Abstract
Papillary thyroid carcinoma (PTC) is the most common type of thyroid cancer. Circular RNAs (circRNAs) are a novel class of RNAs, with higher stability and tissue specificity, which may be of value as novel clinical markers. High-throughput RNA sequencing was used to profile the expression of circRNAs in 5 pairs of cancer and normal tissues, and reverse transcription-quantitative PCR (RT-qPCR) analysis was employed to verify the results of the RNA sequencing in 45 cases of PTC. The dysregulated circRNA expression and clinicopathological characteristics were assessed and the potential roles of circRNAs in the cellular miRNA and mRNA network were predicted using bioinformatics analysis. The results demonstrated that, compared with normal tissues, a total of 53 circRNAs were dysregulated in tumour tissues, and 8 circRNAs were validated at the mRNA level (P<0.001 and P<0.01). Among those, the expression of chr5:161330882-161336769- (P=0.015), chr9:22046750-22097364+ (P=0.041) and chr8:18765448-18804898- (P=0.036) were obviously associated with the BRAFV600E mutation, chr12:129699809-129700698- was associated with capsular invasion (P=0.025) and chr5:38523418-38530666- was associated with pT stage (P=0.037) and lymph node metastasis (P=0.002). Therefore, some dysregulated circRNAs were found to be associated with BRAFV600E mutation, capsular invasion, advanced pT stage and lymph node metastasis of PTC, indicating that circRNAs may be involved in tumourigenesis and cancer progression, and they may be putative biomarkers for the diagnosis and evaluation of progression of PTC.
Introduction
Papillary thyroid carcinoma (PTC) is the most common type of thyroid cancer, accounting for 75–85% of all primary thyroid carcinomas (1). The classical and follicular variants are the most common types of PTC, and they differ markedly regarding diagnosis, prognosis, treatment, recurrence, molecular alterations, genetic alterations and molecular biomarkers (2), such as with the preoperative diagnosis of the follicular variant of PTC being more challenging compared with that of classical PTC (3).
Several markers for the diagnosis or prognosis of PTC have been reported, including thyroglobulin for well-differentiated PTC (4,5), ATP5E for early diagnosis (6), RET/PTC, RAS and B-type Raf kinase (BRAF) mutations for diagnosis or prognosis (7). However, the mechanisms of action of those markers require further study and validation. Therefore, there is a clinical need for more reliable, sensitive and specific markers for PTC.
Most human transcripts are composed of non-coding RNA, and accumulating evidence supports the important regulatory role of non-coding RNAs, such as microRNAs (miRNAs) (8) and long non-coding RNAs (lncRNAs) (9), in several physiological and pathophysiological processes. Recently, circular RNAs (circRNAs) have attracted attention in the field of RNA research (10,11). CircRNAs are a novel class of RNAs that have a closed loop structure and are abundantly present in the eukaryotic transcriptome (12,13). The majority of circRNAs are composed of exon sequences, which are conserved across different species, and have expression specificity for different tissues and developmental stages (10). They are primarily considered to act as sponges that bind competitively with miRNAs to regulate the expression of target genes (10,14,15), and dysregulated expression of circRNAs has been found in several types of human cancer (16–19), such as gastric cancer (20, 21), hepatocellular carcinoma (HCC) (17), colorectal cancer (18) and PTC (22–26). However, the detailed function and molecular mechanism of circRNAs in tumour diagnosis, prognosis and treatment have yet to be fully elucidated.
Therefore, in the present study, the circRNA profile in PTC tissues was screened with high-throughput RNA sequencing (RNA-Seq) and the dysregulated circRNAs were validated with reverse transcription-quantitative PCR (RT-qPCR) analysis. Subsequently, the associations between circRNA expression and clinicopathological characteristics were analysed, and the potential roles of circRNAs were predicted with bioinformatics analysis, with the aim of determining their functions in the tumourigenesis, progression and diagnosis of PTC.
Materials and methods
Patient information
A total of 50 patients who underwent surgical treatment with a final diagnosis of PTC at Peking Union Medical College Hospital (Beijing, China) between January 2017 and December 2018 were enrolled in this study, and the clinicopathological characteristics of the patients are listed in Tables I and III. The recruited patients did not receive any adjunctive treatment prior to surgery, such as radiotherapy, chemotherapy or targeted therapy. Clinical information, including patient age, sex, multifocality, subtype (classical/follicular variant PTC), Ki67, vascular endothelial growth factor (VEGF), pT, pN, metastasis, BRAF mutation, capsular invasion and thyroiditis, was collected from the clinical records and stratified as characteristics according to the 8th edition of the American Joint Committee on Cancer (AJCC) Cancer Staging Manual (27).
Table I.
Clinicopathologic characteristics of the 5 patients for circRNA sequencing.
| Patient no. | Age (years) | Sex | TNM | Subtype | Capsular invasion | Multicentricity |
|---|---|---|---|---|---|---|
| 1 | 46 | Male | T3N0M0 | Classical+follicular variant | Yes | Yes |
| 2 | 78 | Male | T3N0M0 | Follicular variant | Yes | Yes |
| 3 | 38 | Female | T1N1M0 | Classical | Yes | No |
| 4 | 36 | Male | T1N0M0 | Classical | Yes | No |
| 5 | 44 | Male | T1N1M0 | Classical | Yes | No |
circRNA, circular RNAs; TNM, Tumour, Node, Metastasis.
Table III.
Association between the 8 circRNA expression levels and clinicopathological characteristics of the 45 PTC patients.
| chr5:161330882-161336769− | chr12:129699809-129700698− | chr9:22046750-22097364+ | chr20:17456347-17465553+ | chr7:116699070-116700284+ | chr8:18765448-18804898− | chr7:22308338-22318037− | chr5:38523418-38530666− | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinico-pathologic parameters | No. of cases (%) | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value |
| Age (years) | |||||||||||||||||
| <45 | 28 | 1.21E-02± | 0.797 | 2.26E-03± | 0.574 | 4.34E-04± | 0.386 | 1.52E-04± | 0.708 | 2.39E-02± | 0.761 | 2.13E-02± | 0.374 | 8.81E-03± | 0.963 | 8.12E-04± | 0.673 |
| (62.2) | 0.014 | 0.006 | 0.001 | 0.000 | 0.039 | 0.018 | 0.011 | 0.001 | |||||||||
| ≥45 | 17 | 1.25E-02± | 2.12E-03± | 4.45E-04± | 1.54E-04± | 2.32E-02± | 2.21E-02± | 8.51E-03± | 8.01E-04± | ||||||||
| (37.8) | 0.014 | 0.005 | 0.001 | 0.000 | 0.038 | 0.018 | 0.011 | 0.001 | |||||||||
| Sex | |||||||||||||||||
| Female | 33 | 1.29E-02± | 0.209 | 2.06E-03± | 0.259 | 4.47E-04± | 0.472 | 1.57E-04± | 0.590 | 2.37E-02± | 0.505 | 2.26E-02± | 0.228 | 8.61E-03± | 0.878 | 7.99E-04± | 0.342 |
| (73.3) | 0.014 | 0.005 | 0.001 | 0.000 | 0.039 | 0.019 | 0.011 | 0.001 | |||||||||
| Male | 12 | 1.12E-02± | 2.26E-03± | 4.33E-04± | 1.56E-04± | 2.47E-02± | 2.08E-02± | 9.05E-03± | 8.61E-04± | ||||||||
| (26.7) | 0.014 | 0.006 | 0.001 | 0.000 | 0.040 | 0.018 | 0.011 | 0.001 | |||||||||
| Multifocal | |||||||||||||||||
| No | 31 | 1.19E-02± | 0.641 | 2.16E-03± | 0.148 | 4.24E-04± | 0.477 | 1.47E-04± | 0.961 | 2.32E-02± | 0.492 | 2.13E-02± | 0.941 | 8.52E-03± | 0.961 | 8.15E-04± | 0.902 |
| (68.9) | 0.013 | 0.005 | 0.001 | 0.000 | 0.039 | 0.018 | 0.011 | 0.001 | |||||||||
| Yes | 14 | 1.29E-02± | 2.06E-03± | 4.47E-04± | 1.57E-04± | 2.37E-02± | 2.26E-02± | 8.61E-03± | 7.99E-04± | ||||||||
| (31.1) | 0.014 | 0.005 | 0.001 | 0.000 | 0.039 | 0.019 | 0.011 | 0.001 | |||||||||
| Ki67 | |||||||||||||||||
| <3 | 26 | 1.21E-02± | 0.516 | 2.26E-03± | 0.725 | 4.34E-04± | 0.725 | 1.52E-04± | 0.116 | 2.39E-02± | 0.892 | 2.13E-02± | 0.705 | 8.81E-03± | 0.850 | 8.12E-04± | 0.110 |
| (57.8) | 0.014 | 0.006 | 0.001 | 0.000 | 0.039 | 0.018 | 0.011 | 0.001 | |||||||||
| ≥3 | 15 | 1.29E-02± | 2.06E-03± | 4.47E-04± | 1.57E-04± | 2.37E-02± | 2.26E-02± | 8.61E-03± | 7.99E-04± | ||||||||
| (33.3) | 0.014 | 0.005 | 0.001 | 0.000 | 0.039 | 0.019 | 0.011 | 0.001 | |||||||||
| VEGF(+) | |||||||||||||||||
| Yes | 41 | ||||||||||||||||
| (91.1) | |||||||||||||||||
| No | 0 | ||||||||||||||||
| pT-stage | |||||||||||||||||
| x | 1 (2.2) | 2.24E-03 | 0.375 | 2.16E-03 | 0.590 | 4.53E-03 | 0.098 | 1.57E-04 | 0.381 | 2.35E-02 | 0.319 | 2.25E-02 | 0.754 | 8.66E-03 | 0.237 | 7.81E-04 | 0.037a |
| 1 | 29 | 1.25E-02± | 2.12E-03± | 4.45E-04± | 1.54E-04± | 2.32E-02± | 2.21E-02± | 8.51E-03± | 8.01E-04± | ||||||||
| (64.4) | 0.014 | 0.005 | 0.001 | 0.000 | 0.038 | 0.018 | 0.011 | 0.001 | |||||||||
| 2 | 4 | 1.22E-02± | 2.21E-03± | 4.32E-04± | 1.50E-04± | 2.36E-02± | 2.16E-02± | 8.67E-03± | 7.94E-04± | ||||||||
| (8.9) | 0.013 | 0.006 | 0.001 | 0.000 | 0.039 | 0.018 | 0.011 | 0.001 | |||||||||
| 3 | 11 | 1.23E-02± | 2.16E-03± | 4.27E-04± | 1.53E-04± | 2.41E-02± | 2.15E-02± | 8.76E-03± | 8.32E-04± | ||||||||
| (24.4) | 0.014 | 0.005 | 0.001 | 0.000 | 0.040 | 0.018 | 0.011 | 0.001 | |||||||||
| pN stage | |||||||||||||||||
| 0/x | 34 | 1.19E-02± | 0.291 | 2.16E-03± | 0.561 | 4.24E-04± | 0.428 | 1.47E-04± | 0.113 | 2.32E-02± | 0.398 | 2.13E-02± | 0.712 | 8.52E-03± | 0.653 | 8.15E-04± | 0.002a |
| (75.6) | 0.013 | 0.005 | 0.001 | 0.000 | 0.039 | 0.018 | 0.011 | 0.001 | |||||||||
| 1 | 11 | 1.27E-02± | 2.16E-03± | 4.53E-04± | 1.57E-04± | 2.35E-02± | 2.25E-02± | 8.66E-03± | 7.81E-04± | ||||||||
| (24.4) | 0.014 | 0.005 | 0.001 | 0.000 | 0.038 | 0.018 | 0.011 | 0.001 | |||||||||
| BRAF mutation | |||||||||||||||||
| Yes | 36 | 1.27E-02± | 0.015a | 2.16E-03± | 0.865 | 4.53E-04± | 0.041a | 1.57E-04± | 0.955 | 2.35E-02± | 0.496 | 2.25E-02± | 0.036a | 8.66E-03± | 0.670 | 7.81E-04± | 0.444 |
| (80.0) | 0.014 | 0.005 | 0.001 | 0.000 | 0.038 | 0.018 | 0.011 | 0.001 | |||||||||
| No | 9 | 9.71E-03± | 1.95E-03± | 3.96E-04± | 1.38E-04± | 2.14E-02± | 1.75E-02± | 8.59E-03± | 7.24E-04± | ||||||||
| (20.0) | 0.012 | 0.006 | 0.001 | 0.000 | 0.037 | 0.016 | 0.011 | 0.001 | |||||||||
| Capsular invasion | |||||||||||||||||
| Yes | 34 | 1.25E-02± | 0.792 | 2.12E-03± | 0.025a | 4.45E-04± | 0.509 | 1.54E-04± | 0.937 | 2.32E-02± | 0.771 | 2.21E-02± | 0.561 | 8.51E-03± | 0.979 | 8.01E-04± | 0.544 |
| (75.6) | 0.014 | 0.005 | 0.001 | 0.000 | 0.038 | 0.018 | 0.011 | 0.001 | |||||||||
| No | 11 | 1.19E-02± | 2.25E-03± | 4.33E-04± | 1.50E-04± | 2.18E-02± | 2.13E-02± | 8.38E-03± | 8.68E-04± | ||||||||
| (24.4) | 0.014 | 0.006 | 0.001 | 0.000 | 0.037 | 0.018 | 0.011 | 0.001 | |||||||||
| With thyroiditis | |||||||||||||||||
| Yes | 9 | 1.27E-02± | 0.610 | 2.16E-03± | 0.061 | 4.53E-04± | 0.222 | 1.57E-04± | 0.364 | 2.35E-02± | 0.570 | 2.25E-02± | 0.212 | 8.66E-03± | 0.395 | 7.81E-04± | 0.281 |
| (20.0) | 0.014 | 0.005 | 0.001 | 0.000 | 0.038 | 0.018 | 0.011 | 0.001 | |||||||||
| No | 36 | 1.06E-02± | 1.89E-03± | 4.07E-04± | 1.59E-04± | 2.18E-02± | 2.03E-02± | 8.39E-03± | 7.89E-04± | ||||||||
| (80.0) | 0.013 | 0.006 | 0.001 | 0.000 | 0.039 | 0.019 | 0.011 | 0.001 | |||||||||
Associations between the mean ± standard deviation of circRNA expression in the tumor and the clinical characteristics of the 45 PTC patients were analyzed with nonparametric statisticsMann-Whitney U test and Kruskal-Wallis test followed with post-hoc Dunn's multiple comparison test respectively for comparison between two or multiple groups, and P<0.05 was considered to indicate statistically significant differences. circRNA, circular RNA; PTC, papillary thyroid cancer; BRAF, B-type Raf kinase; SD, standard deviation.
A total of 50 paired specimens (tumour and normal tissues) were obtained from the same patients during surgery. Following gross examination, the tissues were resected and washed with PBS (pH 7.2). One part of each specimen was immediately snap-frozen using liquid nitrogen within 30 min and cryopreserved in liquid nitrogen (−196°C) until RNA sequencing and gene expression analysis. The remaining part of the specimen was fixed in formalin at room temperature for 48 h and embedded in paraffin, and then 4-µm sections were cut from each block and stained with haematoxylin and eosin (H&E). The morphology of H&E slides were reviewed by 2 experienced pathologists to confirm the final diagnosis (28).
The present study was approved by the Institutional Review Board of Peking Union Medical College Hospital, and written informed consent was obtained from all patients regarding the use of their tissues for research purposes.
Sample preparation and circRNA sequencing
A total amount of 5 µg RNA per sample was used as input material for RNA sample preparation. First, ribosomal RNAs were depleted using a Ribo-Zero™ rRNA Removal Kit (EPIcentre) to obtain rRNA-depleted RNAs. The rRNA-depleted RNAs were further treated with RNase R (EPIcentre) and were then subjected to TRIzol extraction (Thermo Fisher Scientific, Inc.). Subsequently, sequencing libraries were generated using the rRNA-depleted and RNaseR-digested RNAs by NEBNext Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs) following the manufacturer's recommendations. Then, clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq PE Cluster Kit v4 cBot (Illumina, Inc.) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and 125-bp paired-end reads were generated.
Subsequently, the circRNAs were identified according to the manual (10). Differential expression in normal tissues and PTC was evaluated by using DESeq2 from Dr. Michael Love (version 1.6.3) (29). P-values were adjusted as q (Padj) values by the Benjamini & Hochberg method (30); both q (Padj) <0.05 and fold-change in expression of >2 was set as the threshold for differential expression by default.
Total RNA extraction
Thyroid tissues were processed using the IKA ULTRA-TURRAX T10 basic Disperser/Homogenizer (IKA Works, Inc.), and then total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The total RNA concentrations and quality were determined using Nano Drop One (Thermo Fisher Scientific, Inc.). The RNA integrity was assessed through denaturing agarose gel electrophoresis.
RT-qPCR
The RT-qPCR experiments were conducted as previously described (31,32). Total RNA was digested with RNaseR at 37°C for 30 min and reverse-transcribed to synthesise cDNA using M-MLV reverse transcriptase (Promega Corporation; cat. no. M1701) and random primers at 70°C for 10 min followed by a 60-min incubation at 37°C. Then the cDNA was analysed by qPCR with Fast SYBR® Green Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.; cat. no. 4385612) and a 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). Amplification conditions were 95°C (20 sec), 40 cycles of 95°C (3 sec) and 60°C (30 sec) according to the manufacturer's instructions. GAPDH was used as the internal control. The divergent primers used for circRNA validation are listed in Table SI, the specificity of primers was assessed in NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the backsplice junction of circRNA in the PCR product was verified with Sanger sequencing. The relative expression level of each circRNA with GAPDH was calculated using the 2−ΔΔCq method (24,33).
Immunohistochemistry (IHC)
IHC staining was performed on 4-µm sections using the DAKO Autostainer Link 48 (Dako; Agilent Technologies, Inc.). The tissue epitopes were repaired using the automated water bath heating process in Dako PT Link (Dako; Agilent Technologies, Inc.); the sections were incubated in Tris-EDTA retrieval solution (10 mM Tris, 1 mM EDTA; pH 9.0) at 98°C for 20 min. The sections were subsequently incubated for 50 min at room temperature with the primary anti-Ki67 (cat. no. MIB1; Dako; Agilent Technologies, Inc.) and anti-VEGF (cat. no. VG1; Dako; Agilent Technologies, Inc.) antibodies, diluted at 1:200 and 1:100, respectively, in Dako Envision™ Flex antibody diluents, followed by anti-rabbit immuno-peroxidase polymer (Envision FLEX/HRP) for 20 min at room temperature according to the manufacturer's instructions, and developed with freshly prepared 0.05% 3,3′-diaminobenzidine tetrahydrochloride. Finally, the slides were counterstained with hematoxylin, dehydrated, and mounted.
BRAFV600E mutation detection
Genomic DNA was extracted from fresh-frozen tissues using the TIAamp Genomic DNA Kit (Tiangen Biotech Co., Ltd.; cat. no. DP304). Subsequently, the genomic DNA was used to detect the V600E mutation in the BRAF oncogene through the AmoyDx BRAFV600E Mutations Detection Kit (AmoyDx; cat. no. ADx-BR04).
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (IBM Corp.). The significance of the RT-qPCR validation between the PTC and the normal tissue groups was assessed by the paired Student's t-test (18,34,35). The Mann-Whitney U test was used to analyse the associations between the mean ± standard deviation of circRNA expression and the clinicopathological characteristics of PTC, including age, sex, multifocality, Ki67, pN stage, BRAF mutation status, capsular invasion and thyroiditis, and the Kruskal-Wallis test followed with post-hoc Dunn's multiple comparison test was used to analyse the association between circRNA expression and pT-stage (35,36,37). P<0.05 was considered to indicate statistically significant differences (38). Receiver operating characteristics (ROC) curves were established to evaluate the diagnostic value of circRNAs for PTC. An area under the ROC curve (AUC) of 0.5 was considered to represent a test with no discriminating ability (i.e., no better than chance), while an AUC of 1.0 was considered to represent a test with perfect discrimination (39).
Results
circRNA expression profiles in PTC tissues relative to adjacent tissues
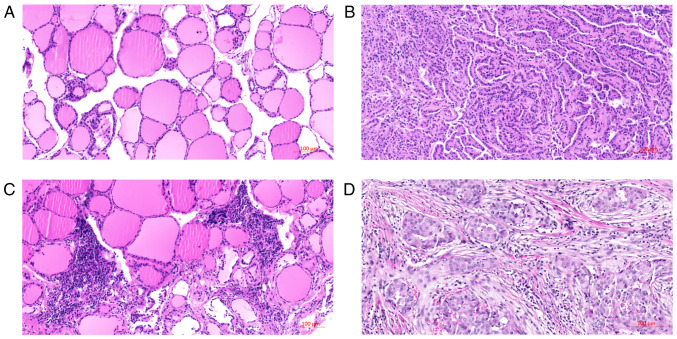
The clinicopathological characteristics of the 5 patients are listed in Table I. The majority were male (4 cases, 75%), 3 cases were classical PTCs, 1 case was follicular PTC and 1 case was mixed classical and follicular PTC, with different tumour-node-metastasis (TNM) stages. Two cases were multifocal and all displayed capsular invasion. The histomorphology of tissues are shown in Fig. 1. Among them, Fig. 1A and C shows the histomorphology of normal tissues and with lymphocyte infiltration. Fig. 1B and represent the classical PTC and follicular variant of PTC, respectively.
Figure 1.
Hematoxylin and eosin staining of papillary thyroid carcinoma (PTC) and normal tissues for RNA sequencing and qPCR validation (magnification, ×100). (A) normal tissues, (B) classical PTC, (C) normal tissues with thyroiditis, and (D) follicular variant of PTC.
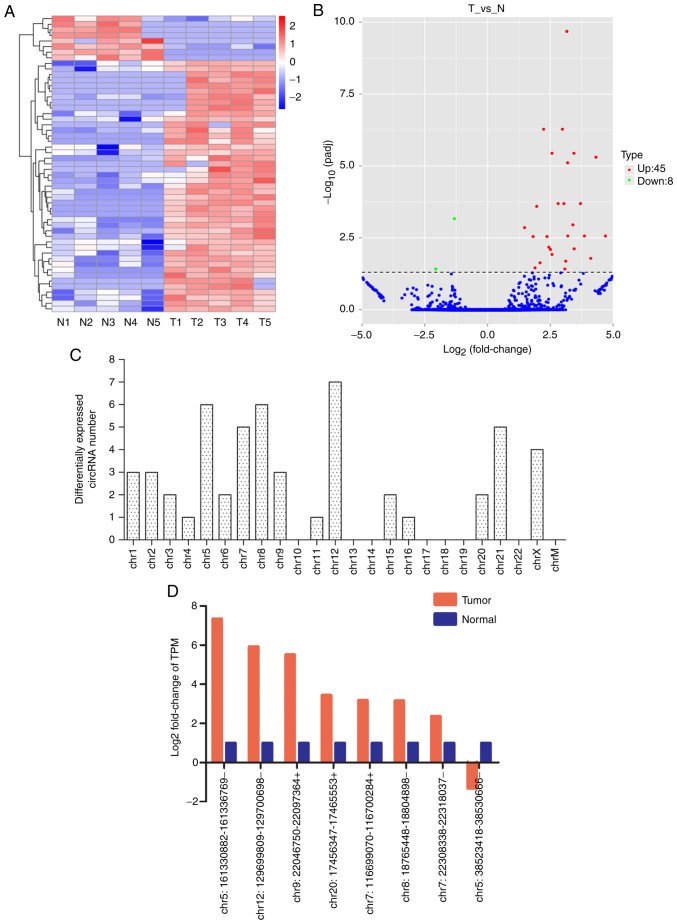
Subsequently, RNA-seq demonstrated that the circRNA expression profile of the 5 paired normal and PTC tissue samples exhibited significant differences. The hierarchical clustering (Fig. 2A) and volcano plots (Fig. 2B) revealed that there were 53 circRNAs differentially expressed in carcinoma tissues, with both q (Padj) <0.05 and fold-change in expression of >2 set as the threshold. Among those, 45 circRNAs were upregulated and 8 were downregulated in tumour tissues.
Figure 2.
Overview of the circRNA profiles (N, normal thyroid tissues; T, tumour thyroid tissues; chr, chromosome). (A) Hierarchical cluster analysis of differentially expressed circRNAs. The red and blue colours indicate high and low expression, respectively. (B) Volcano plot of differentially expressed circRNAs in PTC. The dotted line represents P=0.05. The red and green points represent circRNAs that were significantly upregulated or downregulated, respectively. (C) Chromosomal distributions of differentially expressed circRNAs. (D) RNA sequencing data of 8 circRNAs. PTC, papillary thyroid carcinoma; circRNA, circular RNA.
In the dysregulated circRNAs (Table SII), the Log2 (FC) of abnormally highly expressed circRNAs ranged from 1.48 to 7.34, and the Log2 (FC) of abnormally lowly expressed circRNAs ranged from-1.31 to −5.593. In addition, the magnitude of fold change was highest for chr5:161330882-161336769- in the upregulated circRNAs (fold change, 162; P<0.01), and chr20:20425608-20472956- for downregulated circRNAs (fold changes, 48; P=0.01). As presented in Fig. 2C, the transcription of the differentially regulated circRNAs was found to be widely distributed among all chromosomes, except chr10, chr13, chr14, chr17, chr18, chr19, chr22 and chrM. Moreover, 30 were identified as new circRNAs that have not been annotated in the circBase or circ2 Traits database. Furthermore, multiple circRNAs can be produced from a single gene and alternative splicing events, so circRNAs can arise from exons, introns, intergenic regions, UTRs, ncRNA loci and from locations antisense (40,41). In our research, of the 53 dysregulated circRNAs (Table SII), 29 (54.7%) were from the antisense locus, 10 (18.9%) were formed from exon-intron junction, 8 (15.1%) were from intron, 4 (7.5%) were from exon, and the other 2 were from intergenic regions, respectively.
Validation of circRNA expression
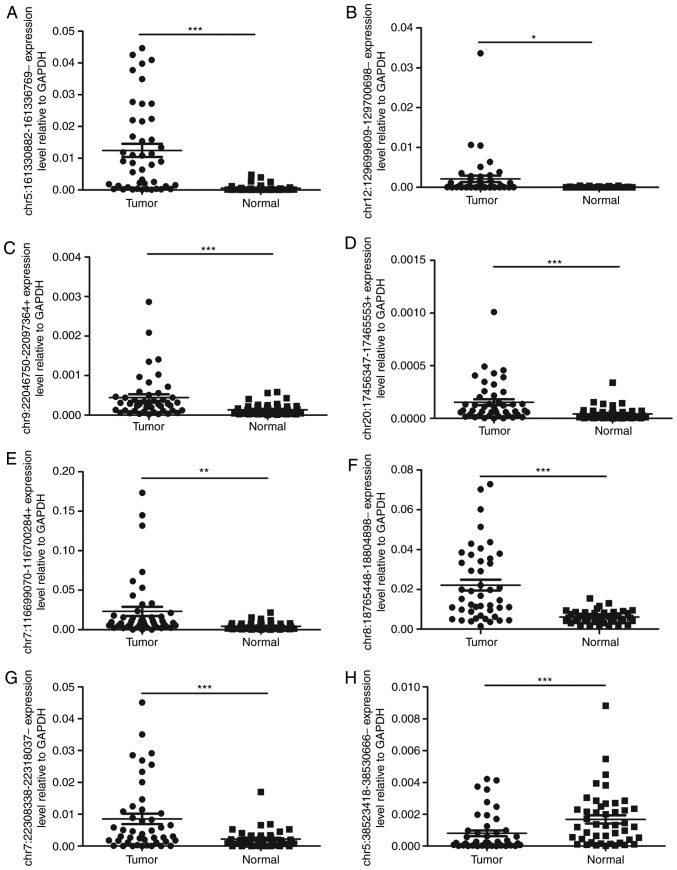
A total of 8 differentially expressed circRNAs were selected according to transcripts per million reads (TPM), fold change, q (Padj) value, junction read, features (arise from exonic/intronic), distribution and length, and then were verified using 45 pairs of tissues by RT-qPCR. The circRNA relative expression level of GAPDH was calculated using the 2−ΔΔCq method. The results confirmed that the upregulation of chr5: 161330882-161336769- (P<0.001), chr12:129699809-129700698- (P<0.05), chr9:22046750-22097364+ (P<0.001), chr20:17456347-17465553+ (P<0.001), chr7:116699070-116700284+ (P<0.01), chr8:18765448-18804898- (P<0.001) and chr7:22308338-22318037- (P<0.001) and the downregulation of chr5:38523418-38530666- (P<0.001) in carcinoma (Fig. 3) were in accordance with the RNA-seq data (Fig. 2D).
Figure 3.
Validation of the differentially expressed circRNAs by reverse transcription-quantitative PCR analysis. Expression levels of (A) chr5:161330882-161336769-, (B) chr12:129699809-129700698-, (C) chr9:22046750-22097364+, (D) chr20:17456347-17465553+, (E) chr7:116699070-116700284+, (F) chr8:18765448-18804898- and (G) chr7:22308338-22318037- in 45 PTC tissues were significantly higher compared with those in their paired normal thyroid tissues (P<0.05). (H) The expression levels of chr5:38523418-38530666- in 45 PTC tissues were significantly higher when compared with those in their paired normal thyroid tissues (P=0.006). ***P<0.001, **P<0.01, *P<0.05.
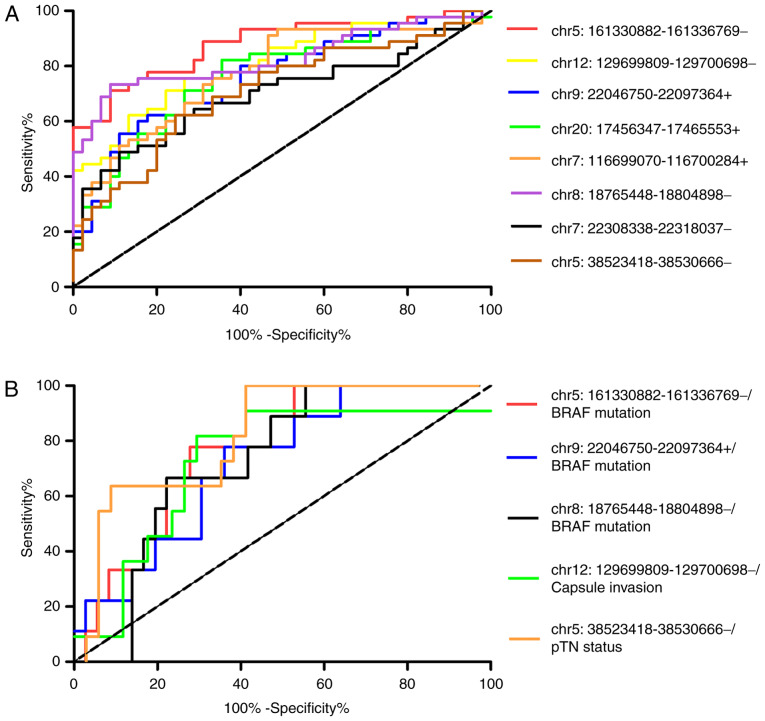
Furthermore, the mRNA expression of these 8 circRNAs was accessed with ROC curve analysis (Fig. 4A), which revealed that all the AUCs of circRNAs were >0.7 (P<0.001), and chr5:161330882-161336769- with the highest AUC (0.878) could be considered as a powerful marker for clinical PTC diagnosis (Table II).
Figure 4.
(A) Receiver operating characteristic (ROC) curves of the 8 reverse transcription-quantitative PCR-validated circRNAs between tumour and normal tissues. (B) The 5 circRNAs with clinicopathological characteristics.
Table II.
Area under the curve (AUC) of RT-qPCR-validated circRNAs in the 45 cases of PTC.
| circRNA | AUC | 95% CI | Sth. Error | P-value |
|---|---|---|---|---|
| chr5: 161330882-161336769- | 0.878 | 0.8068 to 0.9492 | 0.03632 | <0.0001 |
| chr12: 129699809-129700698- | 0.8099 | 0.7213 to 0.8984 | 0.04517 | <0.0001 |
| chr9: 22046750-22097364+ | 0.76 | 0.6610 to 0.8590 | 0.05049 | <0.0001 |
| chr20: 17456347-17465553+ | 0.759 | 0.6583 to 0.8597 | 0.05136 | <0.0001 |
| chr7: 116699070-116700284+ | 0.7763 | 0.6794 to 0.8732 | 0.04945 | <0.0001 |
| chr8: 18765448-18804898- | 0.8277 | 0.7387 to 0.9166 | 0.04538 | <0.0001 |
| chr7: 22308338-22318037- | 0.7017 | 0.5918 to 0.8116 | 0.05607 | 0.0009859 |
| chr5: 38523418-38530666- | 0.7101 | 0.6029 to 0.8174 | 0.05472 | 0.0006003 |
PTC, papillary thyroid carcinoma; circRNAs, circulating RNAs; CI, confidence interval.
circRNA expression and clinical characteristics
The clinical characteristics of 45 PTC patients are summarized in Table III. The median age of the patients at diagnosis was 45 years (range, 17–64 years), while the majority of the patients were female (n=33, 73.3%); 32 cases (71.1%) were classical PTC, 12 cases (26.7%) were the follicular variant of PTC and only 1 case (2%) was a mixture of the classical and follicular variant. Moreover, 31 cases (68.9%) were unifocal, and 14 cases (31.1%) were multifocal. In regards to pT stage, 29 cases (64.4%) had a diameter <2 cm or localized, 4 (8.9%) ranged between 2 and 4 cm, and 11 (24.4%) were >4 cm or invading the soft tissues beyond the thyroid. Only one-quarter of the patients had regional lymph node metastasis. None of the patients had metastasis. The Ki67 index of 33.3% patients was very high (≥3), and VEGF was positive for 91.1% patients. BRAFV600E mutation, capsular invasion and thyroiditis with diffuse lymphocytic infiltration were observed in 80, 75.6 and 20% of the cases, respectively.
Subsequently, we analysed the association between the mean ± standard deviation of circRNA expression in the tumour and the clinical characteristics in 45 PTC patients, including age, sex, multifocality, Ki67, capsular invasion, VEGF, BRAFV600E mutation, TNM stage and thyroiditis (Table III). Among the 8 dysregulated circRNAs, chr5:161330882-161336769- (P=0.015), chr9:22046750-22097364+ (hsa_circ_0008796, P=0.041) and chr8:18765448-18804898- (hsa_circ_0002111, P=0.036) were associated with the BRAFV600E mutation, and chr12:129699809-129700698- was associated with capsular invasion (P=0.025). Additionally, chr5:38523418-38530666- (hsa_circ_0072309) was associated with pT (P=0.037) and pN (P=0.002) stage. This association between circRNA expression and clinicopathological characteristics indicated that those circRNAs may participate in tumourigenesis and progression of PTC. The other three dysregulated circRNAs exhibited no statistically significant association with clinicopathological characteristics.
Furthermore, the association between the mRNA expression of these 5 circRNAs and the clinical characteristics was assessed with ROC curve analysis (Fig. 4B), revealing that the AUCs of all circRNAs were >0.7 (P<0.05), with chr5:38523418-38530666-(associated with pN stage) having the highest AUC (0.8209, P<0.01; Table IV). This result was consistent with the data mentioned above, and indicated that those 5 circRNAs associated with BRAFV600E mutation, capsular invasion, advanced pT stage and lymph node metastasis may be putative biomarkers for the diagnosis and evaluation of the progression of PTC.
Table IV.
Area under the curve (AUC) of the circRNA mRNA expression with the clinical characteristics of the 45 cases of PTC.
| circRNA | Clinical characteristics | AUC | 95% CI | Sth. Error | P-value |
|---|---|---|---|---|---|
| chr5:161330882-161336769- | BRAF mutation | 0.7654 | 0.6142 to 0.9167 | 0.07714 | 0.01471 |
| chr9:22046750-22097364+ | BRAF mutation | 0.7222 | 0.5562 to 0.8882 | 0.08468 | 0.0411 |
| chr8:18765448-18804898- | BRAF mutation | 0.7284 | 0.5757 to 0.8811 | 0.0779 | 0.0358 |
| chr12:129699809-129700698- | Capsular invasion | 0.7273 | 0.5480 to 0.9066 | 0.09146 | 0.02482 |
| chr5:38523418-38530666- | pN Stage | 0.8209 | 0.6905 to 0.9512 | 0.0665 | 0.001538 |
PTC, papillary thyroid cancer; circRNA, circular RNA; CI, confidence interval; BRAF, B-type Raf kinase.
Gene ontology (GO) analysis and pathway analysis of circRNA genes
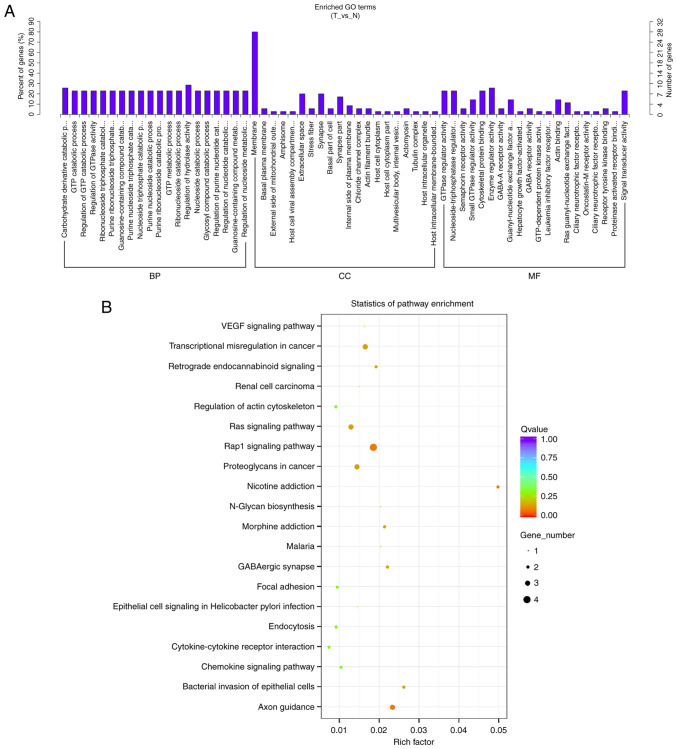
GO enrichment analysis for the host genes of the identified differentially expressed circRNAs was performed, and the significantly enriched GO terms in the biological process, cellular component and molecular function categories, e.g., carbohydrate derivative catabolic process (GO: 1901136, P=0.00021557), membrane (GO: 0016020, P=0.0012195) and GTPase regulator activity (GO: 0030695, P=0.00021632), are presented in Fig. 5A. A total of 28 host genes of the differentially expressed circRNAs were associated with the membrane GO term. The results of the Kyoto Encyclopaedia of Genes and Genomes pathway analysis of the 20 highly related circRNA genes are presented in Fig. 5B. Those circRNA genes were mainly associated with important pathways (Rap1, VEGF and Ras signalling pathways), tumourigenesis (transcriptional misregulation and proteoglycans in cancer), cell signal transduction (axon guidance, retrograde endocannabinoid signalling, cytokine-cytokine receptor interaction), infection and immune defence (bacterial invasion of epithelial cells, chemokine signalling pathway). Furthermore, the Rap1 and Ras signalling pathway contained more related circRNA genes, which suggested that circRNAs may play a key role in PTC tumourigenesis and progression.
Figure 5.
(A) Gene Ontology (GO) terms. N, normal thyroid tissues; T, tumour thyroid tissues. (B) Kyoto Encyclopedia of Genes and Genomes pathways of the differentially expressed circRNA genes in 20 pathways. BP, biological process; CC, cellular component; MF, molecular function.
Prediction for the circRNA-miRNA network
circRNAs have been shown to contain miRNA-binding sites and negatively regulate the inhibitory effects of miRNAs on their target mRNAs, thereby essentially acting as miRNA sponges (14). To evaluate the potential functions of the identified differentially expressed circRNAs, miRNAs that were potentially associated with those circRNAs were investigated using the RegRNA2 platform and Circular RNA Interactome (42,43). The first 5 predicted miRNAs for the dysregulated circRNAs are listed in Table V. For example, circRNA chr5:161330882-161336769- was predicted to harbour hsa-miR-1273g-3p, hsa-miR-498, hsa-miR-1268a, has-miR-363-5p and hsa-miR-566.
Table V.
Predicted miRNAs for the top upregulated or downregulated circRNAs in PTC.
| circRNA | circBase ID | Predicted miRNAs | ||||
|---|---|---|---|---|---|---|
| chr5:161330882-161336769- | / | miR-1273g-3p | miR-498 | miR-1268b | miR-363-5p | miR-566 |
| chr12:129699809-129700698- | / | miR-4726-3p | miR-4793-3p | miR-766-3p | miR-1343 | miR-1226-5p |
| chr9:22046750-22097364+ | hsa_circ_0008796 | miR-490-5p | miR-224 | miR-1228 | miR-576-5p | miR-611 |
| chr20:17456347-17465553+ | / | miR-939 | miR-576-3p | miR-3925-5p | miR-5189 | miR-4732-5p |
| chr7:116699070-116700284+ | hsa_circ_0082002 | miR-512-5p | miR-515-3p | miR-519e | miR-609 | miR-653 |
| chr8:18765448-18804898- | hsa_circ_0002111 | miR-647 | miR-532-3p | miR-526b | miR-1263 | miR-432 |
| chr7:22308338-22318037- | / | miR-5095 | miR-5096 | miR-3159 | miR-4524a-3p | miR-5699 |
| chr5:38523418-38530666- | hsa_circ_0072309 | miR-1277 | miR-331-5p | miR-409-3p | miR-515-5p | miR-1276 |
PTC, papillary thyroid carcinoma; circRNAs, circulating RNAs; miRNAs, microRNAs.
To further explore which miRNAs may be cancer-related, the starBase v2.0 (44) and miRPathDB v1.1 (45) platforms were used, and a network map of circRNA-miRNA interactions was constructed to identify the dysregulated circRNAs in PTC that potentially interacted with cancer-related miRNAs. From the initial list of 40 potential miRNAs identified by the RegRNA2 and circRNA Interactome analysis, several cancer-related miRNAs were identified following a literature research, including hsa-miR-498 (46), hsa-miR-766-3p (47,48) and hsa-miR-661 (49,50), which are associated with cell proliferation in several cancers. The results indicated that those dysregulated circRNAs may act as miRNA sponges, leading to the progression of PTC, but the specific underlying mechanism requires further investigation.
Discussion
The present study profiled the circRNA expression in papillary thyroid carcinoma (PTC) using RNA-seq, and a total of 45 upregulated and 8 downregulated circRNAs were identified in PTC tumours. Among the 53 dysregulated circRNAs, chr3:64594257-64596991-(hsa_circ_0066444) has been reported to be significantly differentially expressed in hepatocellular carcinoma (HCC) tissues by RNA-seq, but it exhibited no significant difference on PCR verification (34). Additionally, chr8:18799294-18804898- (hsa_circ_0004458) was reported to be upregulated in PTC tumours (51) and cells (52), which was consistent with our sequencing results. Another study verified that the silencing of hsa_circ_0004458 inhibited PTC cell proliferation and promoted cell cycle arrest and apoptosis in vitro (52), which may constitute the evidential basis for further research.
Subsequently, the 8 dysregulated circRNAs were screened according to high TPM, fold change, q (Padj) value, junction reads and widespread distribution, and the results were further validated with RT-qPCR. Among the 8 circRNAs, chr5:161330882-161336769- had the highest area under the curve (AUC) (0.878) and may be considered a powerful marker for clinical diagnosis; in addition, chr8:18765448-18804898- (hsa_circ_0002111) was found to be significantly upregulated in PTCs with microarray profiling (51), which was consistent with our results.
The associations between the circRNA expression profile and clinical characteristics in PTC were then explored, and it was observed that chr5:161330882-161336769-(P=0.015), chr9:22046750-22097364+ (hsa_circ_0008796, P=0.041) and chr8:18765448-18804898- (hsa_circ_0002111, P=0.036) were significantly associated with the BRAFV600E mutation. The BRAFV600E mutation is the most common genetic aberration in PTC and has been found in up to 68% of PTCs in adults with aggressive disease characteristics (53). Although it was reported that the major driver mutation of follicular variant PTC and classical PTC was RAS-related mutation and BRAFV600E mutation, respectively (2), other studies indicated that the follicular variant PTC also harbours the BRAFV600E mutation (54,55). In addition to our study, Ren et al reported that hsa_circRNA_047771 is likely to be associated with the BRAFV600E mutation (35). The results indicated that BRAFV600E may be associated with multiple circRNAs, and whether those circRNAs are involve in tumourigenesis should be the focus of further investigation.
Furthermore, the host genes of chr5:161330882-161336769-, chr9:22046750-22097364+ (hsa_circ_0008796) and chr8:18765448-18804898- (hsa_circ_0002111) were γ-aminobutyric acid type A receptor subunit β2 (GABRB2), CDKN2B antisense RNA 1 (CDKN2B-AS1) and pleckstrin and Sec7 domain containing 3 (PSD3), respectively. Among those, GABRB2 was previously reported as being overexpressed in PTC (56), and the hypomethylation and overexpression of GABRB2 were significantly altered in BRAFV600E-mutated PTC (57), further supporting that chr5:161330882-161336769- and GABRB2 may play key roles in the progression of PTC. However, there are yet no published data on this association of CDKN2B-AS1, PSD3 and BRAFV600E mutation.
In addition, the results of the present study also demonstrated that chr12:129699809-129700698- was associated with capsular invasion (P=0.025), which is considered a putative biomarker for the progression of PTC. Furlan et al results suggested that the presence of capsular invasion does not adversely affect the biological behaviour of PTC or patient survival (58), but another study indicated that it is associated with a higher recurrence rate (59) and reduced survival time and quality of life (60). Capsular invasion, advanced pT stage and pathological positive lymph node metastasis were independent predictors of multifocal PTC (61). Additionally, transmembrane protein 132D (TMEM132D), the host gene of chr12:129699809-129700698-, was only reported to be associated with anxiety phenotypes (62) and may serve as a predictor of CD8+T-lymphocyte infiltration and a favourable prognostic marker in early-stage ovarian cancer (63). However, further study is required to determine whether TMEM132D is associated with capsular invasion and whether it may affect T-lymphocyte infiltration in PTC.
Furthermore, the present study demonstrated that the low expression of chr5:38523418-38530666- was associated with advanced pT stage (P=0.037) and lymph node metastasis (P=0.002), and may be considered as a putative biomarker for the progression of PTC. According to the AJCC TNM staging system, T and N stages are independent predictive factors of multifocal and bilateral PTC (61,64). Ren et al reported that low expression of hsa_circRNA_047771 was correlated with lymph node metastasis and advanced TNM stage, whereas higher expression of hsa_circRNA_007148 was correlated with lymph node metastasis (35). In addition, LIF receptor subunit α (LIFR), the host gene of chr5:38523418-38530666-, was found to be a breast cancer metastasis suppressor through the Hippo-YAP pathway, and is considered to have significant prognostic power (65). However, the specific function of chr5:38523418-38530666- in the progression and metastasis of PTC requires further investigation.
In brief, the AUCs of the 8 dysregulated circRNAs indicated that they were specific to PTC, and among them, the 5 circRNAs associated with BRAFV600E, capsular invasion, advanced pT stage and lymph node metastasis, may be putative biomarkers for the diagnosis and evaluation of progression of PTC.
For circRNAs that were found to be associated with the cellular RNA network and negatively regulating the inhibitory effects of miRNAs on their target mRNAs, it was necessary to analyse the downstream miRNAs and mRNA of circRNAs to gain insight into the molecular mechanisms of action and the role of circRNAs in tumourigenesis and cancer progression. Although the literature on the circRNA-miRNA network in regard to other identified circRNAs is limited, there was a considerable amount of literature on those miRNAs. Therefore, in our results, miR-498 was predicted to be a target miRNA of chr5:161330882-161336769-, and the expression of miR-498 was also markedly downregulated in ovarian cancer cells and tumours, acting as a new tumour suppressor by targeting the FOXO3 gene to inhibit cell proliferation in ovarian cancer (46). The present research demonstrated that chr5:161330882-161336769- was significantly upregulated in PTC, although further study is required to determine whether miR-498 could be consequently downregulated in PTC as a result of regulatory disinhibition, and the effects of dysregulation on cancer-related target genes should be the focus of further research.
In addition, miR-766-3p and miR-661, the predicted miRNAs of chr12:129699809-129700698- and chr9:22046750-22097364+ (has_circ_0008796), may also be consequently dysregulated in PTC as a result of regulatory disinhibition. For example, it was previously demonstrated that miR-766-3p inhibits tumour progression by targeting Wnt3a in HCC (47). MiR-661 was shown to inhibit metastatic tumour antigen 1 in breast cancer cells (50), and to inhibit cell proliferation, migration and invasion by targeting hTERT in glioma cells (49). Furthermore, as the predicted miRNA of chr5:38523418-38530666- (hsa_circ_0072309), miR-409-3p may promote epithelial-to-mesenchymal transition and tumourigenesis via the RSU1 or STAG2 pathway in prostate cancer (66,67). However, the specific function and mechanism of action of circRNAs and miRNAs in PTC have not been fully elucidated, and further studies should focus on the circRNA/miRNA/mRNA axis in the regulation of PTC tumourigenesis, in order to conclusively determine cancer-related target genes and facilitate further biomarker identification for PTC.
In general, the function of the parental genes may have some effect on the circRNA, but the effects on tumours are not only associated with the parental genes, as circRNAs may also play several important roles in biological processes, such as regulating gene expression by acting as competitors of pre-mRNA splicing, as decoys for microRNAs, as sponges for RNA-binding proteins, possibly also as substrates for translation (68), and may regulate the transcription of their parental genes (69). Ours is a preliminary study of the expression profile of circRNAs in thyroid cancer. In future studies, we would like to focus more on the mechanisms of action of the dysregulated circRNAs and their sponging miRNAs and potential mRNA targets, which may provide clues to the molecular mechanisms of action of circRNAs in PTC.
Besides, there were certain limitations to the present study, such as the small sample size, different histological subtypes of PTC, and the fact that signaling pathways were not studied. Thus, the results require verification using larger sample sizes and more detailed investigation of the differential expression of circRNAs between the two PTC subtypes and their role in diagnosis, progression and evaluation of prognosis.
In conclusion, the present study demonstrated that several circRNAs are differentially expressed in PTC and were found to be associated with the BRAFV600E mutation, capsular invasion, advanced pT stage and lymph node metastasis, thereby indicating that circRNAs may participate in tumourigenesis and cancer progression. The detailed role of circRNAs as putative biomarkers for the diagnostic and evaluation of progression of PTC should be further explored and validated. Furthermore, those differentially expressed circRNAs may participate in the cellular RNA network with miRNAs and mRNAs, and further research should be performed to gain a better insight into the molecular mechanisms of circRNAs in PTC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine (grant nos. 2017-I2M-1-001 and 2017-I2M-2-001); and The National Key Research and Development program of China: the cluster construction of human genetic resource Bio-bank in north China (grant no. 2016YFC1201703).
Availability of data and materials
All data generated or analysed during the present study are included in this published article.
Authors' contributions
DG carried out the study design and manuscript preparation. FL conducted the manuscript preparation and data analysis/interpretation. XZ carried out the experimental studies and data analysis. BL carried out the manuscript editing and revision. SZ conducted the experimental studies. AW performed the statistical analysis. DC conducted the experimental studies. JS was responsible for the specimen acquisition and diagnosis. BL was responsible for the study conception and clinical studies.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of Peking Union Medical College Hospital (Beijing, China). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lam AK, Lo CY, Lam KS. Papillary carcinoma of thyroid: A 30-yr clinicopathological review of the histological variants. Endocr Pathol. 2005;16:323–330. doi: 10.1385/EP:16:4:323. [DOI] [PubMed] [Google Scholar]
- 2.Abdullah MI, Junit SM, Ng KL, Jayapalan JJ, Karikalan B, Hashim OH. Papillary thyroid cancer: Genetic alterations and molecular biomarker investigations. Int J Med Sci. 2019;16:450–460. doi: 10.7150/ijms.29935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SC, Kuo SF, Hua CC, Huang BY, Chiang KC, Chu YY, Hsueh C, Lin JD. Differentiation of the follicular variant of papillary thyroid carcinoma from classic papillary thyroid carcinoma: An ultrasound analysis and complement to fine-needle aspiration cytology. J Ultrasound Med. 2018;37:667–674. doi: 10.1002/jum.14377. [DOI] [PubMed] [Google Scholar]
- 4.Lin JD. Thyroglobulin and human thyroid cancer. Clin Chim Acta. 2008;388:15–21. doi: 10.1016/j.cca.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Tuttle RM, Leboeuf R, Martorella AJ. Papillary thyroid cancer: Monitoring and therapy. Endocrinol Metabol Clin North Amer. 2007;36:753–778. doi: 10.1016/j.ecl.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Hurtado-Lopez LM, Fernandez-Ramirez F, Martinez-Penafiel E, Carrillo Ruiz JD, Herrera Gonzalez NE. Molecular analysis by gene expression of mitochondrial ATPase subunits in papillary thyroid cancer: Is ATP5E transcript a possible early tumor marker? Med Sci Monit. 2015;21:1745–1751. doi: 10.12659/MSM.893597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang KT, Lee CH. BRAF mutation in papillary thyroid carcinoma: Pathogenic role and clinical implications. J Chin Med Assoc. 2010;73:113–128. doi: 10.1016/S1726-4901(10)70025-3. [DOI] [PubMed] [Google Scholar]
- 8.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 9.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and Cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 18.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, Zheng W. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2017;32:e22281. doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi W, Huang J, Nie C, Liu B, He G, Han J, Pang R, Ding Z, Xu J, Zhang J. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37:275. doi: 10.1186/s13046-018-0936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503:56–61. doi: 10.1016/j.bbrc.2018.05.174. [DOI] [PubMed] [Google Scholar]
- 24.Lan X, Cao J, Xu J, Chen C, Zheng C, Wang J, Zhu X, Zhu X, Ge M. Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. J Clin Lab Anal. 2018;32:e22573. doi: 10.1002/jcla.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem Biophys Res Commun. 2018;504:283–288. doi: 10.1016/j.bbrc.2018.08.175. [DOI] [PubMed] [Google Scholar]
- 26.Lan X, Xu J, Chen C, Zheng C, Wang J, Cao J, Zhu X, Ge M. The landscape of circular RNA expression profiles in papillary thyroid carcinoma based on RNA sequencing. Cell Physiol Biochem. 2018;47:1122–1132. doi: 10.1159/000490188. [DOI] [PubMed] [Google Scholar]
- 27.Doescher J, Veit JA, Hoffmann TK. The 8th edition of the AJCC Cancer Staging Manual. Updates in otorhinolaryngology, head and neck surgery. HNO. 2017;65:956–961. doi: 10.1007/s00106-017-0391-3. (In German) [DOI] [PubMed] [Google Scholar]
- 28.EI-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ. Lyon: 2017. WHO Classification of Head and Neck Tumours. [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D SJ. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Statistics. 2003;31:2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 31.Panda AC, Gorospe M. Detection and analysis of circular RNAs by RT-PCR. Bio Protoc. 2018;8(pii):e2775. doi: 10.21769/BioProtoc.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi: 10.1016/j.biopha.2016.12.097. [DOI] [PubMed] [Google Scholar]
- 33.Sui W, Gan Q, Liu F, Chen H, Liu J, Dai Y. The differentially expressed circular ribonucleic acids of primary hepatic carcinoma following liver transplantation as new diagnostic biomarkers for primary hepatic carcinoma. Tumour Biol. 2018;40:1010428318766928. doi: 10.1177/1010428318766928. [DOI] [PubMed] [Google Scholar]
- 34.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J, Wang D, Yuan G. Profile and clinical implication of circular RNAs in human papillary thyroid carcinoma. PeerJ. 2018;6:e5363. doi: 10.7717/peerj.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP, Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 2018;101:531–538. doi: 10.1016/j.molimm.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 37.Peng Y, Song X, Zheng Y, Cheng H, Lai W. circCOL3A1-859267 regulates type I collagen expression by sponging miR-29c in human dermal fibroblasts. Eur J Dermatol. 2018;28:613–620. doi: 10.1684/ejd.2018.3397. [DOI] [PubMed] [Google Scholar]
- 38.Hu J, Li C, Liu C, Zhao S, Wang Y, Fu Z. Expressions of miRNAs in papillary thyroid carcinoma and their associations with the clinical characteristics of PTC. Cancer Biomark. 2017;18:87–94. doi: 10.3233/CBM-161723. [DOI] [PubMed] [Google Scholar]
- 39.Hoo ZH, Candlish J, Teare D. What is an ROC curve? Emerg Med J. 2017;34:357–359. doi: 10.1136/emermed-2017-206735. [DOI] [PubMed] [Google Scholar]
- 40.Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14:1035–1045. doi: 10.1080/15476286.2016.1271524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Y, He D, Peng Z, Peng W, Shi W, Wang J, Li B, Zhang C, Duan C. Circular RNAs in cancer: An emerging key player. J Hematol Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang TH, Huang HY, Hsu BK, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Backes C, Kehl T, Stöckel D, Fehlmann T, Schneider L, Meese E, Lenhof HP, Keller A. miRPathDB: A new dictionary on microRNAs and target pathways. Nucleic Acids Res. 2017;45:D90–D96. doi: 10.1093/nar/gkw926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R, Liu F, Li L, Sun M, Chen K. MiR-498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed Pharmacother. 2015;72:52–57. doi: 10.1016/j.biopha.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 47.You Y, Que K, Zhou Y, Zhang Z, Zhao X, Gong J, Liu Z. MicroRNA-766-3p inhibits tumour progression by targeting Wnt3a in hepatocellular carcinoma. Mol Cells. 2018;41:830–841. doi: 10.14348/molcells.2018.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Xue S, Zhang J, Chen W, Gong D, Zheng J, Ma J, Xue W, Chen Y, Zhai W, Zheng J. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int J Cancer. 2017;141:1867–1878. doi: 10.1002/ijc.30853. [DOI] [PubMed] [Google Scholar]
- 49.Li Z, Liu YH, Diao HY, Ma J, Yao YL. MiR-661 inhibits glioma cell proliferation, migration and invasion by targeting hTERT. Biochem Biophys Res Commun. 2015;468:870–876. doi: 10.1016/j.bbrc.2015.11.046. [DOI] [PubMed] [Google Scholar]
- 50.Reddy SD, Pakala SB, Ohshiro K, Rayala SK, Kumar R. MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Res. 2009;69:5639–5642. doi: 10.1158/0008-5472.CAN-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS One. 2017;12:e0170287. doi: 10.1371/journal.pone.0170287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin X, Wang Z, Pang W, Zhou J, Liang Y, Yang J, Yang L, Zhang Q. Upregulated hsa_circ_0004458 Contributes to Progression of Papillary Thyroid Carcinoma by Inhibition of miR-885-5p and Activation of RAC1. Med Sci Monit. 2018;24:5488–5500. doi: 10.12659/MSM.911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardee S, Prasad ML, Hui P, Dinauer CA, Morotti RA. Pathologic characteristics, natural history, and prognostic implications of BRAF(V600E) mutation in pediatric papillary thyroid carcinoma. Pediatr Dev Pathol. 2017;20:206–212. doi: 10.1177/1093526616689628. [DOI] [PubMed] [Google Scholar]
- 54.Walts AE, Mirocha JM, Bose S. Follicular variant of papillary thyroid carcinoma (FVPTC): Histological features, BRAF V600E mutation, and lymph node status. J Cancer Res Clin Oncol. 2015;141:1749–1756. doi: 10.1007/s00432-015-1939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H, Kim BH, Kim YK, Kim JM, Oh SY, Kim EH, Lee MJ, Kim JH, Jeon YK, Kim SS, et al. Prevalence of BRAFV600E mutation in follicular variant of papillary thyroid carcinoma and non-invasive follicular tumor with papillary-like nuclear features (NIFTP) in a BRAFV600E prevalent area. J Korean Med Sci. 2018;33:e75. doi: 10.3346/jkms.2018.33.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin Y, Jin W, Zheng Z, Chen E, Wang Q, Wang Y, Wang O, Zhang X. GABRB2 plays an important role in the lymph node metastasis of papillary thyroid cancer. Biochem Biophys Res Commun. 2017;492:323–230. doi: 10.1016/j.bbrc.2017.08.114. [DOI] [PubMed] [Google Scholar]
- 57.Beltrami CM, Reis MBD, Barrosfilho MC, Marchi FA, Kuasne H, Pinto CAL, Ambatipudi S, Herceg Z, Kowalski LP, Rogatto SR. Integrated data analysis reveals potential drivers and pathways disrupted by DNA methylation in papillary thyroid carcinomas. Clin Epigenetics. 2017;9:45. doi: 10.1186/s13148-017-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furlan JC, Bedard YC, Rosen IB. Significance of tumor capsular invasion in well-differentiated thyroid carcinomas. Am Surg. 2007;73:484–491. [PubMed] [Google Scholar]
- 59.Bolognamolina R, Gonzálezgonzález R, Mosquedataylor A, Molinafrechero N, Damiánmatsumura P, Dominguezmalagón H. Expression of syndecan-1 in papillary carcinoma of the thyroid with extracapsular invasion. Arch Med Res. 2010;41:33–37. doi: 10.1016/j.arcmed.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Meng X, Zhu P, Li N, Hu J, Wang S, Pang S, Wang J. Expression of BMP-4 in papillary thyroid carcinoma and its correlation with tumor invasion and progression. Pathol Res Pract. 2017;213:359–363. doi: 10.1016/j.prp.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 61.Genpeng L, Jianyong L, Jiaying Y, Ke J, Zhihui L, Rixiang G, Lihan Z, Jingqiang Z. Independent predictors and lymph node metastasis characteristics of multifocal papillary thyroid cancer. Medicine (Baltimore) 2018;97:e9619. doi: 10.1097/MD.0000000000009619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, Specht M, Kohli MA, Kloiber S, Ising M, et al. TMEM132D, a new candidate for anxiety phenotypes: Evidence from human and mouse studies. Mol Psychiatr. 2011;16:647–663. doi: 10.1038/mp.2010.41. [DOI] [PubMed] [Google Scholar]
- 63.Karapetsas A, Giannakakis A, Dangaj D, Lanitis E, Kynigopoulos S, Lambropoulou M, Tanyi JL, Galanis A, Kakolyris S, Trypsianis G. Overexpression of GPC6 and TMEM132D in early stage ovarian cancer correlates with CD8+ T-lymphocyte infiltration and increased patient survival. Biomed Res Int. 2015;2015:712438. doi: 10.1155/2015/712438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baek HJ, Kim DW, Ryu JH. Association between TNM staging system and histopathological features in patients with papillary thyroid carcinoma. Endocrine. 2015;48:589–594. doi: 10.1007/s12020-014-0362-3. [DOI] [PubMed] [Google Scholar]
- 65.Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, Ma L. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Josson S, Gururajan M, Hu P, Shao C, Chu GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20:4636–4646. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Josson S, Gururajan M, Sung SY, Hu P, Shao C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene. 2015;34:2690–2699. doi: 10.1038/onc.2014.212. [DOI] [PubMed] [Google Scholar]
- 68.Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during the present study are included in this published article.