Abstract
Organic fluorophores, such as Cy3 and Cy5, have been widely used as chemical labels to probe the structure and dynamics of membrane proteins. Although a number of previous studies have reported on the possibility of some of the water-soluble fluorophores to interact with lipid bilayers, detailed fluorophore-lipid interactions and, more importantly, the potential effect of such interactions on the natural dynamics of the labeled membrane proteins have not been well studied. We have performed a large set of all-atom molecular dynamics simulations employing the highly mobile membrane mimetic model to describe spontaneous partitioning of the fluorophores into lipid bilayers with different lipid compositions. Spontaneous membrane partitioning of Cy3 and Cy5 fluorophores captured in these simulations proceeds in two steps. Electrostatic interaction between the fluorophores and the lipid headgroups facilitates the initial, fast membrane association of the fluorophores, followed by slow insertion of hydrophobic moieties into the lipid bilayer core. After the conversion of the resulting membrane-bound systems to full-membrane representations, biased-exchange umbrella sampling simulations are performed for free energy calculations, revealing a higher energy barrier for partitioning into negatively charged (phosphatidylserine or phosphatidylcholine) membranes than purely zwitterionic (phosphatidylcholine or phosphatidylethanolamine) ones. Furthermore, the potential effect of fluorophore-lipid interactions on membrane proteins has been examined by covalently linking Cy5 to single- and multipass transmembrane helical proteins. Equilibrium simulations show strong position-dependent effects of Cy5-tagging on the structure and natural dynamics of membrane proteins. Interactions between the tagged protein and Cy5 were also observed. Our results suggest that fluorophore-lipid interactions can affect the structure and dynamics of membrane proteins to various extents, especially in systems with higher structural flexibility.
Significance
Fluorophores have been extensively used as effective probes to study the structure and dynamics of membrane proteins. However, their potential undesired effects on the native structure and dynamics of the labeled protein, which can arise because of their strong association with lipids, have not been studied. Here, we study at the molecular level membrane interactions of two prototypical fluorophores, Cy3 and Cy5, using advanced simulation and sampling techniques. We present examples of potential effects of membrane-fluorophore interactions in three membrane-protein systems of different complexity. Our results suggest that the structure and dynamics of the labeled proteins can be indeed affected by the fluorophores in a position-dependent manner, thus providing insight for better experimental design of fluorescently labeled membrane proteins.
Introduction
Organic fluorophores have been widely used as fluorescent probes in biological systems to study their structure and dynamics (1), including a large number of membrane proteins. Because some of these largely water-soluble fluorophores have been reported to interact with lipid membranes (2), it can be expected that using them in membrane systems can potentially cause artifacts or false observations in experiments. Systematic studies on a wide variety of the commonly used fluorophores found that most of them interact with membrane and that both hydrophobic and electrostatic lipid-fluorophore interactions contribute to the membrane partitioning of fluorophore molecules (3,4). Although different experiments confirmed that the major determinant for membrane binding is the overall hydrophobicity of the fluorophore molecule, the effects of electrostatics can vary across different fluorophore-lipid pairs (2, 3, 4).
Despite the demonstrated fluorophore-membrane association, the details of fluorophore-lipid interactions and the potential ramifications of such interactions on the natural dynamics of fluorophore-labeled proteins remain elusive. A recent experimental study has reported a reduction in the mobility of a fluorescently labeled soluble protein because of the nonspecific binding of fluorophores to the plasma membrane (2). However, in the case of integral membrane proteins, there is no available experimental information on the fluorophore-induced artifacts. Given the great interest in using fluorophores in fluorescent microscopy, some of the commonly used fluorophores have been parameterized and simulated in the absence of proteins (5). Further characterization of the molecular details of fluorophore-lipid interactions and the effect of such interactions on membrane-protein dynamics using computational approaches could help develop a better understanding of potential artifacts and designing better experiments when using these fluorophores to label membrane proteins.
Here, we study membrane- and lipid-fluorophore interactions using all-atom molecular dynamics (MD) simulations on two amphiphilic, negatively charged fluorophores, Cy3 and Cy5. To allow fast sampling of initial membrane binding and partitioning of the fluorophores, the highly mobile membrane mimetic (HMMM) model (6) was employed. The HMMM model replaces the lipid molecules by short-tailed lipids with the same headgroup and small organic solvent within the membrane interior and, hence, significantly accelerates lateral lipid diffusion (6). This method has proven successful in capturing spontaneous membrane association and insertion in several different peripheral membrane-protein systems (7, 8, 9). The membrane-bound fluorophores captured with the HMMM model were then converted to full-membrane representations. After that, a detailed energetic description of the fluorophores’ membrane partitioning was obtained from bias-exchange umbrella sampling (BEUS) simulations (10,11). The results demonstrate a strong tendency of membrane partitioning of Cy3 and Cy5, governed by both electrostatic and hydrophobic interactions between the fluorophores and the lipid bilayer.
In the second part of the study, the effect of the fluorophore-lipid interactions on membrane proteins was probed by using transmembrane (TM) protein models with various complexity, ranging from a single-pass TM helix to multipass channel proteins, namely 1) glycophorin A (GpA) 2), the β-subunit of a voltage-gated sodium channel (NaVβ), and 3) mechanosensitive channel of large conductance (MscL). Our results indicate that there are nontrivial effects on the structure and dynamics of fluorophore-tagged membrane proteins, with the extent of the effect largely depending on the position of the fluorophore with respect to the lipid bilayer.
Materials and Methods
Spontaneous membrane-binding simulations with HMMM
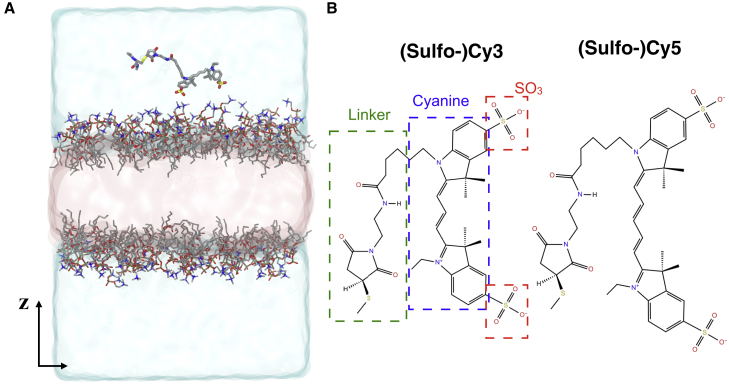
To allow fast sampling for the most likely membrane-binding poses of the fluorophores, the HMMM (6) membrane, in which short-tailed lipids are employed for rapid lateral diffusion, was used for all membrane-binding simulations. The simulations were all carried out with NAMD 2.12 (12), using CHARMM36m protein (13) and CHARMM36 lipid (14) parameters, and the CGenFF parameters for DCLE (15), which is used as an organic solvent for the HMMM membrane. HMMM membranes with pure phosphatidylcholine (PC), pure phosphatidylethanolamine (PE), and 1:1 mixture of phosphatidylserine (PS) and PC were generated using CHARMM-GUI HMMM Builder (16). Each bilayer was then minimized and equilibrated using the default six-step protocol generated by CHARMM-GUI. A 5-ns NPAT (with constant membrane area) equilibrium simulation was carried out for each lipid bilayer following the six-step protocol, in which the C21 and C31 carbonyl carbons of lipid tails were harmonically restrained in z-direction (membrane normal) with a force constant of 0.05 kcal/mol/Å2 (this restraint was applied to all subsequent HMMM simulations). Cy3 and Cy5 fluorophore structures (Fig. 1 B) and parameters were taken from CHARMM-GUI Membrane Builder (17). Methylated neutral chain termini (ACE and CT3 patches) were applied to the end of the linker. Five independent copies of each lipid composition were generated, and in each copy a single fluorophore (Cy3 or Cy5) was placed initially ∼5 Å above the surface of pre-equilibrated HMMM bilayers (Fig. 1 A) with different lipid compositions (PC, PE, or PS:PC 50:50). Each copy was solvated with TIP3P water (18) and ionized with 150 mM of NaCl using VMD (19), resulting in 30 independent copies each with ∼50,000 atoms and dimensions of 70 × 70 × 110 Å3. Here, the aim of carrying out multiple independent, short HMMM simulations is to sample efficiently possible binding poses of the fluorophores, to be used in the subsequent full-tailed membrane simulations.
Figure 1.
Membrane-binding simulations of Cy3 and Cy5 with HMMM bilayers. (A) The simulation box for a single Cy5 molecule and HMMM PC bilayer system. Explicit water is shown in a blue transparent surface, and the DCLE organic solvent between the short-tailed lipids is shown in a pink transparent surface. Ions are not shown for clarity. The z direction is defined to be parallel to the membrane normal. (B) The molecular structures of Cy3 and Cy5 fluorophores. The different components of a Cy3 molecule are highlighted, namely the sulfonate groups (SO3), cyanine, and the linker. To see this figure in color, go online.
Each copy was first subjected to 5000 steps of minimization and 1 ns of NPAT equilibration followed by 200 ns of NVT production run. The convergence of the HMMM-membrane-bound configurations of the fluorophores is shown in Figs. S1 and S2. Simulation parameters were the same in all simulations unless specified otherwise. These include a nonbonded cutoff of 12 Å (with a switching distance of 10 Å), the inclusion of long-range electrostatics using the particle mesh Ewald method (20) with 1-Å grid spacing and 2-fs time steps with the RATTLE algorithm (21) (SETTLE (22) for water hydrogen atoms) to constrain hydrogen bond lengths. A constant temperature of 310 K was maintained by Langevin thermostat (23) with a damping coefficient of 1 ps−1. Nosé-Hoover Langevin piston (24) with a period of 100 ps and a decay of 50 ps was employed to maintain constant pressure at 1 atm. Periodic boundary conditions in all directions were applied to all simulations.
Full-tailed lipid BEUS
To obtain an accurate quantitative description of the interactions between the fluorophores and the membranes, the membrane-binding free energies for the fluorophores in full-tailed membranes were calculated using BEUS (10,11) with the weighted histogram analysis method (WHAM) (25,26,27). For each type of membrane, all HMMM-membrane-bound configurations of the fluorophores were clustered using three features, namely the z position of the two SO3 groups and the center of mass (COM) of the fluorophore. The centroid of the largest cluster was selected as the representative membrane-bound configuration. This representative configuration for each membrane system was then converted to full-tailed membranes using VMD. The full-tailed lipids used were 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine (POPS). BEUS has proven effective in characterizing the free energy profiles for large-scale conformational changes of membrane transporters (11) and membrane-binding processes (9).
The z component of the COM separation (ξz) between the fluorophore molecule, and the membrane was used as the reaction coordinate. 35 umbrella windows were used for each calculation, extending from the membrane core (ξz = 5 Å) to the bulk solution (ξz = 39 Å), with equal spacing of 1 Å. The initial configuration of each window was generated by steered molecular dynamics (SMD) (28), starting from the membrane-bound configuration. For SMD simulations, the full-tailed, lipid-bound systems were first simulated under equilibrium NPT conditions for 15 ns to allow the membrane area and lipid tails to relax while the fluorophore heavy atoms were harmonically restrained with a force constant of 10 kcal/mol/Å2. Then constant-velocity SMD was conducted to drive the unbinding of the fluorophore with a force constant of 10 kcal/mol/Å2 restraining ξz at a speed of 0.67 Å/ns. As shown in Fig. S3, during these full-tailed simulations, the area per lipid of each membrane converges after 7–8 ns to the values consistent with experimental measurements (29, 30, 31).
For BEUS simulations, the fluorophore was first allowed to relax for 5 ns at the center of each umbrella window while restrained with a force constant of 10 kcal/mol/Å2, followed by 20 ns (POPE) or 25 ns (POPC and POPS:POPC) of simulations with replica exchange protocol. The first 5 ns of each window was discarded as equilibration, and the rest of the trajectories was used in calculating the free energy using WHAM. The convergence of the potential of mean force curves were gauged by a change of less than 0.5 kcal/mol in the membrane-binding free energy. As shown in Fig. S4, the potential of mean force of POPE membrane converged after 15 ns, whereas the POPC membrane and POPS:POPC mixture membrane required slightly longer simulations to achieve convergence (20–25 ns). The force constant of each window was chosen such that the exchange ratio for each window falls between 0.2 and 0.4 (11). The resulting magnitude of force constants ranges from 0.6 to 1.7 kcal/mol/Å2.
Equilibrium MD simulations on membrane proteins
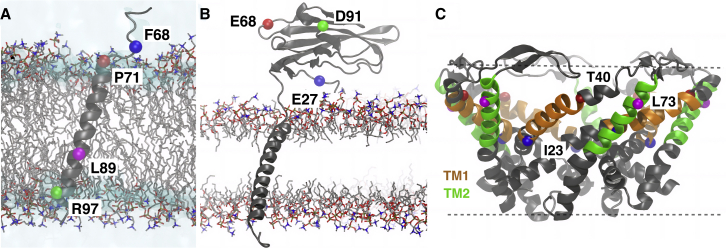
Three TM proteins, namely GpA, NaVβ, and MscL (Fig. 2), were used as benchmark systems to showcase the effect of fluorescent labels and their membrane interaction on TM proteins of different complexity. The target residue on the protein of interest was first mutated to cysteine, and the linker of the fluorophore was covalently attached to the sulfur atom on the cysteine to mimic how proteins are fluorescently labeled in experiments (1).
Figure 2.
The initial structures of different membrane-protein systems used to study the effect of fluorophore tagging. (A) A monomeric GpA TM domain inserted in a POPC bilayer. The blue, red, magenta, and green spheres indicate the positions of F68, P71, L89, and R97, respectively, to which a Cy5 molecule was attached in different simulations (one site per simulation). (B) NaVβ embedded in a PC HMMM lipid bilayer. The E27, E68, and D91 positions on the Ig domain, which were used for Cy5 tagging, are shown in blue, red, and green spheres, respectively. (C) The initial structure of an open-state MscL, with TM1 and TM2 helices colored orange and green, respectively. The POPC lipid bilayer is not shown for clarity. The membrane boundary is indicated by dashed lines. Positions of I23, T40 (TM1), and L73 (TM2), which were used in Cy5 tagging in different simulations, are labeled by blue, red, and magenta spheres, respectively. To see this figure in color, go online.
For GpA (Protein Data Bank, PDB: 1afo (32)), the wild-type (WT) monomeric helix was embedded in a full-tailed POPC bilayer and subjected to 100 ns of equilibrium simulation. The last frame was then used as the initial structure for another 100-ns production run. Cy5 molecules were attached at four different positions (F68, P71, L89, or R97 in different simulation systems; Fig. 2 A) to the same initial structure and simulated for 100 ns each.
NaVβ (PDB: 5XSY (33)) was first embedded in an HMMM PC bilayer and subjected to 50 ns of initial equilibrium simulation. The last frame of the equilibrated NaVβ structure in HMMM bilayer was used as the starting point of both unlabeled NaVβ (control) and Cy5-tagged NaVβ systems. For the control system, four independent 50-ns simulations were performed. For the Cy5-tagged systems, a Cy5 molecule was attached at three different positions (E27, E68, or D91; Fig. 2 B), and four independent 100-ns equilibrium simulations were carried out for each tagged system. Contacts between the Cy5 molecule and lipid bilayer were observed to form within the first 50 ns of each Cy5-tagged NaVβ simulation. Only the last 50 ns of the simulations was used for the analysis to exclude transient behavior.
For MscL (PDB: 2oar (34)), 100 ns of equilibrium simulation was carried out on the open-state MscL embedded in a full-tailed POPC bilayer. The last frame of the initial 100-ns simulation was used as the initial structure for production runs in both unlabeled MscL (control) and Cy5-tagged MscL systems (I23, T40, and L73; Fig. 2 C). Two independent 150-ns simulations were carried out for each of the four systems, and the last 75 ns of each trajectory was included in the analysis. Surface tension of 30 dyn/cm was applied to all simulations of MscL to help maintain the open-pore conformation.
Results and Discussion
To investigate the effect of membrane binding and lipid interaction of fluorophores Cy3 and Cy5 on the dynamics of protein inside a lipid bilayer, membrane-fluorophore interactions are first studied. The lipid-fluorophore contacts, membrane insertion depths, as well as membrane-binding free energies are characterized. Then, the effects of fluorescent labeling on the natural dynamics of different membrane-protein systems (GpA, NaVβ, and MscL) are investigated by MD simulations.
Spontaneous membrane binding of Cy3 and Cy5
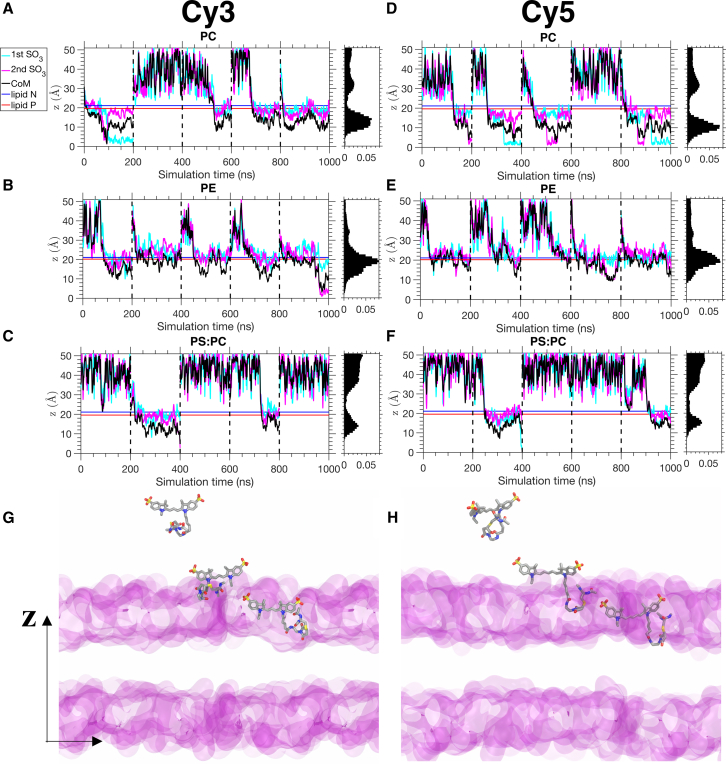
As Cy3 and Cy5 fluorophores have been widely used in studying protein dynamics in membrane environments, it is of interest to study the lipid-dependence of membrane-fluorophore interactions. Five independent simulations for a Cy3-Cy5 fluorophore were carried out with three different lipid compositions, namely PC (major component of eukaryotic membranes), PE (major component of prokaryotic membranes), and PS:PC 1:1 mixture (representing negatively charged membranes). Out of five 200-ns simulations, spontaneous membrane insertion events are observed in the majority of the trajectories with pure PC and PE membrane (Fig. 3, A, B, D, and E). However, stable binding of Cy3 and Cy5 is observed in only two out of five simulations in the presence of PS:PC membranes (Fig. 3, C and F). The negatively charged PS lipid headgroups appear to oppose the formation of initial fluorophore-membrane contact. As a result, Cy3 and Cy5 spend around 63% of the simulation time in the bulk solvent (Table S1). The distribution of COM positions (Fig. 3, A–F, right panels) of the fluorophores show a slightly deeper membrane insertion in PC and PS:PC membranes than PE membranes. The shallower insertion is probably due to the fact that the ethanolamine headgroup in PE allows better packing of the lipid bilayer, resulting in a higher lipid density (17) and shallower insertion depth.
Figure 3.
Spontaneous fluorophore insertion into HMMM membranes. (A–C) The z position of different components of Cy3 as a function of simulation time for PC, PE, and PS:PC 1:1 membrane, respectively. The two SO3 functional groups of Cy3 are represented by cyan and magenta lines, and the center of mass (COM) is represented by black lines. Blue and red horizontal lines represent the average positions of nitrogen and phosphorus atoms, respectively, of the lipid molecules. The histograms of the COM positions are shown on the right to each panel. (D–F) The z position of different components of Cy5 with the same notations as in (A)–(C). Periodic boundary wrapping was applied to all plots to show interactions with only one side of the membrane. For (A)–(F), all five independent trajectories (200 ns each) are concatenated into a single trajectory (0–1000 ns). Independent runs are separated by the dashed vertical lines. (G and H) Snapshots from MD simulation of Cy3 (G) and Cy5 (H) spontaneous insertion to the HMMM PC membrane (transparent magenta surface). For each case, three snapshots representing the positions of the fluorophore are shown relative to the membrane position. The insertion processes are similar for PE and PS:PC membranes. To see this figure in color, go online.
To quantify the lipid contacts of the fluorophores, the total contact C was defined as follows:
| (1) |
where xi is the distance between a heavy atom pair of a fluorophore and a lipid molecule in angstroms and the summation is over all possible atom pairs. C is effectively a weighted sum of all contacts between the fluorophores and lipid molecules, with weaker contacts (xi > 4 Å) downweighted. A similar contact function has been used in a previous study to characterize protein-lipid interactions (9).
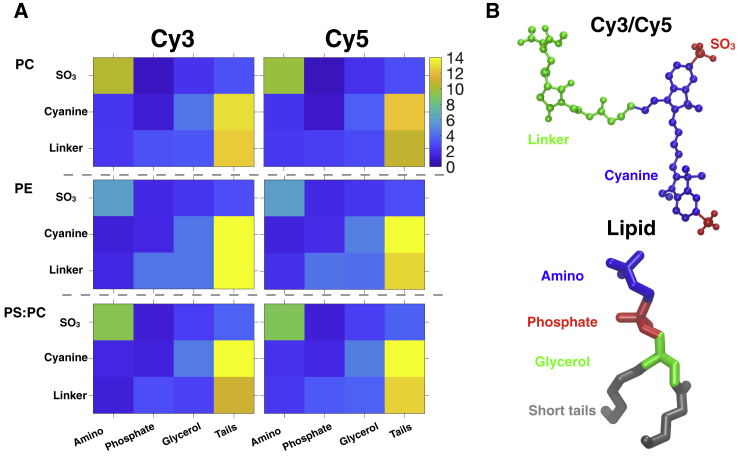
Fig. 4 shows the breakdown of contact counts C between different moieties in the fluorophore and lipid molecules after membrane binding (C > 20). The contact maps for Cy3 and Cy5 are very similar, which is not surprising given the close structural similarity of the two fluorophores (Fig. 1). The SO3 groups on Cy3 and Cy5 preferably interact with the lipid headgroups for all three lipid compositions. This is expected from the strong electrostatic attraction between the lipid headgroup and the negatively charged SO3 group of the fluorophores. On the other hand, the cyanine and linker moieties of the fluorophores are often in contact with the lipid tails. Both electrostatic and hydrophobic interactions with the lipid headgroup and tail help stabilize the fluorophore binding to the lipid bilayer, which is consistent with the observations in fluorescence spectroscopy that both charge and hydrophobicity of a fluorophore affect its membrane partitioning (3,4).
Figure 4.
Contact between Cy3 and Cy5 and the membrane. (A) Contact maps between different moieties of Cy3 and Cy5 and the lipid molecules in the HMMM simulations analyzed after the binding of the fluorophore to the membrane was complete. For each lipid composition, the number of contacts (shown in color scale) is characterized by the lipid-fluorophore contact (C; Eq. 1) in all five trajectories shown in Fig. 3. The value of C is averaged over all membrane-bound snapshots, defined by a total lipid-fluorophore contact Ctotal > 20. (B) The breakdown of heavy atoms into different moieties of Cy5 (also representative of Cy3) and a PC lipid molecule. To see this figure in color, go online.
Binding free energy
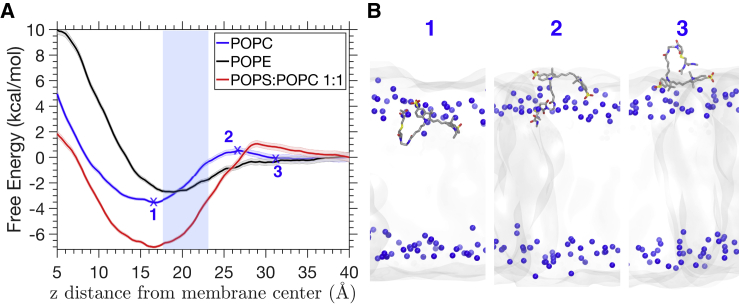
For a more quantitative description of the fluorophore-membrane binding, representative snapshots of the HMMM-membrane-bound Cy5 systems were converted to full-tailed lipid bilayers, and BEUS was used to calculate the free energy profile associated with the membrane-binding process. Fig. 5 A shows the free energy of the membrane-binding process with respect to the z component of the COM of Cy5. The free energy difference between unbound (in bulk water at z = 39 Å) and bound states (at energy minimum), ΔGbind, of Cy5 is about −3.5 kcal/mol for POPC and −2.7 kcal/mol for POPE, whereas the POPS:POPC membrane has a stronger ΔGbind of approximately −7.0 kcal/mol. All lipids show fairly stable binding with Cy5.
Figure 5.
Computed free energy of Cy5 binding to different full-tailed membranes. (A) The blue, black, and red thick lines show the free energy of Cy5 at different z positions for POPC, POPE, and POPS:POPC membranes, respectively. The shaded area indicates the error determined by 1000-step of Monte-Carlo bootstrapping in the WHAM algorithm. The z position is measured from the COM of the Cy5 molecule relative to the membrane center. The headgroup nitrogen atom layer is highlighted by the blue shaded area. (B) Representative structures from the BEUS simulations corresponding to the three labeled positions in (A) for Cy5 binding to the POPC membrane. POPC lipids are shown in gray transparent surface, and the headgroup nitrogen atoms are shown in blue spheres. Positions 1–3 correspond to Cy5 at the energy minimum, energy barrier, and membrane-water interface, respectively. Binding processes of Cy5 to POPE and POPS:POPC membranes are similar to that of POPC. To see this figure in color, go online.
The free energy barrier for Cy5 to insert into the membrane is ∼1.1 kcal/mol for POPS:POPC, which is slightly higher than that of POPC (0.6 kcal/mol). For POPE, however, there is virtually no energy barrier for membrane binding. Moreover, the depth of insertion of Cy5 at the energy minimum is slightly different in different lipid compositions. Both POPC and POPS:POPC membranes show an energy minimum at z = 16.5 Å, whereas POPE shows a slightly shallower binding at z = 18.5 Å.
The BEUS simulations also reveal that the main energy barrier for membrane partitioning of Cy5 corresponds to the translation of the cyanine and linker moieties across the headgroup region (Fig. 5 B). During the insertion, the lipid headgroups have to make room to accommodate Cy5; hence, the smaller ethanolamine headgroup of POPE membrane allows a barrier-free membrane insertion of Cy5.
All the results in full-tailed lipid simulations are consistent with the observations in HMMM binding simulations. In both types of simulations, the fluorophores are found to be more easily bound to POPC and POPE than POPS:POPC, and the binding to POPE is shallower than the other two cases.
Role of electrostatic and hydrophobic interactions
The results from HMMM membrane binding and full-tailed lipid BEUS simulations indicate that both electrostatic and hydrophobic interactions between the Cy3 and Cy5 fluorophores and lipid molecules play important roles in the membrane-binding process. The long-range electrostatic forces determine the affinity of the fluorophores to the membrane surface. As shown in Figs. 3 and 5 A, the COM of negatively charged fluorophores are transiently stable around z = 30 Å for zwitterionic lipid POPC and POPE, allowing more time for the cyanine and linker of the fluorophore to insert into the membrane. In the presence of the anionic lipid POPS, this transient state is not energetically favorable, and the fluorophores are hence repelled away from the membrane surface (Figs. 3, C and F and 5 A). Once the cyanine and linker have inserted into the membrane, hydrophobic interactions start to dominate and stabilize the binding. This observation is consistent with experimental findings that an increase in hydrophobicity of the fluorophore can lead to stronger fluorophore-lipid interactions (2, 3, 4).
Although a POPS:POPC-mixed membrane has a higher energy barrier to fluorophore binding, it also shows more stable binding for Cy5 molecules. The binding free energy in the presence of POPS is ∼3.5–4.3 kcal/mol stronger than that of POPC and POPE (Fig. 5). Close examination of the BEUS trajectories reveals more extensive contacts between water molecules and Cy5 in POPS:POPC bilayer (Fig. S5). The presence of negatively charged POPS headgroups allows more water molecules to penetrate deeper in the membrane and hence allow a more favorable environment for Cy5 to stay inside the lipid bilayer.
Strong effects on structure and dynamics of TM helix
It has been shown experimentally (3) and here using MD simulation that Cy3 and Cy5 can directly interact with lipid bilayers. However, the effect of tagging membrane proteins with these fluorophores can be intricate to quantify. In this section, the equilibrium properties of a simple TM system, GpA monomeric helix (PDB: 1afo (32)) tagged with Cy5, are studied in a POPC bilayer to showcase the effect of the fluorophore on the protein. GpA is an experimentally well-studied TM protein, and because of its simplicity, it has also been studied by MD simulation (35) and used as a benchmark system for membrane simulations (36).
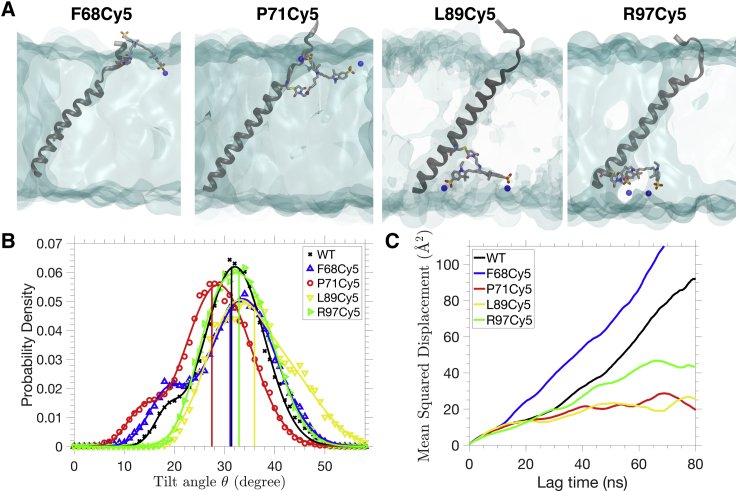
To investigate the effect of Cy5 labeling on a single-pass TM helix, simulations are performed for a single-pass, monomeric GpA TM domain and its mutants tagged with Cy5 at four different locations (F68, P71, L89, and R97; Fig. 6 A). All simulations (duplicate independent 150-ns simulations for each of the systems) are performed with full-tailed POPC to represent an accurate membrane environment. Tagging at all positions allows the attached Cy5 to insert deeply in the membrane, except for F68, in which Cy5 is observed to stay near the membrane-water interface (Fig. 6 A).
Figure 6.
Effect of Cy5 labeling on equilibrium properties of GpA in a POPC lipid bilayer. (A) Representative structures of a monomeric GpA TM helix with Cy5 labeled at different positions. The POPC membrane is shown in cyan transparent surface, and the headgroup nitrogen atoms that are in contact with Cy5 are shown in blue spheres. (B) The distribution of tilt angle θ for unlabeled (WT) and Cy5-tagged GpA systems. θ is defined as the angle between the helical axis of GpA and the normal membrane. Solid curves represent the fitting to a double-Gaussian distribution. The mean tilt angle of each distribution is indicated by the vertical straight line. (C) The mean-squared displacement of a GpA TM helix within the plane of a bilayer against lag time. To see this figure in color, go online.
Fig. 6 B shows the distribution of the tilt angle (θ) of the TM domain for different simulations. All distributions overlap significantly with each other, but the mean tilt angle for different system ranges from θ = 27 to 36°. Labeling at positions closer to the membrane center causes more significant shifts of the mean tilt angle as compared with the control system. In particular, a left shift of 4° and a right shift of 5° are observed for P71Cy5 and L89Cy5, respectively, which is comparable with the difference in the tilt angle of a dimeric GpA TM helix simulated in different environments (lipid bilayer and micelle) (35). The results indicate that, depending on the position, Cy5-lipid interactions can perturb the helical tilt angle in a lipid bilayer.
Fig. 6 C shows the lateral diffusion of GpA within the membrane plane. P71Cy5, L89Cy5, and R97Cy5 mutants show a lower diffusion rate compared with WT. The mean-squared displacement curve plateaus at a longer lag time, indicating that the diffusion is somewhat restricted by Cy5-lipid interactions. However, F68Cy5 (with the mutation site closer to the surface of the membrane) shows smaller effects on the lateral diffusion. The results suggest that the effect of Cy5-tagging on protein diffusion in a membrane is position dependent. Tagging the protein at P71, L89, and R97 allows Cy5 to fully insert into the middle of the lipid bilayer (Fig. 6 A), strengthening hydrophobic interactions and hence hindering the diffusion motion of the GpA TM helix. This observation is consistent with the negative correlation between hydrophobicity of the fluorophore and the diffusivity of the labeled protein reported experimentally (2).
Perturbation of the juxtamembrane domain dynamics
A large number of membrane proteins include in their structure functional or auxiliary juxtamembrane domains that may interact directly with the lipid bilayer. To investigate the effect of Cy5 tagging and its membrane interaction on a juxtamembrane domain of a membrane protein, we use NaVβ (PDB: 5XSY (33)) as a test system. NaVβ is an auxiliary subunit of a voltage-gated sodium channel complex that can modulate the channel’s ion conduction properties (37). NaVβ consists of a single-pass TM helix and a juxtamembrane immunoglobulin (Ig) domain (Fig. 2 B). In a previous study employing Cy dyes for single-molecule imaging (38), the tagged (E27Cy3) NaVβ was shown to render a functional sodium channel complex. Here, we investigate the effect of attaching a Cy5 molecule to the three different sites occupied by negatively charged residues in the WT Ig domain of a NaVβ (E27, E68, and D91) to test the effect of fluorophore tagging on the dynamics of the protein as compared with its unlabeled counterpart. To allow better sampling of the interactions between the Ig domain and lipids, NaVβ was embedded in an HMMM PC lipid bilayer for all the simulations described in this section.
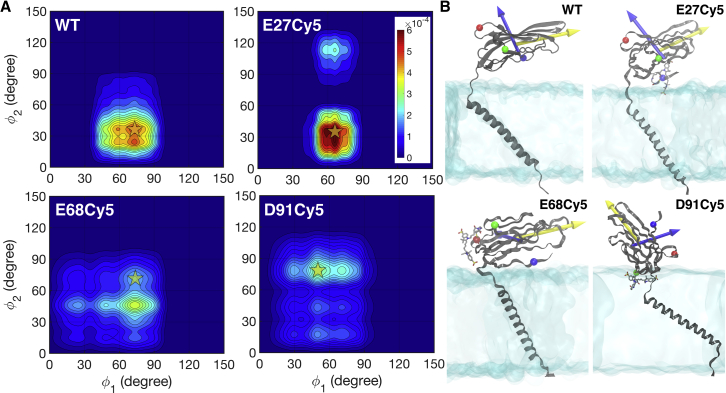
Fig. 7 A shows the dynamics of the Ig domain in the state space formed by its two principal direction angles (ϕ1 and ϕ2). The dynamics of the Ig domain in E27Cy5 remain largely unchanged compared with the unlabeled protein, which is consistent with the observations in single-molecule experiments (38). The distribution of ϕ1 and ϕ2 is much broader for E68Cy5 and D91Cy5, indicating that the Ig domain is more dynamical because of the influence of Cy5-lipid interactions. Among the three positions, E27 was the closest to the lipid headgroups in the equilibrium position of the unlabeled Ig domain (Fig. 7 B); hence, the Cy5-lipid interactions seem to preserve the lipid-protein interactions in the Ig domain. However, the Cy5-lipid interactions are strong enough to perturb the dynamics of the Ig domain in the other two positions such that significant changes in the orientations of the Ig domain are observed (Fig. 7, A and B). The most significant change on the NaVβ structure is observed in D91Cy5, in which Cy5 fully inserts into the membrane to form extensive contacts with the lipid tails, causing the whole Ig domain to flip from its natural orientation and the TM helix to tilt (Fig. 7 B).
Figure 7.
Dynamics of the extracellular immunoglobulin (Ig) domain of a sodium channel β-(NaVβ)-subunit. (A) Dynamics of the Ig domain shown in the state space formed by two principle direction angles ϕ1 and ϕ2 and defined by the angles between the z axis and the first two principal axes of the Ig domain. Color scale gives the normalized probability density in the state space. (B) Snapshots of the conformation highlighted by the stars in the state space plots in (A) for each simulated system. The first and second principal axes are shown by yellow and blue arrows, respectively. The E27, E68, and D91 positions on the Ig domain are shown in blue, red, and green spheres, respectively. To see this figure in color, go online.
Interaction with interhelical loops
Interhelical loops are common to all α-helical TM proteins and known to contribute to the structural organization and stability of TM proteins (39). These loops are often hydrophilic and expose to the extramembrane solution, serving to connect and stabilize TM α-helices in the membrane (40). Because of the hydrophilic nature of the loops, they can interact with soluble fluorophores and potentially affect the structure of TM helical proteins.
Here, we investigate the potential effects of fluorophore labeling on a pentameric channel protein MscL (PDB: 2oar (34)). MscL is a nonselective ion channel (41,42), with each subunit consisting of two TM helices, namely TM1 and TM2 (Fig. 8 A). Functionally, MscL opens under high membrane surface tension to protect bacterial cells from hypo-osmotic shock (43). A previous study on the MscL gating mechanism using single-molecule fluorescence energy transfer technique has proposed that MscL opens with a helix-tilting motion (44). Hence, lipid-protein interactions and helical conformation in the membrane are crucial to the function of MscL, and it is of interest to investigate how fluorophore-lipid and fluorophore-loop interactions may affect the structure of MscL. We attach Cy5 molecules to three positions (I23 and T40 on TM1 and L73 on TM2) in all five monomers that have been used in the aforementioned experiment (44) to investigate the effect of fluorophore-lipid interactions on the structure and dynamics of an open-pore MscL in a full-tailed POPC lipid bilayer.
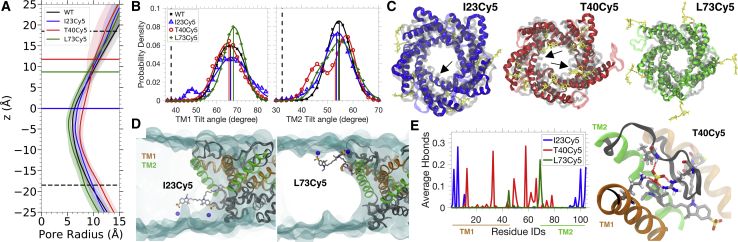
Figure 8.
Effect of Cy5-membrane interaction on the equilibrium properties of an open MscL channel in a POPC lipid bilayer. (A) The mean pore radius of MscL along the channel axis (z) calculated during the last 75 ns of the two independent simulations. The shaded area represents the SD of the pore radius. Black dashed lines represent the average position of the headgroup nitrogen atoms of the POPC bilayer. Colored thick lines (same color code shown in the legend) represent the positions to which Cy5 is attached to the protein. (B) Tilt angles of TM1 and TM2 during the equilibrium simulations. Colored vertical lines indicate the mean tilt angles of TM1 and TM2 for the corresponding systems. Black dashed lines represent the reference tilt angles for TM1 and TM2 measured using the closed state crystal structure. (C) Representative snapshots of MscL showing the structural symmetry in the pore. Structures of I23Cy5, T40Cy5, and L73Cy5 are colored in blue, red, and green, respectively. Cy5 is shown in yellow licorice representation. The control system (WT) is shown as a transparent overlay. The key differences that correspond to the secondary peaks in the tilt angle shown in (B) are highlighted by the arrows. (D) Representative snapshots of MscL with Cy5 attached to I23 and L73. POPC membranes are shown in cyan transparent surface, and the headgroup nitrogen atoms of POPC lipids that are in contact with Cy5 are shown in blue spheres. (E) Hydrogen bonds formed between the SO3 groups of Cy5 and MscL. A representative snapshot depicting the hydrogen bond network formed by T40Cy5, and the protein is shown on the right panel. To see this figure in color, go online.
The pore radius along the channel axis (z axis) is shown in Fig. 8 A. The minimal pore radius of the unlabeled MscL is 6.0 ± 0.5 Å. Compared with the minimal pore radius of I23Cy5 (6.5 ± 0.7 Å) and L73Cy5 (5.2 ± 0.5 Å), there is no significant change in the pore size, but the pore seems to be slightly wider in T40Cy5 (7.4 ± 0.6 Å). The differences in the mean tilt angles (averaged over all five monomers) between the three Cy5-tagged MscL systems and the control system (WT) is less than 2° (Fig. 8 B). However, asymmetry in the helical conformation is observed in the TM1 of I23Cy5 and TM2 of T40Cy5 (Fig. 8 C) because the helical tilt angles show a double-Gaussian distribution instead of a single-Gaussian distribution. The emerged peaks in the tilt angle distribution are significantly different (>10°) from the WT conformations, which leads to a wider pore in T40Cy5. Persistent fluorophore-lipid interactions are observed in both I23Cy5 and L73Cy5 (Fig. 8 D), but only the tilt angle of TM1 of I23Cy5 is affected. Both the pore size and tilt angles of TM helices remain unchanged in L73Cy5.
Cy5 at T40 cannot reach out to the lipid bilayer. Instead, the T40Cy5 mutant forms a persistent hydrogen bonding network with the interhelical loop between TM1 and TM2 (Fig. 8 E) that can otherwise interact with the lipid headgroups or TM helices of the protein. As a result of this fluorophore-loop interaction, fewer contacts are formed between the loops and lipid headgroups in T40Cy5 (Fig. S6), which may destabilize the structure and contribute to the structural asymmetry.
Position-dependent effects of fluorophore labeling
As demonstrated by the three membrane-protein systems, the effects of fluorophore-lipid interactions on the dynamics of membrane proteins are strongly position dependent. As a general observation, the nonspecific hydrophobic interactions between the fluorophore and lipids cause more dramatic effects on the structural and dynamical properties of various membrane proteins. At positions where the attached fluorophores are allowed to make extensive contacts with the lipid tails, the equilibrium structure and diffusion dynamics can be significantly altered, as demonstrated by the GpA simulations (Fig. 6).
Our simulations also suggest that a single fluorophore molecule can alter the surface electrostatic properties of the juxtamembrane Ig domain of NaVβ and, hence, the orientation of the Ig domain with respect to the membrane. The orientation of the unlabeled and E27Cy5 Ig domain in MD simulations is consistent with that captured in a eukaryotic sodium channel complex (33); however, the orientations of the Ig domain in the E68Cy5 and D91Cy5 simulations are quite different from that of the channel complex (Fig. 7). The altered orientation observed in E68Cy5 and D91Cy5 may suggest that the introduced fluorophore label could disrupt the interface between the β-subunit and the α-subunit of a sodium channel. In this case, structural information of the native conformation of a protein becomes crucial to determine the position for fluorescent labeling that can preserve the native structure and dynamics of the protein in an experiment.
The effects of fluorophore-lipid interactions on helical channel protein MscL is less explicit than in the case of single-pass TM helix. There are no significant large-scale structural changes in MscL, such as the pore radius, a result that might be changed in more extended simulations in which long-timescale changes can be captured better. However, after labeling the protein near the center of the membrane (e.g., I23Cy5) where extensive hydrophobic interaction can be formed between the fluorophore and the lipids, the structure is locally perturbed, as shown by the changes in the tilt angles of the TM helices (Fig. 8 B). Besides, the fluorophore can also form a hydrogen bond network with the interhelical loops (T40Cy5) and perturb the lipid-loop interactions (Fig. 8 E; Fig. S6). As a result of the fluorophore-lipid and fluorophore-loop interactions, asymmetry emerges in the pentameric structure of MscL. Among the three tested sites, L73Cy5, which is closer to the lipid headgroup layer, displays the least amount of perturbation to the structure of MscL. Based on our observations, it is advised to avoid labeling a channel protein at a site near the membrane center or exposed interior of the channel pore so that the effect on channel gating will be minimized.
Conclusions
Cy3 and Cy5 fluorophores have been and continue to be extensively used in probing the membrane-protein structure and dynamics in experiments. Our MD simulations, studied at a microscopic level in fluorophore-lipid interactions, show that Cy3 and Cy5 interact directly and strongly with lipid bilayers of common lipid compositions. The two water-soluble fluorophores can interact with lipid bilayers through electrostatic and hydrophobic interactions, resulting in their strong tendency of membrane partitioning.
The MD simulations of various membrane-protein systems suggest that the fluorophore-lipid interaction can cause measurable perturbations on the structural and dynamical properties of membrane proteins, depending on the position of fluorophore labeling and how sensitive the protein structure to these effects might be. We show that when the attached fluorophore is exposed to the membrane core, it forms extensive contacts with the lipid tails and causes more significant effects on the structure and dynamics of the proteins. In addition to the fluorophore-lipid interaction, the fluorophores can also interact with hydrophilic interhelical loops through their charged functional groups and introduce defects in the structure. Taken together, our results suggest that when labeling a TM protein with fluorophores, positions that are exposed to the membrane core or hydrophilic loops of the protein should be avoided to minimize potential experimental artifacts.
Author Contributions
K.L. and E.T. conceived the project. K.L. carried out the simulations and analyzed the data. All the authors wrote the manuscript.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01-GM123455, R01-GM067887, R01-GM122420, and P41-GM104601 (to E.T.)). Simulations were performed using computer time provided by the Extreme Science and Engineering Discovery Environment (grant TG-MCA06N060) and Blue Waters (National Science Foundation Petascale Resource Allocation Committee (PRAC) award ACI-1440026), which is supported by the Blue Waters sustained-petascale computing project (National Science Foundation OCI-0725070 and ACI-1238993) and the State of Illinois.
Editor: Alemayehu Gorfe.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.05.027.
Supporting Material
References
- 1.Toseland C.P. Fluorescent labeling and modification of proteins. J. Chem. Biol. 2013;6:85–95. doi: 10.1007/s12154-013-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanetti-Domingues L.C., Tynan C.J., Martin-Fernandez M. Hydrophobic fluorescent probes introduce artifacts into single molecule tracking experiments due to non-specific binding. PLoS One. 2013;8:e74200. doi: 10.1371/journal.pone.0074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes L.D., Rawle R.J., Boxer S.G. Choose your label wisely: water-soluble fluorophores often interact with lipid bilayers. PLoS One. 2014;9:e87649. doi: 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z., Yomo D., Gradinaru C. Choosing the right fluorophore for single-molecule fluorescence studies in a lipid environment. Biochim. Biophys. Acta Biomembr. 2017;1859:1242–1253. doi: 10.1016/j.bbamem.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Graen T., Hoefling M., Grubmüller H. AMBER-DYES: characterization of charge fluctuations and force field parameterization of fluorescent dyes for molecular dynamics simulations. J. Chem. Theory Comput. 2014;10:5505–5512. doi: 10.1021/ct500869p. [DOI] [PubMed] [Google Scholar]
- 6.Ohkubo Y.Z., Pogorelov T.V., Tajkhorshid E. Accelerating membrane insertion of peripheral proteins with a novel membrane mimetic model. Biophys. J. 2012;102:2130–2139. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baylon J.L., Lenov I.L., Tajkhorshid E. Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J. Am. Chem. Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylon J.L., Vermaas J.V., Tajkhorshid E. Atomic-level description of protein-lipid interactions using an accelerated membrane model. Biochim. Biophys. Acta. 2016;1858:1573–1583. doi: 10.1016/j.bbamem.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermaas J.V., Tajkhorshid E. Differential membrane binding mechanics of synaptotagmin isoforms observed in atomic detail. Biochemistry. 2017;56:281–293. doi: 10.1021/acs.biochem.6b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moradi M., Tajkhorshid E. Mechanistic picture for conformational transition of a membrane transporter at atomic resolution. Proc. Natl. Acad. Sci. USA. 2013;110:18916–18921. doi: 10.1073/pnas.1313202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moradi M., Tajkhorshid E. Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. J. Chem. Theory Comput. 2014;10:2866–2880. doi: 10.1021/ct5002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanommeslaeghe K., Hatcher E., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y., Cheng X., Im W. CHARMM-GUI HMMM builder for membrane simulations with the highly mobile membrane-mimetic model. Biophys. J. 2015;109:2012–2022. doi: 10.1016/j.bpj.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu E.L., Cheng X., Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 19.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 20.Darden T., York D., Pedersen L.G. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 21.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 22.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 23.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 24.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 25.Kumar S., Bouzida D., Rosenberg J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 26.Souaille M., Roux B. Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Comput. Phys. Commun. 2001;135:40–57. [Google Scholar]
- 27.Grossfield A. WHAM: the weighted histogram analysis method, version 2.0.9. 2013. http://membrane.urmc.rochester.edu/wordpress/?page_id=126
- 28.Izrailev S., Crofts A.R., Schulten K. Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc(1) complex. Biophys. J. 1999;77:1753–1768. doi: 10.1016/S0006-3495(99)77022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kučerka N., Nieh M.-P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Kučerka N., van Oosten B., Katsaras J. Molecular structures of fluid phosphatidylethanolamine bilayers obtained from simulation-to-experiment comparisons and experimental scattering density profiles. J. Phys. Chem. B. 2015;119:1947–1956. doi: 10.1021/jp511159q. [DOI] [PubMed] [Google Scholar]
- 31.Pan J., Cheng X., Katsaras J. The molecular structure of a phosphatidylserine bilayer determined by scattering and molecular dynamics simulations. Soft Matter. 2014;10:3716–3725. doi: 10.1039/c4sm00066h. [DOI] [PubMed] [Google Scholar]
- 32.MacKenzie K.R., Prestegard J.H., Engelman D.M. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 33.Yan Z., Zhou Q., Yan N. Structure of the Nav1.4-β1 complex from electric eel. Cell. 2017;170:470–482.e11. doi: 10.1016/j.cell.2017.06.039. [DOI] [PubMed] [Google Scholar]
- 34.Steinbacher S., Bass R.B., Rees D.C. Structures of the prokaryotic mechanosensitive channels MscL and MscS. In: Hamill O.P., editor. Mechanosensitive Ion Channels, Part A. Volume 58. Elsevier Inc.; 2007. pp. 1–24. [Google Scholar]
- 35.Cuthbertson J.M., Bond P.J., Sansom M.S.P. Transmembrane helix-helix interactions: comparative simulations of the glycophorin a dimer. Biochemistry. 2006;45:14298–14310. doi: 10.1021/bi0610911. [DOI] [PubMed] [Google Scholar]
- 36.Qi Y., Cheng X., Im W. CHARMM-GUI PACE CG builder for solution, micelle, and bilayer coarse-grained simulations. J. Chem. Inf. Model. 2014;54:1003–1009. doi: 10.1021/ci500007n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isom L.L., De Jongh K.S., Catterall W.A. Auxiliary subunits of voltage-gated ion channels. Neuron. 1994;12:1183–1194. doi: 10.1016/0896-6273(94)90436-7. [DOI] [PubMed] [Google Scholar]
- 38.Leisle L., Chadda R., Ahern C.A. Cellular encoding of Cy dyes for single-molecule imaging. eLife. 2016;5:e19088. doi: 10.7554/eLife.19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popot J.L., Engelman D.M. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 40.Ulmschneider M.B., Tieleman D.P., Sansom M.S.P. The role of extra-membranous inter-helical loops in helix-helix interactions. Protein Eng. Des. Sel. 2005;18:563–570. doi: 10.1093/protein/gzi059. [DOI] [PubMed] [Google Scholar]
- 41.Blount P., Sukharev S.I., Kung C. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 1996;15:4798–4805. [PMC free article] [PubMed] [Google Scholar]
- 42.Blount P., Moe P.C. Bacterial mechanosensitive channels: integrating physiology, structure and function. Trends Microbiol. 1999;7:420–424. doi: 10.1016/s0966-842x(99)01594-2. [DOI] [PubMed] [Google Scholar]
- 43.Booth I.R., Blount P. The MscS and MscL families of mechanosensitive channels act as microbial emergency release valves. J. Bacteriol. 2012;194:4802–4809. doi: 10.1128/JB.00576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Liu Y., Selvin P.R. Single molecule FRET reveals pore size and opening mechanism of a mechano-sensitive ion channel. eLife. 2014;3:e01834. doi: 10.7554/eLife.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.