Summary
Soybean cyst nematode (SCN, Heterodera glycines) is the most devastating pest affecting soybean production worldwide. SCN resistance requires both the GmSHMT08 and the GmSNAP18 in ‘Peking’‐type resistance. Here, we describe the molecular interaction between GmSHMT08 and GmSNAP18, which is potentiated by a pathogenesis‐related protein GmPR08‐Bet VI. Like GmSNAP18 and GmSHMT08, GmPR08‐Bet VI expression was induced in response to SCN and its overexpression decreased SCN cysts by 65% in infected transgenic soybean roots. Overexpression of GmPR08‐Bet VI did not have an effect on SCN resistance when the two cytokinin‐binding sites in GmPR08‐Bet VI were mutated, indicating a new role of GmPR08‐Bet VI in SCN resistance. GmPR08‐Bet VI was mapped to a QTL for resistance to SCN using different mapping populations. GmSHMT08, GmSNAP18 and GmPR08‐Bet VI localize to the cytosol and plasma membrane. GmSNAP18 expression and localization hyper‐accumulated at the plasma membrane and was specific to the root cells surrounding the nematode in SCN‐resistant soybeans. Genes encoding key components of the salicylic acid signalling pathway were induced under SCN infection. GmSNAP18 and GmPR08‐Bet VI were also induced under salicylic acid and cytokinin exogenous treatments, while GmSHMT08 was induced only when the resistant GmSNAP18 was present, pointing to the presence of a molecular crosstalk between SCN‐resistant genes and defence genes. Expression analysis of GmSHMT08 and GmSNAP18 identified the need of a minimum expression requirement to trigger the SCN resistance reaction. These results provide insight into a new response mechanism towards plant nematode resistance involving haplotype compatibility, gene dosage and hormone signalling.
Keywords: SCN‐R model, SCN mechanism, protein–protein interaction, molecular crosstalk, gene dosage effect, molecular trafficking, salicylic acid, cytokinins
Introduction
The serine hydroxymethyltransferase (SHMT; E.C. 2.1.2.1) plays a key role in one‐carbon metabolism. It is involved in the interconversion of serine/glycine and tetrahydrofolate (THF)/5,10‐methylene THF, impacting the de novo purine pathway cellular methylation reactions, redox homeostasis maintenance and methionine and thymidylate synthesis (Appaji Rao et al., 2003; Mouillon et al., 1999; Schirch, 1982; Stover, 1990). Consequently, in humans, mutations in the SHMT have been linked to a wide range of diseases (Lim et al., 2005; Maddocks et al., 2016; Skibola et al., 2002). shmt knockdown mutants induce apoptosis in lung cancer cells by causing uracil misincorporation (Paone et al., 2014). In plants, SHMTs play an essential role in the metabolic reactions of photorespiration, which is primordial for C3 plants (McClung et al., 2000; Somerville and Ogren, 1981; Wei et al., 2013). SHMTs play a role in the maintenance of redox homeostasis, involving glutathione synthase and peroxidases (Maddocks et al., 2016). In plant–pathogen resistance, GmSHMT08 was identified as the gene conferring resistance to soybean cyst nematode (SCN) (Kandoth et al., 2017; Lakhssassi et al., 2019; Liu et al., 2012). The identification of an SHMT as a plant resistance gene is unique (Liu et al., 2012). It has been suggested that two Gmshmt08 mis‐sense mutants with altered enzymatic properties may have negative effects leading to a hypersensitive response resulting in necrosis and cell death in the nematode feeding cell, the syncytium (Liu et al., 2012; Mahalingam and Skorupska, 1996).
Soluble NSF attachment proteins, SNAPs, are characterized by the presence of a tetratricopeptide repeat (TPR) domain that is shared by a large number of proteins in diverse species including human, yeast, bacteria and plants (D'Andrea and Regan, 2003). In recent years, plant proteins containing TPRs have been found to be essential for responses to hormones such as ethylene, cytokinin, gibberellin, salicylate and auxin in Arabidopsis (Wang et al., 2004; Yoshida et al., 2005). In addition, many cellular functions such as protein folding, cycle regulation, neurogenesis, gametophytic viability, root growth and integrity, mitochondrial and peroxisomal protein transport and protein–protein interactions have been assigned to TPR proteins (Blatch and Lassle, 1999; Lakhssassi et al., 2012a). Mutations in TPR proteins have been found to cause several human diseases, indicating essential roles in cell function (Sohoki et al., 2000). Mutations at the TPR domain of the 17p and p67phox proteins can cause several diseases such as Leber congenital amaurosis and chronic granulomatous, respectively (Grizot et al., 2001; Sohoki et al., 2000). Because of their role in protein–protein interactions, the identification of binding partners is a common strategy for understanding the molecular function of TPR proteins (D'Andrea and Regan, 2003). α‐SNAP proteins are conserved across yeast, animals and plants and are key components of the cellular fusion machinery being involved in sustaining membrane trafficking by disassembling soluble NSF attachment protein receptor (SNARE) complexes that form during membrane fusion (Clary et al., 1990; Marz et al., 2003). In soybean, it has been suggested that expression of SCN resistance‐type rhg1 α‐SNAPs depleted the abundance of SNARE‐recycling 20S complexes, disrupting vesicle trafficking and causing cytotoxicity (Bayless et al., 2016), but the expression of other loci encoding a canonical wild‐type α‐SNAP counteracted the cytotoxicity of resistance‐type rhg1 α‐SNAP (Bayless et al., 2018). However, as of today, the molecular partners and the mechanism of action involving cytotoxicity remain elusive.
Recently, GmSNAP18 and the GmSHMT08 were identified as the Peking‐type rhg1‐a and Rhg4 genes conferring resistance to SCN (Liu et al., 2017; Liu et al., 2012), but not necessarily for PI88788‐type resistance, which requires other proteins including an α‐SNAP, a wound‐inducible domain protein (WI12), and an amino acid transporter (Cook et al., 2012). However, the SCN resistance mechanism and the interacting partners of GmSHMT08 and/or GmSNAP18 have not yet been revealed. The identification of the GmSHMT08 and/or GmSNAP18 molecular partners is indeed crucial for understanding its molecular function and revealing the upstream SCN resistance pathway. Recently, we have demonstrated through whole‐genome re‐sequencing of 106 soybean lines, the impact of copy number variants at both the rhg1 and Rhg4 genes on broad‐based resistance to SCN (Patil et al., 2019). These results provided new insight into epistasis, haplotype compatibility, copy number variation, promoter variation and their impact on broad‐based disease resistance to SCN. Thus, the copy number variations of GmSHMT08 and GmSNAP18 play a major role in SCN resistance.
Pathogenesis‐related proteins (PRs) are widely present in plants and are induced following pathogen attack, elicitors, wounding or stress. Non‐induced pathogenesis‐related genes (NPR1) regulate systemic acquired resistance via regulation of the PR in Arabidopsis thaliana. It has been shown that NPR1 interacts with the transcription factor TGA2 to modulate the expression of some plant defence genes, such as PR‐1 and PR5 (Boyle et al., 2009; Matthews et al., 2014a). Although their precise role is unknown, PRs are involved in systematically acquired resistance and stress responses in plants. PR‐10‐silenced plants exhibit lower accumulation of H2O2 and down‐regulation of PR‐1, defensin 1 (Def1), systematic acquired resistance (SAR82) and a peroxidase (PO2). PR‐10 also acts as a reservoir of cytokinin molecules used to combat pathogens (Fernandes et al., 2008; Pasternak et al., 2006). In addition, cytokinins were reported to play a role during plant–pathogen interaction. In Arabidopsis, nematodes have been shown to release cytokinins that control cell division and orchestrate feeding site formation in host plants (Siddique et al., 2015).
The current study not only demonstrates an unprecedented molecular interaction between the GmSNAP18 and GmSHMT08 proteins, which involves a unique gene dosage requirement, but also identifies a pathogenesis‐related protein, GmPR08‐Bet VI, as a novel molecular partner for both GmSNAP18 and GmSHMT08 impacting soybean resistance to SCN.
Results
Subcellular localization and interaction of GmSNAP18 and GmSHMT08 proteins in planta
Soybean cyst nematode resistance has been shown to be bigenic in the ‘Peking’ type of resistance, requiring both the Rhg4 and rhg1 loci (Meksem et al., 2001). Since GmSHMT08 and GmSNAP18 are the genes at these loci that underlie resistance to SCN (Liu et al., 2017; Liu et al., 2012), we hypothesize that the products of these two genes may interact directly or via other intermediators. The physical GmSHMT08/GmSNAP18 association requires both proteins to be present in the same cellular compartment. Recently, we have demonstrated that GmSHMT08c was localized to the cytosol (Lakhssassi et al., 2019). However, subcellular localization of GmSNAP18 remained to be determined. Therefore, we examined the subcellular localization of the GmSNAP18 protein, in addition to the GmSHMT08 (used as positive control) using two systems: the YFP fusion in onion epidermal cells (Figure 1a) and RFP fusion in Nicotiana benthamiana leaves (See Figure S1). Both YFP‐tagged and RFP‐tagged GmSNAP18 showed similar localization patterns to that of GmSHMT08 and were located in the cytosol and plasma membrane of the infiltrated cells, highlighting the possibility of their physical interaction.
Figure 1.
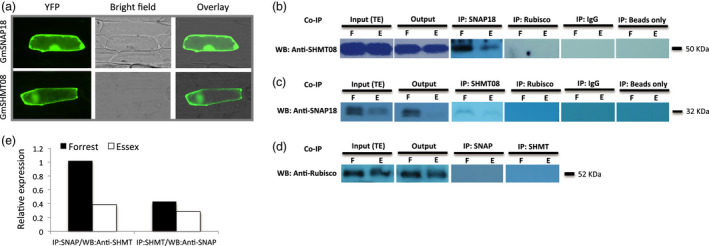
Subcellular localization and interaction analyses of GmSNAP18 and GmSHMT08 proteins by co‐immunoprecipitation (Co‐IP). (a) The coding sequences of GmSNAP18 and GmSHMT08 (used as positive control) were fused to the N‐terminal end of the eYFP and delivered into onion epidermal cells using biolistic bombardment. YFP fluorescence was localized in the cytoplasm and plasma membrane for GmSNAP18 like GmSHMT08. (b) IP: The total protein extracts of soybeans Forrest and Essex from SCN‐infected roots were immunoprecipitated with anti‐SNAP18 polyclonal antibodies (PA) or anti‐Rubisco PA (used as negative control). Blots from the eluted fraction were probed with anti‐SHMT08 PA or anti‐Rubisco PA. (c) The total protein extracts of roots of soybeans Forrest and Essex were immunoprecipitated with anti‐SHMT08 PA or anti‐Rubisco PA (as the control). Blots from the eluted fraction were probed with anti‐SNAP18 PA or anti‐Rubisco PA. Total protein extracts from input (TE) and output were blotted as well. (d) No band observed when using anti‐Rubisco as a negative control from both Co‐IP:SHMT08 and Co‐IP:SNAP18. Only IgG or beads were used for Co‐IP experiments as a negative control and technical control, respectively. The Co‐IP results indicated that GmSNAP18 interacts physically with GmSHMT08 and vice versa in two independent pull‐down experiments. (e) Relative expression of the GmSHMT08 and GmSNAP18 interaction intensity between Forrest and Essex from SDS‐PAGE was measured using ImageJ software and normalized using the Rubisco expression as reference. WB, Western blot.
To further explore this hypothesis, co‐immunoprecipitation experiments were conducted in the SCN‐resistant line Forrest and the SCN‐susceptible line Essex from SCN‐infected roots. The association between GmSNAP18 and GmSHMT08 (and vice versa) in vivo was tested in two independent pull‐down experiments. Immobilized anti‐GmSNAP18 antibodies were used in a chromatography column to capture GmSNAP18 and any GmSNAP18‐interacting proteins from total soybean root protein extract. Under SDS denaturing gel conditions, Western blots of the eluted fraction using anti‐GmSHMT08 antibody showed the presence of strong GmSHMT08 binding at ~50 KDa in Forrest when compared to Essex and the absence of this band in the co‐immunoprecipitation assay using anti‐Rubisco antibodies as a negative control (Figure 1b). In reciprocal pull‐down assays, immobilized anti‐GmSHMT08 antibodies were used, and then, Western blots of the eluted root proteins were performed using anti‐GmSNAP18 antibodies. The results showed a greater presence of GmSNAP18 hybridization at ~32 KDa in Forrest than in Essex (Figure 1c). No bands were detected when the anti‐Rubisco antibodies were used (Figure 1b–d). Thus, GmSHMT08 and GmSNAP18 were correspondingly able to immunoprecipitate in the root protein extract.
Co‐immunoprecipitation of GmSNAP18 and the cytosolic GmSHMT08 was also re‐confirmed in N. benthamiana leaves using agroinfiltration. The coding sequences of GmSNAP18 and GmSHMT08 were amplified from Forrest and Essex soybean cultivars, and the binary p35S‐GmSNAP18 and p35S‐GmSHMT08‐HA plasmids were co‐agroinfiltrated in N. benthamiana leaves. Both anti‐HA and anti‐SNAP18 antibodies showed the presence of GmSHMT08‐HA and GmSNAP18 at ~50 and 32 KDa, respectively, in the immunoprecipitated complex (Figure 2a). The obtained co‐immunoprecipitation data using both the homologous (Soybean) and the heterologous (N. benthamiana) systems further confirm that GmSNAP18 interacts with GmSHMT08 in planta. Most importantly, cell death and necrosis symptoms caused by the GmSNAP18 were observed in N. benthamiana leaves five days after agroinfiltration. The obtained necrosis was similar to what was reported earlier when N. benthamiana leaves were agroinfiltrated with the α‐SNAPRhg1Low Copy (LC‐I289A). Interestingly, necrosis symptoms were intensified when both GmSNAP18 and GmSHMT08 were co‐agroinfiltrated in N. benthamiana, pointing to the possible involvement of both protein partners in the same molecular pathway (Figure 2b).
Figure 2.
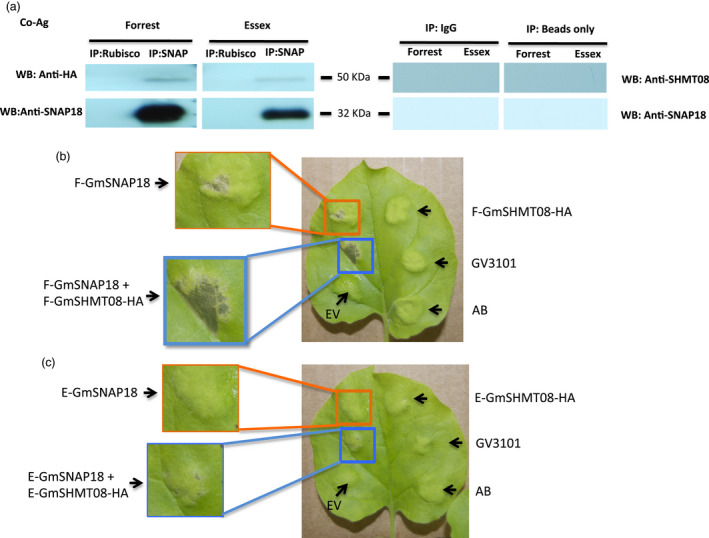
Interaction analyses of GmSNAP18 and GmSHMT08 proteins by co‐agroinfiltration (Co‐Ag) in N. benthamiana. The Agrobacterium mixture containing p35S‐GmSNAP18 and p35S‐GmSHMT08‐HA constructs was mixed with the P19 (suppression of gene silencing) and then incubated for 4 h at 28 ºC before infiltration. (a) After co‐agroinfiltration, total protein of the tobacco (Nicotiana bentamiana) leaves was extracted and co‐immunoprecipitated with anti‐SNAP18 PA or anti‐Rubisco PA (control) and blots from the eluted fractions were probed with anti‐HA, anti‐SHMT08 or anti‐SNAP18 antibodies. Only IgG or beads were used for Co‐IP experiments as a negative control and technical control, respectively. The Co‐Ag results confirmed that GmSNAP18 interacts physically with GmSHMT08. N. benthamiana leaves after 5 days were co‐agroinfiltrated to express either the indicated GmSNAP18 and/or the GmSHMT08 from (b) Forrest or (c) Essex. (b) Cell death and necrosis symptoms caused by the GmSNAP18 were intensified when both GmSNAP18 and GmSHMT08 were co‐agroinfiltrated in N. benthamiana and co‐expressed under the control of the constitutive promoter p35S. (c) Cell death symptoms were very limited in Essex. E, Essex; F, Forrest; EV, empty pGWB vector; AB, agroinfiltration buffer; GV3101, Agrobacterium GV3101 strain, WB, Western blot.
Subcellular localization of the GmSNAP18 in soybean root during SCN infection
It has been demonstrated earlier that the GmSHMT08 was specifically expressed and local to the syncytial feeding cells at 3 days after inoculation with SCN (Liu et al., 2012). The current study has demonstrated that the product of the two genes (GmSNAP18 and GmSHMT08) involved in SCN resistance physically interacts at the molecular level in soybean‐infected root tissue. In order to provide more insight into the role of the GmSNAP18 in SCN resistance, both immunostaining and in situ assays have been carried out in the homologous system (soybean roots infected with SCN) using anti‐GmSNAP18 antibodies and GmSNAP18 probes. Both assays confirmed the presence of a strong GmSNAP18 signal that was specific to the plasma membrane of infected soybean root tissues (Figure 3). Clearly, the GmSNAP18 ‘Forrest’ hyper‐accumulates at SCN infection sites. These data not only confirm the plasma membrane localization and specific response of the GmSNAP18 to SCN infection sites, but clearly demonstrate that the expression is local and specific to the syncytial feeding cells, which is coherent with the GmSHMT08 expression shown earlier (Liu et al., 2012), reinforcing their potential physical interaction and localization within the syncytial feeding cells.
Figure 3.
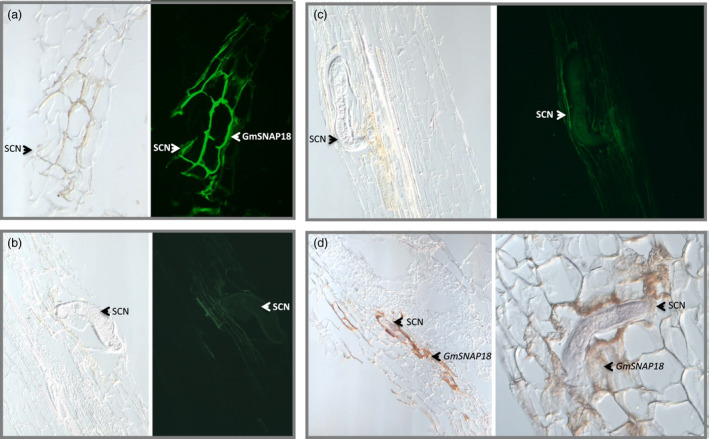
Localization of GmSNAP18 protein at the plasma membrane in soybean in SCN‐infected root. (a, b, c) Immunostaining, (d) In situ hybridization. (a) A specific expression of the GmSNAP18 protein at the plasma membrane was detected in the SCN‐resistant Forrest using anti‐SNAP18 polyclonal antibodies after immunostaining (b) In the SCN‐susceptible Essex, absence of GmSNAP18 expression was seen in the syncytial feeding cells at 3 days after inoculation with SCN. (c) Absence of GmSNAP18 expression at the plasma membrane in negative controls. (d) Expression of GmSNAP18 in the SCN‐infected area was also detected by in situ hybridization using the specific GmSNAP18 probe in ‘Forrest’ soybean roots. The GmSNAP18 expression is induced by SCN infection. GmSNAP18 proteins hyper‐accumulated at the plasma membrane, and their expression was specific to the syncytial feeding cells.
It has been suggested that conserved cysteine residues in HsSNAP25A, HsSNAP23 and other SNAP25 proteins from other organisms including goldfish, Torpedo and Drosophila (Risinger et al., 1993) contribute to stable membrane association (Hess et al., 1992). To determine whether the GmSNAP18 conserved these cysteine residues, in silico structure analysis of the GmSNAP18, its paralog GmSNAP11 (Lakhssassi et al., 2017), in addition to the HsSNAP25A and HsSNAP23 was carried out. Similar to both HsSNAP25A and HsSNAP23 in humans, structural analysis revealed that GmSNAP18 contains the conserved cysteine residues (Figure S2). These data support the possibility of GmSNAP18 binding to the plasma membrane and its role in molecular trafficking.
GmSHMT08 functions downstream of GmSNAP18
We previously demonstrated that both GmSNAP18+ (Lakhssassi et al., 2017) and GmSHMT08+ (Lakhssassi et al., 2019) transcripts were significantly induced in the resistant line Forrest in response to SCN infection, but not in the susceptible line Essex. To investigate the Peking‐type resistance mechanism, expression analysis of GmSHMT08 using an Essex x Forrest (ExF) recombinant inbred line (RIL) population under SCN infection was performed. F5‐derived RILs were haplotyped for GmSNAP18 and GmSHMT08, and then classified into four different genotypes according to their GmSNAP18 and GmSHMT08 allelic combinations (Table S1). Next, the expression profile of both GmSNAP18 and GmSHMT08 in the absence and presence of SCN infection was quantified using qRT‐PCR at 3 days after infection (DAI). Soybean roots infected with SCN revealed significant up‐regulation of GmSNAP18 transcripts in the RILs ExF07 (SHMT08+/SNAP18+ ) and ExF24 (SHMT08−/SNAP18+ ). However, no significant induction was observed in the RILs ExF5 (SHMT08‐/SNAP18− ) and ExF68 (SHMT08+/SNAP18− ) lines. These findings indicate that GmSNAP18 transcripts are induced by SCN infection only when the resistant GmSNAP18+ allele is present, regardless of the GmSHMT08 allele (Figure 4a).
Figure 4.
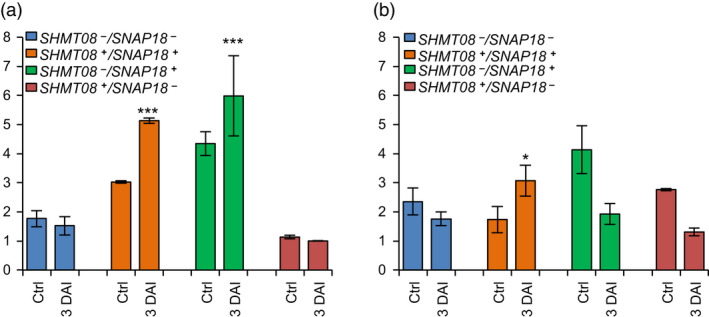
Expression analysis of GmSNAP18 and GmSHMT08 in Forrest, Essex and ExF RIL lines. Quantitative RT‐PCR analysis of (a) GmSNAP18 and (b) GmSHMT08 in F5 ExF RIL lines from infected and non‐infected root tissue with SCN HG‐type 0. F5‐derived RILs from the ExF population were haplotyped for the two genes, GmSNAP18 and GmSHMT08, and then classified into four different genotypes (GmSNAP18 +/−; GmSHMT08 +/−) according to their allelic combinations. Expressions were normalized using ubiquitin as reference as shown; the mean ± standard deviation (SD) is shown. DAI, days after infection; Ctrl, control plant. The experiment was repeated three times, and similar results were obtained. Five plants per line were used for each experiment. Asterisks indicate significant differences between the tested RILs in the presence and absence (Ctr) of SCN infection as determined by ANOVA (*** P < 0.0001, ** P < 0.01, * P < 0.05).
GmSHMT08 transcripts were significantly induced only when both GmSNAP18+ and GmSHMT08+ carried the Forrest haplotype alleles (Figure 4b). However, no significant change in the expression was observed in the three remaining ExF genotypes (SHMT08‐/SNAP18−, SHMT08‐/SNAP18+ and SHMT08+/SNAP18− ) (Figure 4b). This indicates that GmSHMT08 functions downstream of GmSNAP18 and that its induction requires functional GmSNAP18+‐resistant and GmSHMT08+ ‐resistant alleles.
Interaction between resistant and susceptible GmSHMT08 and GmSNAP18 alleles determines reaction specificity to SCN
Soybean lines carrying the resistant GmSNAP18+ and GmSHMT08+ haplotypes develop an SCN resistance response to both HG‐types 0 and 2.7 (Figure 5a, b). However, soybean lines carrying the susceptible GmSNAP18− and GmSHMT08− haplotypes lost resistance to both SCN HG‐types 0 and 2.7 (Figure 5a, b). Interestingly, soybean lines carrying a resistant GmSNAP18+ haplotype but carrying a susceptible GmSHMT08‐ haplotype lost their resistance to SCN HG‐type 0, but maintained resistance to SCN HG‐type 2.7 (Figure 5a, b). This finding was supported by the BiFC assay, where the GmSNAP18 allele from Forrest interacted with the GmSHMT08 allele from Essex (Figure S3b). These data suggest that the interaction between specific allele combinations of GmSNAP18 and GmSHMT08 proteins determines the nematode HG‐type reaction specificity (Figures S3 and S4).
Figure 5.
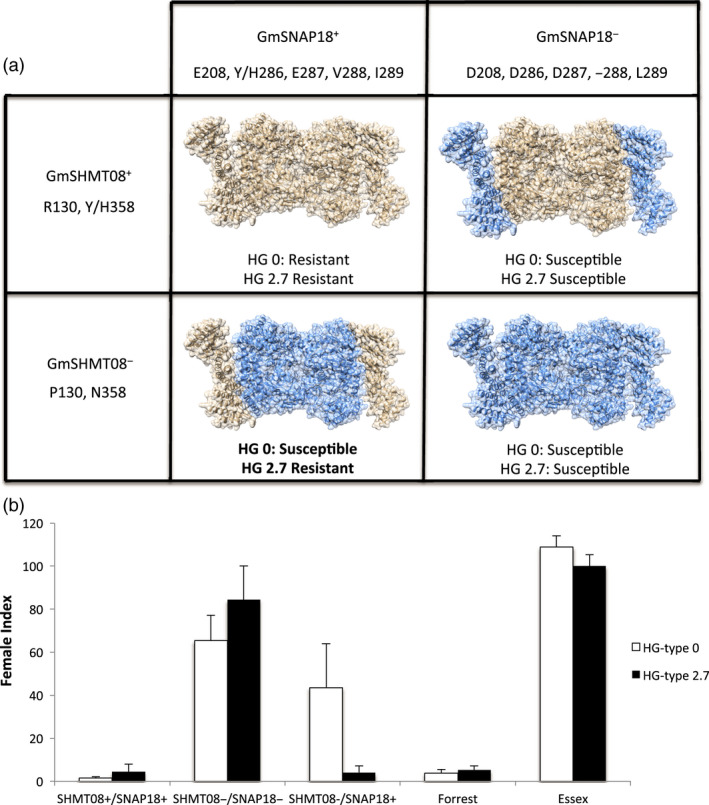
Haplotype specificity and allele combinations of GmSHMT08 and GmSNAP18 determine the SCN HG‐type resistance. (a) Homology modelling showing the predicted interaction model between different haplotypes from Forrest (light brown) and Essex (Blue) of GmSHMT08 and GmSNAP18 predicted proteins. (b) Female index of Essex, Forrest and the soybean PIs, elites and cultivars carrying the different GmSHMT08+/− and GmSNAP18+/− haplotype combinations infected by SCN HG‐type 0 and Hg‐type 2.7. Twelve seeds from each line have been tested to determine SCN resistance to SCN HG‐type 0 and 2.7. FI > 10; lines susceptible to SCN. FI < 10; lines considered resistant to SCN. GmSHMT08+/GmSNAP18+ n = 14, GmSHMT08‐/GmSNAP18‐ n = 14, GmSHMT08‐/GmSNAP18+ n = 10. Soybean PIs, elites and cultivars genotypes are described in Table S1.
GmSHMT08 immunoprecipitates a pathogenesis‐related protein in SCN‐infected roots
To identify interacting partners of GmSHMT08, mass spectrometry analysis of the immunoprecipitated proteins was conducted using anti‐GmSHMT08 antibodies immobilized to beads in a chromatography column. A comparison between non‐infected (Figure S5a) and SCN‐infected (Figure S5b) root eluded fractions showed the presence of several peptides related to SCN infection (Appendix S1). The results obtained from mass spectrometry analysis reveals the presence of a pathogenesis‐related protein belonging to a Bet VI family (PR Bet VI [Pfam: PF00407]) (Figure S6a). Little is known about the PR Bet VI molecular function, other than they are common in many viridiplantae and are reported to bind large hydrophobic compounds such as lipids, hormones and antibiotics (Jain and Kumar, 2015). The soybean genome contains several members belonging to the PR Bet VI family (at least 17 members were found in the phytozome database). In silico analysis of the five fragmented peptides obtained from the LC‐MS analysis (Figure S6b) identified a PR gene on chromosome 08 (Glyma.08G230500) named as the GmPR08‐Bet VI in this study. The predicted protein (GmPR08‐Bet VI) sequence was identical between Essex and Forrest (Figure S6b). The GmPR08‐Bet VI gene contains an open reading frame of 462 nucleotides, encoding a polypeptide of 153 amino acids. The calculated molecular mass of the immunoprecipitated pathogenesis‐related protein is 17.76 KDa (theoretical PI of 5.96). Interestingly, necrosis symptoms were intensified when the GmSNAP18, GmSHMT08 and GmPR08‐Bet VI were co‐agroinfiltrated in N. benthamiana, reinforcing the hypothesis that all three protein partners are involved in the same molecular pathway (Figure S7).
Expression analysis and subcellular localization of the pathogenesis‐related protein GmPR08‐Bet VI
Similar to the GmSNAP18 and GmSHMT08 proteins, GmPR08‐Bet VI was found to localize in the cytosol and plasma membrane of the infiltrated cells (Figure 6a), supporting the possibility of its interaction with both the GmSNAP18 and GmSHMT08 proteins.
Figure 6.
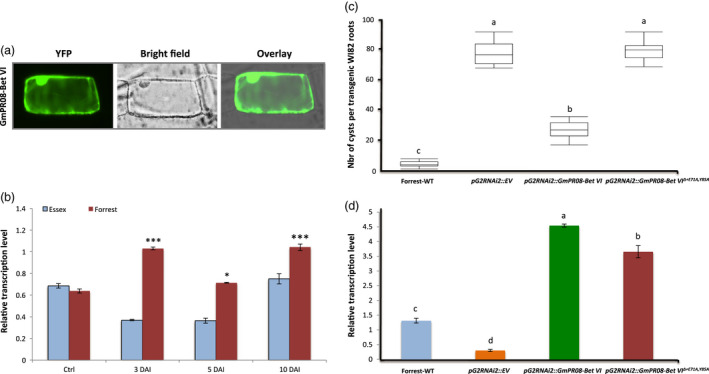
Expression, subcellular localization and overexpression analysis of the GmPR08‐Bet VI. (a) Subcellular localization of the GmPR08‐Bet VI protein. The coding sequences of GmPR08‐Bet VI were fused to the N‐terminal end of the eYFP and delivered into onion epidermal cells using biolistic bombardment. YFP fluorescence was mainly localized in the cytoplasm like the GmSHMT08 and GmSNAP18 proteins. (b) Expression analysis of GmPR08‐Bet VI in Forrest and Essex in response to SCN infection. Quantitative RT‐PCR analysis of GmPR08‐Bet VI in the resistant line, Forrest, and susceptible line, Essex, from infected (3 days, 5 days and 10 days) and non‐infected (control) root tissue with SCN HG‐type 0. Expressions were normalized using ubiquitin as reference; the mean ± standard deviation (SD) is shown. E, Essex; F, Forrest; DAI, days after infection; Ctrl, control plant. (c) Overexpression analysis in transgenic WI82 composite roots transformed by pG2RNAi2::GmPR08‐Bet VI and pG2RNAi2::GmPR08‐Bet VIΔ+E71A,Y85A . The experiments were repeated three times, and similar results were obtained. The data shown represent the averages and SD from all three biological repeats (n = 15). (d) qRT‐PCR of GmPR08‐Bet VI transcript levels in Forrest WT, control, pG2RNAi2::GmPR08‐Bet VI and pG2RNAi2::GmPR08‐Bet VIΔ+E71A,Y85A in the overexpressed soybean transgenic roots. Asterisks and connecting letters indicate significant differences between the tested lines as determined by ANOVA (***P < 0.0001, *P < 0.05).
Expression analysis demonstrates that GmPR08‐Bet VI transcripts were significantly induced in response to SCN infection in Forrest during the incompatible interaction. However, no significant induction was observed in Essex during the compatible interaction (Figure 6b). GmPR08‐Bet VI transcripts were induced at the 3 and 10 DAI in the resistant line Forrest, while its transcripts decreased at 5 DAI. GmPR08‐Bet VI transcripts were 2.86‐, 1.97‐ and 1.4‐fold more abundant in Forrest than in Essex at 3, 5 and 10 DAI, respectively.
Structural analysis of GmPR08‐Bet VI revealed the presence of conserved cytokinin‐binding residues
Three zeatin molecules were shown to bind to PR‐10 in the aqueous environment of the cell. To address structural details regarding the presence of zeatin‐binding sites at the GmPR08‐Bet VI protein sequence, alignment of the immunoprecipitated GmPR08‐Bet VI protein with LIPR‐10.2B/zeatin (2QIM) from yellow lupine (Fernandes et al., 2008) and CSBP/zeatin (2FLH) from mung bean (Pasternak et al., 2006) was carried out. Structural analysis showed that GmPR08‐Bet VI from soybean contains two conserved residues that bind to zeatin via hydrogen bonds (Figure S8). Therefore, the GmPR08‐Bet VI presents common structural features similar to the PR‐10 subfamily. These two conserved residues at the GmPR08‐Bet VI protein sequence correspond to glutamic acid and tyrosine at positions 71 and 85, respectively.
Haplotype analysis of the GmPR08‐Bet VI and SCN resistance
The possible role of the GmPR08‐Bet VI in resistance to SCN was explored using the natural variations within the gene in different soybean germplasms. The correlation with SCN resistance to different SCN races using the whole‐genome re‐sequencing data set (WGRS) including non‐domesticated, semi‐domesticated and elite domesticated introductions belonging to the USDA soybean collection was performed. To infer the allelic variation in the diverse soybean lines, the GmPR08‐Bet VI gene was analysed for synonymous and non‐synonymous SNPs, premature stop codons and indels including the genomic region, promoter, 3’ UTR and 5′ UTR. The haplotyping analysis of the different soybean germplasms showed the presence of three different haplotypes named GmPR08‐Bet VI‐a, GmPR08‐Bet VI‐b and GmPR08‐Bet VI‐c (Figure S9). None of the analysed soybean germplasms presented synonymous and non‐synonymous SNPs at the coding region including the two Exons of the GmPR08‐Bet VI. The GmPR08‐Bet VI‐a haplotype contained several common SNPs at the promoter region and 3′ UTR, in addition to 1 common SNP in the GmPR08‐Bet VI intron, which were different than the GmPR08‐Bet VI‐c. The GmPR08‐Bet VI‐b haplotype contained common SNPs at the 3′ UTR and intron like the GmPR08‐Bet VI‐c haplotype, but SNPs at the promoter region were similar to the SNPs of the GmPR08‐Bet VI‐a haplotype. Interestingly, this clustering correlates significantly with resistance to SCN. Soybean lines belonging to the GmPR08‐Bet VI‐c haplotypes showed resistant to moderate resistant reaction to SCN. However, soybean lines belonging to both the GmPR08‐Bet VI‐a and GmPR08‐Bet VI‐b haplotypes presented susceptible to moderate susceptible resistance reaction to SCN. Clearly, SNPs in the promoter region constituting the GmPR08‐Bet VI‐c haplotype present interesting features that may play a major role in resistance to SCN.
Functional validation of GmPR08‐Bet VI role in SCN resistance
To conduct GmPR08‐Bet VI overexpression analysis, the 462‐bp nucleotide corresponding to the GmPR08‐Bet VI coding sequence was overexpressed in the SCN‐susceptible Williams 82 (WI82) under the control of a soybean ubiquitin promoter using a transgenic hairy root system (Appendix S2). Overexpression of GmPR08‐Bet VI resulted in a significant reduction (65%) in the number of SCN cysts compared with the control plants expressing the empty vector, implying a role of GmPR08‐Bet VI in resistance to SCN (Figure 6c). Interestingly, when soybean roots were transformed with the GmPR08‐Bet VI mutated at both E71A and Y85A residues (GmPR08‐Bet VIΔ+E71A,Y85A) corresponding to the zeatin‐binding sites (Appendix S3), transgenic WI82 soybean lines did not exhibit resistance to SCN and therefore maintained their susceptibility showing an average of 80 cysts per plant (Figure 6c). These results suggest that GmPR08‐Bet VI may mediate resistance to SCN through the cytokinin pathway.
The GmPR08‐Bet VI protein is the molecular partner for both GmSNAP18 and GmSHMT08 proteins
BiFC assay was carried out to test whether GmPR08‐Bet VI interacts with GmSHMT08 and/or GmSNAP18. A combination of different constructs expressing pSAT4‐nEYFP‐C1::GmSHMT08, pSAT4‐cEYFP‐C1‐B::GmSNAP18 and/or pG2RNAi2::GmPR08‐Bet VI (Appendix S4‐S7), in addition to other combinations, was used to examine any possible interaction between the three proteins (Appendix S8‐S10). Since the GmPR08‐Bet VI protein was immunoprecipitated by GmSHMT08, we first tested their direct physical interaction using the BiFC pSAT4‐nEYFP‐C1::GmSHMT08 and pSAT4‐cEYFP‐C1‐B::GmPR08‐Bet VI constructs. As expected, the results obtained confirmed the presence of the interaction between the GmPR08‐Bet VI and GmSHMT08, when the resistant Forrest GmSHMT08 was present (Figure 7). Interestingly, cells co‐transformed with pSAT4‐nEYFP‐C1::GmSNAP18 and pSAT4‐cEYFP‐C1‐B::GmPR08‐Bet VI constructs showed the presence of direct interaction as well. The signal was present at the cytosol and plasma membrane when the resistant GmSNAP18 was present. Almost similar results were obtained when susceptible alleles from GmSNAP18 and GmSHMT08 were used, but with less signal intensity.
Figure 7.
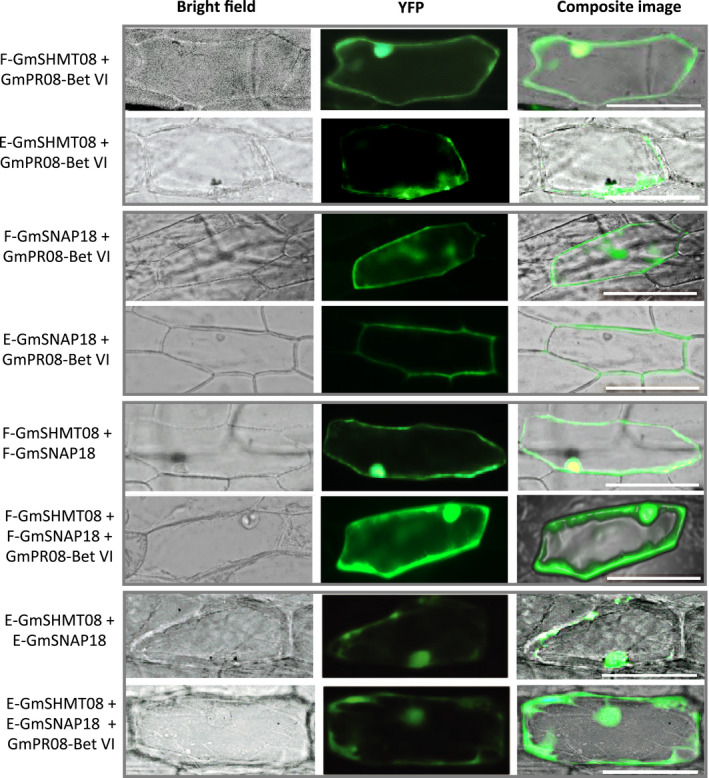
BiFC analysis of GmSNAP18, GmSHMT08 and GmPR08‐Bet VI. The coding sequences of Forrest (f) and Essex (e) wild‐type GmSHMT08 were cloned into pSAT4‐nEYFP‐C1 (E81) to generate nEYFP‐GmSHMT08 fusions. Likewise, GmSNAP18 from Forrest and Essex wild‐type and GmPR08‐Bet VI coding sequences were cloned into pSAT4‐cEYFP‐C1‐B (E82) and pG2RNAi2 to generate cEYFP‐GmSNAP18 and pG2RNAi2‐GmPR08‐Bet VI fusions. Various combinations of cEYFP and nEYFP fusions including controls (Figure S10) were co‐expressed in onion epidermal cells by particle bombardment. Interactions were stronger when the GmPR08‐Bet VI was present, and when both GmSNAP18 and GmSHMT08 were present as resistant alleles. However, weak interactions were observed when at least one of the three partners was missing or present as a susceptible allele. Bar = 200 µm.
BiFC assays also showed the presence of an interaction when the onion epidermal cells were co‐transformed with GmSHMT08 and GmSNAP18 alone (at least five cells), reinforcing their physical interaction. Interestingly, when GmPR08‐Bet VI was present, along with the Forrest variants of GmSHMT08 and GmSNAP18, a strong interaction occurred in a large number of cells (>50 cells), when compared to GmSHMT08 and GmSNAP18 alone (five cells). This interaction was specific to the cytosol and plasma membrane. However, when GmSHMT08 and GmSNAP18 alleles from Essex were used in BiFC assays, a signal with less intensity was observed at the plasma membrane (Figure 7). Similar results were obtained when the constructs were inverted, where combinations of resistant alleles of the GmSHMT08 and GmSNAP18 presented more signals (detected in both cytoplasm and plasma membrane) when compared to the GmSHMT08‐ and GmSNAP18‐carrying susceptible combinations (where the signal was nearly absent and limited to a reduced area at the plasma membrane) (Figure S3b). This includes pSAT4‐nEYFP‐C1::GmSHMT08, pSAT4‐cEYFP‐C1‐B::GmPR08‐Bet VI and pG2RNAi2::GmSNAP18 (Appendix S8‐S10). No signal was detected in all negative controls tested (Figure S10).
Homology modelling of GmSNAP18, GmSHMT08 and GmPR08‐Bet VI predicts a putative interaction site
TPR domains have been reported to facilitate specific interactions with a protein partner (Blatch and Lassle, 1999). Structural analyses show that GmSNAP18 in soybean is characterized by the presence of four tetratricopeptide repeats (TPR1, TPR2, TPR3 and TPR4) in conserved positions along the protein (Lakhssassi et al., 2017). In addition, the four TPR motifs contain a conserved structure and carboxylate clamp residues in both the resistant Forrest and the susceptible Essex cultivars. To predict a putative interaction site, homology modelling was carried out using GmSNAP18, GmSHMT08 and GmPR08‐Bet VI sequences from the resistant cultivar Forrest, followed by protein–protein docking. It has been reported that SHMTs in eukaryotes are found as asymmetric tetramers (Appaji Rao et al., 2003; Lakhssassi et al., 2019; Patil et al., 2019). Thus, we used a tetrameric SHMT protein structure to predict and visualize the interaction site between the three proteins (Figure 8a). The location of the putative interaction site was predicted to be at the GmSNAP18 N‐terminal, the TPR motifs and the tetrameric structure of the GmSHMT08 protein (Figure 8c). Protein homology modelling predicted that the polymorphism E208D (between Essex and Forrest) was located within a pocket between the two interacting proteins GmSHMT08 and GmSNAP18. Moreover, the last 5 amino acids in the GmSNAP18 were predicted to form a surface, where a potential peptide or macromolecule intermediate may interact and form a complex, which in turn may modulate the GmSHMT08 activity (Figure 8c). Interestingly, the homology modelling analysis predicted that the GmPR08‐Bet VI protein fit perfectly in the empty pocket available between the GmSNAP18 and GmSHMT08 proteins (Figure 8d).
Figure 8.
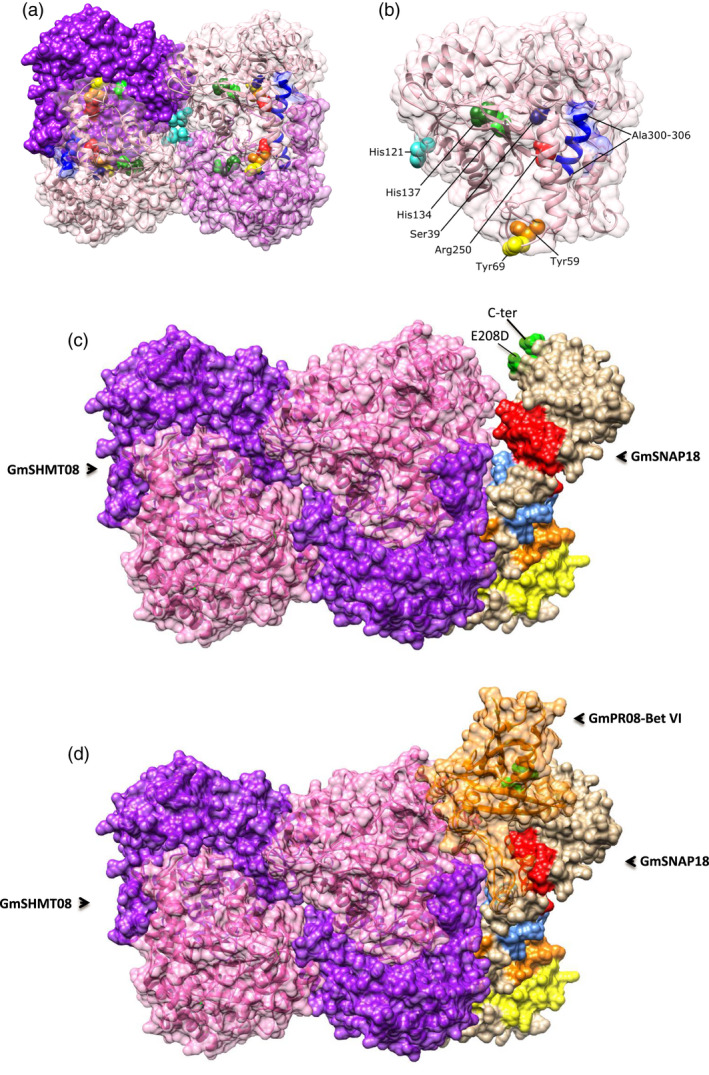
Homology modelling of the GmSNAP18, GmSHMT08 and GmPR08‐Bet VI from Forrest (Peking‐type resistance). (a) The tetrameric GmSHMT08 with important residues highlighted. (b) One SHMT subunit with highlighted catalytic and structural residues labelled. (c) The predicted interaction between GmSHMT08 (left) and GmSNAP18 (right). Locations of the four TPR motifs (TPR1: yellow, TPR2: orange, TPR3: blue, TPR4: red) and polymorphisms (green) at the GmSNAP18 are shown (right). (d) The predicted interaction between the GmSNAP18, GmSHMT08 and GmPR08‐Bet VI protein complex.
Genes encoding key components of salicylic acid signalling pathway were induced under SCN infection
To gain more insight into the possible link between resistance to SCN and the salicylic acid pathway, expression of genes encoding other key components of salicylic acid synthesis and signalling pathways was tested by qRT‐PCR in response to SCN infection. Investigation of the G. max [Williams 82] genome showed the presence of genes encoding key components of the salicylic acid synthesis and signalling pathways such as S‐adenosyl‐L‐methionine‐dependent salicylic acid methyltransferase (GmSAMT02), a transcription factor (GmTGA2‐13), and non‐inducible pathogenesis‐related 1 (GmNPR1.09 and GmNPR1.2.15) genes located on chromosomes 02, 13, 09 and 15, respectively. In order to reveal the possible link of the salicylic acid key gene components in response to SCN infection, we analysed the expression of the five genes above in two lines: the susceptible line, Essex, and the resistant line, Forrest, in the absence and presence of SCN infection at 2, 5 and 10 DAI. The analysis showed that GmSAMT1.02, GmTGA2.13 and GmNPR1.1.15 transcripts were induced in both compatible and incompatible reactions. However, GmNPR1.09 transcripts were induced only in the resistant line Forrest as in the case of GmPR08‐Bet VI and, thus, were specific to the incompatible reaction (Figure S11). Together, these data point to a correlation between the salicylic acid pathway and SCN infection confirming previous reports. It is well known that the salicylic acid pathway induces the expression of PR genes (Kim et al., 2003; Kim et al., 2007), which could explain the induction of transcripts and abundance of GmPR08‐Bet VI after nematode infection.
GmSHMT08, GmSNAP18 and GmPR08‐Bet VI respond to exogenous salicylic acid and cytokinin treatments
Next, we studied the expression of the GmSHMT08, GmSNAP18 and GmPR08‐Bet VI in response to exogenous treatments with two phytohormones, the salicylic acid and cytokinins, at 12, 24 and 72 h. The obtained results show the presence of an early induction of GmSNAP18 transcripts after a 12‐h treatment in the presence of exogenous salicylic acid and cytokinins in Forrest (Figure S12). No significant GmSNAP18 induction was observed in Essex when both hormones were present. GmSNAP18 transcripts were largely (twice) abundant in Forrest under salicylic acid treatment when compared to cytokinin treatments. However, neither of the two phytohormones induced GmSHMT08 transcripts in the two data points tested.
GmPR08‐Bet VI presented different expression profiles. While GmPR08‐Bet VI transcripts were induced in both Essex and Forrest under cytokinin treatments, salicylic acid induces GmPR08‐Bet VI transcripts in the resistant line Forrest, but not in the susceptible line Essex. To bring more insight into the observed differences of GmPR08‐Bet VI expression between both lines, we studied the expression of the two salicylic acid components, GmNPR1.2 and GmTGA2 (Figure S13). Expression analysis revealed that salicylic acid induced GmNPR1.2 transcripts in both lines, whereas transcripts of the transcription factor GmTGA2 were significantly induced in the resistant line only. It is well documented that both components are needed to induce the PR transcripts (Kim et al., 2003; Rahman et al., 2012); therefore, the non‐induction of GmTGA2 transcripts by salicylic acid in the susceptible line may explain the non‐induction of GmPR08‐Bet VI transcripts in Essex.
We also tested the expression of the cytokinin receptor GmARR03 gene. GmARR03 transcripts were significantly induced under exogenous cytokinin treatments, but not by salicylic acid treatments (Figure S13).
Discussion
Haplotype specificity and gene dosage effect of the GmSNAP18/GmSHMT08/GmPR08‐Bet VI complex in SCN resistance
GmSHMT08 and GmSNAP18 are co‐regulated in response to SCN infection, a finding that suggests an interaction at the protein level (Bhardwaj and Lu, 2005). Expression analysis suggests that the presence of the resistant Forrest alleles at GmSNAP18+ and GmSHMT08+ is required to trigger the SCN resistance response to both Hg‐type 0 and Hg‐type 2.7. However, a combination of GmSNAP18+ and GmSHMT08‐ was capable of inducing SCN resistance to Hg‐type 2.7 only. These data indicate the presence of a specific crosstalk between specific rhg1‐a and Rhg4 allele combinations (GmSNAP18 + and GmSHMT08 +/− ) that is needed to determine the nematode Hg‐type reaction specificity in Peking‐type soybeans. Therefore, we conclude that the observed weak interaction between both proteins in the susceptible line is not sufficient to trigger the SCN resistance (Figure 1b, c), but requires the presence of the resistant allele at GmSNAP18+ . These data clearly point to the presence of a gene dosage effect in SCN resistance. The presence of three copies of the GmSNAP18 in the Forrest genome versus one copy in Essex clearly impacted their transcript abundance. Gene dosage is highly related to gene copy numbers and the amount of gene product produced in a cell, which is more commonly dependent on the regulation of gene expression (Lemos et al., 2011; Veitia, 2005). The current study demonstrates that gene dosage effects on the resistant GmSNAP18 allele (three copies) positively impacted its transcript expression and induced the expression of the resistant GmSHMT08 allele, consequently leading to the presence of a strong interaction of the GmSNAP18/GmSHMT08 complex. The induced expression of GmPR08‐Bet VI in response to SCN infection may also play a role in this gene dosage balance for potentiating the GmSNAP18/GmSHMT08/GmPR08‐Bet VI complex’s physical interaction. However, low GmSNAP18 transcripts in Essex could in part explain its susceptible phenotype, which is most likely due to its low copy number and promoter variant as reported earlier (Patil et al., 2019). This is consistent with previous studies showing that alteration of gene dosage could be associated with both quantitative and qualitative phenotype variations (Gardiner, 2004; Veitia, 2004).
GmPR08‐Bet VI is a potential candidate gene for resistance to SCN mapped at the SCN 50‐2 and SCN 37‐4 QTLs
Resistance in Peking‐type soybean requires both the rhg1‐a and Rhg4 alleles (Liu et al., 2017; Liu et al., 2012). These findings revealed an unprecedented resistance gene that has evolved to underlie two types of resistance while ensuring the same function within a plant species (Liu et al., 2017). The current study revealed that unlike the PI88788‐type resistance, Peking‐type resistance requires different molecular interacting partners. Co‐immunoprecipitation, co‐agroinfiltration and BiFC experiments demonstrated the presence of a physical interaction between GmSHMT08 and GmSNAP18. Most importantly, BiFC analysis revealed a new finding involving the identified pathogenesis‐related protein (GmPR08‐Bet VI) that potentiates the interaction between the GmSNAP18 and GmSHMT08 proteins in SCN‐infected soybean roots. Interestingly, GmPR08‐Bet VI was mapped to a novel quantitative trait locus for broad‐based resistance to SCN at the linkage group A2 (chromosome 08) that has been reported earlier using the Magellan x PI 567516C RIL population (Vuong et al., 2010) and the A95‐684043 x LS94‐3207 RIL population (Swaminathan et al., 2018). Therefore, the current study identified GmPR08‐Bet VI as the potential candidate gene for resistance to SCN on chromosome 08 (SCN 50‐2 and SCN 37‐4, Soybase QTL map) (Figure S14).
Structural analysis of the GmSNAP18 points to a possible binding to the plasma membrane
SNAPs have a membrane anchor domain, consisting of a palmitoylation between palmitate and the plasma membrane by a thioester bond at the cysteine residue. This attachment gives the SNAP protein a stable membrane association, a step required for initial membrane targeting (Gonzalo and Linder, 1998a). It has been demonstrated that SNAP‐25 in mouse and chicken contains cysteines organized as a cluster between positions 84 and 92 (Catsicas et al., 1991; Oyler et al., 1989). Clustered cysteines are conserved in similar positions within SNAP‐25 in other organisms including goldfish, Torpedo and Drosophila (Risinger et al., 1993), indicating that these residues are functionally important in order to give the SNAP protein a stable membrane association (Hess et al., 1992). In animals, it has been reported that acylation and deacylation cycles could induce SNAP‐25 conformational changes affecting its affinity with interacting proteins (Gonzalo and Linder, 1998a). Under SCN infection, GmSNAP18 expression and localization hyper‐accumulates at the plasma membrane and was specific to the root cells surrounding the nematode in SCN‐resistant soybean lines, but not in the susceptible ones, indicating the possible role of GmSNAP18 in molecular trafficking.
Interaction between GmSNAP18, GmSHMT08 and GmPR08‐Bet VI proteins and molecular trafficking
Intracellular membrane fusion is mediated by dynamic assembly and disassembly of soluble N‐ethylmaleimide‐sensitive factor (NSF) attachment protein (SNAP) receptors (SNAREs), where α‐SNAP guides NSF to disassemble SNARE complexes after membrane fusion (Ma et al., 2016). In soybean, overexpression of Gm‐α‐SNAP induces Gm‐SYP38 transcription, rescuing susceptible G. max (rhg1−/− ), by suppressing SCN parasitism (Pant et al., 2014). Recently, it has been reported that components of the SNARE‐containing regulon are co‐regulated in root cells undergoing defence against soybean cyst nematodes (Klink et al., 2017).
An atypical resistance‐type PI88788 α‐SNAP protein that is defective in promoting the function of the NSF and is cytotoxic has been reported (Bayless et al., 2018). The unusual resistance‐type α‐SNAP in PI88788 binds to the WT‐NSF less and disrupts the vesicle trafficking and cell death when expressed in Nicotiana benthamiana (Bayless et al., 2016). These findings suggested that the abundance of rhg1 encoding defective α‐SNAP increases in developing syncytial cells by disrupting syncytium viability impacting nematode growth and reproduction in the PI88788 type of resistance (Bayless et al., 2016). Recently, an unusual NSFRAN07 has been reported to bind α‐SNAP in vitro and that its co‐expression in planta was more protective against the rhg1‐b α‐SNAP cytotoxicity in PI88788‐type resistance (Bayless et al., 2018). Therefore, it has been suggested that modulation of vesicle trafficking and cell health at the SCN feeding site is at least one‐core mechanism of rhg1‐mediated SCN resistance involving chromosome 07 (NSFRAN07). Similar to the GmSHMT08 localization following SCN infections, we observed an accumulation of the GmSNAP18 protein in the cells surrounding the nematode developing syncytia in the SCN‐resistant Forrest roots (Peking‐type resistance) (Figure 3a, d). In addition, necrosis symptoms were intensified when the GmSNAP18, GmSHMT08 and GmPR08‐Bet VI complex was present in N. benthamiana co‐agroinfiltration experiments, which explains the hypersensitive response resulting in necrosis and cell death in the nematode feeding cell reported in earlier studies (Liu et al., 2012; Mahalingam and Skorupska, 1996). GmSNAP18 expression was absent in developing syncytia cells in the SCN‐susceptible Essex (Figure 3b). These findings are also congruent with the induced GmSNAP18 transcripts in the resistant Forrest line, but not in the susceptible Essex line.
Exocytosis of the pathogenesis‐related protein occurs by the fusion of vesicles with SNAP proteins at plasma membranes through translocation and docking at the plasma membrane, a process very well studied in animals and plants as well, which involves the SNARE protein complex (Karnik et al., 2013). SNARE complexes involving SNAP, PR, vesicles and other proteins have been shown to contribute to gene for gene resistance against bacteria in N. bentamiana by exocytosis of PR1 in the extracellular space (Kalde et al., 2007; Kwon et al., 2008). Interestingly, we were able to identify, by mass spectrometry, a member of the pathogenesis‐related proteins, GmPR08‐Bet VI, which interacts with both GmSHMT08 and GmSNAP18. Expression analysis demonstrated that GmPR08‐Bet VI transcripts responded to SCN infection in the resistant line only. Moreover, it has been reported that the C‐terminus of SNAPs (last 25 residues) determines its localization and functionality in vesicle trafficking and fusion (Chen et al., 2001). Under SCN infection, the obtained strong signal corresponding to the GmSNAP18 localization at the plasma membrane of infected soybean roots suggests the role of the GmSNAP18 in protein facilitating the trafficking of its molecular partner, the GmSHMT08, in addition to the pathogenesis‐related protein, GmPR08‐Bet VI.
Components of the salicylic acid signalling pathway are co‐regulated in root cells undergoing nematode infection
To combat biotrophic pathogens, plants mainly activate the salicylic acid (SA) signalling pathway (Grant and Lamb, 2006) and the cytokinin (CK) pathway (Albrecht and Argueso, 2017). Studies using the plant model Arabidopsis thaliana, as well as the soybean, Glycine max, investigated the role of salicylic acid as the key hormone triggering the plant defence response against pathogens and demonstrated that salicylic acid reduced soybean cyst nematode infection (Matthews et al., 2014a; Rahman et al., 2012). In tomato, it has been reported that necrotrophs manipulate the salicylic acid signalling pathway to promote their disease (Rahman et al., 2012). Non‐inducible pathogenesis‐related 1 (NPR1) is considered a master regulator of salicylic acid signalling that interacts with the TGA transcription factor, ultimately leading to the activation of SA‐dependent responses (Rahman et al., 2012). Overexpression of the Arabidopsis At‐NPR1, At‐TGA2 and At‐PR5 genes in transgenic soybean roots showed a reduction of SCN cysts by less than 50% (Matthews et al., 2014a). In the current study, we demonstrated that their homologous genes in soybean (Matthews et al., 2014a) were induced during SCN infection. Indeed, transcripts of the soybean salicylic acid signalling genes GmTGA2‐13, GmNPR1‐09 and GmNPR1.2‐09 were induced in response to SCN infection (Figure S11). It has been reported that salicylic acid induces pathogenesis‐related protein 1a (PR1a) in Chinese cabbage at both the protein and mRNA levels (Kim et al., 2003). In the Peking type of resistance, we have shown that GmPR08‐Bet VI transcripts were induced in the SCN‐incompatible reaction. In addition, overexpression of the GmPR08‐Bet VI WT in soybean transgenic hairy roots decreased the number of SCN cysts by over 65%. The observed GmPR08‐Bet VI induction in soybean is most likely due to the activation of the salicylic acid signalling pathway during root wounding caused by nematode infection, which has been tested by the induction of most salicylic acid component genes including the GmPR08‐Bet VI, GmTGA2‐13 and GmNPR1.2‐09 (defence genes) in Forrest, but not in Essex. GmSAMT1, an SCN defence‐related soybean salicylic acid gene, encodes an S‐adenosyl‐L‐methionine‐dependent salicylic acid methyltransferase that uses salicylic acid as a substrate to produce methyl salicylate (Lin et al., 2013). Expression analysis demonstrates that GmSAMT1‐02 transcripts were 61 times more expressed in Forrest than Essex (Figure S11). These results are coherent with the comparative RNA‐Seq analysis that showed a strong induction of genes involved in salicylic acid signalling‐mediated H. glycines resistance, including the GmSAMT1‐02 that was up‐regulated by 54.5‐fold in infected resistant roots (Zhang et al., 2017). Under SCN infection, GmSAMT1‐02 transcripts were significantly induced in Forrest when compared to Essex. Taken together, these data are congruent with previous studies showing that the overexpression of GmSAMT1 modulates both salicylic acid biosynthesis and salicylic acid signal transduction, evidenced by the induced expression of NPR1 genes in GmSAMT1‐overexpressing transgenic hairy roots. Overexpression of GmSAMT1 in soybean roots also reduced the susceptibility of soybeans to nematode infection, indicating that GmSAMT1 plays a role in the soybean’s defence against SCN (Lin et al., 2013). These data point to a correlation between the salicylic acid pathway and SCN infection.
GmPR08‐Bet VI involvement in a crosstalk between the cytokinin and salicylic acid pathways in resistance to SCN
Unlike the GmPR08‐Bet VI WT, GmPR08‐Bet VIΔ+E71A,Y85A carrying mutated cytokinin‐binding sites did not restore resistance to SCN in transgenic WI82 hairy roots. It was not surprising that the inactivation of the two cytokinin‐binding sites in the isolated GmPR08‐Bet VI in this study led to a susceptibility reaction towards SCN infection in soybean. It has been shown in Arabidopsis that the cyst nematode Heterodera schachtii releases exogenous cytokinin that controls cell division and orchestrates feeding site formation in host plants (Siddique et al., 2015). Silencing the cytokinin‐synthesizing isopentenyltransferase gene in Heterodera schachtii was shown to reduce the expansion of feeding sites (Siddique et al., 2015). In a very complex equation, nematodes establish feeding sites by recruiting specific plant developmental pathways involving hormonal crosstalk, while nematodes also need to suppress plant defence and its interacting hormone pathways (Gheysen and Mitchum, 2019). Nematodes have evolved plant peptide hormone effector mimics to facilitate parasitism. These include the CLAVATA3/Embryo surrounding region (CLE)‐like, C‐terminally encoded peptide (CEP)‐like and inflorescence deficient in abscission (IDA)‐like peptides (Gheysen and Mitchum, 2019). The balance of phytohormones is exquisitely controlled in order to maintain plant growth and development; however, this balance is often disturbed following pathogen infection (Albrecht and Argueso, 2017; Denancé et al., 2013). Cytokinins play an important role in plant defence against biotrophic pathogens. Infection by a biotrophic pathogen stimulates oxidative stress and salicylic acid biosynthesis, resulting in salicylic acid‐dependent defence responses to suppress the growth of biotrophic pathogens (Albrecht and Argueso, 2017). Although cytokinins enhance defence activation by salicylic acid processes, this phytohormone can also help pathogen growth, through mechanisms that include suppression of the PTI (cytokinin‐synthesizing isopentenyltransferase) pathway (Albrecht and Argueso, 2017). Increased salicylic acid content/signalling inhibits cytokinin‐regulated processes, causing plant growth inhibition. Data obtained from this study revealed that most components of the salicylic acid pathway including GmPR08‐Bet VI, GmTGA2‐13 and GmNPR1.2 were also induced by cytokinins. In addition, both phytohormones induced GmSNAP18 transcripts, which is coherent with the essential role of plant proteins containing TPR domains in response to hormones including salicylic acid and cytokinins (Wang et al., 2004; Yoshida et al., 2005). While mutagenesis of the two amino acids likely to be involved in ligand binding affected the ability of GmPR08‐Bet VI to enhance resistance of susceptible soybean roots, the particular amino acids involved in binding vary quite considerably between different PR10 group proteins (Fernandes et al., 2008). Those two amino acids could be involved in binding other different hydrophobic ligands (i.e. Brassinosteroid) as it has been shown for other PR10 proteins. Taken together, these data provide evidence about the existence of a molecular crosstalk that may involve the two phytohormones, salicylic acid and cytokinin, and the two major genes for resistance to SCN, GmSNAP18 and GmSHMT08, mediated by the GmPR08‐Bet VI WT.
Deciphering Peking‐type SCN resistance
Collectively, we hypothesize that upon SCN infection, recognition between virulence proteins (i.e. effectors, elicitors) encoded by soybean cyst nematode H. glycines and the GmSNAP18+ (resistant haplotype) will occur, triggering the incompatible interaction (Figure 9). Accordingly, the possible binding of a nematode effector (Bekal et al., 2015) may cause an increase in the transcription of GmSNAP18, consequently triggering GmSHMT08 induction. Most likely, due to the wounding of soybean root by the nematode, in addition to the increase in endogenous cytokinin and the secretion of exogenous cytokinins, the salicylic acid pathway response is being triggered, consequently inducing the GmPR08‐Bet VI.
Figure 9.
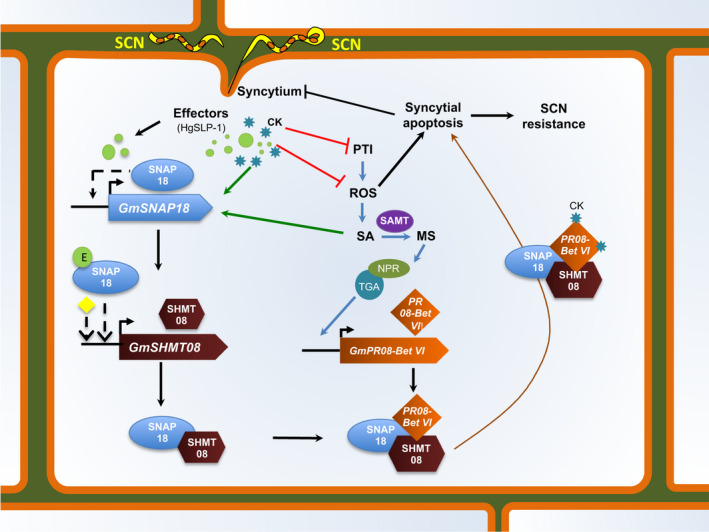
Crosstalk between the GmSNAP18‐, GmSHMT08‐ and GmPR08‐Bet VI‐resistant genes and SA and CK phytohormones to activate defence and resistant response in Peking‐type SCN resistance. In the absence of pathogen attack, endogenous cytokinins (CKs) promote shoot growth. Infection by a biotrophic pathogen (in the case of SCN) stimulates pattern‐triggered immunity (PTI) activation, oxidative stress (ROS) and salicylic acid (SA) biosynthesis, resulting in salicylic acid‐dependent defence responses to suppress pathogen growth (blue) (Albrecht and Argueso, 2017). Endogenous and/or exogenous cytokinins can help pathogen growth, by mechanisms that include suppression of PTI and ROS (cytokinin‐induced susceptibility: red) (Albrecht and Argueso, 2017). Increased salicylic acid content/signalling induces salicylic acid component genes including GmSAMT, GmNPR and GmTGA resulting in the induction of the GmPR08‐Bet VI, potentially promoting SA defence response. QRT‐PCR analysis demonstrates that at ten days post‐SCN inoculation, the expression of GmSNAP18 returns to levels prior to nematode infection. These data point to a model in which GmSNAP18 may directly or indirectly negatively autoregulate itself. Induction of genes in response to stress is commonly associated with rapid inactivation of a negative co‐regulator. Accordingly, the possible binding of a nematode effector (E) like the HgSLP‐1, SA and/or CKs may interrupt the autoregulation of GmSNAP18, causing an increase in the transcription of GmSNAP18, consequently causing an induction of GmSHMT08. An interaction between GmPR08‐Bet VI, GmSNAP18 and the GmSHMT08 protein complex will occur and carry the complex towards the plasma membrane and modulate the activity of the GmSHMT08 in single‐carbon metabolism, methionine synthesis and maintenance of redox homeostasis within the root cells. By binding cytokinins, GmPR08‐Bet VI may hijack the increase of both exogenous and endogenous cytokinins at the feeding site and therefore suppress the cytokinin‐regulated processes (brown). Finally, induction of the reported apoptosis, cell death and necrosis that were intensified when the GmSNAP18/GmSHMT08/GmPR08‐Bet VI protein complex was present, in addition to the degeneration observed in the cells surrounding the syncytia, will occur. Abbreviations are as follows: CKs, endogenous or exogenous cytokinins; E, possible effectors; NPR, non‐inducible pathogenesis‐related; PR, pathogenesis‐related protein; SA, salicylic acid; TGA,transcription factor; SAMT,S‐adenosyl‐L‐methionine‐dependent salicylic acid methyltransferase; SHMT,serine hydroxymethyltransferase; SNAP, soluble NSF attachment protein. Arrows indicate positive interactions; blunt ends indicate negative interactions (inhibition); dashed lines represent unknown possible intermediary steps; (yellow) possibility of the presence of other intermediate(s).
Next, at the protein level, an interaction between GmSNAP18, GmPR08‐Bet VI and GmSHMT08 proteins will occur and may modulate their activity. Consequently, this interaction may result in the trafficking of the GmPR08‐Bet VI towards the infected root tissue, which may increase the cytotoxicity in the cells surrounding the nematode and consequently disrupting syncytium viability, a process that could directly impact nematode growth and reproduction in the Peking type of resistance. The other possible hypothesis may be linked through binding cytokinins. In fact, GmPR08‐Bet VI may be reducing both exogenous and endogenous cytokinins at the feeding site, leading to cytokinin deficiency and therefore stopping syncytia expansion (Figure 9). Interactions with GmSHMT08 may modulate its activity in single‐carbon metabolism, methionine synthesis and the maintenance of redox homeostasis within the root cells. SHMT is involved in the simultaneous interconversion of serine/glycine and THF/5,10‐methylene THF (Mouillon et al., 1999). Accordingly, a potential modulation of SHMT activity may cause a disruption of serine/glycine and/or THF/5,10‐methyleneTHF interconversion. Modulation of the SHMT serine/glycine interconversion may impact important maintenance of redox homeostasis that occurs via glutathione synthase and glutathione peroxidases. Glutathione peroxidase transcription was shown to be significantly modulated in a transcriptomic analysis of SCN infection in syncytia, among a host of other reactive oxygen species (ROS) scavenging enzymes (Kandoth et al., 2011). In fact, it has been reported that maintenance of a certain ROS homeostasis is required for parasitic nematodes to cause and maintain disease (Melillo et al., 2006; Siddique et al., 2014). Disruption of this homeostasis can lead to either the termination of syncytial formation or syncytial apoptosis. It has also been reported that SHMT modulates the salicylic acid (SA) signalling pathway (Rojas et al., 2014). In fact, the shmt1‐1 mutant in Arabidopsis showed a greater accumulation of H2O2, which is known to induce salicylic acid biosynthesis (Leon et al., 1995; Moreno et al., 2005). Collectively, the modulation of the GmSHMT08 and GmPR08‐Bet VI impacts the redox homeostasis pathway, the accumulation of H2O2 and cytokinins in infected soybean roots, which subsequently leads to small syncytia or to its apoptosis. Several reports identify the activation of cytokinin by nematodes to develop syncytia; here, we identified the missing link that plants use to control cytokinin action via the pathogenesis‐related protein in combating nematode growth. Although the initial step of the SCN mechanism in Peking‐type resistance has now been revealed, the following cascade of events needs to be further elucidated.
Experimental procedures
SCN‐infection phenotyping
SCN screening was performed as described by Liu et al. (2011).
SNAP and PR alignments
Alignment of the full‐length amino acid sequences from GmSNAP18, GmSNAP11, GmSNAP25A, HsSNAP23, GmPR08‐Bet VI from G. max, MbPR10 (2FLH) from Mung bean, and YlPR10 (2QIM) from Yellow lupine was performed using the MegAlign (DNASTAR Lasergene 8) software package and the Clustal W algorithm as described earlier (Lakhssassi et al., 2017; Lakhssassi et al., 2019).
Genotyping of ExF RIL population
The ExF RIL population used in this study was developed at Southern Illinois University Carbondale (Lightfoot et al., 2005). The EcoTILLING marker GmSHMT08 (Liu et al., 2012) and GmSNAP18 primers listed in Table S2 were developed and used to identify the genotype of each ExF RIL at the genes GmSHMT08 (Glyma.08g108900) and GmSNAP18 (Glyma.18G022500) as descried earlier (Lakhssassi et al., 2017). The EcoTILLING was conducted as described by Liu et al. (2011).
Haplotype analysis of the GmPR08‐Bet VI
The soybean germplasm lines sequenced at approximately 17X genome coverage were utilized for mapping and detection of allelic variants (Valliyodan et al., 2016). Similar to the published GmSHMT08 and GmSNAP18 haplotyping analysis using a pipeline as previously described (Patil et al., 2019), the haplotype analysis of the GmPR08‐Bet VI gene was performed to study the correlation with SCN resistance using the whole‐genome re‐sequencing data set (WGRS) including non‐domesticated, semi‐domesticated and elite domesticated introductions belonging to the USDA soybean collection.
GmSHMT08 and GmSNAP18 subcellular localization and confocal images in N. benthamiana
The full‐length cDNA of GmSNAP18 and GmSHMT08 cloned previously into the Gateway pDONR221 Vector was cloned behind the cauliflower mosaic virus 35S promoter in translational fusion with RFP at the C‐terminus into a plasmid using the Gateway technology. Those plasmids were transiently expressed by agroinfiltration in N. benthamiana leaves with the viral silencing suppressor p19 (Voinnet et al., 2003). Infected leaves were sectioned 3–5 days after infiltration. Infiltrated N. benthamiana leaves were imaged with a Zeiss LSM 880 microscope (Carl Zeiss Vision GmbH, Le Pecq, France) with a 40× objective.
GmSHMT08, GmSNAP18 and GmPR08‐Bet VI cloning and subcellular localization in onion
The coding sequences of the GmSHMT08 (Glyma.08G108900), GmSNAP18 (Glyma.18G022500) and GmPR08‐Bet VI (Glyma.08G230500) genes were amplified from Forrest cDNA using forward and reverse primers containing EcoRI or HindIII and SalI restriction enzyme sites, respectively. PCR products were digested and then ligated to the N‐terminus of the yellow fluorescent protein (YFP) reporter gene in the pSAT6‐EYFP‐N1 vector. The fusion constructs were verified by sequencing. Gold particles were coated with plasmid DNA and delivered into onion epidermal cells using biolistic bombardment as previously described (Lakhssassi et al., 2019; Liu et al., 2015).
qRT‐PCR analysis
Soybean seedlings from the ExF RILs (Meksem et al., 2001), the susceptible line Essex and from the resistant line Forrest wild types were grown in autoclaved sandy soil in the growth chamber for 1 week, and then infected with infective eggs from SCN HG‐type 0 and Hg‐type 2.7 for soybean elites, PIs and cultivars used for populations. Total RNA was isolated from the root controls and root samples after three, five and ten days following SCN infection as described previously (Lakhssassi et al., 2019). A list of all primers used for qRT‐PCR is found in Table S2. Experiments were repeated threefold with similar results. Results from one biological replicate are shown. Statistical analysis was performed with the analysis of variance (ANOVA), using the JMP Pro V14 software (SAS Institute Inc., Cary, NC).
Soybean Seed Germination and screening of phytohormones
Soybean seeds were surface‐sterilized overnight with 100 mL bleach and 3.5 mL HCl in a dessicator. For in vitro germination, seeds were stratified for 2 days at 4 ºC in the dark and plated onto petri dishes containing Whatman filter paper imbibed with basal MS liquid medium. Hormone screening was conducted as described earlier (Lakhssassi et al., 2012b). Seedlings were grown 4 days in MS medium and then transferred to petri dishes containing different concentrations of the stress agent or hormone. Salicylic acid was used at 5 mm, and cytokinins were used at 10 mg/L. Plates were placed under a long‐day regime (16 h of light/8 h of dark) of 25 mE at 23 ºC (day/night).
p35S‐SNAP18, p35S‐SHMT::3XHA and p35S‐GmPR08‐Bet VI constructs
The p35S‐GmSNAP18, p35S‐GmSHMT08::3xHA and p35S‐GmPR08‐Bet VI full‐length cDNAs were cloned into the Gateway pDONR221 Vector by BP reactions, and then transferred into the Gateway Binary Vector (pGWB) by LR reactions. The pGWB vector was made in the Nakagawa Lab (Research Institute of Molecular Genetics Shimane University, Japan) and obtained from the Cyril Zipfel Lab (Sainsbury, Norwich, UK). The recombinant plasmids were introduced into Agrobacterium tumefaciens strain GV3101. The gene‐specific primers designed to amplify cDNA fragments are detailed in Table S2.
Transient Agrobacterium expression in Nicotiana benthamiana
The A. tumefaciens strain GV3101 containing p35S‐SNAP and p35S‐SHMT‐3XHA, and the suppressor of gene silencing p19 construct (Voinnet et al., 2003), was grown overnight at 28 ºC in 50 μg/mL kanamycin, hygromycin and rifampicin, and 25 μg/mL gentamicin. For co‐immunoprecipitation analysis, strains containing p35S‐SNAP, p35S‐SHMT‐3XHA, pGWB empty vector and p19 mixtures were induced in 10 mm MgCl2, 10 mm Mes (pH 5.6) and 150 μm acetosyringone. Next, at OD600 = 0.5, half of the volume from p19 (OD600 = 0.25) was mixed with 1 volume from each culture containing the previous constructs or an empty vector, and then incubated in the dark for 4 h at 28 ºC before infiltration. Four‐week‐old N. benthamiana plants grown at 25 ºC with a photoperiod of 16‐h light were pressure infiltrated into the abaxial side of a nearly fully expanded leaf using a 1‐mL syringe. Two leaves per plant and three plants per construct were agroinfiltrated. Seventy‐two hours later, agroinfiltrated leaves were collected and ground in liquid nitrogen N2, and total proteins were extracted.
Production of affinity‐purified SNAP antibodies
Total RNA from the soybean Forrest was converted to cDNA and used as a template to amplify GmSNAP18 (Glyma.18G022500) as described previously. XhoI and XmaI restriction sites were added to the forward and reverse primers, XmaI‐SNAP‐Fw and XhoI‐SNAP. The gene‐specific primers designed to amplify cDNA fragments are detailed in Table S2. The resulting PCR product was cloned between the XmaI and XhoI cloning sites in the pGEX‐5x‐1 vector with a GST‐tag in the C‐terminus (pGEX‐5x‐1‐SNAP‐GST). The construct was confirmed by sequencing (Genewiz, South Plainfield, NJ). Next, the plasmid construct was transformed into E. coli BL21 and sent to Rockland Immunochemicals (Gilbertsville, PA), where the protein was produced, purified and then injected into rabbits. The SNAP polyclonal antibody was affinity‐purified and cross‐adsorbed to remove antibodies that might bind to the GST‐tag. Antibody binding was tested through immunoblots using recombinantly produced GmSNAP18 proteins (Figure S15) from root lysates of Essex and Forrest soybean lines, and also from N. benthamiana leaves expressing GmSNAP18 proteins (Figure 1e). Next, the antibody specificity was validated by performing a co‐immunoprecipitation assay using the anti‐GmSNAP18 antibodies, followed by mass spectrometry analysis of the eluted fraction and the obtained band in the polyacrylamide gel at ~32 KDa (Figure S15). Only digested peptides that are specific to the GmSNAP18 protein were obtained. Clearly, anti‐GmSNAP18 antibodies are specific to the GmSNAP18 protein only. The other GmSNAP11 paralog member was not immunoprecipitated by the custom generated anti‐GSNAP18 antibodies in three independent pull‐down experiments.
In situ hybridizations of GmSNAP18
The probe template was prepared by PCR amplification of the pGEX‐5x‐1::GmSNAP18 plasmid with 10 pmol each of GmSNAP18 forward and GmSNAP18 reverse primers (Table S2). The hybridization probe is located within the region between the forward and reverse primers shown in Table S2. The cDNA insert was purified with a Chroma Spin‐200 column. Digoxigenin‐labelled RNA probes were synthesized from 300 ng of purified template with the T7 and T3 AmpliScribe High Yield Transcription Kits (Epicentre Technologies, Madison, WI) with the following nucleotide concentrations: 7.5 mm GTP, 7.5 mm ATP, 7.5 mm CTP, 5 mm UTP, 3 mm dig‐11‐UTP (Roche Molecular Biochemicals, Indianapolis, IN). Labelled RNA was purified with a Chroma Spin‐200 column. The RNA probe was hydrolysed to a length of approximately 150–300 bases (Ruzin, 1999) and then used at a concentration of 1 ng/μL.
Excised nematode‐infected root tips were fixed in FAA (3.7% formaldehyde, 50% ethanol, 5% glacial acetic acid), paraffin‐embedded (Schichnes, Nemson, Sohlberg and Ruzin, 1999) and cut to 10‐μm sections on a rotary microtome. The sections were processed and hybridized to the GmSNAP18 riboprobes according to published protocols (Jackson, 1991). Hybridized probes were detected as previously described (Ruzin, 1999). Labelled cells were examined with a Zeiss Axioskop 2 microscope (Carl Zeiss Microscopy, LLC, White Plains, NY), and images were captured with a Zeiss Axiocam.
Immunostaining of GmSNAP18
To prepare for immunolocalizations, excised nematode‐infected soybean root tips were fixed in AA (50% ethanol, 5% glacial acetic acid), paraffin‐embedded and sectioned as described above. The slides were treated with xylene to remove the paraffin, followed by a one‐minute incubation in cold acetone. The tissue was then rehydrated as previously described (Ruzin, 1999). After treating with 4 μg/mL Proteinase K for 45 min, the tissue was fixed in 4% formalin. The slides were blocked in 1% goat serum and then incubated at room temperature for three hours with purified anti‐GmSNAP18 antibodies diluted 500‐fold. After washing, the slides were incubated for three hours at room temperature with Oregon Green 488 goat anti‐rabbit IgG antibodies (Molecular Probes, Eugene, OR) diluted 1:250. Labelled cells were examined with a Zeiss Axioskop 2 microscope (Carl Zeiss Microscopy, LLC) with a fluorescent light source, and images were captured with a Zeiss Axiocam.
Co‐immunoprecipitation
Total proteins from soybean Forrest c.v. and N. benthamiana were extracted in lysis buffer containing 5mM DTT, 1% (v/v) NP40, 1mM sodium molybdate, 1 mm NaF, 1 mm PMSF, 1.5 mm Na3VO4, 100 mm NaCl, 2 mm EDTA, 50 mm Tris–HCl at pH 7.5, 10% (v/v) glycerol and one tablet from the plant protease and phosphatase inhibitors at 1:100 mL (Thermo Scientific). Protein concentration was quantified using Coomassie Bradford Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). For in planta co‐IP analysis, anti‐SNAP18 and anti‐SHMT08 polyclonal antibodies were immobilized in a column containing Aminolink Plus coupling resin (Pierce Co‐Immunoprecipitation Kit), and then, immunoblot analysis of root protein fraction samples from soybean Forrest, soybeans Essex (homologous system), or of leaf protein from N. benthamiana (heterologous system) was incubated overnight with the immobilized antibodies. After extensive washes, the immunoprecipitated associated proteins were eluted. The eluted fraction was then used for both Western blotting and mass spectrometry analysis.
For Western blotting analysis, anti‐SNAP18 (Rockland Immunochemicals, Limerick, PA), anti‐SHMT (Agrisera #AS05 075) or anti‐HA (Thermo Scientific #RB‐1438) polyclonal antibodies were used. Anti‐Rubisco (Bioss #6988R) polyclonal antibodies were used as a negative control. For native gel analysis, DTT and SDS agents were removed.
Mass spectrometry analysis
Peptide digestion, microsequencing analyses and protein characterization of the SHMT‐associated proteins from non‐infected and SCN‐infected Forrest roots at 5 DAI were carried out in the Charles W Gehrke Proteomics Center at the University of Missouri‐Columbia. The eluted fractions obtained from the Co‐IP experiments using anti‐GmSHMT08 polyclonal antibodies were briefly subjected to lyophilization, and then, all proteins were subsequently digested with trypsin. Furthermore, samples were acidified, lyophilized and re‐suspended in 21 µL, and peptides were analysed by LC‐MS (18 µL injection). BSA was used for quality control on the column. Searches of Swiss‐Prot‐all species and NCBI‐Gmax were conducted using Sorcerer‐Sequest.
BiFC assay
The coding sequence of Forrest and Essex GmSHMT08 wild‐type was cloned into pSAT4‐nEYFP‐C1 to generate nEYFP‐SHMT08 fusions. Likewise, GmPR08‐Bet VI and GmSNAP18 from Forrest and Essex were cloned into pSAT4‐cEYFP‐C1‐B to generate cEYFP‐GmPR08‐Bet VI and cEYFP‐GmSNAP18 fusions. Various combinations of cEYFP and nEYFP fusions including controls were co‐expressed in onion (Allium cepa) epidermal cells by particle bombardment. In order to test interactions among the three genes, the third gene was cloned into the pG2RNAi vector and was then co‐expressed along with cEYFP and nEYFP fusions in onion epidermal cells. Onion tissues co‐transformed with cEYFP and nEYFP fusions were incubated in the dark at 25 °C, and after 16–36 h, the tissues were examined for YFP activity. Fluorescent and bright field images were captured using EVOS® FL Auto Cell Imaging System (Life Technologies, Carlsbad, CA).
Transgenic soybean composite hairy root and SCN screening
A 462‐bp fragment of the GmPR08‐Bet VI (Glyma.08G230500) cDNA sequence was amplified from soybean (cv. Forrest) root cDNA by RT–PCR and cloned into the pG2RNAi2 vector under the control of the soybean ubiquitin (GmUbi) promoter. Cloning was carried out between AscI and AvrII cloning sites in the pG2RNAi2 vector to generate pG2RNAi2::GmPR08‐Bet VI. Williams 82 composite hairy roots transformed with pG2RNAi2::empty vectors were used as a negative control. The pG2RNAi2 vector has a GFP selectable marker in planta. Transgenic Williams 82 composite hairy roots transformed with pG2RNAi2::GmPR08‐Bet VI were produced by injecting agrobacterium bacterial suspensions three times into the hypocotyl directly below soybean cotyledons using a 3‐mL needle (BD#309578). After injection, composite hairy roots from at least 10 independent soybean transgenic plants per construct were grown and propagated in medium vermiculite. Transgenic soybeans were covered with plastic humidity domes sprayed consistently with water and maintained in a growth chamber for 1–2 weeks and fertilized once a week with NPK 20‐20‐20 fertilizer. GFP‐positive composite hairy roots at ~2–3 inches long were transferred to sandy soil before SCN screening. Growth conditions and SCN screenings were performed as mentioned earlier. After 30 days, cysts were counted under a stereomicroscope. The experiment was independently conducted three times with a minimum of 10 independent composite hairy root lines per construct. The results were plotted and analysed for statistical significance by using analysis of variance (ANOVA), using the JMP Pro V12 software as described earlier.
Modelling of GmSNAP18
Homology modelling of a putative GmSNAP18 protein structure was conducted with Deepview and Swiss‐Model Workspace software using the protein sequence from Forrest and an available α‐SNAP crystal structure from Rattus norvegicus as a template; PDB accession 3J96 chain G (Arnold et al., 2006; Guex and Peitsch, 1997; Schwede et al., 2003; Zhao et al., 2015). Residues 6–284 were modelled against this template with a sequence identity of 39% (according to the Protein Data Bank database). TPR domains and haplotype mapping and visualizations were performed using the UCSF Chimera package (Pettersen et al., 2004).
Modelling of GmSHMT08
Homology modelling of a putative tetrameric SHMT protein structure was conducted with the same software as the SNAP, using the SHMT protein sequence from Forrest and the available SHMT crystal structure from Homo sapiens as a template; PDB accession 1BJ4 chain A (Renwick et al., 1998). Residues 11‐462 were modelled against this template with a sequence identity of 60% (according to the Protein Data Bank database). Mutations and haplotypes were then mapped onto the model to elucidate the impact on the predicted interaction and GmSHMT08 tetramerization.
Modelling of GmPR08‐Bet VI
Homology modelling of a putative GmPR08‐Bet VI protein structure was conducted with Deepview and Swiss‐Model Workspace software using the protein sequence from Forrest and an available crystal structure from A. thaliana At1g70830 protein as a template: PDB accession 2I9Y. Residues 2–152 were modelled against this template with a sequence identity of 33.11% (according to the Protein Data Bank database).
Interaction analysis of homology models
After building the GmSNAP18, GmSHMT08 and GmPR08‐Bet VI homology models, structures were submitted to the GRAMM‐X protein docking server (Tovchigrechko and Vakser, 2005). The GRAMM‐X method calculates a fine‐grid projection of a softened Lennard–Jones potential function for each probe atom, followed by a conjugate gradient minimization of the top 4000 grid‐based predictions, accounting for six dimensions: Lennard–Jones potential, evolutionary conservation of predicted interface, statistical residue–residue preference, volume of the minimum, empirical binding free energy and atomic contact energy. A support vector machine filter is then used, and the top ten scoring predictions are returned (Tovchigrechko and Vakser, 2005; Tovchigrechko and Vakser, 2006). Considering that the top two GmSNAP18/GmSHMT08 complex predictions were symmetrical, binding to either side of the tetramer, we chose both as putative interaction sites. Similarly, the obtained top GmPR08‐Bet VI predicted position has been chosen to generate the GmSNAP18/GmSHMT08/GmPR08‐Bet VI protein complex. All three templates used met the minimum requirement of sequence homology (at least 30%) between the target and template (Sensoy et al., 2017).
Conflicts of interest
None.
Author contributions
NL conceived and drafted the manuscript, performed data analysis and interpretation, haplotyping studies, molecular cloning, co‐immunoprecipitation experiments, co‐agroinfiltration in NB, in silico and structural analysis, SCN phenotyping, protein homology modelling, protein–protein docking interaction, and mutational and haplotype mapping analysis. SL and NL carried out the ExF RIL genotyping. SB and NL did qRT‐PCR analysis, immunostaining, in situ analysis and SCN infection. ZZ and NL performed RNA extractions and SCN female index. KL provided for the qRT‐PCR, immunostaining, and in situ analysis, and edited the manuscript. CB and MB did the subcellular localization in Nicotiana. TH and SP did the BiFC analysis. LM assisted with BiFC. Abo, JM, AL, MAK and KJ did data analysis and edited the manuscript. ABe coordinated the SHMT and SNAP localization. KM supervised the work, performed data interpretation, designed the experiment, and edited the manuscript. All authors reviewed and commented on the manuscript.
Consent for publication
Not applicable.
Ethics and consent to participate
This study did not involve humans, human data or animals; no ethics approval or consent is required to publish the results.
Supporting information
Figure S1 Subcellular localization of the GmSNAP18 and GmSHMT08 proteins.
Figure S2 Comparative analysis of the conserved Cysteine Residues in GmSNAP18, GmSNAP11, HsSNAP25 and HsSNAP23.
Figure S3 BiFC analysis between GmSNAP18, GmSHMT08, and GmPR08‐Bet VI from Forrest and Essex.
Figure S4 Haplotype analysis of the soybean PI, Elite, and cultivars used for SCN screening.
Figure S5 Identification of the GmPR08‐Bet VI by mass spectrometry.
Figure S6 Immunoprecipitation of the GmPR08‐Bet VI protein by GmSHMT08.
Figure S7 Cell‐death and necrosis symptoms intensified in N. benthamiana when the three GmSNAP18, GmSHMT08, and GmPR08‐Bet VI genes were co‐agroinfiltrated.
Figure S8 Comparative analysis of the conserved Cytokinin (zeatin) binding site residues at the GmPR08‐Bet VI from Glycine max, MbPR10 (2FLH) from Mung bean, and YlPR10 (2QIM) from Yellow lupine.
Figure S9 GmPR08‐Bet VI haplotype clustering and correlation with SCN female index in sequenced soybean lines.
Figure S10 Negative controls of the BiFC analysis.
Figure S11 Expression analysis of components of the SA signaling pathway reveals that all tested genes are co‐regulated in root cells undergoing nematode infection.
Figure S12 Expression analysis of the GmPR08‐Bet VI, GmSNAP18, and GmSHMT08 under exogenous SA and CKs treatments.
Figure S13 Expression analysis of the (A) GmNPR1.2‐09, (B) GmTGA2‐13, and (C) GmARR03 genes under exogenous SA and CKs treatments.
Figure S14 Physical positions corresponding to GmPR08‐Bet VI and the two identified SCN QTLs at chromosome 08 are shown.
Figure S15 In vivo assays of GmSNAP18 recombinant protein in E. coli, Antibody Anti‐GmSNAP18 production in Rabbit and confirming the specificity of custom‐generated anti‐GmSNAP18 antibodies.
Figure S16 In vivo assays confirming the specificity of custom‐generated anti‐GmSNAP18 antibodies in Soybean.
Table S1 Genotypes of the soybean lines used for expression analysis and SCN screening.
Table S2 The primers used for genotyping, sequencing, subcloning, qRT‐PCR, and in situ analysis.
Appendices S1 Peptide Report LC‐MS.
Appendices S2‐S10 All coding sequences, inserts sizes, vectors and restriction enzyme sites used for cloning (for BiFC analysis and gene overexpression).
Acknowledgements
This research was supported in part from the United Soybean Board to (i) KM and KL, (ii) by a grant from USDA‐NIFA # 2018‐08232 to KM and NL, and (iii) was partially supported by funds from the Tennessee Soybean Promotion Board to the TH. We would like to thank Dr. Gunvant Patil for the haplotyping support and also Dr. Cyril Zipfel for providing the pGWB vectors for agroinfiltration experiments.
Lakhssassi, N. , Piya, S. , Bekal, S. , Liu, S. , Zhou, Z. , Bergounioux, C. , Miao, L. , Meksem, J. , Lakhssassi, A. , Jones, K. , Kassem, M. A. , Benhamed, M. , Bendahmane, A. , Lambert, K. , Boualem, A. , Hewezi, T. and Meksem, K. (2020) A pathogenesis‐related protein GmPR08‐Bet VI promotes a molecular interaction between the GmSHMT08 and GmSNAP18 in resistance to Heterodera glycines . Plant Biotechnol. J., 10.1111/pbi.13343
Data availability statement
The developed cross between the resistant Forrest (+) and the susceptible Essex (−) using both recombinant inbred lines (RILs) and near‐isogenic lines (NILs) seeds, in addition to the soybean EMS mutants, are property of Southern Illinois University (SIU). Access to the germplasm and mutants is subject to a material transfer agreement (MTA).
References
- Albrecht, T. and Argueso, C.T. (2017) Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth‐defence trade‐off. Ann. Bot. 119, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appaji Rao, N. , Ambili, M. , Jala, V.R. , Subramanya, H.S. and Savithri, H.S. (2003) Structure‐function relationship in serine hydroxymethyltransferase. Biochem. Biophys. Acta. 1647, 24–29. [DOI] [PubMed] [Google Scholar]
- Arnold, K. , Bordoli, L. , Kopp, J. and Schwede, T. (2006) The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics (Oxford, England), 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Bayless, A.M. , Smith, J.M. , Song, J. , McMinn, P.H. , Teillet, A. , August, B.K. and Bent, A.F. (2016) Disease resistance through impairment of α‐SNAP–NSF interaction and vesicular trafficking by soybean Rhg1. Proc. Natl Acad. Sci. USA, 113, E7375–E7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless, A.M. , Zapotocny, R.W. , Grunwald, D.J. , Amundson, K.K. , Diers, B.W. and Bent, A.F. (2018) An atypical N‐ethylmaleimide sensitive factor enables the viability of nematode‐resistant Rhg1 soybeans. Proc. Natl Acad. Sci. USA, 115, E4512–E4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal, S. , Domier, L.L. , Gonfa, B. , Lakhssassi, N. , Meksem, K. and Lambert, K.N. (2015) A SNARE‐like protein and biotin are implicated in soybean cyst nematode virulence. PLoS ONE, 10, e0145601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, N. and Lu, H. (2005) Correlation between gene expression profiles and protein‐protein interactions within and across genomes. Bioinformatics (Oxford, England), 21, 2730–2738. [DOI] [PubMed] [Google Scholar]
- Blatch, G.L. and Lassle, M. (1999) The tetratricopeptide repeat: a structural motif mediating protein‐protein interactions. BioEssays, 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Boyle, P. , Le Su, E. , Rochon, A. , Shearer, H.L. , Murmu, J. , Chu, J.Y. , Fobert, P.R. et al. (2009) The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell, 21, 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catsicas, S. , Larhammar, D. , Blomqvist, A. , Sanna, P.P. , Milner, R.J. and Wilson, M.C. (1991) Expression of a conserved cell‐type‐specific protein in nerve terminals coincides with synaptogenesis. Proc. Natl Acad. Sci. USA, 88, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.A. , Scales, S.J. , Jagath, J.R. and Scheller, R.H. (2001) A discontinuous SNAP‐25 C‐terminal coil supports exocytosis. J. Biol. Chem. 276, 28503–28508. [DOI] [PubMed] [Google Scholar]
- Clary, D.O. , Griff, I.C. and Rothman, J.E. (1990) SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell, 61, 709–721. [DOI] [PubMed] [Google Scholar]
- Cook, D.E. , Lee, T.G. , Guo, X. , Melito, S. , Wang, K. , Bayless, A.M. , Wang, J. et al. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science, 338, 1206–1209. [DOI] [PubMed] [Google Scholar]
- D'Andrea, L.D. and Regan, L. (2003) TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662. [DOI] [PubMed] [Google Scholar]
- Denancé, N. , Sánchez‐Vallet, A. , Goffner, D. and Molina, A. (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4, 155–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, H. , Pasternak, O. , Bujacz, G. , Bujacz, A. , Sikorski, M.M. and Jaskolski, M. (2008) Lupinus luteus pathogenesis‐related protein as a reservoir for cytokinin. J. Mol. Biol. 378, 1040–1051. [DOI] [PubMed] [Google Scholar]
- Gardiner, K. (2004) Gene‐dosage effects in Down syndrome and trisomic mouse models. Genome Biol. 5, 244–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen, G. and Mitchum, M.G. (2019) Phytoparasitic nematode control of plant hormone pathways. Plant Physiol. 179, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo, S. and Linder, M.E. (1998a) SNAP‐25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol. Biol. Cell, 9, 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M. and Lamb, C. (2006) Systemic immunity. Curr. Opin. Plant Biol. 9, 414–420. [DOI] [PubMed] [Google Scholar]
- Grizot, S. , Fieschi, F. , Dagher, M.C. and Pebay‐Peyroula, E. (2001) The active N‐terminal region of p67phox. Structure at 1.8 A resolution and biochemical characterizations of the A128V mutant implicated in chronic granulomatous disease. J. Biol. Chem. 276, 21627–21631. [DOI] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. (1997) SWISS‐MODEL and the Swiss‐PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hess, D.T. , Slater, T.M. , Wilson, M.C. and Skene, J.H. (1992) The 25 kDa synaptosomal‐associated protein SNAP‐25 is the major methionine‐rich polypeptide in rapid axonal transport and a major substrate for palmitoylation in adult CNS. J. Neurosci. 12, 4634–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. (1991). in situ hybridization in plants. In Molecular Plant Pathology, A Practical Approach (Gurr S. J., McPherson, Bowles D. J., Eds.), Vol. 1, pp. 163–174. Oxford, England: Oxford University Press. [Google Scholar]
- Jain, S. and Kumar, A. (2015) The pathogenesis related class 10 proteins in plant defense against biotic and abiotic stresses. Adv Plants Agric Res. 2, 305–314. [Google Scholar]
- Kalde, M. , Nuhse, T.S. , Findlay, K. and Peck, S.C. (2007) The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis‐related protein 1. Proc. Natl Acad. Sci. USA, 104, 11850–11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth, P.K. , Ithal, N. , Recknor, J. , Maier, T. , Nettleton, D. , Baum, T.J. and Mitchum, M.G. (2011) The Soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress‐ and defense‐related genes in degenerating feeding cells. Plant Physiol. 155, 1960–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth, P.K. , Liu, S. , Prenger, E. , Ludwig, A. , Lakhssassi, N. , Heinz, R. , Zhou, Z. et al. (2017) Systematic mutagenesis of serine hydroxymethyltransferase reveals essential role in nematode resistance. Plant Physiol. 175, 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik, R. , Grefen, C. , Bayne, R. , Honsbein, A. , Kohler, T. , Kioumourtzoglou, D. , Williams, M. et al. (2013) Arabidopsis Sec1/Munc18 protein SEC11 is a competitive and dynamic modulator of SNARE binding and SYP121‐dependent vesicle traffic. Plant Cell, 25, 1368–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.N. , Cha, J.S. , Cho, T.J. and Kim, H.Y. (2003) Salicylic acid and wounding induce defense‐related proteins in Chinese cabbage. Korean J. Biol. Sci. 7, 213. [Google Scholar]
- Kim, S.T. , Yu, S. , Kang, Y.H. , Kim, S.G. , Kim, J.‐Y. , Kim, S.‐H. and Kang, K.Y. (2007) The rice pathogen‐related protein 10 (JIOsPR10) is induced by abiotic and biotic stresses and exhibits ribonuclease activity. Plant Cell Rep. 27, 593. [DOI] [PubMed] [Google Scholar]
- Klink, V.P. , Sharma, K. , Pant, S.R. , McNeece, B. , Niraula, P. and Lawrence, G.W. (2017) Components of the SNARE‐containing regulon are co‐regulated in root cells undergoing defense. Plant Signal. Behav. 12, e1274481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C. , Neu, C. , Pajonk, S. , Yun, H.S. , Lipka, U. , Humphry, M. , Bau, S. et al. (2008) Co‐option of a default secretory pathway for plant immune responses. Nature, 451, 835–840. [DOI] [PubMed] [Google Scholar]
- Lakhssassi, N. , Doblas, V.G. , Rosado, A. , del Valle, A.E. , Pose, D. , Jimenez, A.J. , Castillo, A.G. et al. (2012a) The Arabidopsis tetratricopeptide thioredoxin‐like gene family is required for osmotic stress tolerance and male sporogenesis. Plant Physiol. 158, 1252–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi, N. , Doblas, V.G. , Rosado, A. , del Valle, A.E. , Posé, D. , Jimenez, A.J. , Castillo, A.G. et al. (2012b) The Arabidopsis TETRATRICOPEPTIDE THIOREDOXIN‐LIKE gene family is required for osmotic stress tolerance and male sporogenesis. Plant Physiol. 158, 1252–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi, N. , Liu, S. , Bekal, S. , Zhou, Z. , Colantonio, V. , Lambert, K. , Barakat, A. et al. (2017) Characterization of the Soluble NSF Attachment Protein gene family identifies two members involved in additive resistance to a plant pathogen. Sci. Rep. 7, 45226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi, N. , Patil, G. , Piya, S. , Zhou, Z. , Baharlouei, A. , Kassem, M.A. , Lightfoot, D.A. et al. (2019) Genome reorganization of the GmSHMT gene family in soybean showed a lack of functional redundancy in resistance to soybean cyst nematode. Sci. Rep. 9, 1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, B. , Hartl, D.L. , Zhou, J. and Dopman, E.B. .(2011) Copy‐number variation: the balance between gene dosage and expression in Drosophila melanogaster . Genome Biol. Evol. 3, 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon, J. , Lawton, M.A. and Raskin, I. (1995) Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol. 108, 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot, D.A. , Njiti, V.N. , Gibson, P.T. , Kassem, M.A. , Iqbal, J.M. and Meksem, K. (2005) Registration of the essex × forrest recombinant inbred line mapping population. Crop Sci. 45, 1678. [DOI] [PubMed] [Google Scholar]
- Lim, U. , Peng, K. , Shane, B. , Stover, P.J. , Litonjua, A.A. , Weiss, S.T. , Gaziano, J.M. et al. (2005) Polymorphisms in cytoplasmic serine hydroxymethyltransferase and methylenetetrahydrofolate reductase affect the risk of cardiovascular disease in men. J. Nutr. 135, 1989–1994. [DOI] [PubMed] [Google Scholar]
- Lin, J. , Mazarei, M. , Zhao, N. , Zhu, J.J. , Zhuang, X. , Liu, W. , Pantalone, V.R. et al. (2013) Overexpression of a soybean salicylic acid methyltransferase gene confers resistance to soybean cyst nematode. Plant Biotechnol. J. 11, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Liu, S. , Jamai, A. , Bendahmane, A. , Lightfoot, D.A. , Mitchum, M.G. and Meksem, K. (2011) Soybean cyst nematode resistance in soybean is independent of the Rhg4 locus LRR‐RLK gene. Funct. Integr. Genomics, 11, 539–549. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Kandoth, P.K. , Warren, S.D. , Yeckel, G. , Heinz, R. , Alden, J. , Yang, C. et al. (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature, 492, 256–260. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Chen, N. , Grant, J.N. , Cheng, Z.M. , Stewart, C.N. Jr and Hewezi, T. (2015) Soybean kinome: functional classification and gene expression patterns. J. Exp. Bot. 66, 1919–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kandoth, P.K. , Lakhssassi, N. , Kang, J. , Colantonio, V. , Heinz, R. , Yeckel, G. et al. (2017) The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat. Commun. 8, 14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Kang, Y. , Jiao, J. , Rebane, A.A. , Cha, H.K. , Xi, Z. , Qu, H. et al. (2016) α‐SNAP enhances SNARE zippering by stabilizing the SNARE four‐helix bundle. Cell Rep. 15, 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks, O.D. , Labuschagne, C.F. , Adams, P.D. and Vousden, K.H . (2016) Serine metabolism supports the methionine cycle and DNA/RNA methylation through De Novo ATP synthesis in cancer cells. Mol. Cell 61, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam, R. and Skorupska, H.T. (1996) Cytological expression of early response to infection by Heterodera glycines Ichinohe in resistant PI 437654 soybean. Genome, 39, 986–998. [DOI] [PubMed] [Google Scholar]
- Marz, K.E. , Lauer, J.M. and Hanson, P.I. (2003) Defining the SNARE complex binding surface of alpha‐SNAP: implications for SNARE complex disassembly. J. Biol. Chem. 278, 27000–27008. [DOI] [PubMed] [Google Scholar]
- Matthews, B.F. , Beard, H. , Brewer, E. , Kabir, S. , MacDonald, M.H. and Youssef, R.M. (2014a) Arabidopsis genes, AtNPR1, AtTGA2 and AtPR‐5, confer partial resistance to soybean cyst nematode (Heterodera glycines) when overexpressed in transgenic soybean roots. BMC Plant Biol. 14, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, C.R. , Hsu, M. , Painter, J.E. , Gagne, J.M. , Karlsberg, S.D. and Salome, P.A. (2000) Integrated temporal regulation of the photorespiratory pathway. Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol. 123, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meksem, K. , Pantazopoulos, P. , Njiti, V.N. , Hyten, L.D. , Arelli, R.P. and Lightfoot, A.D. (2001) ’Forrest’ resistance to the soybean cyst nematode is bigenic: saturation mapping of the Rhg1and Rhg4 loci. Theoret. Appl. Genet. 103, 710–717. [Google Scholar]
- Melillo, M.T. , Leonetti, P. , Bongiovanni, M. , Castagnone‐Sereno, P. and Bleve‐Zacheo, T. (2006) Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato‐root‐knot nematode interactions. New Phytol. 170, 501–512. [DOI] [PubMed] [Google Scholar]
- Moreno, J.I. , Martin, R. and Castresana, C. (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 41, 451–463. [DOI] [PubMed] [Google Scholar]
- Mouillon, J.M. , Aubert, S. , Bourguignon, J. , Gout, E. , Douce, R. and Rebeille, F. (1999) Glycine and serine catabolism in non‐photosynthetic higher plant cells: their role in C1 metabolism. Plant J. 20, 197–205. [DOI] [PubMed] [Google Scholar]
- Oyler, G.A. , Higgins, G.A. , Hart, R.A. , Battenberg, E. , Billingsley, M. , Bloom, F.E. and Wilson, M.C. (1989) The identification of a novel synaptosomal‐associated protein, SNAP‐25, differentially expressed by neuronal subpopulations. J. Cell. Biol. 109, 3039–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, S.R. , Matsye, P.D. , McNeece, B.T. , Sharma, K. , Krishnavajhala, A. , Lawrence, G.W. and Klink, V.P. (2014) Syntaxin 31 functions in Glycine max resistance to the plant parasitic nematode Heterodera glycines. Plant Mol. Biol. 85, 107–121. [DOI] [PubMed] [Google Scholar]
- Paone, A. , Marani, M. , Fiascarelli, A. , Rinaldo, S. , Giardina, G. , Contestabile, R. , Paiardini, A. et al. (2014) SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis. 5, e1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak, O. , Bujacz, G.D. , Fujimoto, Y. , Hashimoto, Y. , Jelen, F. , Otlewski, J. , Sikorski, M.M. et al. (2006) Crystal structure of Vigna radiata cytokinin‐specific binding protein in complex with zeatin. Plant Cell, 18, 2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, G.B. , Lakhssassi, N. , Wan, J. , Song, L. , Zhou, Z. , Klepadlo, M. , Vuong, T.D. et al. (2019) Whole genome re‐sequencing reveals the impact of the interaction of copy number variants of the rhg‐1 and Rhg4 genes on broad‐based resistance to soybean cyst nematode. Plant Biotechnol. J. 17, 1595‐1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F. , Goddard, T.D. , Huang, C.C. , Couch, G.S. , Greenblatt, D.M. , Meng, E.C. and Ferrin, T.E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rahman, T.A. , Oirdi, M.E. , Gonzalez‐Lamothe, R. and Bouarab, K. (2012) Necrotrophic pathogens use the salicylic acid signaling pathway to promote disease development in tomato. MPMI, 25, 1584–1593. [DOI] [PubMed] [Google Scholar]
- Renwick, S.B. , Snell, K. and Baumann, U. (1998) The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure (London, England 1993), 6, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Risinger, C. , Blomqvist, A.G. , Lundell, I. , Lambertsson, A. , Nassel, D. , Pieribone, V.A. , Brodin, L. et al. (1993) Evolutionary conservation of synaptosome‐associated protein 25 kDa (SNAP‐25) shown by Drosophila and Torpedo cDNA clones. J. Biol. Chem. 268, 24408–24414. [PubMed] [Google Scholar]
- Rojas, C.M. , Senthil‐Kumar, M. , Tzin, V. and Mysore, K.S. (2014) Regulation of primary plant metabolism during plant‐pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999) Plant Microtechnique and Microscopy. New York: Oxford University Press. ISBN 0-19-508956-1. [Google Scholar]
- Schichnes, D. , Nemson, J. , Sohlberg, L. and Ruzin, S.E. (1999) Microwave Protocols for Paraffin Microtechnique and In Situ Localization in Plants. Microsc. Microanal. 4, 491–496. [DOI] [PubMed] [Google Scholar]
- Schirch, L. (1982) Serine hydroxymethyltransferase. Adv. Enzymol. Relat. Areas Mol. Biol. 53, 83–112. [DOI] [PubMed] [Google Scholar]
- Schwede, T. , Kopp, J. , Guex, N. and Peitsch, M.C. . (2003) SWISS‐MODEL: an automated protein homology‐modeling server. Nucleic Acids Res. 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensoy, O. , Almeida, J.G. , Shabbir, J. , Moreira, I.S. and Morra, G. (2017) Chapter 16 ‐ Computational studies of G protein‐coupled receptor complexes: Structure and dynamics. In Methods in Cell Biology ( Shukla, A.K. , ed), pp. 205–245. Cambridge, MA: Academic Press. [DOI] [PubMed] [Google Scholar]
- Siddique, S. , Matera, C. , Radakovic, Z.S. , Hasan, M.S. , Gutbrod, P. , Rozanska, E. , Sobczak, M. et al. (2014) Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci. Signal. 7, ra33. [DOI] [PubMed] [Google Scholar]
- Siddique, S. , Radakovic, Z.S. , De La Torre, C.M. , Chronis, D. , Novák, O. , Ramireddy, E. , Holbein, J. et al. (2015) A parasitic nematode releases cytokinin that controls cell division and orchestrates feeding site formation in host plants. Proc. Natl Acad. Sci. USA, 112, 12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola, C.F. , Smith, M.T. , Hubbard, A. , Shane, B. , Roberts, A.C. , Law, G.R. , Rollinson, S. et al. (2002) Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood, 99, 3786–3791. [DOI] [PubMed] [Google Scholar]
- Sohoki, M.M. , Browne, S.J. , Sullivan, L.S. , Blackshaw, S. , Cepko, C.L. , Payne, A.M. , Bhattacharya, S.S. et al. (2000) Mutations in a new photoreceptor‐pineal gene on 17p cause leber congenital amaurosis. Nat gen 2000;24:79–83. Am. J. Ophthalmol. 129, 834–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C.R. and Ogren, W.L. (1981) Photorespiration‐deficient mutants of arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 67, 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, P.V.S. (1990) Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10‐methyltetrahydrofolate to 5‐formyltetrahydrofolate. J. Biol. Chem. 265, 14227–14233. [PubMed] [Google Scholar]
- Swaminathan, S. , Abeysekara, N.S. , Knight, J.M. , Liu, M. , Dong, J. , Hudson, M.E. , Bhattacharyya, M.K. et al. (2018) Mapping of new quantitative trait loci for sudden death syndrome and soybean cyst nematode resistance in two soybean populations. Theor. Appl. Genet. 131, 1047–1062. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko, A. and Vakser, I.A. (2005) Development and testing of an automated approach to protein docking. Proteins, 60, 296–301. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko, A. and Vakser, I.A. (2006) GRAMM‐X public web server for protein‐protein docking. Nucleic Acids Res. 34, W310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan, B. , Dan, Q. , Patil, G. , Zeng, P. , Huang, J. , Dai, L. , Chen, C. et al. (2016) Landscape of genomic diversity and trait discovery in soybean. Sci. Rep. 6, 23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R.A. (2004) Gene dosage balance in cellular pathways: implications for dominance and gene duplicability. Genetics, 168, 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R.A. (2005) Gene dosage balance: deletions, duplications and dominance. Trends Genet. 21, 33–35. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Rivas, S. , Mestre, P. and Baulcombe, D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Vuong, T.D. , Sleper, D.A. , Shannon, J.G. and Nguyen, H.T. (2010) Novel quantitative trait loci for broad‐based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor. Appl. Genet. 121, 1253–1266. [DOI] [PubMed] [Google Scholar]
- Wang, K.L. , Yoshida, H. , Lurin, C. and Ecker, J.R. (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature, 428, 945–950. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Sun, K. , Sandoval, F.J. , Cross, J.M. , Gordon, C. , Kang, C. and Roje, S. (2013) Folate polyglutamylation eliminates dependence of activity on enzyme concentration in mitochondrial serine hydroxymethyltransferases from Arabidopsis thaliana. Arch. Biochem. Biophys. 536, 87–96. [DOI] [PubMed] [Google Scholar]
- Yoshida, H. , Nagata, M. , Saito, K. , Wang, K.L. and Ecker, J.R. (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1‐aminocyclopropane‐1‐carboxylate synthases. BMC Plant Biol 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Kjemtrup‐Lovelace, S. , Li, C. , Luo, Y. , Chen, L.P. and Song, B.H. (2017) Comparative RNA‐Seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja). Sci. Rep. 7, 9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Wu, S. , Zhou, Q. , Vivona, S. , Cipriano, D.J. , Cheng, Y. and Brunger, A.T. (2015) Mechanistic insights into the recycling machine of the SNARE complex. Nature, 518, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Subcellular localization of the GmSNAP18 and GmSHMT08 proteins.
Figure S2 Comparative analysis of the conserved Cysteine Residues in GmSNAP18, GmSNAP11, HsSNAP25 and HsSNAP23.
Figure S3 BiFC analysis between GmSNAP18, GmSHMT08, and GmPR08‐Bet VI from Forrest and Essex.
Figure S4 Haplotype analysis of the soybean PI, Elite, and cultivars used for SCN screening.
Figure S5 Identification of the GmPR08‐Bet VI by mass spectrometry.
Figure S6 Immunoprecipitation of the GmPR08‐Bet VI protein by GmSHMT08.
Figure S7 Cell‐death and necrosis symptoms intensified in N. benthamiana when the three GmSNAP18, GmSHMT08, and GmPR08‐Bet VI genes were co‐agroinfiltrated.
Figure S8 Comparative analysis of the conserved Cytokinin (zeatin) binding site residues at the GmPR08‐Bet VI from Glycine max, MbPR10 (2FLH) from Mung bean, and YlPR10 (2QIM) from Yellow lupine.
Figure S9 GmPR08‐Bet VI haplotype clustering and correlation with SCN female index in sequenced soybean lines.
Figure S10 Negative controls of the BiFC analysis.
Figure S11 Expression analysis of components of the SA signaling pathway reveals that all tested genes are co‐regulated in root cells undergoing nematode infection.
Figure S12 Expression analysis of the GmPR08‐Bet VI, GmSNAP18, and GmSHMT08 under exogenous SA and CKs treatments.
Figure S13 Expression analysis of the (A) GmNPR1.2‐09, (B) GmTGA2‐13, and (C) GmARR03 genes under exogenous SA and CKs treatments.
Figure S14 Physical positions corresponding to GmPR08‐Bet VI and the two identified SCN QTLs at chromosome 08 are shown.
Figure S15 In vivo assays of GmSNAP18 recombinant protein in E. coli, Antibody Anti‐GmSNAP18 production in Rabbit and confirming the specificity of custom‐generated anti‐GmSNAP18 antibodies.
Figure S16 In vivo assays confirming the specificity of custom‐generated anti‐GmSNAP18 antibodies in Soybean.
Table S1 Genotypes of the soybean lines used for expression analysis and SCN screening.
Table S2 The primers used for genotyping, sequencing, subcloning, qRT‐PCR, and in situ analysis.
Appendices S1 Peptide Report LC‐MS.
Appendices S2‐S10 All coding sequences, inserts sizes, vectors and restriction enzyme sites used for cloning (for BiFC analysis and gene overexpression).
Data Availability Statement
The developed cross between the resistant Forrest (+) and the susceptible Essex (−) using both recombinant inbred lines (RILs) and near‐isogenic lines (NILs) seeds, in addition to the soybean EMS mutants, are property of Southern Illinois University (SIU). Access to the germplasm and mutants is subject to a material transfer agreement (MTA).


