Abstract
Mesencephalic astrocyte-derived neurotrophic factor (MANF) is the only human neurotrophic factor with an evolutionarily-conserved C. elegans homolog, Y54G2A.23 or manf-1. MANF is a small, soluble, endoplasmic-reticulum (ER)-resident protein that is secreted upon ER stress and promotes survival of target cells such as neurons. However, the role of MANF in ER stress and its mechanism of cellular protection are not clear and the function of MANF in C. elegans is only beginning to emerge. In this study, we show that depletion of C. elegans manf-1 causes a slight decrease in lifespan and brood size; furthermore, combined depletion of manf-1 and the IRE-1/XBP-1 ER stress/UPR pathway resulted in sterile animals that did not produce viable progeny. We demonstrate upregulation of markers of ER stress in L1 larval nematodes, as measured by hsp-3 and hsp-4 transcription, upon depletion of manf-1 by RNAi or mutation; however, there was no difference in tunicamycin-induced expression of hsp-3 and hsp-4 between wild-type and MANF-deficient worms. Surprisingly, larval growth arrest observed in wild-type nematodes reared on tunicamycin is completely prevented in the manf-1 (tm3603) mutant. Transcriptional microarray analysis revealed that manf-1 mutant L1 larvae exhibit a novel modulation of innate immunity genes in response to tunicamycin. The hypothesis that manf-1 negatively regulates the innate immunity pathway is supported by our finding that the development of manf-1 mutant larvae compared to wild-type larvae is not inhibited by growth on P. aeruginosa. Together, our data represent the first characterization of C. elegans MANF as a key modulator of organismal ER stress and immunity.
Keywords: ARMET, MANF, ER stress, tunicamycin, C. elegans, inflammation
INTRODUCTION
The endoplasmic reticulum (ER) is the organelle where much of the assembly, folding, and post-translational modification occurs for proteins destined to be secreted or localized to vesicles and membranes (Malhotra and Kaufman, 2007). The ER is also the site of synthesis and assembly of the plasma membrane and the major intracellular calcium storage compartment (Appenzeller-Herzog and Simmen, 2016). To facilitate its function, the ER contains molecular chaperones involved in posttranslational modification (Ma and Hendershot, 2004), disulfide bond formation (Sevier and Kaiser, 2002) and quality control mechanisms for ensuring that only properly folded proteins can leave the ER (Malhotra and Kaufman, 2007). Improperly folded proteins are retained in the ER through interactions with chaperone proteins or are actively degraded, either through the proteasome-dependent process known as ER-associated degradation (ERAD) (Olzmann et al., 2013; Pisoni and Molinari, 2016) or through autophagy in the lysosomal pathway (Pisoni and Molinari, 2016).
ER homeostasis is vital for cell function and survival. Environmental insults or genetic deficiencies can compromise the ER protein folding machinery leading to an accumulation of unfolded protein substrates in the lumen (Oakes and Papa, 2015). An increase in the steady-state levels of unfolded or misfolded proteins in the ER (“ER stress”) triggers an adaptive signaling response known as the unfolded protein response (UPR) to restore ER homeostasis by reducing the rate of translation initiation, increasing protein folding capacity, and degradation of misfolded proteins. The UPR is comprised of three pathways controlled by PERK, IRE1 and ATF6 sensor proteins (Bell et al., 2016; Chen and Brandizzi, 2013; Kania et al., 2015). Under normal conditions, the ER chaperone BiP/GRP-78 binds to the luminal domains of the UPR sensor proteins and maintains them in an inactive state. However, when misfolded proteins accumulate in the ER and sequester the BiP chaperone, the stress sensors PERK, IRE1 and ATF6 are then able to initiate downstream signaling (Gardner et al., 2013; Sano and Reed, 2013). If the UPR fails to resolve the protein-folding defect, apoptosis is activated through redundant pathways that include both mitochondrial-dependent (intrinsic) and mitochondrial-independent (extrinsic) apoptotic activation (Sano and Reed, 2013).
Cerebral dopamine neurotrophic factor (CDNF) and mesencephalic astrocyte-derived neurotrophic factor (MANF, previously termed ARMET for Arginine-rich, mutated in early stage of tumors), constitute a unique family of ER-stress-responsive neurotrophic factors. MANF was identified in two independent screens for genes upregulated during ER stress and UPR activation (Apostolou et al., 2008; Mizobuchi et al., 2007). MANF was also identified as a secreted factor from a rat mesencephalic astrocyte cell line that protected dopaminergic neurons from 6-hydroxydopmaine-induced toxicity (Petrova et al., 2003). Later studies reported MANF as a secreted protein that, when upregulated, protected cultured cells against ER stress-induced cell death (Apostolou et al., 2008). CDNF was also shown to be ER-localized and ER stress-induced (Lindholm et al., 2007; Parkash et al., 2009). Both CDNF and MANF have neuroprotective and neurorestorative properties in rodent models of stroke and Parkinson’s Disease (Airavaara et al., 2010; Airavaara et al., 2009; Cordero-Llana et al., 2015; Lindholm et al., 2007; Voutilainen et al., 2009; Yu et al., 2010; Zhang et al., 2018a). In addition to its role as a neuroprotective protein (Voutilainen et al., 2015), MANF also responds to ER stress in the heart to protect cardiomyocytes (Glembotski, 2011) and is essential for the proliferation and survival of mouse pancreatic β cells (Lindahl et al., 2014). In fact, MANF deficiency in mice manifests as severe diabetes due to progressive depletion of the pancreatic β cell mass, an effect which is driven by chronic UPR. Conversely, loss of MANF in zebrafish does not result in any apparent phenotype, although dopamine levels and/or numbers of TH positive cells are decreased by MANF deletion (Chen et al., 2012) and by neurotoxicant exposures that also downregulate MANF (Wang et al., 2015; Wu et al., 2016).
Although mammals have two bifunctional genes CDNF and MANF, the invertebrate genomes of Drosophila melanogaster and Caenorhabditis elegans contain a single ortholog in the CDNF/MANF family with closest homology to MANF (Supplementary Figure S1). In Drosophila melanogaster, DmMANF is expressed in glia and is essential for maintenance of dopamine-positive neurites and dopamine levels (Palgi et al., 2009). Knockdown of DmMANF leads to neurological phenotypes (Palgi et al., 2009) and defects in wing development (Lindstrom et al., 2017). At the cellular level, DmMANF is upregulated in response to ER stress and interacts with UPR and membrane transport pathways (Lindstrom et al., 2016; Palgi et al., 2012) and, surprisingly, with mitochondrial genes (Lindstrom et al., 2017). Very recently, depletion of the C. elegans ortholog, manf-1, was demonstrated to increase ER stress reporters (Bai et al., 2018; Richman et al., 2018). Interestingly, this stress response could be reversed by exposing mutant animals to purified recombinant human MANF protein in a sulfatide-dependent mechanism (Bai et al., 2018). However, the role of MANF-1 in C. elegans biology has not been fully elucidated.
Herein, we characterize a role for C. elegans MANF-1 in modulating the response to ER stress. First, we examined organismal and dopaminergic neuron health following depletion of manf-1 in C. elegans, as well as the effect of MANF-1 depletion on the UPR. We exposed larval nematodes to tunicamycin, a chemical inhibitor of N-linked glycosylation and a known inducer of ER stress, which was previously demonstrated to cause growth delay in wild-type larvae (Richardson et al., 2011). We measured both the UPR response and larval growth after tunicamycin exposure. Finally, we performed a microarray analysis to identify biological pathways that are differentially expressed after wildtype and manf-1 (tm3603) mutants are treated with tunicamycin. This microarray analysis revealed significant changes in genes associated with the innate immunity pathways, which prompted additional experiments to test larval growth upon pathogenic bacterial exposures. Our overall objective was to assess the role of manf-1 in modulating the UPR in order better understand this gene’s mechanism of action.
RESULTS
Depletion of C. elegans manf-1 has minimal effects on total brood size, lifespan, and neuronal phenotypes
In mouse, zebrafish, and the invertebrate Drosophila, deletion of MANF has various detrimental effects on the whole organism (Chen et al., 2012; Lindahl et al., 2014; Lindstrom et al., 2017). In C. elegans, we used complementary approaches of knockdown by RNAi and a mutant allele, tm3603, bearing a deletion in the manf-1 gene (Supplemental Fig. S1) to test organismal effects of manf-1 depletion. MANF has previously been reported to be expressed beginning in post-gastrulating embryos and continuing throughout larval development and into adulthood, with highest expression in intestine, hypodermis, spermatheca, and nervous system based on a transcriptional reporter with GFP being driven by the manf-1 promoter (Bai et al., 2018; Richman et al., 2018). We used CRISPR/Cas9 to generate an endogenously tagged MANF::mKate2 fusion allele and visualized the MANF protein expression throughout development (Supplemental Fig. S2). Using this novel manf-1 reporter allele, we confirmed that the MANF protein is expressed throughout development and is expressed in most somatic cells in L1 larvae, which is the developmental stage that we focused on in the current study.
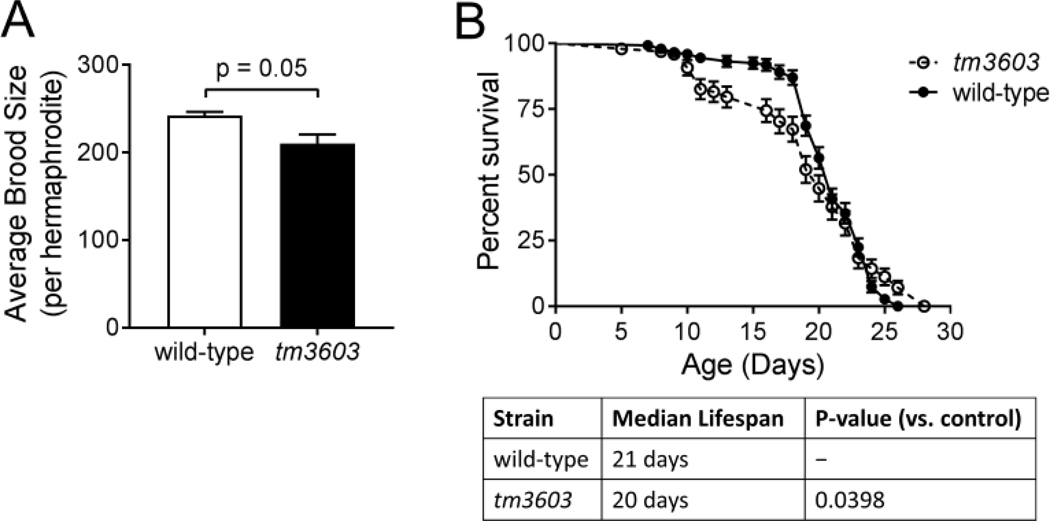
Deletion of MANF in the tm3603 strain showed a trend (p=0.05) to reduced brood size when compared to the wild-type N2 strain (Fig 1A, Table 1) which agrees with recent reported reduction in brood size in this strain (Richman et al., 2018); however, RNAi against manf-1 failed to reduce brood size (Table 1). The mutant lifespan was also significantly different compared to wild-type (Fig. 1B, p=0.0398; individual traces shown in Supplementary Fig. S3), with a slightly shorter median lifespan (21 days vs. 20 days for the wild-type) and the emergence of an increased early die-off from days 10–16.
Figure 1. Depletion of manf-1 has modest effects on worm physiology.
A slight decrease in brood size (Panel A) and lifespan (Panel B) was observed in the manf-1 (tm3603) mutant compared to wild-type. No decrease in brood size was observed in wild-type animals fed RNAi against manf-1 (Table 1). For reproduction experiments, L1 animals were grown on OP50 plates until the L4 stage, at which time three animals were serially moved to new plates each day and followed until they stopped laying embryos (presumably due to sperm exhaustion). The hatched and unhatched progeny on each plate were monitored 24 hours after the mothers were removed. Data shown are the average of three biological replicates. For lifespan experiments, 50 worms were used per biological replicate and the experiment was performed in 4 independent biological replicates. The curve shown is the combined survival from all four replicates. Individual traces from each replicate along with their associated median and maximal lifespans are shown in Supplementary Fig. S3.
Table 1. Combined deficiency for manf-1 and individual components of the IRE-1 UPR pathway results in sterility.
For experiments, L1 animals were grown on the appropriate bacteria (OP50 or HT115(DE3) expressing indicated RNAi (Timmons et al., 2001)) until the L4 stage, at which time three animals were serially moved to new plates each day and followed until they stopped laying embryos (presumably due to sperm exhaustion). The smd-1 RNAi was used as a “negative” control for RNAi phenotypes (Peters et al., 2010). The hatched and unhatched progeny were monitored 24 hours after the mothers were removed.
| Strain | Bacteria/RNAi | Eggs/plate | % Hatch | # replicates |
|---|---|---|---|---|
| N2 (wild-type) | OP50 | 727 | 99.6 | 3 |
| manf-1 (tm3603) | OP50 | 632 | 99.2 | 3 |
| N2 | smd-1 RNAi | 804.5 | 99.6 | 2 |
| N2 | manf-1 RNAi | 872.5 | 99.6 | 2 |
| N2 | hsp-3 RNAi | 782.5 | 99.2 | 2 |
| N2 | hsp-4 RNAi | 671.5 | 92.4 | 2 |
| N2 | xbp-1 RNAi | 766.5 | 99.6 | 2 |
| manf-1 (tm3603) | smd-1 RNAi | 344.5 | 99.1 | 2 |
| manf-1 (tm3603) | manf-1 RNAi | 497 | 99.8 | 2 |
| manf-1 (tm3603) | hsp-3 RNAi | 0 | 0 | 2 |
| manf-1 (tm3603) | hsp-4 RNAi | 21.5 | 37.9* | 2 |
| manf-1 (tm3603) | xbp-1 RNAi | 0 | 0 | 2 |
| hsp-3 (ok1083) | smd-1 RNAi | 886.5 | 99.5 | 2 |
| hsp-3 (ok1083) | manf-1 RNAi | 320.5 | 77.2* | 2 |
| hsp-3 (ok1083) | hsp-3 RNAi | 788.5 | 99.4 | 2 |
| hsp-3 (ok1083) | hsp-4 RNAi | 0 | 0 | 2 |
| hsp-3 (ok1083) | xbp-1 RNAi | 0 | 0 | 2 |
Asterisk () indicates that hatched progeny arrested at L1 stage. Data represent 2 replicate experiments except for N2 and manf-1(tm3603).
Because MANF supports dopaminergic neuronal development and survival in cell culture and in other organisms (Palgi et al., 2009; Petrova et al., 2003; Voutilainen et al., 2009), we hypothesized that dopaminergic neurons would be impacted by manf-1 depletion in the mutant tm3603. However, tm3603 mutant animals developed dopaminergic neurons normally (Supplemental Fig. S4, panel E) and did not show any increased sensitivity to 6-OHDA-induced neurodegeneration (Fig. S4, panel M). Furthermore, tm3603 animals did not have a swimming-induced paralysis (SWIP) dopaminergic phenotype compared to wild-type or dopamine transporter mutant animals (Fig. S4, panel N). We also tested non-dopaminergic neuronal phenotypes such as locomotion (Fig. S4, panels O and P) and chemotaxis (Fig. S4, panel Q) and found no differences compared to wild-type. Together, these data suggest that, at least for young adult animals (day 1–2 of adulthood), there is not a biologically important role for manf-1 in dopaminergic or non-dopaminergic neuronal development or survival.
C. elegans manf-1 depletion increases expression of ER stress markers and does not impact UPR activation by tunicamycin
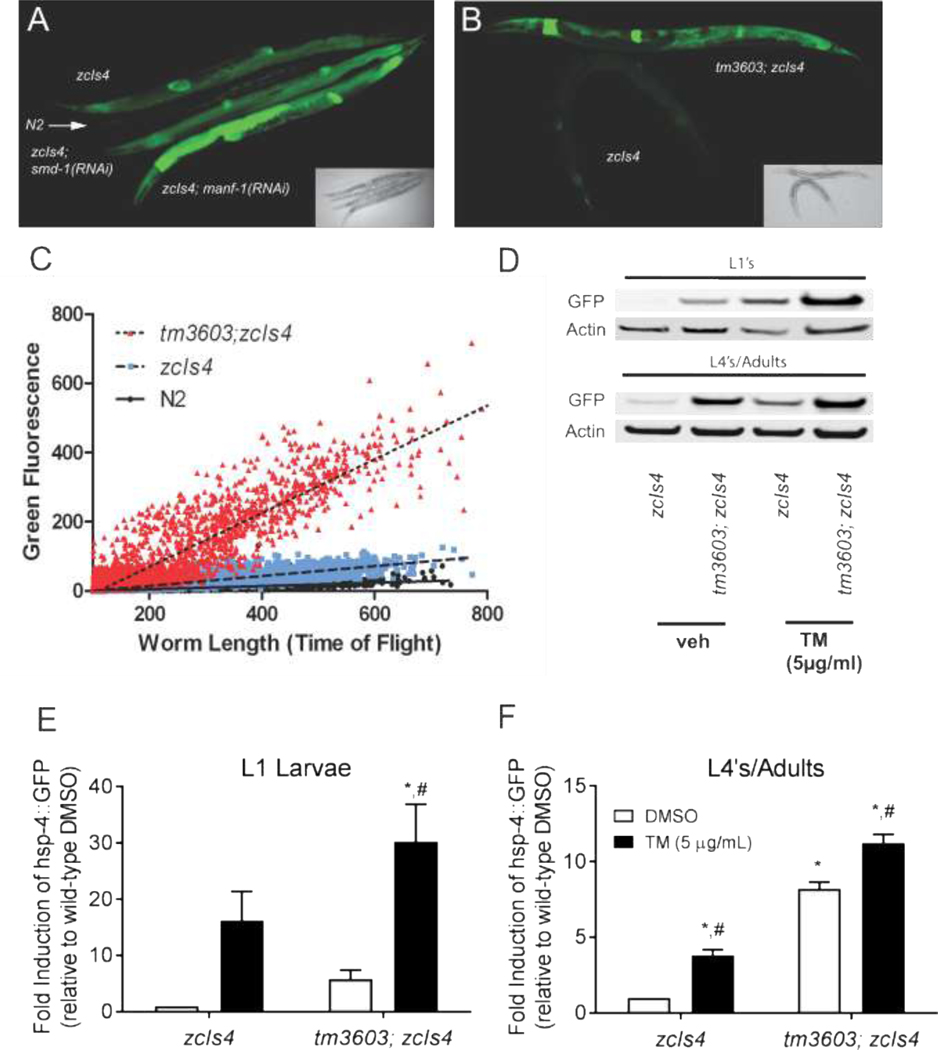
MANF has been implicated as a regulator of and responder to the UPR in mammals and C. elegans (Apostolou et al., 2008; Bai et al., 2018). When the UPR is activated in C. elegans, as in mammalian systems, expression of the ER-resident BiP/Grp78 chaperone protein homolog HSP-4 is upregulated in an IRE-1 and XBP-1 dependent fashion (Kapulkin et al., 2005). A GFP reporter driven by the hsp-4 promoter (zcIs4) allows for monitoring of UPR in live animals (Calfon et al., 2002) We fed zcIs4 transgenic animals RNAi bacteria against manf-1 and observed a significant increase in GFP expression compared to that exhibited by nematodes fed smd-1 RNAi bacteria as a non-UPR control (Fig. 2A). This increase in zcIs4 expression was also detected when the reporter was crossed into the manf-1(tm3603) background (Fig. 2B), an observation first reported in Bai et al (Bai et al., 2018). Using the COPAS fluorescent biosorter, we observed increased GFP fluorescence across all life stages in an asynchronous population of zcIs4; tm3603 mutants compared to the zcIs4 expression in a wild-type background (Fig. 2C). This difference became more dramatic as animals developed, with clear separation between groups beginning at lengths (time of flight) >300. When lysates of these worms were analyzed by immunoblotting with anti-GFP (Fig. 2D, quantification in panels E and F), we observed a 5-fold increase in GFP expression for tm3603 L1s compared to wild-type animals at the same stage, although this difference was not significant (p = 0.068, two-way ANOVA post-hoc test) due to variability in L1 expression. We also observed a significant (p < 0.001) 8-fold increase in adult expression of hsp-4p::GFP on tm3603 compared to wild-type (Fig 2F).
Figure 2. Depletion of manf-1 either by RNAi or mutation induces the expression of an hsp-4 reporter.
(A) A confocal image showing GFP fluorescence from the zcIs4 reporter (hsp-4 promoter::GFP). Age-matched hermaphrodites of several strains and treatments are arranged from top to bottom: zcIs4 (basal expression of the hsp-4::GFP reporter), followed by a wild-type N2 (non-GFP control showing minimal background autofluorescence), a non-UPR related control zcIs4; smd-1(RNAi), and zcIs4; manf-1(RNAi) animals respectively. The inset is a bright field image of the same worms. (B) A confocal image and bright field inset demonstrating the increased expression of the hsp-4::GFP reporter in manf-1(tm3603) mutant animals (top) compared to wildtype N2 worms (bottom). (C) Asynchronous cultures of wildtype (black), zcIs4 (blue), and tm3603; zcIs4 (red) were analyzed by COPAS worm sorter for time of flight (an indicator of body length) and integrated GFP intensity. The manf-1(tm3603) strain has increased expression of the GFP reporter at all stages of development. The dashed and dotted lines denote the best fit line after linear regression analysis for the zcIs4 and zcIs4; manf-1(tm3603) populations respectively. (D) An immunoblot showing the relative GFP expression of synchronized populations before and after exposure to tunicamycin. Worms were treated with tunicamycin (5 micrograms/mL) or DMSO (vehicle) for 5 hours before lysis and separation by electrophoresis. The quantification of tunicamycin-induced GFP expression (relative to actin) for L1 stage animals is shown in (E) and for adult animals in (F), (n=4). While L1 populations exhibited greater levels of inducibility than the adult populations, they also showed greater variability. Animals carrying the manf-1(tm3603) mutation expressed higher levels of GFP expression basally and achieved greater overall levels of GFP expression after tunicamycin, but did not have an overall difference in tunicamycin response (no dose-strain interaction in two-way ANOVA for either L1 or adult animals). Symbols represent significance in twoway ANOVA (strain, dose) with post-hoc analysis: *, p < 0.05 compared to zcIs4 DMSO control; #, p < 0.05 compared to tm3603 DMSO control.
When exposed to ER stress caused by tunicamycin, an N-linked glycosylation inhibitor, both wildtype and tm3603 animals displayed robust induction of the hsp-4p::GFP reporter (Fig 2E, F). Post hoc two-way ANOVA analysis revealed significant drug and strain effects at both L1 (p = 0.048 strain effect, p = 0.0006 tunicamycin effect) and adult (p<0.0001 for strain and drug effects) stages, but no interaction at either stage (p = 0.30 and p = 0.77 for L1 and adults, respectively). Wild-type L1 larvae had a 16-fold increase in GFP following tunicamycin exposure that was trending toward statistical significance (p=0.054), while tm3603 had a significant 5-fold increase (Fig. 2E, p=0.01). Wild-type L4/adult animals showed a significant 3.7-fold increase in GFP after tunicamycin exposure (p=0.0021), while tm3603 L4’s/adults showed a significant 1.5-fold increase (Fig. 2E, p=0.001). Compared to L1 larvae, adult nematodes had far less variability in zcIs4 GFP expression, both with and without tunicamycin exposure. Similar results were observed for mRNA levels of both C. elegans homologues of BiP, hsp-3 and hsp-4 (Supplementary Fig. S5A-B). Furthermore, tunicamycin exposure also increased manf-1 mRNA levels in wild-type animals (Supplementary Fig. S5C).
Deficiency of manf-1 confers larval resistance to ER stress-induced developmental arrest
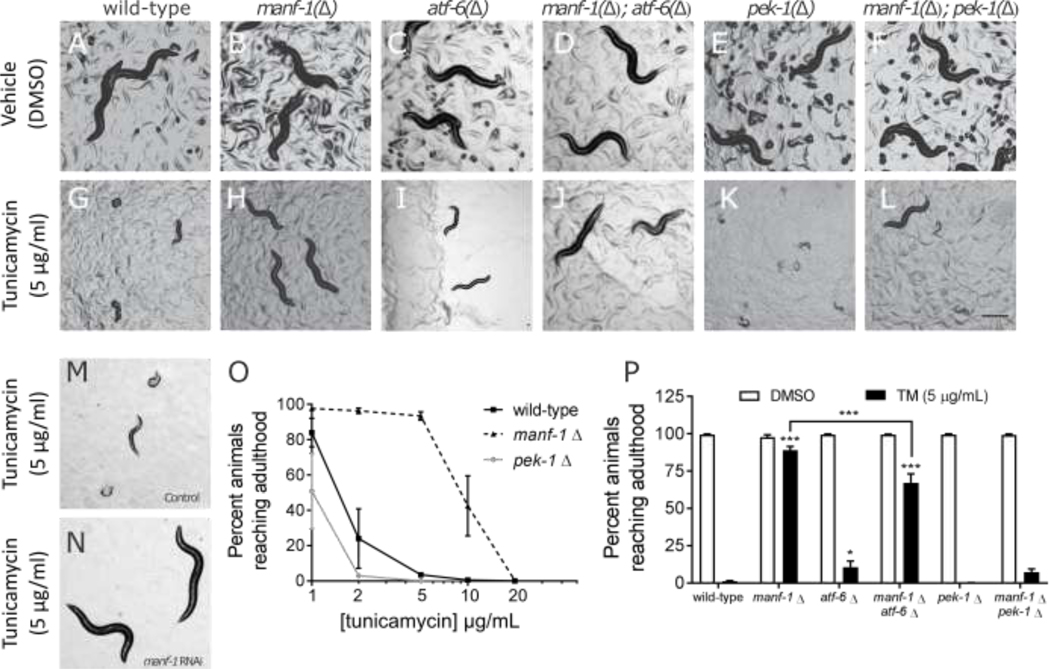
Previous studies have demonstrated that exposure of wild-type larval nematodes to tunicamycin caused a dose-dependent growth arrest and death (Richardson et al., 2011; Struwe et al., 2009). Furthermore, mutants defective in either IRE-1 or PEK-1 (C. elegans PERK homolog) signaling displayed a hypersensitivity to tunicamycin (Richardson et al., 2011). Based on our observation that manf-1 depletion can induce hsp-4::GFP expression, we hypothesized that MANF-1 acts as a negative regulator of IRE-1 and/or XBP-1 activity. If this were true, then the increased expression of XBP-1 transcription targets in the absence of manf-1 may lead to a “pre-conditioning effect” and reduce the worm’s sensitivity to tunicamycin. To test this, we allowed wild-type and tm3603 mutant embryos to hatch on plates containing tunicamycin and assayed their developmental state after 3 days at 20°C (Fig. 3, panels A-B, G-H). We observed that while all wild-type animals grown on 5 μg/mL tunicamycin died or arrested between L1 and L2, most of the tm3603 animals achieved adulthood (developed eggs) within 72 hours. Encouraged by this, we repeated the assay with increasing tunicamycin concentrations and observed that 40% of the tm3603 mutants reached adulthood with 10 μg/mL TM, but the resistance was not sufficient to overcome 20 μg/mL tunicamycin (Fig. 3, panel O). This phenomenon was reproduced when wildtype animals were grown on manf-1(RNAi) plates (Fig. 3, panels M and N). Together, these results demonstrate that manf-1 deficiency allows the animals to overcome tunicamycin-induced growth arrest.
Figure 3. MANF-1 deficiency confers resistance to tunicamycin-mediated developmental arrest.
Worms harboring mutations (indicated by “Δ”) in manf-1 (B), atf-6 (C), and pek-1 (E) have developmental rates similar to wildtype animals (A) when hatched under normal conditions. Similar results were obtained when the manf-1 mutation is paired with either the atf-6 or the pek-1 mutant (D, F). Wild-type (G) and atf-6 mutant (I) larva from embryos hatched in the presence of tunicamycin exhibit delayed or arrested development, while pek-1 mutants die (K). Manf-1 mutant larvae appear unaffected by tunicamycin (H). The tunicamycin-resistant phenotype was also observed in the manf-1; atf-6 double mutant (J), although not to the same extent. The manf-1; pek-1 double mutant was only slightly more resistant to tunicamycin compared to the pek-1 single mutant, and not statistically significant (L). RNAi knockdown of manf-1 also confers resistance to tunicamycin compared to control RNAi (M,N). Dose response to tunicamycin showing the effects on the larval development up to adulthood (O). The percentage of animals (n=100 per condition) reaching adulthood in the presence of tunicamycin (5 μg/mL) for wild-type, manf-1, atf-6, pek-1 as single mutants, and in combination (P). Data shown are the average of four biological replicates. Asterisks represent statistical significance in a two-way ANOVA (dose, strain) compared to the tunicamycin response in wild-type in a Bonferroni post-test unless otherwise indicated.
Combined knockdown of manf-1 and UPR genes reveals genetic interactions in fertility and larval tunicamycin response
To further test our hypothesis that increased resistance to tunicamycin exhibited by tm3603 mutant was dependent on the increased basal activation of the ire-1/xbp-1 signaling, we proceeded to investigate possible genetic interactions between manf-1 and individual UPR genes by combining tm3603 with mutant alleles for UPR genes. In the process of mating the ire-1(v33) allele into the tm3603 background, we observed that an F1 self-fertilizing hermaphrodite with the double heterozygous genotype of (ire-1(v33)/dpy-10(e128) unc-4(e120)); manf-1(tm3603/+)) produced very few non-dumpy non-uncoordinated progeny that were homozygous for tm3603. Those that were recovered developed into sterile adults, even when maintained at 16°C. Thus, combining ire-1 and manf-1 deficiency yields a synthetic sterile (Ste) phenotype, wherein the animals do not produce any progeny. Attempts to bypass this phenomenon by growing single mutant animals (tm3603) on ire-1 RNAi also resulted in sterility (Table 1). Similar results (sterility/reduced brood size) were obtained when we fed tm3603 worms RNAi for hsp-3, hsp-4, and xbp-1, but not a non-UPR control RNAi (smd-1). Conversely, wild-type animals treated with RNAi bacteria for ire-1, hsp-3, hsp-4, and xbp-1 produced no significant changes in brood size or mortality. Impaired fertility was also observed when we grew the hsp-3(ok1083) mutant on RNAi for manf-1. In this case, the total brood size was reduced to 36% of the control, and the embryos exhibited a decreased hatch rate, producing larva that arrested at the L1 stage. Although this failure to produce fertile double-knockouts prevented us from performing the tunicamycin sensitivity assay for these combinations, it is a clear indication that there is a genetic interaction between manf-1 and members of the ire-1/xbp-1 signaling pathway.
Moving to the remaining branches of the UPR, we crossed tm3603 into the backgrounds of pek-1(ok275) or atf-6(ok551). These matings proceeded normally and produced double homozygote animals that were superficially wild-type and had a normal brood size when maintained at 20°C. As expected, the pek-1(ok275) parental strain exhibited a significantly increased sensitivity to tunicamycin compared to wildtype animals in our development assay (Fig 3, panel O), and essentially no pek-1 mutant animals reach adulthood at the 5 μg/mL dose (Fig 3, panel P). Interestingly, the combined depletion of pek-1 and manf-1 rescues this phenotype slightly, with about 8% of animals reaching adulthood. We observed a similar effect for atf-6: depletion of atf-6 alone resulted in only 11% of animals reaching adulthood, while combined depletion of atf-6 and manf-1 resulted in 68% of animals reaching adulthood (Fig 3, panel P). Compared to manf-1 depletion alone, however, which allowed 90% of animals to reach adulthood, combined knockdown of manf-1 with atf-6, and especially pek-1, hindered the ability of manf-1 mutation to confer tunicamycin resistance. Therefore, these results suggest that the resistance to tunicamycin for the manf-1 mutant resistance partially requires the activity of atf-6 and especially that of pek-1.
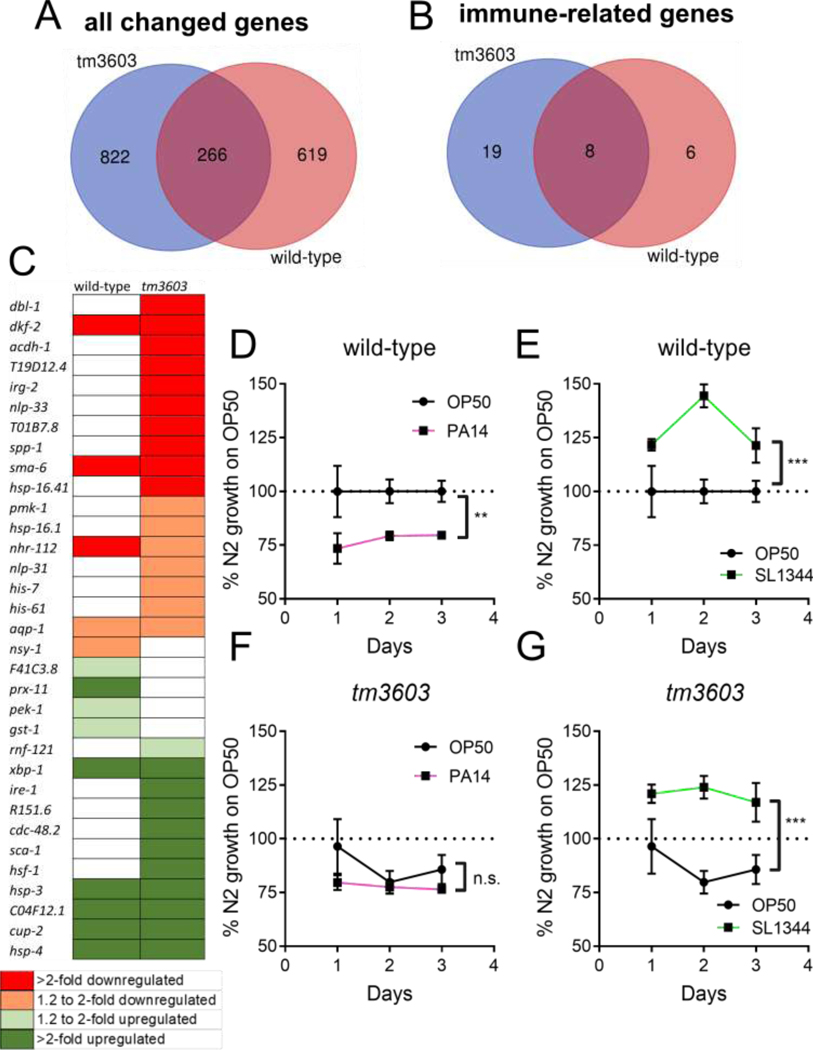
Microarray analysis of wild-type and tm3603 L1 larvae exposed to tunicamycin reveals differentially expressed pathways
The marked difference in the tunicamycin response between tm3603 and wild-type L1 larvae was further examined at the level of the transcriptome. Using Affymetrix microarray analysis, we looked for changes in mRNA levels that could identify pathways that might contribute to the phenotypes described above. Similar to our findings that manf-1 depletion alone had very modest effects on organismal physiology, there were very few pathways enriched at baseline in the tm3603 MANF-deficient mutant compared to wild-type (Table 2). While there were common changes in gene expression in response to tunicamycin (Fig 4, panel A), the majority of significantly changed genes were unique to each strain. The wild-type animals had many more enriched pathways compared to tm3603 (see Table 2), with the majority being related to organismal growth and metabolism. By contrast, tm3603 manf-1-deficient animals had more differently expressed genes, but pathway analysis indicates that these were assignable to fewer enriched pathways. Those pathways that were unique to the tm3603 mutant were mainly involved with innate immunity (Table 2). A closer look at the genes involved in these pathways gives a clear picture that the tm3603 animals had three times more differentially regulated genes related to immune response compared to the wild-type (Fig. 4, panel B). In fact, most of the immune-related genes that are shared between the two strains are upregulated genes related to the UPR (Fig. 4, panel C), which we also observed in earlier experiments to be upregulated in both strains. By contrast, the tm3603 MANF-deficient mutant downregulated many immune genes not directly related to the UPR in response to tunicamycin exposure. It is also worth noting that there was also robust and significant tunicamycin-induced down-regulation of the gene F57F4.4, a putative specific reporter for bacterial infection (Julien-Gau et al., 2014), in the tm3603 MANF-deficient strain (fold-change −5.9; p=0.00033) while the wild-type was not significant (fold-change −1.8, p=0.078). Overall, these findings indicate the tm3603 mutants fail to activate, or are actively repressing several components of the innate immunity system during ER stress.
Table 2.
Pathways enriched in changed genes after exposure to tunicamycin. For experiments, L1 animals were synchronized overnight by liquid starvation and then exposed to tunicamycin (5 μg/mL) for 5 hours. Total RNA was extracted and an Affymetryx microarray analysis was performed. Pathway analysis was performed with GoMiner using a fold-change cutoff of 1.3 and a p-value cutoff of 0.05.
| Go Categories | Description | |
| Enriched pathways shared between wild-type and tm3603, after tunicamycin treatment | ||
| 34976, 30968, 6986, 6984 | ER stress response/UPR | |
| 71445, 51789, 71216 | Response to protein stimulus | |
| 6950 | Stress response | |
| 10033, 71310, 70887 | Response to organic chemical | |
| 51605, 6465 | Protein maturation | |
| 45454, 42592, 19725 | Cell homeostasis | |
| 44281, 6662, 18904 | Small molecule/ether metabolism | |
| 8104, 51641, 15031, 70727, 34613, 46907, 45184, 6886 | Protein localization/transport | |
| 42303, 18988, 18996 | Molting cycle | |
| Enriched pathways only in wild-type animals, after tunicamycin treatment | ||
| 45926, 40015, 48519 | Negative regulation of growth | |
| 30811, 31329, 9154, 46700, 9261, 9894 | Regulation of cellular catabolism | |
| 9166, 9150, 33121, 33124, 9141, 9199, 9205, 9144, 9207 | Nucleoside/nucleotide catabolism | |
| 44270, 44106, 46700 | Cellular heterocyclic amine metabolism | |
| 43087, 32312 | Regulation of GTPase activity | |
| 48870, 46039, 6140, 6520, 42180, 6082, 19752, 16071 | Regulation of metabolism | |
| 40035, 45137, 46546, 46661, 48806, 46660, 46545 | Genitalia development and differentiation | |
| 19098, 18991, 33057 | Reproduction | |
| 40012, 7610 | Locomotion/Behavior | |
| 1703, 7369 | Gastrulation with mouth forming first | |
| 9888, 35121 | Tissue development | |
| 51674, 48870, 16477, 40039 | Cell localization and motility | |
| 9306, 46903 | Protein secretion | |
| 10038, 10035 | Response to inorganic substance/metal | |
| 7167, 23034 | Intracellular signaling pathways | |
| 6888, 33365, 48193, 51649 | Vesicle-mediated transport | |
| Enriched pathways only in tm3603 mutant, after tunicamycin treatment | ||
| 40014, 51704, 35264, 10171 | Multicellular organism growth | |
| 50829, 6952, 9617, 6955, 2376, 51707, 45087, 9607 | Innate immune response | |
| 7592, 40002 | Cuticle development | |
| Enriched pathways in tm3603 mutant compared to wild-type without tunicamycin | ||
| 34504*, 51170, 6606*, 51169 | Protein localization/transport to nucleus | |
| 23060, 23046, 7165 | Signal Transduction | |
| 6996 | Organelle Organization | |
| 6913 | Nucleocytoplasmic transport | |
indicates this term was also found in N2 exposed to tunicamycin
Figure 4. MANF deficiency modulates immune pathway during tunicamycin exposure and ameliorates growth delay from Pseudomonas aeruginosa.
Panel A represents the differentially expressed genes (combined up and down-regulated genes) in wild-type and tm3603 (manf-1 deficient) animals. Panel B, differentially expressed genes related to immune response in each strain, and Panel C, a visual representation of gene expression for the immune-related genes. Red indicates downregulated genes, while green indicates upregulated genes, and white indicates no significant change; the intensity of the shading indicates the fold-change (light color for 1.3–1.9-fold; dark color for >2-fold). For experiments, L1 animals were synchronized overnight by liquid starvation and then exposed to tunicamycin (5 μg/mL) for 5 hours. Following, RNA was extracted and a microarray analysis was performed. Pathway analysis was performed with GoMiner using a fold-change cutoff of 1.3 and a p-value cutoff of 0.05. Venn diagrams were created using the free online webtool at http://bioinformatics.psb.ugent.be/webtools/Venn/. Panels D-G represent larval growth on OP50 (E. coli), PA14 (P. aeruginosa), and SL1344 (S. typhimurium) normalized to wild-type (N2) animals’ growth on OP50. Worm size was determined from bright field images using ImageJ, as described in Methods. Asterisks represent significance in a 2-way ANOVA (bacterial strain vs. time).
MANF-deficient animals grow differently than wild-type animals on Pseudomonas aeruginosa but not on Salmonella typhimurium.
The hypothesis that manf-1 negatively regulates the innate immune response in response to cellular stress was tested by growing the worms on two strains of pathogenic bacteria. Pseudomonas aeruginosa (PA14) and Salmonella typhimurium (SL1344) are gram-negative bacterial strains that infect C. elegans and cause induction of the innate immune response, each through distinct virulence mechanisms (Aballay et al., 2000; Sem and Rhen, 2012; Tan et al., 1999). Larval growth experiments on the pathogenic bacterial P. aeruginosa strain PA14 revealed a PA14-induced growth delay in N2 wild-type animals compared to growth on the mildly pathogenic OP50 E. coli (Fig. 4D). By contrast, wild-type larval growth was significantly accelerated by the pathogenic S. typhimurium strain SL1344 (Fig. 4E). MANF-deficient animals, on the other hand, were delayed on OP50 compared to wild-type, but not further delayed with PA14 (Fig. 4F). However, tm3603 manf-1-deficient larvae had accelerated growth on SL1344 that did not differ from wild-type (Fig. 4G). Together, these findings suggest that C. elegans response to pathogenic bacteria may be altered by MANF deficiency. Further experiments are needed to probe the details of this immune modulation by MANF.
DISCUSSION
The unique and highly conserved CDNF/MANF family of neurotrophic factors has widely been reported to be cytoprotective and ER-stress responsive (Airavaara et al., 2010; Airavaara et al., 2009; Apostolou et al., 2008; Hellman et al., 2011; Henderson et al., 2013; Lindahl et al., 2014; Lindholm et al., 2007; Lindstrom et al., 2013; Palgi et al., 2009; Petrova et al., 2003; Voutilainen et al., 2015; Voutilainen et al., 2009; Yu et al., 2010; Zhang et al., 2018a). These cytoprotective roles are seen in mammals (Lindahl et al., 2014), zebrafish (Chen et al., 2012), and the invertebrate Drosophila (Lindstrom et al., 2013). Herein, we show that depletion of the conserved MANF gene in C. elegans, manf-1, has modest effects on nematode development and reproduction, including a previously reported increase in hsp-4::GFP reporter expression; however, there is a dramatic effect when manf-1-deficient animals are grown on tunicamycin. We report for the first time that while wild-type animals arrest at the L1 stage, nematodes lacking manf-1 expression are very resistant to tunicamycin-induced growth arrest.
We also observed physiological effects when manf-1 was depleted along with components of the IRE-1 pathway. Depletion of manf-1 along with reduction in the IRE-1 pathway components generally led to larval death and sterility. Direct transcriptional regulation of the manf-1 gene at an ER stress response element (ERSE) by spliced xbp-1 in mammalian cells has been recently reported by Wang et al (Wang et al., 2018). Our data supports the idea that this feedback loop (Fig. 5) is also conserved in C. elegans.
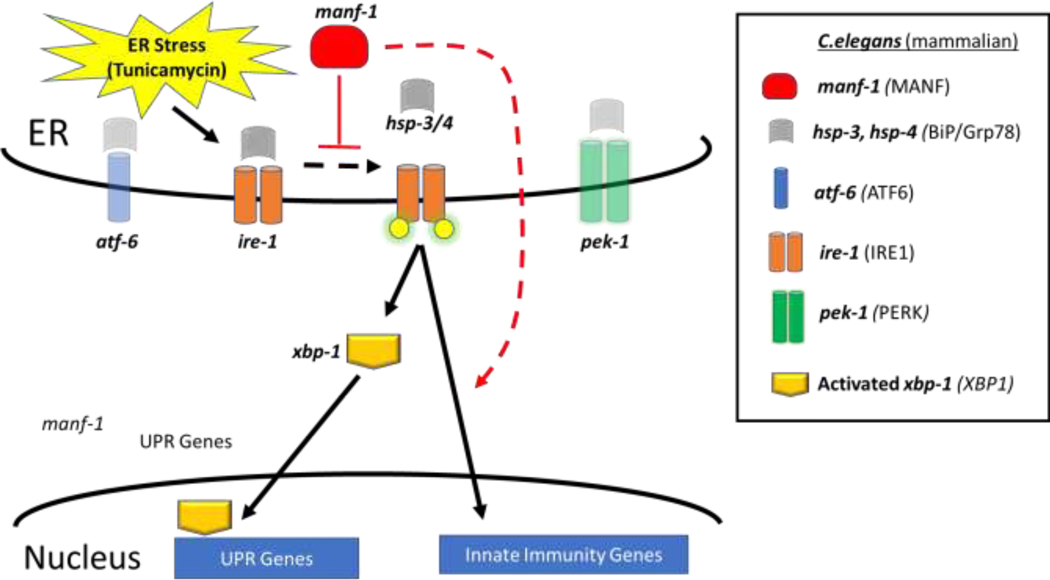
Figure 5. Hypothetical Conserved Pathway for MANF-1 regulation of IRE-1 bifurcation.
ER stress activates the regulators of UPR (atf-6, ire-1 and pek-1) and the data presented herein support a role for manf-1 in the regulation of the ire-1 arm of the unfolded protein response (UPR) by acting as both a negative regulator of xbp-1 transcriptional activation and a positive regulator of innate immunity factors in response to tunicamycin.m. ER stress and protein misfolding activate the ire-1 pathway by dissociating hsp-3/4 from ire-1 allowing ire-1 to become phosphorylate (yellow circles) leading to activation of xbp-1 as well as of other transcriptional factors. We also suggest a conserved feedback loop whereby ER stress and xbp-1 activation results in increased manf-1 expression (as also reported in mammalian cell culture (Wang et al., 2018)), suppressing further xbp-1 activation and acting as a negative regulator of the UPR.
Although manf-1;pek-1 and manf-1;atf-6 double mutants developed into fertile, relatively normal adults in the absence of tunicamycin, both showed a reduction in the resistance to tunicamycin compared to manf-1 mutant alone. While these results demonstrated a genetic interaction between manf-1 and the other branches of the UPR, they do not establish whether the functions supplied by these other branches are required for the manf-1 mutant’s increased resistance to tunicamycin, or if it is the manf-1 mutant’s resistance that partially suppresses the UPR mutants’ increased sensitivity to tunicamycin. Through a transcriptional microarray analysis, we showed that the UPR is upregulated by tunicamycin exposure in both wild-type and manf-1 (tm3603) mutants. However, pek-1 was upregulated only in wild-type and ire-1 was upregulated only in the mutant. Together, this establishes C. elegans MANF as a critical negative regulator of the endoplasmic reticulum UPR stress response (Fig. 5); however, future studies are needed to further establish the interaction of MANF with each arm of the UPR (xbp-1, pek-1, and atf-6).
It is interesting that depletion of MANF in different organisms results in vastly different phenotypes: for example, in mice, MANF deficiency manifests as severe loss of β cells through chronic and sustained UPR activation, resulting in diabetes mellitus (Lindahl et al., 2014). These observations are consistent with our findings that loss of MANF leads to constitutive expression of UPR genes. Conversely, knockdown of MANF in zebrafish manifests in a dopaminergic phenotype (Chen et al., 2012), and in Drosophila causes larval lethality and loss of dopaminergic neurons (Palgi et al., 2009); yet, MANF-deficient worms exhibit a fairly normal physiology with only mild effects on lifespan and brood size, and therefore it seems that, unlike other organisms, there are no cell types in C. elegans for which MANF is essential for development and function under laboratory conditions. This leads to an interesting and unanswered question of the evolutionary pressure to keep manf-1 in C. elegans.
We did not observe any dopaminergic neurodegeneration phenotypes in manf-1-deficient animals. Wild-type and MANF mutant day 1 adult animals had similar sensitivity to the model neurotoxicant 6-OHDA, which selectively damages and kills dopaminergic neurons, and did not display a swimming-induced paralysis (SWIP) phenotype, a behavior previously linked to the function of dopaminergic neurons (McDonald et al., 2007). Recent evidence shows that overexpression of human MANF in C. elegans dopaminergic neurons can protect from alpha-synuclein or 6-OHDA toxicity in aged animals (Zhang et al., 2018b) and that manf-1 mutants have accelerated aging-related dopaminergic neurodegeneration and aggregation of alpha-synuclein (Richman et al., 2018). These studies suggest that, for worms, manf-1 neuroprotection seems to be more important in aged animals, and may not extend to neurodegeneration from 6-OHDA challenge. Further experiments are needed to understand the role of C. elegans MANF in dopaminergic neuronal health.
Our observation that depletion of manf-1 by mutation (tm3603) or RNAi increases the expression of UPR reporter hsp-4 is consistent with recent literature (Bai et al., 2018; Richman et al., 2018) reporting upregulation of hsp-4 in the tm3603 mutant and/or after RNAi against manf-1. In general, these findings and others (Apostolou et al., 2008; Lindahl et al., 2014; Voutilainen et al., 2015) point to a role for MANF as a negative regulator of the UPR. However, the role of MANF in the UPR is likely more complex than simple inhibition, given that depletion of MANF or UPR genes alone did not impact reproduction, but synthetic sterility (lack of progeny) was observed when MANF depletion was combined with depletion of UPR components. Furthermore, it was recently suggested that MANF plays a dual role in alleviating ER stress while activating autophagy pathways (Zhang et al., 2018b).
In MANF-depleted animals, we observed a basal increase in larval expression of hsp-3 and hsp-4, the C. elegans homologues of the BiP protein, a molecular chaperone whose expression is linked with XBP-1 activation (Gardner et al., 2013; Shen et al., 2005). We also observed remarkable larval protection from tunicamycin-induced growth arrest after depletion of manf-1. One possible explanation for this protection is that it is mediated by preconditioning or hormesis (De Haes et al., 2014; Kozlowski et al., 2014; Pinto et al., 2018) from the the elevated expression of hsp-3 and hsp-4. Although we were not able to examine double mutants of UPR genes and MANF due to synthetic sterility, the higher baseline expression of UPR genes with chaperone function in the manf-1 mutant animals provides a plausible means to better accommodate protein misfolding caused by tunicamycin. Further studies are needed to explore this possibility. Similar to MANF, HPL-2 is reported to be a negative regulator of the UPR, and lack of HPL-2 confers protection from ER stress by increasing baseline levels of XBP-1 and ER chaperones (Kozlowski et al., 2014). This paradigm agrees with the larval protection we observe in MANF-deficient animals. However, MANF depletion was previously reported to sensitize cells to tunicamycin-induced cytotoxicity (Apostolou et al., 2008) and was recently demonstrated to sensitize young adult C. elegans nematodes to tunicamycin-induced reproductive defects (Bai et al., 2018). These apparent contradictory roles for MANF in worms could be due to developmental sensitivity to ER stress (larvae vs. young adults) as well as tissue-specific functions (whole organismal development vs. germline effects). As the mechanism of this reproductive defect has not been reported (Bai et al., 2018), it is unclear if there is decreased egg production, impaired spermatogenesis, or both. These details would be important for future studies further exploring the role of MANF in preconditioning or hormesis.
We found that the ability of MANF-deficient tm3603 mutants to overcome larval L1 arrest in the presence of tunicamycin was dependent on atf-6 and pek-1, which agrees with other findings showing that MANF interacts with multiple arms of the UPR (Apostolou et al., 2008; Lindahl et al., 2014; Zhang et al., 2018b). The double mutants (atf-6;manf-1 and pek-1;manf-1) failed to maintain the tunicamycin-resistance phenotype. Thus, the animals must be able to mount a UPR response to tunicamycin to continue through development. However, the hsp-4 induction by tunicamycin did not differ between manf-1 (tm3603) mutants and wild-type animals, as measured by the hsp-4:GFP reporter and confirmed by qPCR. Our microarray analysis clearly showed that the manf-1 (tm3603) mutants failed to change the numerous growth and metabolism pathways that were altered in the wild-type animals by tunicamycin (which have been previously reported (Shen et al., 2005; Urano et al., 2002)), suggesting that other mechanisms are involved in tunicamycin resistance in manf-1-depleted animals.
Furthermore, the analysis of genes that changed in response to tunicamycin revealed significant modulation of components of the innate immune response only in the manf-1 mutant, including downregulation of components of the p38-MAPK (pmk-1) pathway and the TGF-β (dbl-1) innate immunity pathways (Ermolaeva and Schumacher, 2014) and failure to activate antioxidant genes (gst-1 and prx-11) downstream of skn-1 (the C. elegans homolog of NRF-2). Our data supports that MANF is a regulator in the crosstalk between the UPR and bacterial immune response in developing nematodes. These findings are supported by previous studies that have shown that MANF can negatively regulate inflammatory responses via altered NFkB signaling in neural stem cells (Zhu et al., 2016), rat primary astrocytes (Zhao et al., 2013), and human cell lines (Chen et al., 2015). Furthermore, MANF and CDNF have been shown to be protective in animal models where inflammation is a contributing pathogenic mechanism (e.g. ischemia, Parkinson’s and diabetes) (Airavaara et al., 2010; Airavaara et al., 2009; Lindahl et al., 2014; Lindholm et al., 2007; Voutilainen et al., 2009; Yu et al., 2010; Zhang et al., 2018a). Our findings support the growing body of evidence that MANF and the UPR play key roles in immunity and are highly conserved from mammals to C. elegans (Janssens et al., 2014).
Differential regulation of immune genes in response to ER stress in manf-1 mutants likely impacts pathogenic susceptibility, given that infection of C. elegans with Pseudomonas aeruginosa (PA14) induces XBP-1/IRE-1-dependent UPR (Richardson et al., 2010; Singh and Aballay, 2017). Animals deficient in xbp-1 arrest as L1 larvae and die on PA14, while combined loss of xbp-1 and pmk-1 suppresses this larval arrest phenotype (Richardson et al., 2010). Another study implicated the microRNA mir-233 as essential for PA14 innate immune response and UPR, which works by downregulating SCA-1 (C. elegans sarco-endoplasmic reticulum Ca2+ ATPase or SERCA), inducing the UPR (Dai et al., 2015), supporting the involvement of the UPR pathway in PA14 resistance. We observed similar growth of manf-1 mutants on OP50 and PA14 bacteria, whereas wildtype animals were delayed on PA14 compared to OP50. This differential response reveals decreased sensitivity to PA14 in MANF-deficient animals. In comparison to previous study (Mohri-Shiomi and Garsin, 2008), we found showed no difference in growth on SL1344 between wild-type and MANF-deficient animals; in fact, both strains displayed dramatic growth acceleration throughout development. This contrasts with PA14 growth inhibition in wild-type but not MANF-deficient animals, suggesting that SL1344 and PA14 are impacting developing C. elegans larvae differently, which could have contributions both from nutritional availability and toxicity from pathogenic bacteria virulence mechanisms. Additionally, since these studies were carried out in larval stages, pathogenic bacteria had not yet colonized the gut. Thus, if MANF-deficient animals have dysregulated innate immune function, MANF deficiency may also impact adult pathogenic exposures, when animals are susceptible to gut colonization and infection (Aballay et al., 2000; Tan et al., 1999). Future studies are needed to fully characterize the mechanisms of MANF regulation of host-pathogen interactions during development and to expand this knowledge to include pathogen-induced killing of adult animals.
Conclusions.
In mammals, MANF/CDNF can promote cytoprotection and modulate the UPR response, but the mechanisms by which these neurotrophic factors carry out these roles are still poorly understood. Efforts to understand the mechanism of MANF action are critical to support future endeavors to manipulate MANF therapeutically (Lindholm et al., 2016). C. elegans represents an inexpensive, efficient model to identify molecular mechanisms underlying MANF activity. We have shown here that MANF functions as a modifier of the UPR in nematodes and impacts other UPR-sensitive processes such as innate immunity. In fact, many important physiological processes may be affected by MANF modulation of the UPR, including exercise, thermotolerance, and sensitivity to environmental chemicals. Together, our findings represent a new aspect of MANF biology and future research using this model may uncover critical details about MANF biology.
Materials and Methods
Worm maintenance and strains.
A complete list of strains used in this study is provided in Supporting Information, Table S1. The wild-type reference strain N2 (Bristol), and the mutant strains unc-119 (ed3), and dpy-10(e128) unc-4(e120) strains, as well as the bacterial strains OP50 and HT115(DE3), were obtained from Andy Golden (NIDDK, Bethesda, MD). PA14 and SL1344 pathogenic bacterial strains were obtained from Alejandro Aballay (Oregon Health and Science University, Portland, OR). Novel deletions were obtained from Sohei Mitani (Tokyo, Japan). The vtIs1/vsIs33 reporter strain was given to us by Oliver Hobert (Columbia University Medical Center, NY). Other mutants and reporter strains were obtained from the Caenorhabditis Genetic Center (USA). All strains were cultivated on MYOB (Modified Youngren’s, only Bacto-peptone, Eric Lambie, personal communication) media with seeded with OP50 bacteria using standard protocols and maintained at 20°C with the exception of ire-1 (v33) which was maintained at 16°C.
Genotyping.
All deletion alleles were confirmed and followed using polymerase chain reaction (PCR) with allele-specific primer sets (Supplemental Table S2). Single worm lysates and PCR reactions were performed as described (Williams et al., 1992).
Cloning and transgenes.
Multiple Gateway-compatible (ThermoFisher) entry vectors were constructed by PCR amplification from genomic or cDNA templates using attB linkered oligos, followed by BP recombination with appropriate donor vectors (Supplemental Table S2). All entry vectors were confirmed by primer extension sequencing (Seqwright, Houston, TX). Transgenic expression constructs were produced by recombining various entry vectors with a destination vector containing the unc-119(+) rescue cassette, and subsequently introduced into animals by microparticle bombardment (Praitis et al., 2001).
The NK2548 strain containing the fusion allele qy74[manf-1::mKate2] was generated using Cas9-triggered homologous recombination with a self-excising selection cassette (Dickinson et al., 2015). The target sequence was 5’-CTGGTGATTGTCGTCGCCTCGG-3’ with PAM site shown in bold; the guide RNA was cloned into the eft-3p::Cas9+sgRNA expression vector pDD162 using this primer sequence (excluding the PAM). The PAM is 43 nucleotides downstream of the start codon of manf-1, just downstream of the putative signal peptide sequence, which has previously been demonstrated to be an ideal location to insert a tag in the mammalian MANF protein (Norisada et al., 2014). Homology arms were amplified by PCR from N2 genomic DNA (5’ homology arm forward primer manf-1 sequences are 5’-TGGCCGCTGAAACTTATCGG-3’ and reverse primer 5’-TGGAGCAGCAGCTGAGGCG-3’ (with mutation in PAM site shown underlined); 3’ homology arm forward primer manf-1 sequences 5’GAAGGTTTGTAAATTTGAGAAAGC-3’ and reverse primer 5’-TTTTCCTCGAGTTTTCAACG-3’) and cloned into a plasmid modified from (Dickinson et al., 2015) containing an N-terminal linker sequence (with an ATG codon added during assembly), an mKate2 coding gene, and a self-excising cassette (SEC) flanked by LoxP sites in an intron of mKate2. The plasmid was co-injected with sgRNA+Cas9 plasmid into N2 animals, and genome-edited animals were selected by hygromycin B treatment and phenotypic identification (roller). The selection cassette was excised by heat shock as described in (Dickinson et al., 2015).
Lifespan assay.
For lifespan experiments, semi-synchronous gravid adults were obtained by washing the adult and larvae from a plate containing food and eggs, then allowing the eggs to hatch overnight. The following day, L1-L2 animals were transferred to fresh plates seeded with OP50 (300 animals per 10 cm plate) and allowed to develop for 48 hours (until adult day 2). Following, synchronous populations were obtained by bleaching the gravid adults and plating eggs to fresh food plates. After 72 hours, 50 worms were picked from each 6cm plate. Adults were transferred every day during adult days 2–10, then transferred as needed for food depletion. The number of surviving animals was determined by gently prodding each animal with a platinum wire. An animal was considered dead if it did not respond to two repeated prods with the platinum wire. The surviving population was recorded every 1–2 days until all worms were dead.
Brood Size assay.
For brood size experiments, synchronized populations of L4 animals were obtained as described for the Lifespan assay. At the L4 stage, single worms were placed onto 6cm plates and allowed to lay eggs. During the first four days of reproduction, the worms were moved to fresh plates. Offspring were allowed to hatch and were counted after 48 hours. The final plate was counted 48 hours after the worm was placed there (when the adult had reached day 6). Brood size was considered to be the sum of all offspring from a single worm.
Microscopy.
Worms expressing GFP or mCherry reporters were mounted on 1% agarose pads in 15 microliters of M9 buffer containing 25 mM levamisole. Epifluorescent imaging was performed using a Nikon AZ100MS stereomicroscope.
6-OHDA neurotoxicity assay.
A 10mM solution of 6-hydroxydopamine was produced by dissolving 6-OHDA Br salt (Sigma) in a nitrogen-bubbled solution of 2mM ascorbic acid (Sigma). Synchronized L1 populations of the dat-1::GFP reporter strain were fed OP50 bacteria at 20°C for 24 hours at which time they were harvested, rinsed in 2mM ascorbic acid, and exposed to various doses of 6-OHDA for 90 minutes at room temperature with mild agitation. Degeneration of dopaminergic neurons was assayed by fluorescent microscopy in the dat1::GFP reporter strain 24 hours later. A worm was considered to have degeneration of DA neurons if any of the four DAT-positive neurons in the head showed signs of blebbing or breaks(Nass et al., 2002).
SWIP assay.
For determination of swimming-induced paralysis (SWIP), we used a standard protocol (Bermingham et al., 2016) with minor modifications. For each biological replicate, ten wild-type and tm3603 day 2 adult animals were placed into each of 5 wells in a 48-well plate containing 100 μL of molecular biology grade water. The number of animals paralyzed was visually counted after 10, 20, 30, 40, 50, and 60 minutes. The dopamine transporter-deficient strain dat-1 (ok157) was used as a positive control for SWIP.
Locomotion assay.
For determination of adult nematode locomotion, we used a standard worm tracking procedure to measure the speed of wild-type and MANF-deficient animals on food-free plates (Au - Nussbaum-Krammer et al., 2015). For each biological replicate, wild-type and tm3603 day 1 adult animals were transferred to food-free 10 cm plates (around 100 worms per plate) and allowed to acclimate to the plate for 30 minutes. Following, 15 second brightfield videos were collected using the Keyence BZ-X700 All-in-One Microscope (15 videos per plate) with the experimenter blind to strain. The maximal and average velocities were then measured using the wrMTrck plugin in ImageJ (Au - Nussbaum-Krammer et al., 2015).
Chemotaxis assay.
For determination of chemotaxis, a standard four-quadrant assay design was used (Margie et al., 2013). For experiments, 6-cm chemotaxis plates were labeled to divide into equal quadrants, and a 0.5cm circle was drawn in the center. Dots were then placed equidistant (1.5cm) from the center in each quadrant and labeled with alternating “C” for control and “T” for test solution. Synchronized day 1 adult worms (wild-type and tm3603 MANF-deficient strains) were then washed off plates, rinsed three times to remove residual bacteria, and 5μL of the concentrated worm pellet was applied to the center of the plate, which amounted to 100–150 worms. While the worm spot was drying, two microliters of the control and test solutions were applied to the appropriately labeled spots. The control solution was an equal mixture of 0.5M sodium azide and distilled water, while the test solution was an equal mixture of 0.5M sodium azide and either 100% ethanol or 1% benzaldehyde in ethanol. Therefore, the final concentrations of ethanol or benzaldehyde in the test solutions were 50% and 0.5%, respectively. Under these conditions, the plates dried within 3 minutes, allowing the worms to crawl out of the center circle and into the quadrants. Once the plates were dry, they were inverted and placed in the 20ºC incubator for 60 minutes. Following the incubation, plates were immediately placed at 4ºC until they were counted (within 4 hours of the experiment). The chemotaxis index was calculated as: (Number of Worms in Test Quadrants – Number of Worms in Control Quadrants)/Total Number of Worms. Only worms that left the center circle were counted. The experimenter was blind to strain throughout the procedure.
Western Blotting.
Thirty adult hermaphrodites were placed into tubes containing twenty microliters of phosphate-buffered saline. Ten microliters of 4x LDS buffer (Invitrogen) containing beta-mercaptoethanol (Sigma) were added and samples incubated at 95ºC for 10 minutes. The entire volume of each sample was loaded into a single well of a 10% Bis-Tris NUPAGE polyacrylamide gel with MES buffer (Invitrogen). The gels were transferred to PDVF membranes and subsequently probed with mouse anti-Actin (clone C4; Millipore), and rabbit anti-MANF-1 (YZ1272) or rabbit anti-GFP (GN-16469). Anti-MANF-1 is an affinity-purified polyclonal antibody directed against the peptide sequence C-RIEELKPKYVKEEL (amino acids 155–168; YenZyme, Inc.). Anti-GFP is an affinity-purified polyclonal antibody directed against His-tagged EGFP (Sigma-Genosys, Inc., gift from Andy Golden). Primary antibodies were detected using donkey anti-rabbit IRdye 800 and donkey anti-mouse IRdye 700 (LiCor). All blocking and antibody incubations were performed in 1x LiCor Blocking solution. Quantitative measurements of the immunoblots were performed using the Odyssey Infrared Imaging System (LiCor Biosciences). Actin levels were used to normalize sample loading.
Flow-sort analysis.
Asynchronous cultures of worms were grown on MYOB plates with OP50 bacteria and resuspended in M9 and analyzed for body length (time of flight) and GFP fluorescence on a COPAS Biosort (Union Biometrica, Inc.) (Pulak, 2006).
Tunicamycin exposure.
Animals were synchronized at the L1 larval stage using alkaline hypochlorite treatment followed by overnight synchronization on food-free plates. These animals were then suspended in M9 solution (3 g. KH2PO4, 6 g. Na2HPO4, 5 g. NaCl, 1 ml 1M MgSO4 )per L H20) and left untreated or exposed to 5 μg/mL tunicamycin (in DMSO) for 5 hrs. Following treatment, the animals were pelleted for subsequent analyses (immunoblot, qPCR, or microarray).
Quantitative Taqman real-time PCR assay.
Treated and untreated worm pellets (~50,000 worms) were suspended in 300 μl TRIzol reagent (Invitrogen) with 5 μg of linear polyacrylamide (GenElute LPA), extracted with chloroform and the RNA was precipitated with isopropanol. The obtained RNA was then DNase treated (Qiagen) followed with the RNeasy MinElute Cleanup protocol (Qiagen). Total RNA (0.1–0.5 μg) was reverse transcribed following the standard protocol of the iScript Select cDNA Synthesis Kit (Bio-Rad) in 20 μl reactions. The cDNA samples were diluted 1:5 with nuclease-free water. To quantify the gene expression for hsp-4, hsp-3, manf-1 and act-3 (actin), each 25μl reaction contained forward and reverse primers (900 nM each), 50 nM 5’-FAM/3’BHQ1 probe (BioResearch Technologies), 1x of TaqMan Universal Mix (Roche) and 5μl of diluted cDNA. These reactions were performed with the DNA Engine Opticon 2 Thermocycler (MJ Research Inc.) using the following program: 20 cycles of 50°C for 10 sec, plate read; 40 cycles of 95°C for 5 min, 94°C for 20 sec, 60°C for 1 min, plate read. The comparative CT method (ΔΔCT) was used for relative quantitation of gene expression using Actin as a housekeeping gene. Primer and probe sequences are listed in Supplemental Table 2.
Larval tunicamycin resistance assays.
Tunicamycin plates were prepared by adding 40 microliters of (100x of final dose) tunicamycin dissolved in dimethyl sulfoxide (DMSO) directly 30mm (4mL) MYOB plates, which were seeded with OP50 bacteria two hours later. Plates were considered ready for use on the following day. A similar method was also used for tunicamycin+RNAi plates, except that the MYOB plates also contained carbenicillin (25 micrograms/milliliter) (United States Biologicals) and 2 mM isopropyl-β-D-thio-galactoside (IPTG) (Roche Diagnostics). They were seeded with HT115 bacteria harboring various dsRNA-producing plasmids and kept at room temperature for 3 days prior to use (Timmons et al., 2001). The smd-1 RNAi was used as a “negative” control for RNAi phenotypes (Peters et al., 2010). Tunicamycin was obtained from Sigma. Young adult hermaphrodites could lay eggs for 3 hours and then removed. The resulting progeny were incubated at 20°C for three days at which time their developmental state was scored visually using a Nikon SMZ1500 light stereomicroscope with a HR Plan Apo 1x WD54 objective.
Affymetrix microarray analysis.
Following overnight synchronization and 5-hour exposure to tunicamycin, wild-type and tm3603 L1 larvae were pelleted on ice. RNA was extracted from the worm pellets using Trizol, precipitated with isopropanol, and resuspended in RNAase-free water. Microarray analysis was carried out at the Microarray Core at NIDDK. The probes were normalized using Robust Multi-Array Average (RMA) and fold-changes and p-values were calculated for pairwise comparisons (wild-type DMSO vs. tunicamycin; tm3603 DMSO vs. tunicamycin; wild-type DMSO vs. tm3603 DMSO). Microarray data have been deposited in the National Center for Biotechnology Information’s GEO and are accessible through GEO series accession number GSE118294. For each trial, all genes with p < 0.05 and fold change > 1.3 in at least one probe were subjected to Gene Ontology pathway analysis using High-Throughput GoMiner (Zeeberg et al., 2005). Because most pathways involve both positive and negative regulators of the pathway, we chose to focus on pathways enriched from changed genes (both up- and down-regulated genes).
Larval growth assays on pathogenic bacteria.
For larval growth, thin NGM agar plates (2 mL agar/35mm dish) were prepared (3 g/L NaCl, 2.5 g/L bacto-peptone, and 20 g/L bacto-agar); they were autoclaved and then cooled to 55°C before adding to each liter the following: 25 mL 1M potassium phosphate buffer, pH 6.0, 1 mL 1M MgSO4, 1 mL CaCl2, 1 mL cholesterol (10 mg/mL), 5 mL nystatin (1.25 mg/mL in ethanol)). After being allowed to solidify, plates were stored at 4°C until use. For experiments, bacteria from a frozen glycerol stock was spread on a LB plate and allowed to grow overnight. Colony plates were stored at 4°C and used for one week before a new plate was prepared. Fresh overnight cultures were prepared by placing 3–4 colonies into a 4 mL overnight LB culture for 16 hours. After 16 hours, room-temperature NGM plates were seeded with 50 μL bacteria, which was spread over the entire plate, and then placed at 37°C for 24 hours. The plates were equilibrated to room temperature for 1 hour, during which time synchronized eggs were obtained by bleaching 2-day gravid adult hermaphrodites. Egg density in liquid was determined by counting 10 μL drops under the dissecting microscope; 30 eggs were placed on each plate and the plates were kept at 20°C. Images were taken every 24 hours using a Keyence BZ-X700 microscope. Worm area was determined using the Wand selection tool in ImageJ.
Statistical analysis.
All statistical tests were performed in GraphPad Prism 7.04. For lifespan experiments, because we observed an early die-off in every replicate, the Gehan-Breslow-Wilcoxon test was used, which provides extra weight for early time points. For quantified immunoblot and qPCR analysis of hsp-4 induction, tunicamycin growth experiments, and growth on pathogenic bacteria, two-way ANOVA tests were used to compare treatment and strain effects.
Supplementary Material
Acknowledgements.
The manf-1(tm3603) allele was provided by Shohei Mitani at the National BioResource Project (Japan). Other strains used in this study were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the Intramural Research Program at the National Institute on Drug Abuse and by NIEHS F32ES027306 (JHH). The authors would like to thank Andy Golden (NIDDK/NIH) for early support and feedback, Alejandro Aballay for providing the pathogenic bacterial strains, and Luiza Perez for assisting with pathogenic bacterial exposures. The authors would also like to thank Eric Hastie for injecting the CRISPR/Cas9 construct into the wild-type animals.
Abbreviations.
- MANF
mesencephalic astrocyte-derived neurotrophic factor
- CDNF
cerebral dopamine neurotrophic facto
- UPR
unfolded protein response
Footnotes
Conflict of Interest. The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aballay A, Yorgey P, Ausubel FM, 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Current Biology 10, 1539–1542. [DOI] [PubMed] [Google Scholar]
- Airavaara M, Chiocco MJ, Howard DB, Zuchowski KL, Peranen J, Liu C, Fang S, Hoffer BJ, Wang Y, Harvey BK, 2010. Widespread cortical expression of MANF by AAV serotype 7: localization and protection against ischemic brain injury. Experimental neurology 225, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airavaara M, Shen H, Kuo CC, Peranen J, Saarma M, Hoffer B, Wang Y, 2009. Mesencephalic astrocyte-derived neurotrophic factor reduces ischemic brain injury and promotes behavioral recovery in rats. The Journal of comparative neurology 515, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou A, Shen Y, Liang Y, Luo J, Fang S, 2008. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Experimental cell research 314, 2454–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Simmen T, 2016. ER-luminal thiol/selenol-mediated regulation of Ca2+ signalling. Biochemical Society transactions 44, 452–459. [DOI] [PubMed] [Google Scholar]
- Au - Nussbaum-Krammer CI, Au - Neto MF, Au - Brielmann RM, Au - Pedersen JS, Au - Morimoto RI, 2015. Investigating the Spreading and Toxicity of Prion-like Proteins Using the Metazoan Model Organism C. elegans. JoVE, e52321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Vozdek R, Hnizda A, Jiang C, Wang B, Kuchar L, Li T, Zhang Y, Wood C, Feng L, Dang Y, Ma DK, 2018. Conserved roles of C. elegans and human MANFs in sulfatide binding and cytoprotection. Nat Commun 9, 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell MC, Meier SE, Ingram AL, Abisambra JF, 2016. PERK-opathies: An Endoplasmic Reticulum Stress Mechanism Underlying Neurodegeneration. Current Alzheimer research 13, 150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham DP, Hardaway JA, Snarrenberg CL, Robinson SB, Folkes OM, Salimando GJ, Jinnah H, Blakely RD, 2016. Acute Blockade of the C. elegans Dopamine Transporter DAT-1 by the Mammalian Norepinephrine Transporter Inhibitor Nisoxetine Reveals the Influence of Genetic Modifications of Dopamine Signaling In Vivo. Neurochemistry international 98, 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D, 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Chen L, Feng L, Wang X, Du J, Chen Y, Yang W, Zhou C, Cheng L, Shen Y, Fang S, Li J, Shen Y, 2015. Mesencephalic astrocyte-derived neurotrophic factor is involved in inflammation by negatively regulating the NF-kappaB pathway. Scientific reports 5, 8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F, 2013. IRE1: ER stress sensor and cell fate executor. Trends in cell biology 23, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Sundvik M, Rozov S, Priyadarshini M, Panula P, 2012. MANF regulates dopaminergic neuron development in larval zebrafish. Developmental biology 370, 237–249. [DOI] [PubMed] [Google Scholar]
- Cordero-Llana O, Houghton BC, Rinaldi F, Taylor H, Yanez-Munoz RJ, Uney JB, Wong LF, Caldwell MA, 2015. Enhanced efficacy of the CDNF/MANF family by combined intranigral overexpression in the 6-OHDA rat model of Parkinson’s disease. Molecular therapy : the journal of the American Society of Gene Therapy 23, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L-L, Gao J-X, Zou C-G, Ma Y-C, Zhang K-Q, 2015. mir-233 Modulates the Unfolded Protein Response in C. elegans during Pseudomonas aeruginosa Infection. PLOS Pathogens 11, e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haes W, Frooninckx L, Van Assche R, Smolders A, Depuydt G, Billen J, Braeckman BP, Schoofs L, Temmerman L, 2014. Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2. Proc Natl Acad Sci U S A 111, E2501–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B, 2015. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Schumacher B, 2014. Insights from the worm: The C. elegans model for innate immunity. Seminars in immunology 26, 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P, 2013. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harbor perspectives in biology 5, a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glembotski CC, 2011. Functions for the cardiomyokine, MANF, in cardioprotection, hypertrophy and heart failure. Journal of molecular and cellular cardiology 51, 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman M, Arumae U, Yu LY, Lindholm P, Peranen J, Saarma M, Permi P, 2011. Mesencephalic astrocyte-derived neurotrophic factor (MANF) has a unique mechanism to rescue apoptotic neurons. The Journal of biological chemistry 286, 2675–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MJ, Richie CT, Airavaara M, Wang Y, Harvey BK, 2013. Mesencephalic Astrocyte-derived Neurotrophic Factor (MANF) Secretion and Cell Surface Binding Are Modulated by KDEL Receptors. Journal of Biological Chemistry 288, 4209–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Pulendran B, Lambrecht BN, 2014. Emerging functions of the unfolded protein response in immunity. Nature immunology 15, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien-Gau I, Schmidt M, Kurz CL, 2014. f57f4.4p::gfp as a fluorescent reporter for analysis of the C. elegans response to bacterial infection. Developmental & Comparative Immunology 42, 132–137. [DOI] [PubMed] [Google Scholar]
- Kania E, Pajak B, Orzechowski A, 2015. Calcium homeostasis and ER stress in control of autophagy in cancer cells. BioMed research international 2015, 352794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulkin WJ, Hiester BG, Link CD, 2005. Compensatory regulation among ER chaperones in C. elegans. FEBS letters 579, 3063–3068. [DOI] [PubMed] [Google Scholar]
- Kozlowski L, Garvis S, Bedet C, Palladino F, 2014. The Caenorhabditis elegans HP1 family protein HPL-2 maintains ER homeostasis through the UPR and hormesis. Proc Natl Acad Sci U S A 111, 5956–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Danilova T, Palm E, Lindholm P, Voikar V, Hakonen E, Ustinov J, Andressoo JO, Harvey BK, Otonkoski T, Rossi J, Saarma M, 2014. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Rep 7, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Makela J, Di Liberto V, Mudo G, Belluardo N, Eriksson O, Saarma M, 2016. Current disease modifying approaches to treat Parkinson’s disease. Cellular and molecular life sciences : CMLS 73, 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm P, Voutilainen MH, Lauren J, Peranen J, Leppanen VM, Andressoo JO, Lindahl M, Janhunen S, Kalkkinen N, Timmusk T, Tuominen RK, Saarma M, 2007. Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nature 448, 73–77. [DOI] [PubMed] [Google Scholar]
- Lindstrom R, Lindholm P, Kallijarvi J, Palgi M, Saarma M, Heino TI, 2016. Exploring the Conserved Role of MANF in the Unfolded Protein Response in Drosophila melanogaster. PLoS One 11, e0151550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom R, Lindholm P, Kallijarvi J, Yu LY, Piepponen TP, Arumae U, Saarma M, Heino TI, 2013. Characterization of the structural and functional determinants of MANF/CDNF in Drosophila in vivo model. PLoS One 8, e73928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom R, Lindholm P, Palgi M, Saarma M, Heino TI, 2017. In vivo screening reveals interactions between Drosophila Manf and genes involved in the mitochondria and the ubiquinone synthesis pathway. BMC genetics 18, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM, 2004. ER chaperone functions during normal and stress conditions. Journal of chemical neuroanatomy 28, 51–65. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ, 2007. The endoplasmic reticulum and the unfolded protein response. Seminars in cell & developmental biology 18, 716–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margie O, Palmer C, Chin-Sang I, 2013. C. elegans chemotaxis assay. J Vis Exp, e50069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PW, Hardie SL, Jessen TN, Carvelli L, Matthies DS, Blakely RD, 2007. Vigorous motor activity in Caenorhabditis elegans requires efficient clearance of dopamine mediated by synaptic localization of the dopamine transporter DAT-1. J Neurosci 27, 14216–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi N, Hoseki J, Kubota H, Toyokuni S, Nozaki J, Naitoh M, Koizumi A, Nagata K, 2007. ARMET is a soluble ER protein induced by the unfolded protein response via ERSE-II element. Cell structure and function 32, 41–50. [DOI] [PubMed] [Google Scholar]
- Mohri-Shiomi A, Garsin DA, 2008. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. The Journal of biological chemistry 283, 194–201. [DOI] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM 3rd, Blakely RD, 2002. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci U S A 99, 3264–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norisada J, Hirata Y, Amaya F, Kiuchi K, Oh-hashi K, 2014. A sensitive assay for the biosynthesis and secretion of MANF using NanoLuc activity. Biochem Biophys Res Commun 449, 483–489. [DOI] [PubMed] [Google Scholar]
- Oakes SA, Papa FR, 2015. The role of endoplasmic reticulum stress in human pathology. Annual review of pathology 10, 173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC, 2013. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harbor perspectives in biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palgi M, Greco D, Lindstrom R, Auvinen P, Heino TI, 2012. Gene expression analysis of Drosophilaa Manf mutants reveals perturbations in membrane traffic and major metabolic changes. BMC genomics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palgi M, Lindstrom R, Peranen J, Piepponen TP, Saarma M, Heino TI, 2009. Evidence that DmMANF is an invertebrate neurotrophic factor supporting dopaminergic neurons. Proc Natl Acad Sci U S A 106, 2429–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkash V, Lindholm P, Peranen J, Kalkkinen N, Oksanen E, Saarma M, Leppanen VM, Goldman A, 2009. The structure of the conserved neurotrophic factors MANF and CDNF explains why they are bifunctional. Protein engineering, design & selection : PEDS 22, 233–241. [DOI] [PubMed] [Google Scholar]
- Peters N, Perez DE, Song MH, Liu Y, Muller-Reichert T, Caron C, Kemphues KJ, O’Connell KF, 2010. Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. Journal of cell science 123, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova P, Raibekas A, Pevsner J, Vigo N, Anafi M, Moore MK, Peaire AE, Shridhar V, Smith DI, Kelly J, Durocher Y, Commissiong JW, 2003. MANF: a new mesencephalic, astrocyte-derived neurotrophic factor with selectivity for dopaminergic neurons. Journal of molecular neuroscience : MN 20, 173–188. [DOI] [PubMed] [Google Scholar]
- Pinto S, Sato VN, De-Souza EA, Ferraz RC, Camara H, Pinca APF, Mazzotti DR, Lovci MT, Tonon G, Lopes-Ramos CM, Parmigiani RB, Wurtele M, Massirer KB, Mori MA, 2018. Enoxacin extends lifespan of C. elegans by inhibiting miR-34–5p and promoting mitohormesis. Redox biology 18, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisoni GB, Molinari M, 2016. Five Questions (with their Answers) on ER-Associated Degradation. Traffic (Copenhagen, Denmark) 17, 341–350. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, 2006. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol 351, 275–286. [DOI] [PubMed] [Google Scholar]
- Richardson CE, Kinkel S, Kim DH, 2011. Physiological IRE-1-XBP-1 and PEK-1 Signaling in Caenorhabditis elegans Larval Development and Immunity. PLOS Genetics 7, e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, Kim DH, 2010. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman C, Rashid S, Prashar S, Mishra R, Selvaganapathy PR, Gupta BP, 2018. C. elegans MANF Homolog Is Necessary for the Protection of Dopaminergic Neurons and ER Unfolded Protein Response. Frontiers in neuroscience 12, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano R, Reed JC, 2013. ER stress-induced cell death mechanisms. Biochim Biophys Acta 1833, 3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sem X, Rhen M, 2012. Pathogenicity of Salmonella enterica in Caenorhabditis elegans Relies on Disseminated Oxidative Stress in the Infected Host. PLOS ONE 7, e45417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA, 2002. Formation and transfer of disulphide bonds in living cells. Nature reviews. Molecular cell biology 3, 836–847. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ, 2005. Genetic Interactions Due to Constitutive and Inducible Gene Regulation Mediated by the Unfolded Protein Response in C. elegans. PLOS Genetics 1, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Aballay A, 2017. Endoplasmic Reticulum Stress Caused by Lipoprotein Accumulation Suppresses Immunity against Bacterial Pathogens and Contributes to Immunosenescence. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Hughes BL, Osborn DW, Boudreau ED, Shaw KM, Warren CE, 2009. Modeling a congenital disorder of glycosylation type I in C. elegans: a genome-wide RNAi screen for N-glycosylation-dependent loci. Glycobiology 19, 1554–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M-W, Mahajan-Miklos S, Ausubel FM, 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America 96, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. [DOI] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D, 2002. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. The Journal of Cell Biology 158, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutilainen MH, Arumae U, Airavaara M, Saarma M, 2015. Therapeutic potential of the endoplasmic reticulum located and secreted CDNF/MANF family of neurotrophic factors in Parkinson’s disease. FEBS letters 589, 3739–3748. [DOI] [PubMed] [Google Scholar]
- Voutilainen MH, Back S, Porsti E, Toppinen L, Lindgren L, Lindholm P, Peranen J, Saarma M, Tuominen RK, 2009. Mesencephalic astrocyte-derived neurotrophic factor is neurorestorative in rat model of Parkinson’s disease. J Neurosci 29, 9651–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hou C, Cao Y, Cheng Q, Zhang L, Li H, Feng L, Shen Y, 2018. XBP1 activation enhances MANF expression via binding to endoplasmic reticulum stress response elements within MANF promoter region in hepatitis B. The International Journal of Biochemistry & Cell Biology 99, 140–146. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang L, Wu Y, Huang C, Wang Q, Han J, Guo Y, Shi X, Zhou B, 2015. The developmental neurotoxicity of polybrominated diphenyl ethers: Effect of DE-71 on dopamine in zebrafish larvae. Environ Toxicol Chem 34, 1119–1126. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH, 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Yan W, Liu C, Li L, Yu L, Zhao S, Li G, 2016. Microcystin-LR exposure induces developmental neurotoxicity in zebrafish embryo. Environmental pollution (Barking, Essex : 1987) 213, 793–800. [DOI] [PubMed] [Google Scholar]
- Yu YQ, Liu LC, Wang FC, Liang Y, Cha DQ, Zhang JJ, Shen YJ, Wang HP, Fang S, Shen YX, 2010. Induction profile of MANF/ARMET by cerebral ischemia and its implication for neuron protection. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 30, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, Elnekave E, Hari DM, Wynn TA, Cunningham-Rundles C, Stewart DM, Nelson D, Weinstein JN, 2005. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC bioinformatics 6, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GL, Wang LH, Liu XY, Zhang YX, Hu MY, Liu L, Fang YY, Mu Y, Zhao Y, Huang SH, Liu T, Wang XJ, 2018a. Cerebral Dopamine Neurotrophic Factor (CDNF) Has Neuroprotective Effects against Cerebral Ischemia That May Occur through the Endoplasmic Reticulum Stress Pathway. International journal of molecular sciences 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shen Y, Luo H, Zhang F, Jing L, Wu Y, Xia X, Song Y, Li W, Jin L, 2018. b. MANF protects dopamine neurons and locomotion defects from a human alpha-synuclein induced Parkinson’s disease model in C. elegans by regulating ER stress and autophagy pathways. Experimental neurology 308, 59–71. [DOI] [PubMed] [Google Scholar]
- Zhao H, Liu Y, Cheng L, Liu B, Zhang W, Guo YJ, Nie L, 2013. Mesencephalic astrocyte-derived neurotrophic factor inhibits oxygen-glucose deprivation-induced cell damage and inflammation by suppressing endoplasmic reticulum stress in rat primary astrocytes. Journal of molecular neuroscience : MN 51, 671–678. [DOI] [PubMed] [Google Scholar]
- Zhu W, Li J, Liu Y, Xie K, Wang L, Fang J, 2016. Mesencephalic astrocyte-derived neurotrophic factor attenuates inflammatory responses in lipopolysaccharide-induced neural stem cells by regulating NF-kappaB and phosphorylation of p38-MAPKs pathways. Immunopharmacology and immunotoxicology 38, 205–213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.