Abstract
The American lobster, Homarus americanus, is a model for investigating the neuromodulatory control of physiology and behavior. Prior studies have shown that multiple classes of chemicals serve as locally-released/circulating neuromodulators/neurotransmitters in this species. Interestingly, while many neuroactive compounds are known from Homarus, little work has focused on identifying/characterizing the enzymes responsible for their biosynthesis, despite the fact that these enzymes are key components for regulating neuromodulation. Here, an eyestalk ganglia-specific transcriptome was mined for transcripts encoding enzymes involved in neuropeptide, amine, diffusible gas and small molecule transmitter biosynthesis. Using known Drosophila melanogaster proteins as templates, transcripts encoding putative Homarus homologs of peptide precursor processing (signal peptide peptidase, prohormone processing protease and carboxypeptidase) and immature peptide modifying (glutaminyl cyclase, tyrosylprotein sulfotransferase, protein disulfide isomerase, peptidylglycine-α-hydroxylating monooxygenase and peptidyl-α-hydroxyglycine-α-amidating lyase) enzymes were identified in the eyestalk assembly. Similarly, transcripts encoding full complements of the enzymes responsible for dopamine (tryptophan-phenylalanine hydroxylase [TPH], tyrosine hydroxylase and DOPA decarboxylase [DDC]), octopamine (TPH, tyrosine decarboxylase and tyramine β-hydroxylase), serotonin (TPH or tryptophan hydroxylase and DDC) and histamine (histidine decarboxylase) biosynthesis were identified from the eyestalk ganglia, as were those responsible for the generation of the gases nitric oxide (nitric oxide synthase) and carbon monoxide (heme oxygenase), and the small molecule transmitters acetylcholine (choline acetyltransferase), glutamate (glutaminase) and GABA (glutamic acid decarboxylase). The presence and identity of the transcriptome-derived transcripts were confirmed using RT-PCR. The data presented here provide a foundation for future gene-based studies of neuromodulatory control at the level of neurotransmitter/modulator biosynthesis in Homarus.
Keywords: neuromodulator, neurohormone, neurotransmitter, transcriptome mining, Crustacea, Decapoda
1. Introduction
Due to its culinary notoriety, the American lobster, Homarus americanus, is one of the world’s most well-known crustaceans; portions of its nervous system, specifically the cardiac ganglion and the stomatogastric nervous system, are also well-established biomedical models that have contributed greatly to our understanding of the basic principles underlying the generation, maintenance and modulation of rhythmically active motor behaviors across the animal kingdom (e.g., Blitz and Nusbaum, 2011; Christie et al., 2010; Cooke, 2002; Dickinson et al., 2016; Fénelon et al., 2003; Harris-Warrick et al., 1992; Hooper and DiCaprio, Marder and Bucher, 2007; Marder et al., 1995; Nusbaum et al., 2001; Selverston, 2005; Selverston and Ayers, 2006; Selverston and Moulins, 1987; Selverston et al., 1998; Skiebe, 2001; Stein, 2009). One finding derived from studies conducted on the central pattern generator-effector systems of H. americanus and other decapod species is that numerically simple, “hard-wired” neural networks can produce an extensive array of distinct motor outputs via the actions of locally-released and circulating chemical compounds that act to modify the intrinsic properties of the neurons that form these circuits. This modulation of the neural circuitry provides a means by which a single neural network can be converted into a myriad of functionally distinct ones depending on what modulator or combination of modulators are influencing it.
Work from many laboratories over the past several decades has shown that a number of classes of chemical compounds serve as locally-released and/or hormonally-delivered neuromodulators/neurotransmitters in H. americanus and other decapod species (e.g., Christie, 2011). At present, the generally recognized classes of these bioactive agents include peptides, amines, diffusible gases and small molecule transmitters (e.g., Christie, 2011). Of these groups, neuropeptides are by far the largest and most diverse (e.g., Christie et al., 2010), with over 260 distinct peptides, which can be grouped into approximately 35 families, described from the H. americanus nervous system alone (e.g., Christie et al., 2015, 2017; Ma et al., 2008). Four amines (dopamine, octopamine [and/or tyramine, a biosynthetic intermediate; e.g., Jezzini et al., 2014], serotonin and histamine), two diffusible gases (nitric oxide and carbon monoxide) and three small molecule transmitters (acetylcholine, glutamate and γ-aminobutyric acid [GABA]) are also generally recognized to serve as neuromodulators/neurotransmitters in members of the Decapoda (e.g., Christie, 2011).
Regardless of class, neuromodulators are generated via enzymatic pathways of varying complexity. For example, following cleavage from their precursor proteins, many neuropeptides must undergo extensive amino acid modification before they become fully bioactive (e.g., Christie et al., 2010). This is seen for some members of the crustacean hyperglycemic hormone family, which have amino (N)-terminally cyclized glutamine residues, disulfide bridging between internal cysteine residues, and are carboxyl (C)-terminally amidated in their mature, bioactive conformations (e.g., Böcking et al., 2002). While much work has focused on identifying neuromodulators themselves in decapods, little is known about neuromodulator biosynthetic enzymes in this group of crustaceans, despite the fact that knowing the identity, diversity and regulation of these proteins is key to fully understanding the regulatory control of neuromodulation.
Recently, a transcriptome was generated for the eyestalk ganglia of H. americanus (Accession No. GFDA00000000; BioProject No. PRJNA338672; Christie et al., 2017). This assembly consists of 147,542 transcripts and was generated to serve as a resource for future molecular studies of physiological control within and by the eyestalk ganglia. Because the eyestalk ganglia of decapods, which consist of the lamina ganglionaris, medulla externa, medulla interna and medulla terminalis, are well-known sources of multiple classes of neuromodulators (e.g., Christie, 2011), we predicted that this transcriptome could be used to identify the enzymes responsible for modulator biosynthesis. As the data that follow will show, full complements of the enzymes putatively involved in the production of neuropeptides, including those responsible for precursor protein cleavage and amino acid modification of immature peptides, were identified from the eyestalk ganglia assembly. Similarly, full complements of the enzymes putatively responsible for the production of dopamine, octopamine, serotonin and histamine were identified from the eyestalk ganglia transcriptome, as were those putatively responsible for the generation of the gases nitric oxide and carbon monoxide, and the small molecule transmitters acetylcholine, glutamate and GABA. The identified sequences were confirmed using RT-PCR. Taken collectively, the data presented in this report provide a strong foundation from which to initiate gene-based studies of neuromodulatory control at the level of neurotransmitter/modulator biosynthesis in H. americanus and other decapod species.
2. Materials and methods
2.1. In silico identification of putative neuromodulator biosynthetic enzyme transcripts and proteins
2.1.1. Transcriptome mining
Searches of the H. americanus eyestalk ganglia transcriptome were conducted using methods modified from a well-established protocol that has been used previously for the identification of putative neuromodulator biosynthetic enzyme-encoding transcripts in other crustaceans (e.g., Christie et al., 2014a, 2014b; McCoole et al., 2011, 2012a, 2012b). Specifically, the database of the online program tblastn (National Center for Biotechnology Information, Bethesda, MD; http://blast.ncbi.nlm.nih.gov/Blast.cgi) was set to “Transcriptome Shotgun Assembly (TSA)” and restricted to data from eyestalk ganglia assembly, i.e., BioProject PRJNA338672 (Christie et al., 2017). Known Drosophila melanogaster enzymes were used as query sequences. The complete list of proteins searched for in this study, as well as the specific queries used, is provided in Table 1.
Table 1.
Putative Homarus americanus (Homam) neuromodulator biosynthetic enzyme transcripts/proteins identified via in silico mining of an eyestalk ganglia-specific transcriptome
| Modulator class | Target enzyme | Transcript/protein identifications | ||||||
|---|---|---|---|---|---|---|---|---|
| Biosynthetic pathway | Family | Transcript | Deduced protein | |||||
| Accession No. | Trinity No. | Length* | Name | Length† | Type | |||
| Peptide | Neuropeptide | SPP | GFDA01109857 | TR56866:c1_g1_i1 | 2105 | Homam-SPP | 376 | F |
| Neuropeptide | PPP | GFDA01097153 | TR50930:c3_g1_i1 | 5382 | Homam-PPP | 634 | F | |
| Neuropeptide | CP | GFDA01096975 | TR50890:c1_g1_i2 | 4184 | Homam-CP | 1276 | C | |
| Neuropeptide | QC | GFDA01080982 | TR42651:c7_g1_i2 | 2277 | Homam-QC | 364 | F | |
| Neuropeptide | TPST | GFDA01118134 | TR61201:c0_g1_i1 | 3856 | Homam-TPST | 362 | F | |
| Neuropeptide | PDI | GFDA01028451 | TR14457:c0_g2_i1 | 2719 | Homam-PDI-v1 | 502 | F | |
| GFDA01028450 | TR14457:c0_g1_i1 | 2698 | Homam-PDI-v2 | 495 | F | |||
| Neuropeptide | PHM | GFDA01027217 | TR13950:c1_g8_i1 | 1180 | Homam-PHM | 350 | F | |
| Neuropeptide | PAL | GFDA01113631 | TR58721:c0_g5_i2 | 5640 | Homam-PAL-I-v1 | 912 | F | |
| GFDA01113630 | TR58721:c0_g5_i1 | 5868 | Homam-PAL-I-v1 | 912 | F | |||
| GFDA01113634 | TR58721:c0_g5_i5 | 4126 | Homam-PAL-I-v2 | 881 | F | |||
| GFDA01113633 | TR58721:c0_g5_i4 | 4354 | Homam-PAL-I-v2 | 881 | F | |||
| GFDA01107659 | TR56181:c2_g6_i1 | 1853 | Homam-PAL-II | 365 | F | |||
| Amine | Dopamine Octopamine Serotonin |
TPH | GFDA01126226 | TR65054:c1_g3_i10 | 3509 | Homam-TPH-v1 | 460 | F |
| GFDA01126225 | TR65054:c1_g3_i9 | 3480 | Homam-TPH-v1 | 460 | F | |||
| GFDA01126223 | TR65054:c1_g3_i7 | 5139 | Homam-TPH-v2 | 433 | F | |||
| GFDA01126221 | TR65054:c1_g3_i5 | 4601 | Homam-TPH-v2 | 433 | F | |||
| GFDA01126219 | TR65054:c1_g3_i3 | 3218 | Homam-TPH-v3 | 462 | C | |||
| GFDA01126212 | TR65054:c1_g3_i1 | 3247 | Homam-TPH-v4 | 472 | C | |||
| Dopamine | TH | GFDA01027727 | TR13981:c0_g1_i2 | 7109 | Homam-TH | 500 | F | |
| Dopamine Serotonin |
DDC | GFDA01106633 | TR55472:c1_g1_i1 | 2678 | Homam-DDC | 476 | F | |
| Octopamine | TDC | GFDA01037688 | TR19724:c0_g1_i1 | 2971 | Homam-TDC | 652 | F | |
| Octopamine | TBH | GFDA01090865 | TR47480:c0_g1_i3 | 2371 | Homam-TBH | 607 | F | |
| GFDA01090867 | TR47480:c0_g1_i5 | 2403 | Homam-TBH | 607 | F | |||
| GFDA01090863 | TR47480:c0_g1_i1 | 2426 | Homam-TBH | 607 | F | |||
| GFDA01090868 | TR47480:c0_g1_i6 | 2466 | Homam-TBH | 607 | F | |||
| Serotonin | TRH | GFDA01007248 | TR4447:c0_g1_i1 | 2153 | Homam-TRH | 515 | F | |
| Histamine | HDC | GFDA01007849 | TR4952:c0_g1_i1 | 841 | Homam-HDC1 | 256 | N | |
| GFDA01011857 | TR6932:c0_g1_i1 | 357 | Homam-HDC1 | 357 | C | |||
| Gas | Nitric oxide | NOS | GFDA01104849 | TR54971:c4_g3_i2 | 4482 | Homam-NOS-v1 | 1197 | F |
| GFDA01104856 | TR54971:c4_g3_i9 | 4584 | Homam-NOS-v1 | 1197 | F | |||
| GFDA01104855 | TR54971:c4_g3_i8 | 4476 | Homam-NOS-v2 | 1195 | F | |||
| GFDA01104852 | TR54971:c4_g3_i5 | 4578 | Homam-NOS-v2 | 1195 | F | |||
| GFDA01104853 | TR54971:c4_g3_i6 | 4440 | Homam-NOS-v3 | 1183 | F | |||
| GFDA01104857 | TR54971:c4_g3_i10 | 4542 | Homam-NOS-v3 | 1183 | F | |||
| GFDA01104848 | TR54971:c4_g3_i1 | 1806 | Homam-NOS-v4 | 539 | F | |||
| GFDA01104854 | TR54971:c4_g3_i7 | 1838 | Homam-NOS-v4 | 539 | F | |||
| GFDA01104850 | TR54971:c4_g3_i3 | 2834 | Homam-NOS-v5 | 623 | F | |||
| GFDA01104851 | TR54971:c4_g3_i4 | 2904 | Homam-NOS-v5 | 623 | F | |||
| Carbon monoxide | HO | GFDA01001264 | TR1036:c0_g1_i1 | 1226 | Homam-HO-v1 | 264 | F | |
| GFDA01001265 | TR1036:c0_g1_i2 | 1498 | Homam-HO-v2 | 249 | F | |||
| Small molecule transmitter | Acetylcholine | CHAT | GFDA01108811 | TR56332:c0_g3_i1 | 2910 | Homam-CHAT | 746 | F |
| Glutamate | GLS | GFDA01055810 | TR28807:c0_g2_i4 | 3275 | Homam-GLS-I-v1 | 618 | F | |
| GFDA01055808 | TR28807:c0_g2_i2 | 3146 | Homam-GLS-I-v2 | 579 | F | |||
| GFDA01055809 | TR28807:c0_g2_i3 | 2823 | Homam-GLS-I-v3 | 517 | F | |||
| GFDA01055811 | TR28807:c0_g2_i5 | 2781 | Homam-GLS-I-v4 | 593 | C | |||
| GFDA01137996 | TR70838:c2_g2_i5 | 4223 | Homam-GLS-II-v1 | 858 | F | |||
| GFDA01137992 | TR70838:c2_g2_i1 | 4157 | Homam-GLS-II-v1 | 858 | F | |||
| GFDA01137998 | TR70838:c2_g2_i7 | 4192 | Homam-GLS-II-v2 | 848 | F | |||
| GFDA01137993 | TR70838:c2_g2_i2 | 4127 | Homam-GLS-II-v2 | 848 | F | |||
| GABA | GAD1 | GFDA01116953 | TR60641:c0_g1_i1 | 2921 | Homam-GAD-I | 532 | F | |
| GAD2 | GFDA01130724 | TR66607:c1_g1_i1 | 2037 | Homam-GAD-II | 501 | F | ||
Length in nucleotides
Length in amino acids.
Protein type abbreviations: F, full-length; N, amino-terminal partial; C, carboxyl-terminal partial.
The two HDC partial sequences are the N- and C-terminus of the same protein that was cloned full-length (see text).
Enzyme family abbreviations: SPP, signal peptide peptidase; PPP, prohormone processing protease; CP, carboxypeptidase; QC, glutaminyl cyclase; TPST, tyrosylprotein sulfotransferase; PDI, protein disulfide isomerase; PHM, peptidylglycine α-hydroxylating monooxygenase; PAL, peptidyl-α-hydroxyglycine-α-amidating lyase; TPH, tryptophan-phenylalanine hydroxylase; TH, tyrosine hydroxylase; DDC, DOPA decarboxylase; TDC, tyrosine decarboxylase; TBH, tyramine β-hydroxylase; TRH, tryptophan hydroxylase; HDC, histidine decarboxylase; NOS, nitric oxide synthase; HO, heme oxygenase; CHAT, choline acetyltransferase; GLS, glutaminase; GAD, glutamic acid decarboxylase.
BLAST search query proteins: SPP, Drosophila melanogaster signal peptide peptidase (Accession No. ABS20121; Casso et al., 2005); PPP, D. melanogaster prohormone and neuropeptide processing protease (Accession No. AAD49105; Siekhaus and Fuller, 1999); CP, D. melanogaster silver isoform B (Accession No. AAF45514; Adams et al., 2000); QC, D. melanogaster glutaminyl cyclase (Accession No. AAF50733; Adams et al., 2000); TPST, D. melanogaster tyrosylprotein sulfotransferase (Accession No. AAM94031; Moore, 2003); PDI, D. melanogaster protein disulfide isomerase isoform A (Accession No. AAF49659; Adams et al., 2000); PHM, D. melanogaster peptidylglycine α-hydroxylating monooxygenase (Accession No. AAB61676; Kolhekar et al., 1997); PAL, D. melanogaster peptidyl-α-hydroxyglycine-α-amidating lyase 1 isoform A (Accession No AAF58870; Adams et al., 2000); TPH, D. melanogaster phenylalanine hydroxylase (Accession No. CAA66797; RuizVazquez et al., 1996); TH, D. melanogaster tyrosine hydroxylase (Accession No. CAA53802; Neckameyer and Quinn, 1998); DDC, D. melanogaster DOPA decarboxylase isoform B (Accession No. AAF53763; Adams et al., 2000); TDC, D. melanogaster tyrosine decarboxylase 1 (Accession No. AAM70810; Adams et al., 2000); TBH, D. melanogaster tyramine β-hydroxylase isoform B (Accession No. AAO41640; Adams et al., 2000); TRH, D. melanogaster tryptophan hydroxylase (Accession No. AAF47444; Adams et al., 2000); HDC, D. melanogaster histidine decarboxylase isoform A (Accession No. AAF58823; Adams et al., 2000); NOS, D. melanogaster nitric oxide synthase isoform A (Accession No. AAF53014; Adams et al., 2000); HO, D. melanogaster heme oxygenase (Accession No. AAF54680; Adams et al., 2000); CHAT, D. melanogaster choline acetyltransferase isoform A (Accession No AAF55588; Adams et al., 2000); GLS, D. melanogaster glutaminase isoform A (Accession No. AAM68643; Adams et al., 2000); GAD, D. melanogaster glutamic acid decarboxylase 1 isoform A (Accession No. AAF47834; Adams et al., 2000) and D. melanogaster black isoform A (Accession No. AAF53337; Adams et al., 2000).
2.1.2. Protein prediction and confirmation of protein attributions
A workflow developed to vet the identification of a variety of proteins, including those involved in neuromodulator/transmitter biosynthesis (e.g., Christie et al., 2014a, 2014b; McCoole et al., 2011, 2012a, 2012b), was used to characterize the sequences deduced from the H. americanus transcripts. In brief, nucleotide sequences were translated using the “Translate” tool of ExPASy (http://web.expasy.org/translate/) and assessed for completeness. Each protein listed as “full-length” exhibits a “start” methionine and is flanked on its C-terminus by a stop codon. Proteins described here as “partial” lack either a start methionine (referred to as C-terminal partial proteins) or a stop codon (referred to as N-terminal partial proteins); the amino acid sequences of all proteins deduced from the lobster eyestalk transcriptome can be found in Supplemental Figure 1. To confirm that each of the proteins identified here is most similar to the D. melanogaster sequence used to identify the transcript encoding it, each Homarus protein was used as the input query in a BLAST search of the annotated Drosophila protein dataset present in FlyBase (version FB2016_05; Gramates et al., 2017). The arthropod protein most similar to each Homarus sequence was subsequently determined by conducting a BLAST search of the non-redundant arthropod proteins curated at NCBI (taxid:6656). Finally, protein structural motifs were analyzed for each of the H. americanus proteins using the online program Pfam version 29.0 (http://pfam.xfam.org/; Finn et al., 2016). This workflow was conducted for the deduced Homarus proteins on or before January 5, 2018.
All protein alignments were done using the online program MAFFT version 7 (http://mafft.cbrc.jp/alignment/software/; Katoh and Standley, 2013). Amino acid identity/similarity between proteins was determined using MAFFT-generated alignments. Specifically, percent identity was calculated as the number of identical amino acids divided by the total number of residues in the longest sequence (x100), while amino acid similarity was calculated as the number of identical and similar amino acids divided by the total number of residues in longest sequence (x100).
2.2. PCR confirmation of sequences
2.2.1. Animals
H. americanus (n=3) were purchased from seafood retailers in Brunswick, Maine. All lobsters were housed in recirculating natural seawater aquaria at 10–12°C and were fed chopped shrimp approximately weekly.
2.2.2. Dissection of eyestalk ganglia
To isolate eyestalk ganglia, animals were cold-anesthetized by packing in ice for approximately 20–30 min, after which the eyestalks were removed and the eyestalk ganglia were dissected from the overlying carapace and surrounding musculature in chilled (approximately 4°C) physiological saline (composition in mM/l: 479.12 NaCl, 12.74 KCl, 13.67 CaCl2, 20.00 MgSO4, 3.91 Na2SO4, 11.45 Trizma base, and 4.82 maleic acid [pH = 7.45]).
2.2.3. RNA isolation
Freshly dissected eyestalk ganglia pairs (n=3) were placed into sterile RNase-free 1.5 ml microfuge tubes containing 300 μl of TRIzol Reagent (catalog no. 15596018; Thermo Fisher Scientific Inc., Waltham, MA, USA) and manually homogenized using a sterile RNase-free disposable pestle (catalog no. 9950–901; Argos Technologies Inc., Elgin, IL, USA). RNA was isolated from the resulting homogenate using a Direct-zol RNA MiniPrep spin column system (catalog no. R2052; Zymo Research, Irvine, CA, USA) according to the manufacturer-supplied protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). All RNA samples were stored at −80°C until used for the production of cDNA.
2.2.4. cDNA synthesis
Total RNA was treated with DNase I (New England Biolabs, Ipswich, MA, USA) for 10 min at 37°C to remove contaminating genomic DNA. cDNAs were synthesized from 500 ng of total RNA using a SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, USA) according to the recommended manufacturer protocols with custom-made random pentadecamers (IDT, San Diego, CA, USA).
2.2.4. RT-PCR profiling of transcript expression in the eyestalk ganglia
Fragments of selected transcripts, approximately 500 base pair (bp) in length, were amplified from eyestalk ganglia cDNA samples using SapphireAmp Fast PCR Master Mix (Takara Bio USA, Inc., Mountain View, CA, USA) in a 20 μl reaction volume with 0.4 μl cDNA and oligonucleotide primers (Supplemental Table 1) designed to the respective transcriptomic sequences using Primer3 v2.3.7 (Rozen and Skaletsky, 2000) implemented in Geneious v10.1.3 (Biomatters Ltd., Auckland, New Zealand; Kearse et al., 2012). PCR conditions consisted of: 95°C for 2 min, then 35 cycles of 95°C for 20 s, 56°C for 20 s, and 72°C for 20 s, with a final extension at 72°C for 5 min. The resulting PCR products were electrophoresed on 1.5% agarose gels stained with SYBR Safe (Life Technologies), cloned into pCR2.1TOPO TA vector (Life Technologies), and sequenced at the Arizona State University DNA Core laboratory (Tempe, AZ, USA).
2.2.5. Amplification of complete open reading frames
Oligonucleotide primers designed to facilitate RT-PCR amplification of the complete open reading frame (ORF), or the longest possible fragment, of selected enzyme-encoding transcripts were designed with Primer 3 based on the transcriptome assembly data (Supplemental Table 1). Transcripts were amplified as described above in a 20 μl reaction volume with 0.4 μl cDNA and PCR conditions consisting of: 95°C for 2 min, then 35 cycles of 95°C for 20 s, 56°C for 20 s, and 72°C for 105 s, with a final extension at 72°C for 5 min. PCR products were cloned and sequenced as described in the preceding section, with resulting sequence data compared with the in silico assembled transcripts. Given the relative sizes of the putative nitric oxide synthase (3594 bp), choline acetyltransferase (2241 bp), and peptidyl-α-hydroxyglycine-α-amidating lyase I (2739 bp), full-length ORFs were generated via overlap extension PCR using primers encompassing the respective start and stop sites and templates consisting of the amplified transcript fragments. Amplification was performed in a 20 μl reaction volume with SapphireAmp Fast PCR Master Mix and 0.4 μl of the initial PCR products. PCR conditions consisted of: 95°C for 2 min, then 30 cycles of 95°C for 20 s, 56°C for 20 s, and 72°C for 165 s, with a final extension at 72°C for 5 min. Products were cloned and sequenced as described above.
2.3. Generation of cladograms
Cladograms were constructed to examine the phylogenetic relationship of putative H. americanus hydroxylase and decarboxylase sequences with those from a diverse set of arthropods. Phylogenetic analyses (minimum evolution, UPGMA, neighbor joining, and maximum-likelihood) were performed in MEGA 6.06 (Tamura et al., 2013) with bootstrap support based on 1000 iterations, using protein sequences either previously described/annotated (e.g., Adams et al., 2000) or identified here for the first time from publicly accessible transcriptome shotgun assembly data (Supplemental Tables 2 and 3). Analyses were performed using the maximum-likelihood method based on the Whelan and Goldman model (Whelan and Goldman, 2001). Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a Jones-Taylor-Thornton model (Jones et al., 1992). A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, hydroxylase parameter = 1.5256; decarboxylase parameter = 1.1622]). For each analysis, the tree with the highest log likelihood was shown with the percentage of trees in which the associated taxa clustered together in 1000 bootstrap iterations indicated next to the branches. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The hydroxylase analysis involved 40 amino acid sequences, whereas the decarboxylase analysis involved 50 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. The final hydroxylase dataset consisted of a total of 420 nucleotides; the final decarboxylase dataset included 456 positions. Although only the maximum-likelihood analyses have been described in the text, those results were consistent with topologies generated by the other three analyses (minimum evolution, UPGMA, and neighbor joining).
3. Results
3.1. In silico identification of neuromodulator biosynthetic enzyme-encoding transcripts/proteins
The production of the H. americanus eyestalk ganglia-specific transcriptome (Accession No. GFDA00000000) used here for neuromodulator biosynthetic enzyme transcript/protein discovery is described in detail in a previous publication (Christie et al., 2017), and is deposited in GenBank under BioProject No. PRJNA338672. In brief, the assembly was generated from eyestalk ganglia pairs collected from four lobsters, and consists of 147,542 transcripts, 87% of which are singletons. The average length of the transcripts present in the eyestalk ganglia transcriptome is 1,241 bp, with the assembly N50 length being 2,160 bp. Mapping for the RNA-Seq reads used to generate the transcriptome against the complete assembly revealed an overall alignment rate of 91%, with the majority of reads mapping only one time. As the eyestalk ganglia transcriptome was used previously, and highly successfully, for both neuropeptide (Christie et al., 2017) and circadian signaling system (Christie et al., 2018a) transcript/protein discovery, we had confidence that it would also be useful for identifying transcripts encoding the enzymes involved in neuropeptide, amine, diffusible gas and small molecule transmitter biosynthesis. The results of our searches for each of these modulator groups are presented below.
3.1.1. Neuropeptides
Peptides are by far the largest and most diverse single class of modulators present in crustacean nervous systems. In H. americanus, over 260 distinct neuropeptides, encompassing approximately 35 families, have been identified using biochemical techniques, molecular cloning, mass spectrometry, and/or in silico transcriptome mining (e.g., Christie et al., 2015, 2017; Ma et al., 2008). Mature, bioactive neuropeptides are produced from larger precursor proteins via one or more enzymatic cleavage events (e.g., Christie et al., 2010), and often, subsequent enzymatic processing of the immature peptides that results in N-terminal cyclization of glutamine/glutamic acid residues, C-terminal amidation, sulfation of tyrosine residues, and/or disulfide bridging between cysteines (e.g., Christie et al., 2010). Enzymes involved in the cleavage of peptides from their precursor proteins include signal peptide peptidase (SPP; signal peptide cleavage), prohormone processing protease (PPP; cleavage at dibasic target sites within prohormones) and carboxypeptidase (CP; removal of the C-terminal basic residues, which are exposed primarily when the prohormones are cleaved by PPP) (e.g., Christie et al., 2010). Cyclization of N-terminal glutamine/glutamic acid residues in immature peptides is achieved via the action of glutaminyl cyclase (QC)-related proteins (e.g., Christie et al., 2010). Peptides are C-terminally amidated in a two-step process involving peptidylglycine α-hydroxylating monooxygenase (PHM)- and peptidyl-α-hydroxyglycine-α-amidating lyase (PAL)-related enzymes, at least in arthropods such as Drosophila (Kolhekar et al., 1997). Sulfation of tyrosine residues is achieved via the enzymatic actions of tyrosylprotein sulfotransferase (TPST)-related proteins, while disulfide bridge formation between cysteine residues is catalyzed by protein disulfide isomerase (PDI)-like enzymes (e.g., Christie et al., 2010).
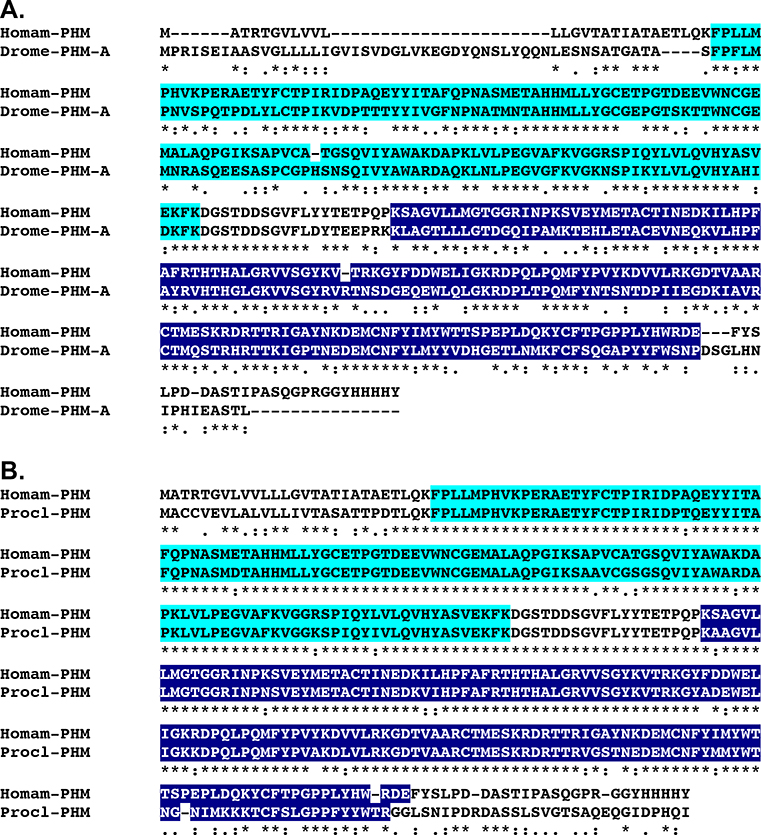
BLAST searches of the eyestalk ganglia transcriptome using known D. melanogaster proteins as the query sequences identified transcripts encoding putative homologs of each of the abovementioned neuropeptide precursor processing and immature peptide modifying enzymes (Table 1). Translation of these sequences revealed one SPP (Homam-SPP; Fig. 1), one PPP (Homam-PPP), one CP (Homam-CP), one QC (Homam-QC), one TPST (Homam-TPST), two PDIs (Homam-PDI-v1 and v2), one PHM (Homam-PHM; Fig. 2) and three PALs (Homam-PAL-I-v1 and v2 and Homam-PAL-II; Fig. 3). All of the proteins deduced from the eyestalk ganglia transcriptome appear to be full-length sequences, with the exception of Homam-CP, which is a C-terminal partial protein. However, when the portion of Homam-CP identified from the eyestalk ganglia was used to search a publicly accessible mixed neural tissue assembly (BioProject No. PRJNA300643; Northcutt et al., 2016), a transcript encoding a full-length protein was identified (Accession No. GEBG01008403), providing the missing amino acid sequence data. The two PDIs appear to be the products of a single gene, likely generated by alternative splicing in a single variable region. In contrast, the three PALs appear to represent products of two different genes (Fig. 3), with Homam-PAL-I-v1 and Homam-PAL-I-v2 differing from one another by the presence/absence of a single insertion/deletion. The presence of two distinct PAL genes has been reported previously in insects, for example in D. melanogaster (Han et al., 2004).
Figure 1.
Alignment of Homarus americanus and related signal peptide peptidases. (A) MAFFT alignment of H. americanus signal peptide peptidase (Homam-SPP) and Drosophila melanogaster signal peptide peptidase (Drome-SPP; Accession No. AAF51486; Adams et al., 2000). (B) MAFFT alignment of Homam-SPP and Daphnia pulex signal peptide peptidase (Dappu-SPP; Accession No. EFX80723; Colbourne et al., 2011). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, signal peptide peptidase domains identified by Pfam analyses are highlighted in yellow.
Figure 2.
Alignment of Homarus americanus and related peptidylglycine α-hydroxylating monooxygenases. (A) MAFFT alignment of H. americanus peptidylglycine α-hydroxylating monooxygenase (Homam-PHM) and Drosophila melanogaster peptidylglycine α-hydroxylating monooxygenase isoform A (Drome-PHM; Accession No. AAF47127; Adams et al., 2000). (B) MAFFT alignment of Homam-PHM and Procambarus clarkii peptidylglycine α-hydroxylating monooxygenase (Procl-PHM; Accession No. BAF64529; Yasuda-Kamatani and Yasuda, unpublished direct GenBank submission). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, copper type II ascorbate-dependent monooxygenase N-terminal and copper type II ascorbate-dependent monooxygenase C-terminal domains identified by Pfam analyses are highlighted in light blue and dark blue, respectively.
Figure 3.
MAFFT alignment of Homarus americanus peptidyl-α-hydroxyglycine-α-amidating lyase I variant 1 (Homam-PAL-I-v1) and peptidyl-α-hydroxyglycine-α-amidating lyase II (Homam-PAL-II). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, copper type II ascorbate-dependent monooxygenase N-terminal and copper type II ascorbate-dependent monooxygenase C-terminal domains identified by Pfam analyses are highlighted in light blue and dark blue, respectively, with NHL repeats highlighted in dark gray.
To provide increased confidence that the Homarus proteins described above represent actual members of the SPP, PPP, CP, QC, TPST, PDI, PHM and PAL families, each sequence was used to search the annotated D. melanogaster proteins in FlyBase, as well as the non-redundant arthropod proteins curated in NCBI, for its most similar homolog. Our expectations for these reciprocal BLAST searches were that each Homarus sequence would return an isoform of the relevant protein family as the top hit from each dataset. As expected, this was indeed the case for each of the FlyBase searches (Table 2). For example, D. melanogaster peptidyl-alpha-hydroxyglycine-alpha-amidating lyase 1 isoform A (Accession No. AAF58870; Adams et al., 2000) was returned as the top hit for both Homam-PAL-I-v1 and v2, with peptidyl-alpha-hydroxyglycine-alpha-amidating lyase 2 isoform A (Accession No. AAF47043; Adams et al., 2000) returned as the top hit for Homam-PAL-II. Similarly, the BLAST searches of the NCBI non-redundant arthropod protein dataset support each of the putative H. americanus SPP, PPP, CP, QC, TPST, PDI, PHM and PAL proteins deduced from the eyestalk assembly as being members of the protein families for which they were named (Table 3), e.g., peptidyl-glycine alpha-amidating monooxygenase B from the amphipod Hyalella azteca (Accession No. XP_018014827; unpublished direct GenBank submission) was returned as the top BLAST hit for both Homam-PAL-I-v1 and v2, while peptidyl-alpha-hydroxyglycine alpha-amidating lyase 2 from the springtail Orchesella cincta (Accession No. ODN01798; Faddeeva-Vakhrusheva et al., 2016) was returned as the most similar non-redundant arthropod protein to Homam-PAL-II.
Table 2.
Most similar Drosophila melanogaster protein to each predicted Homarus americanus neuromodulator biosynthetic enzyme sequence
| Homarus americanus protein | Top FlyBase D. melanogaster annotated protein hit | ||||
|---|---|---|---|---|---|
| Modulator class | Query protein | Accession No. | Name | BLAST statistics | |
| Score | E-value | ||||
| Peptide | Homam-SPP | AAF51486 | Signal peptide peptidase | 463 | 2e-130 |
| Homam-PPP | AAF56615 | Amontillado | 966 | 0.0 | |
| Homam-CP* | AAF45514 | Silver, isoform B | 832 | 0.0 | |
| Homam-QC | AAO41279 | Iso glutaminyl cyclase, isoform A | 287 | 2e-77 | |
| Homam-TPST | AAF48286 | Transport and golgi organization 13, isoform B | 473 | 2e-133 | |
| Homam-PDI-v1 | AAF49659 | Protein disulfide isomerase, isoform A | 616 | 2e-176 | |
| Homam-PDI-v2 | AAF49659 | Protein disulfide isomerase, isoform A | 614 | 1e-175 | |
| Homam-PHM | AAF47127 | Peptidylglycine-α-hydroxylating monooxygenase, isoform A | 371 | 1e-102 | |
| Homam-PAL-I-v1 | AAF58870 | Peptidyl-α-hydroxyglycine-α-amidating lyase 1, isoform A | 297 | 4e-80 | |
| Homam-PAL-I-v2 | AAF58870 | Peptidyl-α-hydroxyglycine-α-amidating lyase 1, isoform A | 298 | 3e-80 | |
| Homam-PAL-II | AAF47043 | Peptidyl-α-hydroxyglycine-α-amidating lyase 2, isoform A | 270 | 2e-72 | |
| Amine | Homam-TPH-v1 | AAF50517 | Henna, isoform A | 669 | 0.0 |
| Homam-TPH-v2 | AAF50517 | Henna, isoform A | 657 | 0.0 | |
| Homam-TPH-v3 | AAF50517 | Henna, isoform A | 666 | 0.0 | |
| Homam-TPH-v4 | AAF50517 | Henna, isoform A | 665 | 0.0 | |
| Homam-TH | AAN12080 | Pale, isoform A | 641 | 0.0 | |
| Homam-DDC | AAF53762 | DOPA decarboxylase, isoform C | 640 | 0.0 | |
| Homam-TDC | AAM70812 | Tyrosine decarboxylase 2 | 779 | 0.0 | |
| Homam-TBH | AAO41640 | Tyramine β-hydroxylase, isoform B | 471 | 1e-132 | |
| Homam-TRH | AAF47444 | Tryptophan hydroxylase | 639 | 0.0 | |
| Homam-HDC* | AAF58823 | Histidine decarboxylase, isoform A | 854 | 0.0 | |
| Gas | Homam-NOS-v1 | AAF53014 | Nitric oxide synthase, isoform A | 1361 | 0.0 |
| Homam-NOS-v2 | AAF53014 | Nitric oxide synthase, isoform A | 1364 | 0.0 | |
| Homam-NOS-v3 | AAF53014 | Nitric oxide synthase, isoform A | 1368 | 0.0 | |
| Homam-NOS-v4 | AAZ66454 | Nitric oxide synthase, isoform F | 806 | 0.0 | |
| Homam-NOS-v5 | AAF53014 | Nitric oxide synthase, isoform A | 560 | 3e-159 | |
| Homam-HO-v1 | AAF54680 | Heme oxygenase | 163 | 3e-40 | |
| Homam-HO-v2 | AAF54680 | Heme oxygenase | 162 | 5e-40 | |
| Small molecule transmitter | Homam-CHAT | AAS65177 | Choline acetyltransferase, isoform B | 522 | 7e-148 |
| Homam-GLS-I-v1 | ACZ94404 | Glutaminase, isoform H | 698 | 0.0 | |
| Homam-GLS-I-v2 | AAM68643 | Glutaminase, isoform A | 708 | 0.0 | |
| Homam-GLS-I-v3 | AAM68647 | Glutaminase, isoform B | 663 | 0.0 | |
| Homam-GLS-I-v4 | ACZ94404 | Glutaminase, isoform H | 705 | 0.0 | |
| Homam-GLS-II-v1 | AHN56158 | Glutaminase, isoform K | 617 | 2e-176 | |
| Homam-GLS-II-v2 | AHN56158 | Glutaminase, isoform K | 617 | 2e-176 | |
| Homam-GAD-I | AAF47834 | Glutamic acid decarboxylase 1, isoform A | 680 | 0.0 | |
| Homam-GAD-II | AAF53337 | Black, isoform A | 570 | 1e-162 | |
Enzyme family abbreviations: SPP, signal peptide peptidase; PPP, prohormone processing protease; CP, carboxypeptidase; QC, glutaminyl cyclase; TPST, tyrosylprotein sulfotransferase; PDI, protein disulfide isomerase; PHM, peptidylglycine α-hydroxylating monooxygenase; PAL, peptidyl-α-hydroxyglycine-α-amidating lyase; TPH, tryptophan-phenylalanine hydroxylase; TH, tyrosine hydroxylase; DDC, DOPA decarboxylase; TDC, tyrosine decarboxylase; TBH, tyramine β-hydroxylase; TRH, tryptophan hydroxylase; HDC, histidine decarboxylase; NOS, nitric oxide synthase; HO, heme oxygenase; CHAT, choline acetyltransferase; GLS, glutaminase; GAD, glutamic acid decarboxylase.
Full-length rather than the partial proteins predicted from the transcriptome used as the query sequence (see text)
Table 3.
Most similar non-redundant arthropod protein to each predicted Homarus americanus (Homam) neuromodulator biosynthetic enzyme sequence
| Homarus americanus protein | Top NCBI non-redundant arthropod protein hit | |||||
|---|---|---|---|---|---|---|
| Modulator class | Query protein | Accession No. | Species | Name | BLAST statistics | |
| Score | E-value | |||||
| Peptide | Homam-SPP | EFX80723 | Daphnia pulex | Hypothetical protein DAPPUDRAFT_303875 | 521 | 0.0 |
| Homam-PPP | AAK28328 | Faxonius limosus | PC2-like protein | 1209 | 0.0 | |
| Homam-CP* | XP_021913626 | Zootermopsis nevadensis | Carboxypeptidase D-like isoform X2 | 1304 | 0.0 | |
| Homam-QC | XP_018023992 | Hyalella azteca | Glutaminyl-peptide cyclotransferase | 355 | 7e-120 | |
| Homam-TPST | EFX85433 | Daphnia pulex | Hypothetical protein DAPPUDRAFT_209116 | 485 | 2e-170 | |
| Homam-PDI-v1 | ACN89260 | Litopenaeus vannamei | Protein disulfide isomerase | 813 | 0.0 | |
| Homam-PDI-v2 | AEE36486 | Fenneropenaeus chinensis | Protein disulfide isomerase 2 | 789 | 0.0 | |
| Homam-PHM | BAF64529 | Procambarus clarkii | Peptidylglycine α-hydroxylating monooxygenase | 587 | 0.0 | |
| Homam-PAL-I-v1 | XP_018014827 | Hyalella azteca | Peptidyl-glycine α-amidating monooxygenase B | 726 | 0.0 | |
| Homam-PAL-I-v2 | XP_018014827 | Hyalella azteca | Peptidyl-glycine α-amidating monooxygenase B | 724 | 0.0 | |
| Homam-PAL-II | ODN01798 | Orchesella cincta | Peptidyl-α-hydroxyglycine α-amidating lyase 2 | 295 | 5e-95 | |
| Amine | Homam-TPH-v1 | XP_020720433 | Bombus terrestris | Henna isoform X3 | 707 | 0.0 |
| Homam-TPH-v2 | XP_018007048 | Hyalella azteca | Henna-like | 700 | 0.0 | |
| Homam-TPH-v3 | XP_018007048 | Hyalella azteca | Henna-like | 702 | 0.0 | |
| Homam-TPH-v4 | XP_018007048 | Hyalella azteca | Henna-like | 703 | 0.0 | |
| Homam-TH | ANA78296 | Litopenaeus vannamei | Tyrosine hydroxylase | 909 | 0.0 | |
| Homam-DDC | XP_022191934 | Nilaparvata lugens | Aromatic-L-amino-acid decarboxylase isoform X1 | 708 | 0.0 | |
| Homam-TDC | XP_013789180 | Limulus polyphemus | Tyrosine decarboxylase | 852 | 0.0 | |
| Homam-TBH | XP_018012883 | Hyalella azteca | Dopamine β-hydroxylase | 749 | 0.0 | |
| Homam-TRH | XP_018008045 | Hyalella azteca | Tryptophan 5-hydroxylase 1 | 761 | 0.0 | |
| Homam-HDC* | XP_018021454 | Hyalella azteca | Histidine decarboxylase | 1013 | 0.0 | |
| Gas | Homam-NOS-v1 | ACZ60615 | Panulirus argus | Nitric oxide synthase | 2137 | 0.0 |
| Homam-NOS-v2 | ACZ60615 | Panulirus argus | Nitric oxide synthase | 2100 | 0.0 | |
| Homam-NOS-v3 | ACZ60615 | Panulirus argus | Nitric oxide synthase | 2109 | 0.0 | |
| Homam-NOS-v4 | ACZ60615 | Panulirus argus | Nitric oxide synthase | 1019 | 0.0 | |
| Homam-NOS-v5 | ACZ60615 | Panulirus argus | Nitric oxide synthase | 1041 | 0.0 | |
| Homam-HO-v1 | XP_014249133 | Cimex lectularius | Heme oxygenase 1 | 265 | 7e-87 | |
| Homam-HO-v2 | XP_014249133 | Cimex lectularius | Heme oxygenase 1 | 264 | 1e-86 | |
| Small molecule transmitter | Homam-CHAT | XP_018026302 | Hyalella azteca | Choline O-acetyltransferase | 875 | 0.0 |
| Homam-GLS-I-v1 | XP_018025541 | Hyalella azteca | Glutaminase | 791 | 0.0 | |
| Homam-GLS-I-v2 | XP_018025541 | Hyalella azteca | Glutaminase | 806 | 0.0 | |
| Homam-GLS-I-v3 | XP_018025541 | Hyalella azteca | Glutaminase | 723 | 0.0 | |
| Homam-GLS-I-v4 | XP_018025541 | Hyalella azteca | Glutaminase | 781 | 0.0 | |
| Homam-GLS-II-v1 | XP_018006525 | Hyalella azteca | Glutaminase | 810 | 0.0 | |
| Homam-GLS-II-v2 | XP_018006525 | Hyalella azteca | Glutaminase | 810 | 0.0 | |
| Homam-GAD-I | XP_018021504 | Hyalella azteca | Glutamate decarboxylase | 808 | 0.0 | |
| Homam-GAD-II | XP_021937855 | Zootermopsis nevadensis | Cysteine sulfinic acid decarboxylase isoform X1 | 652 | 0.0 | |
Enzyme family abbreviations: SPP, signal peptide peptidase; PPP, prohormone processing protease; CP, carboxypeptidase; QC, glutaminyl cyclase; TPST, tyrosylprotein sulfotransferase; PDI, protein disulfide isomerase; PHM, peptidylglycine α-hydroxylating monooxygenase; PAL, peptidyl-α-hydroxyglycine-α-amidating lyase; TPH, tryptophan-phenylalanine hydroxylase; TH, tyrosine hydroxylase; DDC, DOPA decarboxylase; TDC, tyrosine decarboxylase; TBH, tyramine β-hydroxylase; TRH, tryptophan hydroxylase; HDC, histidine decarboxylase; NOS, nitric oxide synthase; HO, heme oxygenase; CHAT, choline acetyltransferase; GLS, glutaminase; GAD, glutamic acid decarboxylase.
Full-length rather than the partial proteins predicted from the transcriptome used as the query sequence (see text)
As a final means of increasing our confidence that the Homarus SPP, PPP, CP, QC, TPST, PDI, PHM and PAL proteins reported here are actual members of the families for which they were named, each lobster protein was analyzed for structural domains using Pfam, and the identified domains were compared to those predicted by the program for the protein’s top FlyBase and NCBI arthropod non-redundant protein hits (see above). As expected, the domain complements predicted by Pfam for each Homarus sequence (Table 4) and its D. melanogaster and non-redundant arthropod protein top hit are essentially identical. For example, a single signal peptide peptidase domain was identified in Homam-SPP (Fig. 1), which was the only domain identified in the D. melanogaster (Accession No. AAF51486; Adams et al., 2000; Fig. 1A) and water flea Daphnia pulex (Accession No. EFX80723; Colbourne et al., 2011; Fig. 1B) SPPs identified as the top BLAST hits for the Homarus sequence in FlyBase and the NCBI non-redundant arthropod datasets, respectively (Fig. 1). Similarly, one copper type II ascorbate-dependent monooxygenase N-terminal domain and one copper type II ascorbate-dependent monooxygenase C-terminal domain were predicted by Pfam in Homam-PHM (Fig. 2), which is the same domain complement predicted by the program for both the D. melanogaster (Accession No. AAF47127; Adams et al., 2000; Fig. 2A) and crayfish Procambarus clarkii (Accession No. BAF64529; Yasuda-Kamatani and Yasuda, unpublished direct GenBank submission; Fig. 2B) PHMs identified as the most similar proteins in FlyBase and NCBI to the lobster sequence. Taken collectively, the protein structural domain predictions and comparisons, in conjunction with the FlyBase and non-redundant arthropod protein BLAST search results, strongly support the H. americanus SPP, PPP, CP, QC, TPST, PDI, PHM and PAL proteins identified here as being true members of their respective enzyme families.
Table 4.
Structural domains/regions identified by Pfam in predicted Homarus americanus (Homam) neuromodulator biosynthetic enzymes
| Homarus americanus protein | Identified domains/regions (amino acid coordinates) | |
|---|---|---|
| Modulator class | Name | |
| Peptide | Homam-SPP | Signal peptide peptidase (67–355) |
| Homam-PPP | Peptidase S8 pro-domain (32–108); Subtilase family (167–455); Proprotein convertase P-domain (514–601) | |
| Homam-CP* | Zinc carboxypeptidase (54–335; 489–764; 904–1001; 1242–1515); Carboxypeptidase regulatory-like domain (346–422; 775–850; 1527–1599) | |
| Homam-QC | Peptidase family M28 (130–356) | |
| Homam-TPST | Sulfotransferase family (57–250) | |
| Homam-PDI-v1 | Thioredoxin (29–133; 369–473); Thioredoxin-like domain (162–346) | |
| Homam-PDI-v2 | Thioredoxin (29–133; 369–473); Thioredoxin-like domain (162–346) | |
| Homam-PHM | Copper type II ascorbate-dependent monooxygenase, N-terminal domain (28–155); Copper type II ascorbate-dependent monooxygenase, C-terminal domain (175–324) | |
| Homam-PAL-I-v1 | Copper type II ascorbate-dependent monooxygenase, N-terminal domain (43–160); Copper type II ascorbate-dependent monooxygenase, C-terminal domain (179–331); NHL repeat (527–554; 580–608; 638–666) | |
| Homam-PAL-I-v2 | Copper type II ascorbate-dependent monooxygenase, N-terminal domain (43–160); Copper type II ascorbate-dependent monooxygenase, C-terminal domain (179–331); NHL repeat (496–523; 549–577; 607–635) | |
| Homam-PAL-II | NHL repeat (182–210; 234–260) | |
| Amine | Homam-TPH-v1 | ACT domain (44–110); Biopterin-dependent aromatic amino acid hydroxylase (127–457) |
| Homam-TPH-v2 | ACT domain (17–83); Biopterin-dependent aromatic amino acid hydroxylase (100–430) | |
| Homam-TPH-v3 | ACT domain (46–112); Biopterin-dependent aromatic amino acid hydroxylase (129–459) | |
| Homam-TPH-v4 | ACT domain (56–122); Biopterin-dependent aromatic amino acid hydroxylase (139–469) | |
| Homam-TH | Biopterin-dependent aromatic amino acid hydroxylase (165–496) | |
| Homam-DDC | Pyridoxal-dependent decarboxylase conserved domain (35–412) | |
| Homam-TDC | Pyridoxal-dependent decarboxylase conserved domain (60–438) | |
| Homam-TBH | DOMON domain (42–163); Copper type II ascorbate-dependent monooxygenase, N-terminal domain (205–334); Copper type II ascorbate-dependent monooxygenase, C-terminal domain (353–509) | |
| Homam-TRH | ACT domain (58–122); Biopterin-dependent aromatic amino acid hydroxylase (156–485) | |
| Homam-HDC* | Pyridoxal-dependent decarboxylase conserved domain (65–443) | |
| Gas | Homam-NOS-v1 | Nitric oxide synthase, oxygenase domain (55–417); Flavodoxin (467–673); FAD binding domain (727–956); Oxidoreductase NAD-binding domain (988–1100) |
| Homam-NOS-v2 | Nitric oxide synthase, oxygenase domain (55–417); Flavodoxin (467–671); FAD binding domain (725–954); Oxidoreductase NAD-binding domain (986–1098) | |
| Homam-NOS-v3 | Nitric oxide synthase, oxygenase domain (55–417); Flavodoxin (467–659); FAD binding domain (713–942); Oxidoreductase NAD-binding domain (974–1086) | |
| Homam-NOS-v4 | Nitric oxide synthase, oxygenase domain (55–417); Flavodoxin (467–539) | |
| Homam-NOS-v5 | Flavodoxin (10–99); FAD binding domain (153–382); Oxidoreductase NAD-binding domain (414–526) | |
| Homam-HO-v1 | Heme oxygenase (7–214) | |
| Homam-HO-v2 | Heme oxygenase (7–214) | |
| Small molecule transmitter | Homam-CHAT | Choline/Carnitine O-acyltransferase (87–738) |
| Homam-GLS-I-v1 | Glutaminase (192–482); Ankyrin repeats (505–605) | |
| Homam-GLS-I-v2 | Glutaminase (153–443); Ankyrin repeats (446–566) | |
| Homam-GLS-I-v3 | Glutaminase (91–381); Ankyrin repeats (383–504) | |
| Homam-GLS-I-v4 | Glutaminase (167–457); Ankyrin repeats (480–580) | |
| Homam-GLS-II-v1 | Glutaminase (439–725); Ankyrin repeats (753–848) | |
| Homam-GLS-II-v2 | Glutaminase (429–715); Ankyrin repeats (743–838) | |
| Homam-GAD-I | Pyridoxal-dependent decarboxylase conserved domain (86–456) | |
| Homam-GAD-II | Pyridoxal-dependent decarboxylase conserved domain (58–425) | |
Enzyme family abbreviations: SPP, signal peptide peptidase; PPP, prohormone processing protease; CP, carboxypeptidase; QC, glutaminyl cyclase; TPST, tyrosylprotein sulfotransferase; PDI, protein disulfide isomerase; PHM, peptidylglycine α-hydroxylating monooxygenase; PAL, peptidyl-α-hydroxyglycine-α-amidating lyase; TPH, tryptophan-phenylalanine hydroxylase; TH, tyrosine hydroxylase; 1DDC, DOPA decarboxylase; TDC, tyrosine decarboxylase; TBH, tyramine β-hydroxylase; TRH, tryptophan hydroxylase; HDC, histidine decarboxylase; NOS, nitric oxide synthase; HO, heme oxygenase; CHAT, choline acetyltransferase; GLS, glutaminase; GAD, glutamic acid decarboxylase.
Full-length rather than the partial proteins predicted from the transcriptome used as the query sequence (see text)
3.1.2. Amines
In H. americanus, four amines are generally recognized to serve as neurotransmitters/modulators, i.e., dopamine (e.g., Ballo and Bucher, 2009; Ballo et al., 2012; Bucher et al., 2003; Pulver et al., 2003), octopamine (e.g., Heinrich et al., 2000; Schneider et al., 1993, 1996), serotonin (e.g., Beltz et al., 1984; Benton et al., 2008; Ma et al., 1992) and histamine (e.g., Kwiatkowski et al., 2013; Mulloney and Hall, 1991; Pulver et al., 2003). The amino acid phenylalanine serves as the initial substrate for the production of both dopamine and octopamine (e.g., Coleman and Neckameyer, 2004; Monastirioti, 1999). In the first stage of the biosynthesis of both amines, phenylalanine is converted to tyrosine via the action of tryptophan-phenylalanine hydroxylase (TPH). For the generation of dopamine, tyrosine is converted to L-3,4-dihydroxyphenylalanine (L-DOPA) by tyrosine hydroxylase (TH), and L-DOPA is subsequently converted to dopamine by DOPA decarboxylase (DDC), alternatively known as aromatic amino acid decarboxylase. To produce octopamine, tyrosine is converted to tyramine (which itself can serve neuromodulatory roles) by tyrosine decarboxylase (TDC), with tyramine subsequently converted to octopamine by tyramine β-hydroxylase (TBH). Tryptophan is the initial substrate for the production of serotonin, with this amino acid converted to 5-hydroxytryptophan via the action of either TPH or tryptophan hydroxylase (TRH), and 5-hydroxytryptophan converted to serotonin by DDC (e.g., Coleman and Neckameyer, 2004, 2005; Monastirioti, 1999). Finally, histidine serves as the initial substrate for the production of histamine, with the former decarboxylated by the enzyme histidine decarboxylase (HDC) to produce the latter (e.g., Monastirioti, 1999; Stuart, 1999).
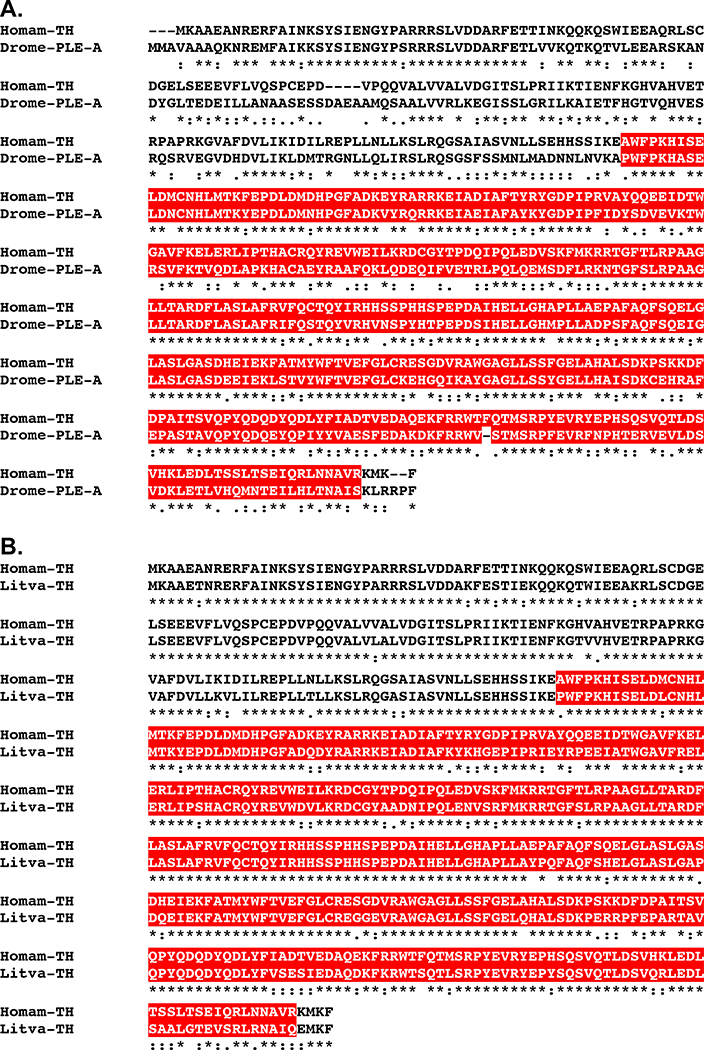
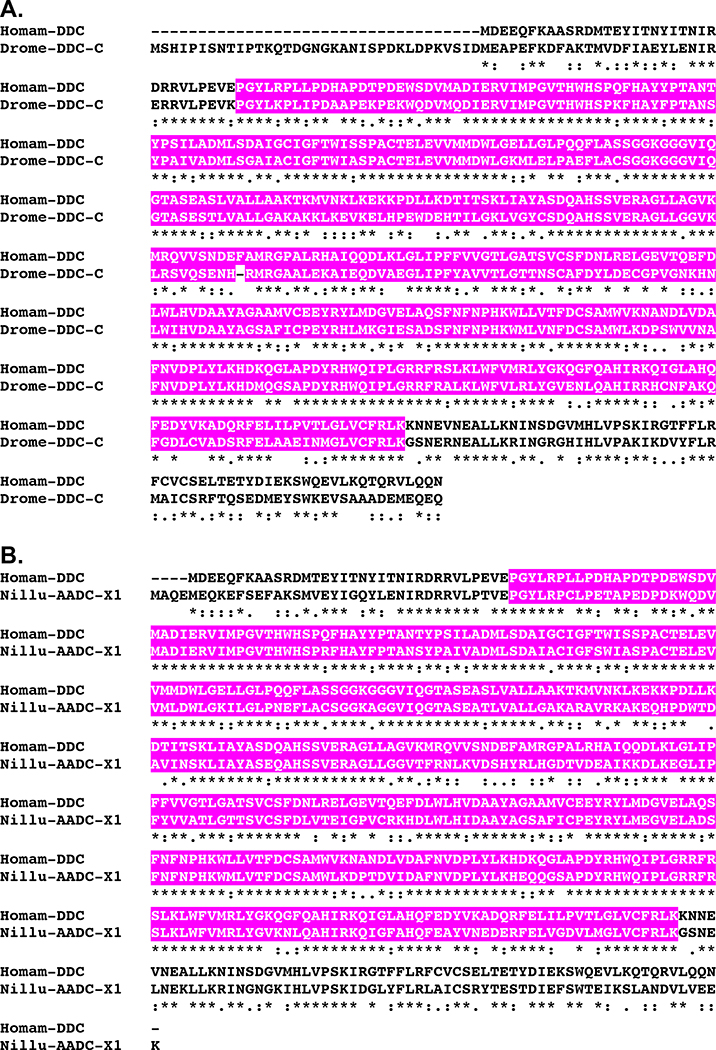
BLAST searches of the H. americanus eyestalk ganglia transcriptome using D. melanogaster proteins as the query sequences identified transcripts encoding putative homologs of each of the abovementioned amine biosynthetic enzymes (Table 1). Translation of these transcripts suggests the presence of four TPHs (Homam-TPH-v1 through v4; Fig. 4), one TH (Homam-TH; Fig. 5), one DDC (Homam-DDC; Fig. 6), one TDC (Homam-TDC), one TBH (Homam-TBH), one TRH (Homam-TRH) and one HDC (Homam-HDC) in the lobster eyestalk ganglia. The deduced proteins all appear to be full-length sequences, with the exceptions of Homam-TPH-v3 and v4, which are both C-terminal partial proteins, and Homam-HDC, for which N-terminal and C-terminal partial proteins were identified. As described below, a cDNA encoding a full-length Homam-HDC was cloned using the two fragments as templates. The four Homarus TPHs have distinct N-termini and are likely generated by alternative splicing of a single gene (Fig. 4).
Figure 4.
MAFFT alignment of Homarus americanus tryptophan-phenylalanine hydroxylase (TPH) variants. In the line immediately below each sequence grouping, “*” indicates identical amino acid residues. In this figure, ACT and biopterin-dependent aromatic amino acid hydroxylase domains identified by Pfam analyses are highlighted in light gray and red, respectively. Homam-TPH-v1 and v2 are full-length sequences, while Homam-TPH-v3 and v4 are C-terminal partial proteins.
Figure 5.
Alignment of Homarus americanus and related tyrosine hydroxylases. (A) MAFFT alignment of H. americanus tyrosine hydroxylase (Homam-TH) and Drosophila melanogaster pale, a synonym for TH, isoform A (Drome-PLE-A; Accession No. AAN12080; Adams et al., 2000). (B) MAFFT alignment of Homam-TH and Litopenaeus vannamei tyrosine hydroxylase (Litva-TH; Accession No. ANA78296; Cheng, unpublished direct GenBank submission). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, biopterin-dependent aromatic amino acid hydroxylase domains identified by Pfam analyses are highlighted in red.
Figure 6.
Alignment of Homarus americanus and related DOPA decarboxylases. (A) MAFFT alignment of H. americanus DOPA decarboxylase (Homam-DDC) and Drosophila melanogaster DOPA decarboxylase isoform C (Drome-DDC-C; Accession No. AAF53762; Adams et al., 2000). (B) MAFFT alignment of Homam-DDC and Nilaparvata lugens aromatic-L-amino-acid decarboxylase, a synonym for DDC, isoform X1 (Nillu-AADC-X1; Accession No. XP_022191934; unpublished direct GenBank submission). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, pyridoxal-dependent decarboxylase conserved domains identified by Pfam analyses are highlighted in pink.
As was the case for the neuropeptide biosynthetic enzymes, searches of FlyBase confirmed an isoform of the expected protein family as the top D. melanogaster BLAST hit for each of the H. americanus amine biosynthetic enzymes predicted from the eyestalk assembly (Table 2). For example, pale, a synonym for TH, isoform A (Accession No. AAN12080; Adams et al., 2000; Fig. 5A) was returned as the top BLAST hit for Homam-TH, while DOPA decarboxylase isoform C (Accession No. AAF53762; Adams et al., 2000; Fig. 6A) was returned as the top hit for Homam-DDC. Similarly, the top non-redundant arthropod protein hit for each of the lobster amine biosynthetic enzymes deduced from the eyestalk ganglia assembly supports the peptide family attribution ascribed to it (Table 3), e.g., tyrosine hydroxylase from the shrimp Litopenaeus vannamei (Accession No. ANA78296; Cheng, unpublished direct GenBank submission; Fig. 5B) was returned as the top hit for Homam-TH, while an aromatic-L-amino-acid decarboxylase, a synonym for DDC, from the Brown planthopper Nilaparvata lugens (Accession No. XP_022191934; unpublished direct GenBank submission; Fig. 6B) was returned as the top hit for Homam-DDC. Lastly, Pfam analyses identified structural domains in each of the putative H. americanus amine biosynthetic enzymes that further support their classification as members of their respective protein families (Table 3), e.g., a biopterin-dependent aromatic amino acid hydroxylase domain in Homam-TH and a pyridoxal-dependent decarboxylase conserved domain in Homam-DDC, domains also predicted by the program for their top D. melanogaster (Figs. 5A and 6A) and non-redundant arthropod protein (Figs. 5B and 6B) BLAST hits, respectively.
3.1.3. Gases
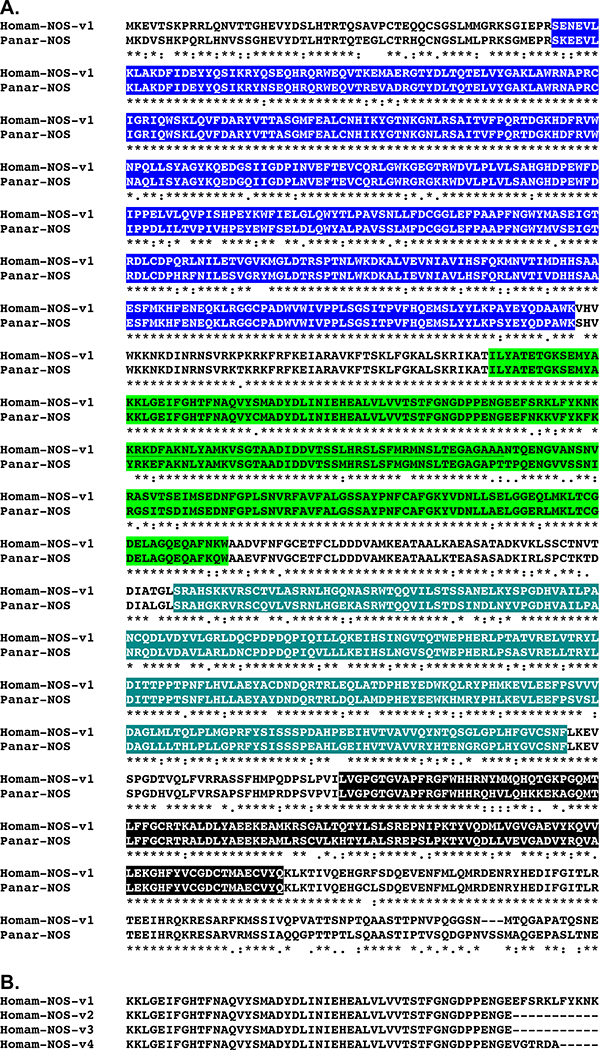
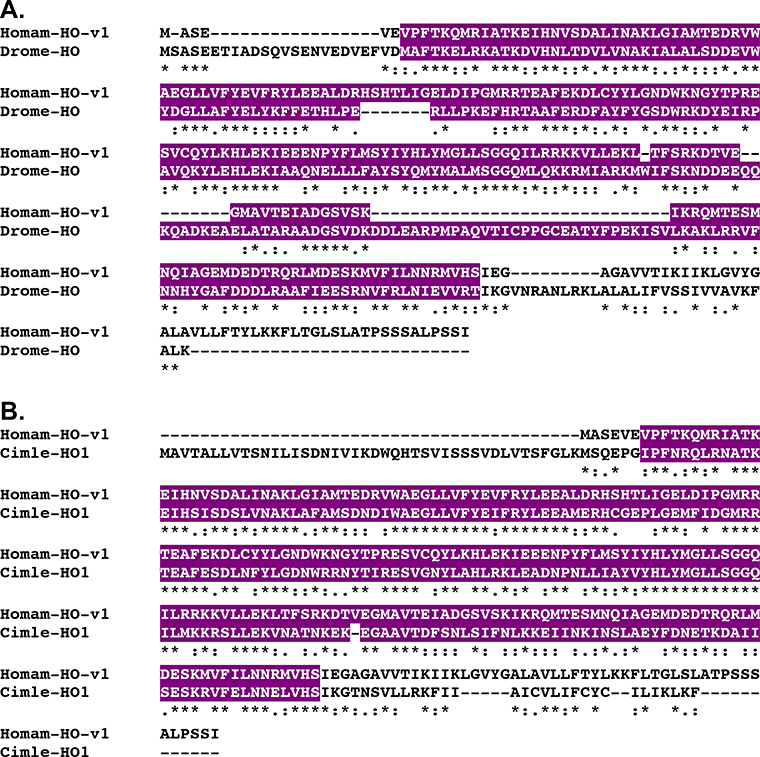
At least two diffusible gas transmitters have been shown, or are hypothesized, to exist in Astacideans, i.e., nitric oxide (e.g., Benton et al., 2007; Mahadevan et al., 2004; Scholz et al., 1998) and carbon monoxide (e.g., Christie et al., 2003). As has been reviewed extensively (e.g., Barañano et al., 2001; Barañano and Snyder, 2001; Boehning and Snyder, 2003; Mustafa et al., 2009), nitric oxide is produced during the conversion of L-arginine to L-citrulline by the enzyme nitric oxide synthase (NOS), while carbon monoxide is generated during the conversion of heme to biliverdin by heme oxygenase (HO)-like enzymes. Transcripts putatively encoding NOS- and HO-like proteins were identified from the Homarus eyestalk ganglia assembly using known D. melanogaster proteins as the query sequences (Table 1). Five full-length isoforms of NOS (Homam-NOS-v1 through v5; Fig. 7) and two isoforms of HO (Homam-HO-v1 and v2; Fig. 8) were predicted from the identified Homarus transcripts; all are full-length proteins and appear to be splice variants of single genes. Isoform diversity in Homam-NOS-v1 through v5 appears to be derived from alternative splicing that results in alternative exon usage (Homam-NOS-v1 through v4) and both N- (Homam-NOS-v5) and C-terminal (Homam-NOS-v4) truncation (Fig. 7B). Homam-HO-v2 is a C-terminally truncated version of Homam-HO-v1, missing the last 15 amino acids of the latter protein. BLAST searches of FlyBase confirmed an isoform of NOS to be the most similar D. melanogaster sequence to each of the Homarus NOSs, isoform A (Accession No. AAF53014; Adams et al., 2000) for Homam-NOS-v1 through v3 and v5, and isoform F (Accession No. AAZ66454; Adams et al., 2000) for Homam-NOS-v4 (Table 2); D. melanogaster heme oxygenase (Accession No. AAF54680; Adams et al., 2000; Fig. 8A) was returned as the top FlyBase hit for both Homam-HO-v1 and v2 (Table 2). BLAST searches of the non-redundant arthropod proteins in NCBI identified nitric oxide synthase from the spiny lobster Panulirus argus (Accession No. ACZ60615; Rodríguez-Ramos et al., 2010; Fig. 7A) as the most similar sequence to each of the Homarus NOS isoforms, while heme oxygenase 1 from the bed bug Cimex lectularius (Accession No. XP_014249133; unpublished direct GenBank submission; Fig. 8B) was returned as the top non-redundant arthropod BLAST hit for both Homam-HO-v1 and v2 (Table 3). Pfam identified one nitric oxide synthase oxygenase, one flavodoxin, one FAD binding, and one oxidoreductase NAD-binding domain in Homam-NOS-v1 (Fig. 7A), v2 and v3 (Table 4), which is the same domain complement predicted by the program for the D. melanogaster and P. argus (Fig. 7A) NOSs identified as the top FlyBase and NCBI non-redundant arthropod BLAST hits for these lobster proteins; due to their truncated nature, a subset of the domains predicted for Homam-NOS-v1 through v3 were identified by Pfam in Homam-v4 and v5 (Table 4). A single heme oxygenase domain was predicted by Pfam for both Homam-HO-v1 and v2 (Table 4), a domain also predicted by the program for their D. melanogaster (Fig. 8A) and C. lectularius (Fig. 8b) counterparts. The protein structural domain predictions/comparisons and reciprocal BLAST search results strongly support the H. americanus NOS and HO proteins identified here as being true members of their respective gas transmitter-producing enzyme families.
Figure 7.
Alignment of Homarus americanus nitric oxide synthase variants and a nitric oxide synthase from Panulirus argus. (A) MAFFT alignment of H. americanus nitric oxide synthase variant 1 (Homam-NOS-v1) and P. argus nitric oxide synthase (Panar-NOS; Accession No. ACZ60615; Rodríguez-Ramos et al., 2010). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. (B) MAFFT alignment of the variable region of the Homarus NOS variants (underlined in A). In panel A of this figure, nitric oxide synthase oxygenase, flavodoxin, FAD binding and oxidoreductase NAD-binding domains identified by Pfam analyses are highlighted in blue, light green, teal and black, respectively.
Figure 8.
Alignment of Homarus americanus and related heme oxygenases. (A) MAFFT alignment of H. americanus heme oxygenase variant 1 (Homam-HO-v1) and Drosophila melanogaster heme oxygenase (Drome-HO; Accession No. AAF54680; Adams et al., 2000). (B) MAFFT alignment of Homam-HO-v1 and Cimex lectularius heme oxygenase 1 (Cimle-HO1; Accession No. XP_014249133; unpublished direct GenBank submission). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, heme oxygenase domains identified by Pfam analyses are highlighted in violet.
3.1.4. Small molecule transmitters
Three small molecules are generally recognized to serve as neurotransmitters/modulators in H. americanus and related species: acetylcholine (e.g., Newkirk et al., 1976), glutamate (e.g., McBride et al., 1975) and GABA (e.g., Cournil et al., 1990; Ducret et al., 2007; Gutovitz et al., 2001; Le Feuvre et al., 2001). Acetylcholine is produced from choline and acetyl-CoA via the enzymatic action of choline acetyltransferase (CHAT) family members (e.g., Itoh et al., 1986), while glutamate is produced from glutamine via the action of glutaminase (GLS)-like proteins (e.g., Márquez et al., 2016). The production of GABA from glutamate is achieved by glutamic acid decarboxylase (GAD)-related proteins (e.g., Martin and Rimvall, 1993).
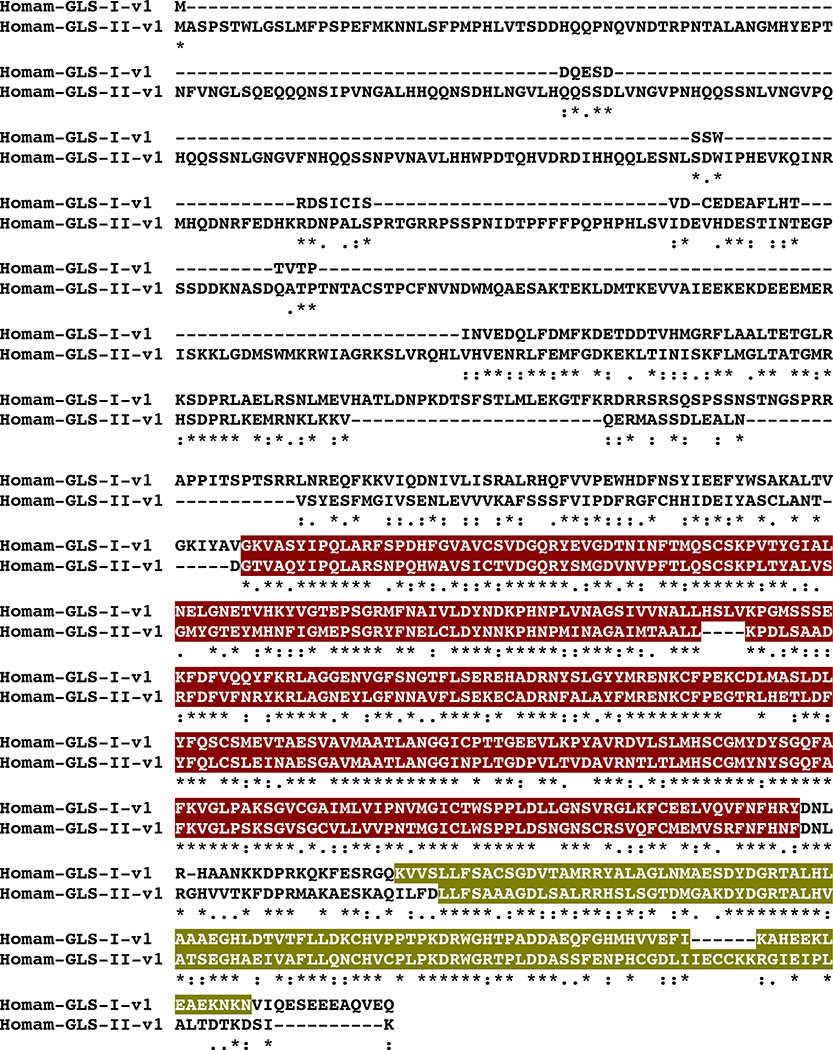
BLAST searches using known D. melanogaster isoforms of the abovementioned proteins as the query sequence identified transcripts encoding putative homologs of each enzyme in the H. americanus eyestalk ganglia transcriptome (Table 1). Translation of the identified lobster transcripts revealed one CHAT (Homam-CHAT; Fig. 9), six GLSs (Homam-GLS-I-v1 through v4 and Homam-GLS-II-v1 and v2; Fig. 10), which are likely the products of two different genes, both showing probable alternative splicing, and two GADs (Homam-GAD-I and Homam-GAD-II; Fig. 11), which are, in all likelihood, the products of different genes. With the exception of Homam-GLS-I-v4, which is a C-terminal partial protein, the deduced H. americanus small molecule transmitter biosynthetic enzymes all appear to be full-length sequences.
Figure 9.
MAFFT alignment of Homarus americanus choline acetyltransferase (Homam-CHAT) and Drosophila melanogaster choline acetyltransferase isoform B (Drome-CHAT-B; Accession No. AAS65177; Adams et al., 2000). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, choline/carnitine o-acyltransferase domains identified by Pfam analyses are highlighted in green.
Figure 10.
MAFFT alignment of Homarus americanus glutaminase I variant 1 (Homam-GLS-I-v1) and glutaminase II variant 1 (Homam-GLS-II-v1). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, glutaminase domains and ankyrin repeat regions identified by Pfam analyses are highlighted in dark red and dark yellow, respectively.
Figure 11.
MAFFT alignment of Homarus americanus glutamic acid decarboxylase I (Homam-GAD-I) and glutamic acid decarboxylase II (Homam-GAD-II). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, pyridoxal-dependent decarboxylase conserved domains identified by Pfam analyses are highlighted in pink.
BLAST searches of FlyBase identified choline acetyltransferase isoform B (Accession No. AAS65177; Adams et al., 2000; Fig. 9) as the Drosophila protein most similar to Homam-CHAT (Table 2), while isoforms of GLS were identified as the most similar Drosophila protein to each of the Homarus GLS-I and GLS-II variants (Table 2), e.g., glutaminase isoform H (Accession No. ACZ94404; Adams et al., 2000) for Homam-GLS-I-v1 and v4, and glutaminase isoform K (Accession No. AHN56158; Adams et al., 2000) for both Homam-GLS-II-v1 and v2. Glutamic acid decarboxylase 1 isoform A (Accession No AAF47834; Adams et al., 2000) and black, a synonym for GAD2, isoform A (Accession No. AAF53337; Adams et al., 2000) were returned as the top FlyBase hits for Homam-GAD-I and Homam-GAD-II, respectively (Table 2). BLAST searches of the non-redundant arthropod protein dataset curated in NCBI also returned top BLAST hits for each of the putative Homarus small molecule transmitter biosynthetic enzymes that were consistent with the protein family attributions ascribed to them (Table 3). For example, H. azteca choline O-acetyltransferase (Accession No. XP_018026302; unpublished direct GenBank submission) was identified as the most similar non-redundant arthropod protein to Homam-CHAT, while an H. azteca glutaminase (Accession No. XP_018025541; unpublished direct GenBank submission) was found to be the most similar sequence to each of the Homam-GLS-I and GLS-II variants. Finally, Pfam analyses identified domain complements in each of the putative small molecule transmitter biosynthetic enzymes that are consistent with their inclusion as true members of the CHAT, GLS or GAD families (Table 4), e.g., a choline/carnitine o-acyltransferase domain in Homam-CHAT, which is the same domain predicted by the program for its D. melanogaster counterpart (Fig. 9).
3.2. RT-PCR amplification of transcripts encoding eyestalk ganglia neuromodulatory biosynthetic enzymes
To validate the transcriptome assembly and increase our confidence that transcripts encoding the various neuromodulatory biosynthetic enzymes described above are expressed in the eyestalk ganglia, fragments (~ 500 bp) of the respective transcripts were amplified using RT-PCR and compared against the transcriptome sequences. Each of the 23 transcripts assessed was consistently amplified with varying degrees of efficiency from each of the three independent eyestalk ganglia cDNA replicates (Fig. 12; representative image). Demonstration that the PCR products exhibited >99% nucleotide identity with their respective transcriptome-derived sequences further supports eyestalk expression and accuracy of the de novo assembly.
Figure 12.
Amplification of Homarus americanus neuromodulator biosynthetic enzyme-encoding genes from eyestalk ganglia. Fragments (~ 500 bp) of the neuropeptide, amine, gas and small molecule transmitter transcripts were amplified using eyestalk ganglia cDNAs. Data shown are representative of amplification from three biological replicates. A 500-bp fragment of H. americanus actin was amplified as a positive control for cDNA template quality. Abbreviations are as follows: SPP (signal peptide peptidase); PPP (prohormone processing protease); CP (carboxypeptidase); QC (glutaminyl cyclase); TPST (tyrosylprotein sulfotransferase); PDI (protein disulfide isomerase); PHM (peptidylglycine α-hydroxylating monooxygenase); PAL-I (peptidyl-α-hydroxyglycine-α-amidating lyase I); PAL-II (peptidyl-α-hydroxyglycine-α-amidating lyase II); TPH (tryptophan-phenylalanine hydroxylase); TH (tyrosine hydroxylase); DDC (DOPA decarboxylase); TDC (tyrosine decarboxylase); TBH (tyramine β-hydroxylase); TRH (tryptophan hydroxylase); HDC (histidine decarboxylase); NOS (nitric oxide synthase); HO (heme oxygenase); CHAT (choline acetyltransferase); GLS-I (glutaminase I); GLS-II (glutaminase II); GAD-I (glutamic acid decarboxylase I); GAD-II (glutamic acid decarboxylase II); ACT (actin).
3.3. Full-length cloning of selected enzyme-encoding transcripts
As an additional means of confirming the transcripts identified from the eyestalk ganglia transcriptome, and by proxy, the proteins deduced from the transcripts, full-length ORFs for six of the transcripts were amplified from eyestalk ganglia cDNA. The transcripts targeted included those encoding the neuropeptide biosynthetic enzymes PPP, TPST and PAL-I, the amine biosynthetic enzyme TH, the gas biosynthetic enzyme NOS, and the small molecule transmitter biosynthetic enzyme CHAT. For all targets, identity between the PCR-generated consensus and transcriptome-derived nucleotide sequences exceeded 99%. The cloned sequences have been submitted to GenBank under Accession Nos. MH673284 (Homam-PPP), MH673285 (Homam-TPST), MH673286 (Homam-PAL-I), MH673287 (Homam-TH), MH673288 (Homam-NOS) and MH673289 (Homam-CHAT). RT-PCR-based ORF validation resulted in the amplification of multiple NOS variants characterized by deletions/insertions within the putative flavodoxin domain (i.e., residues 467–673) and/or deletion of a significant portion of the C-terminal FAD binding domain. Similar diversity among NOS transcripts was also observed in the transcriptome assembly and is supported by the presence of multiple mammalian NOS-encoding mRNAs (Wang et al., 1999; Alderton et al., 2001). The biological significance of multiple NOS variants within the Homarus eyestalk remains to be determined.
As stated earlier, transcripts encoding putative N-terminal and C-terminal fragments of a H. americanus histidine decarboxylase were obtained from the eyestalk ganglia assembly. We assumed that these sequences represented part of a common sequence and thus primers were generated from the partial transcripts to amplify the presumptive full-length ORF from eyestalk ganglia cDNA. The resulting full-length ORF consisted of 2,142 nt and contained 288 nt of new sequence information that spanned the gap between the 5’ and 3’ transcriptomic fragments. As above, the identity between the PCR-generated consensus and partial transcriptome-derived nucleotide sequences exceeded 99%; the cloned ORF encoding Homam-HDC has been submitted to GenBank under Accession No. MH673290.
3.4. Clade-specific clustering of Homarus hydroxylases and decarboxylases
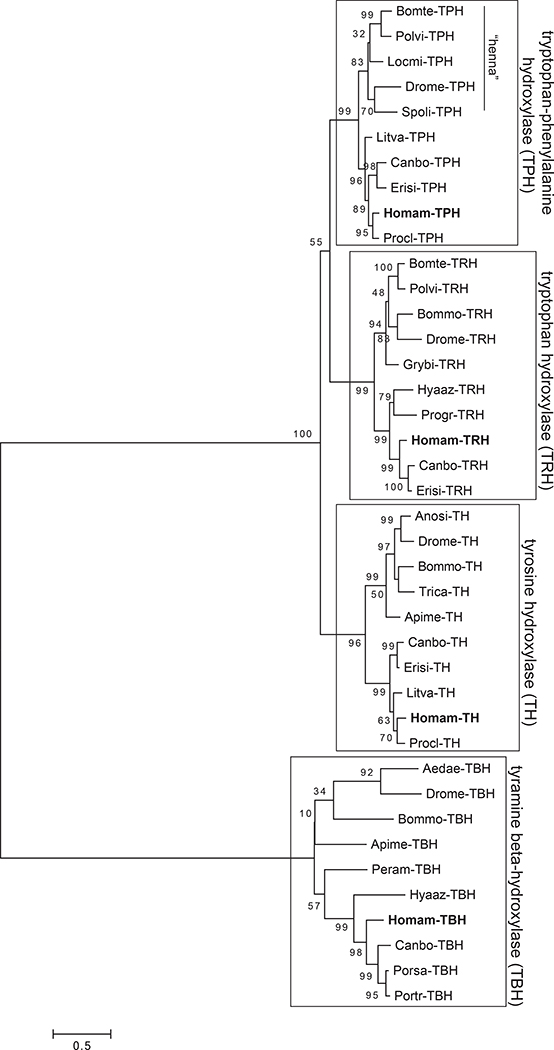
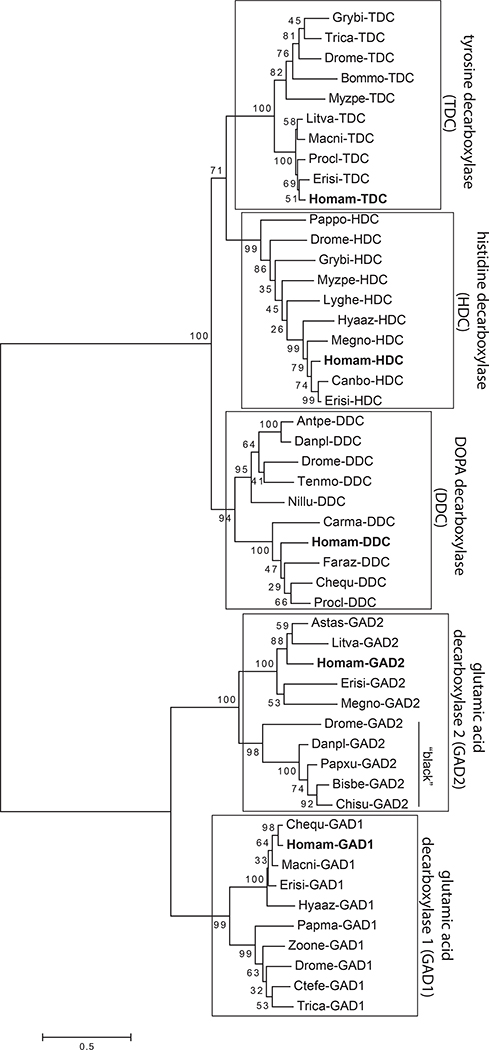
Multiple hydroxylases and decarboxylases representing diverse protein families with significant amino acid conservation were identified in the eyestalk ganglia assembly (Table 5). The initial hydroxylase/decarboxylase substrate-specific family annotations were made based on reciprocal BLAST searches of FlyBase annotated D. melanogaster proteins and non-redundant arthropod proteins in NCBI. To provide further validation for these annotations, we examined the phylogenetic relationship of the deduced H. americanus proteins with crustacean and hexapod sequences (see Supplemental Tables 2 and 3) representative of neuromodulator-associated hydroxylase families (four classes of substrate) and decarboxylase families (five classes of substrate). The respective maximum-likelihood cladograms revealed high bootstrap support for clade-specific clustering of the enzyme families, with each of the Homarus protein sequences sorting as initially annotated (Figs. 13 and 14).
Table 5.
Matrix of amino acid identity/similarity (%) among putative Homarus americanus hydroxylase and decarboxylase proteins
| DDC | TDC | HDC | GAD1 | GAD2 | ||
|---|---|---|---|---|---|---|
| 36.7/62.1 | 36.4/56.7 | 22.9/55.1 | 23.2/60.0 | DDC | ||
| TPH* | 43.4/73.9 | 20.1/47.9 | 20.2/47.1 | TDC | ||
| TH | 41.0/73.8 | 19.3/44.7 | 18.9/45.8 | HDC | ||
| TBH | 12.7/38.4 | 15.2/38.4 | 41.9/74.2 | GAD1 | ||
| TRH | 45.4/72.6 | 38.6/72.6 | 16.5/44.2 | GAD2 | ||
| TPH* | TH | TBH | TRH |
Hydroxylase abbreviations: TPH, tryptophan-phenylalanine hydroxylase; TH, tyrosine hydroxylase; TBH, tyramine β-hydroxylase; TRH, tryptophan hydroxylase.
Decarboxylase abbreviations: DDC, DOPA decarboxylase; TDC, tyrosine decarboxylase; HDC, histidine decarboxylase; GAD, glutamic acid decarboxylase.
Percent identity = number of identical amino acids shared by the two proteins/the length of the longest protein (x100); percent similarity = number of identical and similar amino acids shared by the two proteins/the length of the longest protein (x100).
TPH comparisons were done using TPH variant 1.
Figure 13.
Maximum-likelihood tree of putative tryptophan-phenylalanine hydroxylase (TPH), tyrosine hydroxylase (TH), tyramine β-hydroxylase (TBH) and tryptophan hydroxylase (TRH) sequences from diverse arthropods. The evolutionary history was inferred by using the Maximum-likelihood method based on the Whelan and Goldman model (Whelan and Goldman, 2001). The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together across 1000 bootstrap iterations is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (scale bar at bottom of figure). The Homarus americanus isoforms of TPH, TH, TBH and TRH used in the cladogram sorted with other proteins from the correct hydroxylase family. The hydroxylase sequences used to generate the cladogram, and their associated accession numbers, can be found in Supplemental Table 2. Crustacean species abbreviations: Canbo, Cancer borealis; Erisi, Eriocheir sinensis; Homam, Homarus americanus; Hyaaz, Hyalella azteca; Litva, Litopenaeus vannamei; Porsa, Portunus sanguinolentus; Portr, Portunus trituberculatus; Progr, Proasellus grafi; Procl, Procambarus clarkii. Hexapod species abbreviations: Aedae, Aedes aegypti; Anosi, Anopheles sinensis; Apime, Apis mellifera; Bomte, Bombus terrestris; Bommo, Bombyx mori; Drome, Drosophila melanogaster; Grybi, Gryllus bimaculatus; Locmi, Locusta migratoria; Peram, Periplaneta americana; Polvi, Polyrhachis vicina; Spoli, Spodoptera litura; Trica, Tribolium castaneum.
Figure 14.
Maximum-likelihood tree of putative DOPA decarboxylase (DDC), tyrosine decarboxylase (TDC), histidine decarboxylase (HDC), and glutamic acid decarboxylase (GAD) sequences from diverse arthropods. The evolutionary history was inferred by using the Maximum-likelihood method based on the Whelan and Goldman model (Whelan and Goldman, 2001). The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together across 1000 bootstrap iterations is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (scale bar at bottom of figure). The Homarus americanus isoforms of DDC, TDC, HDC, GAD used in the cladogram sorted with other proteins from the correct decarboxylase family. The decarboxylase sequences used to generate the cladogram, and their associated accession numbers, can be found in Supplemental Table 3. Crustacean species abbreviations: Astas, Astacus astacus; Canbo, Cancer borealis; Carma, Carcinus maenas; Chequ, Cherax quadricarinatus; Erisi, Eriocheir sinensis; Faraz, Farfantepenaeus aztecus; Homma, Homarus americanus; Hyaaz, Hyalella azteca; Litva, Litopenaeus vannamei; Macni, Macrobrachium nipponense; Megno, Meganyctiphanes norvegica; Procl, Procambarus clarkii. Hexapod species abbreviations: Antpe, Antheraea pernyi; Bisbe, Biston betularia; Bommo, Bombyx mori; Chisu, Chilo suppressalis; Ctefe, Ctenocephalides felis; Danpl, Danaus plexippus; Drome, Drosophila melanogaster; Grybi, Gryllus bimaculatus; Lyghe, Lygus hesperus; Myzpe, Myzus persicae; Nillu, Nilaparvata lugens; Pappo, Papilio polytes; Papma, Papilio machaon; Papxu, Papilio xuthus; Tenmo, Tenebrio molitor; Trica, Tribolium castaneum; Zoone, Zootermopsis nevadensis.
4. Discussion
4.1. Homology-based transcriptome mining as a means for rapid protein discovery in crustaceans
New technology and decreasing costs for high-throughput nucleotide sequencing has allowed for expansion of the number of deep transcriptomes for a variety of organisms, including decapod crustaceans (e.g., Cao et al., 2017; Christie et al., 2017, 2018a, 2018b; Gu et al., 2017; Li and Qian, 2017; Lu et al., 2016; Lv et al., 2017; Northcutt et al., 2016; Perina et al., 2016; Toullec et al., 2017; Waiho et al., 2017). The American lobster is one decapod for which significant molecular resources exists, as it has been the subject of several expressed sequence tag (EST) projects (Stepanyan et al., 2006; Towle and Smith, 2006) and extensive deep transcriptome sequencing (Christie et al., 2017, 2018a, 2018b; Northcutt et al., 2016). Collectively, over 500,000 nucleotide (EST and TSA) sequences for H. americanus are now publicly accessible, and hence searchable, on NCBI, providing a rich resource for gene and protein discovery in this species. While many of the H. americanus EST and TSA sequences are derived from mixed tissue RNA (e.g., gill, epipodite, branchiostegite, heart, ovary, testis, antennal gland, skeletal muscle, hepatopancreas, and brain for the ESTs described in Towle and Smith [2006]), others are tissue-specific (e.g., the nervous system for the transcriptome assembly described in Northcutt et al. [2016]) or even specific for a particular region of a tissue (e.g., the mature zone of the olfactory organ, a nervous system-associated structure, for the ESTs described in Stepanyan et al. [2006] and the brain or cardiac ganglion for the TSA assemblies described in two recent studies by Christie et al. [2018a, 2018b]). The transcriptome mined in our study was generated solely from eyestalk ganglia (Christie et al., 2017), one region of the lobster central nervous system.
Recently, we used homology-based BLAST searches of the H. americanus eyestalk ganglia transcriptome (Accession No. GFDA00000000; Christie et al., 2017) to mine for transcripts encoding putative neuropeptide precursor proteins (Christie et al., 2017) and the proteins putatively involved in the establishment of circadian pacemaking systems (Christie et al., 2018a). Both searches were highly successful, with the former resulting in the prediction of a 262-sequence peptidome for the eyestalk ganglia (Christie et al., 2017), and the latter revealing a complete set of circadian signaling system proteins (Christie et al., 2018a), including those likely involved in establishing the core circadian clock, was well as a variety of clock-associated, clock input pathway and clock output pathway proteins. Given their conservation across taxa, and the success of our earlier mining studies, we had confidence that homology-based BLAST searching of the eyestalk ganglia assembly would allow for the identification of transcripts encoding the enzymes responsible for the biosynthesis of neuromodulators, specifically those involved in the generation of neuropeptides, biogenic amines, diffusible gases and small molecule transmitters, some of which, the small molecule transmitters in particular, also are responsible for fast synaptic transmission. The templates used for our BLAST analyses were modulator biosynthetic enzymes from the fruit fly, D. melanogaster, which is one of the most thoroughly investigated arthropods in terms of its neuromodulators and their biosynthetic pathways, and a species for which extensive and well-vetted genetic, transcriptomic and proteomic datasets are readily accessible in FlyBase (Gramates et al., 2017).
4.2. Neuromodulator biosynthetic enzyme discovery in Homarus americanus
Using known Drosophila proteins as input queries, the H. americanus eyestalk transcriptome was mined for transcripts encoding putative homologs of the peptide precursor processing enzymes SPP, PPP and CP, and the immature peptide modifying enzymes QC, TPST, PDI, PHM and PAL; the enzymes responsible for the production of the biogenic amines dopamine (TPH, TH and DDC), octopamine (TPH, TDC and TBH), serotonin (TPH or TRH and DDC) and histamine (HDC) were also searched for in the eyestalk ganglia assembly. The transcriptome was also mined for transcripts encoding the enzymes responsible for the production of the gas transmitters nitric oxide (NOS) and carbon monoxide (HO), and the small molecule transmitters acetylcholine (CHAT), glutamate (GLS) and GABA (GAD). All searches returned one or more positive hits, with the identified transcripts allowing for the prediction of one or more putative protein homologs for each Drosophila query. From the peptide precursor processing and immature peptide modifying enzyme transcripts, one SPP, one PPP, one CP, one QC, one TPST, two PDIs, one PHM and three PALs were identified. From the amine biosynthetic enzyme encoding transcripts, four TPHs, one TH, one DDC, one TDC, one TBH, one TRH and one HDC were deduced. Finally, from the gas transmitter and small molecule transmitter biosynthetic enzyme transcripts, five NOSs, two HOs, one CHAT, six GLSs and two GADs were identified. Each lobster protein was vetted via reciprocal BLAST searches of the annotated D. melanogaster proteins present in FlyBase and the extant non-redundant arthropod proteins curated in NCBI; each Homarus protein was also subjected to structural domain analysis using Pfam. The collective results of these vetting steps strongly support the hypothesis that the H. americanus proteins described here are true members of their respective enzyme families. All transcripts (and, by proxy, proteins) were confirmed using RT-PCR.
4.3. Alternative splicing and multiple genes appear to contribute to protein diversity for several Homarus enzyme families
While a single H. americanus protein was predicted from the eyestalk ganglia for 13 of the 20 neuromodulator biosynthetic enzyme families searched for in our study (i.e., SPP, PPP, CP, QC, TPST, PHM, TH, DDC, TDC, TBH, TRH, HDC and CHAT), multiple proteins were predicted from this portion of the lobster CNS for the remaining seven peptide groups (i.e., PDI, PAL, TPH, NOS, HO, GLS and GAD). For PDI, TPH, NOS and HO, protein diversity is likely due to alternative splicing of single genes. In contrast, for PAL, GLS and GAD, the presence of multiple genes also appears to contribute to protein diversity, with two genes likely to encode members of each of these enzyme families in the lobster. For both PAL and GAD, two genes have also been identified in Drosophila (e.g., Han et al., 2004; Phillips et al., 2005), and searches of publicly accessible TSA datasets suggest that two genes for PAL- and GAD-like enzymes are standard for member of the Decapoda (A.E. Christie, unpublished). Although multiple isoforms of GLS are known from the fruit fly, all appear to be derived from the same gene rather than from multiple ones, which is different from the situation reported here for H. americanus. However, searches of publicly accessible, decapod TSA datasets suggest that the presence of two GLS-encoding genes is standard for members of the Decapoda (A.E. Christie, unpublished).
4.4. Biosynthetic enzyme genes/proteins: a new resource for investigating the regulation of neuromodulation/neurotransmission in Homarus americanus
Due to their numerical simplicity and the large size of their component neurons, the cardiac and stomatogastric nervous systems of decapod crustaceans, including those of the lobster H. americanus, are arguably the best understood rhythmically active neural circuits. As such, these neural networks have long served as models for investigating the basic principles underlying the generation, maintenance and modulation of rhythmically active motor behavior generally (e.g., Blitz and Nusbaum, 2011; Christie et al., 2010; Cooke, 2002; Dickinson et al., 2016; Fénelon et al., 2003; Harris-Warrick et al., 1992; Hooper and DiCaprio, Marder and Bucher, 2007; Marder et al., 1995; Nusbaum et al., 2001; Selverston, 2005; Selverston and Ayers, 2006; Selverston and Moulins, 1987; Selverston et al., 1998; Skiebe, 2001; Stein, 2009), including walking, chewing and breathing in humans. A key finding made using the cardiac and stomatogastric systems is that “hard-wired” neural networks, even very simple ones, can produce an almost infinite array of motor outputs via the actions of locally-released and circulating neuromodulators/neurotransmitters, including peptides, amines, diffusible gases and small molecule transmitters.
While much work has focused on assessment of the physiological actions of neuromodulators/neurotransmitters in the lobster, particularly on the neural networks contained within its stomatogastric and cardiac systems, essentially nothing is known about the mechanisms by which the production of these neuroactive compounds is controlled. Clearly, regulation of the enzymes involved in the biosynthesis of neuromodulators is one area that is likely to be key to understanding the regulation of neuromodulator/neurotransmitter production, and, in all likelihood, the process of neuromodulation/neurotransmission more broadly. The nucleotide and deduced amino acid sequences reported here from the eyestalk ganglia provide a powerful resource for initiating gene-based studies of neuromodulator/neurotransmitter production in H. americanus. For example, probes can be developed for in situ hybridization studies directed at determining which neurons express the various enzymes produced here, and whether or not neurons express one or both of the genes for protein groups such as PAL, GLS and GAD, for which multiple genes were identified. Similarly, reagents for knocking down the genes involved in neuromodulator/neurotransmitter production can be generated from the nucleotide sequences reported here, and the deduced protein sequences can be used to guide the production of antibodies against neuromodulator/neurotransmitter biosynthetic enzymes for use in immunohistochemical mapping studies. Finally, the nucleotide and deduced amino acid sequences reported here can be used as templates for neuromodulator/neurotransmitter biosynthetic enzyme discovery in other crustacean species, as well as members of more distantly related taxa (both invertebrate and vertebrate), potentially providing important insights into the evolution of neuromodulator/neurotransmitter biosynthetic pathways.
Supplementary Material
Amino acid sequences of putative neuromodulator biosynthetic enzymes deduced from Homarus americanus eyestalk ganglia-specific transcripts. (A) Homarus americanus signal peptide peptidase (Homam-SPP). (B) Homarus americanus prohormone processing protease (Homam-PPP). (C) Homarus americanus carboxypeptidase (Homam-CP). (D) Homarus americanus glutaminyl cyclase (Homam-QC). (E) Homarus americanus tyrosylprotein sulfotransferase (Homam-TPST). (F1-2) Homarus americanus protein disulfide isomerase (Homam-PDI) variants. (G) Homarus americanus peptidylglycine α-hydroxylating monooxygenase (Homam-PHM). (H1-3) Homarus americanus peptidyl-α-hydroxyglycine-α-amidating lyase (Homam-PAL) proteins and variants. (I1-4) Homarus americanus tryptophan-phenylalanine hydroxylase (Homam-TPH) variants. (J) Homarus americanus tyrosine hydroxylase (Homam-TH). (K) Homarus americanus DOPA decarboxylase (Homam-DDC). (L) Homarus americanus tyrosine decarboxylase (Homam-TDC). (M) Homarus americanus tyramine β-hydroxylase (Homam-TBH). (N) Homarus americanus tryptophan hydroxylase (Homam-TRH). (O1-2) Homarus americanus histidine decarboxylase (Homam-HDC) partial proteins. (P1-5) Homarus americanus nitric oxide synthase (Homam-NOS) variants. (Q1-2) Homarus americanus heme oxygenase (Homam-HO) variants. (R) Homarus americanus choline acetyltransferase (Homam-CHAT). (S1-6) Homarus americanus glutaminase (Homam-GLS) proteins and variants. (T1-2) Homarus americanus glutamic acid decarboxylase (Homam-GAD) proteins. A “+” symbol on the amino- or carboxyl-terminus indicates the presence of additional unknown amino acids.
Acknowledgements
This study was funded by the National Science Foundation (IOS-1353023 and IOS-1354567), the National Institutes of Health (5P20RR016463–12 and 8P20GM103423–12), base CRIS funding (Project #2020–22620-022–00D), the Arnold and Mabel Beckman Foundation, the Cades Foundation of Honolulu, Hawaii, and a gift from the Henry L. and Grace Doherty Charitable Foundation to Bowdoin College. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. USDA is an equal opportunity provider and employer.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Alderton WK, Cooper CE, Knowles RG (2001) Nitric oxide synthases: structure, function and inhibition. Biochemical J 357:593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballo AW, Bucher D (2009) Complex intrinsic membrane properties and dopamine shape spiking activity in a motor axon. J Neurosci 29:5062–074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballo AW, Nadim F, Bucher D (2012) Dopamine modulation of Ih improves temporal fidelity of spike propagation in an unmyelinated axon. J Neurosci 32:5106–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barañano DE, Ferris CD, Snyder SH (2001) Atypical neural messengers. Trends Neurosci 24:99–106. [DOI] [PubMed] [Google Scholar]
- Barañano DE, Snyder SH (2001) Neural roles for heme oxygenase: contrasts to nitric oxide synthase. Proc Natl Acad Sci USA 98:10996–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz B, Eisen JS, Flamm R, Harris-Warrick RM, Hooper SL, Marder E (1984) Serotonergic innervation and modulation of the stomatogastric ganglion of three decapod crustaceans (Panulirus interruptus, Homarus americanus and Cancer irroratus). J Exp Biol 109:35–54. [DOI] [PubMed] [Google Scholar]
- Benton JL, Goergen EM, Rogan SC, Beltz BS (2008) Hormonal and synaptic influences of serotonin on adult neurogenesis. Gen Comp Endocrinol 158:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton JL, Sandeman DC, Beltz BS (2007) Nitric oxide in the crustacean brain: regulation of neurogenesis and morphogenesis in the developing olfactory pathway. Dev Dyn 236:3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz DM, Nusbaum MP (2011) Neural circuit flexibility in a small sensorimotor system. Curr Opin Neurobiol 21:544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcking D, Dircksen H, Keller R (2002) The crustacean neuropeptide of the CHH/MIH/GIH family: structures and biological activities In: The Crustacean Nervous System. Wiese K (Ed.). Springer, Heidelberg: pp. 84–97. [Google Scholar]
- Boehning D, Snyder SH (2003) Novel neural modulators. Annu Rev Neurosci 26:105–131. [DOI] [PubMed] [Google Scholar]
- Bucher D, Thirumalai V, Marder E (2003) Axonal dopamine receptors activate peripheral spike initiation in a stomatogastric motor neuron. J Neurosci 23:6866–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wu L, Jin M, Li T, Hui K, Ren Q (2017) Transcriptome profiling of the Macrobrachium rosenbergii lymphoid organ under the white spot syndrome virus challenge. Fish Shellfish Immunol 67:27–39. [DOI] [PubMed] [Google Scholar]
- Casso DJ, Tanda S, Biehs B, Martoglio B, Kornberg TB (2005) Drosophila signal peptide peptidase is an essential protease for larval development. Genetics 170:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE (2011) Crustacean neuroendocrine systems and their signaling agents. Cell Tissue Res 345:41–67. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chi M, Lameyer TJ, Pascual MG, Shea DN, Stanhope ME, Schulz DJ, Dickinson PS (2015) Neuropeptidergic signaling in the American lobster Homarus americanus: new insights from high-throughput nucleotide sequencing. PLoS One 10:e0145964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Edwards JM, Cherny E, Clason TA, Graubard K (2003) Immunocytochemical evidence for nitric oxide- and carbon monoxide-producing neurons in the stomatogastric nervous system of the crayfish Cherax quadricarinatus. J Comp Neurol 467:293–306. [DOI] [PubMed] [Google Scholar]
- Christie AE, Fontanilla TM, Roncalli V, Cieslak MC, Lenz PH (2014a) Identification and developmental expression of the enzymes responsible for dopamine, histamine, octopamine and serotonin biosynthesis in the copepod crustacean Calanus finmarchicus. Gen Comp Endocrinol 195:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Fontanilla TM, Roncalli V, Cieslak MC, Lenz PH (2014b) Diffusible gas transmitter signaling in the copepod crustacean Calanus finmarchicus: identification of the biosynthetic enzymes of nitric oxide (NO), carbon monoxide (CO) and hydrogen sulfide (H2S) using a de novo assembled transcriptome. Gen Comp Endocrinol 202:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Cieslak MC, Pascual MG, Yu A, Lameyer TJ, Stanhope ME, Dickinson PS (2017) Prediction of a neuropeptidome for the eyestalk ganglia of the lobster Homarus americanus using a tissue-specific de novo assembled transcriptome. Gen Comp Endocrinol 243:96–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Stemmler EA, Dickinson PS (2010) Crustacean neuropeptides. Cell Mol Life Sci 67:4135–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Yu A, Pascual MG, Roncalli V, Cieslak MC, Warner AN, Lameyer TJ, Stanhope ME, Dickinson PS, Hull JJ (2018a) Circadian signaling in Homarus americanus: region-specific de novo assembled transcriptomes show that both the brain and eyestalk ganglia possess the molecular components of a putative clock system. Mar Genomics In press. [DOI] [PubMed] [Google Scholar]
- Christie AE, Yu A, Roncalli V, Pascual MG, Cieslak MC, Warner AN, Lameyer TJ, Stanhope ME, Dickinson PS, Hull JJ (2018b) Molecular evidence for an intrinsic circadian pacemaker in the cardiac ganglion of the American lobster, Homarus americanus - Is diel cycling of heartbeat frequency controlled by a peripheral clock system? Mar Genomics In press. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Cáceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Fröhlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kültz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL (2011) The ecoresponsive genome of Daphnia pulex. Science 331:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CM, Neckameyer WS (2004) Substrate regulation of serotonin and dopamine synthesis in Drosophila. Invert Neurosci 5:85–96. [DOI] [PubMed] [Google Scholar]
- Coleman CM, Neckameyer WS (2005) Serotonin synthesis by two distinct enzymes in Drosophila melanogaster. Arch Insect Biochem Physiol 59:12–31. [DOI] [PubMed] [Google Scholar]
- Cooke IM (2002) Reliable, responsive pacemaking and pattern generation with minimal cell numbers: the crustacean cardiac ganglion. Biol Bull 202:108–136. [DOI] [PubMed] [Google Scholar]
- Cournil I, Meyrand P, Moulins M (1990) Identification of all GABA-immunoreactive neurons projecting to the lobster stomatogastric ganglion. J Neurocytol 19:478–493. [DOI] [PubMed] [Google Scholar]
- Dickinson PS, Qu X, Stanhope ME (2016) Neuropeptide modulation of pattern-generating systems in crustaceans: comparative studies and approaches. Curr Opin Neurobiol 41:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret E, Le Feuvre Y, Meyrand P, Fénelon VS (2007) Removal of GABA within adult modulatory systems alters electrical coupling and allows expression of an embryonic-like network. J Neurosci 27:3626–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faddeeva-Vakhrusheva A, Derks MF, Anvar SY, Agamennone V, Suring W, Smit S, van Straalen NM, Roelofs D (2016) Gene Family Evolution Reflects Adaptation to Soil Environmental Stressors in the Genome of the Collembolan Orchesella cincta. Genome Biol Evol 8:2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénelon V, Le Feuvre Y, Bem T, Meyrand P (2003) Maturation of rhythmic neural network: role of central modulatory inputs. J Physiol Paris 97:59–68. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P; the FlyBase Consortium (2017) FlyBase at 25: looking to the future. Nucleic Acids Res 45:D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Fu H, Sun S, Qiao H, Zhang W, Jiang S, Xiong Y, Jin S, Gong Y, Wu Y (2017) Dietary cholesterol-induced transcriptome differences in the intestine, hepatopancreas, and muscle of Oriental River prawn Macrobrachium nipponense. Comp Biochem Physiol Part D Genomics Proteomics 23:39–48. [DOI] [PubMed] [Google Scholar]
- Gutovitz S, Birmingham JT, Luther JA, Simon DJ, Marder E (2001) GABA enhances transmission at an excitatory glutamatergic synapse. J Neurosci 21:5935–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Park D, Vanderzalm PJ, Mains RE, Eipper BA, Taghert PH (2004) Drosophila uses two distinct neuropeptide amidating enzymes, dPAL1 and dPAL2. J Neurochem 90:129–141. [DOI] [PubMed] [Google Scholar]
- Harris-Warrick RM, Marder E, Selverston AI, Moulins M (1992) Dynamic biological networks: the stomatogastric nervous system. MIT press; Cambridge. [Google Scholar]
- Heinrich R, Bräunig P, Walter I, Schneider H, Kravitz EA (2000) Aminergic neuron systems of lobsters: morphology and electrophysiology of octopamine-containing neurosecretory cells. J Comp Physiol A 186:617–629. [DOI] [PubMed] [Google Scholar]
- Hooper SL, DiCaprio RA (2004) Crustacean motor pattern generator networks. Neurosignals 13:50–69. [DOI] [PubMed] [Google Scholar]
- Itoh N, Slemmon JR, Hawke DH, Williamson R, Morita E, Itakura K, Roberts E, Shively JE, Crawford GD, Salvaterra PM (1986) Cloning of Drosophila choline acetyltransferase cDNA. Proc Natl Acad Sci USA 83:4081–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezzini SH, Reyes-Colón D, Sosa MA (2014) Characterization of a prawn OA/TA receptor in Xenopus oocytes suggests functional selectivity between octopamine and tyramine. PLoS One 9:e111314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolhekar AS, Roberts MS, Jiang N, Johnson RC, Mains RE, Eipper BA, Taghert PH (1997) Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J Neurosci 17:1363–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski MA, Gabranski ER, Huber KE, Chapline MC, Christie AE, Dickinson PS (2013) Coordination of distinct but interacting rhythmic motor programs by a modulatory projection neuron using different co-transmitters in different ganglia. J Exp Biol 216:1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Feuvre Y, Fenelon VS, Meyrand P (2001) Ontogeny of modulatory inputs to motor networks: early established projection and progressive neurotransmitter acquisition. J Neurosci 21:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GL, Qian H (2017) Transcriptome using Illumina sequencing reveals the traits of spermatogenesis and developing testes in Eriocheir sinensis. PLoS One 12:e0172478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Kong J, Luan S, Dai P, Meng X, Cao B, Luo K (2016) Transcriptome Analysis of the Hepatopancreas in the Pacific White Shrimp (Litopenaeus vannamei) under Acute Ammonia Stress. PLoS One 11:e0164396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Zhang L, Liu P, Li J (2017) Transcriptomic variation of eyestalk reveals the genes and biological processes associated with molting in Portunus trituberculatus. PLoS One 12:e0175315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, Goy MF, Christie AE, Li L (2008) Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen Comp Endocrinol 156:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PM, Beltz BS, Kravitz EA (1992) Serotonin-containing neurons in lobsters: their role as gain-setters in postural control mechanisms. J Neurophysiol 68:36–54. [DOI] [PubMed] [Google Scholar]
- Mahadevan A, Lappé J, Rhyne RT, Cruz-Bermúdez ND, Marder E, Goy MF (2004) Nitric oxide inhibits the rate and strength of cardiac contractions in the lobster Homarus americanus by acting on the cardiac ganglion. J Neurosci 24:2813–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Bucher D (2007) Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol 69:291–316. [DOI] [PubMed] [Google Scholar]
- Marder E, Christie AE, Kilman VL (1995) Functional organization of cotransmission systems: lessons from small nervous systems. Invert Neurosci 1:105–112. [DOI] [PubMed] [Google Scholar]
- Márquez J, Matés JM, Campos-Sandoval JA (2016) Glutaminases. Adv Neurobiol 13:133–171. [DOI] [PubMed] [Google Scholar]
- Martin DL, Rimvall K (1993) Regulation of gamma-aminobutyric acid synthesis in the brain. J Neurochem 60:395–407. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Freeman AR, Graham LT Jr, Aprison MH (1975) Content of amino acids in axons from the CNS of the lobster. J Neurobiol 6:321–328. [DOI] [PubMed] [Google Scholar]
- McCoole MD, Atkinson NJ, Graham DI, Grasser EB, Joselow AL, McCall NM, Welker AM, Wilsterman EJ Jr, Baer KN, Tilden AR, Christie AE (2012a) Genomic analyses of aminergic signaling systems (dopamine, octopamine and serotonin) in Daphnia pulex. Comp Biochem Physiol Part D Genomics Proteomics 7:35–58. [DOI] [PubMed] [Google Scholar]
- McCoole MD, Baer KN, Christie AE (2011) Histaminergic signaling in the central nervous system of Daphnia and a role for it in the control of phototactic behavior. J Exp Biol 214:1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoole MD, D’Andrea BT, Baer KN, Christie AE (2012b) Genomic analyses of gas (nitric oxide and carbon monoxide) and small molecule transmitter (acetylcholine, glutamate and GABA) signaling systems in Daphnia pulex. Comp Biochem Physiol Part D Genomics Proteomics 7:124–160. [DOI] [PubMed] [Google Scholar]
- Monastirioti M (1999) Biogenic amine systems in the fruit fly Drosophila melanogaster. Microsc Res Tech 45:106–121. [DOI] [PubMed] [Google Scholar]
- Moore KL (2003) The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem 278:24243–24246. [DOI] [PubMed] [Google Scholar]
- Mulloney B, Hall WM (1991) Neurons with histamine-like immunoreactivity in the segmental and stomatogastric nervous systems of the crayfish Pacifastacus leniusculus and the lobster Homarus americanus. Cell Tissue Res 266:197–207. [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Snyder SH (2009) Signaling by gasotransmitters. Sci Signal 2:re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckameyer WS, Quinn WG (1989) Isolation and characterization of the gene for Drosophila tyrosine hydroxylase. Neuron 2:1167–1175. [DOI] [PubMed] [Google Scholar]
- Newkirk RF, Ballou EW, Vickers G, Whittaker VP (1976) Comparative studies in synaptosome formation: preparation of synaptosomes from the ventral nerve cord of the lobster (Homarus americanus). Brain Res 101:103–111. [DOI] [PubMed] [Google Scholar]
- Northcutt AJ, Lett KM, Garcia VB, Diester CM, Lane BJ, Marder E, Schulz DJ (2016) Deep sequencing of transcriptomes from the nervous systems of two decapod crustaceans to characterize genes important for neural circuit function and modulation. BMC Genomics 17:868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E (2001) The roles of co-transmission in neural network modulation. Trends Neurosci 24:146–154. [DOI] [PubMed] [Google Scholar]
- Perina A, González-Tizón AM, Meilán IF, Martínez-Lage A (2016) De novo transcriptome assembly of shrimp Palaemon serratus. Genom Data 11:89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AM, Smart R, Strauss R, Brembs B, Kelly LE (2005) The Drosophila black enigma: the molecular and behavioural characterization of the black1 mutant allele. Gene 351:131–142. [DOI] [PubMed] [Google Scholar]
- Pulver SR, Thirumalai V, Richards KS, Marder E (2003) Dopamine and histamine in the developing stomatogastric system of the lobster Homarus americanus. J Comp Neurol 462:400–414. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ramos T, Carpio Y, Bolívar J, Espinosa G, Hernández-López J, Gollas-Galván T, Ramos L, Pendón C, Estrada MP (2010) An inducible nitric oxide synthase (NOS) is expressed in hemocytes of the spiny lobster Panulirus argus: cloning, characterization and expression analysis. Fish Shellfish Immunol 29:469–479. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- Ruiz-Vázquez P, Moulard M, Silva FJ (1996) Structure of the phenylalanine hydroxylase gene in Drosophila melanogaster and evidence of alternative promoter usage. Biochem Biophys Res Commun 225:238–242. [DOI] [PubMed] [Google Scholar]
- Scholz NL, Chang ES, Graubard K, Truman JW (1998) The NO/cGMP pathway and the development of neural networks in postembryonic lobsters. J Neurobiol 34:208–226. [DOI] [PubMed] [Google Scholar]
- Schneider H, Budhiraja P, Walter I, Beltz BS, Peckol E, Kravitz EA (1996) Developmental expression of the octopamine phenotype in lobsters, Homarus americanus. J Comp Neurol 371:3–14. [DOI] [PubMed] [Google Scholar]
- Schneider H, Trimmer BA, Rapus J, Eckert M, Valentine DE, Kravitz EA (1993) Mapping of octopamine-immunoreactive neurons in the central nervous system of the lobster. J Comp Neurol 329:129–142. [DOI] [PubMed] [Google Scholar]
- Selverston AI (2005) A neural infrastructure for rhythmic motor patterns. Cell Mol Neurobiol 25:223–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selverston AI, Ayers J (2006) Oscillations and oscillatory behavior in small neural circuits. Biol Cybern 95:537–554. [DOI] [PubMed] [Google Scholar]
- Selverston A, Elson R, Rabinovich M, Huerta R, Abarbanel H (1998) Basic principles for generating motor output in the stomatogastric ganglion. Ann NY Acad Sci 860:35–50. [DOI] [PubMed] [Google Scholar]
- Selverston AI, Moulins M (1987) The Crustacean Stomatogastric System. Springer; Berlin Heidelberg. [Google Scholar]
- Siekhaus DE, Fuller RS (1999) A role for amontillado, the Drosophila homolog of the neuropeptide precursor processing protease PC2, in triggering hatching behavior. J Neurosci 19:6942–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiebe P (2001) Neuropeptides are ubiquitous chemical mediators: Using the stomatogastric nervous system as a model system. J Exp Biol 204:2035–2048. [DOI] [PubMed] [Google Scholar]
- Stein W (2009) Modulation of stomatogastric rhythms. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 195:989–1009. [DOI] [PubMed] [Google Scholar]
- Stepanyan R, Day K, Urban J, Hardin DL, Shetty RS, Derby CD, Ache BW, McClintock TS (2006) Gene expression and specificity in the mature zone of the lobster olfactory organ. Physiol Genomics 25:224–233. [DOI] [PubMed] [Google Scholar]
- Stuart AE (1999) From fruit flies to barnacles, histamine is the neurotransmitter of arthropod photoreceptors. Neuron 22:431–433. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toullec JY, Corre E, Mandon P, Gonzalez-Aravena M, Ollivaux C, Lee CY (2017) Characterization of the neuropeptidome of a Southern Ocean decapod, the Antarctic shrimp Chorismus antarcticus: Focusing on a new decapod ITP-like peptide belonging to the CHH peptide family. Gen Comp Endocrinol 252:60–78. [DOI] [PubMed] [Google Scholar]
- Towle DW, Smith CM (2006) Gene discovery in Carcinus maenas and Homarus americanus via expressed sequence tags. Integr Comp Biol 46:912–918. [DOI] [PubMed] [Google Scholar]
- Waiho K, Fazhan H, Shahreza MS, Moh JH, Noorbaiduri S, Wong LL, Sinnasamy S, Ikhwanuddin M (2017) Transcriptome analysis and differential gene expression on the testis of orange mud crab, Scylla olivacea, during sexual maturation. PLoS One 12:e0171095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Newton DC, Marsden PA (1999) Neuronal NOS: gene structure, mRNA diversity, and functional relevance. Crit Rev Neurobiol 13:21–43. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences of putative neuromodulator biosynthetic enzymes deduced from Homarus americanus eyestalk ganglia-specific transcripts. (A) Homarus americanus signal peptide peptidase (Homam-SPP). (B) Homarus americanus prohormone processing protease (Homam-PPP). (C) Homarus americanus carboxypeptidase (Homam-CP). (D) Homarus americanus glutaminyl cyclase (Homam-QC). (E) Homarus americanus tyrosylprotein sulfotransferase (Homam-TPST). (F1-2) Homarus americanus protein disulfide isomerase (Homam-PDI) variants. (G) Homarus americanus peptidylglycine α-hydroxylating monooxygenase (Homam-PHM). (H1-3) Homarus americanus peptidyl-α-hydroxyglycine-α-amidating lyase (Homam-PAL) proteins and variants. (I1-4) Homarus americanus tryptophan-phenylalanine hydroxylase (Homam-TPH) variants. (J) Homarus americanus tyrosine hydroxylase (Homam-TH). (K) Homarus americanus DOPA decarboxylase (Homam-DDC). (L) Homarus americanus tyrosine decarboxylase (Homam-TDC). (M) Homarus americanus tyramine β-hydroxylase (Homam-TBH). (N) Homarus americanus tryptophan hydroxylase (Homam-TRH). (O1-2) Homarus americanus histidine decarboxylase (Homam-HDC) partial proteins. (P1-5) Homarus americanus nitric oxide synthase (Homam-NOS) variants. (Q1-2) Homarus americanus heme oxygenase (Homam-HO) variants. (R) Homarus americanus choline acetyltransferase (Homam-CHAT). (S1-6) Homarus americanus glutaminase (Homam-GLS) proteins and variants. (T1-2) Homarus americanus glutamic acid decarboxylase (Homam-GAD) proteins. A “+” symbol on the amino- or carboxyl-terminus indicates the presence of additional unknown amino acids.