Abstract
Mounting evidence indicates that there exists an association between heparanase (HPSE) and several physiological and pathological mechanisms in humans. However, the dynamics of the mechanisms involved in the regulation of HPSE expression in pancreatic cancer (PC) remain unclear. The aim of the present study was to assess the levels of HPSE in PC tissues and cell lines by western blotting and reverse transcription-quantitative PCR (RT-qPCR) analysis. Wound healing and Transwell assays were conducted to examine the effects of HPSE on migration and invasion in sh-NC and sh-HPSE PC cell lines. In addition, tumor growth was assessed in a mouse xenograft model in vivo. The expression levels of epithelial-to-mesenchymal transition (EMT)-related biomarkers and the involvement of the Wnt/β-catenin pathway were assessed by analyzing the results of western blot and RT-qPCR assays. The results indicated that the expression of HPSE was substantially higher in PC tissues and cell lines, whereas experimental knockdown of HPSE suppressed the rates of migration and invasion of PC cells. Western blotting was used to assess the expression of EMT biomarkers and determine the function of HPSE in EMT. Furthermore, our results indicated that downregulation of HPSE expression decreased the expression of Wnt/β-catenin associated proteins. In conclusion, HPSE appears to be a good candidate as a molecular target for the treatment of PC based on the finding of the present study.
Keywords: pancreatic cancer, heparanase, migration, invasion, epithelial-to-mesenchymal transition
Introduction
Pancreatic cancer (PC) is characterized by high malignant potential, insidious onset and typically a poor patient prognosis (1). Due to its high rate of metastasis, ~80% of patients cannot receive radical treatment (1,2). Despite intensive postoperative interventions, including drug treatment and other methods, the 5-year survival rate is ≤7%, and PC has been identified as one of the most serious types of malignant tumors that endanger human health (3). Thus, understanding the dynamics of the molecular mechanisms underlying metastasis is crucial for designing better treatment options and improving the outcomes of this fatal disease.
Heparanase (HPSE) is recently discovered important functional enzyme and it is the only member of the endoglycosidase family that can degrade the heparan sulfate (HS) chain in glycosaminoglycans, and the only that specifically recognizes HS side chains. As the most biologically active of the various proteoglycans, HS participates in the multiple stages of the cell adhesion reaction process and interacts with adhesion molecules, cytokines and intercellular signaling molecules, thereby affecting cell proliferation, differentiation, migration and morphology. HS proteoglycan (HSPG) also plays an important role in various dynamic interactions and pathways associated with inflammation and ischemia-reperfusion injury, thrombosis and tumor metastasis (4,5). HPSE can promote cell infiltration, metastasis, tumor cell division, chemotaxis and microvascular formation (6,7). HPSE is the only hydrolase found in mammals that can cleave the HS side chain on HSPG and is thus an ideal candidate for use as antitumor treatment. Therefore, the main goal of the present study was to investigate the role of HPSE in pancreatic cancer, the effects of HPSE silencing, and the mechanisms underlying the observed dynamics of HPSE.
Materials and methods
Clinical tissue samples
Overall, PC samples and corresponding adjacent non-cancer tissues were obtained from 6 patients (3 women and 3 men; mean age, 60.50±9.42 years; range, 48–72 years) who were diagnosed at Anhui Provincial Hospital (Hefei, China) between June 2013 and June 2016, and the mRNA levels of HPSE were detected. Detailed pathological and clinical data (including patient sex and age, tumor size and location, vascular invasion, degree of differentiation and TNM stage) from 128 patients between June 2013 and June 2016 were obtained from the medical records. Samples were included in the present study based on the 8th edition of the Union for International Cancer Control TNM staging system (8). Patients who had received radiotherapy or chemotherapy prior to surgery were not included. The specimens were fixed in 4% formalin at 37°C for 2 h and embedded in paraffin for pathological analysis and confirmation of the diagnosis. The clinical follow-up data of the patients were obtained from the PC database of Anhui Provincial Hospital. The protocol of the present study was approved by the Ethics Committee of the Anhui Provincial Hospital (certification no. 2019-P-032) and all patients provided written informed consent.
Cell lines and culture
The human HPDE6-C7, Capan-2, PANC-1, AsPC-1 and BxPC-3 pancreatic cancer cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were maintained in DMEM or RPMI-1640 (Biological Industries) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured in an atmosphere of 5% CO2 and at a temperature of 37°C. All cells were authenticated by STR profiling before the experiment.
Stable transfection of pancreatic cancer cell
HPSE-shRNA and control sh-NC were synthesized by Sigma-Aldrich; Merck KGaA. For the construction of the β-catenin vector, β-catenin cDNA was cloned into Flag-tagged-pcDNA3.1 (GenePharma). As regards the transfection of HPSE-shRNA, the transfection of control sh-NC and β-catenin-vector in PANC-1 and BxPC-3 cells was performed by Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the instructions of the manufacturer. The established stable cell lines were verified by RT-qPCR and western blot analyses and used for the subsequent experiments.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from cells using TRIzol® reagent following the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). A cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to synthesize the first-strand cDNA with the following profile: 25°C for 10 min, 37°C for 120 min, 85°C for 5 min and held at 4°C. qPCR was performed with SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) to detect the levels of HPSE transcription as follows: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min and held at 4°C. The following primer sets were used for analysis of the levels of HPSE expression: Forward 5′-ATGCTGCTGCGCTCGAA-3′ and reverse 5′-AGATGCAAGCAGCAACTTTGGC-3′. For GAPDH, the primer set was as follows: Forward 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse 5′-GGGGTCGTTGATGGCAACA-3′. The relative levels of expression were quantified using the 2−ΔΔCq method (9).
Western blot analysis
For the detection of levels of protein expression, western blotting was performed as previously described (10). Briefly, the cells were lysed in RIPA cell lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology, Inc.) with 1 mM phenylmethylsulfonyl fluoride at 4°C for 30 min, with vortexing every 10 min, followed by centrifugation at 13,800 × g for 10 min at 4°C. A BCA kit (cat. no. P0009; Beyotime Institute of Biotechnology, Inc.) was used to quantify the protein concentration. A total of 30 µg of denatured protein for each sample was separated on a 10% SDS PAGE gel and transferred onto a PVDF membrane. The membranes were blocked with 5% non-fat milk in 0.05% Tween-20 in TBS (TBST) for 2 h at room temperature and blotted with the following primary antibodies: HPSE (cat. no. ab85543, 1:500, Abcam), E-cadherin (cat. no. SAB4503751, 1:500, Sigma-Aldrich; Merck KGaA); vimentin (cat. no. SAB1305433, 1:1,000, Sigma-Aldrich; Merck KGaA); Snail (cat. no. SAB4502825, 1:800, Sigma-Aldrich; Merck KGaA); β-catenin (cat. no. SAB4500541, 1:800, Sigma-Aldrich; Merck KGaA); glycogen synthase kinase (GSK)-3β (cat. no. sc-81462, 1:1,000, Santa Cruz Biotechnology, Inc.); phosphorylated (p-)GSK-3β (cat. no. 9336, 1:800, Cell Signaling Technology, Inc.); and/or GAPDH (cat. no. 10494-1-AP, 1:5,000, ProteinTech Group, Inc.). After washing with TBST, the membranes were incubated with anti-rabbit or anti-mouse secondary antibodies, conjugated with horseradish peroxidase (cat. nos. A0208 and A0216; 1:1,000, Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. Following 3 washes, the membrane was visualised with an enhanced chemiluminescence system (cat. no. P0018AM; Beyotime Institute of Biotechnology, Inc.).
Immunohistochemistry (IHC)
IHC was performed using a standard streptavidin-biotin-peroxidase complex-based methodology according to the manufacturer's guidelines (SA2010; Boster Biological Technology). The tissue sections were incubated at 4°C in a moist chamber overnight with the addition of anti-HPSE antibody (cat. no. ab85543, 1:200, Abcam). The expression levels of HPSE were also quantified by determining the percentage of positive tumor cells and the intensity of positive staining. Staining intensity was scored as follows: Negative, 0; bordering, 1, weak, 2; moderate, 3; and strong, 4. Additionally, staining was scored in accordance with the percentage of stained tumor cells in the field as follows: Negative, 0; 0–25%, 1; 26–50%, 2; 51–75%, 3; and 76–100%, 4. The product of the intensity score and percentage of stained cells was considered as the overall IHC score (range, 0–16). Two independent pathologists observed and evaluated the staining process and results.
Cell migration assay
A total of 2.0×105 cells were seeded in 24-well plates and grown to >80% confluence. Following overnight culture of cells in 5% CO2 at 37°C, a 200-µl pipette tip was used to create a longitudinal scratch in the middle of the bottom of the sample well. Detached cells were washed away using PBS and serum-free medium was added. Images were captured at 0 and 24 h after wounding to monitor healing using a light microscope (Ti-S, Nikon Corporation) at a magnification of ×4.
Cell invasion assay
The effect of HPSE on metastatic ability of cancer cells was evaluated by using a 24-well Transwell assay (8 µm pore size; EMD Millipore) as previously described (10). In brief, 2.0×105 cells in serum-free medium were added to the upper culture chambers that had been precoated with Matrigel at 37°C. The bottoms of culture chambers were filled with DMEM supplemented with 10% FBS by volume. After incubation in 5% CO2 at 37°C for 48 h, staining of the invading cells was performed using 0.5% crystal violet solution for 10 min at room temperature and examined with a light microscope (Ti-S, Nikon Corporation) at a magnification of ×4.
Tumor metastasis assay
A total of 100 male BALB/c nude mice (aged 6 weeks and weighing ~18.30 g) were purchased from the Institute for Experimental Animals of the Chinese Academy of Medical Sciences (Beijing, China). The nude mice were maintained under pathogen-free conditions, at 20–26°C, 40–70% humidity and a 12/12 light/dark cycle, with access to food and water ad libitum. PANC-1 cells (2.0×106) transfected with sh-HPSE-luciferase or sh-NC-luciferase were injected into the tail vein of mice (n=6) after 1 month of acclimation. At 50 days after the injection, the mice were sacrificed and their lungs were removed. Observation of metastatic nodules in lung tissues was performed using the IVIS Lumina II high-sensitivity imaging system (PerkinElmer, Inc.). The duration of the experiment was 57 days and the health and behavior of the mice were monitored daily. All mice were sacrificed by cervical dislocation after inhaling 40% carbon dioxide. Death was verified by cardiac arrest (monitoring was continued for 5–6 min after breathing stopped). All animal experiments were approved by the Ethics Committee of Anhui Provincial Hospital (certification no. 2019-P-032).
Hematoxylin and eosin (HE) staining
The tissue samples were cut into 4-µm sections and mounted on silanized glass slides. Following deparaffinization and hydration, the sections were stained with hematoxylin solution for 3 min in 35°C followed by 5 dips in 0.5% acid ethanol (1% HCl in 70% ethanol) and then rinsed in distilled water. Subsequently, the sections were stained with eosin solution for 1 min in 35°C and followed by dehydration with graded alcohol and clearing in xylene. The sections were then examined under a light microscope (Ti-S, Nikon Corporation) at a magnification of ×20.
Statistical analysis
SPSS version 19.0 (SPSS Inc.) software was used to perform analysis of statistical data. Determination of levels of variation and significance of differences for values among cohorts was determined using a paired two-tailed Student's t-test and Dunnett's t-test. Values for P<0.05 were considered to be of statistical significance.
Results
Association of HPSE expression with PC clinicopathological parameters and prognosis
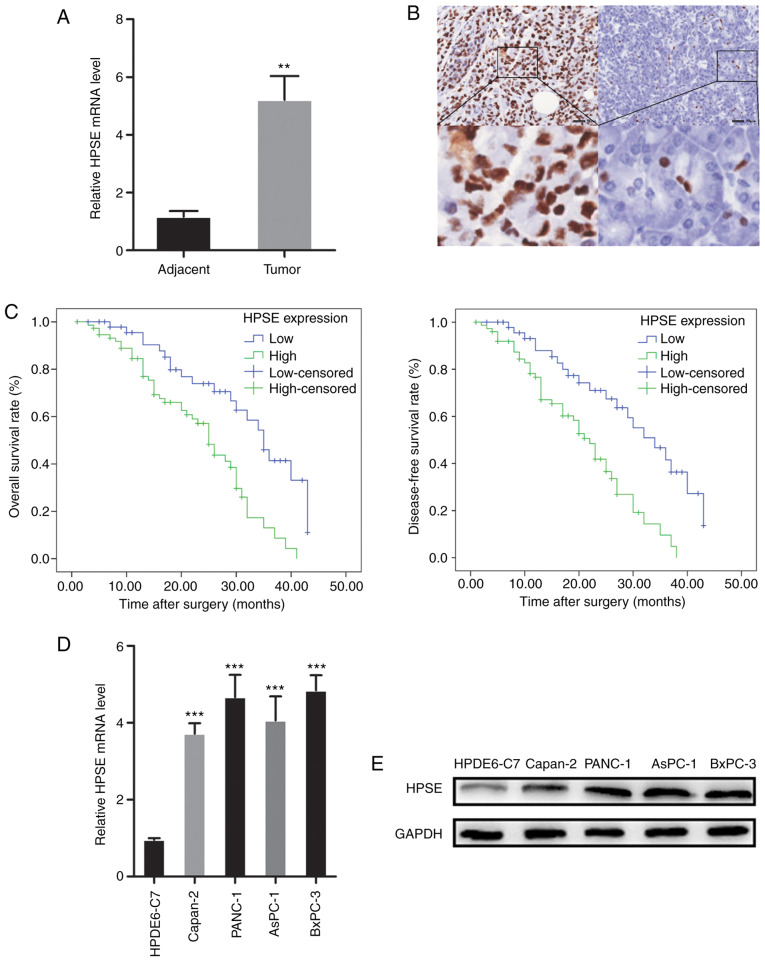
RT-qPCR analysis and IHC were employed to evaluate the expression of HPSE in PC tissue samples. The mRNA and protein expression of HPSE were also examined in pancreatic cancer cell lines. High expression of HPSE was observed in PC tissues and cancer cell lines (Fig. 1). In order to evaluate the biological significance of HPSE in PC, the associations between PC tissue HPSE levels and clinicopathological parameters were analyzed. As shown in Table I, increased HPSE expression was significantly associated with the presence of vascular invasion (P=0.025), poor differentiation (P=0.012) and higher TNM stage (P=0.024). Kaplan-Meier survival curves were plotted to compare the overall survival (OS) and disease-free survival (DFS) of PC patients according to HPSE expression (Fig. 1). Patients with high HPSE expression had poorer prognosis compared with those with low HPSE expression (OS, P=0.001; DFS, P=0.000). Multivariate survival analysis further revealed that intratumoral HPSE expression (OS, P=0.009; DFS, P=0.004) was an independent poor prognostic marker for OS and DFS (Tables II and III).
Figure 1.
High expression of HPSE in PC tissues and cell lines and association with survival. (A) mRNA level of HPSE in PC and adjacent normal tissues. (B) Immunohistochemical staining (magnification, ×4 and ×20) for HPSE in PC (left panel) and adjacent normal tissue (right panel). (C) Kaplan-Meier analysis of overall and disease-free survival of patients with PC according to intra-tumoral HPSE expression. (D and E) mRNA and protein levels of HPSE in normal pancreatic ductal epithelial cells (HPDE6-C7) and four PC cells (Capan-2, PANC-1, AsPC-1 and BxPC-3). Data are presented as mean ± standard deviation. GAPDH served as an internal reference. All experiments were performed three times. **P<0.01, ***P<0.001, compared with adjacent tissue or HPDE6-C7 cells, respectively. HPSE, heparanase; PC, pancreatic cancer.
Table I.
Association between HPSE protein expression (immunohistochemical staining) in pancreatic cancer and clinicopathological variables.
| HPSE expression | |||||
|---|---|---|---|---|---|
| Variables | Total | Low (n=53) | High (n=75) | χ2 | P-value |
| Sex | 0.760 | ||||
| Male | 85 | 36 | 49 | 0.093 | |
| Female | 43 | 17 | 26 | ||
| Age (years) | 0.391 | ||||
| ≤60 | 54 | 20 | 34 | 0.735 | |
| >60 | 74 | 33 | 41 | ||
| Size (cm) | 0.373 | ||||
| ≤4 | 104 | 45 | 59 | 0.793 | |
| >4 | 24 | 8 | 16 | ||
| Tumor location | 0.493 | ||||
| Head and neck | 85 | 37 | 48 | 0.470 | |
| Body and tail | 43 | 16 | 27 | ||
| Vascular invasion | 0.025 | ||||
| Negative | 61 | 19 | 42 | 5.055 | |
| Positive | 67 | 34 | 33 | ||
| Differentiation | 0.012 | ||||
| High/moderate | 58 | 31 | 27 | 6.339 | |
| Poor and undifferentiated | 70 | 22 | 48 | ||
| TNM stage | 0.023 | ||||
| I–II | 62 | 32 | 30 | 5.163 | |
| III–IV | 66 | 21 | 45 | ||
HPSE, heparanase.
Table II.
Univariate and multivariate analysis of the correlation between clinicopathological parameters and overall survival of patients with pancreatic cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex | 0.868 | 0.525–1.432 | 0.579 | |||
| Male | ||||||
| Female | ||||||
| Age (years) | 0.768 | 0.468–1.260 | 0.296 | |||
| ≤60 | ||||||
| >60 | ||||||
| Size (cm) | 1.149 | 0.633–2.088 | 0.648 | |||
| ≤4 | ||||||
| >4 | ||||||
| Tumor location | 1.221 | 0.724–2.059 | 0.455 | |||
| Head and neck | ||||||
| Body and tail | ||||||
| Vascular invasion | 1.812 | 1.109–2.962 | 0.018a | |||
| Negative | ||||||
| Positive | ||||||
| Differentiation | 2.397 | 1.455–3.947 | 0.001a | 1.919 | 1.220–3.916 | 0.014a |
| High/moderate | ||||||
| Poor and undifferentiated | ||||||
| TNM stage | 1.671 | 1.027–2.721 | 0.039a | |||
| I–II | ||||||
| III–IV | ||||||
| HPSE expression | 2.682 | 1.536–4.684 | 0.001a | 2.186 | 1.140–3.229 | 0.009a |
| Low | ||||||
| High | ||||||
Statistically significant. HR, hazard ratio; CI, confidence interval; HPSE, heparanase.
Table III.
Univariate and multivariate analysis of the correlation between clinicopathological parameters and disease-free survival of patients with pancreatic cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex | 0.961 | 0.584–1.579 | 0.874 | |||
| Male | ||||||
| Female | ||||||
| Age (years) | 0.762 | 0.463–1.254 | 0.286 | |||
| ≤60 | ||||||
| >60 | ||||||
| Size (cm) | 1.177 | 0.650–2.134 | 0.590 | |||
| ≤4 | ||||||
| >4 | ||||||
| Tumor location | 1.293 | 0.759–2.202 | 0.344 | |||
| Head and neck | ||||||
| Body and tail | ||||||
| Vascular invasion | 1.685 | 1.034–2.746 | 0.036a | |||
| Negative | ||||||
| Positive | ||||||
| Differentiation | 2.493 | 1.506–4.126 | 0.000a | 2.052 | 1.220–3.452 | 0.007a |
| High/moderate | ||||||
| Poor and undifferentiated | ||||||
| TNM stage | 1.598 | 0.983–2.597 | 0.059 | |||
| I–II | ||||||
| III–IV | ||||||
| HPSE expression | 2.762 | 1.581–4.825 | 0.000a | 2.309 | 1.296–4.112 | 0.004a |
| Low | ||||||
| High | ||||||
Statistically significant. HR, hazard ratio; CI, confidence interval; HPSE, heparanase.
Establishment of HPSE silencing in PC cells
In order to investigate the function of HPSE in PC, the levels of HPSE expression were knocked down in PANC-1 and BxPC-3 with the use of shRNA interference. The successful construction of PANC-1 and BxPC-3 cells with HPSE silencing was verified by RT-qPCR and western blot analyses (Fig. S1).
HPSE modulates the migration and invasion of PC cells
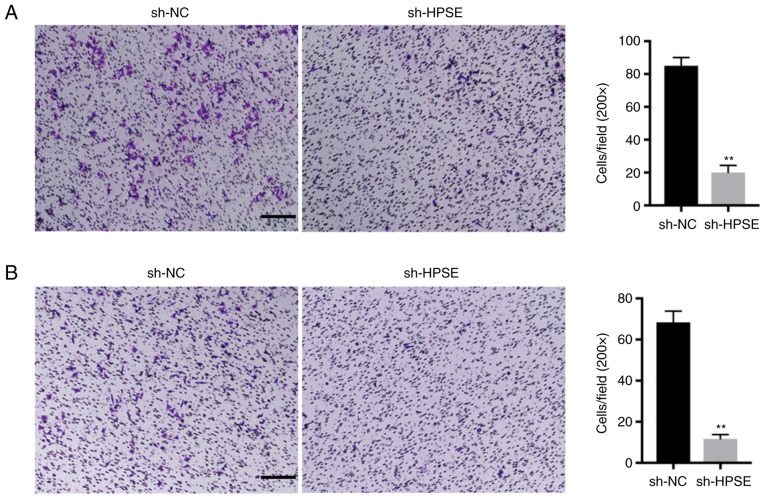
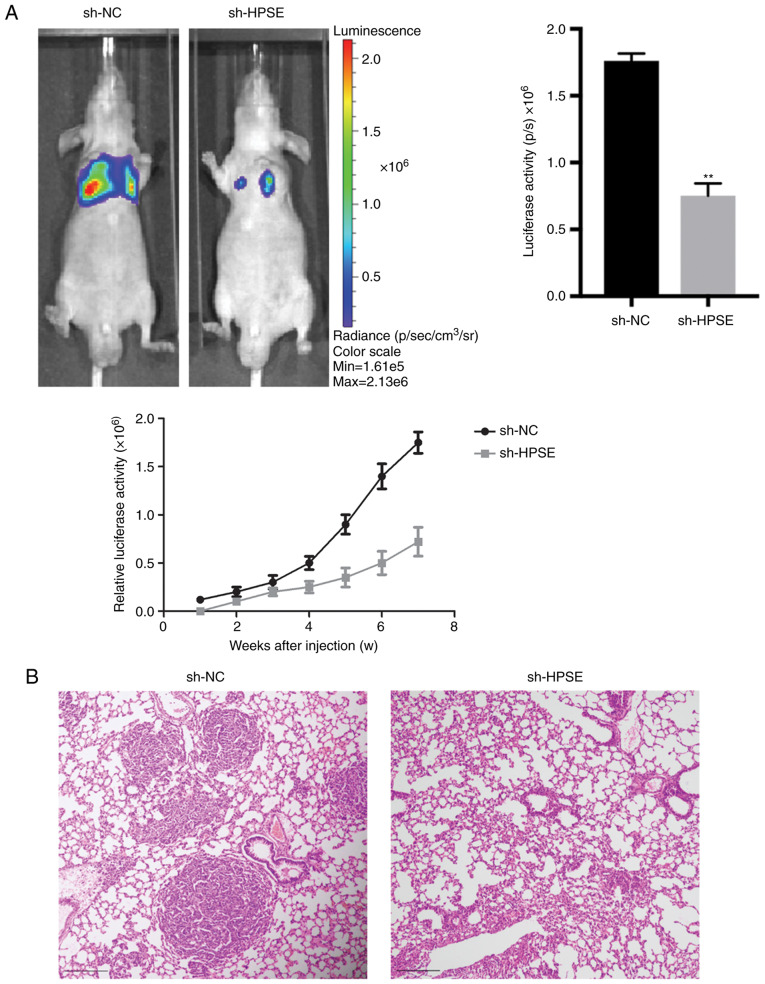
The inhibition of HPSE expression in PANC-1 and BxPC-3 cells markedly delayed wound closure compared with controls (Fig. 2). Additionally, the impact of changes in the levels of expression of HPSE on the invasion potential of the PANC-1 and BxPC-3 PC cell lines was analyzed. In the Transwell invasion assay, PANC-1 and BxPC-3 cells with suppressed HPSE expression exhibited reduced invasion and migration ability in comparison with the control (Fig. 3). In order to validate the role of HPSE in the dynamics of tumorigenesis, a lung metastasis model in nude mice was developed using PANC-1 cells and the effect of endogenous HPSE on metastasis was evaluated in vivo. The results indicated that cells with downregulated HPSE had a significantly reduced ability of inducing tumor formation in the lungs compared with the controls (Fig. 4A). A similar result was also observed based on the results of HE staining (Fig. 4B). Furthermore, the luciferase activity of lung metastatic nodules in the sh-NC group was significantly higher compared with that in the sh-HPSE group at 50 days after injection with PANC-1 cells (P=0.027; Fig. 4A). Taken together, these findings indicate that the downregulated expression of HPSE markedly suppressed cell migration and invasion in vitro and in vivo.
Figure 2.
Representative images of wound healing assays for (A) PANC-1 and (B) BxPC-3 cells transfected with sh-NC or sh-HPSE at 0 and 24 h post-transfection. Wound healing was quantified by measurement of the mean linear speed of movement of the wound edges. All experiments were performed three times. Data are presented as mean ± standard deviation. **P<0.01. HPSE, heparanase.
Figure 3.
Representative images and bar graphs depicting the invasion ability of (A) PANC-1 and (B) BxPC-3 cells after sh-NC or sh-HPSE transfection. Data are presented as mean ± standard deviation. Bar, 100 µm, **P<0.01. HPSE, heparanase.
Figure 4.
Representative luciferase activity images of (A) tumors in vivo and (B) lung metastatic nodules by hematoxylin and eosin staining (magnification, ×20) from six BALB/c nude mice at 50 days after injection with PANC-1 cells transfected with sh-NC or sh-HPSE. HPSE, heparanase. **P<0.01.
HPSE promotes epithelial-to-mesenchymal transition (EMT) in PC cells
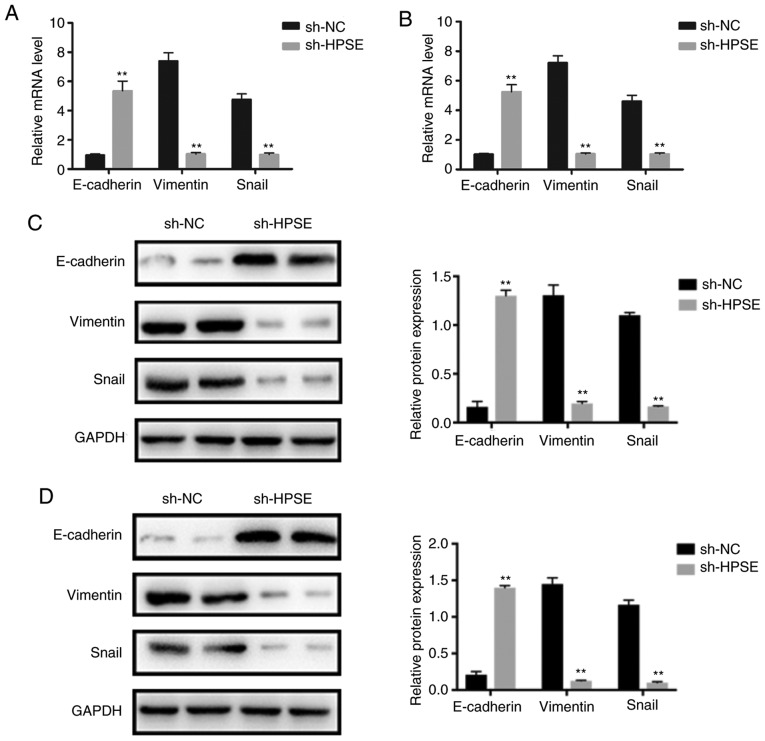
Accumulating evidence has indicated that the migration and invasion of PC cells is regulated by the EMT process. In order to investigate whether HPSE is associated with EMT, the expression levels of epithelial (E-cadherin) and mesenchymal (vimentin, Snail) markers were detected in PANC-1 and BxPC-3 cells with HPSE knockdown. Western blot and RT-qPCR analyses indicated an increase in the expression levels of E-cadherin compared with the controls. By contrast, there was a marked decline in the expression levels of vimentin and Snail in PANC-1 and BxPC-3 cells with HPSE knockdown (Fig. 5). These results indicate that HPSE likely promotes EMT in PC cells.
Figure 5.
Relative mRNA levels of epithelial- and mesenchymal-related markers in (A) PANC-1 and (B) BxPC-3 cells. Representative western blots and summarized data showing the protein levels of epithelial and mesenchymal markers in (C) PANC-1 and (D) BxPC-3 cells. **P<0.01 vs. sh-NC group.
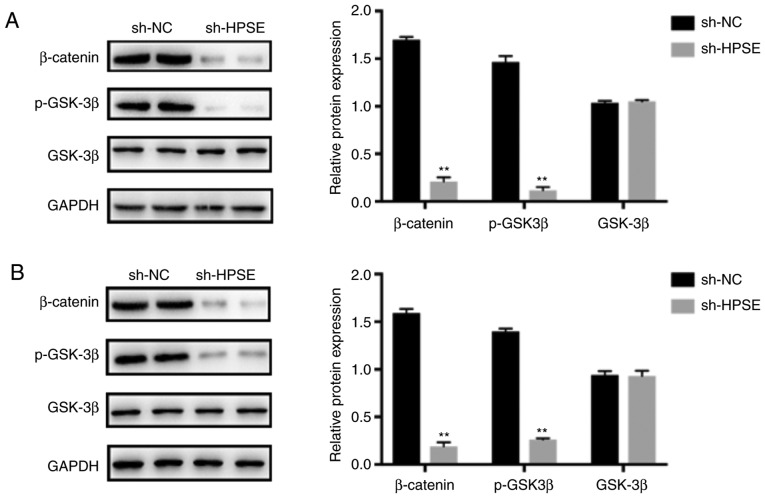
HPSE regulates EMT by activating the Wnt/β-catenin signaling pathway
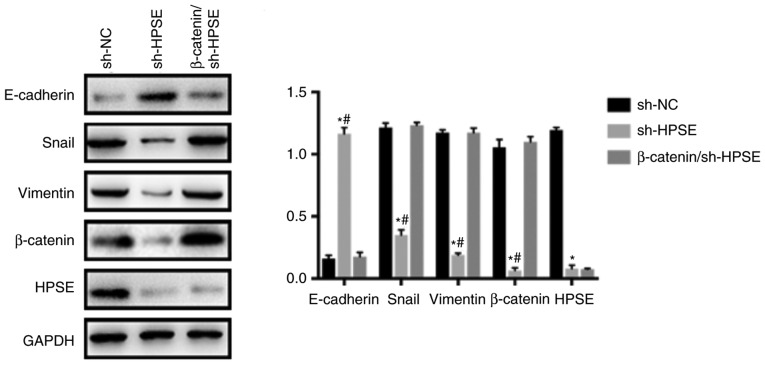
Wnt/β-catenin is among the main contributors to EMT-related signaling pathways that are crucial for the growth and progression of various cancers. To explore the association between HPSE and the Wnt/β-catenin pathway, we first examined the activation of GSK3β and β-catenin. As indicated by the results of western blotting, HPSE interference affected the expression of Wnt/β-catenin-associated proteins. The results demonstrated that the expression levels of p-GSK3β and β-catenin were markedly downregulated when HPSE was inhibited and also in comparison with samples from the control group (Fig. 6). To determine whether the Wnt/β-catenin pathway plays an important role in the pathway that links HPSE and EMT, overexpression of β-catenin was induced in sh-HPSE PANC-1 cells (Fig. S2). As verified by the results of western blotting, β-catenin neutralized the effect of HPSE on EMT, as demonstrated by changes in the expression levels of EMT-related proteins (Fig. 7). Collectively, the results of the present study suggest that HPSE promotes EMT through activating the Wnt/β-catenin signaling pathway, thereby promoting metastasis of PC.
Figure 6.
The effect of HPSE on PC progression is mediated by activating the Wnt/β-catenin signaling pathway. Representative western blots and summarized data showing the expression levels of β-catenin protein and total and phosphorylated GSK3β protein in (A) PANC-1 and (B) BxPC-3 cells. **P<0.01 vs. sh-NC group. HPSE, heparanase; PC, pancreatic cancer; GSK, glycogen synthase kinase.
Figure 7.
Overexpression of β-catenin counteracted the effects of HPSE on epithelial-to-mesenchymal transition. Representative western blots and summarized data showing the protein expression levels of E-cadherin, Snail, vimentin, β-catenin and HPSE in PANC-1 cells. *P<0.05 vs. sh-NC group, #P<0.05 vs. β-catenin/sh-HPSE group. HPSE, heparanase.
Discussion
PC is a malignancy characterized by rapid progression, poor prognosis and extremely low patient survival rates. The biological characteristics of PC include a high propensity for invading nerves, blood vessels and lymph nodes, a high metastatic rate, a deep anatomical location that makes it difficult to detect, lack of specific markers and lack of specific symptoms at the early stages (11). Over 75% of the patients clinically diagnosed with PC have advanced-stage diseased. The surgical resection rate for patients with PC is low, and PC is typically not sensitive to chemotherapy and/or radiotherapy, with a mean 5-year OS rate of ~4% (11). Comprehensive treatment, including surgery, chemotherapy and radiotherapy, the ability to predict tumor recurrence and metastasis and evaluation of the prognosis of PC are key to improving the survival rate of patients. To achieve such a comprehensive treatment approach, it is necessary to establish a specific index that predicts outcome for patients with PC and determine the best course of treatment and the potential for recurrence and metastasis. Thus far, domestic research on primary PC has mainly focused on early diagnosis and achieving the best treatment outcomes. There are currently no clearly identified and accepted specific diagnostic markers for PC and there is no clear clinical-based path for the evaluation of the prognosis of patients with PC. Furthermore, research on metastasis, recurrence rates and prognosis for patients with PC is in its early stages. However, there is an emerging understanding of the value of research on tumor markers related to metastasis and recurrence of PC and on accurate determination of patient prognosis.
The HPSE gene (Hpa) encodes a mammalian β-D glucose endoglycosidase, of which two isomeric isomers are known, namely Hpa1 and Hpa2. The extracellular matrix (ECM) consists of core proteins with HS side chains. The HPSE gene encodes a endoglycosidase that degrades HS, resulting in ECM reconstitution and plays a crucial role in important biological processes such as angiogenesis and tumor metastasis (12). HPSE activates plasminogen and matrix metalloproteinases by degrading HSPG and other proteolytic enzymes, and promotes tumor angiogenesis and lymphangiogenesis through auxiliary receptors such as basic fibroblast growth factor and vascular endothelial growth factor, thereby promoting tumor metastasis and recurrence (13). This indicates that the level of expression of HPSE is closely associated with the potential for tumor infiltration and metastatic ability. Previous studies have also demonstrated that HPSE can promote vascularization of tumor cells and can induce formation of lymphoid tissue, ultimately promoting an increase in rates of tumor invasion and metastasis (6,14,15). In addition, HPSE appears to be of value in assessing tumor invasiveness and metastatic ability (16,17). In the present study, HPSE expression and the correlation with clinicopathological data was examined in PC, in order to determine the effects of HPSE on tumor size, location, vascular invasion, differentiation and recurrence, and uncover the possible mechanism of action. Our investigation may provide a new theoretical basis for further exploration of indicators predicting PC recurrence and prognosis.
The process of EMT involves the transdifferentiation of epithelial cells to mesenchymal cells after the cells have been subjected to particular physiological and pathological conditions, which is accompanied by variations in cell morphology and the levels of expression of associated genes, and affects the rates of development and metastasis of PC (18). The changes in the levels of expression of mesenchymal proteins increases the motility, invasiveness and metastatic potential of PC cells. Cell surface HS is mainly localized in lipid rafts. It reduces transforming growth factor (TGF)-β responsiveness in these cells by facilitating caveolae/lipid raft-mediated endocytosis and rapid degradation of TGF-β bound to TGF-β receptors, thereby diminishing non-lipid raft-mediated endocytosis and TGF-β-stimulated signaling (19). TGF-β is a potent stimulator of EMT in cancer cells, such as PC cells. In addition, HPSE is an enzyme that acts on the cell surface and within the ECM and are responsible for degrading polymeric HS molecules into shorter-chain oligosaccharides. This suggests that high levels of HPSE expression, which reduce the expression of cell surface HS, may activate TGF-β-stimulated non-Smad-dependent and Smad-dependent signaling pathways that are involved in EMT (proliferation and migration) of cancer cells. Furthermore, TGF-β activates the Wnt/β-catenin pathway.
Furthermore, there are a number of critically important pathways that contribute to the promotion of mesenchymal protein expression, including RAS/RAF/MEK/ERK, PI3K/AKT/mTOR and Wnt/β-catenin (20). The downstream activity of these pathways incudes stimulating the expression of EMT transcription determinants, such as Snail, Slug, Twist and Zeb, ultimately promoting inhibition of the epithelial phenotype and acquisition of mesenchymal characteristics (18). The growing evidence base has uncovered that different types of small-molecule inhibitors and phytochemicals can affect the progression of EMT and reversing the underlying mechanisms, thereby inducing re-expression of epithelial markers. The understanding of the association between EMT and PC will likely contribute to the identification of novel therapeutic targets for PC. Our future aim is to use RNAseq to assess the total pathways in HPSE interference cell lines in order to determine whether HPSE is an essential gene in PC cells in vivo, and carry out follow-up research on the role of HPSE in PC.
In conclusion, the present study investigated whether HPSE promotes EMT through the activation of the Wnt/β-catenin signaling pathway in PC cells, and demonstrated that downregulation of HPSE expression inhibited migration and invasion of PC cells in vitro and in vivo. HPSE interference increased the expression of E-cadherin and reduced the expression of vimentin. Furthermore, the overexpression of β-catenin neutralized the effect of HPSE on EMT, as indicated by the observed changes in the protein levels of EMT-related molecular markers in PC cells. Collectively, these findings indicate that HPSE likely acts as an EMT inducer and may be of value as a target for antimetastatic treatment in patients with PC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the Anhui Provincial Natural Science Foundation (grant no. 1708085MH228).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
GW conceived the study. CW, YW, YZ and GW performed the experiments. JZ and KX supervised the experiments and edited the images. CW, YW and GW reviewed the article before submission for spelling and grammar, as well as intellectual content. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Human Research Ethics Committee of the Anhui Provincial Hospital, Anhui Medical University (Hefei, China).
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH, Neoptolemos JP. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 2.Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. doi: 10.1016/j.ccell.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Hessmann E, Johnsen SA, Siveke JT, Ellenrieder V. Epigenetic treatment of pancreatic cancer: Is there a therapeutic perspective on the horizon? Gut. 2017;66:168–179. doi: 10.1136/gutjnl-2016-312539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masola V, Bellin G, Gambaro G, Onisto M. Heparanase: A multitasking protein involved in extracellular matrix (ECM) remodeling and intracellular events. Cells. 2018;7:236. doi: 10.3390/cells7120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masola V, Zaza G, Gambaro G, Franchi M, Onisto M. Role of heparanase in tumor progression: Molecular aspects and therapeutic options. Semin Cancer Biol. 2020;62:86–98. doi: 10.1016/j.semcancer.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Caruana I, Savoldo B, Hoyos V, Weber G, Liu H, Kim ES, Ittmann MM, Marchetti D, Dotti G. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat Med. 2015;21:524–529. doi: 10.1038/nm.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran VM, Wade A, McKinney A, Chen K, Lindberg OR, Engler JR, Persson AI, Phillips JJ. Heparan sulfate glycosaminoglycans in glioblastoma promote tumor invasion. Mol Cancer Res. 2017;15:1623–1633. doi: 10.1158/1541-7786.MCR-17-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley J, Gospodarowicz MK, Wittekind C, editors. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2017. TNM Classification of Malignant tumours. [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017;50:e12389. doi: 10.1111/cpr.12389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlodavsky I, Singh P, Boyango I, Gutter-Kapon L, Elkin M, Sanderson RD, Ilan N. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist Updat. 29:54–75. doi: 10.1016/j.drup.2016.10.001. 20165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabelink TJ, van den Berg BM, Garsen M, Wang G, Elkin M, van der Vlag J. Heparanase: Roles in cell survival, extracellular matrix remodelling and the development of kidney disease. Nat Rev Nephrol. 2017;13:201–212. doi: 10.1038/nrneph.2017.6. [DOI] [PubMed] [Google Scholar]
- 14.Lv Q, Wu K, Liu F, Wu W, Chen Y, Zhang W. Interleukin17A and heparanase promote angiogenesis and cell proliferation and invasion in cervical cancer. Int J Oncol. 2018;53:1809–1817. doi: 10.3892/ijo.2018.4503. [DOI] [PubMed] [Google Scholar]
- 15.Lv B, Zhang B, Hu XY, Zeng QD. Heparanase regulates in vitro VEGF-C expression and its clinical significance to pancreatic ductal cell adenocarcinoma. Oncol Lett. 2016;11:1327–1334. doi: 10.3892/ol.2016.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Zhou ZH, Li W, Zhang SK, Li J, Zhou MJ, Song JW. Heparanase promotes tumor growth and liver metastasis of colorectal cancer cells by activating the p38/MMP1 Axis. Front Oncol. 2019;9:216. doi: 10.3389/fonc.2019.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Jiang W, Yue C, Zhang W, Tong C, Dai D, Cheng B, Huang C, Lu L. Heparanase contributes to trans-endothelial migration of hepatocellular carcinoma cells. J Cancer. 2017;8:3309–3317. doi: 10.7150/jca.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Chen CL, Huang SS, Huang JS. Cellular heparan sulfate negatively modulates transforming growth factor-beta1 (TGF-beta1) responsiveness in epithelial cells. J Biol Chem. 2006;281:11506–11514. doi: 10.1074/jbc.M512821200. [DOI] [PubMed] [Google Scholar]
- 20.Pearlman RL, Montes de Oca MK, Pal HC, Afaq F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017;391:125–140. doi: 10.1016/j.canlet.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.