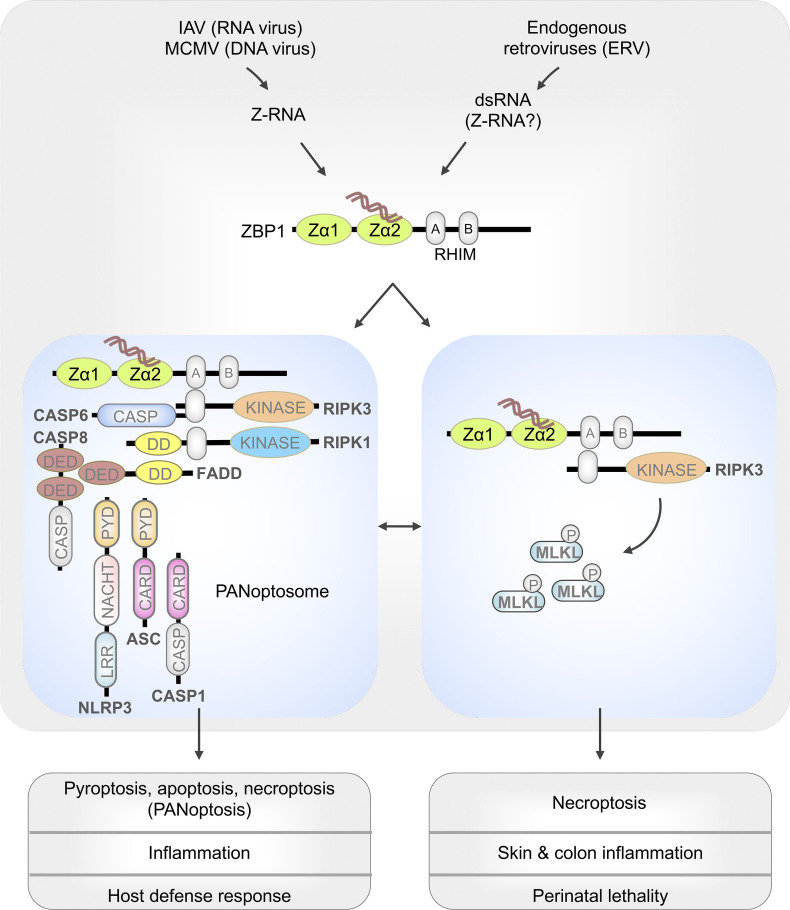
Deciphering ZBP1 activation and its role in sterile or viral-induced cell death.
Abstract
ZBP1 triggers NLRP3 inflammasome activation/pyroptosis, apoptosis, and necroptosis; the specific ligand for ZBP1 activation remains ambiguous. Recent studies, including Devos et al. in this issue of JEM (https://doi.org/10.1084/jem.20191913), collectively suggest that ZBP1 sensing Z-nucleic acids is critical for cell death/inflammatory disease.
Z-DNA–binding protein 1 (ZBP1), also known as DNA-dependent activator of IFN regulatory factors, is an innate nucleic acid sensor that has emerging roles in regulating host defense and inflammatory diseases by orchestrating cell death and inflammation (Kuriakose and Kanneganti, 2018). ZBP1 contains a RIP-homotypic interaction motif (RHIM) through which it interacts with other RHIM-containing proteins, RIPK1 and RIPK3 (Kuriakose and Kanneganti, 2018). ZBP1 is distinct among the RHIM-containing proteins in that it includes nucleic acid–sensing Zα domains. Unlike other nucleic acid sensors, the Zα domains of ZBP1 show high affinity for double-stranded nucleic acids in the Z-conformation. Right-handed DNA (B-form) and RNA (A-form) are conformationally distinct. However, left-handed DNA and RNA, which adopt the Z-conformation, exhibit similar structural features (Rich and Zhang, 2003). Thus, Zα domains of ZBP1 show affinity for both Z-DNA and Z-RNA in vitro. Initial studies proposed that ZBP1 acts as a B-DNA sensor to mount type I interferon responses (Table 1). ZBP1 was also shown to complex with RIPK3 to mediate M45 mutant murine cytomegalovirus (DNA virus)–induced necroptosis (Upton et al., 2012). An independent study in search of an upstream NLRP3 inflammasome regulator identified ZBP1 as an innate immune sensor of influenza A virus (IAV), an RNA virus, that triggers the execution of pyroptosis, apoptosis, and necroptosis (PANoptosis; Kuriakose et al., 2016). Discovery of this unanticipated role for ZBP1 in sensing RNA virus infection prompted researchers to characterize RNAs as endogenous or pathogen-associated Z-nucleic acid (Z-NA) ligands that activate ZBP1 (Kesavardhana et al., 2017; Kuriakose et al., 2016; Thapa et al., 2016). However, current understanding of whether viral or endogenous RNAs attain the Z-conformation and the biological significance of nucleic acids being in the Z-conformation is still lacking.

Insights from Sannula Kesavardhana and Thirumala-Devi Kanneganti.
Schematic representation of ZBP1 activation promoting cell death and inflammation. Z-RNAs from IAV or murine cytomegalovirus (MCMV) or dsRNAs formed by ERVs stimulate Zα domain–dependent activation of ZBP1. When RIPK1 is deleted in keratinocytes or the RHIM domain of RIPK1 is mutated, ERV-mediated activation of ZBP1 promotes RIPK3-MLKL–mediated necroptosis. Activation of ZBP1 by Z-RNAs from IAV triggers assembly of the PANoptosome, which engage pyroptosis, apoptosis, and necroptosis (PANoptosis). CARD, caspase activation and recruitment domain; CASP, caspase; DD, death domain; DED, death effector domain; LRR, leucine-rich repeat; PYD, pyrin domain.
ZBP1-mediated cellular functions are associated with the regulation of RIPK1 and RIPK3. Disrupting the function of RIPK1, a cell survival–regulating signaling kinase, promotes RIPK3 phosphorylation to induce mixed lineage kinase domain–like pseudokinase (MLKL)–dependent necroptosis, an inflammatory form of cell death. Deletion of RIPK1 in keratinocytes (Ripk1EKO) or mutations in the RIPK1 RHIM sequence (Ripk1mRHIM or Ripk1mR) promote spontaneous activation of ZBP1 and subsequent induction of RIPK3-MLKL–driven necroptosis and skin inflammation in mice (Lin et al., 2016; Newton et al., 2016). Moreover, spontaneous activation of ZBP1 drives perinatal lethality in Ripk1mRHIM mice (Lin et al., 2016; Newton et al., 2016). Discovery of these dynamics uncovered a previously unknown homeostatic regulatory function for RIPK1 in restricting endogenous, spontaneous ZBP1 activation. Several groups have recently explored how ZBP1 is activated in sterile conditions and proposed that Z-RNA acts as the ligand to activate ZBP1-mediated cell death and inflammation (Devos et al., 2020; Jiao et al., 2020; Kesavardhana et al., 2020; Wang et al., 2020). Understanding this pathway is essential to unlock key mechanisms, perhaps the sensing of Z-NAs, that prime skin inflammation and autoinflammatory diseases and identify specific signaling cascades for therapeutic targeting.
Devos et al. (2020) and other recent studies have identified that endogenous nucleic acid sensing is critical for ZBP1 activation to induce necroptosis, skin and colon inflammation, and perinatal lethality (Jiao et al., 2020; Kesavardhana et al., 2020; Wang et al., 2020). To understand ZBP1 activation mechanisms, Devos et al. (2020) crossed Ripk1EKO mice with ZBP1 knock-in mice, which contain mutations disrupting the nucleic acid binding of the ZBP1 Zα domains (N46A, Y50A, N122A, Y126A; Maelfait et al., 2017). Ripk1EKO mice developed skin lesions soon after their birth, which resulted in skin inflammation and pathology. Importantly, Ripk1EKO-ZBP1 knock-in mice did not develop lesions until 12 wk of age when they developed mild skin lesions, much less severe than those of Ripk1EKO mice. These observations show that the recognition of nucleic acids, which might exist in the Z-conformation, by ZBP1 drives its activation in Ripk1EKO mice.
Up-regulation of ZBP1 expression via interferon treatment in RIPK1-deficient cells is sufficient to engage RIPK3-MLKL–mediated necroptosis (Devos et al., 2020). Thus, ZBP1 activation is cell intrinsic under sterile conditions, and endogenous nucleic acids may potentially be the ligands engaging its activation. Other recent studies also have shown that the Zα domains of ZBP1 are critical for triggering skin and bowel inflammation, the perinatal lethality of Ripk1mRHIM mice, and IAV-induced PANoptosis via the assembly of a PANoptosome, suggesting a role for Z-NA sensing in regulating physiological functions (Table 1; Christgen et al., 2020; Jiao et al., 2020; Kesavardhana et al., 2020; Samir et al., 2020; Wang et al., 2020; Zhang et al., 2020). Under sterile conditions, double-stranded RNAs (dsRNAs) from endogenous retrovirus (ERV) elements bind ZBP1 and are implicated in its activation (Jiao et al., 2020; Wang et al., 2020). During infection, ZBP1 senses defective IAV RNA genomes that attain the Z-conformation in the nucleus (Zhang et al., 2020). These findings provide evidence for the activation of ZBP1 by sensing Z-NAs. Importantly, deleting or mutating only the Zα2 domain of ZBP1 is sufficient to abolish its activation in both endogenous and infectious conditions (Jiao et al., 2020; Kesavardhana et al., 2020). Overall, these new studies provide genetic evidence that endogenous or viral RNAs which attain Z-NA conformation are sensed by ZBP1, leading to cell death and inflammation.
Table 1. Studies that have proposed an activating ligand for ZBP1 and the timeline for their initial publication.
| Study | Proposed ligand for ZBP1 | Pathogen |
|---|---|---|
| Takaoka et al., 2007 (July 8, 2007) | B-DNA | N/A |
| Kuriakose et al., 2016 (August 5, 2016) | RNA virus (IAV) and RNA genome-associated proteins | IAV |
| Thapa et al., 2016 (October 13, 2016) | IAV genomic RNA | IAV |
| Sridharan et al., 2017 (June 12, 2017) | MCMV RNA transcripts | MCMV |
| Kesavardhana et al., 2017 (June 20, 2017) | IAV RNA genome (RNA associated with nucleoprotein) | IAV |
| Maelfait et al., 2017 (July 17, 2017) | MCMV RNA, endogenous RNA, and Z-RNAs | MCMV |
| Zhang et al., 2020 (March 19, 2020) | IAV viral Z-RNAs | IAV |
| Jiao et al., 2020 (March 25, 2020) | Endogenous Z-nucleic acids, ERV RNAs | N/A |
| Wang et al., 2020 (March 25, 2020) | Endogenous Z-nucleic acids, ERV RNAs | N/A |
| Devos et al., 2020 (April 21, 2020) | Endogenous Z-nucleic acids | N/A |
| Kesavardhana et al., 2020 (April 29, 2020) | IAV Z-RNAs and endogenous Z-nucleic acids | IAV |
N/A, not applicable.
To delineate ZBP1-mediated immune responses that drive skin inflammation and disease in Ripk1EKO mice, Devos et al. (2020) characterized immune cells recruited to the inflamed skin. They observed elevated numbers of IL-17–secreting immune cells, CD4+ T cells and type 3 innate lymphoid cells (ILC3s), in the inflamed epidermis of Ripk1EKO mice. Infiltration of these IL-17–producing immune cells was not seen in Ripk1EKO mice with ZBP1 knock-in mutations at initial stages, but their infiltration was seen in 10–12-wk-old mice. However, it is not clear whether IL-17–mediated inflammation is the crucial driver of detrimental skin inflammation in Ripk1EKO mice. Increased neutrophil infiltration was also observed in the epidermis of Ripk1EKO mice; this infiltration was inhibited when ZBP1 knock-in mutations were introduced in Ripk1EKO mice (Devos et al., 2020). Neutrophil recruitment into damaged or infected tissues generally is known to be promoted by pyroptosis-driven inflammatory immune responses. However, caspase-1/11 deficiency, which abolishes inflammasome-driven pyroptosis, in Ripk1EKO mice did not protect from skin inflammatory disease (Devos et al., 2020). These findings indicate the specific involvement of necroptosis in this pathology. ZBP1 also promotes the inflammasome-independent release of IL-1α and neutrophil inflammation. This suggests a plausible association between ZBP1 activation, release of IL-1α, and neutrophilic inflammation in skin pathology; however, this needs to be experimentally tested.
New studies indicate that the deletion or mutation of only the Zα2 domain of ZBP1 is sufficient to protect Ripk1EKO mice from skin inflammation and Ripk1mRHIM mice from perinatal lethality (Jiao et al., 2020; Kesavardhana et al., 2020). In addition, mutating the Zα domains of ZBP1 did not compromise their interaction with RIPK3 (Devos et al., 2020). This suggests that the mutations in the Zα domains specifically abrogate Z-NA sensing but retain intact RHIM activity. Similar to the protective role of mutating the Zα2 domain, mutations disrupting RHIM activity in ZBP1 also abolish the development of skin inflammation in Ripk1EKO mice and rescue Ripk1mRHIM mice from lethality (Jiao et al., 2020). Thus, both nucleic acid sensing via Zα domains and assembly of signaling complexes via RHIM domains are critical for driving ZBP1-triggered cell death, inflammation, and disease progression.
While ZBP1 was discovered two decades ago, its role in regulating cellular biology has started to unfold only recently. Finding RHIM sequences in the ZBP1 protein hinted at a possible role for ZBP1 in sensing Z-NAs to regulate cell death and inflammation. Intact Zα domains in ZBP1 are critical to sense endogenous or viral RNAs. During viral infections, RNA genomes or RNA transcripts generated in infected cells trigger the activation of ZBP1 and induction of PANoptosis (Table 1; Christgen et al., 2020; Kesavardhana et al., 2017; Maelfait et al., 2017; Thapa et al., 2016; Zhang et al., 2020). Further confirming previous reports on the role of ZBP1 in IAV infection, a recent study demonstrated that the Z-RNAs generated during IAV infection could promote ZBP1-mediated necroptosis (Zhang et al., 2020). In skin or colon inflammatory pathologies, endogenous dsRNAs of ERVs are proposed to activate ZBP1-mediated necroptosis and inflammation, suggesting ERVs might mimic viral infections in activating ZBP1 (Table 1; Jiao et al., 2020; Wang et al., 2020). However, it is still unclear whether endogenous RNAs attain the Z-conformation.
Inhibition of nuclear export in interferon-treated cells promotes ZBP1-mediated cell death (both necroptosis and apoptosis), perhaps by facilitating the accumulation of Z-NAs in the nucleus (Jiao et al., 2020). IAV replicates in the nucleus, and ZBP1 also translocates to the nucleus, which might be essential for sensing IAV Z-RNAs there (Kesavardhana et al., 2017; Kuriakose et al., 2016; Zhang et al., 2020). However, restricting ZBP1 localization to the cytosol does not completely abolish IAV-induced cell death (Zhang et al., 2020). This suggests that the activation of ZBP1 and assembly of signaling complexes can occur in the nucleus as well as in the cytosol. ZBP1 staining is also observed in the cytosol of keratinocytes in the inflamed epidermis, further suggesting cytosolic activation of ZBP1 is occurring (Devos et al., 2020). Additionally, ZBP1 promotes activation of the IAV infection–induced NLRP3 inflammasome, which is located in the cytosol (Kesavardhana et al., 2017; Kuriakose et al., 2016). Further identification of the complete ZBP1 interacting proteome may provide additional clues about its regulation in the nucleus versus cytosol.
Despite having two functional Zα domains, disrupting only the Zα2 domain is sufficient to inhibit activation of ZBP1 during viral infections and pathological inflammation. Notably, several studies have now found that up-regulation of ZBP1 expression, mostly via interferon signaling, follows ZBP1 activation by Z-NAs (Devos et al., 2020; Jiao et al., 2020; Kesavardhana et al., 2017; Kuriakose et al., 2016; Wang et al., 2020; Zhang et al., 2020). Ectopic expression of ZBP1 also drives necroptosis without stimulation (Maelfait et al., 2017). These findings led us to speculate that the presence of two Zα domains may facilitate cooperative binding and sensing of Z-RNAs by ZBP1 to unlock its active conformation. The Zα2 domain might be a regulatory switch for nucleating this cooperative binding to Z-RNAs and subsequent exposure of RHIMs to assemble signaling complexes. It is important for future studies to focus on unraveling the precise activation mechanism of ZBP1 by identifying specific conditions that stimulate the formation of endogenous Z-NAs, conditions that stabilize endogenous nucleic acids in the Z-conformation, and how Z-NAs regulate physiological function and pathogenesis in disease. Addressing these questions will identify the potential of Z-NAs to modulate biological functions and also provide a mechanistic and molecular basis for therapeutic modulation of ZBP1 in cancer and infectious and inflammatory diseases.
Acknowledgments
Research in the Kanneganti laboratory is supported by the National Institutes of Health grants CA163507, AR056296, AI124346, and AI101935 and by the American Lebanese Syrian Associated Charities to T.-D. Kanneganti. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no conflicts of interest.
References
- Christgen, S., et al. 2020. Front. Cell. Infect. Microbiol. 10.3389/fcimb.2020.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, M., et al. 2020. J. Exp. Med. 10.1084/jem.20191913 [DOI] [Google Scholar]
- Jiao, H., et al. 2020. Nature. 10.1038/s41586-020-2129-8 [DOI] [Google Scholar]
- Kesavardhana, S., et al. 2017. J. Exp. Med. 10.1084/jem.20170550 [DOI] [Google Scholar]
- Kesavardhana, S., et al. 2020. J. Biol. Chem. 10.1074/jbc.RA120.013752 [DOI] [Google Scholar]
- Kuriakose, T., and Kanneganti T.D.. 2018. Trends Immunol. 10.1016/j.it.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose, T., et al. 2016. Sci. Immunol. 10.1126/sciimmunol.aag2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., et al. 2016. Nature. 10.1038/nature20558 [DOI] [Google Scholar]
- Maelfait, J., et al. 2017. EMBO J. 10.15252/embj.201796476 [DOI] [Google Scholar]
- Newton, K., et al. 2016. Nature. 10.1038/nature20559 [DOI] [Google Scholar]
- Rich, A., and Zhang S.. 2003. Nat. Rev. Genet. 10.1038/nrg1115 [DOI] [PubMed] [Google Scholar]
- Samir, P., et al. 2020. Front. Cell. Infect. Microbiol. 10.3389/fcimb.2020.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan, H., et al. 2017. EMBO Rep. 10.15252/embr.201743947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka, A., et al. 2007. Nature. 10.1038/nature06013 [DOI] [Google Scholar]
- Thapa, R.J., et al. 2016. Cell Host Microbe. 10.1016/j.chom.2016.09.014 [DOI] [Google Scholar]
- Upton, J.W., et al. 2012. Cell Host Microbe. 10.1016/j.chom.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., et al. 2020. Nature. 10.1038/s41586-020-2127-x [DOI] [Google Scholar]
- Zhang, T., et al. 2020. Cell. 10.1016/j.cell.2020.02.050 [DOI] [Google Scholar]