Abstract
Background
ZDHHC2 is a member of the DHHC protein family, mediating palmitoylation of postsynaptic density‐95 (PSD‐95) and A‐kinase‐anchoring protein 79/150 (AKAP79/150). Genome‐wide association studies (GWASs) have identified ZDHHC2 as a candidate gene for schizophrenia (SCZ). We aimed to fine‐map variants of ZDHHC2 conferring risk to SCZ in the Han Chinese population.
Methods
Targeted sequencing of whole‐exome sequences including untranslated regions (UTRs) along with neighboring regions in 1,827 schizophrenic patients and 1,004 normal controls of Han Chinese origin.
Results
A total of 123 variants, including five common and 118 rare variants, were identified. Among common variants, rs73198534, rs530313445, and rs74406481 were significantly associated with SCZ. Nine nonsynonymous rare variants, p.Glu96fs, p.Arg127X, p.Val145Ile, p.Ala177Thr, p.Arg269Gln, p.Asn312His, p.Glu319Lys, p.Gln340X, and p.Ile347Val, identified only in patients; eight are located in the important domains, including two stop‐gain variants. The 3D structural analysis and functional prediction revealed that all these eight variants may affect AMPAR expression or function, and influence the synaptic plasticity by regulating the palmitoylation of PSD95 and AKAP79/150.
Conclusion
Our results first show strong supportive evidences of the association between the ZDHHC2 and SCZ, and also provide a fine‐mapping of variants of this gene in Han Chinese SCZ patients.
Keywords: polymorphism, schizophrenia, ZDHHC2 domains, ZDHHC2 variant
In the present study, we performed targeted sequencing of the ZDHHC2 in 1,827 schizophrenic patients and 1,004 normal controls of Han Chinese origin. Our results first show strong supportive evidences of the association between the ZDHHC2 and SCZ, and also provide a fine‐mapping of variants of this gene in Han Chinese SCZ patients.
![]()
1. INTRODUCTION
Schizophrenia (SCZ) is a complex and highly heritable polygenic disorder. Genome‐wide association studies (GWASs) have identified numerous SCZ‐associated loci in different populations (Ripke et al., 2014, 2013, 2011; Shi et al., 2011; Yue et al., 2011). These linked single‐nucleotide polymorphisms (SNPs) can define disease‐associated genomic regions, which may span hundreds of genes involved in synaptic function and plasticity, neuronal calcium signaling, glutamatergic neurotransmission, dendritic spines, immune responses, and neurodevelopmental signaling pathways (Thyme et al., 2019). Studies also imply that there is a substantial contribution of common causal variants, rare alleles and de novo single nucleotides to genetic factors in SCZ susceptibility (Lee et al., 2012; Purcell et al., 2014). Follow‐up of GWAS risk genes through targeted sequencing is a promising approach for the detection of SCZ‐associated variants. The identification of novel risk variants and risk mechanisms can accelerate the process of understanding the pathophysiology of SCZ and discovering potential drug targets.
The gene encoding Zinc Finger DHHC‐type Palmitoyltransferase 2 (ZDHHC2, OMIM:618,621) is located on chromosome 8p21.3‐22 and consists of 17 exons; the protein is a member of the DHHC family of protein palmitoyl acyltransferases (PATs) and contains 367 amino acids (Oyama et al., 2000). As the largest transcription factor family in humans, zinc‐finger proteins play a key role in multiple biological processes, including autophagy, metabolism, development and differentiation, and cancer progression (Jen & Wang, 2016). Although commonly referred to as “DHHC PATs,” the official gene names are denoted “ZDHHC” (e.g., ZDHHC2) (Young, Butland, Sanders, Sutton, & Hayden, 2012). PATs are required for protein palmitoylation, one of the most prominent lipid modifications, which can regulate the localization, trafficking, targeting, and function of proteins (Linder & Deschenes, 2007). Emerging evidence indicates that palmitoylation also participates in the regulation of neuronal development (Hess, Patterson, Smith, & Skene, 1993), neuronal differentiation (Chamoun et al., 2001), synaptic transmission, and synaptic plasticity (El‐Husseini & Bredt, 2002). At least 23 members of the DHHC PAT gene family are found in mammals, and the highly conserved DHHC domain is necessary for catalytic activity (Mitchell, Vasudevan, Linder, & Deschenes, 2006). Recently, aberrant palmitoylation of several DHHC family members (ZDHHC12, ZDHHC9, ZDHHC15, ZDHHC13, ZDHHC8) has been demonstrated to be involved in neuropsychiatric diseases, including Alzheimer's disease (Mizumaru et al., 2009), X‐linked mental retardation (Mansouri et al., 2005), Huntington disease (Yanai et al., 2006), and SCZ (Mukai et al., 2004).
ZDHHC2 is highly expressed in the hippocampus, mediating palmitoylation of postsynaptic density‐95 (PSD‐95) and A‐kinase anchoring protein 79/150 (AKAP79/150). The palmitoylation of PSD‐95 is key for clustering synaptic proteins, including α‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazole propionic acid (AMPA) and N‐methyl‐d‐aspartate (NMDA)‐type glutamate receptors (El‐Husseini et al., 2002), while palmitoylation of the AMPAR‐linked scaffold protein A‐kinase anchoring protein (AKAP) 79/150 is required for AMPAR trafficking and synaptic potentiation (Keith et al., 2012; Woolfrey, Sanderson, & Dell'Acqua, 2015). Additionally, synaptosomal‐associated proteins (SNAP) SNAP25 and SNAP23 were identified as important DHHC2‐substrate interactions. Studies have shown that disturbances of synaptic function might result in abnormalities of neuronal connectivity, and the synaptic hypothesis of SCZ has already attracted much attention (Fromer et al., 2014; Owen, O'Donovan, & Harrison, 2005; Owen, Sawa, & Mortensen, 2016).
In the present study, we performed targeted sequencing of the ZDHHC2, a candidate gene for SCZ reported in our prior GWAS (Li et al., 2017). We aimed to detect risk variants in the ZDHHC2 in a Han Chinese population and investigate a possible association with SCZ.
2. MATERIALS AND METHODS
2.1. Ethical compliance
This study was approved by the Institutional Ethical Committee of Human Genetics Resources in Shanghai Jiao Tong University and conformed to the principles of the Declaration of Helsinki (World Medical, 2013). All participants were informed about this study and provided written informed consent before study inclusion.
2.2. Patient
A total of 2,831 blood‐derived DNA samples were collected for targeted sequencing, involving 1,827 patients (mean age ± SD, 44.47 ± 12.13; 1,124 men and 703 women) with schizophrenia and 1,004 (mean age ± SD, 43.17 ± 17.52; 442 men and 562 women) control subjects. All of these subjects were recruited from the Han Chinese population. The clinical diagnoses were made strictly in accordance with the DSM‐IV criteria based on the Structured Clinical Interview for DSM‐IV Axis I Disorders (SCID‐I). Healthy controls were randomly selected from the general Han Chinese population.
2.3. Targeted gene sequencing
Genomic DNA was extracted from peripheral blood samples using the Quick Gene DNA Whole Blood Kit (Tokyo, Japan). Sequencing primers covered the UTRs and all exons of ZDHHC2 (Table S1). Two‐staged PCR amplifications were performed with 20 ng DNA to generate a sequence library according to the manufacturer's instructions (Shanghai DynastyGene Co. Ltd). We barcoded the libraries using the unique 8 bp index for each sample. After purification using microbeads (Shanghai DynastyGene Co. Ltd), libraries were quantified by a Qubit 2.0 Fluorometer and 2100 Bioanalyzer (Agilent Technologies). Sequencing was performed on the Illumina Xten platform following the manufacturer's protocol.
2.4. Data processing and quality control
The quality of the sequence reads was checked by FastQC, and the adaptors were removed followed by low complexity read trimming with Trimmomatic. We used Burrows‐Wheeler Aligner (BWA) to map the sequencing reads to the reference human genome (UCSC, hg38) for ZDHHC2 (NC_000008.11). PCR duplicates were flagged using Picard, and the outputs were locally realigned using the Indel Realignment tool of the Genome Analysis Toolkit (GATK). After local realignment, recalibration was performed according to GATK best practice guidelines. GATK HaplotypeCaller was utilized to call variants, and Annovar was used to annotate variants. The transcript NM_016353.4 was used for gene variant annotation. The mean sequencing depth was 220× and variants with a minimum of 10 reads per base.
2.5. Bioinformatics analysis
Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/) and 1000 Genomes Project (http://www.1000genomes.org) were used to evaluate the frequencies of variants in the whole population and the healthy Eastern Asian population. All variants with minor allele frequencies (MAF) lower than 1% in the above public exome databases were defined as rare variants. The potential deleterious impacts of the candidate variants at the protein level were predicted using LRT (Chun & Fay, 2009), Polyphen‐2 (Adzhubei et al., 2010), and Mutation Taster (Schwarz, Cooper, Schuelke, & Seelow, 2014). Additional clinical variant annotations were obtained from NCBI ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). Localization of the protein domain was based on the UniProt Database (https://www.uniprot.org). Amino acid position is based on the NCBI reference sequences NP_057437.1. Conservation analysis was performed with the aid of ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Although tertiary structure information is not available for ZDHHC2, the structure of the protein can be predicted by using the homology modeling program: SWISS MODEL (Schwede, Kopp, Guex, & Peitsch, 2003). PyMOL was used to evaluate protein tertiary structure and to predict the potential functional effect of a missense mutation on the protein. Nonsynonymous rare variants were selected for Sanger sequencing to determine if the targeted sequencing is correct.
2.6. Statistical analysis
Association analysis was performed on the SHEsisPlus online software platform (http://shesisplus.bio-x.cn/SHEsis.html) (Li et al., 2009; Shi & He, 2005), which was developed by our research group. The χ2 test was used to identify the discrepancies of allele and genotype frequency between patients and controls. Odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were calculated for each association. Hardy‒Weinberg equilibrium (HWE) was analyzed in the healthy controls with χ2 test. All tests were two‐tailed, and p‐value <.05 was considered statistically significant. p‐values were adjusted using the Bonferroni correction method for both allelic, genotypic and haplotype associations tests. The pairwise linkage disequilibrium (LD) values among the common variants were calculated, and SNPs with D’ > 0.95 in both sample sets were categorized in the same block.
3. RESULTS
3.1. Identification of variations
To identify genetic variants, we sequenced the UTRs and exons including the exon–intron boundaries of the ZDDH2 isoform using genomic DNA isolated from 1,827 SCZ patients and 1,004 unaffected controls. A total of 123 variants were identified, 5 of which were defined as common (MAF > 1%) and 118 as rare (MA ≦ 1%) variants. Among the 118 rare variants, 49 are located in introns, 52 in the 3’UTR, and 17 in exons (Table S2).
3.2. Association analysis of the variants with SCZ
The distributions of the detected 123 variants did not deviate from HWE (p > .05). All five common variants are located in noncoding regions, and four common variants showed significance with SCZ. The alleles and genotype frequencies of the common variants are summarized in Table 1. Rs73198534 (corrected P allele = .006, OR = 1.29, P genotype = 0.014), rs530313445 (corrected P allele = 2.37E‐9, OR = 0.113, P genotype = 1.97E‐9), and rs74406481 (corrected P allele = 4.33E‐8, OR = 0.329, P genotype = 4.26E‐7) were significantly associated with SCZ in both allele and genotype distributions after Bonferroni correction. Rs79366093 showed allelic and genotypic significance with SCZ before correction, but the association was eliminated after Bonferroni correction. The T allele of rs79366093 and T allele of rs73198534 conferred risk for SCZ, whereas the T allele of rs530313445 and G allele of rs74406481 were protective against SCZ.
Table 1.
Genotype and allele frequencies of common variants in ZDHHC2
| Position | Location | SNP ID | Allele frequency | OR | 95% CI | P allele | Corrected P | Genotype frequency | P genotype | Corrected P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr8:17208102 | Intron | rs79366093 | C | T | 2.825 | 1.476–5.405 | .001 | .123 | CC | CT | 9.74E−4 | .12 | ||
| SCZ | 3,598 (0.984) | 56 (0.015) | 1771 (0.969) | 56 (0.03) | ||||||||||
| Control | 1997 (0.994) | 11 (0.005) | 993 (0.989) | 11 (0.01) | ||||||||||
| chr8:17210065 | Intron | rs73198534 | C | T | 1.29 | 1.14–1.459 | 4.85E−5 | .006 | CC | TT | CT | 1.11E−4 | .014 | |
| SCZ | 2,549 (0.697) | 1,105 (0.302) | 873 (0.477) | 151 (0.082) | 803 (0.439) | |||||||||
| Control | 1503 (0.748) | 505 (0.251) | 563 (0.56) | 64 (0.063) | 377 (0.375) | |||||||||
| chr8:17215372 | Intron | rs530313445 | — | T | 0.113 | 0.052–0.244 | 1.93E−11 | 2.37E−9 | — | ‐T | 1.60E−11 | 1.97E−9 | ||
| SCZ | 3,646 (0.997) | 8 (0.002) | 1819 (0.995) | 8 (0.004) | ||||||||||
| Control | 1970 (0.981) | 38 (0.018) | 966 (0.962) | 38 (0.037) | ||||||||||
| chr8:17222146 | 3'UTR | rs74406481 | A | G | 0.329 | 0.228–0.473 | 3.52E−10 | 4.33E−8 | AA | GA | GG | 3.46E−9 | 4.26E−7 | |
| SCZ | 3,606 (0.986) | 48 (0.013) | 1,780 (0.974) | 46 (0.025) | 1 (5.47e−04) | |||||||||
| Control | 1930 (0.961) | 78 (0.038) | 928 (0.924) | 74 (0.073) | 2 (0.001) | |||||||||
| chr8:17221849 | 3'UTR | rs3750249 | G | A | 0.952 | 0.846–1.072 | .421 | GA | GG | AA | .679 | |||
| SCZ | 2,556 (0.699) | 1,098 (0.3) | 768 (0.42) | 894 (0.489) | 165 (0.09) | |||||||||
| Control | 1,384 (0.689) | 624 (0.31) | 424 (0.422) | 480 (0.478) | 100 (0.099) | |||||||||
p < .05 as statistical significance and significant p‐values in bold. Genomic position is based on NCBI builds GRCh38 of ZDHHC2 (NC_000008.11, region: 17156000..17230706).
Abbreviations: CHR, chromosome; CI, confidence interval; OR, odds ratio; SCZ, schizophrenia; SNP, single‐nucleotide polymorphism; UTR, untranslated region.
Pairwise LD analysis was performed for the five common SNPs (rs79366093, rs73198534, rs530313445, rs3750249, and rs74406481). Two blocks were identified: rs79366093 and rs73198534 existed in one block, and the other haplotype block was composed of rs530313445, rs3750249, and rs74406481 (Figure 1). We performed haplotype analysis for the two blocks (Table 2). In block 1, one protective haplotype, CC, and two risk haplotypes, CT and TC, were found to be significantly associated with SCZ. In block 2, two protective haplotypes, ‐AG and TGA, and one risk haplotype, ‐GA, were found to be significantly associated with SCZ.
Figure 1.
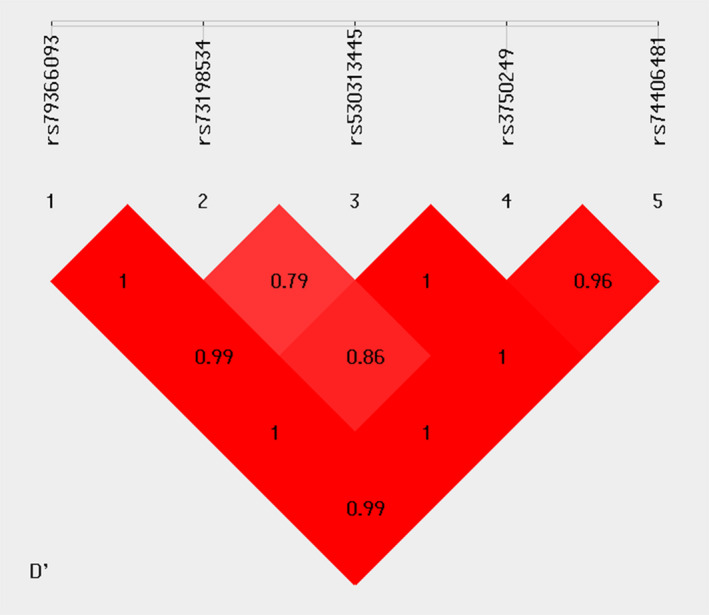
Pairwise linkage disequilibrium (LD) plot for the five common SNPs in ZDHHC2 investigated in healthy controls depicted by D’
Table 2.
Haplotype association analysis
| SCZ(freq) | Control(freq) | P | Corrected P | OR [95% CI] | ||
|---|---|---|---|---|---|---|
| Block1:rs79366093‐rs73198534 | ||||||
| CC | 2,493 (0.682) | 1,492 (0.743) | 1.66E−6 | 4.98E−6 | 0.742 [0.657–0.838] | |
| CT | 1,105 (0.302) | 505 (0.251) | 4.85E−5 | 1.46E−4 | 1.29 [1.14–1.459] | |
| TC | 56 (0.015) | 11 (0.005) | 0.001 | 0.003 | 2.825 [1.476–5.405] | |
| Block2:rs530313445‐rs3750249‐rs74406481 | ||||||
| ‐AA | 1,051 (0.287) | 548 (0.272) | 0.239 | 1.195 | 1.075 [0.952–1.214] | |
| ‐GG | 1 (2.74e‐04) | 2 (9.96e‐04) | 0.258 | 1.29 | 0.274 [0.024–3.03] | |
| ‐GA | 2,547 (0.697) | 1,344 (0.669) | 0.031 | 0.155 | 1.136 [1.011–1.277] | |
| ‐AG | 47 (0.012) | 76 (0.037) | 6.83E−10 | 3.415E−9 | 0.331 [0.229–0.478] | |
| TGA | 8 (0.002) | 38 (0.018) | 1.93E−11 | 9.65E−11 | 0.113 [0.052–0.244] | |
p < .05 as statistical significance and significant p‐values in bold.
Abbreviations: CI, confidence interval; OR, odds ratio.
We next focused on the 12 nonsynonymous (nine missense, one frameshift and two stop‐gain) variants. All of the nonsynonymous variants were validated by Sanger sequencing (Figure 2). In single‐variant association analysis, no significant differences in allele and genotype frequencies of the rare nonsynonymous variants between the patient and control groups were detected (data not shown), which is possibly due to the limited statistical power. Furthermore, we performed gene‐based association analysis using these 12 variants. The frequency of rare nonsynonymous variants in SCZ and control individuals was comparable; 0.657% of SCZ patients (12/1,827) and 0.299% of controls (3/1,004) were identified as carriers, indicating a higher rate of rare nonsynonymous variants in SCZ, although the difference was not statistically significant (p > .05, χ2 test with Yates correction; Table 3).
Figure 2.
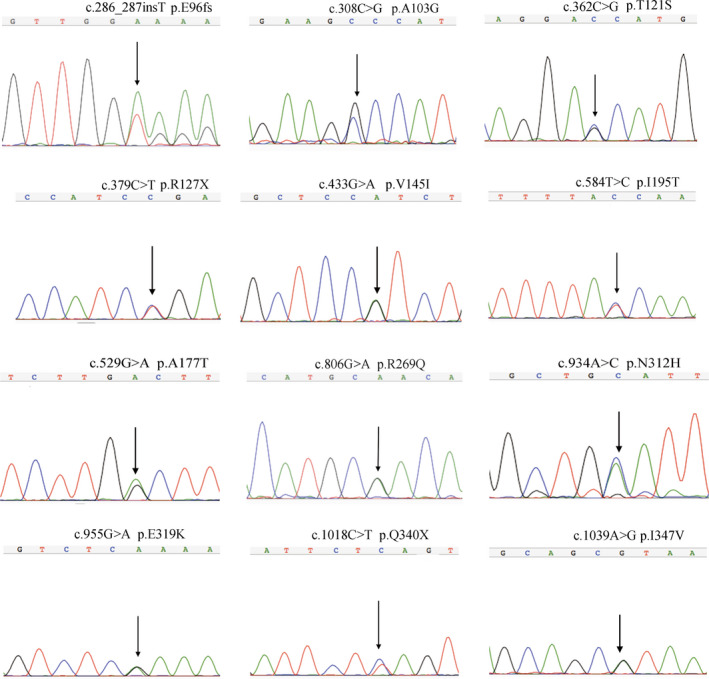
Validation of ZDHHC2 rare nonsynonymous variants via Sanger sequencing. Genomic positions are given according to NCBI builds GRCh38 (NC_000008.11)
Table 3.
Rare ZDHHC2 nonsynonymous variants found in SCZ patients and in controls
| Position | dbSNP | Location | Transcript variant | Amino Acid changes | SCZ | Con | Domains | Exonic Func | ExAC EASa | Polyphen‐2 | LRT | Mutationtaster |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr8:17195537 | Novel | Exon4 | c.286_287insT | p.Glu96fs | 1 M | — | — | Frameshift insertion | — | — | — | ‐ |
| chr8:17195559 | rs567986978 | Exon4 | c.308C>G | p.Ala103Gly | — | 1 M | — | Missense SNV | 0.0001 | B benign | Deleterious | Disease causing |
| chr8:17195613 | rs765056308 | Exon4 | c.362C>G | p.Thr121Ser | — | 1 M | — | Missense SNV | 0.0006 | Probably damaging | Deleterious | Disease causing |
| chr8:17197587 | rs747838463 | Exon5 | c.379C>T | p.Arg127X | 1 M | — | DHHC | Stop—gain | 0 | — | Deleterious | Disease causing automatic |
| chr8:17197641 | rs374897759 | Exon5 | c.433G>A | p.Val145Ile | 2 M | — | DHHC | Missense SNV | 0.0006 | Possibly damaging | Neutral | Disease causing |
| chr8:17205707 | Novel | Exon7 | c.529G>T | p.Ala177Thr | 1 F | — | DHHC | Missense SNV | — | Probably damaging | Deleterious | Disease causing |
| chr8:17205762 | rs758145586 | Exon7 | c.584T>C | p.Ile195Thr | — | 1 M | — | Missense SNV | 0.0002 | B benign | Neutral | Disease causing |
| chr8:17210007 | rs567641948 | Exon9 | c.806G>A | p.Arg269Gln | 1 F,1 M | — | C—terminal | Missense SNV | 0 | Probably damaging | Deleterious | Disease causing |
| chr8:17210464 | Novel | Exon10 | c.934A>C | p.Asn312His | 1 M | — | C—terminal | Missense SNV | — | Possibly damaging | Neutral | Disease causing |
| chr8:17215241 | rs529170689 | Exon11 | c.955G>A | p.Glu319Lys | 1 M, 1 F | — | C—terminal | Missense SNV | 0 | Probably damaging | Deleterious | Disease causing |
| chr8:17215304 | Novel | Exon11 | c.1018C>T | p.Gln340X | 1 M | — | C—terminal | Stop—gain | — | — | Neutral | Disease causing automatic |
| chr8:17215325 | rs752377762 | Exon11 | c.1039A>G | p.Ile347Val | 1 F | — | C—terminal | Missense SNV | 0.0011 | Benign | Neutral | Polymorphism |
| Totle | 12 | 3 |
Genomic positions are given according to NCBI builds GRCh38 (NC_000008.11). Transcript variants and amino acid changes are given according to reference sequences of ZDHHC2(NM_016353.4; NP_057437.1).
Abbreviations: chr, chromosome; ExAC, Exome Aggregation Consortium (http://exac.broadinstitute.org); F, female; M, male; SNV, single nucleotide variants
minor allele count/ total allele count.
3.3. 3D structural and functional predictions for the nonsynonymous variants
The DHHC2 protein comprises different functional domains, including a short intracellular N‐terminal domain, four transmembrane domains, a conserved DHHC (Asp‐His‐His‐Cys) domain and cytoplasmic C‐terminal tail. For the 12 nonsynonymous rare variants, nine variants exclusively occurred in the SCZ group but not in any of the controls, including four novel rare variants: one insertion frameshift variant, p.Glu96fs; one stop‐gain variant, p.Arg127X; and another two missense variants, p.Asn312His and p.Ala177Thr. Three of the nine variants (p.Arg127X, p.Val145Ile, and p.Ala177Thr) are located in the DHHC domain, and five of the nine variants (p.Arg269Gln, p.Asn312His, p.Glu319Lys, p.Gln340X, and p.Ile347Val) are located in the cytoplasmic C‐terminal tail (Figure 3). Three rare missense variants, p.Val145Ile, p.Arg269Gln, and p.Glu319Lys, were identified in more than one SCZ patient with a high expected probability for a functional effect by three different in silico prediction tools (Mutation Taster, PolyPhen2 and LRT; Table 3). Conservation analysis revealed that all nine rare variants are located in highly conserved regions in several species (Figure 3).
Figure 3.
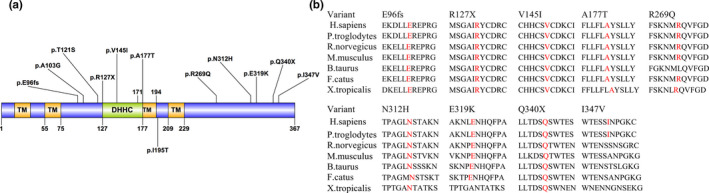
ZDHHC2 rare nonsynonymous variants detected in this study. (a) Protein domain structure of ZDHHC2 and localization of the rare nonsynonymous variants observed in patients and controls. (b) ZDHHC2 protein conservation at mutated positions that occurred only in patients. Localization of a protein domain is based on the UniProt Database (https://www.uniprot.org). Amino acid position is based on the NCBI reference sequences NP_057437.1
Furthermore, we performed 3D structural modeling of ZDHHC2 to determine the potential correlation of these missense mutations on protein structure. Among the six missense mutations, p.Ile347Val could not be predicted due to the length limitation of the modeled protein. The models reveal that p.Val145Ile and p.Ala177Thr mutations increase the side chain size and that p.Ala177Thr abolishes interaction with Leu181 after substitution, which may decrease the stability of this domain. Mutations located at the C‐terminus, that is, p.Arg269Gln, p.Asn312His, and p.Glu319Lys, showed great structural deviation in comparison with wild‐type ZDHHC2, leading to misfolded conformations of the ZDHHC2 protein (Figure 4). As the DHHC domain and C‐terminal tail are key for DHHC2 activity, mutations in these domains may directly impair the protein function consistent with the in silico predictions. In particular, the two stop‐gain variants, p.Arg127X and p.Gln340X, can result in a truncated protein and lead to more severe impacts.
Figure 4.
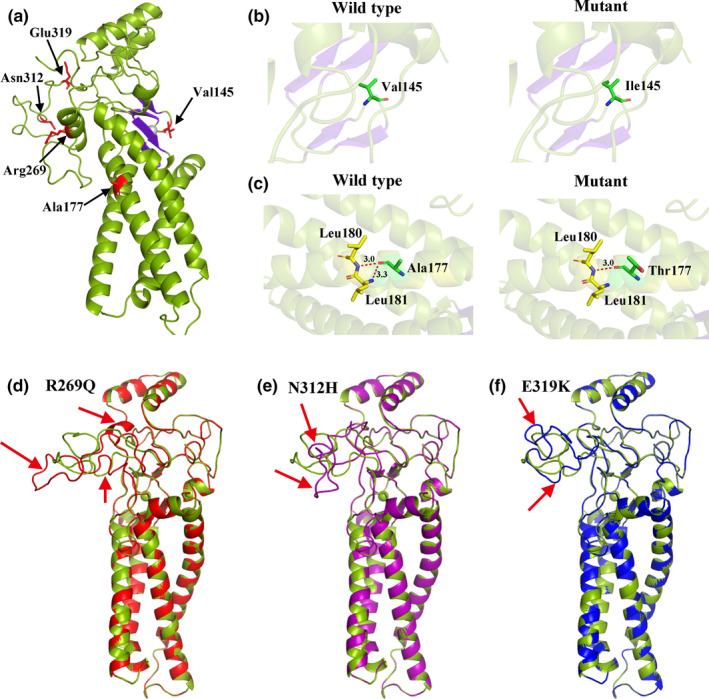
3D model structure of the wildtype and mutated ZDHHC2 protein. (a) Homology modeling of the wildtype structure of ZDHHC2. Five mutated sites (Val145, Ala177, Arg269, Asn312 and Glu319) are shown in red. (b) Wildtype V145 and V145I mutation modeling. The V145I mutation model shows increased side chain size. (c) Wildtype A177 and A177T mutation modeling. The left shows the interaction (red dashed line) of wildtype A177 with neighboring amino acids Leu180 and Leu181. The right shows loss of interaction with Leu181 for the pathogenic variant A177T. (d)Local structural changes for R269Q‐mutated ZDHHC2 protein (red) in comparison with wildtype (green). (e) Local structural changes for N312H‐mutated ZDHHC2 protein (purple) in comparison with wildtype (green). (f)Local structural changes for E319K‐mutated ZDHHC2 protein (blue) in comparison with wildtype (green)
4. DISCUSSION
In this study, we performed targeted sequencing of the ZDHHC2 in 1,827 SCZ patients and 1,004 controls. Among the five common variants, rs73198534, rs530313445, and rs74406481 were significantly associated with SCZ in both allele and genotype distributions. rs79366093 showed allelic and genotypic significance with SCZ before Bonferroni correction, but the association was eliminated after correction. Haplotype analysis identified two haplotype blocks, rs79366093‐rs73198534 and rs530313445‐rs3750249‐rs74406481, that were significantly associated with SCZ. Although the common risk variants are both located in the 3’UTR and intron region, they have the potential to affect miRNA binding and gene splicing, contributing to SCZ susceptibility. The rather small samples (1,827 SCZ patients and 1,004 controls) that generate these genome‐wide significant associations further indicating that ZDHHC2 is a strong candidate gene in the development of SCZ. Investigations with more samples and strict study design are required for a better understanding of the relationship between ZDHHC2 and SCZ.
ZDHHC2 is a member of the “ZDHHC” (zinc finger aspartate‐histidine‐histidine‐cysteine) family of palmitoyl transferases. Recently, the ZDHHC family has received attention for its vital role in regulating synaptic plasticity and the dynamic regulation of synaptic strength in response to neuronal activity (Matt, Kim, Chowdhury, & Hell, 2019). In the neuronal system, neuronal targets for palmitoylation include presynaptic proteins, scaffold proteins (such as AKAP5, GRIP1b, PSD‐95) (Kang et al., 2008; Topinka & Bredt, 1998), neurotransmitter‐receptors and receptor‐associated proteins (such as AMPAR, NMPAR) (Pickering, Taverna, Salter, & Hampson, 1995). There is evidence that mutations of palmitoylation sites can lead to diseases and disorders of the nervous system (Sanders et al., 2015).
ZDHHC2 contains a short intracellular N‐terminal domain, four transmembrane domains, a conserved DHHC (Asp‐His‐His‐Cys) domain (residues 127–177) located in the intracellular loop between transmembrane domains 2 and 3, and a relatively long cytoplasmic C‐terminal tail (residues 229–367). In our present study, there were no significant differences in the allele and genotype frequencies of the DHHC2 rare nonsynonymous variants identified between patients and controls, which may be limited by the size of our testing sample. All identified rare coding variants in SCZ patients were heterozygous, and nine variants were found only in patients, including four novel rare variants. Three rare variants (p.Arg127X, p.Val145Ile, and p.Ala177Thr) are located in the conserved DHHC domain, which binds two zinc ions as necessary structural components, is considered to be the defining feature of palmitoyl transferases and is required for catalytic activity. In addition, the integrity of the DHHC motif is also necessary for autopalmitoylation (Greaves & Chamberlain, 2011). The p.Arg127X variant creates a premature stop codon, altering the DHHC domain, the transmembrane segments and the C‐terminus. Hence, nonsynonymous variants in the DHHC domain may directly influence palmitoyl transferase activity. Furthermore, five rare variants (p.Arg269Gln, p.Asn312His, p.Glu319Lys, p.Gln340X, and p.Ile347Val) are located in the cytoplasmic C‐terminal tail, including two novel variants, p.Asn312His and p.Gln340X. The cytoplasmic C‐terminal domain of ZDHHC2 is responsible for intracellular localization in neurons. When localized to recycling endosomes, ZDHHC2 regulates the exocytosis and palmitoylation of AKAP79/150 (Woolfrey et al., 2015). After translocation and integration at the spine membrane, ZDHHC2 mediates palmitoylation of PSD95 (Noritake et al., 2009). AKAP79/150 and PSD95 are important AMPAR‐interacting proteins that control surface insertion of AMPARs. The palmitoylation of these proteins is pivotal for AMPAR trafficking and recruitment, contributing to the increase in AMPAR expression and the associated long‐term potentiation (LTP) (Han, Wu, Wang, & Chen, 2015). Thus, variants in the ZDHHC2 C‐terminus may impact synaptic plasticity through regulation of the palmitoylation of PSD95 and AKAP79/150. The novel rare variant p.Gln340X results in a premature stop codon that leads to removal of the remaining amino acids and can yield more severe consequences. The 3D structural modeling of ZDHHC2 protein revealed that the p.Ala177Thr mutation abolishes interaction with Leu181, which may decrease the stability of the DHHC domain. Another three mutations, p.Arg269Gln, p.Asn312His, and p.Glu319Lys, showed great structural deviation in comparison with wild‐type ZDHHC2, leading to misfolded conformations. Taken together, we postulate that variants located in the DHHC domain and C‐terminal domain might affect protein function. Further functional experiment is necessary to confirm this hypothesis.
In conclusion, our targeted sequencing of the ZDHHC2 identified four common variants and three rare variants associated with SCZ in the Han Chinese population. The exact molecular mechanisms and networks affected by ZDHHC2 variants in SCZ remain unclear, but the alteration in AMPAR function may be involved. Based on the functional analysis of ZDHHC2 variants, selective regulation of the DHHC domain and C‐terminal domain might become new therapies for SCZ. A much larger sample size and further functional experiments are needed to support our findings.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
We are deeply grateful to all the patients, healthy volunteers, and psychiatrists for their help in recruiting patients and diagnosing SCZ in our patient cohort. This work was supported in part by Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), the National Key R&D Program of China (2017YFC0908105, 2019YFA0905401), the Natural Science Foundation of China (U1804284, 81421061, 81701321, 31571012, 91849104, 31770800, 81571329 and 81501154), Shanghai Hospital Development Center (SHDC12016115), Shanghai Science and Technology Committee (17JC1402900 and 17490712200), Shanghai Municipal Health Commission (ZK2015B01 and 201540114).
Zhang H, Li X, Ma C, et al. Fine‐mapping of ZDHHC2 identifies risk variants for schizophrenia in the Han Chinese population. Mol Genet Genomic Med. 2020;8:e1190 10.1002/mgg3.1190
Han Zhang and Xiuli Li contributed equally to this work.
Funding information
Shanghai Municipal Science and Technology Major Project, Grant Number: 2017SHZDZX01; the National Key R&D Program of China, Grant Number: 2017YFC0908105, 2019YFA0905401; the Natural Science Foundation of China, Grant Number:U1804284, 81421061, 81701321, 31571012, 91849104, 31770800, 81571329, 81501154; Shanghai Hospital Development Center, Grant Number:SHDC12016115; Shanghai Science and Technology Committee, Grant Number:17JC1402900, 17490712200; Shanghai Municipal Health Commission, Grant Number: ZK2015B01, 201540114.
Contributor Information
Yonggang Wang, Email: w100yg@163.com.
Yongyong Shi, Email: shiyongyong@gmail.com.
REFERENCES
- Adzhubei, I. A. , Schmidt, S. , Peshkin, L. , Ramensky, V. E. , Gerasimova, A. , Bork, P. , … Sunyaev, S. R. (2010). A method and server for predicting damaging missense mutations. Nature Methods, 7(4), 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoun, Z. , Mann, R. K. , Nellen, D. , von Kessler, D. P. , Bellotto, M. , Beachy, P. A. , & Basler, K. (2001). Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science, 293(5537), 2080–2084. 10.1126/science.1064437 [DOI] [PubMed] [Google Scholar]
- Chun, S. , & Fay, J. C. (2009). Identification of deleterious mutations within three human genomes. Genome Research, 19(9), 1553–1561. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Husseini, A. E. D. , & Bredt, D. S. (2002). Protein palmitoylation: A regulator of neuronal development and function. Nature Reviews Neuroscience, 3(10), 791–802. 10.1038/nrn940 [DOI] [PubMed] [Google Scholar]
- El‐Husseini, A.‐D. , Schnell, E. , Dakoji, S. , Sweeney, N. , Zhou, Q. , Prange, O. , … Bredt, D. S. (2002). Synaptic strength regulated by palmitate cycling on PSD‐95. Cell, 108(6), 849–863. 10.1016/S0092-8674(02)00683-9 [DOI] [PubMed] [Google Scholar]
- Fromer, M. , Pocklington, A. J. , Kavanagh, D. H. , Williams, H. J. , Dwyer, S. , Gormley, P. , … O'Donovan, M. C. (2014). De novo mutations in schizophrenia implicate synaptic networks. Nature, 506(7487), 179–184. 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, J. , & Chamberlain, L. H. (2011). DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends in Biochemical Sciences, 36(5), 245–253. 10.1016/j.tibs.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Han, J. , Wu, P. F. , Wang, F. , & Chen, J. G. (2015). S‐palmitoylation regulates AMPA receptors trafficking and function: A novel insight into synaptic regulation and therapeutics. Acta Pharmaceutica Sinica B, 5(1), 1–7. 10.1016/j.apsb.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, D. T. , Patterson, S. I. , Smith, D. S. , & Skene, J. H. (1993). Neuronal growth cone collapse and inhibition of protein fatty acylation by nitric oxide. Nature, 366(6455), 562–565. 10.1038/366562a0 [DOI] [PubMed] [Google Scholar]
- Jen, J. Y. , & Wang, Y. C. (2016). Zinc finger proteins in cancer progression. Journal of Biomedical Science, 23, 10.1186/s12929-016-0269-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, R. , Wan, J. , Arstikaitis, P. , Takahashi, H. , Huang, K. , Bailey, A. O. , … El‐Husseini, A. (2008). Neural palmitoyl‐proteomics reveals dynamic synaptic palmitoylation. Nature, 456(7224), 904–909. 10.1038/nature07605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, D. J. , Sanderson, J. L. , Gibson, E. S. , Woolfrey, K. M. , Robertson, H. R. , Olszewski, K. , … Dell'Acqua, M. L. (2012). Palmitoylation of A‐kinase anchoring protein 79/150 regulates dendritic endosomal targeting and synaptic plasticity mechanisms. Journal of Neuroscience, 32(21), 7119–7136. 10.1523/Jneurosci.0784-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. H. , DeCandia, T. R. , Ripke, S. , Yang, J. , Sullivan, P. F. , Goddard, M. E. , … Schizophrenia, M. G. (2012). Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs (vol 44, pg 247, 2012). Nature Genetics, 44(7), 831–831. 10.1038/ng0712-831a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Chen, J. , Yu, H. , He, L. , Xu, Y. , Zhang, D. , … Shi, Y. (2017). Genome‐wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nature Genetics, 49(11), 1576‐+ 10.1038/ng.3973 [DOI] [PubMed] [Google Scholar]
- Li, Z. , Zhang, Z. , He, Z. , Tang, W. , Li, T. , Zeng, Z. , … Shi, Y. (2009). A partition‐ligation‐combination‐subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis (http://analysis.bio‐x.cn). Cell Research, 19(4), 519‐523 10.1038/cr.2009.33 [DOI] [PubMed] [Google Scholar]
- Linder, M. E. , & Deschenes, R. J. (2007). Palmitoylation: Policing protein stability and traffic. Nature Reviews Molecular Cell Biology, 8(1), 74–84. 10.1038/nrm2084 [DOI] [PubMed] [Google Scholar]
- Mansouri, M. R. , Marklund, L. , Gustavsson, P. , Davey, E. , Carlsson, B. , Larsson, C. , … Dahl, N. (2005). Loss of ZDHHC15 expression in a woman with a balanced translocation t(X;15)(q13.3;cen) and severe mental retardation. European Journal of Human Genetics, 13(8), 970–977. 10.1038/sj.ejhg.5201445 [DOI] [PubMed] [Google Scholar]
- Matt, L. , Kim, K. , Chowdhury, D. , & Hell, J. W. (2019). Role of palmitoylation of postsynaptic proteins in promoting synaptic plasticity. Frontiers in Molecular Neuroscience, 12, 10.3389/fnmol.2019.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D. A. , Vasudevan, A. , Linder, M. E. , & Deschenes, R. J. (2006). Protein palmitoylation by a family of DHHC protein S‐acyltransferases. Journal of Lipid Research, 47(6), 1118–1127. 10.1194/jlr.R600007-JLR200 [DOI] [PubMed] [Google Scholar]
- Mizumaru, C. , Saito, Y. , Ishikawa, T. , Yoshida, T. , Yamamoto, T. , Nakaya, T. , & Suzuki, T. (2009). Suppression of APP‐containing vesicle trafficking and production of beta‐amyloid by AID/DHHC‐12 protein. Journal of Neurochemistry, 111(5), 1213–1224. 10.1111/j.1471-4159.2009.06399.x [DOI] [PubMed] [Google Scholar]
- Mukai, J. , Liu, H. , Burt, R. A. , Swor, D. E. , Lai, W. S. , Karayiorgou, M. , & Gogos, J. A. (2004). Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nature Genetics, 36(7), 725–731. 10.1038/ng1375 [DOI] [PubMed] [Google Scholar]
- Noritake, J. , Fukata, Y. , Iwanaga, T. , Hosomi, N. , Tsutsumi, R. , Matsuda, N. , … Fukata, M. (2009). Mobile DHHC palmitoylating enzyme mediates activity‐sensitive synaptic targeting of PSD‐95. Journal of Cell Biology, 186(1), 147–160. 10.1083/jcb.200903101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, M. J. , O'Donovan, M. C. , & Harrison, P. J. (2005). Schizophrenia: A genetic disorder of the synapse? BMJ, 330(7484), 158–159. 10.1136/bmj.330.7484.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, M. J. , Sawa, A. , & Mortensen, P. B. (2016). Schizophrenia. Lancet, 388(10039), 86–97. 10.1016/S0140-6736(15)01121-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama, T. , Miyoshi, Y. , Koyama, K. , Nakagawa, H. , Yamori, T. , Ito, T. , Nakamura, Y. (2000). Isolation of a novel gene on 8p21.3‐22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes Chromosomes and Cancer, 29(1), 9–15. . [DOI] [PubMed] [Google Scholar]
- Pickering, D. S. , Taverna, F. A. , Salter, M. W. , & Hampson, D. R. (1995). Palmitoylation of the GluR6 kainate receptor. Proceedings of the National Academy of Sciences of the United States of America, 92(26), 12090–12094. 10.1073/pnas.92.26.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. M. , Moran, J. L. , Fromer, M. , Ruderfer, D. , Solovieff, N. , Roussos, P. , … Sklar, P. (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506(7487), 185–190. 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke, S. , Neale, B. M. , Corvin, A. , Walters, J. T. R. , Farh, K. H. , Holmans, P. A. , … Consor, W.‐T.‐C.‐C. (2014). Biological insights from 108 schizophrenia‐associated genetic loci. Nature, 511(7510), 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke, S. , O'Dushlaine, C. , Chambert, K. , Moran, J. L. , Kähler, A. K. , Akterin, S. , … Sullivan, P. F. (2013). Genome‐wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics, 45(10), 1150–1159. 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke, S. , Sanders, A. R. , Kendler, K. S. , Levinson, D. F. , Sklar, P. , Holmans, P. A. , … Genome‐Wide, S. P. (2011). Genome‐wide association study identifies five new schizophrenia loci. Nature Genetics, 43(10), 969–976. 10.1038/ng.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, S. S. , Martin, D. D. O. , Butland, S. L. , Lavallée‐Adam, M. , Calzolari, D. , Kay, C. , … Hayden, M. R. (2015). Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Computational Biology, 11(8), e1004405 10.1371/journal.pcbi.1004405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, J. M. , Cooper, D. N. , Schuelke, M. , & Seelow, D. (2014). MutationTaster2: Mutation prediction for the deep‐sequencing age. Nature Methods, 11(4), 361–362. 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- Schwede, T. , Kopp, J. , Guex, N. , & Peitsch, M. C. (2003). SWISS‐MODEL: An automated protein homology‐modeling server. Nucleic Acids Research, 31(13), 3381–3385. 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. Y. , & He, L. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research, 15(2), 97–98. 10.1038/sj.cr.7290272 [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Li, Z. , Xu, Q. I. , Wang, T. I. , Li, T. , Shen, J. , … He, L. (2011). Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nature Genetics, 43(12), 1224–U1279. 10.1038/ng.980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme, S. B. , Pieper, L. M. , Li, E. H. , Pandey, S. , Wang, Y. , Morris, N. S. , … Schier, A. F. (2019). Phenotypic landscape of schizophrenia‐associated genes defines candidates and their shared functions. Cell, 177(2), 478–491.e20. 10.1016/j.cell.2019.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topinka, J. R. , & Bredt, D. S. (1998). N‐terminal palmitoylation of PSD‐95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron, 20(1), 125–134. [DOI] [PubMed] [Google Scholar]
- Woolfrey, K. M. , Sanderson, J. L. , & Dell'Acqua, M. L. (2015). The palmitoyl acyltransferase DHHC2 regulates recycling endosome exocytosis and synaptic potentiation through palmitoylation of AKAP79/150. Journal of Neuroscience, 35(2), 442–456. 10.1523/Jneurosci.2243-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, A. , Huang, K. , Kang, R. , Singaraja, R. R. , Arstikaitis, P. , Gan, L. U. , … Hayden, M. R. (2006). Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nature Neuroscience, 9(6), 824–831. 10.1038/nn1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, F. B. , Butland, S. L. , Sanders, S. S. , Sutton, L. M. , & Hayden, M. R. (2012). Putting proteins in their place: Palmitoylation in Huntington disease and other neuropsychiatric diseases. Progress in Neurobiology, 97(2), 220–238. 10.1016/j.pneurobio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Yue, W.‐H. , Wang, H.‐F. , Sun, L.‐D. , Tang, F.‐L. , Liu, Z.‐H. , Zhang, H.‐X. , … Zhang, D. (2011). Genome‐wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nature Genetics, 43(12), 1228–U1284. 10.1038/ng.979 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


