Abstract
Objectives:
To analyze survival outcomes in patients with oropharygeal cancer treated with primary intensity modulated radiotherapy (IMRT) using decision tree algorithms.
Methods:
A total of 273 patients with newly diagnosed oropharyngeal cancer were identified between March 2010 and December 2016. The data set contained nine predictor variables and a dependent variable (overall survival (OS) status). The open-source R software was used. Survival outcomes were estimated by Kaplan–Meier method. Important explanatory variables were selected using the random forest approach. A classification tree that optimally partitioned patients with different OS rates was then built.
Results:
The 5 year OS for the entire population was 78.1%. The top three important variables identified were HPV status, N stage and early complete response to treatment. Patients were partitioned in five groups on the basis of these explanatory variables.
Conclusion:
The proposed classification tree could help to guide future research in oropharyngeal cancer field.
Advances in knowledge:
Decision tree method seems to be an appropriate tool to partition oropharyngeal cancer patients.
Introduction
In recent years, machine learning approaches for predicting treatment outcomes, including survival and toxicity, have received growing attention in the oncology field, primarily to support decision systems.1–6 At present, the appropriate treatment strategy is mainly based on prognostic stage group, which combines information on primary tumour (T), regional lymph nodes (N) and distant metastasis (M) categories.7 In oropharyngeal cancer, concurrent chemoradiotherapy (CRT) represents the standard treatment, but, over the years, survival improvements have been modest, especially in locally advanced stage disease.7 Identifying factors that are able to predict clinical outcomes could potentially contribute to develop more efficacious therapeutic strategies. Prior attempts at defining “new” survival predictors have shown the importance of factors such as human papillomavirus (HPV) status and tobacco smoking pack-years.8 Confirming HPV prognostic value, the current TNM system has created a separate staging algorithm for HPV-related oropharyngeal cancer and HPV-negative disease, without altering clinical treatment decision-making.9 Apart from these well-established predictors (T stage, N stage and HPV status), research on the impact of novel clinical factors on survival should be encouraged. In response to this need, we analyzed data from our oropharyngeal cancer patients, staged according to eighth edition TNM and who underwent CRT using classification trees and random forests. The aim was to contribute to the existing literature on survival predictors using machine learning. We hope to provide a first step towards personalized care in oropharyngeal cancer.
Methods and patients
Study population
After institutional review board approval, we retrospectively identified a cohort of 273 consecutive patients with histologically confirmed squamous cell carcinoma of the oropharynx treated between March 2010 and December 2016 at our (Guy’s and St Thomas’ Hospital NHS Trust) institution with CRT. Based on clinical examinations (complete medical history, physical examination, flexible nasopharyngoscopy and dental evaluation) and radiologic imaging (head–neck–chest contrast-enhanced CT and head–neck MRI or 18-fludeoxyglucose positron emission tomography-CT (18FDG PET-CT)), all patients were retrospectively re-stage using the eighth edition TNM classification.7 HPV status was determined on p16 immunohistochemistry. p16 immunohistochemistry (clone E6H4, CINtec; Roche mtm labsAG, Basel, Switzerland) was performed on an automated platform (Benchmark Ultra; Ventana Medical Systems, Tucson, AZ) according to the manufacturer’s instructions. Lesions showing strong and diffuse nuclear and cytoplasmic positivity in >70% of tumour cells were then subjected to high-risk HPV testing by the use of DNA in-situ hybridisation (INFORM Family III; Roche) according to the manufacturer’s instructions. The radiological assessment of extracapsular extension and soft tissue invasion of involved nodes was performed by a radiologist and/or a radiation oncologist based on MRI scans. Exclusion criteria were the presence of a synchronous tumour and a non-curative intent approach.
Treatment strategy
All cases were referred to a multidisciplinary head and neck meeting before treatment initiation and written informed consent was obtained from all patients. RT was delivered with an intensity modulated technique (IMRT) to a total dose of 65 Gy (2.17 Gy/fraction) and concomitant chemotherapy consisted of high-dose cisplatin (100 mg/m2 every 3–4 weeks). In case of inadequate renal function or contraindication to platinum, carboplatin (AUC5) or cetuximab (400 mg/m2 loading dose, then 250 mg/m2 weekly during RT) were administered, respectively. Induction chemotherapy was recommended in case of bulky primary or nodal disease. The detailed CRT protocol has been described previously.10,11
Follow-up
After treatment, all patients were monitored by physical examination every 6 weeks for 1 year, then every 3 months for the next 2 years, and every 6 months thereafter. To evaluate treatment response, 18FDG PET-CT (and head-neck CT or MRI where indicated) was recommended 3 months after CRT. Repeat imaging exam was carried out where clinically appropriate.
Statistical analysis
Statistical analysis was performed using the R 0.98.1091 software. Standard descriptive statistics were used to evaluate the distribution of each variable. Continuous variables were reported as mean ± standard deviation (SD) and categorical variables as frequencies or percentages.
Overall survival (OS) and disease-free survival (DFS) were calculated in months from the date of the end of CRT to the first event, including date of the last follow-up or death (OS) and/or relapse (DFS). Complete response was defined as the total disappearance of tumour with a normal oropharyngeal mucosa (T), negative neck (N) and without evidence of distant metastasis (M) determined by clinical (T and N) and/or diagnostic examinations (T, N and M). Patients with a complete response within 3 months from the end of CRT were classified as early responders, whereas patients who never attained local control within 6 months from the end of CRT were classified as persistent disease cases. Living patients without an event corresponding to any end point were censored at the date of their last follow-up.
OS and DFS were estimated by the Kaplan–Meier method and survival curves were compared by the log-rank test for the HPV variable analysis.
In addition to these standard statistical methods, a machine learning-based methodology was applied to define significant clinical predictors of survival rate.12,13 The randomForest and rpart packages in R were used to define important explanatory variables and identify a corresponding optimal decision tree, respectively. Firstly, we applied the randomForest algorithm to build a random forest of a fixed number of classification trees based on a large number of explanatory variables. The following predictor variables were used: gender, age, primary tumour site, alcohol, tobacco smoking, HPV status, clinical T classification, clinical N classification and early responders. The dependent variable referred to death event (no or yes). Then, using the importance() function, we evaluated the importance of each explanatory variable. Variables associated with a mean decrease in accuracy >1% were then included in the classification tree construction. After the most important explanatory variables were identified, the rpart algorithm was used. The rpart algorithm splits a group into two groups that are as different from each other as possible. It was applied to decide which of these variables to split and which splitting value to take at each step of the tree’s construction.12 To define the optimal tree size, the tree was pruned using the cross-validation error criterion. The minimum error rule (size producing the minimum cross-validation error) or the one-standard-error rule (minimal size producing error lower than minimum error plus one standard deviation) was applied.
Results
Patient characteristics
Patient and tumour characteristics are listed in Table 1. Most patients had regional lymph node involvement at presentation (n = 226; 82.8%). Locally advanced disease (>Stage II) was recorded in 110 cases (40.3%). Median follow-up was 54 months. All but 42 patients (15.4%) received concomitant treatment. The median dose was 65 Gy delivered in 30 fractions, with a median overall treatment time of 39 days. Overall, 231 patients (84.6%) received systemic therapy. Cisplatin-based chemotherapy was administered to 209 patients (90.5%), whereas 22 patients (9.5%) received cetuximab. Induction chemotherapy was delivered to 133 patients (48.7%). Induction chemotherapy consisted of a combination of cisplatin and 5-fluorouracil in 75.9% (n = 101), carboplatin and 5-fluorouracil in 8.3% (n = 11) and docetaxel, cisplatin and 5-fluorouracil in 15.8% (n = 21). After induction chemotherapy, all patients received concomitant chemoradiotherapy.
Table 1.
Description of patient and tumour characteristics
| Characteristic | Value (%) | |||
|---|---|---|---|---|
| Age (years) | ||||
| Mean (range) | 59 (35–83) | |||
| Gender | ||||
| Male | 207 (75.8) | |||
| Female | 66 (24.2) | |||
| Tobacco smoking | ||||
| Never | 58 (21.2) | |||
| Former/currenta | 215 (78.8) | |||
| Alcohol habitude | ||||
| Never | 16 (5.9) | |||
| Formerb/currentc | 257 (94.1) | |||
| Primary tumour site | ||||
| Tonsil | 161 (59) | |||
| Soft palate | 11 (4) | |||
| Base of tongue | 95 (34.8) | |||
| Pharyngeal wall | 2 (0.7) | |||
| Vallecula | 4 (1.5) | |||
| HPVstatus | ||||
| Negative | 73 (26.7) | |||
| Positive | 200 (73.3) | |||
| cT (eighth edition) | ||||
| HPV-negative | HPV-positive | |||
| 1 | 1 | 4 (5.5) | 55 (27.5) | |
| 2 | 2 | 32 (43.8) | 74 (37) | |
| 3 | 3 | 21 (28.8) | 31 (15.5) | |
| 4a | 4 | 15 (20.5) | 40 (20) | |
| 4b | 1 (1.4) | |||
| cN (eighth edition) | ||||
| HPV-negative | HPV-positive | |||
| 0 | 0 | 25 (34.2) | 22 (11) | |
| 1 | 1 | 8 (11) | 128 (64) | |
| 2a | 2 | 1 (1.4) | 36 (18) | |
| 2b | 3 | 19 (26) | 14 (7) | |
| 2c | 3 (4.1) | |||
| 3a | 0 (0) | |||
| 3b | 17 (23.3) | |||
| Stage (eighth edition) | ||||
| HPV-negative | HPV-positive | |||
| I | I | 3 | 103 (51.5) | |
| II | II | 12 | 46 (23) | |
| III | III | 14 | 51 (25.5) | |
| IVa | 26 | |||
| IVb | 18 | |||
| Early responders | ||||
| No | 113 (41.4) | |||
| Yes | 160 (58.6) |
Includes ≤10 pack-year (n = 31) and >10 pack-year (n = 124) current smokers.
Includes ≤21 units/week (n = 2) and >21 units/week (n = 17).
Includes ≤21 units/week (n = 159) and >21 units/week (n = 79).
Clinical outcomes
In total, complete response was recorded in 243 patients (89%). 66% (n = 160) of these patients were early responders and 80% (n = 128) had HPV-related oropharyngeal squamous cell carcinoma. Overall, 63 patients died. The 5 year OS and DFS rates for the entire population were 78.1% (95% confidence interval, CI, 0.72–0.83) and 73.9% (95% CI, 0.68–0.79), respectively.
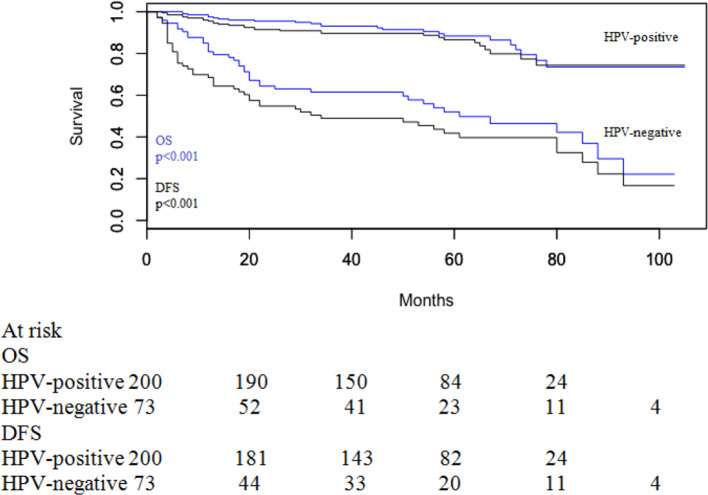
5-year OS in HPV-related oropharyngeal cancers was 88.4% (95% CI 0.82–0.93) and 5 year DFS was 86.6% (95% CI 0.80–0.91). For patients with HPV-negative disease, 5 year OS was 52.0% (95% CI 0.40–0.63) and 5 year DFS was 41.8% (95% CI 0.30–0.53). Survival details according to HPV status are shown in Figure 1. There were significant differences in both OS (p < 0.001) and DFS (p < 0.001) regarding HPV-positive and HPV-negative cancers.
Figure 1.
OS and DFS according to HPV status. DFS, disease-free survival; OS, overall survival.
Random forest
The following variables were investigated with randomForest: gender, age, primary tumour site, alcohol, tobacco smoking, HPV status, clinical T classification, clinical N classification and early responders. The dependent variable, i.e. the response, referred to death event (no or yes). All predictor variables, as well as their values and proportions are listed in Table 1. We applied randomForest using the ntree (number of simulated decision trees)=500 to analyze these data and the top three important predictors were HPV status, N classification and early responders, with a mean decrease accuracy of 4.29, 2.49 and 1.11%, respectively (Figure 2).
Figure 2.
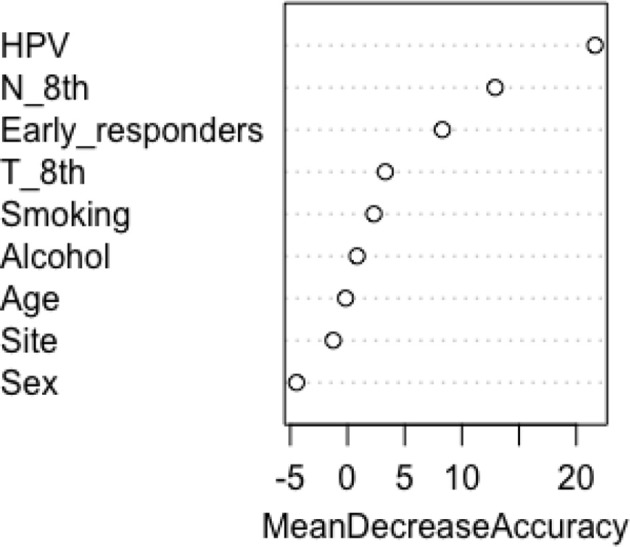
Important variables
Classification tree
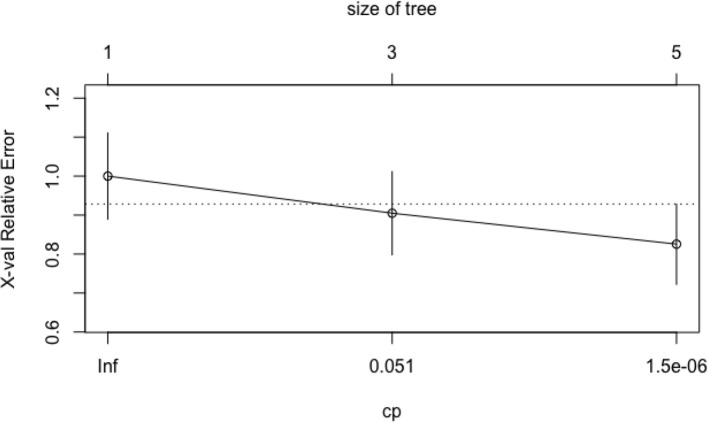
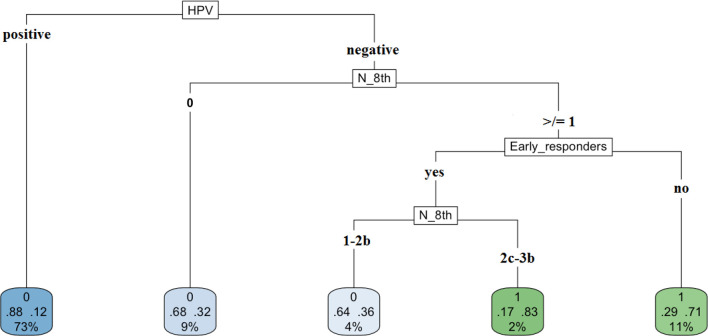
The most important covariates – HPV status, N classification and early responders – were used in rpart to grow an optimal classification tree. Consistent with the categorical nature of the response variable survival, we applied the rpart algorithm with the option method = ”class”, which provides a classification tree. To control the length of the tree before pruning, we used the parameter settings complexity parameter (cp) = 10−9 and minbucket (number of observations in any terminal node)=1. The cross-validation error was used to determine the optimal level of tree complexity and, based on the complexity plot provided in Figure 3, the minimum-error rule was applied. The plot of the final classification tree is displayed in Figure 4. The decision tree predicts the survival rate of oropharyngeal patients, based on the HPV status, the N involvement at diagnosis and the evidence of complete response within 3 months from the end of treatment. The split at the top of the tree resulted in two large branches: the left-hand branch included patients with HPV-related oropharyngeal cancer (73% of the overall sample, with 12% probability of death); the right-hand branch corresponded to HPV-negative disease cancer (27% of the overall sample, with 53% probability of death). The right branch is further subdivided, in three steps, by N classification and early responders. Overall, the tree had five terminal nodes, which resulted in partitioning oropharyngeal patients in five groups: (i) patients with HPV-related oropharyngeal cancer (73% of the overall sample, with 12% probability of death); (ii) patients who had HPV-negative disease without nodal involvement at diagnosis (9% of the overall sample, with 32% probability of death); (iii) patients who had HPV-negative cancer, who had metastasis in single or multiple ipsilateral nodes, none larger than 6 cm in greatest dimension without extranodal extension, and were early responders (4% of the overall, 36% death probability); (iv) patients who had HPV-negative cancer, N stage ≥2c, and were early responders (2% of the overall, 83% death probability); (v) patients who had HPV-negative cancer, with nodal involvement at diagnosis and were not early responders (11% of the overall, 71% death probability).
Figure 3.
The cost–complexity plot. The cost–complexity plot illustrates the relative cross-validation error (y-axis) for various complexity parameter (cp) values (x-axis). The cp is used to define the optimal tree size. To prune the tree, the cp value with the lowest cross-validation error was selected (minimum-error rule).
Figure 4.
Classification tree for patients with locally advanced head and neck cancer.
Discussion
This study used machine learning methods to analyze follow-up data in oropharyngeal patients treated with curative IMRT and concomitant ( ± induction) chemotherapy. A decision tree approach showed that the OS probability for the five groups in which our entire sample had been partitioned was 88%, 68%, 64%, 17% and 29%, respectively. The partition was dependent on HPV status, N classification and early responders. HPV status was of the greatest importance in the classification tree, reflecting and confirming its role in oropharyngeal cancer staging. Interestingly, complete response within 3 months from the end of treatment appeared to be an important variable, more commonly selected in randomForest than T stage and smoking history. As is well-known, randomForest algorithms form many deep decision trees based on randomized versions of the data and/or the predictor set, and average them. However, in this application we were not interested in averaging, since we were looking for a single partition of the predictor set, i.e. a single deterministic tree. We were only interested in the importance scoring, a by-product of random forest algorithms which provides a robust assessment of the predictors having the best explanatory power in the problem at hand. Patients with HPV-negative oropharyngeal cancer either with no evidence of complete tumour regression within 3 months and or early responders but with cN ≥2c (TNM 8) were at higher risk of death, defined as worse survival probability (29 and 17%, respectively), compared toHPV-negative patients without nodal involvement (68%), early responders with N ≤ 2b (64%) and HPV-related cases (88%). Identifying these risk groups has important implications for potential treatment strategies as well as follow-up schedules. This partition seems to be appropriate and clinically applicable, especially when we look at clinical trial design in which randomization could be stratified by response to definitive local therapy, in addition to HPV status and stage disease.14
In the published oropharyngeal cancer literature, it is difficult to compare results on these factors. To date, prior analyses attempted to categorize patients into low-, intermediate- and high-risk groups.8 Survival oropharyngeal data from the Radiation Therapy Oncology Group (RTOG) 0129 trial were retrospectively examined to generate a predictive model.8,15 In total, 266 patients with oropharyngeal squamous cell carcinoma were included in a recursive-partitioning analysis. According to our results, HPV status was the major OS determinant. In addition, tobacco smoking and N stage for HPV-positive cancers or T stage for HPV-negative disease were proposed as independent factors with influential predictive significance.8 Patients were then classified into low-, intermediate- or high-risk groups, with respect to the risk of death (7%, 30% and 54%, respectively). In this context, when building our random forest across all of the variables considered, we confirmed the importance of those predictors (HPV status, N stage, T stage, smoking status) and we defined a new variable to be considered in the oropharyngeal cancer prognosis (early response). Actually, T stage and smoking status displayed a lower importance function than early responders (Figure 2). Although T stage and smoking status had a mean decrease in accuracy <1%, we tried to extend the classification tree to these five covariates. But, after adding the variables T stage and smoking status, the resulting tree was too complex (many terminal leaves), leading to poor performance in reducing prediction accuracy. Therefore, the final data were translated into a simplified classification tree with higher accuracy, based on the top three variables (HPV status, N stage and early responders). For practicality's sake, we merged the information of terminal nodes with similar death probability to define the three categories, showing that low-risk patients had improved survival (88%) compared with both intermediate- (64–68%) and high-risk (12–27%) patients. But a direct comparison between studies was not reliable, except for the objective comment concerning the relative efficacy in subgroup definition.
The main limitation of this study is its retrospective nature, thus hypothesis-generating rather than confirmatory. Biases related to the retrospective design include patient heterogeneity (age, comorbidities, primary tumour location, clinical N classification, HPV status and chemotherapy regimen), treatment selection bias and relatively limited sample size. But, to our knowledge, this is the first study that has investigated the potential decision tree application in oropharyngeal cancer patients in a modern context, including eighth TNM staging system and IMRT technique. Further studies are necessary to confirm our assumptions. The next logical steps should be to conduct an external validation of the model and design a prospective trial based on these partitions.
Conclusion
We introduced for the first time the decision tree approach in oropharyngeal cancer data analysis. The proposed classification tree confirmed the importance of HPV status and N stage and included early response to CRT as survival predictors. Predictive information of our classification tree should be confirmed in further studies to provide an effective and accurate decision-making in the management of patients with oropharyngeal cancer.
Contributor Information
Francesca De Felice, Email: fradefelice@hotmail.it.
Laia Humbert-Vidan, Email: laia.humbert-vidan@kcl.ac.uk.
Mary Lei, Email: mary.lei@gstt.nhs.uk.
Andrew King, Email: andrew.king@kcl.ac.uk.
Teresa Guerrero Urbano, Email: Teresa.GuerreroUrbano@gstt.nhs.uk.
REFERENCES
- 1.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 2015; 13: 8–17. doi: 10.1016/j.csbj.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luna JM, Chao H-H, Diffenderfer ES, Valdes G, Chinniah C, Ma G, et al. . Predicting radiation pneumonitis in locally advanced stage II-III non-small cell lung cancer using machine learning. Radiother Oncol 2019; 133: 106–12. doi: 10.1016/j.radonc.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Ganggayah MD, Taib NA, Har YC, Lio P, Dhillon SK. Predicting factors for survival of breast cancer patients using machine learning techniques. BMC Med Inform Decis Mak 2019; 19: 48. doi: 10.1186/s12911-019-0801-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deist TM, Dankers FJWM, Valdes G, Wijsman R, Hsu I-C, Oberije C, et al. . Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med Phys 2018; 45: 3449–59. doi: 10.1002/mp.12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol 2019; 110: 12–22. doi: 10.1016/j.jclinepi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 6.Fisher A, Rudin C, Dominici F. Model Class Reliance: Variable importance measures for any machine learning model class, from the “Rashomon”. Perspective 2018;. [Google Scholar]
- 7.National Comprehensive Cancer Network (NCCN). Guidelines Head and Neck Cancers, Version 1. 2019. Available from: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf. [DOI] [PubMed]
- 8.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. . Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010; 363: 24–35. doi: 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lydiatt WM, Patel SG, O'Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, et al. . Head and neck cancers-major changes in the American joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin 2017; 67: 122–37. doi: 10.3322/caac.21389 [DOI] [PubMed] [Google Scholar]
- 10.Bird T, De Felice F, Michaelidou A, Thavaraj S, Jeannon J-P, Lyons A, et al. . Outcomes of intensity-modulated radiotherapy as primary treatment for oropharyngeal squamous cell carcinoma - a European singleinstitution analysis. Clin Otolaryngol 2017; 42: 115–22. doi: 10.1111/coa.12674 [DOI] [PubMed] [Google Scholar]
- 11.De Felice F, Thomas C, Barrington S, Pathmanathan A, Lei M, Urbano TG. Analysis of loco-regional failures in head and neck cancer after radical radiation therapy. Oral Oncol 2015; 51: 1051–5. doi: 10.1016/j.oraloncology.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 12.Efron B, Hastie T. Computer age statistical inference. 2017;; 8: 125–8chapter. [Google Scholar]
- 13.Breiman L. Random forests. Mach Learn 2001; 45). : : 5–32Springer. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 14. https://clinicaltrials.gov/ct2/show/NCT03452137?term=NCT03452137&draw=2&rank=1
- 15.Nguyen-Tan PF, Zhang Q, Ang KK, Weber RS, Rosenthal DI, Soulieres D, et al. . Randomized phase III trial to test accelerated versus standard fractionation in combination with concurrent cisplatin for head and neck carcinomas in the radiation therapy Oncology Group 0129 trial: long-term report of efficacy and toxicity. J Clin Oncol 2014; 32: 3858–67. doi: 10.1200/JCO.2014.55.3925 [DOI] [PMC free article] [PubMed] [Google Scholar]