Abstract
SARS-CoV-2 infection is required for COVID-19, but many signs and symptoms of COVID-19 differ from common acute viral diseases. Currently, there are no pre- or post-exposure prophylactic COVID-19 medical countermeasures. Clinical data suggest that famotidine may mitigate COVID-19 disease, but both mechanism of action and rationale for dose selection remain obscure. We explore several plausible avenues of activity including antiviral and host-mediated actions. We propose that the principal famotidine mechanism of action for COVID-19 involves on-target histamine receptor H2 activity, and that development of clinical COVID-19 involves dysfunctional mast cell activation and histamine release.
Keywords: COVID-19, Famotidine, Histamine, Mast Cell, GPCR
Introduction
SARS-CoV–2 is a highly infectious and pathogenic betacoronavirus first detected in human infections during December 2019 1–3. COVID–19 is a disease spectrum causally associated with infection by SARS-CoV–2. Definitive COVID–19 diagnosis requires the presence of the virus, which can be isolated, grown, or otherwise detected as unique SARS-CoV–2 viral nucleic acid sequences. There are SARS-CoV–2 virus shedding or nucleic acid positive patients that do not manifest clinical COVID–19 4–9. 13–20% of patients with symptoms develop severe respiratory compromise requiring oxygenation, with radiological findings of ground glass opacities and consolidation 10–12. Between 5 and 46% of SARS-CoV–2 positive patients are asymptomatic and do not appear to progress to COVID–19 13–16. Therefore, SARS-CoV–2 infection is necessary but not sufficient for development of clinical COVID–19 disease.
Patients with COVID–19 disease can present with a range of mild to severe non-specific clinical signs and symptoms which develop two to fourteen days after exposure to SARS-CoV–2. These symptoms include cough or shortness of breath, and at least two of the following; fever, chills, repeated rigor, myalgia, headache, oropharyngitis, anosmia and ageusia 17,18. More severe symptoms warranting hospital admission include difficulty breathing, a persistent sense of chest pain or pressure, confusion or difficulty to arouse, and central cyanosis. Of hospitalized patients, 20–42% develop ARDS, the most common cause for admission to the ICU. 39–72% of patients admitted to the ICU will die 19.
Early clinical data from a variety of sources indicate that famotidine treatment may reduce morbidity and mortality associated with COVID–19. A retrospective cohort study of 1,620 hospitalized COVID–19 patients indicates that 84 propensity score matched patients receiving famotidine during hospitalization (oral or IV, 20mg or 40mg daily) had a statistically significant reduced risk for death or intubation (adjusted hazard ratio (aHR) 0.42, 95% CI 0.21–0.85) and also a reduced risk for death alone (aHR 0.30, 95% CI 0.11–0.80) 20. In contrast, proton pump inhibitor use was not associated with reduced risk for these outcomes. A preceding anecdotal report from Wuhan, China is purported to have indicated that famotidine may be partially protective for COVID–19, but that neither cimetidine nor proton pump inhibitors were protective 21. Together, these data have been interpreted as indicating that this increased survival pattern is due to an off-target, non-histamine receptor-mediated property of famotidine that is not shared with cimetidine. Famotidine is currently being tested under an IND waiver for treating COVID–19 in a double blind randomized clinical trial at high intravenous doses in combination with either hydroxychloroquine or remdesivir (ClinicalTrials.gov Identifier: NCT04370262).
Herein we aim to investigate how famotidine may act to relieve early phase COVID–19 clinical symptoms. The most likely mechanisms of actions include: via antiviral activity, via novel human targets, or via the on-target mechanism described in the current FDA market authorization—famotidine is a histamine receptor H2 antagonist (and inverse agonist).
Results
Antiviral activity
Famotidine does not bind to SARS-CoV–2 proteases
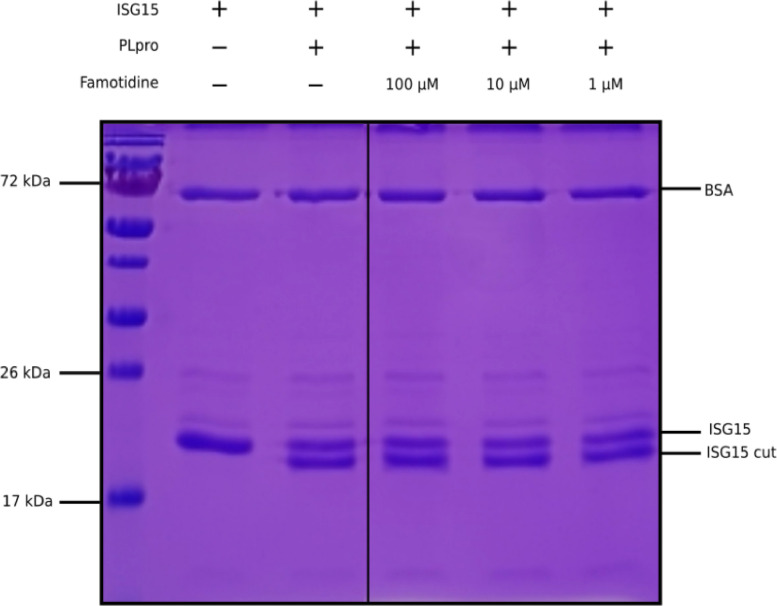
The idea to test the usefulness of famotidine as a medical countermeasure for COVID–19 emerged from a computational molecular docking effort aimed at identifying inhibitors of the papain-like protease (PLpro) of SARS-CoV–2 22,23. In addition to processing the viral polyprotein, the papain-like protease from coronaviruses (PLpro) is known to remove the cellular substrates ubiquitin and the interferon stimulated gene 15 (ISG15) from host cell proteins by cleaving the C-terminal end of the consensus sequence LXGG, a process termed deISGylation 24,25. Here, we used the enzymatic reaction of SARS-CoV–2 PLpro on ISG15 to assess the potential inhibition of PLpro by famotidine. The cleavage of the 8 C-terminal amino acids of ISG15 by PLpro is clearly detected by SDS-PAGE (Figure 1, lanes 2 and 3). However, the addition of 1 to 100 μM famotidine to the reaction does not significantly reduce the amount of ISG15 cleaved during the assay (Figure 1, lanes 4 to 6), thus suggesting that famotidine does not inhibit SARS-CoV–2 PLpro. A previous virtual screening report 26 suggested that famotidine might bind to the 3 chymotrypsin-like protease (3CLpro), more commonly referred to as the main protease (Mpro), however this mechanism was recently discounted 27.
Figure 1.
Cleavage of ISG15 C-terminal 8 amino acids by SARS-CoV-2 PLpro purified from E. coli. ISG15 was incubated with SARS-CoV-2 PLpro (lanes 3 to 6). SARS-CoV-2 PLpro was present at 4 nM, ISG15 was present at 10 μM. For lane 4 to 6, famotidine was present at 100 μM, 10 μM and 1 μM respectively. Control was without enzyme (lane 2). Proteins were resolved by 15% SDS-PAGE and revealed by Coomassie blue staining. The molecular weights of the marker proteins are indicated on the left of the gel.
Famotidine does not directly inhibit SARS-CoV–2 infection
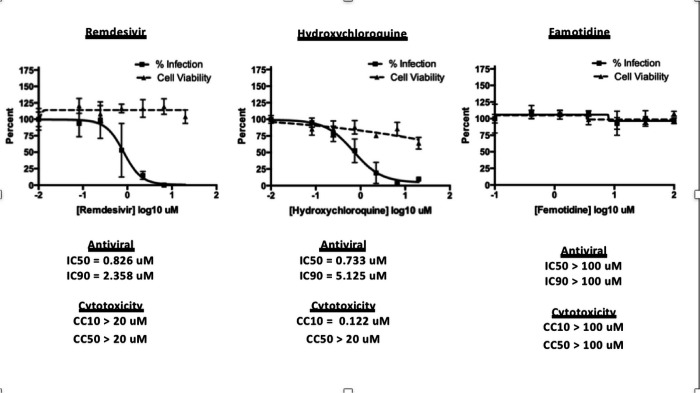
To assess the possibility that famotidine may inhibit SARS-CoV–2 infection by other routes, a Vero E6 cell-based assay was performed to compare median tissue culture infectious doses (TCID50/mL) of famotidine, remdesivir, and hydroxychloroquine (Figure 2). While both remdesivir and hydroxychloroquine demonstrated antiviral activity, no inhibition of SARS-CoV–2 infection was observed with famotidine.
Figure 2.
Famotidine does not directly inhibit SARS-CoV-2 infection
Human receptors
Famotidine does not act via sigma–1 or –2 receptor binding
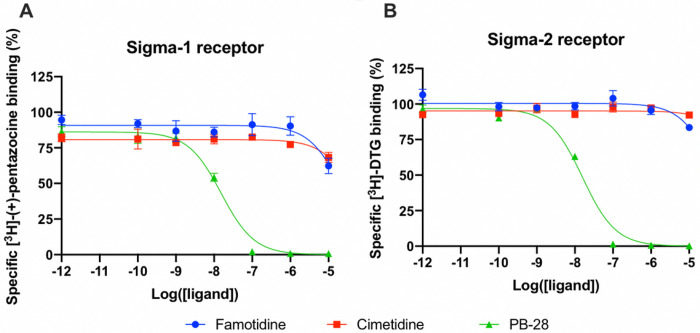
A wide-ranging study recently presented a map of interactions between viral and host proteins 28. It was shown that regulation of the sigma–1 and sigma–2 receptors had antiviral effects. Sigma and histamine receptors share several ligands in common, like the antipsychotic haloperidol, the antihistamines astemizole and clemastine, the antidepressive clomipramine, and many more. As such, we tested for possible interaction between famotidine and sigma–1 or sigma–2 receptors (Figure 3). We performed radioligand competition binding experiments using cloned sigma receptors, following established procedures 29 30. In these assays, famotidine showed no detectable displacement of radioligand probes for either sigma–1 or sigma–2 receptors at famotidine concentrations up to 10 μM. Hence, famotidine’s binding to sigma–1 and sigma–2 receptors is likely negligible at physiologically relevant concentrations.
Figure 3.
Competition binding curves of Famotidine (blue circles), Cimetidine (red squares), and PB-28 (green triangle), a potent sigma receptor ligand as positive control. (A) [3H](+)-pentazocine competition curves in Expi293 membranes expressing sigma-1. (B) [3H]DTG competition curves in Expi293 membranes expressing sigma-2 (TMEM97).
Famotidine is selective for receptor H2
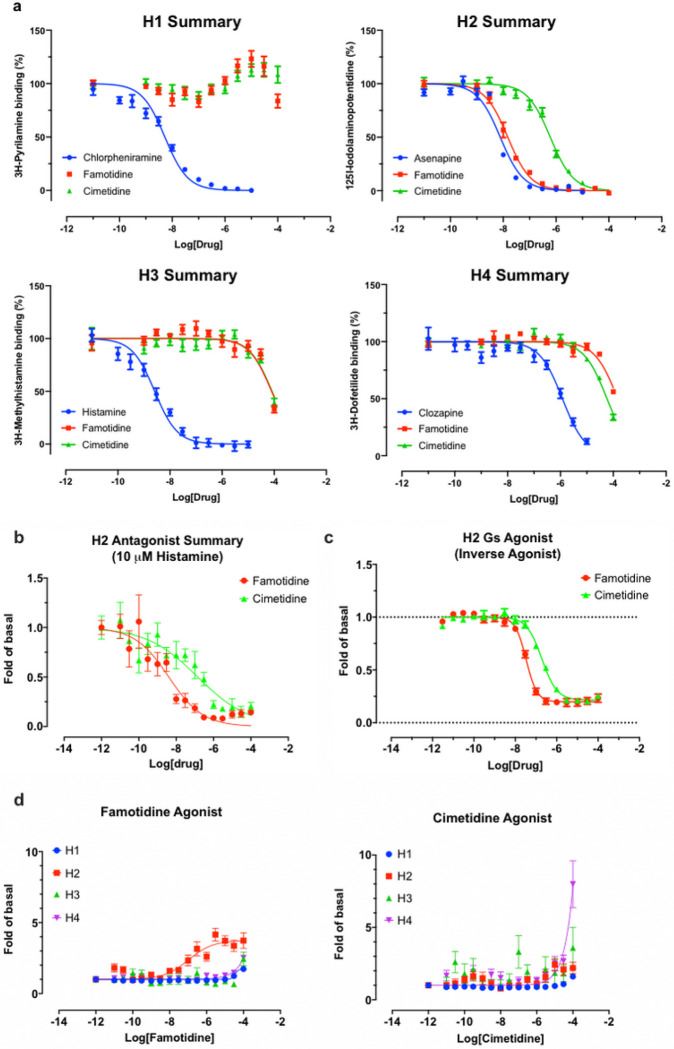
As is well-known 31, famotidine is a selective blocker of the histamine H2 receptor with affinity of approximately 14 nM, substantially more active than the 590 nM cimetidine (Figure 4A). Here we find it to have highly efficacious inverse agonist activity (reducing basal activity by 75%) with a potency of 33 nM (Figure 4C). Intriguingly, and unlike cimetidine, while famotidine acts to block Gs protein signaling it actually acts as a partial agonist of arrestin recruitment, with an efficacy of about 15% that of histamine, and an EC50 of 105 nM (Figure 4D), suggesting that the molecule promotes arrestin-scaffolded signaling—such as through the ERK pathway, 32 and promotes internalization of the receptor and further non-canonical signaling once internalized 33,34 through an arrestin-biased mechanism. These features distinguish famotidine certainly from cimetidine, and potentially from other H2 blockers, as such biased activation of arrrestin recruitment for GPCR antagonists, while not unprecedented, is not common.
Figure 4.
Famotidine and cimetidine activity on histamine receptors. Experiments performed in duplicate. A. Competitive binding dose-response curves for famotidine and cimetidine on four histamine receptors with reference compounds. B. The partial agonist, famotidine, shows antagonist activity of H2 in the presence of potent endogenous agonist, histamine. C. Inverse agonism of famotidine and cimetidine on H2, whereas histamine stimulated cAMP production by 20-fold of basal (N=2). D. Arrestin recruitment by famotidine (left) and cimetidine (right) upon interaction with histamine receptors.
Famotidine may activate other GPCRs
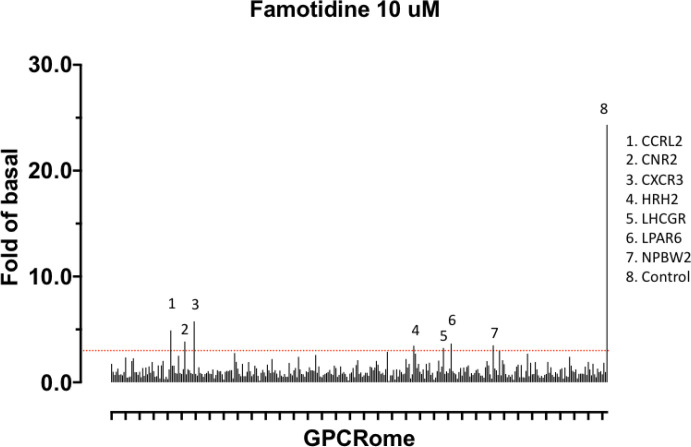
Finally, we note that a screen for activation of 318 receptors of the GPCR-ome reveals only seven receptors with an average fold of basal increase above 3.0, including H2 (Figure 5). In all cases, the quadruplicate replicates were not in agreement and require follow-up studies. Chief among these are the CCR2L and CXCR3 chemokine receptors 35–38. Such activity would be intriguing because these receptors would be expected to activate immune cell mobilization and may plausibly have a role in famotidine’s beneficial activities, especially at the high systemic concentrations it is expected to reach in the clinical studies. This would also be consistent with famotidine’s lack of direct anti-viral activity in the Vero cell direct infectivity assays, where immune cells are not present.
Figure 5.
Screen for activation of 318 receptors of the GPCR-ome.
Famotidine reaches functionally relevant systemic concentrations, whereas cimetidine does not
We calculated predicted steady state concentrations of famotidine and cimetidine at different doses based on published pharmacokinetic and biodistribution data 39–41. This modeling demonstrated that the different clinical outcomes exhibited by COVID–19 patients taking famotidine vs. cimetidine could be readily explained by the distinctive pharmacokinetic and pharmacodistribution properties of the two agents.
Therapeutic efficacy of a pharmacological antagonist requires that it achieves a steady-state concentration that substantially exceeds the half maximal inhibitory concentration (IC50) for its target. Thus, in order to evaluate the relative systemic effects of famotidine and cimetidine, the IC50 values of each agent for the H2 receptor were compared to the steady-state plasma concentrations (Css) predicted at standard clinical doses. As demonstrated above, famotidine binds to H2 with Ki of 14 nM, whereas cimetidine binds to H2 with Ki 586 nM. Previous reports suggest functional IC50s are approximately 3x higher, and these data were used for the current analyses 39,41. In these reports, the IC50 for the H2 receptor were reported as 13 μg/L (0.039 μM) for famotidine and 400–780 μg/L (1.59–3.09 μM) for cimetidine. Css values were calculated using pharmacokinetic data for dosing, clearance, bioavailability, and volume of distribution as summarized previously 41. Table 1 lists the Css values for both famotidine and cimetidine.
Table 1:
Steady-state concentrations (Css) of Famotidine and Cimetidine at standard doses compared to the half maximal inhibitory concentration (IC50) value of Famotidine or Cimetidine for histamine H2 receptor antagonism
| IC50 or Css | Concentration (mass/volume) | Concentration (molarity) | ||
| Famotidine | ||||
| IC50 Histamine H2 | 13 mg/L | 0.039 mM | ||
| IC50 Neutrophil H2 O2− assay | 67 mg/L | 0.201 mM | ||
| IC50 Neutrophil H2 cAMP assay | 8 mg/L | 0.024 mM | ||
| IC50 Eosinophil H2 cAMP assay | 53.6 mg/L | 0.158 mM | ||
| IC50 Mast Cell H2 cAMP increase | Not determined | |||
| Total Concentration (mass/volume) | Total Concentration (mass/volume) | Free drug Concentration3 (mass/volume) | Free drug Concentration3 (molarity) | |
| Css (20 mg tablet p.o. q.d.) | 17.7 mg/L | 0.053 mM | 14.2 mg/L | 0.042 mM |
| Css (20 mg capsule p.o. q.d.) | 18.4 mg/L | 0.055 mM | 14.7mg/L | 0.044 mM |
| Css (20 mg tablet p.o. b.i.d.) | 35.4 mg/L | 0.105 mM | 28.3 mg/L | 0.084 mM |
| Css (20 mg capsule p.o. b.i.d.) | 36.8 mg/L | 0.109 mM | 29.4 mg/L | 0.087 mM |
| Css (20 mg tablet p.o. t.i.d.) | 53.1 mg/L | 0.157 mM | 42.5 mg/L | 0.126 mM |
| Css (20 mg capsule p.o. t.i.d.) | 55.3 mg/L | 0.164 mM | 44.2 mg/L | 0.131 mM |
| Css (40 mg tablet p.o. t.i.d.)1 | 55.4 mg/L | 0.164 mM | 44.3 mg/L | 0.131 mM |
| Css (40 mg tablet p.o. t.i.d.)2 | 80.8 mg/L | 0.239 mM | 64.6 mg/L | 0.192 mM |
| Css (60 mg tablet p.o. t.i.d.) | 144.3 mg/L | 0.425 mM | 115.4 mg/L | 0.340mM |
| Css (120mg IV every 8 hours) | 1,290 mg/L | 1.092 mM | 1,032 mg/L | 0.874 mM |
| Cimetidine | ||||
| IC50 Histamine H2 | 400–780 mg /L | 1.59–3.09 mM | ||
| Css (200 mg tablet p.o. q.d.) | 175 mg /L | 0.69 mM | 140 mg/L | 0.055 mM |
| Css (300 mg tablet p.o. q.d.) | 226 mg /L | 0.90 mM | 180.1 mg/L | 0.720 mM |
In primary human neutrophils and eosinophils, H2 activation by histamine inhibits neutrophil effector functions including O2− release 42,43, platelet-activating-factor induced chemotaxis 44 and leukotriene biosynthesis 45. Eosinophil functions are also inhibited by H2 activation; histamine binding diminishes eosinophil peroxidase release 46 and, at high concentrations, inhibits eosinophil chemotaxis 47,48. Famotidine is one of the most effective antagonists of these H2-mediated histamine effects on neutrophils and eosinophils49. IC50 for two measures that relate to these phenotypes are also listed in Table 1. Mast cells express histamine H2 and H4 receptors, and histamine-induced increase of cAMP in mast cells is inhibited by famotidine 50. 10μM famotidine pre-incubation blocks histamine-induced cAMP increase in human skin mast cells, however, the IC50 for this effect has not been determined 50.
At all dosing regimens, the Css for famotidine exceeds the general IC50 value for the H2 receptor, and at the twice daily (b.i.d.) and thrice daily (t.i.d.) dosing of 20 mg and 40 mg, the Css for unbound famotidine is 2–5 fold greater than the H2 IC50. Also calculated and summarized is the Css for the intravenous dosage currently being administered in clinical trial NCT04370262 and that dose exceeds famotidine IC50 by greater than 20-fold. In contrast, unbound cimetidine levels at standard doses of 200 or 300 mg daily (q.d.), achieve a Css that is a fraction of the reported IC50 range of 400–780 μg/L.
Case history, Severe COVID–19 Outpatient Treatment with Famotidine
Patient JM is a 47 year old male who received PCR diagnosis of COVID–19 after 8 days of complaints of diarrhea, abdominal cramping, eructation, low energy, dry cough, arthralgia, myalgia, anosmia and ageusia. The patient has a history of hypertension (10y), Type II diabetes (4y), hypercholesterolemia (3y) and gout (10y). Current medications included Metformin, Allopurinol, Lisinopril, and Atorvastatin. He is employed as a hospital maintenance worker in the hospital to which he presented.
Contact tracing revealed that his son (same household) had developed COVID–19 symptoms 12 days prior. Receipt on day 8 of positive PCR diagnosis (from a prior outpatient intranasal swab sample) coincided with onset of fever (102oF), night sweats, shortness of breath and a feeling of chest pressure. Famotidine (“PEPCID AC ®” 60mg p.o. t.i.d. = 2.24mg/ft2 t.i.d) was initiated upon receiving the PCR diagnosis due to symptoms meeting FDA criteria for severe COVID–19, combined with high risk pre-existing conditions. The famotidine drug regime was continued for 30 days. After initiating famotidine in the evening, the patient was able to sleep through the night and reported complete relief from the chest pressure sensation, reduction in cough, but continued to be febrile (101.6 °F).
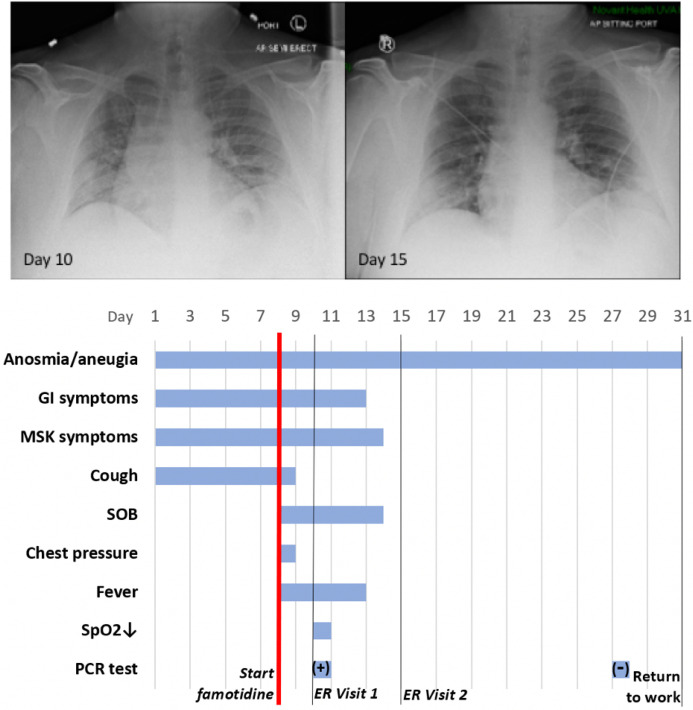
On day 10, he presented to the emergency room (ER) with continuing complaints of diarrhea, abdominal cramping, eructation, low energy, dry cough, arthralgia, myalgia, anosmia and ageusia and shortness of breath on exertion. Day 10 ER physical examination, including the chest, was unremarkable and vital signs were normal. The patient BMI was 36 (Du Bois BSA 26.78 ft2). SpO2 was 93%, rising to 97% and 99% on 3 L/min by nasal cannula over the next 30 minutes. An intranasal sample was obtained for SARS-CoV–2 rtPCR diagnostic analysis. Comprehensive metabolic panel showed a mild decrease in serum sodium and chloride with hyperglycemia (260 mg/dL). Complete blood count (CBC) was normal, specifically including the lymphocyte count. Urinalysis showed a specific gravity of 1.025 but was otherwise normal. A portable chest X-ray had poor inspiration but was interpreted as showing “bibasilar areas of airspace disease” consistent with COVID–19 (Figure 6, CXR day 10). The patient was diagnosed as dehydrated, given ondansetron IV, 1 L IV of normal saline and discharged home with a hospital pulse oximeter. At the time of departure, he had an SpO2 of 94% on room air that did not drop with ambulation.
Figure 6.
Case Study JM: CXR and Timeline Famotidine (60 mg PO tid) was started on Day 8 from start of symptoms. It was continued for 30 days. The anosmia and aneugia are still present at Day 50.
The patient again presented to the emergency room on day 15 after experiencing near-syncope during showering. Physical examination was unremarkable. Vital signs were normal. SpO2 showed values of 98%, 93% and 97% on room air over the 2 hour period. Basic metabolic panel showed only hyperglycemia (266 mg/dL). CBC was normal except for a mild lymphopenia (0.96; reference range 1.00–3.00 ×103/μL) and mild monocytosis (0.87; 0.20–0.80 ×103/μL). Chest X-ray was interpreted as showing “Faint patchy consolidation of lung bases bilaterally, similar to perhaps minimally improved at the lower left lung base compared to prior” (Figure 6 CXR day 15). The patient was placed on azithromycin and discharged to home.
On days 27 and 28 after initial symptoms, he tested negative (2x, successive days) for SARS-CoV–2 nucleic acid by PCR (intranasal swab) and returned to his work at the local hospital 31 days after initial symptoms. 47 days after first developing COVID–19 symptoms he continues to note a lack of ability to taste or smell, but otherwise considers himself largely recovered from COVID–19 (Figure 6 timeline).
Use of famotidine in this patient was recommended due to meeting FDA criteria for severe COVID–19 and his COVID–19 risk factors: male, 47yo, hypertension, obesity and diabetes mellitus Type 2. Although this is an anecdotal example, the patient experienced relief of symptoms overnight upon initiating use of famotidine. While not sufficient to demonstrate proof of cause and effect, this case does provide context for typical COVID–19 presentation and symptoms, as well as support for additional well-controlled famotidine therapeutic clinical trials in an outpatient setting.
Discussion
Famotidine is an off-patent drug available as either branded (“PEPCID®”) or generic medicines in tablet, capsule or intravenous forms. The general pharmacology of famotidine is well-characterized, with an excellent absorption, distribution, metabolism, excretion and toxicology profile 53. Famotidine is unique among the drugs currently being tested for treatment of COVID–19, in that it is an H2 receptor antagonist (and inverse agonist). Famotidine is currently being tested for treating COVID–19 in a double blind randomized clinical trial at high intravenous doses in combination with either hydroxychloroquine or remdesivir (ClinicalTrials.gov Identifier: NCT04370262). A recent retrospective cohort study of 1,620 hospitalized COVID–19 patients indicates that 84 propensity score matched patients receiving famotidine during hospitalization (oral or IV, 20mg or 40mg daily) had a statistically significant reduced risk for death or intubation (adjusted hazard ratio (aHR) 0.42, 95% CI 0.21–0.85) and also a reduced risk for death alone (aHR 0.30, 95% CI 0.11–0.80) 20. In contrast, proton pump inhibitor use was not associated with reduced risk for these outcomes. Anecdotal reports and undisclosed data indicating that famotidine provided protection from COVID–19 mortality while neither cimetidine nor proton pump inhibitors were similarly protective lead to an initial inference that the beneficial effects of famotidine were not related to the known on-target activity of the drug 21. Studies detailed in this report and others, however, indicate that famotidine does not act by directly inhibiting either of the principal SARS-CoV–2 proteases (PLpro or Mpro) 27. Vero E6-based cell assays also indicate that famotidine has no direct antiviral activity in this cell line, although antiviral activity in cells that express H2 has not been tested. Additional hypotheses that famotidine may act via binding either the sigma–1 or –2 receptors have not been supported by the studies summarized herein.
The most straightforward explanation of the apparent famotidine activity as a COVID–19 therapy is that the drug acts via its antagonism or inverse-agonism of histamine signaling and via its arrestin biased activation—all a result of its binding to histamine receptor H2. If true, then it is reasonable to infer that a SARS-CoV–2 infection that results in COVID–19 is at least partially mediated by pathologic histamine release. The anecdotal lack of protection provided by oral administration of the H2 antagonist cimetidine can be accounted for by insufficient systemic drug levels after oral administration and does not contradict potential benefit provided by famotidine H2 binding. Intravenous cimetidine at sufficient doses may achieve levels high enough for clinical benefit and would further support this hypothesis. Failure to achieve clinical COVID–19 responses with cimetidine may indicate that inverse agonism or other GPCR-mediated effects of famotidine may play an important role in the (preliminary) observed clinical benefits. Analysis of famotidine activity in histamine receptor competition assays indicate that, over the range of clinical steady state famotidine drug levels being tested, famotidine is specific for H2. Therefore, off-target antagonism of histamine H1 receptor, H3 receptor, or H4 receptor is unlikely to contribute to famotidine-mediated effects.
Steady state famotidine concentrations sufficient to elicit H2 antagonism (and inverse agonism) are readily achieved using inexpensive oral tablets and safe dosage levels. The famotidine dosage employed in the only retrospective hospital study currently available examining famotidine effects on COVID–19 outcomes appears to have employed dosages (20mg to 40mg daily) which are unlikely to fully inhibit histamine-mediated effects at the H2 receptor 20. In contrast, study NCT04370262 administers intravascular famotidine doses that are more than 20-fold greater than the IC50 for antagonism of H2. The data presented herein provides a rationale for famotidine dose selection to maintain a steady state concentration at a reasonable multiple of the IC50 for systemic antagonism of H2 and indicate that oral tablet dosages of between 40mg every eight hours to 60mg every eight hours should be sufficient to insure maximal H2 target effects. As famotidine is primarily cleared by the kidney, adequate renal function is required for higher dosages 53.
In addition to H2 antagonism, famotidine may also act as an inverse agonist thereby lowering the concentration of cyclic-Adenosine Monophosphate (c-AMP) 32. Endothelial cell permeability has been attributed to histamine H2 activation and is blunted by famotidine pretreatment 54. Histamine, bradykinin and des-arg-bradykinin receptor engagements can lead to increased endothelial permeability through a common pathway that results in AKT–1 activation 55. The H2 receptor also signals through Gq/11 proteins, resulting in inositol phosphate formation and increases in cytosolic Ca2+ concentrations which may account for the increased endothelial cell fluid permeability 56.
One alternative hypothesis is that famotidine may not only inhibit signaling through the H2 receptor but may also engage in cross talk with the kinin B1 receptor, which moderates the response of endothelial cells to DABK and DAKD ligands. Data provided here in are not consistent with this hypothesis; no activation of bradykinin receptor B1 or B2 were observed in quadruplicate replicate TANGO assay.
While COVID–19 symptoms affect multiple organ systems, respiratory failure due to acute respiratory distress syndrome (ARDS) is the most common cause of death. Examination of RNA expression profiles of the cells which contribute to lung anatomy and function demonstrate the presence of multiple ACE2/TMPRSS2 positive cell types susceptible to SARS-CoV–2 infection in the lung. In addition, these and other associated lung cells that are positive for histamine receptors H1 and H2 could respond to local histamine release following mast cell degranulation 57, and therefore those cells positive for H2 may be responsive to famotidine effects.
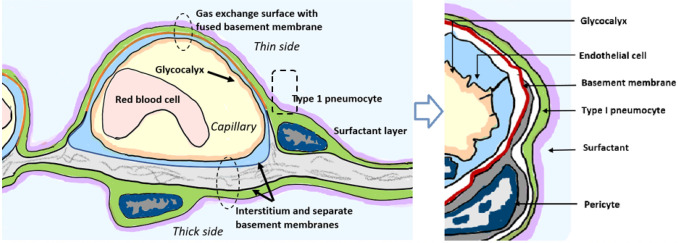
To understand how famotidine may act to reduce pulmonary COVID–19 symptoms requires an understanding of COVID–19 lung pathophysiology, which appears to have two principal disease phases. In turn, this requires an appreciation of pulmonary tissue and cell types. Pulmonary edema results from loss of a regulation of fluid transfer that occurs at several levels in the alveolus, as diagrammed in Figure 7. In the capillary wall, there are the glycocalyx, the endothelial cell with associated tight junctions, and the basement membrane. In the epithelium there is a surfactant layer on the alveolar lining fluid, manufactured and secreted by the Type II pneumocyte, and the Type I pneumocyte itself with its tight junctions and negatively charged basement membrane which restricts albumin. The pulmonary pericytes located in the terminal conducting airway region play a critical role in synthesizing the endothelial basement membrane and regulating blood flow in the precapillary arteriole, the capillary and the postcapillary venule. Disruption of any of these cells or layers can lead to edema. This edema fluid may be a transudate in milder dysfunctions or an exudate when inflammation or necrosis develop. Two possible pathologies that could result in edema of the alveolar wall and space include infection of cells by SARS-CoV–2 and mast cell degranulation with release of hundreds of compounds that can impact on cellular and basement membrane functions, glycocalyx and tight junction integrity. These compounds include histamine, bradykinin, heparin, tryptase and cytokines.
Figure 7.
Lung alveolus cell interactions and gas exchange
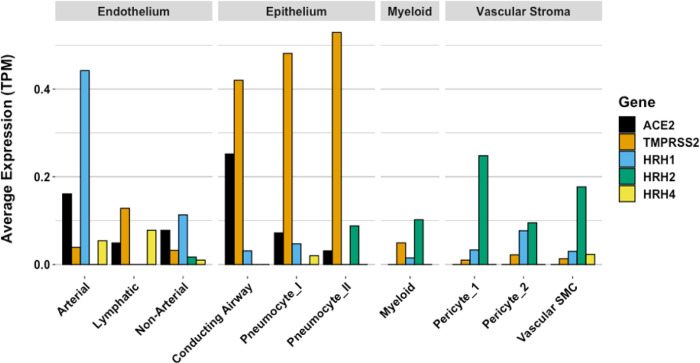
Gene expression patterns of these pulmonary cells provide insight into which cells are likely to be infected, and which express the H2 receptor that could be directly impacted by famotidine treatment and resulting H2 antagonism or inverse agonism (Figure 8). These patterns suggest that epithelial cells and endothelial cells are more likely to be infected based on ACE2 and TMPRSS2 expression patterns in those cell types. The cells most likely to show a famotidine effect include Type 2 pneumocytes, smooth muscle cells, pericytes, and myeloid granulocytes (which includes mast cells, neutrophils and eosinophils).
Figure 8.
Human single cell lung gene expression normalized to transcripts per million (TPM) from LunGENS web portal 58. Single cell lung gene expression patterns from the Dropseq PND1 experiment for angiotensin-converting enzyme 2 (ACE2: black), transmembrane protease, serine 2 (TMPRSS2; orange), and histamine receptors H1 (blue), H2 (green), and H4 (yellow).
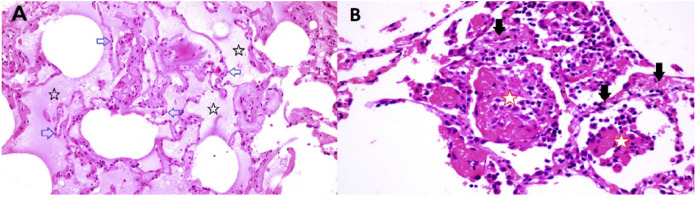
The limited tissue pathology available from early COVID–19 cases seems to support both viral infection as well as histamine effects in the lung. In a singular study of early COVID–19, Sufang Tian et al 59 describe the viral lung pathology of early COVID–19 in tissue resected for cancer. Their photomicrographs show two different patterns of disease. As shown in Figure 9 panel B, some samples of this lung tissue demonstrate the usual mononuclear inflammatory pattern of interstitial pneumonitis and fibrinous exudate that one would associate with a viral infection. It is striking that no neutrophils or eosinophils are observed in the inflammatory infiltrate. One explanation is that H2 activation of neutrophils inhibits neutrophil effector functions including O2− release 42,43, platelet-activating-factor induced chemotaxis 44 and leukotriene biosynthesis 45. Eosinophil functions are also inhibited by H2 activation; histamine binding diminishes eosinophil peroxidase release 46 and, at high concentrations, inhibits eosinophil chemotaxis 47,48.
Figure 9.
Lung pathology of early COVID-19 84 year old female undergoing right middle lobe (RML) resection for adenocarcinoma. On Day 6 of hospitalization a CT scan showed a ground glass opacity (GGO) in the RML in addition to the tumor mass. Lobectomy was performed on Day 12. On Day 13 (Day 1 post-operation), CT scan showed bilateral bibasilar GGO. On Day 16, she developed typical COVID-19 symptoms with cough, dyspnea and chest tightness. Capillary O2 saturation ranged from 77–88%. Death ensued on Day 29. SARS-CoV-2 was confirmed by nasal swab. (Tian et al 59) Panel A (RML). There is extensive pulmonary edema consistent with a transudate (open black stars). Alveolar septae appear normal and there is no inflammation (open blue arrows). Features are not suggestive of an infection. Panel B (RML) There is fibrinous exudate in the alveolar spaces (open red stars). Alveolar septae show edema and a mononuclear infiltrate (solid black arrows). No neutrophils are identified. There is no significant diffuse alveolar damage of ARDS. Features are typical of an interstitial viral pneumonia.
The reports of Tian et al 59 and Zeng et al 60 also include images in which there is interstitial and alveolar edema while the alveolar septae retain normal architecture (Figure 9 panel A). This is not a pattern typically observed in viral infection, as there is no inflammation, and the fluid appears to be a transudate. It is consistent with dysregulation of the fluid barrier due to the effect of histamine or other mast cell products on endothelial cells, pericytes or Type II pneumocytes. Increased endothelial permeability due to histamine is driven by H1 receptor activation, and so if any potential famotidine treatment effect on these cells occurs it would most likely be indirect by inhibition of mast cell degranulation. Forskolin activates the enzyme adenylyl cyclase and increases intracellular levels of cAMP, and can be used to inhibit the release of histamine from human basophils and mast cells 61. Histamine may act as an autocrine regulator of mast cell cytokine and TNF-a release in a PGE2-dependent fashion. Based on in vitro studies, this autocrine feedback appears to be mediated by H2 and H3 (88). Endothelial cells are also susceptible to infection by SARS-CoV–2. Mast cell degranulation-related pulmonary edema could correlate with the early phase silent hypoxia and the high compliance non-ARDS ventilation pattern associated with shortness of breath 62. The image in Figure 9 panel B does not permit evaluation for microvascular thrombi.
These findings are supported in a separate autopsy case report of a patient dying 5 days after onset of COVID–19 symptoms. In this case, photomicrographs also show a non-inflammatory transudative-type edema 63. In both of these studies, the observed non-inflammatory edema in early-stage COVID–19 pulmonary disease is consistent with histamine release by mast cells.
Mast cell degranulation correlates with the COVID–19 natural history that progresses through functionally and clinically different early and later phases. Most SARS-CoV–2 infections follow the typical early phase pattern of any lower respiratory virus, in which a majority of patients have asymptomatic or minimal disease, while a minority go on to later phase acute respiratory distress syndrome (ARDS). Within this spectrum typical of any severe viral disease, COVID–19 has a number of distinctive features. In the out-patient setting, early COVID–19 is usually indistinguishable from other “influenza-like illnesses”, presenting with various non-specific symptoms ranging from sore throat, headache and diarrhea to fever, cough, and myalgias. In these first few days however, COVID–2 may also be associated with anosmia, a unique feature 64. It is towards the end of the first week of symptoms that COVID–19 patients develop shortness of breath (SOB). This follows cough and fever by several days, a feature not typical of other viruses 65. On physical examination of COVID–19 patients with SOB, the oxygen saturation drops dramatically on exertion. CT scan will usually show bilateral bibasilar ground glass opacifications consistent with pulmonary edema. Nasopharyngeal swabs test positive for SARS-CoV–19. This SOB correlates with a distinctive clinical phenotype of hypoxia with near normal compliance (i.e. >50 mLcmH2O). Some authors attribute this to a loss of pulmonary vasoconstriction, one cause of which could be histamine effect on the H2 receptors of pericytes and/or vascular smooth muscle. H1-related edema and microthrombosis of lung vessels could also be causes. These are the patients that PEEP ventilation will not help, as there are no recruitable alveoli. These patients are helped by lying prone 66. It is at this stage that the patient is at greatest risk to progress onto the serious complications of later disease, especially ARDS with its 60–80% mortality if ventilation is required. Patients may also present with additional neurological symptoms and complications including ischemic stroke 67–69. Cardiac complications of later COVID–19 include myocarditis, acute myocardial infarction, heart failure, dysrhythmias, and venous thromboembolic events 70,71.
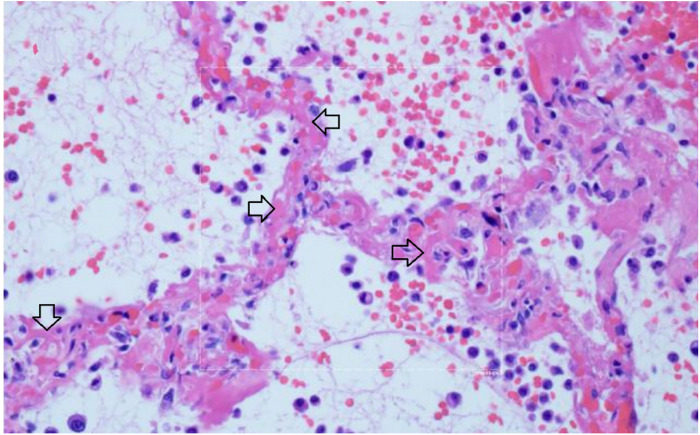
Multiple studies have demonstrated a hypercoagulable state in COVID–19 patients requiring hospitalization. Results from a recent large autopsy study suggests that there is also a novel lung-centric coagulopathy that manifests as a small vessel microthrombosis. Based on this study, there are indications that over 50% of patients dying of COVID–19 have pulmonary microthrombosis 72. This thrombosis is not only in arterial vessels, but also can be found in alveolar capillaries in the absence of inflammation and ARDS, as seen in Figure 10 73.
Figure 10.
Microthrombosis in the pulmonary microvasculature in COVID-19 at autopsy 73 There is widening of the alveolar septae by extensive fibrinous occlusion of capillaries (open black arrows). There is alveolar space edema with red blood cell extravasation. Septae show a mild mononuclear infiltrate. Alveolar edema shows neutrophils in proportion to the blood.
There is widening of the alveolar septae by extensive fibrinous occlusion of capillaries (open black arrows). There is alveolar space edema with red blood cell extravasation. Septae show a mild mononuclear infiltrate. Alveolar edema shows neutrophils in proportion to the blood.
Capillary wall disruption accompanied by fibrin deposition and red cell extravasation, with neutrophils in the septa and within the alveolar spaces. (Hematoxylin and eosin, 1000x). For further discussion of microvascular coagulation associated with COVID–19, see 73.
Because small microthrombi are difficult to identify on CT scan even with iodinated contrast 74, pre-mortem diagnosis is difficult. Laboratory coagulation tests have typically shown normal or mildly prolonged Prothrombin time (PT) and activated partial thromboplastin time (aPTT), normal to increased or slightly decreased platelet counts, elevated fibrinogen levels and very elevated D-dimers 75. Although referred to by some authors as a DIC-like state, this pulmonary microthrombosis does not appear as a typical coagulation factor consumptive bleeding condition typical of overt DIC, but instead more closely resembles hypercoagulable thrombosis. This coagulopathy appears to be a core pathophysiology of COVID–19 as rising D-dimer levels, correlate with a poor prognosis, as do rising levels of IL–6 and CRP. IL–6 levels have been correlated to fibrinogen levels in one study, possibly through the acute phase reactant response 76. The pathogenesis of microthrombosis of the lung in COVID–19 is not known. There are multiple working hypotheses concerning this finding currently being assessed 77. Damage to the vascular endothelial glycocalyx can be caused by TNF-α, ischemia and bacterial lipopolysaccharide. As well, activated mast cells release cytokines, proteases, histamine, and heparinase, which degrade the glycocalyx 78 and may thereby contribute to microthrombosis. Disruption of the glycocalyx exposes endothelial cell adhesion molecules, triggering further inflammation, rolling and adhesion of white blood cells and platelets 79. Glycocalyx components measured in serum positively correlate with increased mortality in septic patients 80. Other causes of hypercoagulability include direct damage to ACE2 positive endothelial cells by viral invasion or secondary damage from the inflammatory response to the infection. Mast cells release heparin which activates the contact system, producing plasmin and bradykinin. Plasmin activation could account for the singular rise in D-dimer levels. Activation of platelets also seems likely as part of the thrombo-inflammatory response but their precise role in thrombus formation remains to be elucidated 81. A more complete understanding awaits further study.
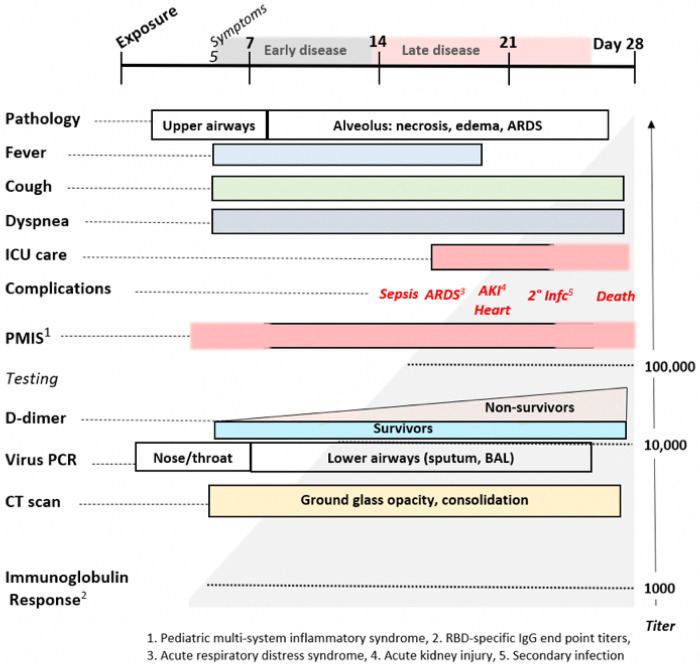
In addition to the usual features of a viral infection, early COVID–19 often presents with anosmia, ageusia, skin rashes including pruritis and urticaria, neuropsychiatric symptoms (including altered dream states), and silent hypoxia. These symptoms are all consistent with histamine signaling. Anosmia, ageusia, and other symptoms relating to cachexia are often reported in both COVID–19 and mast cell degranulation syndrome, and the potential role of histamine signaling in driving the pathophysiology of cachexia has been reviewed 82,83. As summarized in Figure 11, the distinctive later findings of abnormal coagulation, involvement of other organ systems and ARDS occur in the second week after the appearance of symptoms. This is coincidental with a rising immunoglobulin response to SARS-CoV–2 antigens. For a subset of patients, disease progress may suddenly worsen at days 7–10, and this correlates with the onset of SARS-CoV–2 spike protein neutralizing antibody titers 84. In this study, it was shown that IgG starts to rise within 4 days post-symptoms, inconsistent with a first antigenic exposure 84. Rapid onset of specific neutralizing antibody responses beginning less than seven days after exposure to SARS-CoV–2 implies a recall rather than primary B cell response, and therefore the response is being driven by a pre-existing memory cell population. These memory cells may have been educated by prior exposure to another coronavirus (e.g. circulating alphanumeric coronaviruses), raising concerns that this second phase of COVID–19 disease progression could share an immunologic basis with Dengue hemorrhagic fever 85. Antibodies produced from this early rapid humoral response may drive further mast cell degranulation. During this phase rising D-dimer levels correlate with poor prognosis, as do measured levels of CRP and IL–6.
Figure 11.
The Natural History of COVID-19. Modified from Oudkerk et al 74.
Current reviews seek to explain COVID–19 clinical and pathologic findings based on standard models of antiviral innate and adaptive immune responses which do not consider the potential role of mast cell activation and degranulation. Reviews emphasize the inflammatory cell response cascade associated with monocytes, macrophages 86, and adaptive T and B cell helper and effector responses 87. These types of immune responses are also invoked to explain the novel microvascular pulmonary intravascular coagulopathy associated with COVID–19 88.
We propose an alternative paradigm; SARS-CoV–2 infection-induced mast cell activation could account for some of the core pathologic cascade and much of the unusual symptomatology associated with COVID–19 89. Many of the unique clinical symptoms observed during the early phase of COVID–19 are consistent with known effects of histamine release. Histamine may act as an autocrine regulator of mast cell cytokine and TNF-a release in a PGE2-dependent fashion and based on in vitro studies the autocrine feedback appears to be mediated by H2 and H3 90. This model is consistent with the histopathologic findings seen at surgery, autopsies, and is supported by clinical pharmacologic findings suggesting potential benefits of histamine H2 receptor blockade using famotidine. This model is also supported by the significant overlap in the clinical signs and symptoms of the initial phase of COVID–19 disease and those of mast cell activation syndrome (MCAS) 91–94 as well similarities to Dengue hemorrhagic fever and shock syndrome (including T cell depletion) during the later phase of COVID–19 85,95,96. The cardiac events, stroke, and related outcomes associated with COVID–19 also appear consistent with the Kounis syndrome 97–99.
If COVID–19 is partially driven by dysfunctional mast cell degranulation, then a variety of medical interventions employing marketed drugs useful for treating mast cell-related disorders may help to reduce death and disease associated with SARS-CoV–2 infection. Examples include drugs with mast cell stabilizing activity, other histamine antagonists (for example H1 and H4 types), leukotriene antagonists and leukotriene receptor antagonists 100, anti-inflammatory agents such as those developed for inflammatory bowel diseases, and mast cell activation inhibitors 101. If such repurposed drugs are used in combination with pharmaceuticals that directly inhibit SARS-CoV–2 infection or replication, it may be possible to rapidly develop potent, safe and effective outpatient treatments for preventing or treating COVID–19 until such time as a safe and effective SARS-CoV–2 vaccine becomes available.
Online Methods
Analysis of the mechanism of action of famotidine
Famotidine was originally selected by the authors for advancement as a potential repurposed drug candidate therapeutic for COVID–19 based on molecular docking data to PLpro. Based on this analysis the FDA granted an IND waiver for the subsequent double blinded randomized clinical trial currently in progress (ClinicalTrials.gov Identifier: NCT04370262). Briefly, a ranked list of licensed compounds with predicted binding activity in the PLpro catalytic site was computationally generated, and the PLpro catalytic site binding pose of each of the top compounds was examined and ranked by a team of pharmaceutical chemists. Package inserts or product monographs for the licensed compounds which generated high computational binding scores and passed inspection were then reviewed and used to rank compounds based on adverse events, FDA warnings, drug interactions on-target mechanisms, pharmacokinetic and absorption, metabolism, excretion and toxicity (ADMET), protein binding and available therapeutic window considerations. Famotidine (“PEPCID®”), a histamine H2 antagonist widely available in tablet form over-the-counter, as well as in solution form for intravenous administration, was repeatedly computationally ranked as among the most promising of the compounds tested and was associated with the most favorable pharmacokinetic and safety profile. Other compounds considered at this stage of docking model optimization included camostat mesylate and isoquercitrin. Camostat was rejected for further development due to US regulatory status, lack of suitability for outpatient use, and metabolism issues. Isoquercitrin was rejected due to poor oral bioavailability and lack of prior FDA authorization as a therapeutic (including lack of drug master file). A series of analogs of famotidine were generated using PubChem, and many of these scored even higher as potential candidates.
Recognizing that computational docking predictions are typically associated with about a 20% success rate, we applied the method of multiple working hypotheses 77 to assess the mechanism of action of famotidine as a potential treatment for COVID–19. Hypotheses tested included 1) direct binding and action as an inhibitor of SARS-CoV–2 PLpro; 2) action as a direct acting inhibitor of SARS-CoV–2 infection or replication; 3) off-target binding of a non-histamine H2 G-coupled protein receptor 4) histamine H2 receptor inhibition.
1). Famotidine does not appear to directly bind and act as an inhibitor of SARS-CoV–2 PLpro
Production of recombinant SARS-CoV–2 Plpro
An expression plasmid containing the sequence for (His)6-TEVsite-SARS-CoV–2 PLpro (nsp3 from Wuhan-Hu–1 isolate, polyprotein 1ab 1564–1878) was obtained commercially from ATUM. The plasmid was transformed into E. coli BL21(DE3) pLysS. The expression and purification protocols were adapted from 102.
Production of recombinant ISG15
The expression plasmid for proISG15 (2–165) was a gift from David Komander (Addgene plasmid # 110762 ;http://n2t.net/addgene:110762; RRID:Addgene_110762) 103. Expression and purification protocols were adapted from 103. A size exclusion chromatography step on a Superdex 75 column (GE Healthcare) was added as a final step.
PLpro activity assays
Cleavage of ISG15 by SARS-CoV–2 PLpro was tested by incubating 4 nM of PLpro in 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, 2 mM DTT, 0.1 mg.mL–1 BSA, with 10 μM of ISG15 in a final volume of 20 μL for 1 h at room temperature. Control was incubated without enzyme. Samples were subjected to SDS-PAGE.
2). Famotidine does not appear to directly inhibit SARS-CoV–2 infection or replication in Vero cells
Viral Growth and Cytotoxicity Assays in the Presence of Inhibitors
2,000 Vero E6 cells were seeded into 96-well plates in DMEM (10% FBS) and incubated for 24 h at 37C, 5% CO2. Two hours before infection, the medium was replaced with 100ul of DMEM (2% FBS) containing the compound of interest at concentrations 50% greater than those indicated, including a DMSO control. Plates were then transferred into the BSL3 facility and 100 PFU (MOI 0.025) was added in 50ul of DMEM (2% FBS), bringing the final compound concentration to those indicated. Plates were then incubated for 48 h at 37C. After infection, supernatants were removed and cells were fixed with 4% formaldehyde for 24 hours prior to being removed from the BSL3 facility. The cells were then immunostained for the viral NP protein with a DAPI counterstain. Infected cells (488nM) and total cells (DAPI) were quantified using the Celigo (Nexcelcom) imaging cytometer. Percent infection was quantified as ((Infected cells/Total cells) - Background) *100 and the DMSO control was then set to 100% infection for analysis. The IC50 and IC90 for each experiment were determined using the Prism (GraphPad Software) software. For select inhibitors, infected supernatants were assayed for infectious viral titer using the TCID50 method. Cytotoxicity was also performed using the MTT assay (Roche), according to the manufacturer’s instructions. Cytotoxicity was performed in uninfected VeroE6 cells with same compound dilutions and concurrent with viral replication assay.
TCID50 Assay
Infectious supernatants were collected at 48h post infection and frozen at −80 °C until later use. Infectious titers were quantified by limiting dilution titration on Vero E6 cells. Briefly, Vero E6 cells were seeded in 96-well plates at 20,000 cells/well. The next day, SARS-CoV2-containing supernatant was applied at serial 10-fold dilutions ranging from 10−1 to 10−6 and, after 5 days, viral CPE was detected by staining cell monolayers with crystal violet. Median tissue culture infectious doses (TCID50)/mL were calculated using the method of Reed and Muench.
3). Does famotidine bind and interact with the Sigma 1 or 2 receptors?
Sigma–1 and sigma–2 competition binding assays
Sigma–1 receptor [3H](+)-pentazocine competition curves testing the binding of Famotidine, Cimetidine, and PB–28 (as positive control), were performed with Expi293 cells (Thermo Fisher) overexpressing the human sigma–1 receptor. Membranes were incubated in a 100 μL reaction buffered with 50 mM Tris (pH 8.0), with 10 nM [3H](+)-pentazocine, 0.1% BSA, and seven concentrations (ranging from 10 μM to 0.1 nM) of the competing cold ligand. Reactions were incubated for 2 hours at 37 °C and then were terminated by filtration through a glass fiber filter using a Brandel cell harvester. Glass fiber filters were soaked in 0.3% polyethylenimine for at least 30 min at room temperature before harvesting. All reactions were performed in triplicate using a 96-well block. After the membranes were transferred to the filters and washed, the filters were soaked in 5 mL Cytoscint scintillation fluid overnight, and radioactivity was measured using a Beckman Coulter LS 6500 scintillation counter. Data were analyzed using GraphPad Prism software. Ki values were computed by directly fitting the data and using the experimentally determined probe Kd to calculate a Ki value, using the GraphPad Prism software. This process implicitly uses a Cheng–Prusoff correction, so no secondary correction was applied.
Sigma–2 competition curves were performed in a similar manner, using Expi293 cells overexpressing the human sigma–2 (TMEM97) and using [3H] DTG as the radioactive probe.
4). Does famotidine act via an off-target activity involving a G-coupled protein receptor (GPCR) other than the histamine H2 GPCR?
GPCRome screening was carried out according to published procedure (PMID25895059) with minor modifications. In brief, HTLA cells were subcultured into poly-L-lysine coated clear bottom 384-well white plates at a density of 6000 cells/well in 40 ul of DMEM supplemented with 1% dialyzed FBS for overnight, transfected with 20 ng/well Tango constructs for 24 hrs, received drug stimulation (10 uM final) for another 24hrs. Medium and drug solutions were removed and Bright-Glo Reagents (Promega) were added for luminescence counting. For concentration response assays, HTLA cells were transfected with Tango constructs for 24hrs, plated in poly-L-Lysine coated clear bottom 384-well white plates at a density of 10,000 cells/well in 40 ul of DMEM supplemented with 1% dialyzed FBS for 6 hrs before receiving drugs for overnight stimulation. Plates were then counted as above.
5). Famotidine could act by blocking histamine receptor(s)
The known on-target activity of famotidine considered the known primary mechanism of action is as an antagonist of the histamine H2 receptor. This hypothesis was originally rejected due to unverified reports that clinical researchers in PRC (Wuhan) had observed that famotidine use was associated with protection from COVID–19 mortality, while the histamine H2R antagonist cimetidine was not. Positing that this difference in clinical effectiveness for the two different H2R antagonists may reflect absorption, pharmacokinetic and pharmacodistribution differences between famotidine and cimetidine, steady state concentrations were calculated for both drugs when administered at standard oral doses as well as the elevated doses of famotidine which are being prescribed off-label for outpatient clinical use to treat COVID–19 or are being used in the ongoing inpatient clinical trial (NCT04370262), and these were compared to the published H2R IC50 for each drug.
GloSensor cAMP assays: cAMP production was determined in transiently transfected HEK293T cells (PMID 31019306, recent MT1 paper for the method) with minor modifications. In brief, HEK293 T cells were co-transfected with H2 and GloSensor cAMP reporter DNA (Promega) overnight and plated in poly-l-lysine coated clear bottom 384-well white plate at a density of 15,000 cells/well in 40 ul of DMEM supplemented with 1% dialyzed FBS for 6 hrs. Medium was removed and cells were loaded with 3 mM luciferin in drug buffer (20 ul/well, 1x HBSS, 20 mM HEPES, pH 7.40) for 30 min. Test compounds were prepared in the same drug buffer supplemented with 1 mg/ml BSA and added (20 ul/well at 2x) to cells. Luminescence was counted after 20 min and results were analyzed in Prism 8.4.
Radioligand binding assays with human histamine receptors. Radioligand binding assays with human H1, H2, H3, and H4 receptors were conducted according to the NIMH PDSP assay protocol (https://pdsp.unc.edu/pdspweb/?site=assays) and published procedures (PMID 23235874, Besnard et al., 2012, Automated drug design paper).
Famotidine and cimetidine pharmacokinetic analyses
Steady-state values of famotidine and cimetidine at various doses were calculated as follows using standard pharmacokinetic calculations. Bioavailability, and clearance values were obtained from data reported by Lin (1991) for tablet and capsule dosing of famotidine and cimetidine; for 60 mg famotidine bid, kinetic data were obtained from a report by Yeh et al. (1987). For intravenous administration of famotidine (IV 120 mg q8 hours), kinetic values were obtained from the famotidine New Drug Applications (NDAs 19–510/S–029, 20–249/S–012).
Calculations:
Area under the curve (AUC) determinations were made as follows: AUC (mg h/L) = (F x Dose)/Cl; where F = bioavailability, Cl = clearance. Steady (Css) state values were calculated as follows: Css (μg/L) = AUC/T; where T = dosing interval (h). Css levels were converted to μM using the molecular weights of famotidine (337.45) and cimetidine (252.34), respectively.
Acknowledgments
The authors acknowledge the Department of Defense (DoD), Defense Threat Reduction Agency (DTRA), and the Joint Science and Technology Office (JSTO) of the Chemical and Biological Defense Program (CBDP) for funding under the Discovery of Medical countermeasures Against Novel Entities (DOMANE) initiative. This work has also benefitted from advice, guidance, information and comments provided by Drs. Revell Phillips, Howard Haimes, David Hone, and Roland Seifert. We appreciate the valuable input provided by Dr. Frank Weichold of the Office of Regulatory Science and Innovation (ORSI), Office of the Chief Scientist (OCS), Office of the Commissioner (OC), FDA/HHS, and Dr. Lawrence Callahanof the Office of Health Informatics, Office of the Chief Scientist (OCS), Office of the Commissioner (OC), FDA/HHS. We also thank Dr. Anton Yuryev from Elsevier for his assistance in literature mining and reconstruction of the histamine signaling model. D. R. gratefully acknowledges the support of the MIT SuperCloud team.
DISTRIBUTION STATEMENT A. Approved for public release. Distribution is unlimited.
This material is based upon work supported under Air Force Contract No. FA8702–15-D–0001. Any opinions, findings, conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the U.S. Air Force. Funding was also provided by grants from the Defense Advanced Research Projects Agency HR0011–19–2–0020 (to A. G-S.); by CRIP (Center for Research for Influenza Pathogenesis), a NIAID supported Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C) and by supplements to NIAID grant U19AI135972 and DoD grant W81XWH–20–1–0270 to A. G.-S.
Contributor Information
Robert W. Malone, RW Malone MD LLC, Madison, VA
Philip Tisdall, Medical School Companion LLC, Marco Island, FL.
Philip Fremont-Smith, MIT Lincoln Laboratory, Lexington, MA.
Yongfeng Liu, Department of Pharmacology, University of North Carolina, Chapel Hill, Chapel Hill, NC.
Xi-Ping Huang, Department of Pharmacology, University of North Carolina, Chapel Hill, Chapel Hill, NC.
Kris M. White, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY
Lisa Miorin, Global Health and Emerging Pathogens Institute, Icahn School of Medicine at Mount Sinai, New York, NY.
Elena Moreno Del Olmo, Global Health and Emerging Pathogens Institute, Icahn School of Medicine at Mount Sinai, New York, NY.
Assaf Alon, Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA.
Elise Delaforge, McGill University, Department of Chemistry, Montreal, Quebec, Canada.
Christopher D. Hennecker, McGill University, Department of Chemistry, Montreal, Quebec, Canada
Guanyu Wang, McGill University, Department of Chemistry, Montreal, Quebec, Canada.
Joshua Pottel, Molecular Forecaster Inc, Montreal, Quebec, Canada.
Nora Smith, MIT Lincoln Laboratory, Lexington, MA.
Julie M. Hall, Frank H. Netter MD School of Medicine – Quinnipiac University, Hamden, CT
Gideon Shapiro, Pharmorx LLC, Gainesville, FL.
Anthony Mittermaier, McGill University, Department of Chemistry, Montreal, Quebec, Canada.
Andrew C. Kruse, Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA
Adolfo García-Sastre, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY,Global Health and Emerging Pathogens Institute, Icahn School of Medicine at Mount Sinai, New York, NY, Department of Medicine, Division of Infectious Diseases, Icahn School of Medicine at Mount Sinai, New York, NY, The Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY.
Bryan L. Roth, Department of Pharmacology, University of North Carolina, Chapel Hill, Chapel Hill, NC
Jill Glasspool-Malone, RW Malone MD LLC, Madison, VA.
Darrell O. Ricke, MIT Lincoln Laboratory, Lexington, MA
References
- 1.Zhu N., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z. & McGoogan J. M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID–19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA (2020). [DOI] [PubMed] [Google Scholar]
- 3.Wu D., Wu T., Liu Q. & Yang Z. The SARS-CoV–2 outbreak: What we know. Int J Infect Dis 94, 44–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L., et al. SARS-CoV–2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med 382, 1177–1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danis K., et al. Cluster of coronavirus disease 2019 (Covid–19) in the French Alps, 2020. Clin Infect Dis (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai C. C., et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV–2): Facts and myths. J Microbiol Immunol Infect (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan X., et al. Asymptomatic cases in a family cluster with SARS-CoV–2 infection. Lancet Infect Dis 20, 410–411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa N. W., Brooks J. T. & Sobel J. Evidence Supporting Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 While Presymptomatic or Asymptomatic. Emerg Infect Dis 26(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ki M. & Task Force for -nCo, V. Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Korea. Epidemiol Health 42, e2020007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H., et al. Radiological findings from 81 patients with COVID–19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 20, 425–434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He G., et al. The clinical feature of silent infections of novel coronavirus infection (COVID–19) in Wenzhou. J Med Virol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian S., et al. Characteristics of COVID–19 infection in Beijing. J Infect 80, 401–406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z., et al. Clinical characteristics of 24 asymptomatic infections with COVID–19 screened among close contacts in Nanjing, China. Sci China Life Sci 63, 706–711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizumoto K., Kagaya K., Zarebski A. & Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID–19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill 25(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giacomelli A., et al. Self-reported olfactory and taste disorders in SARS-CoV–2 patients: a cross-sectional study. Clin Infect Dis (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lechien J. R., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID–19): a multicenter European study. Eur Arch Otorhinolaryngol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC. Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID–19). Vol. 2020 (2020). [Google Scholar]
- 20.Freedberg D. E., et al. Famotidine Use is Associated with Improved Clinical Outcomes in Hospitalized COVID–19 Patients: A Retrospective Cohort Study. medRxiv, 2020.2005.2001.20086694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borrell B. New York clinical trial quietly tests heartburn remedy against coronavirus. Vol. 2020 (Science Magazine, 2020). [Google Scholar]
- 22.Daczkowski C. M., et al. Structural Insights into the Interaction of Coronavirus Papain-Like Proteases and Interferon-Stimulated Gene Product 15 from Different Species. J Mol Biol 429, 1661–1683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baez-Santos Y. M., St John S. E. & Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res 115, 21–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mielech A.M., Chen Y., Mesecar A.D. & Baker S. C. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Research 194, 184–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y. S., et al. Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry 44, 10349–10359 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Wu C., et al. Analysis of therapeutic targets for SARS-CoV–2 and discovery of potential drugs by computational methods. Acta Pharm Sin B (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anson B. J., et al. Broad-spectrum inhibition of coronavirus main and papain-like proteases by HCV drugs,. Research Square (2020). [Google Scholar]
- 28.Gordon D. E., et al. A SARS-CoV–2 protein interaction map reveals targets for drug repurposing. Nature (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt H. R., et al. Crystal structure of the human σ1 receptor. Nature 532, 527–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alon A., et al. Identification of the gene that codes for the sigma2 receptor. Proc Natl Acad Sci U S A 114, 7160–7165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertaccini G., Coruzzi G., Poli E. & Adami M. Pharmacology of the novel H2 antagonist famotidine: in vitro studies. Agents Actions 19, 180–187 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Alonso N., et al. Physiological implications of biased signaling at histamine H2 receptors. Front Pharmacol 6, 45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irannejad R. & von Zastrow M. GPCR signaling along the endocytic pathway. Curr Opin Cell Biol 27, 109–116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jean-Charles P. Y., Kaur S. & Shenoy S. K. G Protein-Coupled Receptor Signaling Through beta-Arrestin-Dependent Mechanisms. J Cardiovasc Pharmacol 70, 142–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao M., De Clercq E. & Li G. Clinical significance of chemokine receptor antagonists. Expert Opin Drug Metab Toxicol 16, 11–30 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Fantuzzi L., Tagliamonte M., Gauzzi M. C. & Lopalco L. Dual CCR5/CCR2 targeting: opportunities for the cure of complex disorders. Cell Mol Life Sci 76, 4869–4886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y. Q., et al. The Role of CXCR3 in Neurological Diseases. Curr Neuropharmacol 17, 142–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nasi A. & Chiodi F. Mechanisms regulating expansion of CD8+ T cells during HIV–1 infection. J Intern Med 283, 257–267 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Lin J. H., et al. Effects of antacids and food on absorption of famotidine. Br J Clin Pharmacol 24, 551–553 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh K. C., et al. Single-dose pharmacokinetics and bioavailability of famotidine in man. Results of multicenter collaborative studies. Biopharm Drug Dispos 8, 549–560 (1987). [DOI] [PubMed] [Google Scholar]
- 41.Lin J. H. Pharmacokinetic and pharmacodynamic properties of histamine H2-receptor antagonists. Relationship between intrinsic potency and effective plasma concentrations. Clin Pharmacokinet 20, 218–236 (1991). [DOI] [PubMed] [Google Scholar]
- 42.Burde R., Seifert R., Buschauer A. & Schultz G. Histamine inhibits activation of human neutrophils and HL–60 leukemic cells via H2-receptors. Naunyn Schmiedebergs Arch Pharmacol 340, 671–678 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Gespach C. & Abita J. P. Human polymorphonuclear neutrophils. Pharmacological characterization of histamine receptors mediating the elevation of cyclic AMP. Mol Pharmacol 21, 78–85 (1982). [PubMed] [Google Scholar]
- 44.Rabier M., et al. Inhibition by histamine of platelet-activating-factor-induced neutrophil chemotaxis in bronchial asthma. Int Arch Allergy Appl Immunol 89, 314–317 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Flamand N., Plante H., Picard S., Laviolette M. & Borgeat P. Histamine-induced inhibition of leukotriene biosynthesis in human neutrophils: involvement of the H2 receptor and cAMP. Br J Pharmacol 141, 552–561 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ezeamuzie C. I. & Philips E. Histamine H(2) receptors mediate the inhibitory effect of histamine on human eosinophil degranulation. Br J Pharmacol 131, 482–488 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadee A. A., Anderson R. & Sher R. In vitro effects of histamine on eosinophil migration. Int Arch Allergy Appl Immunol 63, 322–329 (1980). [DOI] [PubMed] [Google Scholar]
- 48.Clark R. A., Gallin J. I. & Kaplan A. P. The selective eosinophil chemotactic activity of histamine. J Exp Med 142, 1462–1476 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reher T. M., Brunskole I., Neumann D. & Seifert R. Evidence for ligand-specific conformations of the histamine H(2)-receptor in human eosinophils and neutrophils. Biochem Pharmacol 84, 1174–1185 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Lippert U., et al. Human skin mast cells express H2 and H4, but not H3 receptors. J Invest Dermatol 123, 116–123 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Somogyi A. & Gugler R. Clinical pharmacokinetics of cimetidine. Clin Pharmacokinet 8, 463–495 (1983). [DOI] [PubMed] [Google Scholar]
- 52.Echizen H. & Ishizaki T. Clinical pharmacokinetics of famotidine. Clin Pharmacokinet 21, 178–194 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Administration, U. F.a.D. PEPCID® (famotidine) tablets, for oral use. Vol. 2020 (1986). [Google Scholar]
- 54.Luo T., et al. Histamine H2 receptor activation exacerbates myocardial ischemia/reperfusion injury by disturbing mitochondrial and endothelial function. Basic Res Cardiol 108, 342 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Di Lorenzo A., Fernandez-Hernando C., Cirino G. & Sessa W. C. Akt1 is critical for acute inflammation and histamine-mediated vascular leakage. Proc Natl Acad Sci U S A 106, 14552–14557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panula P., et al. International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors. Pharmacol Rev 67, 601–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krystel-Whittemore M., Dileepan K. N. & Wood J. G. Mast Cell: A Multi-Functional Master Cell. Front Immunol 6, 620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du Y., Guo M., Whitsett J. A. & Xu Y. ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax 70, 1092–1094 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian S., et al. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID–19) Pneumonia in Two Patients With Lung Cancer. J Thorac Oncol 15, 700–704 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng Z., et al. Pulmonary Pathology of Early Phase COVID–19 Pneumonia in a Patient with a Benign Lung Lesion. Histopathology n/a(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marone G., Columbo M., Triggiani M., Vigorita S. & Formisano S. Forskolin inhibits the release of histamine from human basophils and mast cells. Agents Actions 18, 96–99 (1986). [DOI] [PubMed] [Google Scholar]
- 62.Couzin-Frankel J. The mystery of the pandemic’s ‘happy hypoxia’. Science 368, 455–456 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Schweitzer W., et al. Implications for forensic death investigations from first Swiss post-mortem CT in a case of non-hospital treatment with COVID–19. Forensic Imaging 21, 200378 (2020). [Google Scholar]
- 64.Eliezer M., et al. Sudden and Complete Olfactory Loss Function as a Possible Symptom of COVID–19. JAMA Otolaryngology–Head & Neck Surgery (2020). [DOI] [PubMed] [Google Scholar]
- 65.Cohen P. A., Hall L., Johns J. N. & Rapoport A. B. The Early Natural History of SARS-CoV–2 Infection: Clinical Observations From an Urban, Ambulatory COVID–19 Clinic. Mayo Clinic Proceedings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gattinoni L., Chiumello D. & Rossi S. COVID–19 pneumonia: ARDS or not? Critical Care 24, 154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mao L., et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filatov A., Sharma P., Hindi F. & Espinosa P. S. Neurological Complications of Coronavirus Disease (COVID–19): Encephalopathy. Cureus 12, e7352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qureshi A. I., et al. Management of acute ischemic stroke in patients with COVID–19 infection: Report of an international panel. Int J Stroke, 1747493020923234 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Long B., Brady W. J., Koyfman A. & Gottlieb M. Cardiovascular complications in COVID–19. Am J Emerg Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahmud E., et al. Management of Acute Myocardial Infarction During the COVID–19 Pandemic. J Am Coll Cardiol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carsana L., et al. Pulmonary post-mortem findings in a large series of COVID–19 cases from Northern Italy. medRxiv, 2020.2004.2019.20054262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magro C., et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID–19 infection: a report of five cases. Transl Res (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oudkerk M., et al. Diagnosis, Prevention, and Treatment of Thromboembolic Complications in COVID–19: Report of the National Institute for Public Health of the Netherlands. Radiology, 201629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panigada M., et al. Hypercoagulability of COVID–19 patients in Intensive Care Unit. A Report of Thromboelastography Findings and other Parameters of Hemostasis. J Thromb Haemost (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ranucci M., et al. The procoagulant pattern of patients with COVID–19 acute respiratory distress syndrome. J Thromb Haemost (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.The Method of Multiple Working Hypotheses. Science 15, 92–96 (1890). [DOI] [PubMed] [Google Scholar]
- 78.Alphonsus C. S. & Rodseth R. N. The endothelial glycocalyx: a review of the vascular barrier. Anaesthesia 69, 777–784 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Becker B. F., Chappell D., Bruegger D., Annecke T. & Jacob M. Therapeutic strategies targeting the endothelial glycocalyx: acute deficits, but great potential. Cardiovasc Res 87, 300–310 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Nelson A., Berkestedt I., Schmidtchen A., Ljunggren L. & Bodelsson M. Increased levels of glycosaminoglycans during septic shock: relation to mortality and the antibacterial actions of plasma. Shock 30, 623–627 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Jackson S. P., Darbousset R. & Schoenwaelder S. M. Thromboinflammation: challenges of therapeutically targeting coagulation and other host defense mechanisms. Blood 133, 906–918 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Zwickl H., Zwickl-Traxler E. & Pecherstorfer M. Is Neuronal Histamine Signaling Involved in Cancer Cachexia? Implications and Perspectives. Front Oncol 9, 1409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Becker S., et al. Olfactory dysfunction in seasonal and perennial allergic rhinitis. Acta Otolaryngol 132, 763–768 (2012). [DOI] [PubMed] [Google Scholar]
- 84.Suthar M. S., et al. Rapid generation of neutralizing antibody responses in COVID–19 patients. medRxiv, 2020.2005.2003.20084442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mongkolsapaya J., et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med 9, 921–927 (2003). [DOI] [PubMed] [Google Scholar]
- 86.Merad M. & Martin J. C. Pathological inflammation in patients with COVID–19: a key role for monocytes and macrophages. Nature Reviews Immunology (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vabret N., et al. Immunology of COVID–19: current state of the science. Immunity (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGonagle D., O’Donnell J. S., Sharif K., Emery P. & Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID–19 pneumonia. The Lancet Rheumatology (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kritas S., et al. Mast cells contribute to coronavirus-induced inflammation: New anti-inflammatory strategy. Journal of biological regulators and homeostatic agents 34(2019). [DOI] [PubMed] [Google Scholar]
- 90.Bissonnette E. Y. Histamine inhibits tumor necrosis factor alpha release by mast cells through H2 and H3 receptors. Am J Respir Cell Mol Biol 14, 620–626 (1996). [DOI] [PubMed] [Google Scholar]
- 91.Afrin L. B., et al. Diagnosis of mast cell activation syndrome: a global “consensus–2”. Diagnosis (Berl) (2020). [DOI] [PubMed] [Google Scholar]
- 92.Weiler C. R. Mast Cell Activation Syndrome: Tools for Diagnosis and Differential Diagnosis. J Allergy Clin Immunol Pract 8, 498–506 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Weinstock L. B., Pace L. A., Rezaie A., Afrin L. B. & Molderings G. J. Mast Cell Activation Syndrome: A Primer for the Gastroenterologist. Dig Dis Sci (2020). [DOI] [PubMed] [Google Scholar]
- 94.Butterfield J. H. Survey of Mast Cell Mediator Levels from Patients Presenting with Symptoms of Mast Cell Activation. Int Arch Allergy Immunol 181, 43–50 (2020). [DOI] [PubMed] [Google Scholar]
- 95.Guzman M. G. & Harris E. Dengue. Lancet 385, 453–465 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Redoni M., et al. Dengue: Status of current and under-development vaccines. Rev Med Virol, e2101 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Kounis N. G., et al. Anaphylaxis-induced atrial fibrillation and anesthesia: Pathophysiologic and therapeutic considerations. Ann Card Anaesth 23, 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kounis N. G. Kounis syndrome: an update on epidemiology, pathogenesis, diagnosis and therapeutic management. Clin Chem Lab Med 54, 1545–1559 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Gonzalez-de-Olano D., et al. Mast cell activation disorders presenting with cerebral vasospasm-related symptoms: a “Kounis-like” syndrome? Int J Cardiol 150, 210–211 (2011). [DOI] [PubMed] [Google Scholar]
- 100.Fidan C. & Aydogdu A. As a potential treatment of COVID–19: Montelukast. Med Hypotheses 142, 109828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Theoharides T. C., Tsilioni I. & Ren H. Recent advances in our understanding of mast cell activation - or should it be mast cell mediator disorders? Expert Rev Clin Immunol 15, 639–656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lindner H. A., et al. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79, 15199–15208 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Swatek K. N., et al. Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies. Proc Natl Acad Sci U S A 115, 2371–2376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]