Abstract
Background
Ubiquitin‐Specific Peptidase 26 (USP26), located on the X chromosome, encodes a deubiquitinating enzyme expressed mainly in testis, where it regulates protein turnover during spermatogenesis and modulates the ubiquitination levels of the Androgen Receptor (AR), and as a consequence, affects AR signaling.
Methods
The patient was thoroughly characterized clinically. He was genetically tested by chromosome analysis and whole exome sequencing (WES).
Results
The patient was diagnosed with Sertoli cell‐only syndrome pattern (SCOS). The WES analysis revealed only the variation in USP26: causing p.P469S in a highly evolutionary conserved amino acid as the possible cause for SCOS. The literature search identified 34 single variations and 14 clusters of variations in USP26 that were associated with male infertility. Only one of the 22 variations and of one cluster of three mutations tested for ubiquitination activity was found as damaging. Only one out of six variations tested for effect on AR function was found as damaging. Thus, the association of USP26 with male fertility was questioned.
Conclusions
The finding in our patient and the discussion on the reviewed literature support a possible role for USP26 in male fertility.
Keywords: AR, Azoospermia, mutation, SCOS, USP26
Many mutations in USP26 were reported to be causing azoospermia. However, the role of inactivation of this gene as causing azoospermia has been controversial. We present a novel mutation in a Sertoli cell‐only syndrome patient and critically review the previous reports, supporting the role of functional USP26 for male fertility.
1. INTRODUCTION
Nonobstructive azoospermia (NOA) accounts for approximately 60% of men with azoospermia. NOA is histologically characterized by the absence of sperm due to either a Sertoli cell‐only syndrome pattern, maturation arrest, hypospermatogenesis or mixed patterns (Hamada, Esteves, & Agarwal, 2013).
USP26 (MIM: 300,309) encodes a deubiquitinating enzyme which plays an important role in the regulation of protein turnover during spermatogenesis (Amerik & Hochstrasser, 2004) and modulates the ubiquitination levels of the androgen receptor (Dirac & Bernards, 2010) which is essential for male sexual differentiation and maturation, maintenance of spermatogenesis (Collins & Chang, 2002). It is expressed during sperm development in Leydig and Sertoli cells (Wosnitzer et al., 2014) and thus appears to be a good candidate for causing male infertility. However, very recently it was reported that USP26 is not essential for mouse gametogenesis and infertility (Felipe‐Medina et al., 2019). In addition, several papers reported a lack of association between USP26 genetic polymorphisms and male infertility (Christensen, Griffin, & Carrell, 2008; Luddi et al., 2016; Ravel et al., 2006; Ribarski et al., 2009; Shi et al., 2011; Stouffs, Lissens, Tournaye, Steirteghem, & Liebaers, 2006; Zhang et al., 2015). Supporting the observation in mice, was a report on a normozoospermic man carrying a nonsense mutation in USP26 (c.882 C > A) that generated a premature STOP codon which would truncate the protein and eliminate the catalytic Cys domain (Luddi et al., 2016).
In this study we present a NOA patient with a novel missense mutation in USP26 identified by whole exome sequencing (WES), review the controversial information regarding the role of variations in USP26 in male infertility, and discuss the contribution of the finding in our patient in supporting a possible role for USP26 in male fertility.
2. MATERIAL AND METHODS
2.1. Ethical approval and consent to participate
The study was approved by the Soroka Medical Center institutional review board, and the participant had signed a written informed consent prior to participation.
2.1.1. Patient
A detailed history as well as standardized physical, clinical, and laboratory examinations were carried out to record details of the lifestyle, occupation, family history, aside from physical examination and hormone level assessment. Chromosome analysis and Y microdelitions were carried out. Multiple semen analyses (1‐month interval) were according to World Health Organization (WHO) guidelines. Azoospermia was diagnosed when the absence of sperm was observed after centrifugation at 3,000g for 15 min and screening at a magnification of X400 magnification.
2.1.2. Genetic analysis
Genomic DNA (gDNA) was extracted from white blood cells by standard procedures. WES analysis was carried at Theragene, and SureSelect XTHuman All Exon V6 kit was used for library preparation. The target size was 58Mb and the coverage uniformity at X10 coverage was ≥90%. Results were analyzed using QIAGEN’s Ingenuity® Variant Analysis™ software (www.qiagen.com/ingenuity, QIAGEN Redwood City). The variant identified in the USP26 was ascertained by PCR of the genomic region using primers and Sanger sequencing of the PCR product,
PCR amplification of exon 1 of the USP26 gene (NM__031907.1) was performed using the forward: TGTTGCACTCCATTGCTTGT and reverse: GTGCACTCCAACGGAAGTCT (annealing temperature 62°C) primers. Direct sequencing of the PCR products was performed after cleaning the products using ExoSap (ThermoScientific) on an ABI PRISM 3,100 DNA Analyzer with the BigDye Terminator v.1.1 cycle sequencing kit (Applied Biosystems, Carlsbad, California, USA) according to the manufacturer's protocol.
3. RESULTS
3.1. Clinical findings
The patient presented the primary infertility that had lasted for longer than 19 years. His medical history was unremarkable. His BMI was 32.3. Karyotype: 46, XY, no Y chromosome microdeletion were identified. Hormonal profile: FSH: 19.1 mIU/ml; LH: 4.9 mIU/ml; Testosterone: 6.6 ng/ml; Prolactin 12 ng/ml. Testicular volume right ~14 ml, left 11 ~ ml. Among multiple ejaculated semen samples, the volume was in the normal range between 1.4 and 2.2 ml with no sperm. No sperm was found in his testicular sperm extraction and his histopathological diagnosis was Sertoli cell only (Figure 1a).
Figure 1.
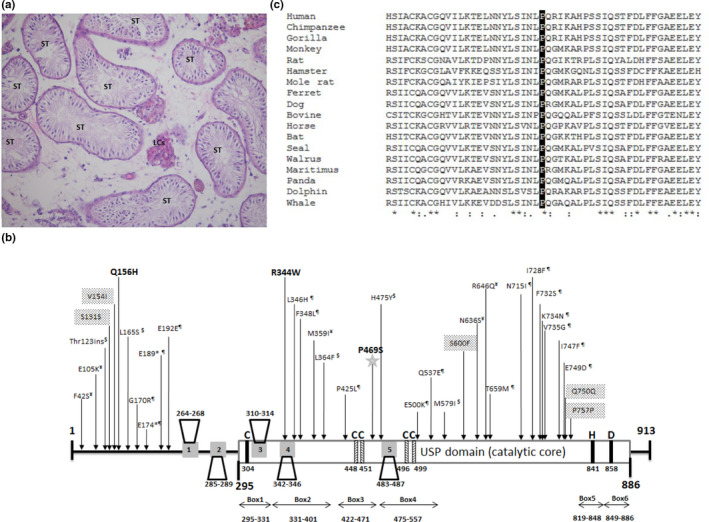
(a) Histopathological section from a biopsy of the left testis of the patient. Hematoxylin eosin staining of formalin‐fixed paraffin embedded sections show seminiferous tubules (STs) with Sertoli cell only. (b) The positions of the variations on USP26 reported to associate with male infertility. Schematic presentation of the catalytic domain (USP domain), the five binding motifs of USP26 protein to AR according to (Dirac & Bernards, 2010) are the numbered gray boxes, the residues of the catalytic triad are shown in black and the residues forming the zinc‐binding motif in the Fingers subdomain are displayed in hatched bars according to ( Ye, Scheel, Hofmann, & Komander, 2009). The mutations reported not to affect enzymatic activity are marked by $ and ¶ according to (Zhang et al., 2015) and (Liu et al., 2018) respectively and the mutations reported not to affect AR signaling are marked by ¥ according to (Ma et al., 2016). The mutation R344W affects AR function according to (Ma et al., 2016), mutation Q156H affects enzymatic activity according to (Liu et al., 2018) and variations V154I, S600F, and synonymous S131S, Q750Q, and P757P were not tested. Our newly identified mutation is marked by star. (c) Evolutionary conservation in the region of the USP domain containing the mutation
3.1.1. Identification of the Mutation
Since the patient's parents are family‐related, we assumed homozygosity by descent of a recessive mutation as the likely cause of the disorder; we analyzed the exome sequence of the patient for homozygous mutations. Thirty five homozygous variations with allele frequencies of less than 1% in the public databases (gnomad, ExAc browser, 1,000 Genomes and dbSNP) were identified. Following bioinformatics predictions and analysis of prevalence in the Bedouin population (Online Resources tables 1 + 2), we found a most likely candidate variation in chromosome X:132,160,844, c.1405C > T (NM_031907.1)(GRCh37/ hg19), in coding exon 1 of the Ubiquitin‐Specific Peptidase 26 (USP26) gene, causing p.P469S. The bioinformatic prediction of this variation is : CADD score: 20.3 (CADD above 15 is predicted damaging), polyphen 2: probably damaging, and the sift score is: tolerated. The exome result of this variation was ascertained by PCR of the genomic region and Sanger sequencing of the PCR product. The mutation is in the USP domain (Figure 1b), with complete conservation in mammals (Figure 1c). According to the 3D structure of USP21 in complex with suicide‐substrate Ub moiety that forms a covalent bond between its C terminus and the catalytic cysteine of the enzyme (PDB: 3MTN), the proline at position 469 resides in the midst of a hydrophobic core preceding a surface loop. The change of proline, which confers stability to the conformation of the hydrophobic core, into serine which enables flexibility of the structure, may destabilize the loop.
Table 1.
Variations in the USP26 gene reported in association with male infertility
| No. | variation | Freq.in patients | Freq. in controls | Ref. | gnomAD | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele Freq. (Total) | East Asia | African | South Asian | Other | Ashkenazi Jewish | Latino | European (non‐Finnish) | European (Finnish) | |||||||||||||
| Hemi. d | Freq. e | Hemi. d | Freq. e | Hemi. d | Freq. e | Hemi. d | Freq. e | Hemi. d | Freq. e | Hemi. d | Freq. e | Hemi. d | Freq. e | Hemi. d | Freq. e | ||||||
| 1 a | F42S | 1/776 | 0/709 | Ma et al. (2016) | 0.00002446 | 2 | 0.0003369 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 a | E105K | 1/776 | 0/709 | Ma et al. (2016) | 0.00002942 | 1 | 0.0003366 | 0 | 0.00005249 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 b | Thr 123 ins | 16/200 | 0/200 | Lee et al. (2008) | 0.02615 | 464 | 0.09628 | 462 | 0.09197 | 696 | 0.06680 | 29 | 0.01772 | 48 | 0.01627 | 49 | 0.006974 | 184 | 0.005174 | 5 | 0.001679 |
| 19/221 | 7/101 | Shi et al. (2011) | |||||||||||||||||||
| 0/78 | 2/72 | Luddi et al. (2016) | |||||||||||||||||||
| 4 | S131S | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 5 | V154I | ‐ | ‐ | Zhang et al. (2007) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 6 | Q156H | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 7 b | L165S | 54/200 | 36/200 | Lee et al. (2008) | 0.02663 | 491 | 0.09787 | 484 | 0.09293 | 751 | 0.06798 | 29 | 0.01761 | 54 | 0.01676 | 52 | 0.007075 | 201 | 0.005345 | 7 | 0.001778 |
| 0/78 | 2/72 | Luddi et al. (2016) | |||||||||||||||||||
| 19/221 | 7/101 | Shi et al. (2011) | |||||||||||||||||||
| 8c | G170R | 2/221 | 0/101 | Shi et al. (2011) | 0.0002600 | 0 | 0 | 1 | 0.0002097 | 26 | 0.001881 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0.0001521 | 0 | 0 |
| 9c | E174ter | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 10c | E189ter | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 11c | E192E | 120/200 | 172/200 | Lee et al. (2008) | |||||||||||||||||
| 108/221 | 79/101 | Shi et al. (2011) | |||||||||||||||||||
| 47/78 | 17/72 | Luddi et al. (2016) | |||||||||||||||||||
| 47/96 | 53/96 | Christensen et al. (2008) | |||||||||||||||||||
| 12 | R344W | 2/776 | 0/709 | Ma et al. (2016) | 0.00006628 | 2 | 0.0005059 | 0 | 0 | 0 | 0 | 1 | 0.0004516 | 0 | 0 | 0 | 0 | 0 | 0.00003707 | 0 | 0 |
| 13 c | L346H | 1/188 | 0/17 | Paduch et al. (2005) | 0.00001973 | 3 | 0.0002703 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 c | F348L | 1/44 | 0/56 | Zhang et al. (2007) | 0.0006696 | 49 | 0.007454 | 0 | 0.0001656 | 13 | 0.0007561 | 1 | 0.0001923 | 0 | 0 | 0 | 0.00007218 | 2 | 0.00005478 | 0 | 0 |
| 3/221 | 2/101 | Shi et al. (2011) | |||||||||||||||||||
| 15 a | M359I | 1/776 | 0/709 | Ma et al. (2016) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 16 b | L364F | 7/188 | 0/17 | Paduch et al. (2005) | 0.03788 | 0 | 0 | 36 | 0.007211 | 61 | 0.005298 | 59 | 0.04045 | 81 | 0.03241 | 150 | 0.02303 | 1987 | 0.05790 | 366 | 0.05737 |
| 7/96 | 7/96 | Christensen et al. (2008) | |||||||||||||||||||
| 14/300 | 10/287 | Ribarski et al. (2009) | |||||||||||||||||||
| 2/166 | 0/60 | Asadpor et al. (2013) | |||||||||||||||||||
| 7/78 | 10/72 | Luddi et al. (2016) | |||||||||||||||||||
| 17 c | P425L | ‐ | ‐ | Paduch et al. (2005) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 18 | P469S | Our mutation | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ||
| 19 b | H475Y | 1/188 | 0/17 | Paduch et al. (2005) | 0.02666 | 491 | 0.09795 | 481 | 0.09293 | 787 | 0.06823 | 28 | 0.01730 | 52 | 0.01648 | 54 | 0.007072 | 201 | 0.005337 | 6 | 0.001724 |
| 14/200 | 8/200 | Lee et al. (2008) | |||||||||||||||||||
| 24/221 | 5/101 | Shi et al. (2011) | |||||||||||||||||||
| 20 c | E500K | 1/188 | 0/17 | Paduch et al. (2005) | 0.0009104 | 0 | 0.00006749 | 0 | 0 | 115 | 0.009440 | 2 | 0.0005655 | 0 | 0 | 0 | 0 | 1 | 0.00002171 | 0 | 0 |
| 21 c | Q537E | 1/96 | 0/96 | Christensen et al. (2008) | 0.00008916 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0001911 | 0 | 0 | 0 | 0 | 5 | 0.0001856 | 0 | 0 |
| 22 b | M579I | 2/188 | 0/17 | Paduch et al. (2005) | 0.004432 | 2 | 0.0001354 | 4 | 0.0005362 | 127 | 0.01043 | 10 | 0.003797 | 4 | 0.001709 | 9 | 0.001292 | 181 | 0.005224 | 61 | 0.007826 |
| 80/200 | 28/200 | Lee et al. (2008) | |||||||||||||||||||
| 3/300 | 2/287 | Ribarski et al. (2009) | |||||||||||||||||||
| 3/166 | 0/60 | Asadpor et al. (2013) | |||||||||||||||||||
| 7/78 | 2/72 | Luddi et al. (2016) | |||||||||||||||||||
| 23 | ss6202791C>T(S600F) | 56/200 | 38/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 24 a | N636S | 1/776 | 0/709 | Ma et al. (2016) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 25 a | R646Q | 1/776 | 0/709 | Ma et al. (2016) | 0.00001968 | 0 | 0 | 0 | 0.00005405 | 0 | 0 | 0 | 0.0001887 | 0 | 0 | 0 | 0 | 0 | 0.00001087 | 0 | 0.00005543 |
| 26 c | T659M | 1/188 | 0/17 | Paduch et al. (2005) | 0.001253 | 1 | 0.00006755 | 0 | 0.0001599 | 9 | 0.0008911 | 0 | 0.0005658 | 30 | 0.01188 | 1 | 0.0002858 | 47 | 0.001430 | 0 | 0.00005442 |
| 1/200 | 0/200 | Lee et al. (2008) | |||||||||||||||||||
| 27 c | N715I | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 28 c | I728F | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 29 c | F732S | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 30 c | K734N | ‐ | ‐ | Paduch et al. (2005) | 0.0003037 | 0 | 0 | 0 | 0 | 1 | 0.0002098 | 1 | 0.0001893 | 0 | 0 | 7 | 0.001001 | 12 | 0.0003142 | 0 | 0 |
| 6/200 | 4/200 | Lee et al. (2008) | |||||||||||||||||||
| 31 c | V735G | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 32 c | I747F | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 33 c | E749D | 1/200 | 0/200 | Lee et al. (2008) | 0.000005474 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00001227 | 0 | 0 |
| 34 | Q750Q | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 35 | P757P | 1/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 36 b | Thr 123 ins+ L165S+ H475Y | 8/143 | 0/142 | Stouffs et al. (2005) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 4/188 | 0/17 | Paduch et al. (2005) | |||||||||||||||||||
| 6/200 | 0/200 | Lee et al. (2008) | |||||||||||||||||||
| 9/300 | 6/287 | Ribarski et al. 2009 | |||||||||||||||||||
| 19/221 | 5/101 | Shi et al. (2011) | |||||||||||||||||||
| 4/166 | 1/60 | Asadpor et al. (2013) | |||||||||||||||||||
| 37 | M579I+ K734N | 1/188 | 0/17 | Paduch et al. (2005) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 38 | Thr 123 ins+ L165S | 1/188 | 0/17 | Paduch et al. (2005) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | |
| 8/200 | 0/200 | Lee et al. (2008) | |||||||||||||||||||
| 0/221 | 2/101 | Shi et al. (2011) | |||||||||||||||||||
| 39 | L364F+ P425L | 1/188 | 0/17 | Paduch et al. (2005) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 40 | Thr 123 ins+ V154I | 8/44 | 0/56 | Zhang et al. (2007) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 41 | Thr 123` ins+ H475Y | 4/166 | 0/60 | Asadpor et al. (2013) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 42 | E192E+ H475Y | 3/221 | 0/101 | Shi et al. (2011) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 43 | E192E+G170R | 2/221 | 0/101 | Shi et al. (2011) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 44 | L165S+ E192E | 2/200 | 14/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 45 | M579I+ S600F+ E192E | 10/200 | 2/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 46 | M579I+ S600F+ E192E+ L165S | 10/200 | 2/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 47 | K734N + S600F+ E192E+ L165S | 8/200 | 0/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 48 | M579I+ E192E | 16/200 | 8/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| 49 | S600F+E192E+ L165S | 8/200 | 12/200 | Lee et al. (2008) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
The variation was not present in our collection of Bedouin exomes from 69 individuals (30 female, 39 male). Thus its prevalence in the Bedouin population is less than 1/99.
3.1.2. Review of the mutations in the USP26 associated with male infertility and the prevalence of the mutations in the gnomAd database
In order to verify the genotype–phenotype correlation of mutations in USP26, we reviewed the literature by a thorough search in PubMed. We found 10 papers that reported mutations associated with male infertility (Asadpor et al., 2013; Christensen et al., 2008; Lee et al., 2008; Luddi et al., 2016; Ma et al., 2016; Paduch, Mielnik, & Schlegel, 2005; Ribarski et al., 2009; Shi et al., 2011; Stouffs, Lissens, Tournaye, Steirteghem, & Liebaers, 2005; Zhang et al., 2007). Additionally, we aimed to find the frequency of the reported mutations in various populations in order to suggest the possibility of further screening for mutations in this gene for patients. and thus assessed the prevalence of the reported mutations in the specific populations in the gnomAD database. In total we report 34 mutations/variations appearing individually including our present mutation and 14 clusters of mutations/variations. The results are presented in Figure 1b and Table 1. Zhang et al. 2015 demonstrated, by enzymatic and meta‐analyses of the literature, that five reported mutations are not causative for male infertility either individually or in a cluster which was associated to infertility by a literature meta‐analysis (Xia et al., 2014). This cluster was later to be reported as a polymorphism among sub‐Saharan African and North African populations (Ravel et al., 2006). Dirac & Bernards reported that the regulatory function of USP26 on AR signaling depends on its deubiquinating activity, but none of the four USP26 mutations associated with fertility affected AR signaling (Dirac & Bernards, 2010). Another study verifying deubiquitinating activity of USP26 toward AR of six variations found in Han Chinese, demonstrated that only one variation, R344W, significantly reduced this activity (Ma et al., 2016). Finally, Liu et al., tested the effect of 19 variations associated with male infertility on the enzymatic activity of ubiquitination, only Q156H caused abrogation of the activity. Unexpectedly, even the nonsense mutations E174ter and E189ter had no effect on the enzyme activity when the variation was included in the entire coding region, only termination of transcription at these sites could abrogate the activity. They reasoned that an initiation methionine is present after the stop codons in both cases that could start the translation and production of active enzymes (Liu et al., 2018).
4. DISCUSSION
We have identified a novel mutation in USP26 which further expands the spectrum of mutations seen in azoospermic patients and may support the possible role of USP26 in male fertility.
Mutated USP26 appears to be a good candidate for causing male infertility considering its unique expression pattern in spermatogonia (Wosnitzer et al., 2014), Sertoli and Leydig cells (Asadpor et al., 2013; Paduch et al., 2005) and for its role in the ubiquitination of AR (Dirac & Bernards, 2010; Ma et al., 2016). However, its role in male fertility was debated because two mouse models lacking the gene were recently reported to be fertile (Felipe‐Medina et al., 2019). This contradiction for the role of USP26 can be argued by claiming that the functional relevance of this protein could differ between mice and men, showing a critical role in human that does not recapitulate in the mouse.
Additional arguments against the role of USP26 in male fertility were that mutations initially reported affecting a high percentage of patients (Paduch et al., 2005; Stouffs et al., 2005) were later reported as polymorphisms appearing also in normo‐spermic males (Ravel et al., 2006) and most of the mutations associated with male infertility did not affect the enzymatic activity (Liu et al., 2018; Zhang et al., 2015). Probably, the most compelling argument against the role of USP26 is the finding of Luddi et al. who reported a normo‐zoospermic man carrying a nonsense mutation in USP26 (c.882 C > A) that generates a premature STOP codon which would eliminate the whole catalytic domain (Luddi et al., 2016). We argue that this finding can be explained by the study of Liu et al. which demonstrated that only termination of transcription abrogates enzymatic activity (Liu et al., 2018).
We conclude that USP26 has a role in male fertility. This is based first on the literature demonstrating that some mutations identified in infertile males affect the enzymatic activity and the deubiquitinating activity of USP26 to AR. The second support for this conclusion is our finding of a single mutation in the WES analysis in a highly evolutionary conserved amino acid residue in the catalytic domain that is predicted to affect the stability of the catalytic domain structure.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
MA: Performed the genetic study and prepared the manuscript; AZ: Diagnosed and recruited the patient and contributed to the manuscript preparation; ElLev: Participated in the diagnosis and recruitment of the patient and contributed to the manuscript preparation; IHV: Performed the sperm analyses and contributed to the manuscript preparation; B S: performed the histology study; RS‐L: contributed to the histological analysis; SD: contributed to the genetic study; EL: participated in the diagnosis and recruitment of the patient, contributed to manuscript preparation; MH: contributed to the histological analysis; RP: supervised the genetic study and wrote the manuscript.
Supporting information
ACKNOWLEDGMENTS
We thank Prof. Raz Zarivach (Ben‐Gurion University of the Negev) for his help in the prediction of the effect of the mutation on the protein structure. This project was partly supported by The Israeli Ministry of Science, Technology and Space and partly by an internal grant of the Reproductive hub, Faculty of Health Sciences, Ben‐Gurion University of the Negev to AZ and RP.
Arafat M, Zeadna A, Levitas E, et al. Novel mutation in USP26 associated with azoospermia in a Sertoli cell‐only syndrome patient. Mol Genet Genomic Med. 2020;8:e1258 10.1002/mgg3.1258
Maram Arafat and Atif Zeadnae contributed equally.
REFERENCES
- Amerik, A. Y. , & Hochstrasser, M. (2004). Mechanism and function of deubiquitinating enzymes. Biochimica et Biophysica Acta, 1695, 189–207. 10.1016/j.bbamcr.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Asadpor, U. , Totonchi, M. , Sabbaghian, M. , Hoseinifar, H. , Akhound, M. R. , Zari Moradi, S. H. , … Mohseni, M. A. (2013). Ubiquitin‐specific protease (USP26) gene alterations associated with male infertility and recurrent pregnancy loss (RPL) in Iranian infertile patients. Journal of Assisted Reproduction and Genetics, 30(7), 923–931. 10.1007/s10815-013-0027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, G. L. , Griffin, J. , & Carrell, D. T. (2008). Sequence analysis of the X‐linked USP26 gene in severe male factor infertility patients and fertile controls. Fertility and Sterility, 90(3), 851–852. [DOI] [PubMed] [Google Scholar]
- Collins, L. L. , & Chang, C. (2002). Androgens and the androgen receptor in male sex development and fertility In Chang C. (Ed.), Androgens and Androgen Receptor (pp. 299–323). Boston, MA: Springer. [Google Scholar]
- Dirac, A. M. , & Bernards, R. (2010). The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Molecular Cancer Research, 8(6), 844–854. 10.1158/1541-7786.MCR-09-0424 [DOI] [PubMed] [Google Scholar]
- Felipe‐Medina, N. , Gómez‐H, L. , Condezo, Y. B. , Sanchez‐Martín, M. , Barbero, J. L. , Ramos, I. , … Pendás, A. M. (2019). Ubiquitin‐specific protease 26 (USP26) is not essential for mouse gametogenesis and fertility. Chromosoma, 128, 237–247. 10.1007/s00412-019-00697-6 [DOI] [PubMed] [Google Scholar]
- Hamada, A. J. , Esteves, S. C. , & Agarwal, A. (2013). A comprehensive review of genetics and genetic testing in azoospermia. Clinics (Sao, Paulo), 68(S1), 39–60. 10.6061/clinics/2013(Sup01)06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I. W. , Kuan, L. C. , Lin, C. H. , Pan, H. A. , Hsu, C. C. , Tsai, Y. C. , … Teng, Y. N. (2008). Association of USP26 haplotypes in men in Taiwan, China with severe spermatogenic defect. Asian Journal of Andrology, 10(6), 896–904. [DOI] [PubMed] [Google Scholar]
- Liu, Y. L. , Zheng, J. , Mi, Y. J. , Zhao, J. , & Tian, Q. B. (2018). The impacts of nineteen mutations on the enzymatic activity of USP26. Gene, 641, 292–296. 10.1016/j.gene.2017.10.074 [DOI] [PubMed] [Google Scholar]
- Luddi, A. , Crifasi, L. , Quagliarello, A. , Governini, L. , De Leo, V. , & Piomboni, P. (2016). Single nucleotide polymorphisms of USP26 in azoospermic men. System Biology in Reproductive Medicine, 62(6), 372–378. [DOI] [PubMed] [Google Scholar]
- Ma, Q. , Li, Y. , Guo, H. , Li, C. , Chen, J. , Luo, M. , … Gui, Y. (2016). A Novel Missense Mutation in USP26 Gene Is Associated With Nonobstructive Azoospermia. Reprod Sci, 23(10), 1434–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduch, D. A. , Mielnik, A. , & Schlegel, P. N. (2005). Novel mutations in testis‐specific ubiquitin protease 26 gene may cause male infertility and hypogonadism. Reproductive BioMedicine Online, 10(6), 747–754. 10.1016/S1472-6483(10)61119-4 [DOI] [PubMed] [Google Scholar]
- Ravel, C. , El Houate, B. , Chantot, S. , Lourenço, D. , Dumaine, A. , Rouba, H. , … McElreavey, K. (2006). Haplotypes, mutations and male fertility: The story of the testis‐specific ubiquitin protease USP26. Molecular Human Reproduction, 12, 643–646. 10.1093/molehr/gal063 [DOI] [PubMed] [Google Scholar]
- Ribarski, I. , Lehavi, O. , Yogev, L. , Hauser, R. , Bar‐Shira Maymon, B. , Botchan, A. , … Kleiman, S. E. (2009). USP26 gene variations in fertile and infertile men. Human Reproduction, 24(2), 477–484. 10.1093/humrep/den374 [DOI] [PubMed] [Google Scholar]
- Shi, Y. C. , Wei, L. , Cui, Y. X. , Shang, X. J. , Wang, H. Y. , Xia, X. Y. , … Huang, Y. F. (2011). Association between ubiquitin‐specific protease USP26 polymorphism and male infertility in Chinese men. Clinica Chimica Acta, 412(7–8), 545–549. 10.1016/j.cca.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Stouffs, K. , Lissens, W. , Tournaye, H. , van Steirteghem, A. , & Liebaers, I. (2005). Possible role of USP26 in patients with severely impaired spermatogenesis. European Journal of Human Genetics, 13, 336–340. [DOI] [PubMed] [Google Scholar]
- Stouffs, K. , Lissens, W. , Tournaye, H. , van Steirteghem, A. , & Liebaers, I. (2006). Alterations of the USP26 gene in Caucasian men. International Journal of Andrology, 29, 614–617. 10.1111/j.1365-2605.2006.00708.x [DOI] [PubMed] [Google Scholar]
- Wosnitzer, M. S. , Mielnik, A. , Dabaja, A. , Robinson, B. , Schlegel, P. N. , & Paduch, D. A. (2014). Ubiquitin Specific Protease 26 (USP26) Expression analysis in human testicular and extra gonadal tissues indicates diverse action of USP26 in cell differentiation and tumorigenesis. PLoS ONE, 9(6), e98638 10.1371/journal.pone.0098638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J. D. , Chen, J. , Han, Y. F. , Chen, H. , Yu, W. , Chen, Y. , & Dai, Y. T. (2014). Association of 370–371insACA, 494T>C, and 1423C>T haplotype in ubiquitin‐specific protease 26 gene and male infertility: A meta‐analysis. Asian Journal of Andrology, 16, 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y. , Scheel, H. , Hofmann, K. , & Komander, D. (2009). Dissection of USP catalytic domains reveal five common insertion points. Molecular BioSystems, 5, 1797–1808. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Qiu, S. D. , Li, S. B. , Zhou, D. X. , Tian, H. , Huo, Y. W. , … Zhang, Q. Y. (2007). Novel mutations in ubiquitin‐specific protease 26 gene might cause spermatogenesis impairment and male infertility. Asian Journal of Andrology, 9(6), 809–814. 10.1111/j.1745-7262.2007.00305.x [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Liu, T. , Mi, Y. J. , Yue, L. D. , Wang, J. M. , Liu, D. W. , … Tian, Q. B. (2015). Evidence from enzymatic and meta‐analyses does not support a direct association between USP26 gene variants and male infertility. Andrology, 3, 271–279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


