Clarifying conflicting previous reports, Tenthorey et al. demonstrate, in vitro and in vivo, that phosphorylation of NLRC4 is neither necessary nor sufficient for NAIP/NLRC4 inflammasome activation. Further, NLRC4 plays no role in a model of melanoma.
Abstract
The NAIP/NLRC4 inflammasome is a cytosolic sensor of bacteria that activates caspase-1 and initiates potent immune responses. Structural, biochemical, and genetic data demonstrate that NAIP proteins are receptors for bacterial ligands, while NLRC4 is a downstream adaptor that multimerizes with NAIPs to form an inflammasome. NLRC4 has also been proposed to suppress tumor growth, though the underlying mechanism is unknown. Further, NLRC4 is phosphorylated on serine 533, which was suggested to be critical for its function. In the absence of S533 phosphorylation, it was proposed that another inflammasome protein, NLRP3, can induce NLRC4 activation. We generated a new Nlrc4-deficient mouse line and mice with S533D phosphomimetic or S533A nonphosphorylatable NLRC4. Using these models in vivo and in vitro, we fail to observe a requirement for phosphorylation in NLRC4 inflammasome function. Furthermore, we find no role for NLRP3 in NLRC4 function, or for NLRC4 in a model of melanoma. These results clarify our understanding of the mechanism and biological functions of NAIP/NLRC4 activation.
Introduction
Inflammasomes are innate immune multi-protein complexes that assemble to activate pro-inflammatory caspases following detection of cytosolic pathogen- or stress-associated stimuli (Martinon et al., 2002; Lamkanfi and Dixit, 2014; Rathinam and Fitzgerald, 2016). The neuronal apoptosis inhibitory protein (NAIP)/NLR family caspase activation and recruitment domain–containing protein 4 (NLRC4) inflammasome is one of the best characterized inflammasomes. NLRC4 was identified in 2001 as a caspase activation and recruitment domain–containing protein that activates caspase-1 (Poyet et al., 2001). NLRC4 activates caspase-1 after cytosolic invasion by flagellated or type 3 secretion system (T3SS)–expressing bacterial pathogens (Franchi et al., 2006; Miao et al., 2006, 2010; Mariathasan et al., 2004; Zamboni et al., 2006). Genetic, biochemical, and structural studies established that activation of NLRC4 requires NAIP proteins (NAIP1, NAIP2, NAIP5, and NAIP6 in C57BL/6 mice), acting as sensors to detect bacterial flagellin or T3SS needle or rod proteins (Kofoed and Vance, 2011; Zhao et al., 2011; Rauch et al., 2016; Zhao et al., 2016; Tenthorey et al., 2017). Ligand binding leads to structural changes in NAIPs that allow recruitment and oligomerization of NLRC4, which acts as an adaptor to recruit and activate caspase-1 (Hu et al., 2015; Zhang et al., 2015; Tenthorey et al., 2017). Activated caspase-1 cleaves and thereby matures the cytokines IL-1β and IL-18 as well as the pore-forming protein gasdermin D (Rathinam and Fitzgerald, 2016; Lamkanfi and Dixit, 2014; Shi et al., 2015; Kayagaki et al., 2015). Mature gasdermin D causes pyroptotic host cell death to release active IL-1β and IL-18, leading to inflammatory responses to control infection.
Despite two decades of study, several aspects of NLRC4 activation and function remain unclear. NLRC4 may have roles outside of pathogen sensing. Several inflammasomes, including NLRP3, NLRP1, and AIM2, have been reported to protect against tumor growth (Karki et al., 2017). Implicating NLRC4 in tumor progression, NLRC4 and caspase-1 were found to promote metastasis of breast and colon cancer in obese mice (Kolb et al., 2016; Ohashi et al., 2019). In contrast, NLRC4 and caspase-1 were reported to protect mice from chemically induced colon cancer (Hu et al., 2010), although others found that caspase-1 but not NLRC4 was protective in the same model (Allen et al., 2010). Intriguingly, a recent paper found that Nlrc4−/− but not Caspase1−/− mice are susceptible to implanted B16 melanomas (Janowski et al., 2016), suggesting that NLRC4 controls tumors independently of its prototypical caspase-1 pathway. Some of the differences in these results might derive from the use of varying tumor models. It is currently unclear what might drive activation of NLRC4 in tumors or tumor-associated macrophages, as the only known agonists of NLRC4 are bacterial proteins.
Another unresolved question is the role of phosphorylation in NLRC4 inflammasome activity. Phosphorylation of NLRP3 by JNK1 is essential for NLRP3 inflammasome assembly (Song et al., 2017), providing precedent for phosphorylation as a checkpoint on inflammasomes. NLRC4 is phosphorylated at serine 533 (S533) during Salmonella enterica serovar Typhimurium (S. Typhimurium) infection, and Nlrc4−/− macrophages reconstituted with nonphosphorylatable NLRC4 (alanine mutant S533A) failed to induce an inflammasome response to S.Typhimurium (Qu et al., 2012). Conversely, it was reported to be difficult to reconstitute Nlrc4−/− macrophages with a phosphomimetic NLRC4 (aspartic acid mutant S533D), suggesting NLRC4-S533D might be constitutively active and induce pyroptosis without infection. Thus, NLRC4 phosphorylation was proposed to be both necessary and sufficient for inflammasome activation (Qu et al., 2012). This group subsequently generated mice with a homozygous S533A mutation in the endogenous Nlrc4 gene (Qu et al., 2016). In contrast to the prior claim that phosphorylation was critical for activation, macrophages from Nlrc4S533A mice exhibited only a partial defect in NLRC4 activation. However, the residual inflammasome signaling in Nlrc4S533A cells was eliminated by deletion of Nlrp3 (Qu et al., 2016), another inflammasome that participates in S. Typhimurium sensing (Broz et al., 2010). It was proposed that NLRP3 is recruited to NLRC4 to mediate inflammasome activation. It is currently unclear why phosphorylation might be required for optimal NLRC4 signaling, as S533 is not near the NLRC4 oligomerization or caspase-1–recruitment interfaces (Tenthorey et al., 2017; Hu et al., 2015; Zhang et al., 2015). Nor is it clear how NLRP3 is recruited and contributes to NLRC4 signaling.
Critically, these studies did not address whether S533 phosphorylation acts in concert with or independently of NAIPs to mediate NLRC4 activation. The S. Typhimurium T3SS was required (Qu et al., 2012), and transfected flagellin was sufficient, to induce NLRC4 phosphorylation (Qu et al., 2016). Since NAIPs are required for flagellin and T3SS proteins to activate NLRC4 (Kofoed and Vance, 2011; Zhao et al., 2011; Lightfield et al., 2008; Zhao et al., 2016; Rauch et al., 2016), the simplest model to explain these results would be a linear pathway in which NAIP activation leads to NLRC4 phosphorylation and activation. However, NLRC4 was phosphorylated in flagellin-transfected Naip5−/− cells, implying the existence of a NAIP-independent pathway for NLRC4 phosphorylation (Matusiak et al., 2015). This lack of requirement for NAIP5 could be due to redundancy between NAIP5 and NAIP6 for detection of flagellin (Kofoed and Vance, 2011). Regardless, Naip5−/− cells did not activate caspase-1 in response to flagellin, despite NLRC4 phosphorylation (Matusiak et al., 2015). Further, NLRC4 was phosphorylated in an inactive, monomeric crystal structure of NLRC4 (Hu et al., 2013). These results imply that S533 phosphorylation is not sufficient to mediate NLRC4 activation, in contrast to the prior suggestion (Qu et al., 2012) that a phosphomimetic S533D NLRC4 mutant is constitutively active.
In an effort to clarify the role of S533 phosphorylation in NLRC4 activation, we used CRISPR/Cas9 to generate three new lines of NLRC4 mutant mice: NLRC4 null mice (Nlrc4−/−), mice harboring a phosphomimetic mutation (Nlrc4S533D/S533D), and mice with nonphosphorylatable NLRC4 (Nlrc4S533A/S533A), all at the endogenous Nlrc4 locus. These mouse lines are on a pure C57BL6/J background, in contrast to another widely used Nlrc4−/− mouse line generated on a C57BL6/N background (Mariathasan et al., 2004). Comparing our new Nlrc4 mutant mice to C57BL6/J wild-type controls, we could only observe a modest and nonessential role for NLRC4 phosphorylation, and no role for NLRP3, in NLRC4 activation. In addition, we found no role for NLRC4 in control of B16 melanoma progression. Our results clarify the function and mechanism of activation of NLRC4 and establish NAIP proteins as the only known upstream activators of NLRC4.
Results and discussion
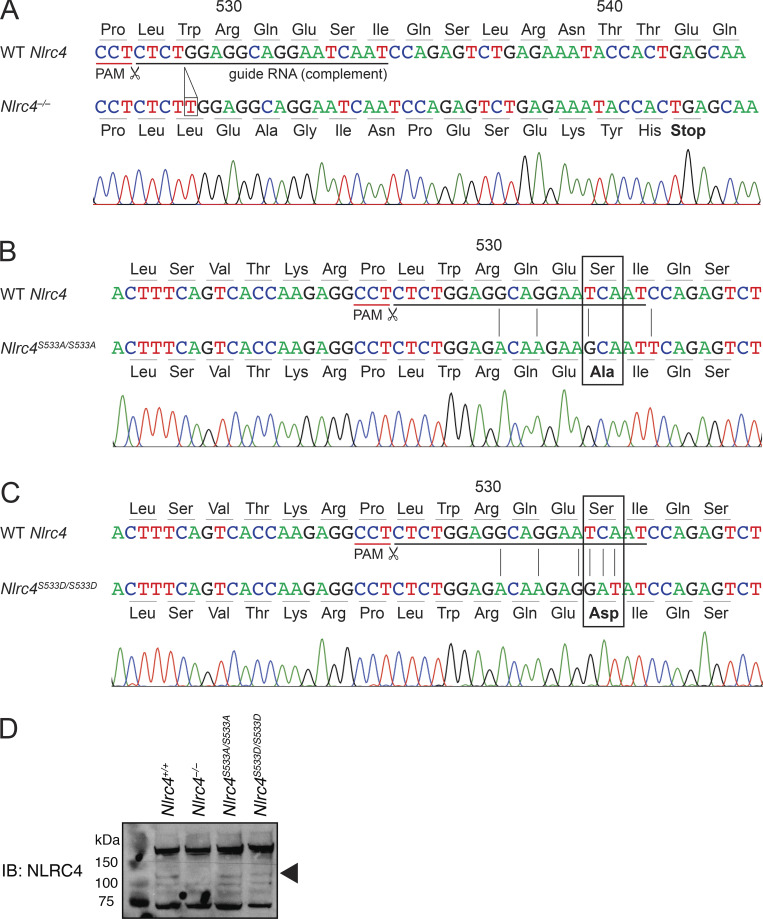
To study the role of phosphorylation in NLRC4 activation, we generated multiple mutant mice via CRISPR/Cas9. We targeted Cas9 to cleave near the S533 codon of Nlrc4, and we repaired the allele with oligonucleotides containing S533A or S533D mutations (Fig. 1). From this targeting, we established three mouse lines with the following mutations: (1) a 1-basepair insertion in codon 529 that leads to a premature stop at codon 542 (Fig. 1 A, official name Nlrc4em1Vnce, here Nlrc4−/−); (2) a homozygous nonphosphorylatable S533A mutation (Fig. 1 B, official name Nlrc4em2(S533A)Vnce, here Nlrc4S533A/S533A); and (3) a homozygous phosphomimetic S533D mutation (Fig. 1 C, official name Nlrc4em3(S533D)Vnce, here Nlrc4S533D/S533D). All mice were viable and fertile, and we did not observe any obvious abnormalities.
Figure 1.
CRISPR/Cas9 targeting of Nlrc4. (A–C) The sequence of the Nlrc4 locus in WT C57BL6/J compared with CRISPR/Cas9-generated (A) Nlrc4−/−, (B) Nlrc4S533A/S533A, and (C) Nlrc4S533D/S533D mice. Sanger sequences traces are shown for mutant mice. Guide RNA sequence and protospacer-adjacent motif (PAM) for CRISPR/Cas9 targeting, as well as nonsilent base exchanges, are indicated. (D) Immunoblot (IB) of NLRC4 levels in bone marrow–derived macrophages. Representative of two independent experiments.
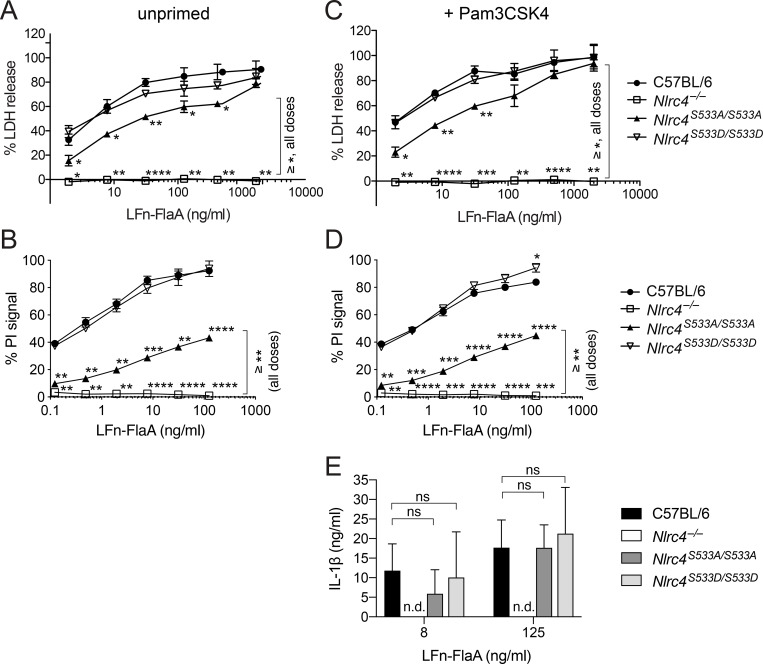
To confirm successful inactivation of Nlrc4 and to test functionality of the nonphosphorylatable andphosphomimetic alleles, we generated bone marrow–derived macrophages. Western blot confirmed the absence of NLRC4 protein in Nlrc4−/− macrophages and the presence of protein in Nlrc4S533A/S533A and Nlrc4S533D/S533D cells (Fig. 1 D). We used FlaTox, a reagent to deliver Legionella pneumophila flagellin into the cytosol of cells (Zhao et al., 2011; Rauch et al., 2016; von Moltke et al., 2012; Ballard et al., 1996), to activate the NAIP/NLRC4 inflammasome. We measured release of the cytosolic enzyme lactate dehydrogenase (LDH) into the supernatant to assay for pyroptosis downstream of NLRC4-mediated caspase-1 activation. As expected, we detected increasing levels of LDH release by wild-type C57BL/6J macrophages treated with increasing amounts of FlaTox, while macrophages from our newly generated Nlrc4−/− mice did not release LDH at any dose of FlaTox (Fig. 2 A). Macrophages from Nlrc4S533A/S533A mice showed about a 50% decrease in LDH release at lower concentrations of FlaTox, but they exhibited no defect at the highest FlaTox doses. Macrophages from phosphomimetic Nlrc4S533D/S533D mice displayed LDH release kinetics comparable to wild-type controls and did not show evidence of spontaneous inflammasome activation in the absence of FlaTox. We confirmed these results with propidium iodide (PI) uptake, a highly sensitive measure of cell death, using nonsaturating doses of FlaTox (Fig. 2 B). Again, we observed normal inflammasome activation in Nlrc4S533D/S533D macrophages, no activation in Nlrc4−/− cells, and a threefold defect in activation in Nlrc4S533A/S533A cells.
Figure 2.
NLRC4 phosphorylation is neither sufficient nor strictly required for response to cytosolic flagellin. (A–E) Macrophages were either left unprimed (A and B) or primed 4 h with 1 µg/ml Pam3CSK4 (C–E) and then treated with 4 µg/ml PA and the indicated dose of LFn-FlaA to activate NLRC4. LDH release (A and C), PI uptake (B and D), or IL-1β release (E) was measured after 4 h. All data are representative of three independent experiments (three biological replicates per experiment). Mean ± SD; n.d., not detectable. Repeated measures two-way ANOVA with Dunnett’s multiple comparisons post-test, all genotypes vs. C57BL6/J (or Nlrc4S533A/S533A vs. Nlrc4−/− as indicated), *, P < 0.05, **, P < 0.005, ***, P < 0.0005, ****, P < 0.00001; ns, not significant.
We repeated our experiments in macrophages pretreated with the TLR2 ligand Pam3CSK4, as it was suggested that TLR priming may help reveal a function for NLRC4 phosphorylation (Qu et al., 2016). There was no difference between primed and unprimed Nlrc4S533A/S533A macrophages in LDH release (Fig. 2 C) or PI uptake (Fig. 2 D) after FlaTox treatment. Furthermore, release of IL-1β into the supernatant was similar for primed wild-type and Nlrc4S533A/S533A macrophages at higher doses of FlaTox (Fig. 2 E). We also did not observe increased pyroptosis or IL-1β secretion in primed Nlrc4S533D/S533D macrophages compared with controls. As the antibody against IL-1β used in ELISAs also recognizes full-length IL-1β, there is a theoretical possibility that we missed a change in cleaved IL-1β (sometimes semiquantitatively assessed by Western blot). However, as we assessed IL-1β levels in combination with LDH release and PI uptake, we are confident that this is a true reflection of the extent of pyroptosis in the respective genotypes.
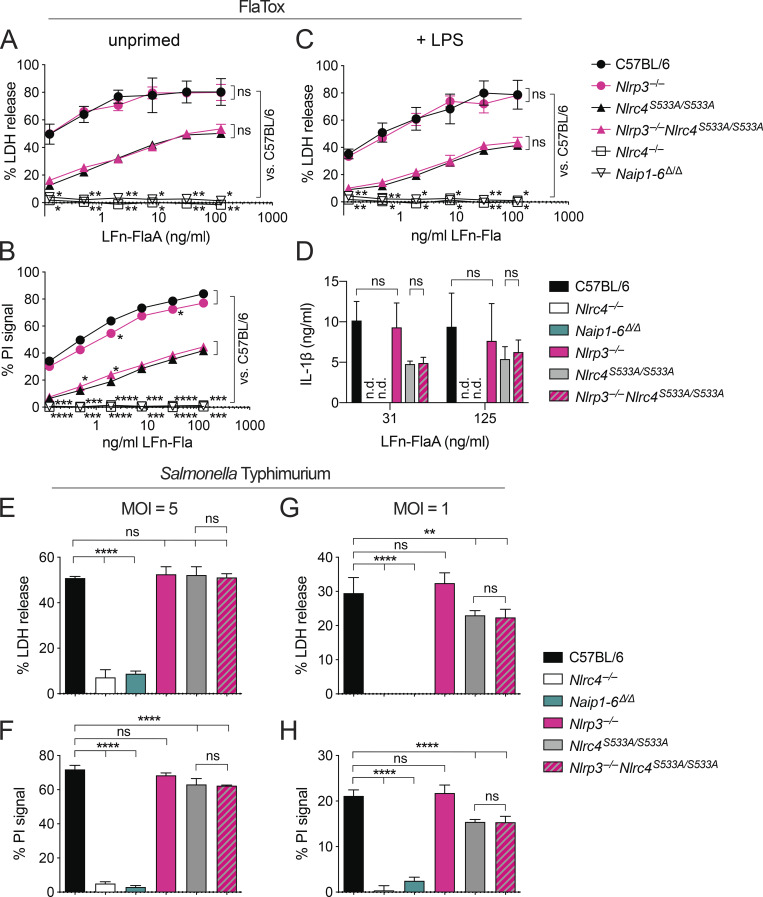
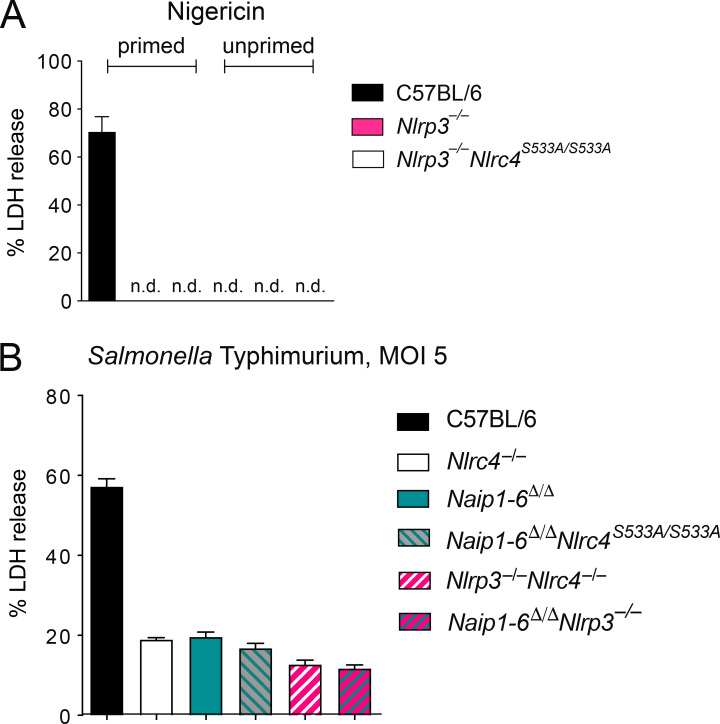
We next addressed whether activation of nonphosphorylatable NLRC4-S533A requires NAIPs, or whether, as previously suggested (Qu et al., 2016), NLRP3 partially compensates for the loss of NLRC4 phosphorylation. We crossed Nlrc4S533A/S533A to Nlrp3−/− mice to generate Nlrc4S533A/S533A;Nlrp3−/− animals. Whether primed or unprimed, Nlrc4S533A/S533A;Nlrp3−/− macrophages exhibited the same amount of cell death and IL-1β secretion as Nlrc4S533A/S533A cells, even with nonsaturating doses of FlaTox (Fig. 3, A–D). In contrast, Nlrc4−/− and Naip1-6Δ/Δ cells were completely protected, confirming the crucial role of NAIPs in NLRC4 activation (Rauch et al., 2016; Zhao et al., 2016). As a control, treatment with the NLRP3 agonist nigericin confirmed that we were able to prime NLRP3 activity and that Nlrp3−/− macrophages lacked NLRP3 activity (Fig. S1 A).
Figure 3.
NLRP3 is not required for signaling by NLRC4-S533A. (A–D) Macrophages were left unprimed (A and B) or primed for 4 h with 2 µg/ml LPS (C and D) and then treated with the indicated dose of LFn-FlaA and 4 µg/ml PA. LDH release (A and C), PI uptake (B), or IL-1β release (D) was measured after 4 h. (E–H) Unprimed macrophages were infected with S. Typhimurium at an MOI of 5 (E and F) or 1 (G and H). LDH release (E and G) and PI uptake (F and H) were measured 4 h after infection. (A–H) All data are representative of three independent experiments (three biological replicates per experiment). Mean ± SD; n.d., not detectable. Repeated measures two-way ANOVA (A–D) or one-way ANOVA (E–H) vs. C57BL6/J or Nlrc4S533A/S533A with Dunnett’s multiple comparisons post-test, *, P < 0.05, **, P < 0.005, ***, P < 0.0005, ****, P < 0.00001; ns, not significant.
Figure S1.
NAIPs, not NLRP3, are required for NLRC4 signaling. (A) Macrophages were left untreated or primed for 4 h with 2 µg/ml LPS and then treated with 10 µM nigericin. LDH release was measured at 4 h after treatment. (B) Unprimed macrophages were infected with S. Typhimurium at an MOI of 5, and LDH release was measured after 4 h. (A and B) All data are representative of three independent experiments (three biological replicates per experiment). Mean ± SD; n.d., not detectable.
Previous studies of NLRC4 phosphorylation used infection of macrophages with S. Typhimurium to induce NLRC4 activation, as this pathogen both primes cells via TLR stimuli and activates the NAIP/NLRC4 inflammasome. The use of S. Typhimurium as an NLRC4 agonist is complicated because it can also activate NLRP3, independently of NLRC4 (Broz et al., 2010). Nevertheless, we also performed S. Typhimurium infections of our mutant macrophages, closely adhering to the published protocol (Qu et al., 2016). However, in contrast to previous results, we failed to detect a significant difference in cell death between S. Typhimurium–infected Nlrc4S533A/S533A;Nlrp3−/− and Nlrc4S533A/S533A macrophages (Fig. 3, E and F). Furthermore, we found no difference between wild-type C57BL6/J and Nlrc4S533A/S533A macrophages in S. Typhimurium–induced pyroptosis. Because we had observed a partial defect for Nlrc4S533A/S533A macrophages at lower doses of FlaTox, we repeated our S. Typhimurium infections at a lower multiplicity of infection (MOI). At an MOI of 1, we observed a slight defect in cell death for Nlrc4S533A/S533A macrophages, while the additional loss of Nlrp3 had no effect (Fig. 3, G and H). As S. Typhimurium induced some macrophage cell death at an MOI of 5 even in the absence of NLRC4, we crossed mice to obtain Nlrc4−/−;Nlrp3−/−, Naip1-6Δ/Δ;Nlrp3−/−, and Naip1-6Δ/Δ; Nlrc4S533A/S533A macrophages. Upon infection with S. Typhimurium, no major differences in cell death were observed in any of these cells compared with Nlrc4−/− cells (Fig. S1 B). Thus, we conclude there is only a modest role for S533 phosphorylation and no role for NLRP3 in NLRC4 activation in macrophages in vitro, whereas we confirm that NAIPs are essential.
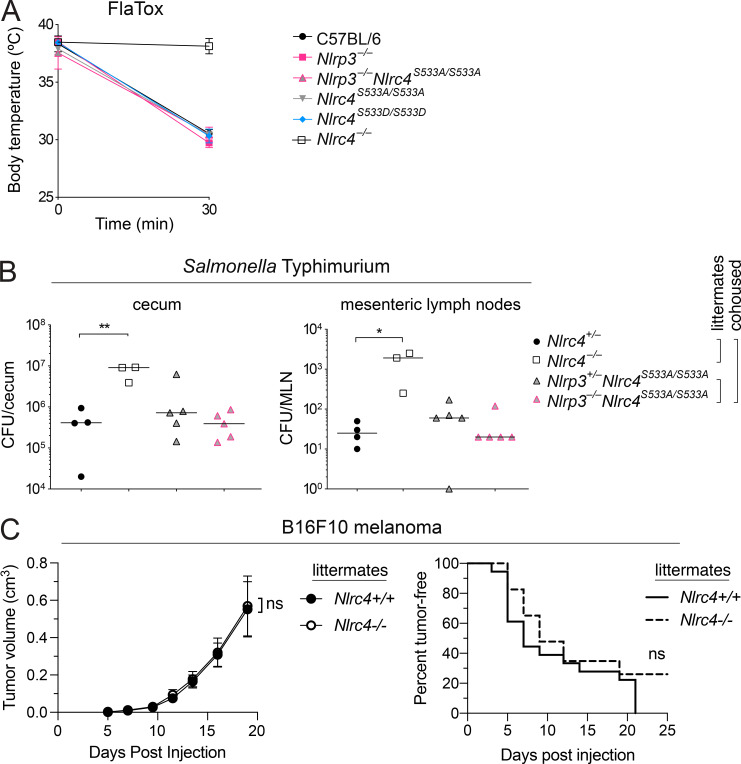
Macrophage generation in vitro relies on conditioned medium that can vary from batch to batch, which may lead to differences in differentiation. We therefore performed in vivo experiments to address the role of NLRC4 phosphorylation. As previously described (von Moltke et al., 2012; Rauch et al., 2016), wild-type C57BL/6J mice experience acute severe Naip/Nlrc4-dependent hypothermia when injected systemically with FlaTox. This phenotype was not mitigated in Nlrc4S533A/S533A, Nlrp3−/−, or Nlrc4S533A/S533A;Nlrp3−/− mice, nor was it exacerbated in Nlrc4S533D/S533D animals (Fig. 4 A). We also performed in vivo S. Typhimurium infections. Streptomycin-pretreated mice were orally infected with 2 × 107 bacteria via gavage. As previously reported with a different Nlrc4−/− mouse line (Rauch et al., 2017; Sellin et al., 2014), our newly generated Nlrc4−/− animals show significantly increased numbers of S. Typhimurium in their cecal tissue and mesenteric lymph nodes, as compared with Nlrc4+/− littermates (Fig. 4 B). We failed to detect any significant difference between Nlrc4S533A/S533A;Nlrp3+/− and Nlrc4S533A/S533A;Nlrp3−/− (littermate) or Nlrc4+/− (cohoused) animals. Collectively, these experiments confirm that our newly generated Nlrc4-deficient mouse line exhibits no NLRC4 activity in response to L. pneumophila flagellin or S. Typhimurium infection. However, we failed to detect a major role for NLRC4 phosphorylation during in vitro or in vivo infection.
Figure 4.
In vivo disease susceptibility of Nlrc4 mutant mice. (A) Rectal temperatures of WT C57BL6/J, Nlrp3−/−, Nlrc4S533A/ S533A;Nlrp3−/−, Nlrc4S533A/S533A, Nlrc4S533D/S533D, and Nlrc4−/− mice (n = 3 per group) injected retroorbitally with 0.8 µg/g body weight of PA and 0.4 µg/g LFn-FlaA. Mean ± SD. (B) CFU in cecum and mesenteric lymph node (MLN) 18 h after oral S. Typhimurium infection of Nlrc4S533A/S533A;Nlrp3−/− and Nlrc4S533A/S533A;Nlrp3−/− littermate mice cohoused with Nlrc4+/− and Nlrc4−/− littermate mice (n = 3–5 per group). Median; Mann–Whitney test, *, P < 0.01, **, P < 0.005. (A and B) Data are representative of three independent experiments. (C) Tumor size and incidence of B16F10 melanoma injected subcutaneously into Nlrc4+/+ and Nlrc4−/− littermate mice (n = 18 or 23, respectively). Mean ± SEM. Data are pooled from three independent experiments. Repeated measures two-way ANOVA with Sidak’s multiple comparisons test (tumor volume) or log-rank Mantel–Cox test (tumor incidence); ns, not significant.
Our new Nlrc4-deficient mice on a genetic background that is isogenic to C57BL/6J provided the opportunity to address another proposed role for NLRC4: suppression of cancer progression. A previous study (Janowski et al., 2016) found that Nlrc4−/− mice on the C57BL/6N background failed to control subcutaneous growth of B16F10 melanomas. However, this study did not compare littermates, leaving open the possibility that differences between wild-type and Nlrc4−/− mice were due to alterations in the microbiota (Sivan et al., 2015) or other genetic differences between these mice. We therefore challenged our new Nlrc4-deficient mice, and their Nlrc4-sufficient littermates, subcutaneously with B16F10 melanoma cells (originally derived from C57BL/6J mice) and tracked tumor development (Fig. 4 C). We did not find significant differences in tumor size or tumor incidence between mice lacking or expressing NLRC4. Thus, we are unable to detect a role for NLRC4 in melanoma suppression.
In conclusion, we do not observe a necessity for NLRC4 phosphorylation or NLRP3 recruitment for NLRC4 inflammasome activity, either in vitro or in vivo. Nlrc4S533A/S533A macrophages did show a two- to threefold decrease in pyroptosis upon low-level ligand stimulation. It is difficult to determine whether this modest defect is due to lack of phosphorylation or, alternatively, to mild destabilization of the protein due to the S533A mutation. However, it is consistent with a slight reduction in inflammasome activity upon deletion or inhibition of LRRK2, the kinase reported to phosphorylate this residue (Liu et al., 2017). Regardless, since there was no difference observed upon stronger stimulation and the phosphomimetic mutant behaved indistinguishably from wild-type, we conclude that phosphorylation is neither strictly required nor sufficient for NLRC4 activation. We can only speculate on the difference in results with our new Nlrc4 mutant mice and previous observations. The importance of NLRC4 phosphorylation might only be revealed in a specific genetic context, as the original Nlrc4−/− mouse line was generated in C57BL/6N embryonic stem cells (Mariathasan et al., 2004). Recently, immunological differences have been described between C57BL/6N and J lines (Simon et al., 2013), explained partially by C57BL/6J carrying a mutation in Nlrp12, leading to defects in neutrophil recruitment (Ulland et al., 2016). The original Nlrc4−/− line might also carry further passenger mutations or have acquired a de novo mutation. Such differences might also explain a previous report that Nlrc4−/− but not Caspase1−/− fails to control implanted tumors (Janowski et al., 2016), a finding that we could not replicate with our Nlrc4−/− mice. Alternately, this discrepancy could be due to microbiota differences between independently maintained wild-type, Nlrc4−/−, and Caspase1−/− lines, which we avoided using cohoused littermate siblings. Regardless, our results clarify our understanding of NLRC4 by confirming that NAIPs—rather than NLRP3 or phosphorylation—are the main upstream activators leading to inflammasome activation.
Materials and methods
Mice
C57BL/6J (B6) mice were purchased from The Jackson Laboratory and bred at University of California, Berkeley. Animal experiments were approved by the University of California, Berkeley, Animal Care and Use Committee. To generate Nlrc4 genetically modified mice, fertilized embryos from C56BL/6J mice were injected with Cas9 mRNA and sgRNA (single-guide RNA), as described (Rauch et al., 2016), along with two DNA oligonucleotides (single-stranded oligodeoxynucleotide; ssODN) for homology-directed repair. We used an sgRNA designed to induce Cas9 cleavage close to the codon for S533 (Fig. 1). The sgRNA was designed using Massachusetts Institute of Technology and Benchling CRISPR design tools and chosen to optimize targeting efficiency relative to efficiency of off-target sites. The following guide was selected: 5′-ATTGATTCCTGCCTCCAGAG-3′, noncoding protospacer-adjacent motif, Massachusetts Institute of Technology targeting score = 48, highest off-target score = 5.2. S533 sgRNA was coinjected with S533A (5′-TGCAATGGTTTATCAGCACGGCAGCCTACAAGGACTTTCAGTCACCAAGAGGCCTCTCTGGAGaCAaGAAgCAATtCAGAGTCTGAGAAATACCACTGAGCAAGATGTTCTGAAAGCCATCAATGTAAATTCCTTC-3′) and S533D (5′-TGCAATGGTTTATCAGCACGGCAGCCTACAAGGACTTTCAGTCACCAAGAGGCCTCTCTGGAGaCAaGAggatATCCAGAGTCTGAGAAATACCACTGAGCAAGATGTTCTGAAAGCCATCAATGTAAATTCC-3′) ssODNs (lowercase or bolded lowercase indicate silent or coding changes). ssODNs contained the indicated coding change and several silent point mutations to prevent continued targeting of the repaired allele. ssODNs were synthesized with a terminal 5′ and 3′ phosphorothioate bond for stability and were PAGE-purified. Founder mice were genotyped by PCR amplification of Nlrc4 exon 4 using Ipaf-GenoF (5′-ATGGGTCCAGCATGAACGAG-3′) and Ipaf-GenoR primers (5′-TCTGAGAACAAATTGATGCCACAC-3′). PCR products were digested with fast alkaline phosphatase (Thermo Fisher Scientific) and exonuclease I (NEB) and sequenced with Ipaf-GenoF or Ipaf-GenoR primers. To separate alleles, and to remove potential off-target mutations, founders were backcrossed at least three times to C57BL/6J mice before crossing heterozygotes to generate homozygous mutant mice. Littermate animals were used wherever possible, and if not possible, animals were cohoused upon weaning for a minimum of 3 wk.
Toxins
His-tagged Bacillus anthracis protective antigen (PA) and the N terminus of lethal factor (LF) fused to L. pneumophila flagellin (LFn-FlaA) were purified from Escherichia coli using Nickel NTA resin, as described previously (von Moltke et al., 2012). Endotoxin was removed with Detoxi-Gel (Pierce). Toxin doses were 0.8 µg/g body weight of PA combined with 0.4 µg/g LFn-Fla for intravenous (retroorbital) delivery, and as indicated in figure legends for in vitro experiments. Rectal temperature was measured at baseline and at 30 min after injection using a MicroTherma 2T thermometer (Braintree Scientific).
Tissue culture
For melanoma, the B16.F10 melanoma cells used were from American Type Culture Collection (CRL-6475). The cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and 1 mM sodium pyruvate.
For macrophages, bone marrow was harvested from mouse femurs, and cells were differentiated into macrophages by culturing in RPMI supplemented with 5% L929 cell supernatant, glutamine, penicillin-streptomycin, and 10% FBS in a humidified incubator (37°C, 5% CO2).
Bacterial strains
S. Typhimurium strain SL1344 (a gift from G. Barton, University of California, Berkeley, Berkeley, CA) was grown overnight in Luria–Bertani broth and was then diluted 1:40 and grown shaking at 37°C to mid-exponential phase (3 h) to induce expression of the Salmonella pathogenicity island 1 T3SS. CFU was calculated after measuring OD600 of a 1:10 dilution, with OD = 1 equivalent to 1.2 × 109; CFU calculations were confirmed by plating.
Cytotoxicity assays
Cytotoxicity was measured via the activity of LDH released from macrophages as described (Lightfield et al., 2008) and via uptake of PI. Macrophages were seeded in 96-well clear-bottom white-walled tissue culture–treated plates (Corning) at 105 cells/well. Where indicated, cells were primed with 1 µg/ml Pam3CSK4 or 2 µg/ml LPS for 4 h. For FlaTox treatments, media were replaced with media containing 4 µg/ml PA and the indicated concentrations of LFn-FlaA as well as 10 µg/ml PI (Sigma-Aldrich). For NLRP3 priming control experiments, primed cells were treated with 10 µM nigericin. Cells were incubated 4 h at 37°C, and then PI fluorescence was assessed and supernatants were analyzed for LDH activity.
For Salmonella infections, S. Typhimurium was added at an MOI of 1 or 5 to a 96-well plate containing 105 unprimed macrophages per well. Medium was replaced with medium containing gentamicin (20 µg/ml), to kill extracellular bacteria, and 10 µg/ml PI (Sigma-Aldrich) 30 min after infection with S. Typhimurium. Propidium iodide fluorescence was assessed and culture supernatants were assayed for LDH activity after 4 h. For both assays, infection-specific lysis was calculated as the percentage of detergent-lysed macrophages.
IL-1β ELISA
Supernatants were collected 4 h after treatment and diluted 1:5 in PBS containing 1% bovine serum albumin. ELISA was performed using anti–IL-1β monoclonal antibody (B122, eBioscience) in 0.1 M sodium phosphate buffer for capture and biotinylated anti–IL-1β polyclonal antibody (13–7112-81, eBioscience) with streptavidin-HRP (Invitrogen) in PBS containing 1% bovine serum albumin and o-phenylenediamine dihydrochloride (Sigma-Aldrich) for detection.
In vivo Salmonella infections
S. Typhimurium infections were done as previously described (Barthel et al., 2003). Briefly, 10- to 15-wk-old mice deprived of food and water for 4 h were gavaged with 25 mg streptomycin sulfate in H2O. After 20 h, mice were again deprived of food and water for 4 h and were then gavaged with 2 × 107 CFU S. Typhimurium SL1344. After 18 h of infection, the animals were sacrificed, tissue was harvested, and ceca were flushed and incubated in PBS containing 400 mg/ml gentamicin for 30 min to kill luminal bacteria before washing six times in PBS. Cecum and mesenteric lymph node were homogenized in sterile PBS and plated on MacConkey agar containing 50 µg/ml streptomycin.
Western blot
Macrophages were grown in tissue culture–treated 24-well plates, medium was removed, and cells were lysed in plate in radioimmunoprecipitation assay buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and 1× Roche protease inhibitor tablet [no EDTA], pH 8.0) for 20 min at 4°C. Lysates were clarified by centrifugation at 16,000 g for 10 min at 4°C, and supernatants were separated by 4–12% SDS-PAGE in morpholinoethanesulfonic acid buffer (Invitrogen). Proteins were transferred to Immobilon-FL polyvinylidene fluoride membranes (Millipore). Membranes were blocked with 5% milk. Anti-NLRC4 antibody (gift of S. Mariathasan and V. Dixit, Genentech, South San Francisco, CA) was detected with a secondary anti-rabbit IgG conjugated to HRP (GE Healthcare).
Tumor model
Heterozygous Nlrc4+/−, newly generated on the C57Bl/6J background, were crossed to generate cohoused littermate Nlrc4+/+ and Nlrc4−/− mice. Mice were subcutaneously injected in the right abdominal flank with 1 × 105 B16F10 cells. The tumors were then measured every 2–3 d with digital calipers. Mice were sacrificed 16–19 d after injection.
Online supplemental material
Fig. S1 demonstrates that NAIPs, not NLRP3, are required for NLRC4 signaling, and that Nlrp3−/− macrophages lack nigericin response.
Acknowledgments
We thank D. Raulet for assistance with B16F10 melanoma experiments and G. Barton for providing S. Typhimurium.
I. Rauch is supported by Oregon Health and Science University. R. Vance is an Investigator of the Howard Hughes Medical Institute, and research in his laboratory is supported by National Institutes of Health grants AI075039 and AI066302.
Author contributions: J.L. Tenthorey, T.W. Thompson, R. Vance, and I. Rauch designed experiments. J.L. Tenthorey, R.A. Chavez, K.A. Deets, and I. Rauch collected and analyzed data. J.L. Tenthorey, R. Vance, and I. Rauch wrote the manuscript.
References
- Allen I.C., TeKippe E.M.E., Woodford R.M.T., Uronis J.M., Holl E.K., Rogers A.B., Herfarth H.H., Jobin C., and Ting J.P.Y.. 2010. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207:1045–1056. 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J.D., Collier R.J., and Starnbach M.N.. 1996. Anthrax toxin-mediated delivery of a cytotoxic T-cell epitope in vivo. Proc. Natl. Acad. Sci. USA. 93:12531–12534. 10.1073/pnas.93.22.12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M., Hapfelmeier S., Quintanilla-Martínez L., Kremer M., Rohde M., Hogardt M., Pfeffer K., Rüssmann H., and Hardt W.D.. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858. 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., and Monack D.M.. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755. 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Hu B., Elinav E., Huber S., Booth C.J., Strowig T., Jin C., Eisenbarth S.C., and Flavell R.A.. 2010. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc. Natl. Acad. Sci. USA. 107:21635–21640. 10.1073/pnas.1016814108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yan C., Liu P., Huang Z., Ma R., Zhang C., Wang R., Zhang Y., Martinon F., Miao D., et al. 2013. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 341:172–175. 10.1126/science.1236381 [DOI] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.W., Sui S.F., and Chai J.. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 350:399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Janowski A.M., Colegio O.R., Hornick E.E., McNiff J.M., Martin M.D., Badovinac V.P., Norian L.A., Zhang W., Cassel S.L., and Sutterwala F.S.. 2016. NLRC4 suppresses melanoma tumor progression independently of inflammasome activation. J. Clin. Invest. 126:3917–3928. 10.1172/JCI86953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Man S.M., and Kanneganti T.D.. 2017. Inflammasomes and Cancer. Cancer Immunol. Res. 5:94–99. 10.1158/2326-6066.CIR-16-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb R., Phan L., Borcherding N., Liu Y., Yuan F., Janowski A.M., Xie Q., Markan K.R., Li W., Potthoff M.J., et al. 2016. Obesity-associated NLRC4 inflammasome activation drives breast cancer progression. Nat. Commun. 7:13007 10.1038/ncomms13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., and Dixit V.M.. 2014. Mechanisms and functions of inflammasomes. Cell. 157:1013–1022. 10.1016/j.cell.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Lightfield K.L., Persson J., Brubaker S.W., Witte C.E., von Moltke J., Dunipace E.A., Henry T., Sun Y.H., Cado D., Dietrich W.F., et al. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9:1171–1178. 10.1038/ni.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu X., Li Y., Zhao J., Liu Z., Hu Z., Wang Y., Yao Y., Miller A.W., Su B., et al. 2017. LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella Typhimurium infection. J. Exp. Med. 214:3051–3066. 10.1084/jem.20170014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Martinon F., Burns K., and Tschopp J.. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Matusiak M., Van Opdenbosch N., Vande Walle L., Sirard J.C., Kanneganti T.D., and Lamkanfi M.. 2015. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc. Natl. Acad. Sci. USA. 112:1541–1546. 10.1073/pnas.1417945112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E., Leaf I.A., and Aderem A.. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 107:3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K., Wang Z., Yang Y.M., Billet S., Tu W., Pimienta M., Cassel S.L., Pandol S.J., Lu S.C., Sutterwala F.S., et al. 2019. NOD-like receptor C4 Inflammasome Regulates the Growth of Colon Cancer Liver Metastasis in NAFLD. Hepatology. 70:1582–1599. 10.1002/hep.30693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet J.L., Srinivasula S.M., Tnani M., Razmara M., Fernandes-Alnemri T., and Alnemri E.S.. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309–28313. 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- Qu Y., Misaghi S., Izrael-Tomasevic A., Newton K., Gilmour L.L., Lamkanfi M., Louie S., Kayagaki N., Liu J., Kömüves L., et al. 2012. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 490:539–542. 10.1038/nature11429 [DOI] [PubMed] [Google Scholar]
- Qu Y., Misaghi S., Newton K., Maltzman A., Izrael-Tomasevic A., Arnott D., and Dixit V.M.. 2016. NLRP3 recruitment by NLRC4 during Salmonella infection. J. Exp. Med. 213:877–885. 10.1084/jem.20132234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A.K., and Fitzgerald K.A.. 2016. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell. 165:792–800. 10.1016/j.cell.2016.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I., Tenthorey J.L., Nichols R.D., Al Moussawi K., Kang J.J., Kang C., Kazmierczak B.I., and Vance R.E.. 2016. NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo. J. Exp. Med. 213:657–665. 10.1084/jem.20151809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I., Deets K.A., Ji D.X., von Moltke J., Tenthorey J.L., Lee A.Y., Philip N.H., Ayres J.S., Brodsky I.E., Gronert K., and Vance R.E.. 2017. NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and -8. Immunity. 46:649–659. 10.1016/j.immuni.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F.. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526:660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Simon M.M., Greenaway S., White J.K., Fuchs H., Gailus-Durner V., Wells S., Sorg T., Wong K., Bedu E., Cartwright E.J., et al. 2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14:R82 10.1186/gb-2013-14-7-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.L., et al. 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 350:1084–1089. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N., Liu Z.S., Xue W., Bai Z.F., Wang Q.Y., Dai J., Liu X., Huang Y.J., Cai H., Zhan X.Y., et al. 2017. NLRP3 Phosphorylation Is an Essential Priming Event for Inflammasome Activation. Mol. Cell. 68:185–197.e6. 10.1016/j.molcel.2017.08.017 [DOI] [PubMed] [Google Scholar]
- Tenthorey J.L., Haloupek N., López-Blanco J.R., Grob P., Adamson E., Hartenian E., Lind N.A., Bourgeois N.M., Chacón P., Nogales E., and Vance R.E.. 2017. The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science. 358:888–893. 10.1126/science.aao1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland T.K., Jain N., Hornick E.E., Elliott E.I., Clay G.M., Sadler J.J., Mills K.A.M., Janowski A.M., Volk A.P.D., Wang K., et al. 2016. Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat. Commun. 7:13180 10.1038/ncomms13180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N.J., Moayeri M., Kintzer A.F., Wang S.B., van Rooijen N., Brown C.R., Krantz B.A., Leppla S.H., Gronert K., and Vance R.E.. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 490:107–111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni D.S., Kobayashi K.S., Kohlsdorf T., Ogura Y., Long E.M., Vance R.E., Kuida K., Mariathasan S., Dixit V.M., Flavell R.A., et al. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318–325. 10.1038/ni1305 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 350:404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.-N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Shi J., Shi X., Wang Y., Wang F., and Shao F.. 2016. Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. J. Exp. Med. 213:647–656. 10.1084/jem.20160006 [DOI] [PMC free article] [PubMed] [Google Scholar]