Abstract
Accurately predicting the protein thermostability changes upon single point mutations in silico is a challenge that has implications for understanding diseases as well as industrial applications of protein engineering. Free energy perturbation (FEP) has been applied to predict the effect of single point mutations on protein stability for over 40 years and emerged as a potentially reliable prediction method with reasonable throughput. However, applications of FEP in protein stability calculations in industrial settings have been hindered by a number of limitations, including the inability to model mutations to and from prolines in which the bonded topology of the backbone is modified and the complexity in modeling charge-changing mutations. In this study, we have extended the FEP+ protocol to enable the accurate modeling of the effects on protein stability from proline mutations and from charge-changing mutations. We also evaluated the influence of the unfolded model in the stability calculations using increasingly longer peptides with native sequence and conformations. With the abovementioned improvements, the accuracy of FEP predictions of protein stability over a data set of 87 mutations on five different proteins has drastically improved compared with previous studies, with a mean unsigned error of 0.86 kcal/mol and root mean square error of 1.11 kcal/mol, comparable with the accuracy of previously published state-of-the-art small-molecule relative binding affinity calculations, which have been shown to be capable of driving discovery projects.
Significance
The structure and the function of the protein are tightly coupled. Single point mutations can drastically change the stability of a protein structure. The ability to predict such changes is valuable in understanding diseases but also in designing therapeutics or industrial proteins. Free energy perturbation is an accurate method for predicting free energy changes upon mutations. In this work, we have implemented a new, to our knowledge, method for proline mutations. We show free energy perturbation can very accurately predict protein thermal stability changes of single point mutations for all 20 natural amino acids. We believe the accuracy of the predictions is sufficient to drive protein engineering projects.
Introduction
The function of a protein is tightly coupled with its structure and dynamic behavior. Therefore, understanding the thermostability of proteins can provide fundamental insight in how proteins fold and how they work (1,2). This structure-function relationship is clearly manifested in protein mutations. Disease-causing single point missense mutations may directly affect protein function, conformational dynamics, and protein-protein interactions. Many of these mutations have been found to be related to protein thermostability (3, 4, 5, 6, 7, 8). From a practical perspective, protein stability engineering has wide applications in biotech industries. For example, industrial enzymes for food, detergents, paper, or fuel may need to be designed to be stable and functional at desired environments (9, 10, 11). The stability of therapeutic antibodies needs to be engineered to have longer shelf-life and prevent aggregation (12, 13, 14, 15, 16). In addition, protein stability engineering is frequently used in crystallography, e.g., crystallization of the transmembrane domain of G-protein-coupled receptors (17).
Significant efforts have been devoted to computationally predicting the stability change upon mutations. Some approaches rely on machine learning, and other methods use empirical energy potentials or statistical potentials (18). These approaches depend to varying extents on existing structural and mutational data and therefore are potentially biased. For example, the vast majority of existing mutational data contain destabilizing mutations, and hence, machine-learning models may be biased toward negative predictions (19), whereas in real applications, we are often more interested in stabilizing mutations. Furthermore, these models are generally based on static protein structures and do not consider potential structural reorganization or dynamic changes and the solvent effects are generally estimated implicitly.
Free energy perturbation (FEP) (20), based on molecular dynamics simulations with an explicit solvent model, is a rigorous method to calculate the change of free energy upon residue mutation or ligand structural modification without the need for any training sets. In the applications of FEP to calculate the effect of residue mutation on protein stability, the native residue is alchemically “morphed” to the mutant residue in both folded and unfolded states. The free energy changes for the folded and unfolded states are estimated from the simulations and the difference, ΔΔG, between the folded and unfolded transformation corresponds to the change of protein thermostability upon mutation. The same principle with slight modifications in the thermodynamic cycle can also be used to calculate the relative binding affinities between two structurally similar ligands to the same protein receptor. The FEP method implemented in FEP+ software (Schrödinger, New York, NY) has been successfully applied in retrospective and prospective predictions of relative protein-ligand binding affinities in which the average root mean square error (RMSE) was found to be generally around 1 kcal/mol (21, 22, 23, 24).
The application of FEP and thermodynamic integration (25) for the calculation of protein stability was first pioneered in the late 1980s, and a number of studies have been published over the years (26, 27, 28, 29, 30, 31, 32, 33, 34, 35). Because of the intensive computational resources required for the FEP and thermodynamic integration calculations, the early applications for protein stability simulations were typically very short and with limited solvation around mutation sites (26, 27, 28, 29, 30). Although the initial results were promising, the number of mutations studied was few, providing little statistical significance. Later works have expanded the number of mutations. However, they only targeted one protein, or only mutations to alanine were attempted (34,36). Gapsys et al. applied a more recently developed alchemical free energy method, the Crooks fluctuation theorem (37,38), to predict protein thermostability for 143 single point mutations in barnase and staphylococcal nuclease using up to six different force fields (33). They found the best average unsigned error for each target to be 0.91 and 0.84 kcal/mol, respectively, with multiple force fields. Two recent studies in 2017 by Steinbrecher et al. (39) and Ford et al. (40), respectively, covered many more mutations on different proteins. The results showed that FEP+ could clearly predict the direction of stability change of proteins upon single point mutations, but overall, the errors were still relatively large, especially for mutations in which the formal charge was altered, suggesting there is room to improve.
In addition to the large errors in modeling charge-changing mutations, none of the earlier studies, as far as we are aware, included mutations involving prolines. Proline is unique among the 20 amino acids because its side chain cyclizes with the backbone to form a covalent bond. Mutations to or from proline involve the formation or breaking of a covalent bond, which can lead to numerical instability problems during the FEP simulations (41). Furthermore, proline residue, because of its cyclization, has restricted flexibility and hence reduced entropy loss upon folding. It has been hypothesized that mutations to proline can stabilize proteins, and indeed, a number of studies that introduced prolines in strategic locations such as loops and β-turns were found to increase stability of the protein (42, 43, 44, 45, 46, 47).
Since the initial study of FEP+ on protein stability in 2017, a number of improvements have been made in the FEP+ program. In particular, an alchemical water method was introduced for perturbations involving net charge changes in the two physical end states (48), and this method was applied successfully to both protein-ligand binding and protein-protein relative binding affinity predictions, with an overall RMSE of 1.2 kcal/mol (49). Also, the numerical instability problem in perturbations involving covalent bond formation or breaking was addressed by introducing a soft bond-stretch potential (50) and has been applied in the pharmaceutical industry, including scaffold hopping mutations (51) and macrocycle formation (24,52). With these recent advances in FEP+, we revisited the question of how accurate FEP+ prediction on protein stability can be, with the focus on charge-changing mutations and proline mutations. For that purpose, we used a carefully curated data set by Pucci et al. (19) and additional data from the literature and performed FEP+ stability calculations on 87 mutations on five different protein targets. We explored the effects of different models of the unfolded state in the simulation and the accuracy of FEP+ prediction on both charge-changing and proline mutations. The overall accuracy was found to be comparable to that of small molecule-protein binding affinity FEP+ predictions.
Materials and Methods
Data set selection and preparation
The protein stability data set in Pucci et al. (19) covers 15 different protein structures associated with 342 mutations in which all wild-types and mutants have crystal structures with resolution below 2.5 Å. We extracted only the experimental data points measured at pH 7 ± 1, resulting in 13 structures with 96 mutations. To gain statistically meaningful results, only the proteins with more than five mutations were kept for this study. In total, there are four protein structures with 69 mutations (Protein Data Bank, PDB: 1EY0, 1BN1, 2LZM, and 1L63). Two additional systems were used to further validate the effect of proline mutations on overall protein stability (PDB: 1RGG and 1PGA) (43,53).
The PDB structures were downloaded and prepared using Protein Preparation Wizard in Maestro (Schrödinger Release 2019-2; Schrödinger), retaining all resolved water molecules in the crystal structures. The hydrogen atoms were added, and hydrogen-bonding networks were sampled. The ionization states of the residues were predicted using Propka at pH 7.0, and the final structures were minimized with restraints on the heavy atom using the OPLS3e force field. The minimization was terminated when the heavy-atom root mean square deviation reached 0.3 Å.
FEP+ simulation
The FEP+ simulations for nonproline mutations were carried out using the 2019-2 release of Schrödinger Suite (Schrödinger) and OPLS3e force field (54,55). FEP+ for mutations involving proline was only fully implemented in more recent 2019-4 release; therefore, all FEP+ predictions for proline mutations in the Pucci set and the complete proline set were from the most recent version of the software. The prepared protein structures were solvated in a box of water molecules with 5 Å buffer width with no counterions added to the system. As unfolded models, we used capped monopeptide, tripeptide, pentapeptide, or heptapeptide with the mutation site in the center of the peptides. The capping groups were acetyl on the N-terminus and an N-methyl group on the C-terminus. Both the sequences and coordinates of the peptides were directly extracted from the crystal structures of the corresponding native proteins. The solvation buffer width for unfolded models was set to 10 Å. When the mutation involves a charged residue, the buffer width was chosen to be 8 Å for the folded protein. Further, both the folded and unfolded systems were neutralized by adding an appropriate number of sodium or chloride ions in addition to a NaCl solution at physiological concentration (0.15 M). The ions were randomly placed in the simulation box. The mutated residue is included in the replica exchange solute tempering (REST) region (56,57), which effectively increases the temperature for the residue. During the outlier analysis of 2LZM, the side chains of Leu36, Leu103, and Val23 for mutations involving Thr62 and side chains of Val23 and Ile72 for the mutations involving Val66 were included in the REST region.
The solvated systems were relaxed and equilibrated using the default Desmond (Desmond Molecular Dynamics System; D. E. Shaw Research, New York, NY) (58) relaxation protocol implemented in Maestro, which consists of a series of minimizations and short simulations with restraints. Each perturbation was performed over 12 λ windows for charge conserving mutations and 24 λ windows for charge altering mutations. Each λ window was simulated for 5 ns by default or up to 100 ns. For charge mutations, a co-alchemical water approach was employed to maintain an overall neutral system charge. The protocol essentially mutates an alchemical ion, sodium or chloride, to a water molecule at the same time as the charge-changing residue mutation (48). For mutations involving a proline amino acid, a core-hopping protocol was deployed with 16 λ windows. In this protocol, a “CG-CD” bond within the pyrrolidine ring is replaced by a softcore bond, allowing bond breaking to accommodating a noncyclic side-chain mutation.
WaterMap simulation
WaterMap (59, 60, 61) simulations on Staphylococcus nuclease (S. nuclease) structures were also carried out using the Schrödinger 2019-2 release (Schrödinger). WaterMap was initially designed for analysis of protein-ligand binding pockets; hence, the analysis region is identified by the ligand. A probe, dimethylpropane, was manually placed close to Thr62 and Val66, and a region with 10 Å within the probe was analyzed. Before the simulations, all mutant structures were aligned on the wild-type structure, PDB: 1EY0. The probe was merged into the mutant structures ensuring that the same region was analyzed.
Residue scanning
Residue scanning calculation (62), as implemented in BioLuminate (Schrödinger Release 2019-2; Schrödinger), samples only the side-chain rotamers of the mutated residue followed by minimization. The stability ΔΔG is defined by ΔGunfolded − ΔGfolded, where each individual term is the energy difference between the mutant and the wild-type in unfolded or folded states. The protein backbone and the neighboring side chains are kept fixed. The input structures are identical to those for FEP+, and because the method uses the implicit solvation model VSGB (63), all crystallographic water molecules were removed before the calculations.
Results
Data sets
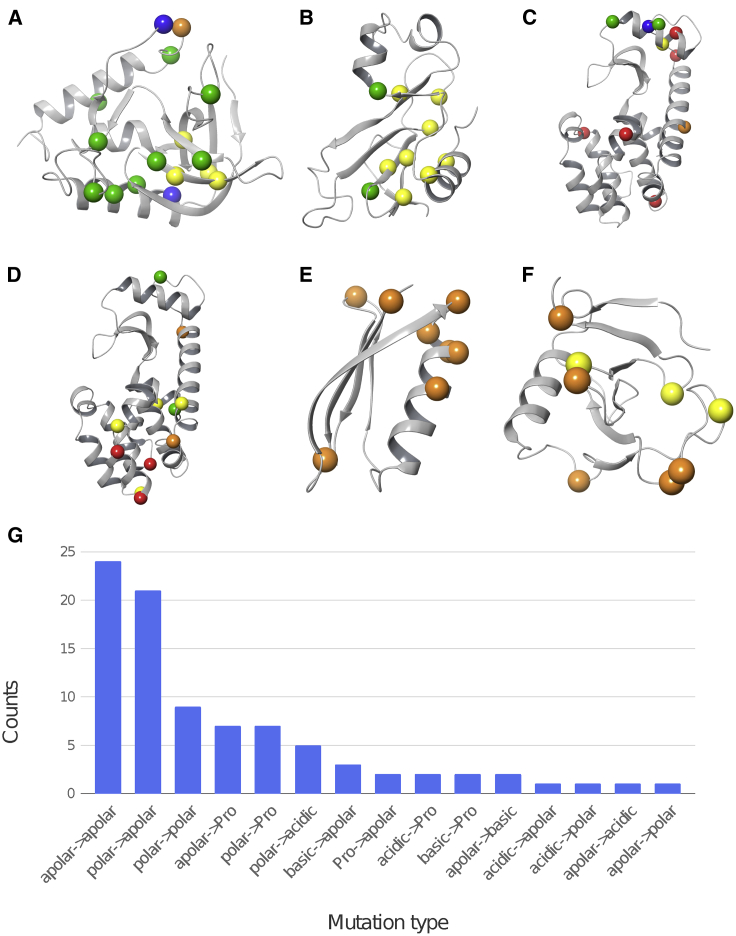
Pucci et al. (19) collected a set of protein stability data from the ProTherm database (64), in which high-resolution crystal structures are available for both wild-type proteins and their mutants. The experimental conditions such as pH and temperatures were also recorded, as well as the relative protein stability in ΔΔG. In this work, we focused on the experiments conducted at pH 7 ± 1, resulting in four sets of data with 69 mutations. Two of the mutation sets were measured using thermal unfolding assays, and two were from protein unfolding measurements at room temperature (20–25°C) using chemical denaturants. The two thermal unfolding assay sets were both from T4 lysozyme but with different wild-type crystal structures (PDB: 2LZM and 1L63). The chemical denaturation assay sets were from S. nuclease and barnase (PDB: 1EY0 and 1BNI). This data set will be referred to as the “Pucci set.” To collect sufficient data on the accuracy of proline mutations, we included an additional eight mutations, of which five involved prolines from ribonuclease Sa (43) and 10 involved proline mutations from protein G (53), and the stabilities of these mutations were all from thermal unfolding experiments. This data set will be referred to as the “proline set.” Information about the mutations, their location, and type is shown in Fig. 1. The data set includes a large number of combinations of residue types, and, not surprisingly, the apolar-to-apolar mutations are the most common ones. Curiously, apolar residues mutated to other residue types are rare. In fact, there were only four cases in which apolar residues were mutated to polar, acidic, or basic residues. Of the total 87 mutations, 20 mutations involved proline, 20 alanine, nine acidic, and seven basic residue types.
Figure 1.
Crystal structures of the wild-type proteins with colored spheres indicating the mutation sites: (A) S. nuclease (PDB: 1EY0), (B) barnase (PDB: 1BNI), (C) T4 lysozyme (PDB: 1L63), (D) T4 lysozyme (PDB: 2LZM), (E) protein G (PDB: 1PGA), and (F) ribonuclease (PDB: 1RGG). The color codes indicate whether the mutations involve apolar residues (yellow), polar residues (green), basic residues (blue), acidic residues (red), or proline (orange). Polar residues include T, S, C, N, and Q; basic residues are K and R; and acid residues are D and E. The remaining nonproline residues, including Y, are apolar residues. In cases in which the mutations change residue types, we color based on the following rank: proline > charge > polar > apolar, e.g., the color of a mutation from a polar residue to proline or reverse will be colored orange (proline). (G) The number of mutations for each mutation type is shown. To see this figure in color, go online.
FEP+ predictions of the Pucci set: approximating an unfolded state of the protein
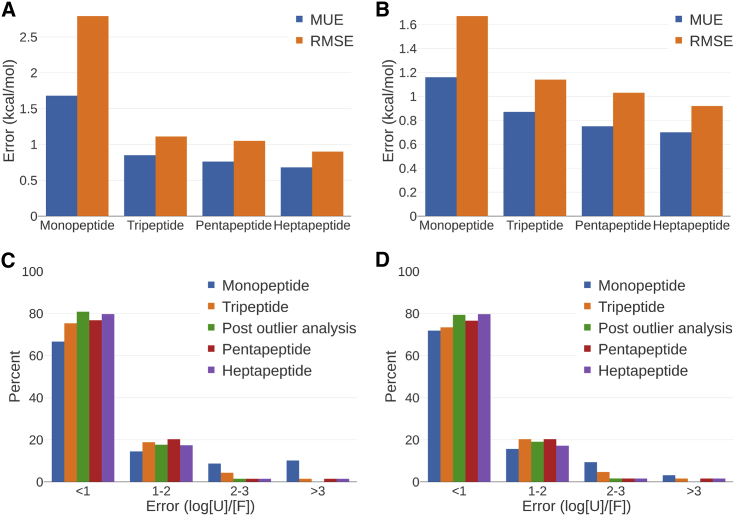
We examined four different unfolded models based on short peptides with the native sequence flanking the mutation sites. The simplest one is a single capped amino acid (monopeptide), which was used in previous studies (39,40). The other three models are capped tri-, penta-, and heptapeptide. In each of these models, the residue that is being mutated is flanked by one, two, or three neighboring amino acids, respectively. The input conformations for the unfolded models were from the corresponding wild-type crystal structures. Across the different systems in the Pucci set, the error reduced significantly (p < 0.016) when tripeptide was used instead of monopeptide. Mean unsigned error (MUE) dropped from 1.71 (monopeptide) to 1.05 (tripeptide) and RMSE from 2.80 to 1.63. Extending the peptide further slightly reduced the MUE further, to 0.95 (pentapeptide) and 0.89 (heptapeptide). We noticed that although there are only five mutations in the Pucci set that involved proline, their absolute prediction errors for the monopeptide unfolded model can be as high as 10 kcal/mol. By removing these proline mutations, the monopeptide MUE dropped down to 1.20 kcal/mol, whereas the MUE for tripeptide, pentapeptide, and heptapeptide remained the same. It seems that mutations involving prolines is the chief beneficiary of using tripeptide as an unfolded model. As the largest benefit seems to come from using tripeptide instead of the monopeptide, we focused our additional analysis on predictions using tripeptide as an unfolded model.
There were a number of clear prediction outliers with absolute prediction error larger than 2.8 kcal/mol, which correspond to a 100-fold difference in the predicted folded/unfolded population ratio compared to that of experiment. These include Thr62Val, Val66Lys, and Val66Leu in 1EY0 and Ile3Cys and Gly156Asp in 2LZM. Thr62Ser in 1EY0 was also included in the analysis, although the absolute error was 2.44 kcal/mol. We will describe each of the outliers in detail in the next section. After addressing the outliers, the MUE and RMSE for the tripeptide unfolded model reduced to 0.85 and 1.11 kcal/mol, respectively (Fig. 2, A and B; Tables S1 and S2), which is comparable with FEP+ prediction accuracy of small molecule-protein relative binding affinities (22,23). If we disregard the proline mutations, the improvement in accuracy using the tripeptide unfolded model compared to the monopeptide is smaller. MUE and RMSE for monopeptide are 1.16 and 1.67 kcal/mol and for tripeptide are 0.87 and 1.14 kcal/mol. However, the difference is still significant (p = 0.04).
Figure 2.
(A) MUE and RMSE of predictions in the Pucci set after outlier analysis, (B) MUE and RMSE for nonproline mutations, (C) error distribution for Pucci set mutations, and (D) error distribution for nonproline mutations. The orange bar corresponds to predictions using the tripeptide unfolded model before outlier analysis, and the green bar is after outlier analysis. Error distribution is expressed as log[U]/[F], where [U] is the concentration of the unfolded state and [F] is the concentration of folded state. To see this figure in color, go online.
Further, we analyzed the error distribution of the predictions in the Pucci set, which is expressed as log of the unfolded/folded concentration ratio (Fig. 2, C and D). An error of 1 log unit means that the prediction is off by 10-fold, corresponding to RTln (10) in free energy, which is ∼1.37 kcal/mol. After addressing the outlier as discussed in the following section, ∼80% of all the predictions were accurate within 1 log unit, and none were greater than 3 log units.
Outlier analysis of the Pucci set
Understanding these apparent “failures” is paramount to the characterization of the domain of applicability. Without the knowledge of the “truth” in the form of mutant crystal structures, outlier analysis can be speculative. Fortunately, in this study, the outlier mutants all have crystal structures. This section is organized by the type of outliers.
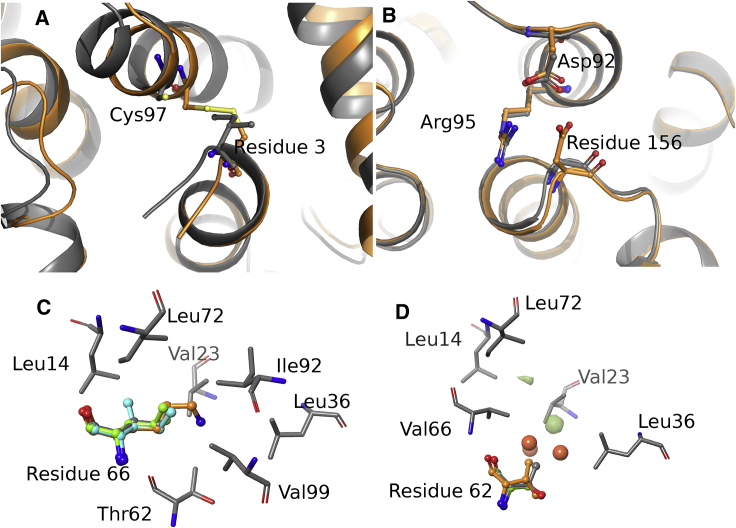
One of the outliers in 2LZM, Ile3Cys, is a stability neutral mutant that FEP+ predicted to be destabilizing by 2.8 kcal/mol. The crystal structure of the Ile3Cys (PDB: 172L) revealed a disulfide bond between Cys3 and Cys97, and the mutant structure is markedly different compared to the native fold (Fig. 3 A). Because the formation of the disulfide bond could not be modeled by FEP+, this mutation falls outside the domain of applicability, and the poor prediction is expected.
Figure 3.
(A) An overlay of the Ile3Cys mutant structure in orange with wild-type, PDB: 2LZM, in gray. (B) An overlay of the Gly156Asp mutant structure is shown in orange with wild-type, PDB: 2LZM, in gray. (C) An overlay of S. nuclease mutant structures in position 66 is shown with wild-type, PDB: 1EY0, in gray. The residues Val66Lys are in orange, Val66Leu in green, and Val66Ile in cyan. (D) An overlay of mutant structures in position 62 with wild-type, PDB: 1EY0, in gray is given. The residues Thr62Ser are in green and Thr62Val in orange. The red spheres are Thr62Ser WaterMap hydration sites with ΔΔG >5 kcal/mol, and the green surface is a cavity map representing areas lacking hydration in the Thr62Ser mutant structure. Cavity maps of similar size were also found in analyzed S. nuclease structures but are not shown for clarity reason. To see this figure in color, go online.
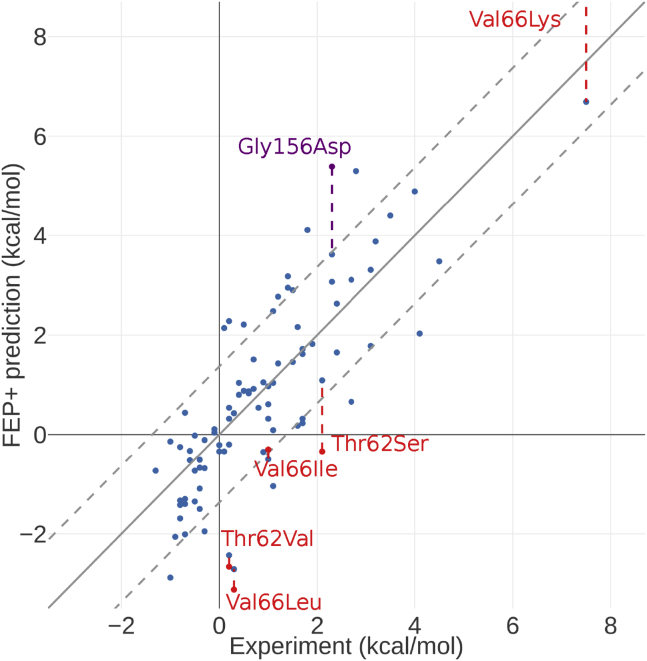
The second outlier in 2LZM, Gly156Asp, is an excellent example of uncertainty in protonation states of ionizable residues. The mutant was destabilizing by 2.3 kcal/mol, whereas FEP+ predicted it to be 5.39 kcal/mol. The environment surrounding residue 156 is highly polar and solvent exposed. It is located next to a salt bridge between Arg95 and Asp92. The crystal structure of mutant (PDB: 1L16) showed a surprising head-to-head interaction between Asp92 and Asp156, and this particular interaction is not an artifact due to crystal packing (Fig. 3 B). The pKa of Asp156 was predicted to be 5.3 by Propka (65) as implemented in Maestro, and the stability measurement was done at pH 6.5 (66), which suggests that a small fraction of the residue would be protonated in the folded state. A similar phenomenon has been observed for small molecule-protein and protein-protein binding and is a common source of prediction error. It is particularly acute when the pKa of the ionizable group and the experimental pH are close. A pKa correction has been developed requiring FEP+ simulations of both neutral and ionized forms and with the intrinsic pKa of the ionizable group and experimental pH as input (67). This approach provides a rigorous treatment of the ionization equilibria in unfolded and folded forms. By following the free energy changes as a function of time, the convergence of the simulations could be tracked (68). To achieve convergence, the FEP+ calculations of Gly156Asp in both forms were extended to 25 ns. The intrinsic pKa of aspartic acid is 3.9 (69), and after pKa correction, the prediction is 3.62 kcal/mol (Fig. 4). A closer examination of the FEP+ trajectories showed agreement with the mutant crystal structure. A protonated Asp156 fluctuates between interacting directly with Asp92 and stacking with Arg95, whereas the deprotonated form appears to interact with Arg95 only.
Figure 4.
Correlation between the FEP+ prediction and experiment for both the Pucci and proline sets. The dashed gray lines show the boundary of 1 log unit error. The red points are outliers in 1EY0, and the purple point is the outlier in 2LZM. The colored dashed line shows how the predictions changed after outlier analysis. The FEP+ prediction of Val66Lys mutant before outlier analysis was 16.28 kcal/mol and therefore off the chart. To see this figure in color, go online.
There are four outliers in the 1EY0 set: Thr62Ser, Thr62Val, Val66Lys, and Val66Leu. The largest outlier, Val66Lys, is a destabilizing mutation with experimental ΔΔG of 7.5 kcal/mol. FEP+ correctly identified the destabilizing effect but overestimated by more than 5 kcal/mol independent of the unfolded model we used. A close examination of the Val66Lys mutation crystal structure (PDB: 2SNM) revealed that the protein structure maintained native fold, and lysine ε-amine is entirely buried in the hydrophobic core without salt bridge or hydrogen-bonding partners (Fig. 3 C). This suggests that the amine should be neutral to avoid the desolvation penalty and the structure must be solved at high pH. Indeed, Stites et al. found that Val66Lys is highly unstable at pH 7; therefore, it was only possible to crystalize the structure at high pH (70). Because FEP+ by default mutated to a positively charged Lys, it is conceivable that the additional desolvation penalty for charged Lys in the predicted ΔΔG was a key reason for the overestimation. We carried out FEP+ calculations for Val66Lys in both neutral and ionized forms and applied the pKa correction scheme (67). The experimentally determined pKa of lysine residue at 26°C in a GGKGG peptide is 10.5 (69), and the pKa-corrected FEP+ prediction for Val66Lys was 6.69 kcal/mol, which is in agreement with the experimental value of 7.5 kcal/mol.
At the same position, the Val66Leu mutation was slightly destabilized by 0.3 kcal/mol in experiment, but the FEP+ predicted it to be stabilized by −3.12 kcal/mol. Although the FEP+ error for Val66Ile mutation was not as large as that for Val66Leu mutation, the stability of the mutant was also overpredicted by 1.3 kcal/mol (−0.3 kcal/mol by FEP+ and 1 kcal/mol by experiment). Furthermore, two mutations on the Thr62 position, Thr62Val and Thr62Ser, were also predicted to be more stable in FEP+ than in the experiments. Val66 and Thr62 are located on the same helix and belong to the same hydrophobic core formed by Leu36, Val23, Thr62, and Val66. At first glance, the crystal structures of the mutants and the wild-type are very similar. Upon closer inspection, a number of side chains have observed partial occupancies. These changes were observed in the mutant crystal structures, but not in the wild-type structure, suggesting increased flexibility. These include Leu36, Leu103, and Val23 in the Thr62Ser and Thr62Val crystal structures (PDB: 2EYH and 2EYJ) and Val23 and Ile72 in the Val66Ile mutant structure (PDB: 2F0G). We also analyzed the structure and energetics of the water molecules in the hydrophobic core for both the wild-type and mutant structures using WaterMap (59), which revealed a conserved cavity (71) in this hydrophobic core. Three additional high-energy hydration sites with estimated free energies of 5.23, 6.25, and 8.46 kcal/mol, respectively, were observed in the Thr62Ser mutant (Fig. 3 D), suggesting a destabilizing effect. Indeed, the experiment measured a destabilization of 2.1 kcal/mol for Thr62Ser mutation. We hypothesized that the water molecules and side-chain flexibilities were insufficiently sampled by the default 5 ns per λ window. To address this group of outliers, we extended the FEP+ calculation for all four mutants to 100 ns, adding the abovementioned residues with partial occupancies in the REST (56,57) region, “heating up” these side chains in the intermediate λ windows to improve their sampling. With the extended simulations, the prediction error for the Thr62Ser mutant reduced substantially from 2.44 to 1.01 kcal/mol, and the errors for Thr62Val and Val66Leu also reduced marginally, from 2.86 to 2.63 kcal/mol and from 3.42 to 3.01 kcal/mol, respectively (Fig. 4).
In a separate FEP+ study on protein-protein binding (49), it was observed that mutations can cause slight reorganization of the side chains and allow water molecules to penetrate the binding interface, but the simulations were difficult to converge; hence, the prediction accuracies were low. Running FEP+ for hundreds of nanoseconds would require days of simulations for one mutation and it is simply impractical for large numbers of mutations. However, for mutations in a tightly packed hydrophobic core that can trigger complex side-chain and water reorganization, longer simulations should be considered.
After addressing these outliers by pKa corrections or more extensive sampling, the MUE was significantly reduced (Table 1).
Table 1.
Summary of the System Studied in this Work
| Protein | PDB | ΔΔG Range (kcal/mol) | # Mutations | Monopeptide MUE (kcal/mol) | Tripeptide MUE (kcal/mol) |
|---|---|---|---|---|---|
| S. nuclease | 1EY0 | −1.0 to 2.1 | 27 | 2.01 | 0.78 (1.14) |
| Barnase | 1BNI | 0.5 to 4.5 | 13 | 0.93 | 1.11 |
| T4 lysozyme | 1L63 | −0.6 to 2.7 | 16 | 1.13 | 0.64 |
| T4 lysozyme | 2LZM | −0.8 to 2.3 | 13 | 2.51 | 0.91 (1.19) |
| Protein G | 1PGA | −0.8 to 3.5 | 10 | NA | 1.11 |
| Ribonuclease | 1RGG | −0.8 to 1.3 | 8 | NA | 0.58 |
The ΔΔG refers to the experimental free energy difference between the wild-type and the mutants. A negative value indicates stabilization and positive value destabilization. The numbers in parenthesis are before outlier analysis.
FEP+ predictions of additional proline mutations
To collect sufficient data on the accuracy of proline mutation predictions, another 18 mutations, including 15 involving prolines on protein G (1PGA) and ribonuclease (1RGG), were predicted using FEP+. The results were in agreement with what was observed for the Pucci set. The MUE for 1PGA was 1.11 kcal/mol and for 1RGG is 0.58 kcal/mol (Table 1). No outlier predictions were found for this set.
Overall performance
For both the Pucci and proline sets, the coefficient of determination (R2) for FEP+ prediction using tripeptide model was 0.66, the slope was 1.03, and the intercept −0.12 (Fig. 4). The MUE and RMSE were 0.86 and 1.11 kcal/mol. This shows that FEP+-predicted ΔΔG is directly in line with experimental measurements. The experimental data were derived using either thermal unfolding (2LZM, 1L63, 1RGG, and 1PGA) or chemical unfolding (1EY0 and 1BNI) assays. There is no clear difference in FEP+ prediction accuracy compared with each of the types of assay. In this study, the proline set and proline mutations in the Pucci set were predicted using a more recent release (2019-4 release) that has the full support of proline mutations for FEP calculation, whereas the other nonproline mutations were performed using an earlier version of the FEP+ product. To confirm that the two releases are consistent with each other for the nonproline mutations, we also rerun FEP+ using the 2019-4 release on nonoutlier mutations in 1L36 and 2LZM, including most of the charge-changing and proline mutations. The mean unassigned deviation and root mean square deviation between the predictions from the two releases were 0.20 and 0.29 kcal/mol, respectively. In addition, the MUE and RMSE of the predictions as compared with the experimental measurements from the two releases differed by no more than 0.02 kcal/mol.
Another way to look at the prediction performance is to calculate the sensitivity (true positive rate), specificity (true negative rate), and accuracy. True positive is defined as both the predicted and experimentally measured ΔΔG of a mutation being below 0 kcal/mol. The accuracy, sensitivity, and specificity across both sets were 0.86, 0.88, and 0.85.
The performance of FEP+ predictions can be compared with the following two null models. 1) There are 69.4% destabilizing mutations in both sets, and if we predict all mutations to be destabilizing, the accuracy would be 0.69, sensitivity 0, and specificity 1. Clearly such a null model is not useful at all because it cannot drive improvement in the prediction. 2) Randomly predict 69.4% of the mutations to be destabilizing and the remaining to be stabilizing. We repeated the experiment 1000 times and found the average accuracy to be 0.57, average sensitivity 0.31, and average specificity 0.69 with this null model. The performance of FEP+ is indisputably better than both the null models.
We also compared the accuracy of FEP+ predictions with a less computationally expensive MM-GBSA-based stability predictions, as implemented in the Residue scanning (62) panel that is part of BioLuminate. Unlike FEP+, the stability energies of Residue scanning in BioLuminate cannot be directly compared with the experimental free energies; hence, the MUE and RMSE are not at all meaningful. The R2 for Residue scanning is much lower at 0.33 for the entire data set. We classified a mutation as stabilizing if the predicted stability energy is below 0 or 3 kcal/mol. The choice of 3 kcal/mol is based on earlier observation of a shift in Residue scanning predicted protein-protein affinity energies versus the experimental energies (62). With 0 kcal/mol as cutoff, the sensitivity is 0.46, the specificity is 0.86, and the resulting accuracy is 0.74. Lifting the cutoff to 3 kcal/mol improved the sensitivity drastically to 0.81; specificity, however, was reduced to 0.71, and the accuracy remained the same.
Discussion
Significant improvement of FEP+ prediction accuracy
Previous works on large-scale protein stability predictions using FEP+ technology have shown relatively large error. For example, Steinbrecher et al. found that the RMSE for over 700 mutations were 2.27 or 2.07 kcal/mol if the subset of charge-changing mutations were removed (39). The MUE was slightly lower at 1.58 or 1.38 kcal/mol. Nevertheless, these errors are significantly larger than the small molecule-protein relative binding affinity prediction errors by FEP+, in which RMSE is estimated to be 1.1 kcal/mol (22) over a large data set. Ford et al. (40) also benchmarked FEP+ predictions. Unfortunately, their data set consists of mutations with ΔTm-values, and it was not possible to calculate average prediction errors (40). In both studies, FEP+ was compared with many other methods using static structures only, including MM-GBSA-based methods, and the authors found that FEP+ indeed performed better. In this work, our aim is not to provide yet another comparison between different methods, but rather to assess how we can improve FEP+’s prediction power compared with earlier work and whether it is possible to speed up the screening process without sacrificing the performance too much.
As a starting point, we analyzed possible reasons for why the reported FEP+ prediction performance for protein stability appeared to be much worse than that of small-molecule relative binding affinity to proteins. The main reason relates to the quality of the data set. The selected data set in Steinbrecher et al. (39) was very large, and it was impractical to ensure that all experimental conditions were consistent. For example, the pH at which the measurements were performed may have an especially large impact on the prediction results. At extreme pH conditions, the protonation states of the ionizable residues are likely to be very different compared to neutral pH. It was also unclear whether the mutants would adapt different conformations compared to the wild-type. For example, the mutant structure could be partially or fully unfolded. Because the simulations lasted only 5 ns per λ window, the sampling is insufficient to capture large structural transitions, which may lead to incorrect predictions. For small-molecule FEP+ studies, this would correspond to molecules adopting different binding modes in the same chemical series, a known cause of large prediction errors (72). Pucci et al. collected a set of protein stability mutations from the ProTherm database (64) in which the experimental pH was noted (19), enabling us to focus only on the stability measurements done at pH 7 ± 1. There are additional data points that were measured at pH between 6 and 2. At those pH ranges, the protonation state of the acidic residues may be dynamic, and the current version of FEP+ is not able to handle them properly. More valuably, the authors have identified all crystal structures of the protein mutants, which allows us to confirm that the wild-type and mutant structures are conserved.
A second issue that Steinbrecher et al. (39) identified is the large errors associated with charge-changing mutations. A subset of these mutations may very well have been measured at extreme pH, whereas they were simulated at pH 7. Changing charges is in itself a challenge associated with long-range electrostatic effects, polarization, salt concentration, etc., and it was largely addressed and applied on small molecule-protein and protein-protein relative affinity predictions (48,49). The nonproline mutations in our data set have 20% charge-changing mutations, but our FEP+ predictions for these mutations and with the monopeptide unfolded model achieved an MUE of 1.09 kcal/mol, showing clear improvement compared to the 1.58 kcal/mol reported by Steinbrecher et al. (39). Although our data set is much smaller, the improvement in MUE can presumably be attributed to both improved treatment of charge perturbations and a clean data set. This suggests that the true performance of FEP+ is likely better than what has been reported before.
A third possible source of prediction error is the representation of the unfolded state, for which we do not have experimental structures. The extreme opposite of the folded state would be a random-coil model in which the backbone torsion angles of each amino acid are independent of its neighbors. In effect, this means that the unfolded model has no native structure characteristics (73,74). If this is true, then a single capped amino acid should suffice as an unfolded model, which was used in earlier studies (26,32,39,40). The benefit of such a model is a significant reduction of the simulation system size and sampling needed for convergence. However, numerous studies showed that the unfolded form of a protein may still have residual secondary structure elements and local interactions (73, 74, 75, 76, 77). An alternative model is an extended and capped tripeptide with an Ala-X-Ala or Gly-X-Gly sequence in which X is the mutation site (27,31,33,34), but such a model would not account for neighboring side-chain interactions. We rationalized that using the native sequence around the mutation site in its native conformation should be ideal for preserving the native interactions that may occur in an unfolded state. Using an extended starting conformation may require much longer simulations to sample native contacts. Furthermore, a turn in the α-helix is ∼3.4 amino acids, and a β-turn is four residues; hence, it seemed prudent to include capped penta- and heptapeptides as well.
The FEP+ predictions clearly showed that there is significant improvement by adding just one flanking residue on each side to the mutation site. The MUE dropped from 1.68 to 0.85 kcal/mol, and RMSE reduced from 2.79 to 1.11 kcal/mol. If we remove the five mutations involving proline from the Pucci set, there is still a small but significant improvement. It appears most of the improvements are from proline mutations. We sought out an additional 15 proline mutations from two different proteins in the literature, and the prediction accuracy agreed very well. Adding additional flanking residues to the tripeptide model reduced both MUE and RMSE further but less pronounced. Across both the Pucci and proline sets, the RMSE and MUE are 1.11 and 0.86 kcal/mol, respectively. The linear correlation between the FEP+ predictions and the experimental energies has a slope of 1.03 and intercept of −0.12 kcal/mol. Overall, the results clearly show that FEP+-predicted protein stability energies can be meaningfully compared with the experiments.
With a carefully curated data set, implementation of charge-changing mutations, cyclization for proline mutation, and capped peptides with three to seven residues as unfolded model, the performance of FEP+ for protein stability prediction is on par with relative binding affinity predictions of small molecule-protein complexes. Small molecule-protein relative binding affinity FEP+ predictions have been extensively used in drug discovery programs, with tens of thousands of predictions made at Schrödinger alone. It has made remarkable impact on project progression (22,23). Our results imply that FEP+ could be accurate enough to drive the design of more stable (in at least a single chain) proteins.
Comparison to other protein thermostability prediction methods
In addition to the MM-GBSA-based Residue scanning protocol, there are a plethora of different protein thermostability prediction methods. Pucci et al. reported the performance of 15 predictors (19), and we chose to compare them with commonly used ones such as FoldX (78), Rosetta (79), MUPRO (80), and PoPMuSiCsym (81) using the raw data kindly provided by Pucci (Table 2; (78)). FEP+ compares favorably to all methods except MUPRO, which is a machine-learning model using only sequence information and all mutations as part of its training set. Pucci et al. observed that MUPRO suffered from training set biases (19), and indeed, our analysis also showed that the RMSE increased drastically for the reverse mutations (Table S4). Because of its implementation of replica exchange technology, FEP+ does not have any directionality. Furthermore, FEP+ does not rely on any fitting to training sets, and therefore, it does not suffer from similar biases. FEP+ also outperforms the other predictors as a classification tool, assuming that the appropriate cutoff for stabilizing mutations is 0 kcal/mol. Interestingly, MUPRO has almost perfect specificity but poor sensitivity for forward mutation, which confirms that it is strongly biased by the abundance of destabilizing mutations in the training sets.
Table 2.
Performance Comparison with Commonly Used Protein Thermostability Prediction Methods
| FEP+ | MM-GBSA | PoPMuSiC_sym | MUPRO | FoldX | Rosetta | |
|---|---|---|---|---|---|---|
| MUE | 0.85 | NA | 1.08 | 0.70 | 1.20 | 1.65 |
| RMSE | 1.11 | NA | 1.46 | 1.21 | 1.79 | 2.19 |
| R2 | 0.68 | 0.39 | 0.27 | 0.36 | 0.22 | 0.39 |
| Accuracy | 0.85 | 0.74 | 0.65 | 0.77 | 0.70 | 0.64 |
| Sensitivity | 0.89 | 0.46 | 0.68 | 0.32 | 0.68 | 0.53 |
| Specificity | 0.84 | 0.86 | 0.64 | 0.94 | 0.70 | 0.68 |
Using the Crooks fluctuation theorem, Gapsys et al. also reported excellent MUE of 0.89 kcal/mol for 143 unique mutations in barnase and S. nuclease (33), which is very similar to our study. The RMSE and R2 are slightly worse at 1.22 kcal/mol and 0.47. Interestingly, none of the outliers at positions Thr62 and Val66 in S. nuclease were in their data set, and none of the mutations involved proline. The same study also predicted the thermostability of five mutants of a G-protein coupled receptor with ΔTm data, and the correlation was outstanding, R2 = 0.74 (33). Curiously, the use of any membrane models was not mentioned.
Efficient screening cascade by combining Residue scanning and FEP+
The FEP+ simulation for our data set typically takes 6 h on a single Nvidia Pascal architecture graphics card. Because the number of possible mutations on a protein can be very large, FEP+ simulation as the sole filter may take too long time unless a very large cluster is available. The idea of using a faster filter before FEP+ simulation is not new (40). Residue scanning in BioLuminate uses static structure and only predicts the side-chain rotamer of the mutant residue, followed by a short minimization by default in which the solvent effect is treated using a modified MM-GBSA solvent model (63). It has been established that similar fast methods perform worse compared to FEP+ (40), which agrees with our observation. Nevertheless, Residue scanning calculation for each mutation takes minutes compared with hours for FEP+. We observed that Residue scanning tends to introduce false positives, which can be filtered out by FEP+ before experimental testing. Here, we explored how Residue scanning can be used as a filter before FEP+ in a screening cascade to drastically reduce the simulation time without sacrificing performance.
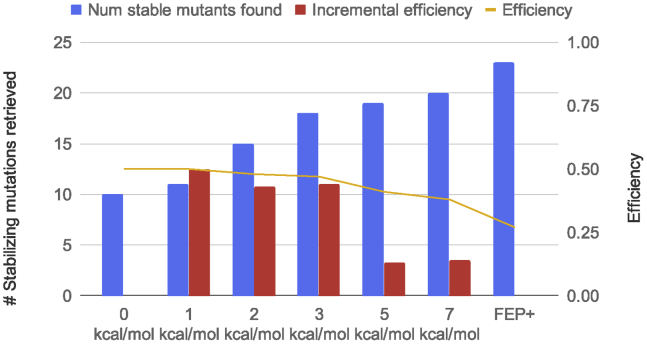
Unfortunately, unlike FEP+ prediction, the MM-GBSA-based energies are not comparable with the experimental energies, and hence, the exact cutoff for the filter is an unknown factor. In retrospect, one can compare the distribution of the experimental energy with that of the predicted energy to derive the cutoff, but such knowledge is not possible a priori. To identify the best energy cutoff for the Residue scanning, we retained mutations with predicted stability energies below 0, 1, 2, 3, 5 and 7 kcal/mol, and we plotted the number of true positives that would come out of the entire cascade (Fig. 5). We also examined how efficient such a workflow would be compared to using FEP+ simulations alone. Because the time needed for a Residue scanning calculation is negligible compared with that for FEP+, we define efficiency as a number of retrieved actives per FEP+ simulations. The efficiency of FEP+ alone was therefore 23 actives divided by 86 simulations, i.e., 0.27. Interestingly, the efficiency of the screening cascade using different MM-GBSA energy cutoffs was flat at ∼0.5 up to 3 kcal/mol before it declined to ∼0.4. The efficiency of 0.5 suggests that for every two FEP+ simulations, we found one active for this data set. We know that by using higher cutoff value, more false positives were introduced, and hence, more nonproductive FEP+ simulations were carried out. We then defined incremental efficiency, which is a number of additional actives found per additional FEP+ simulations. The incremental efficiency tells us the extra effort needed to find new actives if we increase the cutoff. As an illustrative example, when the Residue scanning cutoff is raised from 3 to 5 kcal/mol, to find one additional stabilizing mutant, we have to screen eight additional mutations using FEP+. Hence, the incremental efficiency would be 0.125. The incremental efficiency was between 0.4 and 0.5 up to 3 kcal/mol cutoff before declining down to 0.125 at 5 kcal/mol cutoff. Therefore, 3 kcal/mol seems to be a good compromise between speed and performance.
Figure 5.
The performance of a computational screening cascade in which Residue scanning is used as a first filter, followed by FEP+ as a second filter. The blue bar shows the number of stabilizing mutations retrieved by the cascade using different cutoff values for Residue scanning compared to using FEP+ only. The yellow line (right y axis) shows the efficiency, which is defined as the ratio of number of true positives retrieved per FEP+ simulation. The red bar (right axis) shows the incremental efficiency, which is defined as the number of additional true positives found per additional FEP+ simulation when a higher cutoff value is used. To see this figure in color, go online.
In the envisioned screening cascade, we would filter the initial 86 mutations through Residue scanning with 3 kcal/mol as a cutoff. Of these 86 mutations, 38 would be evaluated by FEP+, and only 22 would be passed on to experimental measurement, resulting in 18 true stabilizing mutations. Compared with using FEP+ alone, the cascade will save 55% of FEP+ computational time, and compared with using Residue scanning at 3 kcal/mol cutoff alone, it will save 39% of the experiments. In the scenario of thousands of mutation designs to be evaluated, this screening cascade can significantly reduce both computational cost as well as experimental cost. Based on the availability of the GPU cluster and the project requirement, the cutoff can be easily adjusted. In an era with easy accessibility of GPU resources on the cloud, we think efficient screening of stabilizing mutations using free energy calculations is realistically possible with sufficient accuracy to impact the projects.
Conclusion
In this study, we introduced improvements in FEP+ prediction of protein stability, including more accurate modeling of the unfolded states and methods for dealing with proline mutations and charge-changing mutations. We found that modeling the unfolded state as polypeptides with native sequence and conformation improved the accuracy for stability prediction. The outlier analysis is particularly valuable because it highlighted the potential pitfalls when running FEP+ prediction on protein stability. This shows that blindly running screening on all residues in a protein to all 19 amino acids is probably ill-advised. Careful thoughts are desired to avoid obvious cases outside the applicability domain. Charge-changing mutations close to charge clusters need to be treated with care regarding the protonation state, and possibly pKa corrections are needed. Likewise, mutations in the tight hydrophobic core may demand extended simulations to permit sampling of water molecules and side-chain reorganization. Encouragingly, the predictions for these challenging cases often correctly classify or rank the mutations. Within the applicability domain, FEP+ is clearly able to accurately predict the protein stability change upon mutations. The MUE was found to be 0.86 kcal/mol, and RMSE is slightly higher at 1.11 kcal/mol, which is comparable with FEP+ prediction accuracy for small molecule-protein relative binding affinity. FEP+’s ability to identify stabilizing mutations is substantially higher than randomly guess with prior knowledge of fraction of stabilizing mutations.
With the simulation time of a few hours on a GPU and access to large GPU farms on the cloud, it is possible to screen hundreds or thousands of mutations for stabilizing mutations within a few days. Combined with a fast Residue scanning method in a screening cascade, both computational and experimental resources can be reduced. FEP+ technology for small molecule-protein binding affinity prediction is already being applied at a large scale in the pharmaceutical industry and has been found to be sufficient to drive discovery projects. The results in this work demonstrate a similarly high level of accuracy in the prediction of change in protein thermostability upon mutations and position free energy calculations to play a guiding role in protein engineering projects.
Author Contributions
J.D., D.L., and L.W. designed the research, analyzed the data, and wrote the manuscript. D.L. and L.W. implemented the software algorithms. J.D. and D.L. performed the experiments.
Acknowledgments
We thank Dr. Fabrizio Pucci for kindly sharing the raw data.
J.D. is an employee of Schrödinger GmbH. D.L. and L.W. are employees of Schrödinger Inc.
Editor: Rohit Pappu.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.05.020.
Supporting Material
References
- 1.Dill K.A., MacCallum J.L. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 2.Mannige R.V. Dynamic new world: refining our view of protein structure, function and evolution. Proteomes. 2014;2:128–153. doi: 10.3390/proteomes2010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan J., Nilsson L., Lambert B. Structural and functional analysis of mutations at the human hypoxanthine phosphoribosyl transferase (HPRT1) locus. Hum. Mutat. 2004;23:599–611. doi: 10.1002/humu.20047. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Moult J. SNPs, protein structure, and disease. Hum. Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 5.Ferrer-Costa C., Orozco M., de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 2002;315:771–786. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 6.Kucukkal T.G., Petukh M., Alexov E. Structural and physico-chemical effects of disease and non-disease nsSNPs on proteins. Curr. Opin. Struct. Biol. 2015;32:18–24. doi: 10.1016/j.sbi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petukh M., Kucukkal T.G., Alexov E. On human disease-causing amino acid variants: statistical study of sequence and structural patterns. Hum. Mutat. 2015;36:524–534. doi: 10.1002/humu.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefl S., Nishi H., Alexov E. Molecular mechanisms of disease-causing missense mutations. J. Mol. Biol. 2013;425:3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold F.H. Enzyme engineering reaches the boiling point. Proc. Natl. Acad. Sci. USA. 1998;95:2035–2036. doi: 10.1073/pnas.95.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pantazes R.J., Grisewood M.J., Maranas C.D. Recent advances in computational protein design. Curr. Opin. Struct. Biol. 2011;21:467–472. doi: 10.1016/j.sbi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Rigoldi F., Donini S., Gautieri A. Review: engineering of thermostable enzymes for industrial applications. APL Bioeng. 2018;2:011501. doi: 10.1063/1.4997367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wörn A., Plückthun A. Stability engineering of antibody single-chain Fv fragments. J. Mol. Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 13.Honegger A. Engineering antibodies for stability and efficient folding. Handb. Exp. Pharmacol. 2008;(181):47–68. doi: 10.1007/978-3-540-73259-4_3. [DOI] [PubMed] [Google Scholar]
- 14.Jung S., Plückthun A. Improving in vivo folding and stability of a single-chain Fv antibody fragment by loop grafting. Protein Eng. 1997;10:959–966. doi: 10.1093/protein/10.8.959. [DOI] [PubMed] [Google Scholar]
- 15.Tiller K.E., Tessier P.M. Advances in antibody design. Annu. Rev. Biomed. Eng. 2015;17:191–216. doi: 10.1146/annurev-bioeng-071114-040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McConnell A.D., Zhang X., Bowers P.M. A general approach to antibody thermostabilization. MAbs. 2014;6:1274–1282. doi: 10.4161/mabs.29680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaidehi N., Grisshammer R., Tate C.G. How can mutations thermostabilize G-protein-coupled receptors? Trends Pharmacol. Sci. 2016;37:37–46. doi: 10.1016/j.tips.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan S., Vihinen M. Performance of protein stability predictors. Hum. Mutat. 2010;31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 19.Pucci F., Bernaerts K.V., Rooman M. Quantification of biases in predictions of protein stability changes upon mutations. Bioinformatics. 2018;34:3659–3665. doi: 10.1093/bioinformatics/bty348. [DOI] [PubMed] [Google Scholar]
- 20.Zwanzig R.W. High-temperature equation of state by a perturbation method. I. Nonpolar gases. J. Chem. Phys. 1954;22:1420–1426. [Google Scholar]
- 21.Kuhn B., Tichý M., Hert J. Prospective evaluation of free energy calculations for the prioritization of cathepsin L inhibitors. J. Med. Chem. 2017;60:2485–2497. doi: 10.1021/acs.jmedchem.6b01881. [DOI] [PubMed] [Google Scholar]
- 22.Abel R., Wang L., Friesner R.A. A critical review of validation, blind testing, and real- world use of alchemical protein-ligand binding free energy calculations. Curr. Top. Med. Chem. 2017;17:2577–2585. doi: 10.2174/1568026617666170414142131. [DOI] [PubMed] [Google Scholar]
- 23.Abel R., Wang L., Friesner R.A. Advancing drug discovery through enhanced free energy calculations. Acc. Chem. Res. 2017;50:1625–1632. doi: 10.1021/acs.accounts.7b00083. [DOI] [PubMed] [Google Scholar]
- 24.Wagner V., Jantz L., Christ C.D. Computational macrocyclization: from de novo macrocycle generation to binding affinity estimation. ChemMedChem. 2017;12:1866–1872. doi: 10.1002/cmdc.201700478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkwood J.G. Statistical mechanics of fluid mixtures. J. Chem. Phys. 1935;3:300–313. [Google Scholar]
- 26.Bash P.A., Singh U.C., Kollman P.A. Free energy calculations by computer simulation. Science. 1987;236:564–568. doi: 10.1126/science.3576184. [DOI] [PubMed] [Google Scholar]
- 27.Dang L.X., Merz K.M., Kollman P.A. Free energy calculations on protein stability: Thr-157. fwdarw. Val-157 mutation of T4 lysozyme. J. Am. Chem. Soc. 1989;111:8505–8508. [Google Scholar]
- 28.Prevost M., Wodak S.J., Karplus M. Contribution of the hydrophobic effect to protein stability: analysis based on simulations of the Ile-96----Ala mutation in barnase. Proc. Natl. Acad. Sci. USA. 1991;88:10880–10884. doi: 10.1073/pnas.88.23.10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tidor B., Karplus M. Simulation analysis of the stability mutant R96H of T4 lysozyme. Biochemistry. 1991;30:3217–3228. doi: 10.1021/bi00227a009. [DOI] [PubMed] [Google Scholar]
- 30.Yamaotsu N., Moriguchi I., Hirono S. Molecular dynamics study of the stability of staphylococcal nuclease mutants: component analysis of the free energy difference of denaturation. Biochim. Biophys. Acta. 1993;1163:81–88. doi: 10.1016/0167-4838(93)90282-v. [DOI] [PubMed] [Google Scholar]
- 31.Sun Y.C., Veenstra D.L., Kollman P.A. Free energy calculations of the mutation of Ile96-->Ala in barnase: contributions to the difference in stability. Protein Eng. 1996;9:273–281. doi: 10.1093/protein/9.3.273. [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Veenstra D.L., Kollman P.A. Can one predict protein stability? An attempt to do so for residue 133 of T4 lysozyme using a combination of free energy derivatives, PROFEC, and free energy perturbation methods. Proteins. 1998;32:438–458. [PubMed] [Google Scholar]
- 33.Gapsys V., Michielssens S., de Groot B.L. Accurate and rigorous prediction of the changes in protein free energies in a large-scale mutation scan. Angew. Chem. Int. Ed. Engl. 2016;55:7364–7368. doi: 10.1002/anie.201510054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeliger D., de Groot B.L. Protein thermostability calculations using alchemical free energy simulations. Biophys. J. 2010;98:2309–2316. doi: 10.1016/j.bpj.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mooney S.D., Huang C.C., Klein T.E. Computed free energy differences between point mutations in a collagen-like peptide. Biopolymers. 2001;58:347–353. doi: 10.1002/1097-0282(200103)58:3<347::AID-BIP1011>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 36.Jespers W., Isaksen G.V., Gutiérrez-de-Terán H. QresFEP: an automated protocol for free energy calculations of protein mutations in Q. J. Chem. Theory Comput. 2019;15:5461–5473. doi: 10.1021/acs.jctc.9b00538. [DOI] [PubMed] [Google Scholar]
- 37.Crooks G.E. Nonequilibrium measurements of free energy differences for microscopically reversible markovian systems. J. Stat. Phys. 1998;90:1481–1487. [Google Scholar]
- 38.Crooks G.E. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1999;60:2721–2726. doi: 10.1103/physreve.60.2721. [DOI] [PubMed] [Google Scholar]
- 39.Steinbrecher T., Zhu C., Sherman W. Predicting the effect of amino acid single-point mutations on protein stability-large-scale validation of MD-based relative free energy calculations. J. Mol. Biol. 2017;429:948–963. doi: 10.1016/j.jmb.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Ford M.C., Babaoglu K. Examining the feasibility of using free energy perturbation (FEP+) in predicting protein stability. J. Chem. Inf. Model. 2017;57:1276–1285. doi: 10.1021/acs.jcim.7b00002. [DOI] [PubMed] [Google Scholar]
- 41.Liu S., Wang L., Mobley D.L. Is ring breaking feasible in relative binding free energy calculations? J. Chem. Inf. Model. 2015;55:727–735. doi: 10.1021/acs.jcim.5b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor T.J., Vaisman I.I. Discrimination of thermophilic and mesophilic proteins. BMC Struct. Biol. 2010;10(Suppl 1):S5. doi: 10.1186/1472-6807-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trevino S.R., Schaefer S., Pace C.N. Increasing protein conformational stability by optimizing beta-turn sequence. J. Mol. Biol. 2007;373:211–218. doi: 10.1016/j.jmb.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe K., Hata Y., Suzuki Y. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 A resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 1997;269:142–153. doi: 10.1006/jmbi.1997.1018. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe K., Kitamura K., Suzuki Y. Analysis of the critical sites for protein thermostabilization by proline substitution in oligo-1,6-glucosidase from Bacillus coagulans ATCC 7050 and the evolutionary consideration of proline residues. Appl. Environ. Microbiol. 1996;62:2066–2073. doi: 10.1128/aem.62.6.2066-2073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe K., Suzuki Y. Protein thermostabilization by proline substitutions. J. Mol. Catal., B Enzym. 1998;4:167–180. [Google Scholar]
- 47.Zhu G.P., Xu C., Wang Y.Z. Increasing the thermostability of D-xylose isomerase by introduction of a proline into the turn of a random coil. Protein Eng. 1999;12:635–638. doi: 10.1093/protein/12.8.635. [DOI] [PubMed] [Google Scholar]
- 48.Chen W., Deng Y., Wang L. Accurate calculation of relative binding free energies between ligands with different net charges. J. Chem. Theory Comput. 2018;14:6346–6358. doi: 10.1021/acs.jctc.8b00825. [DOI] [PubMed] [Google Scholar]
- 49.Clark A.J., Negron C., Friesner R.A. Relative binding affinity prediction of charge-changing sequence mutations with FEP in protein-protein interfaces. J. Mol. Biol. 2019;431:1481–1493. doi: 10.1016/j.jmb.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., Deng Y., Abel R. Accurate modeling of scaffold hopping transformations in drug discovery. J. Chem. Theory Comput. 2017;13:42–54. doi: 10.1021/acs.jctc.6b00991. [DOI] [PubMed] [Google Scholar]
- 51.Keränen H., Pérez-Benito L., Tresadern G. Acylguanidine beta secretase 1 inhibitors: a combined experimental and free energy perturbation study. J. Chem. Theory Comput. 2017;13:1439–1453. doi: 10.1021/acs.jctc.6b01141. [DOI] [PubMed] [Google Scholar]
- 52.Yu H.S., Deng Y., Wang L. Accurate and reliable prediction of the binding affinities of macrocycles to their protein targets. J. Chem. Theory Comput. 2017;13:6290–6300. doi: 10.1021/acs.jctc.7b00885. [DOI] [PubMed] [Google Scholar]
- 53.Choi E.J., Mayo S.L. Generation and analysis of proline mutants in protein G. Protein Eng. Des. Sel. 2006;19:285–289. doi: 10.1093/protein/gzl007. [DOI] [PubMed] [Google Scholar]
- 54.Harder E., Damm W., Friesner R.A. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 55.Roos K., Wu C., Harder E.D. OPLS3e: extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 2019;15:1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- 56.Wang L., Berne B.J., Friesner R.A. On achieving high accuracy and reliability in the calculation of relative protein-ligand binding affinities. Proc. Natl. Acad. Sci. USA. 2012;109:1937–1942. doi: 10.1073/pnas.1114017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Deng Y., Abel R. Modeling local structural rearrangements using FEP/REST: application to relative binding affinity predictions of CDK2 inhibitors. J. Chem. Theory Comput. 2013;9:1282–1293. doi: 10.1021/ct300911a. [DOI] [PubMed] [Google Scholar]
- 58.Bowers K.J., Chow E., Shaw D.E. ACM; 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), B. Horner-Miller, conference chair; pp. 84–96. [Google Scholar]
- 59.Abel R., Young T., Friesner R.A. Role of the active-site solvent in the thermodynamics of factor Xa ligand binding. J. Am. Chem. Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cappel D., Sherman W., Beuming T. Calculating water thermodynamics in the binding site of proteins - applications of waterMap to drug discovery. Curr. Top. Med. Chem. 2017;17:2586–2598. doi: 10.2174/1568026617666170414141452. [DOI] [PubMed] [Google Scholar]
- 61.Young T., Abel R., Friesner R.A. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc. Natl. Acad. Sci. USA. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beard H., Cholleti A., Loving K.A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS One. 2013;8:e82849. doi: 10.1371/journal.pone.0082849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J., Abel R., Friesner R.A. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bava K.A., Gromiha M.M., Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32:D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olsson M.H., Søndergaard C.R., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 66.Gray T.M., Matthews B.W. Structural analysis of the temperature-sensitive mutant of bacteriophage T4 lysozyme, glycine 156----aspartic acid. J. Biol. Chem. 1987;262:16858–16864. doi: 10.2210/pdb1l16/pdb. [DOI] [PubMed] [Google Scholar]
- 67.de Oliveira C., Yu H.S., Wang L. Rigorous free energy perturbation approach to estimating relative binding affinities between ligands with multiple protonation and tautomeric states. J. Chem. Theory Comput. 2019;15:424–435. doi: 10.1021/acs.jctc.8b00826. [DOI] [PubMed] [Google Scholar]
- 68.Bennett C.H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976;22:245–268. [Google Scholar]
- 69.Thurlkill R.L., Grimsley G.R., Pace C.N. pK values of the ionizable groups of proteins. Protein Sci. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stites W.E., Gittis A.G., Shortle D. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J. Mol. Biol. 1991;221:7–14. doi: 10.1016/0022-2836(91)80195-z. [DOI] [PubMed] [Google Scholar]
- 71.Wang L., Berne B.J., Friesner R.A. Ligand binding to protein-binding pockets with wet and dry regions. Proc. Natl. Acad. Sci. USA. 2011;108:1326–1330. doi: 10.1073/pnas.1016793108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L., Wu Y., Abel R. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J. Am. Chem. Soc. 2015;137:2695–2703. doi: 10.1021/ja512751q. [DOI] [PubMed] [Google Scholar]
- 73.Smith L.J., Fiebig K.M., Dobson C.M. The concept of a random coil. Residual structure in peptides and denatured proteins. Fold. Des. 1996;1:R95–R106. doi: 10.1016/S1359-0278(96)00046-6. [DOI] [PubMed] [Google Scholar]
- 74.Baldwin R.L., Zimm B.H. Are denatured proteins ever random coils? Proc. Natl. Acad. Sci. USA. 2000;97:12391–12392. doi: 10.1073/pnas.97.23.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basharov M.A. Residual ordered structure in denatured proteins and the problem of protein folding. Indian J. Biochem. Biophys. 2012;49:7–17. [PubMed] [Google Scholar]
- 76.Bowler B.E. Residual structure in unfolded proteins. Curr. Opin. Struct. Biol. 2012;22:4–13. doi: 10.1016/j.sbi.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camilloni C., De Simone A., Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 78.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 79.Kellogg E.H., Leaver-Fay A., Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 81.Pucci F., Bernaerts K., Rooman M. Symmetry principles in optimization problems: an application to protein stability prediction. IFAC-PapersOnLine. 2015;48:458–463. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.