Abstract
Testis-specific protein Y-encoded-like 5 (TSPYL5), a member of the nucleosome assembly protein (NAP) superfamily, functions as a tumor suppressor in ovarian and lung cancer, yet its clinical significance and molecular mechanism in colorectal cancer (CRC) remain unclear. TSPYL5 expression was analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) database. CRC cell lines HCT116 and HT29 were forced to overexpress TSPYL5 by transfection with pcDNA3.1-TSPYL5. Cell proliferation, apoptosis, migration, and invasion were examined by EdU proliferation assays, flow cytometry, and Transwell assays, respectively. Endoplasmic reticulum stress (ERS) was examined by transmission electron microscopy. Western blot analyses were performed to assess the expression of ERS-associated proteins. GEPIA database analysis showed that CRC patients had lower levels of TSPYL5 expression in their tumor tissues when compared with their para-carcinoma tissues. In vitro experiments indicated that TSPYL5 overexpression significantly suppressed cell proliferation, migration, and invasion, and induced apoptosis and ERS in HCT116 and HT29 cells. Furthermore, the levels of caspase-1, caspase-3, Bax, ATF4, and CHOP protein expression were upregulated after TSPYL5 was overexpressed. In conclusion, our data suggest that TSPYL5 can activate an ERS response that suppresses the proliferation, migration, and invasion of tumor cells. This mechanism may represent a promising therapeutic strategy for CRC.
Keywords: colorectal cancer, TSPYL5, endoplasmic reticulum stress, apoptosis, cell proliferation
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, and its incidence has significantly increased during the past 20 years (1,2). While surgical treatments have significantly increased the 5-year survival rate of patients with most types of primary tumors, late-stage CRC patients still have a 5-year overall survival rate of <10% (3). Radiotherapy and chemotherapy can be used for patients with different stages of CRC, but are not recommended for patients with functional organs and for extended treatment of >6 months (4). Recently, molecular-targeting agents have emerged as promising treatments for prolonging the overall survival of CRC patients, (5). This has created an urgent need to identify new targets that can be used for the early diagnosis and personalized treatment of CRC.
Testis-specific protein Y-encoded-like 5 (TSPYL5), located on chromosome 8q22.1, is a member of the TSPY-L gene family (6), and is also a member of the nucleosome assembly protein (NAP) superfamily (7). TSPYL5 has been shown to interact with ubiquitin-specific protease to reduce the tumor-suppressor activity of p53 (8). Accumulating evidence suggests a critical role for TSPYL5 in tumor progression. For example, TSPYL5 was shown to modulate the growth of A549 cells and their sensitization to the detrimental effects of toxic agents via regulation of p21(WAF1/Cip1) and the PTEN/AKT pathway (9). Restoration of TSPYL5 by a DNA methyltransferase inhibitor was demonstrated to suppress the growth of gastric cancer cells (10). Furthermore, TSPYL5 functions as a tumor suppressor in ovarian cancer (11) and an oncogene in breast cancer (12).
Endoplasmic reticulum (ER) is made up of membranous tubules and vesicles. An accumulation of unfolded and misfolded proteins usually leads to ER stress (ERS) (13,14). ERS is mediated by pancreatic endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6) for the purpose of maintaining protein homeostasis (15). As a major signal-transducing event, ERS can induce apoptosis to enhance the cytotoxicity of various chemotherapeutic drugs (16,17). It is now well-established that targeting the ERS response is an effective strategy for suppressing the growth of human hepatocellular carcinoma (18), breast cancer (19), and ovarian cancer cells (20). Although some investigators have focused on the role of ERS in CRC, few studies have examined the mechanism by which TSPYL5 affects ERS and CRC progression.
In the present study, we investigated the expression patterns and clinical significance of TSPYL5 in CRC patients via a GEPIA database analysis and an analysis of clinical samples. Furthermore, we explored the biological function of TSPYL5 and its effects on ERS-associated factors for the purpose of identifying molecular pathways involved in the malignant behaviors of CRC cells.
Materials and methods
GEPIA database analysis
The levels of TSPYL5 expressed in CRC tumors and normal tissues were identified using the online Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html), which is an interactive website that includes information for 9,736 tumor samples and 8,587 normal tissue samples obtained from TCGA and GTEx projects. The GEPIA database was also used to generate survival curves based on the levels of TSPYL5 gene expression in CRC tissues, as determined by the log-rank test.
Clinical tissues
Thirty pairs of CRC and para-carcinoma tissue samples were collected from CRC patients who underwent surgical resection of their tumors at the Renmin Hospital of Wuhan University from February 2017 to December 2018 (age range, 45–86 years; Females, 41%). None of the patients had received any radiotherapy or chemotherapy prior to surgery, and each patient provided a written informed consent. All the tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until use. The protocol for this study was approved by the Ethics Committee of The Renmin Hospital of Wuhan University (Wuhan, Hubei, China). All procedures involving human subjects were performed in accordance with the 1964 Helsinki declaration.
Quantitative real time PCR
Total RNA was isolated from frozen tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reversed transcribed into cDNA with an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc.). Quantitative real-time PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Inc.) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The PCR reaction conditions consisted of 95°C for 3 min, followed by 40 cycles of 95°C for 30 sec and 60°C for 30 sec. The primer sequences used were as follows: TSPYL5 forward, GGTTGTTTTTTGTGTAGTTGTAGT and reverse, CATCACAAACATACAACTATACCA. TSPYL5 expression was normalized to that of GAPDH, and analyzed using the 2−ΔΔCq method (21).
Cell culture and transfection
Human CRC cell lines HCT116 and HT29 were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). Both cell lines were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
For TSPYL5 overexpression, the cDNA for TSPYL5 without its 3′-UTR was inserted into a pcDNA3.1 vector using Lipofectamine 2000 transfection reagent (both from Invitrogen; Thermo Fisher Scientific, Inc.) to generate the recombinant vector, pcDNA3.1-TSPYL5. Subsequently, HCT116 and HT29 cells were seeded into 6-well plates and transfected with pcDNA3.1-TSPYL5 or empty pcDNA3.1, to produce TSPYL5 and negative control (NC) groups, respectively. Non-transfected cells served as a blank group.
EdU proliferation assay
A Cell Light™ EDU Apollo®488 In Vitro Imaging Kit (Guangzhou RiboBio) was used to detect the proliferation rates of the transfected CRC cells, according to instructions provided by the manufacturer. Briefly, HCT116 and HT29 cells were fixed with 4% paraformaldehyde for 15 min, washed three times with PBS, and then stained with 200 µl of 1X Apollo solution for 30 min. After another wash with PBS, the cells with EdU-positive signals were detected by flow cytometry (BD Biosciences).
Flow cytometry
Cell apoptosis was detected with an Annexin V-fluorescein Isothiocyanate Propidium Iodide (FITC/PI) apoptosis detection kit (cat. no. 70-AP101-100; Hangzhou MultiSciences Biotech Co., Ltd.). Briefly, HCT116 and HT29 cells were harvested, washed twice in PBS, and then stained with Annexin V-FIFC/PI for 15 min; after which, they were analyzed by flow cytometry (BD Biosciences). The total apoptotic rate, including early apoptosis and late apoptosis, was calculated and averaged for three experiments.
Hoechst 33342 staining
HCT116 and HT29 cells in their logarithmic growth phase were plated into 6-well plates and incubated for 48 h. The cells were then washed twice with PBS, incubated in the dark for 10 min with Hoechst 33342 nucleic acid stain (Sigma-Aldrich; Merck KGaA), and subsequently examined for their nuclear morphology under a fluorescence microscope (Olympus Corp.).
Transwell assay
Cell migration and invasion abilities were examined using Transwell assays. Briefly, ~2×104 HCT116 or HT29 cells suspended in 1 ml of serum-free medium were added into the upper chamber (without Matrigel for migration or with Matrigel for invasion) of a Transwell plate (Costar, cat. no. 3422; Corning Life Sciences). The lower chamber was filled with 500 µl of culture medium containing 10% FBS. After a 48-h incubation, the migrated cells in the lower chamber were fixed with formaldehyde for 5 min, and then stained with 0.5% crystal violet solution (Sigma-Aldrich; Merck KGaA). The stained cells were observed under a phase-contrast microscope (Olympus Corp.) at a magnification of ×200, and various visual fields were randomly selected for cell counting.
Transmission electron microscopy
ERS was examined by transmission electron microscopy as previously described (22). In brief, transfected HCT116 and HT29 cells were harvested, fixed in glutaraldehyde, and then dehydrated in serial dilutions of acetone (30, 80 and 90%). Next, ultrathin sections were produced by embedding cells in Ultracut (Leica, Germany) and cutting into 60-nm sections. After staining with uranyl acetate, the ultrathin sections were examined with a JEM-1230 transmission electron microscope (JEOL Ltd.).
Western blot analysis
Total cellular protein was obtained by lysing CRC cells in RIPA buffer (Pierce; Thermo Fisher Scientific, Inc.) that contained a protease inhibitor cocktail and phosphatase inhibitors (Sigma-Aldrich; Merck KGaA). Protein concentrations were detected using a BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.). Samples containing ~30 µg of protein were separated by 10% SDS-PAGE, and the protein bands were transferred onto PVDF membranes (Invitrogen; Thermo Fisher Scientific, Inc.). The membranes were then blocked with 5% skim milk and subsequently incubated overnight with primary antibodies against TSPYL5 (cat. no. ab203657, dilution: 1:800; Abcam), caspase-1 (cat. no. ab62698, dilution: 1:800; Abcam), caspase-3 (cat. no. ab49822, dilution: 1:500; Abcam), Bax (cat. no. M00183-2, dilution: 1:1,000, Boster), ATF4 (cat. no. A00371, dilution: 1:500; Boster), CHOP (cat. no. A00311, dilution: 1:500; Boster), and GAPDH (cat. no. ab9485, dilution: 1:800; Abcam) at 4°C. On the following day, the membranes were incubated with an HRP conjugated secondary antibody (cat. no. ab97080, dilution: 1:30,000; Abcam). The immunostained proteins were detected using an enhanced chemiluminescence kit (Thermo Fisher Scientific, Inc.). Quantification of western blot bands was performed using Image-Pro Plus (Version 6; Media Cybernetics, Inc.).
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics for Windows, version 21.0 (IBM Corp.). Quantitative results are expressed as the mean ± SD of data obtained from at least three experiments. Differences between two groups were analyzed using Student's t-test and differences among groups were assessed by one-way analysis of variance (ANOVA). A P-value <0.05 was considered to indicate a statistically significant difference.
Results
Expression of TSPYL5 was downregulated in CRC tissues
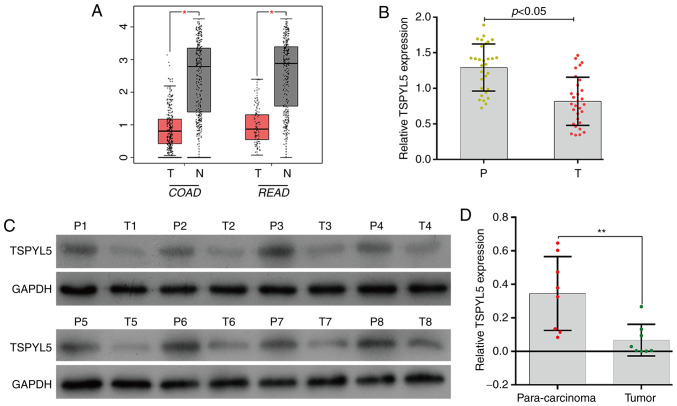
To determine differences in TSPYL5 expression in CRC tumor tissues vs. normal tissues, the TSPYL5 mRNA levels in COAD (colon adenocarcinoma) and READ (rectum adenocarcinoma) were analyzed using the GEPIA database. As shown in Fig. 1A, TSPYL5 expression was significantly lower in the COAD and READ tissues when compared to levels in the respective adjacent normal tissues (P<0.05). To verify the expression results obtained from the GEPIA database, we determined the levels of TSPYL5 expression in 30 pairs of CRC tumor and para-carcinoma tissues by quantitative real-time PCR. As shown in Fig. 1B, TSPYL5 expression was significantly downregulated in the tumor tissues when compared with that noted in the para-carcinoma tissues (P<0.05). Western blot analyses of 8 pairs of representative tissues showed a result similar to that obtained by quantitative real-time PCR (Fig. 1C and D, P<0.01). These results suggest the role of TSPYL5 as a possible tumor suppressor in CRC.
Figure 1.
Expression of TSPYL5 was downregulated in CRC tissues. (A) TSPYL5 expression levels in COAD (colon adenocarcinoma) T, tumor [n=275], N, normal [n=349] and READ (rectum adenocarcinoma) T [n=92], N [n=318] were investigated via the GEPIA database. (B) The levels of TSPYL5 mRNA expression in 30 paired tumor tissues and para-carcinoma tissues were investigated by quantitative real-time PCR analysis. (C) The levels of TSPYL5 protein expression in 8 paired tumor tissues (T1-T8) and para-carcinoma tissues (P1-P8) were investigated by western blotting. (D) Quantitative analysis of TSPYL5 protein levels in 8 paired tumor tissues and para-carcinoma tissues. *P<0.05, **P<0.01. P, para-carcinoma tissues; T, CRC tumor tissues. CRC, colorectal cancer; TSPYL5, testis-specific protein Y-encoded-like 5; GEPIA, Gene Expression Profiling Interactive Analysis. T, Tumor tissues; N, non-tumor tissues; n, number of samples used.
Overexpression of TSPYL5 suppresses the proliferation and promotes the apoptosis of CRC cells
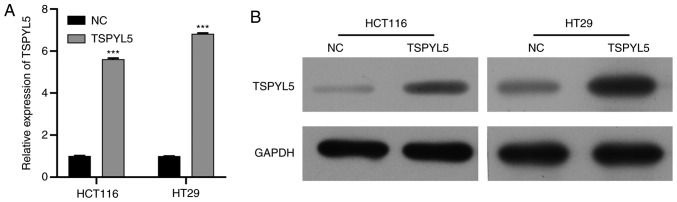
To further confirm that TSPYL5 acts as a tumor suppressor in CRC in vitro, two CRC cell lines, HCT116 and HT29, were transfected with pcDNA3.1-TSPYL5, and then used in a series of functional experiments. Firstly, RT-qPCR and western blot analysis were performed to validated the overexpression of TSPYL5 in HCT116 and HT29 cell lines. Results showed that expression of TSPYL5 was enhanced after cell transfection (Fig. 2A and B).
Figure 2.
Validation of expression of TSPYL5 in CRC HCT116 and HT29 cells lines after transfection with the overexpression vector. (A) RT-qPCR was used to measure the mRNA expression of TSPYL5. (B) Western blot analysis was used to detect the expression of TSPYL5 protein. ***P<0.001, compared to the negative control (NC) group. CRC, colorectal cancer; TSPYL5, testis-specific protein Y-encoded-like 5.
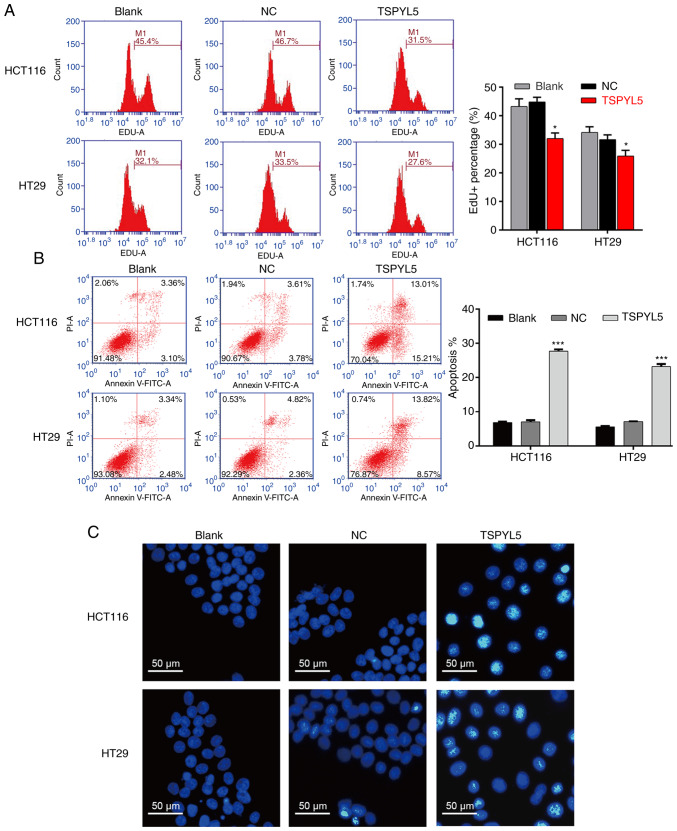
As shown in Fig. 3A, flow cytometry of EdU-positive cells revealed that TSPYL5 overexpression decreased the percentage of EDU-positive HCT116 cells from 46.7 to 31.5% and the percentage of EDU-positive HT29 cells from 33.5 to 27.6%. In addition, flow cytometry with Annexin V/PI double staining was performed to evaluate cell apoptosis. As illustrated in Fig. 3B, the apoptotic rate of the TSPYL5-overexpressing HCT116 cells was significantly increased (27.69±0.52%) when compared to the apoptotic rates of cells in the blank (6.81±0.30%) and NC (7.05±0.49%) groups (P<0.001). Similarly, overexpression of TSPYL5 promoted the apoptosis of HT29 cells (Fig. 3B, P<0.001). Next, HCT116 and HT29 cells stained with a DNA-specific dye (Hoechst 33342) were examined for morphologic changes. The results showed that TSPYL5-overexpressing HCT116 and HT29 cells exhibited bright fluorescence and characteristic features of apoptosis, including chromatin condensation when compared with cells in the NC and blank groups (Fig. 3C).
Figure 3.
Effects of TSPYL5 overexpression on the proliferation and apoptosis of CRC cells. HCT116 and HT29 cells were transfected with pcDNA3.1-TSPYL5 or empty pcDNA3.1, and subsequently divided into TSPYL5 and negative control (NC) groups, respectively. (A) EdU flow cytometry was used to analyze the percentage of EdU-positive cells. (B) Flow cytometric analysis of cell apoptosis rates using Annexin V-FITC/PI double labeling. Data represent the mean ± SD of results from three individual experiments; ***P<0.001 vs. NC. (C) Fluorescence assay of Hoechst 33342 staining. HCT116 and HT29 cells were stained with Hoechst 33342 staining solution and visualized under a fluorescence microscope. CRC, colorectal cancer; TSPYL5, testis-specific protein Y-encoded-like 5.
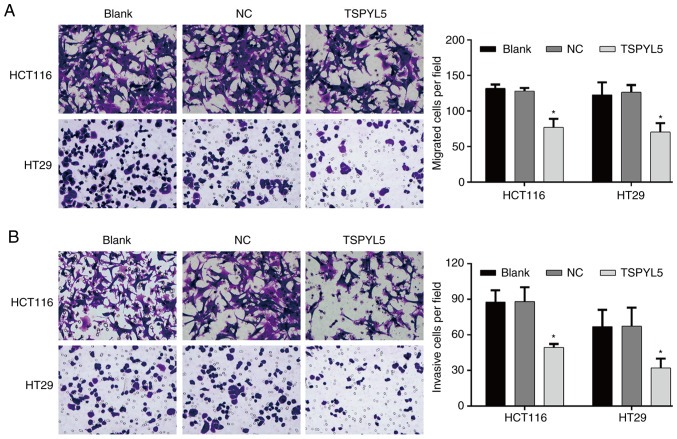
Overexpression of TSPYL5 suppresses the migration and invasion abilities of CRC cells. Transwell assays were performed to investigate the effect of TSPYL5 on the migration and invasion abilities of CRC cells. As shown in Fig. 4A, overexpression of TSPYL5 significantly reduced the numbers of migrated HCT116 cells when compared to the numbers of migrated control cells (TSPYL5-overexpressing HCT116 cells vs. NC cells: 77.00±12.12 vs. 128.00±4.36; P<0.05), and similar results were obtained for the TSPYL5-overexpressing HT29 cells (TSPYL5-overexpressing HT29 cells vs. NC cells: 70.33±12.66 vs. 126.33±10.26, P<0.05). Moreover, after transfection with pcDNA3.1-TSPYL5, the numbers of invading HCT166 and HT29 (Fig. 4B) cells were also decreased when compared to the numbers of invading NC cells (TSPYL5-overexpressing HCT116 cells vs. NC cells: 49.33±3.06 vs. 88.00±12.12, P<0.05 and TSPYL5-overexpressing HT29 cells vs. NC cells: 32.00±7.94 vs. 67.33±15.57, P<0.05).
Figure 4.
Effects of TSPYL5 overexpression on the migration and invasion abilities of CRC cells. HCT116 and HT29 cells were transfected with pcDNA3.1-TSPYL5 or empty pcDNA3.1, and subsequently divided into TSPYL5 and negative control (NC) groups, respectively. Transwell assays were performed to investigate cell migration (A) and invasion (B) Representative images were captured after transfection for 48 h (left panel). A quantification of migrated or invasive cells is shown in the right panel. Data represent the mean ± SD of results from three individual experiments; *P<0.05 vs. the NC group. CRC, colorectal cancer; TSPYL5, testis-specific protein Y-encoded-like 5.
Effects of TSPYL5 overexpression on ERS and associated proteins
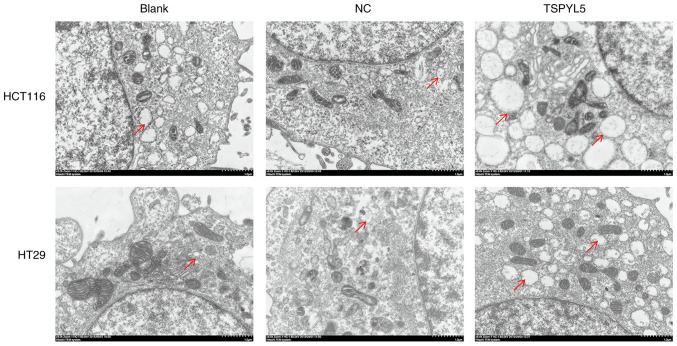
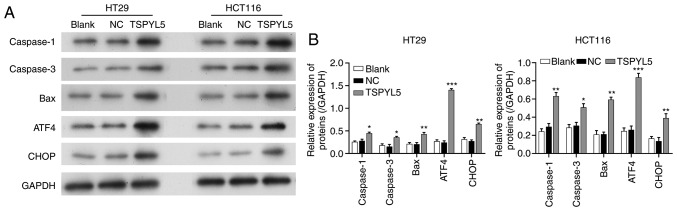
As the effect of TSPYL5 on ERS in CRC cells remains unknown, we next investigated whether TSPYL5 overexpression could induce ERS. First, ERS was detected using a transmission electron microscope. As shown in Fig. 5, larger amounts of swollen ER were observed in the cytoplasm of HCT116 and HT29 cells that overexpressed TSPYL5, and that change was not observed in the control or NC cells. Next, western blot analyses were performed to examine the direct effects of TSPYL5 on ERS and apoptosis. As shown in Fig. 6, the levels of caspase-1, caspase-3, bcl-2-like protein 4 (Bax), activating transcription factor 4 (ATF4) and CCAAT-enhancer-binding protein homologous protein (CHOP) proteins were upregulated after TSPYL5 overexpression. These observed changes indicated that TSPYL5 overexpression-induced apoptosis may be correlated with activated ERS in CRC cells.
Figure 5.
TSPYL5 overexpression induces endoplasmic reticulum stress (ERS) in CRC cells. HCT116 and HT29 cells were transfected with pcDNA3.1-TSPYL5 or empty pcDNA3.1, and subsequently divided into TSPYL5 and negative control (NC) groups, respectively. Photomicrographs of HCT and HT29 cells collected by transmission electron microscopy (magnification ×12,000; red arrow, endoplasmic reticulum). CRC, colorectal cancer; TSPYL5, testis-specific protein Y-encoded-like 5.
Figure 6.
Effects of TSPYL5 overexpression on ERS and its associated proteins. HCT116 and HT29 cells were transfected with pcDNA3.1-TSPYL5 or empty pcDNA3.1, and subsequently divided into TSPYL5 and negative control (NC) groups, respectively. (A) Western blot analysis of the protein levels of caspase-1, caspase-3, bcl-2-like protein 4 (Bax), activating transcription factor 4 (ATF4) and CCAAT-enhancer-binding protein homologous protein (CHOP) in the HT29 and HCT116 cells in the TSPYL5 overexpression and NC groups. (B) Quantification of the levels of caspase-1, caspase-3, Bax, ATF4, and CHOP protein expression as determined by western blot analysis. *P<0.05, **P<0.01 and ***P<0.001 vs. the NC group. CRC, colorectal cancer; TSPYL5, testis-specific protein Y-encoded-like 5.
Discussion
Several studies have reported that TSPYL5 acts as a tumor-suppressor gene in gastric (10) and ovarian cancer (11), yet its clinical significance and biological role in CRC remain unclear. In the present study, we first found that TSPYL5 expression was significantly lower in CRC tissues when compared with that noted in adjacent normal tissues. The methylation levels of TSPYL5 were found to be significantly increased in HCC tissues when compared with those levels in adjacent non-tumor tissues, and that may be an independent unfavorable factor affecting disease-free survival (6). Aberrant methylation of the TSPYL5 promoter has been reported to be associated with high-risk oral leukoplakia (23). In addition, TSPYL5 gene expression has been correlated with the survival of patients with all grades of endometrial cancer (24).
Next, we further demonstrated that overexpression of TSPYL5 significantly suppressed cell proliferation, migration and invasion, and induced apoptosis in two human CRC cell lines. Our findings were consistent with those reported by Shao et al (11), who observed that ovarian cancer inhibition ability could be elevated by miR-629 inhibitor-mediated upregulation of TSPYL5. Lakshmanan et al (25) showed that MUC16 regulates TSPYL5 via the JAK2/STAT3/GR signaling axis in regards to lung cancer cell growth and metastasis. Kumar et al (26) revealed that TSPYL5 overexpression in prostate cancer cells increased the sensitivity of those cells to chemotherapy drugs. Strikingly, a recent study by Huang and Luo (27) demonstrated that overexpression of TSPYL5 promoted apoptosis in HT29 cells and reduced cell proliferation, migration and invasion, which further validates our results (27).
Recently, interest has developed in determining how to utilize the endoplasmic reticulum stress (ERS) response as a method for treating cancer. In the present study, larger amounts of swollen ER were observed in the cytoplasm of HCT116 and HT29 cells that exhibited overexpression of TSPYL5, indicating an increase in ERS. We also found that TSPYL5 overexpression triggered ERS that significantly increased the levels of ATF4, CHOP, caspase-1, caspase-3, and Bax proteins in CRC cells. These findings suggest that ER stress can trigger apoptosis via ER stress-specific cell-death signals, including CHOP and caspase, as previously described (28). ERS-induced apoptosis in CRC was previously described in another report (29,30). Shikonin was found to induce apoptotic cell death by activating ERS, accompanied by increases in Bax and CHOP protein levels (30). Moreover, interleukin-1 receptor associated kinase 2 (IRAK2), as a potential tumor suppressor to counterbalance oncogenic Smad ubiquitylation regulatory factor 1 (Smurf1) in response to ERS, also induced cell death (31). To the best of our knowledge, the effects of TSPYL5 on ERS activation have not been previously investigated. Our present findings suggest that TSPYL5 overexpression suppresses cell proliferation, migration, and invasion via activation of ERS. While knockdown experiments of TSPYL5 must also be performed in further in vitro and in vivo investigations.
Overall, the data presented here demonstrated that TSPYL5 exerted anticancer effects on HCT116 and HT29 cells by activating ERS-induced apoptosis, as evidenced by an accumulation of caspase-1, caspase-3, and Bax proteins, induction of ERS markers, and induction of ATF4 and CHOP. These findings indicate that targeting the ERS response using TSPYL5 may be a promising strategy for treating CRC. This approach should be investigated in future clinical trials.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Data will be provided based on requirement from the corresponding author upon reasonable request.
Authors' contributions
CH and RZ conceived and designed the study. CH and CH performed the experiments. CH and PR wrote the manuscript. CH and PR collected the data. PR and RZ reviewed, revised it critically for important intellectual content and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The protocol for this study was approved by the Ethics Committee of The Renmin Hospital of Wuhan University (Wuhan, Hubei, China). All procedures involving human subjects were performed in accordance with the 1964 Helsinki declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declared no competing interest.
References
- 1.Smeby J, Sveen A, Merok MA, Danielsen SA, Eilertsen IA, Guren MG, Dienstmann R, Nesbakken A, Lothe RA. CMS-dependent prognostic impact of KRAS and BRAFV600E mutations in primary colorectal cancer. Ann Oncol. 2018;29:1227–1234. doi: 10.1093/annonc/mdy085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somers E. International Agency for Research on Cancer. Encyclopedia of Toxicology. 2014;133:1067–1069. [Google Scholar]
- 3.Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97–119. doi: 10.1002/1878-0261.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JP. Chinese standard for the diagnosis and treatment of colorectal cancer (2010) Zhonghua Wei Chang Wai Ke Za Zhi. 2011;14:1–4. (In Chinese) [PubMed] [Google Scholar]
- 5.Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, et al. Molecular biomarkers for the evaluation of colorectal cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J Clin Oncol. 2017;35:1453–1486. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 6.Qiu X, Hu B, Huang Y, Deng Y, Wang X, Zheng F. Hypermethylation of ACP1, BMP4, and TSPYL5 in hepatocellular carcinoma and their potential clinical significance. Dig Dis Sci. 2016;61:149–157. doi: 10.1007/s10620-015-3878-3. [DOI] [PubMed] [Google Scholar]
- 7.Vogel T, Dittrich O, Mehraein Y, Dechend F, Schnieders F, Schmidtke J. Murine and human TSPYL genes: Novel members of the TSPY-SET-NAP1L1 family. Cytogenet Cell Genet. 1998;81:265–270. doi: 10.1159/000015042. [DOI] [PubMed] [Google Scholar]
- 8.Epping MT, Meijer LA, Krijgsman O, Bos JL, Pandolfi PP, Bernards R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol. 2011;13:102–108. doi: 10.1038/ncb2142. [DOI] [PubMed] [Google Scholar]
- 9.Kim EJ, Lee SY, Kim TR, Choi SI, Cho EW, Kim KC, Kim IG. TSPYL5 is involved in cell growth and the resistance to radiation in A549 cells via the regulation of p21(WAF1/Cip1) and PTEN/AKT pathway. Biochem Biophys Res Commun. 2010;392:448–453. doi: 10.1016/j.bbrc.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 10.Jung Y, Park J, Bang YJ, Kim TY. Gene silencing of TSPYL5 mediated by aberrant promoter methylation in gastric cancers. Lab Invest. 2008;88:153–160. doi: 10.1038/labinvest.3700706. [DOI] [PubMed] [Google Scholar]
- 11.Shao L, Shen Z, Qian H, Zhou S, Chen Y. Knockdown of miR-629 Inhibits ovarian cancer malignant behaviors by targeting testis-specific Y-like protein 5. DNA Cell Biol. 2017;36:1108–1116. doi: 10.1089/dna.2017.3904. [DOI] [PubMed] [Google Scholar]
- 12.Lyu JH, Park DW, Huang B, Kang SH, Lee SJ, Lee C, Bae YS, Lee JG, Baek SH. RGS2 suppresses breast cancer cell growth via a MCPIP1-dependent pathway. J Cell Biochem. 2015;116:260–267. doi: 10.1002/jcb.24964. [DOI] [PubMed] [Google Scholar]
- 13.Ansari SS, Sharma AK, Soni H, Ali DM, Tews B, König R, Eibl H, Berger MR. Induction of ER and mitochondrial stress by the alkylphosphocholine erufosine in oral squamous cell carcinoma cells. Cell Death Dis. 2018;9:296. doi: 10.1038/s41419-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khaket TP, Singh MP, Khan I, Bhardwaj M, Kang SC. Targeting of cathepsin C induces autophagic dysregulation that directs ER stress mediated cellular cytotoxicity in colorectal cancer cells. Cell Signal. 2018;46:92–102. doi: 10.1016/j.cellsig.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Luo J. Endoplasmic reticulum stress and ethanol neurotoxicity. Biomolecules. 2015;5:2538–2553. doi: 10.3390/biom5042538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra RR, Belder N, Ansari SA, Kayhan M, Bal H, Raza U, Ersan PG, Tokat ÜM, Eyüpoğlu E, Saatci Ö, et al. Reactivation of cAMP pathway by PDE4D inhibition represents a novel druggable axis for overcoming tamoxifen resistance in ER-positive breast cancer. Clin Cancer Res. 2018;24:1987–2001. doi: 10.1158/1078-0432.CCR-17-2776. [DOI] [PubMed] [Google Scholar]
- 17.Bakhtou H, Olfatbakhsh A, Deezagi A, Ahangari G. The expression of dopamine receptors gene and their potential role in targeting breast cancer cells with selective agonist and antagonist drugs. Could it be the novel insight to therapy? Curr Drug Discov Technol. 2019;16:184–197. doi: 10.2174/1570163815666180130101421. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Zhang L, Gao M, Han M, Liu K, Zhang Z, Gong Z, Xing L, Shi X, Lu K, Gao H. Endoplasmic reticulum stress triggers Xanthoangelol-induced protective autophagy via activation of JNK/c-Jun Axis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:8. doi: 10.1186/s13046-018-1012-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Ma Z, Yang Y, Di S, Feng X, Liu D, Jiang S, Hu W, Qin Z, Li Y, Lv J, et al. Pterostilbene exerts anticancer activity on non-small-cell lung cancer via activating endoplasmic reticulum stress. Sci Rep. 2017;7:8091. doi: 10.1038/s41598-017-08547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Gong W, Yang ZY, Zhou XS, Gong C, Zhang TR, Wei X, Ma D, Ye F, Gao QL. Quercetin induces protective autophagy and apoptosis through ER stress via the p-STAT3/Bcl-2 axis in ovarian cancer. Apoptosis. 2017;22:544–557. doi: 10.1007/s10495-016-1334-2. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Ba-Omar TA, Al-Jardani S, Victor R. Effects of pesticide temephos on the gills of Aphanius dispar (Pisces: Cyprinodontidae) Tissue Cell. 2011;43:29–38. doi: 10.1016/j.tice.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Abe M, Yamashita S, Mori Y, Abe T, Saijo H, Hoshi K, Ushijima T, Takato T. High-risk oral leukoplakia is associated with aberrant promoter methylation of multiple genes. BMC Cancer. 2016;16:350. doi: 10.1186/s12885-016-2371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witek Ł, Janikowski T, Bodzek P, Olejek A, Mazurek U. Expression of tumor suppressor genes related to the cell cycle in endometrial cancer patients. Adv Med Sci. 2016;61:317–324. doi: 10.1016/j.advms.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Lakshmanan I, Salfity S, Seshacharyulu P, Rachagani S, Thomas A, Das S, Majhi PD, Nimmakayala RK, Vengoji R, Lele SM, et al. MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing p53. Clin Cancer Res. 2017;23:3906–3917. doi: 10.1158/1078-0432.CCR-16-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar SR, Bryan JN, Esebua M, Amos-Landgraf J, May TJ. Testis specific Y-like 5: Gene expression, methylation and implications for drug sensitivity in prostate carcinoma. BMC Cancer. 2017;17:158. doi: 10.1186/s12885-017-3134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Luo H. miR-19-5p enhances tumorigenesis in human colorectal cancer cells by targeting TSPYL5. DNA Cell Biol. 2018;37:23–30. doi: 10.1089/dna.2017.3804. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 29.Kim IY, Shim MJ, Lee DM, Lee AR, Kim MA, Yoon MJ, Kwon MR, Lee HI, Seo MJ, Choi YW, Choi KS. Loperamide overcomes the resistance of colon cancer cells to bortezomib by inducing CHOP-mediated paraptosis-like cell death. Biochem Pharmacol. 2019;162:41–54. doi: 10.1016/j.bcp.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Han X, Kang KA, Piao MJ, Zhen AX, Hyun YJ, Kim HM, Ryu YS, Hyun JW. Shikonin exerts cytotoxic effects in human colon cancers by inducing apoptotic cell death via the endoplasmic reticulum and mitochondria-mediated pathways. Biomol Ther (Seoul) 2019;27:41–47. doi: 10.4062/biomolther.2018.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Chen Y, Huang Q, Liu W, Ji X, Hu F, Zhu Y, Zhang L, Dong G. IRAK2 counterbalances oncogenic Smurf1 in colon cancer cells by dictating ER stress. Cell Signal. 2018;48:69–80. doi: 10.1016/j.cellsig.2018.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be provided based on requirement from the corresponding author upon reasonable request.