Abstract
Reuterin is an antimicrobial compound produced by Lactobacillus reuteri, and has been proposed to mediate, in part, the probiotic health benefits ascribed to this micro-organism. Despite 20 years of investigation, the mechanism of action by which reuterin exerts its antimicrobial effects has remained elusive. Here we provide evidence that reuterin induces oxidative stress in cells, most likely by modifying thiol groups in proteins and small molecules. Escherichia coli cells subjected to sublethal levels of reuterin expressed a set of genes that overlapped with the set of genes composing the OxyR regulon, which senses and responds to various forms of oxidative stress. E. coli cells mutated for oxyR were more sensitive to reuterin compared with wild-type cells, further supporting a role for reuterin in exerting oxidative stress. The addition of cysteine to E. coli or Clostridium difficile growth media prior to exposure to reuterin suppressed the antimicrobial effect of reuterin on these bacteria. Interestingly, interaction with E. coli stimulated reuterin production or secretion by L. reuteri, indicating that contact with other microbes in the gut increases reuterin output. Thus, reuterin inhibits bacterial growth by modifying thiol groups, which indicates that reuterin negatively affects a large number of cellular targets.
Keywords: DTNB, 5,5′-dithiobis-(2-nitrobenzoic acid); 3-HPA, 3-hydroxypropionaldehyde; TNB, 5-thio-2-nitrobenzoic acid
INTRODUCTION
The use of probiotic bacteria to improve human and animal health has enjoyed a renaissance in the past decade. Companies in the food and dietary supplement industries have increasingly been adding probiotic strains to several different products and touting a variety of health benefits associated with these foods. Recent clinical trials have shown that probiotics are effective in treating a variety of ailments, including diarrhoea, colic, eczema and pouchitis (Floch et al., 2008; Savino et al., 2007; Wickens et al., 2008). However, most of these trials are small, and to date few well-controlled large-scale clinical trials have been conducted, so the efficacy of probiotics in many diseases remains to be proven.
A number of mechanisms by which probiotics exert their beneficial effects have been proposed, including the modulation of the immune system, the alteration of the intestinal microbiota, and the production of antimicrobial compounds that inhibit the growth of pathogens (Marco et al., 2006; O'Hara & Shanahan, 2007). Understanding the mechanisms that are actively contributing to the beneficial effects of probiotics will be an important step in tapping the true potential of these organisms in benefiting health.
Strains of Lactobacillus reuteri have been shown to be effective against a variety of ailments, including diarrhoea and colic (Rosenfeldt et al., 2002a, b; Savino et al., 2007; Weizman et al., 2005). One of the proposed mechanisms of action that L. reuteri uses to effect probiosis is the production of the antimicrobial compound 3-hydroxypropionaldehyde (3-HPA), also referred to as reuterin (Talarico et al., 1988). Reuterin is produced as an intermediate step in the conversion of glycerol to 1,3-propanediol, a pathway proposed to regenerate NAD+ from NADH and to contribute to improved growth yield (Luthi-Peng et al., 2002). Reuterin is metabolized in a specialized bacterial compartment called the metabolosome, perhaps due to its toxicity (Sriramulu et al., 2008). For reasons that are unclear, L. reuteri secretes high levels of reuterin when grown or incubated in the presence of excess amounts of glycerol. Reuterin has been shown to be bioactive against bacteria, viruses and fungi (Chung et al., 1989; Cleusix et al., 2007), and has been studied as a possible additive to prevent food spoilage and pathogen growth in food (Arques et al., 2004, 2008). Recent work using germ-free mice mono-associated with a reuterin-producing strain of L. reuteri has demonstrated that reuterin can be produced by L. reuteri in vivo, indicating that the in vitro antimicrobial activity occurs in the gastrointestinal tract (Morita et al., 2008).
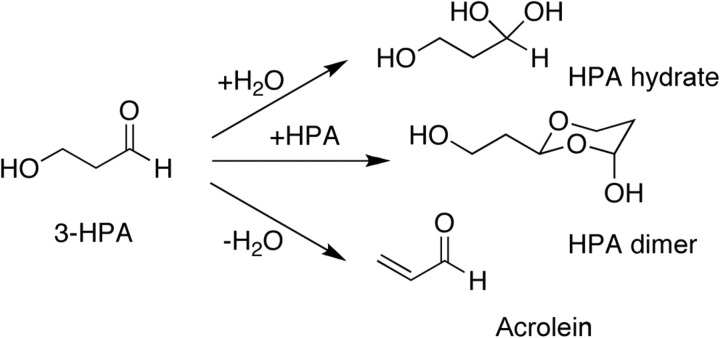
The aldehyde group of reuterin is highly reactive and thus reuterin can form additional compounds in aqueous solution (Vollenweider & Lacroix, 2004). Reuterin can dimerize, forming HPA dimer, or can be hydrated to form HPA hydrate (Fig. 1). Reuterin can also be dehydrated into the toxic compound acrolein. Because reuterin can be converted into these various compounds, the mechanism by which reuterin exerts its antimicrobial effects has been difficult to determine. Two main hypotheses have been proposed. First, the aldehyde group of reuterin is proposed to be highly reactive with thiol groups and primary amines, and therefore reuterin could inactivate proteins and small molecules containing these groups (Vollenweider & Lacroix, 2004). This would explain the broad spectrum of bacteria, fungi and viruses affected by reuterin. Alternatively, the dimeric form of reuterin, HPA dimer, which is structurally similar to a ribose sugar, could specifically block the enzyme ribonucleotide reductase by acting as a competitive inhibitor. This enzyme is required for the generation of deoxynucleotides, which are required for DNA synthesis. An earlier report has suggested that reuterin could indeed inhibit ribonucleotide reductase and possibly exert its broad-spectrum effects through this inhibition (Talarico & Dobrogosz, 1989). Because the active site of this enzyme contains a thiol group, it is impossible to determine which proposed mode of action is correct from earlier experiments.
Fig. 1.
3-HPA (reuterin) can adopt a number of different forms, as indicated in the main text. 3-HPA can react with water to form HPA hydrate or interact with itself to form HPA dimer (normally seen only at very high 3-HPA concentrations). 3-HPA can also undergo dehydration to form acrolein.
We were interested in further exploring the expression of reuterin in L. reuteri as well as its mode of action. Our work demonstrates that reuterin is produced solely by the enzyme glycerol dehydratase and that direct contact with other bacteria can stimulate reuterin production or secretion. Several lines of in vivo and in vitro evidence indicate that the aldehyde form of reuterin is the bioactive agent and that this causes an oxidative stress response by modifying thiol groups inside cells.
METHODS
Bacterial strains, media, and growth conditions.
Strains used in this study are listed in Table 1. L. reuteri was grown in liquid culture under hypoxic conditions (2 % O2, 5 % CO2, balanced with N2) in de Man, Rogosa and Sharpe (MRS) broth (BD Difco). L. reuteri was also grown on plates under anaerobic conditions using the GasPak EZ anaerobe Container System (BD Difco) on MRS agar (BD Difco) at 37 °C for 24–48 h. L. reuteri mutants were grown in the presence of 10 μg erythromycin ml−1. Bacillus subtilis, Citrobacter rodentium and Escherichia coli strains were grown in liquid culture in LB broth (BD Difco) at 37 °C under aerobic conditions. Clostridium difficile strain CD630 was grown in brain heart infusion media (Fluka) supplemented with yeast extract (5 mg ml−1, BD Difco) under anaerobic conditions in a Coy Laboratory anaerobic chamber.
Table 1.
Strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| L. reuteri strains | ||
| ATCC 23272 | Type strain, isolated from human faeces | BioGaia |
| ATCC PTA-6475 | Isolated from human breast milk | BioGaia |
| ATCC 55730 | Isolated from human breast milk | BioGaia |
| PRB90 | ΔpduC mutant in ATCC 23272 background | This study |
| PRB94 | ΔpduC mutant in ATCC PTA-6475 background | This study |
| PRB81 | ΔdhaT mutant in ATCC 23272 background | This study |
| PRB104 | ΔdhaT mutant in ATCC PTA-6475 background | This study |
| E. coli strains | ||
| DH5α | Strain used for MIC assays | Laboratory stock |
| TW6375 | Enteropathogenic strain | T. Whittam, Michigan State University, East Lansing, MI |
| TW951 | Enterohaemorrhagic strain | T. Whittam |
| TW116 | Enterohaemorrhagic strain | T. Whittam |
| JW3933-3 | ΔoxyR mutant | Coli Genetic Stock Center, Yale University, New Haven, CT (Baba et al., 2006) |
| BW25113 | Background strain for JW3933-3 | Coli Genetic Stock Center (Datsenko & Wanner, 2000) |
| Other strains | ||
| Clostridium difficile CD630 | Wild-type | A. L. Sonenshein, Tufts University, Medford, MA |
| B. subtilis RB247 (JH642) | Wild-type | Dean et al. (1977) |
| C. rodentium DBS120 | Wild-type | V. Young, University of Michigan, Ann Arbor, MI (Schauer & Falkow, 1993) |
Reuterin production and purification.
Reuterin was produced from L. reuteri cultures using methods described elsewhere (Talarico & Dobrogosz, 1989; Vollenweider et al., 2003). Briefly, cells were harvested from liquid cultures of L. reuteri by centrifugation and washed twice with 50 mM sodium phosphate buffer (pH 7.4). Approximately 150 mg cells (wet weight) were resuspended in 15 ml 250 mM glycerol and transferred to 15 ml screw-capped tubes; if needed, the appropriate amount of other bacteria was added as described below. The cells in glycerol were incubated at 37 °C for 2 h. Reuterin was collected by pelleting the cells and filtering the supernatants through 0.22 μm pore-size filters (Millipore). The resulting cell-free supernatant was shown by HPLC to contain only reuterin and glycerol (see Methods, HPLC, below).
Reuterin quantification and activity.
Reuterin was quantified using an adapted colorimetric assay described by Doleyres et al. (2005); reuterin samples were diluted 10-fold in water and the A560 was determined on a Spectramax model M5 microplate reader (Molecular Devices). The antimicrobial activity of reuterin was quantified using a MIC assay described by Chung et al. (1989) and adapted for use in a 96-well format. Relative reuterin concentration (units reuterin ml−1) was defined as the reciprocal of the sample dilution preceding the dilution that allowed cell growth. E. coli DH5α cells were grown overnight at 37 °C in LB broth and diluted to approximately 104 cells ml−1. Twofold serial dilutions of reuterin-containing cell-free supernatant samples were prepared with water in 96-well 0.4 ml deep-well flat-bottomed plates (Nalge Nunc) in a total volume of 150 μl per well. A 150 μl volume of the E. coli suspension was added to each well and the plates were incubated aerobically at 37 °C overnight. Water was assayed in place of reuterin as a positive growth control. OD600 values were measured in a Spectramax model M5. The absence of a turbid culture after overnight growth was interpreted as growth inhibition by reuterin. For the experiments assaying reuterin activity in the presence of cysteine, valine or serine, each amino acid (Sigma-Aldrich) was added to each well to a final concentration of 600 μM.
Microarray experiments.
Oligonucleotide microarrays for E. coli were obtained from T. Whittam (Michigan State University, East Lansing, MI) (Bergholz et al., 2007). The microarrays were printed onto Corning UltraGap slides at the Research Technology Support Facility at Michigan State University using E. coli oligonucleotide set version 1.0.1 (Operon), which contained probes specific for three E. coli strains, K-12 (MG1655), O157 : H7 Sakai and O157 : H7 EDL-933. Overnight cultures of E. coli were diluted 1 : 100 in 25 ml LB broth and grown to mid-exponential phase (OD600 0.25) under hypoxic conditions (2 % O2) with shaking at 250 r.p.m. Reuterin-containing cell-free supernatant was added to cultures to yield a final concentration of approximately 5 units reuterin ml−1. This concentration is sufficient to slow growth of E. coli without being lethal. LB broth (2×) was added at the same time to maintain the same nutrient concentration. Glycerol (250 mM) was added to identical cultures as controls. Cells were allowed to grow for one doubling before samples were taken for analysis. Sample preparation, RNA isolation using RNeasy RNA isolation kits (Qiagen), indirect cDNA labelling with Cy3- or Cy5-labelled aminoallyl dUTP, and slide hybridization were performed as described previously (Britton et al., 2002; Britton, 2003). Slides were scanned on a GenePix 4000B scanner (Axon Instruments). Five biological replicates were analysed. Microarray data were analysed using iterative outlier analysis with three iterations, as previously described (Britton et al., 2002; Uicker et al., 2006). Genes that had expression values three standard deviations or more away from the mean of the population were considered statistically significant.
Sensitivity of E. coli oxyR mutant strains to reuterin and hydrogen peroxide.
E. coli strains JW3933-3 and BW25113 were grown overnight at 37 °C in LB broth and diluted to approximately 104 cells ml−1. MIC assays were performed to test sensitivity of the cells to reuterin and hydrogen peroxide. To test sensitivity to reuterin, twofold serial dilutions of reuterin-containing cell-free supernatant samples were generated and combined with E. coli suspension as described above. To test sensitivity to hydrogen peroxide, twofold serial dilutions of hydrogen peroxide (Sigma-Aldrich) were prepared with water in 96-well 0.4 ml deep-well flat-bottomed plates (Nalge Nunc) in a total volume of 150 μl per well; the final concentration of hydrogen peroxide ranged from 1.2 mM to 1.2 μM. A 150 ml volume of the E. coli suspension was added to each well as described above. The 96-well plates were incubated aerobically at 37 °C overnight. Five biological replicates were analysed.
Mutant construction.
Mutant L. reuteri ATCC 23272 and L. reuteri ATCC PTA-6475 strains for the glycerol dehydratase subunit γ gene (pduC, also known as gupC) and the 1,3-propanediol oxidoreductase gene (dhaT) were constructed using the system developed by Russell & Klaenhammer (2001) and modified for use in L. reuteri (Walter et al., 2005; Whitehead et al., 2008). To create the ΔpduC mutant strains, a 356 bp internal fragment (+37 to +392) was PCR-amplified from the pduC gene in ATCC 23272 using primers LS74 (5′-TGACTGGATCCTAACGGCCAATTCATCAAGATAC-3′) and LS87 (5′-TGACTGAATTCTTGATAGCCATAATCATTTCACC-3′), and a 367 bp internal fragment (+43 to +409) was PCR-amplified from ATCC PTA-6475 using primers LS67 (5′-TGACTGGATCCTAATTTGGTCCTGGTGTTATTGC-3′) and LS89 (5′-TGACTGAATTCTCAATGGTGGAAGTGGATTC-3′). The fragments were cloned into pORI28 to create plasmids pLS46 for ATCC 23272 and pLS47 for ATCC PTA-6475; these plasmids were then used to generate ΔpduC mutant L. reuteri strains PRB90 and PRB94, as previously described (Whitehead et al., 2008). To create the ΔdhaT mutant strains, a 194 bp fragment (+43 to +236) was PCR-amplified from the dhaT gene in L. reuteri ATCC 55730 using primers LS67 (5′-TGACTGGATCCTAATTTGGTCCTGGTGTTATTGC-3′) and LS68 (5′-TGACTGAATTCTTCCGGATCTTAGGGTTAGG-3′) and cloned into pORI28 to create plasmid pLS39. The dhaT gene sequence is 99 % identical among L. reuteri ATCC 23272, L. reuteri ATCC PTA-6475 and L. reuteri ATCC 55730, and therefore pLS39 could be used to generate ΔdhaT mutants in both L. reuteri ATCC 23272 and L. reuteri ATCC PTA-6475 backgrounds to create strains PRB81 and PRB104, respectively.
Inhibition of free thiol groups.
The reduction of Ellman's reagent, or 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) by free thiol groups releases 5-thio-2-nitrobenzoic acid (TNB; Amax 412 nm) and can be monitored spectrophotometrically (Ellman, 1959). Increasing amounts of reuterin produced from ATCC PTA-6475 were prepared in water in a final volume of 125 μl and then mixed with 8.3 μl 3 mM glutathione (GSH) and 108.3 μl 100 mM phosphate buffer, pH 7.0, in 96-well plates. After 1 h at room temperature, 8.3 μl 15 mM DTNB was added and A412 was measured. Samples and standard curves were done in duplicate. Control reactions with no GSH had no absorbance. Iodoacetamide, which is an irreversible thiol-modifying compound, was assayed as a positive control. PRB94, the glycerol dehydratase (ΔpduC) mutant of ATCC PTA-6475, was used as a negative control.
HPLC.
Reuterin was produced, and reuterin-containing cell-free supernatant from early stationary phase cultures of ATCC 55730 was prepared and concentrated fivefold under nitrogen. H2SO4 was added to a final concentration of 10 mM. The samples were injected using a 200 μl loop and run through a 300×7.8 mm Bio-Rad Aminex HPX-87H ion exclusion column at a flow rate of 0.6 ml min−1 at room temperature using 10 mM de-gassed H2SO4 as eluent. Components were detected with Breeze software using model 2487 UV absorbance (210 nm) and 2414 refractive index detectors (Waters), before being collected in a 96-well plate (300 μl per well).
Co-incubation of L. reuteri with other bacteria.
E. coli, B. subtilis and C. rodentium cells were grown in liquid culture for 24 h, pelleted, washed twice with 50 mM potassium phosphate buffer (pH 7.4), and resuspended in potassium buffer at approximately 50 mg cells ml−1. The appropriate amounts of bacteria were combined with 150 mg L. reuteri cells in potassium buffer in a final volume of 15 ml in 15 ml screw-capped tubes. Cell mixtures were then pelleted, resuspended in 15 ml 250 mM glycerol, and incubated at 37 °C for 2 h. Reuterin was collected from the supernatants by pelleting the cells and filtering the supernatants through 0.22 μm pore-size filters (Millipore).
In order to determine whether the viability of E. coli was required for the increase in reuterin levels, E. coli cells were killed via heat or UV irradiation. For heat-killed E. coli, cells were boiled for 10 min. For UV-killed E. coli, cells were washed in 50 mM potassium phosphate buffer (pH 7.4) and diluted to approximately 108 cells ml−1. Diluted cells in a Petri dish were then exposed to short-wave UV light at a distance of 1 cm for 60 s. Loss of viability of E. coli was confirmed by plating cells onto LB agar and incubating overnight at 37 °C. E. coli (50 mg) and L. reuteri (150 mg) were then combined, resuspended in 250 mM glycerol in a final volume of 15 ml, and incubated at 37 °C for 2 h as described above.
RESULTS
Exposure of E. coli cells to reuterin results in the increased expression of genes that encode proteins containing thiol groups and genes involved in the oxidative stress response
We were interested in identifying the mechanism of action by which reuterin exerts its antimicrobial effects. Using DNA microarrays to monitor E. coli gene expression we found that many genes involved in the oxidative stress response were induced in response to reuterin exposure. E. coli cells were grown to mid-exponential phase under hypoxic conditions (2 % O2) and exposed to a sublethal concentration of reuterin that caused a mild reduction in growth rate. Gene expression profiles of cells exposed to reuterin compared with those of untreated cells were determined using DNA microarrays. Table 2 lists the 33 genes whose expression was most strongly induced in response to reuterin exposure. A full list of the genes induced by reuterin is available in Supplementary Table S1. Many of the genes that were significantly overexpressed in reuterin-treated cells encode proteins that require thiol groups for their function (e.g. reductases). In addition, several genes that were induced by H2O2 in E. coli were upregulated and are highlighted in Table 3, including thioredoxin, glutaredoxin and hydroperoxide reductase (Zheng et al., 2001). The gene most highly overexpressed in response to reuterin was bhs, which is also highly induced by exposure to H2O2; bhs has been shown to be involved in stress resistance and biofilm formation in E. coli (Zhang et al., 2007). yqhD, another gene highly expressed as a result of reuterin exposure, encodes an aldehyde reductase involved in protecting against harmful aldehydes produced by lipid peroxidation. Overexpression of yqhD increases the resistance of E. coli to the effects of reactive oxygen molecules (Perez et al., 2008). Overall, genes regulated by hydrogen peroxide were eightfold over-represented in the genes that were induced by reuterin compared with what would be expected if 33 genes were selected randomly from the E. coli genome. These results indicate that reuterin is exerting an oxidative stress response upon cells, most likely by acting as a thiol-modifying agent in the aldehyde form.
Table 2.
The most highly overexpressed genes in E. coli cells treated with sublethal concentrations of reuterin
| Gene | Fold induction* | Function |
|---|---|---|
| bhs† | 35.7 | Protein involved in stress resistance and biofilm formation |
| marA | 23.3 | Transcriptional activator of multiple antibiotic resistance |
| yqhD | 21.9 | Putative oxidoreductase |
| nemR | 20.5 | Putative transcriptional regulator inactivated by Cys-modification reagents |
| marR | 15.5 | Repressor of multiple antibiotic resistance |
| grxA† | 14.3 | Glutaredoxin 1 |
| ybiJ | 13.6 | Putative periplasmic protein of unknown function |
| nemA | 13.1 | N-Ethylmaleimide reductase |
| yqhC | 10.3 | Putative ARAC-type regulatory protein |
| gloA | 8.6 | Lactoylglutathione lyase |
| ahpF† | 8.5 | Alkyl hydroperoxide reductase, F52a subunit |
| yhcN | 7.4 | Putative periplasmic protein of unknown function |
| mdaB | 5.2 | Modulator of drug activity B |
| ahpC† | 5.0 | Alkyl hydroperoxide reductase, C22 subunit |
| yjbM | 4.8 | Function unknown |
| yjgI | 4.6 | Putative oxidoreductase |
| emrR | 4.6 | Regulator of plasmid mcrB operon (microcin B17 synthesis) |
| katG† | 4.6 | Catalase hydrogen peroxidase I |
| frmR | 4.5 | Negative regulator of frmRAB operon; expression is induced by formaldehyde |
| marB | 4.5 | Multiple antibiotic resistance protein |
| adhC | 4.1 | Glutathione-dependent alcohol dehydrogenase |
| cysD | 4.1 | Sulfate adenylyltransferase, subunit 2 |
| ibpB | 4.1 | Heat-shock protein |
| rnt | 4.0 | RNase T |
| nfnB | 3.9 | Oxygen-insensitive NAD(P)H nitroreductase |
| dkgA | 3.9 | 2,5-Diketo-d-gluconate reductase A |
| yafB | 3.8 | Putative aldose reductase |
| yegQ | 3.6 | ORF, hypothetical protein |
| iscR† | 3.6 | Transcriptional repressor for isc operon |
| trxC† | 3.5 | Thioredoxin 2 |
| iscS† | 3.4 | Putative aminotransferase |
| adiY | 3.3 | Putative ARAC-type regulatory protein |
| ibpA† | 3.3 | Heat-shock protein |
*Increased level of expression in reuterin-treated cells versus untreated cells.
†Genes overexpressed in response to hydrogen peroxide (Zheng et al., 2001).
Table 3.
Overlap between genes overexpressed in response to hydrogen peroxide treatment and in response to reuterin treatment
| Gene | Fold induction* | Function |
|---|---|---|
| bhs | 35.7 | Protein involved in stress resistance and biofilm formation |
| grxA | 14.3 | Glutaredoxin 1 |
| ahpF | 8.5 | Alkyl hydroperoxide reductase, F52a subunit |
| ahpC | 5.0 | Alkyl hydroperoxide reductase, C22 subunit |
| katG | 4.6 | Catalase hydrogen peroxidase I |
| iscR | 3.6 | Transcriptional repressor for isc operon |
| trxC | 3.5 | Thioredoxin 2 |
| iscS | 3.4 | Putative aminotransferase |
| ibpA | 3.3 | Heat-shock protein |
| iscU | 2.8 | Iron–sulfur cluster assembly scaffold protein |
| iscA | 2.8 | Iron–sulfur cluster assembly protein |
| iscX | 2.6 | Iron–sulfur cluster assembly protein |
| yeeD | 2.1 | Function unknown |
*Increased level of expression in reuterin-treated cells versus untreated cells.
An E. coli strain defective in the OxyR-mediated oxidative stress response is more sensitive to reuterin
Exposure of E. coli to reuterin induced the expression of several genes that are under the transcriptional control of the positive transcriptional regulator OxyR, which upregulates many genes in response to oxidative stress (including hydrogen peroxide). Disruption of the oxyR gene results in increased sensitivity to agents causing oxidative stress, notably hydrogen peroxide (Christman et al., 1985). The expression profiles from E. coli exposed to reuterin overlapped significantly with profiles from E. coli exposed to H2O2, leading us to investigate whether ΔoxyR mutant cells are also more sensitive to reuterin. We found that an E. coli ΔoxyR mutant (JW3933-3) was four times more sensitive to reuterin than the isogenic wild-type parental strain as determined by MIC assay, further supporting the suggestion that reuterin is exerting oxidative stress. The same ΔoxyR mutant strain was eight times more sensitive to H2O2 than wild-type cells, indicating that the degree of sensitivity to reuterin and H2O2 is similar. These results suggest that reuterin induces oxidative stress in E. coli.
Addition of an exogenous thiol group can suppress the antimicrobial effects of reuterin
To further test if reuterin could interact with thiol groups we asked whether the addition of excess cysteine to media could inhibit the antimicrobial activity of reuterin. MIC assays for reuterin against E. coli were conducted in the presence of 600 μM cysteine. With the addition of cysteine, only the most concentrated level of reuterin (32 units ml−1) was able to suppress the growth of E. coli, representing a 16-fold increase in the MIC value for reuterin with respect to E. coli without the amino acid added (2 units ml−1). The addition of the amino acids valine or serine at 600 μM concentrations had no effect on reuterin activity, indicating that reuterin was interacting with the thiol group of cysteine and not the free amino group. We also confirmed the inhibition of reuterin activity by cysteine in a MIC assay using Clostridium difficile strain CD630. The ability of cysteine to detoxify reuterin further supports the hypothesis that reuterin induces oxidative stress through interaction with thiol groups.
Glycerol dehydratase is required for reuterin production
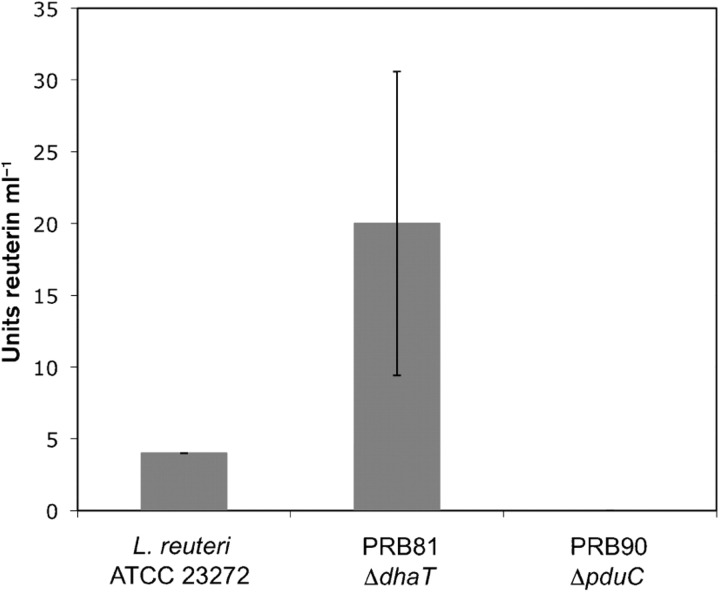
The glycerol dehydratase of L. reuteri is a cobalamin-dependent enzyme composed of three subunits (α, β and γ) and catalyses the conversion of glycerol to reuterin (3-HPA). To assess whether this is the sole pathway for reuterin production in L. reuteri, we created a disruption in the pduC gene, which encodes the large subunit γ of glycerol dehydratase in two L. reuteri strains, ATCC 23272, which is the type strain originally isolated from human faeces (Kandler et al., 1980), and ATCC PTA-6475, which was isolated from breast milk and has been shown to possess immunomodulatory activity (Lin et al., 2008). After overnight growth in MRS broth, ΔpduC mutant cells and wild-type cells were incubated in aqueous glycerol solution, and reuterin production was measured using both an adapted colorimetric assay (Doleyres et al., 2005) and a MIC bioassay (Chung et al., 1989). The mutant strains did not produce any detectable reuterin according to either assay, demonstrating that glycerol dehydratase is required for reuterin production in L. reuteri (Fig. 2). These results confirm and extend those published by Morita et al. (2008), in which the entire genomic region containing the genes encoding the three glycerol dehydratase subunits was deleted in L. reuteri strain JCM 1112T, resulting in elimination of reuterin production capability.
Fig. 2.
Reuterin production in L. reuteri strains mutant for enzymes involved in glycerol metabolism. Inactivation of 1,3-propanediol oxidoreductase in strain PRB81 (ΔdhaT) produced higher levels of reuterin as compared with wild-type strain L. reuteri ATCC 23272 during late stationary phase, as measured by MIC assay in three replicate experiments. Inactivation of glycerol dehydratase (ΔpduC) in strain PRB90 resulted in no detectable reuterin production. Units reuterin ml−1 were determined from the fold dilution of the reuterin supernatant that was still able to inhibit the growth of E. coli in the MIC assay. Error bars, sd. These results were independently confirmed using the colorimetric assay (data not shown).
We were also interested in determining if disruption of the dhaT gene, encoding 1,3-propanediol oxidoreductase, would lead to overproduction of reuterin. Reuterin is normally converted to 1,3-propanediol by 1,3-propanediol oxidoreductase. We created deletion mutants in strains ATCC 23272 and ATCC PTA-6475 by disrupting the gene encoding this enzyme and measured their ability to produce reuterin compared with wild-type cells. An increase in reuterin levels two- to eightfold over wild-type was observed for stationary-phase cells (Fig. 2); the difference in reuterin production between mutant and wild-type cells increased as cells moved into later stationary phase. Little or no difference in reuterin levels was observed between the ΔdhaT mutant and wild-type cells in exponential phase and early stationary phase.
Reuterin can inhibit free thiol groups in an in vitro assay
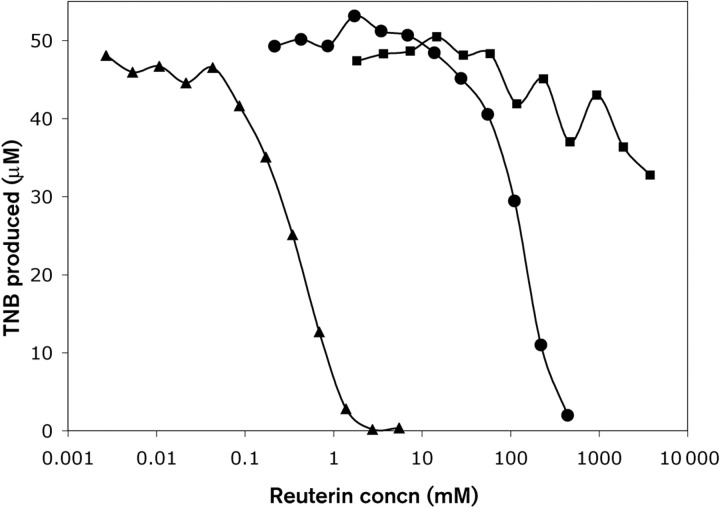
One proposed mechanism by which aldehydes cause stress in cells is via their ability to react with thiol and primary amine groups of proteins (Vollenweider & Lacroix, 2004). Using an assay that detects the presence of free thiol groups, we investigated whether reuterin can directly modify thiol groups. Ellman's reagent (DTNB) contains a disulfide bond that can be reduced by free thiol groups, and has been used extensively to quantify free thiol levels in a variety of samples (Ellman, 1959). DTNB reduction releases TNB, which can be monitored spectrophotometrically at 412 nm. Increasing amounts of reuterin produced from ATCC PTA-6475 were mixed with glutathione for 1 h prior to adding this mixture to Ellman's reagent and monitoring TNB levels. Iodoacetamide, a compound that irreversibly interacts with thiol groups, was used as a positive control for thiol inhibition. The glycerol dehydratase (ΔpduC) mutant of ATCC PTA-6475 was used to control for any compounds other than reuterin that may affect this assay. Reuterin was able to inhibit the reduction of Ellman's reagent in a concentration-dependent manner, similar to iodoacetamide (Fig. 3). The need for increased levels of reuterin to inhibit the reaction compared with iodoacetamide likely reflects the fact that the reaction of reuterin with thiol groups is reversible (see Discussion). As expected, the ΔpduC mutant did not produce any compound capable of inhibiting TNB production, demonstrating that the effect was specific to the production of reuterin.
Fig. 3.
Effect of reuterin on thiol-mediated reduction of Ellman's reagent. Increasing amounts of iodoacetamide (▴), reuterin (•) or control supernatant (▪) (derived from L. reuteri PRB94 cells, which have deletions of the pduC gene) were incubated in phosphate buffer with glutathione for 1 h, after which time Ellman's reagent was added and the amount of TNB produced was determined. There was no detectable TNB production when glutathione was omitted.
Antimicrobial activity corresponds to the presence of reuterin and not acrolein
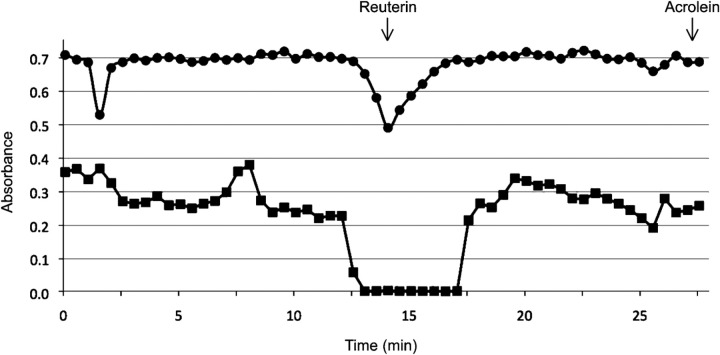
Dehydration of reuterin generates the highly toxic compound acrolein (Vollenweider & Lacroix, 2004). Although the conditions that we used to produce reuterin should result in a 3 : 1 ratio of HPA hydrate to reuterin, we wanted to rule out the possibility that small amounts of acrolein were producing the observed killing effect. Resting L. reuteri cells were incubated in aqueous glycerol to produce reuterin; the reuterin-containing supernatant was then subjected to HPLC to separate the components. Fractions were collected and tested for antimicrobial activity using the MIC bioassay and the ability to inhibit the reduction of DTNB by glutathione. The only two peaks detected in the supernatant corresponded to glycerol and reuterin. Antimicrobial activity and the ability to inhibit DTNB reduction were found only in the HPLC fractions that contained reuterin; we did not detect these activities with the fraction that would contain acrolein if present (Fig. 4). This indicates that under our reuterin production conditions, L. reuteri cells are not producing levels of acrolein that are deleterious to cell growth.
Fig. 4.
HPLC fractionation of L. reuteri supernatants. HPLC fractions of reuterin supernatant produced from L. reuteri ATCC PTA-6475 were assayed for inhibition of DTNB reduction by free thiol groups (•, A412) and inhibition of E. coli growth (▪, OD600; from modified MIC assay). Pure reuterin and acrolein were run on the HPLC column and detected at 14 and 27.25 min, respectively (indicated by arrows), in agreement with earlier reports by others. Antimicrobial activity and thiol inhibition corresponded with reuterin but not acrolein.
Reuterin production is increased by interaction with a variety of bacteria
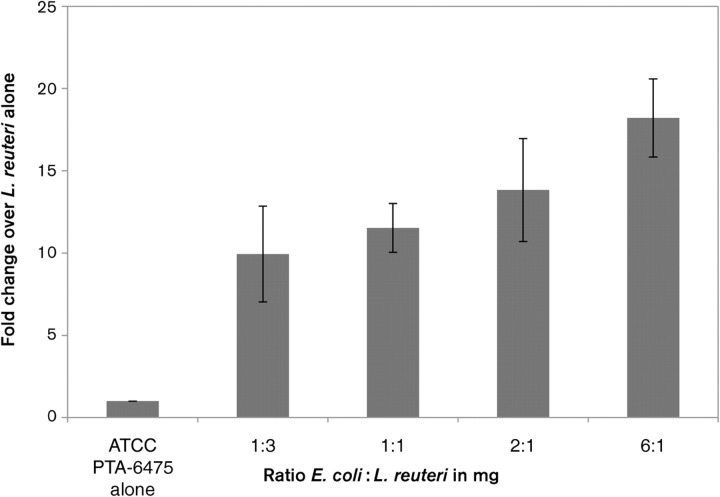
Reuterin production in L. reuteri is stimulated when cells are co-cultured with a variety of other microbes, including bacterial, protozoan and fungal species (Chung et al., 1989). This indicates that interaction with other bacteria in the gastrointestinal tract may trigger reuterin production in vivo. To address the mechanism by which other bacteria stimulate reuterin production we asked whether or not co-culturing of the bacteria was required for this stimulation. We found that co-culturing is not necessary; direct interaction with other micro-organisms during incubation of L. reuteri in aqueous glycerol solution is sufficient to produce this stimulation. L. reuteri ATCC 23272 and E. coli DH5α were grown overnight to stationary phase, spun down to remove culture supernatant, and then mixed together in aqueous glycerol solution. Increasing amounts of E. coli DH5α incubated with L. reuteri produced a corresponding increase in reuterin levels (Fig. 5). This increase was observed with up to six times the amount (in milligrams) of E. coli DH5α compared with that of L. reuteri. E. coli DH5α has no ability to produce reuterin on its own (data not shown). This stimulation was observed for all L. reuteri strains tested, including ATCC 23272, ATCC PTA-6475 and ATCC 55730. In addition to E. coli DH5α, stimulation of reuterin production was achieved by co-incubation with a variety of bacteria, including B. subtilis, the mouse pathogen C. rodentium, enterohaemorrhagic E. coli and enteropathogenic E. coli (data not shown).
Fig. 5.
Reuterin production is increased by interaction with E. coli. Increasing amounts of E. coli DH5α incubated with L. reuteri ATCC PTA-6475 in glycerol solution produced increased reuterin levels. Reuterin levels were monitored by colorimetric change at A560, as described by Circle et al. (1945), in three replicate experiments. The wild-type strain ATCC PTA-6475 was incubated with the increasing amounts of E. coli cells indicated on the x axis. The fold change over the reuterin levels produced by ATCC PTA-6475 in the absence of E. coli is represented on the y axis. Error bars, sd.
The viability of E. coli is not required for stimulation of reuterin production
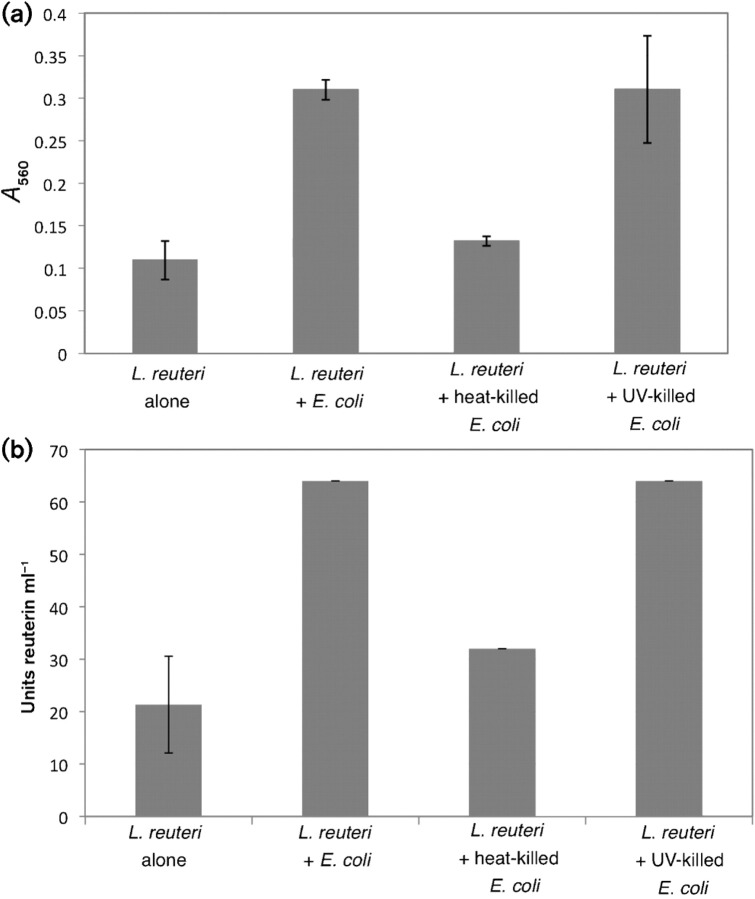
To address the mechanism through which E. coli increases reuterin production in L. reuteri, we tested whether the viability of the E. coli cells was necessary for stimulation of reuterin synthesis. E. coli DH5α cells were heat-killed either by boiling or by prolonged exposure to UV irradiation before incubation with L. reuteri in glycerol solution. Reuterin levels were measured by both MIC and colorimetric assays (Fig. 6). Full stimulation was observed using UV-killed E. coli but not the heat-killed cells (Fig. 6), indicating that although viability is not a requirement, treatment with extreme heat eliminates the ability of the E. coli to increase reuterin production.
Fig. 6.
Viability of E. coli is not required to induce higher levels of reuterin accumulation. Wild-type L. reuteri ATCC PTA-6475 cells were incubated in the presence of UV-killed or heat-killed E. coli cells and the levels of reuterin production were measured using the colorimetric assay (a) and MIC assay (b). Units reuterin ml−1 were determined from the fold dilution of the reuterin supernatant that was still able to inhibit the growth of E. coli in the MIC assay. The data presented are from three replicate experiments. Error bars, sd.
DISCUSSION
The antimicrobial compound reuterin has been proposed to be important for the ability of certain probiotic L. reuteri strains to exert beneficial properties on their hosts. Here we present evidence that the broad-spectrum antimicrobial activity of reuterin is due to 3-HPA, which is predicted to be highly reactive towards thiol groups and primary amines. Microarray analysis of cells treated with reuterin indicated that cells were undergoing oxidative stress. Reuterin assays conducted with oxyR mutant strains demonstrated that the OxyR oxidative stress response was required for E. coli to most effectively resist the effects of reuterin. Exogenously applied cysteine, but not other amino acids, could counteract the effects of reuterin. This indicates that the interaction of reuterin with thiol groups but not with primary amines was responsible for the growth inhibition. These results all indicate that the bioactive component of reuterin is the aldehyde form interacting with thiol groups of small molecules and proteins, rather than the HPA dimer inhibiting ribonucleotide reductase as a competitive inhibitor. Previous work had suggested that the HPA dimer was not favoured as the bioactive component, since it is only found in concentrations of reuterin greater than 1 M and is a minor component at relevant antimicrobial concentrations (Vollenweider et al., 2003). In addition, the inhibition of ribonucleotide reductase would activate a strong SOS response in E. coli, which was not observed in our global gene expression experiments.
One concern regarding the use of reuterin as an antimicrobial agent is its ability to create the highly toxic compound acrolein by dehydration of reuterin. We verified that the antimicrobial activity that we were observing using our reuterin production conditions was in fact due to reuterin and not to other compounds such as acrolein. Using HPLC we determined that the bioactive component of reuterin was always associated with a peak corresponding to reuterin. Antimicrobial activity was not detected with the fraction that would contain acrolein. This result eliminates any concern that low levels of acrolein were contaminants of the reuterin preparations tested in this study.
The discovery that reuterin production or secretion can be stimulated by interaction with other bacteria suggests that L. reuteri utilizes reuterin to gain a competitive advantage in the intestinal tract. While previous work had shown that co-culturing produces this stimulatory effect (Chung et al., 1989), we have demonstrated that simply mixing stationary phase L. reuteri with a number of other types of bacteria in a concentrated glycerol solution was sufficient. Because the bacteria were washed prior to being mixed together and viability is not required, we can mostly rule out the accumulation of a secondary metabolite as the mechanism of stimulation. The fact that heat-killed bacteria are not able to support reuterin stimulation while UV-killed bacteria retain this ability suggests that a heat-labile component is involved in the stimulation response. At this time we do not know if additional reuterin is being synthesized or the increase is due to more reuterin being released from the cell.
One issue concerning reuterin as an antimicrobial agent that would be bioactive against pathogens is the relatively high levels of reuterin required for killing activity in vitro. We estimate that approximately 1 mM reuterin is required to prevent bacterial growth in vitro, similar to concentrations that have been reported elsewhere (Cleusix et al., 2007; Spinler et al., 2008). NMR analysis of the forms of reuterin present at different concentrations indicates that a 1 mM solution of reuterin would contain 750 μM HPA hydrate and only 250 μM of the bioactive aldehyde 3-HPA (Vollenweider et al., 2003). The requirement for high levels of reuterin is likely due to the fact that reactions between thiol groups and aldehydes forming hemithioacetals are reversible. The 3-hydroxy substituent of reuterin also accentuates the electrophilicity of the aldehyde and therefore its propensity towards hydration, which explains, in part, why the HPA hydrate form predominates in vitro under aqueous conditions. Thus, the environment in which 3-HPA exists will greatly affect how much of the bioactive aldehyde form exists. At this time we do not know how conditions within the intestinal tract would influence the balance of the aldehyde and HPA hydrate forms.
It seems unlikely that L. reuteri would be able to produce such high levels throughout the intestinal tract and have a systemic killing effect on a pathogen. A more likely function of reuterin is perhaps to provide a localized antimicrobial effect in which production of reuterin could provide a competitive advantage in the intestinal tract. L. reuteri itself is highly resistant to the effects of reuterin and therefore could create a zone of growth inhibition to compete with other organisms in the gastrointestinal tract. Future studies using the mutants described in this manuscript will address the ability of reuterin to enhance colonization of L. reuteri as well as its role in evoking beneficial effects in animal models.
Supplementary Data
Acknowledgments
We thank Scott Mulrooney and Piotr Grzyska for HPLC assistance, and Cathy Robinson for analysing the effects of reuterin on Clostridium difficile. We appreciate the support of Eamonn Connolly and BioGaia for access to probiotic L. reuteri strains and helpful research discussions. We also thank Abraham L. Sonenshein (Tufts University, Medford, MA) for providing us with Clostridium difficile CD630. This work was supported by grants from The Gerber Foundation and the Michigan State Center for Microbial Pathogenesis.
Footnotes
The microarray data discussed in this paper are available from the Gene Expression Omnibus (GEO) under accession number GSE19760.
A supplementary table, listing E. coli genes induced and repressed upon exposure to reuterin, is available with the online version of this paper.
Edited by: P. W. O'Toole
References
- Arques J. L., Fernandez J., Gaya P., Nunez M., Rodriguez E., Medina M. Antimicrobial activity of reuterin in combination with nisin against food-borne pathogens. Int J Food Microbiol. 2004;95:225–229. doi: 10.1016/j.ijfoodmicro.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Arques J. L., Rodriguez E., Nunez M., Medina M. Antimicrobial activity of nisin, reuterin, and the lactoperoxidase system on Listeria monocytogenes and Staphylococcus aureus in cuajada, a semisolid dairy product manufactured in Spain. J Dairy Sci. 2008;91:70–75. doi: 10.3168/jds.2007-0133. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz T. M., Wick L. M., Qi W., Riordan J. T., Ouellette L. M., Whittam T. S. Global transcriptional response of Escherichia coli O157 : H7 to growth transitions in glucose minimal medium. BMC Microbiol. 2007;7:97. doi: 10.1186/1471-2180-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton R. A. DNA microarrays and bacterial gene expression. Methods Enzymol. 2003;370:264–278. doi: 10.1016/S0076-6879(03)70023-8. [DOI] [PubMed] [Google Scholar]
- Britton R. A., Eichenberger P., Gonzalez-Pastor J. E., Fawcett P., Monson R., Losick R., Grossman A. D. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Morgan R. W., Jacobson F. S., Ames B. N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- Chung T. C., Axelsson L., Lindgren S. E., Dobrogosz W. J. In vitro studies on reuterin synthesis by Lactobacillus reuteri. Microb Ecol Health Dis. 1989;2:137–144. [Google Scholar]
- Circle S. J., Stone L., Boruff C. S. Acrolein determination by means of tryptophan. Ind Eng Chem Res. 1945;17:259–262. [Google Scholar]
- Cleusix V., Lacroix C., Vollenweider S., Duboux M., Le Blay G. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol. 2007;7:101. doi: 10.1186/1471-2180-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. R., Hoch J. A., Aronson A. I. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol. 1977;131:981–987. doi: 10.1128/jb.131.3.981-987.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doleyres Y., Beck P., Vollenweider S., Lacroix C. Production of 3-hydroxypropionaldehyde using a two-step process with Lactobacillus reuteri. Appl Microbiol Biotechnol. 2005;68:467–474. doi: 10.1007/s00253-005-1895-4. [DOI] [PubMed] [Google Scholar]
- Ellman G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Floch M. H., Walker W. A., Guandalini S., Hibberd P., Gorbach S., Surawicz C., Sanders M. E., Garcia-Tsao G., Quigley E. M., other authors Recommendations for probiotic use – 2008. J Clin Gastroenterol. 2008;42(Suppl. 2):S104–S108. doi: 10.1097/MCG.0b013e31816b903f. [DOI] [PubMed] [Google Scholar]
- Kandler O., Setter K. O., Kohl R. Lactobacillus reuteri sp. nov., a new species of heterofermentative lactobacilli. Zentralbl Bakteriol Mikrobiol Hyg Abt. 1980;1 Orig C1:264–269. [Google Scholar]
- Lin Y. P., Thibodeaux C. H., Pena J. A., Ferry G. D., Versalovic J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis. 2008;14:1068–1083. doi: 10.1002/ibd.20448. [DOI] [PubMed] [Google Scholar]
- Luthi-Peng Q., Dileme F. B., Puhan Z. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl Microbiol Biotechnol. 2002;59:289–296. doi: 10.1007/s00253-002-1002-z. [DOI] [PubMed] [Google Scholar]
- Marco M. L., Pavan S., Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 2006;17:204–210. doi: 10.1016/j.copbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Morita H., Toh H., Fukuda S., Horikawa H., Oshima K., Suzuki T., Murakami M., Hisamatsu S., Kato Y., other authors Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 2008;15:151–161. doi: 10.1093/dnares/dsn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara A. M., Shanahan F. Mechanisms of action of probiotics in intestinal diseases. ScientificWorldJournal. 2007;7:31–46. doi: 10.1100/tsw.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J. M., Arenas F. A., Pradenas G. A., Sandoval J. M., Vasquez C. C. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem. 2008;283:7346–7353. doi: 10.1074/jbc.M708846200. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt V., Michaelsen K. F., Jakobsen M., Larsen C. N., Møller P. L., Pedersen P., Tvede M., Weyrehter H., Valerius N. H., Paerregaard A. Effect of probiotic Lactobacillus strains in young children hospitalized with acute diarrhea. Pediatr Infect Dis J. 2002a;21:411–416. doi: 10.1097/00006454-200205000-00012. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt V., Michaelsen K. F., Jakobsen M., Larsen C. N., Moller P. L., Tvede M., Weyrehter H., Valerius N. H., Paerregaard A. Effect of probiotic Lactobacillus strains on acute diarrhea in a cohort of nonhospitalized children attending day-care centers. Pediatr Infect Dis J. 2002b;21:417–419. doi: 10.1097/00006454-200205000-00013. [DOI] [PubMed] [Google Scholar]
- Russell W. M., Klaenhammer T. R. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl Environ Microbiol. 2001;67:4361–4364. doi: 10.1128/AEM.67.9.4361-4364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino F., Pelle E., Palumeri E., Oggero R., Miniero R. Lactobacillus reuteri (American Type Culture Collection Strain 55730) versus simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics. 2007;119:e124–e130. doi: 10.1542/peds.2006-1222. [DOI] [PubMed] [Google Scholar]
- Schauer D. B., Falkow S. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun. 1993;61:4654–4661. doi: 10.1128/iai.61.11.4654-4661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinler J. K., Taweechotipatr M., Rognerud C. L., Ou C. N., Tumwasorn S., Versalovic J. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14:166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramulu D. D., Liang M., Hernandez-Romero D., Raux-Deery E., Lunsdorf H., Parsons J. B., Warren M. J., Prentice M. B. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol. 2008;190:4559–4567. doi: 10.1128/JB.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarico T. L., Dobrogosz W. J. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1989;33:674–679. doi: 10.1128/aac.33.5.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarico T. L., Casas I. A., Chung T. C., Dobrogosz W. J. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1988;32:1854–1858. doi: 10.1128/aac.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker W. C., Schaefer L., Britton R. A. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol Microbiol. 2006;59:528–540. doi: 10.1111/j.1365-2958.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Vollenweider S., Lacroix C. 3-Hydroxypropionaldehyde: applications and perspectives of biotechnological production. Appl Microbiol Biotechnol. 2004;64:16–27. doi: 10.1007/s00253-003-1497-y. [DOI] [PubMed] [Google Scholar]
- Vollenweider S., Grassi G., Konig I., Puhan Z. Purification and structural characterization of 3-hydroxypropionaldehyde and its derivatives. J Agric Food Chem. 2003;51:3287–3293. doi: 10.1021/jf021086d. [DOI] [PubMed] [Google Scholar]
- Walter J., Chagnaud P., Tannock G. W., Loach D. M., Dal Bello F., Jenkinson H. F., Hammes W. P., Hertel C. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl Environ Microbiol. 2005;71:979–986. doi: 10.1128/AEM.71.2.979-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman Z., Asli G., Alsheikh A. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics. 2005;115:5–9. doi: 10.1542/peds.2004-1815. [DOI] [PubMed] [Google Scholar]
- Whitehead K., Versalovic J., Roos S., Britton R. A. Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl Environ Microbiol. 2008;74:1812–1819. doi: 10.1128/AEM.02259-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens K., Black P. N., Stanley T. V., Mitchell E., Fitzharris P., Tannock G. W., Purdie G., Crane J. A differential effect of 2 probiotics in the prevention of eczema and atopy: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2008;122:788–794. doi: 10.1016/j.jaci.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Zhang X. S., Garcia-Contreras R., Wood T. K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol. 2007;189:3051–3062. doi: 10.1128/JB.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.