Summary
Increasing drought resistance without sacrificing grain yield remains an ongoing challenge in crop improvement. In this study, we report that Oryza sativa CCCH‐tandem zinc finger protein 5 (OsTZF5) can confer drought resistance and increase grain yield in transgenic rice plants. Expression of OsTZF5 was induced by abscisic acid, dehydration and cold stress. Upon stress, OsTZF5‐GFP localized to the cytoplasm and cytoplasmic foci. Transgenic rice plants overexpressing OsTZF5 under the constitutive maize ubiquitin promoter exhibited improved survival under drought but also growth retardation. By introducing OsTZF5 behind the stress‐responsive OsNAC6 promoter in two commercial upland cultivars, Curinga and NERICA4, we obtained transgenic plants that showed no growth retardation. Moreover, these plants exhibited significantly increased grain yield compared to non‐transgenic cultivars in different confined field drought environments. Physiological analysis indicated that OsTZF5 promoted both drought tolerance and drought avoidance. Collectively, our results provide strong evidence that OsTZF5 is a useful biotechnological tool to minimize yield losses in rice grown under drought conditions.
Keywords: OsTZF5, transgenic, zinc finger protein, drought, rice, confined field
Introduction
Rice comprises 27% of total cereal utilization, with a global output of 738.2 million tons (FAO, 2016: http://fao.org/faostat/en/#home). To feed an increasing population, global rice production is forecast to increase by 26% by 2035 (Cassman et al., 2003; Seck et al., 2012). Water scarcity is a limiting factor in rice production (Manavalan et al., 2012), and drought alone is estimated to reduce worldwide rice production by 18 million tons annually (O’Toole, 2004). Drought affects approximately 23 million ha devoted to rice production under rain‐fed conditions (Huke and Huke, 1997). Not surprisingly, continued efforts have been made to generate drought‐resistant genotypes and thereby reduce the impact of water availability on rice production.
Transgenic technologies represent a promising tool for the development of stress‐tolerant crop varieties (Younis et al., 2014). Despite considerable efforts to develop drought‐resistant rice, very few attempts have managed to improve grain yield under actual field conditions (Gaudin et al., 2013; Jeong et al., 2010; Jeong et al., 2013; Selvaraj et al., 2017). This is likely because drought resistance is a complex trait not governed by a single gene, but involves a network of many genes and multiple mechanisms. Drought resistance could be enhanced if the multiple and complex mechanism crops have evolved to deal with water scarcity (Pennisi, 2008) could be introduced into commercial rice cultivars as a single trait. Unfortunately, in most cases, a specific gene is linked to a particular drought mechanism (Yue et al., 2006) and combining several mechanisms into one trait is often difficult (Zhou et al., 2016).
So far, transgenes providing increased drought resistance have been tested mostly on a single model rice variety (e.g. Nipponbare) under greenhouse conditions. By contrast, only a few have been tested in a natural hotspot and in different rice genetic backgrounds (Selvaraj et al., 2017). For an improved commercial rice variety to be accepted by consumers, it has to adapt to the target environment and fulfil local grain quality and taste preferences. These requirements should be of primary importance during transgenic studies, when the recipient genetic background is often selected based on its ability to be transformed rather than its agronomic or sensory characteristics (Gaudin et al., 2013).
Zinc finger proteins are involved in various aspects of plant growth and development, cellular functions, transcriptional regulation, RNA binding and protein–protein interactions (Ciftci‐Yilmaz and Mittler, 2008). Moreover, they play a vital role in plants’ response to biotic and abiotic stress (Ciftci‐Yilmaz et al., 2000; Mukhopadhyay et al., 2004; Sakamoto et al., 2004). Zinc finger proteins containing a tandem zinc finger (TZF) domain (two CCCH zinc fingers separated by 18 amino acids) have been documented in humans, mice and yeasts, and many of them have been associated with RNA metabolism (Al‐Souhibani et al., 2010; Carballo et al., 1998; Jeong et al., 2010; Shimada et al., 2002; Varnum et al., 1991). However, the function of most TZF genes in plants remains unknown. In Arabidopsis, PEI1 has been shown to function in embryo formation (Li and Thomas, 1998). AtTZF1 has been reported to be involved in abscisic acid (ABA)/gibberellic acid‐mediated growth and abiotic stress responses (Lin et al., 2011; Pomeranz et al., 2010), AtSZF1/AtSZF2 have been shown to confer salt stress tolerance (Sun et al., 2007), and SOMNUS has been shown to mediate light‐dependent seed germination (Kim et al., 2008). Rice OsDOS (also known as OsTZF2) has been reported to localize to nuclei and delay leaf senescence (Kong et al., 2006). Induction of OsTZF1 under abiotic stress conditions, such as drought and salt, results in delayed leaf senescence in rice (Jan et al., 2013).
In this study, we identified and characterized a novel gene, OsTZF5 (OsC3H33; Os05g0128200), which encodes a CCCH‐TZF protein and is a homolog of AtSZF1/AtSZF2 and oxidative stress 2 (OXS2) in Arabidopsis. AtOXS2 is involved in the transition from vegetative growth to the flowering stage (Blanvillain et al., 2011). OsTZF5 was first expressed during different types of controlled abiotic stress conditions in rice. We then introduced OsTZF5 into two popular upland rice cultivars in Latin America and Africa, namely Curinga and NERICA4, to evaluate the functions of this gene under actual field conditions.
To our knowledge, this is the first report whereby the newly identified OsTZF5 gene is expressed in two different transgenic rice genetic backgrounds under two different environments (managed stress and natural rain‐fed). We provide extensive evidence that expression of OsTZF5 under control of the rice stress‐responsive OsNAC6 promoter confers multiple drought resistance mechanisms in different rice genetic backgrounds.
Results
Expression profiles, sub‐cellular localization and RNA binding of OsTZF5
To identify essential genes involved in abiotic stress responses, the 44K rice microarray system was applied and the expression profile of the Nipponbare cultivar was acquired (Maruyama et al., 2012). The microarray analysis identified OsTZF5, whose expression was induced in response to different abiotic stresses, such as ABA, dehydration, NaCl and cold stress. To determine the induction patterns under different stress conditions, we applied time‐course RNA gel‐blot analysis (Figure 1a). The OsTZF5 transcript accumulated in rice seedlings within 1 h following ABA, dehydration, NaCl and cold treatments. In dehydration and NaCl stress treatment, the expression of OsTZF5 peaked after 2 h and then decreased gradually over the next 24 h (Figure 1a). In contrast, no significant accumulation of OsTZF5 transcript was detected in seedlings treated with hydrogen peroxide (H2O2) or water (H2O) (Figure 1a). To determine the organ‐specific expression of OsTZF5, total RNA was isolated from roots, shoot bases and young leaves at seedling stage, as well as mature leaves at heading stage and two different stages of the rice panicle (before or after heading). RNA gel‐blot analysis revealed that expression of OsTZF5 was high in young and mature leaves and moderate in roots, shoot bases and panicles after heading, but low in panicles before heading (Figure 1b). Transgenic rice containing a 1533‐bp OsTZF5 promoter fragment (POsTZF5) fused to the β‐glucuronidase (GUS) reporter gene exhibited GUS activity in the leaves and roots of 2‐week‐old rice seedlings, mature leaves, leaf sheath, nodes, anthers and panicles after heading (Figure 1c–h). The intensity of GUS activity was higher in response to dehydration and ABA (Figure 1i).
Figure 1.
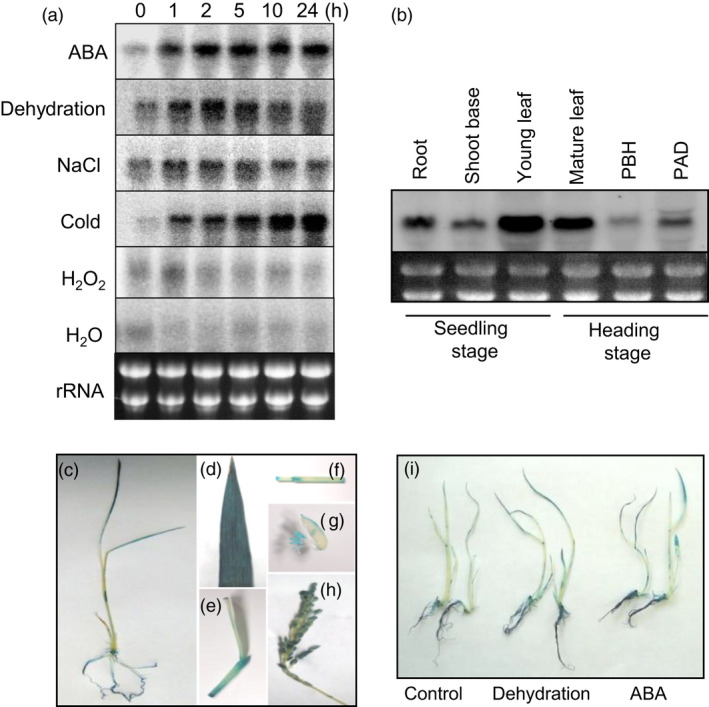
Expression profiles of OsTZF5. (a) Expression profiles of OsTZF5 under different stress and hormone treatments. Hydroponically grown 2‐week‐old rice plants were exposed to the following treatments: ABA (10 µm), dehydration, NaCl (250 mm), cold, H2O2 (10 mm) and H2O. The times for each treatment are indicated above the panels. (b) Expression profiles of OsTZF5 in different rice tissues based on RNA gel blots. For organ‐specific expression, total RNA was extracted from roots and shoot bases at seedling stage, as well as young leaves, mature leaves and panicles before heading (PBH), and panicles after heading (PAH) at heading stage. (c–h) Histochemical analysis of GUS activity provided by the POsTZF5:GUS construct in rice seedlings and different rice tissues: (c) 2‐week‐old seedlings, (d) mature leaves, (e) leaf joints, (f) longitudinal sections of node and internode, (g) panicles before heading and (h) panicle after heading. (i) Histochemical activity of POsTZF5:GUS transgenic rice plants under dehydration and ABA (10 μm) treatments.
We then determined the sub‐cellular localization of OsTZF5 using the transgenic Nipponbare plants expressing OsTZF5 fused to synthetic GFP (sGFP) under control of the OsTZF5 promoter (POsTZF5:OsTZF5‐sGFP). OsTZF5‐sGFP in untreated transgenic rice seedlings localized predominantly to the cytoplasm of root meristem cells and occasionally to cytoplasmic foci (Figure S1a), while being hardly observed in the nucleus at the young seedling stage. Incubation of transgenic seedlings with 10 µm ABA for 24 h enhanced the formation of OsTZF5‐GFP cytoplasmic foci in root cells, indicating that these foci might be stress‐related (Figure S1a). Co‐localization of marker proteins DCP2 and PABP8 in rice protoplasts confirmed the strong resemblance of these cytoplasmic foci with processing bodies (PBs) and stress granules (SGs), respectively (Figure S1b).
Given that plant TZFs have been shown to bind DNA or RNA in vitro (Pomeranz et al., 2010), we performed in vitro binding assays. The assay showed that OsTZF5 protein could only form a complex with U homopolymers and the ARE sequence, whereas GST alone could not bind to any homopolymer sequence (Figure S1c). These results demonstrated that OsTZF5 could bind to U‐rich regions as well as ARE, possibly in the 3' untranslated region of RNA.
Identification of genes regulated by OsTZF5
To examine the function of OsTZF5 in rice, transgenic rice plants overexpressing OsTZF5 driven by the maize ubiquitin promoter (PUbi:OsTZF5) were generated. The expression of PUbi:OsTZF5 in five transgenic rice lines (T2) was confirmed by RNA gel blots (Figure S2a). 44K rice microarray analysis of genes regulated by OsTZF5 in transgenic PUbi:OsTZF5 Nipponbare relative to non‐transgenic (NT) Nipponbare lines revealed 609 up‐regulated and 196 down‐regulated‐genes (Figure S2b). Among these, 171 up‐regulated genes and 43 down‐regulated genes were associated also with dehydration treatment. Up‐regulation of the representative genes PR1, salT and GolS2, and down‐regulation of OsSAM3 and flavin monooxygenase following both overexpression of OsTZF5 and dehydration treatment were confirmed by quantitative reverse transcription (qRT)‐PCR (Figure S2c).
Overexpression of OsTZF5 improves survival in a drought‐stressed environment
Given that elevated expression of OsTZF5 resulted in extremely stunted growth and poor seed setting, we selected two lines (PUbi:OsTZF5 #3 and PUbi:OsTZF5 #5) with moderate level of transgene expression (Figure S3a) for further analyses. The PUbi:OsTZF5 transgenic rice seedlings grew normally when transplanted in the soil; however, upon maturity, transgenic PUbi:OsTZF5 Nipponbare plants showed stunted growth, lower biomass (Figure S3a), reduced seed setting and underdeveloped panicles compared to NT Nipponbare. The above results highlighted how constitutive overexpression of OsTZF5 negatively affected plant growth.
We then evaluated the survival of PUbi:OsTZF5 transgenic rice plants under severe drought conditions. PUbi:OsTZF5 transgenic seedlings of lines #3 and #5 showed a survival rate of 63.6% (28/44) and 59.1% (26/44), respectively (Figure S3b), whereas NT plants exhibited a survival rate of only 16.7% (8/48), confirming the positive effect of the transgene on survival of seedlings exposed to drought.
OsTZF5 under the control of the stress‐inducible OsNAC6 promoter improves survival of rice during drought without negatively affecting growth
As mentioned above, overexpression of OsTZF5 adversely affected growth of rice plants under unstressed conditions. The stress‐inducible promoter of OsNAC6 is known to regulate expression of genes related to stress tolerance without adversely affecting rice growth (Nakashima et al., 2007, 2014). Therefore, we generated a construct, whereby the OsTZF5 gene was under the control of the OsNAC6 promoter (POsNAC6:OsTZF5) and introduced it into two commercial upland rice cultivars, NERICA4 and Curinga. No significant difference was observed in shoot dry biomass between transgenic lines and NT NERICA4 when they were grown for 2 months under unstressed (well‐watered) condition in a greenhouse (Figure S4). When the plants were subjected to survival test under drought conditions, two out of five NERICA4 transgenic lines showed greater survival rate than NT plants (Table S1). These results confirmed that OsTZF5:OsNAC6 conferred drought tolerance to rice without compromising growth during unstressed conditions.
OsNac6:OsTZF5 improves growth during drought and post‐drought recovery in the vegetative stage
To understand the effect of OsTZF5 on drought resistance, we performed field target environment (TE) trials in Santa Rosa, Colombia, and managed drought stress environment (MDSE) trials in Palmira, Colombia, using transgenic OsNAC6:OsTZF5 Curinga rice. First, we conducted a vegetative MDSE rainout shelter drought trial (vegetative MDSE‐2011) to evaluate the ability of plants at a vegetative stage to grow during drought stress period (up to 3 weeks) and recover afterwards. Soil moisture was measured at two different depths (0–20 and 20–40 cm) to confirm that uniform stress treatments were applied to all plants (Figure S5a). Six out of 13 independent POsNAC6:OsTZF5 transgenic lines displayed significantly higher shoot dry biomass than NT Curinga (Figure S5b). The plants were then rewatered to restore growth for 1 week. Five transgenic lines exhibited significantly higher biomass after recovery than NT Curinga. Enhanced and drought‐inducible expression of OsTZF5 in six representative lines (#3135, #3229, #3474, #3480, #3507 and #3508) was verified by reverse transcription PCR (RT‐PCR) (Figure S6).
Agronomic traits of transgenic OsNac6:OsTZF5 rice plants
Next, we evaluated the agronomic traits of the transgenic Curinga lines against NT lines under field conditions. In the well‐watered (WW) 2012 condition trial, we found no significant difference between transgenic and NT Curinga lines in terms of single‐plant grain yield (Figure S7), revealing that POsNAC6:OsTZF5 had no adverse effect on grain yield under well‐watered conditions. To further evaluate the agronomic yield traits of the selected six transgenic lines under drought conditions, we conducted three MDSE and two TE field trials. Drought stress was recorded with an AquaPro soil moisture sensor (Figure 2a), and the values were converted to soil matric potential (SMP) (Figure S8 and S9). In MDSE‐2013 and MDSE‐2014 trials, drought progressed rapidly in the top 20 cm of soil. Specifically, SMP fell below the permanent wilting point (PWP; −1.6 MPa) at 25 days in MDSE‐2013 and at 13 days in MDSE‐2014, after termination of irrigation (Figure S8a, b). In TE‐2012–2013 and TE‐2014–2015 trials, SMP reached even lower values (−2.35 and −3.63 MPa, respectively) at 14 and 16 days after the beginning of the rain‐free period (Figure S9a, b). In the MDSE‐2013 trial, five out of six transgenic lines flowered significantly earlier than NT Curinga (Figure 2b, Figure S10). No significant difference in panicle length was observed between transgenic and NT Curinga (Figure 2c). The single‐plant grain yield of four transgenic lines (#3229, #3474, #3507 and #3508) was significantly higher than for NT Curinga (Figure 2d, e). Interestingly, lines #3474 and #3507 also showed significantly higher grain yield than NT Curinga in MDSE‐2012 dry and TE 2012–2013 trials (Figure 3a, c). Except for MDSE‐2014 (Figure 3b), at least one or more transgenic lines produced significantly higher grain yield than NT Curinga in each trial. In particular, line #3474 exhibited consistently higher grain yield than NT Curinga in both trial environments (Figure 3a, c, d). Our drought stress experiments conducted under different environmental conditions confirmed that grain yield was elevated in transgenic plants at both single‐plant level (g/plant) and per unit grain yield (kg/ha).
Figure 2.
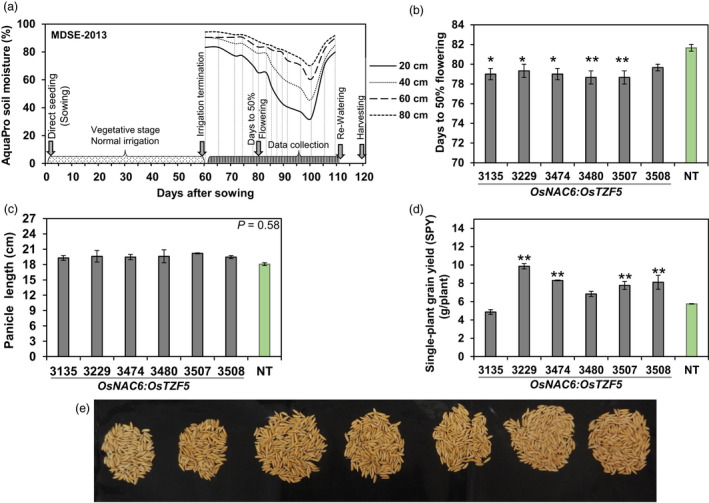
POsNAC6:OsTZF5 expressed in Curinga plants improves rice grain yield in the MDSE‐2013 trial using a rainout shelter. (a) AquaPro soil moisture profile during the drought stress period. Lines denote soil moisture at four different depths (20, 40, 60 and 80 cm). Arrowheads indicate drought stress scheduling and sampling times. (b) Variation in flowering time. (c) Panicle length. (d) Single‐plant grain yield (SPY) of Curinga transgenic lines. (e) Images showing single‐plant grain yield obtained in the MDSE trial. Each flowering time value represents the mean ± SE from three replications (at plot level). Each panicle length and single‐plant yield value represents the mean ± SE from three replications (eight plants per each replication). Asterisks denote values different from NT Curinga plants by Dunnett’s test: *P < 0.05, **P < 0.01.
Figure 3.
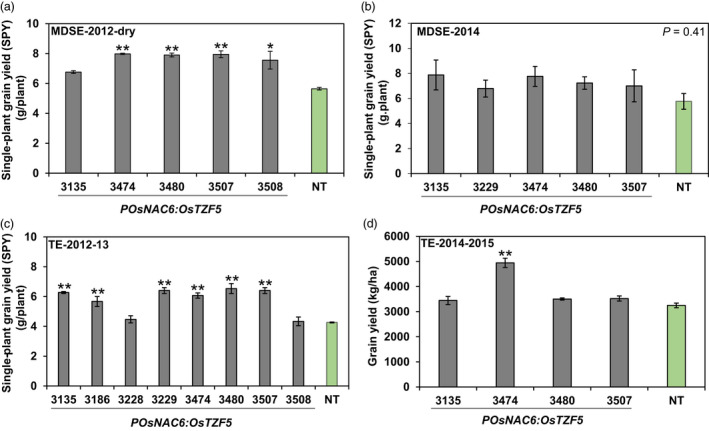
Effect of overexpressing POsNAC6:OsTZF5 on grain yield of Curinga plants under field drought conditions. The plants were grown in a MDSE trial under a rainout shelter in Palmira or in a TE trial in Santa Rosa, Colombia. Each figure indicates single‐plant grain yield in (a) MDSE‐2012, (b) MDSE‐2014 and (c) TE‐2012–2013. (d) Grain yield per area in TE‐2014–2015. Each single‐plant grain yield value represents the mean ± SE from three replications (4–8 plants per each replication). Grain yield values represent the mean ± SE from three replications (three plants per each replication). Asterisks denote values different from NT Curinga plants by Dunnett’s test: *P < 0.05, **P < 0.01.
Effect of OsTZF5 on NERICA4 under field drought conditions
To evaluate the performance of transgenic POsNAC6:OsTZF5 NERICA4 lines under drought conditions, we conducted the TE‐2015–2016 and MDSE‐2016 trials. Based on the survival rate of the transgenic lines under drought, five representative lines (#4890, #4926, #4927, #5035 and #5036) were selected for further evaluation of yield parameters (Table S1). Except for panicle length and biomass of line #4890 (MDSE‐2016), no significant differences were observed between transgenic lines and NT NERICA4 with respect to the selected traits (Table S2). Nevertheless, grain yield of two lines (#4890 and #4926) was significantly higher than that of NT NERICA4 in both trials (Figure 4a, b). Interestingly, the same lines also showed greater survival under drought stress in the greenhouse experiments (Table S1). These results indicated that the transgenic NERICA4 lines showed greater survival at seedling stage and higher grain yield under field drought conditions.
Figure 4.
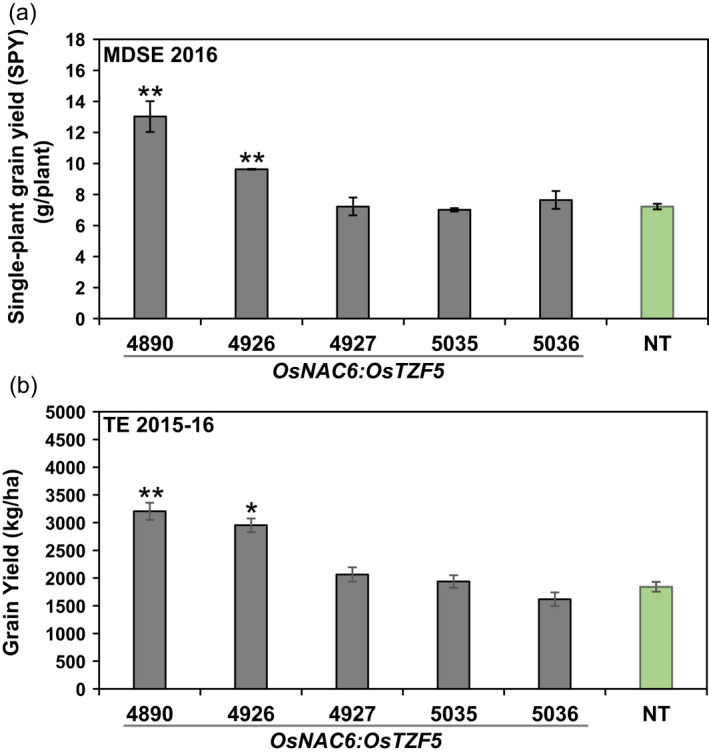
Effect of overexpressing POsNAC6:OsTZF5 on grain yield of NERICA4 plants under field drought conditions. The plants were grown in a MDSE trial under a rainout shelter in Palmira or in a TE trial in Santa Rosa, Colombia. (a) Single‐plant grain yield in MDSE‐2016. (b) Grain yield per area in TE‐2015–2016. Each single‐plant grain yield value represents the mean ± SE from three replications (five plants per each replication). Grain yield values represent the mean ± SE from three replications (three plants per each replication). Asterisks denote values different from NT NERICA4 plants by Dunnett’s test: *P < 0.05, **P < 0.01.
Physiological traits of transgenic POsNAC6:OsTZF5 Curinga lines
To evaluate the physiological traits of POsNAC6:OsTZF5 plants in response to drought, we used five promising transgenic Curinga lines (T4). The plants were subjected to a week‐long time‐course of drought stress treatments, after which relative water content (RWC), maximum photochemical efficiency of photosystem II (Fv/Fm) and relative chlorophyll content were assessed at one, two or 3 weeks. In transgenic lines, RWC decreased slowly at one and 2 weeks after stress, whereas in NT Curinga, RWC dropped much faster (Figure 5a). After 3 weeks of drought stress, the transgenic lines retained significantly higher RWC (72.6–79.3%) than the corresponding NT Curinga (63.2%).
Figure 5.
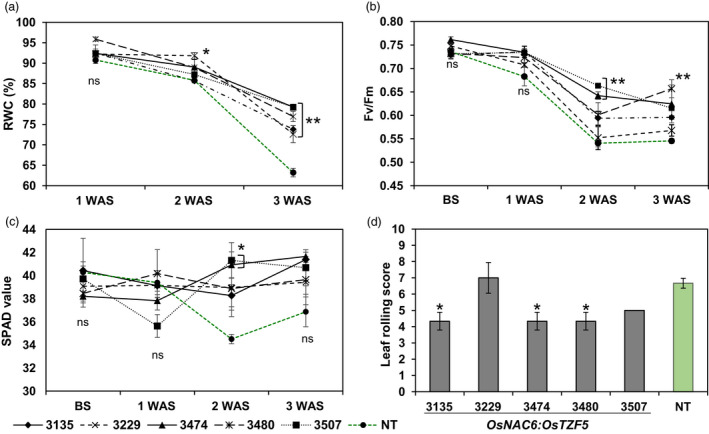
Variation in physiological parameters among transgenic POsNAC6:OsTZF5 Curinga lines in the MDSE‐2014 trial. (a) Percentage relative water content (RWC) of leaves in transgenic and NT lines at first, second and third weeks after stress (WAS). (b) Changes in chlorophyll fluorescence (Fv/Fm ) of transgenic and NT Curinga lines before stress (BS), and at first, second and third weeks after stress. (c) SPAD chlorophyll values of transgenic and NT Curinga lines before stress (BS), and at first, second and third weeks after stress. (d) Variation in leaf rolling score among transgenic lines at peak stress (4 weeks after stress). The value of leaf rolling score of #3507 was eliminated from statistical analysis, since no variant was found in this line. Each RWC and leaf rolling score parameter represents the mean ± SE from three replications (three plants per each replication). Each Fv/Fm and SPAD values represents the mean ± SE from three replications (nine plants per each replication). Asterisks in (a), (b) and (c) denote values different from NT Curinga plants by Dunnett’s test: *P < 0.05, **P < 0.01. P‐value indicated in (d) was calculated with one‐factor ANOVA. Dunnett's test detected no significant differences in leaf rolling score of transgenic lines from NT Curinga.
To further estimate the stress status of transgenic plants, Fv/Fm values were measured before and during the stress period (Figure 5b). The Fv/Fm values of transgenic POsNAC6:OsTZF5 and NT Curinga showed no significant difference at 1 week after stress, and values were generally lower at 2 weeks. Only one transgenic line (#3480) showed significantly higher Fv/Fm values compared to NT Curinga at 3 weeks after stress (Figure 5b). Under normal growth conditions, Fv/Fm values showed no significant differences between transgenic lines and NT Curinga, and they ranged from 0.73 to 0.76.
The relative chlorophyll content in transgenic and NT Curinga plants was measured at one, two and 3 weeks after stress treatments (Figure 5c). At 2 weeks, chlorophyll content was significantly higher in two transgenic lines (#3135 and #3474) than in NT plants. Before and after that point, however, no significant differences could be detected between transgenic and NT Curinga plants. The transgenic lines tended to display lower leaf rolling scores compared to NT Curinga under stress conditions (Figure 5d).
Discussion
Several reports have demonstrated that zinc finger proteins are involved in plant growth, development and stress responses through transcriptional regulation, RNA binding and protein–protein interactions (Bogamuwa and Jang, 2014; Ciftci‐Yilmaz and Mittler, 2008). In this study, we identified a novel stress‐responsive CCCH‐tandem zinc finger protein, OsTZF5, and generated transgenic rice plants overexpressing OsTZF5 with improved performance under field drought conditions. We report the molecular, agronomic and physiological characteristics of drought‐resistant OsTZF5 overexpressing transgenic rice.
Characterization of rice plants overexpressing OsTZF5
RNA gel‐blot analysis and histochemical GUS activity assay of transgenic POsTZF5:GUS plants confirmed OsTZF5 expression in all examined tissues (Figure 1) and revealed that OsTZF5 was induced by multiple abiotic stresses. Microscopic observation revealed that OsTZF5 localized to cytoplasmic foci resembling PBs and SGs (Figure S1b), which are known to participate in mRNA turnover and translational repression (Balagopal and Parker, 2009; Parker and Sheth, 2007). Other CCCH‐TZF proteins also localize to PBs and SGs, where they play critical roles in mRNA metabolism, regulation of plant growth, and development and stress response (Bogamuwa and Jang, 2014). To determine whether OsTZF5 bound to RNA, we performed an RNA electrophoretic mobility shift assay and the results revealed that OsTZF5 could bind to U homopolymers and ARE motives, possibly in the 3' untranslated region of RNA (Figure S1c). This observation suggests that, like other CCCH‐TZF proteins, OsTZF5 is also an RNA regulator.
Gene expression profiling of transgenic PUbi:OsTZF5 Nipponbare plants revealed that overexpression of OsTZF5 up‐ and down‐regulated many stress‐related genes (Figure S2). In addition, the same plants exhibited greater survival under severe drought stress (Figure S3). Accordingly, overexpression of OsTZF5 appears to facilitate the regulation of drought tolerance in rice plants, probably through RNA metabolism of stress‐related genes. Concurrently, transgenic Nipponbare plants showed also reduced growth, suggesting that overexpression of OsTZF5 conferred not only higher tolerance to stress but caused also growth retardation on plants. Increased survival but reduced growth under drought conditions has been reported previously in transgenic rice plants overexpressing candidate genes for drought tolerance, such as CCCH‐TZF protein OsTZF1 (Jan et al., 2013), dehydration‐responsive element‐binding protein 1 (DREB1s) (Ito et al., 2006) or the NAC‐type transcription factor OsNAC6 (Nakashima et al., 2007).
OsTZF5 under the stress‐inducible promoter confers higher drought resistance in rice without compromising growth
Given the above findings, we selected the stress‐inducible promoter of the OsNAC6 gene to drive the expression of OsTZF5. The resulting construct (POsNAC6:OsTZF5) was then introduced into two commercial upland rice cultivars, Curinga and NERICA4. Transgenic NERICA4 lines carrying POsNAC6:OsTZF5 exhibited greater survival under severe drought (Table S1), as observed previously for the transgenic PUbi:OsTZF5 Nipponbare plants (Figure S3a, b) but, unlike the latter, no significant reduction in plant growth (Figure S4). These results suggest that POsNAC6:OsTZF5 can improve survival of rice during drought conditions without any adverse effect on growth. Previous reports showed that transgenic rice plants carrying DREB1s or OsNAC6 genes with stress‐inducible promoters exhibited higher tolerance to drought without negatively impacting plant growth. This was likely made possible by the activation of transcriptional factors involved in regulating the expression of other stress‐related genes (Ito et al., 2006; Nakashima et al., 2007).
The transgenic Curinga and NERICA4 POsNAC6:OsTZF5 plants were then subjected to field trials. Grain yield was similar under well‐watered conditions for transgenic and NT Curinga (Figure S7), suggesting that the stress‐inducible OsNAC6 promoter eliminated the adverse effect of OsTZF5 on growth also under field conditions. Transgenic POsNAC6:OsTZF5 Curinga showed higher growth recovery than NT Curinga after vegetative stage drought stress (Figure S5b). In addition, several transgenic lines showed increased grain yield per plant and per unit area than NT Curinga in repeated field drought trials (Figures 2 and 3).
Interestingly, transgenic Curinga exhibited early flowering (Figure 2, Figure S10), which helps plants to effectively avoid drought through an ‘escape mechanism’ (Yue et al., 2006). However, it remains unclear whether early flowering affected somehow the agronomic traits of transgenic plants. The OsTZF5 gene, an ortholog of OXS2 from Arabidopsis, is involved in the transition from vegetative growth to flowering stage under stress conditions (Blanvillain et al., 2011). Thus, it is not surprising that induction of OsTZF5 in transgenic lines exposed to drought stress in the field triggered flowering. Transgenic POsNAC6:OsTZF5 NERICA4 also showed higher yield than NT NERICA4 under field drought conditions (Figure 4). Collectively, our findings indicate that introduction of POsNAC6:OsTZF5 into rice cultivars affords greater survival when exposed to severe drought at seedling stage, as well as faster recovery after being exposed to severe drought during vegetative growth or early flowering. Importantly, it also leads to higher grain yield than NT cultivars.
It should be noted that not all POsNAC6:OsTZF5 transgenic lines showed higher performance under drought conditions than NT cultivars. This might be due to position effect and differences in the level of expression of the transgene. Curinga line #3474 (Figure 3) and NERICA4 lines #4890 and #4926 (Figure 4) showed consistently increased grain yields. Selection of elite lines to be evaluated under actual field conditions may be essential to breed drought‐resistant rice varieties through transgenic approaches.
POsNAC6:OsTZF5 offers a two‐in‐one mechanism against drought
Drought tolerance and avoidance are two key mechanisms for drought resistance in rice. Physiological analyses of transgenic plants grown in confined field trials revealed that transgenic POsNAC6:OsTZF5 Curinga lines had higher leaf RWC, higher Fv/Fm values and lower leaf rolling scores than NT Curinga (Figure 5). Leaf RWC and leaf rolling scores are associated with drought avoidance strategies, whereas Fv/Fm values are associated with drought tolerance mechanisms. Our results suggest that, unlike NT cultivars, our transgenic POsNAC6:OsTZF5 lines have a unique capability of regulating two different drought mechanisms at both seedling and reproductive stages under real field conditions. Additionally, they favour early flowering, which acts as a drought escape mechanism in rice plants. Zhou et al. (2016) reported that rice plants overexpressing OsAHL1 exhibited both drought avoidance and drought tolerance, suggesting that the two strategies might be genetically controlled either through the same molecular pathway or similar gene regulation and are not, consequently, two separate mechanisms. Altered RNA regulation of POsNAC6:OsTZF5 in transgenic rice plants might contribute to high yields and improved drought tolerance as manifested by higher RWC, higher Fv/Fm values and lower leaf rolling scores.
POsNAC6:OsTZF5 improves drought resistance across different stages of rice growth, genetic background, drought intensity and environments
The phenotypes of plants exposed to drought are controlled by genetic factors (genes, transgenes and genotype of transgenic plants), environmental parameters (phenology, intensity, duration and frequency of the stress) and diverse plant–soil–environmental interactions (Saint Pierre et al., 2012). In this study, we evaluated the agronomic traits of the same gene construct, POsNAC6:OsTZF5, in two different rice genetic backgrounds (Curinga and NERICA4), against two different seasons (dry and rainy), and in two different environments (rainout shelter and TE).
Conclusions and prospects
Our study is the first to report extensive field evaluation of transgenic rice plants expressing the newly identified OsTZF5 gene under drought stress and natural rain‐fed conditions. The study provides strong evidence supporting the use of stress‐responsive POsNAC6:OsTZF5 to reduce yield losses under field drought conditions in different environments and rice genetic backgrounds. Improved grain yield was associated with two different genetic mechanisms (drought tolerance and avoidance) contributed by the same transgene expressed under field drought stress. Because this gene was tested in two commercial rice varieties already used in Latin America (Curinga) and Africa (NERICA4), it is very easy to include this transgene into ongoing transgenic rice breeding programs of participating countries. Development of drought‐resistant rice may offer significant economic and environmental benefits in low‐input agricultural systems in Asia, Africa and Latin America.
Experimental procedures
Plant material and growth conditions
Nipponbare transgenic lines were used for basic analysis including gene expression and microscopic observation. Two‐week‐old seedlings grown in a growth chamber at 28˚C under 12‐h light/12‐h dark cycles were used for studies on responses to abiotic stresses. For dehydration treatment, plants were removed from hydroponic basal Yoshida nutrient solution (Yoshida et al., 1976) and were kept in a dehydrated state in the same growth chamber. For other stress treatments, plants were transferred into Yoshida nutrient solution supplemented with 250 mm NaCl, 10 mm H2O2 or 10 μm ABA. For H2O stress conditions, plants grown in hypotonic Yoshida solution were transferred to water. For cold treatment, plants were maintained in hydroponic solution and transferred to 4 °C.
Expression analysis by RNA gel blot, qRT‐PCR and microarray
Isolation of total RNA, RNA gel blot, qRT‐PCR and 44K microarray was performed as previously described (Maruyama et al., 2014; Nakashima et al., 2006, 2007; Yamaguchi‐Shinozaki et al., 1990) with some modifications. In brief, total RNA was isolated with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). cDNA was synthesized with SuperScript III reverse transcriptase (Thermo Fisher Scientific). Real‐time PCR analyses were performed using an Applied Biosystems 7500 real‐time PCR system (Thermo Fisher Scientific) and SYBR Premix Ex Taq kit (Takara, Shiga, Japan). Probe for RNA gel blots was the 3’ UTR of OsTZF5, which was radio‐labelled using [alpha‐32P] dCTP (GE Healthcare Life Sciences, Buckinghamshire, UK) and BcaBEST Labeling Kit (Takara). For 44K microarray, total RNAs were labelled using Low RNA Input Linear Amplification/Labeling Kit reagents (Agilent Technologies, Santa Clara, CA).
Analysis of OsTZF5 activity and localization using POsTZF5:GUS and OsTZF5‐sGFP constructs
The promoter region of OsTZF5, comprising 1533 bp upstream of the ATG, was amplified from the rice genome by PCR. POsTZF5 was then inserted in the pGreenII 0129 vector to generate POsTZF5:GUS (Hellens et al., 2000) as previously described by Jan et al. (2013). GUS histochemical activity in POsTZF5:GUS transgenic plant organs was detected according to Jefferson et al. (1987) following a 12‐h incubation in GUS staining solution.
The OsTZF5 coding region was fused to sGFP (Chiu et al., 2002; Nakashima et al., 2007) using the pGreenII 0129 vector (Hellens et al., 2000) with the OsTZF5 native promoter and then introduced into Nipponbare plants by Agrobacterium‐mediated transformation (Toki et al., 2006). Cellular localization of OsTZF5‐sGFP and co‐localization of OsTZF5 with cytoplasmic foci markers were assessed as described by Jan et al. (2013).
RNA‐binding assay
The RNA electrophoretic mobility shift assay was conducted as described by Brewer et al. (2004).
Generation of PUbi:OsTZF5 plants
We used the pBIG‐ubi vector (Becker, 1990) to generate transgenic rice overexpressing OsTZF5. The construct was introduced into Nipponbare plants by Agrobacterium‐mediated transformation (Toki et al., 2006), and T2 and T3 seeds were used in further experiments.
Generation of constructs and transformation of Curinga and NERICA4 plants
The promoter region of OsNAC6 (1501 bp) (Nakashima et al., 2007) was inserted into the HindIII and BamHI sites of the pBIG‐HYG vector (Becker, 1990), while the OsTZF5 ORF was inserted into the SmaI site, generating pBIH‐OsNAC6:OsTZF5. Successful transformation of transgenic Curinga and NERICA4 plants was confirmed by PCR and Southern blot analysis as described previously (Ishizaki and Kumashiro, 2008; Uga et al., 2013).
Survival under severe drought stress
For drought stress treatment, 2‐week‐old seedlings of transgenic Nipponbare (PUbi:OsTZF5) and 3‐week‐old seedlings of transgenic NERICA4 (POsNac6:OsTZF5) were grown in pots containing soil and subjected to drought stress by withholding water for 3–7 days. Following the drought stress treatment, plants were allowed to recover for 2 weeks under normal growth conditions and the number of surviving plants was counted. To evaluate the growth of NERICA4 transgenic lines of POsNac6:OsTZF5, uniform transgenic seedlings were transplanted into 4‐L pots containing soil and allowed to grow for 2 months. The dry weight of shoots was measured.
Vegetative drought stress experiment in a confined field
To evaluate the drought tolerance of transgenic rice plants at the vegetative stage, single‐copy independent homozygous transgenic POsNAC6:OsTZF5 Curinga lines, together with NT Curinga controls, were direct‐seeded in confined field conditions under rainout shelter at the CIAT, Palmira, in the dry season (September–November 2011). Drought stress was imposed by withholding irrigation at initial tillering stage (21 days after direct sowing) and rewatered after 21 days. The experiments followed the same field design and drought characterization protocols as described previously by Selvaraj et al. (2017).
MDSE trials with rainout shelter at reproductive stage
All rainout shelter reproductive stress experiments were carried out at our confined field facility at CIAT, Palmira. For Curinga, three MDSE trials (from 2012 to 2014) over two contrasting seasons (2012 dry and 2013, 2014 rainy) were conducted under a movable semi‐automatic rainout shelter facility. For NERICA4, one MDSE trial was conducted during the rainy season in 2016 to confirm the agronomic performance of the most promising lines selected by the greenhouse survival test.
Drought was imposed by withholding irrigation when panicle initiation was around 10 mm long (60–68 days after sowing for both Curinga and NERICA4) for 3–4 weeks or until severe leaf rolling and drying appeared in the NT control. Then, the plants were rewatered to 90% field capacity until physiological maturity. All four experiments followed the same design and field drought and physiological and yield characterization (SPAD, RWC, Fv/Fm, leaf rolling and grain yield‐related traits) protocols as described previously (Selvaraj et al., 2017).
Well‐watered experiment in confined field conditions
To evaluate the yielding performance of transgenic Curinga lines under normal well‐watered field conditions, the independent T4 homozygous lines of transgenic POsNAC6:OsTZF5 rice plants together with NT controls were transplanted to a confined field at the CIAT, Palmira, and grown during the dry season (September–January 2013) as described previously (Selvaraj et al., 2017).
RT‐PCR analysis of field samples and rainout shelter drought trials
Expression analysis by RT‐PCR followed the same sampling and timing procedure as described previously by Selvaraj et al. (2017).
TE trials
To evaluate yield performance of transgenic plants grown under rain‐fed (TE) upland conditions, the most promising T4 homozygous Curinga lines from the previous year drought experiments were subjected to TE at the CIAT rain‐fed upland station in Santa Rosa from 2012 to 2015 (two consecutive field trials). Promising NERICA4 lines were selected based on the survival test, and rainout shelter trials were also evaluated under TE‐2015–2016. The plants were established in dry soil, and irrigation was provided until 50 days after sowing via sprinklers to establish the crop. After plant establishment, irrigation was stopped and plants were totally dependent on rainfall. The rain‐free periods in trials for Curinga ranged from 3 to 4 weeks, and the rain‐free period in trial for NERICA4 was around 4 weeks. The rain‐fed experiments design and grain yield characterization were conducted as described previously (Selvaraj et al., 2017).
Soil moisture measurement and SMP calculation
The development of drought‐induced stress was monitored by measuring changes in soil moisture at three different depths (20, 40 and 60 cm) using AquaPro sensors (Reno, NV). Humidity data were converted to SMP values using soil water retention curves constructed for each trial location. AquaPro soil moisture values (%) were converted to volumetric soil water content using our previously published protocol (Ghneim‐Herrera et al., 2017).
Data analysis
All the data were analysed by one‐factor ANOVA, chi‐square test, Dunnett's test or Tukey's test.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
A.J. and K.N. of JIRCAS and K.Y.‐S. of the University of Tokyo planned experiments and A.J. and K.M. of JIRCAS and D.T. of the University of Tokyo conducted construction, transformation and phenotypic analysis of transgenic rice (Nipponbare) plants. T.I planned experiments in TARF and JIRCAS and conducted construction, transformation and phenotypic analysis of transgenic rice (NERICA) plants. M.G.S. and M.I planned the experiments in CIAT, and M.G.S., M.O.V. and B.D. conducted transformation and evaluation experiments of transgenic rice (Curinga) plants. T.O. of JIRCAS conducted expression analysis of the transgenic rice plants. M.G.S., T.I., A.J., K.Y.‐S. M. I. and K. N wrote the manuscript, and all other authors checked it.
Supporting information
Figure S1 Sub‐cellular localization and RNA‐binding ability of OsTZF5.
Figure S2 Transcriptome analysis of genes regulated in Nipponbare plants overexpressing OsTZF5.
Figure S3 Phenotypic analyses of transgenic PUbi:OsTZF5 and NT Nipponbare plants.
Figure S4 Dry biomass of transgenic NERICA4. Shoot dry biomass of two‐month‐old transgenic OsTZF5 NERICA4 under control of the OsNAC6 promoter grown without stress treatment.
Figure S5 Vegetative drought stress experiment on transgenic POsNAC6:OsTZF5 Curinga plants conducted at CIAT, Palmira, from September to November 2011.
Figure S6 Semi‐quantitative PCR analysis of transgenic POsNAC6:OsTZF5 and NT Curinga lines under different drought stress conditions during the MDSE trial.
Figure S7 Transgenic POsNAC6:OsTZF5 rice yield performance in the field under well‐watered (WW) conditions.
Figure S8 Progress of drought stress measured as changes in soil matric potential (SMP) under rainout shelter trials.
Figure S9 Progress of drought‐induced stress measured as changes in soil matric potential (SMP) under upland rain‐fed trials.
Figure S10 Images of transgenic Curinga line #3474 and NT Curinga during flowering.
Table S1 Survival rates of NT NERICA4 and transgenic POsNAC6:OsTZF5 NERICA4 lines under drought stress.
Table S2 Agronomic data captured from the TE‐2015–2016 trial and from the MDSE‐2016 trial of transgenic POsNAC6:OsTZF5 NERICA4 lines.
Acknowledgements
We thank Y. Fujita (JIRCAS) for plasmid construction; T. Urao, K. Suenaga, T. Kumashiro (JIRCAS) and K. Shinozaki (RIKEN) for their valuable advice; M. Kishimoto, K. Amano, E. Kishi, K. Murai and K. Yoshiwara (JIRCAS) for their technical support; and S. Ogawa and M. Recio (CIAT) for help with data collection. This work was supported by the Ministry of Agriculture, Forestry and Fisheries (MAFF); the Program for Promotion of Basic and Applied Researches for Innovations in Bio‐oriented Industry (BRAIN); Grants‐in‐Aid for Scientific Research by the Ministry of Education, Culture, Sports, Science and Technology (MEXT); and the Japan Society for the Promotion of Science (JSPS).
Selvaraj, M. G. , Jan, A. , Ishizaki, T. , Valencia, M. , Dedicova, B. , Maruyama, K. , Ogata, T. , Todaka, D. , Yamaguchi‐Shinozaki, K. , Nakashima, K. and Ishitani, M. (2020) Expression of the CCCH‐tandem zinc finger protein gene OsTZF5 under a stress‐inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotechnol. J., 10.1111/pbi.13334
References
- Al‐Souhibani, N. , Al‐Ahmadi, W. , Hesketh, J.E. , Blackshear, P.J. and Khabar, K.S. (2010) The RNA‐binding zinc‐finger protein tristetraprolin regulates AU‐rich mRNAs involved in breast cancer‐related processes. Oncogene, 29, 4205–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal, V. and Parker, R. (2009) Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D. (1990) Binary Vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18, 203–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain, R. , Wei, S. , Wei, P. , Kim, J. and Ow, D. (2011) Stress tolerance to stress escape in plants: role of the OXS2 zinc‐finger transcription factor family. EMBO J. 30, 3812–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa, S.P. and Jang, J.C.C. (2014) Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 55, 1367–1375. [DOI] [PubMed] [Google Scholar]
- Brewer, B.Y. , Malicka, J. , Blackshear, P.J. and Wilson, G.M. (2004) RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au‐rich mRNA‐destabilizing motifs. J. Biol. Chem. 279, 27870–27877. [DOI] [PubMed] [Google Scholar]
- Carballo, E. , Lai, W.S. and Blackshear, P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor‐alpha production by tristetraprolin. Science, 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Cassman, K.G. , Dobermann, A. , Walters, D.T. and Yang, H. (2003) Meeting cereal demand while protecting natural resources and improving environmental quality. Annu. Rev. Environ. Resour. 28, 315–358. [Google Scholar]
- Chiu, W.L. , Niwa, Y. , Zeng, W. , Hirano, T. , Kobayashi, H. and Sheen, J. (2002) Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Ciftci‐Yilmaz, S. and Mittler, R. (2008) The zinc finger network of plants. Cell Mol. Life Sci. 65, 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci‐Yilmaz, S. , Morsy, M.R. , Song, L. , Coutu, A. , Krizek, B.A. , Lewis, M.W. , Warren, D. et al. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- FAO (2016) Rice Market Monitor, April 2016, Volume XIX Issue No. 1. Available at: http://www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Rice/Images/RMM/RMM_APR16.pdf [Google Scholar]
- Gaudin, A.C. , Henry, A. , Sparks, A.H. and Slamet‐Loedin, I.H. (2013) Taking transgenic rice drought screening to the field. J. Exp. Bot. 64, 109–117. [DOI] [PubMed] [Google Scholar]
- Ghneim‐Herrera, T. , Selvaraj, M.G. , Meynard, D. , Fabre, D. , Peña, A. , Ben, R.W. , Ben, S.R. et al. (2017) Expression of the Aeluropus littoralis AlSAP gene enhances rice yield under field drought at the reproductive stage. Front. Plant Sci. 8, 994. 10.3389/fpls.2017.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P. , Edwards, E.A. , Leyland, N.R. , Bean, S. and Mullineaux, P.M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Huke, R.E. and Huke, E.H. .(1997) Rice area by type of culture. South, Southeast, and East Asia. A revised and updated data base. Los Baños, Philippines: IRRI. [Google Scholar]
- Ishizaki, T. and Kumashiro, T. (2008) Genetic transformation of NERICA, interspecific hybrid rice between Oryza glaberrima and O. sativa, mediated by Agrobacterium tumefaciens . Plant Cell Rep. 27, 319–327. [DOI] [PubMed] [Google Scholar]
- Ito, Y. , Katsura, K. , Maruyama, K. , Taji, T. , Kobayashi, M. , Seki, M. , Shinozaki, K. et al. (2006) Functional analysis of rice DREB1/CBF‐type transcription factors involved in cold‐responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. [DOI] [PubMed] [Google Scholar]
- Jan, A. , Maruyama, K. , Todaka, D. , Kidokoro, S. , Abo, M. , Yoshimura, E. , Shinozaki, K. et al. (2013) OsTZF1, a CCCH‐tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress‐related genes. Plant Physiol. 161, 1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, M.S. , Kim, E.J. and Jang, S.B. (2010) Expression and RNA‐binding of human zinc‐finger antiviral protein. Biochem. Biophys. Res. Commun. 396, 696–702. [DOI] [PubMed] [Google Scholar]
- Jeong, J.S. , Kim, Y.S. , Redillas, M.C. , Jang, G. , Jung, H. , Bang, S.W. , Choi, Y.D. et al. (2013) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotech. J. 11, 101–114. [DOI] [PubMed] [Google Scholar]
- Kim, D.H. , Yamaguchi, S. , Lim, S. , Oh, E. , Park, J. , Hanada, A. , Kamiya, Y. et al. (2008) SOMNUS, a CCCH‐type zinc finger protein in Arabidopsis, negatively regulates light‐dependent seed germination downstream of PIL5. Plant Cell, 20, 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Z. , Li, M. , Yang, W. , Xu, W. and Xue, Y. (2006) A novel nuclear‐localized CCCH‐type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. and Thomas, T.L. (1998) PEI1, an embryo‐specific zinc finger protein gene required for heart‐stage embryo formation in Arabidopsis. Plant Cell, 10, 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, P.C. , Pomeranz, M.C. , Jikumaru, Y. , Kang, S.G. , Hah, C. , Fujioka, S. , Kamiya, Y. et al. (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA‐ and GA‐mediated growth, stress and gene expression responses. Plant J. 65, 253–268. [DOI] [PubMed] [Google Scholar]
- Manavalan, L.P. , Chen, X. , Clarke, J. , Salmeron, J. and Nguyen, H.T. (2012) RNAi‐mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 63, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K. , Todaka, D. , Mizoi, J. , Yoshida, T. , Kidokoro, S. , Matsukura, S. , Takasaki, H. et al. (2012) Identification of cis‐acting promoter elements in cold‐ and dehydration‐induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2014) Gene expression profiling using DNA microarrays. In Methods in Molecular Biology( Sanchez‐Serrano, J. and Salinas, J. , eds), pp. 381–391. Totowa: Humana Press. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, A. , Vij, S. and Tyagi, A.K. (2004) Overexpression of a zinc‐finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101, 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, K. , Fujita, Y. , Katsura, K. , Maruyama, K. , Narusaka, Y. , Seki, M. , Shinozaki, K. et al. (2006) Transcriptional regulation of ABI3‐ and ABA‐responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. , Tran, L.S. , Van Nguyen, D. , Fujita, M. , Maruyama, K. , Todaka, D. , Ito, Y. et al. (2007) Functional analysis of a NAC‐type transcription factor OsNAC6 involved in abiotic and biotic stress‐responsive gene expression in rice. Plant J. 51, 617–630. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole, J.C. .(2004) Rice and water: The final frontier. In First International Conference on Rice for the Future, pp. 1–26. Bangkok, Thailand: Rockefeller Foundation. [Google Scholar]
- Parker, R. and Sheth, U. (2007) P bodies and the control of mRNA translation and degradation. Mol. Cell, 25, 635–646. [DOI] [PubMed] [Google Scholar]
- Pennisi, E. (2008) The blue revolution, drop by drop, gene by gene. Science, 320, 171–173. [DOI] [PubMed] [Google Scholar]
- Pomeranz, M.C. , Hah, C. , Lin, P.C. , Kang, S.G. , Finer, J.J. , Blackshear, P.J. and Jang, J.C. (2010) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 152, 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Pierre, C. , Crossa, J.L. , Bonnett, D. , Yamaguchi‐Shinozaki, K. and Reynolds, M.P. (2012) Phenotyping transgenic wheat for drought resistance. J. Exp. Bot. 63, 1799–1808. [DOI] [PubMed] [Google Scholar]
- Sakamoto, H. , Maruyama, K. , Sakuma, Y. , Meshi, T. , Iwabuchi, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2004) Arabidopsis Cys2/His2‐type zinc‐finger proteins function as transcription repressors under drought, cold, and high‐salinity stress conditions. Plant Physiol. 136, 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seck, P.A. , Diagne, A. , Mohanty, S. and Wopereis, M.C. (2012) Crops that feed the world 7: rice. Food Secur. 4, 7–24. [Google Scholar]
- Selvaraj, M.G. , Ishizaki, T. , Valencia, M. , Ogawa, S. , Dedicova, B. , Ogata, T. , Yoshiwara, K. et al. (2017) Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 15, 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, M. , Kawahara, H. and Doi, H. (2002) Novel family of CCCH‐type zinc‐finger proteins, MOE‐1, ‐2 and ‐3, participates in C. elegans oocyte maturation. Genes Cells, 7, 933–947. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Jiang, H. , Xu, Y. , Li, H. , Wu, X. , Xie, Q. and Li, C. (2007) The CCCH‐type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis . Plant Cell Physiol. 48, 1148–1158. [DOI] [PubMed] [Google Scholar]
- Toki, S. , Hara, N. , Ono, K. , Onodera, H. , Tagiri, A. , Oka, S. and Tanaka, H. (2006) Early infection of scutellum tissue with Agrobacterium allows high‐speed transformation of rice. Plant J. 47, 969–976. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. , Kitomi, Y. et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Varnum, B.C. , Ma, Q.F. , Chi, T.H. , Fletcher, B. and Herschman, H.R. (1991) The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys‐His repeat. Mol. Cell Biol. 11, 1754–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. , Mundy, J. and Chua, N.H. (1990) Four tightly linked rab genes are differentially expressed in rice. Plant Mol. Biol. 14, 29–39. [DOI] [PubMed] [Google Scholar]
- Yoshida, S.D. , Forno, D. , Cock, J.K. and Gomez, K.A. .(1976) Laboratory Manual for Physiological Studies of Rice. Manila: International Rice Research Institute. [Google Scholar]
- Younis, A. , Siddique, M.I. , Kim, C.‐K. and Lim, K.‐B. (2014) RNA interference (RNAi) induced gene silencing: a promising approach of hi‐tech plant breeding. Int. J. Biol. Sci. 10, 1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, B. , Xue, W. , Xiong, L. , Yu, X. , Luo, L. , Cui, K. , Jin, D. et al. (2006) Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics, 172, 1213–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Liu, Z. , Liu, Y. , Kong, D. , Li, T. , Yu, S. , Mei, S. et al. (2016) A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice. Sci. Rep. 6, 30264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sub‐cellular localization and RNA‐binding ability of OsTZF5.
Figure S2 Transcriptome analysis of genes regulated in Nipponbare plants overexpressing OsTZF5.
Figure S3 Phenotypic analyses of transgenic PUbi:OsTZF5 and NT Nipponbare plants.
Figure S4 Dry biomass of transgenic NERICA4. Shoot dry biomass of two‐month‐old transgenic OsTZF5 NERICA4 under control of the OsNAC6 promoter grown without stress treatment.
Figure S5 Vegetative drought stress experiment on transgenic POsNAC6:OsTZF5 Curinga plants conducted at CIAT, Palmira, from September to November 2011.
Figure S6 Semi‐quantitative PCR analysis of transgenic POsNAC6:OsTZF5 and NT Curinga lines under different drought stress conditions during the MDSE trial.
Figure S7 Transgenic POsNAC6:OsTZF5 rice yield performance in the field under well‐watered (WW) conditions.
Figure S8 Progress of drought stress measured as changes in soil matric potential (SMP) under rainout shelter trials.
Figure S9 Progress of drought‐induced stress measured as changes in soil matric potential (SMP) under upland rain‐fed trials.
Figure S10 Images of transgenic Curinga line #3474 and NT Curinga during flowering.
Table S1 Survival rates of NT NERICA4 and transgenic POsNAC6:OsTZF5 NERICA4 lines under drought stress.
Table S2 Agronomic data captured from the TE‐2015–2016 trial and from the MDSE‐2016 trial of transgenic POsNAC6:OsTZF5 NERICA4 lines.


