Abstract
Purpose:
Response criteria of glioblastoma after chemoradiation do not account for metabolic changes that occur after treatment. The purpose of this study is to evaluate the utility of positron emission tomography (PET) imaging with C11 methionine (MET) (MET-PET) for detecting changes that occur after chemoradiation therapy and the value of molecular biomarkers for predicting the magnitude of metabolic response.
Methods and Materials:
Patients with newly diagnosed glioblastoma undergoing standard chemoradiation treatment were enrolled in this prospective imaging study, with MET-PET scan performed within 3 days after surgical resection and again at 4 weeks after completion of chemoradiation. Near contemporaneous contrast-enhanced magnetic resonance imaging was performed within 2 weeks of each MET-PET scan. MET-PET imaging was analyzed for maximum standardized uptake value (SUV), SUVmean, and SUVvolume on a multimodality workstation.
Results:
A total of 18 patients underwent baseline postoperative MET-PET imaging, 14 of whom underwent postchemoradiation MET-PET imaging. Among those who showed residual MET-avid disease on immediate postoperative MET-PET scans and underwent postchemoradiation MET-PET imaging (n = 10), mean ΔSUVmax was −40% (range −100% to 0%), mean ΔSUVmean was −35% (range −100% to 0%), and mean ΔSUV volume was −64% (range −100% to 0%). The Δtumor/brain reference was −40% (range −100% to 0%) using SUVmax and −35% (range −100% to 0%) using SUVmean. In contrast, none of the T2-weighted images on contrast-enhanced magnetic resonance imaging showed a >25% reduction in abnormal T2/fluid-attenuated inversion recovery signal on visual assessment. ΔSUVmax, ΔSUVmean, and ΔSUVvolume correlated with O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status (P = .01), but not with epidermal growth factor receptor or c-MET amplification status. All patients were IDH-1 wildtype.
Conclusions:
MET-PET scanning shows a significant decrease in metabolic signal at 1 month after chemoradiation compared with the immediate postoperative period, even when T2/fluid-attenuated inversion recovery changed little. MGMT promoter methylation status further predicts differential metabolic responses. MET-PET may be a useful tool for delineation of radiation targets and assessment of response.
Summary
C11 methionine positron emission tomography may be a useful tool for delineation of radiation targets and assessment of response in glioblastoma. C11 methionine positron emission tomography scanning can show a significant decrease in extent of methionine-avid glioblastoma at 1 month after completion of chemoradiation compared with the immediate postoperative period, even when T2/ fluid-attenuated inversion recovery changes little. O6-methylguanine-DNA methyltransferase promoter methylation status further predicts differential metabolic responses.
Introduction
Contrast-enhanced magnetic resonance imaging (ceMRI) is the standard modality for diagnosis and monitoring of glioblastoma; however, it is limited by an inability to show the full extent of tumor, irrespective of the burden of macroscopic disease at pathology and variable biologic activity (1). Emerging molecular imaging techniques offer the potential to assess metabolic tumor status, which is increasingly important because molecular profiling and individualized risk-adapted treatment considerations have become the standard of care. Positron emission tomography (PET) imaging with 11C methionine (MET) (MET-PET) has shown promise for delineating tumor margins, localizing residual tumor sites, and differentiating residual tumor from reactive changes (2-5). We hypothesize that serial MET-PET imaging can be used as an early time point biomarker to predict treatment response not evident on ceMRI. We further assert that molecular biomarkers previously established to be predictive of outcome, including O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation, epidermal growth factor receptor (EGFR) amplification, c-MET amplification status, and IDH-1 gene mutations can potentially predict the level of metabolic response on 11C MET-PET (6-9).
Methods and Materials
Adult patients with newly diagnosed glioblastoma and intent to undergo standard chemotherapy with temozolomide and fractionated radiation therapy were enrolled on this prospective, institutional review board—approved study.
A baseline postoperative MET-PET scan was performed within 3 days after intended maximal surgical resection of the primary glioblastoma, and a second postchemoradiation MET-PET scan was performed within 4 weeks after standard radiation therapy and chemotherapy.
For PET scanning, 10 ± 2 mCi of 11C MET was injected. After approximately 15 minutes, imaging was performed for 15 minutes on a Seimens Biograph 64 PET/ CT scanner.
CeMRI imaging, performed within 2 weeks of MET-PET scanning, included sagittal T1, axial T2, axial T2 fluid-attenuated inversion recovery (FLAIR), axial T1, and triplanar T1 weighted postcontrast images.
MET-PET images were analyzed by using a dedicated multimodality viewing workstation (Siemens SyngoVia 24B). PET and magnetic resonance imaging (MRI) data sets were registered manually using rigid anatomic landmarks. A well-defined auto-contouring threshold volume of interest (VOI) was drawn around any abnormal 11C MET uptake suspicious for residual glioma in the resection cavity, as described by Kracht et al (8). We recorded tumor mean standardized uptake value (SUV), SUVmax, and SUVvolume from the threshold VOI. A reference VOI of at least 3 cm3 was drawn in a region of contralateral cerebrum without correlative enhancement and/or T2/FLAIR abnormalities on ceMRI; SUVmean_reference was recorded. We subsequently calculated tumor/reference brain ratios. Kracht et al proposed a tumor/reference brain threshold of 1.3 as producing optimal differentiation between tumor uptake and nontumor tissue changes for their data set. This threshold appears applicable to our data set because the mean SUVmean of postoperative uptake in our cohort was 1.3 (as determined by drawing a manual VOI around the craniotomy site superficial to the resection cavity and exclusive of any abnormal brain parenchyma) whereas the mean SUVmean_reference was 1.0.
On ceMRI, residual enhancing tumor was defined as any region of nodular enhancement. At the early postoperative time point, categories of resection are as follows: gross total resection (GTR), defined as the absence of gross residual enhancing tumor; subtotal resection (STR), defined as the presence of residual enhancing tumor; and biopsy only, defined as tissue sampling with no apparent removal of additional enhancing tissue. Potential nonenhancing tumor was defined as peritumoral regions of T2/FLAIR signal abnormality. Because the extent of nonenhancing tumor is often diffuse and difficult to measure precisely, change in nonenhancing tumor burden was estimated visually as <25%, between 25% and 50%, and >50% on T2/FLAIR weighted images.
Molecular biomarker data were obtained from histopathology reports of the primary tumor at initial biopsy or resection.
Radiation therapy to a total dose of 60 Gy in 2-Gy fractions was initiated within 6 weeks after surgery. Temozolomide was given concurrently and sequentially, per Stupp et al (10).
Statistical analysis was performed using SAS 9.4 software (Cary, NC). Comparison of the postchemoradiation versus baseline SUV and tumor/brain reference data was performed with the Mann-Whitney U test. Statistically significant differences in metabolic response indicated by molecular biomarker status was determined by Wilcoxon rank sum testing.
Results
Eighteen patients were enrolled (Table 1). All study patients were IDH-1 wildtype.
Table 1.
Patient characteristics
| Characteristics | No (%) |
|---|---|
| Demographic characteristics | |
| Total patients | 18 |
| Grade 4 glioblastoma | 18 (100%) |
| Age, y | 55 (range 48-70) |
| Sex | |
| Male | 10 (56%) |
| Female | 8 (44%) |
| Evidence for enhancing tumor on preoperative ceMRI | 18 (100%) |
| MET-PET scans | |
| Scan 1: Postoperative | 18 (100%) |
| Scan 2: Postchemoradiation | 14 (78%) |
| Tumor characteristics | |
| MGMT hypermethylation status | |
| Methylated | 8 (44%) |
| Not methylated | 8 (44%) |
| Unknown | 2 (12%) |
| EGFR amplification | |
| Positive | 5 (28%) |
| Negative | 11 (61%) |
| Unknown | 2 (12%) |
| MET amplification | |
| Positive | 2 (12%) |
| Negative | 14 (78%) |
| Unknown | 2 (12%) |
| IDH-1 gene mutation status | |
| Wildtype | 18 (100%) |
| Mutated | 0 (0%) |
Abbreviations: ceMRI = contrast-enhanced magnetic resonance imaging; EGFR = epidermal growth factor receptor; IDH-1 = isocitrate dehydrogenase 1; MET = methionine; MGMT = O6-methylguanine-DNA methyltransferase; PET = positron emission tomography.
Postoperative MRI results are shown in Table 2. All patients had T2/FLAIR signal abnormalities on the early postoperative ceMRI that were stable (<25% change) on postchemoradiation ceMRI, irrespective of the status of nodular enhancement.
Table 2.
Summary of baseline postsurgical ceMRI and 11C MET-PET status and reduction of MET-avid disease at postchemoradiation
| MET-PET | 11C MET- PET (−) |
11C MET- PET (+) |
Total |
|---|---|---|---|
| Postoperative | |||
| Total scans | 6 | 12 | 18 |
| GTR by ceMRI | 5 | 2 | 7 (39%) |
| STR by ceMRI | 1 | 8 | 9 (50%) |
| Biopsy only by ceMRI | 0 | 2 | 2 (11%) |
| SUVmax (range) | N/A | 6.9 (3.2-8.6) | |
| SUVmean (Range) | N/A | 2.6 (1.9-4.5) | |
| SUV volume (range) | N/A | 3.0 (1.0-128) | |
| Postchemo radiation | |||
| Total scans | 7 | 7 | 14 |
| ΔSUVmax (range) | N/A | 40% (0%-100%) | N/A |
| ΔSUVmean (range) | N/A | 35% (0%-100%) | N/A |
| ΔSUVvolume (range) | N/A | 64% (0%-100%) | N/A |
| ΔSUVmax/brain reference (range) | N/A | 40% (0%-100%) | N/A |
| ΔSUVmean/brain reference (range) | N/A | 35% (0%-100%) | N/A |
Abbreviations: ceMRI = contrast-enhanced magnetic resonance imaging; GTR = gross total resection; MET = methionine; N/A = not available; PET = positron emission tomogrpahy; STR = subtotal resection; SUV = standardized uptake value.
Postoperative MET-PET results are shown in Table 2. Postoperative MET-PET and ceMRI scans were concordant in 15 patients (83%) and discordant in 3 patients (17%). In 2 patients, focal and intensely increased 11C MET uptake suspicious for tumor remnant was observed when ceMRI showed no enhancing tumor (Fig. 1). Areas of increased 11C MET uptake represented a subset of T2/FLAIR signal abnormalities and therefore would have been included within standard radiation therapy treatment volumes. In a third patient, 11C MET-PET was negative when postoperative MRI showed subtotal resection.
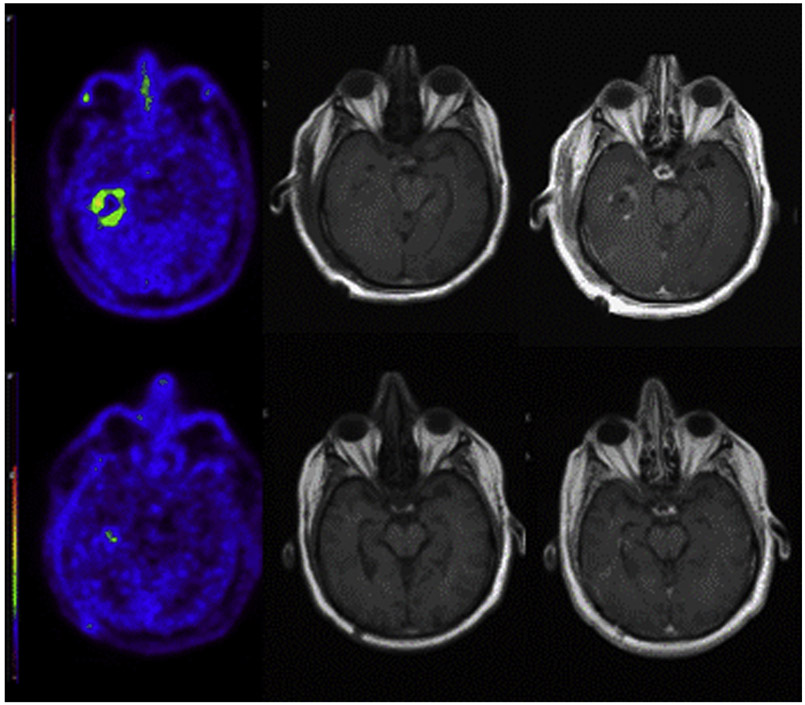
Fig. 1.
Postoperative positron emission tomography imaging with C11 methionine and contrast-enhanced magnetic resonance imaging showing intensely abnormal uptake (left) in right parietal lobule with only minimal contrast enhancement (T1 precontrast, left center; T1 postcontrast, right center) but with T2/fluid-attenuated inversion recovery signal abnormality (right).
Four patients did not complete a postchemoradiation MET-PET scan because of intercurrent illness (n = 2), non—health-related logistical challenges preventing MET-PET scanning (n = 1), and transfer of care to a different institution (n = 1).
All 5 patients considered as having GTR by ceMRI and a single patient considered as having STR by ceMRI with negative 11C MET-PET scans at baseline underwent subsequent MET-PET imaging that was also negative. Eight patients considered to have STR or biopsy only by ceMRI with MET-avid disease at baseline underwent follow-up MET-PET imaging, which showed reduced uptake and extent of MET uptake abnormality (Fig. 2, Table 2).
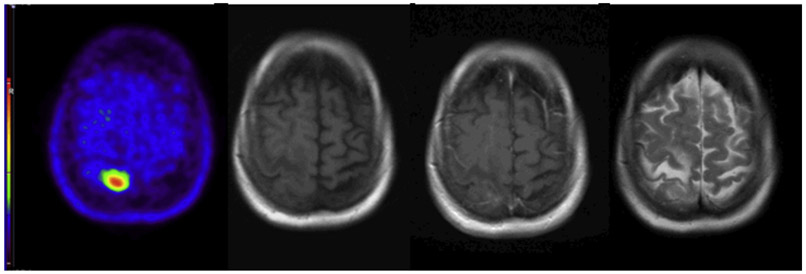
Fig. 2.
Postoperative scans (upper row) show residual intense uptake abnormality and abnormal enhancement in the right temporal lobe that is nearly resolved on postchemoradiation scans (bottom row). T2/fluid-attenuated inversion recovery signal abnormality is essentially unchanged at follow-up (not shown). Positron emission tomography imaging with C11 methionine, left; precontrast T1, center, postcontrast T1, right.
Molecular profiling status is shown in Table 1. MGMT methylation status was correlated significantly with greater reductions in ΔSUVmax, ΔSUVmean, ΔSUVvolume, and tumor/brain reference ratio (P = .01) (Fig. 3). In contrast, EGFR and c-MET status did not correlate significantly with any quantitative metabolic parameters.
Fig. 3.
Treatment response by MGMT promoter methylation status with standard error bars. Abbreviations: MGMT = O6-methylguanine-DNA methyltransferase; SUV = standardized uptake value.
Discussion
We found that MGMT promoter—hypermethylated tumors have a higher magnitude of metabolic response compared with MGMT promoter—unmethylated tumors. This result is not surprising because MGMT methylation status is known to be a strong independent predictor of improved survival in glioblastoma. We were not able to demonstrate a relationship between EGFR or c-MET amplification status and response; however, our sample size was small, and the status of these markers was weighted toward negativity in a larger subset of our cohort. All of the tumors in our cohort were IDH-1 wild-type, which is not surprising because IDH-1 mutations are uncommon in de novo high-grade glioblastomas and in an older study population.
Our data support MET-PET imaging as a platform for classifying glioblastoma into metabolic phenotypes predictive of response to chemoradiation therapy. MET-PET imaging successfully demonstrated treatment response in the majority (all except 1) of patients with evidence of 11C MET—avid residual disease postoperatively, in the setting of little change in nonenhancing tumor burden by ceMRI. This suggests that MET-PET imaging is potentially useful for assessing the degree of response and defining subsets of complete and partial response, even among those broadly considered to be responders by standard clinical and ceMRI imaging criteria. Long-term follow-up studies with clinical correlation will be required to validate this hypothesis.
Footnotes
This study was funded by a grant through the Federal Proton Share Program.
Conflict of interest: none.
References
- 1.Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Br J Radiol 2011;84(Spec No 2):S107–S111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosu AL, Weber WA, Riedel E, et al. L-(methyl-11c) methionine positron emission tomography for target delineation in resected high-grade gliomas before radiotherapy. Int J Radiat Oncol Biol Phys 2005; 63:64–74. [DOI] [PubMed] [Google Scholar]
- 3.Lee IH, Piert M, Gomez-Hassan D, et al. Association of 11C-methionine PET uptake with site of failure after concurrent temozolomide and radiation for primary glioblastoma multiforme. Int J Radiat Oncol Biol Phys 2009;73:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miwa K, Shinoda J, Yano H, et al. Discrepancy between lesion distributions on methionine PET and MR images in patients with glioblastoma multiforme: Insight from a PET and MR fusion image study. J Neurol Neurosurg Psychiatry 2004;75: 1457–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terakawa Y, Tsuyuguchi N, Iwai Y, et al. Diagnostic accuracy of 11C-methionine PET for differentiation of recurrent brain tumors from radiation necrosis after radiotherapy. J Nucl Med 2008;49: 694–699. [DOI] [PubMed] [Google Scholar]
- 6.Frederick L, Wang XY, Eley G, et al. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res 2000;60:1383–1387. [PubMed] [Google Scholar]
- 7.Kong DS, Song SY, Kim DH, et al. Prognostic significance of c-met expression in glioblastomas. Cancer 2009;115:140–148. [DOI] [PubMed] [Google Scholar]
- 8.Kracht LW, Miletic H, Busch S, et al. Delineation of brain tumor extent with [11c]L-methionine positron emission tomography: Local comparison with stereotactic histopathology. Clin Cancer Res 2004; 10:7163–7170. [DOI] [PubMed] [Google Scholar]
- 9.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–996. [DOI] [PubMed] [Google Scholar]