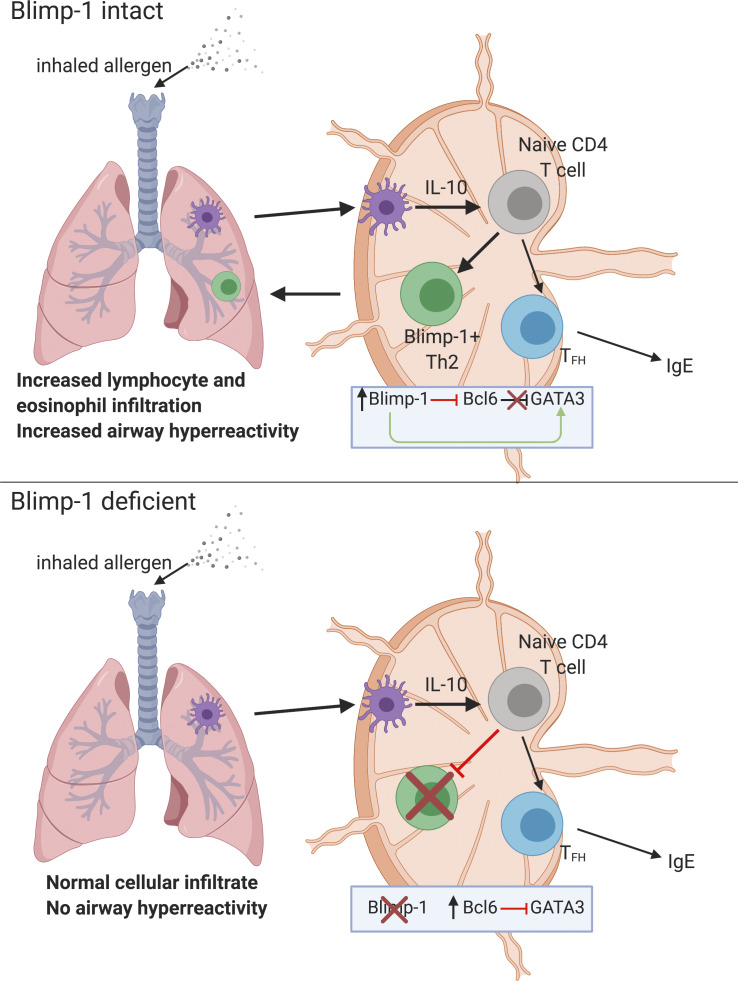
The transcriptional repressor Blimp-1, known to constrain autoimmunity, is an unexpected and critical positive regulator of Th2 cells in the lung acting via an IL-10–STAT3 axis in response to inhaled allergens, driving pathophysiology associated with asthma disease.
Abstract
A Th2 immune response is central to allergic airway inflammation, which afflicts millions worldwide. However, the mechanisms that augment GATA3 expression in an antigen-primed developing Th2 cell are not well understood. Here, we describe an unexpected role for Blimp-1, a transcriptional repressor that constrains autoimmunity, as an upstream promoter of GATA3 expression that is critical for Th2 cell development in the lung to inhaled but not systemically delivered allergens but is dispensable for TFH function and IgE production. Mechanistically, Blimp-1 acts through Bcl6, leading to increased GATA3 expression in lung Th2 cells. Surprisingly, the anti-inflammatory cytokine IL-10, but not the pro-inflammatory cytokines IL-6 or IL-21, is required via STAT3 activation to up-regulate Blimp-1 and promote Th2 cell development. These data reveal a hitherto unappreciated role for an IL-10–STAT3–Blimp-1 circuit as an initiator of an inflammatory Th2 response in the lung to allergens. Thus, Blimp-1 in a context-dependent fashion can drive inflammation by promoting rather than terminating effector T cell responses.
Graphical Abstract
Introduction
Asthma is a complex, chronic inflammatory disease of the airways. House dust mite (HDM) is a major indoor allergen that is globally ubiquitous in living environments and is capable of inducing allergic lung inflammatory diseases (Calderón et al., 2015). Immune cell infiltration, including eosinophils and IgE-mediated sensitization, are hallmarks of allergic airway disease, which is primarily driven by strong type 2 cytokine responses (such as IL-4, IL-5, and IL-13) predominantly produced by activated CD4+ T cells of a helper T (Th) type 2 cell phenotype (Lambrecht and Hammad, 2015; Licona-Limón et al., 2013; Pascual and Peters, 2005; Ray and Cohn, 1999; Zhang et al., 1999). Th2 cells are differentiated following the activation of naive CD4+ T cells in the presence of IL-4, and the master transcription factor GATA3 (Kopf et al., 1993; Zhang et al., 1997; Zheng and Flavell, 1997). However, the signals that support this process in vivo are still not well understood (Lambrecht and Hammad, 2015; Pulendran et al., 2010). Several IL-4–secreting cells have been proposed to promote Th2 cell development such as natural killer T cells, basophils, or early-activated CD4 T cells (Croft and Swain, 1995; Seder et al., 1991; Yoshimoto et al., 1995). However, there is evidence that IL-4–independent Th2 cell differentiation can occur, suggesting additional cytokines may play an important role in initiating or supporting Th2 cell differentiation in response to allergens (Dent et al., 1998; Oliphant et al., 2011; Ouyang et al., 2000; Stritesky et al., 2011). As evidence, both STAT3 signaling and cytokines such as thymic stromal lymphopoietin can promote Th2 cell differentiation (Rochman et al., 2018; Stritesky et al., 2011). Thus, additional regulators of Th2 cells beyond the IL-4–STAT6–GATA3 circuit play a role in type 2 immune responses.
B lymphocyte–induced maturation protein-1 (Blimp-1) is a transcriptional repressor required for plasma cell development and function (Minnich et al., 2016; Turner et al., 1994). However, Blimp-1 also has important functions in T cells to regulate effector responses (Crotty et al., 2010; Fu et al., 2017). Conditional deletion of Blimp-1 in T cells causes spontaneous accumulation of effector T cells and systemic autoimmunity, suggesting that Blimp-1 constrains T cell–mediated autoimmunity (Kallies et al., 2006; Martins et al., 2006). In CD4 T cells, Blimp-1 can repress Bcl6 to antagonize T follicular helper (TFH) cell differentiation, control IL-10 expression in effector (Th1 and Th17) and regulatory T (T reg) cells, and regulate the differentiation and function of effector T cells (Cretney et al., 2011; Heinemann et al., 2014; Johnston et al., 2009; Neumann et al., 2014; Parish et al., 2014). Furthermore, we previously found that overexpression of Blimp-1 could lead to cell death, suggesting Blimp-1 also controls effector responses by limiting effector cell numbers directly (Poholek et al., 2016).
Our previous studies showed that disrupting Blimp-1 in T cells increased Th2 cell responses in a worm antigen model delivered via s.c. injection of the footpad. Therefore, we hypothesized that T cell–specific deficiency of Blimp-1 in an allergic airway inflammation model would lead to increased expansion of effector cells and more severe disease due to increased Th2 cell responses. Unexpectedly, we found that T cell–specific Blimp-1 deficiency protected mice from the development of allergic lung inflammation in a model of inhaled allergen delivery, and Th2 cells in the lung were severely reduced. STAT3 via IL-10 was required for Blimp-1 expression and Th2 cell development in this model, suggesting IL-10 may play an unexpected role in supporting Th2 cell differentiation. Mechanistically, our data support an intrinsic role for Blimp-1–mediated repression of Bcl6, which in turn can repress GATA3 in the context of responses to allergens. Thus, Blimp-1 can indirectly support Th2 cell differentiation by promoting GATA3 expression. These data identify a new context-dependent role for Blimp-1 in T cells that is essential for the full development of allergic lung disease, highlighting a previously unappreciated pathway with potential therapeutic targets for the treatment of asthma disease.
Results
Blimp-1 in T cells promotes allergic airway inflammation
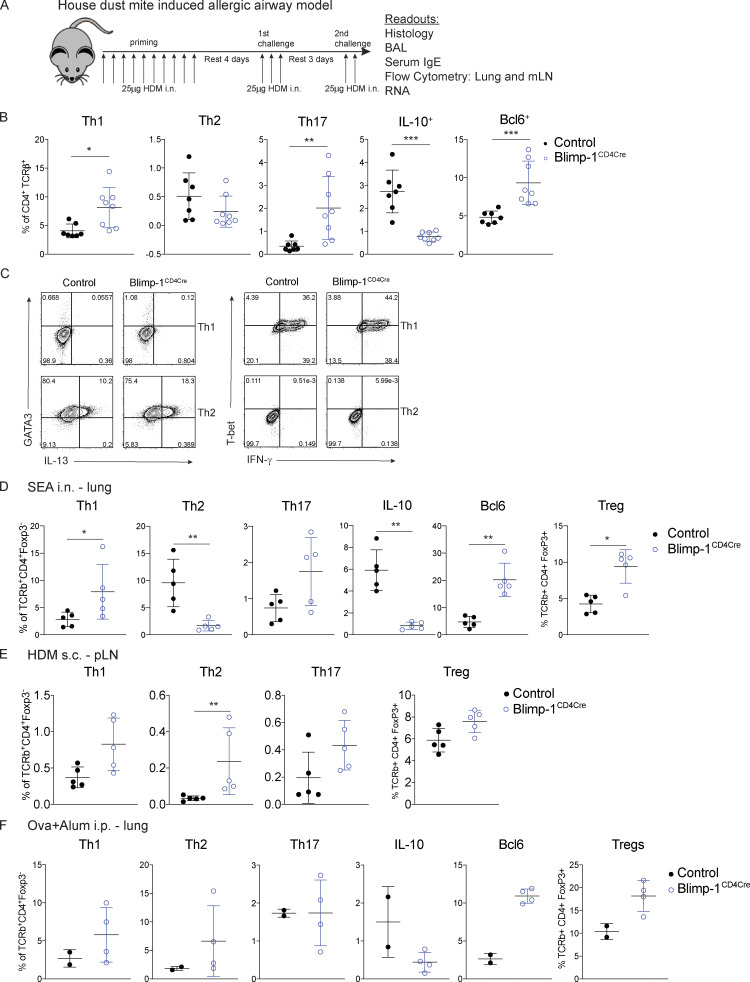
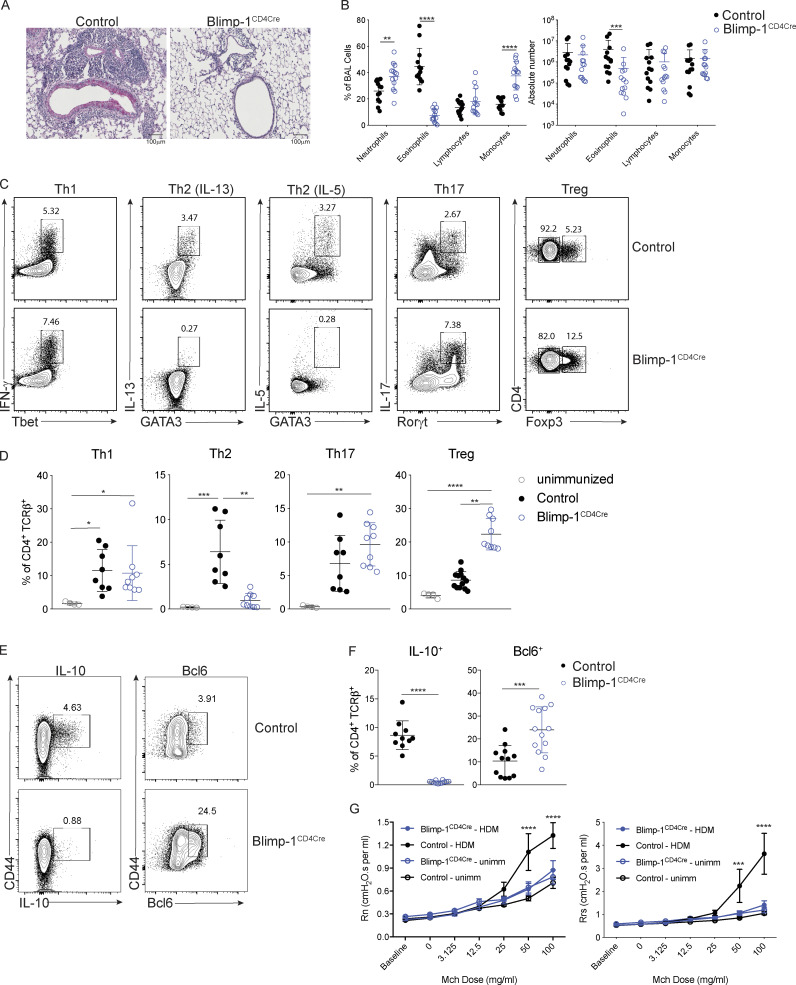
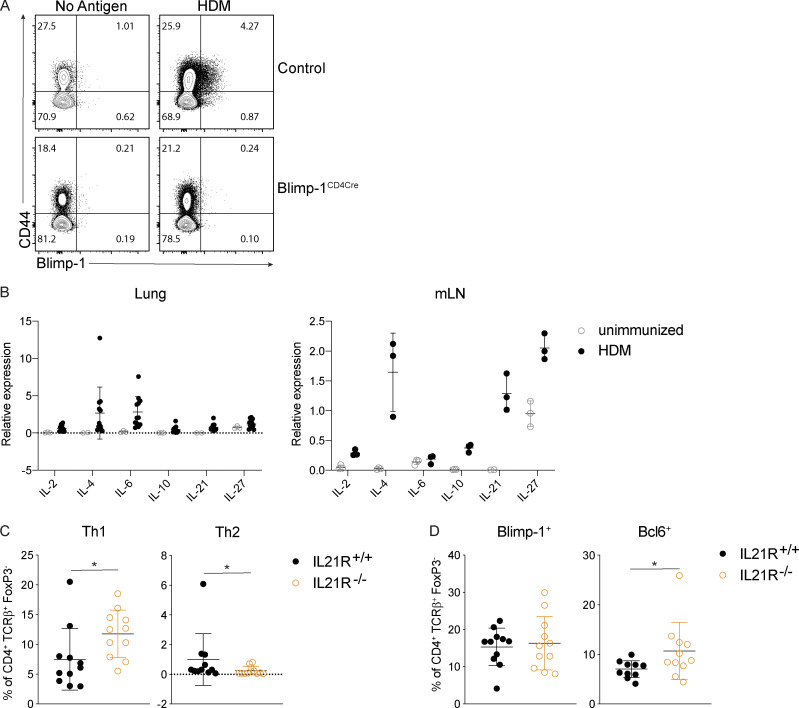
Blimp-1 controls effector T cell responses and constrains autoimmunity (Crotty et al., 2010; Poholek et al., 2016). We reasoned that during a HDM-induced model of allergic lung disease, the absence of Blimp-1 in T cells would drive increased effector T cells and exacerbate disease. To test this, we compared mice in which Blimp-1 was conditionally deleted in all T cells (Blimp-1f/f × CD4-cre, referred to as Blimp-1CD4Cre) to Cre-negative littermate controls (Blimp-1f/f). Allergic inflammation in the airways was induced by repeated i.n. administration of HDM antigen for 10 d (priming) followed by two rechallenges of 2–3 d each after rests of 3–4 d (Fig. S1 A). As expected, histological assessment of lung tissue following HDM administration of control animals revealed substantial lymphocyte infiltration to the tissue and mucus in the airways, indicating a robust allergic inflammatory response to HDM had occurred (Fig. 1 A). In addition, neutrophils, eosinophils, lymphocytes, and monocytes were readily found in the bronchioalveolar lavage (BAL) fluid of control animals (Fig. 1 B). Contrary to our hypothesis, we found that Blimp-1CD4Cre animals had no signs of inflammation based on histological analysis of lung tissue (Fig. 1 A). Analysis of BAL identified a specific loss of eosinophils, suggesting type 2 immunity was impaired (Fig. 1 B). Eosinophils are recruited by Th2 cells that produce IL-5, while mucus in the airways is due to the presence of IL-13+ cells (Erle and Sheppard, 2014; McBrien and Menzies-Gow, 2017). To determine if there were alterations in the CD4+ T cell response to HDM, we isolated lymphocytes from the lung and mediastinal LN (mLN) and assessed CD4 T cell subsets by flow cytometry. HDM is known to induce a mixed response with differentiation/expansion of Th1, Th2, and Th17 cells. We observed robust increases of Th1 (Tbet+ IFN-γ+), Th2 (GATA3+ IL-13+ or GATA3+ IL-5+), and Th17 (Rorγt+ IL-17+) cells as well as T reg cells (FoxP3+) in the lungs of control animals compared with unimmunized, naive animals (Fig. 1, C and D). In contrast, Blimp-1CD4Cre animals were specifically lacking Th2 cells in the lung, while Th1 and Th17 cells were largely unaffected (Fig. 1, C and D). In contrast to the lungs, Blimp-1CD4Cre mice had increased Th1 and Th17 cells in the mLN, but Th2 cells (GATA3+ IL-13+) were largely absent, constituting less than 1% of all CD4+ T cells (Fig. S1 B; Bao and Reinhardt, 2015). Consistent with previous publications, T reg cells were increased in Blimp-1CD4Cre mice, likely due to Blimp-1’s role in repressing IL-2, and T reg cells’ capability to expand in conditions with excess IL-2 (Fig. 1, C and D; Cretney et al., 2011; D’Cruz and Klein, 2005; Fontenot et al., 2005; Malek et al., 2002; Martins et al., 2008; Webster et al., 2009). Blimp-1 is well known to be a critical driver of IL-10, and indeed Blimp-1CD4Cre mice were specifically lacking IL-10 production from T cells (Fig. 1, E and F; and Fig. S1 B; Kallies et al., 2006; Martins et al., 2006; Neumann et al., 2014; Parish et al., 2014). Bcl6 and Blimp-1 are transcriptional repressors known to regulate one another in both T and B cells; thus, we explored the levels of Bcl6 in CD4+ T cells (Crotty et al., 2010). Blimp-1CD4Cre mice had increased expression of Bcl6 in both the lung and mLN (Fig. 1, E and F; and Fig. S1 B), suggesting that the absence of Blimp-1 in T cells resulted in a concomitant increase in Bcl6. We next assessed airway hyperreactivity (AHR) to determine if Blimp-1–dependent loss of Th2 cells in the lung was sufficient to alter lung function. Control mice repeatedly immunized and rechallenged with HDM had increased AHR in response to methylcholine; however, Blimp-1CD4Cre were protected from AHR and looked similar to unimmunized mice (Fig. 1 G). Thus, Blimp-1 in T cells played a significant role in promoting allergic asthma in response to inhaled allergens.
Figure S1.
Lung-specific Blimp-1–dependent Th2 development in vivo. (A) Schematic for HDM-induced allergic lung inflammation model used throughout this study. (B) Percent of indicated subsets of T cells isolated from mLN gated on live CD4+ TCRβ+. Data in B are pooled from two or three experiments with 8–13 total mice per group, mean ± SD. (C) Flow staining of Th1 (IFN-γ+ T-bet+) and Th2 (IL-13+ GATA3+) in vitro polarization of T cells isolated from control or Blimp-1CD4Cre animals. (D–F) Analysis of lung or pLN tissue isolated from control (Blimp-1f/f CD4Cre−) or Blimp-1CD4Cre (Blimp-1f/f CD4Cre+) animals (D) i.n. immunized with SEA, (E) s.c. immunized with HDM, and (F) i.p. immunized with OVA + alum followed by 1% nebulizer treatment. Percent of Th1, Th2, Th17, IL-10, and Bcl6 cells isolated from lungs gated on live CD4+ TCRβ+ FoxP3-. T reg cells are gated on live CD4+ TCRβ+ FoxP3+. Data in D–F are pooled from two experiments with four or five total mice per group, mean ± SD. Mann–Whitney t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 1.
T cell expression of Blimp-1 promotes allergic lung inflammation. Analysis of lung tissue isolated from control (Blimp-1f/f CD4Cre−) or Blimp-1CD4Cre (Blimp-1f/f CD4Cre+) animals i.n. immunized with HDM. (A) PAS staining (scale bar, 100 µm). (B) Percent and absolute number of infiltrating inflammatory cells in the BAL. (C–F) Flow analysis (C and E) and (D and F) percent of Th1 (IFN-γ+ T-bet+), Th2 (GATA3+ IL-13+ or GATA3+ IL-5+) Th17 (Rorγt+ IL-17+) cells and T regulatory cells (FoxP3+) isolated from lungs (gated on Live, CD4+ TCRβ+). (G) Assessment of AHR in unimmunized or HDM-immunized mice. Rn, central airway resistance (Newtonian resistance). Rrs, respiratory system resistance. Mch, methylcholine. Data in A–G are pooled from two or three experiments with 8–13 total mice per group, mean ± SD. Kruskal–Wallis one-way ANOVA (D), Mann–Whitney t test (B and F), or two-way ANOVA (G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
These results were surprising as prior examination of Blimp-1–deficient T cells had increased Th2 cells in a Th2 cell model using a worm antigen (Poholek et al., 2016), and in vitro, Blimp-1 is not required for Th2 cell differentiation (Fig. S1 C; Cimmino et al., 2008). To determine if the requirement for Blimp-1 to drive Th2 cells was specific to HDM or to a lung-specific pathway in response to inhaled allergens, we i.n. immunized control and Blimp-1CD4Cre mice with soluble egg antigen (SEA) derived from Schistosoma mansoni worms. In our previous study (Poholek et al., 2016), we found s.c. injection of SEA in Blimp-1CD4Cre mice resulted in a modest increase in Th2 cell responses. In contrast, i.n. immunization of SEA resulted in a Blimp-1–dependent requirement for Th2 cell generation in the lung, similar to our results with HDM (Fig. S1 D). We next immunized mice s.c. with HDM in the footpad and looked at Th2 cell responses in the popliteal LN (pLN). Similar to our previously published results with SEA in the footpad, loss of Blimp-1 led to an increase in Th2 cells after s.c. injection of HDM (Fig. S1 E). These data suggest that Blimp-1 plays a surprising and critical role in driving a lung-specific pathway driving the differentiation of Th2 cells in response to inhaled antigens but may constrain effector Th2 cell responses when antigens are primed s.c. We next asked if Th2 cells in the lung require Blimp-1 in a model of allergic lung inflammation that first primes T cells systemically followed by i.n. challenge. Control or Blimp-1CD4Cre mice were immunized i.p. with OVA in alum twice, followed by challenge via nebulizer treatment of 1% OVA for 3 d. In contrast to when antigen is delivered i.n., we found the Blimp-1 was dispensable for Th2 cells in the lung when antigen is introduced systemically (Fig. S1 F). These data suggest Blimp-1 can have contrasting functions in T cell differentiation and function depending on the route of immunization and location of T cell priming, which has not previously been appreciated.
IgE responses are Blimp-1 independent
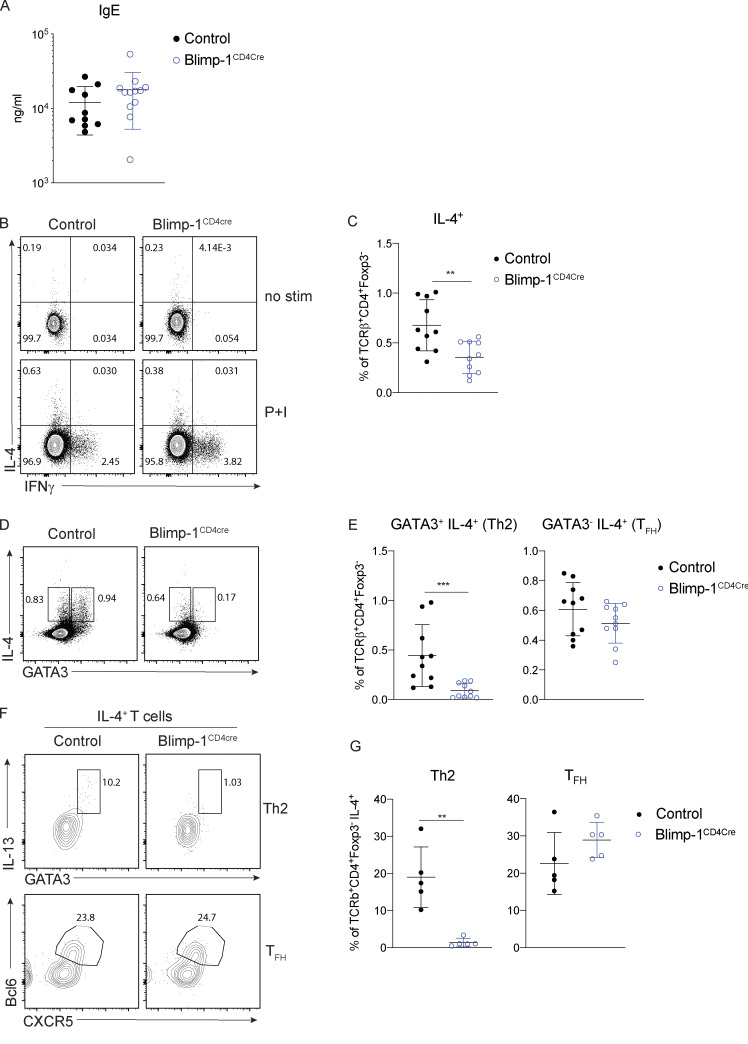
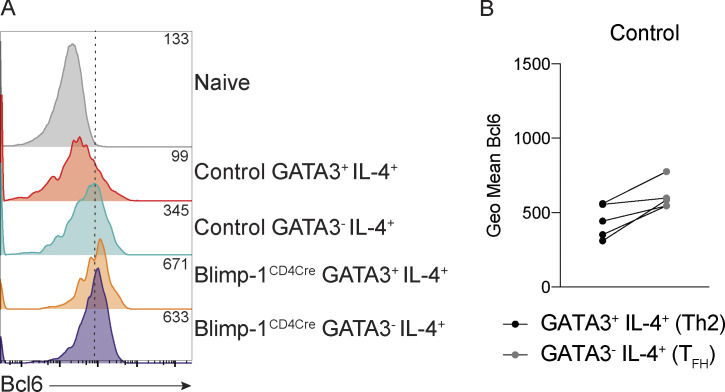
A hallmark of type 2–driven allergic responses is the presence of high levels of circulating IgE. In contrast to the decrease in type 2 immunity observed in the lungs of Blimp-1CD4Cre animals (reduced Th2 cells and eosinophils in BAL), IgE levels were unchanged (Fig. 2 A). Isotype switch to IgE is dependent on IL-4 (Kopf et al., 1993). However, similar to loss of Th2 cells in the lung, IL-4+ T cells in the mLN were significantly reduced in Blimp-1CD4Cre animals (Fig. 2, B and C). Antibody responses are largely driven by TFH cells, and Blimp-1 antagonizes TFH responses. Thus, we reasoned that TFH cell responses would be intact in Blimp-1CD4Cre and thus capable of driving robust IgE responses. IL-4 can be produced by both Th2 and TFH cells (Zhu, 2015); therefore, we determined the percentage IL-4+ cells in both subsets (Th2, GATA3+ and TFH, GATA3−) of cells in the mLNs of control and Blimp-1CD4Cre animals. Despite total IL-4+ cells being decreased in mLNs of Blimp-1CD4Cre animals, analysis of IL-4+ cells within the TFH cell population (GATA3−) was similar to controls (Fig. 2, D and E). In contrast, IL-4+ cells within the Th2 cell population (GATA3+) were significantly reduced (Fig. 2, D and E). As expected, expression of Bcl6 was higher in TFH cells (GATA3−) than Th2 cells (GATA3+) of control animals, but still reduced compared with Blimp-1CD4Cre animals (Fig. S2, A and B). Indeed, IL-4+ cells from control animals comprised both Th2 (GATA3+ IL-13+) and TFH (Bcl6+ CXCR5+) cells (Fig. 2 F). Blimp-1CD4Cre animals, however, had similar IL-4+ TFH cell percentages, but Th2 cells were absent (Fig. 2, F and G). These data suggest that although Blimp-1 is a master regulator of Th2 cell development in the lungs, it is not required for IL-4+ TFH cell development and subsequent type 2–mediated IgE responses. Thus, there are unique molecular pathways that can drive type 2 humoral immunity for IgE production (IL-4–producing TFH cells) distinct from those that drive eosinophil recruitment to the lung (IL-13 and IL-5–producing Th2 cells), and Blimp-1 is a major regulator that molecularly dissects these two independent pathways.
Figure 2.
IgE responses are Blimp-1 independent. (A) Total IgE in serum from animals in Fig. 1. (B–E) Flow analysis (B and D) and (C and E) percent of IL-4, IFN-γ, and GATA3 expression in mLN T cells (gated on live CD4+ TCRβ+ FoxP3−). P+I, PMA, ionomycin. (F and G) Flow analysis and percentage of IL-4+ cells shown in B. Gated on live CD4+ TCRβ+ FoxP3− IL-4+. Data in A–G are pooled from two or three experiments with 9–13 total mice per group, mean ± SD. Mann–Whitney t test (A, C, E, and G). **, P < 0.01; ***, P < 0.001.
Figure S2.
Bcl6 expression in TFH and Th2 cells in the mLN. (A) Expression of Bcl6 in indicated populations. Geometric mean shown in top right corner of each track. (B) Geometric (Geo) mean of Bcl6 in indicated samples. Wilcoxon matched-pairs signed rank test.
Blimp-1 is intrinsically required in T cells to promote Th2 cells
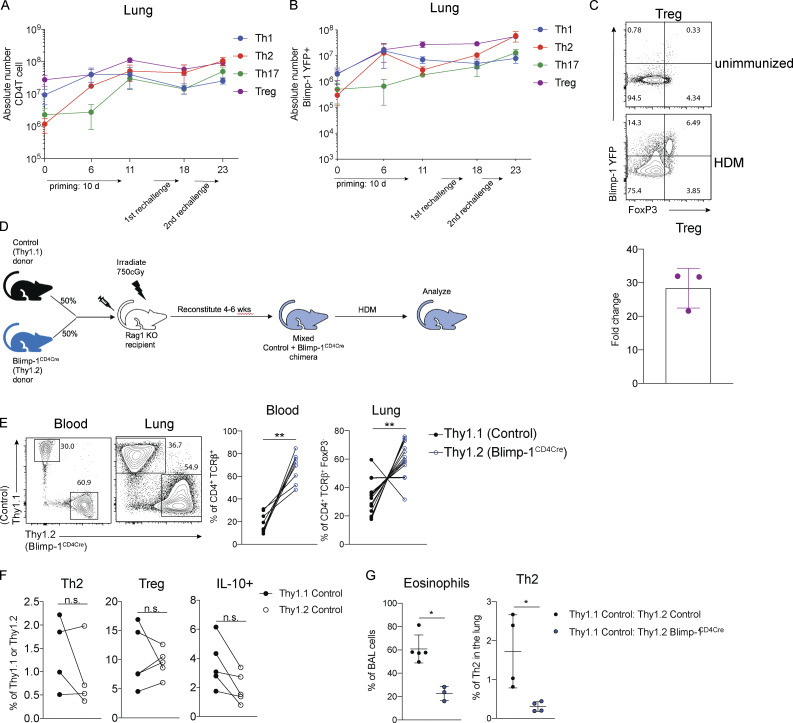
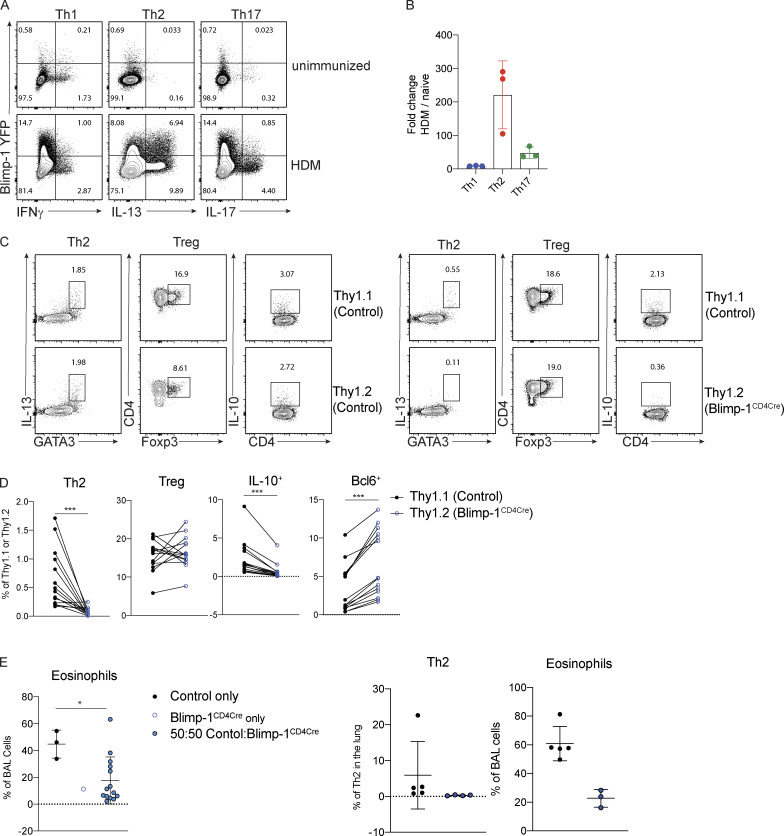
We next sought to determine if Blimp-1 was expressed by all CD4 T cell subsets in the lung during the course of allergic airway inflammation, or if expression was limited to Th2 cells. To assess Blimp-1 expression, we induced allergic inflammation in Blimp-1 YFP reporter mice, which reliably tracks Blimp-1 transcription by YFP (Rutishauser et al., 2009). We assessed each CD4 T cell subset and Blimp-1 expression within each subset at four time points: day 6 and day 10, which are during and after priming, day 18, after the first challenge, and day 23, after the second challenge. Th1, Th2, Th17, and T reg cells were present at each time point in the lung in naive unimmunized animals and after HDM challenges (Fig. S3 A). When looking at the percentage of Blimp-1 YFP+ cells among each subset, we found that all subsets initially up-regulated Blimp-1 but that Th2 cells did so to a greater degree compared with Th1 and Th17 cells (Fig. 3, A and B; and Fig. S3 B). T reg cells have been reported to express high levels of Blimp-1 in effector tissues; thus, as a positive control, we assessed Blimp-1 in T reg cells and found nearly all T reg cells expressed Blimp-1 after HDM (Fig. S3 C; Cretney et al., 2011). Blimp-1 continued to increase in Th2, T reg, and Th17 cell populations, while the percentage of Blimp-1+ cells in Th1 cells remained fairly constant over time after the initial increase from day 0 to day 5 (Fig. S3 B).
Figure S3.
Blimp-1 expression dynamics in HDM-induced allergic lung inflammation. (A) Absolute number of CD4 T cell subsets during the course of HDM-induced allergic lung inflammation model. (B) Absolute number of Blimp-1 YFP+ cells of each CD4 T cell subset shown in A. (C) Flow analysis and fold change of Blimp-1 YFP+ FoxP3+ T reg cells in the lung of naive unimmunized animals and after HDM. Fold change calculated as percent of Blimp-1 YFP+ T regs after HDM over average of Blimp-1 YFP+ T reg cells in unimmunized mice. Data in A–C are from two experiments with three mice per group. (D) Schematic of mixed BM chimera generation. (E) Reconstitution 4–6 wk after irradiation and injection of donor marrow isolated from blood (4 wk) or lungs after HDM-induced lung inflammation. Flow cytometry and percent of Thy1.1+ (control) and Thy1.2+ (Blimp-1CD4Cre) in both tissues shown. Data in E are pooled from two experiments with 14 mixed BM chimeras, mean ± SD. Wilcoxon matched-pairs signed rank test. (F) Percent of Th2, T reg, and IL-10+ cells in 50:50 mixed Thy1.1 control: Thy1.2 control BM chimeras in the lung after HDM immunization. Th2 and IL-10+ cells gated on CD4+ TCRb+ FoxP3- Thy1.1+ or Thy1.2+. T reg cells gated on CD4+ TCRb+ FoxP3+ Thy1.1+ or Thy1.2+. (G) Percent of eosinophils and total Th2 cells in the lungs of 50:50 mixed Thy1.1 control: Thy1.2 control or 50:50 mixed Thy1.1 control: Thy1.2 Blimp-1CD4Cre BM chimeras. Data in F and G are pooled from two experiments with five mixed BM chimeras, mean ± SD. Wilcoxon matched-pairs signed rank test. **, P < 0.01.
Figure 3.
Blimp-1 is intrinsically required in T cells to promote Th2 cells. (A and B) Expression (A) and (B) fold changes of Blimp-1 YFP+ cells in T cell subsets (gated on live CD4+ TCRβ+ FoxP3−) in lungs of naive unimmunized mice or after HDM. Fold change calculated as percent of Blimp-1 YFP+ in each subset after HDM over average of Blimp YFP+ in each subset in unimmunized naive animals. Data in A and B are from two experiments with three mice per group. (C) Flow analysis of lungs from 50:50 mixed Thy1.1 control: Thy1.2 control (left panels) or 50:50 mixed Thy1.1 control: Thy1.2 Blimp-1CD4Cre BM chimeras gated on live CD4+ TCRβ+ (T regs) and FoxP3− (Th2 and IL-10+) and Thy1.1 (control); or Thy1.2 (Blimp-1CD4Cre) after HDM. Plots are representative of two mice from each group (three to five per group). (D) Percentage of Th2 (Gata3+ IL-13+), T reg (FoxP3+), IL-10+, and Bcl6+ cells isolated from lungs BM chimeras in C. (E) Eosinophil percentages in BAL of control BM chimeras (control only or Blimp-1CD4Cre only) or mixed BM chimeras. Data in C–E are pooled from two experiments with 14 mixed BM chimeras and one to three control only BM chimera mice per group, mean ± SD. Wilcoxon matched-pairs signed rank test (D) or Mann–Whitney t test (E). *, P < 0.05; ***, P < 0.001.
It was possible that the increase in T reg cells (Fig. 1, C and D) might be responsible for suppressing Th2 cells. Therefore, we wondered if the expression of Blimp-1 was required intrinsically in T cells to drive Th2 cells or if an extrinsic mechanism such as T reg cell–mediated control may be at play. To test the intrinsic function of Blimp-1, we generated mixed bone marrow (BM) chimeras in Rag-deficient hosts where 50% of donor BM was from congenically marked B6 control mice (Thy1.1) and 50% from Blimp-1CD4Cre (Thy1.2). 6 wk after reconstitution, allergic lung inflammation was induced with HDM as previously described (Fig. S3 D). Although irradiated hosts received equal percentages of donor BM, Blimp-1CD4Cre BM reconstituted at greater than 50%, supporting its role in negatively regulating T cell survival (Poholek et al., 2016; Fig. S3 E). Nevertheless, Th2 cells were significantly reduced in Blimp-1CD4Cre T cells after HDM-induced allergic lung inflammation in contrast to 50:50 mixed control chimeras, where Th2 cells arose equally from both Thy1.1 control or Thy1.2 control cells (Fig. 3, C and D; and Fig. S3 F). T reg cells, however, were equivalently derived from control or Blimp-1CD4Cre BM, suggesting that Blimp-1 is intrinsically required in Th2 cells to promote Th2 cell differentiation, and that increases in T reg cells were not responsible for this effect. The reduction in Th2 cells was significant enough to result in a slight but significant decrease in eosinophils in the BAL compared with animals that only received control BM or a 50:50 mix of control Thy1.1:control Thy1.2 BM, in accordance with a reduction in the overall Th2 cells due to the lack of Blimp-1–deficient T cells forming Th2 cells (Fig. 3 E and Fig. S3 G). Blimp-1CD4Cre T cells (Thy1.2+) had significant decreases in IL-10 and increases in Bcl6 compared with control cells in the same animal, consistent with Blimp-1’s function in driving IL-10 and repressing Bcl6 (Fig. 3, C and D). Taken together, these data suggest that the mechanism of Blimp-1–mediated Th2 cell differentiation is cell intrinsic.
STAT3 is required for Blimp-1 expression in T cells
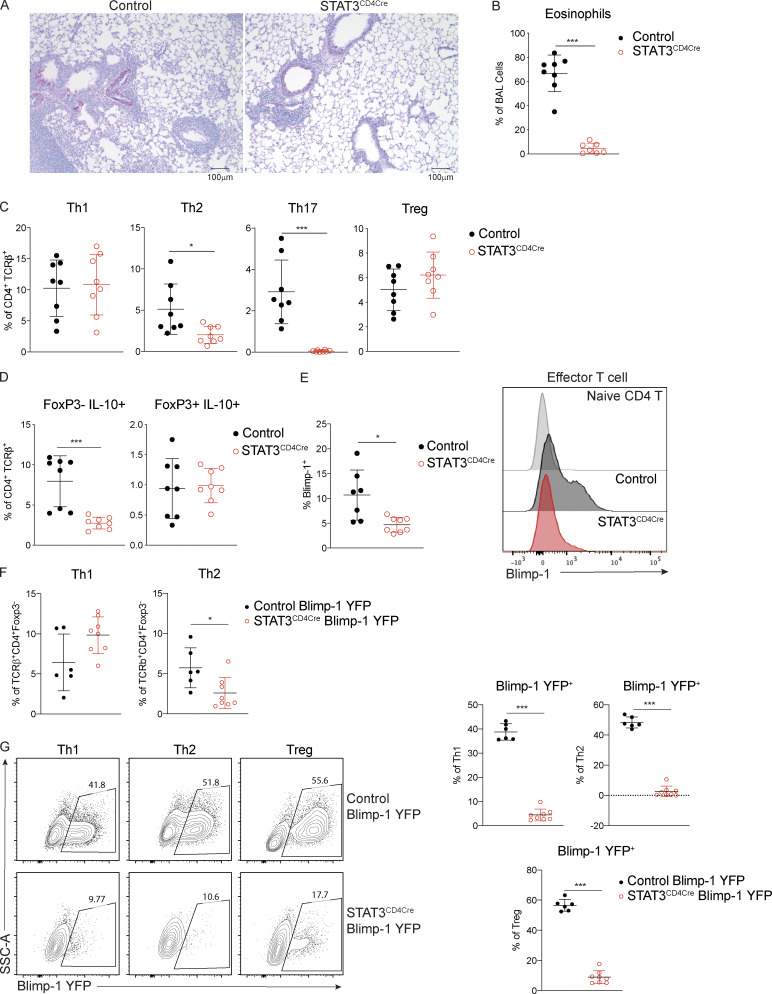
We next explored the upstream drivers of Blimp-1. Our previous examination of Th2 cells found that STAT3, but not STAT6, was a critical regulator of Blimp-1. In vitro, STAT3 can bind directly to the Prdm1 locus when stimulated with a strong STAT3-activating cytokine such as IL-10 (Poholek et al., 2016). Furthermore, STAT3 can support Th2 cell differentiation in murine allergic lung models (Stritesky et al., 2011). Therefore, we wondered whether STAT3 could be promoting Th2 cell development in our HDM-induced allergic model through up-regulation of Blimp-1. To explore the role of STAT3, we used T cell–specific STAT3–deficient animals (STAT3f/f CD4-Cre, referred to as STAT3CD4Cre) and assessed T cell subsets in the lung. After immunization with HDM, STAT3CD4Cre lung tissue had reduced lymphocytic infiltration as assessed by periodic acid-Schiff (PAS) staining, and there was a significant reduction in eosinophils found in the BAL (Fig. 4, A and B). Consistent with STAT3’s role in driving Th17 differentiation (Zhou et al., 2007), STAT3CD4Cre had a complete absence of Th17 cells in the lungs (Fig. 4 C). Intriguingly, we found a significant reduction in Th2 cells, similar to our results in Blimp-1CD4Cre, confirming previous studies that STAT3 could support Th2 cell development in the context of allergic lung inflammation (Stritesky et al., 2011; Fig. 4 C). In contrast, Th1 and T reg cell percentages were similar in control and STAT3CD4Cre mice. IL-10 was reduced in effector T cells (FoxP3−), consistent with a role for STAT3 in driving IL-10 expression. In contrast, IL-10 was unaffected in STAT3–deficient T reg cells (Fig. 4 D; Stumhofer et al., 2007). Thus, T reg cell percentages and effector function appeared intact in the absence of STAT3 expression in T cells. To determine if STAT3 was required for Blimp-1 expression, we assessed Blimp-1 protein expression in effector T cells. We first confirmed the specificity of our Blimp-1 intracellular staining with Blimp-1CD4Cre mice (Fig. S4 A). In agreement with our previous study, Blimp-1 protein was significantly reduced in the absence of STAT3, suggesting that STAT3 is required for Blimp-1 expression and subsequent Th2 cell differentiation in allergic lung inflammation (Fig. 4 E). To determine if all T cells required STAT3 for Blimp-1 expression or just Th2 cells, we crossed the STAT3CD4Cre to Blimp-1 YFP reporter mice. Consistent with STAT3CD4Cre animals, Th2 cells were specifically reduced in STAT3CD4Cre Blimp-1 YFP after HDM (Fig. 4 F). STAT3CD4Cre Blimp-1 YFP mice had significantly reduced Blimp-1 expression (assessed by YFP) in Th1, Th2, and T reg cells compared with control Blimp-1 YFP mice (Fig. 4 G). Although Th1 cells also lacked Blimp-1, it did not affect Th1 cell percentages in the lung, suggesting that Blimp-1 and STAT3 are not necessary for Th1 cell differentiation in response to allergens. Taken together, these data confirm the critical role of intrinsic STAT3 in driving Blimp-1 in Th2 cells in vivo in an allergic lung inflammation model.
Figure 4.
STAT3 is required for Blimp-1 expression in T cells. Analysis of lung tissue isolated from control (STAT3f/f CD4Cre−) or STAT3CD4Cre (STAT3f/f CD4Cre+) animals i.n. immunized with HDM. (A) PAS staining (scale bar, 100 µm). (B) Percent of eosinophils in the BAL. (C) Percent of T cell subsets in lung (gated on live CD4+TCRβ+). (D) Percent of IL-10 producing effector T cells (FoxP3−) and T reg cells (FoxP3+). (E) Percent of Blimp-1+ effector T cells (gated on CD4+TCRβ+FoxP3−CD44+) in mLN and representative histogram. Data from B–E are pooled from two experiments with eight mice total per group, mean ± SD. (F and G) Percent of Th1 (IFN-γ+ T-bet+) and Th2 (IL-13+ GATA3+) cells (gated on CD4+TCRβ+FoxP3−; F), and (G) representative flow plots and percent of Blimp-1 YFP+ cells within Th1, Th2, and T reg (FoxP3+) cells in the lung after HDM. Data from F and G are pooled from two experiments with six to eight total mice per group, mean ± SD. Mann–Whitney t test (B–G). *, P < 0.05; ***, P < 0.001.
Figure S4.
IL-21R signaling is required for T cell responses but not Blimp-1 in the mLN. (A) Representative flow plots of cells isolated from the mLN of control or Blimp-1CD4Cre after HDM immunization stimulated in vitro for 36 h with 50 μg of HDM or PBS alone, then stained for Blimp-1. (B) qPCR of indicated cytokines in lungs and mLN of unimmunized or HDM-immunized animals. Data in B are from two experiments with 10–12 mice per group. (C) Percent of Th1 (IFN-γ+ T-bet+) and Th2 (IL-13+ GATA3+) cells in mLN gated on live CD4+TCRβ+FoxP3−. (D) Expression of Blimp-1 and Bcl6 in mLN in T cells gated on live CD4+TCRβ+FoxP3−. Data in C and D are pooled from three experiments with 9 or 10 mice total per group, mean ± SD, Mann–Whitney t test. *, P < 0.05.
IL-6 and IL-21 are not required for expression of Blimp-1 in Th2 cells
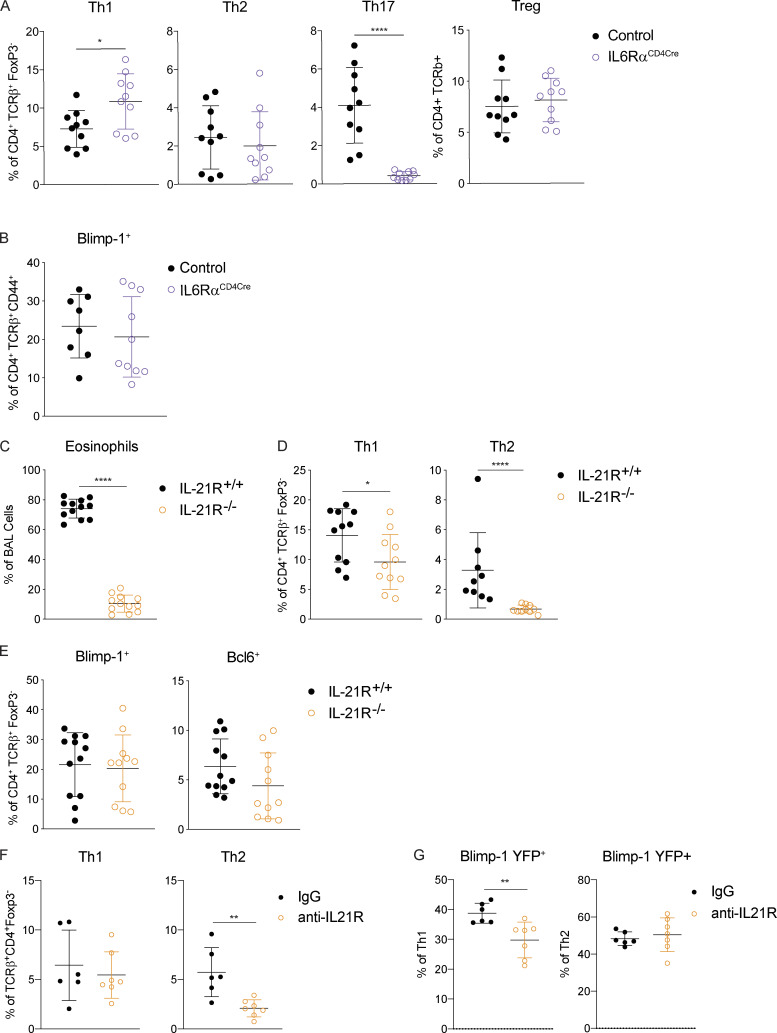
We next sought to determine what environmental factors were driving Blimp-1 expression in Th2 cells in HDM-induced lung inflammation. We focused our analysis on known STAT3-activating cytokines. As IL-6 has previously been shown to support Th2 cells in allergic lung disease (Doganci et al., 2005) and is a strong STAT3 activator, we first tested whether IL-6 signaling on T cells was required for Blimp-1 induction and Th2 cell formation in this model. Using IL-6Rα–deficient T cells (IL-6Rαf/f CD4-Cre, referred to as IL-6RαCD4Cre), we found no significant difference in Th2 cells in the lung after HDM; however, Th17 cells in the lung were absent, in line with the well-known role of IL-6 to drive Th17 cell development via STAT3 (Fig. 5 A). Importantly, Blimp-1 levels were unchanged in effector (CD44+) T cells (Fig. 5 B), suggesting IL-6 was not required to drive Blimp-1 or Th2 cells in response to HDM.
Figure 5.
IL-6 and IL-21 are not required for Blimp-1–mediated Th2 cell differentiation. (A and B) Percent of T cell subsets (A; gated on live CD4+ TCRβ+ FoxP3− [non-T reg] or FoxP3+ [T reg]) or (B) percent of Blimp-1+ cells (gated on Live, CD4+ TCRβ+ FoxP3− CD44+) isolated from lungs of control (IL-6Rαf/f CD4Cre−) or IL6RαCD4Cre (IL-6Rαf/f CD4Cre+) animals i.n. immunized with HDM. Data from A and B are pooled from two experiments with 10 mice per group, mean ± SD. (C–E) Percent of eosinophils in the BAL (C), (D) Th1 (IFN-γ+, T-bet+), and Th2 (IL-13+ GATA3+) cells (gated on live CD4+TCRβ+FoxP3−) and (E) percent of Blimp-1+ or Bcl6+ (gated on live CD4+TCRβ+FoxP3−) T cells in lung isolated from IL-21R–intact or IL-21R–deficient animals immunized with HDM. Data from C–E are pooled from two or three experiments with 9 or 10 total mice per group, mean ± SD. (F and G) Percent of Th1 (IFN-γ+ T-bet+) and Th2 (IL-13+ GATA3+) cells (gated on live CD4+TCRβ+FoxP3−; F) and (G) Blimp-1 YFP+ cells within Th1 and Th2 cells isolated from the lung from control (IgG) or anti–IL-21R–treated animals immunized with HDM. Data from F and G are pooled from two experiments with six or seven mice total per group, mean ± SD, Mann–Whitney t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
To identify other possible STAT3-driven cytokines, we performed whole-tissue quantitative real-time PCR (qPCR) of both lung and mLN and found IL-10, IL-21, and IL-27 were all up-regulated in response to HDM, particularly in the mLN (Fig. S4 B). IL-21 is known to play a critical role in Th2 cell generation in HDM-induced allergic lung inflammation; however, the mechanism is unclear (Coquet et al., 2015). Furthermore, IL-21 is known to drive Blimp-1 in B cells (Ozaki et al., 2004). Using IL-21R–deficient mice (IL-21R−/−), we found a significant reduction in eosinophils in the lung, consistent with previous reports (Coquet et al., 2015; Fig. 5 C). We also found a significant reduction in Th2 cells in the lung and mLN (Fig. 5 D and Fig. S4 C). Surprisingly, although IL-21R−/− displayed a loss of Th2 cells similar to Blimp-1CD4Cre and STAT3CD4Cre, Blimp-1 levels were similar in effector T cells in both control and IL-21R−/− animals (Fig. 5 E and Fig. S4 D). We wondered if IL-21 could play an important role in driving Blimp-1 only in Th2 cells, and the absence of Th2 cells limited our ability to detect this. To test this, we treated Blimp-1 YFP mice with anti–IL-21R antibody (or control IgG antibody) during the course of HDM-induced allergic lung inflammation and tracked Blimp-1 expression by YFP. Similar to IL-21R−/− mice, Th2 cells were significantly reduced in the lung of anti–IL-21R–treated mice (Fig. 5 F). However, Blimp-1 levels were intact in the few remaining Th2 cells found (Fig. 5 G). While Blimp-1 levels were slightly reduced in Th1 cells, this had no effect on Th1 cell development (Fig. 5, F and G). These data suggest that while IL-21 is critical for Th2 cell development and eosinophil recruitment during HDM-induced allergic lung inflammation as has previously been shown, the role of IL-21 in Th2 cell formation is independent of the function of Blimp-1 in driving Th2 cells (Coquet et al., 2015).
IL-10 is required for Blimp-1 expression in Th2 cells
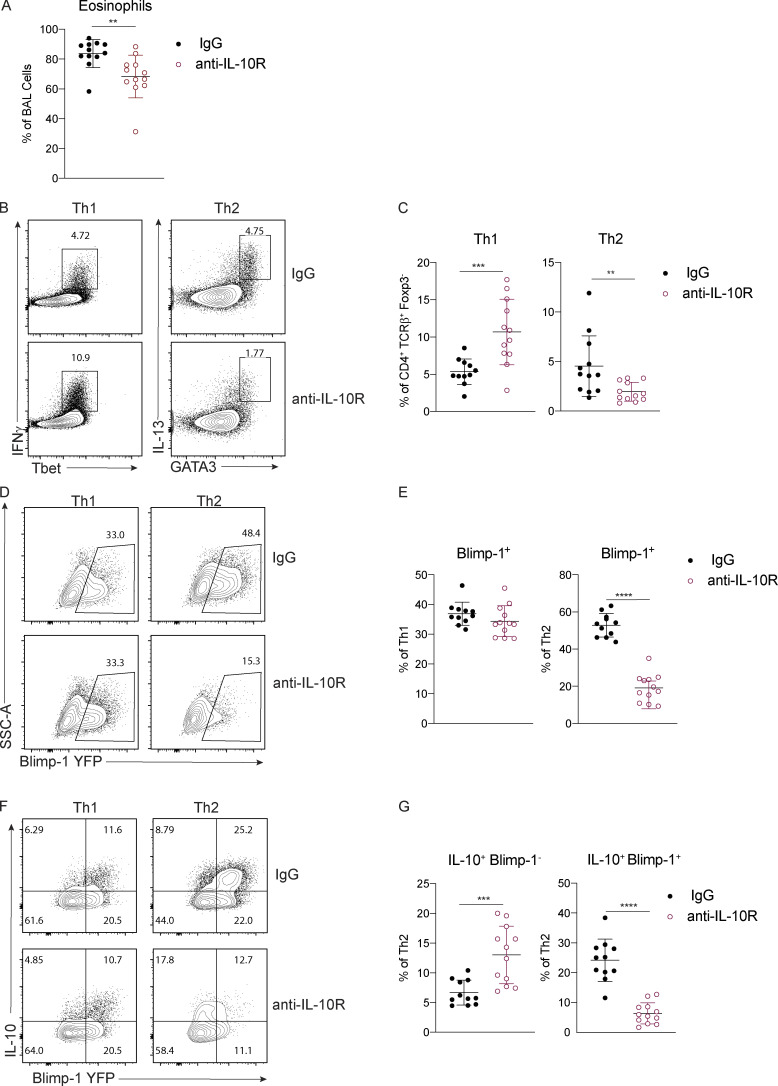
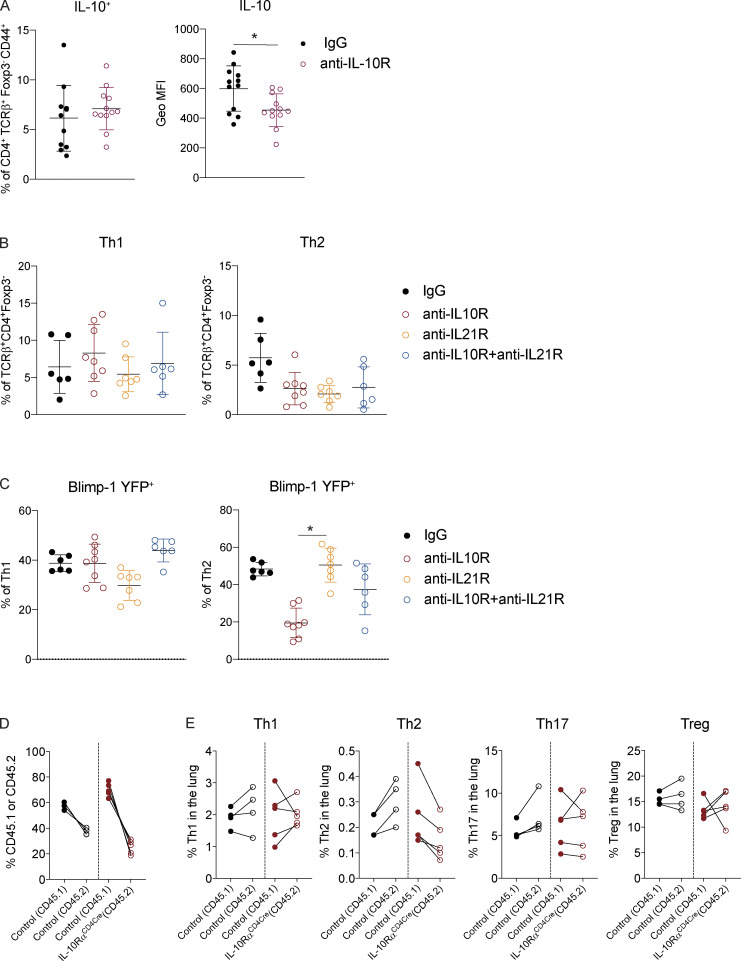
Our previous studies using worm-derived antigens demonstrated that IL-10 is a critical factor driving Blimp-1 expression in Th2 cells (Poholek et al., 2016). Although IL-10 is generally thought to be anti-inflammatory, there have been contradictory findings in allergic asthma, with both anti-inflammatory and pro-Th2 cell roles described (Bandukwala et al., 2007; Coomes et al., 2016; Hammad et al., 2001; Igietseme et al., 2000; Laouini et al., 2003; Pulendran et al., 2010; Wilson et al., 2007). To test if IL-10 might be a driver of Blimp-1 in HDM-induced allergic lung inflammation, we treated Blimp-1 YFP mice with anti–IL-10R antibody or IgG control during HDM immunization. Surprisingly, we found a consistent reduction in eosinophils recruited to the lungs (Fig. 6 A), and a concomitant decrease in Th2 cells in the lung (Fig. 6, B and C). Th1 cells were increased, while Th17 and T reg cells were unaffected (Fig. 6 C and data not shown). Intriguingly, Blimp-1 was specifically and significantly reduced in Th2 cells but unaffected in Th1 cells (Fig. 6, D and E), suggesting IL-10 was an important regulator of Blimp-1 in Th2 cells in the context of allergic antigen stimulation. As Blimp-1 is an intrinsic regulator of IL-10 expression (Fig. 3 D), we wondered if IL-10 was also affected by loss of Blimp-1. Within the total effector (CD44+) T cell population, the percentage of IL-10+ cells was unchanged; however, the total amount of IL-10 per cell was slightly reduced (mean fluorescence intensity [MFI]; Fig. S5 A). Specifically within Th2 cells, expression of IL-10 shifted from a Blimp-1+ population in IgG-treated control mice to a Blimp-1–negative population in anti–IL-10R–treated mice (Fig. 6, F and G). These data suggest that Blimp-1 expression may not be continually required for IL-10 expression in T cells. To address the possibility that IL-10 and IL-21 act in a redundant fashion to regulate Blimp-1 expression, we performed double antibody blockade of both IL-10R and IL-21R in Blimp-1 YFP animals. Blockade of IL-10R and IL-21R together did not have a synergistic effect on Blimp-1 expression, suggesting the effects of IL-10 and IL-21 are independent of one another (Fig. S5, B and C).
Figure 6.
IL-10 is required for Blimp-1 expression in Th2 cells. Analysis of lungs isolated control (IgG) or anti–IL-10R treated animals i.n. immunized with HDM. (A) Percent of eosinophils in the BAL. (B and C) Flow analysis (B) and (C) percent of Th1 and Th2 cells in lungs (gated on live CD4+TCRβ+FoxP3−). (D and E) Flow analysis (D) and (E) percent of Blimp-1 YFP+ cells in Th1 and Th2 cells in the lungs. (F and G) Flow analysis (F) and (G) percent of IL-10+ and Blimp-1 YFP+ expression in Th1 and Th2 cells in lungs. Data from A–G are pooled from three experiments with 11 to 12 mice total per group, mean ± SD. Mann–Whitney t test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S5.
IL-10 and IL-21 do not cooperatively regulate Blimp-1 expression. (A) Percent and geometric (Geo) MFI of IL-10–expressing cells isolated from the lung gated on live CD4+ TCRβ+ FoxP3− CD44+. Data in A are pooled from three experiments with 11 or 12 mice total per group. Mann–Whitney t test. (B) Percent of Th1 (IFN-γ+ T-bet+) and Th2 (IL-13+ GATA3+) cells in the lung isolated from Blimp-1 YFP animals treated as control (IgG) or treated with anti–IL-10R, anti–IL-21R, or both anti–IL-10R and anti–IL-21R for the duration of the HDM-induced allergic lung inflammation model. (C) Percent of Blimp-1 YFP+ Th1 and Th2 cells in B. Data in B and C are pooled from two or three experiments with 6–10 total mice per group. Kruskal–Wallis one-way ANOVA. (D) Reconstitution 4–6 wk after irradiation and injection of donor marrow isolated from the lungs post HDM-induced lung inflammation. Percent of CD45.1+ (control) and CD45.2+ (IL-10RαCD4Cre) are shown in both 50:50 control:control and 50:50 control:IL-10RαCD4Cre. (E) Percentages of T cell subsets isolated from lungs of mixed BM chimeras gated on live CD4+ TCRβ+ FoxP3− (Th1, Th2, Th17) or FoxP3+ (T reg) and CD45.1 (control); or CD45.2 (control or IL-10RαCD4Cre) after HDM. Data in D and E are pooled from two experiments with five total mice per group. Wilcoxon matched-pairs signed rank test. *, P < 0.05.
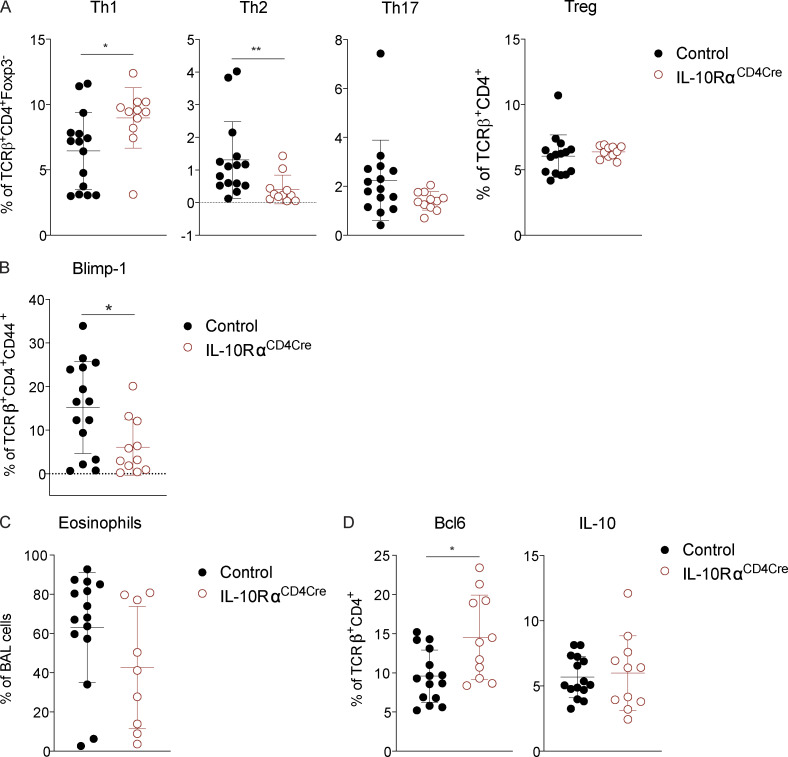
IL-10 is a pleotropic cytokine that acts on many cell types; thus, it was unclear if blockade of IL-10 signaling in vivo acted by directly inhibiting signaling to T cells, or if it was through an accessory cell such as dendritic cells. To assess the role of IL-10R on T cells directly, we generated mice lacking IL-10Rα on T cells (IL-10Rαf/f crossed to CD4-Cre, herein referred to as IL-10RαCD4Cre). In contrast to published studies showing IL-10 signaling on T cells was required to suppress Th2 responses (Coomes et al., 2016), we found that loss of IL-10Rα on T cells led to a reduction in Th2 cells in the lung, while Th1 cells were elevated (Fig. 7 A). Th17 and T reg cells were unchanged compared with controls. In addition, Blimp-1 was significantly reduced in effector T cells lacking IL-10Rα (Fig. 7 B). The reduction in Th2 cells led to a slight decrease in eosinophils in the lung (Fig. 7 C). Similar to our results in Blimp-1CD4Cre mice, Bcl6 was elevated (Fig. 7 D). In contrast, IL-10 levels were subtly increased in effector T cells, despite decreases in Blimp-1 (Fig. 7 D). These data were highly similar to our results with IL-10R blocking antibody, suggesting IL-10 signaling during responses to HDM directly to T cells leads to Blimp-1 up-regulation and Th2 cell differentiation, but is not sufficient to alter overall IL-10 production from T cells (Fig. S5 A). Finally, to determine if IL-10 signaling was required intrinsically on T cells, we generated 50:50 mixed BM chimeras of IL-10Rα–intact and –deficient BM (control [CD45.1]:control [CD45.2] or control [CD45.1]:IL-10RaCD4Cre[CD45.2]) and assessed T cell responses in the lung after HDM inhalation. Th2 cells were reproducibly lower when derived from IL-10RαCD4Cre BM than from control BM, whereas Th1, Th17, and T reg cells were similar or increased (Fig. S5, D and E). Taken together, these data suggest that IL-10 is an important driver of Blimp-1 in T cells, supporting Th2 cells in the lung during allergic inflammation in response to inhaled allergens. These data are consistent with a pro-inflammatory role for IL-10 that promotes Th2 cells, similar to prior studies showing a pro–Th2 cell role for IL-10 in allergic asthma (Bandukwala et al., 2007; Hammad et al., 2001; Igietseme et al., 2000; Laouini et al., 2003; Pulendran et al., 2010; Williams et al., 2013).
Figure 7.
IL-10R is required on T cells for Blimp-1 expression and Th2 cell development. Analysis of lung tissue isolated from control (IL-10Rαf/f CD4Cre−) or IL10RαCD4Cre (IL-10Rαf/f CD4Cre+) animals i.n. immunized with HDM. (A) Percent of T cell subsets isolated from lungs (gated on live CD4+ TCRβ+ FoxP3− [non-T reg] or FoxP3+ [T reg]). (B) Percent of Blimp-1+ cells (gated on live CD4+ TCRβ+ FoxP3− CD44+). (C) Percent of eosinophils in the BAL. (D) Percent of Bcl6+ and IL-10+ T cells isolated from lungs (gated on live CD4+ TCRβ+). Data in A–D are from two experiments with 11 or 12 mice per group, mean ± SD. Mann–Whitney t test. *, P < 0.05; **, P < 0.01.
Blimp-1 supports GATA3 expression and Th2 cell development in the lung through suppression of Bcl6
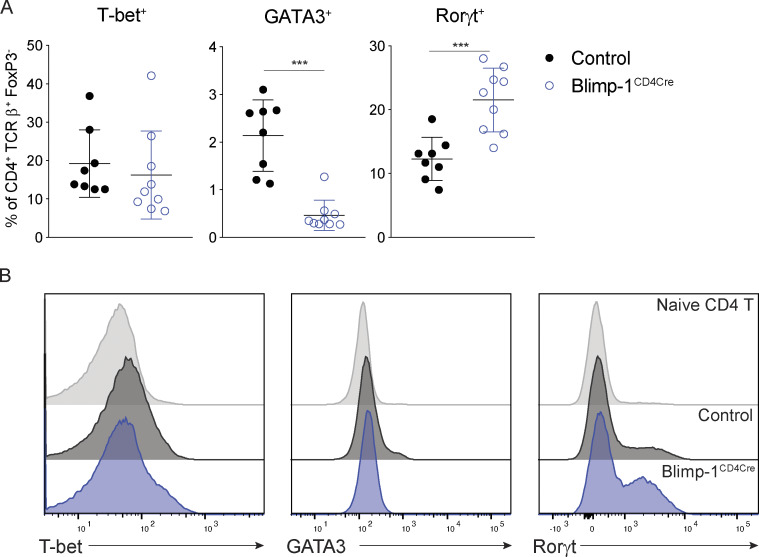
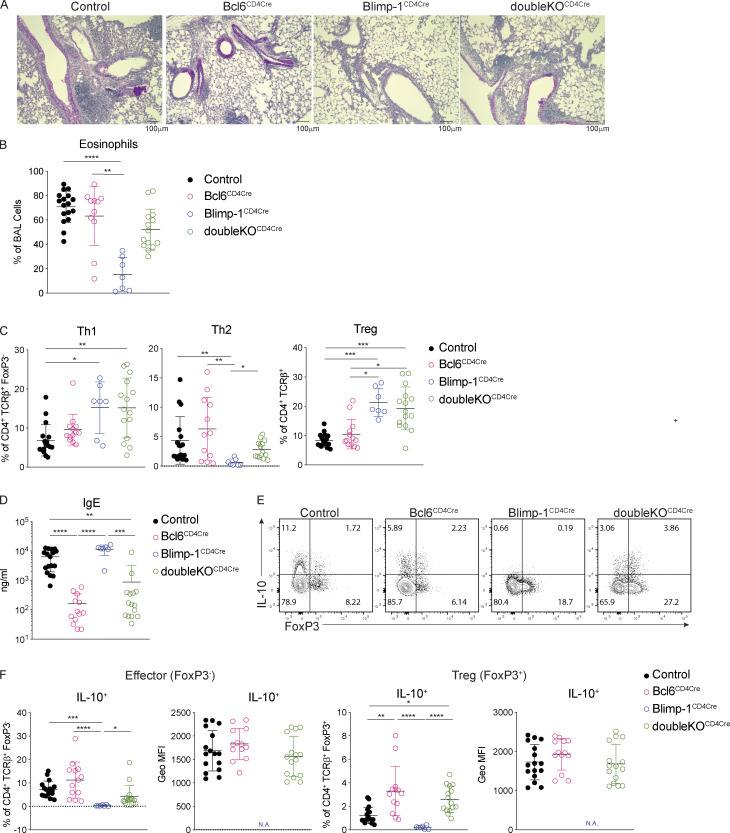
We next sought to understand how Blimp-1 was functioning to control Th2 cell differentiation. Since GATA3 is the master regulator of Th2 cell development (Zhang et al., 1997; Zheng and Flavell, 1997), we determined more specifically the expression of master regulators in T cell subsets in the absence of Blimp-1. The loss of Blimp-1 led to a specific and robust decrease in GATA3 in lung Th2 cells in response to HDM, while T-bet was unaffected and Rorγt was increased (Fig. 8, A and B). Bcl6 is known to be a repressor of GATA3 under some circumstances (Kusam et al., 2003; Sawant et al., 2012), and Blimp-1–deficient T cells had a robust increase in Bcl6 expression (Fig. 1 F). We hypothesized that in wild-type cells, Blimp-1 may be suppressing Bcl6, which in turn can promote GATA3 expression, but that in Blimp-1CD4Cre animals, increased expression of Bcl6 might suppress GATA3 and Th2 cell development. To test this, we induced allergic lung inflammation in mice that had a T cell–specific deficiency in both Blimp-1 and Bcl6 (Blimp-1f/f Bcl6f/f CD4Cre+, referred to as doubleKOCD4Cre; Xie et al., 2017). Control (Bcl6f/f or Blimp-1f/f, CD4Cre−), Bcl6-deficient (Bcl6f/f CD4Cre+, Bcl6CD4Cre), Blimp-1CD4Cre, or doubleKOCD4Cre mice were immunized with HDM and assessed for lung inflammation as well as development of T cell subsets in the lung and mLN. As before, examination of lung tissue found a consistent reduction in inflammation in Blimp-1CD4Cre lungs compared with control or Bcl6CD4Cre (Fig. 9 A). DoubleKOCD4Cre mice had lymphocyte infiltration and mucus that was similar to controls, suggesting inflammation was present when Bcl6 and Blimp-1 were both absent in T cells (Fig. 9 A). Consistent with this finding, we saw no difference in the recruitment of eosinophils to the BAL in the doubleKOCD4Cre lungs compared with control or Bcl6CD4Cre; however, Blimp-1CD4Cre mice had a consistent reduction in eosinophils as seen previously (Fig. 9 B). In the lung, Th1 cells were increased in both Blimp-1CD4Cre and doubleKOCD4Cre mice compared with control and Bcl6CD4Cre animals (Fig. 9 C). Interestingly, Th2 cells were increased in doubleKOCD4Cre compared with Blimp-1CD4Cre mice, suggesting the loss of Bcl6 in Blimp-1CD4Cre was sufficient to restore Th2 cell differentiation (Fig. 9 C). T reg cells in doubleKOCD4Cre were increased compared with controls, similar to Blimp-1CD4Cre, suggesting that Blimp-1’s repression of IL-2 is Bcl6 independent (Fig. 9 C). As expected, IgE levels were dramatically reduced in Bcl6CD4Cre and doubleKOCD4Cre mice, likely due to the loss of TFH cells in the absence of Bcl6 expression (Fig. 9 D). Finally, we explored the expression of IL-10 in both effector and T reg cells. Intriguingly, the absence of Bcl6 restored IL-10 expression in doubleKOCD4Cre, suggesting the function of Blimp-1 in driving IL-10 is via suppression of Bcl6 expression, and thus may be indirect rather than direct, as previously reported (Fig. 9, E and F; Minnich et al., 2016; Xie et al., 2017). This effect was greater in T reg cells than in effector T cells, although the amount of IL-10 per cell was similar (Fig. 9 F). Taken together, these data indicate that the mechanism that drives a loss of Th2 cells in the absence of Blimp-1 in response to inhaled allergens is mediated by increased Bcl6 expression, which can repress GATA3. Thus, in this context, Blimp-1 functions intrinsically in T cells to suppress Bcl6 expression, which is permissive for GATA3 expression, subsequent development of Th2 cells, and expression of IL-10 in T cells in the context of an HDM-induced allergic lung inflammation model.
Figure 8.
Transcription factor expression in effector T cells in the absence of Blimp-1. (A) Percent of effector cells expressing the transcription factors shown isolated from lung after HDM. Gated on live CD4+ TCRβ+ FoxP3−. (B) Histograms of data from A comparing naive, control effector (CD44+) or Blimp-1CD4Cre effector (CD44+) T cells. Data are pooled from two or three experiments with 8–10 total mice per group. Mann–Whitney t test. ***, P < 0.001.
Figure 9.
Blimp-1 supports Th2 development through suppression of Bcl6. Analysis of lungs isolated from control, Bcl6CD4Cre (Bcl6f/f CD4Cre+), Blimp-1CD4Cre (Blimp-1f/f CD4Cre+), or doubleKOCD4Cre (Bcl6f/f Blimp-1f/f CD4Cre+) animals i.n. immunized with HDM. (A) PAS staining (scale bar, 100 µm). (B) Percent of eosinophils in the BAL. (C) Percent of Th1 (IFN-γ+ T-bet+), Th2 (IL-13+ GATA3+), and T reg (FoxP3+) cells isolated from lungs (gated on live CD4+TCRβ+ [T reg], and FoxP3− [non-T reg]). (D) Total IgE in serum. (E) Flow analysis of IL-10 and FoxP3 T cells isolated from lungs of indicated mice after HDM-induced allergic asthma. Gated on live TCRβ+ CD4+. (F) Percent and geometric (Geo) MFI of IL-10+ cells in effector (CD4+TCRβ+FoxP3−) or T reg (CD4+TCRβ+FoxP3+) cells in lung. Data from B–F are pooled from three experiments with 7–17 total mice per group, mean ± SD. Kruskal–Wallis one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Discussion
In this study, we have uncovered a previously unappreciated role for Blimp-1 in driving lung-specific Th2 cell development in the context of an allergic asthma model. HDM antigen is a widely used model of allergic lung inflammation owing to its physiological relevance to human health as dust mites are a well-known allergic trigger in many individuals with asthma disease (Thomas, 2016). Based on our previous study, we expected Blimp-1 to suppress inflammatory effector responses to HDM. On the contrary, we uncovered an unexpected role for Blimp-1 in the specific control of Th2 cell differentiation, which has not been previously demonstrated in in vitro assays or our prior in vivo Th2 cell model (Cimmino et al., 2008; Poholek et al., 2016). Previous studies exploring the role of Blimp-1 in T cells have elucidated many functions for Blimp-1 that collectively could be described as controlling effector functions of T cells that mediate disease states. In CD8 T cell responses to viral infection, Blimp-1 supports effector cell differentiation while repressing memory cells (Kallies et al., 2009; Rutishauser et al., 2009; Shin et al., 2009). Blimp-1 also plays a critical role in supporting IL-10 expression in T reg cells and effector CD4 T cells, the absence of which can have important consequences for controlling infections by pathogens such as toxoplasma or chronic viral infection (Cretney et al., 2011; Neumann et al., 2014; Parish et al., 2014). Blimp-1 also controls differentiation of an inflammatory Th17 cell that produces IFN-γ and GM-CSF and subsequent autoimmune disease in an experimental autoimmune encephalomyelitis model (Jain et al., 2016). In each of these systems, Blimp-1 primarily controls effector properties of the T cells, supporting cytokines such as IL-10, IFN-γ, and GM-CSF, suggesting that Blimp-1 is part of a cell program to further drive a terminal effector differentiation state. Finally, Blimp-1 is known to repress the differentiation of TFH cells (Johnston et al., 2009; Nurieva et al., 2009). Here, for the first time, we have unexpectedly shown that Blimp-1 is essential for supporting the differentiation of a lineage of helper cells, Th2 cells, suggesting Blimp-1 has a more complex role than previously appreciated.
To better understand the difference for Blimp-1 function in Th2 cell differentiation between responses to inhaled (i.n.) HDM and s.c. SEA explored in our prior publication, we immunized mice with SEA i.n. and HDM s.c. and tracked Th2 cell responses in the lung. We found that Blimp-1 was required for Th2 cells in the lung in response to SEA (Fig. S1 D), while there was an increase in Th2 cells to HDM when delivered s.c. (Fig. S1 E). Furthermore, a model of allergic asthma that uses OVA + alum priming via i.p. followed by rechallenge via nebulizer treatment with 1% OVA did not require Blimp-1 for Th2 cells in the lung. This points to a lung-specific role for Blimp-1 in driving Th2 cells in response to inhaled but not systemically delivered antigens, suggesting that lung-specific factors or pathways control type 2 immunity in response to inhaled allergens, which may have important therapeutic implications as allergens are introduced via inhalation.
Th2 cells are primary producers of several important type 2 cytokines that drive allergic lung disease, including IL-4, IL-5, and IL-13 (Licona-Limón et al., 2013; Ray and Cohn, 1999). Each of these cytokines has a unique function in asthma disease, but collectively they enforce several hallmarks of asthma, including IgE production, recruitment of eosinophils, and mucus production in the airways. Intriguingly, we found that Blimp-1 controlled only GATA3+ Th2 cells but was not required for all IL-4+–producing cells. TFH cells are a critical population of effector T cells that are superior at initiating and supporting B cell responses such as IgE (Crotty, 2011). Furthermore, they can express IL-4, which is required for isotype switch to IgE (Zhu, 2015). Although the loss of Blimp-1 did not result in an increase in TFH cells after HDM, as it has in other systems, IL-4+ TFH cells were intact, resulting in normal IgE responses to HDM despite the absence of Th2 cells. These results suggest that the pathways that drive TFH and Th2 cells in response to allergens can be molecularly dissected, and that Blimp-1 is a major regulator distinguishing these two inflammatory cell types. This may enable therapeutic targeting of these populations distinctly and supports further exploring the upstream pathways of each cell population.
Multiple STATs including STAT3, STAT4, and STAT5 have been implicated in driving Blimp-1 in effector T cells (Jain et al., 2016; Neumann et al., 2014; Xin et al., 2016). In line with previous reports, we have found a central role for STAT3 in up-regulating expression of Blimp-1 (Jain et al., 2016; Poholek et al., 2016). Although we had previously found that IL-10 was required for Blimp-1 expression in Th2 cells, we did not initially expect IL-10 to promote allergic disease based on its known anti-inflammatory roles. There are several well-known STAT3-activating cytokines such as IL-6 and IL-21 that had already been implicated in supporting Th2 cells in allergic lung disease models (Rincón et al., 1997; Leonard and O’Shea, 1998; Doganci et al., 2005; Licona-Limón et al., 2013; Coquet et al., 2015). In addition, some studies had shown IL-10 suppresses Th2 cell responses in an HDM-driven model (Coomes et al., 2016; Tournoy et al., 2000). However, upon finding that neither IL-6 nor IL-21 signaling to T cells had any significant function on Blimp-1 expression in T cells, we pursued IL-10 as a potential regulator of Blimp-1 in this context. Our studies using both IL-10R antibody blockade and genetic deletion of IL-10R from T cells demonstrated a clear role for IL-10 acting directly on T cells to drive Blimp-1 and subsequent Th2 cells in response to inhaled allergens. A previous study analyzed the responses of IL-10Rα−/− CD4+ T cells during HDM-induced allergic lung inflammation and found that IL-10 acts directly on T cells to suppress Th2 cells, in contrast to our findings here (Coomes et al., 2016). We believe a difference in location of antigen priming may explain this discrepancy, as our immunizations were all i.n.; however, in Coomes et al., (2016), mice were first primed i.p. with HDM in alum followed by i.n. challenge. Together with our data using SEA i.n., this suggests that the route of immunization plays a major role in driving unique Th2 cell differentiation pathways. Several reports have suggested a STAT6- and IL-4–independent mechanism exists to drive Th2 cells (Dent et al., 1998; Ouyang et al., 2000). Furthermore, IL-10 from IRF4+ dendritic cells, the main Th2 cell–driving dendritic cell population, has been implicated in supporting Th2 cells during allergic responses (Cook and MacDonald, 2016; Gao et al., 2013; Krishnaswamy et al., 2017; Kumamoto et al., 2013; Williams et al., 2013). Given that many cells can secrete IL-10, it will be interesting to determine the pertinent cell types involved in this process (Gabryšová et al., 2014). We postulate that dendritic cells can secrete IL-10 after sensing allergens such as HDM via the lung, and then promote Blimp-1 expression and subsequent Th2 cell development through STAT3 activation. IL-10 can also act in a paracrine or autocrine manner from T cells to further drive Blimp-1 expression and IL-10 expression, which may further support Th2 cells. However, it is likely additional factors may act in concert with IL-10 to support Blimp-1, such as thymic stromal lymphopoietin, which can directly limit Bcl6 via STAT5 (Rochman et al., 2018). Full understanding of how a lung-specific pathway via IL-10 and Blimp-1 can promote Th2 cells will be of interest for future studies.
Blimp-1 mainly acts as a transcriptional repressor by recruiting histone methyltransferases such as G9a, which results in down-regulation of gene expression (Gyory et al., 2004). Recent studies have identified Blimp-1 binding near the IL-10 gene, suggesting Blimp-1 may also have a role as an activator of gene expression (Minnich et al., 2016). However, our study is more in line with an indirect mechanism of activation whereby Blimp-1 represses Bcl6, which represses IL-10 (Hollister et al., 2013; Xie et al., 2017). Loss of Bcl6 and Blimp-1 together restored IL-10 expression in T cells, and blockade of IL-10R signaling, resulted in robust IL-10+ Blimp-1− populations, suggesting T cells can make IL-10 in the absence of Blimp-1. We postulate that there may be more than one way for Blimp-1 to control gene expression of IL-10; however, our studies clearly point to a more complex mechanism beyond Blimp-1 directly activating the IL-10 gene, at least in CD4 effector cells responding to allergens in the lung. More studies carefully exploring Blimp-1 and IL-10 expression in T cells are required to fully understand this complex interaction.
As Blimp-1 has effects in almost every T cell subset, it was important to determine the intrinsic function of Blimp-1 in Th2 cells in this system. As Th2-specific deletion of Blimp-1 is challenging, we opted to use mixed BM chimeras, which supported an intrinsic role for Blimp-1. We found that intrinsic loss of Blimp-1 led to an increase in Bcl6, which could suppress GATA3, thus limiting Th2 cells. Although it is known that Bcl6 can repress GATA3 (Kusam et al., 2003; Sawant et al., 2012), it has not been appreciated that Blimp-1 can play an important role in suppressing Bcl6 in Th2 cells. Although STAT5 can also directly suppress Bcl6, our data suggest that this may be insufficient in some contexts, and that STAT3-driven Blimp-1 is also required to limit Bcl6 and indirectly support GATA3 and subsequent Th2 cell differentiation in the context of inhaled allergens (Johnston et al., 2012; Oestreich et al., 2012). However, as Blimp-1 is not required for in vitro differentiation of Th2 cells or Th2 cell formation after s.c. antigen administration, we suspect that down-regulation of Bcl6 is not required for GATA3 expression in all contexts. Further studies that explore the complex interplay of STAT3- and STAT5-activating cytokines in the control of Blimp-1, Bcl6, and GATA3 may reveal exactly how Th2 cells differentiate in response to allergens, which has long been not fully understood.
In summary, our study exploring the T cell–intrinsic role of Blimp-1 in HDM-induced allergic airway inflammation has uncovered several findings relevant to the differentiation of Th2 cells in allergen-induced lung inflammation. We found an important role of STAT3 in promoting Blimp-1 expression and subsequent Th2 cell differentiation. We additionally determined a potential pro-inflammatory role of IL-10 in driving Blimp-1 expression and promoting inflammation of lung via Th2 cells. Finally, we have described a new role for Blimp-1 in Th2 cell differentiation by repressing Bcl6 and indirectly supporting GATA3 expression. Taken together, our study elucidates a new context-dependent role for Blimp-1 in T cells that promotes, rather than constrains, allergen-induced airway disease.
Materials and methods
Mice
C57BL/6NJ (005304), B6.Cg-Tg(Prdm1-EYFP)1Mnz/J (008828), Tg(Cd4-cre)1Cwi/BfluJ (017336), B6.129-Prdm1tm1Clme/J (008100), B6.129S(FVB)-Bcl6tm1.1Dent/J (023727), B6.129S7-Rag1tm1Mom/J (002216), B6N.129-Il21rtm1Kopf/J (019115), B6;SJL-Il6ratm1.Drew/J (012944), and B6(SJL)-Il10ra tm1.1Tlg/J (028146) were purchased from Jackson Laboratories. STAT3flox were from D. Levy (New York University, New York, NY; Mouse Genome Informatics: 2384272; Lee et al., 2002). Animals were housed in specific pathogen–free enclosures at the John G. Rangos Sr. Research Center of University of Pittsburgh Medical Center Children’s Hospital of Pittsburgh. All experiments were approved by the University of Pittsburgh Animal Care Committee and the American Veterinary Medical Association. All mice were over 6 wk of age, and a mixture of male and female mice was used. All in vivo experiments were performed independently two to three times, with at least four mice per group.
Chemicals and reagents
Flow cytometry antibodies were purchased from BD Biosciences or Thermo Fisher Scientific (eBioscence). Blocking antibodies (IL-21R, IL-10R, and IgG control) were purchased from BioXCell. CD90 Microbeads were from Miltenyi Biotec. DNase I and collagenase A were purchased from Sigma-Aldrich. qPCR reagents (Trizol, qScript, and qPCR FastMix II) were purchased from Thermo Fisher Scientific or Quanta Biosciences. IgE ELISA Kit was purchased from Thermo Fisher Scientific.
In vivo immunizations
25 µg of LPS-low HDM (Stallergenes-Greer Pharmaceuticals) or SEA (provided by Dr. Dragana Jankovic, National Institutes of Health/National Institute of Allergy and Infectious Diseases, Bethesda, MD) was resuspended in 25 µl of sterile PBS and given i.n. to mice anesthetized with isoflurane daily for 10 d (priming) and rechallenged two times for 2–3 d per rechallenge with periods of 3–4 d rest between challenges (Fig. S1 A). For an OVA + alum model of driving allergy, mice were sensitized with i.p. injections of OVA (Invivogen, 10 µg) emulsified in alum (2 mg) in 200 µl PBS on day 1 and day 6. Mice were challenged by exposure to aerosolized 1% OVA twice a day for 20 min on days 12, 13, and 14. For s.c. immunizations, mice were injected in the footpad with 100 µg of HDM. 9 d after immunization, draining and nondraining pLNs were isolated for detection.
BAL
After the final rechallenge, BAL was isolated from anesthetized mice by infusion and recovery of ∼0.9 ml of sterile PBS into the lungs. Total BAL cell numbers were determined using automated cell counters (Nexcelom Bioscience LLC), and 4 × 105 BAL fluid cells were cytospun onto glass slides for staining using Richard-Allan Scientific Three-Step Stain (Thermo Fisher Scientific) following the manufacturer’s protocol. Neutrophils, eosinophils, monocytes, and lymphocytes were determined out of a total of 300 cells.
Histology
Lungs fixed in Safefix II (Thermo Fisher Scientific) were paraffin-embedded, sectioned, and stained with H&E and PAS stains. The pathological changes were visualized under a microscope.
Flow cytometry
Lungs of anesthetized mice were perfused with sterile PBS, removed, and digested. Briefly, the lung tissues were digested in a collagenase-DNase suspension for 45 min at 37°C, then processed on a gentleMACS dissociator (Miltenyi Biotec) according to the manufacturer’s protocol. Single cell suspensions were obtained by passing the dissociated tissue through a 70-µm cell strainer (Thermo Fisher Scientific) and washed with PBS containing 2% FBS and 0.5M EDTA. Red cell lysis was performed (ACT lysing buffer, Thermo Fisher Scientific) and washed with PBS containing 2% FBS. The mediastinal draining LN and pLN was dissociated in 10 ml of complete RPMI 1640 medium and passed through 40-µm filters. Cells were stimulated for 2.5 h with PMA (50 ng/ml) and ionomycin (1 µg/ml), with the addition of brefeldin A (GolgiPlug, BD) and monensin. Cells were processed for staining with fluorochrome-labeled antibodies. Cells were resuspended in Hanks’ balanced salt solution and stained for live cells using LIVE/DEAD Fixable Dead Cell Stain and cell surface markers (see Table S1 for antibodies). Cells were fixed overnight with Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) and stained with intracellular cytokine or transcription factor staining for 45 min. For intracellular Blimp-1 staining, cells from lung or mediastinal draining LN were stimulated with 10 µg/ml anti-CD3/anti-CD28 or 100 µg/ml HDM for 36 h, respectively, then restimulated with PMA and ionomycin, with the addition of brefeldin A and monensin for 1.5 h, and processed for staining as above. Cells were collected using a five-laser BD Fortessa and analyzed with FlowJo.
BM chimeras
BM cells were isolated from femurs of mice, and T cells were depleted using Thy1.1- or Thy1.2-positive selection and a magnetic separation kit (Miltenyi Biotec). Cells were counted, mixed in a ratio of 50:50, and injected intravenously into Rag1−/−-deficient hosts. Rag1−/−-deficient host mice were lethally irradiated at 750 cGy for 10 min, followed by 4 h rest, and a subsequent at 750 cGy for 10 min 1 d before injection with cells. A total of 20 × 106 cells was injected per mouse. 4 wk after reconstitution, mice were bled to check reconstitution efficiency, and 6 wk after reconstitution, they were used for HDM experiment, as described above.
Antibody blockade
Mice were injected i.p. with 100 µl of sterile PBS containing 0.2 mg of anti–IL-10R mAb (1B1.3A), anti–IL-21R mAb (4A9), or both anti–IL-10R and anti–IL-21R mAbs on days 1, 3, 5, 7, 9, 15, 17, and 21 during the HDM immunization as described above. Control mice were injected with 0.2 mg IgG1 isotype control (TNP6A7) mAb.
ELISA
The total IgE in the serum was assessed by commercial ELISA kit according to the manufacturer’s protocol.
Quantitative polymerase chain reaction
Total RNA was isolated from lung tissue and mediastinal draining LN using TRIzol Reagent (Thermo Fisher Scientific). About 1 µg RNA was converted into cDNA using a qScript cDNA Synthesis Kit (Quanta BioSciences). qPCR was performed with the following TaqMan probes (mouse IL-2 [Mm00434256_m1], mouse IL-4 [Mm00445259_m1], mouse IL-6 [Mm0046190_m1], mouse IL-10 [Mm01288386_m1], mouse IL-21 [Mm00517640_m1], mouse IL-27 [Mm00469294_m1], and mouse β-actin [Mm02619580_g1]; Thermo Fisher Scientific) and TaqMan PerfeCTa qPCR FastMix II mix (Quanta BioSciences) and detected by a CFX96 Real-time PCR Detection Machine (Bio-Rad). ΔΔCt was calculated to determine the relative expression normalized to β-actin.
In vitro T cell polarization
Naive CD4 T cells were isolated from spleens and LNs by magnetic enrichment and flow sorting (CD4+ CD25− CD44lo CD62L+) and activated with plate-bound anti-CD3 and anti-CD28 (10 μg/ml each) for 3 d in complete RPMI in Th1 conditions (IL-12 [10 ng/ml], anti–IL-4 [10 μg/ml]) or Th2 conditions (IL-4 [10 ng/ml], anti–IFN-γ [10 μg/ml]). Cells were stimulated with PMA, ionomycin, and brefeldin A for 2 h before flow staining as above.
AHR
Assessment of AHR was made essentially as previously described (Raundhal et al., 2015). Briefly, mice were anesthetized using a regimen of xylazine (12 mg/kg)/sodium pentobarbital (90 mg/kg)/pancuronium bromide (0.8 mg/kg) and subjected to the forced oscillation technique using flexiVent (SCIREQ). Measurements of lung function were made following perturbation with increasing doses of methacholine.
Statistical analysis
For statistical analysis, an unpaired Mann–Whitney t test, one-way ANOVA, or Kruskal–Wallis test was performed using GraphPad Prism (version 8.02, GraphPad), unless otherwise specified to calculate statistical significance (reported as means ± SD). A P value of 0.05 was considered significant.
Online supplemental material
Fig. S1 shows lung-specific Blimp-1–dependent Th2 development in vivo, related to Fig. 1. Fig. S2 shows Bcl6 expression levels in TFH and Th2 cells in mLN, related to Fig. 2. Fig. S3 shows Blimp-1 expression dynamics in HDM-induced allergic lung inflammation, related to Fig. 3. Fig. S4 shows IL-21R signaling is required for T cell responses but not Blimp-1 in the mLN, related to Fig. 5. Fig. S5 shows IL-10 and IL-21 do not cooperatively regulate Blimp-1 expression, related to Fig. 6 and Fig. 7. Table S1 lists antibodies for cell surface markers and intracellular cytokines/transcription factors.
Supplementary Material
lists antibodies for cell surface markers and intracellular cytokines/transcription factors.
Acknowledgments
We gratefully thank T. Oriss, R. Huff, E. Schmitz, S. Pandya, K. Scholl, H. Yun, J. Chen, and J. Woodall for technical help with lung isolation, preparation for histology, and BAL counting. We also thank J. Alcorn and M. Manni for expertise, discussions, and use of cytospin and microscope, and C. Byersdorfer and A. Dobbs for help and expertise with chimera studies. We also thank D. Jankovic for SEA. We also thank M. Mulkeen and the Rangos Histology Core for histology support, the Division of Laboratory Animal Resources (for support related to animal husbandry, J. Michael and the Rangos Flow Cytometry Core, and the Unified Flow Core at the University of Pittsburgh for flow cytometry support. We also thank T. Hand, S. Canna, and M. McGeachy for helpful discussions and comments on the manuscript.
This work benefitted from the BDFortessa funded by National Institutes of Health grant 1S10OD011925-01. This work was supported by an American Lung Association Biomedical Research Grant (A.C. Poholek), and grants from the National Institutes of Health: AI132771 (A.L. Dent), AI106684 and HL113956 (A. Ray), and AI135027 (A.C. Poholek).
Author contributions: Conceptualization, K. He, A. Ray, and A.C. Poholek; Methodology, S.L. Kale, A. Ray, K. He, and A.C. Poholek; Investigation and validation, K. He, A. Hettinga, S.L. Kale, S. Hu, and A.C. Poholek; Formal analysis, K. He, A. Hettinga, S.L. Kale, and A.C. Poholek; Resources, M.M. Xie, A.L. Dent, and A. Ray; Writing (original draft), K. He and A.C. Poholek; Writing (review & editing), K. He, A. Ray, and A.C. Poholek; Supervision, A. Ray and A.C. Poholek; Funding acquisition, A.C. Poholek.
References
- Bandukwala H.S., Clay B.S., Tong J., Mody P.D., Cannon J.L., Shilling R.A., Verbeek J.S., Weinstock J.V., Solway J., and Sperling A.I.. 2007. Signaling through Fc gamma RIII is required for optimal T helper type (Th)2 responses and Th2-mediated airway inflammation. J. Exp. Med. 204:1875–1889. 10.1084/jem.20061134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K., and Reinhardt R.L.. 2015. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine. 75:25–37. 10.1016/j.cyto.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón M.A., Linneberg A., Kleine-Tebbe J., De Blay F., Hernandez Fernandez de Rojas D., Virchow J.C., and Demoly P.. 2015. Respiratory allergy caused by house dust mites: What do we really know? J. Allergy Clin. Immunol. 136:38–48. 10.1016/j.jaci.2014.10.012 [DOI] [PubMed] [Google Scholar]
- Cimmino L., Martins G.A., Liao J., Magnusdottir E., Grunig G., Perez R.K., and Calame K.L.. 2008. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J. Immunol. 181:2338–2347. 10.4049/jimmunol.181.4.2338 [DOI] [PubMed] [Google Scholar]
- Cook P.C., and MacDonald A.S.. 2016. Dendritic cells in lung immunopathology. Semin. Immunopathol. 38:449–460. 10.1007/s00281-016-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes S.M., Kannan Y., Pelly V.S., Entwistle L.J., Guidi R., Perez-Lloret J., Nikolov N., Muller W., and Wilson M.S.. 2016. CD4+ Th2 cells are directly regulated by IL-10 during allergic airway inflammation. Mucosal Immunol. [DOI] [PubMed] [Google Scholar]
- Coquet J.M., Schuijs M.J., Smyth M.J., Deswarte K., Beyaert R., Braun H., Boon L., Karlsson Hedestam G.B., Nutt S.L., Hammad H., et al. . 2015. Interleukin-21-Producing CD4(+) T Cells Promote Type 2 Immunity to House Dust Mites. Immunity. 43:318–330. 10.1016/j.immuni.2015.07.015 [DOI] [PubMed] [Google Scholar]
- Cretney E., Xin A., Shi W., Minnich M., Masson F., Miasari M., Belz G.T., Smyth G.K., Busslinger M., Nutt S.L., et al. . 2011. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 12:304–311. 10.1038/ni.2006 [DOI] [PubMed] [Google Scholar]
- Croft M., and Swain S.L.. 1995. Recently activated naive CD4 T cells can help resting B cells, and can produce sufficient autocrine IL-4 to drive differentiation to secretion of T helper 2-type cytokines. J. Immunol. 154:4269–4282. [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty S., Johnston R.J., and Schoenberger S.P.. 2010. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11:114–120. 10.1038/ni.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz L.M., and Klein L.. 2005. Development and function of agonist-induced CD25+Foxp3+ regulatory T cells in the absence of interleukin 2 signaling. Nat. Immunol. 6:1152–1159. 10.1038/ni1264 [DOI] [PubMed] [Google Scholar]
- Dent A.L., Hu-Li J., Paul W.E., and Staudt L.M.. 1998. T helper type 2 inflammatory disease in the absence of interleukin 4 and transcription factor STAT6. Proc. Natl. Acad. Sci. USA. 95:13823–13828. 10.1073/pnas.95.23.13823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganci A., Sauer K., Karwot R., and Finotto S.. 2005. Pathological role of IL-6 in the experimental allergic bronchial asthma in mice. Clin. Rev. Allergy Immunol. 28:257–270. 10.1385/CRIAI:28:3:257 [DOI] [PubMed] [Google Scholar]
- Erle D.J., and Sheppard D.. 2014. The cell biology of asthma. J. Cell Biol. 205:621–631. 10.1083/jcb.201401050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Gavin M.A., and Rudensky A.Y.. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6:1142–1151. 10.1038/ni1263 [DOI] [PubMed] [Google Scholar]
- Fu S.-H., Yeh L.-T., Chu C.-C., Yen B.L.-J., and Sytwu H.-K.. 2017. New insights into Blimp-1 in T lymphocytes: a divergent regulator of cell destiny and effector function. J. Biomed. Sci. 24:49 10.1186/s12929-017-0354-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabryšová L., Howes A., Saraiva M., and O’Garra A.. 2014. The regulation of IL-10 expression. Curr. Top. Microbiol. Immunol. 380:157–190. [DOI] [PubMed] [Google Scholar]
- Gao Y., Nish S.A., Jiang R., Hou L., Licona-Limón P., Weinstein J.S., Zhao H., and Medzhitov R.. 2013. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 39:722–732. 10.1016/j.immuni.2013.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I., Wu J., Fejér G., Seto E., and Wright K.L.. 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5:299–308. 10.1038/ni1046 [DOI] [PubMed] [Google Scholar]
- Hammad H., Charbonnier A.S., Duez C., Jacquet A., Stewart G.A., Tonnel A.B., and Pestel J.. 2001. Th2 polarization by Der p 1--pulsed monocyte-derived dendritic cells is due to the allergic status of the donors. Blood. 98:1135–1141. 10.1182/blood.V98.4.1135 [DOI] [PubMed] [Google Scholar]
- Heinemann C., Heink S., Petermann F., Vasanthakumar A., Rothhammer V., Doorduijn E., Mitsdoerffer M., Sie C., Prazeres da Costa O., Buch T., et al. . 2014. IL-27 and IL-12 oppose pro-inflammatory IL-23 in CD4+ T cells by inducing Blimp1. Nat. Commun. 5:3770 10.1038/ncomms4770 [DOI] [PubMed] [Google Scholar]
- Hollister K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D.V., and Dent A.L.. 2013. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 191:3705–3711. 10.4049/jimmunol.1300378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igietseme J.U., Ananaba G.A., Bolier J., Bowers S., Moore T., Belay T., Eko F.O., Lyn D., and Black C.M.. 2000. Suppression of endogenous IL-10 gene expression in dendritic cells enhances antigen presentation for specific Th1 induction: potential for cellular vaccine development. J. Immunol. 164:4212–4219. 10.4049/jimmunol.164.8.4212 [DOI] [PubMed] [Google Scholar]
- Jain R., Chen Y., Kanno Y., Joyce-Shaikh B., Vahedi G., Hirahara K., Blumenschein W.M., Sukumar S., Haines C.J., Sadekova S., et al. . 2016. Interleukin-23-Induced Transcription Factor Blimp-1 Promotes Pathogenicity of T Helper 17 Cells. Immunity. 44:131–142. 10.1016/j.immuni.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Choi Y.S., Diamond J.A., Yang J.A., and Crotty S.. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209:243–250. 10.1084/jem.20111174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hawkins E.D., Belz G.T., Metcalf D., Hommel M., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2006. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 7:466–474. 10.1038/ni1321 [DOI] [PubMed] [Google Scholar]
- Kallies A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kopf M., Le Gros G., Bachmann M., Lamers M.C., Bluethmann H., and Köhler G.. 1993. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 362:245–248. 10.1038/362245a0 [DOI] [PubMed] [Google Scholar]
- Krishnaswamy J.K., Gowthaman U., Zhang B., Mattsson J., Szeponik L., Liu D., Wu R., White T., Calabro S., Xu L., et al. . 2017. Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol. 2 eaam9169 10.1126/sciimmunol.aam9169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y., Linehan M., Weinstein J.S., Laidlaw B.J., Craft J.E., and Iwasaki A.. 2013. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 39:733–743. 10.1016/j.immuni.2013.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusam S., Toney L.M., Sato H., and Dent A.L.. 2003. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J. Immunol. 170:2435–2441. 10.4049/jimmunol.170.5.2435 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2015. The immunology of asthma. Nat. Immunol. 16:45–56. 10.1038/ni.3049 [DOI] [PubMed] [Google Scholar]
- Laouini D., Alenius H., Bryce P., Oettgen H., Tsitsikov E., and Geha R.S.. 2003. IL-10 is critical for Th2 responses in a murine model of allergic dermatitis. J. Clin. Invest. 112:1058–1066. 10.1172/JCI18246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.K., Raz R., Gimeno R., Gertner R., Wistinghausen B., Takeshita K., DePinho R.A., and Levy D.E.. 2002. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 17:63–72. 10.1016/S1074-7613(02)00336-9 [DOI] [PubMed] [Google Scholar]
- Leonard W.J., and O’Shea J.J.. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16:293–322. 10.1146/annurev.immunol.16.1.293 [DOI] [PubMed] [Google Scholar]
- Licona-Limón P., Kim L.K., Palm N.W., and Flavell R.A.. 2013. TH2, allergy and group 2 innate lymphoid cells. Nat. Immunol. 14:536–542. 10.1038/ni.2617 [DOI] [PubMed] [Google Scholar]
- Malek T.R., Yu A., Vincek V., Scibelli P., and Kong L.. 2002. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 17:167–178. 10.1016/S1074-7613(02)00367-9 [DOI] [PubMed] [Google Scholar]
- Martins G.A., Cimmino L., Liao J., Magnusdottir E., and Calame K.. 2008. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J. Exp. Med. 205:1959–1965. 10.1084/jem.20080526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G.A., Cimmino L., Shapiro-Shelef M., Szabolcs M., Herron A., Magnusdottir E., and Calame K.. 2006. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 7:457–465. 10.1038/ni1320 [DOI] [PubMed] [Google Scholar]
- McBrien C.N., and Menzies-Gow A.. 2017. The Biology of Eosinophils and Their Role in Asthma. Front. Med. (Lausanne). 4:93 10.3389/fmed.2017.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich M., Tagoh H., Bönelt P., Axelsson E., Fischer M., Cebolla B., Tarakhovsky A., Nutt S.L., Jaritz M., and Busslinger M.. 2016. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 17:331–343. 10.1038/ni.3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C., Heinrich F., Neumann K., Junghans V., Mashreghi M.F., Ahlers J., Janke M., Rudolph C., Mockel-Tenbrinck N., Kühl A.A., et al. . 2014. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med. 211:1807–1819. 10.1084/jem.20131548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Mohn S.E., and Weinmann A.S.. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13:405–411. 10.1038/ni.2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant C.J., Barlow J.L., and McKenzie A.N.. 2011. Insights into the initiation of type 2 immune responses. Immunology. 134:378–385. 10.1111/j.1365-2567.2011.03499.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Löhning M., Gao Z., Assenmacher M., Ranganath S., Radbruch A., and Murphy K.M.. 2000. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 12:27–37. 10.1016/S1074-7613(00)80156-9 [DOI] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Ettinger R., Kim H.P., Wang G., Qi C.F., Hwu P., Shaffer D.J., Akilesh S., Roopenian D.C., et al. . 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173:5361–5371. 10.4049/jimmunol.173.9.5361 [DOI] [PubMed] [Google Scholar]
- Parish I.A., Marshall H.D., Staron M.M., Lang P.A., Brüstle A., Chen J.H., Cui W., Tsui Y.C., Perry C., Laidlaw B.J., et al. . 2014. Chronic viral infection promotes sustained Th1-derived immunoregulatory IL-10 via BLIMP-1. J. Clin. Invest. 124:3455–3468. 10.1172/JCI66108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual R.M., and Peters S.P.. 2005. Airway remodeling contributes to the progressive loss of lung function in asthma: an overview. J. Allergy Clin. Immunol. 116:477–486, quiz :487. 10.1016/j.jaci.2005.07.011 [DOI] [PubMed] [Google Scholar]
- Poholek A.C., Jankovic D., Villarino A.V., Petermann F., Hettinga A., Shouval D.S., Snapper S.B., Kaech S.M., Brooks S.R., Vahedi G., et al. . 2016. IL-10 induces a STAT3-dependent autoregulatory loop in TH2 cells that promotes Blimp-1 restriction of cell expansion via antagonism of STAT5 target genes. Sci. Immunol. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., Tang H., and Manicassamy S.. 2010. Programming dendritic cells to induce T(H)2 and tolerogenic responses. Nat. Immunol. 11:647–655. 10.1038/ni.1894 [DOI] [PubMed] [Google Scholar]
- Raundhal M., Morse C., Khare A., Oriss T.B., Milosevic J., Trudeau J., Huff R., Pilewski J., Holguin F., Kolls J., et al. . 2015. High IFN-γ and low SLPI mark severe asthma in mice and humans. J. Clin. Invest. 125:3037–3050. 10.1172/JCI80911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., and Cohn L.. 1999. Th2 cells and GATA-3 in asthma: new insights into the regulation of airway inflammation. J. Clin. Invest. 104:985–993. 10.1172/JCI8204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón M., Anguita J., Nakamura T., Fikrig E., and Flavell R.A.. 1997. Interleukin (IL)-6 directs the differentiation of IL-4-producing CD4+ T cells. J. Exp. Med. 185:461–469. 10.1084/jem.185.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y., Dienger-Stambaugh K., Richgels P.K., Lewkowich I.P., Kartashov A.V., Barski A., Khurana Hershey G.K., Leonard W.J., and Singh H.. 2018. TSLP signaling in CD4+ T cells programs a pathogenic T helper 2 cell state. Sci. Signal. 11 eaam8858 10.1126/scisignal.aam8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant D.V., Sehra S., Nguyen E.T., Jadhav R., Englert K., Shinnakasu R., Hangoc G., Broxmeyer H.E., Nakayama T., Perumal N.B., et al. . 2012. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J. Immunol. 189:4759–4769. 10.4049/jimmunol.1201794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder R.A., Paul W.E., Dvorak A.M., Sharkis S.J., Kagey-Sobotka A., Niv Y., Finkelman F.D., Barbieri S.A., Galli S.J., and Plaut M.. 1991. Mouse splenic and bone marrow cell populations that express high-affinity Fc epsilon receptors and produce interleukin 4 are highly enriched in basophils. Proc. Natl. Acad. Sci. USA. 88:2835–2839. 10.1073/pnas.88.7.2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Blackburn S.D., Intlekofer A.M., Kao C., Angelosanto J.M., Reiner S.L., and Wherry E.J.. 2009. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 31:309–320. 10.1016/j.immuni.2009.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky G.L., Muthukrishnan R., Sehra S., Goswami R., Pham D., Travers J., Nguyen E.T., Levy D.E., and Kaplan M.H.. 2011. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 34:39–49. 10.1016/j.immuni.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumhofer J.S., Silver J.S., Laurence A., Porrett P.M., Harris T.H., Turka L.A., Ernst M., Saris C.J., O’Shea J.J., and Hunter C.A.. 2007. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 8:1363–1371. 10.1038/ni1537 [DOI] [PubMed] [Google Scholar]
- Thomas W.R. 2016. House Dust Mite Allergens: New Discoveries and Relevance to the Allergic Patient. Curr. Allergy Asthma Rep. 16:69 10.1007/s11882-016-0649-y [DOI] [PubMed] [Google Scholar]
- Tournoy K.G., Kips J.C., and Pauwels R.A.. 2000. Endogenous interleukin-10 suppresses allergen-induced airway inflammation and nonspecific airway responsiveness. Clin. Exp. Allergy. 30:775–783. 10.1046/j.1365-2222.2000.00838.x [DOI] [PubMed] [Google Scholar]
- Turner C.A. Jr., Mack D.H., and Davis M.M.. 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 77:297–306. 10.1016/0092-8674(94)90321-2 [DOI] [PubMed] [Google Scholar]
- Webster K.E., Walters S., Kohler R.E., Mrkvan T., Boyman O., Surh C.D., Grey S.T., and Sprent J.. 2009. In vivo expansion of T reg cells with IL-2-mAb complexes: induction of resistance to EAE and long-term acceptance of islet allografts without immunosuppression. J. Exp. Med. 206:751–760. 10.1084/jem.20082824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.W., Tjota M.Y., Clay B.S., Vander Lugt B., Bandukwala H.S., Hrusch C.L., Decker D.C., Blaine K.M., Fixsen B.R., Singh H., et al. . 2013. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 4:2990 10.1038/ncomms3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.S., Elnekave E., Mentink-Kane M.M., Hodges M.G., Pesce J.T., Ramalingam T.R., Thompson R.W., Kamanaka M., Flavell R.A., Keane-Myers A., et al. . 2007. IL-13Ralpha2 and IL-10 coordinately suppress airway inflammation, airway-hyperreactivity, and fibrosis in mice. J. Clin. Invest. 117:2941–2951. 10.1172/JCI31546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M.M., Koh B.H., Hollister K., Wu H., Sun J., Kaplan M.H., and Dent A.L.. 2017. Bcl6 promotes follicular helper T-cell differentiation and PD-1 expression in a Blimp1-independent manner in mice. Eur. J. Immunol. 47:1136–1141. 10.1002/eji.201747034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin A., Masson F., Liao Y., Preston S., Guan T., Gloury R., Olshansky M., Lin J.X., Li P., Speed T.P., et al. . 2016. A molecular threshold for effector CD8(+) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17:422–432. 10.1038/ni.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T., Bendelac A., Watson C., Hu-Li J., and Paul W.E.. 1995. Role of NK1.1+ T cells in a TH2 response and in immunoglobulin E production. Science. 270:1845–1847. 10.1126/science.270.5243.1845 [DOI] [PubMed] [Google Scholar]
- Zhang D.H., Cohn L., Ray P., Bottomly K., and Ray A.. 1997. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J. Biol. Chem. 272:21597–21603. 10.1074/jbc.272.34.21597 [DOI] [PubMed] [Google Scholar]
- Zhang D.H., Yang L., Cohn L., Parkyn L., Homer R., Ray P., and Ray A.. 1999. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 11:473–482. 10.1016/S1074-7613(00)80122-3 [DOI] [PubMed] [Google Scholar]
- Zheng W., and Flavell R.A.. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 89:587–596. 10.1016/S0092-8674(00)80240-8 [DOI] [PubMed] [Google Scholar]
- Zhou L., Ivanov I.I., Spolski R., Min R., Shenderov K., Egawa T., Levy D.E., Leonard W.J., and Littman D.R.. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8:967–974. 10.1038/ni1488 [DOI] [PubMed] [Google Scholar]
- Zhu J. 2015. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine. 75:14–24. 10.1016/j.cyto.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists antibodies for cell surface markers and intracellular cytokines/transcription factors.