Abstract
The COVID-19 pandemic caused by the novel SARS-CoV-2 is more contagious than other coronaviruses and has higher rates of mortality than influenza. As no vaccine or drugs are currently approved to specifically treat COVID-19, identification of effective therapeutics is crucial to treat the afflicted and limit disease spread. We deployed a bioinformatics workflow to identify candidate drugs for the treatment of COVID-19. Using an “omics” repository, the Library of Integrated Network-Based Cellular Signatures (LINCS), we simultaneously probed transcriptomic signatures of putative COVID-19 drugs and signatures of coronavirus-infected cell lines to identify therapeutics with concordant signatures and discordant signatures, respectively. Our findings include three FDA approved drugs that have established antiviral activity, including protein kinase inhibitors, providing a promising new category of candidates for COVID-19 interventions.
Keywords: transcriptomic signatures, putative COVID-19 drugs, coronavirus-infected cell lines
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the first global pandemic in a decade, coronavirus disease 2019 (COVID-19)1. Initial reports of a novel SARS-like acute respiratory syndrome emerged in late 2019 from Wuhan, China2. Since then, COVID-19 has spread to over 150 countries and all continents except Antarctica3,4. At the time of writing, over one million people have been infected, over 60,000 deaths have been attributed to this outbreak4, and millions of additional infections are projected to occur globally in upcoming months3,4. COVID-19 is less infectious than SARS-CoV1 but more lethal than the common flu1 with an estimated mortality rate of 3.4%2. The incubation period, on average, is 5.2 days; in severe cases, the median time course from disease onset to death is 14 days5. While fever, cough, fatigue, and myalgias6–10 are common, mild presentations of COVID-19, the disease can fatally evolve into a severe pneumonia, complicated by acute respiratory distress syndrome, hypoxemic respiratory failure, and cytokine storm secondary to prolonged infection8. In addition to the significant medical burden imposed by this outbreak, it is estimated that the global economic cost of COVID-19 will be over $1 trillion in 202011. The emotional toll on individuals will be incalculable, with prolonged quarantine policies restricting personal freedom and social contacts.
Current treatment is supportive and is focused on managing disease complications and secondary symptoms12–14. Drugs indicated for other infectious diseases, such as antiviral and antiparasitic therapies, have been used for COVID-19 patients, but there is a paucity of evidence supporting their efficacy15.
There is now an immediate need to identify therapeutic compounds that can be rapidly repurposed for COVID-19 treatment. Recent efforts to address this pressing public health concern include a comprehensive network-based approach to identify 16 candidate drugs (and drug combinations) that may be repurposed for the treatment of this disease16. To further expand this area of novel research, in the present study we employ a bioinformatics approach with the goal of datamining an extensive drug signature resource to distill a list of drug therapies that may prove fruitful in the search for COVID-19 therapies.
To accomplish this, we apply a signature-based connectivity analysis17–19 utilizing the extensive chemical perturbagen “omics” datasets deposited in the Library of Integrated Network-based Signatures (LINCS) database17,20,21. LINCS is a repository for systematically generated gene signatures based on the L1000 assay22. These gene signatures reflect cellular perturbations in response to pharmacological treatments; LINCS contains datasets for over 40,000 small molecules (drugs) in various cell lines. Different small molecules that produce signatures composed of highly similar patterns of gene expression changes, or “concordant” signatures, reflect shared connections between small molecules.
Here, we apply a two-pronged approach to identify novel compounds for the treatment of COVID-19. First, we identify pharmacologic therapies that are effective in the treatment of pathogens in the coronavirus family, like SARS and Middle East Respiratory Syndrome (MERS), as well as other viral illnesses23–27. We then generate gene signatures for these targets in iLINCS (http://ilincs.com) and highlight connected small molecule signatures to identify which of these candidate drugs are highly concordant with current therapies. Simultaneously, we generate gene signatures from coronavirus infected human cell line transcriptomic datasets. We analyze these data in iLINCS to directly match disease signatures with discordant small molecule signatures, thereby identifying drugs that “reverse” the disease signature. Finally, we compile a list of drugs from these two approaches to identify high-yield candidate drugs that may have therapeutic utility in the treatment of COVID-19.
Results
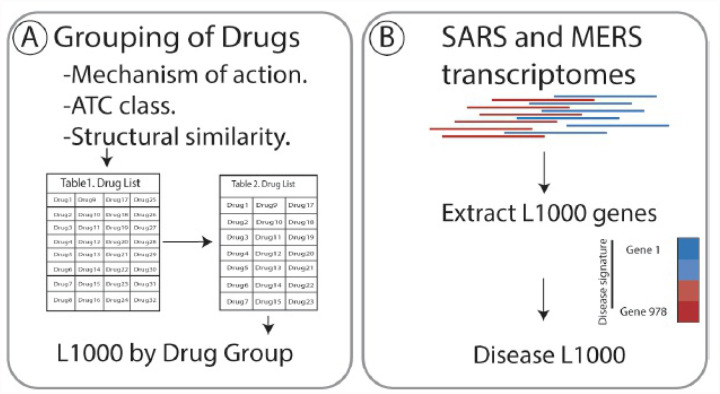
Applying the workflow outlined in Figure 1, we identified nine drugs, with known efficacy in treating coronavirus family pathogens, for which there are gene signatures in iLINCS (Table 1 and extended information in Table S1). These drugs were clustered into five groupings according to their mechanism of action, Anatomical Therapeutic Chemical (ATC) classification and/or structural similarity (Table 1).
Figure 1.
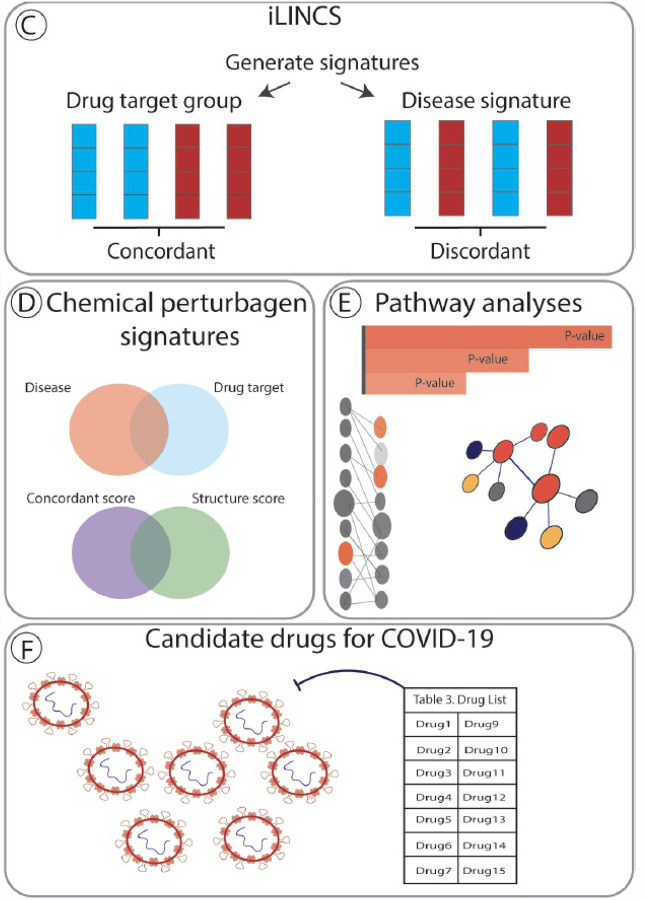
Overview of the workflow to identify candidate repurposable drugs to combat COVID-19. A) Drugs that are currently in use to treat coronavirus and putative COVID-19 treatments were clustered based on mechanism of action, ATC class and structural similarity. B) Gene expression data of the 978 genes that comprise the Library of Integrated Network-Based Cellular Signature (iLINCS) L1000 genes were extracted from severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome- related coronavirus (MERS) (GSE56192) transcriptomic datasets. C) Consensus iLINCS gene signatures were generated for drug groupings and disease. D) Connectivity analysis was conducted and a list of chemical perturbagens that are concordant (≥ 0.321 concordance) to the drug target grouping signatures or discordant (≤ − 0.321 discordance) to the disease signatures was generated. Chemical perturbagens are curated to identify top candidate drugs. E) Biological pathways of drug target groupings, disease signatures and candidate drugs was conducted. F) Fourteen perturbagens with concordance scores ≥ 0.8 in MCF7 and HA1E cell lines were shortlisted as repurposable candidate drugs.
Table 1. Drug target groupings.
Drug targets with iLINCS signatures that are in use or under investigation for the treatment of COVID-19 were grouped together if they met a least two of the three following criteria: Canonical Mechanism of Action, referenced from the database DrugBank (https://www.drugbank.ca/); Anatomic Therapeutic Chemical classification, referenced from https://www.whocc.no/atc ddd index/: structural similarity as determined by intracluster Tanimoto coefficient. F = Febratininb; R = Ruxolinitib; B = Bariticinib
| Drug Cluster | Drug | Canonical Mechanism of Action | Anatomical Therapeutic Chemical First Level |
Tanimoto Coefficient |
|---|---|---|---|---|
| 1 | Chloroquine Hydroxychloroquine |
Toll-like receptor antagonists | Antiparasitic Products, Insecticides and Repellants | 0.84 |
| 2 | Lopinavir Ritonavir |
Protease inhibitors | Anti-infective for Systemic Use | 0.72 |
| 3 | Fedratinib Ruxolinitib Bariticinib |
JAK inhibitors | Antineoplastic and Immunomodulating Agents |
F − R: 0.25 F − B: 0.20 B − R: 0.72 |
| 4 | Azithromycin | Macrolide antibiotic | Anti-Infective for Systemic Use | 1 |
| 5 | Losartan | Angiotensin receptor blocker antagonist | Cardiovascular System | 1 |
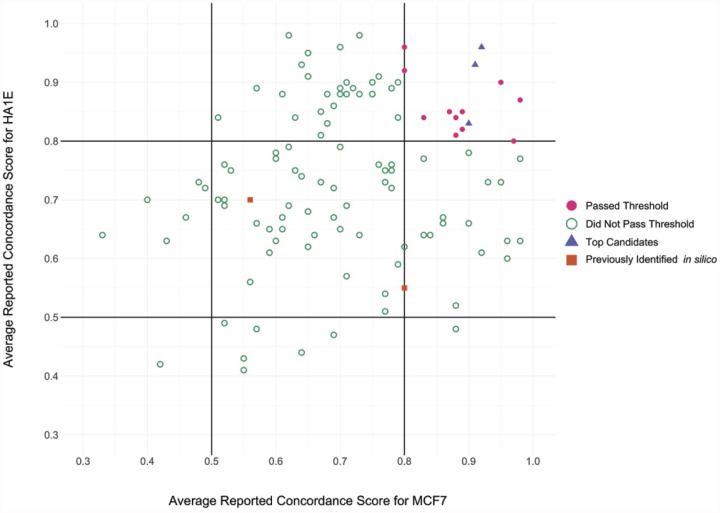
Simultaneously, we extracted differential gene expression data on the 978 genes that comprise the L1000 from a publically available SARS (GSE56192) transcriptomic dataset. Gene signatures composed of genes changed LFC ≥ 0.5 and ≤ −0.5 were generated for the disease signature (Table S2). In iLINCS, we conducted connectivity analysis to identify chemical perturbagens that are highly concordant to the drug target groupings (≥ 0.321) or highly discordant to the disease signatures (≤ −0.321), established minimum iLINCS concordance score cutoffs 22,28. This resulted in identification of 112 chemical perturbagens common to two cell lines, MCF7 and HA1E (Figure 2). Fourteen chemical perturbagens were identified at concordance scores ≥ 0.8 in both cell lines and were considered “candidate” drugs for the treatment of COVID-19 (Table 2). The Tanimoto scores for the candidate drugs and the original 9 drug targets were generated, showing structural similarity between drugs currently in use for the treatment of coronavirus family pathogens and our newly identified candidate drugs (Figure S1).
Figure 2.
Scatter Plot of average reported concordance scores for candidate drugs in the MCF7 cell line and HA1E cell line. A total of 112 drugs with concordance scores > 0.321 were common between both cell lines (open circles). Fourteen drugs were identified with concordance scores ≥ 0.8 in both cell lines (closed circles). Drugs above this threshold are considered “candidate” drugs. Top candidate drugs, those approved for use in humans and with demonstrated antiviral activity in vitro, are also identified (triangles). Drugs identified by other in silico drug screening studies that were also found in our analysis are identified (squares).
Table 2.
Candidate repurposable drugs for the treatment of COVID-19.
| Drug | DrugBank I.D. | MCF7 Concordance | HA1E Concordance | MOA | ATC Drug Class | Indication |
|---|---|---|---|---|---|---|
| Drugs with reported antiviral activity. | ||||||
| Alvocidib | DB03496 | 0.92 | 0.96 | CDK inhibitor | Unclassified | Antineoplastic |
| Genistein | DB01645 | 0.91 | 0.93 | Tyrosine kinase and topoisomerase-II inhibitor | Unclassified | Antineoplastic; nthelmintic |
| Ivermectin | DB00602 | 0.90 | 0.83 | Chloride channel agonist | Anthelmintic | Anthelmintic |
| Drugs with no reported antiviral activity. | ||||||
| Clinically Relevant Drugs | ||||||
| Idebenone | DB09081 | 0.88 | 0.81 | Electron donor to mitochondrial electron transport chain | Psychoanaleptic | Leber’s Hereditary Optic Neuropathy |
| Penicillin V | DB01053 | 0.97 | 0.80 | Binds penicillin binding proteins, inhibits bacterial cell wall synt hesis | Antibacterials for Systemic Use | Antibiotic |
| Idelalisib | DB09054 | 0.88 | 0.84 | Phosphoinositid e 3-kinase inhibitor | Antineoplastic Agent |
| GSK-1059615 | DB11962 | 0.83 | 0.84 | Phosphoinositid e 3-kinase, mTOR inhibitor 46 | Unclassified |
| AT-9283 | DB05169 | 0.89 | 0.82 | kinase inhibitor | Unclassified |
| Experimental Drugs | |||||
| GSK-3 Inhibitor IX | -- | 0.87 | 0.85 | Inhibitor of glycogen synthase kinase-3a/b 47 | Unclassified |
| AC1MJ3VH | -- | 0.89 | 0.82 | RNA synthesis and topoisomerase inhibitor 48 | Unclassified |
| CCT-10B | -- | 0.95 | 0.90 | #Kinase binding, protein kinase binding | Unclassified |
| CHEMBL2136735 | -- | 0.80 | 0.96 | #Oxidoreductas e activity, cadherin binding | Unclassified |
| Broad-Sai-595 | -- | 0.82 | 0.92 | #NAPH binding, cadherin binding | Unclassified |
| BRD-K54343811 | -- | 0.98 | 0.87 | #Protein tyrosine kinase binding, phosphotyrosin e residue binding | Unclassified |
Table 2 Candidate drugs are separated into two cohorts: drugs with reported antiviral activity and those with no reported antiviral activity. These drugs are then subcategorized as clinically relevant (used in human subjects) or experimental (used in research but not yet approved for humans). Concordance scores in the MCF7 and HA1E cell lines represent the average concordance scores between the identified candidate drug and at least 2 of the drug target clusters in that cell line. All 14 candidate drugs have an average concordance ≥ 0.80 in both cell lines. DrugBank I.D. and Mechanism of Action (MOA) is referenced from Drug Bank (https://www.drugbank.ca/). Drug Class is referenced from the second level of the Anatomical Therapeutic Chemical (ATC) classification (https://www.whocc.no/atc_ddd_index/). For “Experimental drugs”, the MOA was cited from iLINCS, or alternatively, the perturbagens top two significant Gene Ontology (GO) Molecular Functions are listed under MOA and denoted with the superscript “#”. “^” Indicated and approved for use only by the European Medicines Agency. HSV, Herpes Simplex Virus; HIV, Human Immunodeficiency Virus; LV, Lassa Virus; ASFV, African Swine Fever Virus; YF, Yellow Fever; DF, Dengue Fever; CV, Chikungunya virus; SARS, Severe Acute Respiratory Syndrome virus; MERS, Middle East Respiratory Syndrome virus.
Unsupervised clustering of L1000 disease gene signatures demonstrates significant differences in patterns of gene expression induced by SARS, MERS and influenza (Figure S2). Influenza is utilized as a control dataset as it represents a non-coronavirus pathogen that also causes respiratory illness. As expected, unsupervised clustering of L1000 gene signatures shows discordance between disease signatures and drug target grouping signatures, which are comprised of drugs utilized to treat SARS and to a lesser extent, MERS (Figure S3–4).
Biological pathways analysis demonstrated a range of perturbations, including those in similar biological pathways (immune system pathways and cell cycle processes) induced by both drug target groupings and disease signatures (Figure S5–6). Unsupervised clustering of L1000 gene signatures also shows discordance between SARS (and MERS) disease signatures and the identified candidate drug signatures (Figure S7–10), including those with antiviral properties.
Biological pathway analysis indicates that the identified candidate drugs also induce changes including in similar biological pathways as disease signatures (immune system and cell cycle related pathways) (Figure S11–12).
Thus, we distilled a list of drugs, derived from pharmacological and disease perturbation signatures that may have therapeutic utility in the treatment of COVID-19. The candidate drugs identified are:
Tyrosine Kinase Inhibitors.
Tyrosine kinases are essential for viral RNA synthesis, viral ribonucleoprotein nuclear export, and virion release. Inhibitors that target this protein class may, therefore, demonstrate activity against viruses49.
Genistein is an isoflavonoid derived from soy-products that has been implicated as an antiparasitic50 and antineoplastic agent51 in humans. Several clinical trials of Genistein are ongoing to treat prostate, breast, and bladder cancers52,53. Genistein also has potent antiviral activity in a number of in vitro models. It has efficacy against RNA viruses from different families, such as filoviridae (Ebola virus), feoviridae (rotavirus), and arenaviridae (Lassavirus, Pichindé virus)34,35,38,39; retroviruses like HIV-136,37 and DNA viruses. Both in vivo and in vitro studies of Genistein demonstrated activity against African swine fever viruses40 and Epstein- Barr virus54. While the pre-clinical evidence is promising, Genistein has yet to be explored as an antiviral therapy in humans.
AT9283 is a broad protein kinase inhibitor55,56. Canonically, it acts as a receptor and nonreceptor tyrosine kinase inhibitor but also effectively inhibits serine/threonine kinases, Aurora A/B kinases, Janus kinases (JAK) 2/3, and ABL kinases57. In a clinical setting, AT9283 has been predominantly studied as an antineoplastic for hematologic malignances and several trials are underway58–60. AT9283 has not been explored directly as an antiviral, although the ABL kinase inhibitor, Imatinib, is efficacious in preventing coronavirus (SARS-CoV and MERS-CoV) viral fusion with endosomes, effectively halting viral activity61. Given its role as a broad-spectrum kinase inhibitor, researching the antiviral properties of AT9283 may prove fruitful.
Serine Threonine Kinase Inhibitors.
Alvocidib (also known as flavopiridol) is a pan-specific cyclin-dependent kinases (CDK) inhibitor that inhibits CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, and CDK962. Alvocidib is under clinical investigation as an antineoplastic for both solid tumors and hematologic malignancies63. Like other CDK kinase inhibitors, Alvocidib has been implicated as a broad antiviral against several DNA virus families64. Alvocidib has been studied as an inhibitor of transcriptional activation and elongation in the infectious lifecycle of DNA viruses (HSV-1, HSV-2) and retroviruses (HIV)29–32 and also suppresses replication of Influenza A33. This suggests that Alvocidib is a strong candidate drug for repurposing.
The antiviral activity of the following serine/threonine kinase candidate drugs have yet to be studied directly and further research is required to determine their potential as repurposable antiviral therapies. However, these candidates are members of drug families with demonstrated antiviral or antimicrobial properties that could be exploited for the treatment of COVID-19.
GSK-1059615 is a reversible, ATP-competitive, thiazolidinedione inhibitor of phosphoinositide 3-kinase (PI3K) and has been studied as an antineoplastic for solid tumors65. Though the antiviral activity of GSK-1059615 has yet to be determined, the thiazolidinedione drug family has a broad range of antibacterial and antiparasitic activity66–68. Idelalisib is a phosphoinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling inhibitor69–71. In vitro experiments show that the downstream target pathways of kinase inhibitors like Idelalisib, extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) and PI3K/AKT/mammalian target of rapamycin (mTOR) signaling responses, are specifically modulated during infection with coronavirus pathogen MERS72. Thus, inhibiting this virulent signaling pathway using kinase inhibitors is potentially an efficacious therapeutic strategy.
Antioxidants and Antimicrobials.
Ivermectin is a promising drug candidate for COVID-19. It is a well-characterized anthelmintic for Onchocerca volvulus, the causative parasitic roundworm of “river blindness” or the “black sight”73–75. Canonically, Ivermectin works as a chloride-channel agonist76. Ivermectin has a well-established safety profile in humans and has been under investigation for repurposing in various parasitic diseases, cancers, neurological disorders, and viral infections75–77. The efficacy of Ivermectin in the treatment of RNA virus families, such as flaviviridae41,43,78,79 and togaviridae43 has been demonstrated in vitro. Of note, Ivermectin has been used as an adjunct therapy in patients with HIV and concomitant parasitic infections80,81.
In vitro, Ivermectin has shown efficacy in targeting HIV41 alone; by inhibiting HIV-1 integrase Ivermectin potentially prevents the viral genetic material from entering the host genome42. Efficacy for Ivermectin as an antiviral in humans warrants further investigation, especially as the global COVID-19 pandemic ensues.
Idebenone is a synthetic derivative of ubiquinone, also known as Coenzyme Q1082,83. This drug acts to increase the production of ATP by enhancing oxidative phosphorylation. As a general antioxidant, Idebenone may prevent lipid peroxidation, reduce membrane oxidative stress, and scavenge free radicals84. Idebenone has been used for a number of human neurodegenerative disorders, and its safety has been validated85,86. Antioxidants such as Idebenone have been hypothesized to mitigate the deleterious effects of a “reactive-oxygen species burst”45 from viruses with a pulmonary and respiratory predilections (Influenza and SARS)44. However, further work is required to determine the utility of antioxidants like Idebenone as adjunct treatments for COVID-19.
Penicillin V is a beta-lactam antibiotic and is indicated primarily for treating gram-positive bacterial infections like Treponema pallidum, the causative organism of syphilis87. Penicillin binds to a family of bacterial transpeptidases, termed penicillin binding proteins, which effectively inhibit cross-linking of peptidoglycan in the bacterial cell wall87. To the best of our knowledge, penicillin V does not have an experimental or clinical indication in the treatment of viruses. In addition, antibiotics such as penicillin should be employed judiciously, given their well-characterized ability to induce hypersensitivity reactions88,89. However, like azithromycin, utilizing penicillin as an adjunct therapy may be advantageous to empirically cover bacterial infections co-morbid with viral infections.
Candidate Drugs with Unknown Utility.
Finally, five drugs (AC1MJ3VH, Broad-Sai-595, CHEMBL2136735, COT-10B and GSK Inhibitor IX) with limited or no known biological function were identified. These drugs do not have identifiers in Drug Bank. They were not considered to be of utility as candidate drugs to repurpose for treatment of COVID-19.
Discussion
The COVID-19 outbreak is an escalating public health concern that requires a swift and comprehensive response. Research is progressing rapidly. There are currently over 200 clinical trials exploring a range of pharmacological and non-pharmacological options for the treatment of COVID-19, but as of yet, there are no vaccines or therapies approved specifically for this disease. In recent months, a number of in silico studies addressing this gap in our knowledge have identified putative repurposable drugs for treating COVID-1916,90–93. Many of these studies exploit the recent finding that SARS-CoV-2 may enter the cell by binding to angiotensin converting enzyme 2 (ACE2)94 and utilize a combination of structural and biomedical data to identify drug candidates. One such study used artificial intelligence algorithms (BenevolentAI) to identify the JAK inhibitor Baricitinib90. Baricitinib may reduce the ability of the virus to enter cells and is currently in clinical use as a treatment for rheumatoid arthritis. To advance therapeutic discovery and the identification of candidate drugs for COVID-19, we employ an alternative, signature-based bioinformatic approach. In this study, we data mine the extensive LINCS database, which acts as a repository of “L1000” gene signatures generated by treating various cell lines with over 20,000 small molecules. The L1000 genes are a reduced representation of the transcriptome, a method by which a select group of genes account for ~82% of the information content of the transcriptome95. The approach involved feature selection/reduction techniques applied to 12,063 gene expression samples profiled on microarrays from GEO96. Benchmarking of the L1000 assay versus RNAseq yielded a cross-platform correlation of 0.8495, suggesting the L1000 assay represents an efficient alternative to RNA-Seq.
Utilizing this resource, our two-pronged connectivity analysis approach identified candidate drugs that are 1) highly concordant to current drugs employed to treat coronavirus family pathogens and 2) highly discordant to SARS disease gene signatures. As there are currently no publically available datasets for human (or other) tissues infected with SARS-CoV-2 virus, we generated a disease signature from an RNAseq dataset of human lung cells infected with SARS; SARS and SARS-CoV-2 are highly homologous, sharing envelope and nucleocapsid protein sequence identities of 96% and 89.6%, respectively16.
The main class of drugs identified from our analyses are kinase inhibitors. Kinase inhibitors are high-yield targets, with new small molecule kinase inhibitors being developed every year and over two dozen small molecule kinase inhibitors already approved for human use97. Their potential as antiviral treatments has also been explored in recent years91,98–100. Viruses depend on host cell protein kinases for every step of their life cycle, including viral entry into the cell, cell cycle processes and cellular stress response101. Thus, targeting these protein kinases using kinase inhibitors will disrupt the virus’s ability to hijack cellular processes. As many host protein kinases are broadly required by different viruses, kinase inhibitors are excellent candidates for broad-spectrum antiviral therapies99. Our study identified two kinase inhibitors, Genistein and Alvocidib. These drugs have demonstrated, extensive, antiviral properties in vitro30,40 and both are approved for use in humans, making them strong candidate therapies for the treatment of COVID-19.
Kinase inhibitors AT9283, GSK1059615 and Idelalisib were identified by our analyses and are approved for use in humans, but their antiviral properties have yet to be tested directly. These drugs inhibit a range of different protein kinases including PI3K/mTOR, GSK3β and ABL kinases. In vitro work demonstrates that inhibitors targeting these kinases are highly effective at treating coronavirus pathogens SARS and/or MERS61,72,102,103, in addition to other viral pathogens104–108. However, further work is required to determine the specific antiviral profiles of these candidate drugs, before they can be considered for repurposing.
Interestingly, we, and another group, identified Ruxolitinib, a JAK kinase inhibitor that is utilized as an antineoplastic91. Although Ruxolitinib was not identified above the same stringent threshold as our top candidate drugs, its discovery by two different in silico approaches suggests that this kinase inhibitor may have utility as a candidate drug for treating COVID-19. Importantly, kinase inhibitor Barcitinib is now undergoing clinical trial in response to the COVID-19 pandemic (NCT04320277). Thus, kinase inhibitors represent an expanding, if underexplored, avenue of research for the treatment of viral illnesses, including coronaviruses. Repurposing kinase inhibitors, many of which are already approved for use in humans, is a time-and cost-effective method to identify new therapeutics in a rapidly evolving situation such as the one posed by the current outbreak of COVID-19.
Another strong candidate drug finding from our analyses is the antimicrobial Ivermectin. Best known for its efficacy in treating “river blindness,” Ivermectin is already in widespread use as an anthelmintic and was recently shown to have potent antiviral characteristics in vitro, in experiments targeting flaviviruses and RNA viruses76. Indeed, Ivermectin is used as an adjuvant therapy in patients with HIV and a concomitant parasitic infections80,81. In phase II/III clinical trial, Ivermectin safely and significantly reduced Dengue viral NS1 protein serum levels 109,110.
Although no clinical benefit was seen, the dosing regimen may yet be optimized based on pharmacokinetic and pharmacodynamic data109. In addition, Ivermectin shows potent antiviral activity for SARS-CoV-2, reducing viral RNA approximately 5000-fold in infected cells at 48 hours, in a recent in vitro study111. Taken together, Ivermectin’s established safety profile and efficacy in reducing SARS-CoV-2 viral material in vitro suggest this drug is worthy of further consideration as a treatment for COVID-19.
A number of our other candidate drug findings may also prove useful as adjuvant treatments for the secondary effects associated with viral infection, including the antioxidant Idebenone, and the antibiotic Penicillin112. During the 2009 H1N1 influenza pandemic, bacterial infection was a suspected, underreported contributor to patient hospitalization and death113.
Interestingly, the antibiotic quinacrine was also identified in a recent network-based COVID-19 drug screen16 and another β-lactam antibody ceftriaxone, is in clinical trial for (adjunct) treatment of COVID-19 (NCT02735707). The immunosuppressant Sirolimus was identified in our study, albeit at a less stringent threshold, as well as in another in silico drug screen16, as a candidate repurposable drug for treating COVID19. Immunosuppressants may address the symptoms resulting from overactivation of the immune system (“cytokine storm”) in response to COVID-19 infection114, and this class of drug is also in clinical trial as adjunct treatments (e.g. Thalidomide NCT04273529).
Limitations
The antimicrobial drugs that comprise our drug target groupings are limited to those that have gene signatures in iLINCS. We analyze gene signatures generated in two cell lines, MCF7 and HA1E, as data was available for all of our drug targets in these cell lines only. Our drug target clusters may induce different patterns of gene expression changes in other cell lines, resulting in different gene signatures, and potentially, the identification of different chemical perturbagens. We utilize a single, representative SARS transcriptomic disease signature to identify discordant chemical perturbagens, as no transcriptomic SARS-CoV-2 datasets are available at the time of writing. Although the two viral genomes are highly similar (envelope and nucleocapsid protein share sequence identities of 96% and 89.6%, respectively), we cannot exclude that chemical perturbagens that are discordant to SARS may not have the same level of discordance to SARS-CoV-2. Finally, as with other in silico screening approaches, the candidate drugs identified here are not necessarily ready for human use. Several of the candidate drugs are used in the treatment of human disease, but not viral infections or COVID-19 specifically, and require further investigation for dosage, efficacy etc. A subset of the drugs were explored in experimental, but not clinical settings, and are not currently approved for use in humans.
In summary, our approach has identified candidate repurposable drugs, from the >20,000 small molecules in the LINCS repository, that may be utilized to combat COVID-19. Several of the candidate drugs are 1) currently approved for use in humans, 2) have demonstrated antiviral efficacy in vitro and 3) a number were also identified in other in silico analyses. Our findings contribute to the relatively novel literature addressing the purported broad-spectrum antiviral efficacy of kinase inhibitors and may offer a novel avenue for investigation in the search for COVID-19 therapies. While there is evolving evidence for kinase inhibitors as antivirals, other antimicrobials could be repurposed as well. Finally, our bioinformatic workflow identified an antioxidant and two known antimicrobial drugs, which are concordant with current therapies being explored to combat SARS-CoV-2.
Methods
Selecting and grouping antimicrobials with known efficacy in treating coronavirus family pathogens
The workflow for this study is outlined in Figure 1. We conducted a PubMed search using search terms “coronavirus” or “COVID-19” and “antiviral” or “drug” or “therapy” and generated a list of compounds utilized to treat coronavirus family pathogens or identified as putative COVID-19 therapeutics. We identified seventeen drugs for potential analysis (Table S1). L1000 gene signature datasets were available for nine of the seventeen drugs (Table 1) using the integrative web platform iLINCS (http://ilincs.com). The iLINCS L1000 hub gene assay assesses genome-wide transcriptional changes following perturbation by one of more than 20,000 small molecules95. Eight drugs without signatures were excluded from further analysis. Gene signatures were generated for all remaining drugs. To standardize our analysis, we used the two cell lines that appeared most frequently in the signatures: MCF7 and HA1E. Hydroxychloroquine has an MCF7 signature only. For further standardization, where possible, signatures for a 24-hour time point and 10 uM concentration conditions were used.
Next, we grouped the nine drug targets based on canonical mechanism of action, the Anatomical Therapeutic Chemical (ATC) classification, and structural similarity. Drugs were grouped together if they were categorized by at least two of the three methods. The database DrugBank (https://www.drugbank.ca/) was used to group the drugs by their canonical mechanisms of actions.
Drug identification was only referenced from Drug Bank I.D. If no Drug Bank I.D. was available, this is indicated in Table 1 and Table S1. If there was no listed MOA from Drug Bank, then the MOA was appropriately cited, referenced from iLINCS, or was referenced from Gene Ontology (GO) Molecular Function 2018 accessed via Enrichr (http://amp.pharm.mssm.edu/Enrichr/enrich). Next, drugs were classified based on the ATC classification system (https://www.whocc.no/atc_ddd_index/). If a particular drug did not have an ATC classification, it was marked as “unclassified.” From DrugBank, we also collected the clinical indications, gene targets, and trade names. In addition, we probed the ATC Index (https://www.whocc.no/atc_ddd_index/) to identify the first- and second-level of drug classifications. The first-level classification was used to confirm drug grouping. Finally, to group drugs based on structural similarity, the structural data files (sdf) for the nine drugs under investigation were downloaded from DrugBank. The package ChemmineR was used to generate 1024-bit binary fingerprints for each compound. The Tanimoto coefficient between all pairs of fingerprints were then computed also using the R package “ChemmineR”. The Tanimoto coefficient, also known as the Jaccard similarity, represents the most popular measure for chemical similarity115 and is the ratio of the intersection of the two fingerprints divided by the union of the two fingerprints. The data were visualized using the “Corrplot” package. With a final list of drug clusters, the individual drug signatures within each grouping were collected and averaged across the L1000.
Generating iLINCS gene signatures
Using the iLINCS portal, we acquired the LINCS chemical perturbagen signatures (978 genes that comprise the L1000) for each drug candidate. Genes with a log fold change (LFC) value of ≥ 0.85 or ≤ −0.85, indicating differential gene expression induced by the drug target compared to a corresponding control cell line, were exported to Microsoft Excel. Gene lists were pooled and averaged such that a master list of differentially expressed genes was generated for each drug candidate family. For example, genes with a LFC ≥ 0.85 or ≤ −0.85 that appeared in both the hydroxychloroquine gene signature and the chloroquine gene signature were averaged to calculate mean values for each differentially expressed gene in drug target grouping 1.
Next, the upregulated genes (LFC ≥ 0.85) were clustered and the downregulated genes (≤ −0.85) were clustered. These clusters were uploaded as user generated signatures into iLINCS. Next, we identified connected chemical perturbagens, utilizing a concordance threshold score of ≥ 0.321, an established minimum concordance score cutoff 22,28, to identify chemical perturbagen signatures that are considered highly correlated with our drug target grouping signatures.
Gene signatures coronavirus-family induced disease datasets
At the time of writing, there are no publically availably Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2; COVID-19) transcriptomic datasets available. Thus, SARS (GSE56192) and Middle East Respiratory Syndrome (MERS; GSE56192) RNAseq datasets were identified in the Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/). SARS and MERS are coronavirus family members that cause severe respiratory illnesses116. We selected these datasets based on the viral type, transcriptomic platform, and infected tissue type. The SARS genome in particular is highly homologous to SARS-CoV-2, the virus responsible for COVID-19117. The envelope and nucleocapsid proteins of SARS-CoV-2 are two evolutionarily conserved regions, with sequence identities of 96% and 89.6%, respectively, to SARS16. The selected datasets focused on evaluating transcriptomic changes post-infection in lung cell lines, the primary tissue affected by the viral infection in humans. Each selected dataset represents a single time point from each study. We selected the most representative time points based on the top 50 differentially expressed genes from each dataset. The time point selected for these datasets is 24h.
We conducted differential gene expression analysis of these datasets comparing virus infected samples to corresponding control samples. RNAseq raw count data were analyzed using edgeR R package v.3.28.1 for differential gene expression118. As a quality control step, we require that a gene have a count of at least 10 in at least some libraries before it is considered to be expressed. Normalization was performed using the default method, trimmed mean of M-values (TMM). This step is performed by using the calcNormFactor function, which returns the DGEList argument with a set of calculated normalization factors, one for each sample, to eliminate composition biases between libraries. org.Hs.eg.db R package v.3.10.0 was used to complement the gene annotation. Following analysis of the disease transcriptomic datasets, the subset of genes that comprise the LINCS L1000 were extracted from the differentially expressed gene list for each dataset. When the L1000 genes were extracted from the SARS and MERS RNAseq datasets, 944 and 947 overlapping genes, respectively, were extracted. We used the existing gene raw counts from GEO and did not remap the genes. Thus, if some of the L1000 genes were not mapped we were unable to access their expression. In addition, we applied a gene raw count cutoff of 10 as a quality control step, which also reduced the number of L1000 genes found in the final differential expression gene lists.
The extracted L1000 genes were uploaded into iLINCS. Genes with LFC in expression within three thresholds, 0.26 LFC, 0.5 LFC and all L1000 genes, were identified with a custom R script for further processing. As described above, upregulated and down regulated disease gene signatures were generated for each disease dataset (within each threshold) and uploaded into iLINCS to identify connected perturbagens. For disease gene signatures, chemical perturbagen signatures that are highly discordant (discordance score ≤ −0.321), indicating these perturbagens may “reverse” the disease signature, were identified. Genes at LFC ≥ 0.5 and ≤ − 0.5 threshold were then carried forward for further analysis. Utilizing this gene threshold generated optimal SARS disease signatures to identify a large number of discordant chemical perturbagens. In addition, we identified a microarray dataset of human airway epithelium cells infected with influenza A of the H1N1 serotype (GSE47963, 36h time point). The microarray data were extracted using GEOquery R package v.2.54.1 119. Specifically, SOFT format files from GEO containing all of the information in the GEO records were parsed. The normalized gene expression values associated with SOFT files for selected samples filewere then analyzed using limma R package v.3.42.2, linear model for microarray120. As described above, the L1000 genes were extracted. Since this dataset was performed on the microarray platform, which limits the gene detection to the probes available in the arrays, we extracted 968 L1000 genes.
In addition to the MERS dataset, which represents a pathogenic coronavirus like SARS, the influenza dataset was used as a comparison dataset in unsupervised clustering and biological pathway analyses (described below). Influenza A represents a non-corona family virus that also causes respiratory disease121. A total of 906 overlapping L1000 genes were used in clustering analysis and heat map generation.
Identification of candidate chemical perturbagens (drugs) to treat COVID-19.
Candidate drugs were identified from the chemical perturbagen connectivity analysis using a custom script in R. The script downloaded the data from the iLINCS API and used the following criteria: Chemical perturbagens had a concordance score ≥ 0.321 compared to drug target grouping signatures or a discordance score ≤ −0.321 compared to disease signatures in cell lines MCF7 (6373/14355) or HA1E (4179/8141). If the same chemical perturbagen is identified multiple times, from different experimental conditions, replicate findings are removed so that only the highest concordance score (or lowest discordance score) for each chemical perturbagen remains. Chemical perturbagens were identified in the SARS disease signature analysis AND at least 2/5 drug target grouping signature analyses. This resulted in 506 chemical perturbagens from cell line MCF7 and 411 chemical perturbagens from cell line HA1E.
In total, 112 chemical perturbagens were common to both cell lines. Chemical perturbagens with concordance scores ≥ 0.8 in the MCF7 AND HA1E cell lines are considered “candidate drugs,” resulting in a final list of 14 candidate drugs. Heat maps of the L1000 gene data for each of the drug target groupings, candidate drugs and disease (SARS and MERS) datasets were generated as described above. In addition, the L1000 gene data for Furosemide (FUR), a drug which is not used to treat viral pathogens or related illnesses, and influenza, a non-coronavirus family pathogen associated with respiratory illness, were included in the heat maps. These datasets acts as controls, suggesting that the drug target groupings and candidate drugs, but not unrelated drugs, are discordant with coronavirus disease signatures specifically.
The sdf for the 9 drugs that comprise the drug target groupings were downloaded from DrugBank. The sdf files for the 14 candidate drugs were downloaded from PubChem and Chembl. ChemmineR was used to convert the sdf files to binary chemical fingerprints. The Tanimoto coefficients of the chemical fingerprints between the 9 drugs that comprise the drug target groupings and the 14 candidate drugs for all pairs were computed using the ChemmineR library. The Tanimoto coefficient, also known as the Jaccard similarity, represents the established measure for chemical similarity. The data were then visualized as a heat map using the “gplots” package.
GitHub repository access
R scripts utilized in data processing can be accessed at https://github.com/AliSajid/Covid19.
Comparing disease and drug target group signatures
Unsupervised clustering analysis was performed on log fold change values of disease and drug target signatures and heat map was generated using “pheatmap” package122 in R programming language.
Biological Pathway analysis
To generate biological pathways associated with the drug targets and disease datasets, the gene list was searched in Reactome v70 (https://reactome.org/). For drug target groupings 1–3, the gene signatures (LFC ≥ 0.85 and ≤ −0.85) were searched and pathways with a minimum of 10 entities (genes), p-value < 0.05, were generated. For drug target groupings 4 and 5, the gene signatures (LFC ≥ 0.85 and ≤ −0.85) were searched and pathways with a minimum of 3 entities (genes), p-value < 0.05, were generated, as fewer genes were input into pathway analysis at this threshold. For disease data, the gene signatures (LFC ≥ 0.5 and ≤ − 0.5) were searched and pathways with a minimum of 6 entities (genes), p-value < 0.05, were generated.
Pathways are organized under their top-level hierarchical identifier. Pathways are then identified at multiple sub-levels. To reduce the number of redundant pathways whilst still allowing for meaningful interpretation of biological pathways, some sub-level pathways are contracted.
Supplementary Material
Acknowledgements:
This work was supported by NIMH R01 MH107487 and MH121102.
Footnotes
Competing Interests: The authors have no conflicts to declare.
Data availability: The datasets analysed during the current study are available in the Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) (GSE56192; GSE47963) and the Library of Integrated Network-Based Cellular Signatures (LINCS) via iLINCS (http://ilincs.com).
Code Availability: R scripts can be accessed via: https://github.com/AliSajid/Covid19
Supplementary Files
This is a list of supplementary files associated with this preprint. Click to download. Supplementalmaterial.pdf
Contributor Information
Sinead M O’Donovan, University of Toledo.
Hunter Eby, University of Toledo.
Nicholas D Henkel, University of Toledo.
Justin Creeden, University of Toledo.
Ali Imami, University of Toledo.
Sophie Asah, University of Toledo.
Xiaolu Zhang, University of Toledo.
Xiaojun Wu, University of Toledo.
Rawan Alnafisah, University of Toledo.
R. Travis Taylor, University of Toledo.
James Reigle, University of Cincinnati College of Medicine.
Alexander Thorman, University of Cincinnati College of Medicine.
Behrouz Shamsaei, University of Cincinnati College of Medicine.
Jarek Meller, University of Cincinnati College of Medicine.
Robert E McCullumsmith, University of Toledo College of Medicine and Life Sciences.
References
- 1.Organization, W.H. Naming the coronavirus disease (COVID-19) and the virus that causes it. (2020).
- 2.Organization, W.H. WHO: Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). (2020).
- 3.Prevention, C.f.D.C.a. Coronavirus (COVID-19). (2020).
- 4.Organisation, W.H. Rolling updates on coronavirus disease (COVID-19). (2020).
- 5.Li Q., et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlos W.G., Dela Cruz C.S., Cao B., Pasnick S. & Jamil S. Novel Wuhan (2019-nCoV) Coronavirus. American journal of respiratory and critical care medicine 201, P7–P8 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Wang W., Tang J. & Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol 92, 441–447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren L.L., et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chinese medical journal (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan W.J., et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Development, U.N.C.o.T.a. Coronavirus (COVID-19): New, analysis and resources. (2020).
- 12.Wang D., et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.She J., et al. 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin Transl Med 9, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y.H., et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res 7, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Administration, F.a.D. Coronvvirus Disease 2019 (COVID-19). (2020).
- 16.Zhou Y., et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keenan A.B., et al. Connectivity Mapping: Methods and Applications. Annual Review of Biomedical Data Science 2, 69–92 (2019). [Google Scholar]
- 18.Stromback L., Jakoniene V., Tan H. & Lambrix P. Representing, storing and accessing molecular interaction data: a review of models and tools. Brief Bioinform 7, 331–338 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Lamb J., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Wang Z., et al. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat Commun 7, 12846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keenan A.B., et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst 6, 13–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilarczyk M. Connecting omics signatures of diseases, drugs, and mechanisms of actions with iLINCS. bioRxiv (2019). [Google Scholar]
- 23.Cao B., et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19 N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L., et al. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019:a retrospective cohort study. J Infect (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao T.T., Qian J.D., Zhu W.Y., Wang Y. & Wang G.Q. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J Med Virol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colson P., Rolain J.M., Lagier J.C., Brouqui P. & Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents, 105932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30, 269–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S. & Ahlquist P. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. The Annals of Applied Statistics 1, 85–106 (2007). [Google Scholar]
- 29.Diwan P., Lacasse J.J. & Schang L.M. Roscovitine inhibits activation of promoters in herpes simplex virus type 1 genomes independently of promoter-specific factors. Journal of virology 78, 9352–9365 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schang L.M., et al. Pharmacological cyclin-dependent kinase inhibitors inhibit replication of wild-type and drug-resistant strains of herpes simplex virus and human immunodeficiency virus type 1 by targeting cellular, not viral, proteins. Journal of virology 76, 7874–7882 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biglione S., et al. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 4, 47 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou M., et al. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol 78, 13522–13533 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S., Zhang J. & Ye X. [Protein kinase inhibitor flavopiridol inhibits the replication of influenza virus in vitro]. Wei Sheng Wu Xue Bao 52, 1137–1142 (2012). [PubMed] [Google Scholar]
- 34.Kolokoltsov A.A., et al. Inhibition of Lassa virus and Ebola virus infection in host cells treated with the kinase inhibitors genistein and tyrphostin. Arch Virol 157, 121–127 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Huang H., Liao D., Liang L., Song L. & Zhao W. Genistein inhibits rotavirus replication and upregulates AQP4 expression in rotavirus-infected Caco-2 cells. Arch Virol 160, 1421–1433 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Sauter D., et al. Genistein as antiviral drug against HIV ion channel. Planta medica 80, 682–687 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Guo J., et al. Genistein interferes with SDF-1- and HIV-mediated actin dynamics and inhibits HIV infection of resting CD4 T cells. Retrovirology 10, 62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vela E.M., Bowick G.C., Herzog N.K. & Aronson J.F. Genistein treatment of cells inhibits arenavirus infection. Antiviral research 77, 153–156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moraz M.L., et al. Cell entry of Lassa virus induces tyrosine phosphorylation of dystroglycan. Cell Microbiol 15, 689–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arabyan E., et al. Genistein inhibits African swine fever virus replication in vitro by disrupting viral DNA synthesis. Antiviral research 156, 128–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S.N.Y., et al. The broad spectrum antiviral ivermectin targets the host nuclear transport importin alpha/beta1 heterodimer. Antiviral research, 104760 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D. & Jans D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J 443, 851–856 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varghese F.S., et al. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral research 126, 117–124 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Yuan S. Drugs to cure avian influenza infection--multiple ways to prevent cell Cell Death Dis 4, e835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saso L., Korkina L. & Zarkovic N. Modulation of Oxidative Stress: Pharmaceutical and Pharmacological Aspects 2017. Oxid Med Cell Longev 2017, 4802824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Institute N.C. PI3K inhibitor GSK1059615. (NCI Drug Dictionary; ). [Google Scholar]
- 47.Meijer L., et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol 10, 1255–1266 (2003). [DOI] [PubMed] [Google Scholar]
- 48.(iLINCS).
- 49.Kumar N., Liang Y., Parslow T.G. & Liang Y. Receptor tyrosine kinase inhibitors block multiple steps of influenza a virus replication. J Virol 85, 2818–2827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toner E., Brennan G.P., Wells K., McGeown J.G. & Fairweather I. Physiological and morphological effects of genistein against the liver fluke, Fasciola hepatica. Parasitology 135, 1189–1203 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Baechler S.A., et al. Topoisomerase poisoning by genistein in the intestine of Toxicol Lett 243, 88–97 (2016). [DOI] [PubMed] [Google Scholar]
- 52.de Ruijter J., et al. Genistein in Sanfilippo disease: a randomized controlled crossover trial. Ann Neurol 71, 110–120 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Messing E., et al. A phase 2 cancer chemoprevention biomarker trial of isoflavone G-2535 (genistein) in presurgical bladder cancer patients. Cancer prevention research 5, 621–630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukuda M. & Longnecker R. Epstein-Barr virus (EBV) latent membrane protein 2A regulates B-cell receptor-induced apoptosis and EBV reactivation through tyrosine phosphorylation. J Virol 79, 8655–8660 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howard S., et al. Fragment-based discovery of the pyrazol-4-yl urea (AT9283), a multitargeted kinase inhibitor with potent aurora kinase activity. J Med Chem 52, 379–388 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Qi W., et al. AT9283, a novel aurora kinase inhibitor, suppresses tumor growth in aggressive B-cell lymphomas. Int J Cancer 130, 2997–3005 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Santo L., et al. Antimyeloma activity of a multitargeted kinase inhibitor, AT9283, via potent Aurora kinase and STAT3 inhibition either alone or in combination with lenalidomide. Clin Cancer Res 17, 3259–3271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vormoor B., et al. A phase I/II trial of AT9283, a selective inhibitor of aurora kinase in children with relapsed or refractory acute leukemia: challenges to run early phase clinical trials for children with leukemia. Pediatric blood & cancer 64(2017). [DOI] [PubMed] [Google Scholar]
- 59.Moreno L., et al. A phase I trial of AT9283 (a selective inhibitor of aurora kinases) in children and adolescents with solid tumors: a Cancer Research UK study. Clin Cancer Res 21, 267–273 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Hay A.E., et al. A phase II study of AT9283, an aurora kinase inhibitor, in patients with relapsed or refractory multiple myeloma: NCIC clinical trials group IND.191. Leukemia & lymphoma 57, 1463–1466 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Coleman C.M., et al. Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion. J Virol 90, 8924–8933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schang L.M., St Vincent M.R. & Lacasse J.J. Five years of progress on cyclin-dependent kinases and other cellular proteins as potential targets for antiviral drugs. Antivir Chem Chemother 17, 293–320 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Drugbank. Alvocidib. (2005).
- 64.Yamamoto M., et al. CDK9 inhibitor FIT-039 prevents replication of multiple DNA viruses. J Clin Invest 124, 3479–3488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pubchem. (Z)-5-((4-(Pyridin-4-yl)quinolin-6-yl)methylene)thiazolidine-2,4-dione.
- 66.Desai N.C., Rajpara K.M. & Joshi V.V. Synthesis of pyrazole encompassing 2-pyridone derivatives as antibacterial agents. Bioorg Med Chem Lett 23, 2714–2717 (2013). [DOI] [PubMed] [Google Scholar]
- 67.D’Ascenzio M., et al. Design, synthesis and biological characterization of thiazolidin-4- one derivatives as promising inhibitors of Toxoplasma gondii. Eur J Med Chem 86, 17–30 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Tomasic T., et al. Novel 2-thioxothiazolidin-4-one inhibitors of bacterial MurD ligase targeting D-Glu- and diphosphate-binding sites. Eur J Med Chem 46, 3964–3975 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Cuneo A., et al. Management of adverse events associated with idelalisib treatment in chronic lymphocytic leukemia and follicular lymphoma: A multidisciplinary position paper. Hematol Oncol 37, 3–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiestner A. Emerging role of kinase-targeted strategies in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program 2012, 88–96 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Falchi L., Baron J.M., Orlikowski C.A. & Ferrajoli A. BCR Signaling Inhibitors: an Overview of Toxicities Associated with Ibrutinib and Idelalisib in Patients with Chronic Lymphocytic Leukemia. Mediterr J Hematol Infect Dis 8, e2016011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kindrachuk J., et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrobial agents and chemotherapy 59, 1088–1099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walker M., et al. Macrofilaricidal Efficacy of Repeated Doses of Ivermectin for the Treatment of River Blindness. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 65, 2026–2034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim Y.E., et al. Control, elimination, and eradication of river blindness: scenarios, timelines, and ivermectin treatment needs in Africa. PLoS Negl Trop Dis 9, e0003664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laing R., Gillan V. & Devaney E. Ivermectin - Old Drug, New Tricks? Trends Parasitol 33, 463–472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crump A. & Omura S. Ivermectin, ‘wonder drug’ from Japan: the human use perspective. Proc Jpn Acad Ser B Phys Biol Sci 87, 13–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonzalez Canga A., et al. The pharmacokinetics and interactions of ivermectin in humans--a mini-review. AAPS J 10, 42–46 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mastrangelo E., et al. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. The Journal of antimicrobial chemotherapy 67, 1884–1894 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tay M.Y., et al. Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral research 99, 301–306 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Liu L.X. & Weller P.F. Antiparasitic drugs. N Engl J Med 334, 1178–1184 (1996). [DOI] [PubMed] [Google Scholar]
- 81.Chou A. & Serpa J.A. Eosinophilia in patients infected with human immunodeficiency virus. Curr HIV/AIDS Rep 12, 313–316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zs-Nagy I. Chemistry, toxicology, pharmacology and pharmacokinetics of idebenone: a review. Archives of gerontology and geriatrics 11, 177–186 (1990). [DOI] [PubMed] [Google Scholar]
- 83.Meier T. & Buyse G. Idebenone: an emerging therapy for Friedreich ataxia. J Neurol 256 Suppl 1, 25–30 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Jaber S. & Polster B.M. Idebenone and neuroprotection: antioxidant, pro-oxidant, or electron carrier? J Bioenerg Biomembr 47, 111–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gillis J.C., Benefield P. & McTavish D. Idebenone. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic use in age-related cognitive disorders. Drugs & aging 5, 133–152 (1994). [DOI] [PubMed] [Google Scholar]
- 86.Montenegro L., Turnaturi R., Parenti C. & Pasquinucci L. Idebenone: Novel Strategies to Improve Its Systemic and Local Efficacy. Nanomaterials (Basel) 8(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phenoxymethylpenicillin. (2005). [Google Scholar]
- 88.de Haan P., Bruynzeel D.P. & van Ketel W.G. Onset of penicillin rashes: relation between type of penicillin administered and type of immune reactivity. Allergy 41, 75–78 (1986). [DOI] [PubMed] [Google Scholar]
- 89.Gruchalla R.S. & Pirmohamed M. Clinical practice. Antibiotic allergy. N Engl J Med 354, 601–609 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Richardson P., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 395, e30–e31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stebbing J., et al. COVID-19: combining antiviral and anti-inflammatory Lancet Infect Dis 20, 400–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci 248, 117477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang D.H., Wu K.L., Zhang X., Deng S.Q. & Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med 18, 152–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Y., et al. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv (2020). [Google Scholar]
- 95.Subramanian A., et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452 e1417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edgar R., Domrachev M. & Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu P., Nielsen T.E. & Clausen M.H. FDA-approved small-molecule kinase Trends Pharmacol Sci 36, 422–439 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Perwitasari O., Yan X., O’Donnell J., Johnson S. & Tripp R.A. Repurposing Kinase Inhibitors as Antiviral Agents to Control Influenza A Virus Replication. Assay and drug development technologies 13, 638–649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schor S. & Einav S. Repurposing of Kinase Inhibitors as Broad-Spectrum Antiviral Drugs. DNA Cell Biol 37, 63–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lesch M., et al. RNAi-based small molecule repositioning reveals clinically approved urea-based kinase inhibitors as broadly active antivirals. PLoS pathogens 15, e1007601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meineke R., Rimmelzwaan G.F. & Elbahesh H. Influenza Virus Infections and Cellular Kinases. Viruses 11(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu C.H., et al. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem 284, 5229–5239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sisk J.M., Frieman M.B. & Machamer C.E. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. The Journal of general virology 99, 619–630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Badia R., et al. Inhibition of herpes simplex virus type 1 by the CDK6 inhibitor PD-0332991 (palbociclib) through the control of SAMHD1. The Journal of antimicrobial chemotherapy 71, 387–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pauls E., et al. Palbociclib, a selective inhibitor of cyclin-dependent kinase4/6, blocks HIV-1 reverse transcription through the control of sterile alpha motif and HD domain- containing protein-1 (SAMHD1) activity. Aids 28, 2213–2222 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Jaw J., Hill P. & Goodman D. Combination of Leflunomide and Everolimus for treatment of BK virus nephropathy. Nephrology 22, 326–329 (2017). [DOI] [PubMed] [Google Scholar]
- 107.Guendel I., Agbottah E.T., Kehn-Hall K. & Kashanchi F. Inhibition of human immunodeficiency virus type-1 by cdk inhibitors. AIDS Res Ther 7, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liacini A., Seamone M.E., Muruve D.A. & Tibbles L.A. Anti-BK virus mechanisms of sirolimus and leflunomide alone and in combination: toward a new therapy for BK virus infection. Transplantation 90, 1450–1457 (2010). [DOI] [PubMed] [Google Scholar]
- 109.Yamasmith E., et al. Efficacy and Safety of Ivermectin against Dengue Infection: A Phase III, Randomized, Double-blind, Placebo-controlled Trial. in The 34th Annual Meeting The Royal College of Physicians of Thailand ‘Internal Medicine and One Health’ (PEACH Royal Cliff Beach Resort, Pattaya, Chonburi, Thailand, 2018). [Google Scholar]
- 110.Avirutnan P. Ivermectin: a promising anti-dengue replication treatment [abstract S634] in 26th European Congress of Clinical Microbiology and Infectious Diseases 9–12 (Amsterdam, Netherlands, 2016). [Google Scholar]
- 111.Caly L., Druce J.D., Catton M.G., Jans D.A. & Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral research, 104787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peterhans E. Oxidants and antioxidants in viral diseases: disease mechanisms and metabolic regulation. The Journal of nutrition 127, 962S–965S (1997). [DOI] [PubMed] [Google Scholar]
- 113.McCullers J.A. Preventing and treating secondary bacterial infections with antiviral agents. Antiviral therapy 16, 123–135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mehta P., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maggiora G., Vogt M., Stumpfe D. & Bajorath J. Molecular similarity in medicinal chemistry. J Med Chem 57, 3186–3204 (2014). [DOI] [PubMed] [Google Scholar]
- 116.Hui D.S., Memish Z.A. & Zumla A. Severe acute respiratory syndrome vs. the Middle East respiratory syndrome. Curr Opin Pulm Med 20, 233–241 (2014). [DOI] [PubMed] [Google Scholar]
- 117.van Doremalen N, B. T., Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. The new england journal of medicine (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen Y., Lun A.T. & Smyth G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 5, 1438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis S. & Meltzer P.S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007). [DOI] [PubMed] [Google Scholar]
- 120.Smyth G.K. Limma: linear models for microarray data. in Bioinformatics and Computational Biology Solutions using R and Bioconductor, R (2005). [Google Scholar]
- 121.Gallaher W.R. Towards a sane and rational approach to management of Influenza H1N1 2009. Virol J 6, 51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kolde R. pheatmap : Pretty Heatmaps, (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.