Tay et al. show that HDAC3 acts early during CD8 T cell activation to inhibit cytotoxicity and effector differentiation gene programs. This study identifies a novel role for HDAC3 as an epigenetic regulator of CD8 T cell cytotoxicity and persistence following activation.
Abstract
Cytotoxic T cells play a key role in adaptive immunity by killing infected or cancerous cells. While the transcriptional control of CD8 T cell differentiation and effector function following T cell activation has been extensively studied, little is known about epigenetic regulation of these processes. Here we show that the histone deacetylase HDAC3 inhibits CD8 T cell cytotoxicity early during activation and is required for persistence of activated CD8 T cells following resolution of an acute infection. Mechanistically, HDAC3 inhibits gene programs associated with cytotoxicity and effector differentiation of CD8 T cells including genes encoding essential cytotoxicity proteins and key transcription factors. These data identify HDAC3 as an epigenetic regulator of the CD8 T cell cytotoxicity program.
Introduction
Following an infection, naive CD8 T cells become activated and develop into effector cytotoxic T lymphocytes that mediate immunity by contact-dependent killing of infected cells and secretion of effector cytokines including IFN-γ and TNF-α. At the peak of the CD8 T cell effector response to infection, CD8 T cells have already committed to one of at least two cell fates, a short-lived, terminally differentiated state with potent effector function (Joshi et al., 2007; Sarkar et al., 2008) or a memory precursor state with higher proliferative potential (Kaech et al., 2003; Schluns et al., 2000). Following antigen clearance, the majority of terminally differentiated effector T cells undergo apoptosis, whereas memory precursor cells differentiate into memory T cells that can mount a robust and rapid response upon reinfection.
The transcriptional regulation of these developmental processes has been extensively studied. Key transcription factors have been identified that regulate effector and memory differentiation, including T-bet (Tbx21 gene), Eomes (Eomes), Blimp-1 (Prdm1), Bcl-6 (Bcl6), Id2 (Id2), and Id3 (Id3; reviewed in Chang et al., 2014; Kaech and Cui, 2012). T-bet and Eomes cooperate in several key aspects of CD8 T cell function, including effector and memory programs (Intlekofer et al., 2005). The T-bet to Eomes ratio is highest in effector cells and lowest in memory cells, consistent with an important role for T-bet in effector differentiation (Intlekofer et al., 2007; Joshi et al., 2007) and for Eomes in memory CD8 T cell differentiation (Banerjee et al., 2010; Intlekofer et al., 2005). Blimp-1 is part of a transcriptional program that favors differentiation of terminal effector T cells (Kallies et al., 2009; Rutishauser et al., 2009; Shin et al., 2013), while Bcl-6 acts as an antagonist of Blimp-1 that promotes memory T cell differentiation (Cui et al., 2011; Ichii et al., 2002, 2004). Id2 favors survival of effector T cells while Id3 is required for the generation of long-lived memory CD8 T cells (Cannarile et al., 2006; Ji et al., 2011; Yang et al., 2011). These studies have been indispensable to our current understanding of CD8 T cell effector and memory differentiation as a process governed by key transcriptional factors that form a complex regulatory network (Best et al., 2013).
However, much less is known about the epigenetic regulation of CD8 T cell effector and memory differentiation (recently reviewed in Henning et al., 2018). The majority of investigations have focused on profiling global patterns of epigenetic modifications or chromatin accessibility in relationship to effector and memory differentiation (Gray et al., 2017; Philip et al., 2017; Russ et al., 2014), whereas other studies have taken a classical reverse genetics approach of characterizing the phenotype of mice deficient for a known epigenetic regulator (Gray et al., 2017; Pace et al., 2018). Two recent studies using combinations of these approaches have uncovered novel roles for the histone methyltransferases EZH2 (Gray et al., 2017; a key component of the polycomb repressive complex) and SUV39H1 (Pace et al., 2018) in regulating CD8 T cell function and differentiation. Here, we used a complementary approach to uncover and characterize novel epigenetic regulators of CD8 T cell effector function by screening small molecule inhibitors covering diverse epigenetic regulatory pathways (recently reviewed in Bennett and Licht, 2018) in order to perturb CD8 T cell function.
Results
HDAC3 is a negative regulator of CD8 T cell cytotoxicity
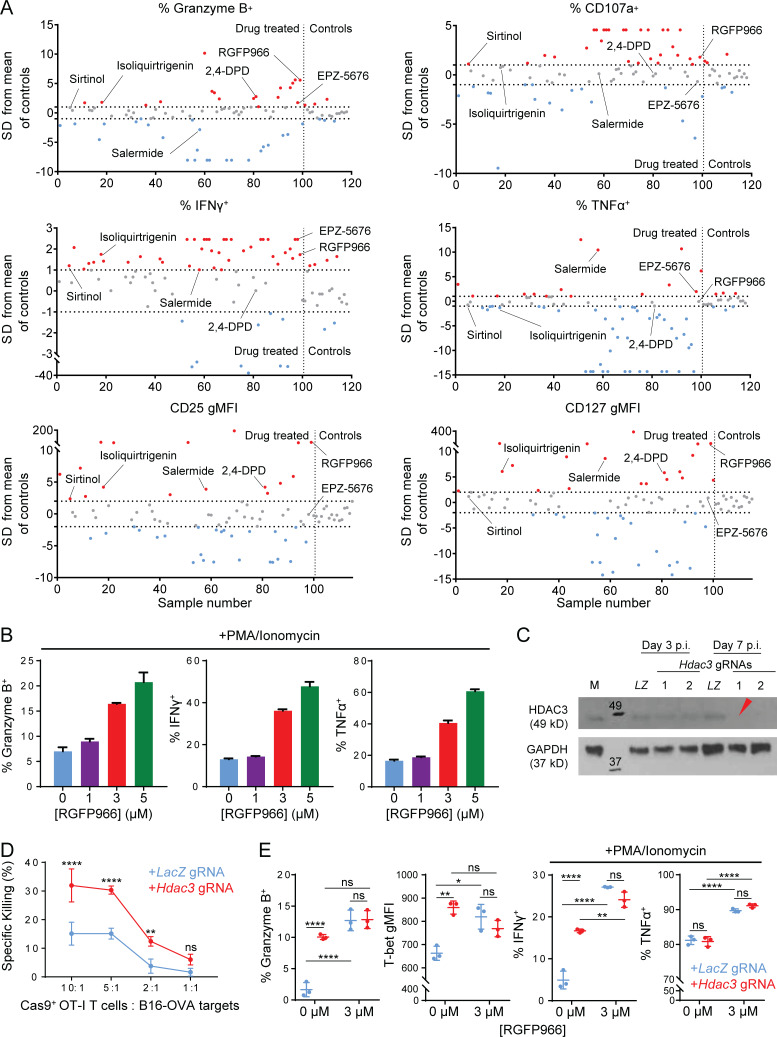
We identified histone deacetylase 3 (HDAC3) as a potential regulator of CD8 T cell cytotoxicity in a functional screen of a library of 100 small molecules targeting epigenetic regulators. We tested these inhibitors by coculturing OVA-specific OT-I+ CD8 T cells (OT-I T cells) with irradiated OVA peptide–pulsed bone marrow–derived dendritic cells (BMDCs) for 7 d in the presence of drugs, and used high-throughput flow cytometry to measure perturbations of markers of CD8 T cell effector function including Granzyme B, IFN-γ, and CD25. The drugs were then ranked based on their ability to modulate these readouts of CD8 T cell effector function (Fig. S1 A and Table S1). The top hit from this screen was the HDAC3-specific inhibitor RGFP966 (Xu et al., 2009), which induced a substantial increase in the cytotoxicity-associated functional markers Granzyme B and CD107a (LAMP-1), the effector cytokine IFN-γ, and the activation-associated surface receptors CD25 (IL-2Rα) and CD127 (IL-7Rα; Fig. S1 A). In subsequent validation experiments, we confirmed that activating CD8 T cells in the presence of RGFP966 resulted in a dose-dependent increase in the percentage of T cells expressing Granzyme B, IFN-γ, and TNF-α (Fig. S1 B). Based on the reported high specificity of RGFP966 in inhibiting HDAC3 relative to other class I HDACs (HDAC1, 2, and 8), we selected HDAC3 for further investigation.
Figure S1.
HDAC3-specific inhibitor RGFP966 augments cytotoxicity function of CD8 T cells. Related to Fig. 1. (A) Summary of flow cytometry screen of 100 epigenetic inhibitor drugs during in vitro CD8 T cell activation for perturbations to effector phenotype. OT-I CD8 T cells were activated in vitro by co-culture with OVA peptide–pulsed BMDCs for 7 d in the presence of indicated epigenetic inhibitor drugs (10 µM) or an equivalent volume of DMSO as vehicle control. Flow-cytometric measurements for each of the six indicated markers were acquired in parallel for all samples. Perturbations to each marker in drug-treated samples were normalized relative to the mean of control samples, and are expressed in terms of SDs from the mean of control samples. Horizontal dashed lines indicate threshold for significance (|Drug perturbation| ≥1 SD for bimodal markers, ≥2 SD for unimodal markers); the vertical dashed line separates drug-treated samples (samples numbers 1–100) from control samples (sample numbers ≥101). Drugs that increased (red) or decreased (blue) a given marker above the significance threshold are indicated. The top six drug hits are labeled. (B) Flow-cytometric measurement of RGFP966-induced changes of CD8 effector functions in OT-I T cells activated in vitro for 7 d. Data are representative of two independent experiments. (C) Evaluation of KO efficiencies for lentiviral gRNA expression vectors targeting Hdac3 in transduced Cas9+ OT-I T cells. Whole cell lysates prepared from 5 × 105 magnetically purified transduced T cells were loaded per lane. LZ, LacZ-targeting gRNA vector; M, mock-transduced (both negative controls); p.i., post-infection. Red arrowhead indicates the gRNA used in subsequent experiments. Molecular weights (kD) are indicated on immunoblot images. (D) Cytotoxicity of in vitro activated Cas9+ OT-I T cells transduced with indicated gRNA sequences against B16-OVA targets. Data are representative of two independent experiments with four technical replicates per condition. (E) Flow-cytometric measurement of markers of CD8 effector function in activated Hdac3- or LacZ-edited Cas9+ OT-I T cells after 7 d of activation in the presence or absence of RGFP966. Data are representative of two independent experiments with three replicates per condition. Means ± SD are indicated (B, D, and E). P values were calculated by two-way ANOVA (D and E). *, P < 0.05, **, P < 0.01, ****, P < 0.0001.
The data from drug screen supported the hypothesis that inactivation of HDAC3 may enhance the cytotoxic function of CD8 T cells. Indeed, we found that RGFP966-treated OT-I T cells exhibited substantially higher cytotoxic activity against OVA-expressing B16F10 melanoma target cells (B16-OVA) relative to vehicle-treated OT-I T cells (Fig. 1 A). To validate HDAC3 as a regulator of CD8 T cell cytotoxicity, we inactivated the Hdac3 gene in OT-I T cells isolated from Cas9 transgenic mice with a lentiviral guide RNA (gRNA) expression vector (Fig. S1 C). Hdac3 gRNA-transduced CD8 T cells showed increased cytotoxicity against B16-OVA target cells compared with CD8 T cells transduced with a control LacZ gRNA (Fig. S1 D). In addition, a larger percentage of Hdac3-inactivated CD8 T cells expressed Granzyme B and the effector phenotype-associated transcription factor T-bet (Lazarevic et al., 2013) compared with control gRNA-transduced CD8 T cells (Fig. S1 E, left panels); addition of RGFP966 to Hdac3 gRNA-transduced cells did not further increase the percentage of Granzyme B+ or T-bet+ cells. Additionally, inactivation of Hdac3 in CD8 T cells did not result in substantial changes in effector cytokine expression as was observed in RGFP966-treated CD8 T cells, except for a small increase in IFN-γ expression (Fig. S1 E, right panels).
Figure 1.
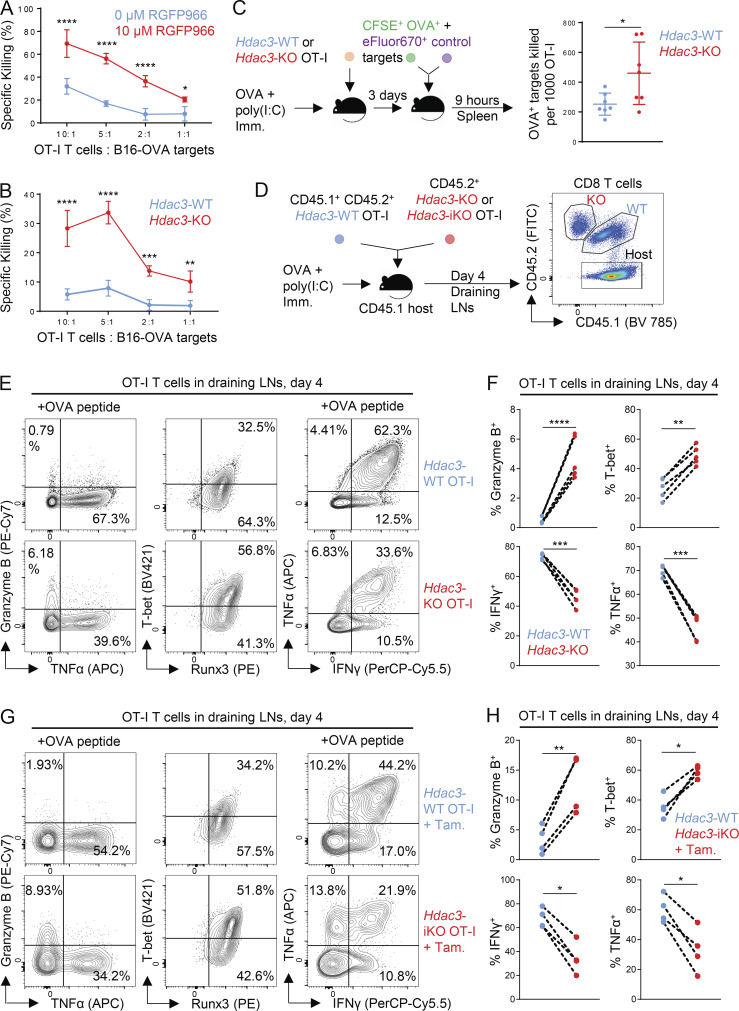
HDAC3 negatively regulates CD8 T cell cytotoxicity. (A and B) Cytotoxicity of in vitro activated RGFP966-treated (A) and Hdac3-KO (B) OT-I T cells against OVA-expressing B16 melanoma targets (B16-OVA). Data are representative of two independent experiments, each with four technical replicates per condition. (C) In vivo cytotoxicity of adoptively transferred Hdac3-KO and Hdac3-WT OT-I T cells against OVA peptide–pulsed target cells. Data are representative of two independent experiments with seven host mice for each genotype of transferred donor OT-I T cells. (D) Experimental scheme of OVA immunization (Imm.) experiments following OT-I transfer. 5 × 105 cells of a 1:1 mix of congenically distinct Hdac3-KO (E8I-Cre+; Hdac3fl/fl) and Hdac3-WT OT-I T cells were transferred into CD45.1+ TCR-polyclonal recipients. Alternatively, 5 × 105 cells of a 1:1 mix of congenically distinct Hdac3-iKO (UBC-Cre-ERT2+; Hdac3fl/fl) and Hdac3-WT OT-I T cells were transferred, and 2 mg of tamoxifen (Tam.) was administered daily for 3 d to induce Cre-mediated inactivation of Hdac3. Mice were immunized subcutaneously with OVA + poly(I:C) in PBS. (E and F) Flow-cytometric analysis (E) and quantification (F) of markers of effector phenotype of Hdac3-WT and Hdac3-KO OT-I T cells in inguinal LNs draining the immunization site harvested 4 d after immunization as in D. Data are representative of two independent experiments with five recipient mice each. Data from cotransferred donor cells of each genotype within the same recipient mouse were analyzed as pairs. (G and H) Flow-cytometric analysis (G) and quantification (H) of markers of effector phenotype of Hdac3-WT and Hdac3-iKO (Hdac3-inducible knockout) OT-I T cells in inguinal LNs draining the immunization site harvested 4 d after immunization as in D. Data are representative of two independent experiments with five recipient mice each. Data from cotransferred donor cells of each genotype within the same recipient mouse were analyzed as pairs. Means ± SD are indicated (A–C). P values were calculated by two-way ANOVA (A and B), two-tailed Student’s t test (C), or two-tailed ratio-paired t test (F and H). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001.
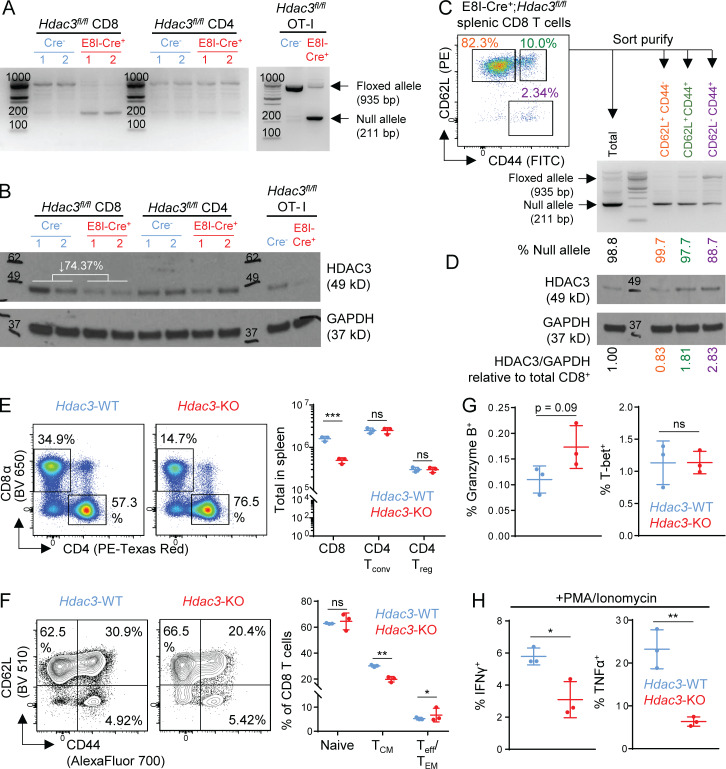
To investigate the potential role of HDAC3 in regulating CD8 T cell cytotoxicity and effector function in vivo, we generated TCR polyclonal and OT-I TCR transgenic mice with CD8 T cell–restricted deletion of Hdac3. Because HDAC3 is essential for T cell development in the thymus (Hsu et al., 2015; Philips et al., 2016; Stengel et al., 2015), Hdac3 deletion was restricted to mature CD8 T cells by crossing a E8I-Cre transgenic strain to mice with a floxed allele of Hdac3 to generate E8I-Cre+; Hdac3fl/fl mice (hereafter referred to as Hdac3-KO mice). E8I-Cre+ mice express Cre recombinase under the control of a regulatory element of Cd8a that is active during the late CD8 single-positive (SP) stage and in mature CD8 T cells (Ellmeier et al., 1997). We did not observe significant changes in thymic development for TCR polyclonal Hdac3-KO compared with Hdac3-WT mice (Fig. S2, A–D), except for a small decrease in the relative frequencies of CD4+ CD8+ double-positive (DP) and CD44hi CD25lo CD4− CD8− double-negative (DN1) cells, and a small increase in CD4+ SP (CD4 SP) thymocytes in Hdac3-KO mice relative to Hdac3-WT littermates (Fig. S2, B–D). Importantly, the numbers of total and CD8 SP thymocytes were similar between Hdac3-KO and Hdac3-WT mice (Fig. S2 B), as were the frequencies of mature CD5+ cells within the CD8 SP thymocyte populations from both genotypes (Fig. S2, C and D, rightmost panels).
Figure S2.
Characterization of thymopoiesis in E8I-Cre+; Hdac3fl/flHdac3-KO mice. Related to Fig. 1. (A) Representative flow cytometry gating of live CD11b− CD11c− thymocyte populations in male 10-wk-old Hdac3-KO (E8I-Cre+; Hdac3fl/fl) mice and Hdac3-WT (Cre−; Hdac3fl/fl) littermates based on CD4 and CD8α expression. (B) Flow-cytometric measurement of thymocyte developmental stages, gated as in A. DD, CD4lo CD8lo double-dull; CD4, CD4+ CD8− single-positive; CD8, CD4− CD8+ single-positive thymocytes. (C and D) Flow-cytometric analysis (C) and quantification (D) of indicated thymocyte developmental stages in Hdac3-KO mice and Hdac3-WT littermates. (From left to right) Distribution of DN subsets, CD69hi MHC Ilo DD cells undergoing negative selection, CD5hi TCRβ+ DP cells undergoing positive selection, CD5+ mature CD4 SP, and CD5+ mature CD8 SP thymocytes. Data are representative of two independent experiments with three to five mice per genotype each. Means ± SD are indicated (B and D). P values were calculated by two-tailed Student’s t test (B and D). *, P < 0.05.
We further confirmed that Hdac3 was inactivated only in mature CD8 but not CD4 T cells of Hdac3-KO mice, both in TCR polyclonal and OT-I TCR transgenic strains, by performing PCR analysis of genomic DNA and Western blot analysis of whole lysates from sorted cell populations (Fig. S3, A and B). The extent of Hdac3 KO in TCR polyclonal mice was highest in CD62L+ CD44− naive CD8 T cells, followed by CD62L+ CD44+ central memory phenotype cells (TCM) and CD62L− CD44+ effector/effector memory phenotype cells (Teff/EM; Fig. S3, C and D). The major peripheral T cell compartments were intact in Hdac3-KO mice, albeit with moderately reduced total CD8 T cell numbers relative to Hdac3-WT littermates (Fig. S3 E). CD8 T cells from Hdac3-KO mice showed a small decrease in the percentage of CD44hi CD62L+ (TCM) and a small increase in the percentage CD44hi CD62L− (Teff/TEM) phenotypes compared with Hdac3-WT mice, while the percentage of CD44lo CD62L+ naive cells was unchanged (Fig. S3 F). Finally, while CD8 T cells from Hdac3-KO and Hdac3-WT mice were not significantly different in terms of Granzyme B or T-bet expression (Fig. S3 G), a smaller percentage of Hdac3-KO T cells produced IFN-γ or TNF-α in response to ex vivo stimulation with PMA and ionomycin (Fig. S3 H). Overall, these results indicated that Hdac3-KO mice indeed possessed a relatively intact peripheral T cell compartment with CD8 T cell–restricted inactivation of Hdac3.
Figure S3.
Characterization of peripheral T cells in E8I-Cre+; Hdac3fl/flHdac3-KO mice. Related to Fig. 1. (A and B) Evaluation of on-target KO efficiency of Hdac3 in E8I-Cre+; Hdac3fl/fl mice. Total CD8 and CD4 T cells were sorted from 6-wk-old TCR-polyclonal E8I-Cre+;Hdac3fl/fl and Cre− Hdac3fl/fl littermates (three mice each) for assessment of KO efficiency by PCR (A) and immunoblot (B) analysis. A parallel analysis was performed for magnetically purified OT-I transgenic E8I-Cre+; Hdac3fl/fl and Cre−; Hdac3fl/fl T cells. 50 ng of purified genomic DNA was loaded as a template for PCR. Whole cell lysates prepared from 5 × 105 cells were loaded for immunoblotting. Molecular weights in basepairs or kilodaltons are indicated on PCR gel and immunoblot images, respectively. The mean percentage reduction in band intensity for HDAC3 in Hdac3-KO relative to Hdac3-WT CD8 T cells is indicated on the Western blot image. (C and D) Evaluation of Cre-mediated loss of Hdac3 in subpopulations of peripheral CD8 T cells in E8I-Cre+; Hdac3fl/fl mice. Sort-purified total, CD62L+ CD44− naive, CD62L+ CD44+ TCM, and CD62L− CD44+ Teff/EM CD8 T cells pooled from 3 E8I-Cre+; Hdac3fl/fl mice were analyzed by PCR (C) and immunoblot (D) as in A and B, respectively. (E) Cell numbers of splenic T cell compartments in 10-wk-old TCR-polyclonal E8I-Cre+ and Cre− Hdac3fl/fl littermates. CD4 Tconv, conventional CD4+ FoxP3− T cells; CD4+ Treg, CD4+ FoxP3+ regulatory T cells. Data are from one experiment with three mice per genotype. (F) Flow-cytometric characterization of surface marker phenotypes of splenic CD8 T cells in Hdac3-KO and Hdac3-WT mice gated as in E. Naive, CD62Lhi CD44lo cells; TCM, CD62Lhi CD44hi central memory cells; Teff/TEM, CD62Llo CD44hi effector or effector memory cells. Data are from one experiment with three mice per genotype. (G and H) Analysis of markers of CD8 effector function in splenic CD8 T cells in Hdac3-KO and Hdac3-WT mice gated as in E. Data are from one experiment with three mice per genotype (E–H). Means ± SD are indicated (E–H). P values were calculated by two-tailed Student’s t test. *, P < 0.05, **, P < 0.01, ***, P < 0.001.
Hdac3-KO OT-I T cells were more cytotoxic against B16-OVA melanoma target cells than their Hdac3-WT counterparts following in vitro activation (Fig. 1 B), consistent with our previous results using gRNA-edited CD8 T cells. We further validated this phenotype in an in vivo model of T cell–mediated cytotoxicity. We first transferred naive OT-I T cells from Hdac3-KO mice or Hdac3-WT littermates into C57BL/6 hosts and immunized recipient mice with a low dose of OVA + polyinosinic:polycytidylic acid (poly[I:C]). 3 d later, we transferred differentially labeled OVA peptide– and control peptide–pulsed splenocyte targets and measured depletion of both populations in the spleen after 9 h. We found that Hdac3-KO OT-I T cells killed a larger number of OVA peptide–pulsed than control target cells on a per cell basis (Fig. 1 C). Overall, our results using three separate approaches to ablate HDAC3 activity in CD8 T cells demonstrated that HDAC3 inhibits CD8 T cell cytotoxicity.
To evaluate cell-intrinsic changes in CD8 T cell effector function caused by loss of HDAC3 in an in vivo setting, we cotransferred congenically distinct Hdac3-KO and Hdac3-WT naive OT-I T cells into TCR-polyclonal hosts, immunized recipient mice with OVA + poly(I:C), and evaluated the effector phenotypes of transferred OT-I T cells on day 4 in draining LNs (Fig. 1 D). In agreement with our previous observations, an increased percentage of Hdac3-KO CD8 T cells expressed Granzyme B and T-bet compared with cotransferred Hdac3-WT cells (Fig. 1, E and F). We also observed that a reduced percentage of Hdac3-KO compared with cotransferred Hdac3-WT CD8 T cells produced IFN-γ or TNF-α following ex vivo restimulation with OVA peptide (Fig. 1, E and F). Finally, to verify that these phenotypes were specific to a loss of Hdac3 during CD8 T cell activation and not due to changes in CD8 T cell development, we repeated the cotransfer and OVA immunization experiments using OT-I T cells derived from Hdac3fl/fl mice bearing a tamoxifen-inducible Cre transgene (Hdac3-iKO mice) and inactivated Hdac3 by administration of tamoxifen beginning 3 d before activation. Hdac3-iKO OT-I T cells with Hdac3 inactivated just before activation showed an increased frequency of Granzyme B+ and T-bet+ cells, with decreased percentages of IFN-γ– and TNF-α–secreting cells, relative to cotransferred Hdac3-WT OT-I T cells (Fig. 1, G and H), recapitulating our observations with Hdac3-KO OT-I T cells.
HDAC3 is required for persistence of CD8 T cells following antigen stimulation
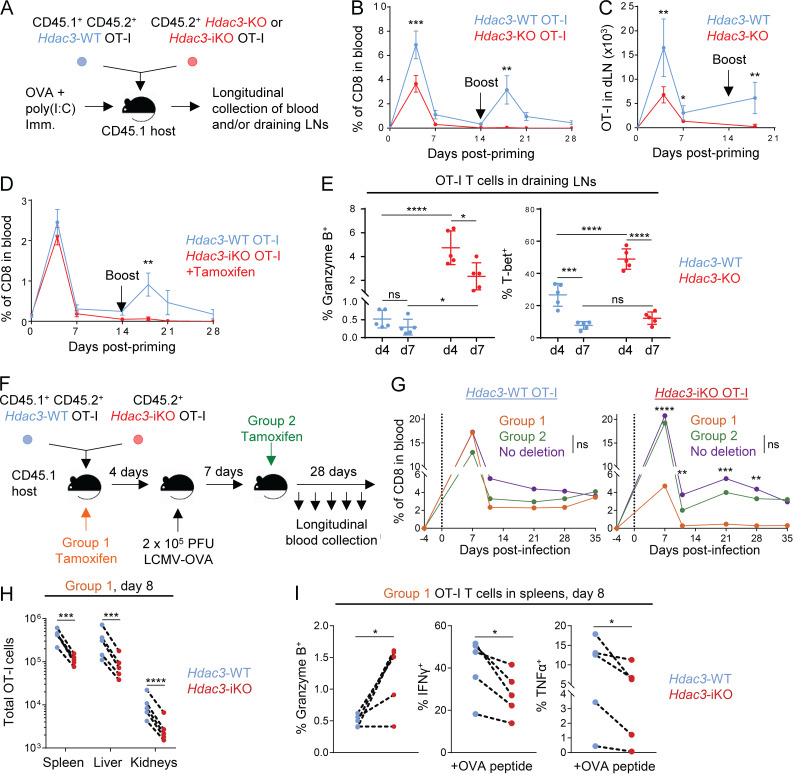
We next investigated whether loss of HDAC3 altered the dynamics of the CD8 T cell response by tracking the relative frequencies of cotransferred Hdac3-KO and Hdac3-WT OT-I CD8 T cells over a time course following OVA + poly(I:C) immunization (Fig. 2 A). In this setting, we found that Hdac3-KO OT-I CD8 T cells did not persist in peripheral blood or draining LNs compared with cotransferred Hdac3-WT cells following contraction of the OT-I response (Fig. 2, B and C). Furthermore, Hdac3-KO OT-I T cells failed to expand following rechallenge with OVA + poly(I:C) (Fig. 2, B and C); we also observed this failure to expand following OVA + poly(I:C) rechallenge in experiments where we cotransferred Hdac3-iKO and Hdac3-WT OT-I T cells and inactivated Hdac3 by tamoxifen injection before immunization (Fig. 2 D). In addition, Granzyme B and T-bet expression in activated OT-I T cells declined between days 4 and 7, such that there was only a small increase in the percentage of Granzyme B+ Hdac3-KO relative to Hdac3-WT cells and no significant differences in the proportions of T-bet+ cells by day 7 after immunization (Fig. 2 E).
Figure 2.
HDAC3 is required during T cell activation for persistence of the CD8 T cell response. (A) Experimental scheme for longitudinal tracking of OT-I T cell persistence following transfer and OVA immunization. 5 × 105 cells of a 1:1 mix of congenically distinct Hdac3-KO and Hdac3-WT OT-I T cells were transferred into CD45.1+ TCR-polyclonal recipients. Alternatively, 5 × 105 cells of a 1:1 mix of congenically distinct Hdac3-iKO and Hdac3-WT OT-I T cells were transferred, and 2 mg of tamoxifen was administered daily for 3 d to induce Cre-mediated inactivation of Hdac3. Mice were immunized subcutaneously with OVA + poly(I:C) in PBS. A secondary boost of OVA + poly(I:C) was administered on day 14. (B and C) Dynamics of Hdac3-KO and Hdac3-WT OT-I T cell responses were tracked longitudinally in peripheral blood (B) and inguinal LNs (C) draining the immunization site following immunization as described in A. Data are representative of two independent experiments with four or five recipient mice each. (D) Dynamics of Hdac3-iKO and Hdac3-WT OT-I T cell responses were tracked longitudinally in peripheral blood following immunization as in A. Data are representative of two independent experiments with five recipient mice each. (E) Quantification of flow-cytometric analysis of Granzyme B+ and T-bet+ OT-I T cells on days 4 and 7 following transfer and immunization as in A. Data are representative of two independent experiments with five recipient mice each. (F) Experimental scheme for longitudinal tracking of OT-I T cell persistence following transfer in an acute viral infection model. 5 × 103 cells of a 1:1 mix of congenically distinct Hdac3-iKO and Hdac3-WT OT-I T cells were transferred into CD45.1+ TCR-polyclonal recipients. Mice were then infected intraperitoneally with 2 × 105 PFU of LCMV Armstrong expressing the OVA SIINFEKL epitope (LCMV-OVA), and Hdac3 deletion was induced at the indicated time points by intraperitoneal injection of tamoxifen on 3 consecutive days. (G) Donor OT-I responses in peripheral blood were monitored longitudinally for the indicated treatment groups (Group 1, n = 4; Group 2, n = 4; No deletion, n = 3) after treatment as in F. Data are representative of two independent experiments with three to five recipient mice per treatment group. (H and I) Analysis of total numbers (H) and markers of effector phenotype (I) of cotransferred Hdac3-iKO (Hdac3-inducible knockout) and Hdac3-WT OT-I T cells by flow cytometry 8 d after infection with LCMV-OVA. Data are representative of two independent experiments with five recipient mice each, treated as in F. Data from cotransferred donor cells of each genotype within the same recipient mouse were analyzed as pairs. Means ± SEM (B–D), means ± SD (E), or means (G) are indicated. P values were calculated by two-way ANOVA (B–E and G) or two-tailed ratio-paired t test (H and I). Error bars for G were omitted for clarity of presentation; statistical analysis for this experiment was also performed on the ratios of transferred Hdac3-KO to Hdac3-WT OT-I T cells and is shown in Fig. S5 B. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001; ns, not significant.
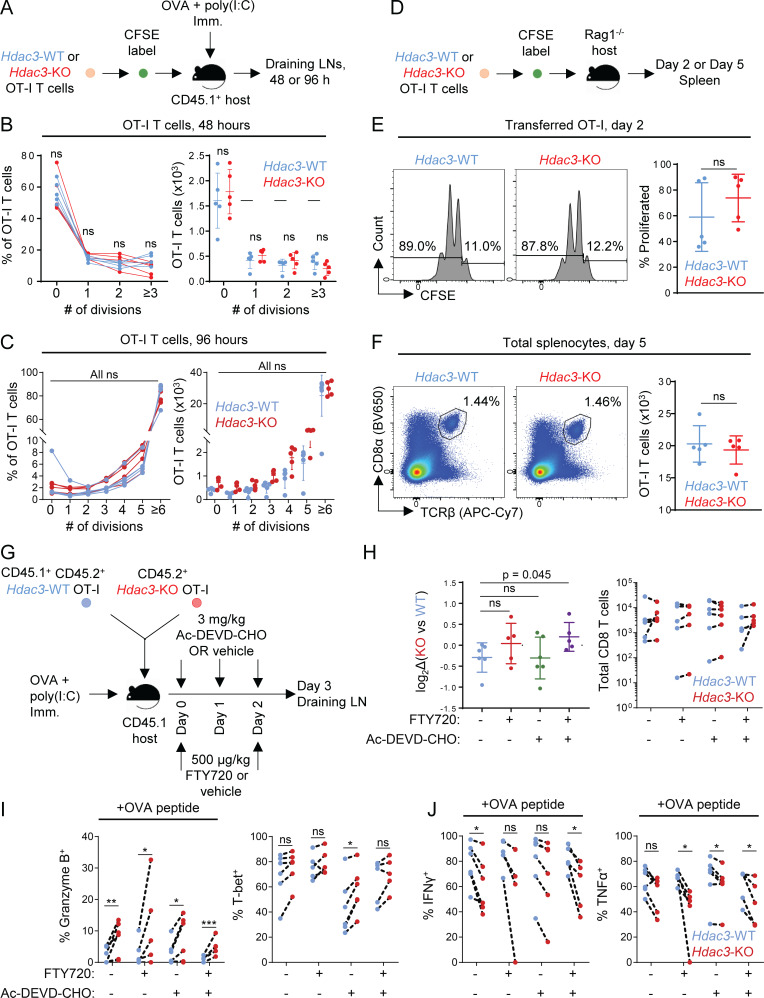
We therefore explored potential explanations for the failure of Hdac3-KO T cells to persist. Both Hdac3-KO and Hdac3-WT OT-I T cells showed similar kinetics of proliferation as assessed by CFSE dilution at 48 and 96 h following adoptive transfer and immunization of recipient hosts (Fig. S4, A–C). Moreover, naive Hdac3-KO OT-I T cells were able to proliferate and accumulate to the same extent as Hdac3-WT cells following transfer to lymphopenic Rag1−/− hosts (Fig. S4, D–F); such homeostatic expansion is thought to be cytokine dependent. Because it did not appear that there was a general defect in survival fitness or proliferation, at least within the first 96 h, we evaluated whether an increase in activation-induced apoptosis or LN egress might be responsible for the reduced persistence of Hdac3-KO CD8 T cells following activation. Following cotransfer of Hdac3-KO and Hdac3-WT OT-I T cells and OVA immunization, we treated recipient host mice with Ac-DEVD-CHO (an inhibitor of apoptosis) and/or the sphingosine 1–phosphate receptor (S1PR) agonist FTY720 (to block LN egress; Fig. S4 G). Inhibition of LN egress or activation-induced apoptosis did not rescue the failure of Hdac3-KO CD8 T cells to accumulate in draining LNs (Fig. S4, H–J).
Figure S4.
Analysis of reduced accumulation of Hdac3-KO CD8 T cells in draining LNs after in vivo activation. Related to Fig. 2 and Fig. 3. (A) Experimental scheme for evaluating TCR activation-induced proliferation of Hdac3-KO and Hdac3-WT CD8 T cells. OT-I T cells from Hdac3-KO mice or Hdac3-WT littermates were labeled with CFSE and adoptively transferred into CD45.1 congenic WT hosts. Mice were immunized subcutaneously with OVA + poly(I:C) in PBS, and transferred OT-I T cells were analyzed in LNs draining the immunization site 48 or 96 h after immunization. (B and C) Analysis of proliferation of Hdac3-KO and Hdac3-WT OT-I T cells after 48 h (B) or 96 h (C) following adoptive transfer and OVA immunization as in A. Data are representative of two independent experiments with five mice per genotype of transferred OT-I T cells. (D) Experimental scheme for evaluating homeostatic proliferation of Hdac3-KO and Hdac3-WT CD8 T cells. OT-I T cells from Hdac3-KO mice or Hdac3-WT littermates were labeled with CFSE and adoptively transferred into lymphopenic Rag1−/− hosts to assess cytokine-mediated homeostatic proliferation. (E) Representative flow cytometry plots and quantification of CFSE dilution in transferred OT-I T cells 48 h after transfer. Gated on live TCRβ+ CD8α+ Kb-SIINFEKL-tetramer+ cells. Data are representative of two independent experiments with five recipient mice per genotype of transferred OT-I T cells. (F) Representative flow cytometry plots and quantification of OT-I T cell numbers in spleens of Rag1−/− recipient mice after 5 d. OT-I T cells were defined as live TCRβ+ CD8α+ Kb-SIINFEKL-tetramer+ events. Data are representative of two independent experiments with five recipient mice for each genotype of OT-I CD8 T cells transferred. (G) Experimental scheme for evaluating the contributions of apoptosis or LN egress to reduced accumulation of Hdac3-KO CD8 T cells in LNs following activation. CD45.1+ TCR-polyclonal mice were adoptively transferred with 106 cells of a 1:1 mix of congenically distinct Hdac3-KO and Hdac3-WT OT-I T cells and immunized with OVA + poly(I:C) in PBS. Recipient mice received S1PR agonist FTY720 and/or pan-caspase inhibitor Ac-DEVD-CHO intraperitoneally at indicated time points after immunization. (n = 5, FTY720-treated groups; n = 6, FTY720-untreated groups). (H) Flow-cytometric quantification of changes in relative KO vs. WT frequencies normalized to pretransfer ratios (left), and absolute numbers of transferred Hdac3-KO and Hdac3-WT OT-I T cells in inguinal LNs of host mice 3 d after immunization (right). Data from cotransferred donor cells of each genotype within the same recipient mouse were analyzed as pairs. Data are representative of two independent experiments with five mice per treatment group. (I and J) Flow-cytometric quantification of changes in effector phenotypic markers in transferred OT-I T cells between different treatment groups as in G. Data from cotransferred donor cells of each genotype within the same recipient mouse were analyzed as pairs. Data are representative of two independent experiments with five mice per treatment group. Means ± SD are indicated (B, C, E, F, and H). P values were calculated by two-tailed Student’s t test (B, C, E, F, and H), or two-tailed ratio-paired t test (I and J). *, P < 0.05, **, P < 0.01, ***, P < 0.001.
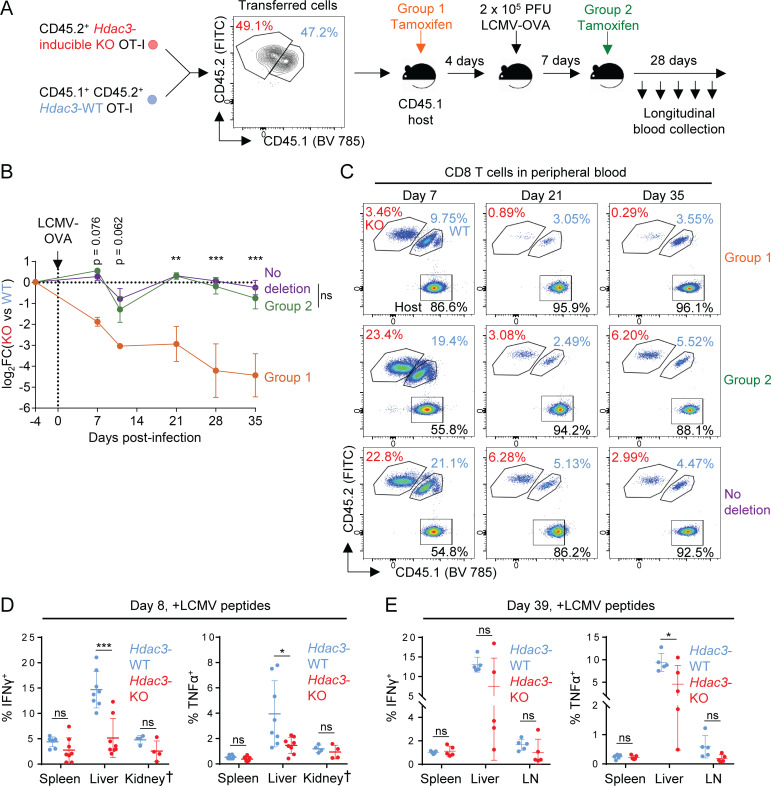
We next examined the persistence of Hdac3-KO CD8 T cells in an acute viral infection model, specifically the Armstrong strain of lymphocytic choriomeningitis virus engineered to express the SIINFEKL epitope recognized by OT-I T cells (LCMV-OVA). Congenically marked Hdac3-iKO and Hdac3-WT OT-I T cells were transferred to WT recipients that were subsequently challenged with 2 × 105 PFU LCMV-OVA. Hdac3 inactivation was induced by tamoxifen injection either before infection or on day 7 (Fig. 2 F and Fig. S5 A), when the virus is typically cleared (Wherry et al., 2003). The frequency of donor Hdac3-iKO and Hdac3-WT OT-I CD8 T cells was tracked over time to examine expansion and persistence. Both Hdac3-iKO and Hdac3-WT OT-I T cells expanded following viral infection, but Hdac3-iKO cells failed to persist in the experimental group receiving tamoxifen before infection (Fig. 2 G and Fig. S5, B and C). Such a failure of Hdac3-iKO T cell persistence was only observed when Hdac3 was inactivated before viral infection, but not when tamoxifen was administered on day 7 following infection (Fig. 2 G and Fig. S5, B and C). We also observed a reduction in the number of Hdac3-iKO OT-I CD8 T cells relative to cotransferred Hdac3-WT OT-I cells across three major organ reservoirs of LCMV infection on day 8 following infection when Hdac3 was deleted before infection (Fig. 2 H). Consistent with data from the immunization model, a higher percentage of Hdac3-iKO OT-I CD8 T cells was Granzyme B+ compared with cotransferred Hdac3-WT T cells, and the percentages of IFN-γ+ and TNF-α+ cells after ex vivo restimulation with OVA peptide were reduced (Fig. 2 I). These data support the conclusion that HDAC3 is required for CD8 T cell persistence following resolution of an acute infection.
Figure S5.
Phenotypes of virus-specific CD8 T cells during acute or chronic LCMV infection. Related to Fig. 2 and Fig. 4. (A) Experimental scheme for longitudinal tracking of OT-I T cell persistence following transfer and acute viral infection with inactivation of Hdac3 at indicated time points. 5 × 103 cells of a 1:1 mix of congenically distinct Hdac3-inducible KO (Hdac3-iKO) and Hdac3-WT OT-I cells were transferred into CD45.1+ TCR-polyclonal recipients. Mice were then infected intraperitoneally with 2 × 105 PFU of LCMV Armstrong expressing the OVA SIINFEKL epitope (LCMV-OVA), and Hdac3 deletion was induced at the indicated time points by intraperitoneal injection of tamoxifen on 3 consecutive days. (B and C) Comparison of frequencies of transferred Hdac3-iKO (Hdac3-inducible knockout) and Hdac3-WT OT-I T cells (B) and representative flow cytometry plots of total CD8 T cells (C) in peripheral blood during and after resolution of acute LCMV-OVA infection. Data are representative of two independent experiments with three to five recipient mice per treatment group. Data from cotransferred donor cells of each genotype within the same recipient mouse were analyzed as pairs. (D and E) Analysis of the IFN-γ and TNF-α response in CD8 T cells in indicated organs on day 8 (D) and day 39 (E) of chronic LCMV clone 13 infection as measured by flow cytometry. Data are pooled from two independent experiments (D) or from one experiment (E), each with four mice per genotype except for kidney data marked †, which is from one experiment with four mice per genotype. Means ± SEM (B) or means ± SD (D and E) are indicated. P values were calculated by two-way ANOVA (B) or two-tailed Student’s t test (D and E). *, P < 0.05, **, P < 0.01, ***, P < 0.001.
HDAC3 inhibits the CD8 T cell cytotoxic response early during activation
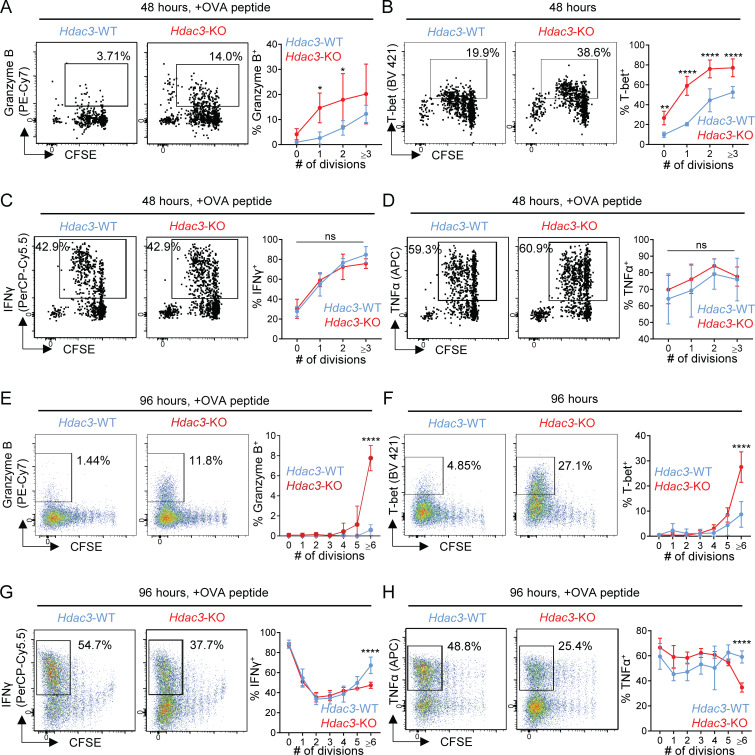
We hypothesized that HDAC3 functioned early during CD8 T cell activation to regulate acquisition of CD8 T cell effector function. We therefore investigated the kinetics of acquisition of CD8 effector functions in Hdac3-KO and Hdac3-WT CD8 T cells by transferring CFSE-labeled OT-I T cells into WT recipients followed by immunization with OVA + poly(I:C). Within 48 h after activation, a larger percentage of Hdac3-KO CD8 T cells expressed Granzyme B and T-bet compared with Hdac3-WT CD8 T cells (Fig. 3, A and B), with a significant increase observed even following one cell division following activation. There was no significant difference in the percentage of IFN-γ or TNF-α–producing cells at this time point (Fig. 3, C and D). The increased frequencies of Granzyme B+ and T-bet+ cells persisted at 96 h after activation (Fig. 3, E and F); a reduced percentage of IFN-γ+ or TNF-α+ cells was only observed by the sixth cell division (Fig. 3, G and H). These data suggest that HDAC3 inhibits the cytotoxicity program at an early time point following activation.
Figure 3.
HDAC3 inhibits the cytotoxic response early during CD8 T cell priming. 5 × 105 CFSE-labeled CD45.2+ OT-I T cells from Hdac3-KO mice or Hdac3-WT littermates were transferred into CD45.1+ TCR polyclonal recipients. Mice were immunized with OVA + poly(I:C) in PBS. Inguinal LNs draining the immunization site were collected for flow-cytometric analysis. (A–D) Kinetics of acquisition for markers of CD8 T cell effector function in OT-I T cells 48 h after immunization. Flow cytometry plots are representative of two independent experiments with five recipient mice per genotype of donor OT-I T cells transferred. (E–H) Kinetics for acquisition of markers of CD8 T cell effector function in OT-I T cells 96 h after immunization. Flow cytometry plots are representative of two independent experiments with five recipient mice per genotype of donor OT-I T cells transferred. Means ± SD are indicated (A–H). P values were calculated by two-way ANOVA. *, P < 0.05, **, P < 0.01, ****, P < 0.0001.
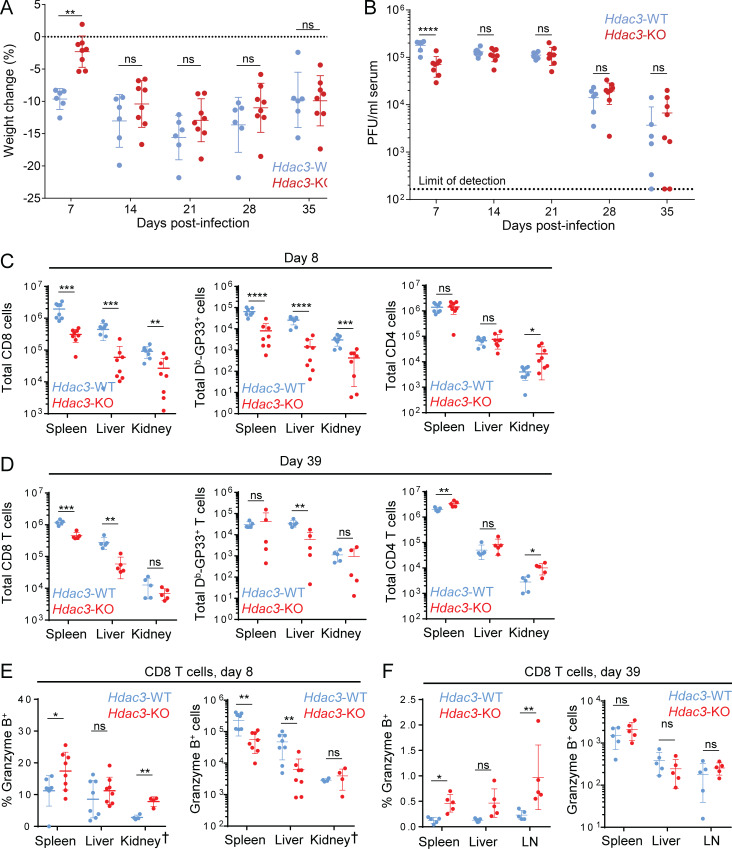
We next investigated how inactivation of the Hdac3 gene affects CD8 T cell responses in a model of chronic viral infection, both at early and late stages of the T cell response. We infected Hdac3-KO (E8I-Cre+; Hdac3fl/fl) and Hdac3-WT littermates with 2 × 106 PFU LCMV Clone 13 to induce chronic infection, and monitored disease progression and CD8 T cell effector phenotypes in infected mice. Hdac3-KO mice showed a moderate delay in disease progression as indicated by delayed weight loss and reduced viremia on day 7, during the establishment stage of chronic viral infection (Zajac et al., 1998), but not at later time points (Fig. 4, A and B). This delay in early disease progression on day 8 was observed despite lower numbers of total and Db-GP33 tetramer+ virus-specific CD8 T cells in Hdac3-KO mice compared with Hdac3-WT littermates (Fig. 4 C, left and center panels); in contrast, CD4 T cell numbers were similar between both groups except for a small increase in kidneys of Hdac3-KO mice (Fig. 4 C, right panel). At the late stage of chronic viral infection (day 39), numbers of total and virus-specific CD8 T cells were generally lower in Hdac3-KO than Hdac3-WT littermates (Fig. 4 D, left and center panels), although these differences were less pronounced than those observed on day 8. CD4+ T cell numbers were slightly increased in the spleens and kidneys of Hdac3-KO mice compared with Hdac3-WT mice (Fig. 4 D, right panel).
Figure 4.
Analysis of Hdac3-deficient CD8 T cells in a model of chronic viral infection. TCR-polyclonal Hdac3-KO mice and Hdac3-WT littermates were infected with 2 × 106 PFU LCMV Clone 13 by intravenous injection. (A and B) Weight change (A) and viral load in sera (B) of infected Hdac3-KO (n = 8) and Hdac3-WT (n = 6) mice. Data are representative of two independent experiments with six to eight mice of each genotype. (C and D) Analysis of T cell numbers on day 8 (C) and day 39 (D) in indicated organs as measured by flow cytometry. (E and F) Analysis of the Granzyme B response in CD8 T cells in indicated organs on day 8 (E) and day 39 (F) as measured by flow cytometry, showing both percentage (left) and number (right) of Granzyme B+ CD8 T cells. Data are pooled from two independent experiments (C and E) or from one experiment (D and F), each with four mice per genotype except for kidney data marked †, which is from one experiment with four mice per genotype. Means ± SEM (A) or means ± SD are indicated (B–F). P values were calculated by two-way ANOVA (A and B) or two-tailed Student’s t test (C–H). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001.
We also observed a larger percentage of Granzyme B+ CD8 T cells in the spleens and kidneys of Hdac3-KO mice on day 8 (Fig. 4 E, left panel), even though the total numbers of Granzyme B+ cells were lower due to reduced total CD8 T cell numbers (Fig. 4 E, right panel). Importantly, a larger percentage of CD8 T cells were Granzyme B+ in the spleens and inguinal LNs of Hdac3-KO compared with Hdac3-WT mice even on day 39 following infection (Fig. 4 F, left panel), even though the total numbers of Granzyme B+ cells were not significantly increased in Hdac3-KO relative to Hdac3-WT mice (Fig. 4 F, right panel). In contrast, the percentage of IFN-γ+ and TNF-α+ CD8 T cells was either unchanged (in spleen and kidneys) or lower (in liver) in Hdac3-KO mice compared with Hdac3-WT mice at both the early (day 8) and late (day 39) time points (Fig. S5, D and E).
Transcriptional and epigenetic signatures of HDAC3
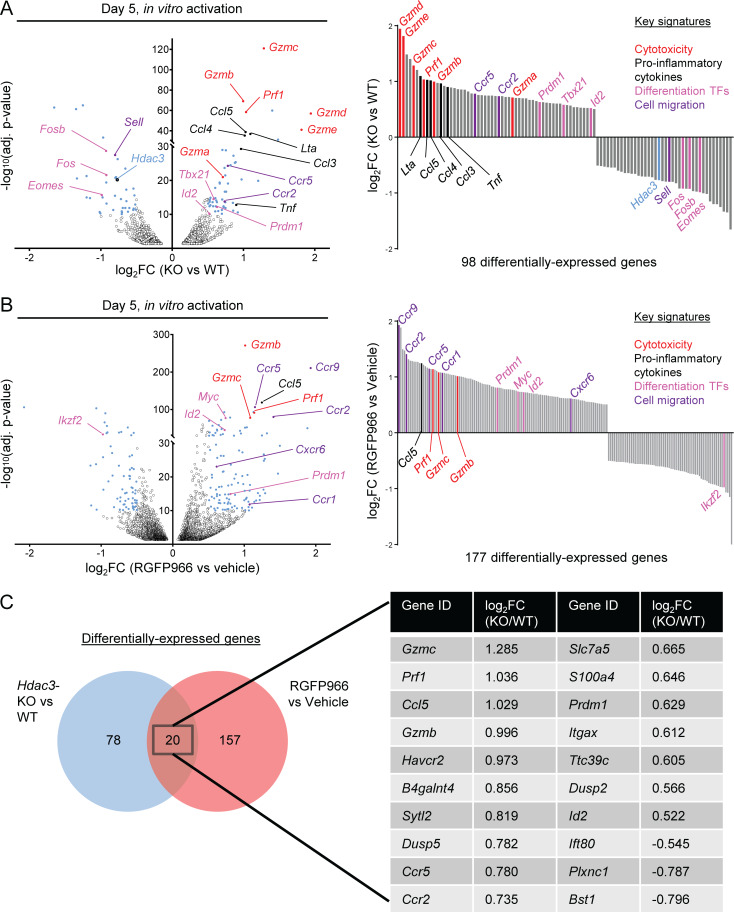
To uncover potential molecular mechanisms of HDAC3-mediated regulation of CD8 T cell cytotoxic function, we examined global transcriptional differences between Hdac3-KO and Hdac3-WT OT-I T cells that had been activated for 5 d by co-culture with irradiated OVA peptide–pulsed BMDCs (Fig. 5 A and Table S2). Key genes of the cytotoxicity program (Gzma, Gzmb, Gzmc, Gzmd, Gzme, Prf1) were more highly expressed in Hdac3-KO compared with Hdac3-WT CD8 T cells, consistent with the in vivo data. In addition, genes encoding transcription factors that promote CD8 T cell effector function and/or terminal effector differentiation (Tbx21, Id2, Prdm1) were also more highly transcribed in Hdac3-KO than Hdac3-WT CD8 T cells. Finally, transcripts of a subset of chemokine receptor genes (Ccr5, Ccr2) and pro-inflammatory chemokine/cytokine genes (Lta, Ccl5, Ccl4, Ccl3, Tnf) were more highly expressed by Hdac3-KO compared with Hdac3-WT cells.
Figure 5.
HDAC3 negatively regulates a cytotoxic effector-associated transcriptional program in CD8 T cells. OT-I T cells were co-cultured with irradiated OVA peptide–pulsed BMDCs in vitro, and sorted to purity for molecular analysis. (A and B) Whole genome RNA-seq of Hdac3-KO and Hdac3-WT (A) or of RGFP966- and vehicle-treated (B) CD8 T cells 5 d after in vitro activation. Significance was determined using thresholds of −log10(adjusted P value) >10 and |log2(fold change)| >0.5. Data are from one experiment with three technical replicates per sample. TFs, transcription factors. (C) Venn diagram showing overlap between genes differentially expressed between Hdac3-KO and Hdac3-WT CD8 T cells, or RGFP966- and vehicle-treated CD8 T cells after 5 d of in vitro activation. Statistical analyses for RNA-seq data are described in the Materials and methods section. RNA-seq data are available through GEO accession no. GSE143644.
We performed a similar RNA sequencing (RNA-seq) analysis for OT-I T cells activated in the presence of the HDAC3 inhibitor RGFP966 or a solvent control (Fig. 5 B and Table S3). We identified 20 genes that were differentially expressed in both Hdac3-KO and HDAC3 inhibitor–treated CD8 T cells (Fig. 5 C). Within this common set of 20 genes, we identified genes belonging to the four functional categories described above: cytotoxicity (Gzmb, Gzmc, Prf1) and transcription factors associated with CD8 T cell effector differentiation (Prdm1, Id2), as well as pro-inflammatory chemokines (Ccl5) and chemokine receptors (Ccr5, Ccr2). Overall, these RNA-seq data indicate that HDAC3 negatively regulates gene programs associated with CD8 T cell cytotoxic effector function, including direct mediators of cytotoxicity (Gzmb, Gzmc, Prf1) and transcription factor genes that promote an effector phenotype in CD8 T cells (Prdm1, Id2).
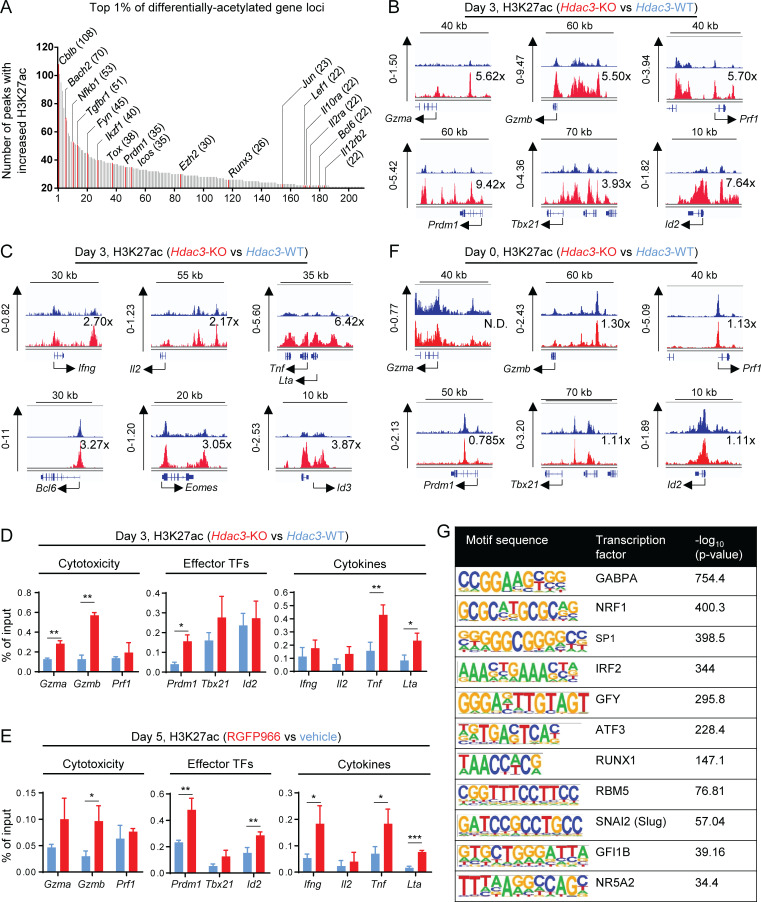
We next examined the impact of Hdac3 gene inactivation on histone acetylation in CD8 T cells, specifically histone 3 lysine 27 acetylation (H3K27ac), an epigenetic modification associated with transcriptionally active genes (Zhang et al., 2015). H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) analysis was performed on Hdac3-KO and Hdac3-WT OT-I T cells activated in vitro by co-culture with irradiated OVA peptide–pulsed BMDCs. Relative differences in peak intensities were calculated for each genomic locus with unique significant peaks present in both Hdac3-KO and Hdac3-WT samples. Consistent with the role of HDAC3 as an HDAC, activated Hdac3-KO CD8 T cells showed a median 3.1-fold increase in global H3K27ac levels relative to Hdac3-WT cells. Genes encoding transcription factors that regulate T cell activation and differentiation (Bach2, Nfkb1, Ikzf1, Tox, Prdm1, Ezh2, Runx3, Jun, Lef1, Bcl6), regulators of T cell receptor signaling (Cblb, Fyn), and surface signaling receptors (Tgfbr1, Icos, Il10ra, Il2ra, Il12rb2) were represented within the top 1% of gene loci–containing peaks with increased H3K27ac signal (≥21 peaks; Fig. 6 A and Table S4). Furthermore, we observed an increase in H3K27ac signal (greater than the median of 3.1×) at the loci of genes that encoded cytotoxic mediators (Gzma, Gzmb, Prf1), transcription factors known to polarize CD8 T cell effector differentiation (Prdm1, Tbx21, Id2), and the cytokines Tnf and Lta in Hdac3-KO cells compared with Hdac3-WT cells (Fig. 6, B and C). H3K27ac levels at the loci of the effector cytokine genes Ifng and Il2 were not increased beyond the global median increase of 3.1 (Fig. 6 C). Similar findings were made for the genetic loci of the transcription factors Bcl6, Eomes, and Id3 (Fig. 6 C), whose transcript levels were not increased in Hdac3-KO cells.
Figure 6.
HDAC3 deacetylates H3K27ac epigenetic marks at gene loci associated with cytotoxic effector function in activated CD8 T cells. (A–C) Hdac3-KO and Hdac3-WT OT-I T cells were co-cultured with irradiated OVA peptide–pulsed BMDCs in vitro for 3 d and sorted to purity. Total chromatin was prepared from fixed cell pellets for immunoprecipitation with antibodies specific to H3K27ac for ChIP-seq analysis. (A) Top 1% of differentially acetylated gene loci in activated Hdac3-KO CD8 T cells with at least 21 differentially acetylated H3K27ac peaks per locus. Genes with described regulatory roles in T cell activation, effector function, or differentiation are highlighted in red and labeled. The number of differentially acetylated H3K27ac peaks mapping to each highlighted gene is indicated in parentheses. (B) H3K27ac ChIP-seq tracks of H3K27ac showing representative genes encoding cytotoxicity genes (top row) or transcription factors polarizing toward an effector phenotype in CD8 T cells (bottom row). Relative increases in H3K27ac signal in Hdac3-KO relative to Hdac3-WT cells for each genomic locus shown are indicated. (C) H3K27ac ChIP-seq tracks of H3K27ac showing representative genes encoding CD8 effector cytokines (top row) or transcription factors polarizing toward a memory phenotype in CD8 T cells (bottom row). Relative increases in H3K27ac signal in Hdac3-KO relative to Hdac3-WT cells for each genomic locus shown are indicated. (D and E) OT-I T cells were co-cultured with irradiated OVA peptide–pulsed BMDCs in vitro for indicated times and sorted to purity. Total chromatin was prepared from fixed cell pellets for immunoprecipitation with antibodies specific to H3K27ac for ChIP-qPCR analysis. Data are representative of two independent experiments each with three or four technical replicates per qPCR reaction. (D) Comparison of H3K27ac at promoters of indicated genes in Hdac3-KO and Hdac3-WT CD8 T cells activated for 3 d in vitro. (E) Comparison of H3K27ac at promoters of indicated genes in RGFP966- and vehicle-treated CD8 T cells activated for 5 d in vitro. (F) H3K27ac ChIP-seq tracks of H3K27ac showing representative genes encoding cytotoxicity genes (top row) or transcription factors polarizing toward an effector phenotype in CD8 T cells (bottom row) in naive Hdac3-KO and Hdac3-WT CD8 T cells. Relative increase in H3K27ac signal in Hdac3-KO relative to Hdac3-WT cells is indicated for each genomic locus. (G) Transcription factor binding motifs enriched in genomic regions with increased H3K27ac in activated Hdac3-KO CD8 T cells as in A–C. Genomic regions containing H3K27ac peaks with increased signal in Hdac3-KO T cells relative to Hdac3-WT cells were analyzed for mammalian (Homo sapiens and Mus musculus) transcription factor binding motifs using the HOMER motif analysis algorithm (Heinz et al., 2010). Means ± SD are indicated (D and E). P values were calculated by two-tailed Student’s t test (D and E). *, P < 0.05, **, P < 0.01, ***, P < 0.001. Statistical analysis for H3K27ac ChIP-seq data are described in the Materials and methods section. H3K27ac ChIP-seq data are available through GEO accession no. GSE143644.
We validated these findings by performing ChIP-quantitative PCR (ChIP-qPCR) to measure H3K27ac at the promoter regions of cytotoxicity, effector transcription factor, and cytokine genes using in vitro activated Hdac3-KO and Hdac3-WT CD8 T cells. Consistent with the ChIP-seq results, higher levels of H3K27ac were observed at promoters of cytotoxicity genes (Gzma, Gzmb) in Hdac3-KO compared with Hdac3-WT cells (Fig. 6 D, left panel). We also observed increased H3K27ac at the Prdm1 promoter, and at the promoters of the cytokine genes Tnf and Lta (Fig. 6 D, center and right panels). Increased H3K27ac was also observed at the promoters of Gzmb, Prdm1, Tnf, and Lta in RGFP966-treated activated CD8 T cells relative to vehicle control-treated cells (Fig. 6 E). In addition, higher levels of H3K27ac were observed at the Ifng and Id2 promoters in activated RGFP966-treated CD8 T cells (Fig. 6 E). We further performed ChIP-seq on freshly isolated naive Hdac3-KO and Hdac3-WT OT-I T cells to verify that the increase in H3K27ac was only observed following activation. We did not observe a substantial increase in H3K27ac signal between naive Hdac3-KO and Hdac3-WT cells at the loci of cytotoxicity genes (Gzma, Gzmb, Prf1) or at genes coding for transcription factors known to polarize CD8 T cells toward effector differentiation (Prdm1, Tbx21, Id2; Fig. 6 F).
Finally, to uncover how HDAC3-mediated deacetylation of H3K27ac epigenetic marks may interact with other regulatory pathways of CD8 T cell effector function and differentiation, we analyzed the set of genomic loci containing peaks with increased H3K27ac signal in Hdac3-KO relative to Hdac3-WT samples for enrichment of transcription factor–binding motif sequences using the Hypergeometric Optimization of Motif EnRichment (HOMER) motif analysis algorithm (Heinz et al., 2010). The binding motif of the transcription factor GA-binding protein α chain (GABPA) was the most significantly enriched in the differentially acetylated regions (Fig. 6 G). GABPA is an erythroblast transformation specific (ETS)–family transcription factor that is coredundant with ETS1 and may colocalize with Runt-related (RUNX) family transcription factors at promoter regions. Our analysis also highlighted a RUNX1 binding motif that had been identified via ChIP-seq in Jurkat cells (Sanda et al., 2012; Fig. 6 G).
Inactivation of Prdm1 increases the frequency of T-bet+ Hdac3-KO CD8 T cells
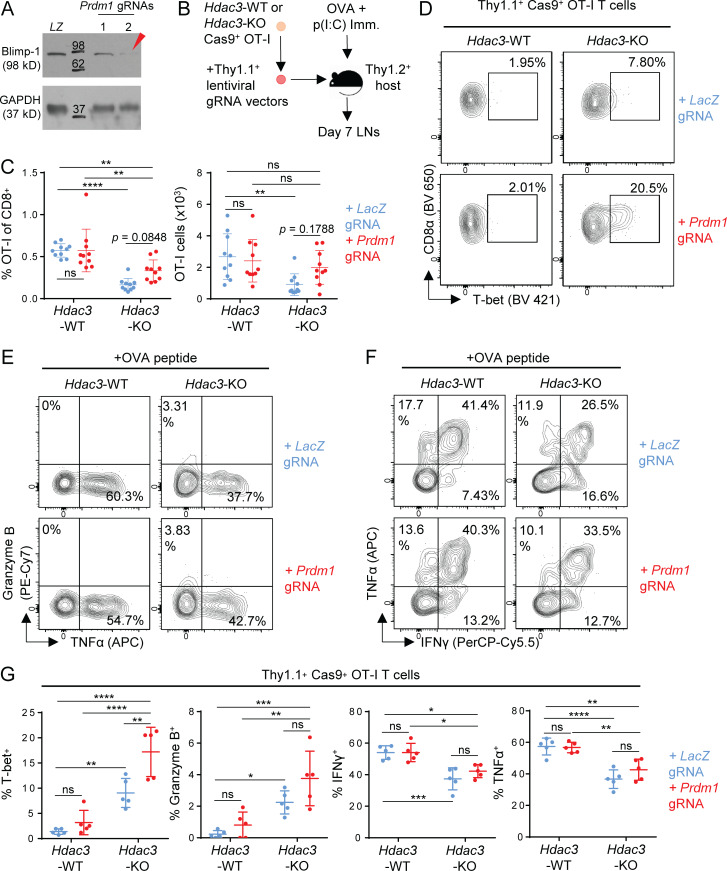
In view of the known role of Blimp-1 (encoded by Prdm1) in driving terminal effector differentiation of activated CD8 T cells (Kallies et al., 2009; Rutishauser et al., 2009), we hypothesized that an increase in Blimp-1 activity (as suggested by our RNA-seq and ChIP-qPCR data) might contribute to the increased frequency of Granzyme B+ and T-bet+ Hdac3-KO cells, or potentially to the reduced persistence of activated Hdac3-KO CD8 T cells. To test these possibilities, we generated Prdm1-Hdac3–double KO OT-I T cells by transducing Cas9+ Hdac3-KO OT-I T cells with lentiviral gRNA expression constructs targeting Prdm1. Prdm1 inactivation was confirmed by Western blot analysis of Blimp-1 (Fig. 7 A). Hdac3-KO or Hdac3-WT OT-I T cells transduced with either Prdm1 or control (LacZ) gRNAs were transferred into congenic hosts that were then immunized with OVA + poly(I:C) (Fig. 7 B). We found that inactivation of Prdm1 in Hdac3-KO CD8 T cells showed a trend toward improved persistence compared with Hdac3-WT CD8 T cells, but this effect did not reach statistical significance in all experiments (pooled data shown in Fig. 7 C). Interestingly, activated Prdm1-Hdac3–double KO CD8 T cells had a higher proportion of T-bet+ cells compared with Hdac3-KO CD8 T cells (Fig. 7 D), but did not show differences in the percentage of Granzyme B+, IFN-γ+, or TNF-α+ cells (Fig. 7, E–G). These data indicate that Prdm1 reduces the frequency of T-bet+ Hdac3-KO CD8 T cells, and may also contribute to the reduced persistence of Hdac3-KO cells following activation.
Figure 7.
Inactivation of Prdm1 increases the frequency of T-bet+Hdac3-KO CD8 T cells. (A) Validation of lentiviral gRNA vectors targeting Prdm1 in Cas9+ OT-I T cells. Whole cell lysates from 2 × 105 magnetically purified transduced T cells were loaded per lane for immunoblot analysis. LZ, LacZ-targeting gRNA vector (negative control). Molecular weights (kD) are indicated on immunoblot images. The red arrow indicates the gRNA used in subsequent experiments. (B) Experimental scheme for phenotype analysis of Prdm1-Hdac3-double KO CD8 T cells following in vivo activation. Hdac3-KO and Hdac3-WT Cas9+ OT-I T cells were transduced with lentiviral vectors expressing Prdm1 or LacZ targeting gRNA sequences, magnetically purified based on Thy1.1 expression, and adoptively transferred into Thy1.2+ C57BL/6 recipients. Mice were immunized subcutaneously with OVA and poly(I:C) in PBS. (C) Persistence of Prdm1-Hdac3-double KO CD8 T cells on day 7 after in vivo activation as in B, measured in terms of percentage of transferred OT-I T cells within the total CD8 T cell population (left) and total numbers of OT-I T cells (right) in LNs draining the immunization site. Data are pooled from two independent experiments with five mice per group. (D–G) Flow-cytometric analysis of T-bet (D), Granzyme B (E), and effector cytokine (F) expression in transferred OT-I T cells in inguinal LNs 7 d after OVA immunization as in B. Quantification of flow-cytometric data are shown in G. Data are representative of two independent experiments with five mice per group. Means ± SD are indicated (C and G). P values were calculated by two-way ANOVA (C and G). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001.
Runx3 is required for the augmented cytotoxic phenotype of Hdac3-KO T cells
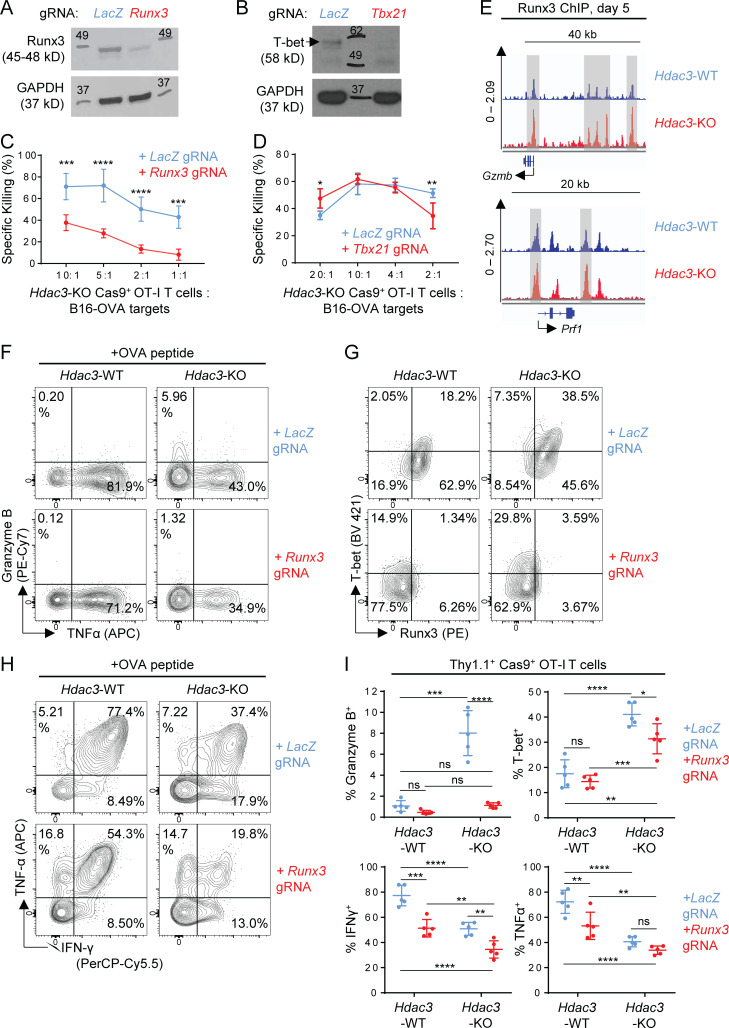
We next assessed whether the transcription factors Runx3 and T-bet (encoded by Tbx21) were required for the augmented cytotoxic phenotype of activated Hdac3-KO CD8 T cells, because both transcription factors were previously shown to be required for cytotoxic function of CD8 T cells (Cruz-Guilloty et al., 2009). We generated Runx3-Hdac3–double KO and Tbx21-Hdac3–double KO CD8 T cells using the approach described above for Prdm1 (Fig. 8, A and B). Inactivation of Runx3 but not Tbx21 abrogated the enhanced cytotoxic function of Hdac3-KO CD8 T cells (Fig. 8, C and D). ChIP-seq analysis of in vitro activated OT-I T cells also revealed an increase in Runx3 binding at regulatory elements near the cytotoxicity genes Gzmb and Prf1 in Hdac3-KO CD8 T cells relative to Hdac3-WT cells (Fig. 8 E).
Figure 8.
Runx3 but not Tbx21 is required for the augmented cytotoxic phenotype of Hdac3-KO T cells. (A and B) Validation of lentiviral gRNA vectors targeting Runx3 (A) and Tbx21 (B) in Cas9+ OT-I T cells. Whole cell lysates from 2 × 105 magnetically purified transduced T cells were loaded per lane for immunoblot analysis. A LacZ-targeting gRNA vector was used as a negative control for Runx3 or Tbx21 editing. Molecular weights (kD) are indicated on immunoblot images. Where multiple bands are present, a black arrow indicates a band of the correct molecular weight. (C and D) Comparison of in vitro cytotoxicity of Hdac3-KO and Runx3-Hdac3–double KO (C) or Hdac3-KO and Tbx21-Hdac3–double KO (D) CD8 T cells. Hdac3-KO Cas9+ OT-I T cells were transduced with indicated gRNA sequences, activated in vitro with irradiated OVA peptide–pulsed BMDCs, and evaluated for cytotoxicity against B16-OVA targets. Data are from one experiment each, with four replicates per condition. (E) Runx3 ChIP-seq tracks of genes coding for representative cytotoxic mediators for in vitro activated OT-I T cells. Hdac3-KO and Hdac3-WT OT-I T cells were co-cultured with irradiated OVA peptide–pulsed BMDCs and sorted to purity after 5 d for chromatin preparation and ChIP-seq. Specific peaks with increased Runx3 binding in Hdac3-KO relative to Hdac3-WT CD8 T cells are highlighted with gray columns. Runx3 ChIP-seq data are available through GEO accession no. GSE143644. (F–I) Hdac3-KO and Hdac3-WT Cas9+ OT-I T cells were transduced with lentiviral vectors expressing Runx3- or LacZ-targeting gRNA sequences, magnetically purified for Thy1.1+ expression, and adoptively transferred into Thy1.2+ C57BL/6 recipients. Mice were immunized subcutaneously with OVA + poly(I:C) in PBS. OT-I T cells in inguinal LNs draining immunization site were analyzed after 4 d. Flow-cytometric analysis of Granzyme B (F), T-bet (G), and effector cytokine (H) expression in transferred OT-I T cells in inguinal LNs. Quantification of flow-cytometric data are shown in I. Data are representative of two independent experiments with five mice per treatment group. Means ± SD are indicated (C, D, and I). P values were calculated by two-way ANOVA (C, D, and I). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001.
We therefore investigated the functional phenotype of Runx3-Hdac3-double KO CD8 T cells by transfer of edited OT-I T cells followed by immunization with OVA. Inactivation of Runx3 in Hdac3-KO CD8 T cells resulted in a substantial reduction in the percentage of Granzyme B+ CD8 T cells and also reduced the percentage of T-bet+ cells (Fig. 8, F and G). Runx3 inactivation also reduced the percentage of IFN-γ+ CD8 T cells both in Hdac3-KO and Hdac3-WT cells (Fig. 8, H and I). Overall, these data indicate that Runx3 plays an important role in the augmented cytotoxic phenotype of Hdac3-KO CD8 T cells.
Discussion
HDAC3 has been shown to be required for proper development of the T cell compartment (Hsu et al., 2015; Stengel et al., 2015). In particular, during the thymic maturation of T cell precursors, HDAC3 is required at the CD4+ CD8+ DP stage for down-regulation of RORγt (Philips et al., 2016) and for maintaining the potential of DP thymocytes to mature into either the CD4 or CD8 T cell lineages by deacetylating CD8-lineage associated gene loci (Philips et al., 2019). However, the role of HDAC3 in peripheral conventional T cells has not been described, partly because previous attempts at generating conditional-KO mice resulted in partial or total loss of T cells in the periphery (Hsu et al., 2015; Philips et al., 2016; Stengel et al., 2015).
Here we describe a novel role for HDAC3 in regulating the effector phenotype of CD8 T cells after activation. Our data demonstrate that HDAC3 inhibits the development of a short-lived, highly cytotoxic state in CD8 T cells. Loss of HDAC3 activity during activation resulted in a gain-of-function phenotype with increased CD8 T cell cytotoxicity, in contrast to the loss-of-function phenotypes described in recent studies of other epigenetic regulators in CD8 T cells (Gray et al., 2017; Pace et al., 2018). Inactivation of the Hdac3 gene or inhibition of the enzyme with a small molecule strongly increased the cytotoxic function of CD8 T cells, and kinetic analyses demonstrated an increased fraction of Granzyme B+ and T-bet+ Hdac3-KO CD8 T cells relative to Hdac3-WT cells even at the first cell division following TCR activation, suggesting that HDAC3 impacts early steps of T cell differentiation. Hdac3-KO CD8 T cells failed to persist following resolution of an acute viral infection or rechallenge after initial immunization with a peptide antigen, consistent with a model in which the absence of HDAC3 biases differentiation toward a short-lived cytotoxic effector state. This lack of persistence was not due to early differences in proliferation, apoptosis, or egress from lymphatics, nor due to reduced survival fitness in the absence of TCR activation. We hypothesize that a loss of proliferative potential later during activation, consistent with an increased propensity toward terminal effector differentiation, may be responsible for the reduced persistence of Hdac3-KO CD8 T cells following activation. Furthermore, our observation that inactivation of Hdac3 before but not after the peak of the effector CD8 response led to a loss in persistence again suggests that HDAC3 may regulate CD8 T cell differentiation and effector function beginning at an early stage, with the consequences of this early regulation being inherited with subsequent cell divisions. Overall, our observations of the role of HDAC3 in inhibiting the cytotoxic effector phenotype and regulating the persistence of CD8 T cells fit classical hallmarks for epigenetic control of cell differentiation (Deans and Maggert, 2015).
To uncover potential molecular pathways by which HDAC3 regulates CD8 T cell effector function and differentiation, we profiled both transcriptional and epigenetic changes in activated CD8 T cells deficient in HDAC3 activity. Our RNA-seq analysis demonstrated increased transcription of cytotoxicity and effector genes in Hdac3-KO T cells or HDAC3 inhibitor–treated T cells, consistent with our previous observations across several experimental settings of CD8 T cell activation. Although we observed that key cytotoxicity genes (including Gzmb and Prf1) and transcription factors (Prdm1 and Id2) were up-regulated in both Hdac3-KO and HDAC3 inhibitor–treated CD8 T cells, there was also a large number of nonoverlapping genes between the two experimental conditions. Contributing factors could be the following: (1) HDAC3 activity was absent in Hdac3-KO T cells but may have only been partially reduced by the inhibitor; (2) even though RGFP966 has been reported to be quite specific for HDAC3 (Xu et al., 2009), it is possible that it also reduced the activity of one or several other HDACs; (3) it is possible that RGFP966 had off-target activity unrelated to HDAC3 or other HDAC enzymes. Nevertheless, both inactivation of the Hdac3 gene and inhibition of HDAC3 enzyme enhanced the cytotoxic function of CD8 T cells and negatively regulated a core set of genes associated with the CD8 cytotoxic effector phenotype. We therefore focused our molecular analyses on these genes.
H3K27ac ChIP-seq analysis of activated Hdac3-KO CD8 T cells revealed that HDAC3 deacetylates H3K27ac at several genes encoding regulators of CD8 T cell activation, effector function, and differentiation, including transcription factors (Bach2, Nfkb1, Ikzf1, Tox, Prdm1, Ezh2, Runx3, Jun, Lef1, Bcl6), regulators of T cell receptor signaling (Cblb, Fyn), and surface receptors (Tgfbr1, Icos, Il10ra, Il2ra, Il12rb2). This increase in H3K27ac was observed following CD8 T cell activation, while there were no substantial changes in H3K27ac between freshly isolated, unstimulated Hdac3-KO and Hdac3-WT CD8 T cells. Whereas a general increase in H3K27ac was present in activated Hdac3-KO compared with Hdac3-WT T cells, we observed an even greater increase in H3K27ac at the loci of cytotoxicity genes (Gzma, Gzmb, Prf1) and genes encoding transcription factors (Prdm1, Tbx21, Id2) that showed increased transcription in Hdac3-KO compared with Hdac3-WT CD8 T cells during activation. While we were able to validate the increase in H3K27ac at the loci of Gzmb and Prdm1 independently by performing ChIP-qPCR on both activated Hdac3-KO and HDAC inhibitor-treated CD8 T cells, we did not observe significantly increased H3K27ac at all of the loci examined by ChIP-qPCR; this could have been due to the limited sensitivity of our ChIP-qPCR assay, which assessed H3K27ac only for segments of proximal promoters (rather than entire gene loci as in ChIP-seq). Also, the overall global increase in H3K27ac signal in Hdac3-KO samples may have made it more difficult to identify such H3K27ac signals after normalization to the input.
To gain further insight into how HDAC3-mediated deacetylation of H3K27ac might interact with transcriptional regulators of CD8 T cell activation and differentiation, we performed motif analysis on differentially acetylated genomic regions in activated Hdac3-KO and Hdac3-WT CD8 T cells to identify transcription factors that might potentially interact with HDAC3. We had previously attempted to perform HDAC3 ChIP-seq experiments to directly address this question but were not able to identify suitable antibodies for this purpose. Our motif analysis suggested that transcription factors from the Runx and Ets families may bind to genomic regions targeted by HDAC3 for H3K27ac deacetylation. However, it remains unknown whether and how HDAC3 interacts with key transcription factors (e.g., Runx3) that regulate CD8 T cell differentiation and effector function during activation. Several mechanisms are possible: (1) HDAC3 may first bind to H3K27ac sites, resulting in chromatin condensation and preventing transcription factor binding to genes regulating CD8 cytotoxic effector function; (2) transcription factors such as Runx3 might be recruited to H3K27ac marks via interactions with epigenetic readers such as bromodomain proteins (which recognize and bind acetylated lysine residues on histones), and might thus compete with HDAC3 for binding to chromatin. Further investigation to identify direct molecular interactions between HDAC3 and transcription factors regulating CD8 T cell activation, as well as analyses of chromatin accessibility changes in activated Hdac3-KO CD8 T cells, will be necessary to elucidate the order of epigenetic and transcriptional events regulated by HDAC3. It is also certainly possible that other histone acetylation marks in addition to H3K27ac (e.g., H3K9ac) could be regulated by HDAC3 activity and contribute to regulation of cytotoxic effector function.
Because our RNA-seq and H3K27ac ChIP-seq data suggested that HDAC3 negatively regulates Prdm1 expression, we focused on investigating the genetic interaction between Hdac3 and Prdm1 (Blimp-1). Blimp-1 is a transcriptional repressor that enhances terminal differentiation of effector CD8 T cells during viral infection, and Prdm1-deficient T cells primarily develop into memory precursor cells with higher proliferative potential (Kallies et al., 2009; Rutishauser et al., 2009). Our data suggest that HDAC3 may regulate the persistence of activated CD8 T cells in part by repressing expression of Blimp-1; however, this is unlikely to be the only molecular mechanism by which HDAC3 activity regulates CD8 T cell persistence. Our data are consistent with a model in which HDAC3-mediated inhibition of terminal effector differentiation (driven in part by Blimp-1) controls the persistence of the CD8 T cell response following activation. At the molecular level, Blimp-1 represses transcription of the gene encoding Fos, which associates with Jun to form the AP-1 transcription factor required for transcriptional activation of the Il2 gene (Martins et al., 2008), and was also recently shown to regulate Granzyme B expression in murine effector CD8 T cells (Kragten et al., 2018). Blimp-1 also inhibits expression of the CD25 chain of the IL-2 receptor by recruiting the histone methyltransferase G9a (encoded by Ehmt2; Shin et al., 2013). Given that Hdac3-KO CD8 T cells express T-bet at a higher frequency than Hdac3-WT cells and also differentiate preferentially into terminally differentiated effector cells, it is likely that HDAC3-regulated expression of Blimp-1 may play a role in the observed cellular phenotype.
We further demonstrated that the increased cytotoxic phenotype of CD8 T cells following T cell activation required the presence of Runx3. Runx3 is a transcription factor with an important role for commitment to the CD8 T cell lineage in the thymus (Taniuchi et al., 2002; Woolf et al., 2003). It plays a critical role in CTL differentiation and regulates expression of a number of key effector genes, including those encoding Granzyme B and perforin. Runx3 is also known to cooperate with the effector function–associated transcription factor T-bet (encoded by Tbx21) during differentiation into cytotoxic effector CD8 T cells (Cruz-Guilloty et al., 2009). Runx3 was recently shown to be required for memory CTL development during acute viral infection and to function as a pioneer transcription factor for cis-regulatory elements beginning early during CD8 T cell activation (Wang et al., 2018). Finally, HDAC3 was recently reported to bind to a super-enhancer regulatory region of the Runx3 locus and was associated with suppression of Runx3 transcription during differentiation of thymocytes into CD4 rather than CD8 T cells (Philips et al., 2019). Together with our data showing an enrichment of potential Runx family transcription factor binding sites within genomic loci targeted by HDAC3 for H3K27ac deacetylation, we suggest that HDAC3 is a potential regulator of the Runx3-dependent CD8 T cell cytotoxicity program.
Taken together, we propose a model in which HDAC3 epigenetically regulates a network of genes including Runx3 and Prdm1 in CD8 T cells during CD8 T cell activation to inhibit differentiation into cytotoxic effector cells. Transient inhibition of HDAC3 with small molecule inhibitors may offer opportunities to enhance CD8 T cell differentiation into cytotoxic effector cells. This work thus identifies an important epigenetic layer of regulation for the differentiation of activated CD8 T cells into cytotoxic effector cells.
Materials and methods
Mice
All mice were maintained in specific pathogen–free conditions and used in accordance to guidelines of the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee. C57BL/6, CD45.1+ congenic, Rag1−/−, and OT-I mice were purchased from The Jackson Laboratory. Cas9+ OT-I mice were generated by crossing the OT-I TCR transgenic strain with mice carrying a Rosa26-targeted knock-in of Cas9 (Jackson strain 024858) for constitutive Cas9 expression; all Cas9+ mice in this study were homozygous for the Cas9 knock-in allele. Hdac3fl/fl mice were developed by S. Hiebert (Vanderbilt University, Nashville, TN; Knutson et al., 2008). Hdac3fl/fl mice were crossed with E8I-Cre transgenic mice (Jackson strain 008766) and OT-I mice to generate TCR-polyclonal and OT-I TCR-transgenic mice with CD8 T cell–restricted deletion of Hdac3; the latter strain was further crossed with Cas9+ OT-I mice to generate Cas9+ E8I-Cre+ Hdac3fl/fl OT-I mice. Hdac3fl/fl mice were also crossed with UBC-Cre-ERT2 mice (Jackson strain 008085) and OT-I mice to generate OT-I TCR-transgenic mice with a tamoxifen-inducible deletion of Hdac3. Hdac3-conditional KO strains were maintained by crossing Cre+ Hdac3fl/fl mice with Cre− Hdac3fl/fl mice to generate Hdac3-WT littermate controls for experiments, and to maintain hemizygous inheritance of the Cre transgene. All mice used as hosts in experiments were male mice 8–9 wk of age. For adoptive transfer experiments, 6–12-wk-old mice of both sexes were used as T cell donors.
Flow cytometry
Fluorochrome-conjugated antibodies against CD5 (53–7.3), CD8α (53–6.7), CD11b (M1/70), CD11c (N418), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD62L (MEL-14), H-2 (M1/42; pan-MHC I), H-2Kb-SIINFEKL (25-D1.16), TCR-β (H57-597), Thy1.1 (OX-7), FoxP3 (FJK-16s), IFN-γ (XMG1.2), TNF-α (MP6-XT22), and T-bet (4B10) were purchased from Biolegend. Antibodies specific for CD4 (RM4-5) and Granzyme B (NGZB) were purchased from Thermo Fisher Scientific. Antibodies specific for Runx3 (R3-5G4) were purchased from BD Biosciences. PE-conjugated Kb-SIINFEKL (OVA peptide) and Db-KAVYNFATC (LCMV GP33 epitope) tetramers were purchased from MBL International Corporation. Antibodies against surface epitopes were used at 1:200 dilution, except α-CD44 and α-CD62L antibodies, which were used at 1:100 dilution. Antibodies against intracellular epitopes were used at 1:50 dilution, except α-Runx3 antibodies, which were used at 1:20 dilution. All flow-cytometry samples were first stained with Zombie UV fixable viability dye (Biolegend) for live/dead exclusion at 1:200 dilution, then treated with unconjugated α-CD16/32 antibodies (93, Biolegend) at 1:50 dilution to block nonspecific binding to Fc receptors before staining. Where required, staining with tetramers was done at the dilution recommended by the manufacturer after live/dead staining and before Fc receptor blockade. For intracellular staining of cytokines and transcription factors, samples were fixed and permeabilized with the eBioscience FoxP3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). Where indicated, samples were restimulated with PMA and ionomycin using eBioscience Cell Stimulation Cocktail (plus protein transport inhibitors; Thermo Fisher Scientific), or with 5 µg/ml of indicated peptide(s) plus eBioscience Protein Transport Inhibitor Cocktail (Thermo Fisher Scientific) for 3–4 h before evaluation by flow cytometry. Unstimulated samples (treated only with Protein Transport Inhibitor Cocktail) were acquired in parallel as gating controls. All flow cytometry data were acquired using an LSRFortessa X-20 (BD Biosciences), and all cell sorting was performed using a FACSAria IIIu (BD Biosciences). Flow cytometry data were analyzed using FlowJo software.
Cell culture and cell lines
Cell culture media and supplements were purchased from Gibco (Thermo Fisher Scientific). All primary cell cultures were grown in RPMI 1640 media (RPMI) supplemented with 10% (vol/vol) FBS, 1× GlutaMAX (Gibco), 100 U penicillin-streptomycin, 1 mM sodium pyruvate, 20 mM Hepes, and 50 µM 2-mercaptoethanol. All other cell lines were grown in DMEM media supplemented with 10% (vol/vol) FBS, 1× GlutaMAX, 100 U penicillin-streptomycin, and 10 mM Hepes. The OVA-expressing B16F10 tumor cell line (B16-OVA) was generated by lentiviral transduction of B16F10 (ATCC) with a pHAGE expression vector bearing an N-terminally truncated variant of chicken OVA that was sub-cloned from pcDNA3-deltaOVA (Addgene plasmid 64595; Diebold et al., 2001). zsGreen+ transduced tumor cells were sorted to purity to establish the cell lines. B16-OVA cells were validated by flow-cytometric measurement of the OVA epitope SIINFEKL in complex with H2-Kb, and by evaluation of their ability to activate OT-I T cells.
Isolation of immune cell populations from mice
For spleens and LNs, organs were mechanically dissociated on a 70-µm cell strainer with unsupplemented RPMI media. For liver and kidney samples, organs were minced with scalpels in RPMI and further dissociated in GentleMACS C tubes using a GentleMACS dissociator (Miltenyi Biotec); resulting cell suspensions were separated at 1,200 g on a 40%/70% discontinuous Percoll gradient (Sigma-Aldrich). Samples from spleens and livers were treated with RBC lysis buffer (Biolegend) after dissociation to remove erythrocytes.
In vitro activation of OT-I T cells with OVA peptide–pulsed BMDCs
1 wk before T cell activation, BMDCs were generated by GM-CSF culture of bone marrow cells isolated from the femora and tibiae of C57BL/6-background mice as previously described (Madaan et al., 2014). Immature BMDCs were activated with 100 ng/ml of lipopolysaccharide (Sigma-Aldrich) overnight before γ-irradiation (3,500 rad) and incubation with 100 ng/ml of SIINFEKL peptide. OT-I T cells were prepared from pooled cell suspensions from spleens and peripheral LNs (inguinal, brachial, and cervical) of OT-I TCR-transgenic mice by negative magnetic selection using an EasySep Mouse CD8+ T Cell Isolation Kit (StemCell Technologies, Inc.). BMDCs and OT-I T cells were co-cultured at a 1:1 ratio at a total density of 106 cells/ml in 96-well, 24-well, or 6-well tissue culture plates (at volumes of 0.2, 0.5, or 2 ml, respectively). For in vitro HDAC3 inhibitor studies, RGFP966 (Selleck Chemicals) was prepared as a 10 mM stock in DMSO (Sigma-Aldrich) and diluted to the indicated concentrations directly into cell culture medium; control cell cultures were treated with an equivalent volume of DMSO vehicle.
In vitro screen of epigenetic regulator drugs on CD8 T cell activation
96-well plates containing BMDC and OT-I T cell co-cultures were set up as described in the preceding section. Immediately after OT-I T cells were added to BMDCs, a custom small molecule library containing aliquots of 100 epigenetic inhibitor drugs in DMSO vehicle (Table S1) was directly diluted into the 96-well plates using 96-well pin applicators (Thermo Fisher Scientific) to a final concentration of 10 µM. Each 96-well plate also contained 10 control wells (equivalent volume of DMSO vehicle added). Fresh media containing 10 µM epigenetic inhibitor drugs or DMSO vehicle was added on days 3 and 5 after activation. On day 7, cells were collected in 96-well plates and fixed for flow-cytometric analysis of a panel of markers of effector CD8 T cell function.
Means and standard distributions of either the geometric mean fluorescence intensities (gMFIs; for markers with unimodal staining patterns, e.g., CD25 and CD127), or of the percentage of positive cells (for markers with bimodal distributions, e.g., Granzyme B and IFN-γ), were computed across the 10 control wells for each plate. Thresholds for significance were set as |drug perturbation| ≥1 SD or ≥2 SD for bimodal and unimodal markers, respectively. Each drug was then scored with +1, 0, or −1 for each marker that was significantly elevated, not significantly changed, or significantly reduced relative to the mean of the control, respectively. The total score for each drug was then used to rank and prioritize small molecules for follow-up analysis.
CRISPR/Cas9-mediated gene KO in CD8 T cells by lentiviral transduction
To efficiently transduce primary CD8 T cells without prior TCR activation, we constructed the Thy1.1-marked lentiviral gRNA expression plasmid vector pLKO-gRNA-Thy1.1 by subcloning the gRNA expression cassette of lentiGuide-Puro (Addgene plasmid 52963; Sanjana et al., 2014) into a pLKO.3G backbone (Addgene plasmid 14748) and replacing the enhanced green fluorescent protein (eGFP) marker with Thy1.1. gRNA sequences targeting specific genes were picked from a list generated by the online single guide RNA (sgRNA) Design Tool (Broad Institute, Cambridge, MA) and inserted into pLKO-gRNA-Thy1.1 using sticky ends generated by BsmBI digestion. Lentivirus production and CD8 T cell transduction were performed as previously described (Zhou et al., 2014). Briefly, Cas9+ OT-I T cells were cultured in 100 ng/ml IL-15 and 5 ng/ml IL-7 for 48 h before spin-infection in retronectin-coated (Takara Bio) 24-well plates with concentrated lentivirus (multiplicity of infection = 15). Spin-infection was done in RPMI media with 5 µg/ml protamine sulfate (Sigma-Aldrich) for 1.5–2 h at 32°C. Cells were cultured for 72 h after transduction with 50 ng/ml IL-15, 2.5 ng/ml IL-7, and 2 ng/ml IL-2 before magnetic enrichment using the Thy1.1 marker using an EasySep Mouse CD90.1 Positive Selection Kit (StemCell Technologies) to ≥93% Thy1.1+ purity. The following gRNA-coding sequences were used to generate lentiviruses targeting the indicated genes for inactivation (all sequences read 5′ to 3′): Hdac3, AGTGGAAGTTGCCCACATCG; Prdm1, GGATAGGATAAACCACCCGA; Runx3, GTTCACCAACCCTACCCAAG; and Tbx21, GCTCTACCCAGGGCCGCGCG.
Western blot analysis of gRNA KO efficiency
Thy1.1+-enriched cells after lentiviral gRNA vector transduction were activated in vitro with 3 µg/ml plate-bound α-CD3ε (145-2C11, Biolegend) and 1 µg/ml of soluble α-CD28 (37.51, Biolegend). Cells were collected after 3–7 d after activation for preparation of whole cell lysates in radioimmunoassay precipitation buffer for subsequent immunoblot analysis to assess the efficiency of gene inactivation. Total cell lysates equivalent to 2–5 × 105 cells were loaded into each lane for SDS-PAGE and subsequent protein transfer to a polyvinylidene difluoride membrane. Antibodies for Western blotting against HDAC3 (2632, 85057), Blimp-1 (9115), Runx3 (9647), and GAPDH (2118) were purchased from Cell Signaling Technology. Membranes were incubated with primary antibodies overnight at 4°C with gentle shaking at a dilution of 1:1,000 in SuperBlock T20 (TBS) (Thermo Fisher Scientific), except for α-GAPDH antibodies, which were diluted 1:5,000. Membranes were then incubated with secondary goat α-rabbit HRP-conjugated antibodies (1:10,000; Cell Signaling Technology) for 1 h at room temperature and then treated with enhanced chemiluminescence reagent (Thermo Fisher Scientific). Protein bands were visualized on autoradiography film (Denville Scientific), and protein KO levels were quantified from scanned film images using FIJI software (National Institutes of Health).
Cytotoxicity assays
For in vitro cytotoxicity assays, B16-OVA cells were treated with 10 ng/ml of IFN-γ overnight and labeled with 51Cr the following day. OT-I T cells were collected from in vitro cultures, resuspended at appropriate densities, and cultured with 2 × 103 B16-OVA target cells per well at effector-to-target ratios ranging from 10:1 to 1:1 with four replicates per condition. Cells were gently centrifuged to accelerate cell–cell interactions. Eight wells of B16-OVA target cells without T cells were left untreated to assess spontaneous 51Cr release, and eight wells of B16-OVA cells were fully lysed with 1% Triton X-100 in PBS to assess maximum 51Cr release. After 4 h of co-culture, cells were pelleted, and the supernatant was collected for quantification of 51Cr radioactivity with a Microbeta2 microplate counter (Perkin Elmer). In vitro cytotoxicity was calculated using the following equation:
For in vivo cytotoxicity assays, target cells were prepared by depleting C57BL/6 splenocytes with biotin-conjugated α-CD3 (17A2, Biolegend) and α-NK1.1 (PK136, Biolegend) antibodies followed by negative magnetic selection with streptavidin beads (StemCell Technologies). Target cells were then incubated with 1 µM of either OVA SIINFEKL peptide (Sigma-Aldrich) or gp100 KVPRNQDWL control peptide (AnaSpec), and labeled with 1 µM CFSE (Biolegend) or eFluor670 (Thermo Fisher Scientific), respectively. 5 × 106 cells of a 1:1 mix of the two differentially labeled target cell populations were intravenously injected into OVA-immunized recipient mice that had previously received Hdac3-KO or Hdac3-WT OT-I T cells. Mice that had received OT-I T cells without OVA immunization were used as controls. 9 h following transfer of cells, spleens from recipient mice were analyzed, and the numbers of CFSE+ OVA-labeled targets, eFluor670+ control peptide–labeled targets, and OVA peptide tetramer+ CD8 T cells were recorded. In vivo cytotoxicity was calculated as a ratio of depleted OVA-labeled targets relative to the numbers of OT-I T cells as follows:
OVA immunization studies
1 d before immunization, OT-I T cells were prepared from pooled cell suspensions from spleens and peripheral LNs as described earlier, and 5 × 105 OT-I T cells in PBS were transferred into recipient mice by intravenous injection. Recipient mice were immunized by subcutaneous injection into the flanks with a dose of 10 µg OVA (Imject Ovalbumin, Thermo Fisher Scientific) + 50 µg poly(I:C) (Sigma-Aldrich) in PBS. For longitudinal studies of transferred OT-I populations, blood lymphocytes from retro-orbital bleeds were analyzed by flow cytometry. For terminal analysis of donor OT-I T cell phenotype, total cell suspensions prepared from inguinal LNs were restimulated with 5 µg/ml OVA SIINFEKL peptide (Sigma-Aldrich) for 3–4 h before analysis by flow cytometry. The contribution of LN egress and/or activation-induced cell death on T cell numbers was examined by intraperitoneal injection of 500 µg/kg FTY720 (Cayman Chemical) in a vehicle of 50% (vol/vol) ethanol in PBS and/or 3 mg/kg pan-caspase inhibitor peptide Ac-DEVD-CHO (Selleck Chemical) at indicated time points.
LCMV infections
LCMV Armstrong strain expressing OVA (LCMV-OVA) was a gift from U. von Andrian (Harvard Medical School, Boston, MA) and generated by Juan de la Torre (The Scripps Research Institute, La Jolla, CA; Gerlach et al., 2016). LCMV Clone 13 was provided by H.-J. Kim. All viruses were propagated and titrated as previously described (Welsh and Seedhom, 2008). For acute virus infection experiments, TCR-polyclonal CD45.1+ congenic mice were adoptively transferred with 5 × 103 cells of a 1:1 mix of CD45.1− CD45.2+ Hdac3-inducible KO and CD45.1+ CD45.2+ Hdac3-WT OT-I T cells isolated from age- and sex-matched donors. Hdac3 deletion at indicated time points was induced by intraperitoneal injection of 100 µl of a 20 mg/ml tamoxifen (Sigma-Aldrich) solution in corn oil daily for 3 consecutive days. 4 d after T cell transfer, host mice were infected with 2 × 105 PFU of LCMV-OVA by intraperitoneal injection. For longitudinal studies of transferred OT-I T cell populations, lymphocytes from retro-orbital bleeds were analyzed by flow cytometry. For terminal analysis of donor OT-I T cell phenotypes, total cell suspensions prepared from indicated organs were restimulated with 5 µg/ml OVA peptide (Sigma-Aldrich) for 3–4 h before flow cytometry analysis.
For chronic virus infection experiments, TCR-polyclonal E8I-Cre+ Hdac3fl/fl mice with a CD8 T cell–restricted deletion of Hdac3 or Cre− Hdac3fl/fl littermate controls of both sexes were infected with 2 × 106 PFU LCMV Clone 13 by intravenous injection. Weight loss was monitored every 2–3 d after infection, and viremia was monitored weekly by titration of serum samples on Vero cell monolayers and counting of plaques, as previously described (Welsh and Seedhom, 2008). For terminal analysis of virus-specific CD8 T cell responses in viral reservoir organs, total cell suspensions prepared from indicated organs were restimulated with 5 µg/ml of LCMV GP33 and GP276 peptides (AnaSpec) for 3–4 h before flow-cytometric analysis.
RNA-seq and analysis
RNA-seq analysis was performed on Hdac3-KO and Hdac3-WT OT-I T cells activated in parallel, and on RGFP966- and vehicle control-treated OT-I T cells activated in parallel, to robustly identify genes that were regulated by HDAC3 activity. OT-I T cells were isolated and activated on irradiated SIINFEKL-pulsed BMDCs as described above and sorted to purity after 5 d. Total RNA was prepared using the RNAeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions and submitted to the Dana-Farber Molecular Biology Core Facility for library preparation and sequencing. A standard mRNA library preparation kit (RS-122-2101, Illumina) was used for library preparation. Single-end 75-bp sequencing was done on an Illumina NextSeq 500. Statistics for differentially expressed genes were calculated by DESeq2 (version 3.5; Love et al., 2014). Thresholds of −log10(adjusted P value) >10 and |log2(fold change)| >0.5 were used to identify genes that were differentially transcribed between Hdac3-KO and Hdac3-WT CD8 T cells (Table S2), or between RGFP966-treated and vehicle control-treated CD8 T cells (Table S3). The HDAC3 activity-regulated list of genes was compiled from genes that were present in both lists of differentially transcribed genes (in the KO vs. WT and RGFP966 vs vehicle datasets), and whose direction of change was also conserved between both datasets. The RNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO; Edgar et al., 2002) under accession no. GSE143644.
Analysis of H3K27ac by ChIP-seq
Hdac3-KO and Hdac3-WT OT-I T cells were isolated and activated on irradiated SIINFEKL-pulsed BMDCs as described above and sorted to purity after 3 d. Sorted cells were fixed using 1% paraformaldehyde (PFA; Sigma-Aldrich) in PBS for 7 min, and the reaction was then quenched by addition of glycine to a final concentration of 125 mM (Sigma-Aldrich) in PBS for 5 min, all with gentle rotation at room temperature. Cells were then washed with ice-cold PBS in the presence of 1× protease inhibitor cocktail (P8340, Sigma-Aldrich), 1 mM PMSF (Sigma-Aldrich), and 10 mM sodium butyrate (Sigma-Aldrich), and dry pellets were stored at −80°C until all samples were collected. Cross-linked material was resuspended in sonication buffer (1% SDS, 50 mM Tris-HCl, pH 8, and 10 mM EDTA) and sonicated in a Covaris E220 instrument (15 min, 5% duty cycle, 75 peak incident power, 200 cycles per burst) using a 1-ml AFA Fiber milliTUBE (Covaris). 10 µl of recombinant barcoded nucleosomes with various acetyl-lysine modifications (Epicypher v0.5 K-AcylStat SNAP-ChIP panel) to 5 µg of soluble chromatin before taking 10% as input and using the remaining for immunoprecipitation with 10 µg of rabbit anti-H3K27ac antibody (Diagenode C15410196, lot A1723-0041D). Libraries were prepared with an Accel-NGS 2S Plus DNA Library Kit (Swift Biosciences) following the manufacturer’s protocol. 36-bp paired end reads were sequenced on a NextSeq 2500 instrument (Illumina).
Reads for the entire barcoded spike-in histone K-acetyl panel were quantified before and after ChIP. We evaluated the reads from all barcoded spike-in acetylation marks to calculate the enrichment of ChIPs from both Hdac3-KO and Hdac3-WT samples, and used the ratio of the barcode signals to normalize the sequencing Bedgraph files. Normalization was done by taking the union set of all peaks from the two H3K27ac ChIP samples (Hdac3-KO and Hdac3-WT), calculating the read depth-normalized ratio of reads at each peak location, and then applying the normalization factor derived from the spike-in panel barcodes to the sequencing results. Finally, the normalized Bedgraph files were converted to Bigwiggle format for viewing in the Integrative Genomics Viewer platform and downstream analysis.
Pre-processing and quality control of normalized ChIP-seq data were performed using the ChiLin pipeline 2.0.0 (Qin et al., 2016). We used the Burrows-Wheeler Aligner (Li and Durbin, 2009) to map reads. Peak-calling was performed using the MACS2 (Model-based Analysis of ChIP-Seq) algorithm (Zhang et al., 2008), with specifications to call only unique peaks to reduce the probabilities of calling false positive peaks. Statistically significant peaks were then selected based on the calculated false discovery rate of reported peaks. The H3K27ac ChIP-seq data are accessible through GEO accession no. GSE143644.
To calculate the changes in H3K27ac signal between Hdac3-KO and Hdac3-WT samples, we first defined a set K of genomic coordinates corresponding to H3K27ac peaks that were present within both Hdac3-KO and Hdac3-KO samples, i.e., . We then compiled a new set R of fold changes in H3K27ac intensities by computing the ratio of H3K27ac signals between Hdac3-KO and Hdac3-WT normalized ChIP-seq traces across the set of genomic loci described by each element of set K. From the distribution of elements within set R, we then determined the median increase of H3K27ac peak intensities in Hdac3-KO relative to Hdac3-WT samples. Peaks with differential H3K27ac signals were also mapped to the nearest gene loci to generate a list of gene loci rank-ordered by the numbers of H3K27ac peaks with increased signal in Hdac3-KO relative to Hdac3-WT samples (Table S4). Relative changes in H3K27ac at selected gene loci were also quantified by calculating the mean increase in signal at each peak present within the genomic regions indicated in the ChIP-seq traces in Fig. 6.
Unbiased identification of mammalian (Homo sapiens or Mus musculus) transcription factor binding motifs within the genomic regions with differentially acetylated H3K27ac peaks in activated Hdac3-KO CD8 T cells relative to Hdac3-WT cells was performed using the HOMER motif analysis algorithm (Heinz et al., 2010).
ChIP-qPCR validation of H3K27 acetylation
OT-I T cells were isolated and activated on irradiated SIINFEKL-pulsed BMDCs as described above and sorted to purity after 3 or 5 d. Dry, PFA-fixed cell pellets were prepared and stored at −80°C as previously described. Total chromatin was prepared using a Zymo-Spin ChIP Kit (Zymo Research) using the mechanical shearing protocol supplied by the manufacturer; chromatin was sheared by direct probe sonication using a Q125 sonicator with a 1/16 inch (1.6 mm) diameter probe tip (Qsonica), using 12 pulsed cycles of 15 s at 10% amplitude followed by 30 s incubation at 4°C. ChIP was performed using reagents from the Zymo-Spin ChIP Kit (Zymo Research), and with a 1:100 dilution of antibodies against H3K27ac (8173) or equivalent amount of normal rabbit IgG (negative control; 2729; Cell Signaling Technology). After reverse cross-linking and chromatin elution, samples were analyzed by qPCR using PowerSYBR Green reagents (Thermo Fisher Scientific) with primers targeting promoter regions (up to −1,000 bp before the transcription start site) of target gene loci. PCR reactions were run on a C1000 thermocycler with a CFX96 optical reaction module (Bio-Rad) for 40 cycles of 15 s denaturation at 95°C followed by 60 s annealing/extension at 60°C. The following primer pairs were used to generate amplicons from the promoters of the indicated genes (all sequences read 5′ to 3′): Gzma, CCCTTCTAAGGTCACGTGGT and ATGTCTGGTTAAGCCCCTCG; Gzmb, AGACGGACCATGGAGGATGT and GGACAGTAGTTCGTGCCGT; Prf1, TCGACTCTTTTCCCGAAGGC and CCGGCCTCCTCTTCTGTTAC; Ifng, TGGAGCATTTGGTTGAGCCT and TGGCTATGGGTGCAGACTTG; Il2, AGCAGACCCAGCACAGTCTA and ACCCCAGGTTAGGTGCTACA; Tnf, GAGGTAGGGATGTTGGCTGC and TCTCTTTGCGGAGGCCTAAAG; Lta, CTGGGAATGTGGGGATGGAC and TATTGGAAGGCCTGGGACAC; Prdm1, GCTGTGCTCTTGTAAGGCGA and CGTCGGAGGAGAAACGAGTC; Tbx21, AAATGCAGGACTTACCGTAGTT and ATACCAACCGGTGTCTGTGT; and Id2, CTAGCGACCTGTGACTTCCC and GTGCCCGGAGCTAGCTATAC.
Runx3 ChIP-sequencing
Hdac3-KO or Hdac3-WT OT-I T cells were isolated and activated on irradiated SIINFEKL-pulsed BMDCs as described above and sorted to purity after 5 d. Sorted cells were double-fixed with di(N-succinimidyl) glutarate (Sigma-Aldrich) and 1% PFA (Sigma-Aldrich) according to a previously described protocol (Tian et al., 2012). Cells were then washed with ice-cold PBS in the presence of 1× protease inhibitor cocktail (P8340, Sigma-Aldrich), 1 mM PMSF (Sigma-Aldrich), and 10 mM sodium butyrate (Sigma-Aldrich), and dry pellets were stored at −80°C until all samples were collected. Cross-linked material was resuspended in sonication buffer (1% SDS, 50 mM Tris-HCl, pH 8, and 10 mM EDTA) and sonicated in a Covaris E220 instrument (15 min, 5% duty cycle, 75 peak incident power, 200 cycles per burst) using a 1 ml AFA Fiber milliTUBE (Covaris). 40 µg of soluble chromatin was immunoprecipitated with 5 µg of rabbit anti-Runx3 antibody (ab11905, Abcam). Libraries were prepared with an Accel-NGS 2S Plus DNA Library Kit (Swift Biosciences) following the manufacturer’s protocol. 36-bp paired end reads were sequenced on a NextSeq 2500 instrument (Illumina). The Runx3 ChIP-seq data are accessible through GEO accession no. GSE143644.
Online supplemental material
Fig. S1 contains an overview of the epigenetic inhibitor drug screen and additional data characterizing the regulation of CD8 T cell cytotoxicity by the HDAC3-specific inhibitor RGFP966. Fig. S2 and Fig. S3 contain data characterizing the phenotypes of Hdac3-KO (E8I-Cre+; Hdac3fl/fl) in developing thymocytes and in peripheral CD8 T cells, respectively. Fig. S4 contains detailed analysis of the reduced accumulation of Hdac3-KO CD8 T cells in draining LNs following in vivo activation. Fig. S5 describes additional aspects of the phenotype of Hdac3-KO virus-specific CD8 T cells during acute or chronic viral infection, related to Fig. 2and Fig. 4, respectively. Table S1 describes the results of the epigenetic inhibitor drug screen in detail. Table S2 and Table S3 list genes that are differentially expressed in HDAC3 inhibitor-treated activated CD8 T cells or in Hdac3-KO activated CD8 T cells relative to control cells, respectively. Table S4 lists gene loci with at least one H3K27ac peak showing increased acetylation in activated Hdac3-KO CD8 T cells relative to Hdac3-WT cells.
Supplementary Material
lists epigenetic inhibitor drugs screened and their perturbations to the expression of markers of CD8 T cell effector function.
lists differentially expressed genes between activated Hdac3-KO and Hdac3-WT CD8 T cells.
lists differentially expressed genes between activated CD8 T cells treated with HDAC3 inhibitor RGFP966 or vehicle control.
lists gene loci with at least one H3K27ac peak showing increased acetylation in activated Hdac3-KO CD8 T cells relative to Hdac3-WT cells.
Acknowledgments
We would like to thank Scott W. Hiebert (Vanderbilt University, Nashville, TN) for providing the Hdac3fl/fl mouse strain generated in his laboratory and Ulrich von Andrian and Olga Barreiro del Rio (Harvard Medical School, Boston, MA) for sharing the LCMV-OVA strain (generated by Juan Carlos de la Torre, The Scripps Research Institute, La Jolla, CA). We are thankful to Sabrina Haag (Dana-Farber Cancer Institute, Boston, MA) for her helpful discussions in developing this project.
This work was supported by grants from the National Institutes of Health (NCI R01 CA238039 to K.W. Wucherpfennig and P01 CA163222, R01 AR072304, R01 CA222871, and R01 AR043369-19 to D.E. Fisher). This work was also supported by The Bridge Project, a collaboration between the Koch Institute of Massachusetts Institute of Technology and the Dana-Farber/Harvard Cancer Center (K.W. Wucherpfennig). R.E. Tay was supported by a graduate fellowship from A*STAR (Singapore). D.E. Fisher also acknowledges generous support from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Author contributions: R.E. Tay, H.-J. Kim, and K.W. Wucherpfennig designed the study and wrote the paper; R.E. Tay and O. Olawoyin performed in vitro and in vivo immunological studies with input from H.-J. Kim and K.W. Wucherpfennig; Y. Ito contributed to the characterization of Hdac3-KO mice; P. Cejas, Y. Xie, C.A. Meyer, H.W. Long, and M. Brown performed ChIP-seq experiments and analysis; Q.Y. Weng and D.E. Fisher provided the small molecule epigenetic inhibitor library for the initial drug screen.
References
- Banerjee A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., and Reiner S.L.. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.L., and Licht J.D.. 2018. Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 58:187–207. 10.1146/annurev-pharmtox-010716-105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best J.A., Blair D.A., Knell J., Yang E., Mayya V., Doedens A., Dustin M.L., and Goldrath A.W.; Immunological Genome Project Consortium . 2013. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14:404–412. 10.1038/ni.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarile M.A., Lind N.A., Rivera R., Sheridan A.D., Camfield K.A., Wu B.B., Cheung K.P., Ding Z., and Goldrath A.W.. 2006. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7:1317–1325. 10.1038/ni1403 [DOI] [PubMed] [Google Scholar]
- Chang J.T., Wherry E.J., and Goldrath A.W.. 2014. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15:1104–1115. 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty F., Pipkin M.E., Djuretic I.M., Levanon D., Lotem J., Lichtenheld M.G., Groner Y., and Rao A.. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206:51–59. 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Liu Y., Weinstein J.S., Craft J., and Kaech S.M.. 2011. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 35:792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans C., and Maggert K.A.. 2015. What do you mean, “epigenetic”? Genetics. 199:887–896. 10.1534/genetics.114.173492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S.S., Cotten M., Koch N., and Zenke M.. 2001. MHC class II presentation of endogenously expressed antigens by transfected dendritic cells. Gene Ther. 8:487–493. 10.1038/sj.gt.3301433 [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W., Sunshine M.J., Losos K., Hatam F., and Littman D.R.. 1997. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 7:537–547. 10.1016/S1074-7613(00)80375-1 [DOI] [PubMed] [Google Scholar]
- Gerlach C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., and von Andrian U.H.. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.M., Amezquita R.A., Guan T., Kleinstein S.H., and Kaech S.M.. 2017. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity. 46:596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning A.N., Roychoudhuri R., and Restifo N.P.. 2018. Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 18:340–356. 10.1038/nri.2017.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F.C., Belmonte P.J., Constans M.M., Chen M.W., McWilliams D.C., Hiebert S.W., and Shapiro V.S.. 2015. Histone Deacetylase 3 Is Required for T Cell Maturation. J. Immunol. 195:1578–1590. 10.4049/jimmunol.1500435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Hatano M., Okada S., Toyama H., Taki S., Arima M., Kuroda Y., and Tokuhisa T.. 2002. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 3:558–563. 10.1038/ni802 [DOI] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Kuroda Y., and Tokuhisa T.. 2004. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J. Immunol. 173:883–891. 10.4049/jimmunol.173.2.883 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Kao C., Banerjee A., Schambach F., Northrop J.K., Shen H., Wherry E.J., and Reiner S.L.. 2007. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 204:2015–2021. 10.1084/jem.20070841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Pos Z., Rao M., Klebanoff C.A., Yu Z., Sukumar M., Reger R.N., Palmer D.C., Borman Z.A., Muranski P., et al. 2011. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12:1230–1237. 10.1038/ni.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., and Ahmed R.. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kallies A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Knutson S.K., Chyla B.J., Amann J.M., Bhaskara S., Huppert S.S., and Hiebert S.W.. 2008. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 27:1017–1028. 10.1038/emboj.2008.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragten N.A.M., Behr F.M., Vieira Braga F.A., Remmerswaal E.B.M., Wesselink T.H., Oja A.E., Hombrink P., Kallies A., van Lier R.A.W., Stark R., et al. 2018. Blimp-1 induces and Hobit maintains the cytotoxic mediator granzyme B in CD8 T cells. Eur. J. Immunol. 48:1644–1662. 10.1002/eji.201847771 [DOI] [PubMed] [Google Scholar]
- Lazarevic V., Glimcher L.H., and Lord G.M.. 2013. T-bet: a bridge between innate and adaptive immunity. Nat. Rev. Immunol. 13:777–789. 10.1038/nri3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaan A., Verma R., Singh A.T., Jain S.K., and Jaggi M.. 2014. A stepwise procedure for isolation of murine bone marrow and generation of dendritic cells. J. Biol. Methods. 1:1 10.14440/jbm.2014.12 [DOI] [Google Scholar]
- Martins G.A., Cimmino L., Liao J., Magnusdottir E., and Calame K.. 2008. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J. Exp. Med. 205:1959–1965. 10.1084/jem.20080526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace L., Goudot C., Zueva E., Gueguen P., Burgdorf N., Waterfall J.J., Quivy J.-P., Almouzni G., and Amigorena S.. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science. 359:177–186. 10.1126/science.aah6499 [DOI] [PubMed] [Google Scholar]
- Philip M., Fairchild L., Sun L., Horste E.L., Camara S., Shakiba M., Scott A.C., Viale A., Lauer P., Merghoub T., et al. 2017. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 545:452–456. 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips R.L., Chen M.W., McWilliams D.C., Belmonte P.J., Constans M.M., and Shapiro V.S.. 2016. HDAC3 Is Required for the Downregulation of RORγt during Thymocyte Positive Selection. J. Immunol. 197:541–554. 10.4049/jimmunol.1502529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips R.L., Lee J.-H., Gaonkar K., Chanana P., Chung J.Y., Romero Arocha S.R., Schwab A., Ordog T., and Shapiro V.S.. 2019. HDAC3 restrains CD8-lineage genes to maintain a bi-potential state in CD4+CD8+ thymocytes for CD4-lineage commitment. eLife. 8 e43821 10.7554/eLife.43821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Mei S., Wu Q., Sun H., Li L., Taing L., Chen S., Li F., Liu T., Zang C., et al. 2016. ChiLin: a comprehensive ChIP-seq and DNase-seq quality control and analysis pipeline. BMC Bioinformatics. 17:404 10.1186/s12859-016-1274-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ B.E., Olshanksy M., Smallwood H.S., Li J., Denton A.E., Prier J.E., Stock A.T., Croom H.A., Cullen J.G., Nguyen M.L., et al. 2014. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 41:853–865. 10.1016/j.immuni.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda T., Lawton L.N., Barrasa M.I., Fan Z.P., Kohlhammer H., Gutierrez A., Ma W., Tatarek J., Ahn Y., Kelliher M.A., et al. 2012. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 22:209–221. 10.1016/j.ccr.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., and Ahmed R.. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns K.S., Kieper W.C., Jameson S.C., and Lefrançois L.. 2000. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 1:426–432. 10.1038/80868 [DOI] [PubMed] [Google Scholar]
- Shin H.M., Kapoor V., Guan T., Kaech S.M., Welsh R.M., and Berg L.J.. 2013. Epigenetic modifications induced by Blimp-1 Regulate CD8+ T cell memory progression during acute virus infection. Immunity. 39:661–675. 10.1016/j.immuni.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel K.R., Zhao Y., Klus N.J., Kaiser J.F., Gordy L.E., Joyce S., Hiebert S.W., and Summers A.R.. 2015. Histone Deacetylase 3 Is Required for Efficient T Cell Development. Mol. Cell. Biol. 35:3854–3865. 10.1128/MCB.00706-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.-C., Komori T., Ito Y., and Littman D.R.. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. 10.1016/S0092-8674(02)01111-X [DOI] [PubMed] [Google Scholar]
- Tian B., Yang J., and Brasier A.R.. 2012. Two-step cross-linking for analysis of protein-chromatin interactions. Methods Mol. Biol. 809:105–120. 10.1007/978-1-61779-376-9_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Diao H., Getzler A.J., Rogal W., Frederick M.A., Milner J., Yu B., Crotty S., Goldrath A.W., and Pipkin M.E.. 2018. The Transcription Factor Runx3 Establishes Chromatin Accessibility of cis-Regulatory Landscapes that Drive Memory Cytotoxic T Lymphocyte Formation. Immunity. 48:659–674.e6. 10.1016/j.immuni.2018.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.M., and Seedhom M.O.. 2008. Lymphocytic choriomeningitis virus (LCMV): propagation, quantitation, and storage. Curr. Protoc. Microbiol. Chapter 15:1 10.1002/9780471729259.mc15a01s8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Blattman J.N., Murali-Krishna K., van der Most R., and Ahmed R.. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911–4927. 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736. 10.1073/pnas.1232420100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Soragni E., Chou C.J., Herman D., Plasterer H.L., Rusche J.R., and Gottesfeld J.M.. 2009. Chemical probes identify a role for histone deacetylase 3 in Friedreich’s ataxia gene silencing. Chem. Biol. 16:980–989. 10.1016/j.chembiol.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.Y., Best J.A., Knell J., Yang E., Sheridan A.D., Jesionek A.K., Li H.S., Rivera R.R., Lind K.C., D’Cruz L.M., et al. 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12:1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac A.J., Blattman J.N., Murali-Krishna K., Sourdive D.J., Suresh M., Altman J.D., and Ahmed R.. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205–2213. 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Cooper S., and Brockdorff N.. 2015. The interplay of histone modifications - writers that read. EMBO Rep. 16:1467–1481. 10.15252/embr.201540945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Shaffer D.R., Alvarez Arias D.A., Nakazaki Y., Pos W., Torres A.J., Cremasco V., Dougan S.K., Cowley G.S., Elpek K., et al. 2014. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature. 506:52–57. 10.1038/nature12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists epigenetic inhibitor drugs screened and their perturbations to the expression of markers of CD8 T cell effector function.
lists differentially expressed genes between activated Hdac3-KO and Hdac3-WT CD8 T cells.
lists differentially expressed genes between activated CD8 T cells treated with HDAC3 inhibitor RGFP966 or vehicle control.
lists gene loci with at least one H3K27ac peak showing increased acetylation in activated Hdac3-KO CD8 T cells relative to Hdac3-WT cells.