Abstract
The aim of the present study was to observe the influence of the small breast epithelial mucin (MUCL1) (also known as SBEM) gene on migration and invasion ability of breast cancer cells and to explore the potentially involved mechanism. SBEM-interference plasmid and SBEM-overexpressing plasmid were constructed. SBEM-knockdown or SBEM- overexpressing MCF-7 and MDA-MB-231 breast cancer cells were established by lentivirus-mediated stable transfection method. The scratch wound-healing assay and Transwell chamber experiment were used to detect the influence of the SBEM gene on the migration and invasion abilities of MCF-7 and MDA-MB-231 cells. Real-time PCR (polymerase chain reaction) and western blotting were used to detect the expression of epithelial-to-mesenchymal transition (EMT)-related markers and regulators. The cell morphology was observed after transfection. The SBEM-knockdown or SBEM-overexpressing MCF-7 and MDA-MB-231 cells were established successfully. The migration and invasion abilities were decreased after SBEM was downregulated, and were increased after SBEM was overexpressed both in MCF-7 and MDA-MB-231 cell lines. The mRNA and protein expressions of N-cadherin, Twist and vimentin were elevated following SBEM overexpression, while the expression of E-cadherin and claudin-1 were found to be decreased following SBEM overexpression. In conclusion, SBEM has the potential to promote migration and invasion ability of breast cancer cells via promoting epithelial-to-mesenchymal transition.
Keywords: breast cancer, mucin-like protein 1, MCF-7 cells, MDA-MB-231 cells, epithelial-to-mesenchymal transition
Introduction
In recent years, the incidence of breast cancer in China has been exhibiting an increasing trend on an annual basis (1). Hematogenous micrometastasis at initial diagnosis is considered to be one of the causes of recurrence that affects the overall survival of breast cancer patients. Therefore, detection of hematogenous micrometastases based on tissue-specific markers may provide valuable information and guidance for the early screening of high-risk breast cancer patients (2). Small breast epithelial mucin (SBEM) (also known as MUCL1) has been identified as a putative breast-specific gene and has been considered to be a promising breast-specific marker (3). In our previous study, we detected SBEM expression in tissues and peripheral blood specimens of breast cancer patients to analyze its correlation with prognostic parameters. SBEM was proposed as a marker for predicting hematogenous micrometastasis and response to neoadjuvant chemotherapy in breast cancer (4). However, although published studies have suggested that SBEM may represent a suitable marker for molecular detection of isolated tumor cells in the bone marrow and targeting bone marrow micrometastasis in breast cancer patients (5,6), studies that have been conducted to date in order to observe the effect of the SBEM gene on breast cancer cells and explore the underlying possible mechanism are sparse.
In the present study, MCF-7 and MDA-MB-231 cells with stable SBEM knockdown or overexpression were first generated. After detecting the effect of SBEM on the migration and invasion abilities of MCF-7 and MDA-MB-231 cells, the expression of EMT-related markers and regulators in SBEM-overexpressing MCF-7 cells was monitored. Among the EMT-related markers and regulators, E-cadherin and claudin-1 are considered to be two important suppressors of invasion. E-cadherin plays a crucial role in the maintenance of epithelial cell polarization, and deficiency of this molecule causes cancer metastasis due to the loss of cell-cell adhesion, with concomitant increased cell motility (7). Breast cancer patients with lower expression of E-cadherin are at a higher risk of recurrence and metastasis, and have a worse prognosis (8). Claudin-1 plays a key role in the formation of tight junctions (9). In several cancers, the loss of claudin-1 expression has been associated with cancer progression, invasiveness, and acquisition of the metastatic phenotype (10,11). Claudin-1 is frequently downregulated in cancer, and its downregulation has been shown to be associated with poor clinical outcome in human invasive breast cancer (12). The aim of the present study was to elucidate the possible mechanisms of action of SBEM by observing its effects on EMT-related markers and regulators, including E-cadherin and claudin-1, in the hope of the results laying an experimental foundation for further exploring the role of SBEM in breast cancer.
Materials and methods
Cell characteristics and culture
MCF-7 is a widely studied epithelial cancer cell line derived from breast adenocarcinoma, with the characteristics of differentiated mammary epithelium. MCF-7 cells express estrogen receptor-α (ER-α), as well as androgen, progesterone and glucocorticoid receptors, which make them valuable tools in medical research. Although MCF-7 cells are easy to propagate, they are generally a slow-growing population, with a doubling time of 30–40 h. MDA-MB-231 is a highly aggressive, invasive and poorly differentiated triple-negative breast cancer (TNBC) cell line. Similar to other invasive cancer cell lines, the invasiveness of the MDA-MB-231 cells is mediated by proteolytic degradation of the extracellular matrix. In a 3D culture, this cell line displays endothelial-like morphology and is distinguished by the stellate projections that often bridge multiple cell colonies.
MCF-7 and MDA-MB-231 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were grown in 25-cm2 cell culture flasks with RPMI-1640 medium (HyClone; GE Healthcare Life Sciences) or L15 (HyClone; GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences), 2 mM L-glutamate, 100 U/ml penicillin G, and 100 U/ml streptomycin at 37°C in 5% CO2 and 95% air. The cells were then seeded into 6- or 24-well culture plates (Corning, Inc.) with the cell confluence of ~70% prior to transfection.
Construction of recombinant shRNA lentivirus vector
According to the design principle of mammalian eukaryotic cell RNA interference, shRNA sequences were designed for the different targets of mucin-like protein 1. Each interference sequence was synthesized into a single chain of sense and antisense, and a double chain was formed after annealing. The primers were as follows: 5′-CCGGGTGTGTCCCTGAGATGGAATCCTCGAGGATTCCATCTCAGGGACACACTTTTTG-3′ (sense) and 5′-AATTCAAAAAGTGTGTCCCTGAGATGGAATCCTCGAGGATTCCATCTCAGGGACACAC-3′ (antisense).
The two chains were annealed into double chains, connected by T4DNA ligase and the AgeI/EcoRI double enzyme cutting carrier PLKO1 (Fig. 1), converted by DH5a competent cells, and single clones were selected. The contrast scramble plasmid was provided by Genesent, and its hairpin structure was as follows: 5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3.
Figure 1.
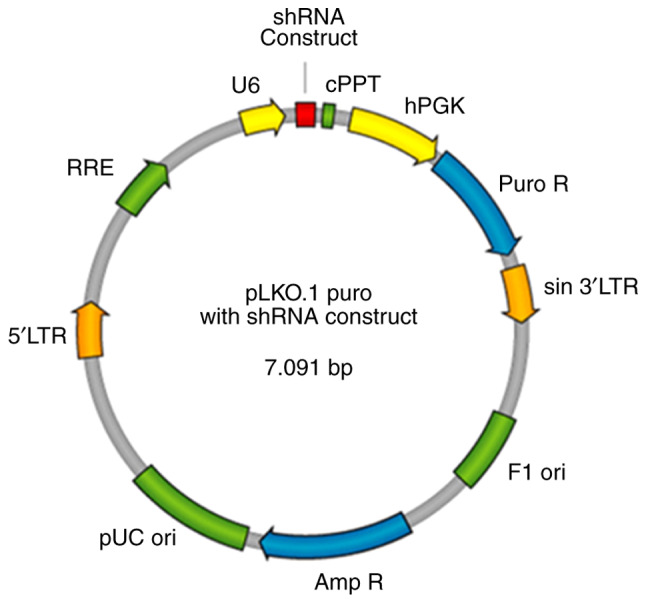
SBEM shRNA plasmid map. SBEM, small breast epithelial mucin; RRE, Rev responsive element; 5′LTR, 5′ long terminal repeat; pUC ori, pUC bacterial origin of replication; Amp R, ampicillin resistance gene for selection of pLKO.1 plasmid in bacterial cells; Puro R, puromycin-resistance gene for selection of pLKO.1 plasmid in mammalian cells; sin 3′LTR, 3′ self-inactivating long terminal repeat; F1 ori, F1 bacterial origin of replication; hPGK, human phosphoglycerate kinase promoter drives expression of puromycin; cPPT, central polypurine tract.
Construction of SBEM lentivirus expression plasmid
SBEM cDNA was provided by Genesent, and the lentivirus overexpression vector was pCDH-CMV-MCS-EF1-Puro. The plasmid map is shown in the Fig. 2. The cloning primers were as follows: 5′-TAGAGCTAGCGAATTATGAAGTTCTTAGCAGTCC-3′ (sense) and 5′-AGATCCTTCGCGGCCTCAGGGACACACTCTACCA-3′ (antisense).
Figure 2.
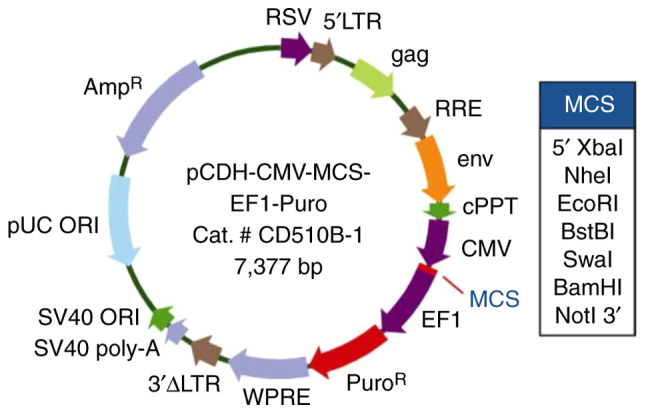
The SBEM-overexpressing plasmid map. SBEM, small breast epithelial mucin. SV40 ORI, SV40 promoter/origin; SV40 poly-A, simian vacuolating virus 40 poly-A; 3′ΔLTR, 3′ self-inactivating long terminal repeat; WPRE, In cis Woodchuck hepatitis virus post & hyphentranscriptional regulatory element; EF1, elongation factor 1; MCS, multiple cloning site; CMV, cytomegalovirus; env, envelope; RSV, Rous sarcoma virus.
Generation of stable SBEM knockdown or overexpression cell lines
MCF-7 and MDA-MB-231 cells with stable SBEM knockdown or overexpression were generated by lentiviral vectors carrying either sh-SBEM, a SBEM overexpression construct, or the respective negative controls (Genesent), in accordance with the manufacturer's instructions. Cells were placed into 60-mm dishes at 3×105/dish and allowed to grow overnight. Lentivirus expression plasmid (8 µg) was mixed with 20 µl Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) and transfected into the cells according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were trypsinized, diluted, and placed into 96-well plates. Transfected cells were then selected with 2 µg/ml puromycin and 200 µg/ml G418. Western blotting was conducted to detect the knockdown and overexpression effects of SBEM.
Scratch wound-healing assay
To determine the regeneration and repair abilities of breast cancer cells, 4×105 MCF-7 and MDA-MB-231 cells were seeded in 6-well plates and incubated at 37°C with 5% CO2 overnight. Artificial wounds were created using a 10-µl pipette tip (0 h) to generate a gap in the confluent cell layer with confluence percentage of ~80%. The cells were washed with PBS twice and incubated with serum-free medium at 37°C with 5% CO2 as a control. At 0 and 8 h, phase-contrast images were captured using a microscope at a magnification of ×400 (CX41-32C02PH, Olympus Corporation).
Cell migration and invasion assays
The detailed procedures of cell migration and invasion assays were conducted as previously reported (13). Briefly, 3×105 cells were suspended in serum-free medium and seeded into the upper layer of Transwell membrane with an 8-µm pore size in a 24-well plate (Corning, Inc.). The membranes were coated with Matrigel (1:8; BD Biosciences) for invasion assays, or left uncoated for migration assays. Medium containing 10% FBS was placed in the bottom chamber as an attractant. After 24 h, the cells were fixed in 4% paraformaldehyde for 15 min at room temperature and stained with 0.1% crystal violet solution for 15 min. The invading cells were then examined and counted in 10 randomly selected fields under a light microscope at a magnification of ×400. The mean number of invading cells was then calculated.
Real-time PCR
Total RNA of 5×106 MCF-7 cells was extracted using the Trizol RNA extraction protocol (cat. no. 10606ES60, Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Reverse transcription of mRNA to cDNA was performed in 20 µl reaction volumes with random priming using an RT-PCR Kit (cat. no. RR047A, Takara Biomedical Technology Co., Ltd.). The sequences of primers used in this study are listed in Table I, and the real-time PCR reaction system is provided in Table II. The primers were synthesized by Beijing AuGCT Biotech Co. Ltd. Quantitative PCR was performed using SYBR Green PCR Kit (cat. no. RR820A, Takara Biomedical Technology Co. Ltd.) and Fast Real-Time PCR System (ABI 7900HT, Applied Biosystems; Thermo Fisher Scientific, Inc.). The detailed process was similar to that previously described (4). The increase of fluorescence was detected due to the exponential accumulation of PCR products and the 2−ΔΔCq method was used to calculate the relative quantity of gene expression in each sample (14).
Table I.
Sequence of the primers for real-time PCR.
| Primer | Sequence (5′ to 3′) |
|---|---|
| N-cadherin forward | GATGTTGAGGTACAGAATCGT |
| N-cadherin reverse | GGTCGGTATGGATGGCGA |
| Twist forward | GGAGTCCGCAGTCTTACGAG |
| Twist reverse | TCTGGAGGACCTGGTAGAGG |
| Vimentin forward | GGACCAGCTAACCAACGACA |
| Vimentin reverse | AAGGTCAAGACGTGCCAGAG |
| E-cadherin forward | ATTCTGATTCTGCTGCTCTTG |
| E-cadherin reverse | AGTAGTCATAGTCCTGGTCTT |
| Claudin-1 forward | CAGCTGTTGGGCTTCATTCTC |
| Claudin-1 reverse | ATCACTCCCAGGAGGATGCC |
Table II.
Real-time PCR reaction system (25 µl).
| Reaction | Volume (µl) |
|---|---|
| cDNA | 2 |
| SYBR Green I | 12.5 |
| Primer F (0.2 µmol/l) | 0.5 |
| Primer R (0.2 µmol/l) | 0.5 |
| Sterile deionized water | 9.5 |
Western blot assay
The proteins were quantified using the BCA method. An amount of 10 µl protein plus 10 µl PBS were loaded and 200 µl working liquid was added in 96-well plates. The absorbance value at 560 nm was measured by enzyme-labeling instrument. The concentration of proteins was calculated on the basis of the standard curve. For western blotting, 1×106 cells were plated in 100-mm Petri dishes for 24 h. The cells were then washed with cold PBS and lysed with 200 µl of cold lysis buffer [150 mmol/l NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 50 mmol/l Tris-HCl (pH 7.2), 0.2 mmol/l sodium vanadate, 1% phenylmethylsulfonyl fluoride and 0.2% aprotinin].
The samples were kept on ice for 20 min and then spun at 12,000 × g at 4°C for 20 min, and the protein concentration of the supernatant was determined. Cell lysates were fractionated on 10% SDS-PAGE, and protein was transferred onto nitrocellulose membranes (Pall Life Sciences). The membranes were blocked with 5% skimmed milk powder dissolved in TBST at room temperature for 2 h. The membranes were then probed with primary antibodies against SBEM (dilution 1:1,000, cat. no. HPA-039093; Sigma-Aldrich; Merck KGaA), N-cadherin (dilution 1:500, cat. no. 13116S; Cell Signaling Technology, Inc.), E-cadherin (dilution 1:500, cat. no. 3195S; Cell Signaling Technology, Inc.), Twist (dilution 1:500, cat. no. 69366S; Cell Signaling Technology, Inc.), vimentin (dilution 1:500, cat. no. 5741S; Cell Signaling Technology, Inc.) and claudin-1 (dilution 1:500, cat. no. 13995S; Cell Signaling Technology, Inc.). The expression of GAPDH as control was determined using anti-GAPDH (dilution 1:500, cat. no. 5174S; Cell Signaling Technology, Inc.). After hybridization at 4°C overnight, HRP-labeled IgG (dilution 1:500, cat. no. 074S; Cell Signaling Technology, Inc.) was added and incubated at 37°C for 2 h. The membrane was washed with TBST (0.1% Tween-20) for 3 times, and ECL chemiluminescence system (Applygen Technologies Inc.) was used for coloration. The immunoreactive bands were detected using an Odyssey Infrared Imaging System (Gene Company Ltd.). The intensity of each band was measured with Odyssey 3.0 software (Li Cor Inc.).
Statistical analysis
All data are presented as mean ± standard deviation. Statistical analysis was performed using Statistical Package for the Social Sciences version 14.0 (SPSS, Inc.). All statistical tests were two-sided. The comparison between two groups of samples adopted the t-test, and P-value ≤0.05 was considered to indicate a statistically significant difference.
Results
Observation of cell morphology after transfection
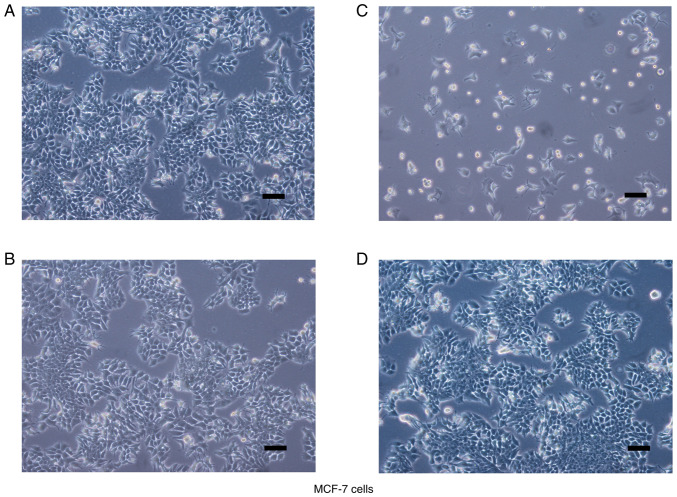
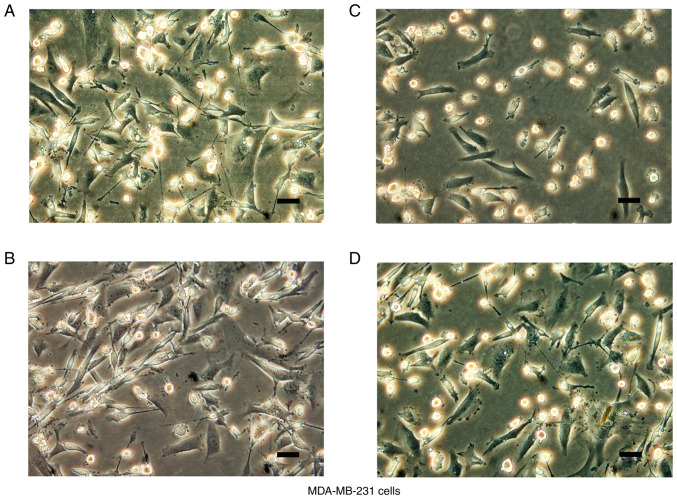
In accordance with the manufacturer's instructions, MCF-7 and MDA-MB-231 cells with stable SBEM knockdown or overexpression were generated by lentiviral vectors carrying sh-SBEM, a SBEM overexpression construct, or the negative controls, respectively. The microscopic observations of cell morphology are presented in Figs. 3 and 4. MCF-7 cells displayed typical epithelioid characteristics. They were small and polygonal in shape. MDA-MB-231 cells are mostly spindle-like with narrow strip in shape, and some of the cells adopted a more rounded morphology.
Figure 3.
Cell morphology after transfection. Stable SBEM knockdown or overexpressing MCF-7 cells were generated by lentiviral vectors carrying (A and B) negative controls, (C) sh-SBEM, or (D) SBEM overexpression construct. Cell morphology is shown. Scale bars represent 100 µm. SBEM, small breast epithelial mucin.
Figure 4.
Cell morphology after transfection. Stable SBEM knockdown or overexpressing MDA-MB-231 cells were generated by lentiviral vectors carrying (A and B) negative controls, (C) sh-SBEM, or (D) SBEM overexpression construct. Cell morphology is shown. Scale bars represent 100 µm. SBEM, small breast epithelial mucin.
SBEM protein expression in MCF-7 and MDA-MB-231 cells after transfection
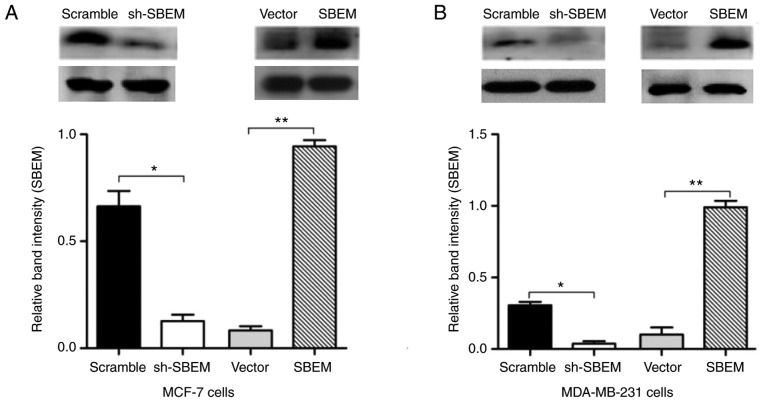
In order to test and verify the knockdown and overexpression of the SBEM gene, western blotting was used to detect SBEM protein expression in MCF-7 and MDA-MB-231 cells. Compared with the scramble group, SBEM protein expression decreased significantly after cells were transfected with sh-SBEM. Compared with the vector group, SBEM protein expression was increased significantly after the cells were transfected with the overexpression plasmid. The difference was statistically significant in both MCF-7 and MDA-MB-231 cells (P<0.05). Therefore, MCF-7 and MDA-MB-231 cell lines with stable SBEM knockdown or overexpression were successfully established (Fig. 5).
Figure 5.
MCF-7 and MDA-MB-231 cell lines with stable SBEM knockdown or overexpression are successfully established. SBEM protein expression was significantly decreased after sh-SBEM was transfected), and was significantly increased after the overexpression plasmid was transfected in (A) MCF-7 and (B) MDA-MB-231 cells. *P<0.05 and **P<0.01, compared with the scramble or vector group. SBEM, small breast epithelial mucin.
SBEM promotes scratch wound healing of breast cancer cells
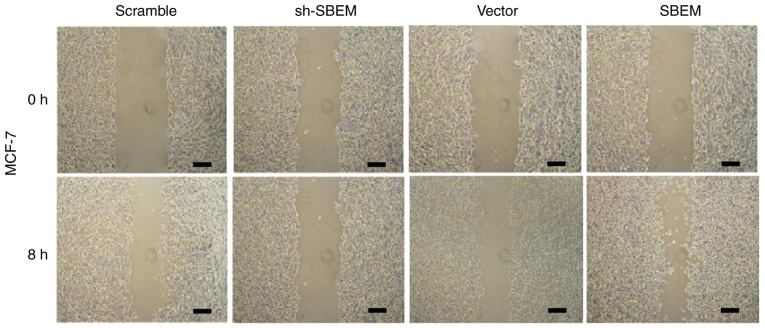
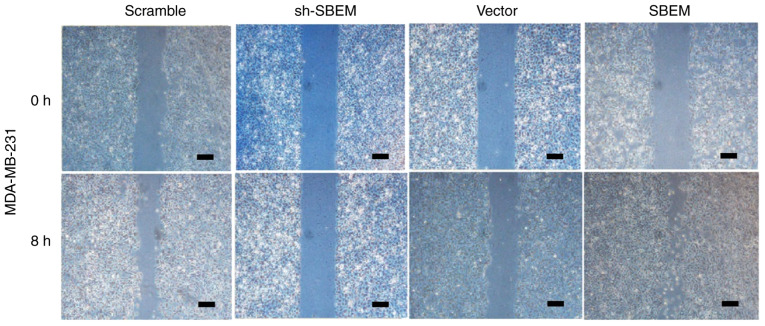
Wound healing was observed after 8 h. The results of the MCF-7 cells demonstrated that the cell repair rate in sh-SBEM group was (6.8%) much lower than that in the scramble group (39.6%). The cell repair rate in the SBEM overexpression group was higher (48.1%) than that in the vector group (27.0%) (Fig. 6 and Table III). The results of the MDA-MB-231 cells demonstrated that the cell repair rate in the sh-SBEM group (4.8%) was markedly lower compared with that in the scramble group (31.0%). The cell repair rate in the SBEM overexpression group was higher (69.9%) compared with that in the vector group (26.2%) (Fig. 7, Table III). These results indicated that SBEM knockdown obviously inhibited and SBEM overexpression obviously promoted the migration ability of the breast cancer cells.
Figure 6.
SBEM promotes the scratch wound healing of MCF-7 cells. The cell repair rate of the sh-SBEM group was much lower than that of the scramble group in the MCF-7 cells. The cell repair rate of the SBEM-overexpression group was higher than of the vector group in the MCF-7 cells. Scale bars represent 200 µm. SBEM, small breast epithelial mucin.
Table III.
Cell repair rate (%) of each group in the scratch wound-healing assay.
| Cell line | ||
|---|---|---|
| Group | MCF-7 | MDA-MB-231 |
| sh-SBEM | 6.8 | 4.8 |
| Scramble | 39.6 | 31.0 |
| SBEM overexpression | 48.1 | 69.9 |
| Vector | 27.0 | 26.2 |
SBEM, small breast epithelial mucin.
Figure 7.
SBEM promotes the scratch wound healing of MDA-MB-231 cells. The cell repair rate of the sh-SBEM group was much lower than that of the scramble group in the MDA-MB-231 cells at 8 h. The cell repair rate of the SBEM overexpression group was higher than of the vector group in the MDA-MB-231 cells at 8 h. Scale bars represent 200 µm. SBEM, small breast epithelial mucin.
SBEM promotes the invasion of breast cancer cells
The numbers of cells crossing the basement membrane were recorded for 24 h. In the MCF-7 cells, the cell numbers were 20±1.15 in the sh-SBEM group, and 75.6±2.01 in the scramble group (P<0.05); the cell numbers were 81.2±1.47 in the SBEM-overexpressing group, and 25.5±1.08 in the vector group. The difference between the two groups was statistically significant (P<0.05; Fig. 8). In MDA-MB-231 cells, the cell numbers were 48.5±1.35 in sh-SBEM group, and 161.7±1.25 in the scramble group; the cell numbers were 241.7±1.15 in the SBEM-overexpressing group, and 118±1.63 in the vector group. The difference between the two groups was statistically significant (P<0.05; Fig. 9). The results demonstrated that SBEM knockdown obviously inhibited and SBEM overexpression obviously promoted the invasion ability of breast cancer cells.
Figure 8.
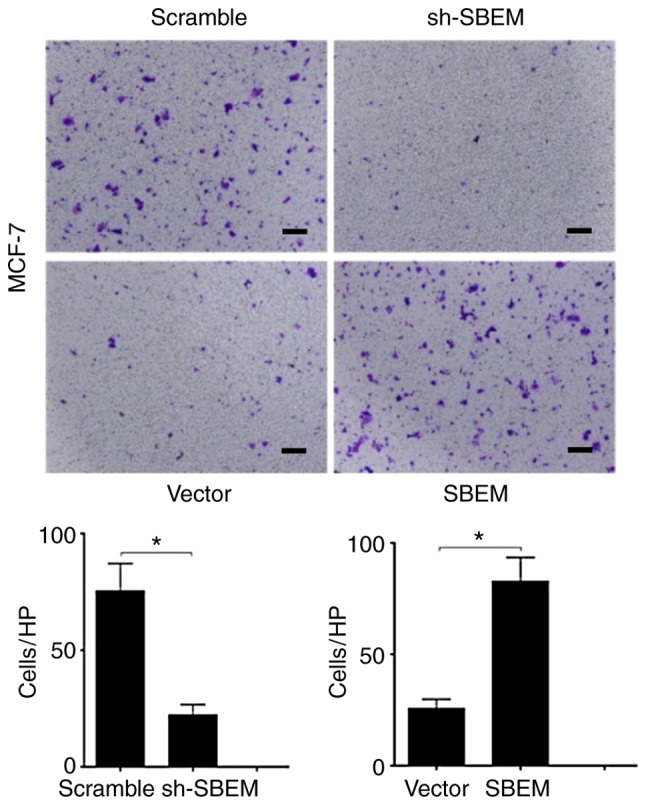
SBEM promotes the invasion of MCF-7 cells. The invasive cell numbers/HPF (high-power field) of the sh-SBEM group were much lower than that of the scramble group (*P<0.05). The invasive cell numbers/HPF of the SBEM-overexpressing group were much higher than that of the vector group (*P<0.05). Scale bars represent 50 µm. SBEM, small breast epithelial mucin.
Figure 9.
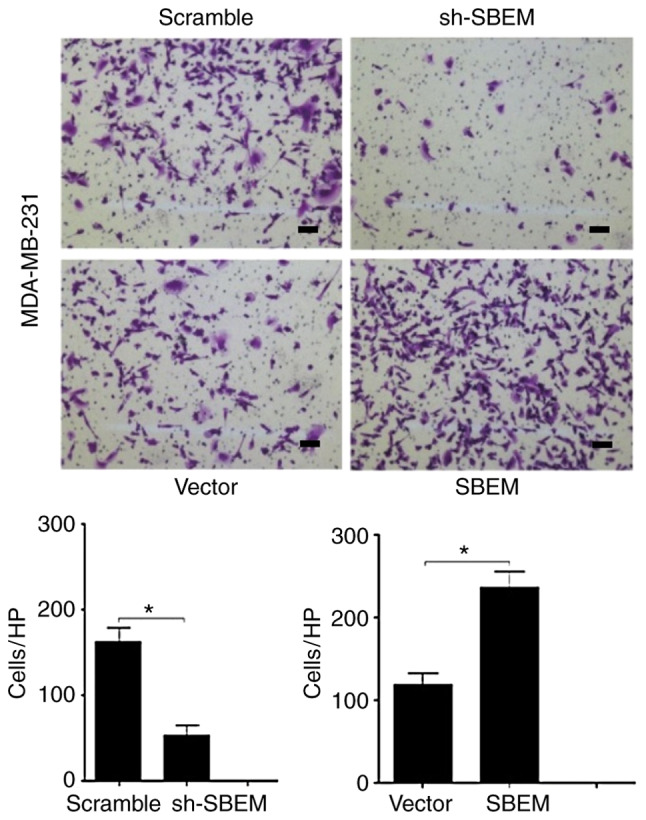
SBEM promotes the invasion of MDA-MB-231 cells. The invasive cell numbers/HPF (high-power field) of the sh-SBEM group were much lower than that of the scramble group (*P<0.05). The invasive cell numbers/HPF of the SBEM-overexpressing group were much higher than that of the vector group (*P<0.05). Scale bars represent 50 µm. SBEM, small breast epithelial mucin.
SBEM affects the expression of EMT-related markers and regulators
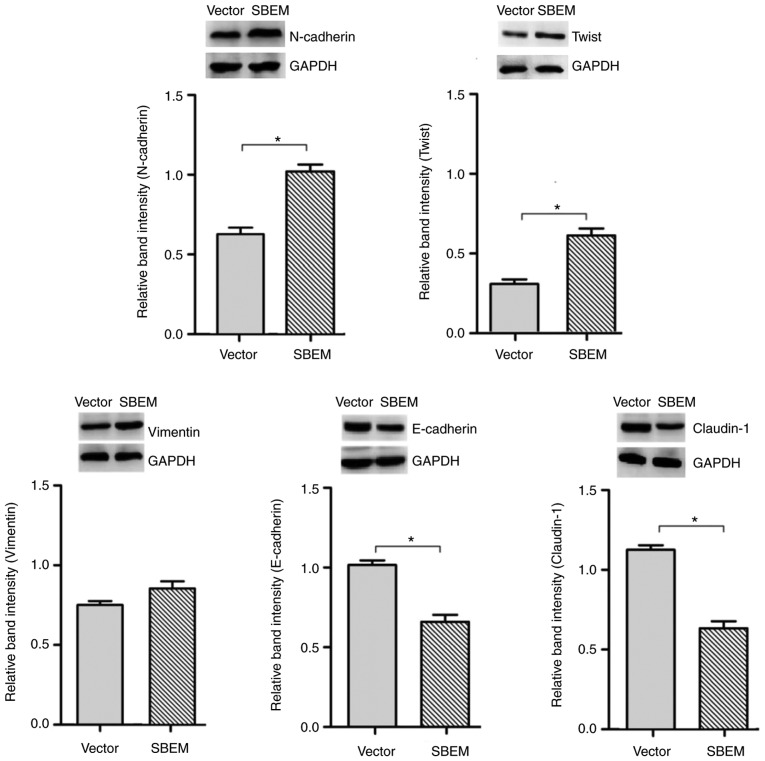
In order to confirm the association of the SBEM gene with the process of EMT, the real-time PCR and western blotting were used to detect the expression of EMT-related markers and regulators in SBEM-overexpressing MCF-7 cells. The results revealed that the levels of N-cadherin, Twist and vimentin were elevated, while those of E-cadherin and claudin-1 were decreased following SBEM overexpression. The relative mRNA expression levels, protein bands and relative band intensities are presented in Figs. 10 and 11.
Figure 10.
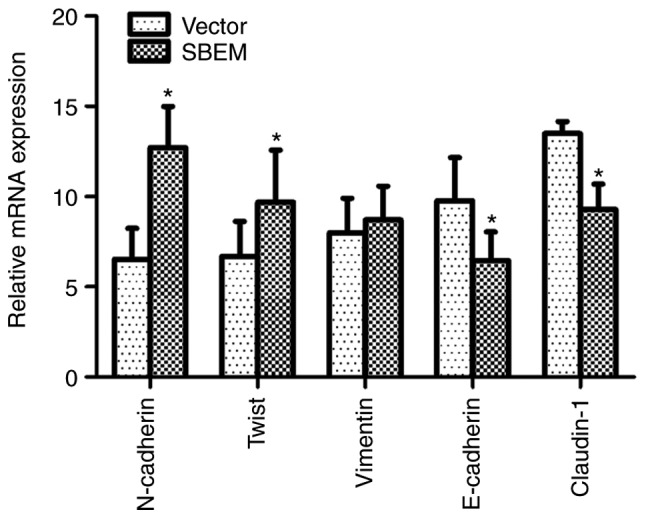
In SBEM-overexpressing MCF-7 cells, SBEM affects the mRNA expression of EMT-related markers and regulators. The relative mRNA expression levels of N-cadherin, Twist and vimentin were elevated), while those of E-cadherin and Claudin-1 were decreased following SBEM overexpression. *P<0.05, compared with the vector group. SBEM, small breast epithelial mucin.
Figure 11.
In SBEM-overexpressing MCF-7 cells, SBEM affects the protein expression of EMT-related markers and regulators. The relative protein band intensities of N-cadherin, Twist and vimentin were elevated, while those of E-cadherin and claudin-1 were decreased following SBEM overexpression. *P<0.05, compared with the vector group. SBEM, small breast epithelial mucin.
Discussion
In recent years, breast cancer has become the primary cause of cancer-related death among women worldwide (15). Patients with isolated tumor cells or micrometastases have a comparably poor 5-year disease-free survival rate (16). Detection of breast cancer micrometastases based on specific molecular markers and exploration of the potential underlying mechanism may provide useful information for clinical research (17). Small breast epithelial mucin (SBEM) is a type of secretory protein, which belongs to the family of MUC (18). The SBEM gene is mainly expressed in the breast and salivary glands (19), and exhibits higher expression in breast cancer tissue and metastatic lymph nodes (20,21). We previously reported that SBEM expression had the potential to serve as a useful and specific marker for hematogenous metastasis of breast cancer (3). Our previous study demonstrated that the expression of SBEM was significantly correlated with the disease-free and overall survival of patients with TNBC, and that it appears to be a promising prognostic biomarker for TNBC diagnosis and treatment (22). Due to its high specificity for breast tissue, SBEM was considered to play an important role in the metastatic process of breast cancer (23,24). However, little is known concerning the potential role and mechanism of action of the SBEM gene in the migration and invasion of breast cancer cells.
In the present study, MCF-7 and MDA-MB-231 cells with stable SBEM knockdown or overexpression were generated by lentiviral vectors. In both cell lines, the cell repair rate in the sh-SBEM group was markedly lower when compared with that in the scramble group, and the cell repair rate in the SBEM overexpression group was markedly higher when compared with that in the vector group. These findings indicated that SBEM knockdown obviously inhibited and SBEM overexpression obviously enhanced the migration ability of breast cancer cells. Similarly, in both MCF-7 and MDA-MB-231 cells, the numbers of cells that invaded through the basement membrane over 24 h were markedly lower in the sh-SBEM group compared with those in the scramble group, whereas they were markedly higher in the SBEM-overexpressing group compared with those in the vector group. These findings indicated that SBEM knockdown obviously inhibited and SBEM overexpression obviously promoted the invasion ability of the breast cancer cells.
Epithelial-mesenchymal transition (EMT) is a cellular process during which epithelial cells acquire mesenchymal phenotypes and behavior following the downregulation of epithelial features (25). The underlying cytological mechanisms include changes in cell morphology, loss of polarity, decreased adhesion, weakening of connections to the basement membrane, and the increase of cell migration and invasion abilities. Approximately 95% of breast cancer cells originate from epithelial cells, whereas the surrounding cells display mesenchymal phenotypes. Mesenchymal cells are characterized by stronger mobility, which enables invasion of blood and lymphatic vessels and metastasis to distant organs (26). The loss of E-cadherin expression has been considered as the key step during EMT in breast cancer. When the expression of E-cadherin is decreased, the intercellular adhesions become weaker, resulting in the loss of cell polarity. The expression of E-cadherin has been shown to be negatively correlated with the migration and invasion abilities of breast cancer cells (27,28). Claudin-1 is the main cytoskeletal protein that constitutes the tight junction chain of epithelial cells. The loss of claudin-1 expression leads to the separation of epithelial cells and an increase of mobility, which facilitates the invasion and metastasis of cancer cells after EMT (9). Vimentin, a type of intermediate filament that is distributed in mesenchymal tissues and cells, helps maintain interstitial cell characteristics (30). When the expression of E-cadherin is decreased, the cytoskeleton mainly composed of keratin is transformed into vimentin-based cytoskeleton proteins, resulting in a change of cell morphology and rendering tumor cells more invasive (31,32). Twist is a basic-helix-loop-helix transcription factor that promotes cell migration and tissue recombination, which may enhance cell invasiveness (33). Overexpression of Twist was also found to induce angiogenesis in breast cancer (34). In the present study, the expression of N-cadherin, Twist and vimentin was found to be increased, while the expression of E-cadherin and claudin-1 was decreased following SBEM overexpression in MCF-7 cells. These findings indicated that overexpression of SBEM downregulated the expression of the epithelial marker E-cadherin, and upregulated the expression of the mesenchymal markers N-cadherin and vimentin. Furthermore, SBEM promoted EMT by upregulating the expression of the transcription factor Twist. Of note, the increase in the expression of vimentin was not as significant compared with that of N-cadherin and Twist following SBEM overexpression. It was hypothesized that this may be associated with the early observational time point, as vimentin elevation is usually a late event during the EMT process.
EMT marks the initiation of the malignant phenotype transformation process, and it is the first step in the invasion-metastasis cascade of breast cancer cells through the basement membrane (35). There are a number of EMT-related signaling pathways in breast cancer, including TGF-β, NF-κB, Notch, Wnt/β-catenin and PI3K/AKT, among others (36–40). Determining which signaling pathways are involved in SBEM-induced EMT constitutes an important research direction. There remain a number of relevant mechanisms and pathways to be investigated.
In summary, the effects of SBEM on the invasion-metastasis cascade suggest that it may be a potential effective target for anti-metastasis treatment in breast cancer. However, there are yet no data from experiments in vivo to validate our in vitro findings, and there are no exact data on EMT-related signaling pathways of SBEM. The findings of the present study may prove helpful as the basis for further research on the mechanisms of action and relevant pathways of SBEM. More focus will be placed on these two aspects of research to acquire relevant data in the future.
Acknowledgements
We thank Professor Guo-Xin Li from The Second Affiliated Hospital of Liaoning University of Traditional Chinese Medicine for his secretarial and organizational support in our experiments.
Funding
The present study was financially supported by Foundation projects: 2019 Subsidization Project of Liaoning Province Natural Foundation (2019-MS-351 and 2019-MS-210); 2018 Guidance Project of Liaoning Province Natural Foundation (20180550359); 2017 Liaoning Province TCM Clinics (Specialized) Branch Capacity Building Project; 2018 Liaoning Province Doctoral Start-up Foundation (20180540043).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
QHL, ZZL and BL contributed to the study conception and design. Material preparation, data collection and analysis were performed by QHL, ZZL, YNG, XL, XDX, ZDZ and YHM. The first draft of the manuscript was written by ZZL. All authors read, revised and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Li T, Mello-Thoms C, Brennan PC. Descriptive epidemiology of breast cancer in China: Incidence, mortality, survival and prevalence. Breast Cancer Res Treat. 2016;159:395–406. doi: 10.1007/s10549-016-3947-0. [DOI] [PubMed] [Google Scholar]
- 2.Schindlbeck C, Andergassen U, Jueckstock J, Rack B, Janni W, Jeschke U. Disseminated and circulating tumor cells in bone marrow and blood of breast cancer patients: Properties, enrichment, and potential targets. J Cancer Res Clin. 2016;142:1883–1895. doi: 10.1007/s00432-016-2118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skliris GP, Hubé F, Gheorghiu I, Mutawe MM, Penner C, Watson PH, Murphy LC, Leygue E, Myal Y. Expression of small breast epithelial mucin (SBEM) protein in tissue microarrays (TMAs) of primary invasive breast cancers. Histopathology. 2008;52:355–369. doi: 10.1111/j.1365-2559.2007.02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu ZZ, Xie XD, Qu SX, Zheng ZD, Wang YK. Small breast epithelial mucin (SBEM) has the potential to be a marker for predicting hematogenous micrometastasis and response to neoadjuvant chemotherapy in breast cancer. Clin Exp Metastasis. 2010;27:251–259. doi: 10.1007/s10585-010-9323-2. [DOI] [PubMed] [Google Scholar]
- 5.Ayerbes MV, Diaz-Prado S, Ayude D, Campelo RG, Iglesias P, Haz M, Medina V, Gallegos I, Quindos M, Aparicio LA. In silico and in vitro analysis of small breast epithelial mucin as a marker for bone marrow micrometastasis in breast cancer. Adv Exp Med Biol. 2008;617:331–339. doi: 10.1007/978-0-387-69080-3_31. [DOI] [PubMed] [Google Scholar]
- 6.Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: Mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Corso G, Pravettoni G, Galimberti V, Veronesi P. Clinical implication of E-cadherin deficiency in lobular breast cancer. Breast Cancer Res Treat. 2019;173:751–752. doi: 10.1007/s10549-018-5051-0. [DOI] [PubMed] [Google Scholar]
- 8.Luo CW, Wu CC, Chang SJ, Chang TM, Chen TY, Chai CY, Chang CL, Hou MF, Pan MR. CHD4-mediated loss of E-cadherin determines metastatic ability in triple-negative breast cancer cells. Exp Cell Res. 2018;363:65–72. doi: 10.1016/j.yexcr.2017.12.032. [DOI] [PubMed] [Google Scholar]
- 9.Zhou B, Blanchard A, Wang N, Ma X, Han J, Schroedter I, Leygue E, Myal Y. Claudin 1 promotes migration and increases sensitivity to tamoxifen and anticancer drugs in luminal-like human breast cancer cells MCF7. Cancer Invest. 2015;33:429–439. doi: 10.3109/07357907.2015.1060996. [DOI] [PubMed] [Google Scholar]
- 10.Upmanyu N, Bulldan A, Papadopoulos D, Dietze R, Malviya VN, Scheiner-Bobis G. Impairment of the Gnα11-controlled expression of claudin-1 and MMP-9 and collective migration of human breast cancer MCF-7 cells by DHEAS. J Steroid Biochem Mol Biol. 2018;182:50–61. doi: 10.1016/j.jsbmb.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Sheehan GM, Kallakury BV, Sheehan CE, Fisher HA, Kaufman RP, Jr, Ross JS. Loss of claudins-1 and −7 and expression of claudins-3 and −4 correlate with prognostic variables in prostatic adenocarcinomas. Hum Pathol. 2007;38:564–569. doi: 10.1016/j.humpath.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Blanchard AA, Ma X, Dueck KJ, Penner C, Cooper SC, Mulhall D, Murphy LC, Leygue E, Myal Y. Claudin 1 expression in basal-like breast cancer is related to patient age. BMC Cancer. 2013;13:268. doi: 10.1186/1471-2407-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Huang S, Zeng L, Ma J, Sun S, Zeng F, Kong F, Cheng X. HMOX-1 inhibits TGF-β-induced epithelial-mesenchymal transition in the MCF-7 breast cancer cell line. Int J Mol Med. 2017;40:411–417. doi: 10.3892/ijmm.2017.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: Burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26:444–457. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 16.de Boer M, van Deurzen CH, van Dijck JA, Borm GF, van Diest PJ, Adang EM, Nortier JW, Rutgers EJ, Seynaeve C, Menke-Pluymers MB, et al. Micrometastases or isolated tumor cells and the outcome of breast cancer. N Engl J Med. 2009;361:653–663. doi: 10.1056/NEJMoa0904832. [DOI] [PubMed] [Google Scholar]
- 17.Hubé F, Mutawe M, Leygue E, Myal Y. Human small breast epithelial mucin: The promise of a new breast tumor biomarker. DNA Cell Biol. 2004;23:842–849. doi: 10.1089/dna.2004.23.842. [DOI] [PubMed] [Google Scholar]
- 18.Park HK, Seov UH. MUC1 from the mucin family as potential tools in breast cancer immunotherapy. J Breast Cancer. 2009;12:125. doi: 10.4048/jbc.2009.12.3.125. [DOI] [Google Scholar]
- 19.Kontani K, Taguchi O, Narita T, Izawa M, Hiraiwa N, Zenita K, Takeuchi T, Murai H, Miura S, Kannagi R. Modulation of MUC1 mucin as an escape mechanism of breast cancer cells from autologous cytotoxic T-lymphocytes. Br J Cancer. 2001;84:1258–1264. doi: 10.1054/bjoc.2000.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heublein S, Mayr D, Egger M, Karsten U, Goletz S, Angele M, Gallwas J, Jeschke U, Ditsch N. Immunoreactivity of the fully humanized therapeutic antibody PankoMab-GEX™ is an independent prognostic marker for breast cancer patients. J Exp Clin Cancer Res. 2015;34:50. doi: 10.1186/s13046-015-0152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber PA, Hevezi P, Buhren BA, Martinez C, Schrumpf H, Gasis M, Grether-Beck S, Krutmann J, Homey B, Zlotnik A. Systematic identification and characterization of novel human skin-associated genes encoding membrane and secreted proteins. PLoS One. 2013;8:e63949. doi: 10.1371/journal.pone.0063949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Liu Z, Qu S, Zheng Z, Liu Y, Xie X, Song F. Small breast epithelial mucin tumor tissue expression is associated with increased risk of recurrence and death in triple-negative breast cancer patients. Diagn Pathol. 2013;8:71. doi: 10.1186/1746-1596-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miksicek RJ, Myal Y, Watson PH, Walker C, Murphy LC, Leygue E. Identification of a novel breast- and salivary gland-specific, mucin-like gene strongly expressed in normal and tumor human mammary epithelium. Cancer Res. 2002;62:2736–2740. [PubMed] [Google Scholar]
- 24.Liu Z, Guo F, Xie X. Individual detection significances of small breast epithelial mucin (SBEM) and human mammaglobin (hMAM) expressions in peripheral blood of breast cancer patients. Chin-Ger J Clin Oncol. 2012;11:716–720. doi: 10.1007/s10330-012-1078-6. [DOI] [Google Scholar]
- 25.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori G, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Bio. 2020 Apr 16; doi: 10.1038/s41580-020-0237-9. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creighton CJ, Gibbons DL, Kurie JM. The role of epithelial-mesenchymal transition programming in invasion and metastasis: A clinical perspective. Cancer Mang Res. 2013;5:187–195. doi: 10.2147/CMAR.S35171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vergara D, Simeone P, Latorre D, Cascione F, Leporatti S, Trerotola M, Giudetti AM, Capobianco L, Lunetti P, Rizzello A, et al. Proteomics analysis of E-cadherin knockdown in epithelial breast cancer cells. J Biotechnol. 2015;202:3–11. doi: 10.1016/j.jbiotec.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 28.Carrizo MN. Loss of E-cadherin expression and epithelial mesenchymal transition (EMT) as key steps in tumor progression. J Cancer Prev Curr Res. 2017;7:00235. doi: 10.15406/jcpcr.2017.07.00235. [DOI] [Google Scholar]
- 29.Bar I, Merhi A, Larbanoix L, Constant M, Haussy S, Laurent S, Canon JL, Delrée P. Silencing of casein kinase 1 delta reduces migration and metastasis of triple negative breast cancer cells. Oncotarget. 2018;9:30821–30836. doi: 10.18632/oncotarget.25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Gu LN, Shan BE, Geng CZ, Sang MX. Biomarkers for EMT and MET in breast cancer: An update. Oncol Lett. 2016;12:4869–4876. doi: 10.3892/ol.2016.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Libring S, Solorio L. Cancer mechanobiology: Interaction of biomaterials with cancer cells. Biomaterials for Cancer Therapeutics. 2020:445–470. doi: 10.1016/B978-0-08-102983-1.00016-8. [DOI] [Google Scholar]
- 32.Trogden KP, Battaglia RA, Kabiraj P, Madden VJ, Herrmann H, Snider NT. An image-based small-molecule screen identifies vimentin as a pharmacologically relevant target of simvastatin in cancer cells. FASEB J. 2018;32:2841–2854. doi: 10.1096/fj.201700663R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, Mo Q, Liao L, Xu J. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36:1157–1166. doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mironchik Y, Winnard PT, Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, et al. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 2005;65:10801–10809. doi: 10.1158/0008-5472.CAN-05-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mali AV, Joshi AA, Hegde MV, Kadam SS. Enterolactone modulates the ERK/NF-κB/Snail signaling pathway in triple-negative breast cancer cell line MDA-MB-231 to revert the TGF-β-induced epithelial-mesenchymal transition. Cancer Biol Med. 2018;15:137–156. doi: 10.20892/j.issn.2095-3941.2018.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pires BR, Mencalha AL, Ferreira GM, de Souza WF, Morgado-Díaz JA, Maia AM, Corrêa S, Abdelhay ES. NF-kappaB is involved in the regulation of EMT genes in breast cancer cells. PLoS One. 2017;12:e0169622. doi: 10.1371/journal.pone.0169622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Tang P, Li S, Qin X, Yang H, Wu C, Liu Y. Notch signaling pathway networks in cancer metastasis: A new target for cancer therapy. Med Oncol. 2017;34:180. doi: 10.1007/s12032-017-1039-6. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Ji L, Jiang G, Liu R, Liu Z, Yang Y, Ma Q, Zhao H. FL118, a novel camptothecin analogue, suppressed migration and invasion of human breast cancer cells by inhibiting epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway. Biosci Trends. 2018;12:40–46. doi: 10.5582/bst.2017.01288. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Fu H, Luo Y, Chen L, Cheng R, Zhang N, Guo H. cPLA2α mediates TGF-β-induced epithelial-mesenchymal transition in breast cancer through PI3k/Akt signaling. Cell Death Dis. 2017;8:e2728. doi: 10.1038/cddis.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.