Abstract
Identifying the native range of invasive species is useful to understand their evolution and natural history, as well as to develop new methods to control potentially harmful introduced organisms. The clonal raider ant, Ooceraea biroi, is an introduced species and an increasingly important social insect model organism, but its native range remains unknown. Here, we report a new series of O. biroi collections from Bangladesh, Singapore, Vietnam and China. We use a molecular phylogeny constructed with five gene fragments from 27 samples to determine that invasive lineages of O. biroi originated in Bangladesh. These lineages may have spread from Bangladesh via the historically significant Bay of Bengal shipping ports. Ooceraea biroi shares multiple features of its biology with other introduced ants, including parthenogenesis, retention of heterozygosity and presence of multiple egg-layers in the colony. Using laboratory rearing and microsatellite markers, we show that colonies collected from disturbed habitat in Bangladesh have these traits in common with colonies from the invasive range. Ancestral populations with sexual reproduction in primary habitats either remain to be discovered or have gone extinct. Our findings advance our understanding of the global spread of the clonal raider ant and highlight a suite of general traits that make certain ants prone to becoming invasive.
Keywords: clonality, Formicidae, invasion history, invasive species, Ooceraea biroi, thelytoky
1. Introduction
A number of tramp ant species have been spread by human commerce throughout the world. Studies of their native populations allow researchers to better understand the circumstances under which these species have evolved, identify general characteristics that predispose them to become invasive and uncover natural biological control agents that could limit their spread. Unfortunately, the exact native range and likely route of invasion remain unknown for the great majority of invasive species [1].
In recent years, genetic data have identified the source populations and likely invasion routes for a handful of invasive ants [2–5]. In the absence of strong confounding factors, the native range of a species is expected to contain more genetic diversity than the invasive range [6]. Furthermore, invasive genotypes will be phylogenetically nested within the diversity of native genotypes and will be more closely related to native genotypes from their source population than from geographically more distant native populations. For most invasive ants, however, these genetic signatures have not been reported, and precise source populations therefore remain unknown (e.g. [7–9]). Challenges arise when the global distribution of a species is poorly known, when species are difficult to collect and when putatively native populations are difficult to pinpoint or reside in inaccessible regions of the planet.
The clonal raider ant, Ooceraea biroi, is queenless, and colonies are composed of a few dozen to a few hundred unmated workers, all of which can reproduce via thelytokous parthenogenesis [10,11]. Ooceraea biroi reproduces via automixis with central fusion, where the two central meiotic products fuse after meiosis II, which, in the absence of recombination, restores the maternal genotype [11]. Parthenogenesis and the presence of several reproductively active females in a colony are overrepresented among introduced ant species and are believed to facilitate the establishment of small founding populations in new habitats [12–14]. Its unusual biology might therefore explain why O. biroi is currently the only known invasive species in the ant subfamily Dorylinae.
The clonal raider ant is known primarily from tropical and subtropical islands worldwide, where it was presumably introduced via human activity [10,15]. Additional localities reported since earlier reviews [10,15] include Pakistan [16], Sri Lanka [17], Macau [18] and Cuba [19].
Based on phylogenetic evidence and microsatellite markers, all assayed invasive colonies belong to one of four clonal lineages, termed Lines A, B, C and D, or to lineages that arose from rare mating events between invasive lines [10]. Four presumably native samples of Ooceraea from India, China and Vietnam have been determined to correspond either to genetically divergent lineages of O. biroi or to closely related species (genotypes E, F, G and H) [10,20,21]. Of these, a colony from Uttarakhand, India was the closest relative of invasive lines (genotype E; [10]). However, the native source population of Lines A, B, C and D remains unknown.
Subterranean invasive ants are mainly spread via soil, such as the ballast of ships [5]. We hypothesized that Bangladesh could be the source of invasive O. biroi populations, as this country neighbours eastern India and is host to the Bay of Bengal, a major Asian shipping port.
2. Methods
For additional details, see electronic supplementary material, Methods. Our final dataset consisted of 16 independent collections of O. biroi from Bangladesh, one colony from Shenzhen, China, one colony from Singapore and one colony from Ba Vì, Vietnam, in conjunction with previously published sequences. Phylogenetic analysis was conducted using five gene fragments (cytochrome oxidase I (COI), cytochrome oxidase II (COII), wingless (wg), elongation factor 1α (EF1α) and long wavelength rhodopsin (LR)) from a total of 27 independent samples (electronic supplementary material table S1).
Five conserved microsatellite loci from nine exemplar colonies (5–7 ants per colony) were used for population genetic analysis. Two criteria were employed to infer clonality in colonies collected from Bangladesh: (i) colonies could be maintained in the laboratory without mating (see details in §3) and (ii) genotypes across five microsatellite loci were consistent with clonal reproduction.
3. Results
From the 16 colonies we collected in Bangladesh, we recovered seven unique mitochondrial haplotypes across COI and COII sequences. Two of these were identical to those of known invasive lines, Lines C and D, and the remaining five had not been previously reported (electronic supplementary material, table S1). All microsatellite data were consistent with clonal reproduction (electronic supplementary material, table S2) [10]. As further support for asexual reproduction, we were able to maintain two of the new lines, I and L, in the laboratory for over five years without any evidence of mating. The maximum lifespan of O. biroi is ca. 1.5 years, implying that all of the individuals collected initially in the field had long died. Colonies that were not maintained in the laboratory were either collected with low numbers of individuals and/or died in captivity (for colony sizes in the field, see electronic supplementary material, table S1). Finally, we visually inspected all ants from each colony immediately upon collection and never observed any morphological queens. Queen production was neither observed in the laboratory.
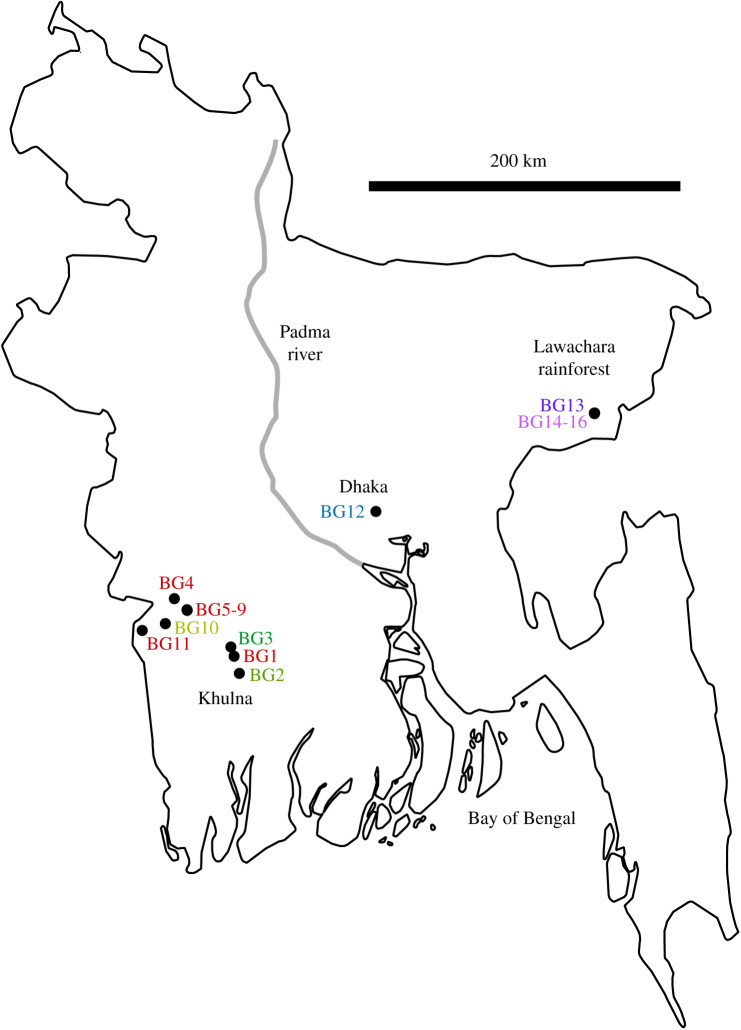
Based on the totality of evidence, we conclude that the seven mitochondrial haplotypes we collected in Bangladesh correspond to at least seven unique clonal lines. Two of these belong to known invasive lines, Lines C and D, and we designate the other five as new lines, Lines I, J, K, L and M (electronic supplementary material, tables S1 and S2). This sample population encompasses the 600 km width of the country, and new lines were found in the vicinity of Khulna, Dhaka and the Lawachara Rainforest (figure 1).
Figure 1.
Collection localities for colonies from Bangladesh. Geographic coordinates and location names are given in electronic supplementary material, table S1.
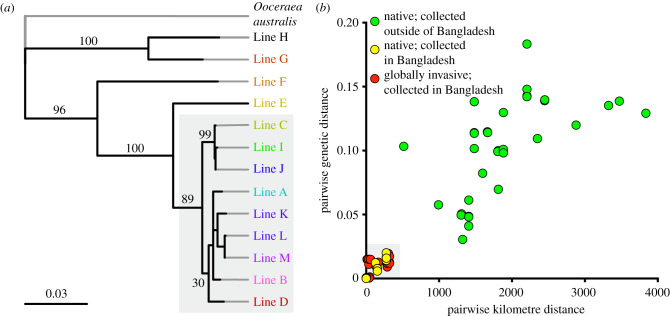
To test whether Bangladesh is the source of invasive lines, we performed a molecular phylogenetic analysis (figure 2a). The five new lines from Bangladesh are more closely related to the four invasive lines than the previously described lines from continental Asia. In fact, the invasive lines are phylogenetically nested within the genetic diversity of Bangladesh lines.
Figure 2.
Phylogenetic analysis. (a) Phylogeny of O. biroi from Bangladesh and the globally invasive range. Numbers indicate bootstrap support; scale bar indicates proportional divergence at informative loci. Globally invasive lines (A–D) are nested within the diversity of Bangladesh lines (I–M; globally invasive lines C and D were also found in Bangladesh) (grey box). Outgroup in grey: O. australis (senior synonym of Cerapachys edentata [22]). (b) Relationship between geographic and genetic distances of Ooceraea collections from the Asian continent. Genetic distances are derived from branch lengths in figure 2a. Green points (native; collected outside of Bangladesh) are pairwise distances between every native Ooceraea collection from India, Vietnam and China with every other such collection, and with every Bangladesh line. Yellow points (native; collected in Bangladesh) are distances between all pairs of Bangladesh lines (I–M). Red points (globally invasive; collected in Bangladesh) represent distances between our new Bangladesh collections of globally invasive lines (C and D) and our new Bangladesh lines (I–M).
The distance matrix of this phylogeny reveals a linear association of genetic and geographic distances in native samples from the Asian continent (figure 2b, green). The globally invasive lines we collected in Bangladesh did not deviate from this relationship and were geographically and genetically close to our newly described Bangladesh lines (figure 2b; red and yellow, respectively). As an example, Lines I, J and C were found within a 50 km radius, are closely related and form a clade, whereas Line E, found 1300 km away in Uttarakhand, is genetically more distant and the sister to the clade of Bangladesh and invasive lines. This concordance of geographic and genetic distances indicates that the invasive lines are almost certainly derived from within Bangladesh.
Finally, we genotyped three additional new collections from Singapore, Shenzhen (China) and Ba Vì (Vietnam). These colonies belong to Lines B, C and D, respectively, confirming that invasive O. biroi lines have become established in Singapore and mainland Asia [15].
4. Discussion
Our results provide a number of new insights into the biology and invasion history of the clonal raider ant. With seven unique mitochondrial haplotypes among 16 sampled colonies, Bangladesh contains by far the most genetically diverse O. biroi population known to date. For example, [10] observed two mitochondrial haplotypes in 22 colonies from Okinawa, Japan, and just one mitochondrial haplotype in 17 colonies from St. Croix, U.S. Virgin Islands. This high diversity suggests that O. biroi is native to Bangladesh, and our phylogenetic data indicate that the globally invasive lines originally stem from Bangladesh.
Our results confirm that O. biroi had already become invasive when it was originally described from Singapore in 1907 [15,23]. These samples were collected by Lajos Bíró following his travels to New Guinea from 1896 to 1902, so O. biroi must have become invasive before 1902 [24]. It is likely that O. biroi spread initially from Bangladesh via the historically important Bay of Bengal shipping ports, Dacca (present-day Dhaka) and Chittagong [25]. The Chittagong port existed at least as early as the second century, and both ports became major sources of international shipping activity from the 1600s beyond the 1800s [25]. Unfortunately, our present data do not provide a precise estimate for which port(s) and time periods might be responsible for the export of O. biroi from Bangladesh.
Invasive lines of O. biroi share a few traits with other invasive ant species that may allow them to thrive in human-modified habitats. These include parthenogenesis, retention of heterozygosity and the presence of multiple egg-layers within a colony [11,13]. Interestingly, we observed these three traits in the new O. biroi lines from the native range in Bangladesh (Lines I, J, K, L and M), which were not phenotypically distinct from the previously described lines from the invasive range (Lines A, B, C and D). Foucaud et al. [3] proposed a two-step model for the evolution of invasive ants: populations may adapt first to human-modified habitats within their native range and then spread to similar anthropogenic habitats around the world. Under this view, parthenogenesis, retention of heterozygosity and the presence of multiple egg-layers in O. biroi may all represent adaptations for survival in human-modified habitats in Bangladesh. Indeed, all three traits are found in invasive, but not native, populations of the little fire ant Wasmannia auropunctata. Evolution of parthenogenesis and polygyny in human-modified habitats possibly also occurred in Mycocepurus smithii and Solenopsis geminata [26–28].
In the light of the above case studies, we propose the following scenario. Prior to major anthropogenic impact, native populations in Bangladesh largely reproduced sexually, as do most ants, but had a propensity for asexual reproduction at some rate. As the native habitat became increasingly modified by humans, a larger subset of asexual genotypes were reproductively successful and became locally prolific in anthropogenic habitats. From that population, four genotypes were transported out of Bangladesh and ultimately became globally invasive. This model predicts that extant populations of O. biroi in undisturbed habitats may still reproduce sexually at some rate. We might also expect to find sexual reproduction in closely related Ooceraea species provided they have not experienced parallel evolution of parthenogenesis. The alternative scenario is that O. biroi was strictly asexual even before humans began transforming their habitat in Bangladesh. This seems unlikely, however, because automixis with central fusion leads to the gradual loss of heterozygosity. The fact that all genotyped samples of O. biroi are still highly heterozygous therefore suggests that they belong to relatively young asexual lineages, as is the case in several other invasive ants [3,13,26,27]. Genetic data from undisturbed habitats in Bangladesh and surrounding areas will be required to distinguish more definitively between these two possibilities.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Lisa Kronauer, Megna Guhathakurta, Hasan Aref, Dilip Datta and Karsten Schroeder for help organizing field research in Bangladesh. We thank Tawhid Hossain and Shamul Biswas for field assistance in Bangladesh, and Jihea Choi for sharing samples from Singapore. We thank Ian Butler for assistance with microsatellite genotyping and Leonora Olivos-Cisneros; Amelia Ritger and Stephany Valdés-Rodríguez for assistance with ant maintenance. This is Clonal Raider Ant Project paper #11.
Ethics
This work has been approved by and complies with the requirements of all relevant institutional and government authorities.
Data accessibility
All raw data for this article are available in electronic supplementary material, table S1 and table S2, and GenBank accession numbers provided therein.
Authors' contributions
W.T., S.K.M. and D.J.C.K. conceived and planned the project. W.T. and S.K.M. performed fieldwork. W.T. collected and analysed molecular data, with input and assistance from S.K.M. and D.J.C.K. W.T. wrote the manuscript, with input, critical insight and revisions from S.K.M. and D.J.C.K. D.J.C.K. supervised the project. All authors have read and approved the final version of the manuscript and agree to be accountable for any questions related to the accuracy or integrity of any part of the work.
Competing interests
We declare we have no competing interests.
Funding
Research reported in this publication was supported by a grant from the Faculty Scholars Program of the Howard Hughes Medical Institute to D.J.C.K.
References
- 1.Global Invasive Species Database. 2020. http://www.issg.org/database . Accessed 29 May 2020.
- 2.Tsutsui ND, Suarez AV, Holway DA, Case TJ. 2001. Relationships among native and introduced populations of the Argentine ant (Linepithema humile) and the source of introduced populations. Mol. Ecol. 10, 2151–2161. ( 10.1046/j.0962-1083.2001.01363.x) [DOI] [PubMed] [Google Scholar]
- 3.Foucaud J, et al. 2010. Worldwide invasion by the little fire ant: routes of introduction and eco-evolutionary pathways. Evol. Appl. 3, 363–374. ( 10.1111/j.1752-4571.2010.00119.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascunce MS, et al. 2011. Global invasion history of the fire ant Solenopsis invicta. Science 331, 1066–1068. ( 10.1126/science.1198734) [DOI] [PubMed] [Google Scholar]
- 5.Gotzek D, Axen HJ, Suarez AV, Helms Cahan S, Shoemaker D. 2015. Global invasion history of the tropical fire ant: a stowaway on the first global trade routes. Mol. Ecol. 24, 374–388. ( 10.1111/mec.13040) [DOI] [PubMed] [Google Scholar]
- 6.Sakai AK, et al. 2001. The population biology of invasive species. Annu. Rev. Ecol. Syst. 32, 305–332. ( 10.1146/annurev.ecolsys.32.081501.114037) [DOI] [Google Scholar]
- 7.Wetterer JK. 2005. Worldwide distribution and potential spread of the long-legged ant, Anoplolepis gracilipes (Hymenoptera: Formicidae). Sociobiology 45, 77–97. [Google Scholar]
- 8.Wetterer JK. 2010. Worldwide spread of the pharaoh ant, Monomorium pharaonis (Hymenoptera: Formicidae). Myrmecol. News 13, 115–129. [Google Scholar]
- 9.Tseng S-P, Darras H, Lee C-Y, Yoshimura T, Keller L, Yang C-CS. 2019. Isolation and characterization of novel microsatellite markers for a globally distributed invasive ant Paratrechina longicornis (Hymenoptera: Formicidae). Eur. J. Entomol. 116, 253–257. ( 10.14411/eje.2019.029) [DOI] [Google Scholar]
- 10.Kronauer DJC, Pierce NE, Keller L. 2012. Asexual reproduction in introduced and native populations of the ant Cerapachys biroi. Mol. Ecol. 21, 5221–5235. ( 10.1111/mec.12041) [DOI] [PubMed] [Google Scholar]
- 11.Oxley PR, Ji L, Fetter-Pruneda I, McKenzie SK, Li C, Hu H, Zhang G, Kronauer DJC. 2014. The genome of the clonal raider ant Cerapachys biroi. Curr. Biol. 24, 451–458. ( 10.1016/j.cub.2014.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suarez AV, McGlynn TP, Tsutsui ND. 2010. Biogeographic and taxonomic patterns of invasive ants. In Ant ecology, pp. 233–244. Oxford, UK: Oxford University Press. [Google Scholar]
- 13.Rabeling C, Kronauer DJC. 2013. Thelytokous parthenogenesis in eusocial Hymenoptera. Annu. Rev. Entomol. 58, 273–292. ( 10.1146/annurev-ento-120811-153710) [DOI] [PubMed] [Google Scholar]
- 14.Wetterer JK. 2015. Geographic origin and spread of cosmopolitan ants (Hymenoptera: Formicidae). Halteres 6, 66–78. [Google Scholar]
- 15.Wetterer J, Kronauer D, Borowiec M. 2012. Worldwide spread of Cerapachys biroi (Hymenoptera: Formicidae: Cerapachyinae). Myrmecol. News 17, 1–4. [Google Scholar]
- 16.Rasheed M, Bodlah I, Fareen A, Wachkoo A, Huang X, Akbar S. 2019. A checklist of ants (Hymenoptera: Formicidae) in Pakistan. Sociobiology 66, 426–439. ( 10.13102/sociobiology.v66i3.4330) [DOI] [Google Scholar]
- 17.Dias RKS, Udayakantha S, Wachkoo AA, Akbar SA. 2018. New records of ants (Hymenoptera: Formicidae) from Sri Lanka, including four tramp species. Sociobiology 65, 449–455. ( 10.13102/sociobiology.v65i3.3180) [DOI] [Google Scholar]
- 18.Leong C, Shiao S, Guénard B. 2017. Ants in the city, a preliminary checklist of Formicidae (Hymenoptera) in Macau, one of the most heavily urbanized regions of the world. Asian Myrmecology 9, e009014 ( 10.20362/am.009014) [DOI] [Google Scholar]
- 19.Fontenla JL, Aolfonso-Simonetti J. 2018. Classification of Cuban ants (Hymenoptera: Formicidae) into functional groups. Poeyana 506, 21–30. [Google Scholar]
- 20.Bharti H, Akbar S. 2013. Taxonomic studies on the ant genus Cerapachys Smith (Hymenoptera, Formicidae) from India. Zookeys 336, 79–103. ( 10.3897/zookeys.336.5719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada A, Luong PTH, Eguchi K. 2018. Description of a new species of the ant genus Ooceraea Roger, 1862 (Hymenoptera: Formicidae; Dorylinae) from the Vietnam's Central Highlands. J. Insect Biodivers. 007, 17–23. ( 10.12976/jib/2018.07.1.2) [DOI] [Google Scholar]
- 22.Borowiec M. 2016. Generic revision of the ant subfamily Dorylinae (Hymenoptera, Formicidae). Zookeys 608, 1–280. ( 10.3897/zookeys.608.9427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forel A. 1907. Formicides du Musée National Hongrois. Ann. Museei Natl. Hungarici 5, 1–42. [Google Scholar]
- 24.Mackellar CD. 1912. Scented isles and coral gardens: Torres Stra its, German New Guinea and the Dutch East Indies. London, England: John Muray. [Google Scholar]
- 25.Amrith SS. 2013. Crossing the Bay of Bengal. Cambridge, MA: Harvard University Press. [Google Scholar]
- 26.Rabeling C, Lino-Neto J, Cappellari SC, Dos-Santos IA, Mueller UG, Bacci M. 2009. Thelytokous parthenogenesis in the fungus-gardening ant Mycocepurus smithii (Hymenoptera: Formicidae). PLoS ONE 4, e6781 ( 10.1371/journal.pone.0006781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabeling C, Gonzales O, Schultz TR, Bacci M, Garcia MVB, Verhaagh M, Ishak HD, Mueller UG. 2011. Cryptic sexual populations account for genetic diversity and ecological success in a widely distributed, asexual fungus-growing ant. Proc. Natl Acad. Sci. USA 108, 12 366–12 371. ( 10.1073/pnas.1105467108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacy KD, Shoemaker D, Ross KG. 2019. Joint evolution of asexuality and queen number in an ant. Curr. Biol. 29, 1–7. ( 10.1016/j.cub.2019.03.018) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data for this article are available in electronic supplementary material, table S1 and table S2, and GenBank accession numbers provided therein.