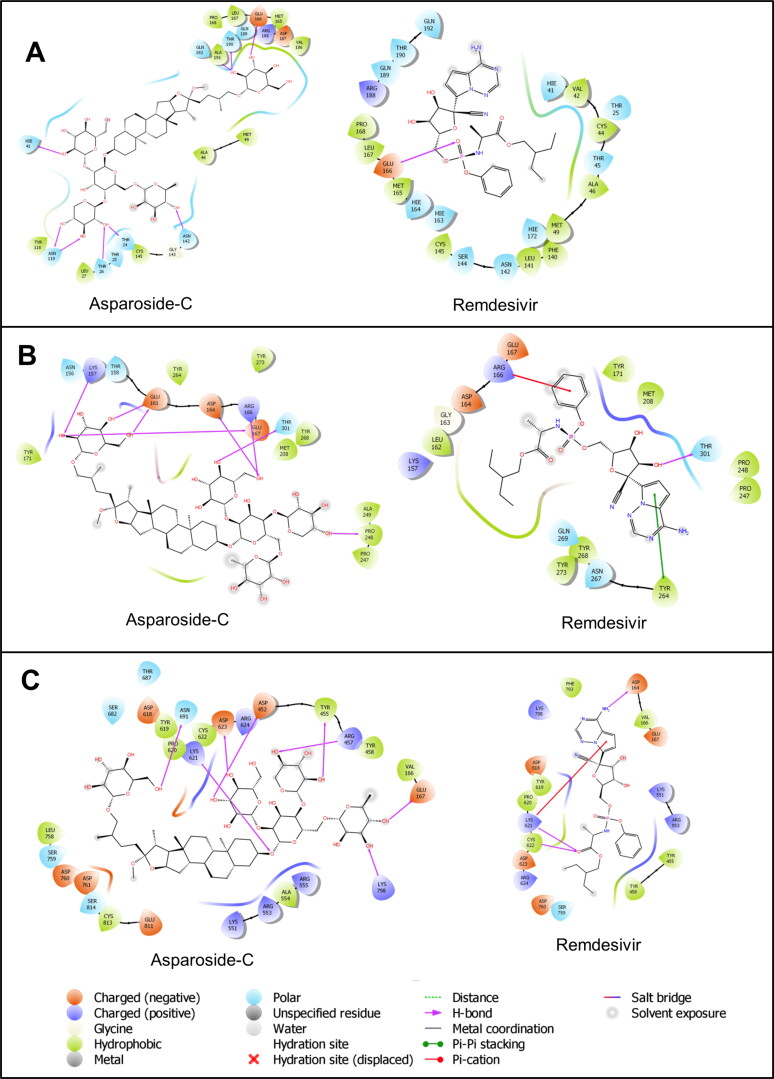
SARS-CoV2 main protease (Mpro), papain like protease (PLpro) are other viral proteins which facilitates viral assembly by cleaving polyproteins (Mengist et al., 2020; Cardenas‐Conejo et al., 2020). RNA dependent RNA polymerase (RdRp) is another nonstructural protein which is vital for viral life cycle (Elfiky, 2020). The docking study against these protein with the most promising bioactive ligand (Asparoside-C) against spike RBD and NSP 15 proteins was also performed. The crystal structure of SARS-Cov2 main protease (PDB ID: 4MDS) with 1.60 Å resolution was used in docking studies. All the previously mentioned protein refinement steps were performed. The aglycone part of Asparoside-C was properly positioned in to the binding site of main protease. The hydroxyl groups of sugar moieties form hydrogen bonds with the residues His41, Thr 190, Ala191, Glu 166, Asn 142, Asn 119, Thr 26 and Thr 24 (Figure 5A). Compared to the reference drug Remdesivir, which forms the hydrogen bond with Glu 166, Asparoside-C binds more favorably at the binding site. The docking score for Asparoside-C and Remdesivir are -7.431 and -6.099 respectively, which also suggests more binding affinity of Asaparoside-C.
Figure 5.
Binding-interaction analysis of Asparoside-C and Remdesivir at the binding site of A. SARS-CoV2 main protease, B. SARS-CoV2 papain like protease and C. SARS-CoV2 RNA dependent RNA polymease.
In case of SARS-CoV2 papain like protease (PLpro), the experimentally solved crystal structure (PDB ID: 6WX4) with resolution 1.66 Å was used in docking study. The Asparoside-C binds at the binding site where the hydroxyl groups of sugar moieties form hydrogen bond with positively charged Lys 157, Arg 166, polar Thr 301, negatively charged Glu 161, Asp164, Glu 167 and hydrophobic Pro 248 (Figure 5B). On the other hand, the phenyl ring in Remdesivir forms π-cation interaction with positively charged Arg 166, while the pyrrole ring forms π-π stacking interaction with Thr 264 and only one hydrogen bond is formed with Thr 301. The docking scores for Asparoside-C and Remdesivir were -5.448 and -4.055 respectively.
RdRp crystal structure with resolution 2.90 Å (PDB ID: 6M71) was used to investigate possible mode of binding of Asparoside-C and compared with the binding mode of RdRp inhibitor, Remdesivir. The aglycone part of Asparoside-C occupies binding site in such a way that the major glycosidic part of this molecule accommodates in a binding cavity surrounded by hydrophobic Tyr 455, Tyr 458 and Val 166 residues. The hydroxyl groups of sugar moieties form hydrogen bond with Lys 798, Glu 167, Tyr 455, Arg 457, Asn 691, Asp 623, Asp 452, while the oxygen atom from glycosidic linkage forms hydrogen bond with positively charged Lys 621 residue (Figure 5C). Remdesivir, on the other hand forms hydrogen bond interactions with Asp 164, Lys 621 and Cys 622 residues. The charged Lys 621 also forms the π-cation interaction with pyrrole ring in Remdesivir. These docking scores for Asparoside-C and Remdesivir are -6.652 and -4.661 respectively. The docking study on these proteins suggest that Asparoside-C has favorable binding interactions and reasobaly good docking scores compared to reference drug Remdesivir.
3.2. Molecular dynamics
Molecular dynamics simulations (MDS) provides with greater insight into binding of ligand molecules with the target protein under investigation. A workflow integrated with molecular docking, molecular dynamics simulations and free energy calculations has been employed by us in understanding the properties of various small molecule inhibitors (Chikhale et al., 2018; Kerzare et al., 2016; Pant et al., 2014). The MDS studies of protein-ligand complex over a sufficiently long period allow exploring a wide space of conformational optimization and binding affinity. In this study a 100 ns MDS was performed to evaluate the conformational stability and binding affinity of the ligands bound to the viral spike receptor-binding domain and NSP15 endoribonuclease proteins. Six phytochemicals, Asparoside-C, Asparoside-D, Shatavarin-I in complex with SARS-CoV-2 spike protein receptor-binding domain and Asparoside-C, Asparoside-D, Asparoside-F in complex with NSP15 endoribonuclease protein were selected for the MDS based on their docking performance. Along with these phytochemicals, standard drug Remdesivir was studied for its interactions with spike protein receptor-binding domain and NSP15 endoribonuclease protein. MDS was performed with stages of three minimization steps, one heating step and two equilibration steps followed by 100 ns of production stage. MDS of these complexes provides for analyzing binding modes, formation of hydrogen bond, pi-pi interactions and van der Waals interactions. The MDS trajectories are analyzed study the following parameters; RMSD, RMSF, Ligand-RMSD and the binding free energies on the complexes with MMGBSA calculations.
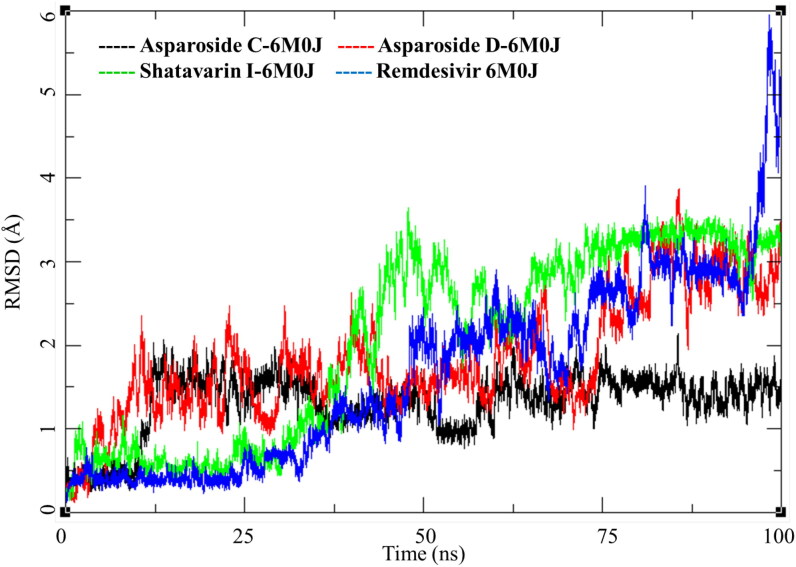
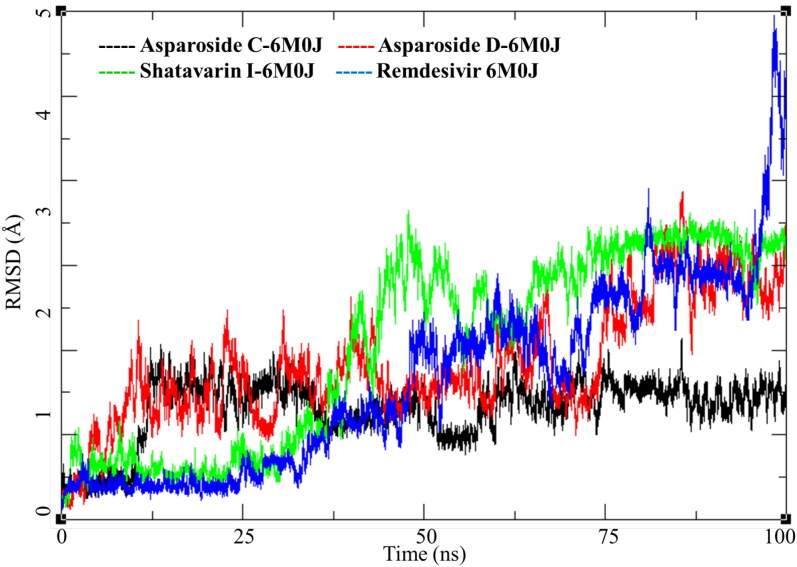
SARS-CoV-2 spike receptor-binding domain binds with the host ACE-2 receptor leading to the viral entry in the host cell. Targeting this protein is a clear and direct approach to inhibit the COVID-19 infection. Three phytochemicals Asparoside-C, Asparoside-D, Shatavarin-I bound to spike protein were obtained from the docking studies. Post-MDS RMSD analysis showed Asparoside-C- spike protein complex achieves the conformational stability 12.5 ns with a RMSD of 2.5 Å to the initial structure and stays in this conformation for rest of the period of time (Figure 6, Black). The Asparoside-D-spike protein complex shows optimum conformational stability around 2.5 Å between 15 to 60 ns of the MDS after which the RMSD increases to about 4 Å (Figure 6, Red). The Shatavarin-I-spike protein complex is less stable conformationally with varying RMSD between 2-4 Å (Figure 6, Green), however, this is not a very large fluctuation. In case of Remdesivir, it had a stable RMSD convergence for first 25 ns around 0.5 Å. However, the RMSD increased gradually throughout the simulation and fluctuated to 6 Å towards the end, this could be attributed to the breaking of some interactions between the ligand and receptor active site residues.
Figure 6.
Root mean square deviation (RMSD) of Asparoside-C, Asparoside-D, Shatavarin-I and Remdesivir bound to SARS-CoV-2 spike receptor-binding domain (PDB: 6M0J).
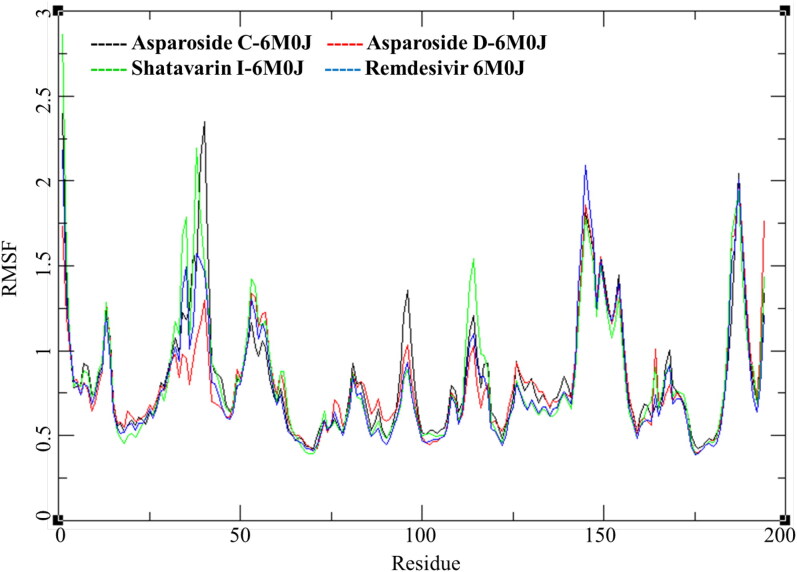
The RMSF for individual amino acid (aa) reflects the trend observed in RMSD of complexes (Figure 7). The RMSF per residue of Shatavarin-I-spike protein complex is higher compared to other tow complex. The RMSF of residue number between aa25 to aa50 is higher followed by residues between aa140 to aa160. To understand the conformational stability of the ligand bound in the active site ligand RMSD was studied for individual ligands; Asparoside-C, Asparoside-D, Shatavarin-I and Remdesivir.
Figure 7.
Root mean square fluctuations per amino acid (aa) residue of Asparoside-C, Asparoside-D, Shatavarin-I and Remdesivir bound to SARS-CoV-2 spike receptor-binding domain (PDB: 6M0J).
The ligand RMSD was calculated to understand the ligand conformations during the period of MDS. The Asparoside-C (Figure 8, Black) equilibrates for first 15 ns below 1 Å and then fluctuates by about 1.5 Å to stabilize for next 20 ns and then again fluctuates between 35 to 60 ns for about 1.5 Å. It equilibrates and stabilizes to 2 Å from the mean position after 60 ns. The Asparoside-D RMSD rises to around 2 Å and equilibrates for about 60 ns and then again increases to 4 Å after 60 ns (Figure 8, Red). The RMSD of Shatavarin-I increases consistently over the MSD time form 0.5 Å to 4.5 and then equilibrates around 4.5 Å towards the end of MDS.
Figure 8.
Root mean square deviation for each individual ligand; Asparoside-C, Asparoside-D, Shatavarin-I and Remdesivir bound to SARS-CoV-2 spike receptor-binding domain (PDB: 6M0J).
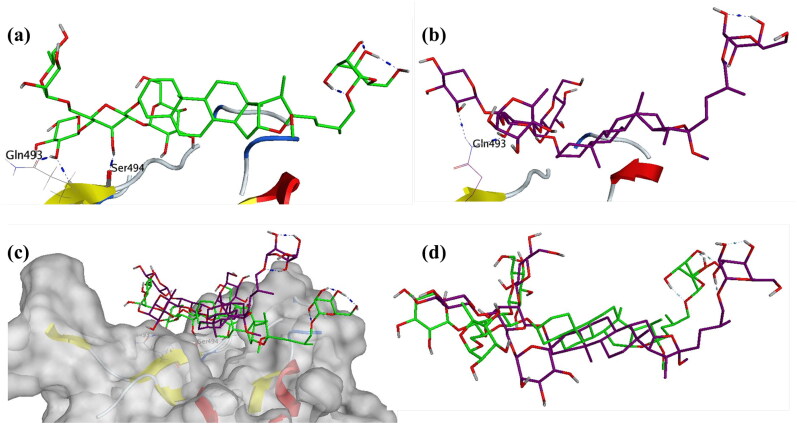
These ligands were studied further for their interactions with the individual residues port MDS. Asparoside-C bound to spike protein has two hydrogen bonds between hydroxyl groups of the Oxane rings and Gln493 and Ser494 (Figure 9a) before the start of production stage of the MDS. The Asparoside-C (O7-Ser494-O, Bond length 2.79 Å) hydrogen bond breaks during the MDS but Asparoside-C (O7-Gln493-NE2, Bond length 6.63 Å) is formed and stabilizes the complex (Figure 9b).
Figure 9.
Interaction analysis for Asparoside C bound to the SARS-CoV-2 viral spike receptor-binding domain during the molecular dynamics simulation; (a) Equilibrated structure of Asparoside-C (green) bound to the SARS-CoV-2 viral spike receptor-binding domain before MDS production phase; (b) Asparoside-C (Purple) bound to the SARS-CoV-2 viral spike receptor-binding domain post-MDS production phase; (c) Receptor surface analysis; (d) Ligand analysis.
The receptor surface analysis of the Asparoside-C bound to spike protein shows that the ligands are well accommodated in the active site during the MDS (Figure 9c), the superposition of ligand conformation at the beginning and end of MDS shed light on the stability of the complex (Figure 9d), this is also supported by the equilibrated ligand RMSD after 15 ns of the MDS. Three other phytochemicals Asparoside-C, Asparoside-D and Asparoside-F bound to SARS-CoV-2 NSP15 Endoribonuclease were selected from the docked complexes for MDS.
The binding mode of Remdesivir with the RBD domain of S protein is elaborated in Figure 10. It was observed from the protein and ligand RMSD that there is gradual rise in RMSDs, on visual inspection of the MDS trajectory and analyzing the bond breaking and formation it was found that there are several conformational changes in Remdesivir during the MDS. These conformational changes contribute to the fluctuations in the RMSD. The hydrogen bond between Remdesivir and RBD domain (N12-Arg71) reduces in bond length from 3.26 to 2.90 Å, which explains the drop in RMSD in the last 5 ns (Figures 6 and 8).
Figure 10.
Interaction analysis for Remdesivir bound to the SARS-CoV-2 viral spike receptor-binding domain during the molecular dynamics simulation; (a) Equilibrated structure of Remdesivir (green) bound to the SARS-CoV-2 viral spike receptor-binding domain before MDS production phase; (b) Remdesivir (Purple) bound to the SARS-CoV-2 viral spike receptor-binding domain post-MDS production phase; (c) Receptor surface analysis; (d) Ligand analysis.
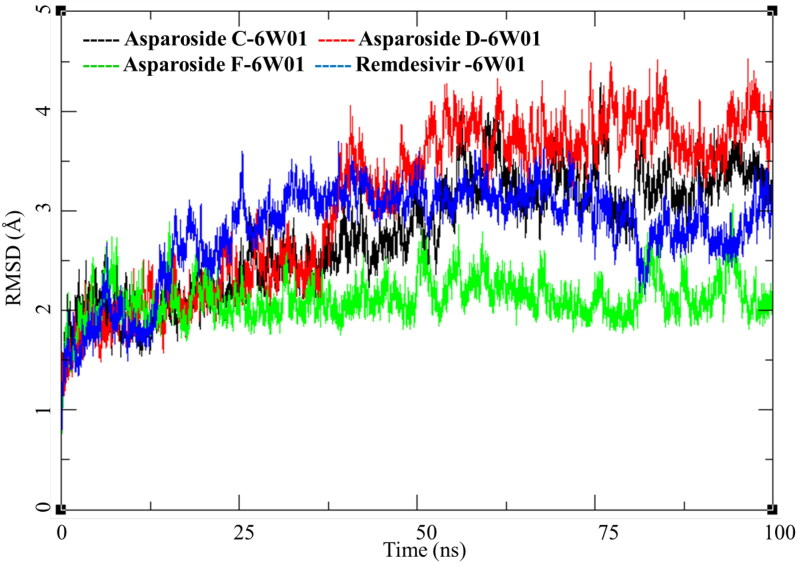
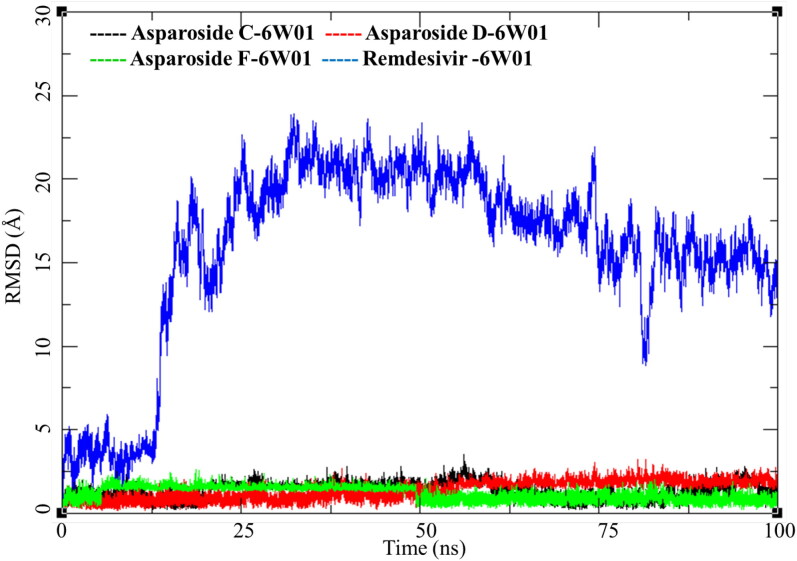
RMSD analysis of these complexes of phytochemicals and one of Remdesivir was performed the results are presented in Figure 11. Asparoside-C and D-NSP15 endoribonuclease complexes (Figure 11, black and Red respectively) shows similar pattern of RMSD with the initial equilibration to 2 Å for 35 ns. After 40 ns both the complexes shows increase in RMSD by another 1 Å and stays equilibrated in the same conformation till the end of MDS. NSP15 bound to Remdesivir also show similar trend and converges between 30 and 100 ns to RMSD between 2.5 to 3.0 Å (Figure 11, Blue) Contrary to this the AsparosideF-NSP15 endoribonuclease complex remains around 2 Å throughout the MDS (Figure 11, Green).
Figure 11.
Root mean square deviation of ligand-protein complexes of Asparoside-C, Asparoside-D, Asparoside-F and Remdesivir bound to SARS-CoV-2 NSP15 Endoribonuclease (PDB: 6W01).
To further understand these three complexes and the effect of ligand binding on the active site of the receptor RMSF of these complexes was studied (Figure 12). The RMSF for Asparoside-C, Asparoside-D, Asparoside-F and Remdesivir bound D-NSP15 endoribonuclease protein amino acid residues had significantly low variations. The residues in the pocket forming the active site for binding of ligands had a low RMSF between 1-2 Å, this could be attributed to the low b-factor in that region owing to its structural confirmation.
Figure 12.
Root mean square fluctuations of Asparoside-C, Asparoside-D, Asparoside-F and Remdesivir bound to SARS-CoV-2 NSP15 Endoribonuclease (PDB: 6W01).
The ligand RMSD for Asparoside-C, Asparoside-D, Asparoside-F and Remdesivir bound D-NSP15 endoribonuclease protein was studied to understand the conformational changes occurring during the MDS, the plot for ligand RMSD is presented in Figure 13.
Figure 13.
Ligand root mean square deviation of Asparoside C, Asparoside D, Asparoside F and Remdesivir bound to SARS-CoV-2 NSP15 Endoribonuclease (PDB: 6W01).
The phytochemical Asparoside-C had several conformational changes during the MDS which is evident from its evolving RMSD which fluctuates between 0 to 3 Å (Figure 13, black). Asparoside-D equilibrated and the RMSD remains low for first 50 ns between 0.5 to 2 Å, after 50 ns the RMSD gradually rises to 2.5 and converges for good till the end of MDS (Figure 13, Red). Asparoside-F, is an interesting case, the ligand RMSD changes from initial 1 Å to a narrow range of 1.75 to 2.0 Å, this reflects the limited access to conformational change due to either strong ligand binding due to hydrogen bonds or van Der Waals interactions or due to structural limitations. However, after 50 ns the molecule regains its original RMSD and fluctuates between 0.5 to 1.5 equilibrating till the end of MDS (Figure 13, Green). More interesting changes were observed with Remdesivir, initially the there were fluctuation between 0-5 Å for first 10 ns and then the RMSD suddenly peaked to 20 Å and later stabilized for rest of the simulation period (Figure 13, Blue). This high fluctuation in RMSD suggest major conformational and binding mode of Remdesivir.
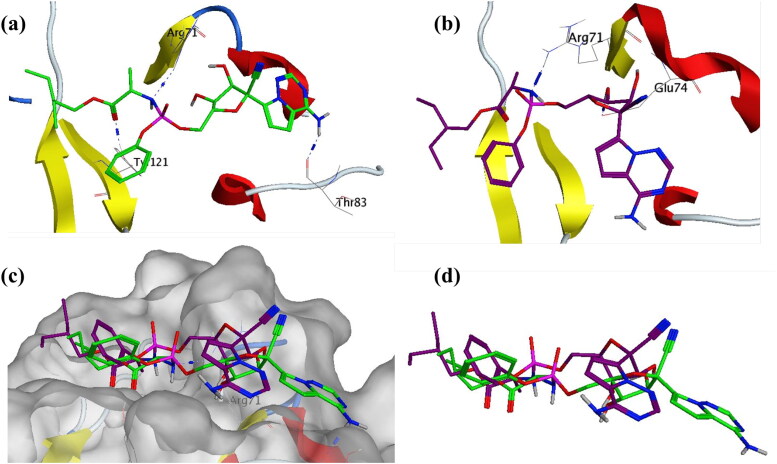
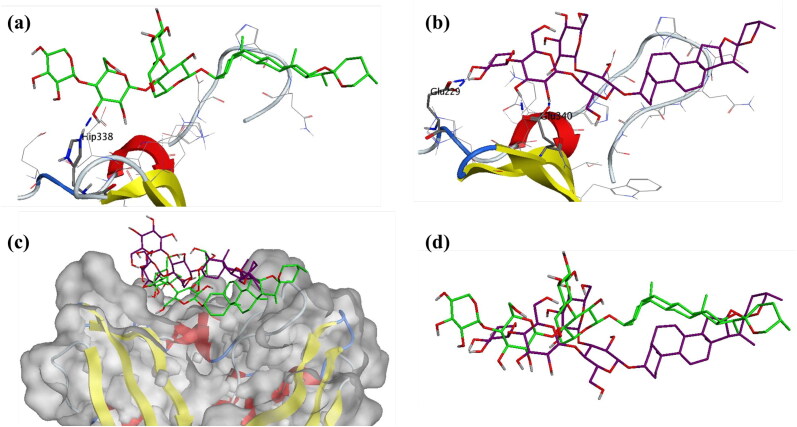
To shed more light into this compound, interaction between receptor and ligand was explored by visualizing three-dimensional structure, given in Figure 14. The initial conformation at the start of the MDS production phase was analyzed, the oxane ring of Asparoside-F formed a hydrogen bond with the His338 residue of the endonuclease protein active site (Asparoside-F-O14-His338-NE2, Bond length 2.91 Å) (Figure 14a). After the completion of the MDS, this interaction is lost and the oxane rings form new bonds between Asparoside-F and Endoribonuclease (Asparoside-F-O12-Glu340-OE2, Bond length 2.65 Å) and (Asparoside-F-O20-Glu229-OE1, Bond length 2.65 Å) (Figure 14b). The newly formed bonds and shorter bond distances, lowered ligand RMSD suggest improved binding of the compounds during MDS. The protein surface interaction and ligand superposition (Figure 14c and d) suggest the improved stability of the molecule.
Figure 14.
Interaction analysis for Asparoside-F bound to the SARS-CoV-2 NSP15 Endoribonuclease (PDB: 6W01) during the molecular dynamics simulation; (a) Equilibrated structure of Asparoside-F (green) bound to the SARS-CoV-2 NSP15 Endoribonuclease before MDS production phase; (b) Asparoside-F (Purple) bound to the SARS-CoV-2 NSP15 Endoribonuclease post-MDS production phase; (c) Receptor surface analysis; (d) Ligand analysis.
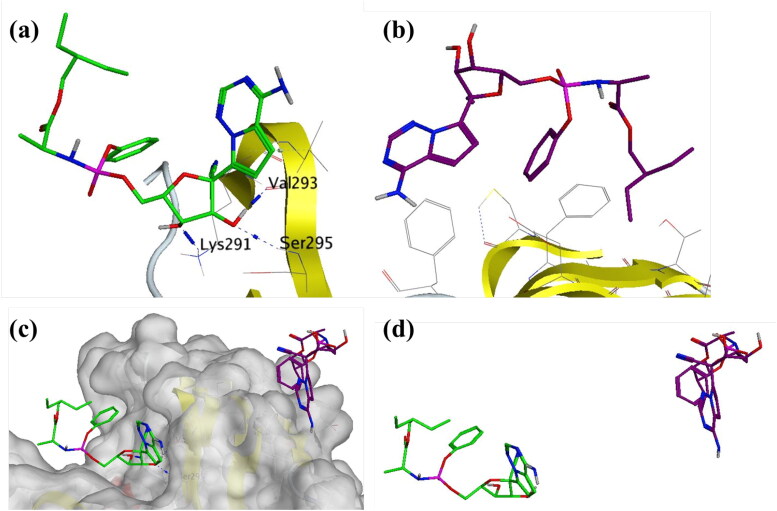
Remdesivir-SARS-CoV-2 NSP15 Endoribonuclease shows high Ligand-RMSD but no major fluctuation in protein RMSD and RMSF. This was studied in detail and the trajectory was analyzed visually for the conformational evolution. It was found that the ligand breaks away from the complex around 12 ns breaking the interactions with Lys291, Val293 and Ser295 (Figure 15a and b). During rest of the MDS, it does not form any further interactions with the receptor. Its positional and conformation changes (Figure 15c and d) contributed to the higher RMSD as observed in Figure 12.
Figure 15.
Interaction analysis for Remdesivir bound to the SARS-CoV-2 NSP15 Endoribonuclease (PDB: 6W01) during the molecular dynamics simulation; (a) Equilibrated structure of Remdesivir (green) bound to the SARS-CoV-2 NSP15 Endoribonuclease before MDS production phase; (b) Remdesivir (Purple) bound to the SARS-CoV-2 NSP15 Endoribonuclease post-MDS production phase; (c) Receptor surface analysis; (d) Ligand analysis.
MM-GBSA analysis was performed on all of the six protein-ligand complexes to evaluate the affinity of ligands to the target protein receptors. The MM-GBSA based binding free energy (ΔGbind) calculations were performed on the 100 ns long MDS trajectories. The binding energies measured by this method are more efficient than the GlideScore values for selection of protein-ligand complexes. The major energy components, such as H-bond interaction energy (ΔGbind_H-bond), coulomb or electrostatics interaction energy (ΔGbind_Coul), covalent interaction energy (ΔGbind_Cov), lipophilic interaction energy (ΔGbind_Lipo), electrostatic solvation free energy (ΔGbind_Solv) and van der Waals interaction energy (ΔGbind_vdW) altogether contribute to the calculation of MM-GBSA based relative binding affinity. The binding energies and the contributing factors calculated for the MDS trajectories are mentioned in the Table 4. Out of the six complexes studied three complexes belonged to the SARS CoV-2 spike receptor-binding domain and other three bound to SARS-CoV-2 NSP15 Endoribonuclease. The Asparoside-D bound to SARS CoV-2 spike receptor-binding domain showed high binding free energies with ΔGbind of -66.49.41 Kcal/mol with high contribution form the ΔGgas (-115.98 kcal/mol) and ΔGGB (54.61 Kcal/mol). Asparoside-C had the second highest binding free energy (ΔGbind of -62.61 Kcal/mol). The ΔGSurf (-5.84 Kcal/mol) for Asparoside-C indicates its better surface accessibility compared to other molecules. Remdesivir showed least binding energy (ΔGbind of -18.45 Kcal/mol) in the studied molecules bound to the S protein. In the second group of compounds binding the SARS-CoV-2 NSP15 endoribonucelase, Asparoside-D shows a very good binding free energy (ΔGbind = -72.46 Kcal/mol), second highest was Asparoside-F with binding free energy (ΔGbind = -55.19 Kcal/mol). The high binding affinity is also justified by the fact that during MDS the molecule optimizes its conformation for better fitting with the receptor active site which is clearly evident with Asparoside-F bound to the SARS-CoV-2 NSP15 Endoribonuclease. The most significant finding was the comparison of Remdesivir binding to the SARS-CoV-2 NSP15 Endoribonuclease, it has a very poor binding free energy (ΔGbind = -0.12 Kcal/mol). These results corroborate with results obtained from ligand RMSD analysis of Remdesivir. It suggests that binding of Remdesivir to SARS-CoV-2 NSP15 Endoribonuclease is weaker compared the phytochemicals under investigation.
Table 4.
Binding free energy components for the protein ligand complexes calculated by MM-GBSA analysis.
| Compounds | MM-GBSA* |
||||||
|---|---|---|---|---|---|---|---|
| ΔEVDW | ΔEELE | ΔGGB | ΔGSurf | ΔGgas | ΔGSol | ΔGbind | |
| 6M0J | |||||||
| AsparosideC | −35.36 (11.09) | −26.07 (9.44) | 54.24 (9.48) | −5.84 (1.60) | −111.00 (15.62) | 48.39 (8.42) | −62.61 (8.95) |
| AsparosideD | −27.58 (7.17) | −41.21 (16.73) | 54.61 (13.72) | −5.12 (1.10) | −115.98 (21.37) | 49.48 (12.89) | −66.49 (10.28) |
| ShatawarinI | −35.45 (4.28) | −24.34 (13.49) | 47.34 (10.97) | −5.34 (0.70) | −97.19 (15.55) | 41.99 (10.49) | −55.19 (6.81) |
| Remdesivir | −29.02 (3.59) | −48.96 (7.82) | 58.92 (6.14) | −4.98 (0.36) | −72.39 (9.14) | 53.94 (5.88) | −18.45 (4.64) |
| 6W01 | |||||||
| AsparosideC | −29.19 (3.63) | −65.14 (16.10) | 72.17 (10.64) | −6.96 (0.53) | −124.62 (10.09) | 65.20 (11.20) | −51.42 (6.11) |
| AsparosideD | −36.70 (4.59) | −50.19 (24.12) | 67.51 (20.59) | −6.13 (0.75) | −133.84 (24.98) | 61.37 (20.02) | −72.46 (7.02) |
| AsparosideF | −30.51 (5.46) | −62.78 (20.97) | 82.24 (16.76) | −5.53 (0.69) | −132.02 (21.22) | 76.71 (16.30) | −55.30 (6.06) |
| Remdesivir | −17.18 (7.29) | −16.34 (11.16) | 28.14 (11.97) | −2.71 (0.94) | −25.55 (16.69) | 25.43 (11.26) | −0.12 (6.89) |
*All energies are in Kcal/mol with standard deviation in parenthesis.
ΔEVDW = van der Waals contribution from MM; ΔEELE = electrostatic energy as calculated by the MM force field; ΔGGB = the electrostatic contribution to the solvation free energy calculated by GB; ΔGSurf = solvent-accessible surface area; ΔGSol= solvation free energy; ΔGgas = gas phase interaction energy; ΔGbind =Binding free energy.
4. Conclusion
The diverse pharmacological activities of Asparagus racemosus are well documented. However, in current outbreak of SARS-CoV-2 its anti-viral activities are immensely important. All together due to its anti-viral, immunomodulatory, anti-inflammatory and antioxidant potentials, this plant and its phytoconstituents could become vital therapeutic alternative in COVID 19. On this line, in the present work, the potential of major phytochemicals from Asparagus racemosus was explored through in silico methods. The docking studies pointed out the phytoconstituents viz. Asparoside-C, Shatavarin-I, Asparoside-D, Shatavarin-X and Racemoside-A having the lowest binding free energies with SARS-CoV-2 spike receptor-binding domain and Asparoside-C, Asparoside-F, Rutin, Asparoside-D, Racemoside-A with NSP15 endoribonuclease. In order to gain deeper insights of binding affinities and binding modes of top phytoconstituents, 100 ns molecular dynamics simulations and MM-GBSA binding free energy calculations were carried out. It was found that the Asparoside-C- spike protein complex achieves the conformational stability at 12.5 ns with comparably low RMSD of 2.5 Å. In case of NSP15 endoribonuclease, Asparoside-F was found better stabilized with RMSD of 2 Å throughout the MDS. Compared to docking studies, MM-GBSA binding free energy calculations and estimates of various ΔG parameters are more accurate. These calculations suggest that the Asparoside-D and Asparoside-C bound to SAR CoV-2 spike receptor-binding domain with ΔGbind of -66.49 and -62.61 Kcal/mol respectively are most favorable while Asparoside-D and Asparoside-F bound to NSP15 endoribonucelase with ΔGbind -72.46 and -55.19 Kcal/mol respectively are most favorable in terms of binding interactions and binding free energies. In this study we also found that the standard drug Remdesivir, which is being used as treatment for COVID-19 has less binding affinity to the S protein compared to other phytochemicals. In case of NSP15 endoribonuclease, it shows very low binding affinity compared to the phytochemicals. The binding affinity of Remdesivir is more towards the SARS CoV-2 S protein compared to NSP15 endoribonuclease which suggest its site of binding to be RBD domain of S protein. The future scope of our findings lies in the experimental verification of these results towards the potential of Asparoside-D, Asparoside-C and Asparoside-F in inhibiting key SAR CoV-2 proteins. These results could be exploited further to design future ligands from natural sources.
Supplementary Material
Glossary
Abbreviations
- 2019-nCoV
2019 Novel Coronavirus
- CoV
Corona Virus
- COVID-19
Coronavirus Disease 2019
- HCQ
Hydroxychloroquine
- MD
Molecular Dynamics
- MMGBSA
Molecular Mechanics-Generalized Born Solvent Accessibility
- NSP
Non-structural Protein
- ORF
Open Reading Frame
- OPLS
Optimized Potentials for Liquid Simulations
- PHEIC
Public Health Emergency of International Concern
- RBD
Receptor Binding Domain
- RMSD
Root Mean Square Deviation
- SARS
Severe Acute Respiratory syndrome
- SARS-COV-2
Coronavirus Disease Strain
- SDF
Structure Data File
- WHO
World Health Organization
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Al-Khafaji K., Al-DuhaidahawiL D., & Taskin Tok T. (2020). Using integrated computational approaches to identify safe and rapid treatment for SARS -CoV- 2. Journal of Biomolecular Structure & Dynamics, 1–11. 10.1080/07391102.2020.1764392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alok S., Jain S. K., Verma A., Kumar M., Mahor A., & Sabharwal M. (2013). Plant profile, phytochemistry and pharmacology of Asparagus racemosus (Shatavari): A review. Asian Pacific Journal of Tropical Disease, 3(3), 242–251. 10.1016/S2222-1808(13)60049-3 [DOI] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J. M. O., Yi L. L., Guarino L., Sacchettini J. C., & Kao C. C. (2008). Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. The Journal of Biological Chemistry, 283(6), 3655–3664. 10.1074/jbc.M708375200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsom T., Waranuch N., Ingkaninan K., Denduangboripant J., & Sukrong S. (2012). Molecular analysis of the genus Asparagus based on matK sequences and its application to identify A. racemosus, a medicinally phytoestrogenic species. Fitoterapia, 83(5), 947–953. 10.1016/j.fitote.2012.04.014 [DOI] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure & Dynamics, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Conejo Y., Linan-Rico A., Garcia-Rodriguez D. A., Centeno-Leija S., & Serrano-Posada H. (2020). An exclusive 42 amino acid signature in pp1ab protein provides insights into the evolutive history of the 2019 novel human‐pathogenic coronavirus (SARS-CoV-2). Journal of Medical Virology, 92, 688–692. 10.1002/jmv.25758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. J., Michaelis M., Hsu H. K., Tsai C. C., Yang K. D., Wu Y. C., Cinatl J. Jr., & Doerr H. W. (2008). Toona sinensis Roem tender leaf extract inhibits SARS coronavirus replication. Journal of Ethnopharmacology, 120(1), 108–111. 10.1016/j.jep.2008.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhale R. V., Sinha S. K., Patil Rb, Prasad S. K., Shrivastava S. K., Shakya A., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). SARS-CoV-2 host entry and replication inhibitors from Indian ginseng: In silico approach. Journal of Biomolecular Structure and Dynamics. ( 10.1080/07391102.2020.1778539). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhale R., Thorat S., Choudhary R. K., Gadewal N., & Khedekar P. (2018). Design, synthesis and anticancer studies of novel aminobenzazolyl pyrimidines as tyrosine kinase inhibitors. Bioorganic Chemistry, 77, 84–100. 10.1016/j.bioorg.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Choudhury C. (2020). Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1771424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Watson S. J., Kellam P., Al-Rabeeah A. A., Makhdoom H. Q., Assiri A., Al-Tawfiq J. A., Alhakeem R. F., Madani H., AlRabiah F. A., Hajjar S. A., Al-Nassir W. N., Albarrak A., Flemban H., Balkhy H. H., Alsubaie S., Palser A. L., Gall A., Bashford-Rogers R., … Memish Z. A. (2013). Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. The Lancet, 382(9909), 1993–2002. 10.1016/S0140-6736(13)61887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., & Baker S. C. (2018). An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology, 517, 157–763. 10.1016/j.virol.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA News Release. (2020, May 1). Coronavirus (COVID-19) update: FDA issues emergency use authorization for potential COVID-19 treatment https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment.
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurav S., & Gurav N. (2014). Herbal drug microscopy In Gurav, S., & Gurav, N. (Eds.), Indian herbal drug microscopy (pp. 15–196). Springer; 10.1007/978-1-4614-9515-4_4 [DOI] [Google Scholar]
- Han Y., & Král P. (2020). Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2. ACS Nano, 14(4), 5143–5147. 10.1021/acsnano.0c02857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G. A., Doerr H. W., & Cinatl J. Jr.(2005). Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. Journal of Medicinal Chemistry, 48(4), 1256–1259. 10.1021/jm0493008 [DOI] [PubMed] [Google Scholar]
- Jayawardena R., Sooriyaarachchi P., Chourdakis M., Jeewandara C., & Ranasinghe P. (2020). Enhancing immunity in viral infections, with special emphasis on COVID-19: A review. Diabetes and Metabolic Syndrome. Diabetes & Metabolic Syndrome, 14(4), 367–382. 10.1016/j.dsx.2020.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure & Dynamics, 1–16. 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzare D., Chikhale R., Bansode R., Amnerkar N., Karodia N., Paradkar A., & Khedekar P. (2016). Design, synthesis, pharmacological evaluation and molecular docking studies of substituted oxadiazolyl-2-oxoindolinylidene propane hydrazide derivatives. Journal of the Brazilian Chemical Society, 27(11), 1998–2010. 10.5935/0103-5053.20160090 [DOI] [Google Scholar]
- Lalert L., Kruevaisayawan H., Amatyakul P., Ingkaninan K., & Khongsombat O. (2018). Neuroprotective effect of Asparagus racemosus root extract via the enhancement of brain-derived neurotrophic factor and estrogen receptor in ovariectomized rats. Journal of Ethnopharmacology, 225, 336–341. 10.1016/j.jep.2018.07.014 [DOI] [PubMed] [Google Scholar]
- Li S.-Y., Chen C., Zhang H.-Q., Guo H.-Y., Wang H., Wang L., Zhang X., Hua S.-N., Yu J., Xiao P.-G., Li R.-S., & Tan X. (2005). TanIdentification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Research, 67(1), 18–23. 10.1016/j.antiviral.2005.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. W., Tsai F. J., Tsai C. H., Lai C. C., Wan L., Ho T. Y., Hsieh C. C., & Chao P. D. (2005). Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Research, 68(1), 36–42. 10.1016/j.antiviral.2005.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S. C., Nandy A., Pal M., & Saha B. P. (2000). Evaluation of antibacterial activity of Asparagus racemosus Willd. root. Phytotherapy Research, 14(2), 118–119. [DOI] [PubMed] [Google Scholar]
- Mengist H. M., Fan X., & Jin T. (2020). Designing of improved drugs for COVID-19: Crystal structure of SARS-CoV-2 main protease Mpro. Signal Transduction and Targeted Therapy, 5(1), 67. 10.1038/s41392-020-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal L., Kumari A., Suri C., Bhattacharya S., & Asthana S. (2020). Insights into structural dynamics of allosteric binding sites in HCV RNA-dependent RNA polymerase . Journal of Biomolecular Structure & Dynamics, 38(6), 1612–1625. 10.1080/07391102.2019.1614480 [DOI] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure & Dynamics, 1–6. 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., & Qian Z. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications, 11(1) 1-12. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant A. M., Chikhale R. V., Menghani S. S., & Khedekar P. B. (2014). LEDGF/p75 IN interaction inhibitors: In silico studies of an old target with novel approach. BMC Infectious Diseases, 14(S3) P18. 10.1186/1471-2334-14-S3-P18 [DOI] [Google Scholar]
- Patwardhan B., Chavan-Gautam P., Gautam M., Tillu G., & Chopra A. (2020). Ayurveda rasayana in prophylaxis of COVID-19. Current Science, 19, 1–3. [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., & Manickam M. (2020). Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today, 25(4), 668–688. 10.1016/j.drudis.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A., Muhammad F., Bashir S., Anwar M. I., Awais M. M., Akhtar M., Aslam B., Khaliq T., & Naseer M. U. (2015). Antiviral and immune boosting activities of different medicinal plants against Newcastle disease virus in poultry. World's Poultry Science Journal, 71(3), 523–532. 10.1017/S0043933915002147 [DOI] [Google Scholar]
- Ryu Y. B., Jeong H. J., Kim J. H., Kim Y. M., Park J. Y., Kim D., Nguyen T. T., Park S. J., Chang J. S., Park K. H., Rho M. C., & Lee W. S. (2010). Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorganic & Medicinal Chemistry, 18(22), 7940–7947. 10.1016/j.bmc.2010.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M. A., Abdullah S. M., Khan M. A., Nasar G., & Saba I. (2014). Antibacterial activity of chemical constituents isolated from asparagus racemosus. Bangladesh Journal of Pharmacology, 9(1), 1–3. 10.3329/bjp.v9i1.16672 [DOI] [Google Scholar]
- Singh R., & Geetanjali (2016). Asparagus racemosus: A review on its phytochemical and therapeutic potential. Natural Product Research, 30(17), 1896–1908. 10.1080/14786419.2015.1092148 [DOI] [PubMed] [Google Scholar]
- Sinha S. K., Prasad S. K., Gurav Ss, Patil Rb…Shakya A. (2020. b). Identification of bioactive compounds from glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: A pharmacoinformatics study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1779132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020. a). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillu G., Chaturvedi S., Chopra A., & Patwardhan B. (2020). Public Health Approach of Ayurveda and Yoga for COVID-19 Prophylaxis. The Journal of Alternative and Complementary Medicine), 26(5), 360–365. 10.1089/acm.2020.0129 [DOI] [PubMed] [Google Scholar]
- Uma B., Prabhakar K., & Rajendran S. (2009). Anticandidal activity of Asparagus racemosus. Indian Journal of Pharmaceutical Sciences, 71(3), 342–343. 10.4103/0250-474X.56017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S., Jeena G. S., Kumar S., & Shukla R. K. (2020). Asparagus racemosus bZIP transcription factor-regulated squalene epoxidase (ArSQE) promotes germination and abiotic stress tolerance in transgenic tobacco. Plant Science : An International Journal of Experimental Plant Biology, 290, 110291 10.1016/j.plantsci.2019.110291 [DOI] [PubMed] [Google Scholar]
- Verma S., Kuhad A., Bhandari R., Prasad S. K., Shakya A., Prasad R. S., & Sinha S. K. (2020). Effect of ethanolic extract of Solanum virginianum Linn. on neuropathic pain using chronic constriction injury rat model and molecular docking studies. Naunyn-Schmiedeberg’s Archives of Pharmacology. 10.1007/s00210-020-01872-8 [DOI] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wen C.-C., Shyur L.-F., Jan J.-T., Liang P.-H., Kuo C.-J., Arulselvan P., Wu J.-B., Kuo S.-C., & Yang N.-S. (2011). Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and Taxillus chinensis inhibit SARS CoV replication. Journal of Traditional and Complementary Medicine, 1(1), 41–50. 10.1016/S2225-4110(16)30055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Hai, Wu K., Lun, Zhang X., Deng S., Qiong., & Peng B. (2020). In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. Journal of Integrative Medicine, 18(2), 152–158. 10.1016/j.joim.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., … Shi Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.