Abstract
Laryngeal carcinoma (LCC) is a common malignant tumor with low radiosensitivity and generally poor response rates. The ubiquitin protein ligase E3 component n-recognin 5 (UBR5) has prognostic implications in several neoplasms; however, its role in LCC and radiotherapy sensitivity remains unknown. Immunohistochemistry and bioinformatics analyses were performed to measure UBR5 protein and mRNA expression in LCC and adjacent non-tumor tissues. The gene and protein expression of UBR5 in LCC and HuLa-PC cell lines were measured using quantitative PCR and western blot analyses. Following transfection with small interfering RNA or UBR5 overexpression plasmid in LCC cells, the proliferation, cell cycle distribution, invasion, migration and radiosensitivity of LCC cells were analyzed. UBR5-related lncRNA, targeted miRNA and protein-protein interaction networks were analyzed using bioinformatics. Finally, the expression of the p38/mitogen-activated protein kinase (MAPK) pathway was evaluated following UBR5 silencing in M2E cells treated with radiation. Increased UBR5 expression was observed in LCC tissues compared with adjacent non-tumor tissues, and it was correlated with poor overall survival of LCC patients. After overexpression or silencing of UBR5 in M2E and M4E LCC cells, cell proliferation and radiosensitivity were significantly increased or decreased, respectively, compared with the control groups. The percentage of S phase cells decreased in the UBR5 si-RNA group compared with that in the control group, while overexpression of UBR5 exerted no effect on the cell cycle. In addition, the expression of Bcl-2 and p38 was decreased in the si-UBR5 combined with radiation groups. The level of phosphorylated p38 expression was increased after combination of si-UBR5 with radiation. The small molecule inhibitor of p38/MAPK signaling, SB203580, decreased the viability of UBR5-overexpressing cells and the survival fraction when cells were exposed to radiation. These findings demonstrated that UBR5 may be involved in regulating cell proliferation and sensitivity to radiotherapy in LCC via the p38/MAPK pathway, thereby highlighting its possible value for the development of new therapeutic strategies and targets for the treatment of this disease.
Keywords: ubiquitin protein ligase E3 component n-recognin 5, laryngeal carcinoma, radiosensitivity, proliferation, p38/mitogen-activated protein kinase
Introduction
Laryngeal carcinoma (LCC) is a common malignant tumor of the head and neck, which accounts for 5.7–7.6% of all malignant tumors (1). Approximately 40% of patients with LCC have late-stage (III–IV) disease at the time of diagnosis (1,2). Due to the basic laryngeal and hypopharyngeal dysfunction, the presenting symptoms, such as hoarseness, difficulty swallowing, or difficulty breathing, lead to a significant decline in the quality of life of the patients (3,4). Therefore, in addition to achieving optimal local control, it is crucial to maintain vocalization and swallowing function. Radiotherapy, one of the main treatment modalities for LCC, may fully preserve the vocal function of the patients and is considered to be a valuable alternative to total laryngectomy for advanced tumors (5,6). A comprehensive treatment strategy has been developed for LCC that includes surgical resection combined with radiotherapy or chemotherapy (7,8). However, despite this comprehensive treatment regimen, tumors often exhibit low sensitivity to radiotherapy and the response rates are generally poor, particularly in patients with advanced and/or recurrent tumors (8). Therefore, improved strategies for the prevention and treatment of LCC, including the exploration of radiotherapy and therapeutic targets, are urgently needed, which represents an important and pressing issue in the field of biomedical research.
The ubiquitin-proteasome system (UPS) is a regulator of protein homeostasis and cellular signaling. Defective UPS may lead to abnormal protein expression, interaction and cellular localization (9). Among the three known components of the UPS, the E3 ubiquitin ligases are primarily responsible for determining substrate specificity and ubiquitin chain topology (10). Recently, targeting E3 ligases has attracted interest as a strategy for cancer treatment (9,10). A previous study demonstrated that inhibition of the E3 ubiquitin ligase CHIP promotes radiosensitivity in human lung cancer cells via the CHIP-HSP70-p21 ubiquitination/degradation axis (11). Increased expression of the E3 ligase cIAP2 resulted in altered MRE11 ubiquitination models and mediated radiosensitization in response to histone deacetylase inhibition (12). In addition, an increasing number of studies have indicated E3 ubiquitin ligases as novel effectors linking the p38/mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase signaling pathways to the cell cycle (13,14). These findings prompt further investigation of E3 ligases as potential regulators of radiosensitivity in cancer.
Ubiquitin protein ligase E3 component n-recognin 5 (UBR5), also referred to as EDD, is a nuclear phosphoprotein involved in the regulation of DNA damage responses, β-catenin activity, metabolic processes and apoptosis (9). UBR5 was also recently identified as a key regulator of the UPS and ciliogenesis, which may have important implications in elucidating cancer pathophysiology (10,15). As a downstream factor of BMI1, UBR5 is enriched to ultraviolet (UV) radiation-induced damage along with the FACT component SPT16 (16). UBR5 was identified as a mediator of the activating phosphorylation of checkpoint kinase 2 in response to DNA damage following exposure to ionizing radiation (IR), revealing its potential importance in cancer (17). Moreover, UBR5 is a key component of ataxia-telangiectasia mutated (ATM) activation in response to IR. Upon stimulation by IR, UBR5 catalyzes the ubiquitination of ATM, which decreases the interaction of the ATM interacting protein with ATM and promotes Mre11/Rad50/NBS1-mediated signaling, thus impairing checkpoint activation and increasing radiosensitivity (18). In addition, increased UBR5 expression has been shown to promote metastasis in triple-negative breast cancer (19), while high UBR5 expression in ovarian cancer is associated with poor prognosis (20). Therefore, UBR5 may play an important role in regulating sensitivity to radiotherapy in these types of cancer. However, the function of UBR5 in LCC and its potential role in radiosensitivity remain unclear. A previous study indicated that p38/MAPK phosphorylation is associated with radiosensitivity in liver cancer (21). The aim of the present study was to analyze the expression and function of UBR5 in LCC cell lines, and to further investigate whether UBR5 regulates radiosensitivity in LCC via p38/MAPK signaling, with the hope of uncovering the mechanism underlying the development of radiosensitivity and identifying a novel target for the clinical treatment of LCC.
Materials and methods
Patients and variables
A total of 171 LCC patients from the First People's Hospital of Foshan were consecutively recruited between August 2011 and May 2018. LCC and adjacent tissue samples were collected during surgery or biopsy. The clinicopathological characteristics of LCC patients, including age, sex, TNM stage and absence of radiotherapy history, were reviewed to identify their association with UBR5 expression. Biopsies from LCC patients were obtained prior to radiotherapy. The present study was approved by the Ethics Committee of the First People's Hospital of Foshan. Written informed consent was obtained from all patients who participated in this study, according to the committee regulations.
Raw biological microarray data and functional enrichment analysis
Raw DNA microarray data of patients with LCC were obtained from Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) (22). Corresponding probes were converted into symbols according to the annotation information in the platform. Three chip datasets GSE51985 (10 LCC and 10 normal samples) were downloaded from GEO (Illumina GPL10558 platform). Back-ground correction of probe data, normalization and summarization were executed by robust multi-array average analysis algorithm17 in the affy package of R. The follow-up duration was estimated using the Kaplan-Meier method with 95% confidence interval (95%CI) and log-rank test in separate curves. In addition, related long non-coding RNAs (lncRNAs), targeted miRNA and protein-protein interaction (PPI) networks were predicted using R software. The biological process from Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis of PPI genes was performed and visualized using ClueGO, version 2.5.3 (23) and CluePedia, version 1.5.3 (24).
Cell lines and culture
The human LCC cell lines M2E and M4E were purchased from Central South University (http://gdyjzx.csu.edu.cn/info/1034/1204_2.htm). The HuLa-PC cell line was a kind gift from Dr Deng (Tongren Hospital of WuHan University). All cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 100 µg/ml penicillin. The cells were cultured in a 37°C humidified incubator with an atmosphere of 5% CO2 and were harvested in the logarithmic growth phase. Both M2E and M4E cell lines were authenticated by STR profiling.
Immunohistochemistry (IHC) staining
The tissue samples were fixed in formalin at room temperature for 24 h, embedded in paraffin, cut into 10-µm sections and deparaffinized. IHC staining was performed using a Dako Envision System (Dako; Agilent Technologies, Inc.) following the manufacturer's protocol at room temperature. The sections were blocked using serum-free protein blocking buffer (Dako; Agilent Technologies, Inc.) for 30 min at room temperature, after which time they were incubated with anti-UBR5 antibody (1:200, cat. no. ab70311, Abcam). Images were captured using a Nikon light microscope (magnification, ×200), and staining intensity was analyzed using Nikon software (NIS-Elements AR 3.2; Nikon Corporation). The IHC score was calculated according to the amount and level of immunoreactivity as follows: The percentage of stained cells was scored on a scale from 0 to 4 [0 (<1%), 1 (1-24%), 2 (25-49%), 3 (50-74%) and 4 (75-100%)], and the staining intensity was scored from 0 to 3 [0 (no staining), 1 (pale yellow), 2 (yellow), and 3 (reddish-brown)]. The final IHC score was determined by multiplying the intensity score and the positivity score to achieve a final score ranging between 0 and 12. Expression scores higher than the median score were categorized as positive.
RNA interference and UBR5 overexpression vector construction
Small interfering RNAs (si-RNAs) targeting UBR5 were synthesized; the sequences were as follows: si-UBR5-1 sense 5′-GCAGUGUUCCUGCCUUCU-3′ and antisense 5′-AGAAGGCAGGAACACUGC-3′; si-UBR5-2 sense 5′-GCGACUCUCCAUGGUUUCU-3′ and antisense 5′-AGAAACCAUGGAGAGUCGC-3′; scrambled control sense 5′-UUCUCCGAACGUGUCACGU-3′; scrambled control antisense 5′-ACGUGACACGUUCGGAGAA-3′. In order to construct a plasmid for overexpressing UBR5, the coding sequence of UBR5 was inserted into the p-Enter vector (Vigene Bio.) according to the manufacturer's instructions. The empty p-Enter vector was used as control. UBR5 si-RNAs or scrambled si-RNA were transfected into cells at a final concentration of 20 nM/ml. UBR5 overexpression plasmid or the empty vector control were transfected in LCC cells at a final concentration of 1 µg/µl with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Successful transfection was confirmed by reverse transcription-quantitative PCR (RT-qPCR) analysis and western blotting. All subsequent experiments were performed 48 h after the transfection.
RT-qPCR analysis
Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNAs were then reverse-transcribed into cDNA using the PrimeScript™ RT Master Mix (Takara Bio, Inc.) at 42°C for 20 min and 90°C for 5 min. qPCR reactions were performed using SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) following standard protocols. Briefly, an initial denaturation at 95°C for 3 min was followed by 40 cycles of denaturation (at 95°C for 20 sec), annealing (at 55°C for 45 sec), and elongation (at 72°C for 30 sec), with a final extension at 72°C for 5 min. The PCR primers used for UBR5 were as follows: Forward 5′-GACGCGAGAACTCTTGGAAC-3′ and reverse 5′-TTCAAATGGATTTGGGGGTA-3′. The PCR primers used for β-actin were as follows: Forward 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse 5′-CTCCTTAATGTCACGCACGAT-3′. The relative UBR5 expression quantity was calculated using the 2−ΔΔCq method (25). Each sample was replicated three times.
Western blotting
Total protein was extracted from cells using RIPA lysis buffer (Takara Bio, Inc.) according to the manufacturer's instructions, and the protein concentration was quantified using a bicinchoninic acid protein assay kit. A total of 15 µg protein per lane was separated by 10% SDS/PAGE and then transferred onto a polyvinylidene fluoride membrane. The membrane was incubated with blocking buffer (1X TBS, 0.1% Tween-20, 5% non-fat dry milk) for 2 h at room temperature and then with the appropriate primary antibody overnight at 4°C. The following primary antibodies were used: Anti-UBR5 (1:1,000, cat. no. ab70311, Abcam), anti-Bax (1:1,000, cat. no. ab32503, Abcam), anti-Bcl-2 (1:1,000, cat. no. ab185002, Abcam), anti-p38 (1:1,000, cat. no. ab170099, Abcam), anti-phosphorylated (p)-p38 (1:1,000, cat. no. ab178867, Abcam), and anti-β-actin (1:1,000, cat. no. ab115777, Abcam) antibody. The membrane was then incubated with secondary antibody for 2 h at room temperature. Protein bands were visualized using ECL plus western blotting detection reagents (BD Biosciences) and detected with an enhanced chemiluminescence kit.
Cell proliferation assays
LCC cells were seeded in a 96-well plate (5×103 cells/well) and cultured in a 5% CO2 incubator at 37°C for 24 h. Next, 10 µl CCK8 solution (KeyGen Bio.) was added to each well, and the cells were incubated for 4 h. The OD value of the medium was detected using a spectrophotometer at 450 nm. In some experiments, 5 µM SB203580 (Selleck Chemicals), a p38/MAPK inhibitor, was added to UBR5-overexpressing cells overnight before analyses.
Cell migration and invasion assays
A Transwell 24-well Boyden chamber with a 8-µm polycarbonate membrane (Corning, Inc.) was used to examine the migration capacity. For invasion assay, the Transwell insert was pretreated with Matrigel (BD Biocoat) at 37°C for 8 h. LCC cells (1×105 cells) were seeded in serum-free medium in the upper chamber of the insert; medium with 20% FBS was included in the lower chamber. After 24 h, cells on the bottom surface of the filter were fixed with 4% polyoxymethylene at room temperature for 15 min and then 1% crystal violet dye was used to stain the cells at room temperature for 10 min. Cells were visualized using a Leica AF6000 LX fluorescence microscope (Meyer Instruments, Inc.) and counted in 5 randomly selected fields of vision at a magnification, ×100.
Cell cycle analysis
Flow cytometry was used to examine the cell cycle distribution of LCC cells using a cell cycle assay kit (KeyGen Bio.) according to the manufacturer's instructions. The cells were washed twice with PBS and assayed on the machine. The percentage of cells in each phase of the cell cycle was then calculated.
Radiosensitivity assays
To investigate the impact of UBR5 on radiosensitivity, M2E and M4E cells transfected with si-RNA, overexpression plasmid or controls were seeded in 96-well plates at a density of 5,000 cells/well. The cells were then exposed to 0, 2, 5 or 8 Gy, and CCK8 assay was performed 48 h later, as described above. Cell numbers of treated and control groups were converted by the CCK8 standard curve method. The cell viability rate was calculated as the cell number of the treated group/5,000 ×100%.
For survival fraction assays, cells seeded in a 6-cm culture plate were exposed to 0, 2, 4, 6 or 8 Gy, after which time the cells were routinely cultured for 2 weeks. Cells were fixed in methanol at room temperature for 15 min, stained with 1% crystal violet-ethanol solution at room temperature for 10 min, and the number of clones was calculated in 5 randomly selected fields of vision at a magnification of ×100, using a Leica DM2500 system microscope (Meyer Instruments, Inc.). The survival rate was calculated as the number of clones in the treated group/the number of clones in the control group. All experiments were replicated three times.
Statistical analysis
The results are expressed as the mean ± standard error of the mean. Statistical analysis was performed using R software (version 3.5.1; http://cran.r-project.org/bin/windows/base/old/3.5.1/), R studio software (version 1.2.1335; http://rstudio.com/products/rstudio/download/) and GraphPad Prism 5 software (version 7.0; GraphPad Software, Inc.). Comparisons between two groups were analyzed using a two-tailed unpaired Student's t-test. One-way ANOVA with Bonferroni correction was used to compare multiple groups. The χ2 test was used for cell cycle distribution analysis. The median mRNA and protein levels were used as cut-off values in the two cohorts of LCC cases. The Kaplan-Meier method with log-rank hazard ratio (95% CI) was used to construct survival plots. P<0.05 was considered to indicate statistically significant differences in all tests.
Results
UBR5 expression pattern and prognostic value in human LCC tissues
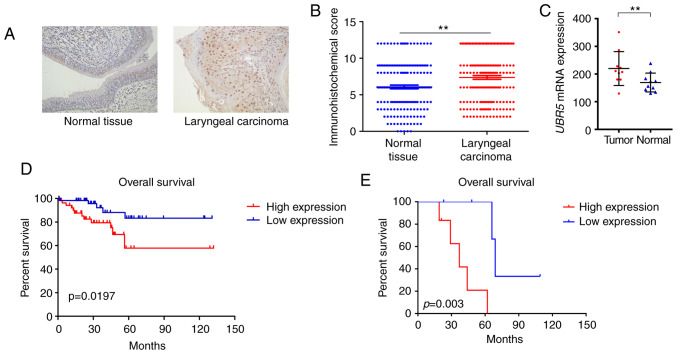
To investigate the potential role of UBR5 in LCC development, the expression pattern of UBR5 was first examined in 171 LCC tissue samples and adjacent non-tumor tissues. IHC demonstrated that UBR5 was significantly overexpressed in LCC tissues compared with its levels in adjacent non-tumor tissues (P<0.05; Fig. 1A and B). Patients positive for UBR5 expression were likely to be male with T3-4, N2-3 or M1 disease, while patients with T1-2, N0-1 or M0 stage tended to be UBR5-negative (Table I).
Figure 1.
UBR5 expression pattern and prognostic value in human LCC tissues. (A) Immunohistochemistry staining results demonstrated that UBR5 was overexpressed in LCC specimens compared with non-tumor tissues (magnification, ×200). (B) Immunohistochemical score of UBR5 in LCC and adjacent non-tumor tissues. (C) Increased UBR5 mRNA expression was observed in LCC tissues from the Gene Expression Omnibus database. (D and E) Survival curves demonstrated that increased UBR5 expression was significantly correlated with poor overall survival in patients with LCC (P<0.05). UBR5, ubiquitin protein ligase E3 component n-recognin 5; LCC, laryngeal carcinoma. **P<0.01.
Table I.
Association of clinicopathological characteristics with UBR5 expression status in patients with LCC (n=171).
| UBR5 expression, n (%) | |||||
|---|---|---|---|---|---|
| Characteristics | Entire cohort, n (%) | IHC negative (n=85) | IHC positive (n=86) | χ2 | P-value |
| Age, years | 0.874 | 0.350 | |||
| <70 | 128 (74.8) | 60 (70.6) | 68 (79.1) | ||
| ≥70 | 43 (25.2) | 25 (29.4) | 18 (20.9) | ||
| Sex | 0.491 | 0.712 | |||
| Male | 131 (76.7) | 62 (72.9) | 69 (80.2) | ||
| Female | 40 (23.3) | 23 (27.1) | 17 (19.8) | ||
| T stagea | 7.754 | <0.001 | |||
| T1-T2 | 101 (59.1) | 64 (75.3) | 37 (43.0) | ||
| T3-T4 | 70 (40.9) | 21 (24.7) | 49 (57.0) | ||
| N stagea | 9.173 | <0.001 | |||
| N0-N1 | 151 (88.3) | 80 (94.1) | 71 (82.6) | ||
| N2-N3 | 20 (11.7) | 5 (5.9) | 15 (17.4) | ||
| M stagea | 4.229 | 0.037 | |||
| M0 | 152 (88.9) | 78 (91.8) | 74 (86.0) | ||
| M1 | 19 (11.1) | 7 (8.2) | 12 (14.0) | ||
| Radiotherapy | 0.917 | 0.203 | |||
| Yes | 43 (25.0) | 18 (21.2) | 25 (29.1) | ||
| No | 128 (75.0) | 67 (78.8) | 61 (70.9) | ||
TNM scoring system: Tumor size, lymph nodes affected, metastases. Bold print indicates statistical significance. UBR5, ubiquitin protein ligase E3 component n-recognin 5; LCC, laryngeal carcinoma; IHC, immunohistochemistry.
In addition, transcriptional expression data from the GEO database indicated that UBR5 expression was markedly increased in tumor samples (213.23±74.1) compared with that in non-tumor tissues (172.4±47.23; Fig. 1C). The median survival time of the high UBR5 expression group was 43 months (interquartile range, 12–57 months) compared with 67 months (interquartile range, 23–79 months) in the low UBR5 expression group. The median survival time of patients in our cohort has not been reached. Survival curves demonstrated that increased UBR5 mRNA and protein expression was significantly correlated with poor overall survival in LCC patients (P<0.05; Fig. 1D and E).
Expression of UBR5 in LCC cell lines
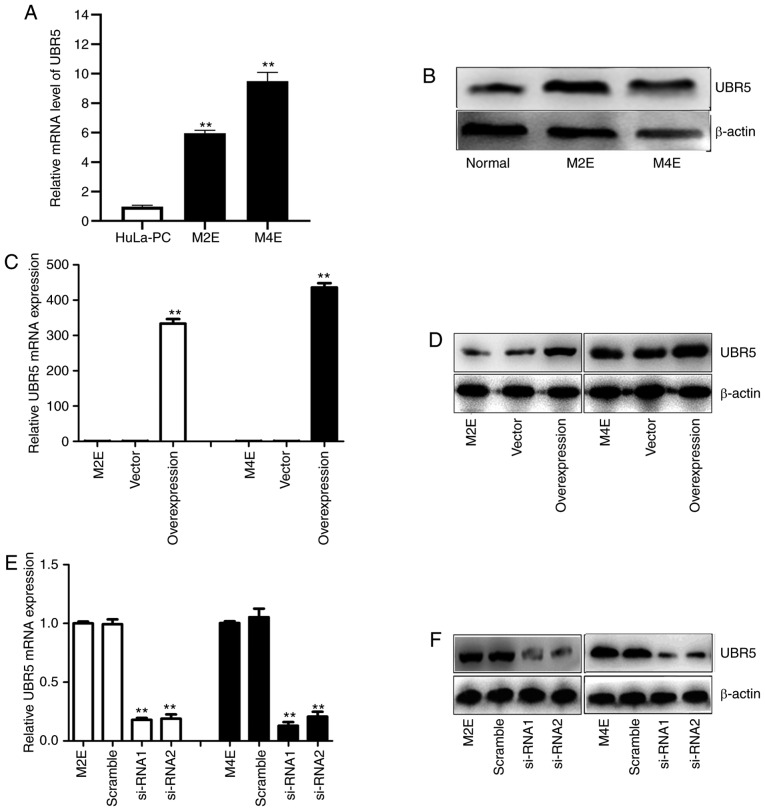
Using RT-qPCR and western blot analyses, UBR5 expression was examined in two LCC cell lines. As illustrated in Fig. 2A and B, the UBR5 mRNA and protein expression levels were significantly higher in M2E and M4E cells compared with HuLa-PC cells.
Figure 2.
Overexpression and silencing of UBR5 in M2E and M4E cells. (A) Compared with the control group, UBR5 expression was significantly increased in M2E and M4E cells. (B) The protein expression of UBR5 was evaluated in M2E and M4E cells. Investigating the expression of UBR5 mRNA and protein in cells transfected with UBR5 overexpression plasmid, UBR5 si-RNA1, UBR5 si-RNA2 or controls, revealed (C and D) significantly increased expression in the UBR5 overexpression group and (E and F) decreased expression in the si-UBR5 groups, respectively. UBR5, ubiquitin protein ligase E3 component n-recognin 5; **P<0.01.
To more closely examine the function of UBR5 in LCC, a UBR5 overexpression construct was generated and si-RNA was used to target UBR5 in LCC cells. Transfection of M2E and M4E cells with the overexpression plasmid resulted in increased UBR5 mRNA expression by 300-400-fold compared with its levels in cells transfected with vector (Fig. 2C). Western blotting revealed a similar trend (Fig. 2D). After transfection of M2E and M4E cells with UBR5 si-RNAs, UBR5 mRNA expression decreased significantly (0.27±0.07 in M2E cells and 0.31±0.09 in M4E cells) compared with its levels in the negative control groups (P<0.05; Fig. 2E). Western blotting similarly confirmed that UBR5 expression was downregulated by transfection of UBR5 si-RNA in both M2E and M4E cells (Fig. 2F).
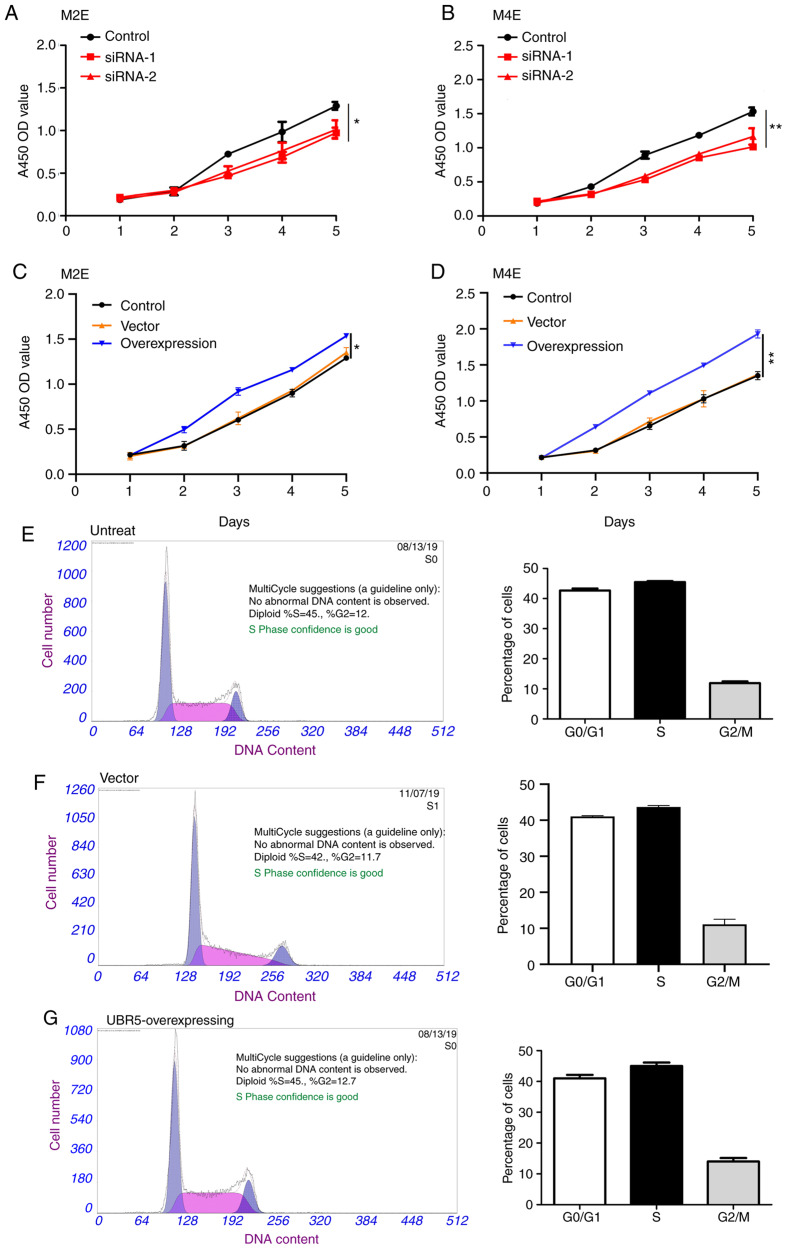
UBR5 regulates the proliferation of M2E and M4E cells
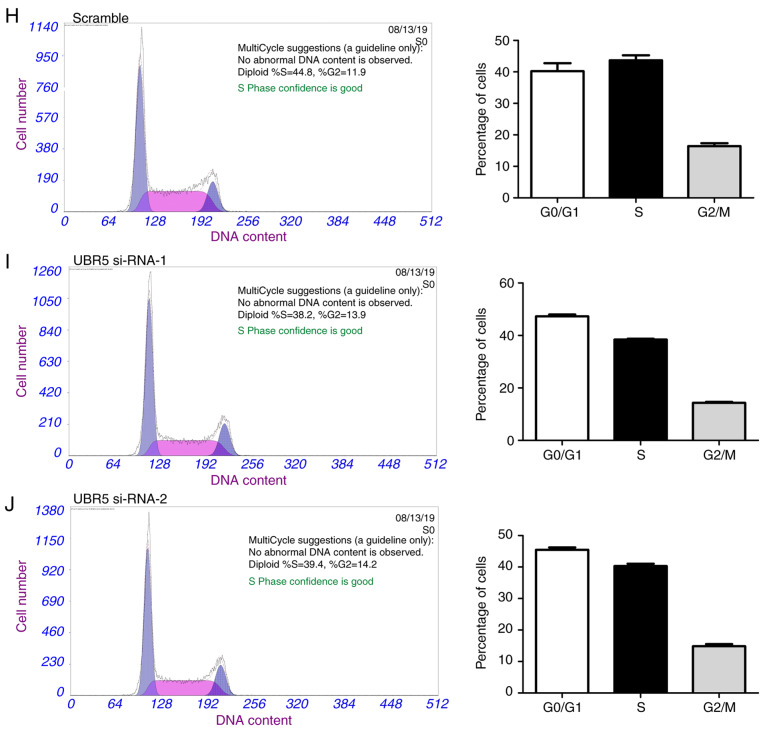
CCK-8 assay was next performed to evaluate the effect of UBR5 on the proliferation capacities of M2E and M4E LCC cells. UBR5 silencing significantly decreased cell proliferation (Fig. 3A and B), whereas UBR5 overexpression significantly increased the proliferation of both cell lines compared with the control groups (Fig. 3C and D). We further analyzed the impact of URB5 expression on the cell cycle, as shown in Fig. 3E-J. The percentage of cells in the S phase was relatively similar in the untreated, vector and UBR5 overexpression groups (45, 42 and 45%, respectively). However, compared to the control group (Fig. 3H), the percentage of S phase cells significantly decreased in both UBR5 si-RNA-1- and UBR5 si-RNA-2-transfected cells (38.2 and 39.4%, respectively; P<0.05; Fig. 3I and J).
Figure 3.
UBR5 increases M2E and M4E cell proliferation. The Cell Counting Kit-8 assay was employed to evaluate the role of UBR5 in the proliferation of M2E and M4E cells. (A and B) Silencing UBR5 significantly decreased cell proliferation, whereas (C and D) overexpression of UBR5 significantly increased cell proliferation compared with the control group. Cell cycle analysis revealed that the percentage of S phase cells was comparable among (E) untreated, (F) vector and (G) UBR5-overexpressing cells. Compared with the (H) control group, the percentage of S phase cells was significantly decreased in the (I) UBR5 si-RNA-1 and (J) UBR5 si-RNA-2 groups (P<0.05). UBR5, ubiquitin protein ligase E3 component n-recognin 5; *P<0.05; **P<0.01.
Effect of UBR5 on the invasion and migration of LCC cells
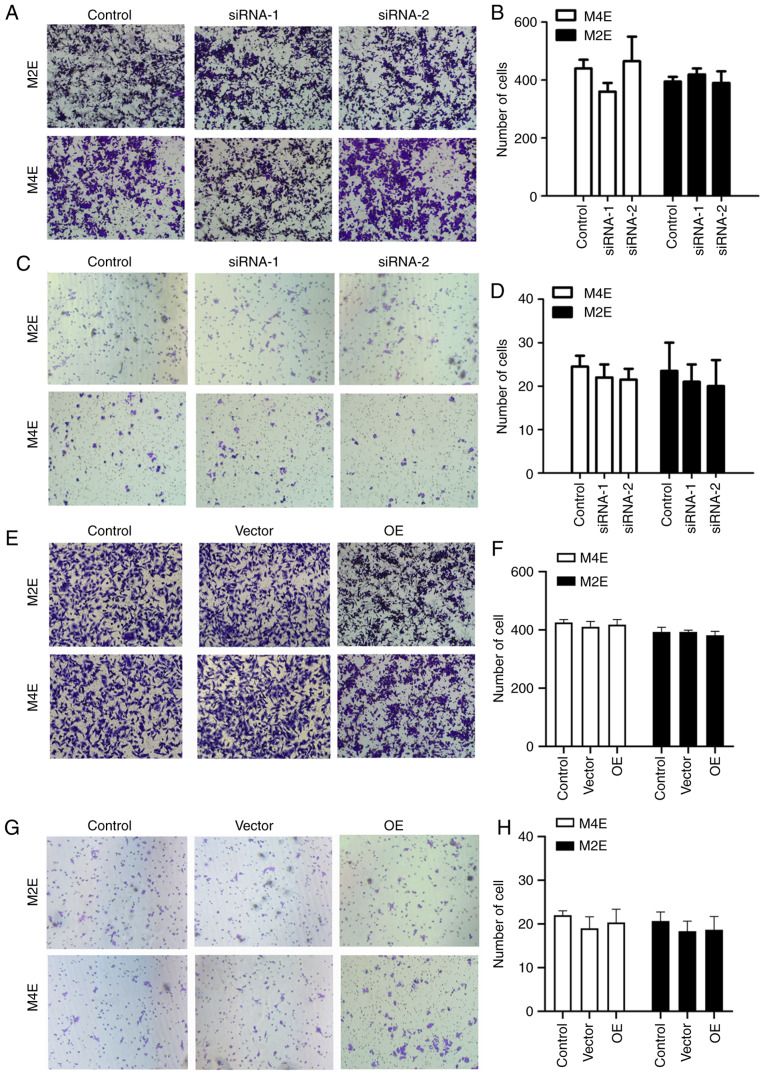
To explore the role of UBR5 in the invasion and migration of LCC cells, invasion and migration assays were performed. As shown in Fig. 4A and B, the number of invading cells slightly decreased in M4E cells transfected with si-RNA-1 (382±14) compared with controls (413±15), but the difference was not statistically significant (P>0.05). Similarly, no statistically significant differences were observed in terms of migration or invasion among the four groups of M2E cells. We also observed no statistically significant difference in the number of migrating cells in the UBR5-overexpressing M2E and M4E cells compared with the control or vector groups (Fig. 4E and F). The invasion assay revealed similar trends in M4E and M2E cells (Fig. 4C, D, G and H), wit up- or downregulation of UBR5 exerting no effect on cell invasion.
Figure 4.
Effect of UBR5 on M2E and M4E cell invasion and migration. In the migration assays, no significant change was observed in the cell numbers of (A and B) the UBR5 si-RNA and (E and F) the UBR5 overexpression groups compared with controls in M2E and M4E cells. Invasion assays revealed no significant differences in invading cell number in (C and D) UBR5 si-RNA or (G and H) overexpression groups. OE, overexpression; UBR5, ubiquitin protein ligase E3 component n-recognin 5.
UBR5 regulates radiosensitivity in M2E and M4E cells
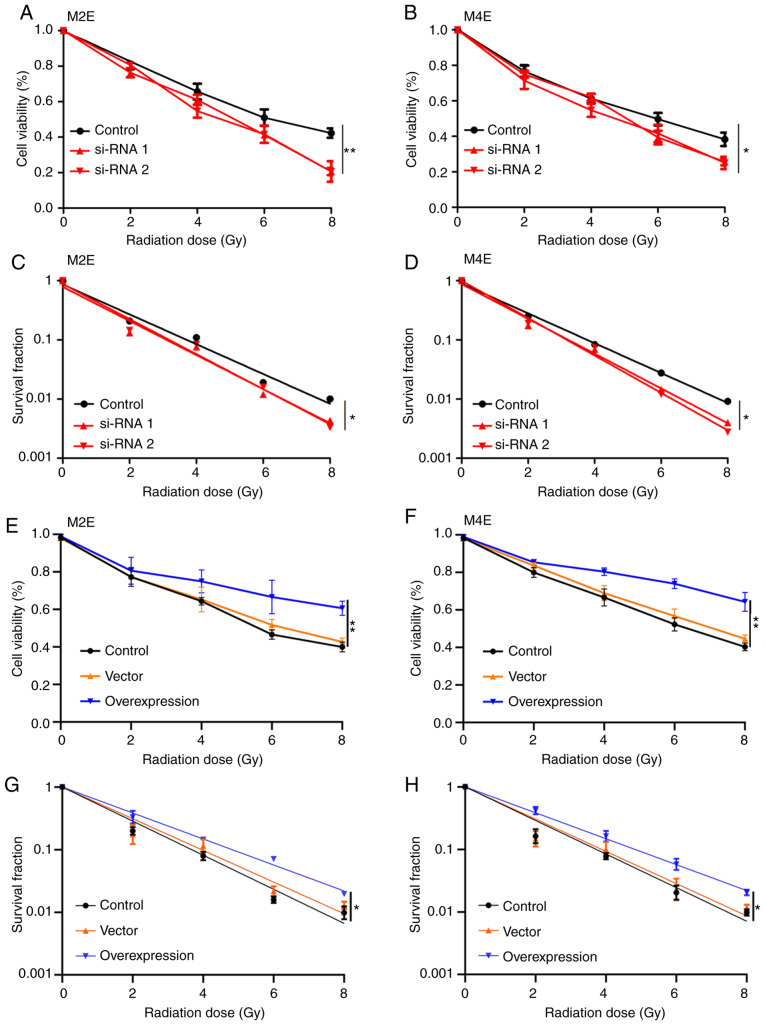
The role of UBR5 in the radiosensitivity of LCC cells was next investigated. As shown in Fig. 5, downregulation of UBR5 expression in M2E and M4E cells significantly suppressed the viability and survival fraction of both cell lines after exposure to radiation (Fig. 5A-D). Conversely, overexpression of UBR5 increased cell viability and the survival fraction of M2E and M4E cells with increasing radiation dose compared with the control or vector groups (Fig. 5E-H).
Figure 5.
UBR5 regulates the radiosensitivity of M2E and M4E cells. (A-D) UBR5 knockdown significantly suppressed the viability and survival fraction of M2E and M4E cells treated with radiation. (E-H) Conversely, UBR5 overexpression increased the viability and survival fraction of M2E and M4E cells with increasing radiation dose (0, 2, 4, 6 and 8 Gy) compared with controls. UBR5, ubiquitin protein ligase E3 component n-recognin 5; *P<0.05; **P<0.01.
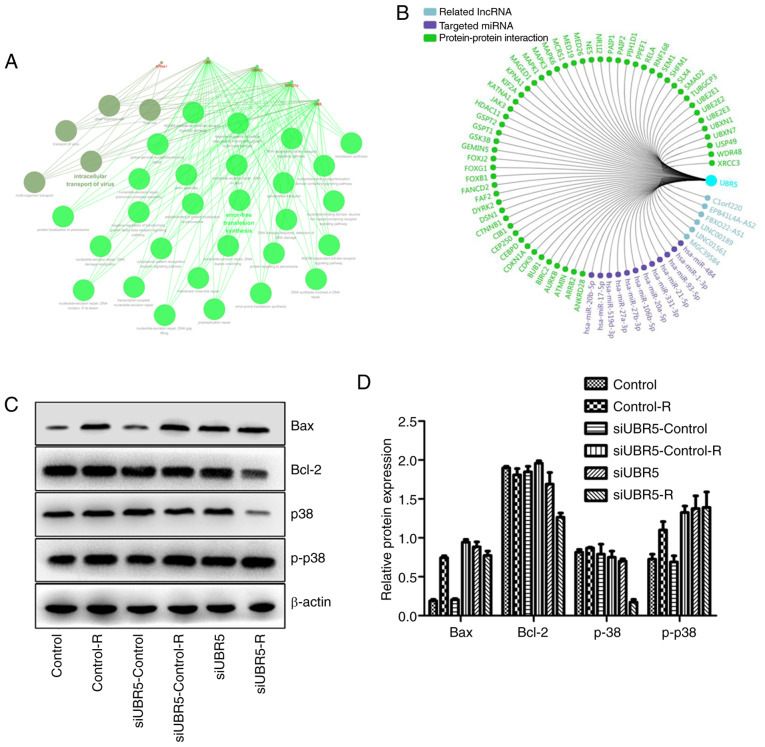
UBR5 silencing increases the Bax/Bcl2 ratio and activates the p38/MAPK signaling pathway following radiotherapy
Using bioinformatics, related lncRNA, targeted miRNA and PPI networks were obtained and a functional enrichment analysis of significant co-regulated nodes of UBR5 was performed, as shown in Fig. 6A and B. Alterations in the expression of p38/MAPK were measured in different groups. Western blot analysis revealed that radiation increased Bax expression in the control-R group compared with the control group, whereas it exerted no effect on the expression of p38. However, when M2E cells transfected with UBR5 si-RNA were exposed to radiation, Bcl-2 expression decreased significantly compared with its levels in cells transfected with si-RNA control and then exposed to radiation. Bax expression was the same in the two groups mentioned above. In addition, the p-p38 protein level was higher in the UBR5 si-RNA combined with radiation group compared with that in the radiation alone group, while total p38 protein was significantly decreased in the UBR5 si-RNA and radiation combination group (P<0.05; Fig. 6C and D).
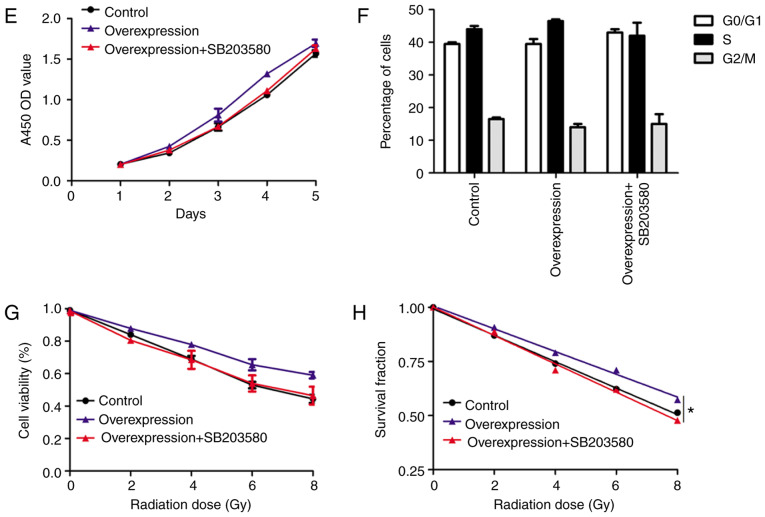
Figure 6.
UBR5 regulates cell proliferation and radiosensitivity via p38/MAPK signaling. (A and B) Related lncRNAs, targeted miRNAs and protein-protein interaction network and functional enrichment analysis of significant co-regulated nodes of UBR5 were obtained using bioinformatics tools. (C and D) The expression of Bcl-2 and p38 was decreased in the si-UBR5 combined with radiation groups, whereas the level of p-p38 was increased after combination of si-UBR5 with radiation. (E) The small molecule inhibitor of p38/MAPK signaling, SB203580, reduced the promotion of proliferation induced by UBR5 overexpression. (F) SB203580 treatment exerted no effect on the percentage of LCC cells in the S phase. (G-H) SB203580 treatment decreased UBR5-overexpressing cell viability and survival fraction when cells were exposed to radiation. UBR5, ubiquitin protein ligase E3 component n-recognin 5; LCC, laryngeal carcinoma; MAPK, mitogen-activated protein kinase; *P<0.05.
As shown in Fig. 6E-H, significantly increased proliferation was observed in the UBR5 overexpression group compared with the normal control. Following radiation exposure, the cell viability and survival fraction significantly increased in cells transfected with the UBR5 overexpression plasmid compared with the vector group. Moreover, UBR5-overexpressing cells were treated with SB203580, an inhibitor of the p38/MAPK signaling pathway, and cell proliferation, viability and survival fraction were rescued to the original levels. However, SB203580 treatment exerted no effect on the percentage of LCC cells in the S phase (Figs. 6F and S1). Taken together, these findings suggest that UBR5 regulates cell proliferation and radiosensitivity via p38/MAPK in LCC cells.
Discussion
The treatment strategy for LCC currently involves a comprehensive treatment based on surgery. However, with the development of molecular biology and technologies in medical research, increasing attention has been focused on developing approaches that improve the quality of life of patients with LCC after surgery, such as preservation of vocal and swallowing functions. Currently, the preservation of laryngeal function while maximally eradicating tumors is a major challenge in LCC (26,27). Radiotherapy can preserve the integrity of the laryngeal stent to the maximum extent, thus maintaining the full function of the larynx. However, the major drawback of radiotherapy for LCC is that individual sensitivity varies greatly (26). The present study demonstrated that survival fraction was decreased when IR was administered in conjunction with downregulation of UBR5 expression, suggesting that downregulation of UBR5 enhances radiosensitivity in LCC cells. These findings also suggest the possibility that increased UBR5 expression may represent a mechanism of radioresistance in LCC, which, to the best of our knowledge, has not been reported to date.
Recent studies indicated that the development of resistance to radiotherapy involves various signaling pathways that regulate DNA repair, cell survival, proliferation and apoptosis (8). Previous reports revealed that IR regulates the expression of Bcl-2 family genes, and its expression depends on the downregulation of Bcl-2 and the upregulation of Bax expression in wild-type p53 cancer (28,29). Nix et al reported that Bcl-2 expression in LCC predicted radioresistance with an accuracy of 71%, suggesting a potential mechanism by which LCC cells avoid the destructive effects of radiotherapy (30). In acute leukemia, regulating the response of cells to apoptotic stimuli affects radiosensitivity (31). IR-induced DNA double-strand breaks are among the most cytotoxic types of DNA damage. In LCC cells, DNA damage checkpoint 1 and p53-binding protein 1 limit tumor cell radiosensitivity via upstream mediators of the ATM pathway (32).
UBR5 is an important nuclear phosphoprotein involved in the regulation of DNA damage response, β-catenin activity, metabolism, transcription and apoptosis (33). The UBR5 gene is localized to chromosome 8q22, a region that is disrupted in a number of cancers. The UBR5 gene encodes a progestin-induced protein that belongs to the homologous to E6-AP carboxyl terminus (HECT) family (34,35). HECT family proteins function as E3 ubiquitin-protein ligases, targeting specific proteins for ubiquitin-mediated proteolysis (36). Recently, targeting E3 ligases as a strategy for radiotherapy treatment in several neoplasms has attracted significant interest (9,10). For example, increased E3 ligase cIAP2 expression resulted in altered MRE11 ubiquitination models and mediated radiosensitization in response to histone deacetylase inhibition (12). The E3 ligase UBR5 is a key regulator of the UPS in cells and in cancer development, and a potential gene that regulates IR sensitivity. In addition, UBR5-knockout cells are hypersensitive to UV radiation, which supports the role of UBR5 in IR-induced lesions in cancers (16). These results indicate that UBR5 may be a new potential independent prognostic marker of outcome or radiosensitivity in cancer. To the best of our knowledge, the present study was the first to demonstrate that UBR5 plays a key role in regulating the malignant behavior and radiosensitivity of LCC cells through the p38/MAPK pathway, thereby highlighting possible approaches to the development of new therapeutic strategies and targets for the treatment of this disease. However, there was a major limitation to the present study: Of all the included LCC patients from a retrospective cohort, only 43 received radiotherapy, and biopsy was performed on these patients prior to radiotherapy. Further studies are required to explore the association between UBR5 expression and response to radiotherapy in clinical biopsies following radiotherapy.
In conclusion, the present study demonstrated that UBR5 is highly expressed in LCC tissues and that downregulation of UBR5 in LCC cells may reduce their proliferation and increase their radiosensitivity. The mechanism underlying reduced proliferation and increased radiosensitivity may be associated with the activation of p38-MAPK signaling and downregulation of the apoptosis-related proteins Bax and Bcl-2. These findings indicate that UBR5 may be a novel treatment target in patients with radiation-resistant LCC and should be further investigated in future clinical studies.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Science and Technology Planning Project of Guangdong Province of China (nos. 2014A020212004 and 2016A02021500), the Natural Science Foundation of Guangdong Province of China (no. 2016A030313245), the Project of Administration of Traditional Chinese Medicine of Guangdong Province of China (no. 20161238), the Medical Scientific Research Foundation of Guangdong Province of China (no. A2015212), and the Capital Health Research and Development of Special (no. 2018-4-2082).
Availability of materials and data
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
RZ and WC conceived and designed the study. KW and JT performed the experiments. KW and XL wrote the paper. YW and RZ reviewed and edited the manuscript. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First People's Hospital of Foshan. Written informed consent was obtained from all patients who participated in this study according to the committee regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.American Society of Clinical Oncology, corp-author. Pfister DG, Laurie SA, Weinstein GS, Mendenhall WM, Adelstein DJ, Ang KK, Clayman GL, Fisher SG, Forastiere AA, et al. American Society of Clinical Oncology clinical practice guideline for the use of larynx-preservation strategies in the treatment of laryngeal cancer. J Clin Oncol. 2006;24:3693–3704. doi: 10.1200/JCO.2006.07.4559. [DOI] [PubMed] [Google Scholar]
- 2.Tobias JS, Monson K, Gupta N, Macdougall H, Glaholm J, Hutchison I, Kadalayil L, Hackshaw A, UK Head and Neck Cancer Trialists' Group Chemoradiotherapy for locally advanced head and neck cancer: 10-year follow-up of the UK Head and Neck (UKHAN1) trial. Lancet Oncol. 2010;11:66–74. doi: 10.1016/S1470-2045(09)70306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langendijk JA, Doornaert P, Verdonck-de Leeuw IM, Leemans CR, Aaronson NK, Slotman BJ. Impact of late treatment-related toxicity on quality of life among patients with head and neck cancer treated with radiotherapy. J Clin Oncol. 2008;26:3770–3776. doi: 10.1200/JCO.2007.14.6647. [DOI] [PubMed] [Google Scholar]
- 4.Duke RL, Campbell BH, Indresano AT, Eaton DJ, Marbella AM, Myers KB, Layde PM. Dental status and quality of life in long-term head and neck cancer survivors. Laryngoscope. 2005;115:678–683. doi: 10.1097/01.mlg.0000161354.28073.bc. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Zhang Y, Li Z, Liu S, Li H, Xu Z. Benefit of salvage total pharyngolaryngoesophagectomy for recurrent locally advanced head and neck cancer after radiotherapy. Radiat Oncol. 2017;12:164. doi: 10.1186/s13014-017-0900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pujo K, Philouze P, Scalabre A, Céruse P, Poupart M, Buiret G. Salvage surgery for recurrence of laryngeal and hypopharyngeal squamous cell carcinoma: A retrospective study from 2005 to 2013. Eur Ann Otorhinolaryngol Head Neck Dis. 2018;135:111–117. doi: 10.1016/j.anorl.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003;349:2091–2098. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- 8.Popovtzer A, Burnstein H, Stemmer S, Limon D, Hili O, Bachar G, Sopov V, Feinmesser R, Groshar D, Shvero J. Phase II organ-preservation trial: Concurrent cisplatin and radiotherapy for advanced laryngeal cancer after response to docetaxel, cisplatin, and 5-fluorouracil-based induction chemotherapy. Head Neck. 2017;39:227–233. doi: 10.1002/hed.24571. [DOI] [PubMed] [Google Scholar]
- 9.Shearer RF, Iconomou M, Watts CK, Saunders DN. Functional roles of the E3 Ubiquitin ligase UBR5 in cancer. Mol Cancer Res. 2015;13:1523–1532. doi: 10.1158/1541-7786.MCR-15-0383. [DOI] [PubMed] [Google Scholar]
- 10.Shearer RF, Frikstad KM, McKenna J, McCloy RA, Deng N, Burgess A, Stokke T, Patzke S, Saunders DN. The E3 ubiquitin ligase UBR5 regulates centriolar satellite stability and primary cilia. Mol Biol Cell. 2018;29:1542–1554. doi: 10.1091/mbc.E17-04-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas K, Sarkar S, Du K, Brautigan DL, Abbas T, Larner JM. The E3 Ligase CHIP Mediates p21 degradation to maintain radioresistance. Mol Cancer Res. 2017;15:651–659. doi: 10.1158/1541-7786.MCR-16-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson J, Jevons SJ, Groselj B, Ellermann S, Konietzny R, Kerr M, Kessler BM, Kiltie AE. E3 Ligase cIAP2 mediates downregulation of MRE11 and radiosensitization in response to HDAC inhibition in bladder cancer. Cancer Res. 2017;77:3027–3039. doi: 10.1158/0008-5472.CAN-16-3232. [DOI] [PubMed] [Google Scholar]
- 13.Dogan T, Gnad F, Chan J, Phu L, Young A, Chen MJ, Doll S, Stokes MP, Belvin M, Friedman LS, et al. Role of the E3 ubiquitin ligase RNF157 as a novel downstream effector linking PI3K and MAPK signaling pathways to the cell cycle. J Biol Chem. 2017;292:14311–14324. doi: 10.1074/jbc.M117.792754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rom O, Kaisari S, Aizenbud D, Reznick A. Involvement of E3 Ubiquitin ligases in Cigarette Smoke associated muscle catabolism. Free Radic Biol Med. 2014;75(Suppl 1):S5. doi: 10.1016/j.freeradbiomed.2014.10.835. [DOI] [PubMed] [Google Scholar]
- 15.Xie Z, Liang H, Wang J, Xu X, Zhu Y, Guo A, Shen X, Cao F, Chang W. Significance of the E3 ubiquitin protein UBR5 as an oncogene and a prognostic biomarker in colorectal cancer. Oncotarget. 2017;8:108079–108092. doi: 10.18632/oncotarget.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez A, De Vivo A, Uprety N, Kim J, Stevens SM, Jr, Kee Y. BMI1-UBR5 axis regulates transcriptional repression at damaged chromatin. Proc Natl Acad Sci USA. 2016;113:11243–11248. doi: 10.1073/pnas.1610735113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson MJ, Munoz MA, Saunders DN, Clancy JL, Russell AJ, Williams B, Pappin D, Khanna KK, Jackson SP, Sutherland RL, Watts CK. EDD mediates DNA damage-induced activation of CHK2. J Biol Chem. 2006;281:39990–40000. doi: 10.1074/jbc.M602818200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Cronshaw J, Kanu N, Snijders AP, Behrens A. UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proc Natl Acad Sci USA. 2014;111:12091–12096. doi: 10.1073/pnas.1400230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alpsoy A, Yasa S, Gündüz U. Gunduz, Etoposide resistance in MCF-7 breast cancer cell line is marked by multiple mechanisms. Biomed Pharmacother. 2014;68:351–355. doi: 10.1016/j.biopha.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien PM, Davies MJ, Scurry JP, Smith AN, Barton CA, Henderson MJ, Saunders DN, Gloss BS, Patterson KI, Clancy JL, et al. The E3 ubiquitin ligase EDD is an adverse prognostic factor for serous epithelial ovarian cancer and modulates cisplatin resistance in vitro. Br J Cancer. 2008;98:1085–1093. doi: 10.1038/sj.bjc.6604281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chun SY, Kim S, Nam KS, Lee KS. Anti-metastatic potential of a proton beam is regulated by p38 MAPK/c-Fos signaling pathway in TPA-treated HepG2 human hepatocellular carcinoma. Biomed Pharmacother. 2018;99:904–912. doi: 10.1016/j.biopha.2018.01.134. [DOI] [PubMed] [Google Scholar]
- 22.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Graboyes EM, Zhan KY, Garrett-Mayer E, Lentsch EJ, Sharma AK, Day TA. Effect of postoperative radiotherapy on survival for surgically managed pT3N0 and pT4aN0 laryngeal cancer: Analysis of the National Cancer Data Base. Cancer. 2017;23:2248–2257. doi: 10.1002/cncr.30586. [DOI] [PubMed] [Google Scholar]
- 27.Dyckhoff G, Plinkert PK, Ramroth H. A change in the study evaluation paradigm reveals that larynx preservation compromises survival in T4 laryngeal cancer patients. BMC Cancer. 2017;17:609. doi: 10.1186/s12885-017-3608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong QS, Zheng LD, Chen FM, Zeng FQ, Wang L, Dong JH, Lu GC. Selection of optimal antisense accessible sites of survivin and its application in treatment of gastric cancer. World J Gastroenterol. 2005;11:634–640. doi: 10.3748/wjg.v11.i5.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bokarewa M, Lindblad S, Bokarew D, Tarkowski A. Balance between survivin, a key member of the apoptosis inhibitor family, and its specific antibodies determines erosivity in rheumatoid arthritis. Arthritis Res Ther. 2005;7:R349–R358. doi: 10.1186/ar1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nix P, Cawkwell L, Patmore H, Greenman J, Stafford N. Bcl-2 expression predicts radiotherapy failure in laryngeal cancer. Br J Cancer. 2005;92:2185–2189. doi: 10.1038/sj.bjc.6602647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrzesień-Kuś A, Smolewski P, Sobczak-Pluta A, Wierzbowska A, Robak T. The inhibitor of apoptosis protein family and its antagonists in acute leukemias. Apoptosis. 2004;9:705–715. doi: 10.1023/B:APPT.0000045788.61012.b2. [DOI] [PubMed] [Google Scholar]
- 32.Gou Q, Xie Y, Liu L, Xie K, Wu Y, Wang Q, Wang Z, Li P. Downregulation of MDC1 and 53BP1 by short hairpin RNA enhances radiosensitivity in laryngeal carcinoma cells. Oncol Rep. 2015;34:251–257. doi: 10.3892/or.2015.3980. [DOI] [PubMed] [Google Scholar]
- 33.Shen Q, Qiu Z, Wu W, Zheng J, Jia Z. Characterization of interaction and ubiquitination of phosphoenolpyruvate carboxykinase by E3 ligase UBR5. Biol Open. 2018;7(pii):bio037366. doi: 10.1242/bio.037366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matta-Camacho E, Kozlov G, Menade M, Gehring K. Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:1158–1163. doi: 10.1107/S1744309112036937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safdar K, Gu A, Xu X, Au V, Taylor J, Flibotte S, Moerman DG, Maine EM. UBR-5, a Conserved HECT-Type E3 Ubiquitin Ligase, negatively regulates Notch-type signaling in Caenorhabditis Elegans. G3 (Bethesda) 2016;6:2125–2134. doi: 10.1534/g3.116.027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flack JE, Mieszczanek J, Novcic N, Bienz M. Wnt-Dependent inactivation of the Groucho/TLE Co-repressor by the HECT E3 Ubiquitin Ligase Hyd/UBR5. Mol Cell. 2017;67:181–193.e5. doi: 10.1016/j.molcel.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.