Abstract
Long Evans rat strains are applied as research models in a broad spectrum of biomedical fields (>15,800 citations, NCBI PubMed). Here, we report an approach to genetically modify the Long Evans rat germline in donor spermatogonial stem cells. Long Evans rat spermatogonial lines were derived from freshly isolated laminin-binding spermatogonia. Laminin-binding spermatogonia were cultured over multiple passages on fibroblast feeder layers in serum-free culture medium containing GDNF and FGF2. Long Evans rat spermatogonial lines were genetically modified by transposon transduction to express a germline, tdTomato reporter gene. Donor rat spermatogonial lines robustly regenerated spermatogenesis after transplantation into testes of busulfan-treated, allogenic, Long Evans rats. Donor-derived spermatogenesis largely restored testis size in the chemically sterilized, recipient Long Evans rats. Recipient Long Evans rats stably transmitted the tdTomato germline marker to subsequent generations. Overall, Long Evans rat spermatogonial lines provided effective donor germline vectors for genetically modifying Long Evans rats.
Keywords: Spermatogonial stem cells, transgenic rats, germline editing, allogeneic transplantation, drug addiction, drug abuse
Introduction
Rat research models have been established to study a broad spectrum of human health issues (NCBI PubMed listings/species: rats ~1.55e6, mice ~1.39e6, rabbits ~0.34e6, guinea pigs ~0.14e6, hamsters ~0.11e6). Hundreds of inbred and outbred rat strains have been generated through selective breeding and genome mutagenesis (Aitman et al. 2008; Atanur et al. 2013; Lazar et al. 2005). The Rat Genome Database (RGD) provides a comprehensive resource on rat genetic diversity, and has accumulated informational resources on rat biology over the last 14 years (Laulederkind et al. 2013; Petri et al. 2014). Most recently, numerous breakthroughs in genome engineering have revolutionized the potential to create transgenic rats (Geurts et al. 2009; Izsvak et al. 2010; Ma et al. 2014; Tong et al. 2010).
Expanding genome editing capabilities into additional rat strains (and additional model organisms) will allow researchers to study gene function on genetic backgrounds most relevant to scientific goals. In a primary example, Long Evans rat strains are becoming popular for studying substance abuse due to their robust discriminative responses to self-administered, addictive compounds (Yamamoto et al. 2013). Accordingly, researchers at the National Institute on Drug Addiction (NIDA) have built the most assorted resource of neuron-specific Cre recombinase and tetracycline-responsive transactivation rats on the Long Evans genetic background (NIDA IRP Rat Project). These unique Long Evans “gene deleting” and “transgene-inducing” rat strains will facilitate controllable gene regulation during studies on addictive behavior, and a diversity of other biological processes.
Here, to advance technologies for genetically modifying Long Evans rats using donor germline stem cells, we derived fully functional, genetically modifiable Long Evans rat spermatogonial stem cell lines and used them to produce transgenic rats.
Results
Deriving and Sub-Culturing Long Evans Rat Spermatogonial Lines
Potent Sprague Dawley rat spermatogonial lines have been successfully derived from laminin-binding spermatogonia using a simplified, serum free spermatogonial culture medium (SG Medium) (Wu et al. 2009). Here, a formulation of the original SG Medium containing reduced concentrations of GDNF and FGF2 (Chapman et al. 2015) was applied to derive Long Evans rat spermatogonial lines from laminin binding spermatogonia.
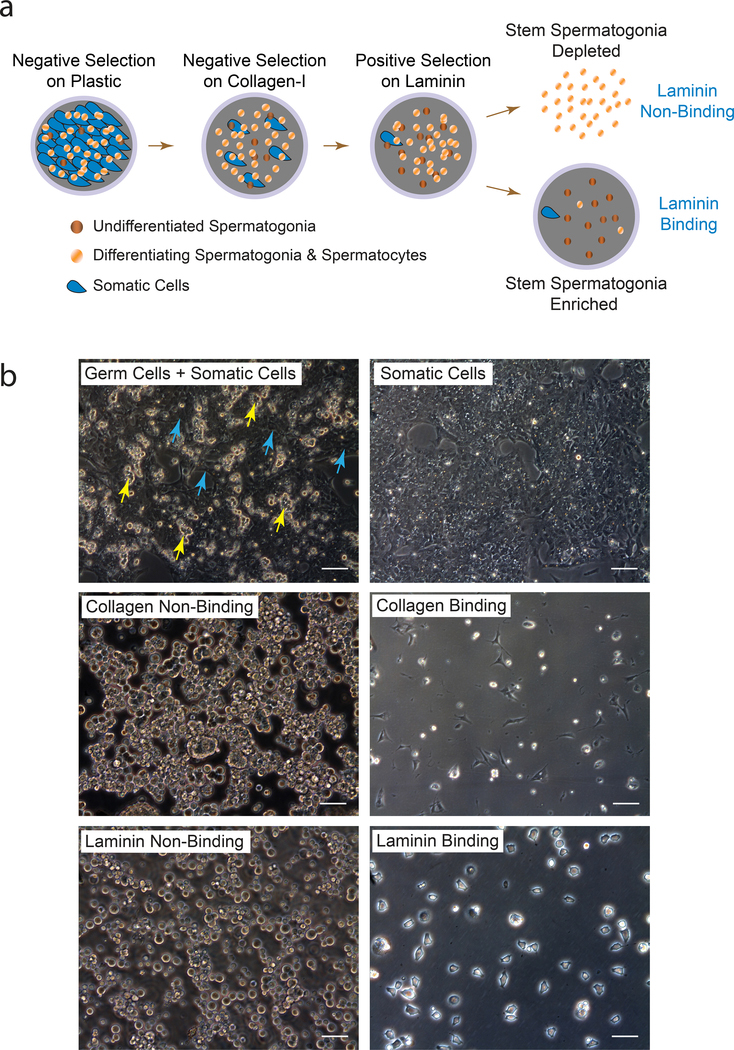
Laminin binding spermatogonia were isolated from postnatal d23 Long Evans rat testes by first disaggregating seminiferous tubules with collagenase, type 1 (pooled testes from 4 rats). The testis cell suspension was then processed by negative selection in culture on plastic and collagen-I, following by positive selection on laminin (Fig. 1a) (Hamra et al. 2002). Accordingly, spermatogenic cells in primary Long Evans rat testis cell suspensions were depleted of somatic cells by negative selection during culture on plastic and collagen-I-coated dishes in serum containing medium (Fig. 1b). The harvested collagen non-binding testis cell fractions were next used to select for cultures of laminin-binding spermatogonia in SG Medium (Fig. 1b).
Figure 1. Isolating Long Evans rat laminin-binding spermatogonia.
a) Matrix selection method used to isolate Long Evans rat spermatogonial stem cells in culture. Germ cells were harvested from Somatic Cells bound to plastic by gentle pipetting, and plated onto collagen-I for ~4 hr. Collagen Non-Binding germ cells were harvested and incubated on laminin ~40 min to collect laminin Non-Binding (LNB) and Laminin-Binding (LB) spermatogenic cell fractions.
b) Primary cultures of Long Evans rat testis cells (Germ Cells + Somatic Cells) isolated from 23 day old rats as described in panel “a”. Yellow and cyan arrows point to Spermatogenic-like (Germ Cells) and somatic-like testis cells (Somatic Cells), respectively. Scale, 40 μm
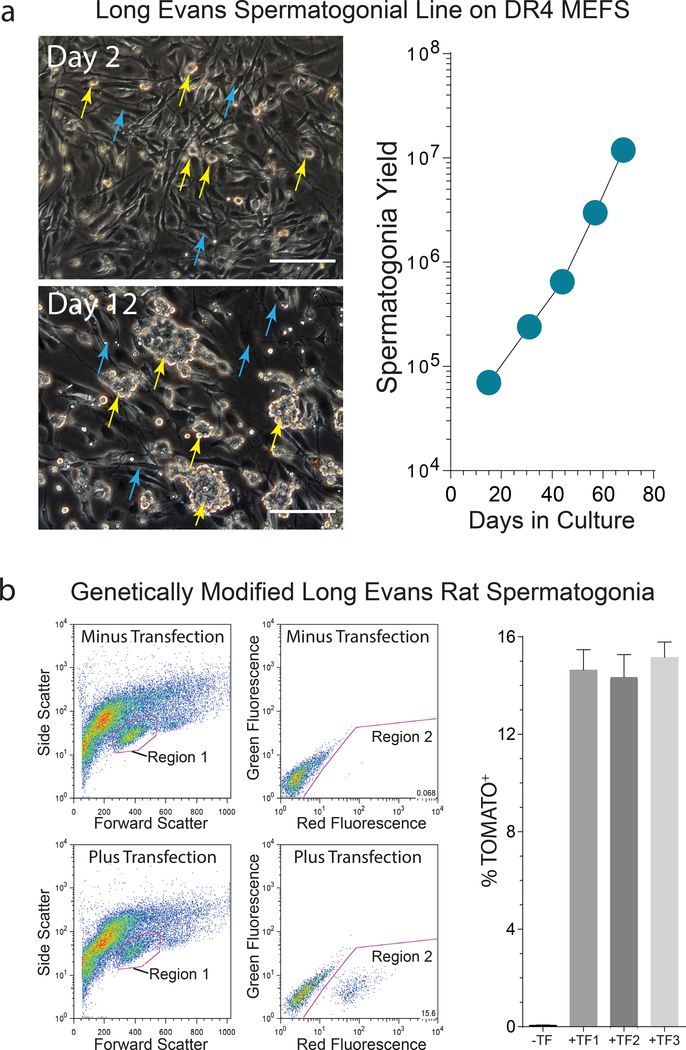
Long Evans rat laminin-binding spermatogonia were further depleted of somatic testis cells by 3 passages on gelatin-coated plates in SG Medium, prior to continuously sub-culturing the derived spermatogonial lines on DR4 mouse embryonic fibroblasts (MEFs) in SG Medium (Fig. 2a). In a subsequent experiment, individual Longs Evans rat spermatogonial lines were derived from separate d23 rats, yielding a mean double time of 6.4±0.7 days between passages 5–8 (±SD, n=4 spermatogonial lines). Thus, the SG Medium effectively supported derivation of Long Evans rat spermatogonial lines.
Figure 2. Long Evans rat spermatogonial sub-culture and genetic modification.
a) Left: Laminin-Binding spermatogenic cells illustrated in Figure 1 were used to derive Long Evans rat spermatogonial lines that develop into colonies (yellow arrows) on MEF feeder layers (cyan arrows). Scale, 40 μm. Right: Propagation of a Long Evans rat spermatogonial line during sub-culture on MEFs. Graph depicts spermatogonial yield following passages 1–5 on MEFs.
b) Flow analysis of a Long Evans rat spermatogonial line following transfection with pSBDazl-tdTomato (SB transposon) + pSBM3A (SB transposase) plasmids. Left: Red Fluorescence intensity (Region 2) in cells gated from Region 1 in adjacent forward and side scatter plots following transfection with pSBDazl-tdTomato + pSBM3A plasmids (Plus Transfection). Untransfected spermatogonia from the same culture were used to define Region 2 (Minus Transfection). Right: percent tdTomato+ cells in Region 2 scatter plots at passage 4 following co-transfection with pSBDazl-tdTomato and pSBM3A (+TF; ±SEM, n=3 transfections/spermatogonial line) -TF, Minus Transfection culture used to determine background fluorescence.
Genetically Modifying Long Evans Rat Spermatogonial Lines
Long Evans rat spermatogonial lines were evaluated for their ability to be transfected and genetically modified with donor DNA constructs. Neon co-transfection (Chapman et al. 2015) was used to deliver a mixture of DNA plasmids into Long Evans rat spermatogonial lines at passage 4 on MEFs. The co-transfection mixture included a selectable marker plasmid conferring G418 resistance (pPGK-Neo), a Sleeping Beauty transposase plasmid (pSB-M3A) and a Sleeping Beauty transposon plasmid harboring a red fluorescent germline reporter transgene as cargo (pSB-Dazl-tdTomato). The same Sleeping Beauty transposon system, harboring a β-galactosidase-neomycin-phosphotransferase fusion gene trap reporter as cargo (pSB-β-Geo trap), was initially reported to effectively stably transduce Sprague Dawley rat spermatogonial stem cell lines (Ivics et al. 2011a; Ivics et al. 2011b; Izsvak et al. 2010). And, the ~1.7 kb mouse Deleted in Azoospermia (Dazl) promoter fragment was originally reported to drive expression of a green fluorescent germline reporter transgene in mice (Nicholas et al. 2009).
Co-transfected spermatogonial lines were maintained for 3 days in SG Medium prior to selection in SG Medium containing 65 μg/ml G418 for 6 days, followed by continuous sub-culture under standard conditions in SG Medium between passages 5–11. Flow analyses on co-transfected spermatogonial lines at passage 7 detected the Tomato germline reporter in 14.6±0.4% of the cultured cells (n=3 transfections/line; n=3 spermatogonial lines, ±SEM) (Fig. 2b). The stably integrated, Dazl-tdTomato germline transgene provided a vital marker for measuring the relative germline stem cell activity of Long Evans rat spermatogonial lines.
Preparing Long Evans Rat Spermatogonial Recipients
Prior to testing the germline stem cell activity of Long Evans rat spermatogonial lines, it was necessary to prepare male-sterile recipient rats with testes that could be effectively colonized by donor stem spermatogonia. In an initial report, the ability of donor Sprague Dawley rat spermatogonial stem cells to colonize recipient testes was largely dependent on clearing endogenous spermatogonia from germline stem cell niches localized within the basal compartment of the seminiferous epithelium (Ogawa et al. 1999).
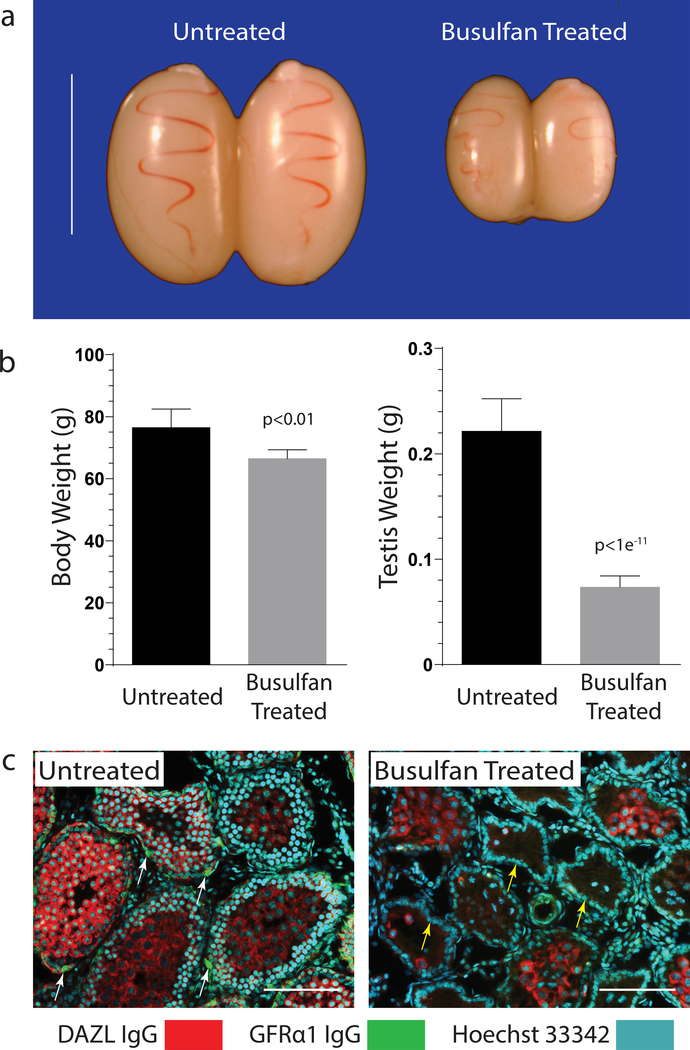
Here, allogeneic recipients for donor Long Evans rat spermatogonial lines were prepared by using busulfan to deplete endogenous spermatogenesis in pre-pubertal Long Evans rats. Groups of Long Evans rats were treated by intra-peritoneal injections with 0 mg/kg (n=4), 10.5 mg/kg (n=4), 11.5 mg/kg (n=4) and 12.5 (n=5) mg/kg busulfan at 12 days of age, prior to analyzing testes and body growth at 26–27 days of age (Fig. 3).
Figure 3. Recipient Long Evans rat preparation.
a) Testes from 24 day old Long Evans rats treated at 12 days of age with 12.5 mg/kg busulfan (Busulfan Treated) compared to untreated littermate control rats (Untreated).
b) Body and testis weights in Untreated and Busulfan Treated Long Evans rats described in panel “a”. ±SD, n=4 rats/group.
c) Immunofluorescence labeling for total spermatogenic cells (DAZL IgG, red cytoplasmic marker), type A spermatogonia (GFRα1 IgG, green cytoplasmic marker; white arrows) and total cell nuclei (Hoescht 33342, cyan nuclear marker) in testis cross sections from representative Untreated and Busulfan Treated 24 day old rats described in panel “a”. Note: yellow arrows point to Sertoli cell nuclei lining the basal compartment of the seminiferous epithelium. Scale, 200 μm
All rats treated with busulfan survived until the time of analysis. Mean testis weight/rat (±SD) was reduced 65–70% in busulfan-treated groups (10.5, 11.5, 12.5 mg/kg busulfan = 93±7, 83±7, 72±8 mg testis wt), as compared to an untreated group (0 mg/kg busulfan = 233±39 mg testis wt) (Fig. 3a). The highest busulfan dose tested selectively reduced testis weight in Long Evans rats (p<1e−11), as compared to a smaller % reduction in body weight (p<0.01) (Fig. 3b).
Reduced testis weight in busulfan treated groups was associated with loss of most GFRα1+ and GFRα1− spermatogenic cells (DAZL+) within seminiferous tubules. Notably, GFRα1+ type A spermatogonia containing the undifferentiated spermatogonial populations were depleted from the seminiferous epithelium (Fig. 3c). In contrast, untreated rat testes displayed normal populations of type A spermatogonia (DAZL+, GFRα1+), plus more differentiated spermatogonial and spermatocyte types (DAZL+, GFRα1−) (Fig. 3c). Several tubules in busulfan-treated rats contained residual populations of pachytene spermatocytes that presumably developed from busulfan-resistant, differentiating spermatogonia/early spermatocytes present in d12 rats (Fig. 3c).
The basal compartment of the seminiferous epithelium remained circumferentially lined by Sertoli cells in the busulfan-treated rats (Fig. 3c). Sertoli cells are a critical component of germline stem cell niches in rodents (Oatley et al. 2011). Overall, busulfan treatment effectively depleted spermatogenic cells, including GFRα1+ spermatogonia, from pre-pubertal Long Evans rat testes.
Donor germline transmission from genetically modified Long Evans spermatogonial lines
Based on the ability to genetically modify Long Evans rat spermatogonial lines with a germline transgene marker (Fig. 2), and the effectiveness of busulfan for depleting spermatogenesis in Long Evans rats (Fig. 3), we next tested the sperm-forming potential of donor Long Evans rat spermatogonial lines following genetic modification in culture and transplantation into allogeneic hosts (Fig. 4, Table 1).
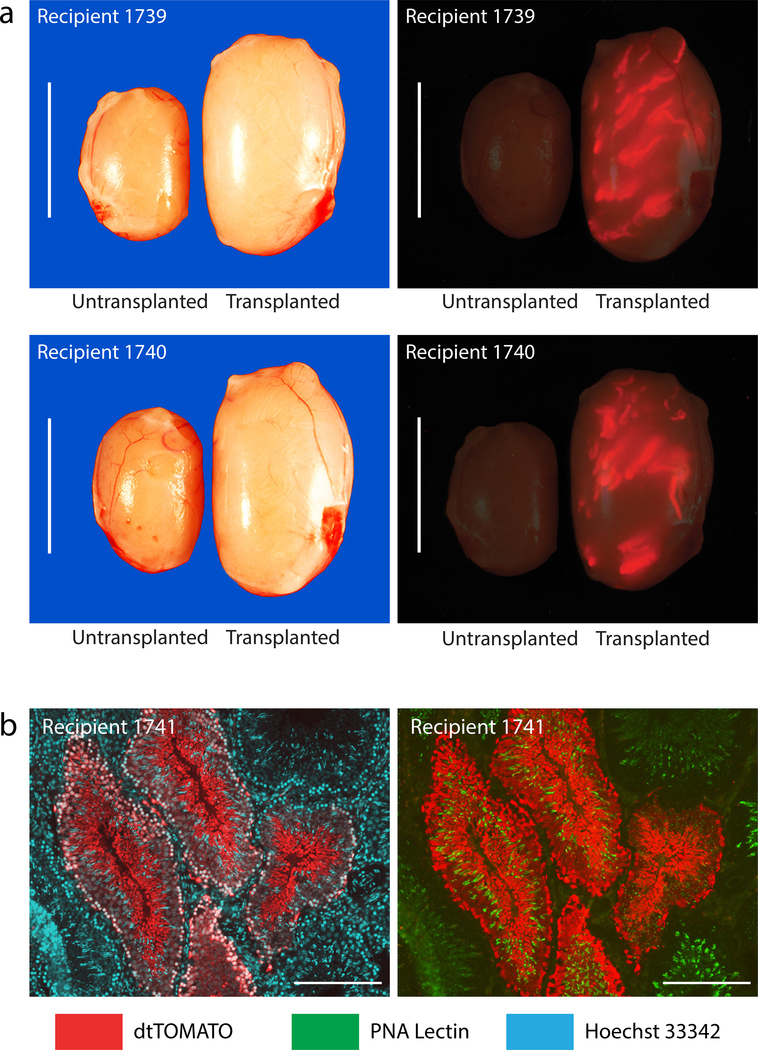
Figure 4. Donor derived spermatogenesis in allogenic Long Evans rat recipients.
a) Testes from Long Evans rats at 7–8 mo after being transplanted with a Long Evans rat spermatogonial line at passage 5 following genetic modification with pSBDazl-tdTomato (donor spermatogonia population contained ~14.5% tdTomato+ cells). Note: Red, tdTomato+ seminiferous tubules in Transplanted testes, but not in Untransplanted testes from Recipients 1739–1740. Scale, 1 cm.
b) Donor derived spermatogenesis in a testis section prepared from Recipient 1741 at ~8 mo post-transplantation (also see Supplementary Figure 1). Note the development of elongated tdTomato+ spermatids (Red, cytoplasmic germ cell marker) co-labeled with Hoeschet 33342 (Cyan, nuclear marker; left panel) and PNA (Green, acrosome marker; right panel). Also note full spermatogenesis in seminiferous tubules colonized with tdTomato− cells. Scale 200 μm.
Table 1.
Transgenic Long Evans rats produced using donor spermatogonial stem cell lines
| Generation | Breeders† | Litters | Pups | F:M | DTB | # Tg∥ pups | % Tg pups | Estimated Donor Derived Pups (%)‡ |
|---|---|---|---|---|---|---|---|---|
| F0 Recipient Founder Males | R1739 | 2 | 19 | 58:42 | 125 | 1 | 5.3 | 54–73 |
| R1740 | 2 | 27 | 63:37 | 122 | 2 | 7.4 | 76–100 | |
| R1741 | 2 | 13 | 51:49 | 147 | 1 | 7.7 | 79–100 | |
| Totals | 6 | 59 | 57:43* | 131* | 4 | 6.8 | 70–93 | |
| F1 Transgenic Females | tg1739-1-4 | 1 | 11 | 54:46 | 218 | 5 | 45 | n.a. |
| tg1740-1-2 | 2 | 24 | 42:58 | 210 | 19 | 79 | n.a. | |
| tg1741-2-6 | 1 | 16 | 56:44 | 241 | 9 | 56 | n.a. | |
| Totals | 4 | 51 | 51:49* | 223* | 33 | 65 | n.a. | |
DTB, days from transplantation to first litter born containing transgenic animals; F, female progeny; M, male progeny; Tg, Transgenic
Number tgSB-Dazl-dtTomato transgenic pups. Note: all 4 F1 transgenic pups were female.
Average value; n.a., not applicable
R(n), Recipient Founder male identifier; tg(n-n-n), F1 transgenic female identifier; F0 Recipient Founder × wildtype female and transgenic F1 female × wildtype male rat crosses were conducted on Long Evans rat genetic backgrounds.
Assumes 1–2 transposon copies inherited/transgenic F1 pup. Estimated donor derived progeny = (% Tg pups × 2 meiotic dilution factor) / %TOMATO+ donor cells when assuming 1 transposon copy integrated/transmitting transgenic spermatozoon. Meiotic dilution factor = 1.5 when assuming 2 independently segregating transposon copies integrated/transmitting transgenic spermatozoon.
A Long Evans rat spermatogonial culture (~14.5% Tomato+ cells) derived from the transfection experiment described in Figure 3b was transplanted into busulfan-treated Long Evans rat seminiferous tubules at passage 11 (n=5 recipients receiving 12.5 mg/kg dose). Three of 5 busulfan-treated rats were successfully transplanted with greater than 50% of seminiferous tubule surface area/testis filled by the donor spermatogonial suspension in a least one testis/rat (monitored by trypan blue dye). The 3 recipients receiving the most successful spermatogonial injections (R1739, R1740, R1741) were paired with wildtype female Long Evans rats at D60 post-transplantation, yielding transgenic progeny between D122-D147 post-transplantation (Table 1).
Transgenic F1 progeny (tgDazl-tdTomato rats) were produced from each recipient-founder (F0) at rates consistent with the abundance of tdTomato+ cells in the donor spermatogonial population (Fig. 3, Table 1). Recipient breeders were maintained for up to 7–8 months prior to analyzing their testes (Fig. 4). Transplanted testes weighed 2–3 times more than contralateral, un-transplanted testes (Fig. 4, Table 1), and robustly supported spermatozoa production by the donor cell population (Fig. 4, Supplementary Fig. S1, Table 1).
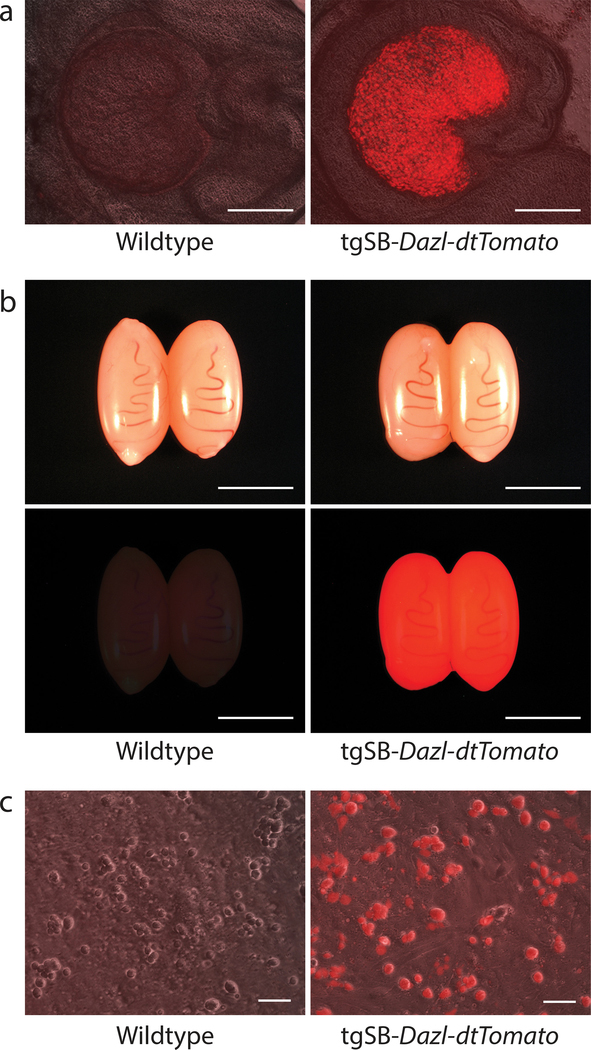
Newly produced transgenic rat strains stably transmitted the tgSB-Dazl-tdTomato transgene to F2 progeny at Mendelian rates, consistent with 1–2 transposon copies/transgenic strain (Table 1). Male and female F2 progeny born from transgenic F1 strains displayed vibrant tdTomato fluorescence in their gonads (Fig. 5a, b). Primary testis cell cultures prepared from F2 progeny born from each transgenic strain selectively expressed the red fluorescent tdTomato reporter in spermatogenic cells (Fig. 5c). Thus, the tgSB-Dazl-tdTomato transgene stably integrated into Long Evans rat germlines during spermatogonial culture in vitro, providing a robust marker to measure germline transmission rates achieved by donor Long Evans rat spermatogonial lines.
Figure 5. Transgenic Long Evans Rat Testes Express tgSB-Dazl-tdTomato.
a) Ovaries from F2 wildtype and transgenic (tgSB-Dazl-tdTomato) Long Evans rat littermates generated by mating a F1 tgSBDazl-tdTomato female rat with a wildtype male Long Evans rat. Scale, 500 μm
b) Testes from F2 wildtype and transgenic (tgSBDazl-tdTomato) Long Evans rat littermates generated by mating a F1 tgSBDazl-tdTomato female rat with a wildtype male Long Evans rat. Scale, 0.5 cm
c) Primary testis cell cultures derived from F2 wildtype and transgenic tgSBDazl-tdTomato Long Evans rats. Note, tdTomato fluorescence is selectively detected in spermatogenic-like cells. Scale, 50 μm
Discussion
Here, Long Evans rat spermatogonial lines were derived from primary cultures of laminin-binding spermatogonia that displayed long-term sperm forming potential following sub-culture and genetic manipulation on MEFs in SG Medium. Long Evans rat spermatogonial lines were propagated with doubling times ~2 days faster than reported for Sprague Dawley rat spermatogonial lines initially derived in the original SG Medium (Wu et al. 2009).
Long Evans rat spermatogonial stem cell lines effectively transmitted donor germline haplotypes from recipients to F1 progeny by natural mating. Given estimates that ~14–15% of the donor Long Evans rat spermatogonial population stably expressed the tgSB-Dazl-tdTomato germline marker (Fig. 2b), and that ~6.8% of F1 progeny fathered by F0 recipient-founders inherited the tgSB-Dazl-tdTomato genetic marker (4 of 59 F1 pups; n=8 litters), it is estimated that between 50–100% of the F1 progeny were derived from donor germlines (i.e. estimating a 2-fold allelic dilution by meiosis and 1–2 transposon copies/transgenic rat strain).
Estimated donor germline transmission rates achieved by Long Evans rat spermatogonial stem cell lines are consistent with their ability to robustly regenerate testis growth in busulfan treated rats (Fig. 4a). Similarly, Sprague Dawley rat spermatogonial lines that were sub-cultured on MEFs in SG Medium and clonally expanded in G418-containing SG Medium were previously reported to display robust regenerative potential in busulfan-treated, Sprague Dawley rat testes (i.e. >70% donor germline chimerism) (Chapman et al. 2015). In future studies, the tgSB-Dazl-tdTomato Long Evans rat strain can be used to study donor spermatogonial colonization efficiency and donor germline transmission rates by providing genetically pure spermatogonial cultures that uniformly express the tdTomato germline marker.
The ability to produce transgenic Long Evans rats from genetically modified donor spermatogonia reported herein, complements a previous report demonstrating donor spermatogonial stem cells within Long Evans rat total testis cell suspensions (Zhang et al. 2003). Stem spermatogonia in Long Evans rat testis cell suspensions developed into functional donor-derived spermatozoa after being transplanted into busulfan-treated, adult, Sprague Dawley rat testes (Zhang et al. 2003). However, adult recipient Sprague Dawley rats in the former report required extensive protocols that included a series of cyclosporine, testosterone and follicle stimulating hormone treatments before and after donor cell transplantation (Zhang et al. 2003). Here, highly pure Long Evans rat spermatogonial lines were more simply transplanted into young, busulfan-treated, allogenic recipients that robustly regenerated spermatogenesis and produced new transgenic Long Evans rat strains.
Transgenic Long Evans rat production efficiency using donor spermatogonial stem cells and transposon transgenes is similar to efficiencies for generating Long Evans transgenic rats by pronuclear injection methods (Cifani et al. 2012), and to methods that introduce transgenes into rodent early embryos using transposon systems and site-specific integrases (Furushima et al. 2012; Katter et al. 2013; Kitada et al. 2007; Lu et al. 2007; Ohtsuka et al. 2012). Based on several reported target alleles, site-specific transgenic knockin mice and rats can also be generated at similar efficiencies by CRISPR/Cas9-mediated homology directed repair in donor early embryos or pluripotent cell lines (Jung et al. 2016; Li et al. 2013; Qiu et al. 2013; Yang et al. 2013).
A primary advantage of donor spermatogonia-mediated gene transfer is that it directly modifies the germline, and therefore, eliminates production, genotyping and breeding of mosaic and/or chimeric transgenic progeny (Hamra et al. 2002; Nagano et al. 2001; Yen et al. 2014). Recently, targeted CRISPR/Cas9-mediated genomic modifications in donor spermatogonial stem cell lines were effectively transmitted to Sprague-Dawley rats (Chapman et al 2015). With the successful production of Long Evans spermatogonial stem cells described herein, future studies will employ CRISPR/Cas9 to modify genes in Long Evans rats, including targeted insertion of human gene variants linked to disease processes. Long Evans rat spermatogonial lines will also be used to create more complex, cell type-dependent recombinase expressing rats by targeting endogenous genes with RNA-linked (via internal ribosomal entry sequence, IRES; Pelletier and Sonenberg 1988, Nature) or protein-linked (2A peptide linkers; Ryan MD 1991 J Gen Virol) recombinases.
Transgenic rats on the Long Evans background will provide broad support for neuroscience applications, including studies on complex behaviors to further our understanding of mechanisms underlying cognition and diseases of the brain, such as addiction, schizophrenia and neurodegeneration.
Materials and Method
Animal Care and Use
Protocols for the use of rats in this study were approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center in Dallas, as certified by the Association for Assessment and Accreditation of Laboratory Animal Care International. Rats used for this study were housed in individually ventilated, Lab Products 2100 cages in a dedicated room with atmosphere controls set to 72°F, 45–50% humidity during a 12 hr light/dark cycle (i.e. Light cycle = 6:00am-6:00pm, Central Standard Time adjusted for daylight savings time). Rats were fed Harlan Teklad Irradiated 7912, LM-485 Mouse/Rat Diet, 5% fat Diet and a continuous supply of reverse osmosis water. Wildtype Long Evans rats were form Charles Rivers, Inc.
Isolating Undifferentiated Type A Spermatogonia
Seminiferous tubules were isolated from testes of 23 day old Long Evans rats. The most advanced germ cell types in 23-day old male rats are spermatocytes (Clermont and Perey 1957). Tubules were enzymatically and mechanically dissociated into a cellular suspension to generate cultures of testis cells in serum-containing medium, as described (Hamra et al. 2008; Hamra et al. 2002), but using 1.2 mg/ml Clostridium histolyticum Collogenase (Sigma; 2.1 units/mg FLGPA) in place of dispase. Testis cell cultures were then used to isolate enriched populations of laminin-binding spermatogonia by established methods (Hamra et al. 2008; Hamra et al. 2002) that first deplete somatic testis cells from the germ cell population by negative selection on plastic and collagen, before positive selection for the spermatogonial stem cells based on their ability to bind to laminin (Hamra et al. 2002). Laminin binding spermatogonia were then sub-cultured every 2 days for 3 passages on fresh gelatin-coated dishes prior to passaging onto irradiated DR4 MEFs (Chapman et al. 2011).
Spermatogonial Culture
Long Evans rat spermatogonial lines were derived from freshly isolated laminin binding spermatogonia, as previously described in detail (Chapman et al. 2011; Wu et al. 2009). Here, a SG Medium formulation was used to sub-culture Long Evans rat spermatogonial stem cell lines: Dulbecco’s Modified Eagle’s Medium:Ham’s F12 Nutrient Mixture (1:1), 6 ng/ml GDNF, 6 ng/ml FGF2, 100 μM 2-mercaptoethanol, 6 mM L-glutamine and a 1x concentration of the B27-Minus Vitamin-A Supplement solution (Chapman et al. 2015). Spermatogonia were sub-cultured in SG Medium on feeder layers of DR4 MEFs (Chapman et al. 2011; Wu et al. 2009). Prior to transplantation, spermatogonia were harvested from MEFs and incubated for 2 hr on gelatin-coated plates in SG Medium to deplete feeder cells.
Spermatogonial Transfection and G418 Selection
Long Evans rat spermatogonial lines were co-transfected using a Neon (Life technologies) transfection apparatus with a 10 μl plasmid mixture containing 2 μg pPGK-Neo, 4 μg pM3A (Sleeping Beauty transposase plasmid) and 4 μg pSB-Dazl-tdTomato (Sleeping Beauty transposon plasmid) at passage 4, as previously described (Chapman et al. 2015).
DR4 Fibroblast Feeder Layer Preparation
Primary stocks of DR4 mouse embryonic fibroblasts (MEFs) were from ATCC, Inc. and expanded after plating into Dulbecco’s modified Eagle’s medium supplemented with 1.5 g/l sodium bicarbonate, and 15% heat-inactivated FBS (MEF medium) at 37°C/5% CO2 for up to 4 passages after their thawing and initial plating from the manufacturer’s vial. Following expansion, secondary stocks of MEFs are irradiated (100 Gy) and then cryo-preserved in liquid nitrogen using Recovery Cell Culture Freezing Medium (Invitrogen) according to manufacturer’s protocol. Tissue culture dishes were pre-coated with a solution of sterile 0.1% gelatin for 1 hr at room temperature and rinsed once with sterile PBS before plating MEFs. Prior to use for culture with spermatogonia, irradiated MEFs were plated into gelatin-coated dishes (~6.8 × 104 cells/cm2) in MEF medium for 16–48 hr, rinsed 1x with PBS and then pre-incubated in SG medium for an additional 16–48 hr. The SG medium used for pre-incubation is then discarded and spermatogonia are passaged onto the MEFs in fresh SG medium.
Transgenic Rat Production
Long Evans rats were injected (i.p.) with 12.5 mg/kg busulfan (4 mg/mL in 50% DMSO) at 12 days of age and then used as recipient males at 24 days of age. Busulfan is a spermatogonial toxin commonly used to kill spermatogonia in recipient rat testes prior to transplantation because it increases donor spermatogonial stem cell colonization efficiency (Hamra et al. 2002; Ogawa et al. 1999; Ryu et al. 2003). Donor spermatogonia were loaded into injection needles fashioned from 100 μl glass capillary tubes at a concentration of 3×105 cells/65 μl SG Medium containing 0.032% (wt/vol) trypan blue, and then transplanted into rat seminiferous by retrograde injection through the rete testes (Dym 1976; Ogawa et al. 1999). Recipient testes were analyzed 7–8 months post-transplantation, and their seminiferous tubules expressing the tgSBDazl-tdTomato transgene were imaged in whole testes using a Nikon SMZ1500 fluorescence stereomicroscope equipped with ACT-1 imaging software (Nikon Instruments, Inc. NY) (Hamra et al. 2004). Gonads from F1 and F2 generation tgSB-Dazl-tdTomatofkh rats were imaged similarly.
Supplementary Material
Acknowledgments
We thank Karen M. Chapman, Jaideep Chaudhary, Priscilla Jaichander, Gerardo Medrano and Andrew Syvyk for their help with, respective, cell culture and transfection, flow cytometry and histology, PNA-labeling techniques, recipient rat preparation and transplantation, and pSB-Dazl-tdTomato transposon construction and validation experiments. We thank Zoltán Ivics and Zsuzsanna Izsvák for providing the Sleeping Beauty transposon and transposase plasmids. This work was supported by the NIH Intramural Research Program (IRP) of the National Institute on Drug Abuse (NIDA) including the NIDA-IRP, Scientific Director’s Innovators Partnership Program to BKH/FKH.
Abbreviations
- SB
Sleeping Beauty transposon
- GDNF
glial cell line derived neurotropic factor
- FGF2
fibroblast growth factor 2
- F1
familial generation 1
- F2
familial generation 2
- SG Medium
Spermatogonial Medium
- MEFs
mouse embryonic fibroblasts
- FBS
fetal bovine serum
- HS
horse serum
- PNA
peanut agglutinin lectin
References
- Aitman TJ et al. (2008) Progress and prospects in rat genetics: a community view Nat Genet 40:516–522 doi: 10.1038/ng.147 [DOI] [PubMed] [Google Scholar]
- Atanur SS et al. (2013) Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat Cell 154:691–703 doi: 10.1016/j.cell.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KM et al. (2015) Targeted Germline Modifications in Rats Using CRISPR/Cas9 and Spermatogonial Stem Cells Cell Rep 10:1828–1835 doi: 10.1016/j.celrep.2015.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KM, Saidley-Alsaadi D, Syvyk AE, Shirley JR, Thompson LM, Hamra FK (2011) Spermatogonial Stem Cell Mediated Gene Transfer Transgenic Technology vol 1, Eds. Pease S & Saunders T Springer Press. doi:DOI: 10.1007/978-3-642-20792-1_12 [DOI] [Google Scholar]
- Cifani C et al. (2012) Medial prefrontal cortex neuronal activation and synaptic alterations after stress-induced reinstatement of palatable food seeking: a study using c-fos-GFP transgenic female rats J Neurosci 32:8480–8490 doi: 10.1523/JNEUROSCI.5895-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont Y, Perey B (1957) Quantitative study of the cell population of the seminiferous tubules in immature rats The American journal of anatomy 100:241–267 doi: 10.1002/aja.1001000205 [DOI] [PubMed] [Google Scholar]
- Dym M (1976) The mammalian rete testis--a morphological examination Anat Rec 186:493–523 [DOI] [PubMed] [Google Scholar]
- Furushima K, Jang CW, Chen DW, Xiao N, Overbeek PA, Behringer RR (2012) Insertional mutagenesis by a hybrid piggyBac and sleeping beauty transposon in the rat Genetics 192:1235–1248 doi: 10.1534/genetics.112.140855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM et al. (2009) Knockout rats via embryo microinjection of zinc-finger nucleases Science 325:433 doi: 10.1126/science.1172447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra FK, Chapman KM, Wu Z, Garbers DL (2008) Isolating highly pure rat spermatogonial stem cells in culture Methods in molecular biology 450:163–179 doi: 10.1007/978-1-60327-214-8_12 [DOI] [PubMed] [Google Scholar]
- Hamra FK, Gatlin J, Chapman KM, Grellhesl DM, Garcia JV, Hammer RE, Garbers DL (2002) Production of transgenic rats by lentiviral transduction of male germ-line stem cells Proc Natl Acad Sci U S A 99:14931–14936 doi: 10.1073/pnas.222561399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, Garbers DL (2004) Defining the spermatogonial stem cell Developmental biology 269:393–410 doi: 10.1016/j.ydbio.2004.01.027 [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z, Chapman KM, Hamra FK (2011a) Sleeping Beauty transposon mutagenesis of the rat genome in spermatogonial stem cells Methods 53:356–365 doi: 10.1016/j.ymeth.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z, Medrano G, Chapman KM, Hamra FK (2011b) Sleeping Beauty transposon mutagenesis in rat spermatogonial stem cells Nature protocols 6:1521–1535 doi: 10.1038/nprot.2011.378 [DOI] [PubMed] [Google Scholar]
- Izsvak Z et al. (2010) Generating knockout rats by transposon mutagenesis in spermatogonial stem cells Nature methods 7:443–445 doi: 10.1038/nmeth.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CJ, Zhang J, Trenchard E, Lloyd KC, West DB, Rosen B, de Jong PJ (2016) Efficient gene targeting in mouse zygotes mediated by CRISPR/Cas9-protein Transgenic Res doi: 10.1007/s11248-016-9998-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katter K et al. (2013) Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits FASEB J 27:930–941 doi: 10.1096/fj.12-205526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K et al. (2007) Transposon-tagged mutagenesis in the rat Nature methods 4:131–133 doi: 10.1038/nmeth1002 [DOI] [PubMed] [Google Scholar]
- Laulederkind SJ et al. (2013) The Rat Genome Database 2013--data, tools and users Brief Bioinform 14:520–526 doi: 10.1093/bib/bbt007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar J, Moreno C, Jacob HJ, Kwitek AE (2005) Impact of genomics on research in the rat Genome Res 15:1717–1728 doi: 10.1101/gr.3744005 [DOI] [PubMed] [Google Scholar]
- Li D et al. (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system Nat Biotechnol 31:681–683 doi: 10.1038/nbt.2661 [DOI] [PubMed] [Google Scholar]
- Lu B, Geurts AM, Poirier C, Petit DC, Harrison W, Overbeek PA, Bishop CE (2007) Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system Mamm Genome 18:338–346 doi: 10.1007/s00335-007-9025-5 [DOI] [PubMed] [Google Scholar]
- Ma Y et al. (2014) Generating rats with conditional alleles using CRISPR/Cas9 Cell research 24:122–125 doi: 10.1038/cr.2013.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Brinster CJ, Orwig KE, Ryu BY, Avarbock MR, Brinster RL (2001) Transgenic mice produced by retroviral transduction of male germ-line stem cells Proc Natl Acad Sci U S A 98:13090–13095 doi: 10.1073/pnas.231473498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas CR, Xu EY, Banani SF, Hammer RE, Hamra FK, Reijo Pera RA (2009) Characterization of a Dazl-GFP germ cell-specific reporter Genesis 47:74–84 doi: 10.1002/dvg.20460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley MJ, Racicot KE, Oatley JM (2011) Sertoli cells dictate spermatogonial stem cell niches in the mouse testis Biology of reproduction 84:639–645 doi: 10.1095/biolreprod.110.087320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Dobrinski I, Brinster RL (1999) Recipient preparation is critical for spermatogonial transplantation in the rat Tissue Cell 31:461–472 doi: 10.1054/tice.1999.0060 [DOI] [PubMed] [Google Scholar]
- Ohtsuka M, Miura H, Sato M, Kimura M, Inoko H, Gurumurthy CB (2012) PITT: pronuclear injection-based targeted transgenesis, a reliable transgene expression method in mice Exp Anim 61:489–502 [DOI] [PubMed] [Google Scholar]
- Petri V et al. (2014) Disease pathways at the Rat Genome Database Pathway Portal: genes in context-a network approach to understanding the molecular mechanisms of disease Hum Genomics 8:17 doi: 10.1186/s40246-014-0017-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z et al. (2013) High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases Nucleic Acids Res 41:e120 doi: 10.1093/nar/gkt258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie CT et al. (2017) Near-infrared fluorescent protein iRFP713 as a reporter protein for optogenetic vectors, a transgenic Cre-reporter rat, and other neuronal studies J Neurosci Methods doi: 10.1016/j.jneumeth.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Avarbock MR, Brinster RL (2003) Stem cell and niche development in the postnatal rat testis Dev Biol 263:253–263 doi:S0012160603004615 [pii] [DOI] [PubMed] [Google Scholar]
- Tong C, Li P, Wu NL, Yan Y, Ying QL (2010) Production of p53 gene knockout rats by homologous recombination in embryonic stem cells Nature 467:211–213 doi: 10.1038/nature09368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Falciatori I, Molyneux LA, Richardson TE, Chapman KM, Hamra FK (2009) Spermatogonial culture medium: an effective and efficient nutrient mixture for culturing rat spermatogonial stem cells Biology of reproduction 81:77–86 doi: 10.1095/biolreprod.108.072645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto DJ et al. (2013) Rats classified as low or high cocaine locomotor responders: a unique model involving striatal dopamine transporters that predicts cocaine addiction-like behaviors Neurosci Biobehav Rev 37:1738–1753 doi: 10.1016/j.neubiorev.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering Cell 154:1370–1379 doi: 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen ST et al. (2014) Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes Developmental biology 393:3–9 doi: 10.1016/j.ydbio.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Renfree MB, Short RV (2003) Successful intra- and interspecific male germ cell transplantation in the rat Biology of reproduction 68:961–967 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.