Summary
Though the temporal precision of neural computation has been studied intensively, a data-driven determination of this precision remains a fundamental challenge. Reproducible spike patterns may be obscured on single trials by uncontrolled temporal variability in behavior and cognition, and may not be time locked to measurable signatures in behavior or local field potentials (LFP). To overcome these challenges, we describe a general-purpose time warping framework that reveals precise spike-time patterns in an unsupervised manner, even when these patterns are decoupled from behavior or are temporally stretched across single trials. We demonstrate this method across diverse systems: cued reaching in nonhuman primates, motor sequence production in rats, and olfaction in mice. This approach flexibly uncovers diverse dynamical firing patterns, including pulsatile responses to behavioral events, LFP-aligned oscillatory spiking, and even unanticipated patterns, like 7 Hz oscillations in rat motor cortex that are not time-locked to measured behaviors or LFP.
Introduction
The role of spike time precision in neural computation has been widely examined from both experimental and theoretical perspectives, engendering intense debates in systems neuroscience over the last several decades (Softky and Koch 1993; London et al. 2010; Bruno 2011; Brette 2015; Denève and Machens 2016). Assessing the degree of temporal precision in multi-neuronal spike trains is challenging since variations in behavioral and cognitive variables can mask precise spike time patterns. For example, the temporal precision of olfactory coding may be underestimated by factors of two to four when spike times are aligned to stimulus delivery instead of inhalation onset (Shusterman et al. 2011; Cury and Uchida 2010; Shusterman et al. 2018).
Thus, experimental estimates of spike time precision hinge on the choice of an alignment point, which defines the origin of the time axis on each trial. This choice can often be challenging and subjective. Even relatively simple tasks often involve a sequence of stimuli, actions, and rewards that occur with varying latencies, thus presenting multiple choices for temporal alignment. Moreover, in addition to choosing an origin of time, we must also choose its units. Should spike times be measured in absolute clock time relative to some measured event, or in units of fractional time between two events? Could any one of these choices unmask spike-timing precision that is otherwise invisible? These questions are most challenging to answer in systems far from the sensory or motor periphery, where neural responses may not be locked to any measurable behavior, and instead reflect internal decisions or changes-of-mind.
Past studies have addressed these challenges in a number of ways: grouping trials together with similar durations before averaging spike counts (Murakami et al. 2014; Starkweather et al. 2017; Wang et al. 2018), manually stretching or compressing time units between measured task events (Leonardo and Fee 2005; Shusterman et al. 2011; Kobak et al. 2016; Aronov et al. 2017), or repeating statistical analyses around different choices of alignment point (Feierstein et al. 2006; Harvey et al. 2012; Jazayeri and Shadlen 2015; Shushruth et al. 2018).
These complications, and the diversity of heuristic approaches used to address them, underscore a need for statistical frameworks to assess the temporal precision of neural computation. Of particular interest are unsupervised statistical methods that reveal precise patterns in multi-neuronal spike trains without reference to behavioral measurements. Such methods would be broadly applicable, as they make few restrictive assumptions about the experimental design. Furthermore, by moving beyond pre-conceived alignments designed by human experts, these data-driven methods may discover novel and unexpected results.
While time series and image alignment methods are a well-studied topic in signal processing (Marron et al. 2015; Pnevmatikakis and Giovannucci 2017), these techniques have rarely been applied to large-scale neural recordings (but see recent work by Poole et al. 2017; Lawlor et al. 2018; Duncker and Sahani 2018). Neuroscientists have historically utilized simple alignment operations—namely, translating (Baker and Gerstein 2001; Ventura 2004) and potentially stretching/compressing activity traces between pairs of behavioral events (Shusterman et al. 2011; Leonardo and Fee 2005; Perez et al. 2013; Kobak et al. 2016). In contrast, popular statistical methods, such as Dynamic Time Warping (DTW; Berndt and Clifford 1994), allow signals to be non-uniformly compressed and dilated on each trial. While such nonlinear warping models can be useful, we demonstrate that they can be difficult to interpret and sensitive to the high level of noise that is typical of neural data.
To identify interpretable alignments for high-dimensional spike trains, we developed a framework for linear and piecewise linear time warping that encompasses existing human-annotated procedures (Leonardo and Fee 2005; Kobak et al. 2016). We applied these methods to multielectrode recordings collected from three experiments spanning different animal models (rodents and primates), brain regions (olfactory and motor cortex), and behavioral tasks (sensation and motor production). In each case, time warping revealed precise spike patterns that were imperceptible in the raw data. Moreover, some of these results were not easily captured by any common behavioral alignment. For example, in rodents performing a motor timing task, we uncovered ~7 Hz oscillations in spike times that were not aligned to the LFP or any measured behavior.
Results
Time Warping Framework
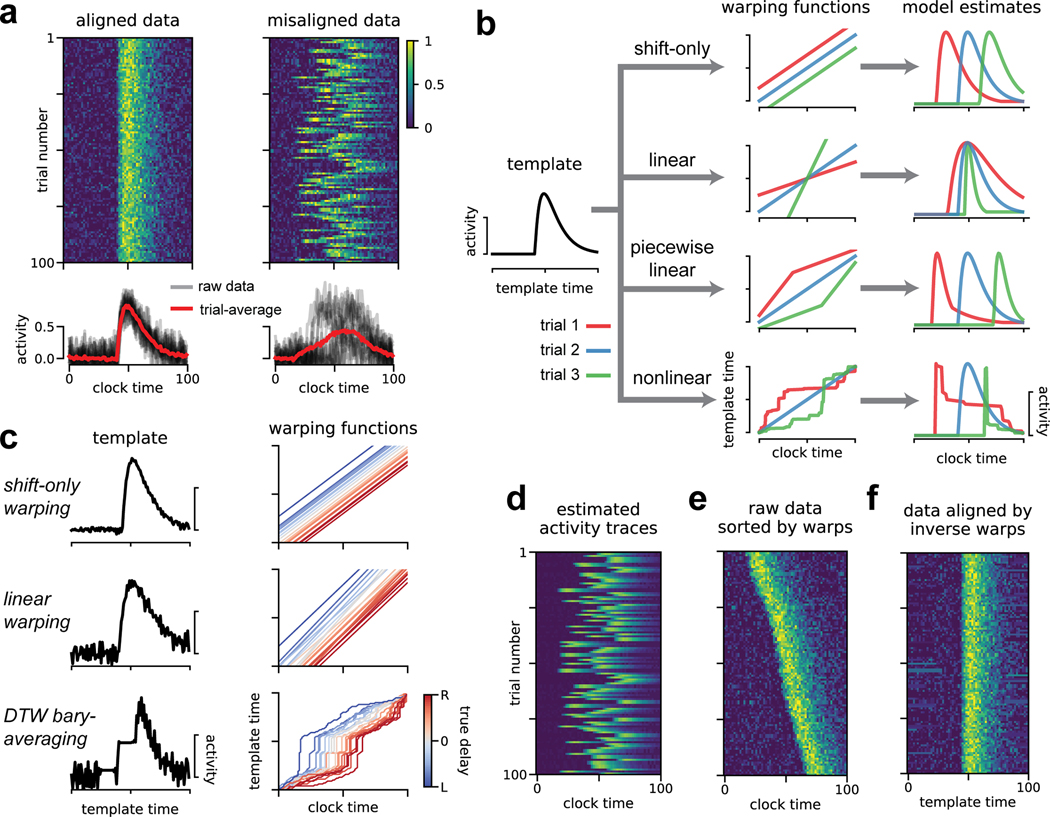
Our ultimate goal is to identify firing patterns that reliably occur on a trial-by-trial basis. If these activity patterns are tightly time-locked to a sensory or behavioral event, then we can characterize the neural response by averaging over trials. This is illustrated in Figure 1a (left), which shows 100 trials of simulated neural activity with additive Gaussian noise. The average activity trace (red; bottom) extracts the neural response from noisy single-trial instantiations. This simulated example loosely resembles calcium fluorescence traces, but the methods we describe can be flexibly applied to any multi-dimensional time series including spike trains, fMRI data, or LFP traces.
Fig 1. Illustration of time warping models.
(A) Single-neuron synthetic data from 100 trials. When data are aligned (top left) the trial-average provides a good description of activity (bottom left). When the data are misaligned by introducing jitter (right), the trial-average does not produce a useful description (bottom right). (B) Time warping models estimate a template (black line, left) that is transformed on a trial-by-trial basis by warping functions (middle column) producing single-trial estimates of neural activity (right column). Colored lines represent warping functions and estimated firing rates on three example trials. Top row illustrates shifting the template activity (shift-only warping), second row illustrates stretching and compressing the template in addition to shifting (linear warping), third row illustrates piecewise-linear warping with two line segments, the bottom row illustrates a fully nonlinear warping. (C) Results of shift-only warping (top), linear warping (middle), and nonlinear warping (bottom; Petitjean et al. 2011) fit to the synthetic data from panel A, right. For each model, we show the warping templates (left, black lines) and warping functions (right, lines colored by ground truth shift; red indicates rightward shift, blue indicates leftward shift). (D) The predicted firing rates of shift-only warping provide a denoised trial-by-trial estimate of neural activity. (E) The raw data sorted by the learned per-trial shift parameter. (F) Data aligned by the shift-only model; the per-trial shift is applied in the opposite direction to the raw data.
More formally, if N neurons are measured at T timepoints over K trials, the trial-average is given by:
| (1) |
Here, Xk is a T × N matrix denoting the activity of all neurons on trial k. In the context of spiking data, each column of corresponds to a peri-stimulus time histogram (PSTH) of a recorded neuron. Trial averaging is also a common step in population-level statistical analyses (Kobak et al. 2016).
Despite its widespread use, trial averaging can produce inaccurate and misleading results when neural activity is misaligned across trials. For example, introducing a random shift to each simulated trial produces a less informative result (Fig 1a, right). Such jitter commonly arises in practice, leading many research groups to develop custom-built alignment procedures. For example, in songbirds it is common to manually segment and cluster song syllables and warp intervening spike times on a per-syllable basis (Leonardo and Fee 2005). In olfaction, detailed measurements and fluid dynamics modeling of the sniff cycle have been pursued to understand the accuracy of sensory responses (Shusterman et al. 2011; Shusterman et al. 2018).
Time warping methods address these challenges through a data-driven, statistical approach. The key idea is to fit a response template time series that is shifted and stretched—i.e., warped—on a trial-by-trial basis to match the data. The response template, denoted , is an T × N matrix of activity traces that captures the average activity across trials after correcting for variability in timing.
The time axis of the response template is transformed by a warping function on each trial. Formally, we denote the warping function for trial k as ωk(t); this function maps each timebin t (clock time) to a new index ωk(t) (template time). If ωk(t) is an integer between 1 and T, then the warping transformation for every neuron n on trial k amounts to the transformation . If ωk(t) is not an integer then time warping is implemented by linear interpolation (see Methods). Note that this model assumes all recorded neurons share the same warping function on a trial-by-trial basis, though this assumption could be relaxed by future work.
Figure 1b illustrates how different classes of warping functions account for single-trial variability in timing. We focus on three main model classes: shift-only warping, linear warping, and piecewise linear warping (Fig 1b, top three models). Shift-only warping is the simplest model: the warping functions are constrained to be linear with slope equal to one, and only a single parameter (the y-intercept of ωk(t)) is fit on each trial. As its name suggests, the shift-only model can only account for trial-to-trial differences in response latency. In contrast, a linear warping model, which fits the slope in addition to the intercept of ωk(t), can account for variable latencies as well as uniform stretches and compressions of the response template. A piecewise linear warping model adds further complexity by adding one or more knots (points where the slope of ωk(t) can change). Most generally, nonlinear warping functions may be used, which non-uniformly stretch and compress portions of the template on each trial (Fig 1b, bottom). In all cases, we constrain the time warping functions to be monotonically increasing. Loosely speaking, this ensures that the model cannot go backwards in time while making a prediction. This constraint also implies that the warping functions are invertible, which we later exploit to align spike times across trials.
The model parameters are fit to minimize reconstruction error over all neurons, trials, and timepoints. For simplicity, we chose the mean squared error to quantify model performance. Ignoring the interpolation step of time warping for the sake of clarity (see Methods), the objective function is:
| (2) |
This expression is minimized with respect to the warping functions on each trial, ωk(t), and the response template, . Other loss functions may be substituted for the mean squared error. In particular, a loss function based on Poisson noise is popular in neural modeling (Paninski 2004) and our accompanying Python package supports this option.
For illustration, we fit shift-only, linear, and nonlinear warping models to the misaligned synthetic data shown Figure 1a. By design, the shift-only model is sufficient to capture the ground truth variability. As expected, this model identifies a highly accurate template firing pattern (Fig 1c, top left), along with warping functions that tightly correlate with the ground truth delay (Fig 1c, top right). The linear warping model is a gentle extension of the shift-only warping model, which only introduces one additional parameter on each trial—the slope of each warping function. Yet, even this minor extension produces a slightly worse estimate, due to overfitting (Fig 1c, middle).
To demonstrate a more severe case of overfitting, we fit a nonlinear warping model using Dynamic Time Warping (DTW; Berndt and Clifford 1994), combined with a standard barycenter averaging procedure (Petitjean et al. 2011). This method can be highly effective on datasets with low levels of noise and complex temporal deformations. However, as we will soon see, neural datasets often exhibit the opposite—high levels of noise and simple temporal deformations. In this regime, DTW barycenter averaging identifies a noisy template (Fig 1c, bottom left), and unnecessarily complex warping paths (Fig 1c, bottom right).
These results demonstrate that time warping models, particularly with complex warping functions, are susceptible to overfitting. We adopted four strategies to mitigate this possibility. First, as mentioned above, we assume that each trial’s warping function is shared across neurons, thus mitigating the possibility of overfitting to noise in any single neuron. Second, as illustrated by the progression of models in Figure 1c, we always compare the estimates of complex warping models (e.g. with piecewise linear warping functions) to the performance of simpler models (e.g. shift-only warping). Third, we include a roughness penalty on the template, which directly encourages the model to identify smooth firing rates. Fourth, we place a penalty on the area between each warping function and the identity line, which penalizes the magnitude of warping on each trial. We include these roughness and warp-magnitude penalties in subsequent results, but show the results of unregularized time warping in Figure 1c for the sake of illustration.
These models enable several strategies for visualizing and understanding neural data. First, one can directly inspect the model parameters (Fig 1c): the response template for each neuron captures the shape of the neural response, while the warping functions capture trial-to-trial variability in timing. Second, one can view the model prediction as a denoised estimate of firing rates on a single-trial basis (Fig 1d). Third, one can re-sort the trials by the slope or the intercept of the warping function, producing a multi-trial raster plot that is easier to visually digest (Fig 1e). Finally, one can invert the warping functions on each trial to transform the raw data into an aligned time domain (Fig 1f). This alignment procedure simply entails plotting each activity trace as a function of instead of raw clock time, t. Intuitively, this amounts to reversing the flow diagram shown Figure 1b, which is possible as long as the warping functions are monotonically increasing and thus invertible.
Extraction of precise, ground truth spike patterns on synthetic data
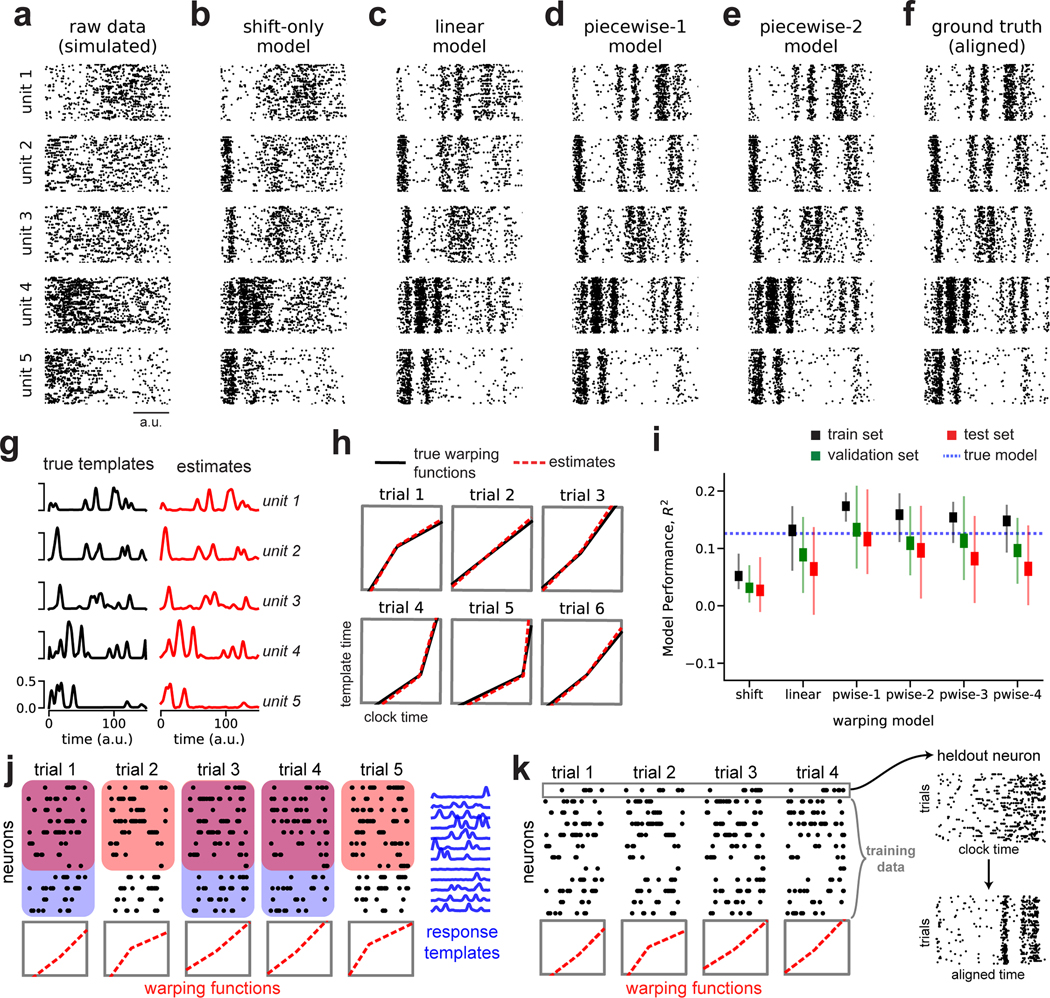
Before proceeding to biological data, we examined a more challenging simulated dataset consisting of spike trains from N = 5 neurons, T = 150 timebins, and K = 75 trials. On each trial, the neural firing rates were time warped by randomized piecewise linear functions with one knot (the “ground truth” model). This resulted in spike trains that appear highly variable in their raw form (Fig 2a).
Fig 2. Recovery of ground truth warping functions in synthetic data.
(A) Synthetic spiking data from N = 5 units, T = 150 timebins, and K = 75 trials. Data were simulated from a ground truth model with piecewise linear warping functions with 1 knot. (B) Data re-aligned by a shift-only warping model. (C) Data re-aligned by a linear warping model. (D) Data re-aligned by a piecewise-linear (1-knot) warping model. (E) Data re-aligned by a piecewise-linear (2-knots) warping model. (F) Data re-aligned by the ground truth model. Note similarity with panels D & E. (G) Ground truth neural response templates (black) and estimated response templates (red) from the piecewise-linear (1-knot) model. Y-axis denotes the probability of spiking in each time bin. (H) Ground truth warping functions (black) on six representative trials, and estimated warping functions (red) from the piecewise-linear (1-knot) model. (I) Model performance (R2) in training, validation, and test partitions of various warping models. Thin lines represent maximum and minimum values; thick lines represent mean ± standard error. Results were computed over 40 randomized cross-validation runs. The horizontal dashed blue line reflects the performance ceiling on held out data achieved by the ground-truth model. (J) Cross-validation procedure (see Methods). On each trial, the warping functions are fit to a subset of neurons (red boxes). For each neuron, the response templates are fit to a subset of the trials (blue boxes). The training set refers to the overlap of these two regions (purple overlap). The validation and test sets are subsets of the remaining data (uncolored white regions). (K) Validation procedure for single-neuron raster plots. The visualized neuron is held out; the warping functions are fit to the remaining neurons and then applied to the held out cell.
Time warping successfully reveals the spike patterns corresponding to the ground truth process. Figures 2b–e show model-aligned spike trains (as in Fig 1f) across warping models of increasing complexity. The patterns evident in the ground truth data (Fig 2f) are partially revealed by shift-only and linear time warping (Fig 2b–c), but these models are too simple to capture the fine-scale temporal structure in the data. A piecewise linear warping model with one knot (piecewise-1 model; Fig 2d) accurately captures these details, and represents a parsimonious and “correct” model since it matches the data generation process. Furthermore, the parameters this model closely matched the ground truth response template (Fig 2g) and warping functions (Fig 2h). Using a piecewise linear model with 2 knots did not result in substantial overfitting and indeed closely matched the result of the piecewise-1 model (Fig 2e).
Identifying a parsimonious warping model is challenging in real-world applications where there is no observable ground truth. To select the appropriate model and regularization strengths we developed a nested cross-validation scheme, which correctly identified the piecewise-1 model as having optimal performance on a held out test set (Fig 2i). This approach fits the neural response templates to a subset of trials (Fig 2j; shaded blue regions), and fits the warping functions to a subset of the neurons (Fig 2j; shaded red regions). We refer to the intersection of these two sets as the training set (Fig 2j; shaded purple regions), while the intersection of the complement sets are used for validation and testing (Fig 2j; unshaded regions). This procedure is then repeated many times with different randomized partitions of the data.
We used the proportion of variance explained (R2) to quantify model performance. Unlike in trial-averaged analyses, R2 values will typically be very low because spike trains exhibit substantial single-trial variability. In the case of this synthetic dataset in Figure 2, the ground truth model achieves an R2 of approximately 0.131 (blue dashed line, Fig 2i). This R2 value, while low due to unexplainable trial-to-trial spiking variability, nevertheless constitutes an upper bound on the R2 value achievable by any model on a held out test set. The piecewise-1 model closely matches this performance, achieving an average R2 of 0.113 on the test set (Fig 2i). However, in real experimental data it is not possible to know how close we are to this performance ceiling. Thus, while R2 can be useful as a relative measure when comparing two different models, the absolute magnitude should be interpreted cautiously.
After using cross-validation to select the number of warping function knots and the regularization parameters, we are often interested in using time warping to visualize single neuron raster plots (as in Fig 2a–f). To further validate these raster plots, we can hold out single neurons, fit the model to the remaining data, and apply the inverse warping functions to align the held out neuron (Fig 2k). All model-aligned raster plots shown in subsequent results are generated using this procedure.
Alignment of olfactory responses to sniff cycle
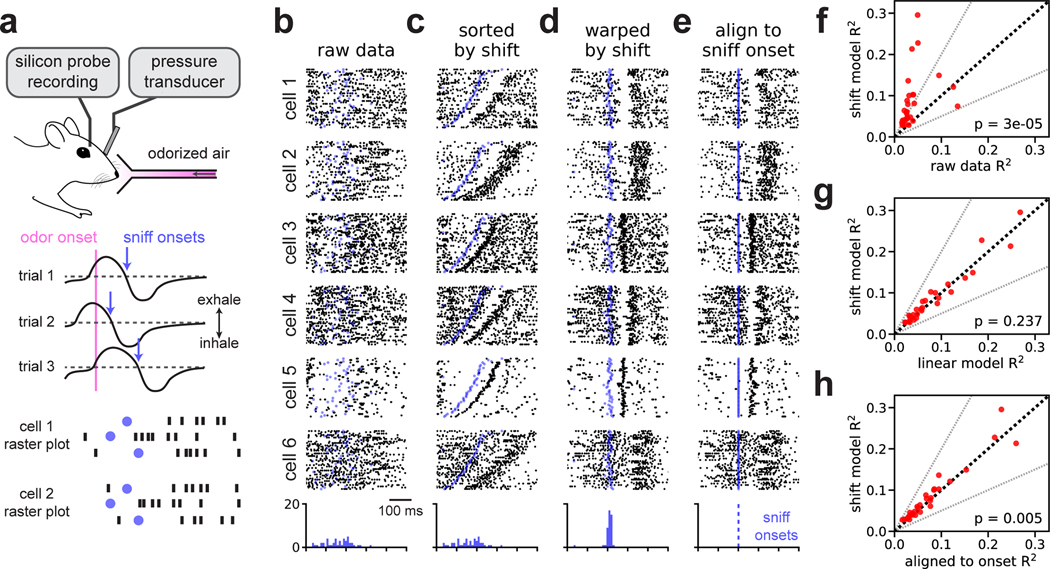
Mitral/tufted cells in the mouse olfactory bulb exhibit variable firing patterns across trials when naïvely aligned to odor delivery (Fig 3a). If spike times are instead aligned to the onset of inhalation neural responses are drastically more reliable (Shusterman et al. 2011).
Fig 3. Time warping of mitral/tufted cell activity recovers sniff-locked activity patterns.
(A) A head-fixed mouse sampled odorized air (α-pinene, 10−2 M). Variability in inhalation onset from trial-to-trial caused jitter in the olfactory response. (B) Spike raster plots for six representative cells over all K = 45 trials with spike times aligned to odor delivery. Black dots denote spike times, blue dots denote sniff onsets. Blue histogram indicates the distribution of sniff onsets. (C) Same as (B), but with trials re-ordered by the magnitude of shift warping. (D) Same as (B), but with the time axis aligned by shift-only warping. (E) Same as (B), but with the time axis aligned to sniff onset. (F) Trial-to-trial reliability (R2) for all N = 30 cells before (x-axis) and after (y-axis) alignment by the shift-only model. Dashed black line indicates the identity line. Dashed grey lines indicate a two-fold increase or decrease in R2. The p-value is computed using a Wilcoxon signed rank test. (G) Same as (F), but comparing the alignment of linear warping to shift-only warping. (H) Same as (F), but comparing the alignment of shift-only warping to sniff onset alignment.
We reasoned that simple time warping models could be used to align mitral/tufted cell activity using purely neural activity, bypassing the need to measure inhalation directly. We tested this hypothesis on a multielectrode recording from N = 30 neurons over K = 45 trials of odor presentation at a fixed concentration (α-pinene, 10−2 M). We experimentally measured intra-nasal pressure to detect sniff onset and offset, but these measurements were not provided to the model and spike times were instead aligned to odor presentation. As expected, this initial alignment strategy produced highly disordered spike rasters (Fig 3b).
We found that a shift-only time warping model captured precise sensory responses from these raw data, as revealed by re-sorting the trials based on the model’s shifts (Fig 3c) or by applying these shifts to align the raw spike times (Fig 3d). Here, as well as in all subsequent results, we use the leave-one-out validation procedure shown in Figure 2k; thus, any temporal structure seen in Figure 3d is unlikely to arise as an artifact of overfitting.
As expected, aligning spike times to inhalation onset revealed similar patterns in these data (Fig 3e). Indeed, the shifts learned by the model correlated very tightly with the onset of sniffing (see blue dots and histograms, Fig 3b–e). These results are nonetheless a useful demonstration, since the model inferred these precise responses from the neural data directly. Furthermore, closer examination suggested that the unsupervised, shift-only model may enjoy slight performance advantages relative to the simple supervised alignment method. For example, when aligned to sniff onset, cells 4 and 5 in Figure 3 exhibit subtle, but perceptible, jitter in their responses (Fig 3e), and this variability is visibly corrected by time warping (compare to Fig 3d).
To explore these effects on a neuron-by-neuron basis, we quantified trial-to-trial variability before and after time warping by computing the coefficient of determination (R2) of the neuron’s PSTH. Larger R2 values imply that the trial-averaged PSTH better approximates single trial firing rates. We fit time warping models while holding out neurons one at a time and computed R2 on the held out neuron before and after warping. Relative to the raw spike times, shift-only time warping improved R2 in nearly all neurons, with many increasing over two-fold (Fig 3f; average 107% increase in R2, geometric mean; Wilcoxon signed rank test, p < 10−4, n = 30). Linear warping did not produce consistent improvements at the level of single neurons (Fig 3g; average 6.6% decrease in R2, geometric mean; Wilcoxon signed rank test, p = 0.24, n = 30). Relative to sniff onset alignment, shift-only time warping improved the R2 criterion mildly (Fig 3h; average 11% increase in R2, geometric mean; Wilcoxon signed rank test, p = 0.005, n = 30). Overall, we conclude that timing variability in these olfactory responses is well-described by a per-trial shift. Further, the optimal shift correlates tightly with, but may not coincide exactly with, sniff onset (Shusterman et al. 2018).
Alignment of motor cortex dynamics during reaching in nonhuman primates
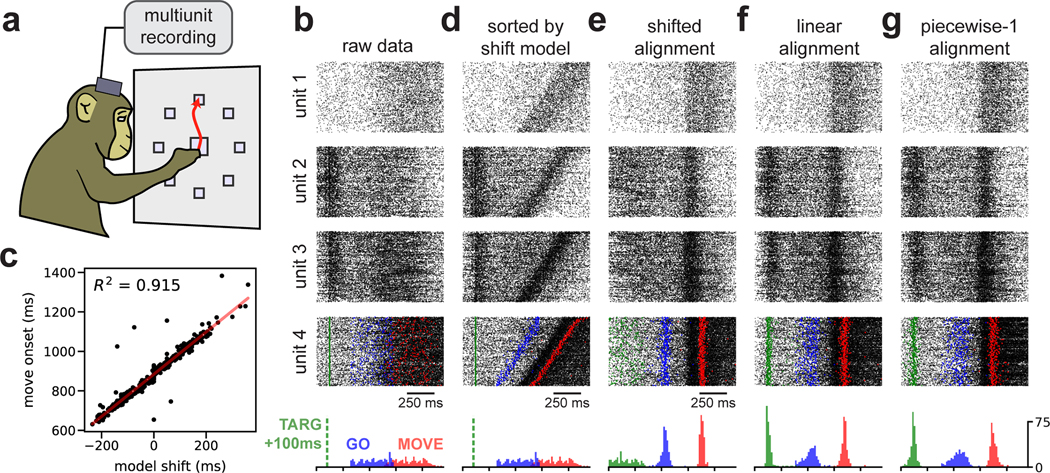
Neural dynamics underlying motor control exhibit variable time courses due to differences in reaction times and muscle kinematics. To investigate the benefits of time warping in this setting, we examined data from a canonical reaching experiment in a nonhuman primate (Fig 4a). On each trial, the subject (Monkey J) moved its arm to one of several target locations after a delay period that randomly varied between 300 and 700 ms. In addition to this inherent timing variability due to task design, the monkey exhibited variable reaction time ranging from 293–442 ms (5th and 95th percentiles). We limited our analysis to upward reaches (90° from center) with the target placed at 40, 80, or 120 cm from the center. We observed similar results on other reach angles (Fig S1), as well as when data was pooled across all reach angles (data not shown). Multiunit activity was collected from N =191 electrodes across two Utah multielectrode arrays placed in primary motor (M1) and premotor (PMd) cortices (see Methods).
Fig 4. Time warping of reach dynamics in a nonhuman primate.
(A) Reaches towards a 90° target were analyzed. (B) Spike data from four example multiunits over all trials. Units 1 & 4 were from primary motor cortex; units 2 & 3 were from pre-motor cortex. At the bottom, the temporal distribution of task events is shown: 100 ms after target onset (TARG, green), end of delay period (GO, blue), and movement onset (MOVE, red). (C) Scatterplot showing correlation between the hand movement onset on each trial and the learned shift parameter in the shift-only warping model. (D) Same as (B), but with trials re-ordered by the magnitude of shift warping. (E) Same as (B), but with the time axis aligned by shift-only warping. (F) Same as (B), but with the time axis aligned by linear warping. (G) Same as (B), but with the time axis aligned by piecewise-linear warping with one knot.
The most dramatic changes in neural firing rates are closely time-locked to movement (Kaufman et al. 2016). Thus, it is common to track hand position on a moment-by-moment basis and use these measurements to align spike times to the onset of movement or the peak hand velocity on each trial. We instead examined spike trains aligned to the beginning of the delay period (Fig 4b), and used time warping to infer an alignment without any reference to the animal’s behavior.
As expected, a shift-only warping model closely aligned spike times with the onset of movement. The model’s learned shift parameter on each trial correlated very tightly with movement onset (Fig 4c), achieving a comparable level of performance (R2 = 0.9) to what was recently reported for a complex, nonlinear warping method (Duncker and Sahani 2018). Furthermore, the shift-only warping model enabled the visualization of movement-related firing rate changes in single-neuron rasters, either by re-sorting the trial order of the raw data (Fig 4d) or by re-aligning the spike times (Fig 4e).
Thus, learning a single per-trial shift was sufficient to align neural spike times to movement, without any reference to hand tracking data. However, shifting spike times in this manner also destroyed other structure in the data. Namely, a subset of multiunits, mostly in PMd, showed increased firing around ~100ms into the delay period—i.e. shortly after the reach target was visually presented to the animal (see units 2 & 3 in Fig 4). Due to the variable delay between target onset and movement onset, a shift-only warping model is incapable of simultaneously aligning spikes across these two events.
A linear time warping model more appropriately captures this structure in the data. On each trial, the model utilizes its two free parameters—the slope and intercept of the warping function—to precisely align these two task events (Fig 4f). Thus, these results provide strong evidence, via an unsupervised time warping method, that reliable neural dynamics occur around the time of movement onset and shortly after target onset. Piecewise-linear warping functions did not provide substantial benefits over linear warping; however, linear warping provided a reproducible benefit over the shift-only warping model in cross-validated performance (Fig S1).
Detection of ~13–40 Hz spike-time oscillations in primate pre-motor cortex
Thus far, we have shown that the temporal alignments learned by simple warping models can closely correlate with behaviors (e.g. movement or sniffing) and sensory cues (e.g. reach target presentation). This agreement demonstrates that time warping models can converge to reasonable and human-interpretable solutions, and, conversely, suggests that established alignment practices in these systems are well-justified from a statistical perspective. We now show that time warping can also uncover more subtle and unexpected features in spike train data.
LFP signals in primate premotor cortex show oscillations in the beta frequency range (13–40 Hz) during movement preparation, which are correlated with spike timing (Murthy and Fetz 1992; Sanes and Donoghue 1993; Reimer and Hatsopoulos 2010). While recent work has investigated the relationships between LFP and behavior (Khanna and Carmena 2015; Khanna and Carmena 2017; Chandrasekaran et al. 2019), the impact of beta oscillations on population-level spiking activity is still poorly understood. Recent work used a complex, black box model of neural dynamics to detect oscillatory structure in high-dimensional spike trains (Pandarinath et al. 2018). Here, we show that shift-only or linear time warping models can recover similar oscillations.
We examined premotor cortical data collected from two different monkey subjects (Monkey J and Monkey U) performing point-to-point reaches; one animal performed these reaches with an unrestrained hand, while the other used a manipulandum (see Methods). Since the oscillations are strongest during the pre-movement delay period, we focused on a time window beginning 400 ms prior to and 100 ms after go cue presentation. We found that having a larger number of trials was beneficial, so we pooled trials from all reach angles for this analysis. We analyzed multiunit data for each monkey from N = 96 electrodes placed in PMd.
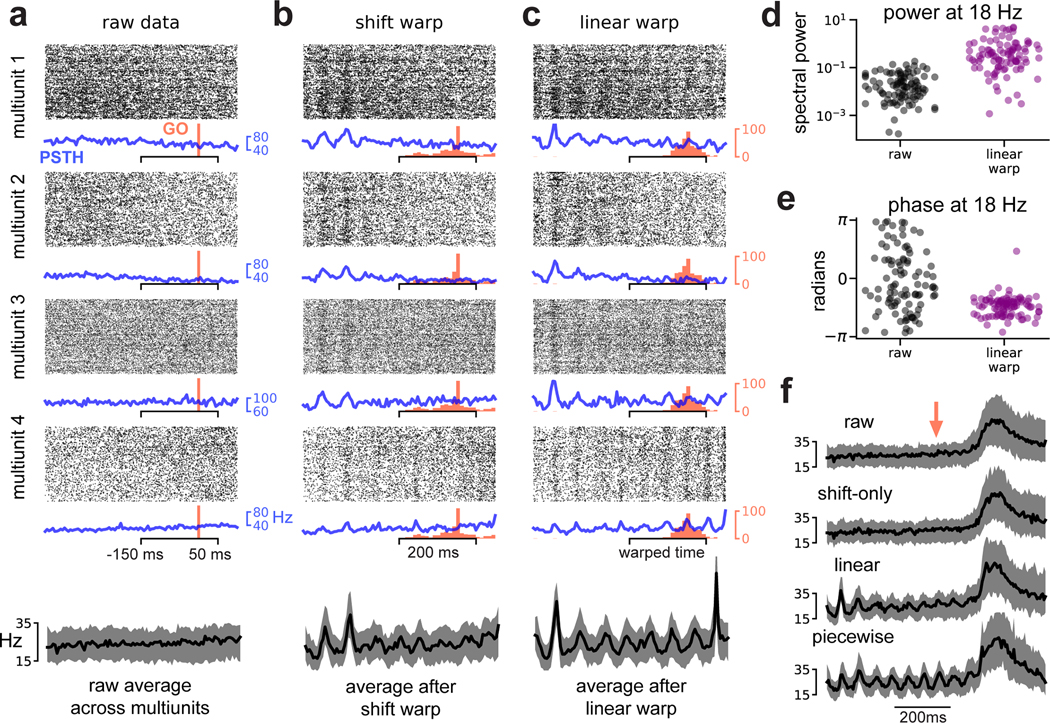
No oscillations were visible in spike rasters aligned to go cue (Fig 5a; data from Monkey U). However, re-aligning these spike trains based on a shift-only warping model revealed oscillations at ~18 Hz in Monkey U (shown in Fig 5b) and at ~40 Hz in Monkey J (Fig S2); these results are within previously reported frequency ranges (Murthy and Fetz 1992; Sanes and Donoghue 1993). In Monkey U, these oscillations were more apparent after linear warping (Fig 5c), suggesting that the frequency (in addition to the phase) of the oscillations may be variable. Further, these oscillations were roughly in-phase across multiunits—as a result, averaging spike counts across all multiunits and trials after time warping produced cleaner estimates (Fig 5a–c, bottom).
Fig 5. Time warping reveals spike-level oscillations in primate premotor cortex.
(A) Multiunit activity aligned to go cue. Four representative multiunits are shown; spike rasters (black) trial-average PSTH (blue) are shown for each multiunit. Go cue onset is shown in red. The average firing rate across all multiunits is shown on the bottom (black trace). Shaded grey region denotes upper and lower quartiles. (B) Same as (A) but with spike times aligned by shift-only warping model. Each displayed multiunit was held out from the model fit. (C) Same as (A) but with spike times aligned by linear warping model. Each displayed multiunit was held out from the model fit. (D) Oscillatory power at 18Hz in trial-average multiunit activity aligned to go cue (black dots) and after linear warping (purple dots). Each dot represents one of N = 96 multiunits. (E) Same as (D) but showing oscillation phase for each multiunit. (F) Multiunit activity averaged across electrodes and trials in a larger time window (800 ms) around go cue (red arrow). Oscillations are not visible in the raw data (top) or after shift warping (second from top). Oscillations are recovered by either linear (second from bottom) or piecewise linear warping (bottom). Shaded grey region denotes upper and lower quartiles.
To quantify these effects across all multiunits, we compared the PSTHs computed from raw data (blue traces; Fig 5a) to PSTHs computed from data aligned by linear time warping (blue traces; Fig 5c). We used Fourier analysis to estimate the amplitude and phase of the oscillation at 18 Hz, and found that time warping increased the strength of the oscillation by 1–2 orders of magnitude in most multiunits (Fig 5d). Furthermore, in the raw PSTHs the oscillation phases were widely spread across multiunits, consistent with there being no detectable oscillations above background noise (Fig 5e; gray dots); in the aligned PSTHs, the phases were tightly clustered, reflecting that oscillations were detectable and in-phase across nearly all multiunits (Fig 5e; purple dots).
We confirmed that the spike-level oscillations were in-phase with LFP oscillations in Monkey U. To do this, we applied the time warping models fit on spike train data to align bandpass-filtered LFP signals (10–30 Hz). The LFP signal was misaligned across trials in raw data, but was accurately aligned by the spike-level time warping models (Fig S3), suggesting that the two signals are coherently time-warped (in this case, temporally shifted and/or stretched) on a trial-by-trial basis. On a methodological level, this demonstrates that time warping models can generalize and make accurate predictions about other time series (e.g., LFP) with qualitatively distinct statistics from the training data (e.g., spike times).
We wondered whether time warping would fail to recover these oscillations if the movement-related spiking, which occurs at a much higher firing rate than pre-movement activity, was included in the analysis. To examine this, we fit warping models to a larger time window (±400 ms around go cue presentation). Time warping was still able to extract oscillations under these more challenging circumstances (Fig 5f). Interestingly, a shift-only model was no longer sufficient to capture oscillatory activity, suggesting that the oscillations were not phase-locked to movement onset. In contrast, linear or piecewise linear warping functions were able to recover the oscillations (Fig 5f; bottom). Thus, while the shift-only model is simplest to interpret, it may be insufficient to capture certain results. This demonstrates the utility of exploring a range of time warping models, with varying levels of complexity, on the same dataset.
Detection of ~6–7 Hz oscillations in rat motor cortex
We have seen that time warping can reveal interpretable structure, even under well-controlled experimental conditions. Discrete reaching, for example, is arguably the simplest volitional motor behavior that one can study, and yet straightforward behavioral alignments obscure salient spike-time oscillations (Fig 5). To study a more complex behavior, in a different animal model, we analyzed motor cortical activity in rats trained to produce a timed motor sequence (Kawai et al. 2015; Dhawale et al. 2017). Rats were trained to press a lever twice with a target time interval of 780 ms, and were rewarded if the sequence was completed within ±80 ms of this target (Fig 6a). While rats produce stereotyped motor sequences in this setting, the duration between lever presses and the timing of intermediate motor actions is variable from trial-to-trial. We examined a dataset consisting of N = 30 neurons and K = 1265 trials (3 recording sessions, collected over a 2 day period). In these sessions, the interval between lever presses ranged from 521–976 ms (5th- and 95th-percentiles) across trials.
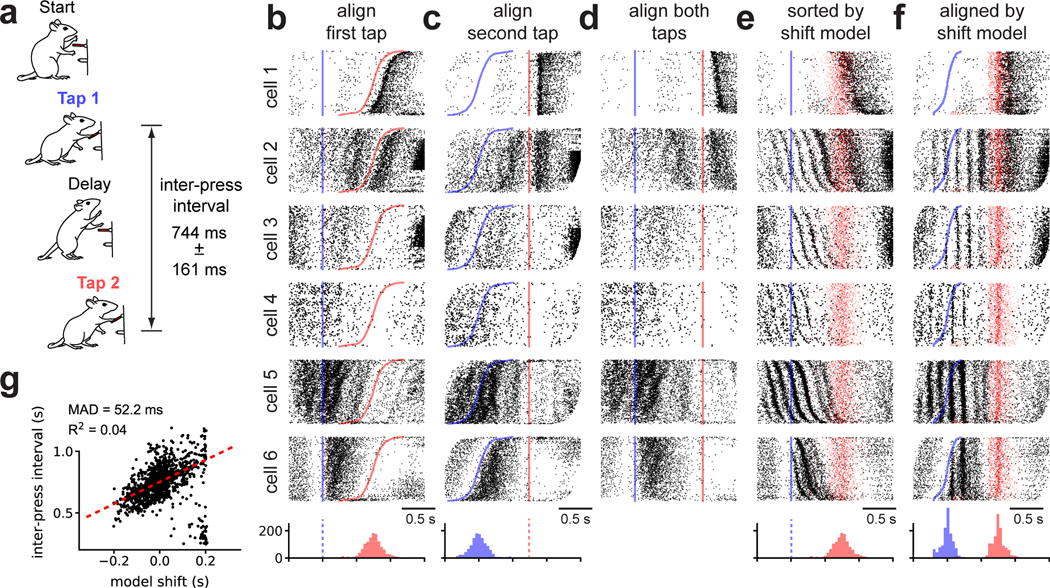
Fig 6. Shift-only time warping reveals temporally precise theta-locked oscillations in rat motor cortex.
(A) Rats were trained to press a lever twice with a prescribed delay. The range of inter-press-intervals across trials is listed (median ± inter-quartile range). (B) Raster plots for six representative cells with spike times aligned to the first lever press (blue line) and trials sorted by second lever press time (red line). Histograms at the bottom denote the distribution of the lever press times. (C) Same as (B) but with time aligned to the second lever press. (D) Same as (B) but with time linearly re-scaled to align both lever presses. The missing scale bar reflects the fact that time units are no longer preserved across trials. (E) Same as (B) but with trials re-ordered by the magnitude of shift warping. The time of the second lever press on each trial is denoted by a semi-transparent red dot. (F) Same as (E) but with the time axis aligned by shift-only warping. (G) Relationship between the magnitude of shift warping and inter-press interval on each trial. Dashed red line denotes a robust linear regression fit (Huber loss function, ϵ = 1.001); the Median Absolute Deviation (MAD) and R2 are listed as measures of fit.
This experiment has three obvious alignment procedures: align spike times to the first lever press, align spike times to the second lever press, or linearly stretch/compress the spike times to align both lever presses across trials (i.e. human-supervised time warping). Figure 6b–d shows the activity of six example neurons under these alignment strategies. At a high level, these raster plots demonstrate that neurons preferentially respond to different behavioral events within a trial. For example, cell 1 in Figure 6 fires after the second lever press, while cell 6 in Figure 6 fires after the first press. Thus, it is not obvious which alignment is preferable and indeed different insights may be gained from analyzing each.
Unsupervised time warping revealed structure in the data that is hidden in all three behavioral alignments. A shift-only warping model uncovered strong oscillations in many neurons, as visualized either by re-sorting trials based on the learned shift (Fig 6e, same alignment as Fig 6b), or by using the model to re-align spike times (Fig 6f). These findings are not due to spurious alignments produced by an overfit model; each spike raster in Figure 6f was generated on held out data as diagrammed in Fig 2k. Linear and piecewise linear time warping recovered qualitatively similar oscillations (data not shown), and were not superior to the shift-only model in terms of cross-validated performance (Fig S4).
These results reveal a partial decoupling of behavioral events (lever presses) with neural firing patterns. After alignment, both the first and second lever presses occur at variable times within each trial (Fig 6f, histograms at bottom), and the learned shift on each trial only loosely correlated with inter-press interval (Fig 6g). Furthermore, while multiple neurons exhibited spike oscillations, these oscillations were out-of-phase across neurons and therefore not visible in single-trial raster plots (Fig S4). Taken together, these features of the data suggest that it would be difficult to discover this oscillatory structure by manual alignment, demonstrating the power of unsupervised time warping.
While the uncovered oscillations are not phase locked with lever press times, they are nonetheless correlated with certain aspects of the animal’s behavior. In particular, some cells only exhibit the ~6–7 Hz oscillation following the first lever press, with remarkable temporal precision (see cells 3 and 6 in Fig 6f). Indeed, multiple cells exhibit non-oscillatory firing prior to the first lever press, but rapidly switch to an oscillatory behavior following the lever press (see cell 3 in Fig 6f and Fig S5). Other cells exhibit oscillations prior to the first lever press, but the amplitude and precision of the oscillations appears to improve following the first lever press (see cell 4 in Fig 5F). Still other cells either do not exhibit oscillations (cell 1 in Fig 6f) or exhibit strong oscillations both prior to and following the first lever press (cell 5 in Fig 6f). Time warping enables us to discover and visualize this full spectrum of functional cell types, which are otherwise difficult to detect and characterize. The presence of oscillations in single neurons can be confirmed by plotting the distribution of inter-spike-intervals (Fig S5); the shift-only model goes beyond this method by demonstrating that a large population of neurons are coherently phase-shifted on a trial-by-trial basis, and by enabling characterization of the full population dynamics and behavioral events in an aligned temporal space.
We then examined whether these spike-level oscillations were aligned with oscillations in LFP. The average frequency spectrum of the LFP did display a prominent peak at ~6–7 Hz—a very similar frequency range to the spike-level oscillations identified in Figure 6. To characterize the relationship between these two oscillatory signals, we bandpassed filtered the LFP between 5–9 Hz on each trial and fit a separate shift-only time warping model to the LFP traces. The time warping functions learned on LFP data did not uncover the spike-time oscillations shown in Figure 6, and likewise the LFP signals were not well-aligned by the time warping functions fit to spike times (Fig S6). Thus, unlike the oscillations identified in primate premotor cortex, the oscillations in rat motor cortex were not aligned with the LFP.
Discussion
While the temporal precision of neural coding has been a matter of intense debate, few studies have leveraged statistical alignment methods to investigate this problem. Earlier work incorporated time warping into single neuron encoding and decoding models (Aldworth et al. 2005; Gollisch 2006; Smith and Paninski 2013; Lawlor et al. 2018), as well as dimensionality reduction methods (Poole et al. 2017; Duncker and Sahani 2018). Here, we decoupled time warping from these other modeling objectives to achieve a flexible and simplified framework. We surveyed a broader range of datasets than past work, spanning multiple model organisms, brain areas, and sensory/motor tasks. In all cases, we found that the simplest and most interpretable models—often those with shift-only or linear warping functions—matched the performance of more complex models, while uncovering striking and sometimes unanticipated dynamics.
Our results suggest that shift-only and linear warping models can match or even outperform more complex nonlinear warping methods. These simpler models have three attractive properties. First, they manipulate model estimates of single-trial firing rates in a more interpretable manner (see Fig 1), enabling exploratory data analysis and visualization. Second, we developed efficient and scalable optimization methods for this class of models. On a modern laptop, these models can typically be fit to data from 1000 neurons, 100 timepoints, and 1000 trials in one minute or less. This scalability is of great practical importance given the increasing scale of neural recordings (Stevenson and Kording 2011), and the growing need for rigorous cross-validation and model comparison methods (Chandrasekaran et al. 2018), which are often computationally intensive if not prohibitive. Indeed, another contribution of this work is the development of cross-validation procedures to compare the performance of time warping models ranging from simple (shift-only) to more complex (piecewise linear; see Fig 2i–k). In essence, this enables a data-driven determination of the questions we posed in the introduction of this manuscript—should spike times be reported in absolute clock time, relative time between two behavioral events, or something else? Though the answer to this question will depend on the particulars of the experimental context, time warping may be used to rigorously compare these possibilities on a case-by-case basis.
Time warping also uncovered firing patterns that were not aligned to any stimulus or measured behavior. For example, we observed ~13–40Hz spike time oscillations in primate premotor cortex during movement preparation (see Fig 5), which we then verified were phase-aligned with LFP (see Fig S3). Similar non-phase-locked oscillations have been previously observed in functional neuroimaging experiments (Makeig et al. 2002; David et al. 2006). Notably, the time warping models we used did not assume any oscillatory structure in the data, and thus provide strong evidence that spike-level oscillations are a salient feature of the dynamics. When an oscillation is uncovered, a linear time warping model considers the activity of the full neural population to estimate the changes in the phase (y-intercept) and frequency (slope) of the oscillation on a trial-by-trial basis. This population-level approach can be contrasted with popular frequency-domain statistical measures like coherence, which measures the degree of phase synchronization between two spike trains or between a single spike train and LFP (Fries et al. 1997). These pairwise analysis methods are likely more sensitive to noise in single neurons than the population-level models we investigated here; future work could explore the relative advantages and disadvantages of these approaches and develop new methods that combine desirable elements from both.
Oscillatory spike patterns may not always be synchronized to LFP, as we observed in rat motor cortex (see Fig 6). These ~7 Hz oscillations were sometimes gated by a motor action—specifically, the first lever press—thus suggesting a potential relevance to timed motor production (Fig S5). Another possibility is that orofacial behaviors such as whisking and licking are the primary driver of these oscillations (Hill et al. 2011). Other work has shown that persistent ~7 Hz LFP oscillations may be locked to the respiration cycle (Tort et al. 2018); however, the transient, spike-level oscillations we observed were decoupled from LFP, and are thus potentially distinct from this phenomenon. Regardless of their root cause, this result demonstrates the ability of time warping to extract unexpected features of scientific interest from high-dimensional spike trains. Further work is needed to confirm and fully elucidate the properties of these oscillations.
Time warping is only one form of variability exhibited by single-trial neural dynamics. For simplicity, we examined time warping in the absence of other modeling assumptions, such as trial-to-trial variation in amplitude (Bollimunta et al. 2007; Williams et al. 2018), or condition-specific changes in dynamics (Duncker and Sahani 2018). Nonetheless, the models described here could be extended in several ways to capture more complex single-trial dynamics. First, we made the restrictive assumption that all neurons share the same time warping function on each trial. Under more complex experimental conditions—e.g., in large-scale recordings of multiple brain regions—it may be fruitful to fit multiple time warping functions associated with different neural sub-populations. Second, we fit only a single response template for each neuron. Relaxing this assumption may be useful in situations where neurons exhibit more than one canonical response pattern. Finally, we assumed that neural data was collected over repeated trials. To accommodate more unstructured experimental designs, future work could incorporate time warping into state space models (Macke et al. 2015) or sequence extraction algorithms (Mackevicius et al. 2019). Despite these exciting prospects for future statistical methodology, our work demonstrates that even a simple time warping framework can provide a rich and practical set of tools for the modern neuroscientist.
While our results already show that averaging over short, stereotyped trials can obscure fine temporal oscillations and firing events, these shortcomings are undoubtedly more severe in behaviors that have longer temporal extents and exhibit more variability. Thus, we expect time warping methods to play an increasingly crucial role in neural data analysis as the field moves to study more complex and unstructured animal behaviors (e.g. under more naturalistic settings; Krakauer et al. 2017). Furthermore, in complex experimental tasks involving large numbers of conditions and exploratory behaviors, the same motor act or sensory percept may present itself only a small number of times. In this trial-limited regime, precise data alignment may be critical to achieve the necessary statistical power to make scientific claims. We expect simple models, such as linear and piecewise linear warping, to perform best on these emerging datasets due to their interpretability, computational efficiency, and robustness to overfitting.
STAR Methods
Contact for Reagent and Resource Sharing
Further requests for resources should be directed to and will be fulfilled by the Lead Contact, Alex H. Williams (ahwillia@stanford.edu).
Experimental Model and Subject Details
Mouse Olfactory Task
Experimental subjects were male C57B/6 mice (Jackson Laboratories). All procedures were approved by the Institutional Animal Care and Use Committee of New York University Langone Medical Center.
Primate Motor Task
Experimental subjects were two male rhesus macaque monkeys (Macacca mulatta), denoted monkey J and monkey U. The monkeys were 13 (J) and 7 (U) years old and weighed 16 kg (J) and 13 kg (U) at the time of these experiments. All procedures and experiments were reviewed and approved by the Stanford University Institutional Animal Care and Use Committee.
Rat Motor Task
Experimental subjects were female Long Evans rats, 3–8 months old at the start of the experiment (Charles River). All procedures and experiments were reviewed and approved by the Harvard Institutional Animal Care and Use Committee.
Method Details
Mathematical Notation
We follow the same notation introduced in the main text. Matrices are denoted in bold, uppercase fonts, e.g. M, while vectors are denoted in bold, lowercase fonts, e.g. v. Unless otherwise specified, non-boldface letters specify scalar quantities, e.g. S or s. We use MT and M−1 to denote the transpose and inverse of a matrix, respectively.
We consider a dataset consisting of N features over K trials with T timesteps per trial. For simplicity, we refer to N as the number of neurons in the dataset; however, N could also refer to the number of fMRI voxels, multiunits, or regions of interest in imaging data. The full dataset is a third-order tensor (a three-dimensional data array) with dimensions K × T × N. The kth slice of the data tensor is a T × N matrix Xk, which represents the activity of the neural population on trial k. We denote a single element of the tensor as Xk,t,n, which specifies the activity of neuron n at timebin t on trial k.
The time warping model produces an estimate of population activity on each trial. Mirroring standard notation in linear regression, we denote the model estimate on trial k as (a T × N matrix).
Model Estimate and Template Interpolation Scheme
The main idea behind time warping is to approximate each trial, Xk, as a warped version of a T × N template, , that is shared across all trials. For neuron n, at time bin t, on trial k, the spirit behind the model is:
| (3) |
However, this expression is only valid when the warping function, ωk(t), produces integer values. To allow the warping functions to produce non-integer values, we adopt a standard linear interpolation scheme. Let ωk(t) denote the time warping function for trial k. The inputs to the warping function are integer-valued time indices t (clock time), and the outputs are any real number (aligned time). We assume that ωk(t) is monotonically increasing and thus invertible. Then, the model estimate for neuron n, at time bin t, on trial k is given by:
| (4) |
where ⎿·⏌ represents the “flooring” operation, and ⎾·⏋ represents the “ceiling” operation. Because the model estimate (Eq 4) is a linear combination of and , the warping transformation can be represented as a matrix W with nonzero elements given by:
| (5) |
For each trial, the warping matrix Wk can be uniquely determined from the warping function ωk. Thus, the model estimate on each trial is given by:
| (6) |
Optimization Strategy
The model template and warping functions are optimized to minimize an objective function, which we denote as . We assume that this objective function decomposes across trials as follows:
| (7) |
Here fk is a function defining the model loss on trial k, and ρ1 is a regularization term, penalizing the roughness and size of the template (described in the next section). Our online code package supports least-squares and Poisson loss functions; we adopted the least-squares criterion for the purposes of this paper due to its computational efficiency and its ability to be adapted to non-spike time data (e.g. fMRI or calcium imaging). Under this choice, the per-trial loss function is:
| (8) |
Here, ρ2 is a regularization term that penalizes the magnitude of warping (described in the next section), and denotes the squared Frobenius norm, which is simply the sum of squared residuals, .
To minimize F, we adopt an alternating optimization (block coordinate descent) approach (Wright 2015). First, each warping function is initialized to be the identity, ωk(t) = t, and the template and warping functions are cyclically updated according to the following sequence of optimization subproblems:
| (9) |
Here, an underlined variable denotes a dummy variable that is optimized over in each subproblem. This sequence of parameter updates is cyclically repeated until the objective value ceases to improve; by construction, the objective monotonically decreases at each step of the algorithm so convergence is guaranteed under mild assumptions (Wright 2015).
Partitioning the parameter updates in this manner is useful because each subproblem can be solved very efficiently. When the template is considered a fixed variable, the objective function decouples across trials (Eq 8), which simplifies the warping function updates considerably:
| (10) |
These parameter updates are entirely independent, with each update only depending on the raw data for trial k, Xk, and the current warping template . Our code package executes them efficiently in parallel across CPU threads. Furthermore, each warping function is controlled by a small number of parameters in our framework—at best a single parameter (shift-only warping) and at worst only a few parameters (piecewise linear warping). Thus, we perform these updates by a brute force random search (see Warping Function Regularization and Update Rule).
The response template is also very simple to update, especially under a least-squares loss criterion. Assume for the moment that the model is not regularized; i.e., and ρ2(Wk) = 0. Then, because each Wk is held constant, updating the template amounts to a least-squares problem that can be solved in closed form:
| (11) |
Furthermore the matrix is a symmetric, tridiagonal matrix. Intuitively, this tridiagonal structure arises from the constraint that each warping function is monotonically increasing, and the local structure of the linear interpolation scheme: for any warping matrix W, with an associated warping function ω, Equation 4 implies that Wi,tWj,t = 0 if |ω(t) − i| > 1 or if |ω(t) − j| > 1 and thus [WTW]i,j = 0 if |i – j| > 1.
The tridiagonal structure of enables the template parameters to be updated extraordinarily fast for practical applications. We use a specialized solver for systems of linear equations with banded, symmetric structure (scipy.linalg.solveh_banded). This allows the optimization problem in Equation 11 to be solved with O(TN) operations. If were naïvely treated as a dense matrix, the solution would instead require O(T3 + T2N) operations.
Template Regularization and Update Rule
We found that introducing regularization (penalties on the magnitude or complexity of model parameters) can improve the interpretability of the model and its ability to predict held out data. First, we found in some datasets that the warping template could exhibit rapid, high-frequency changes in firing rate (see, e.g., the template in Fig 1C, which was fit without regularization). These irregularities likely correspond to the model overfitting to noisy neuronal data, and can be discouraged by penalizing the magnitude of the second finite differences along the temporal dimension of the template (Grosenick et al. 2013; Maheswaranathan et al. 2018). We refer to this term as a roughness penalty or smoothness regularization. Second, it is possible that the matrix appearing in eq. (11) would become non-invertible or ill-conditioned during optimization. To prevent this, and to discourage the template firing rates from becoming too large, we added a penalty on the squared Frobenius norm of the template. Formally, the regularization on the template is given by:
| (12) |
where λ > 0 controls the strength of the roughness penalty and γ > 0 controls the strength of the Frobenius norm penalty. The matrix D is a (T − 2) × T matrix that computes second-order finite differences:
| (13) |
Incorporating this regularization term into the update of the warping template (eq. (11)), we get:
| (14) |
Which yields the template update rule:
| (15) |
Thus, the solution is the same as before except a term λDTD + γI is added to the inverted matrix (left-hand side of linear system). These modifications hardly affect the computational complexity of the parameter update since λDTD + γI is also a symmetric, banded matrix. Furthermore, as long as γ > 0 the overall matrix is positive definite and therefore guaranteed to be invertible.
In practice, we have found that it is simple to hand-tune the regularization strengths for exploratory analysis (though cross-validation procedures, described below, should always be used to monitor for overfitting). We typically set the L2 regularization (γ) to be zero or very small (e.g., 1e-4) and do not tune it further. A reasonable value for the roughness penalty scale can be found by visually inspecting the template for various neurons (columns of ) and increasing λ if these time series appear noisy.
Warping Function Regularization and Update Rule
We found that the optimization landscape of linear and piecewise linear warping functions is complex and full of local minima. Thus, gradient-based optimization methods can be ineffective. Thankfully, the warping functions are (a) low-dimensional and (b) entirely decoupled across trials. Thus, when updating the warping functions, we perform a brute force parameter search for each trial in parallel. For shift-only warping models, we perform a dense grid search over the parameter (the magnitude of the shift).
For piecewise linear warping models we perform an annealed random search as follows. Consider a warping function ωk(t) for any arbitrary trial. We parameterize the warping function as:
| (16) |
Where is a piecewise linear function mapping the unit interval [0, 1] to any real number, and denotes clipping any real number z to have a value between zero and one.
The piecewise linear function intuitively defines the “unclipped” warping function for trial k. We define these functions by a series of M x-y coordinates, {(α1, β1),(α2, β2), …(αM, βM)}, where 0 = α1 < α2 < … < αM = 1 and β1 ≤ β2 ≤ … ≤ βM. We refer to these coordinates as the knots of the warping function.
| (17) |
Note that the warping function for each trial has a different set of knots; however, we have suppressed the trial index variable k on the knot coordinates (αm and βm) for notational brevity.
To optimize the warping functions we perform a random search over these coordinates/knots. Let α = [α1, α2, …, αM] and β = [β1, β2, …, βM] denote the current coordinates for any particular trial. We form a new proposed warping function by:
| (18) |
where Q > 0 is a scalar parameter tuning the amount of exploration, and η is a vector of random normal variables with mean zero and unit variance. The procedure “sort(v)” re-orders the elements of a vector so that they are in ascending order. If the proposed warping function improves the objective function, we accept the new parameters:
For each round of optimization we exponentially relax Q from 1.0 to 0.01 over a fixed number of iterations.
We also found that penalizing the warping functions based on their distance from the identity line was helpful in some cases. Intuitively, this encourages the warping functions to be minimal—as the penalty strength increases the warping functions will approach ω(t) = t, resulting in no warping at all in this extreme limit. Similar penalties or hard constraints on time warping have been examined in prior literature (see e.g., Zhang et al. 2017). We chose the penalty to be the area between the unclipped warping function and the identity line:
| (19) |
which, for piecewise linear functions with relatively small M, can be efficiently computed as the sum of triangular and trapezoidal regions. Here, μ ≥ 0 is a scalar hyperparameter controlling the strength of the penalty. In practice we start with μ = 0 and increase it if, upon visual inspection, the warping functions are highly deviant from the identity line. Increasing μ in these cases can result in more sensible and interpretable templates. Again, cross-validation procedures can be used to asses whether μ is too low (resulting in overfitting) or too high (resulting in underfitting).
Synthetic data examples
In Figure 1 data from a single neuron was simulated as a difference of two exponential curves. The activity at T = 100 equally spaced time points between [−8, +8] was given by:
Where sk was a random shift parameter drawn uniformly on the interval [−5.5,3.5), and η was randomly drawn zero-mean gaussian noise with a standard deviation of 0.15. Unregularized shift-only, linear, and piecewise linear (with 1 knot) models were fit to K = 100 simulated trials. DTW-Barycenter Averaging (DBA; Petitjean et al. 2011) was fit to the same data using an open-source package called tslearn (Tavenard 2017).
In Figure 2 we simulated random warping warping functions following the procedure listed in Equation 18, with Q = .12. The firing rate template of each neuron was given by a smoothed, sparse sequence of heavy-tailed random variables:
where et were randomly drawn from an exponential distribution (with scale parameter equal to one) and bt were binary random variables drawn from a Bernoulli distribution (with probability of 0.92 that bt = 0). The conv(·) procedure denotes convolution with a Gaussian smoothing kernel with a standard deviation of 2. Truncated Poisson random variables were then drawn in each timebin; any bins with more than two spikes were truncated to one spike.
Mouse Olfactory Task
We analyzed data from a single recording session collected as part of a previously published study (Wilson et al. 2017). Subjects were implanted with a RIVETS headbar for head-fixation 7 days prior to the experiment (see description in Arneodo et al. 2018). Subjects were water deprived prior to the experiment and were administered water during random odor presentations to acclimate animals to the experimental apparatus.
On the day of experiment, subjects were anesthetized using isoflurane and a ~0.3 mm craniotomy was preformed to gain access to the dorsal olfactory bulb. NeuroNexus A2×32 probes were inserted approximately 500 μm into the dorsal bulb to record from the mitral-tufted cell layer. After probe insertion, subjects were allowed to recover from anesthesia for 30 minutes prior to recording. Electrophysiological and respiration signals were recorded using the HHMI Janelia Whisper recording system at 25000 Hz. Respiration (sniff) was monitored non-invasively using a pressure sensor sampling from the airflow in front of the nose. Action potentials from the recording were identified and classified into units offline using Spyking Circus template-matching software (Yger et al. 2018).
Two odors at 3 concentrations were presented in randomly interleaved trials. Subjects were passively sampling odor during trials. Concentrations covered a range of 2 orders of magnitude of molarity in carrier air. Odorants were diluted in mineral oil, stored in amber volatile organic analysis vials, and delivered via a 8-odor olfactometer. Odorant concentrations were controlled using a combination of gas- and liquid-phase dilution. We restricted our analysis to subsets of trials with odorant concentrations of 10−2 M—the highest concentration analyzed in (Wilson et al. 2017).
Primate Motor Task
Monkeys performed a standard center-out delayed reach task described previously in (Gilja et al. 2012; Ames et al. 2014). Targets were presented at (40, 80, 120) cm and 90 cm from the central starting location for monkeys J and U respectively. For monkey J, delay periods were evenly distributed between 300 and 700 ms (monkey J) with ~4.5% non-delay trials randomly interleaved. For monkey U, delays were randomly distributed between 350 and 600 ms on 87% trials, between 5 and 350 ms on 10% of trials, with 3% non-delay trials. Monkeys received a liquid reward upon touching and holding the cursor on the target. Movement during the delay period caused a trial failure and provided a brief automated time out (~1 s). Non-delay trials were not analyzed.
For monkey J, the virtual cursor and targets were presented in a three-dimensional environment (MusculoSkeletal Modeling Software, Medical Device Development Facility, University of Southern California). Hand-position data were measured at 60 Hz with an infrared reflective bead–tracking system (Polaris, Northern Digital). Behavioral control and neural decode were run on separate PCs using the Simulink/xPC platform (Mathworks) with communication latencies of less than 3ms. This system enabled millisecond timing precision for all computations. Visual presentation was provided via two LCD monitors in a Wheatstone stereotax configuration, with refresh rates at 120 Hz, yielding frame updates of 7 ± 4 ms. Two mirrors visually fused the displays into a single three-dimensional percept for the user, as described previously in (Gilja et al. 2012).
For monkey U, the virtual cursor and targets were presented on a standard 2D display. The monkey controlled the position of an onscreen cursor using a haptic manipulandum which applied no additional forces applied to the arm and was only used for positional cursor control. The haptic device was constrained to move within a 2D vertical workspace and cursor position tracks hand position 1:1 without perceptible lag.
Neural recordings were obtained via implanted 96-electrode Utah Microelectrode arrays (Blackrock Microsystems) using standard neurosurgical techniques. Two arrays were implanted in the left hemisphere of Monkey J, one in dorsal premotor cortex (PMd) and one in primary motor cortex (M1). Three arrays were implanted in the left hemisphere of Monkey U, one in PMd, one in medial M1, and one in lateral M1. For both monkeys, implantation location was estimated visually from local anatomical landmarks.
Neural data were band-pass filtered between 250–7500 Hz, and processed to obtain multiunit ‘threshold crossings’ spikes, defined as any time the signal crosses −3.5 times RMS voltage. We did not perform spike sorting, and instead grouped together the multiple neurons present on each electrode. As such, we anticipate that these population recordings contain both single and multiunit activity.
For Figure 4 and Figure S2 (Monkey J), each trial was defined as the 1200 ms following the reach target onset. Spike times were binned in 5 ms increments. For Figure 5 (Monkey U) and Figure S3 (Monkey J), we aligned spike times to the go cue instead of target onset. To highlight oscillatory spiking activity, we defined each trial as the period occurring 400 ms prior to go cue and 100 ms after go cue. Spike times were binned in 2.5 ms increments for Monkey J and 5 ms increments for Monkey U; similar results were found for smaller bin sizes, and stronger smoothness regularization. In Figure 5F (Monkey U), we extended each trial duration to ±400 ms around the go cue, but otherwise kept the same parameters.
Tuning the regularization strength of on template smoothness (λ) and warp magnitude (μ) was important to uncover the oscillations in premotor cortex. We used the cross-validation procedure described above to determine roughly appropriate values for these parameters. To be conservative and to be confident that these results were not due to overfitting, we increased the regularization strength further for the purposes of visualization.
Rat Motor Task
We analyzed data that was collected as part of a previously published study (Dhawale et al. 2017), which describes all experimental procedures and data collection protocols in greater detail. Extracellular recordings were obtained from 16 chronically implanted tetrodes in the motor cortex. Signals were amplified and digitized on a customized head-stage, and sampled at 30 kHz. The head stage was attached to a custom-designed tethering system that allowed the animal to move freely within its cage. Before implantation, an automated behavioral training framework (described in Poddar et al. 2013) was used to train the rats on a timed lever-pressing task (described in Kawai et al. 2015) until asymptotic performance was achieved.
The tetrode drive was then surgically implanted and targeted to motor cortex, through a 4–5 mm diameter craniotomy made 2 mm anterior and 3 mm lateral to bregma. The tetrode array was lowered to a target depth of 1.85 mm. At the end of the experiments, the position of the electrodes was verified by standard histological methods—brains were fixed via transcardial perfusion (4% paraformaldehyde in phosphate-buffered saline, Electron Microscopy Sciences) and the location of the electrodes was reconstructed by viewing mounted coronal sections (60 mm).
After 7 days of recovery post-surgery, training on the task resumed in the animal’s home cage. Neural and behavioral data was recorded continuously during this time (12–16 weeks) with only brief interruptions (median time of 0.2 hr). Spikes were sorted using Fast Automated Spike Tracker (FAST), a custom algorithm designed for parsing long-term continuous neural recordings (for details, see Dhawale et al. 2017). We examined K = 1265 trials, collected over a two day period.
Each trial was defined as the period starting 500 ms prior to the first lever press and 1500 ms after the first lever press. Spike times were binned in 10 ms increments for each unit. Raw spike counts were provided to the time warping algorithm; however, we observed similar results under various normalization schemes, such as soft-normalization (Churchland et al. 2012). All analyses of these data used a shift-only time warping model. The per-trial shift was constrained to be less than 10% of the total trial duration.
Quantification and Statistical Analysis
Cross-validation
As with any statistical method, one must be very careful that time warping does not reveal spurious structure and features of the data. In Fig 1, we saw that even a simple linear warping model can result in noticeable overfitting on a simple synthetic time series. An important technical contribution of our work is a rigorous cross-validation framework for time warping models. This framework, described in detail below, enables us to fine-tune all regularization terms—i.e. the hyperparameters {γ, λ, μ}—across all warping models. That is, we can rigorously compare the performance of shift-only, linear, and piecewise-linear time warping models on an even footing, and thus critically examine the degree of nonlinearity in time warping. While cross-validation is a common procedure in statistical modeling and in modern neuroscience, there are subtle pitfalls that must be avoided in unsupervised learning models (Bro et al. 2008; Perry 2009), and in models with temporally correlated noise (Opsomer et al. 2001).
To properly compare the performance of different warping models, it is important to perform nested cross-validation, so that regularization terms are separately tuned for each model. For example, a piecewise linear warping model will often require stronger smoothness and warp regularization terms, compared to a simpler, shift-only warping model. Thus, on each cross-validation run we split the data in three partitions: a training set, a validation set, and a test set. For each model class (shift-only, linear warping, piecewise-linear warping, etc.) we fit 100 models with randomized regularization strengths to the training set; we then evaluated all 100 models on the held out validation set; finally, the best-performing model was evaluated on the test set. The test set performance is then compared across model classes. Model performance was measured by computing the coefficient of determination (R2), averaged across neurons. Formally, for any partition of neurons N ⊆ {1, 2, …, N} and trials K ⊆ {1, 2, …, K} we define the coefficient of determination as:
| (20) |
Where is the average activation for neuron n.
Recall that our dataset consists of N neurons, T timebins, and K trials. The question then arises, should we hold out neurons, timepoints, or trials during cross-validation? Since the warping functions are assumed to be shared across all neurons, these model parameters can be fit on a subset of neurons (training set), and then evaluated on held out neurons (validation/test sets). However, if we hold out individual neurons entirely, then it is impossible to fit the response template matrix for those cells. Conversely, the response template can be fit to a subset of trials (training set) and evaluated on the remaining trials (validation/test sets). However, if we hold out individual trials entirely, then it is impossible to fit the warping functions associated with those held out trials.
To circumvent this problem we adopt a bi-cross-validation hold out pattern (Owen and Perry 2009). This entails separately and independently partitioning neurons and trials. Thus, we randomly choose training neurons (~73% of all cells), validation neurons (~13% of all cells), and testing neurons (the remaining ~13%). Additionally, we randomly choose training trials, validation trials, and testing trials, according to these same ratios. The model warping functions are fit to all trials, but only on the training neurons; the response template is fit for all neurons, but only on the training trials. When reporting the training performance, we compute the reconstruction loss on the intersection of the training neurons and training trials. Likewise, when evaluating models on the validation (or test) set, we compute the the reconstruction loss on the intersection of the validation (or test) neurons and validiation (or test) trials.
Temporal dependencies in model errors can complicate proper cross-validation (Opsomer et al. 2001; Latimer 2018). To avoid these complications, we leave out entire trials for individual neurons, rather than leaving out a subset of time bins.
Null models and other sanity checks
The cross-validation procedure described above is fully rigorous, but computationally expensive to perform. Even if each optimization run only takes a few seconds to complete, comparing M warping models over P random samples of the regularization parameters, and repeating the whole process over Q randomized folds leads to long run times; for example, ~26 hours for M = 5, P = 100, Q = 50 and each model taking ~20 seconds to optimize. This rather unfavorable scaling underscores why our attention to performance enhancements—e.g. by exploiting banded matrix structure when updating the model template—is critical for practical applications. On the other hand, a full cross-validation run is often unnecessary for exploratory data analysis and visualization. Here we briefly outline two simple procedures for validating time warping visualizations in a more interactive manner. Our online code package also supports both of these options.
First, one can create a very simple null dataset of neural activity that, by construction, contains no warping. By comparing the results of time warping on this null dataset to those achieved on the real data, we gain an informative reference point. For spiking data, we simulate null data by computing the trial-average firing rate of each neuron and then drawing Poisson i.i.d. random spike trains on every trial. That is, on each trial, the spike train for a neuron is drawn from an inhomogeneous Poisson process, with a rate function given by the trial-average firing rate. Similar baselines could be developed for calcium imaging and fMRI studies after specifying an appropriate noise model. We did not apply this method directly in this manuscript, but nonetheless mention it here for completeness.
Second, a key visualization tool enabled by time warping is the alignment of neural activity across trials. This alignment is achieved by applying the inverse warping functions to re-scale the time axis on the raw data; it does not directly rely on the response template, . Thus, one can visualize the aligned activity of an individual neuron in a held out manner—the model is fit to all trials and all other neurons, and the warping functions are applied to the held out cell (see Fig 2k). This can then be repeated for each neuron in the full population. All spike raster plots in the main paper were produced using this procedure.
While these two approaches do not supplant the need for careful cross-validation, they can provide a quick validation for visualizations and presented results.
Data and Software Availability
Our code for fitting linear and piecewise linear time warping models is distributed as a GitHub repository (under an MIT license): https://github.com/ahwillia/affinewarp. This repository also contains pre-processed versions of all three experimental datasets detailed in this paper, alongside step-by-step tutorials which reproduce the major results in Figures 2, 3, 5, and 6. Our Python implementation relies on the standard SciPy scientific computing libraries (Jones et al. 2001–; Hunter 2007). Additionally, we achieved substantial performance enhancements by leveraging numba, a Python library that enables just-in-time (JIT) compilation (Lam et al. 2015).
Supplementary Material
Acknowledgements
We thank Isabel Low and Chandramouli Chandrasekaran for feedback on the manuscript. This work received support from the following research grants and foundations: Department of Energy CSGF fellowship (A.H.W.), NIH NRSA 1F31NS089376-01 (E.M.T.), Stanford Graduate Fellowship (E.M.T.), NSF IGERT 0734683 (E.M.T.), NIH NINDS T-R01NS076460 (K.V.S.), NIH NIMH T-R01MH09964703 (K.V.S.), NIH Director’s Pioneer Award 8DP1HD075623 (K.V.S.), DARPA BTO ”REPAIR” N66001-10-C-2010 and ”NeuroFAST” W911NF-14-2-0013 (K.V.S.), the Simons Foundation Collaboration on the Global Brain awards 325380 and 543045 (K.V.S.), ONR N000141812158 (K.V.S.), the Howard Hughes Medical Institute (K.V.S.), Charles A. King Trust (A.K.D.), the Life Sciences Research Foundation (A.K.D.), NIH NINDS R01-NS099323-01 (B.P.O.), NIH R01-DC013797 (D.R.), NIH R01-DC014366 (D.R.), Burroughs Wellcome (S.G.), Alfred P. Sloan Foundation (S.G.), Simons Foundation (S.G.), McKnight Foundation (S.G.), James S. McDonell Foundation (S.G.), and the Office of Naval Research (S.G.).
Declaration of Interests
K.V.S. is a consultant to CTRL-Labs and Neuralink Inc. and on the Scientific Advisory Boards of Cognescent and Heal; these entities in no way influenced or supported this work.
References
- Aldworth Zane N., Miller John P., Gedeon Tomás, Cummins Graham I., and Dimitrov Alexander G. (2005). “Dejittered Spike-Conditioned Stimulus Waveforms Yield Improved Estimates of Neuronal Feature Selectivity and Spike-Timing Precision of Sensory Interneurons”. Journal of Neuroscience 25.22, pp. 5323–5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames K. Cora, Ryu Stephen I., and Shenoy Krishna V. (2014). “Neural Dynamics of Reaching following Incorrect or Absent Motor Preparation”. Neuron 81.2, pp. 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneodo Ezequiel M, Penikis Kristina B, Neil Rabinowitz, Angela Licata, Annika Cichy, Jingji Zhang, Thomas Bozza, and Dmitry Rinberg (2018). “Stimulus dependent diversity and stereotypy in the output of an olfactory functional unit”. Nature Communications 9.1, p. 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov Dmitriy, Nevers Rhino, and Tank David W (2017). “Mapping of a non-spatial dimension by the hippocampal–entorhinal circuit”. Nature 543, p. 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Stuart N and Gerstein George L (2001). “Determination of response latency and its application to normalization of cross-correlation measures”. Neural Computation 13.6, pp. 1351–1377. [DOI] [PubMed] [Google Scholar]
- Berndt Donald J. and Clifford James (1994). “Using Dynamic Time Warping to Find Patterns in Time Series”. Proceedings of the 3rd International Conference on Knowledge Discovery and Data Mining. AAAIWS’94 Seattle, WA: AAAI Press, pp. 359–370. [Google Scholar]
- Bollimunta Anil, Knuth Kevin H, and Mingzhou Ding (2007). “Trial-by-trial estimation of amplitude and latency variability in neuronal spike trains”. Journal of neuroscience methods 160.1, pp. 163–170. [DOI] [PubMed] [Google Scholar]
- Brette Romain (2015). “Philosophy of the Spike: Rate-Based vs. Spike-Based Theories of the Brain”. Frontiers in Systems Neuroscience 9, p. 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro R, Kjeldahl K, Smilde AK, and Kiers HAL (2008). “Cross-validation of component models: A critical look at current methods”. Analytical and Bioanalytical Chemistry 390.5, pp. 1241–1251. [DOI] [PubMed] [Google Scholar]
- Bruno Randy M (2011). “Synchrony in sensation”. Current Opinion in Neurobiology 21.5. Networks, circuits and computation, pp. 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran Chandramouli, Bray Iliana E., and Shenoy Krishna V. (2019). “Frequency Shifts and Depth Dependence of Premotor Beta Band Activity during Perceptual Decision-Making”. Journal of Neuroscience 39.8, pp. 1420–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran Chandramouli, Joana Soldado-Magraner, Diogo Peixoto, Newsome William T, Krishna Shenoy, and Sahani Maneesh (2018). “Brittleness in model selection analysis of single neuron firing rates”. bioRxiv. [Google Scholar]
- Churchland Mark M., Cunningham John P., Kaufman Matthew T., Foster Justin D., Nuyujukian Paul, Ryu Stephen I., and Shenoy Krishna V. (2012). “Neural population dynamics during reaching”. Nature 487, 51 EP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cury Kevin M. and Uchida Naoshige (2010). “Robust Odor Coding via Inhalation-Coupled Transient Activity in the Mammalian Olfactory Bulb”. Neuron 68.3, pp. 570–585. [DOI] [PubMed] [Google Scholar]
- David Olivier, Kilner James M, and Friston Karl J (July 2006). “Mechanisms of evoked and induced responses in MEG/EEG”. en. Neuroimage 31.4, pp. 1580–1591. [DOI] [PubMed] [Google Scholar]
- Denève Sophie and Machens Christian K (2016). “Efficient codes and balanced networks”. Nature Neuroscience 19, p. 375. [DOI] [PubMed] [Google Scholar]
- Dhawale Ashesh K, Rajesh Poddar, Wolff Steffen BE, Normand Valentin A, Evi Kopelowitz, and Ölveczky Bence P (2017). “Automated long-term recording and analysis of neural activity in behaving animals”. eLife 6 Ed. by Andrew J King, e27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker Lea and Sahani Maneesh (2018). “Temporal alignment and latent Gaussian process factor inference in population spike trains”. bioRxiv. [Google Scholar]
- Feierstein Claudia E., Quirk Michael C., Uchida Naoshige, Sosulski Dara L., and Mainen Zachary F. (2006). “Representation of Spatial Goals in Rat Orbitofrontal Cortex”. Neuron 51.4, pp. 495–507. [DOI] [PubMed] [Google Scholar]
- Fries Pascal, Roelfsema Pieter R., Engel Andreas K., Peter König, and Wolf Singer (1997). “Synchronization of oscillatory responses in visual cortex correlates with perception in interocular rivalry”. Proceedings of the National Academy of Sciences 94.23, pp. 12699–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilja Vikash, Nuyujukian Paul, Chestek Cindy A, Cunningham John P, Yu Byron M, Fan Joline M, Churchland Mark M, Kaufman Matthew T, Kao Jonathan C, Ryu Stephen I, and Shenoy Krishna V (2012). “A highperformance neural prosthesis enabled by control algorithm design”. Nature Neuroscience 15, p. 1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollisch Tim (2006). “Estimating receptive fields in the presence of spike-time jitter”. Network: Computation in Neural Systems 17.2, pp. 103–129. [DOI] [PubMed] [Google Scholar]
- Grosenick Logan, Klingenberg Brad, Katovich Kiefer, Knutson Brian, and Taylor Jonathan E (2013). “Interpretable whole-brain prediction analysis with GraphNet”. NeuroImage 72, pp. 304–321. [DOI] [PubMed] [Google Scholar]
- Harvey Christopher D., Coen Philip, and Tank David W. (2012). “Choice-specific sequences in parietal cortex during a virtual-navigation decision task”. Nature 484.7392, pp. 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill Daniel N., Curtis John C., Moore Jeffrey D., and Kleinfeld David (2011). “Primary Motor Cortex Reports Efferent Control of Vibrissa Motion on Multiple Timescales”. Neuron 72.2, pp. 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD (2007). “Matplotlib: A 2D graphics environment”. Computing In Science & Engineering 9.3, pp. 90–95. [Google Scholar]
- Jazayeri Mehrdad and Shadlen Michael N. (2015). “A Neural Mechanism for Sensing and Reproducing a Time Interval”. Current Biology 25.20, pp. 2599–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones Eric, Oliphant Travis, Peterson Pearu, et al. (2001. –). SciPy: Open source scientific tools for Python. [Google Scholar]
- Kaufman Matthew T., Seely Jeffrey S., Sussillo David, Ryu Stephen I., Shenoy Krishna V., and Churchland Mark M. (2016). “The Largest Response Component in the Motor Cortex Reflects Movement Timing but Not Movement Type”. eNeuro 3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Risa, Markman Timothy, Poddar Rajesh, Ko Raymond, Fantana Antoniu L., Dhawale Ashesh K., Kampff Adam R., and Ölveczky Bence P. (2015). “Motor Cortex Is Required for Learning but Not for Executing a Motor Skill”. Neuron 86.3, pp. 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna Preeya and Carmena Jose M (2017). “Beta band oscillations in motor cortex reflect neural population signals that delay movement onset”. eLife 6 Ed. by Pascal Fries, e24573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- — (2015). “Neural oscillations: beta band activity across motor networks”. Current Opinion in Neurobiology 32. Large-Scale Recording Technology (32), pp. 60–67. [DOI] [PubMed] [Google Scholar]
- Kobak Dmitry, Brendel Wieland, Constantinidis Christos, Feierstein Claudia E, Adam Kepecs, Mainen Zachary F, Xue-Lian Qi, Romo Ranulfo, Uchida Naoshige, and Machens Christian K (2016). “Demixed principal component analysis of neural population data”. eLife 5 Ed. by Mark CW van Rossum, e10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer John W., Ghazanfar Asif A., Alex Gomez-Marin, MacIver Malcolm A., and Poeppel David (2017). “Neuroscience Needs Behavior: Correcting a Reductionist Bias”. Neuron 93.3, pp. 480–490. [DOI] [PubMed] [Google Scholar]
- Lam Siu Kwan, Pitrou Antoine, and Seibert Stanley (2015). “Numba: A LLVM-based Python JIT Compiler”. Proceedings of the Second Workshop on the LLVM Compiler Infrastructure in HPC. LLVM ‘15 Austin, Texas: ACM, 7:1–7:6. [Google Scholar]
- Latimer Kenneth (2018). Simplicity Is Good, But Only When It’s Correct. http://www.kennethlatimer.com/blog/simplicity-is-good-but-only-when-its-correct.Blog.
- Lawlor Patrick N, Perich Matthew G, Lee E Miller, and Kording Konrad P (2018). “Linear-Nonlinear-Time-Warp-Poisson models of neural activity”. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo Anthony and Fee Michale S. (2005). “Ensemble Coding of Vocal Control in Birdsong”. J Neurosci 25.3, pp. 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London Michael, Roth Arnd, Beeren Lisa, Michael Häusser, and Latham Peter E (2010). “Sensitivity to perturbations in vivo implies high noise and suggests rate coding in cortex”. Nature 466, p. 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke JH, Buesing L, and Sahani M (2015). “Estimating state and parameters in state space models of spike trains”. Advanced State Space Methods for Neural and Clinical Data. Ed. by ZheEditor Chen. Cambridge University Press, pp. 137–159. [Google Scholar]
- Mackevicius Emily L, Bahle Andrew H, Williams Alex H, Shijie Gu, Denisenko Natalia I, Goldman Mark S, and Fee Michale S (2019). “Unsupervised discovery of temporal sequences in high-dimensional datasets, with applications to neuroscience”. eLife 8 Ed. by Laura Colgin and Timothy E Behrens, e38471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaranathan Niru, Kastner David B., Baccus Stephen A., and Ganguli Surya (2018). “Inferring hidden structure in multilayered neural circuits”. PLOS Computational Biology 14.8, pp. 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeig S, Westerfield M, Jung TP, Enghoff S, Townsend J, Courchesne E, and Sejnowski TJ (Jan. 2002). “Dynamic brain sources of visual evoked responses”. en. Science 2955555, pp. 690–694. [DOI] [PubMed] [Google Scholar]
- Marron JS, Ramsay James O., Sangalli Laura M., and Anuj Srivastava (2015). “Functional Data Analysis of Amplitude and Phase Variation”. Statist. Sci 30.4, pp. 468–484. [Google Scholar]
- Murakami Masayoshi, Vicente M Inês, Costa Gil M, and Mainen Zachary F (2014). “Neural antecedents of self-initiated actions in secondary motor cortex”. Nature Neuroscience 17, p. 1574. [DOI] [PubMed] [Google Scholar]
- Murthy VN and Fetz EE (1992). “Coherent 25- to 35-Hz oscillations in the sensorimotor cortex of awake behaving monkeys.” Proceedings of the National Academy of Sciences 89.12, pp. 5670–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsomer Jean, Wang Yuedong, and Yang Yuhong (2001). “Nonparametric Regression with Correlated Errors”. Statistical Science 16.2, pp. 134–153. [Google Scholar]
- Owen Art B. and Perry Patrick O. (2009). “Bi-cross-validation of the SVD and the nonnegative matrix factorization”. Ann. Appl. Stat 3.2, pp. 564–594. [Google Scholar]
- Pandarinath Chethan, O’Shea Daniel J., Jasmine Collins, Rafal Jozefowicz, Stavisky Sergey D., Kao Jonathan C., Trautmann Eric M., Kaufman Matthew T., Ryu Stephen I., Hochberg Leigh R., Henderson Jaimie M., Shenoy Krishna V., Abbott LF, and David Sussillo (2018). “Inferring single-trial neural population dynamics using sequential auto-encoders”. Nature Methods 15.10, pp. 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski Liam (2004). “Maximum likelihood estimation of cascade point-process neural encoding models”. Network: Computation in Neural Systems 15.4, pp. 243–262. [PubMed] [Google Scholar]
- Perez Oswaldo, Kass Robert E., and Merchant Hugo (2013). “Trial time warping to discriminate stimulus-related from movement-related neural activity”. Journal of Neuroscience Methods 212.2, pp. 203–210. [DOI] [PubMed] [Google Scholar]
- Perry Patrick O. (2009). “Cross-Validation for Unsupervised Learning”. PhD thesis Stanford University. [Google Scholar]
- Petitjean François, Alain Ketterlin, and Pierre Gançarski (2011). “A global averaging method for dynamic time warping, with applications to clustering”. Pattern Recognition 44.3, pp. 678–693. [Google Scholar]
- Pnevmatikakis Eftychios A. and Giovannucci Andrea (2017). “NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data”. Journal of Neuroscience Methods 291, pp. 83–94. [DOI] [PubMed] [Google Scholar]
- Poddar Rajesh, Kawai Risa, and Ölveczky Bence P. (2013). “A Fully Automated High-Throughput Training System for Rodents”. PLOS ONE 8.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole Ben, Williams Alex H., Maheswaranathan Niru, Yu Byron, Santhanam Gopal, Ryu Stephen, Baccuss Stephen A., Shenoy Krishna V., and Ganguli Surya (2017). “Time-warped PCA: simultaneous alignment and dimensionality reduction of neural data”. Cosyne Abstracts. Salt Lake City, UT, USA. [Google Scholar]
- Reimer Jacob and Hatsopoulos Nicholas G. (2010). “Periodicity and Evoked Responses in Motor Cortex”. J Neurosci 30.34, pp. 11506–11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JN and Donoghue JP (1993). “Oscillations in local field potentials of the primate motor cortex during voluntary movement.” Proc Natl Acad Sci U S A 90.10, pp. 4470–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shushruth S, Mark Mazurek, and Shadlen Michael N. (2018). “Comparison of Decision-Related Signals in Sensory and Motor Preparatory Responses of Neurons in Area LIP”. Journal of Neuroscience 38.28, pp. 6350–6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shusterman Roman, Sirotin Yevgeniy B., Smear Matthew C., Ahmadian Yashar, and Rinberg Dmitry (2018). “Sniff Invariant Odor Coding”. eNeuro 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shusterman Roman, Smear Matthew C, Koulakov Alexei A, and Dmitry Rinberg (2011). “Precise olfactory responses tile the sniff cycle”. Nat Neurosci 14.8, pp. 1039–1044. [DOI] [PubMed] [Google Scholar]
- Smith Carl and Paninski Liam (2013). “Computing loss of efficiency in optimal Bayesian decoders given noisy or incomplete spike trains”. Network: Computation in Neural Systems 24.2, pp. 75–98. [DOI] [PubMed] [Google Scholar]
- Softky WR and Koch C (1993). “The highly irregular firing of cortical cells is inconsistent with temporal integration of random EPSPs”. Journal of Neuroscience 13.1, pp. 334–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkweather Clara Kwon, Babayan Benedicte M, Naoshige Uchida, and Gershman Samuel J (2017). “Dopamine reward prediction errors reflect hidden-state inference across time”. Nature Neuroscience 20, p. 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson Ian H and Kording Konrad P (2011). “How advances in neural recording affect data analysis”. Nature Neuroscience 14, p. 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavenard Romain (2017). tslearn: A machine learning toolkit dedicated to time-series data. https://github.com/rtavenar/tslearn. [Google Scholar]
- Tort Adriano B L, Simon Ponsel, Jakob Jessberger, Yevgenij Yanovsky, Jurij Brankačk, and Andreas Draguhn (2018). “Parallel detection of theta and respiration-coupled oscillations throughout the mouse brain”. Scientific Reports 8.1, p. 6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura Valérie (2004). “Testing for and estimating latency effects for Poisson and non-Poisson spike trains”. Neural Computation 16.11, pp. 2323–2349. [DOI] [PubMed] [Google Scholar]
- Wang Jing, Narain Devika, Hosseini Eghbal A., and Jazayeri Mehrdad (2018). “Flexible timing by temporal scaling of cortical responses”. Nature Neuroscience 21.1, pp. 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Alex H., Kim Tony Hyun, Forea Wang, Vyas Saurabh, Ryu Stephen I., Shenoy Krishna V., Schnitzer Mark, Kolda Tamara G., and Ganguli Surya (2018). “Unsupervised Discovery of Demixed, Low-Dimensional Neural Dynamics across Multiple Timescales through Tensor Component Analysis”. Neuron 986, 1099–1115.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Christopher D., Serrano Gabriela O., Koulakov Alexei A., and Rinberg Dmitry (2017). “A primacy code for odor identity”. Nature Communications 8.1, p. 1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright Stephen J. (2015). “Coordinate descent algorithms”. Mathematical Programming 151.1, pp. 3–34. [Google Scholar]
- Yger Pierre, Spampinato Giulia LB, Elric Esposito, Lefebvre Baptiste, Stéphane Deny, Christophe Gardella, Marcel Stimberg, Florian Jetter, Guenther Zeck, Serge Picaud, Jens Duebel, and Olivier Marre (2018). “A spike sorting toolbox for up to thousands of electrodes validated with ground truth recordings in vitro and in vivo”. eLife 7 Ed. by David Kleinfeld, e34518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Zheng, Tavenard Romain, Bailly Adeline, Tang Xiaotong, Tang Ping, and Corpetti Thomas (2017). “Dynamic Time Warping under limited warping path length”. Information Sciences 393, pp. 91–107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our code for fitting linear and piecewise linear time warping models is distributed as a GitHub repository (under an MIT license): https://github.com/ahwillia/affinewarp. This repository also contains pre-processed versions of all three experimental datasets detailed in this paper, alongside step-by-step tutorials which reproduce the major results in Figures 2, 3, 5, and 6. Our Python implementation relies on the standard SciPy scientific computing libraries (Jones et al. 2001–; Hunter 2007). Additionally, we achieved substantial performance enhancements by leveraging numba, a Python library that enables just-in-time (JIT) compilation (Lam et al. 2015).