In the title Schiff base compound, C13H9IN2O3, the hydroxy group forms a intramolecular hydrogen bond to the imine N atom and generates an S(6) ring motif. The 4-iodobenzene ring is inclined to the phenol ring by 39.1 (2)°. The configuration about the C=N bonds is E. The crystal structure features C—H⋯O hydrogen-bonding interactions.
Keywords: crystal structure, salicylaldehyde derivative, 4-iodoaniline, 2-hydroxy-5-nitrobenzaldehyde, hydrogen bonding
Abstract
The title compound, C13H9IN2O3, was synthesized by a condensation reaction between 2-hydroxy-5-nitrobenzaldehyde and 4-iodoaniline, and crystallizes in the orthorhombic space group Pna21. The 4-iodobenzene ring is inclined to the phenol ring by a dihedral angle of 39.1 (2)°. The configuration about the C=N double bond is E. The crystal structure features C—H⋯O hydrogen-bonding interactions. A Hirshfeld surface analysis of the crystal structure indicates that the most important contributions for the packing arrangement are O⋯H/H⋯O (26.9%) and H⋯H (22.0%) interactions.
Chemical context
Over the past 25 years, extensive research has been directed towards the synthesis and use of Schiff base compounds in organic and inorganic chemistry as they have important medicinal and pharmaceutical applications. These compounds exhibit biological activities, including antibacterial, antifungal, anticancer and herbicidal properties (Desai et al., 2001 ▸; Singh & Dash, 1988 ▸; Karia & Parsania, 1999 ▸). They may also show useful photochromic properties, leading to applications in various fields such as the measurement and control of radiation intensities in imaging systems and optical computers, electronics, optoelectronics and photonics (Iwan et al., 2007 ▸). Schiff bases derived from 2-hydroxy-5-nitrobenzaldehyde are widely used either as materials or as intermediates in explosives, dyestuffs, pesticides and organic synthesis (Yan et al., 2006 ▸). Intramolecular hydrogen-atom transfer (tautomerism) from the o-hydroxy group to the imine-N atom is of prime importance with respect to the solvato-, thermo- and photochromic properties of o-hydroxy Schiff bases (Filarowski, 2005 ▸; Hadjoudis & Mavridis 2004 ▸). Such proton-exchanging materials can be utilized for the design of various molecular electronic devices (Alarcón et al., 1999 ▸). The present work is a part of an ongoing structural study of Schiff bases and their utilization in the synthesis of quinoxaline derivatives (Faizi et al., 2018 ▸), fluorescence sensors (Faizi et al., 2016 ▸; Mukherjee et al., 2018 ▸; Kumar et al., 2017 ▸; 2018 ▸) and non-linear optical properties (Faizi et al., 2020 ▸). We report herein the synthesis (from 2-hydroxy-5-nitrobenzaldehyde and 4-iodoaniline) and crystal structure of the title compound (I), along with the findings of a Hirshfeld surface analysis.
Structural commentary
The molecular structure of compound (I) is shown in Fig. 1 ▸. An intramolecular O—H⋯N hydrogen bond is observed (Table 1 ▸ and Fig. 1 ▸). This is a relatively common feature in analogous imine–phenol compounds (see Database survey section). The imine group displays a C8—C7—N1—C4 torsion angle of 174.5 (6)°. The 4-iodobenzene ring (C1–C6) is inclined by a dihedral angle of 39.1 (2)° to the phenol ring (C8–C13), which renders the molecule non-planar. The configuration of the C7=N1 bond of this Schiff base is E, and the intramolecular O1—H1⋯N1 hydrogen bond forms an S(6) ring motif (Fig. 1 ▸ and Table 1 ▸). The 4-nitro group is slightly tilted away from co-planarity with the benzene ring to which it is attached [O2—N2—C10—C9 = −7.4 (10)° and O3—N2—C10—C11= −7.4 (10)°]. The C13—O1 distance [1.330 (7) Å] is close to normal for values reported for single C—O bonds in phenols and salicylideneamines (Ozeryanskii et al., 2006 ▸). The N1=C7 bond is short at 1.264 (8) Å, indicative of double-bond character, while the long C7—C8 bond [1.444 (8) Å] implies a single bond. All these data support the existence of the phenol–imine tautomer for (I) in its crystalline state. These features are similar to those observed in related 4-dimethylamino-N-salicylideneanilines (Filipenko et al., 1983 ▸; Aldoshin et al., 1984 ▸; Wozniak et al., 1995 ▸; Pizzala et al., 2000 ▸). The C—N, C=N and C—C bond lengths are normal and close to the values observed in related structures (Faizi et al., 2017a ▸,b ▸).
Figure 1.
The molecular structure of the title compound, with the atom labelling. Displacement ellipsoids are drawn at the 40% probability level. The intramolecular N—H⋯O hydrogen bond (see Table1), forming an S(6) ring motif, is shown as a dashed line.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.82 | 1.86 | 2.591 (6) | 148 |
| C7—H7⋯O2i | 0.93 | 2.45 | 3.309 (8) | 154 |
Symmetry code: (i) 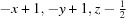 .
.
Supramolecular features
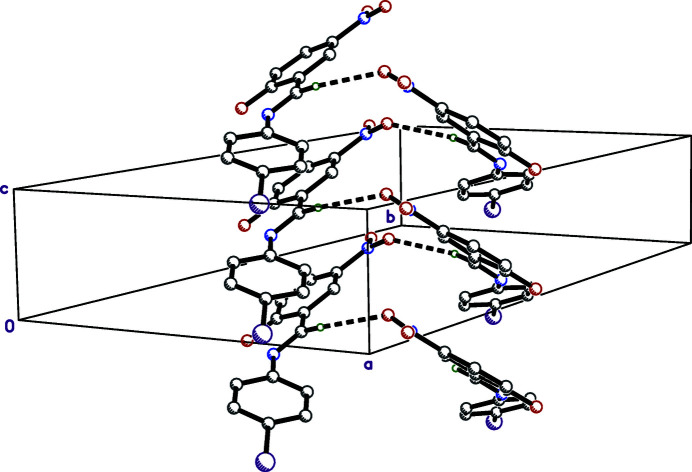
In the crystal packing of (I), the most important intermolecular interactions are weak C7—H7⋯O2i [symmetry code: (i) 1 − x, 1 − y, − + z] hydrogen bonds between screw-related molecules, which form helical chains propagating along the crystallographic screw axis parallel to c (Fig. 2 ▸, Table 1 ▸). The shortest intermolecular contact involving the iodine is 3.351 (5) Å, between glide-related molecules, I1⋯O1ii [symmetry code: (ii) x +
+ z] hydrogen bonds between screw-related molecules, which form helical chains propagating along the crystallographic screw axis parallel to c (Fig. 2 ▸, Table 1 ▸). The shortest intermolecular contact involving the iodine is 3.351 (5) Å, between glide-related molecules, I1⋯O1ii [symmetry code: (ii) x +  ,
,  − y, −1 + z)], which makes a zigzag tape motif (Fig. 3 ▸). There are no other significant intermolecular interactions present in the crystal. The Hirshfeld surface analysis confirms the role of the C—H⋯O interactions in the packing arrangement.
− y, −1 + z)], which makes a zigzag tape motif (Fig. 3 ▸). There are no other significant intermolecular interactions present in the crystal. The Hirshfeld surface analysis confirms the role of the C—H⋯O interactions in the packing arrangement.
Figure 2.
A partial packing plot showing the C—H⋯O hydrogen-bonded (thick dashed lines) helical chains about the crystallographic 21 screw axis parallel to c.
Figure 3.
A partial packing plot showing close contacts (dashed lines) between iodine and the phenolic oxygen of glide-related (x +  ,
,  − y, −1 + z) molecules.
− y, −1 + z) molecules.
Hirshfeld surface analysis
In order to visualize the intermolecular interactions in the crystal packing of (I), a Hirshfeld surface (HS) analysis (Hirshfeld, 1977 ▸; Spackman & Jayatilaka, 2009 ▸) was carried out using Crystal Explorer 17.5 (Turner et al., 2017 ▸). In the HS plotted over d norm (Fig. 4 ▸), white surfaces indicate contacts with distances equal to the sum of van der Waals radii, and the red and blue colours indicate distances shorter (i.e., in close contact) or longer than the van der Waals radii sum, respectively (Venkatesan et al., 2016 ▸). The two-dimensional finger print plots are depicted in Fig. 5 ▸. The O⋯H/H⋯O (26.9%) interactions form the majority of contacts, with H⋯H (22.0%) interactions representing the next highest contribution. The percentage contributions of other interactions are: I⋯H/H⋯I (16.3%), C⋯H/H⋯C (10.5%), C⋯C (8.7%), O⋯C/C⋯O (4.7%), N⋯C/C⋯N (3.8%), I⋯C/C⋯I (2.3%), H⋯N/N⋯H (1.4%), I⋯O/O⋯I (2.0%), I⋯N/N⋯I (0.6%), I⋯I (0.5%), O⋯N/N⋯O (0.2%), N⋯N (0.1%) and O⋯O (0.1%).
Figure 4.
Hirshfeld surface of the title compound plotted over d norm.
Figure 5.
Two-dimensional fingerprint plots of the crystal with the relative contributions of the atom pairs to the Hirshfeld surface along with d norm full.
Database survey
A search of the Cambridge Structural Database (CSD, version 5.39; Groom et al., 2016 ▸) gave 26 hits for the (E)-2-{[(4-iodophenyl)imino]methyl}-phenol fragment. Of these 26, the most similar to (I), are as follows. In p-iodo-N-(p-cyanobenzylidene)aniline (LALMEQ; Ojala et al., 1999 ▸), the OH group is absent and the NO2 group is replaced by a cyano group. In (E)-5-(diethylamino)-2-[(4-iodophenylimino)methyl]phenol (VEFPED; Kaştaş et al., 2012 ▸), the NO2 is replaced by an N,N diethyl group. In N-(3,5-di-tert-butylsalicylidene)-4-iodobenzene; (MILFET; Spangenberg et al., 2007 ▸), the NO2 group is absent but a pair of tBu groups occupy the 3,5 positions of the salicylidene group. In 2-{[(4-iodophenyl)imino]methyl}-6-methoxyphenol (SEDBIP; Carletta, et al., 2017 ▸), the NO2 group is absent and a methoxy group is ortho to the hydroxyl. Lastly, in N-(2-cyanobenzylidene)-4-iodoaniline (XOXKIF; Ojala et al., 1999 ▸) the NO2 is absent and the OH is replaced by cyano. All these compounds have an E configuration about the C=N bond and form the S(6) ring motif.
Synthesis and crystallization
The title compound was synthesized by condensation of 2-hydroxy-5-nitrobenzaldehyde (11.0 mg, 0.066 mmol) and 4-iodoaniline (14.4 mg, 0.066 mmol) in ethanol (15 ml). After the mixture had refluxed for about 15 h, the orange product was washed with ether and dried at room temperature (yield 60%, m.p. 484–486 K). Crystals suitable for X-ray analysis were obtained by slow evaporation of an ethanol solution.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The OH hydrogen atoms and the C-bound H atoms were included in calculated positions and allowed to ride on the parent atoms: O—H = 0.82 Å, C—H = 0.93–0.96 Å with U iso(H) = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C13H9IN2O3 |
| M r | 368.12 |
| Crystal system, space group | Orthorhombic, P n a21 |
| Temperature (K) | 296 |
| a, b, c (Å) | 12.8022 (4), 24.4556 (9), 4.1459 (1) |
| V (Å3) | 1298.02 (7) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 2.47 |
| Crystal size (mm) | 0.42 × 0.34 × 0.21 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.944, 0.981 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 15403, 2508, 2231 |
| R int | 0.084 |
| (sin θ/λ)max (Å−1) | 0.617 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.037, 0.094, 1.05 |
| No. of reflections | 2508 |
| No. of parameters | 173 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.81, −0.25 |
| Absolute structure | Flack x determined using 814 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.00 (4) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020008191/pk2635sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020008191/pk2635Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020008191/pk2635Isup3.cml
CCDC reference: 1922980
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful to the Department of Chemistry, Langat Singh College, B. R. A. Bihar University, Muzaffarpur, India, for providing laboratory facilities.
supplementary crystallographic information
Crystal data
| C13H9IN2O3 | Dx = 1.884 Mg m−3 |
| Mr = 368.12 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 25449 reflections |
| a = 12.8022 (4) Å | θ = 1.7–29.9° |
| b = 24.4556 (9) Å | µ = 2.47 mm−1 |
| c = 4.1459 (1) Å | T = 296 K |
| V = 1298.02 (7) Å3 | Prism, colorless |
| Z = 4 | 0.42 × 0.34 × 0.21 mm |
| F(000) = 712 |
Data collection
| STOE IPDS 2 diffractometer | 2508 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 2231 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.084 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 26.0°, θmin = 1.8° |
| rotation method scans | h = −15→15 |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | k = −30→30 |
| Tmin = 0.944, Tmax = 0.981 | l = −5→4 |
| 15403 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.037 | w = 1/[σ2(Fo2) + (0.0632P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.094 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.81 e Å−3 |
| 2508 reflections | Δρmin = −0.25 e Å−3 |
| 173 parameters | Absolute structure: Flack x determined using 814 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 1 restraint | Absolute structure parameter: 0.00 (4) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| I1 | 0.50544 (3) | 0.16672 (2) | −0.1482 (5) | 0.06914 (19) | |
| O1 | 0.1141 (3) | 0.43513 (17) | 0.4285 (13) | 0.0721 (13) | |
| H1 | 0.148899 | 0.407604 | 0.393280 | 0.108* | |
| N1 | 0.2784 (3) | 0.37325 (18) | 0.4176 (13) | 0.0605 (13) | |
| C8 | 0.2751 (4) | 0.4584 (2) | 0.6892 (16) | 0.0575 (13) | |
| C1 | 0.4310 (5) | 0.2352 (2) | 0.0459 (14) | 0.0578 (12) | |
| C9 | 0.3293 (4) | 0.4960 (2) | 0.875 (2) | 0.0614 (12) | |
| H9 | 0.397472 | 0.488588 | 0.939016 | 0.074* | |
| C13 | 0.1701 (4) | 0.4702 (2) | 0.6025 (17) | 0.0564 (12) | |
| C11 | 0.1808 (4) | 0.5566 (2) | 0.870 (2) | 0.0688 (15) | |
| H11 | 0.151047 | 0.589917 | 0.926484 | 0.083* | |
| C6 | 0.3291 (5) | 0.2321 (2) | 0.1458 (18) | 0.0689 (16) | |
| H6 | 0.293084 | 0.199177 | 0.130244 | 0.083* | |
| C10 | 0.2824 (4) | 0.5444 (2) | 0.9636 (16) | 0.0618 (15) | |
| O3 | 0.3034 (5) | 0.6282 (2) | 1.207 (2) | 0.120 (3) | |
| N2 | 0.3421 (4) | 0.5833 (2) | 1.1596 (16) | 0.0751 (16) | |
| C7 | 0.3254 (4) | 0.4086 (2) | 0.5862 (18) | 0.0592 (12) | |
| H7 | 0.394439 | 0.402274 | 0.645609 | 0.071* | |
| C3 | 0.4358 (5) | 0.3296 (2) | 0.1943 (18) | 0.0654 (15) | |
| H3 | 0.472183 | 0.362411 | 0.211171 | 0.079* | |
| C12 | 0.1245 (4) | 0.5192 (3) | 0.6944 (17) | 0.0655 (15) | |
| H12 | 0.055800 | 0.526809 | 0.636953 | 0.079* | |
| C2 | 0.4847 (4) | 0.2839 (3) | 0.067 (2) | 0.0679 (16) | |
| H2 | 0.553487 | 0.286056 | −0.003196 | 0.082* | |
| C4 | 0.3331 (4) | 0.3266 (2) | 0.2959 (17) | 0.0575 (15) | |
| C5 | 0.2798 (4) | 0.2777 (2) | 0.2694 (15) | 0.0660 (18) | |
| H5 | 0.210481 | 0.275532 | 0.335185 | 0.079* | |
| O2 | 0.4261 (4) | 0.5694 (2) | 1.2640 (15) | 0.0946 (19) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| I1 | 0.0821 (3) | 0.0597 (3) | 0.0656 (3) | 0.01321 (13) | −0.0044 (3) | −0.0083 (2) |
| O1 | 0.0590 (19) | 0.063 (2) | 0.094 (4) | 0.0028 (16) | −0.007 (2) | −0.007 (2) |
| N1 | 0.061 (2) | 0.051 (2) | 0.069 (4) | 0.0033 (17) | −0.004 (2) | 0.002 (2) |
| C8 | 0.054 (3) | 0.052 (3) | 0.066 (4) | 0.002 (2) | 0.007 (3) | 0.004 (2) |
| C1 | 0.071 (3) | 0.049 (3) | 0.054 (3) | 0.009 (2) | −0.006 (3) | 0.002 (2) |
| C9 | 0.054 (2) | 0.060 (3) | 0.070 (4) | 0.0001 (18) | −0.001 (4) | −0.001 (3) |
| C13 | 0.056 (3) | 0.049 (3) | 0.064 (3) | 0.001 (2) | 0.004 (3) | 0.003 (3) |
| C11 | 0.067 (3) | 0.058 (3) | 0.081 (4) | 0.007 (2) | 0.022 (4) | 0.000 (4) |
| C6 | 0.070 (3) | 0.054 (3) | 0.083 (5) | −0.002 (2) | −0.006 (3) | −0.007 (3) |
| C10 | 0.064 (3) | 0.056 (3) | 0.066 (4) | −0.006 (2) | 0.010 (2) | −0.001 (2) |
| O3 | 0.111 (4) | 0.077 (3) | 0.171 (8) | 0.001 (3) | −0.007 (4) | −0.049 (4) |
| N2 | 0.074 (3) | 0.066 (3) | 0.085 (5) | −0.015 (2) | 0.014 (3) | −0.015 (3) |
| C7 | 0.057 (3) | 0.052 (3) | 0.068 (3) | 0.004 (2) | −0.001 (3) | 0.008 (3) |
| C3 | 0.068 (3) | 0.049 (3) | 0.079 (4) | −0.004 (2) | 0.001 (3) | −0.004 (2) |
| C12 | 0.054 (3) | 0.061 (3) | 0.081 (4) | 0.006 (2) | 0.003 (3) | −0.001 (3) |
| C2 | 0.059 (3) | 0.069 (4) | 0.075 (5) | 0.002 (2) | 0.005 (3) | 0.003 (4) |
| C4 | 0.062 (2) | 0.046 (2) | 0.064 (5) | 0.0059 (19) | −0.005 (3) | 0.002 (2) |
| C5 | 0.060 (3) | 0.060 (3) | 0.078 (5) | 0.003 (2) | −0.003 (3) | −0.005 (3) |
| O2 | 0.072 (2) | 0.088 (3) | 0.124 (6) | −0.012 (2) | −0.008 (3) | −0.028 (3) |
Geometric parameters (Å, º)
| I1—C1 | 2.089 (5) | C11—H11 | 0.9300 |
| O1—C13 | 1.330 (7) | C6—C5 | 1.381 (8) |
| O1—H1 | 0.8200 | C6—H6 | 0.9300 |
| N1—C7 | 1.264 (8) | C10—N2 | 1.466 (8) |
| N1—C4 | 1.430 (7) | O3—N2 | 1.220 (8) |
| C8—C9 | 1.385 (9) | N2—O2 | 1.208 (8) |
| C8—C13 | 1.421 (7) | C7—H7 | 0.9300 |
| C8—C7 | 1.444 (8) | C3—C4 | 1.383 (8) |
| C1—C6 | 1.371 (9) | C3—C2 | 1.385 (10) |
| C1—C2 | 1.377 (9) | C3—H3 | 0.9300 |
| C9—C10 | 1.377 (7) | C12—H12 | 0.9300 |
| C9—H9 | 0.9300 | C2—H2 | 0.9300 |
| C13—C12 | 1.388 (8) | C4—C5 | 1.381 (8) |
| C11—C12 | 1.373 (10) | C5—H5 | 0.9300 |
| C11—C10 | 1.389 (9) | ||
| C13—O1—H1 | 109.5 | C11—C10—N2 | 120.2 (5) |
| C7—N1—C4 | 120.5 (5) | O2—N2—O3 | 123.8 (6) |
| C9—C8—C13 | 118.7 (5) | O2—N2—C10 | 118.8 (5) |
| C9—C8—C7 | 120.1 (5) | O3—N2—C10 | 117.4 (6) |
| C13—C8—C7 | 121.2 (5) | N1—C7—C8 | 121.9 (5) |
| C6—C1—C2 | 120.2 (6) | N1—C7—H7 | 119.1 |
| C6—C1—I1 | 120.4 (4) | C8—C7—H7 | 119.1 |
| C2—C1—I1 | 119.4 (4) | C4—C3—C2 | 120.2 (5) |
| C10—C9—C8 | 120.0 (5) | C4—C3—H3 | 119.9 |
| C10—C9—H9 | 120.0 | C2—C3—H3 | 119.9 |
| C8—C9—H9 | 120.0 | C11—C12—C13 | 120.0 (5) |
| O1—C13—C12 | 118.6 (5) | C11—C12—H12 | 120.0 |
| O1—C13—C8 | 121.1 (5) | C13—C12—H12 | 120.0 |
| C12—C13—C8 | 120.2 (5) | C1—C2—C3 | 119.8 (6) |
| C12—C11—C10 | 119.8 (5) | C1—C2—H2 | 120.1 |
| C12—C11—H11 | 120.1 | C3—C2—H2 | 120.1 |
| C10—C11—H11 | 120.1 | C5—C4—C3 | 119.4 (5) |
| C1—C6—C5 | 120.1 (6) | C5—C4—N1 | 118.5 (5) |
| C1—C6—H6 | 119.9 | C3—C4—N1 | 122.1 (5) |
| C5—C6—H6 | 119.9 | C6—C5—C4 | 120.2 (5) |
| C9—C10—C11 | 121.2 (6) | C6—C5—H5 | 119.9 |
| C9—C10—N2 | 118.6 (5) | C4—C5—H5 | 119.9 |
| C13—C8—C9—C10 | −1.8 (10) | C4—N1—C7—C8 | 174.5 (6) |
| C7—C8—C9—C10 | 177.8 (7) | C9—C8—C7—N1 | 179.8 (6) |
| C9—C8—C13—O1 | −179.1 (6) | C13—C8—C7—N1 | −0.5 (10) |
| C7—C8—C13—O1 | 1.3 (9) | C10—C11—C12—C13 | −1.9 (11) |
| C9—C8—C13—C12 | 2.0 (9) | O1—C13—C12—C11 | −179.1 (7) |
| C7—C8—C13—C12 | −177.7 (6) | C8—C13—C12—C11 | −0.1 (10) |
| C2—C1—C6—C5 | −0.2 (10) | C6—C1—C2—C3 | 0.9 (11) |
| I1—C1—C6—C5 | −179.0 (5) | I1—C1—C2—C3 | 179.7 (6) |
| C8—C9—C10—C11 | −0.1 (10) | C4—C3—C2—C1 | −0.7 (12) |
| C8—C9—C10—N2 | −179.9 (6) | C2—C3—C4—C5 | 0.0 (11) |
| C12—C11—C10—C9 | 2.1 (11) | C2—C3—C4—N1 | −177.7 (7) |
| C12—C11—C10—N2 | −178.2 (7) | C7—N1—C4—C5 | 146.2 (7) |
| C9—C10—N2—O2 | −7.4 (10) | C7—N1—C4—C3 | −36.1 (10) |
| C11—C10—N2—O2 | 172.8 (7) | C1—C6—C5—C4 | −0.6 (10) |
| C9—C10—N2—O3 | 172.4 (7) | C3—C4—C5—C6 | 0.7 (10) |
| C11—C10—N2—O3 | −7.4 (10) | N1—C4—C5—C6 | 178.5 (6) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.82 | 1.86 | 2.591 (6) | 148 |
| C7—H7···O2i | 0.93 | 2.45 | 3.309 (8) | 154 |
Symmetry code: (i) −x+1, −y+1, z−1/2.
Funding Statement
This work was funded by University Grants Commission grant . University of Science and Technology, Ibb Branch, Yemen grant .
References
- Alarcón, S. H., Pagani, D., Bacigalupo, J. & Olivieri, A. C. (1999). J. Mol. Struct. 475, 233–240.
- Aldoshin, S. M., Atovmyan, L. O. & Ponomarev, V. I. (1984). Khim. Fiz. 3, 787–791.
- Carletta, A., Spinelli, F., d’Agostino, S., Ventura, B., Chierotti, M. R., Gobetto, R., Wouters, J. & Grepioni, F. (2017). Chem. Eur. J. 23, 5317–5329. [DOI] [PubMed]
- Desai, S. B., Desai, P. B. & Desai, K. R. (2001). Heterocycl. Commun. 7, 83–90.
- Faizi, M. S. H., Ahmad, M., Kapshuk, A. A. & Golenya, I. A. (2017a). Acta Cryst. E73, 38–40. [DOI] [PMC free article] [PubMed]
- Faizi, M. S. H., Alam, M. J., Haque, A., Ahmad, S., Shahid, M. & Ahmad, M. (2018). J. Mol. Struct. 1156, 457–464.
- Faizi, M. S. H., Dege, N., Haque, A., Kalibabchuk, V. A. & Cemberci, M. (2017b). Acta Cryst. E73, 96–98. [DOI] [PMC free article] [PubMed]
- Faizi, M. S. H., Gupta, S., Mohan, V. K., Jain, K. V. & Sen, P. (2016). Sens. Actuators B Chem. 222, 15–20.
- Faizi, M. S. H., Osório, F. A. P. & Valverde, C. (2020). J. Mol. Struct. 1210, 128039–464.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Filarowski, A. (2005). J. Phys. Org. Chem. 18, 686–698.
- Filipenko, O. S., Ponomarev, V. I., Bolotin, B. M. & Atovmyan, L. O. (1983). Kristallografiya, 28, 889–895.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hadjoudis, E. & Mavridis, I. M. (2004). Chem. Soc. Rev. 33, 579–588. [DOI] [PubMed]
- Hirshfeld, H. L. (1977). Theor. Chim. Acta, 44, 129–138.
- Iwan, A., Kaczmarczyk, B., Janeczek, H., Sek, D. & Ostrowski, S. (2007). Spectrochim. Acta A Mol. Biomol. Spectrosc. 66, 1030–1041. [DOI] [PubMed]
- Karia, F. D. & Parsania, P. H. (1999). Asian J. Chem. 11, 991–995.
- Kaştaş, G., Albayrak, C., Odabaşoğlu, M. & Frank, R. (2012). Spectrochim. Acta A Mol. Biomol. Spectrosc. 94, 200–204. [DOI] [PubMed]
- Kumar, M., Kumar, A., Faizi, M. S. H., Kumar, S., Singh, M. K., Sahu, S. K., Kishor, S. & John, R. P. (2018). Sens. Actuators B Chem. 260, 888–899.
- Kumar, S., Hansda, A., Chandra, A., Kumar, A., Kumar, M., Sithambaresan, M., Faizi, M. S. H., Kumar, V. & John, R. P. (2017). Polyhedron, 134, 11–21.
- Mukherjee, P., Das, A., Faizi, M. S. H. & Sen, P. (2018). Chemistry Select, 3, 3787–3796.
- Ojala, C. R., Ojala, W. H., Gleason, W. B. & Britton, D. (1999). J. Chem. Crystallogr. 29, 27–32.
- Ozeryanskii, V. A., Pozharskii, A. F., Schilf, W., Kamieński, B., Sawka-Dobrowolska, W., Sobczyk, L. & Grech, E. (2006). Eur. J. Org. Chem. pp. 782–790.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Pizzala, H., Carles, M., Stone, W. E. E. & Thevand, A. (2000). J. Chem. Soc. Perkin Trans. 2, pp. 935–939.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Singh, W. M. & Dash, B. C. (1988). Pesticides, 22, 33–37.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spangenberg, A., Sliwa, M., Métivier, R., Dagnélie, R., Brosseau, A., Nakatani, K., Pansu, R. & Malfant, I. (2007). J. Phys. Org. Chem. 20, 992–997.
- Stoe & Cie (2002). X-AREA, X-RED32 and X-SHAPE. Stoe & Cie GmbH, Darmstadt, Germany.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. The University of Western Australia.
- Venkatesan, P., Thamotharan, S., Ilangovan, A., Liang, H. & Sundius, T. (2016). Spectrochim. Acta Part A, 153, 625–636. [DOI] [PubMed]
- Wozniak, K., He, H., Klinowski, J., Jones, W., Dziembowska, T. & Grech, E. (1995). J. Chem. Soc. Faraday Trans. 91, 7–85.
- Yan, X. F., Xiao, H. M., Gong, X. D. & Ju, X. H. (2006). J. Mol. Struct. Theochem, 764, 141–148.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020008191/pk2635sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020008191/pk2635Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020008191/pk2635Isup3.cml
CCDC reference: 1922980
Additional supporting information: crystallographic information; 3D view; checkCIF report