Abstract
Purpose:
We evaluated strategies for identifying disease-causing variants in genetic testing for dilated cardiomyopathy (DCM).
Methods:
Cardiomyopathy gene panel testing was performed in 532 DCM patients and 527 healthy elderly subjects. Rare variants in 41 genes were stratified using variant-level and gene-level characteristics.
Results:
A majority of DCM cases and controls carried rare protein-altering cardiomyopathy gene variants. Variant-level characteristics alone had limited discriminative value. Differentiation between groups was substantially improved by addition of gene-level information that incorporated ranking of genes based on literature evidence for disease association. The odds of DCM were increased to nearly 9-fold for truncating variants or high-impact missense variants in the subset of 14 genes that had the strongest biological links to DCM (P < 0.0001). For some of these genes, DCM-associated variants appeared to be clustered in key protein functional domains. Multiple rare variants were present in many family probands, however, there was generally only one “driver” mutation that co-segregated with disease.
Conclusions
Rare variants in cardiomyopathy genes can be effectively stratified by combining variant-level and gene-level information. Prioritization of genes based on their a priori likelihood of disease causation is a key factor in identifying clinically-actionable variants in cardiac genetic testing.
Keywords: Dilated cardiomyopathy, mutation, genetic testing, next-generation sequencing
INTRODUCTION
Determining variant pathogenicity is the major challenge in cardiomyopathy genetic testing, spawning debate in clinics worldwide. Data analysis has been biased by a focus on affected patients without a full appreciation of the normal spectrum of variation in cardiomyopathy “disease genes”. Rare protein-altering variants, many of which were initially reported to be pathogenic, are now known to occur in apparently healthy individuals in the general population.1-4 These disturbing findings raise doubts about the discriminative efficacy of variant annotation pipelines used in literature reports and the reliability of genetics results provided to patients. Given the increasing role of personal genome sequencing in clinical practice, there is a critical need for an improved strategy for identifying the subset of rare variants that are truly disease-causing.
Dilated cardiomyopathy (DCM) is the most common cardiomyopathy and frequently has a genetic etiology.5-8 More than 100 genes have been implicated to date but the weight of evidence in DCM causation varies widely. Genetic testing panels generally contain putative DCM-associated genes and have expanded over time to include genes with direct and indirect links to numerous other cardiac and skeletal myopathies. This inclusive approach potentially reduces negative screening results but magnifies the problem of rare variant interpretation.
The aim of this study was to investigate parameters that might improve discrimination between rare cardiomyopathy gene variants in patients with DCM and apparently healthy individuals. We evaluated elderly (>70 years) control subjects to account for the recognized age-related penetrance associated with genetic causes of DCM. Our analysis included evaluation of variant-level and gene-level characteristics, as well as assessment of the total numbers of rare variants in each individual. This type of information about personal burden of rare variants is unable to be determined from studies of single genes or small gene panels in DCM patients and is not available in population databases where single variants in de-identified subjects are listed separately. Our findings provide a new framework for variant prioritization and highlight the key role of a subset of genes in DCM pathogenesis.
MATERIALS AND METHODS
Study subjects
Participants provided informed written consent and protocols were approved by the institutional human ethics committees. Patients with familial or sporadic idiopathic DCM (aged 47 ± 19 years, 69% males) and healthy subjects without cardiovascular disease (aged 49 ± 15 years, 38% males) were recruited from Brigham and Women’s Hospital, Boston Children’s Hospital, Victor Chang Cardiac Research Institute, Royal Brompton & Harefield NHS Foundation Trust, and the London Institute of Medical Sciences, Imperial College, London. All subjects had self-reported European ancestry and significant population stratification was excluded using a principal component analysis (Supplementary Figure S1). The DCM replication cohort was comprised of 101 Australian familial DCM probands (aged 45 ± 16 years, 67% males). Replication control cohorts included elderly (>70 years) subjects drawn from the Alzheimer’s Disease Sequencing Project (n=2971) (www.niagads.org/adsp/) and the Medical Genome Reference Bank (n=1144) (https://sgc.garvan.org.au/initiatives/mgrb).
Gene sequencing
Genomic DNA libraries were constructed using standard library preparation protocols. Fragments were ligated to adaptors, amplified, purified, then hybridized to custom arrays enriched for coding regions of genes associated with DCM and other inherited cardiac disorders: 69-gene panel (n=203 DCM cases, 208 controls), 64-gene panel (n=320 DCM cases, 319 controls). The replication familial DCM cohort was also tested using the 69-gene panel. Sequencing was performed using the Illumina HiSeq 2000 or SOLiD 5500x1 platforms. Selected variants were evaluated in probands and family members using Sanger sequencing.
Data analysis
Sequence data were processed and aligned to the hg19 (GRCh37) human genome reference using Novoalign and the Genome Analysis Toolkit, or Lifescope v2.5.1. Data for the 41 genes that were in represented on both the 69-gene and 64-gene panels and for which variants were detected in cases, were included in this study. Coding-sequence variants that were truncating (stop gain, splice donor or acceptor site gain or loss, frame-shift indels) or missense, and that met quality metrics for mapping, read depth, and allelic balance were evaluated. Population data for variant minor allele frequency (MAF) were obtained from the Exome Sequencing Project (ESP) and Exome Aggregation Consortium (ExAC) v1 databases. In silico pathogenicity predictions for missense variants were made using PolyPhen2, SIFT, PROVEAN, and MetaSVM.9-11 Statistical comparisons between groups were made using Fisher’s exact or chi-squared (2 x 2, 2 x 4) tests. To identify the cells that contributed most to 2 x 4 chi-squared tests, standardized residuals were calculated and deviations of absolute value >2 were considered significant. To evaluate variant distribution in annotated domains of group A genes, missense variants identified in DCM cases were pooled with those of Walsh et al.12 and compared with ExAC missense variants (MAF <0.1 %) within the same protein domains using Fisher’s exact test. P < 0.05 was used as the threshold for statistical significance.
RESULTS
Rare variants in cardiomyopathy gene are common in cases and controls
Genetic screening was performed in 532 DCM patients and 527 healthy control subjects. Variants that were protein-altering (truncating, missense) and rare (defined here by MAF < 0.1% in the European [EA] subgroup in the ESP database) in a set of 41 cardiomyopathy- associated genes were evaluated. Variants that met these criteria were found in 407 (77%) DCM cases and in 348 (66%) control subjects (P = 0.0002), with the number of rare variants per person ranging from 0 to 13 (mean 1.63) in DCM cases and from 0 to 8 (mean 1.24) in controls (P < 0.0001). To explore strategies for identifying disease-associated variants, we compared 770 different variants found in DCM cases and 589 variants in control subjects (Supplementary Table S1).
TTN variants
There was an excess of truncating TTN variants (TTNtv) in DCM cases (83 [11%] variants vs controls, 6 [1%] variants; P < 0.0001; Supplementary Table S2). TTN missense variants comprised ~ 40% of all variants found in cases and controls. Unlike missense variants in other genes (see below), TTN missense variants were not differentiated between groups by assessment of novelty or in silico functional predictions (Supplementary Table S3). All TTN variants were excluded from subsequent variant-level and gene-level analyses.
Limited discriminative value of variant characteristics
Unlike TTN, there were relatively few truncating variants in other cardiomyopathy genes and these were equally seen in DCM cases (25 [3%] variants) and controls (17 [3%] variants; P = 0.75). To start to stratify the larger numbers of missense variants found in both groups, we looked at novelty and in silico functional predictions.
Variant novelty.
The absence of a variant in healthy subjects has been used as a criterion of pathogenicity in many DCM mutation reports.13-15 Assessment of novelty varies with the size of the reference cohort evaluated and many disease-associated variants initially deemed to be novel have subsequently been detected in population sequence databases.1-4 A majority of the missense variants present in DCM cases and controls were absent from the EA subgroup (>4,250 subjects) of the ESP database (P = 0.87; Supplementary Table S3). Using the larger ExAC database (>60,000 subjects), differences between groups became apparent with a greater proportion of novel variants in DCM cases (P < 0.0001; Supplementary Table S3).
Variant functional predictions.
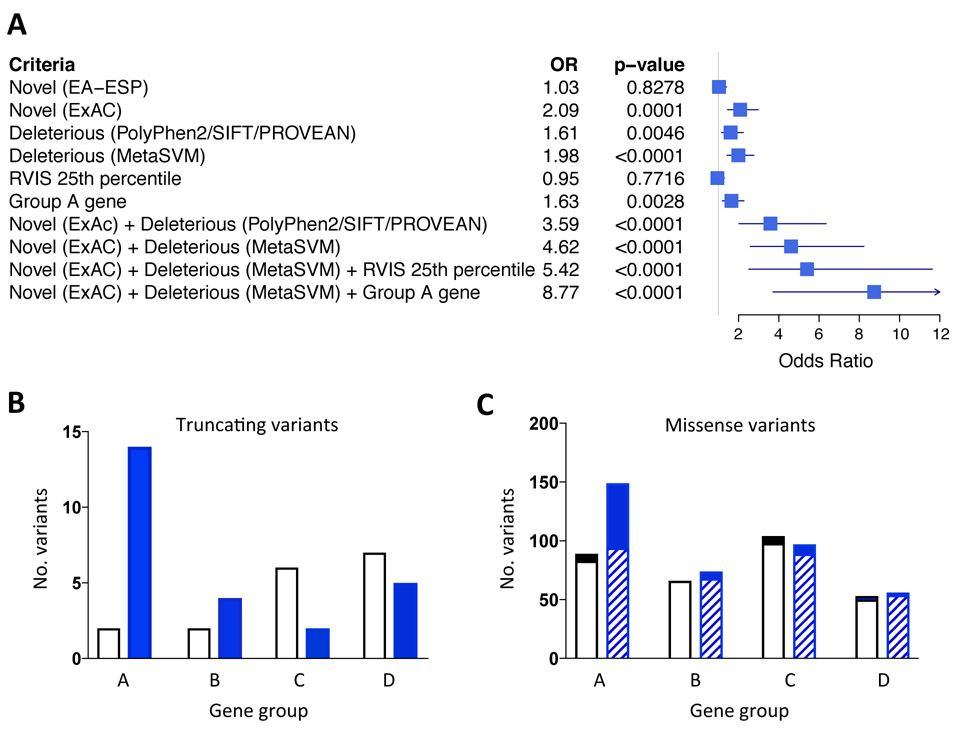
To assess variant function, we evaluated subsets of variants that were (i) uniformly predicted to be deleterious in PolyPhen2, SIFT, and PROVEAN,9,10 or (ii) predicted deleterious by MetaSVM, which provides an ensemble score derived from 10 separate prediction algorithms.11 Both of these approaches yielded significant differences between DCM cases and controls, with modestly better results for MetaSVM’s ensemble score than for the three-program consensus score (Supplementary Table S3). When novelty and functional predictions were combined, discrimination between groups was increased. DCM cases were twice as likely as controls to have variants that were absent from ExAC or deleterious (MetaSVM), and nearly five-fold more likely to have variants that were absent from ExAC and deleterious (MetaSVM) (Figure 1A).
Figure 1.
Criteria for variant prioritization. (A) Missense rare variants in cardiomyopathy genes in DCM cases and controls were compared using a number of criteria including novelty, defined as absence from the European subgroup of the Exome Sequencing Project (EA-ESP) or Exome Aggregation Consortium (ExAC), in silico functional predictions (PolyPhen2, SIFT, PROVEAN, MetaSVM), RVIS score and gene group. The odds ratios (OR) for DCM are displayed in a forest plot. (B) Distribution of truncating variants across gene groups in controls (white bars) and DCM cases (blue bars). (C) Distribution of missense variants across gene groups in controls (white bars) and DCM cases (blue bars). The subset of “novel (ExAC) + deleterious (MetaSVM)” missense variants are denoted in the solid sections of bars. There were significant effects of clinical status and gene group for truncating variants (B, P = 0.01; 2 x 4 chi-squared test) and missense variants (C; P = 0.018); see Supplementary Table S6 for statistical analysis. For all panels, TTN truncating and missense variants were excluded.
Importance of gene-based parameters
Since variant level parameters incompletely differentiated DCM cases from controls, we next sought to determine whether there might be a hierarchy for pathogenicity between genes.
ExAC metrics of expected genetic variation.
Two recently-devised constraint metrics in the ExAC database provide information on a per gene basis for the probability of variation in the general population.16 Cardiomyopathy genes had pLI (probability of being loss-of-function intolerant) scores ranging from 0 to 1, with only 10 genes assessed as extremely intolerant (pLI ≥ 0.9; Table 3). DCM-associated truncating variants were enriched in genes with high pLI scores (11 of 23 [48%] variants vs controls, 1 of 17 [6%] variants, P = 0.005). However, over half of the truncating variants in DCM cases were in genes with intermediate or low pLI scores. Twenty-six genes had positive Z-scores, indicating intolerance to missense variants (Table 1). There was a statistically-significant but relatively modest excess of missense variants in these genes in DCM cases (182 [52%] variants vs controls, 129 [43%] variants; P = 0.018).
Table 1.
Gene characteristics and evidence for role in DCM pathogenesis
| Gene | Protein | Gene group |
RVIS | pLI score |
Z score |
Genetic data* |
In vitro defects |
Animal model† |
Other phenotypes |
|---|---|---|---|---|---|---|---|---|---|
| MYH7 | Myosin heavy chain 7 | A | 0.28 | 0.00 | 6.54 | +++ | Yes | +++ | HCM, RCM, LVNC, CHD, DM, MSM, SPM |
| DSP | Desmoplakin | A | 1.32 | 1.00 | 0.91 | +++ | Yes | +++ | ARVC, LVNC, palmoplantar keratoderma, skin-fragility woolly hair syndrome |
| SCN5A | Cardiac sodium channel | A | 1.99 | 1.00 | 2.53 | +++ | Yes | +++ | CD, AF, LQTS, BS, VF, LVNC |
| LMNA | Lamin A/C | A | 9.96 | 0.99 | 3.37 | +++ | Yes | +++ | CD, EDMD, >10 additional phenotypes/overlap syndromes. |
| LDB3 | LIM domain binding protein 3 | A | 10.16 | 0.00 | 0.32 | +++ | Yes | +++ | HCM, LVNC, ARVC, MM |
| DMD | Dystrophin | A | 11.28 | 1.00 | −4.82 | +++ | Yes | +++ | DMD, BMD |
| DES | Desmin | A | 21.56 | 0.00 | 2.34 | +++ | Yes | +++ | RCM, ARVC, HCM, CD, MM, LGMD-2R, NSS |
| TPM1 | Tropomyosin α-1 chain | A | 25.15 | 0.80 | 3.42 | +++ | Yes | +++ | HCM, RCM, LVNC |
| TNNC1 | Troponin C | A | 39.68 | 0.51 | 2.22 | +++ | Yes | +++ | HCM |
| TNNT2 | Troponin T | A | 41.64 | 0.01 | 1.54 | +++ | Yes | +++ | HCM, RCM, LVNC |
| BAG3 | BCL-2 associated athanogene | A | 62.14 | 0.53 | −1.01 | +++ | Yes | +++ | MM |
| PLN | Phospholamban | A | 62.38 | 0.11 | 0.57 | +++ | Yes | +++ | HCM, ARVC |
| RBM20 | RNA-binding protein 20 | A | 90.98 | NA | NA | +++ | Yes | + | |
| TTN | Titin | A | 98.04 | 0.00 | −5.48 | +++ | Yes | ++ | HCM, ARVC, RCM, LVNC, CM, CNM, HMERF, LGMD-2J, TMD |
| VCL | Vinculin | B | 2.23 | 0.99 | 3.11 | ++ | Yes | ++ | HCM |
| LAMA4 | Laminin subunit α-4 | B | 4.68 | 0.00 | −1.14 | + | Yes | ++ | |
| ILK | Integrin-linked protein kinase | B | 14.97 | 0.04 | 0.96 | + | Yes | ++ | |
| MYBPC3 | Cardiac myosin binding protein 3 | B | 20.54 | 0.00 | 0.69 | ++ | Yes | ++ | HCM, LVNC |
| CSRP3 | Muscle LIM protein | B | 28.93 | 0.00 | −0.66 | ++ | Yes | ++ | HCM |
| CRYAB | αβ-crystallin | B | 35.42 | 0.01 | 0.38 | ++ | Yes | +++ | MM, DM, cataracts |
| ACTC1 | Cardiac actin | B | 38.28 | 0.95 | 5.25 | ++ | Yes | +++ | HCM, RCM, LVNC, CHD |
| NEXN | Nexilin | B | 50.34 | 0.00 | −1.32 | ++ | NA | ++ | HCM, CHD |
| TAZ | Tafazzin | B | 59.76 | 0.97 | 2.42 | +++ | Yes | ++ | Barth syndrome (may present as infantile X-linked DCM), LVNC, EFE |
| LAMP2 | Lysosome-associated membrane glycoprotein 2 | B | 61.73 | 0.95 | 0.41 | ++ | Yes | + | Danon disease (may present as HCM or DCM) |
| TNNI3 | Troponin I | B | 64.11 | 0.17 | 1.88 | ++ | Yes | + | HCM, RCM, AF |
| ANKRD1 | Cardiac ankyrin repeat protein | B | 65.76 | 0.00 | −0.01 | ++ | Yes | No | HCM, CHD |
| DSG2 | Desmoglein-2 | B | 98.32 | 0.00 | −1.20 | ++ | NA | +++ | ARVC |
| MYH6 | Myosin heavy chain 6 | C | 0.66 | 0.00 | 2.87 | ++ | NA | ++§ | HCM, CHD, SSS |
| ABCC9 | KATP channel SUR2A subunit | C | 1.82 | 0.00 | 4.89 | + | Yes | ++ | AF, BS, Cantu syndrome, coronary vasospasm |
| ACTN2 | α-actinin | C | 2.47 | 1.00 | 1.76 | + | Yes | + | HCM |
| PDLIM3 | PDZ and LIM domain protein 3 | C | 20.70 | 0.00 | −0.04 | + | NA | ++ | |
| SGCD | δ - sarcoglycan | C | 34.32 | 0.00 | −0.23 | ++ | Yes | ++ | LGMD-2F |
| SYNE1 | Nesprin-1 | C | 41.65 | 0.00 | −0.95 | + | Yes | + | EDMD, cerebellar ataxia, autism, arthrogryposis |
| FXN | Frataxin | C | 76.67 | 0.82 | 0.47 | + | NA | ++ | Friedreich’s ataxia (may develop LVH + late DCM) |
| CAV3 | Caveolin-3 | C | 77.70 | 0.34 | 1.19 | + | Yes | ++ | HCM, LQTS, SIDS, LGMD-1C, RMD-2, DM, HCK |
| TCAP | Telethonin | C | 78.28 | 0.08 | 0.45 | + | Yes | + | HCM, LGMD-2G |
| FKTN | Fukutin | C | 82.25 | 0.00 | −0.64 | ++ | Yes | NA | LGMD-2M (may present as DCM) |
| SDHA | Succinate dehydrogenase flavoprotein subunit | D | 10.95 | 0.00 | 2.32 | ++ | Yes | NA | Leigh syndrome (rarely associated with pediatric recessive DCM) |
| DTNA | α-dystrobrevin | D | 21.65 | 0.92 | 1.17 | + | NA | No | LVNC, CHD |
| LAMA2 | Laminin subunit α-2 | D | 79.64 | 0.00 | −1.58 | + | NA | NA | Merosin-deficient congenital MD/LGMD (rarely includes DCM) |
| SYNM | Synemin | D | NA | 0.00 | −0.15 | NA | NA | NA | Accumulates in skeletal myopathy |
Genetic evidence: +++ co-segregation of variants in multiple (3+) families, with at least one family showing variant co-segregation in 5 or more affected individuals; ++ co-segregation of variant in 1 or more small families (<5 affected); + variant demonstrated in single cases (with/without positive family history), or SNPs associated with DCM susceptibility.
Animal models: +++ Spontaneous development of DCM in animal model with a heterozygous gene loss-of-function or expressing a human disease-associated non-synonymous variant; ++ spontaneous DCM present in homozygous gene loss-of-function model, or stress-induced DCM in model of a human non-synonymous variant; + No DCM but other relevant phenotypes, eg. isolated ventricular dilation or contractile dysfunction, evidence of reduced contractile reserve, skeletal myopathy. See Supplemental Table X for more details.
α-MHC is major myosin isoform in murine ventricle: human MYH7 mutations modelled in murine myh6 gene.
AF, atrial fibrillation; ARVC, arrhythmogenic right ventricular cardiomyopathy; BMD, Becker muscular dystrophy; BS, Brugada syndrome; CD, conduction-system defects; CHD, congenital heart disease; CM, congenital myopathy; CNM, centronuclear myopathy; DCM, dilated cardiomyopathy; DM, distal myopathy; DMD, Duchenne muscular dystrophy; EDMD, Emery-Dreifuss muscular dystrophy; EFE, endomyocardial fibroelastosis; HCK, high creatine kinase; HCM, hypertrophic cardiomyopathy; HMERF, hereditary myopathy with early respiratory failure; LGMD, limb girdle muscular dystrophy; LQTS, long QT syndrome; LVH, left ventricular hypertrophy; LVNC, left ventricular non-compaction; MD, muscular dystrophy; MM, myofibrillar myopathy; MSM, myosin storage myopathy; NA, not available; NSS, neurogenic scapuloperoneal syndrome; RCM, restrictive cardiomyopathy; RMD, rippling muscle disease; SIDS, sudden infant death syndrome; SPM, scapuloperoneal myopathy; SSS, sick sinus syndrome; TMD, tibial muscular dystrophy; VF, ventricular fibrillation.
Residual Variation Intolerance Score (RVIS).
Another metric used to assess the intolerance of genes to sequence variation is RVIS.17 Genes that cause Mendelian disorders are expected to show little variation in the general population (25th percentile), with those associated with complex disorders being highly variable (100th percentile). The RVIS model predicts that disease-causing rare variants would be relatively enriched in 25th percentile genes, and indeed, 18 genes of 40 cardiomyopathy genes were in this percentile bin (Table 1). However, while DCM-associated truncating variants were preferentially distributed in 25th percentile genes (P = 0.01, 2 x 4 chi-squared test; Supplementary Table S4), this was not the case for missense variants (P = 0.67).
Variant burden per gene.
There were only three genes, RBM20, MYH7, and LMNA, in which there were significantly more rare variants in DCM cases than in controls (Supplementary Table S1). These findings strongly suggest that these genes harbor pathogenic rare variants, but do not directly inform the interpretation of any single variant. For most genes, the relatively small numbers of variants in both cases and controls limited statistical comparisons.
DCM gene ranking.
To focus more specifically on DCM pathogenesis, we undertook a literature search and allocated grades of A (strong) to D (weak) for the strength of genetic, in vitro, and in vivo evidence of disease association (Table 1, Supplementary Table S5). Only 14 of 41 genes were classified as group A. There was a prominent peak of DCM-associated truncating variants in these genes, with control-associated truncating variants mostly found in gene groups C and D (P = 0.01, 2 x 4 chi-squared test; Supplemental Table S6; Figure 1B). DCM-associated missense variants also showed a peak in group A genes (P = 0.018), with marked enrichment of variants that were novel (ExAC) + deleterious (MetaSVM) (P = 0.001, Figure 1C). Adding the “group A” gene parameter to these variant parameters increased the odds ratio for DCM from approximately 5-fold to nearly 9-fold (Figure 1A).
Yield of variants per person
We next looked at the yield per person of prioritized variants (Table 2). For this analysis, truncating variants and “novel (ExAC) + deleterious (MetaSVM)” missense variants are referred to as “damaging” group A gene variants (Supplementary Table S7). Damaging variants were present in 65 (12.2%) DCM cases and in 8 (1.5%) control subjects (P < 0.0001). Similar patterns for the prevalence of damaging group A variants were seen in an independent cohort of familial DCM cases (17 probands [16.8%]), and in two replication control cohorts from the Alzheimer’s Disease Sequencing project (54 subjects [1.8%]) and the Medical Genome Reference Bank (33 subjects [2.9%]).
Table 2.
Yield per person of TTNtv and damaging* group A gene variants in the discovery cohort and in replication cohorts.
| Variants | DCM cases | Control subjects | ||||
|---|---|---|---|---|---|---|
| Discovery (n=532) |
FDCM (n=101) |
Discovery (n=527) |
Alzheimer’s (n=2971) |
MGRB (n=1144) |
||
| Single TTNtv | 85 | 15 | 6 | 6 | 13 | |
| Two TTNtv | 1 | 0 | 0 | 0 | 0 | |
| TTNtv + group A trunc | 1 | 0 | 0 | 0 | 0 | |
| TTNtv + group A missense | 5 | 1 | 0 | 0 | 1 | |
| Group A trunc only | 13 | 6 | 2 | 9 | 3 | |
| Group A trunc + missense | 0 | 1 | 0 | 0 | 0 | |
| Group A missense only | 46† | 9 | 6 | 46 | 29 | |
| None | 381 | 69 | 513 | 2910 | 1098 | |
Damaging variants included truncating (trunc) or “novel (ExAC) + deleterious (MetaSVM)” missense variants in group A genes (TTN excluded).
Three cases had 2 variants. ADSP, Alzheimer’s Disease Sequencing Project; FDCM, familial dilated cardiomyopathy; MGRB, Medical Genome Reference Bank.
When TTNtv were also considered, 151 (28.4%) of our DCM cases were positive for damaging variants and/or TTNtv compared to 14 (2.7%) controls (P < 0.0001). Damaging variants and/or TTNtv were present in 32 (31.7%) probands in the familial DCM replication cohort, and in 61 (2.1%) and 46 (4.0%) subjects, respectively in the two control replication cohorts. It has been questioned whether TTNtv are sufficient alone to cause DCM or require “second hit” genetic and/or acquired factors for DCM manifestation. We found no differences in the background burden of variants in TTNtv+ DCM cases (1.58 ± 1.74 variants, range 0-9) when compared with TTNtv-/damaging+ DCM cases (1.17 ± 1.22 variants, range 0-5), TTNtv-/damaging- DCM cases (1.30 ± 1.55 variants, range 0-13), or TTNtv-/damaging-control subjects (1.20 ± 1.29, range 0-8; P = 0.3819, Kruskal-Wallis test). These data suggest that TTNtv carriers generally do not show an excess of second damaging variants.
Family segregation
To further test our criteria for rare variant prioritization, we performed co-segregation analysis in 28 DCM families in which DNA samples from 3 or more informative individuals were available. All rare variants that were identified in each family proband were evaluated in the respective family members.
Truncating group A gene variants.
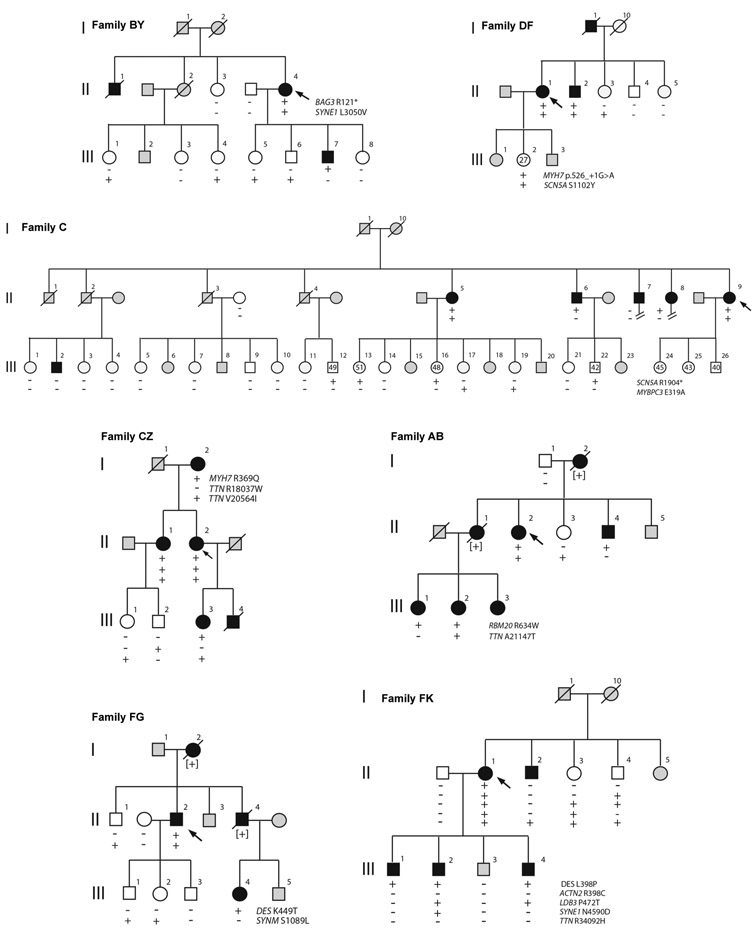
Truncating variants in group A genes were evaluated in 3 families (Figure 2). In Family BY, the proband carried a BAG3 stop codon with segregation analysis consistent with linkage (odds ratio 1/18, LOD score 1.3). The truncated protein would lack the signature BAG domain that binds to HSP70 and is required for chaperone activity.18 Numerous BAG3 truncating mutations have been reported in DCM patients, and reduced levels of BAG3 protein have been seen in ventricular tissues from patients with heart failure.19-21
Figure 2.
Family co-segregation analysis. Pedigrees showing genotype-phenotype correlations for rare variants identified in family probands. The presence (+) or absence (−) of variants in relatives is shown. DCM status is denoted as affected (solid symbols), unaffected (open symbols), or unknown (gray symbols); probands are indicated by arrows.
The Family DF proband and her affected brother carried a MYH7 splice acceptor site variant. This variant was absent from 3 unaffected individuals aged >40 years, but the family size was insufficient to show statistically-significant linkage (odds ratio 1/6, LOD score 0.8). MYH7 truncating variants were found in 2 additional sporadic DCM cases in our cohort (Supplementary Table S7) but have rarely been reported in primary DCM cases,12,22 and there has been uncertainty about their clinical significance.
The Family C proband carried a SCN5A stop codon that would truncate the C-terminus with loss of a PY-motif (binding site for Nedd4 ubiquitin ligase) and a PDZ-domain binding motif (interacts with the cytoskeletal adapter protein, syntrophin), potentially giving rise to protein degradation or trafficking defects 23,24 The cardiac phenotype associated with SCN5A mutations often includes arrhythmias and conduction-system abnormalities as well as DCM,25,26 and atrial fibrillation was notably present in all affected individuals in Family C. This SCN5A truncation did not segregate with disease (LOD score −2.5) but this may be confounded by two affected genotype-negative individuals who had other possible acquired causes of DCM.
Missense variants.
Segregation of 30 missense variants was evaluated in 14 kindreds (Supplementary Table S8). Only 4 of the 30 variants had LOD scores >1 (suggestive of linkage relative to family size) and all of these were damaging group A gene variants (Figure 2). Affected individuals in Family CZ carried a p.Arg369Gln MYH7 variant and were noted to have DCM as well as left ventricular non-compaction. This variant, in the myosin motor domain, has previously been associated with both phenotypes.6,27 The p. Arg634Trp RBM20 variant, present in all affected individuals in Family AB, lies within the arginine-serine-rich (RS) domain which is a putative DCM mutation hotspot.28,29 There were also two co-segregating DES variants, p.Leu398Pro (Family FK) and p.Lys449Thr (Family FG), the latter associated previously with myofibrillar myopathy.30 Four additional kindreds (Families BG, BK, FR, KS) had damaging group A gene variants with LOD scores <1 (Supplementary Figure 2). Despite these low scores, the damaging variants were present in 15 of 16 affected individuals in these families. Apparent genotype-phenotype discordance was mainly attributable to unaffected variant carriers and might be reflective of age and sex effects on penetrance. In all families tested, there was no evidence for co-segregation of any variants did not meet the criteria for being “damaging” or that were in genes other than group A.
Multiple variants.
With expanded genetic testing panels, it is not uncommon to find several rare variants in DCM cases, prompting hypotheses of multiple mutations.7,31 In an extension of the current analysis, we selected 5 of the DCM kindreds and evaluated rare variants in all of the genes in the original 69-gene testing panel as well as increasing the threshold level of MAF to < 1%. In 4 families (Families BY, FK, AB, BA), although the proband carried 5-7 variants, only one of these segregated with DCM in each kindred (Supplementary Figure S3). In the remaining family (Family GX), the proband had 5 variants, none of which segregated with disease. Four of these rare variants were inherited from the proband’s unaffected father, and none of these were damaging group A variants. These studies demonstrate the value of family analysis and support the expectation that a single “driver” variant will be present when DCM appears as a Mendelian trait.
Location of variants in group A genes
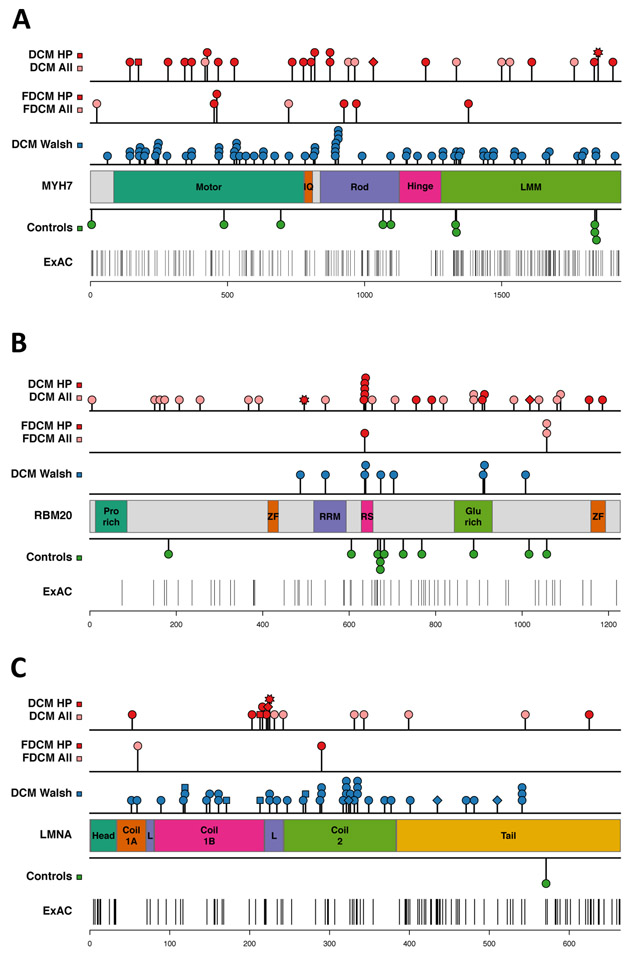
To explore potential effects of variant location within group A genes, we compared the distribution of missense variants identified in DCM cases (derived from our discovery and replications and two clinical laboratories12) with rare (MAF < 0.1%) missense variants in the ExAC database (Figure 3, Supplementary Figure S3, Supplementary Table S9). In MYH7, DCM variants occurred in all protein regions but were preferentially located in the myosin motor (Fig. 3A). DCM-associated variants in RBM20 were significantly enriched in the RS domain that has been associated with changes in titin splicing and myocardial compliance (Figure 3B).28,29 More than half of the damaging RBM20 variants resided in the RS domain or in a glutamate-rich region that is also associated with titin splicing defects.32 DCM-associated variants in LMNA were mostly located in the coiled-coil rod domain, particularly in coil 2, which is critical for dimer formation (Figure 3C),33 and there was a cluster of DCM-associated variants in the S4 voltage-sensor, repeat I, of SCN5A (Supplementary Figure S4B). Variants in the S4 transmembrane segments cause arrhythmic forms of DCM and have been associated with gain-of-function effects and gating pore currents25,26,34-36 Clustering of DCM variants was also apparent in the α-tropomyosin binding domain of TNNT2, and several damaging variants in TPMI resided in the cardiac troponin T binding domain (Supplementary Figure S4D and S4E).
Figure 3.
Rare variant distribution in DCM cases and control subjects. The location of variants in different protein domains is shown for the group A genes: (A) MYH7, (B) RBM20, (C) LMNA. Variants identified in DCM patients in the discovery cohort (top row) and in the familial DCM (FDCM) replication cohort (second row) are shown: all variants (pink, “DCM all”), damaging variants (red, “DCM HP”). Variant types are indicated by: circle =missense, square = splice site change, diamond = frameshift, star = stop codon. The third row shows DCM variants identified by two clinical diagnostic laboratories (“DCM Walsh”)12. Missense variants identified in control subjects are shown below the protein schematic: control subjects in this study (Controls), Exome Aggregation Consortium (ExAC) database. See Supplementary Table S9 for protein domain coordinates and statistical analysis.
DISCUSSION
Rare protein-altering variants in cardiomyopathy genes that are discovered in affected patients have been considered potentially disease-causing. Primary evaluation of disease cohorts is subject to ascertainment bias however, and our data confirm that rare variants in cardiomyopathy genes are highly prevalent in control cohorts.1-4 Here we show the importance of combining variant-level and gene-level information and provide a new strategy for assessment of rare variants in the clinical context of DCM.
Metrics such as pLI, Z scores, and RVIS provide gene-level information about the expected frequency of sequence variation in the general population.16,17 A limitation of these metrics is that they are unable to take each gene’s specific role in cardiomyocyte biology into account and cannot be used to infer disease mechanisms. For example, a truncating variant in a gene with a high pLI score might be directly relevant to DCM if that gene is known to be critical for myocardial contraction, or have modest, if any, relevance for genes with non-essential or redundant functions. Similar arguments can be made for missense variants in genes with high Z scores. For some genes, loss-of-function may be a recognized mechanism of disease, while for others, dominant negative effects of missense variants might be relatively more important. It is worth bearing in mind that function-altering variants may not always have deleterious effects and that some might be protective. Genes with high intolerance scores may be subject to extreme selective constraint, hence loss-of-function variants may be more likely to manifest as severe pediatric-onset disease rather than adult-onset DCM. In our data, there was no consistent relationship between metrics of expected genetic variation and DCM, findings that help to underscore the need to consider the fundamental roles of individual genes in disease causation.
We ranked the 41 genes in our dataset based on published genetic and functional data and found robust evidence for DCM causation for only 14 of these genes. There are a number of factors, however, that influence gene scoring, including the relative richness of the knowledge base. Genetic data can provide a compelling case to support disease association and genes in which variants co-segregated with DCM status in large kindreds (≥5 affected) scored highly in our system. For many genes, only small families or single cases have been studied, raising the possibility that variants, even if function-altering, might co-segregate by chance or be incidental findings. Assessment of genetic evidence for DCM is intrinsically biased by the size of the kindreds evaluated, the frequency in which specific genes have been screened, and available literature reports. Although positive data are useful, no conclusions can be drawn if there is insufficient information. Similarly, in vitro or in vivo functional data for human variants are helpful if these are available but are often lacking from the literature. Most of the genes that achieved group A status had animal data showing spontaneous development of DCM in models expressing heterozygous loss-of-function alleles or human missense mutations. This level of evidence was missing from many studies in which only homozygous loss-of-function animal models have been looked at. While such models can implicate genes in normal cardiac function, it cannot be assumed that there is s direct correlation with gene “dose” and that heterozygous counterparts will have a similar, albeit less severe, phenotype. In homozygous loss-of-function animals, there may also be profound effects on cardiac development that independently predispose to contractile impairment. Despite these limitations, incorporation of gene group proved to be a powerful discriminator of rare variants.
The group A genes MYH7, RBM20, and LMNA, showed significant differences in rare variant numbers between DCM cases and controls, and had a predilection for damaging rare variants. MYH7 and LMNA have consistently appeared on lists of the “most frequently mutated” genes in DCM genetics studies, and were the top 2 genes (after TTN) showing an excess of rare variants (MAF <0.0001) in a recent analysis of 1315 DCM patients referred to two diagnostic genetic testing laboratories.6,7,12,22 RBM20 is a relatively more recent addition to the DCM disease gene list and has been less frequently screened. Despite their overall enrichment in DCM cases, not all rare variants in MYH7, RBM20, and LMNA are necessarily deleterious and additional information is required for clinical interpretation of any specific variant. Interestingly, variants in these genes had a non-random distribution in DCM cases: MYH7 variants were more likely to reside in the myosin motor, RBM20 variants in the RS domain, and LMNA variants in the coiled-coil rod. As the numbers of reported DCM-associated variants expands, further evaluation of patterns of variant distribution should be informative.
Our method for stratifying rare variants provides a framework for future studies and ongoing refinements can be anticipated. For missense variants, the criterion of absence from ExAC may underestimate the yield of potentially pathogenic variants and varying MAF threshold levels need to be evaluated. Gene group is a dynamic variable and the status of genes is likely to change as more families are studied and more animal models generated. It can be expected that a number of genes previously associated with DCM but not included in our current analysis would also achieve group A status. Although pLI, Z-scores and RVIS did not provide incremental information over gene group for differentiating variants in genes already associated with disease, more nuanced interpretations may be possible with consideration of protein subdomains. These metrics may also prove to be useful in whole-exome or whole-genome analysis pipelines for prioritizing variants that occur in genes with unknown roles in heart function. In assessing individual burden of genetic variation, only rare cardiomyopathy gene variants were considered. The extent to which rare, low-frequency and common variants in a broad range of cardiac genes might cumulatively contribute to a myopathic substrate is unknown.
Understanding which genes are important in DCM pathogenesis is a vital starting point for interpretation of genetic testing results and for identification of clinically-actionable variants. Efforts by authoritative bodies such as the American College of Medical Genetics to refine and standardize variant annotation methods are underway and will play a important role in establishing international guidelines for genetic testing in DCM and other inherited human disorders.37 The cost-efficacy of first-line screening of a core set of key DCM disease genes (e.g. group A genes) vs extended panels or genome-wide testing warrants further analysis. As genome sequencing is poised to become part of mainstream healthcare, there will be an ongoing need for curation of clinical and genetic information in databases such as ClinGen,38 functional evaluation of variants and clinical trials in genotyped patients. Advancements in these areas should expedite implementation of personalized medicine.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erica Mazaika, Monique Ohanian, Santiago Pineda, Celine Santiago. Magdalena Soka, and Gunjan Trivedi for assistance with data collection, collaborating physicians who referred patients to the study and participating families. Data for healthy elderly subjects were obtained from the Medical Genome Research Ban (see Supplementary Acknowledgments).
FUNDING SOURCES
This study was supported by grants from the National Health and Medical Research Council of Australia (D.F., P.S.M.), Harvard Club of Australia Foundation (D.F.), Estate of the Late RT Hall (D.F.), Simon Lee Foundation (D.F.), British Heart Foundation (S.A.C.), National Institute for Health Research Cardiovascular BRU at the Brompton and Harefield & Harefield NHS Foundation Trust and Imperial College London (S.A.C., P.J.B.), Howard Hughes Medical Institute (C.E.S.), National Institutes of Health (D.S.H., J.G.S.), and the Leducq Foundation (S.A.C., J.G.S., C.E.S.).
Footnotes
DISCLOSURE
The authors declare no conflict of interest.
REFERENCES
- 1.Pan S, Caleshu CA, Dunn KE, et al. Cardiac structural and sarcomere genes associated with cardiomyopathy exhibit marked intolerance of genetic variation. Circ Cardiovasc Genet 2012;5:602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton N, Robertson PD, Rieder MK, et al. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet 2012;5:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet 2012;5:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haggerty CM, James CA, Calkins H, et al. Electronic health record phenotype in subjects with genetic variants associated with arrhythmogenic right ventricular cardiomyopathy: a study of 30,716 subjects with exome sequencing. Genet Med 2017;19:1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatkin D, Seidman CE, Seidman JG. Genetics and disease of ventricular muscle. Cold Spring Harb Perspect Med 2014;4;a021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med 2014:16:601–608. [DOI] [PubMed] [Google Scholar]
- 7.Haas J, Frese KS, Peil B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J 2015;36:1123–1135. [DOI] [PubMed] [Google Scholar]
- 8.McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res 2017;121:731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acid Res 2012;40:W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 2015;24:2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh R, Thomson KL, Ware JS, et al. Exome Aggregation Consortium, MacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 1998;280:750–752. [DOI] [PubMed] [Google Scholar]
- 14.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 1999;341:1715–1724. [DOI] [PubMed] [Google Scholar]
- 15.Kamisago M, Sharma SD, dePalma SR, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med 2000;343:1688–1696. [DOI] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013;9:e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hishiya A, Kitazawa T, Takayama S. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ Res 2010;107:1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villard E, Perret C, Gary F, et al. A genome-wide association study identified two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J 2011;32:1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton N, Li D, Rieder MJ, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet 2011;88:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldman AM, Begay RL, Knezevic T, et al. Decreased levels of BAG3 in a family with a rare variant and in idiopathic dilated cardiomyopathy. J Cell Physiol 2014;229:1697–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayvanpour E, Sedaghat-Hamedani F, Amr A, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol 2017;106:127–139. [DOI] [PubMed] [Google Scholar]
- 23.Van Bemmelen MX, Rougier JS, Gavillet B, et al. Cardiac voltage-gated sodium channel Nav1.5 is regulated by Nedd4-2 mediated ubiquitination. Circ Res 2004;95:284–291. [DOI] [PubMed] [Google Scholar]
- 24.Gavillet B, Rougier JS, Domenighetti AA, et al. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res 2006;99:407–414. [DOI] [PubMed] [Google Scholar]
- 25.McNair WP, Ku L, Taylor MR, et al. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation 2004;110:2163–2167. [DOI] [PubMed] [Google Scholar]
- 26.Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 2005;293:447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dellefave LM, Pytel P, Mewborn S, et al. Sarcomere mutations in cardiomyopathy with left ventricular hypertrabeculation. Circ Cardiovasc Genet 2009;2:442–449. [DOI] [PubMed] [Google Scholar]
- 28.Brauch KM, Karst ML, Herron KJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol 2009;54:930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo W, Schafer S, Greaser ML, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med 2012;18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selcen D, Ohno K, Engel AG. Myofibrillar myopathy: clinical, morphological and genetic studies in 63 patients. Brain 2004;127:439–451. [DOI] [PubMed] [Google Scholar]
- 31.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. [DOI] [PubMed] [Google Scholar]
- 32.Beqqali A, Bollen IA, Rasmussen TB, et al. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc Res 2016;112:452–463. [DOI] [PubMed] [Google Scholar]
- 33.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 2015;84:131–164. [DOI] [PubMed] [Google Scholar]
- 34.McNair WP, Sinagra G, Taylor MR, et al. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol 2011;57:2160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann SA, Castro ML, Ohanian M, et al. R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J Am Coll Cardiol 2012;60:1566–1573. [DOI] [PubMed] [Google Scholar]
- 36.Moreau A, Gosselin-Badaroudine P, Boutjdir M, Chahine M. Mutations in the voltage sensors of domain I and II of Nav1.5 that are associated with arrhythmias and dilated cardiomyopathy generate gating pore currents. Front Pharmacol 2015;6:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rehm HL, Berg JS, Brooks LD, et al. ClinGen – the clinical genome resource. N Engl J Med 2015;372:2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.