SUMMARY
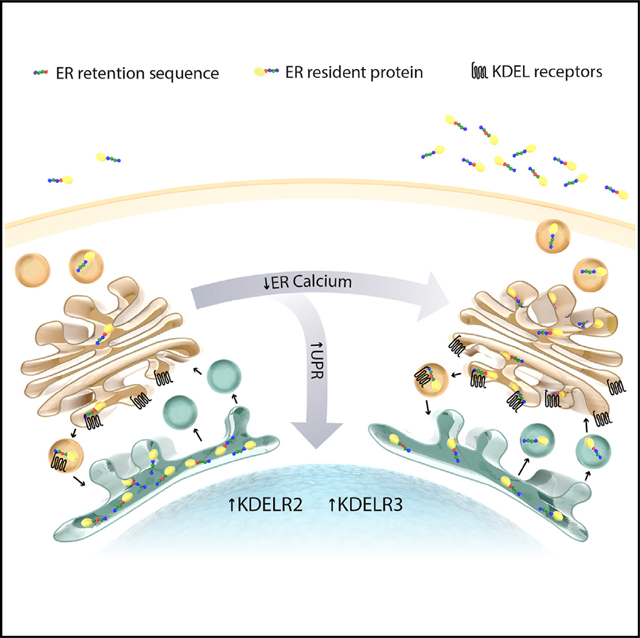
Retention of critical endoplasmic reticulum (ER) luminal proteins needed to carry out diverse functions (e.g., protein synthesis and folding, lipid metabolism) is mediated through a carboxy-terminal ER retention sequence (ERS) and its interaction with KDEL receptors. Here, we demonstrate that depleting ER calcium causes mass departure of ERS-containing proteins from cells by overwhelming KDEL receptors. In addition, we provide evidence that KDELR2 and KDELR3, but not KDELR1, are unfolded protein response (UPR) genes upregulated as an adaptive response to counteract the loss of ERS-containing proteins, suggesting previously unknown isoform-specific functions of the KDEL receptors. Overall, our findings establish that decreases in ER calcium change the composition of the ER luminal proteome and secretome, which can impact cellular functions and cell viability. The redistribution of the ER proteome from inside the cell to the outside has implications for dissecting the complex relationship of ER homeostasis with diverse disease pathologies.
In Brief
Trychta et al. identify an exodus of resident proteins from the endoplasmic reticulum (ER) in response to the loss of luminal ER calcium. They show that the cell works to maintain the ER proteome under calcium depletion by upregulating KDEL receptors, which act as unfolded protein response genes.
Graphical Abstract
INTRODUCTION
The endoplasmic reticulum (ER) lumen is a site of essential cellular functions including lipid metabolism, calcium storage, and protein folding and trafficking (Schwarz and Blower, 2016). Luminal ER resident proteins execute these important cellular functions and are maintained within the ER under homeostatic conditions by carboxy-terminal ER retention sequences (ERSs) that are proposed to interact with KDEL receptors (Munro and Pelham, 1987; Mei et al., 2017). KDEL receptors are located in the cis-Golgi and retrogradely transport ERS-containing proteins to the ER through COPI vesicle-mediated retrograde transport (Orci et al., 1997; Munro and Pelham, 1987). The mammalian genome encodes three closely related KDEL receptors: KDELR1 (Lewis and Pelham, 1990), KDELR2 (Lewis and Pelham, 1992; Hsu et al., 1992), and KDELR3 (Collins et al., 2004; Raykhel et al., 2007). Thus far, no isoform-specific functions have been described, and although all three proteins are ubiquitously expressed (http://biogps.org) (Wu et al., 2009), the majority of KDEL receptor studies have focused solely on KDELR1.
The importance of the ERS-KDEL receptor interaction is most apparent in organisms expressing only one KDEL receptor homolog. For example, yeast KDEL receptor homolog (ERD2) is essential for the retention of ER resident proteins, and loss of the receptor leads to growth defects, an accumulation of intracellular membranes and vesicles, and inhibition of the secretory pathway (Semenza et al., 1990). In Drosophila, a loss-of-function mutation in the KDEL receptor homolog results in death at the larval stage (Abrams et al., 2013). There is currently no knockout data for all three KDEL receptors in mammals, but in humans, the loss of an ERS on specific proteins has been linked to pathological conditions such as myeloproliferative disease (Klampfl et al., 2013; Nangalia et al., 2013) and osteogenesis imperfecta (Takagi et al., 2012). Interestingly, an increase in the secretion of some ERS-containing proteins has also been observed upon disruption of ER calcium homeostasis (Henderson et al., 2013, 2014; Booth and Koch, 1989), suggesting a link between ER calcium and the cell’s ability to maintain ERS proteins in the ER lumen. However, no reported studies to date have examined the relationship between putative ERS (Raykhel et al., 2007), KDEL receptors, and ER calcium homeostasis in depth.
Here we show that proteins with an ERS are indeed secreted from the cell specifically in response to ER calcium depletion. Our data suggest that this phenomenon is partly due to a saturation of the KDEL receptors, and a model is proposed in which selective KDEL receptors function as unfolded protein response (UPR)gene products that are differentially upregulated as an adaptive mechanism to counteract the efflux of ERS-containing proteins. This described phenomenon provides an avenue of exploration in diverse diseases linked to ER calcium depletion, such as diabetes, stroke, Alzheimer’s disease, and cardiac diseases.
RESULTS
The ER Retention of GLuc-ERS Reporters Is Impaired by Depletion of ER Calcium
To investigate the connection between ERS, ER calcium depletion, and protein secretion, we created a library of luminescent protein reporters based off our previously described Gaussia luciferase (GLuc) SERCaMP reporter (Henderson et al., 2013, 2014). A bioinformatic analysis was previously performed to identify soluble proteins with a signal peptide and a putative ER localization motif defined as XX[DE][FLM] (Raykhel et al., 2007). Using similar criteria, we fused the last seven amino acids (Alanen et al., 2011) of human proteins to the C terminus of GLuc to create 94 constructs that comprise an ERS reporter library (Figure S1).
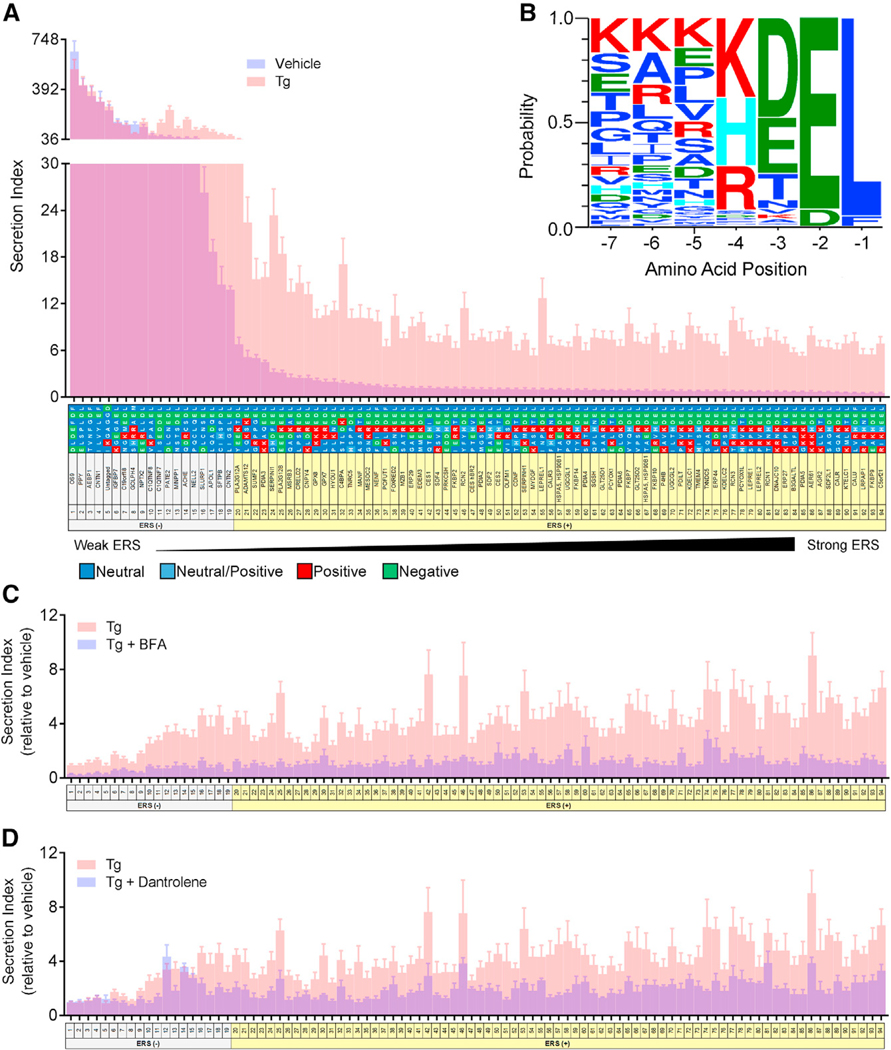
Each construct with a putative ERS was introduced into the human neuroblastoma cell line, SH-SY5Y, and the ratio of extracellular to intracellular GLuc activity was used as the “secretion index” where a high ratio is highly secreted. The tails were sorted by secretion index from highly secreted to highly retained based on GLuc activity under basal conditions (Figure 1A, blue). To evaluate responsiveness to ER calcium depletion, GLuc activity was measured following 24 hr treatment with thapsigargin (Tg), a selective inhibitor of the sarcoplasmic endoplasmic reticulum ATPase (SERCA) (Thastrup et al., 1990) (Figure 1A, pink). Most ERS motifs previously classified as ER localization signals (Raykhel et al., 2007) exhibited a basal secretion index of less than 10 and a ≥3-fold increase in GLuc secretion in response to Tg (Figure S1; Figure 1A). We used these criteria to define an ERS, and 75/94 motifs exhibited this ERS (+) phenotype (Figure S1; Figure 1A). The remaining ERS tails showing high basal secretion and minimal responsiveness to Tg were considered ERS (−) tails. For ERS (+) tails, the increased extracellular GLuc activity observed following Tg treatment was accompanied by a decrease in intracellular levels of GLuc, suggesting GLuc was trafficked out of cells (Figure S2A). Brefeldin A (BFA) blocked the Tg-induced secretion of ERS (+) reporters indicating that trafficking from the ER to Golgi is critical for ERS (+) secretion (Figure 1C) (Helms and Rothman, 1992). As expected, ERS (−) secretion was also inhibited by BFA treatment both in the presence and absence of Tg (Figure 1C; Figure S2D). Additionally, the ryanodine receptor (RyR) antagonist dantrolene, which stabilizes ER calcium following Tg treatment (Wei and Perry, 1996), broadly attenuated Tg-invoked ERS (+) secretion but not ERS (−) secretion (Figure 1D). The secretion of ERS-containing proteins was specific to ER calcium depletion, as other ER stressors (tunicamycin and DTT) caused no change in ERS (+) secretion (Figures S2B and S2C). Taken together, all constructs in the ERS library were secreted via the classical ER to Golgi secretory pathway and a majority responded to ER calcium depletion.
Figure 1. Characterizing the Secretion of ER Resident Proteins with ERSs.
(A) Secretion of GLuc fusion proteins from transiently transfected SH-SY5Y cells under basal conditions (blue) and after 200 nM Tg for 24 hr (pink) (n = 9). Overlap of the two groups is purple. Secretion index refers to the ratio of extracellular to intracellular GLuc.
(B) Sequence logo produced by WebLogo 3.4 forproteins designated as ERS (+).
(C and D) Fold change in secretion of GLuc fusion protein from transiently transfected SH-SY5Y with (blue) or without (pink) (C) 500 nM brefeldin A (BFA) or (D) 50 μM dantrolene prior to an 8 hr exposure to 200 nM Tg (n = 6 for dantrolene, n = 12 for Tg).
See also Figures S1 and S2.
A sequence logo derived from the ERS (+) tails graphically represents the amino acid frequency for C-terminal tails of proteins sensitive to ER calcium depletion (Figure 1B). The logo predicted that KKKKDEL, a tail not present in a human protein, would be the most sensitive to Tg-induced ER calcium depletion, and while our library did not include such a tail, we did observe that KKKHDEL exhibited the greatest fold change in secretion following Tg (Figure 1A, ERS #86).
Endogenous ERS-Containing Proteins Are Secreted in Response to ER Calcium Depletion
We performed pathway analysis of the endogenous proteins corresponding to the ERS (+) tails from the reporter library using Ingenuity Pathway Analysis (IPA) software. The top two canonical pathways involving ERS (+) tails were the UPR and ER stress pathways (Figure S3A). The two most significant functional groups represented by ERS (+) proteins were post-translational modification and protein folding (Figure S3B). These data support that endogenous ERS-containing proteins play critical roles in cellular proteostasis.
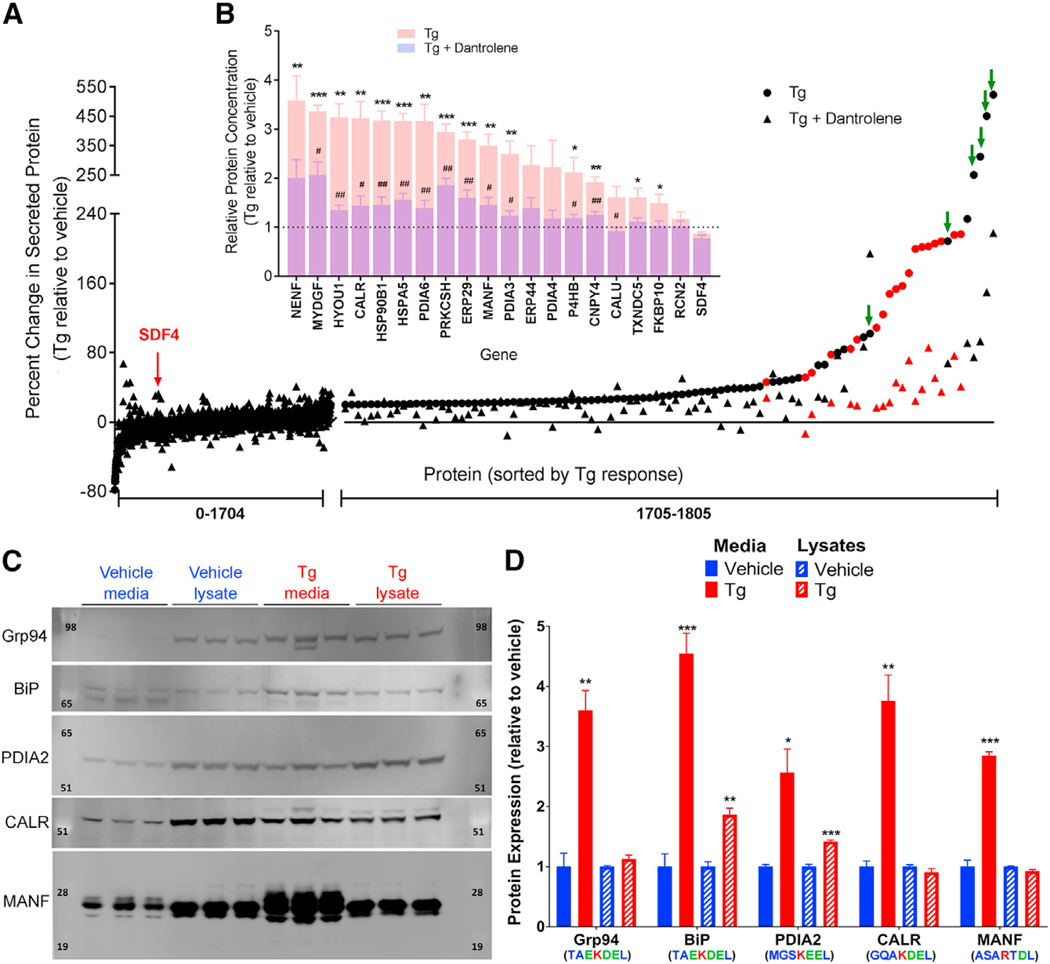
We next examined the secretome of ER calcium depleted cells for the presence of endogenous proteins corresponding to the ERS (+) tails identified by our reporter library. SH-SY5Y cells were treated with vehicle, Tg, or Tg plus dantrolene, and secreted proteins were analyzed by mass spectrometry. Of the 1,805 proteins identified, twenty corresponded to ERS (+) proteins, nineteen of which were in the top 2% of proteins increased following Tg (Figure 2A). Of the nineteen, fifteen were significantly released following Tg treatment (relative to vehicle), and the remaining four exhibited the same trend (Figure 2B). An additional six proteins (GANAB isoforms 2 and 3, NRROS, SPTB, PRDX4, and C9orf78) that are known to interact with ERS (+) proteins were also in the top 2% (Figure 2A, green arrows) (Trombetta et al., 1996; Huttlin et al., 2015; Hermjakob et al., 2004; Tavender et al., 2010). Treatment with dantrolene attenuated the Tg-induced secretion of both ERS (+) proteins and their putative interacting proteins (Figures 2A and 2B). Only one ERS (−) protein from our library, CNTN1, was identified, and as predicted, its extracellular expression was not affected by Tg (ERS #4 in Figure 1A; protein #1538 in Figure 2A). To corroborate our mass spectrometry findings, western blot analysis of a subset of endogenous ERS (+) proteins revealed a significant increase in GRP94, BiP, CALR, and MANF in the media following Tg treatment (Figures 2C and 2D). We also observed an increase in PDIA2, which was predicted from the reporter library but not detected by mass spectrometry (Figures 2C and 2D). The intracellular levels of BiP and PDIA2 also increased (Figures 2C and 2D), consistent with previous reports that they are UPR genes upregulated by Tg (Földi et al., 2013). Together, these data demonstrate a phenomenon in which a departure of ER resident proteins from cells occurs in response to ER calcium depletion.
Figure 2. Endogenous ERS-Containing Proteins Are Secreted in Response to Calcium Dysregulation.
(A) Mass spectrometry identified 1,805 proteins from SH-SY5Y cell culture media following treatment with 200 nM Tg for 8 hr (circles) or with 50 μM dantrolene pre-treatment for 30 min (triangles) (n = 3). Peptides of putative endogenous ERS (+) proteins are identified in red. Green arrows indicate proteins known to interact with ERS (+) proteins.
(B) Fold change in peptide fragments identified in Tg versus vehicle treated media for detectable ERS (+) proteins (n = 3; unpaired two-tailed t tests; *p < 0.05, **p < 0.01, ***p < 0.001 for Tg versus vehicle; #p < 0.05, ##p < 0.01 for Tg versus Tg plus dantrolene).
(C) western blots of cell culture media and lysates for a selected set of putative endogenous ERS (+) proteins following treatment with 200 nM Tg or vehicle for 8 hr.
(D) Densitometry analysis of western blots (n = 3; unpaired two-tailed t test; *p < 0.05, **p < 0.01, ***p < 0.001).
See also Figure S3.
KDEL Receptors Identified as Primary Modulators of ERS Secretion in Response to ER Calcium Depletion
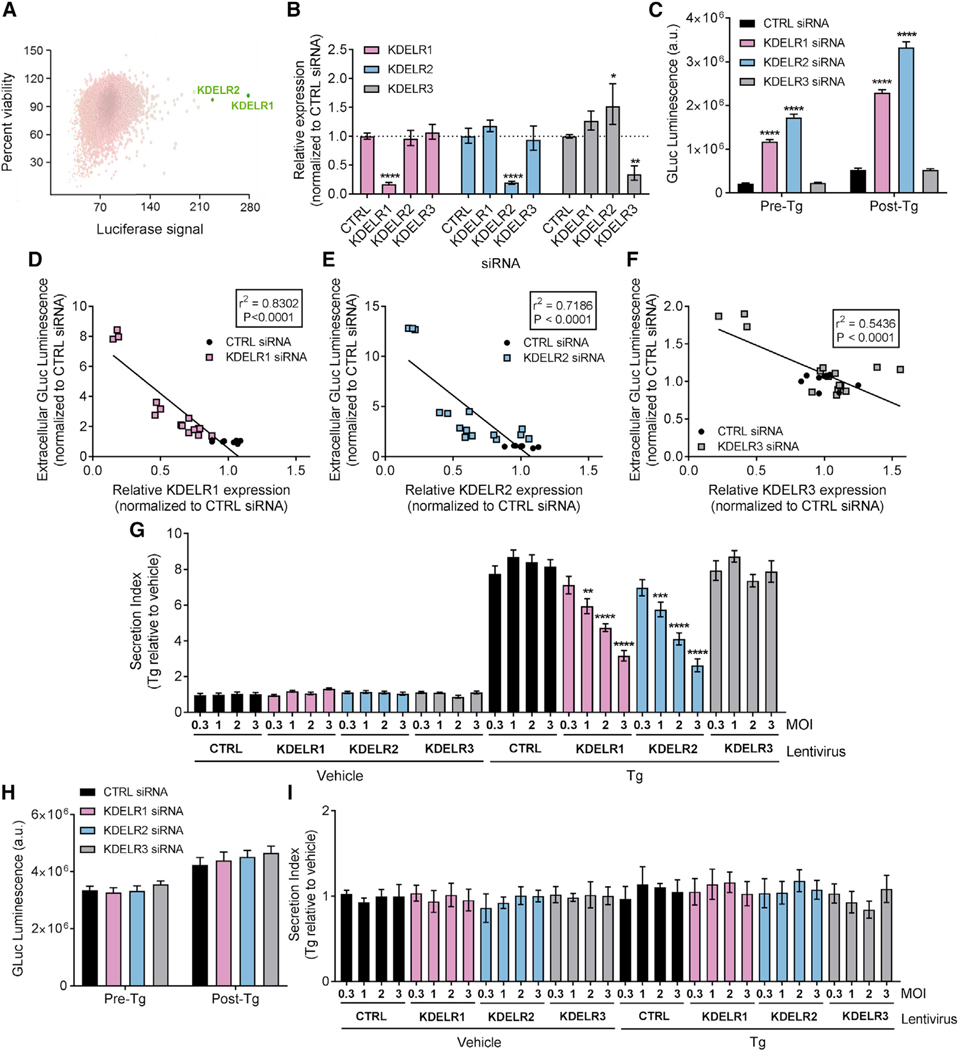
The similarity of the identified putative ERS to the canonical ERS, KDEL, implies that KDEL receptors likely regulate GLuc-ERS secretion (Raykhel et al., 2007). A high-throughput RNAi screen including over 20,000 genes identified KDELR1 and KDELR2 in the top 0.1% of genes that when knocked down caused the largest increase in Tg-induced GLuc-ASARTDL secretion in SH-SY5Y cells, without affecting cell viability (Figure 3A). Validation of the RNAi screen confirmed that the small interfering RNAs (siRNAs) were KDEL receptor isoform-specific (Figure 3B) and that knockdown of KDELR1 and KDELR2 led to a robust increase in GLuc-ASARTDL secretion both under basal conditions (pre-Tg) and following Tg treatment (post-Tg; Figure 3C), with the increase in secretion being highly dependent on the degree of KDEL receptor knockdown (Figures 3D and 3F). The lack of a significant effect of KDELR3 knockdown on the secretion of GLuc-ASARTDL may be due to the low basal expression of KDELR3 in SH-SY5Y cells (Figure S4A). Based on the variation in relative expression of the three receptors between cell lines (Figures S4A–S4D) and mammalian tissues (Figures S4E–S4P), the effect of a knockdown of different isoforms is likely to result in different cell- or tissue-specific phenotypes. In contrast to knockdown, overexpression of KDELR1 and KDELR2 attenuated GLuc-ASARTDL secretion following Tg treatment in a dose-dependent manner (Figure 3G). Importantly, neither knockdown nor overexpression of single KDEL receptors influenced general secretion as measured by extracellular levels of the constitutively secreted GLuc-Untagged, which lacks an ERS (Figures 3H and 3I).
Figure 3. KDEL Receptors Regulate the Secretion of GLuc-ASARTDL.
(A) A human RNAi library was screened for effects of gene knockdown on Tg-induced GLuc-ASARTDL secretion. The plot shows the effect of each gene on cell-viability versus Tg-induced GLuc-ASARTDL secretion compared to control siRNA for each gene in the library. Positive hits are colored for the GLuc signal effect (maximal GLuc is green) and sized by seed-corrected Z score (maximum Z score is the largest sphere).
(B) KDEL receptor mRNA levels estimated with real-time RT-qPCR in SH-SY5Y cells transfected with siRNAs specific for KDELR1, KDELR2, or KDELR3 (n = 3; *p < 0.05, ****p < 0.0001 versus control siRNA; two-way ANOVA and Tukey’s test).
(C) GLuc activity in media under normal conditions (pre-Tg), and after an 8 hr incubation in 100 nM Tg (post-Tg) following RNA interference of the KDEL receptors in SH-SY5Y cells stably expressing GLuc-ASARTDL (n = 9; ****p < 0.0001 versus control siRNA; two-way ANOVA and Tukey’s multiple comparison test).
(D–F) Correlation of extracellular GLuc activity with mRNA expression of (D) KDELR1, (E) KDELR2, and (F) KDELR3 following transfection with 5–20 nM siRNAs.
(G) SH-SY5Y cells transiently transfected with GLuc-ASARTDL and transduced with lentiviral vectors expressing KDEL receptors at different MOIs were exposed to 200 nM Tg or vehicle for 24 hr. Fold change in GLuc secretion is indicated (n = 6; one-way ANOVA; **p < 0.01, ***p < 0.001, ****p < 0.0001 KDEL receptor versus control).
(H) GLuc activity in the media from SH-SY5Y cells stably expressing GLuc-Untagged after transfection with KDEL receptor siRNAs (n = 6).
(I) SH-SY5Y cells overexpressing KDEL receptors at different MOI and transiently transfected with GLuc-Untagged were exposed to 200 nM Tg or vehicle for 24 hr. Fold change in GLuc secretion is indicated (n = 6).
See also Figures S4 and S5.
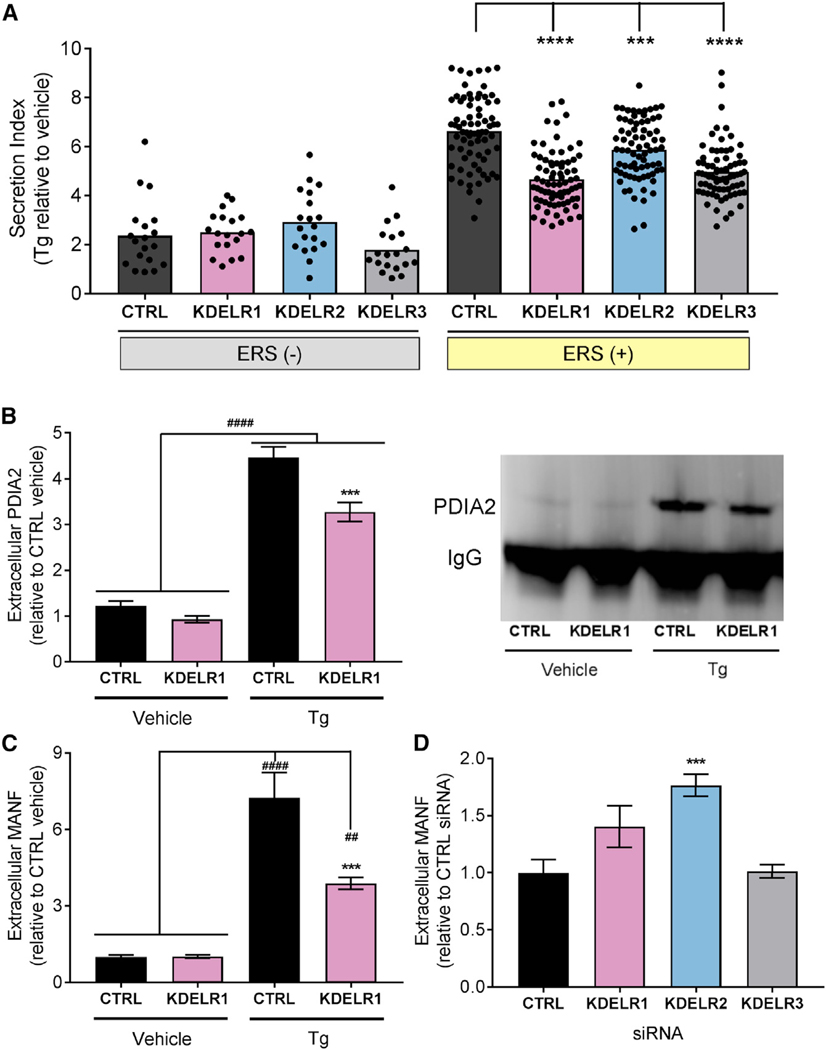
We individually overexpressed the KDEL receptors (KDELR1, KDELR2, and KDELR3) together with the ERS tail library in SH-SY5Y cells, and each KDEL receptor isoform showed a distinct profile for inhibiting the release of GLuc-ERS variants (Figures S5A–S5C). However, when ERS (+) tails are taken as a group, all KDEL receptors significantly attenuated Tg-induced secretion, an effect not seen with ERS (−) tails (Figure 4A). Additionally, KDELR1 and KDELR3 broadly attenuated Tg-induced responses compared to KDELR2, which appeared to affect a smaller subset of ERS (+) tails.
Figure 4. KDEL Receptors Regulate the Secretion of ERS-Containing Proteins.
(A) Average fold change in secretion of ERS (+) and ERS (−) constructs from transiently transfected SH-SY5Y cells overexpressing KDEL receptors following treatment with 200 nM Tg for 24 hr (n = 4; two-way ANOVA with Dunnett’s multiple comparisons test; p < 0.0001 for ERS (+) versus ERS (−), ***p < 0.001 and ****p < 0.0001 for control versus KDEL receptor). Each dot represents a unique GLuc-ERS.
(B and C) Fold change in (B) immunoprecipitated PDIA2 (representative blot is shown to the right) or (C) MANF in media from SH-SY5Y cells overexpressing KDELR1 or control and treated with 200 nM Tg or vehicle for 24 hr (n = 9; **p < 0.01, ####p < 0.0001 Tg versus vehicle, ***p < 0.001 KDELR1 Tg versus CTRL Tg). (C) Extracellular MANF levels in the media of SH-SY5Y cells transfected with KDEL receptor siRNAs (n = 6; one-way ANOVA and Dunnett’s test; ***p < 0.001 versus CTRL).
See also Figures S4 and S5.
Given our results with ERS reporter constructs and KDEL receptors, we proposed that KDEL receptors are capable of attenuating ER calcium-induced secretion of endogenous proteins. Indeed, overexpression of KDELR1 resulted in decreased extracellular levels of PDIA2 and MANF (Figures 4B and 4C), while KDEL receptor knockdown led to an increase in extracellular ERS (+) protein levels (Figure 4D). Taken together, these data support a model in which the localization of ER resident proteins depends both on ER calcium levels and the presence of KDEL receptors.
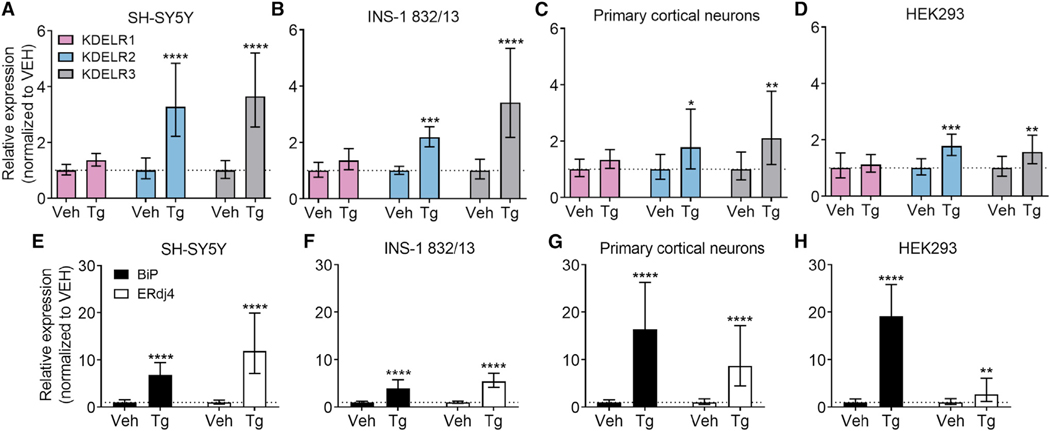
KDELR2 and KDELR3, but Not KDELR1, Are Upregulated by ER Stress
We next examined the influence of ER calcium depletion and ER stress on KDEL receptor expression as a possible adaptive response. Using four cell lines (SH-SY5Y, INS-1 832/13, HEK293, rat primary cortical neurons) with variable relative expression of the KDEL receptor isoforms at baseline (Figures S4A–S4D), we observed an upregulation of KDELR2 and/or KDELR3 mRNA in response to Tg in all cell lines (Figures 5A–5D). An increase in the expression of KDELR2 and KDELR3 was also observed in response to another chemical inducer of ER stress, the N-linked glycosylation-blocking agent tunicamycin (Figures S6A–S6C). The more limited increase in KDEL receptor mRNA following tunicamycin treatment is probably caused by different ER stress onset times due to separate modes of action for Tg and tunicamycin. For example, Tg has been reported to cause a peak increase in spliced XBP1 in HeLa cells already at 4 hr, while the peak is observed at 6 hr with tunicamycin (Yoshida et al., 2001).
Figure 5. KDEL Receptor Expression Is Increased by Thapsigargin-Induced ER Stress.
(A–D) KDEL receptor mRNA levels were analyzed with real-time RT-qPCR after 8 hr treatment with vehicle (Veh) or 100 nM Tg in (A) SH-SY5Y cells, (B) INS-1 832/13 cells, (C) rat primary cortical neurons, or (D) HEK293 cells (n = 8–9).
(E–H) BiP and ERdj4 expression in the same cells. BiP and ERdj4 mRNA levels were analyzed with real-time RT-qPCR after 8 hr treatment with vehicle (Veh) or 100 nM Tg in (E) SH-SY5Y cells, (F) INS-1 832/13 cells, (G) rat primary cortical neurons, or (H) HEK293 cells (n = 8–9). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 Tg versus Veh treatment, two-way ANOVA and Tukey’s post-hoc test.
See also Figures S4 and S6.
To verify a Tg-induced activation of the UPR, the expression of two established UPR genes known to be activated by ATF6 and IRE1α/XBP1, BiP and ERdj4 (Lee et al., 2003; Shoulders et al., 2013), were analyzed from the same samples (Figures 5E–5H). Tg treatment resulted in upregulation of both BiP and ERdj4 in all cell lines studied; however, the relative increase in the expression of the two UPR genes differed among the cell lines.
IRE1α/XBP1-Induced Upregulation of KDELR2 and KDELR3 Affects ERS Retention
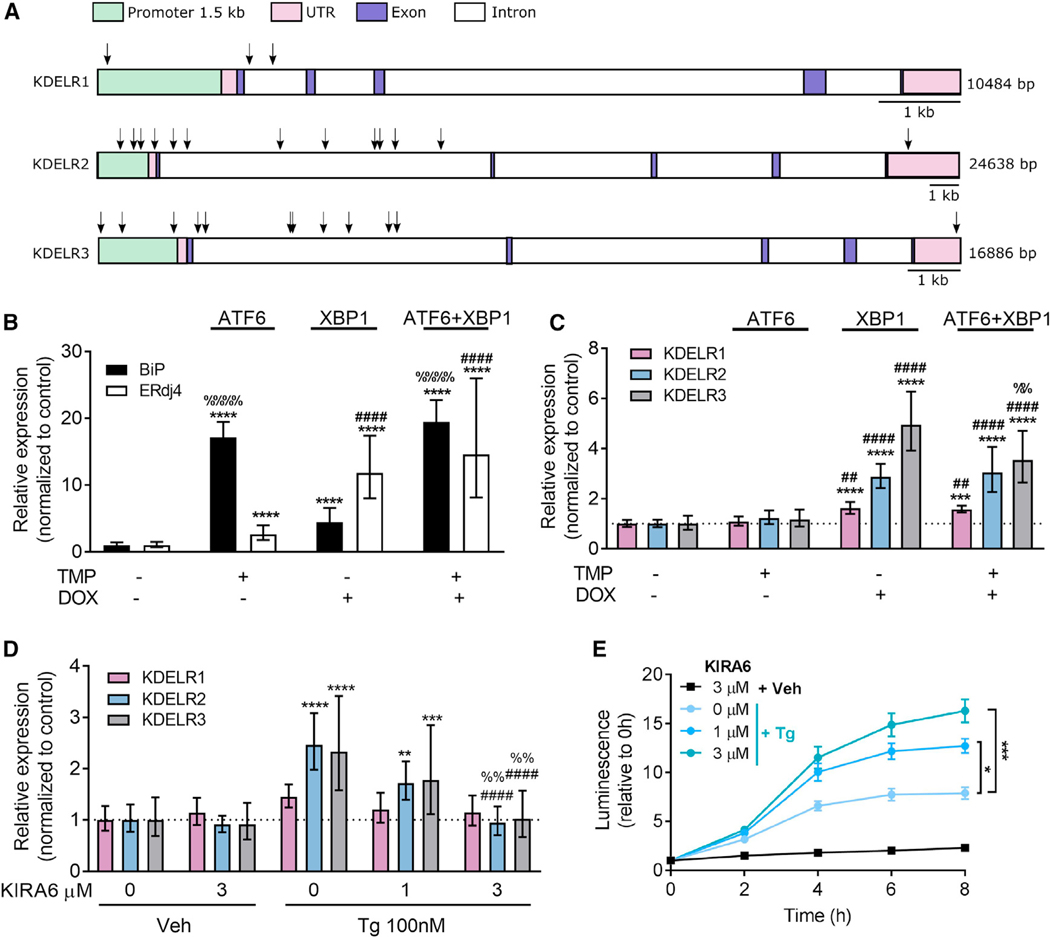
The observed upregulation of KDEL receptors during ER stress prompted us to examine the influence of UPR-related transcription factors on KDEL receptor expression. An explorative in silico analysis of transcription factor binding sites in the promoters of the three KDEL receptor genes using ConTra v2 (Broos et al., 2011) was performed. Putative binding sites for XBP1 were identified in the promoters of KDELR2 and KDELR3 with cumulative scores (based on number of predicted binding sites, phylogenetic conservation, distance from transcription start site, proportion of conserved predicted binding sites, and information content of the predicting position weight matrix (Hooghe et al., 2008)) of 2.322 and 2.884, respectively, for a 1.5 kb region upstream of the transcriptional start site (Table S1, upper table). As a comparison, the known XBP1-regulated gene ERdj4 had a cumulative score of 0.588 for XBP1 binding, while the score for the KDELR1 promoter was 0.000. The scores for ATF6 and ATF4, two other UPR-linked transcription factors, were 0.000 for all four gene promoters. A visualization and list of putative XBP1 binding sites in the KDEL receptor genes are found in Figure 6A and Table S1 (lower table). These data suggest that KDELR2 and KDEL3, but not KDELR1 are likely to respond to the transcription factor XBP1.
Figure 6. XBP1 Upregulates KDEL Receptor Expression.
(A) Schematic figure of putative binding sites for XBP1 in the KDEL receptor genes (arrows).
(B and C) ATF6 and XBP1 activity was induced in HEK293DAX cells by a 16 hr incubation in 10 mM trimethoprim (TMP) and/or 1 μg/mL doxycycline (DOX), and expression levels of (B) BiP and ERdj4 and (C) KDEL receptors were analyzed using real-time RT-qPCR (n = 8–9).
(D) XBP1 splicing was inhibited by a 1hr pretreatment with 1 or 3 μM KIRA6, and changes in KDEL receptor expression were assessed after exposure of SH-SY5Y cells to 100 nM Tg for 8 hr (n = 6–9).
(E) Extracellular GLuc-ASARTDL from cells described in (D) (n = 6).
All transcription results are shown as 2−ddCq (mean ± upper and lower limit) with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 versus control groups using two-way ANOVA followed by Tukey’s test. In (B) and (C): ##p < 0.01, ####p < 0.0001 as compared to TMP only, and %%p < 0.01, %%%%p < 0.0001 as compared to DOX only. In (D): ####p < 0.0001 as compared to 0 mM KIRA6/Tg and %%p < 0.01 as compared to 1 μM KIRA6/Tg. See also Figure S6 and Table S1.
We tested whether XBP1 was sufficient for the transcriptional activation of KDELR2 and KDELR3 using the previously described HEK293DAX cell line (Shoulders et al., 2013). This cell line expresses the ATF6 protein fused to a destabilizing domain from dihydrofolate reductase (ddDHFR) and a Tet-inducible gene for spliced XBP1. Induction of ATF6 activity with 10 mM trimethoprim or XBP1 activity with 1 μg/mL doxycycline allowed for stress-independent activation of these two pathways and resulted in transcriptional activation of BiP or ERdj4, respectively (Figure 6B). Only doxycycline treatment increased KDELR2 and KDELR3 expression, indicating that the genes are responsive to XBP1 but not ATF6 (Figure 6C). The upregulation of KDELR2 and/or KDELR3 in response to XBP1 activity is consistent with the published transcriptional profiling of the HEK293DAX cells (Shoulders et al., 2013) and whole-genome array data from mouse fibroblasts (Sriburi et al., 2007). Applying doxycycline and trimethoprim to parental HEK293FT cells did not affect gene expression, suggesting that the upregulation was indeed a result of XBP1 pathway activation and not an unspecific effect of the chemical inducers (Figures S6D and S6E).
To further examine the XBP1-inducible upregulation of KDELR2 and KDELR3, we used an inhibitor of IRE1α kinase activity, KIRA6, to inhibit ER-stress-induced splicing of XBP1 (Ghosh et al., 2014). One-hour pretreatment with KIRA6 attenuated the Tg-induced upregulation of KDELR2 and KDELR3 in a dose-dependent manner (Figure 6D), further supporting our observation that spliced XBP1 acts as a transcription factor for KDELR2 and KDELR3 during ER stress. We did not observe changes in KDEL receptor expression with KIRA6 treatment only, suggesting that XBP1 is not essential for basal KDEL receptor expression. Importantly, pretreatment of GLuc-ASARTDL stable SH-SY5Y cells with increasing doses of KIRA6 resulted in a dose-dependent increase of luminescence in the media following Tg (Figure 6E), indicating that the inhibition of ER-stress-induced KDEL receptor upregulation potentiated the release of GLuc-ASARTDL. Since inhibition of KDELR3 upregulation with RNAi during ER stress did not affect secretion of GLuc-ASARTDL (Figure 3C; Figure S6F), the effect of KIRA6 treatment on ER-stress-induced GLuc-ASARTDL secretion in SH-SY5Y cells is probably due to a failure of the cell to upregulate KDELR2. The KIRA6-induced increase in GLuc-ASARTDL secretion was further verified to be dependent on the ERS tail, since KIRA6 pretreatment did not affect the secretion of GLuc-Untagged (Figure S6G).
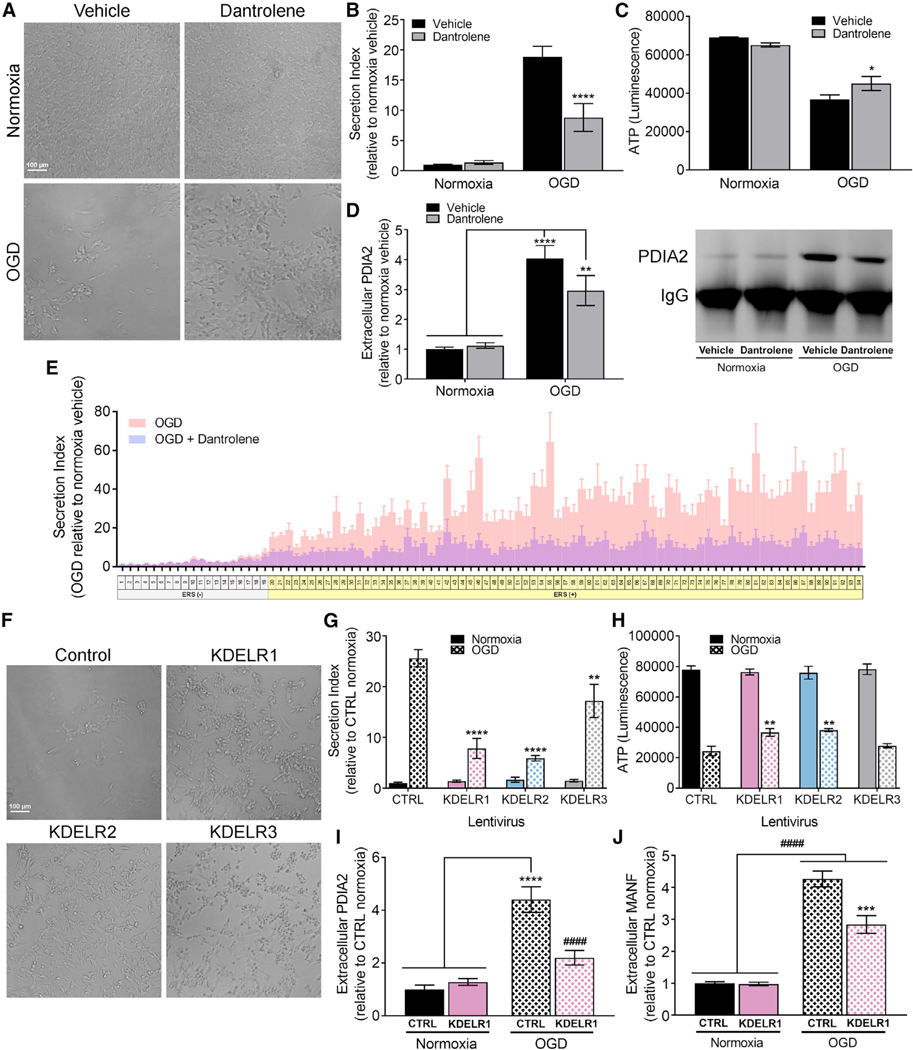
Stabilizing ER Calcium Attenuates ERS Secretion and Toxicity Caused by OGD
Oxygen-glucose deprivation (OGD) is an established in vitro model of ischemia and has been linked to ER stress and reductions in intracellular calcium stores (Badiola et al., 2011; Wang et al., 2002). We exposed SH-SY5Y cells stably expressing GLuc-ASARTDL to OGD and found a marked increase in GLuc-ASARTDL release (Figure 7B) and decrease in cell viability (Figures 7A and 7C; Figure S7A). Treatment with dantrolene attenuated GLuc-ASARTDL release and improved cell viability following OGD (Figures 7A–7C; Figure S7A). Examination of PDIA2, an endogenous ERS-containing protein, found a significant increase in extracellular PDIA2 following OGD, an effect that was partially attenuated by treatment with dantrolene (Figure 7D).
Figure 7. Dantrolene and KDEL Receptor Overexpression Improve Cell Viability and Decrease ER Exodosis in an In Vitro Model of Ischemia.
(A–D) SH-SY5Y cells stably expressing GLuc-ASARTDL were exposed to 16 hr of OGD followed by 24 hr of normoxia with or without 30 mM dantrolene. (A) Representative images of treated cells. Scale bar indicates 100 μm. (B) Fold change in secretion of GLuc-ASARTDL (n = 8; two-way ANOVA with Bonferroni’s multiple comparisons test; p < 0.001 for overall OGD effect, ****p < 0.0001 vehicle versus dantrolene,). (C) ATP viability assay (n = 8; two-way ANOVA with Bonferroni’s multiple comparisons test; p < 0.001 for overall OGD effect, *p < 0.05 vehicle versus dantrolene). (D) Densitometry analysis and representative blot of PDIA2 immunoprecipitated from cell culture media (n = 6; two-way ANOVA with Tukey’s multiple comparisons test; ****p < 0.0001, **p < 0.01 normoxia versus OGD).
(E) Fold change in secretion of GLuc fusion proteins exposed to 16 hr of OGD followed by 24 hr of normoxia with or without 30 μM dantrolene (n = 6 for dantrolene, n = 15 for OGD).
(F–H) SH-SY5Y cells overexpressing KDEL receptors and transiently transfected with GLuc-ASARTDL were exposed to 16 hr of OGD followed by 24 hr of normoxia. (F) Representative images of cells. Scale bar indicates 100 μm. (G) Fold change in secretion of GLuc-ASARTDL (n = 13; two-way ANOVA with Sidak’s multiple comparisons test; p < 0.001 for overall OGD effect, for control OGD versus KDEL receptor OGD ****p < 0.0001, **p < 0.01). (H) Viability ATP assay (n = 13 wells; two-way ANOVA with Dunn’s multiple comparisons test; p < 0.001 for overall OGD effect, **p < 0.01 for control versus KDEL receptors).
(I and J) SH-SY5Y overexpressing KDELR1 or control exposed to 16 hr of OGD followed by 24 hr of normoxia. (I) PDIA2 immunoprecipitation of cell culture media quantified as fold change in extracellular PDIA2 (n = 9; ****p < 0.0001 normoxia versus OGD, ####p < 0.0001 KDELR1 OGD versus CTRL OGD). (J) Fold change in extracellular MANF (n = 6; ####p < 0.0001 normoxia versus OGD, ***p < 0.001 KDELR1 OGD versus CTRL OGD).
See also Figure S7.
We expanded our analysis of OGD-induced secretion to our ERS reporter library and showed that ERS (−) tails exhibited minimal change in response to OGD, whereas the release of ERS (+) tails was significantly elevated when compared to normoxia controls (Figure 7E). In addition, dantrolene attenuated the secretion of ERS (+) reporters, indicating that OGD-induced secretion depends on the levels of ER calcium. Dantrolene did not alter secretion of GLuc reporters under normoxic conditions (Figure S7C). While the relative secretion profile of ERS (+) tails in response to OGD was of greater magnitude than the secretion profile from Tg, there was a positive correlation (linear regression, p < 0.0001, R2 = 0.209) between OGD response and Tg response (Figure S7B).
Despite the robust increase in secretion of KDEL ligand already at 8 hr post-OGD recovery (Figure S7D), we detected a significant upregulation of only KDELR3 and not until 16 hr later (Figure S7E). The upregulation of KDELR3 correlated with a significant increase in mRNA levels of the IRE1α/XBP1-regulated UPR gene ERdj4 (Figure S7F). Because of the relatively low basal KDELR3 expression in SH-SY5Y cells (Figure S4A), the increase in endogenous KDELR3 likely has minimal effect on the secretion of GLuc-ASARTDL. We next examined whether ERS secretion caused by OGD could be altered by an increase in exogenous KDEL receptor expression and found that lentiviral-mediated expression of each KDEL receptor isoform significantly reduced GLuc-ASARTDL secretion (Figure 7G) and improved cell viability (Figures 7F and 7H) with KDELR3 being less effective at attenuating GLuc-ASARTDL release and causing no improvement in viability. Overexpression of KDELR1 reduced extracellular levels PDIA2 and MANF following OGD (Figures 7I and 7J), suggesting that KDEL receptors can modulate the secretion of endogenous as well as exogenous proteins with an ERS. The data collectively support that KDEL receptor overexpression and dantrolene treatment reduce ERS secretion in pharmacological and physiological models of ER calcium depletion.
DISCUSSION
The concentration of calcium within the ER exceeds that of the cytosol, and maintenance of this gradient is important for sustaining proper ER functioning. We discovered that ERS-containing proteins are redistributed from the ER lumen to the extracellular space in response to pathophysiological conditions where ER calcium stores are depleted. We propose classifying the phenomenon of ERS-containing proteins leaving the cell as “ER exodosis,” where exodosis is defined as the departure of a resident protein from its organelle under pathophysiological conditions (exodos [Greek]: “a final scene or departure in a play, especially a tragedy”). Our results show that cells upregulate KDELR2 and KDELR3 as part of the UPR, which counteracts exodosis. Importantly, we provide evidence that both stabilizing ER calcium and augmenting KDEL receptor expression can attenuate ER exodosis and improve cell viability, suggesting that exodosis is a therapeutic target for diseases associated with ER calcium dysregulation.
The library of GLuc reporters containing seven amino acid C-terminal ERS was built on the seminal work carried out by Raykhel et al. (2007) that identified four amino acid sequences representing ER localization motifs. In addition to corroborating ER retention, we found 75 ERS corresponding to endogenous proteins with critical functions in ER homeostasis that were secreted in response to ER calcium depletion. The precise mechanism by which calcium levels within the ER mediate the retention of ERS proteins is not known. Several of the ERS proteins (e.g., BiP, GRP94, PDIA2, PDIA4, and calreticulin) bind calcium with low affinity and interact with both each other and misfolded proteins (Nigam et al., 1994; Sönnichsen et al., 1994; Meldolesi and Pozzan, 1998). A decrease in calcium concentration has been speculated to reduce their interactions and cause them to be secreted (Sönnichsen et al., 1994). Another hypothesis is that the ER environment, with its high concentration of proteins and calcium, promotes the formation of dynamic hydrogels that allows mobility of the luminal, calcium-interacting proteins but limits their access to ER exit sites (Barlowe and Helenius, 2016). A decrease in the calcium concentration would thereby change the luminal matrix, allowing ER resident proteins to escape and possibly overwhelm the KDEL retrieval pathway.
While the mechanism that regulates ER exodosis is not fully understood, we provide evidence that the well-established KDEL retrieval pathway (Munro and Pelham, 1987; Semenza et al., 1990) plays an adaptive, modulatory role in exodosis. Augmenting the expression of KDEL receptors attenuated ER exodosis, supporting a model in which ERS protein efflux from the cell occurs as a result of saturation of endogenous KDEL receptors (Xiao et al., 1999). Our data also support the KDEL retrieval pathway as part of an endogenous UPR mechanism against exodosis and identify XBP1 as a regulator of the ER-stress-induced expression of KDELR2 and KDELR3. The observed upregulation of KDELR2 and KDELR3, but not KDELR1, during ER stress suggests that the isoforms have previously unknown, distinct functions during stress conditions. We also observed differences in the KDEL receptors’ ability to alter secretion of GLuc-ERS reporters. The variable effects on different ERS tails, differential expression of KDEL receptors across tissues, and the differences in activity of each UPR pathway (Harding et al., 2002) likely create cell-specific responses to exodosis.
The departure of ER resident proteins provides mechanistic insight on ER calcium dysregulation associated with numerous disease states (Missiaen et al., 2000), including cellular apoptosis (Bian et al., 1997), inflammation (Peters and Raghavan, 2011), heart failure (Lindner et al., 1998), and both acute (e.g., ischemia) and chronic (e.g., Alzheimer’s disease) neurological diseases (Paschen, 2001; Resende et al., 2008). An abnormal departure of some ER resident proteins has previously been reported following disruption of ER calcium homeostasis (Henderson et al., 2013, 2014; Booth and Koch, 1989) and in several pathophysiological conditions where ER calcium dysregulation has been implicated (Aksoy et al., 2017; Luo and Lee, 2013; Wires et al., 2017; Wu et al., 2017). In our current study, we show that dantrolene, a drug used clinically for malignant hyperthermia and central core disease, can reduce exodosis caused by Tg and OGD, providing further evidence that the retention and secretion of ERS proteins are calcium-dependent events. The prior clinical use of dantrolene also opens the possibility of examining ER calcium stabilization in other diseases where ER calcium dysregulation is implicated.
Taken together, our findings describe a mechanism of cellular pathology linked to ER calcium depletion. Given the complex and diverse functions of ERS-containing proteins, their secretion is likely to have both positive and negative effects on cell viability. Exodosis of ER resident proteins has at least two potential negative consequences on an affected cell. First, there is a loss of proteins involved in critical ER functions. Second, the relocation of such proteins outside of the cell may contribute to pathological changes in the extracellular environment and trigger inflammation. For example, both BiP/Grp78 and calreticulin are suggested to be involved in rheumatoid arthritis (RA) pathology (Bodman-Smith et al., 2004; Corrigall et al., 2001; Tarr et al., 2010). However, the release of ERS proteins in response to ER calcium depletion may also be advantageous and contribute to their physiological roles. For example, extracellular neuroserpin was recently shown to be a modulator of synaptic development and plasticity (Reumann et al., 2017) and have a protective effect in OGD models (Wang et al., 2015). Similarly, MANF has been shown to have protective effects in models of ischemia (Glembotski et al., 2012; Airavaara et al., 2009), Parkinson’s disease (Voutilainen et al., 2009), and diabetes (Lindahl et al., 2014). Based on our observations with neuroserp in- and MANF-based ERS reporters, the secretion of these proteins is likely impacted by ER calcium. Thus, we suggest that studies on proteins with identified or putative ERS should take into consideration their regulation by ER calcium. In summary, we propose a model in which ER calcium depletion leads to (1) the secretion of resident ER luminal proteins containing an ERS and (2) the activation of KDEL receptor retrieval pathway to reduce the loss of ERS containing proteins. Future studies on interventions aimed at stabilizing ER-calcium or augmenting the KDEL receptor pathway may have therapeutic value in diseases characterized by ER calcium dysregulation and exodosis.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brandon Harvey (BHarvey@intra.nida.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal studies
Rat tissue (brain, liver, heart, kidney) was collected from adult Long-Evans rats (three females, three males) housed in 12 h light/dark cycle. Primary cortex samples were collected from Sprague-Dawley E15 rat embryos (three different litters, three samples per litter). All animal procedures were reviewed and approved by the National Institute on Drug Abuse Animal Care and Use Committee.
Cell Lines
SH-SY5Y
Human neuroblastoma cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM + GlutaMAX, 4.5 g/L D-glucose, 110 mg/L sodium pyruvate; Thermo Fisher Scientific, Waltham, MA) supplemented with 10 units/mL penicillin, 10 μg/mL streptomycin (Thermo Fisher Scientific), and 10% bovine growth serum (BGS; GE Life Sciences). Cells were grown at 37C with 5.5% CO2 in a humidified incubator.
HEK293, HEK293FT, HEK293DAX
Embryonic kidney cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM + GlutaMAX, 4.5 g/L D-glucose, 110 mg/L sodium pyruvate; Thermo Fisher Scientific, Waltham, MA) supplemented with 10 units/mL penicillin, 10 μg/mL streptomycin (Thermo Fisher Scientific). For HEK293 cells (a gift from Dr. Xiao Xiao) media contained 5% bovine growth serum (BGS; GE Life Sciences). For HEK293FT and HEK293DAX cells media contained 10% fetal bovine serum (FBS; Sigma, St. Louis, MO). Cells were grown at 37C with 5.5% CO2 in a humidified incubator. The HEK293DAX cells were a generous gift from Dr. Matt Shoulders at MIT (Shoulders et al., 2013).
INS-1 832/13
Rat insulinoma cell line INS-1 832/13 was maintained at 37C with 5.5% carbon dioxide in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS, 10 units/mL penicillin, 10 μg/mL streptomycin, 10 mM HEPES, 2 mM L-glutamine, 1 mM sodium pyruvate, and 0.05 mM 2-mercaptoethanol. Cells were generously provided by Dr. Christopher Newgard, Duke University.
Rat Primary Cortical Neurons
Rat primary cortical neurons were isolated from Sprague-Dawley rats on embryonal day 15 (E15) as previously described (Howard et al., 2008). Cultures are comprised of embryos of both sexes to obtain adequate cell numbers for experiments. Cells were maintained on PEI-coated 24-well plates in neurobasal media (Thermo Fisher Scientific) supplemented with 2% FBS, 2% B-27 (Thermo Fisher Scientific) and 0.5 mM L-glutamine (Sigma) at 37C with 5.5% carbon dioxide. A 50% media exchange was done every fortnight. Treatments were applied to the cells at 13 days in vitro (DIV13) as a 50% media exchange.
METHOD DETAILS
ER retention sequence plasmid construction
ERS tails were cloned into a pLenti6.3-CMV-MANF sigpep-GLuc-MCS plasmid. The coding region of GLuc was amplified from a previously described GLuc reporter with a MANF signal peptide. This region was then cloned into the BamHI and MluI sites of a pLenti6.3-V5-DEST plasmid to create a pLenti6.3-CMV-MANF sigpep-GLuc-MCS plasmid. This plasmid was then linearized using SgrAI and AscI enzymes to create complementary “sticky ends” for ligating annealed oligonucleotide duplexes to create in-frame ERS tails. Custom forward and reverse oligonucleotides coding for each specific seven amino carboxy-terminal tails were synthesized (Integrated DNA Technologies, Coralville, IA) and annealed to form an oligonucleotide duplex by denaturing at 95C for 10 min and cooling to room temperature (25C) for 1–1.25 h. The oligonucleotides used can be found in Table S2. The oligonucleotide duplexes were ligated using Ligate-IT (Affymetrix) in a 10 μL reaction consisting of 20 ng of SgrAI/AscI digested vector, 0.5 μL of Ligate-IT, and duplexes at a final concentration of 10 mM. Immediately following ligation, DNA was transformed and propagated in Stbl3 recombination deficient cells cultured at 30C (Life Technologies). The resulting DNA plasmids encoding each ERS tail fused to GLuc with a MANF signal peptide were sequence verified for proper ligation of the tail.
Transfections
Plasmid transfection
For reverse transfections, 5 × 104 SH-SY5Y cells in 90 μL of growth media containing no antibiotics were plated in opaque 96-well plates containing 0.06 μL Xfect (Takara, Mountain View, CA) and 200 ng plasmid DNA per well. After 28–29h, a full media exchange into growth media containing 1.5% BGS was performed. Drug treatments (e.g., Tg) began 16–17 h after the media exchange and typically lasted for 8 h or 24 h. For pre-treatments (e.g., dantrolene, brefeldin A), drugs were added 30 min prior to Tg. At the end of each experiment, media was removed from the wells and cells were lysed in 75 μL of 50 mM Tris (pH 7.5), 150 mM NaCl, 1% NP40, and protease inhibitors (Sigma) directly in the plate and used for luciferase assay.
siRNA transfection
Using Lipofectamine RNAiMax (Thermo Fisher Scientific), SH-SY5Y cells were reverse transfected with 5–20 nM siRNAs specific for the KDEL receptors (Silencer Select®, assay #548 (KDELR1), #21689 (KDELR2), #21690 (KDELR3), Thermo Fisher Scientific). Media was changed 48 h after transfection, and treatment assays were performed 64–72 h after the transfection.
Lentivirus
Lentiviral vectors expressing Myc-FLAG tagged KDEL receptors 1, 2 and 3 were previously described and titered using the Lenti-X p24 rapid titer kit (Takara) (Henderson et al., 2013). SH-SY5Y were transduced at the indicated multiplicity of infection (MOI), incubated for 48 h, and then re-plated for reverse transfections with the ERS library as described above. Following dose response experiments, a MOI of 2 was used for all further experiments.
Gaussia luciferase secretion assay
Luciferase levels in 5 μL culture medium or 75 μL cell lysate (cells lysed in 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP40, protease inhibitors (Sigma)) were determined using a plate reader with an injector setup (BioTek Synergy II, Winooski, VT) at 25°C with a sensitivity of 100 and a 0.5 s integration time with a 5 s delay following injection of 100 μL of 10 mM coelenterazine (Regis Technologies, Morten Grove, IL) as previously described (Henderson et al., 2015).
Mass spectrometry
SH-SY5Y cells (2.3 × 107) were plated in a 15 cm dish. At 28 h, a full media exchange was done into 1.5% BGS media and 16 h later, media was removed and cells were rinsed with PBS and mass spectrometry assay media (150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 20 mM HEPES, 1 mM CaCl2, and 1.9 g/L glucose). Drugs were added via full media exchanges into mass spectrometry assay media. After 8 h, 20 mL of media was collected and centrifuged at 1000xg for 5 min. 15 mL of supernatant was concentrated using 10,000MW cutoff concentrator tubes (Millipore, Burlington, MA) by centrifuging at 4000xg for 40 min at 4°C. Briefly, 10 μg of protein was analyzed by tandem mass tag (TMT) labeling, basic reverse phase (bRP) fractionation, and liquid chromatography tandem mass spectrometry (LC-MSMS) analysis. Detailed methods follow.
The pH of supernatants containing 10 μg protein in mass spectrometry assay media (pH 7.5) was adjusted to pH 8 by adding 10 μL of 500 mM TEAB (triethyl ammonium bicarbonate). Proteins were reduced by adding 1 μL of 50 mM TCEP (tris(2-carboxyethyl) phosphine) at 60°C for 1 h, and, after cooling to room temperature, alkylated by adding 0.5 μL 200 mM MMTS (methyl methanethiosulfonate) in isopropanol for 15 min at room temperature. Reduced and alkylated proteins were digested overnight at 37°C by adding 0.55 μg of a 27 μg/mL Trypsin/LysC mixture (V5071, Promega, Madison, WT) in 200 mM TEAB. TEAB (35 μL of 200 mM) was added to each protein digest prior to labeling the peptides with a unique TMT 10-plex reagent according to the manufacturer’s protocol. The nine TMT-labeled peptide samples were mixed, and, after the organic phase was evaporated, excess TMT reagents were removed using Pierce Detergent Removal Spin columns (125 mL, #87776, Thermo Fisher Scientific) and the combined TMT sample was fractionated by basic reverse phase (bRP) chromatography on Oasis HLB uElution plates (Waters, Milford, MA). TMT-labeled peptides were bound to HLB resin in 10 mM TEAB buffer, step-eluted with 15%, 25% and 75% acetonitrile in 10 mM TEAB and dried by vacuum centrifugation. Approximately 1 μg of each fraction (calculated based on the original amount of total protein) was analyzed by liquid chromatography interfaced with tandem mass spectrometry (LCMSMS) using a Nano-Acquits HPLC system (Waters) interfaced with a QExactive HF (Thermo Fisher Scientific). Peptides were loaded onto a C18 trap (S-10 μM, 120Å, 75 μm x 2 cm, YMC, Japan) for 5 min at 5 mL/min in 2% acetonitrile/0.1% formic acid in-line with a 75 mm x 150 mm ProntoSIL-120–5-C18 H column (5 mm, 120Å (BISCHOFF Chromatography, Leonberg, Germany). Peptides eluting during the 2%–90% acetonitrile in 0.1% formic acid gradient over 90 min at 300 nL/min were directly sprayed into a QExactive HF mass spectrometer through 1 μm emitter tip (New Objective, Woburn, MA) at 2.0 kV. Survey scans (full ms) were acquired from 350–1700 m/z with data dependent monitoring of up to 15 peptide masses (precursor ions), each individually isolated in a 1.0 Da window and fragmented using HCD activation collision energy at 32 and 30 s dynamic exclusion. Precursor and fragment ions were analyzed at resolutions 120,000 and 60,000, respectively, and automatic gain control (AGC) target values at 3e5 with 50 ms maximum injection time (IT) and 1e5 with 200 ms maximum IT, respectively.
Isotopically resolved masses in precursor (MS) and fragmentation (MS/MS) spectra were extracted from raw MS data without deconvolution and with deconvolution using Xtract or MS2 Processor in Proteome Discoverer (PD) software (v1.4, Thermo Fisher Scientific). All extracted data were searched using Mascot (2.5.1; Matrix Science, Boston, MA) against the 2015RefSeq_72r_human protein database with the added enzymes and BSA, using the following criteria: sample’s species; trypsin as the enzyme, allowing two missed cleavage; cysteine methanethiosulfenylation and TMT 10-plex on N terminus as fixed modifications; methionine oxidation, asparagine and glutamine deamidation and TMT 6plex on lysine as variable modifications. Peptide identifications from Mascot searches were filtered at 1% False Discovery Rate (FDR) confidence threshold, based on a concatenated decoy database search, using the Proteome Discoverer. Proteome Discoverer uses TMT reporter ions for quantification only from the peptide identifications with the highest Mascot score from the three different extraction methods of the same peptide matched spectrum (PSM). Protein quantification is based on the normalized median ratio of all spectra of tagged peptides from the same protein (Herbrich et al., 2013).
Western blot analysis
Equal amounts of total protein from concentrated media or lysates (cells lysed in 50 mM Tris-HCl (pH 7.4), 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1% NP-40, protease inhibitors) were separated on 4%–12% Bis-Tris NuPage gels (Thermo Fisher Scientific) using MOPS SDS running buffer (Thermo Fisher Scientific). Proteins were transferred to 0.20 μm PVDF (Thermo Fisher Scientific) membranes and immunoblotted with antibodies listed in Key Resources table. Blots were scanned using an Odyssey scanner (LI-COR Biosciences, Lincoln, NE).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-BiP | Cell Signaling | Cat#3177S; RRID:AB_2119845 |
| rabbit anti-calreticulin | Cell Signaling | Cat#2891S; RRID:AB_2275208 |
| rabbit anti-MANF | YenZyme, Henderson et al., 2013 | N/A |
| rabbit anti-Grp94 | Cell Signaling | Cat#2104S; RRID:AB_823506 |
| mouse anti-PDI [RL90] | Abcam | Cat#2792; RRID:AB_303304 |
| goat anti-rabbit IR800 | Rockland | Cat#611-132-122; RRID:AB_220152 |
| goat anti-mouse IR700 | Rockland | Cat#610-130-121; RRID:AB_220121 |
| Bacterial and Virus Strains | ||
| One Shot Stbl3 Chemically Competent E. coli | Thermo Fisher Scientific | Cat#C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s Modified Eagle Medium (1X) + GlutaMAX-I | Thermo Fisher Scientific | Cat#10569-010 |
| Dulbecco’s Modified Eagle Medium (1X), no glucose | Thermo Fisher Scientific | Cat#11966-025 |
| Neurobasal medium | Thermo Fisher Scientific | Cat#12348-017 |
| Bovine growth serum (BGS) | GE Life Sciences | Cat#SH30541.03 |
| Fetal bovine serum (FBS) | Millipore Sigma | Cat#F6178 |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat#15140-122 |
| B-27 | Thermo Fisher Scientific | Cat#17504-044 |
| L-glutamine | Millipore Sigma | Cat#G8540 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat#11360-070 |
| 2-mercaptoethanol | Sigma | Cat#M6250 |
| RPMI-1640 | Thermo Fisher Scientific | Cat#61870036 |
| X-fect | Takara | Cat#631318 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | Cat#13778030 |
| SgrAI | New England BioLabs | Cat#R0603S |
| AscI | New England BioLabs | Cat#R0558S |
| Thapsigargin | Sigma | Cat#T9033-1MG |
| Dantrolene | Sigma | Cat# D9175-250MG |
| Brefeldin A | Sigma | Cat#B7651-5mg |
| Tunicamycin | Sigma | Cat# T7765-5mg |
| Dithiothreitol (DTT) | Sigma | Cat#646563-10X.5ML |
| Doxycyline | Sigma | Cat#D9891-1G |
| Trimethoprim | Sigma | Cat# 92131-1G |
| KIRA6 | Cayman Chemicals | Cat#19151 |
| Blocking buffer | Rockland | Cat#MB-070 |
| Tris-HCl pH 7.4 | Quality Biological | Cat#351-006-101 |
| Sodium deoxycholate | Millipore Sigma | Cat#6750 |
| EDTA | Millipore Sigma | Cat#E7889 |
| NP-40 | Thermo Fisher Scientific | Cat#PI-28324 |
| Protease inhibitors | Millipore Sigma | Cat#P8340-5ML |
| 4–12% NuPAGE gels | Thermo Fisher Scientific | Cat#NP0329BOX |
| NuPAGE MOPS SDS running buffer | Thermo Fisher Scientific | Cat#NP0001 |
| PDVF membranes, 0.2 mm | Thermo Fisher Scientific | Cat#LC2002 |
| NuPAGE transfer buffer | Thermo Fisher Scientific | Cat#NP0006-1 |
| Coelenterazine | Regis Technologies | Cat#1-361204.200 |
| Trypsin/LysC Mix | Promega | Cat#V5071 |
| TaqMan Universal PCR Master Mix | Thermo Fisher Scientific | Cat#4364340 |
| Protein A SureBeads | Bio-Rad | Cat#1614013 |
| NuPAGE LDS sample buffer (4x) | Thermo Fisher Scientific | Cat#NP0007 |
| Critical Commercial Assays | ||
| Ligate-IT | Affymetrix | Cat#78410 |
| NucleoSnap Plasmid Midi | Takara | Cat# 740494.10 |
| NucleoSpin RNA kit | Takara | Cat#740955 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#1708891 |
| DC Protein Assay Kit | Bio-Rad | Cat#5000112 |
| Lenti-X p24 rapid titer kit | Takara | Cat#632200 |
| RNeasy Lipid Tissue Mini Kit | QIAGEN | Cat#74804 |
| MANF HTRF | Cisbio | Cat#63ADK056PEG |
| CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS assay) | Promega | Cat#G3580 |
| CellTiter-Glo Luminescent Cell Viability Assay (ATP assay) | Promega | Cat#G7571 |
| Experimental Models: Cell Lines | ||
| SH-SY5Y | Dr. Howard Federoff, University of Rochester, circa 1999 | ATCC®CRL-2266 |
| HEK293 | Dr. Xiao Xiao, UNC | N/A |
| HEK293FT | ThermoFisher Scientific | R70007 |
| HEK293DAX | Shoulders et al., 2013 | |
| INS-1 832/13 | Christopher Newgard | Cell Line 832/13 |
| Experimental Models: Organisms/Strains | ||
| Long-Evans rats, WT | Charles River Laboratories | Strain Code: 006 |
| Sprague-Dawley, WT | Charles River Laboratories | Strain Code: 400 |
| Oligonucleotides | ||
| See Table S2 for oligonucleotide sequences used to create GLuc plasmids with ER retention sequences. | N/A | |
| See Table S3 for nucleotide sequences for all primers and probes. | N/A | |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Proteome Discoverer | Thermo Fisher | v1.4 |
| Odyssey Infrared Imaging System 3.0.30 | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/odyssey/ |
| Mascot 2.5.1 | Matrix Science | |
| WebLogo3.4 | University of California, Berkeley | http://weblogo.threeplusone.com |
| Ingenuity Pathway Analysis | QIAGEN | Version: 33559992 |
| ConTra v2 | Broos et al., 2011 | http://bioit.irc.ugent.be/contrav2/index.php?v2 |
| BioGPS | Wu et al., 2009 | http://biogps.org |
| Other | ||
| 10,000 MWCO cutoff concentrator tubes | Millipore Sigma | UFC901008 |
| Pierce Detergent Removal Spin columns | Thermo Fisher | Cat#87776 |
| Oasis HLB uElution plates | Waters | Cat#186001828BA |
| Nano-Acquits HPLC | Waters | N/A |
| QExactive HF mass spectrometer | Thermo Fisher | Cat#IQLAAEGAAPFALGMBFZ |
| C18 trap | YMC, Japan | N/A |
| ProntoSIL-120-5-C18 H column | Bischoff Chromatography | N/A |
| 1 μm emitter tip | New Objective | N/A |
| C1000 Thermal Cycler CFX96 Real-Time System | Bio-Rad | Cat#1855195 |
| Hypoxia chambers | Billups-Rothenberg | Cat#MIC-101 |
Gene expression analysis
Animal tissue collection
Rats were deeply anesthetized with isoflurane and intracardially perfused with heparinized saline for 4 min after which the organs were quickly removed. Rat brains and samples from the liver (superior right lobe), heart (left ventricle), and kidney were immediately frozen in ice-cold isopentane. Dissection of the rat prefrontal cortex, striatum, midbrain, hippocampus and cerebellum was done in a freezing microtome. Primary cortex was collected from Sprague-Dawley E15 rat embryos and immediately frozen on dry ice.
Real time qPCR
Total RNA was isolated from cells using NucleoSpin RNA kit (Takara) or from rat tissue using RNeasy Lipid Tissue Mini kit including an on-column DNA digest (Qiazol). Using the iScript cDNA Synthesis Kit (Bio-Rad), 0.5 μg of the RNA samples was transcribed into cDNA in a 20 μL reaction mix and diluted 1:20 with DNase-free water. 5.0 μl of the cDNA samples were applied as duplicates in a 20 μl reaction mix consisting of TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), 450 nM primers and 100 nM probe. Real time qPCR was performed with C1000 Thermal Cycler CFX96 Real-Time System (Bio-Rad) with 50 amplification repeats (94°C for 20 s, 60°C or 63°C (rat KDEL receptors) for 1 min). All Cq values were normalized to the geometric mean of the Cq for the reference genes ubiquitin-conjugating enzyme 2i (Ube2i) and RNA polymerase II (PRNAII).
Primers and probes
Probes for KDEL receptors and markers of ER stress signaling activation were labeled with FAM/BHQ1, and probes for reference genes with HEX/BHQ1. KDELR3 oligonucleotides were designed to recognize both KDELR3 isoforms (3a and 3b). KDEL receptor primer and probe efficiency was confirmed by applying a DNA standard curve consisting of five-log dilutions of KDEL receptor DNA (50 to 500,000 copy numbers). Delta Cq (KDEL receptors versus reference genes) remained constant over a three-log dilution of cDNA sample (slopes between 0.1 and +0.1). To exclude isoform cross-reactivity, 500,000 copy numbers of each isoform DNA (plasmid DNA for human, and gene fragments (gBlocks, Integrated DNA Technologies) for rat KDEL receptors) were applied to the PCR reaction mixes. No-template controls (NTC) and no-reverse transcriptase controls (NRT) were used to control for contamination. The nucleotide sequences for all primers and probes can be found in Table S3.
Oxygen glucose deprivation (OGD)
Cells were reverse transfected in 96-well plates 48 h prior to the start of the OGD procedure. The OGD media (culture media without glucose) was deoxygenated by bubbling a gas mixture containing 5% CO2, 10% hydrogen, and 85% nitrogen into the media for 15 min (Goldberg and Choi, 1993). Every plate underwent a full media exchange into standard culture media (normoxia control) or OGD media with or without treatments (30 μM dantrolene or vehicle). OGD plates were placed into hypoxia chambers (BillupsRothenberg, Del Mar, CA) deoxygenated for 10 min with the gas mixture above and placed in a 37C incubator. After 16 h of OGD, cells were removed from the hypoxia chambers, media was replaced with glucose-containing growth media (media was also exchanged in normoxia controls) and returned to the incubator for a further 24 h before collection of samples.
Immunoprecipitation (IP)
IP was performed with magnetic Protein A beads (SureBeads, Bio-Rad) following centrifugation of samples at 4°C for 5 min at 1000x g. Beads were washed with PBS + 0.1% Tween, then incubated with antibodies 1:100 for 10 min. After washing, beads were incubated with 400 μL cell culture media for 1 h. A final wash was followed by elution with 40 μL of 1x LDS (Thermo Fisher Scientific).
MANF Homogeneous Time Resolved Fluorescence assay
The concentration of MANF was determined on media samples diluted 1:2 in dilution buffer using a MANF HTRF assay (Cisbio, Codolet, France) according to the manufacturer’s instructions.
Cell viability
Cell viability assays were performed with the Promega CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS assay) and Promega CellTiter-Glo Luminescent Cell Viability Assay (ATP assay) per the manufacturer’s protocol.
Bioinformatic analyses
WebLogo3.4
WebLogo3.4 (http://weblogo.threeplusone.com) was used to create a sequence logo representing the amino acid alignment associated with Tg responsiveness. For the sequence logo, the amino acids of ERS (+) tails were weighted based on the tail’s response to Tg (fold change in secretion index relative to vehicle) and entered WebLogo. The height of each symbol within the column represents the probability of each amino acid at that position. No information regarding interactions between the different amino acid positions is conveyed.
Ingenuity IPA
Ingenuity IPA version: 33559992 was used to group the 76 proteins with ERS (+) tails. Of the 75 proteins analyzed using Ingenuity IPA, one (CES hBr2, UniProt Q8TDZ9) was not recognized and therefore not present in the dataset. For the canonical pathways analysis, the -log p value was calculated by Fisher’s exact test right-tailed and only those pathways with p < 0.05 are shown. For the functional classification, the disease and biofunction option was chosen to look at downstream effects of the proteins of interest. Z-score algorithm was used to make predictions and a threshold p value of 0.05 was used (as calculated by Fisher’s exact test right tailed).
ConTra v2
Potential transcription factor binding sites within 1.5 kb upstream of the transcriptional start site for human KDELR1 (NM_00681), KDELR2 (NM_006854), KDELR3 (NM_006855), and ERdj4 (NM012328) were explored using ConTra v2 using the TRANSFAC position weight matrix library with stringency criteria set to 0.90 (core matrix) and 0.75 (similarity matrix). For visualization of putative XBP1 binding sites the entire gene sequences including a 1.5-kb region upstream of the transcriptional start site were screened using following TRANSFAC position weight matrices: F$XBP_Q2, F$XBP_01, V$XBP_01 and V$XBP1_02.
QUANTIFICATION AND STATISTICAL ANALYSIS
Results were analyzed using GraphPad Prism 7, with details about tests used found in the figure legends. For cell culture experiments, multiple wells of multi-welled plates were used in varied positions within the plates, and all results have been reproduced in R 2 independent experiments. Cell culture plates were randomly chosen for OGD versus normoxia or drug treatment studies, but the investigator was not blinded for the treatments. Statistical data were analyzed using two-tailed Student’s t tests, One-way ANOVA with multiple comparison’s test (Tukey’s, Dunn’s, Dunnett’s, Holm-Sidak’s), Two-way ANOVA with multiple comparison’s test (Bonferroni’s, Sidak’s, Dunn’s, Tukey’s, Dunnett’s). Data are represented as means ± standard error of the mean (SEM) with error bars representing SEM unless otherwise indicated. For expression data, results are presented as mean for 2−ddCq, with error bars showing the upper and lower limits calculated using the standard deviation (SD) for delta Cq values. Statistical testing of qPCR data was done using delta Cq values. Sample sizes and significance are shown in corresponding figure legends or stated in the results section.
Supplementary Material
Highlights.
A mass departure of ER-resident proteins occurs in response to ER calcium depletion
KDELR2 and KDELR3, but not KDELR1, are unfolded protein response genes
KDELR2 and KDELR3 upregulation by ER calcium loss counteracts ER protein loss
Stabilizing calcium and increasing KDEL receptor expression have therapeutic potential
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Programs at the National Institute on Drug Abuse and the National Center for Advancing Translational Sciences. We thank Dr. Madhu Lal and the RNAi Screening Facility at the National Center for Advancing Translational Sciences for providing data on RNAi screen. We thank Lowella Fortuno, Yaroslav Markov, and Xiaokang Yan for technical assistance. We thank Dr. Christopher Richie for discussion and guidance with molecular biology. We thank the Johns Hopkins Mass Spectrometry and Proteomics Core Facility, who performed the mass spectrometry sample processing and data acquisition. We thank Dr. Matt Shoulders for providing ATF6 and XBP1 transgenic cell lines. We thank Lauren Brick and the Visual Media at NIDA IRP for assistance with the graphical abstract.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and three tables can be found with this article online at https://doi.org/10.1016/j.celrep.2018.10.055.
REFERENCES
- Abrams EW, Cheng YL, and Andrew DJ (2013). Drosophila KDEL receptor function in the embryonic salivary gland and epidermis. PLoS ONE 8, e77618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airavaara M, Shen H, Kuo CC, Perä nen J, Saarma M, Hoffer B, and Wang Y. (2009). Mesencephalic astrocyte-derived neurotrophic factor reduces ischemic brain injury and promotes behavioral recovery in rats. J. Comp. Neurol 515, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy MO, Kim V, Cornwell WD, Rogers TJ, Kosmider B, Bahmed K, Barrero C, Merali S, Shetty N, and Kelsen SG (2017). Secretion of the endoplasmic reticulum stress protein, GRP78, into the BALF is increased in cigarette smokers. Respir. Res 18, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanen HI, Raykhel IB, Luukas MJ, Salo KE, and Ruddock LW (2011). Beyond KDEL: the role of positions 5 and 6 in determining ER localization. J. Mol. Biol 409, 291–297. [DOI] [PubMed] [Google Scholar]
- Badiola N, Penas C, Miñano-Molina A, Barneda-Zahonero B, Fadó R, Sá nchez-Opazo G, Comella JX, Sabriá J, Zhu C, Blomgren K, et al. (2011). Induction of ER stress in response to oxygen-glucose deprivation of cortical cultures involves the activation of the PERK and IRE-1 pathways and of caspase-12. Cell Death Dis. 2, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, and Helenius A. (2016). Cargo capture and bulk flow in the early secretory pathway. Annu. Rev. Cell Dev. Biol 32, 197–222. [DOI] [PubMed] [Google Scholar]
- Bian X, Hughes FM Jr., Huang Y, Cidlowski JA, and Putney JW Jr. (1997). Roles of cytoplasmic Ca2+ and intracellular Ca2+ stores in induction and suppression of apoptosis in S49 cells. Am. J. Physiol 272, C1241–C1249. [DOI] [PubMed] [Google Scholar]
- Bodman-Smith MD, Corrigall VM, Berglin E, Cornell HR, Tzioufas AG, Mavragani CP, Chan C, Rantapää -Dahlqvist S, and Panayi GS (2004). Antibody response to the human stress protein BiP in rheumatoid arthritis. Rheumatology (Oxford) 43, 1283–1287. [DOI] [PubMed] [Google Scholar]
- Booth C, and Koch GL (1989). Perturbation of cellular calcium induces secretion of luminal ER proteins. Cell 59, 729–737. [DOI] [PubMed] [Google Scholar]
- Broos S, Hulpiau P, Galle J, Hooghe B, Van Roy F, and De Bleser P. (2011). ConTra v2: a tool to identify transcription factor binding sites across species, update 2011. Nucleic Acids Res. 39, W74–W78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, Wright CL, Edwards CA, Davis MP, Grinham JA, Cole CG, Goward ME, Aguado B, Mallya M, Mokrab Y, et al. (2004). A genome annotation-driven approach to cloning the human ORFeome. Genome Biol. 5, R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigall VM, Bodman-Smith MD, Fife MS, Canas B, Myers LK, Wooley P, Soh C, Staines NA, Pappin DJC, Berlo SE, et al. (2001). The human endoplasmic reticulum molecular chaperone BiP is an autoantigen for rheumatoid arthritis and prevents the induction of experimental arthritis. J. Immunol 166, 1492–1498. [DOI] [PubMed] [Google Scholar]
- Földi I, Tóth AM, Szabó Z, Mózes E, Berkecz R, Datki ZL, Penke B, and Janáky T. (2013). Proteome-wide study of endoplasmic reticulum stress induced by thapsigargin in N2a neuroblastoma cells. Neurochem. Int 62, 58–69. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, Prado K, Thamsen M, Caswell D, Macias H, et al. (2014). Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158, 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glembotski CC, Thuerauf DJ, Huang C, Vekich JA, Gottlieb RA, and Doroudgar S. (2012). Mesencephalic astrocyte-derived neurotrophic factor protects the heart from ischemic damage and is selectively secreted upon sarco/endoplasmic reticulum calcium depletion. J. Biol. Chem 287, 25893–25904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MP, and Choi DW (1993). Combined oxygen and glucose deprivation in cortical cell culture: calcium-dependent and calcium-independent mechanisms of neuronal injury. J. Neurosci 13, 3510–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Calfon M, Urano F, Novoa I, and Ron D. (2002). Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol 18, 575–599. [DOI] [PubMed] [Google Scholar]
- Helms JB, and Rothman JE (1992). Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature 360, 352–354. [DOI] [PubMed] [Google Scholar]
- Henderson MJ, Richie CT, Airavaara M, Wang Y, and Harvey BK (2013). Mesencephalic astrocyte-derived neurotrophic factor (MANF) secretion and cell surface binding are modulated by KDEL receptors. J. Biol. Chem 288, 4209–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MJ, Wires ES, Trychta KA, Richie CT, and Harvey BK (2014). SERCaMP: a carboxy-terminal protein modification that enables monitoring of ER calcium homeostasis. Mol. Biol. Cell 25, 2828–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MJ, Wires ES, Trychta KA, Yan X, and Harvey BK (2015). Monitoring endoplasmic reticulum calcium homeostasis using a Gaussia luciferase SERCaMP. J. Vis. Exp 103, e53199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrich SM, Cole RN, West KP Jr., Schulze K, Yager JD, Groopman JD, Christian P, Wu L, O’Meally RN, May DH, et al. (2013). Statistical inference from multiple iTRAQ experiments without using common reference standards. J. Proteome Res 12, 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. (2004). The HUPO PSI’s molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol 22, 177–183. [DOI] [PubMed] [Google Scholar]
- Hooghe B, Hulpiau P, van Roy F, and De Bleser P. (2008). ConTra: a promoter alignment analysis tool for identification of transcription factor binding sites across species. Nucleic Acids Res. 36, W128–W132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DB, Powers K, Wang Y, and Harvey BK (2008). Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology 372, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu VW, Shah N, and Klausner RD (1992). A brefeldin A-like phenotype is induced by the overexpression of a human ERD-2-like protein, ELP-1. Cell 69, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. (2015). The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, et al. (2013). Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med 369, 2379–2390. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, and Glimcher LH (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol 23, 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, and Pelham HR (1990). A human homologue of the yeast HDEL receptor. Nature 348, 162–163. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, and Pelham HR (1992). Sequence of a second human KDEL receptor. J. Mol. Biol 226, 913–916. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Danilova T, Palm E, Lindholm P, Võikar V, Hakonen E, Ustinov J, Andressoo JO, Harvey BK, Otonkoski T, et al. (2014). MANF is indispensable for the proliferation and survival of pancreatic b cells. Cell Rep. 7, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner M, Erdmann E, and Beuckelmann DJ (1998). Calcium content of the sarcoplasmic reticulum in isolated ventricular myocytes from patients with terminal heart failure. J. Mol. Cell. Cardiol 30, 743–749. [DOI] [PubMed] [Google Scholar]
- Luo B, and Lee AS (2013). The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 32, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei M, Zhai C, Li X, Zhou Y, Peng W, Ma L, Wang Q, Iverson BL, Zhang G, and Yi L. (2017). Characterization of aromatic residue-controlled protein retention in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem 292, 20707–20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi J, and Pozzan T. (1998). The endoplasmic reticulum Ca2+ store: a view from the lumen. Trends Biochem. Sci 23, 10–14. [DOI] [PubMed] [Google Scholar]
- Missiaen L, Robberecht W, van den Bosch L, Callewaert G, Parys JB, Wuytack F, Raeymaekers L, Nilius B, Eggermont J, and De Smedt H. (2000). Abnormal intracellular ca(2+)homeostasis and disease. Cell Calcium 28, 1–21. [DOI] [PubMed] [Google Scholar]
- Munro S, and Pelham HR (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907. [DOI] [PubMed] [Google Scholar]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, et al. (2013). Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med 369, 2391–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK, Goldberg AL, Ho S, Rohde MF, Bush KT, and Sherman MYu. (1994). A set of endoplasmic reticulum proteins possessing properties of molecular chaperones includes Ca(2+)-binding proteins and members of the thioredoxin superfamily. J. Biol. Chem 269, 1744–1749. [PubMed] [Google Scholar]
- Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sö llner TH, and Rothman JE (1997). Bidirectional transport by distinct populations of COPI-coated vesicles. Cell 90, 335–349. [DOI] [PubMed] [Google Scholar]
- Paschen W. (2001). Dependence of vital cell function on endoplasmic reticulum calcium levels: implications for the mechanisms underlying neuronal cell injury in different pathological states. Cell Calcium 29, 1–11. [DOI] [PubMed] [Google Scholar]
- Peters LR, and Raghavan M. (2011). Endoplasmic reticulum calcium depletion impacts chaperone secretion, innate immunity, and phagocytic uptake of cells. J. Immunol 187, 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, and Ruddock L. (2007). A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol 179, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende R, Ferreiro E, Pereira C, and Resende de Oliveira C. (2008). Neurotoxic effect of oligomeric and fibrillar species of amyloid-beta peptide 1–42: involvement of endoplasmic reticulum calcium release in oligomerinduced cell death. Neuroscience 155, 725–737. [DOI] [PubMed] [Google Scholar]
- Reumann R, Vierk R, Zhou L, Gries F, Kraus V, Mienert J, Romswinkel E, Morellini F, Ferrer I, Nicolini C, et al. (2017). The serine protease inhibitor neuroserpin is required for normal synaptic plasticity and regulates learning and social behavior. Learn. Mem 24, 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, and Blower MD (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci 73, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza JC, Hardwick KG, Dean N, and Pelham HRB (1990). ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357. [DOI] [PubMed] [Google Scholar]
- Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, Yates JR 3rd, Su AI, Kelly JW, and Wiseman RL (2013). Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Füllekrug J, Nguyen Van P, Diekmann W, Robinson DG, and Mieskes G. (1994). Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J. Cell Sci 107, 2705–2717. [DOI] [PubMed] [Google Scholar]
- Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, and Brewer JW (2007). Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J. Biol. Chem 282, 7024–7034. [DOI] [PubMed] [Google Scholar]
- Takagi M, Ishii T, Barnes AM, Weis M, Amano N, Tanaka M, Fukuzawa R, Nishimura G, Eyre DR, Marini JC, and Hasegawa T. (2012). A novel mutation in LEPRE1 that eliminates only the KDEL ER-retrieval sequence causes non-lethal osteogenesis imperfecta. PLoS ONE 7, e36809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr JM, Winyard PG, Ryan B, Harries LW, Haigh R, Viner N, and Eggleton P. (2010). Extracellular calreticulin is present in the joints of patients with rheumatoid arthritis and inhibits FasL (CD95L)-mediated apoptosis of T cells. Arthritis Rheum. 62, 2919–2929. [DOI] [PubMed] [Google Scholar]
- Tavender TJ, Springate JJ, and Bulleid NJ (2010). Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 29, 4185–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, and Dawson AP (1990). Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. USA 87, 2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Simons JF, and Helenius A. (1996). Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem 271, 27509–27516. [DOI] [PubMed] [Google Scholar]
- Voutilainen MH, Bäck S, Pörsti E, Toppinen L, Lindgren L, Lindholm P, Peränen J, Saarma M, and Tuominen RK (2009). Mesencephalic astrocyte-derived neurotrophic factor is neurorestorative in rat model of Parkinson’s disease. J. Neurosci 29, 9651–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Nguyen HN, Maguire JL, and Perry DC (2002). Role of intracellular calcium stores in cell death from oxygen-glucose deprivation in a neuronal cell line. J. Cereb. Blood Flow Metab 22, 206–214. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang Y, Asakawa T, Li W, Han S, Li Q, Xiao B, Namba H, Lu C, and Dong Q. (2015). Neuroprotective effect of neuroserpin in oxygen-glucose deprivation- and reoxygenation-treated rat astrocytes in vitro. PLoS ONE 10, e0123932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, and Perry DC (1996). Dantrolene is cytoprotective in two models of neuronal cell death. J. Neurochem 67, 2390–2398. [DOI] [PubMed] [Google Scholar]
- Wires ES, Trychta KA, Bäck S, Sulima A, Rice KC, and Harvey BK (2017). High fat diet disrupts endoplasmic reticulum calcium homeostasis in the rat liver. J. Hepatol 67, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd, and Su AI (2009). BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Zhang F, Yang Q, Zhang Y, Liu Q, Jiang W, Cao H, Li D, Xie S, Tong N, and He J. (2017). Circulating mesencephalic astrocyte-derived neurotrophic factor is increased in newly diagnosed prediabetic and diabetic patients, and is associated with insulin resistance. Endocr. J 64, 403–410. [DOI] [PubMed] [Google Scholar]
- Xiao G, Chung TF, Pyun HY, Fine RE, and Johnson RJ (1999). KDEL proteins are found on the surface of NG108–15 cells. Brain Res. Mol. Brain Res 72, 121–128. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, and Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.